Abstract
Translation is a central cellular process and is optimized for speed and fidelity. The speed of translation of a single codon depends on the concentration of aminoacyl-tRNAs. Here, we used microarray-based approaches to analyze the charging levels of tRNAs in Escherichia coli growing at different growth rates. Strikingly, we observed a non-uniform aminoacylation of tRNAs in complex media. In contrast, in minimal medium, the level of aminoacyl-tRNAs is more uniform and rises to approximately 60%. Particularly, the charging level of tRNASer, tRNACys, tRNAThr and tRNAHis is below 50% in complex medium and their aminoacylation levels mirror the degree that amino acids inhibit growth when individually added to minimal medium. Serine is among the most toxic amino acids for bacteria and tRNAsSer exhibit the lowest charging levels, below 10%, at high growth rate although intracellular serine concentration is plentiful. As a result some serine codons are among the most slowly translated codons. A large fraction of the serine is most likely degraded by L-serine-deaminase, which competes with the seryl-tRNA-synthetase that charges the tRNAsSer. These results indicate that the level of aminoacylation in complex media might be a competition between charging for translation and degradation of amino acids that inhibit growth.
INTRODUCTION
Transfer RNAs (tRNAs) are fundamental components of the translation machinery and link two chemically distinct entities, the nucleotide mRNA sequence and the growing polypeptide (1). Different tRNAs decode all 61 sense codons, yet wobbling at the first position of some anticodons reduces the total number of tRNAs required for decoding. For example, E. coli employs 46 tRNAs (2). In prokaryotes and single-cell eukaryotes the abundance of each tRNA correlates with its genomic copy number (3) and with the codon usage of highly expressed genes (4–6). Hence, tRNA isoacceptors (that are tRNA species charged with the same amino acids but with different anticodon sequences) pairing to frequent codons have higher concentrations than tRNAs reading rare codons. This asymmetric tRNA abundance leads to variations in the translation rate of each codon (7) and this non-uniform speed of elongation along mRNA provides an important benefit to facilitate protein folding and biogenesis (8).
The total concentration of individual tRNAs in E. coli varies with growth rate, i.e. growth in various media (2,3). Importantly, the abundance of each tRNA species changes non-uniformly with growth rate. For example, the levels of abundant tRNAs increase while minor tRNAs remain relatively unchanged when growth rate increases (2), implying that the adaptability of tRNA pools provides a plasticity to maintain translation balance under different growth conditions. In support of this view, imbalance of the naturally selected tRNA levels alters cell growth and decreases cell fitness (9,10).
tRNAs enter a translation cycle only when charged with their cognate amino acid at its 3′ end. tRNAs are aminoacylated by 20 different aminoacyl-tRNA-synthetases (AARS), each of which is specific for the 20 different amino acids (11). Experimental determination of the charging level of several individual tRNAs in exponentially growing E. coli has suggested that they are aminoacylated to 80–90% with their cognate amino acid (12–14). This result has been widely extrapolated to all tRNAs, thus assuming a nearly uniform charging level for all tRNAs (15). However, the aminoacylation level of the tRNA isoacceptors sensitively reacts to changes in environmental conditions (16,17). Thereby various tRNA isoacceptors within one tRNA isoacceptor family (i.e. all tRNA isoacceptors carrying the same amino acid (1)) respond differently to environmental changes, suggesting regulatory functions of aminoacylation in shaping the growth response of the cell. For example, amino acid limitations trigger preferential charging of minor tRNAs within one isoacceptor family to facilitate specific translation of amino acid biosynthetic genes enriched in codons pairing to those rare tRNAs (16). Furthermore, tandems of rare codons (18) or long consecutive repeats of a codon with even a plentiful cognate tRNA (19) results in transient ribosomal pausing and frameshifting, implying that a paucity of charged tRNA might be a causal factor. In addition, many tRNAs participate in a plethora of processes beyond translation, including N-terminal tagging of proteins for degradation by phenylalanyl-tRNAPhe and leucyl–tRNALeu (1), hence the turnover of aminoacyl–tRNA levels may differ from the turnover of tRNAs involved only in elongation. Together, these observations question the general assumption that tRNAs are uniformly charged. Using the power of microarray technology to determine the charging levels of all tRNA isoacceptor families (17), we set to revisit this arbitrarily established assumption which is based on only a few experimental examples (12–14). We found that the levels of aminoacyl-tRNAs are non-uniform and the low suboptimal charging of some tRNAs is determined by the trade-off between amino acid sequestration into aminoacyl–tRNAs for biosynthesis and degradation for detoxification.
MATERIALS AND METHODS
Media and growth conditions
Escherichia coli MC4100 was grown aerobically at 37°C in LB medium (10 g/l NaCl, 10 g/l tryptone and 5 g/l yeast extract), M9 minimal medium (MM, 12.8 g/l Na2HPO4.7H2O, 3 g/l KH2PO4, 1 g/l NH4Cl, 0.24 g/l MgSO4, 0.011 g/l CaCl2, 4 g/l glucose) or defined rich medium (M9 minimal medium supplemented with each amino acid to the following concentrations as reported in (20): Cys, Trp – 0.1 mM; His, Met, Tyr – 0.2. mM; Arg, Asn, Asp, Ile, Lys, Phe, Pro, Thr, Trp – 0.4 mM; Glu, Gln, Val – 0.6 mM; Ala, Gly, Leu – 0.8 mM; Ser 10 mM). Starter cultures from a single colony were grown overnight at 37°C, diluted 100-fold and used to inoculate fresh medium. For microarray analysis cultures were grown in shake flasks. Growth curves in MM supplemented with one amino acid were performed in 96-well plates sealed with gas permeable foil at 37°C and 2 mm shaking amplitude. OD600 data were collected with TECAN Infinite M200 plate reader and zeroed with a blank of the medium. The intracellular concentration of free amino acids was determined as described (21). Calculation of the concentration from mol/dry cell weight to mM was performed as described for calculating various intracellular metabolites using 9 × 10−13g and 3 × 10−13g for E. coli volume in complex media (LB and MM+AA) and MM, respectively (22). The acetate concentration in the growth medium was measured at OD 0.5 with Acetate Colorimetric Assay kit (Sigma).
Microarray analysis
RNA was isolated under mild acidic conditions as described (13) with slight modifications at later steps. After phenol extraction, samples were precipitated with isopropanol for at least 30 min at −80°C and the centrifugation steps were extended to 30 min at 21 000 xg at 4°C. tRNAs were oxidized with NaIO4 as described (3), e.g. 0.1 μg/μl total RNA in 100 mM NaOAc/AcOH (pH 4.5) was treated with 50 mM NaIO4 at 22°C for 30 min. In parallel, a control non-oxidized sample was treated the same way with NaCl instead of NaIO4. Oxidized sample was quenched by adding 100 mM of glucose and incubated at 22°C for 10 min. Both samples were spiked in with in vitro synthesized transcripts of yeast tRNAPhe, human tRNAHis and human tRNAGln, each of which with a final concentration of 0.08 μM. To remove NaIO4 from oxidized sample, a G25 spin column was used and both oxidized and control RNA samples were precipitated in ethanol. Thereafter, both RNA samples were deacylated into 0.1 M Tris (pH 9) at 37°C for 45 min and neutralized with the same volume of 100 mM NaOAc/AcOH (pH 4.8), 100 mM NaCl. After ethanol precipitation, RNA samples were dissolved into H2O. For labeling, 0.25 μg/μl RNA was mixed with 4.5 μM tagging fluorescent oligonucleotide (3) in reaction containing 20 U/μl T4 DNA Ligase (New England Biolabs), 1x T4 DNA Ligase buffer (New England Biolabs) and 15% (v/v) DMSO and incubated at 16°C for 16 h. The oxidized sample (i.e. aminoacyl–tRNAs) was labeled with Atto647-labeled oligonucleotide and the control samples representing the total tRNA was labeled with Cy3-labeled oligonucleotide.
After phenol/chloroform extraction, RNA was precipitated with ethanol. The ligation efficiency was estimated on denaturing 10% PAGE by comparing the fluorescent signal of the labeled tRNAs (Fujifilm LAS-4000) to the intensity of the total tRNA visualized with SYBR gold (Invitrogen). Two micrograms of each total labeled tRNA sample was loaded on a microarray containing full-length tDNA probes recognizing 40 E. coli tRNA isoacceptors (3) and the three standard spike-ins. On each array, the Atto647-labeled aminoacyl-tRNA and Cy3-labeled total tRNA were loaded and the hybridization was carried out at 60°C for 16 h on Hyb4 microarray hybridization system (Digilab). Each array contained 24 replicates of each tDNA probe. The arrays were normalized to spike-in standards and the signal of Atto647-labeled aminoacyl-tRNA was calculated as a percentage of the cognate Cy3-labeled total tRNA. The quantification was performed with in-house R scripts.
Northern blot analysis
Aminoacylation levels of single tRNAs were determined by acidic denaturing polyacrylamide gel as described (13) with some modifications. Untreated and deacylated RNA samples were resolved on acidic denaturing gel (6.5% polyacrylamide, 8 M urea, 0.1 M NaOAc/AcOH pH 5.0) and electroblotted onto a nylon membrane (GE Healthcare, Amersham HybondTM-N). Hybridization was performed at 60°C for 16 h with a 5′-32P labeled full length tDNA.
Quantitative RT-PCR
Total RNA from three biological replicates of exponentially growing cells (OD600 = 0.5) was isolated with TRI reagent (Sigma, Aldrich) according to the manufacturer's instructions with an additional washing step with 70% ethanol. Precipitated RNA was dissolved in RNAse-free water, its amount adjusted to 1 μg and subjected to DNA depletion with DNase I (Thermo Scientific) followed by cDNA synthesis using RevertAid RT Kit (Thermo Scientific) with random hexamer primers. The mRNA expression of sdaB and sdaC were quantified using QuantiFast SYBR Green PCR Kit (Qiagen) and normalized against the housekeeping gene gapA, which encodes glyceraldehyde-3-phosphate dehydrogenase (23). Primer sequences were as follows: forward, 5′-AATACTCATGGTCGCCTGATG-3′ and reverse,- 5′-ATAAAGCCACCGCCAATAGAG-3′ for sdaB: forward, 5′-ACTCGCTTCGTACTGTCTGG-3′ and reverse, 5′-AGCAGGATCGGGTAGATAGC-3′ for sdaC; forward, 5′-CCTGTTAGACGCTGATTAC-3′ and reverse 5′-CGTCCCATTTCAGGTTAG-3′ for gapA.
Analysis of effective codon usage and ribosomal occupancy from ribosome profiling and RNA-Seq
To determine the codon identities in the ribosomal A site over the whole transcriptome, we used our recently generated ribosome profiling and RNA-Seq data sets from exponentially growing E. coli MC4100 grown either in LB or MM (24); the data were deposited in Gene Expression Omnibus (GEO), GSE68762. Ribosome-protected fragments (RPFs) were binned in groups of equal read length and each group was aligned to the stop codons. We used 3′ calibration of the reads as the MNase cleavage was more variable on the 5′ side of the ribosome-protected fragment, which is consistent with prior studies (25). The calibration was performed over the stop codon that allows precise assignment of the ribosomal A site and was used to determine the distance between the middle nucleotide in the A site and 3′ end of the read for each binned read length. We found that 23–26 nt-long reads showed the most evident 3-nt periodicity in frame. The aggregated A site codons were normalized to the library depth and the effective codon usage that represents the codon usage calculated from all expressed mRNAs from the RNA-seq experiment.
The gene expression examples were calculated by total mapped reads to the corresponding coding sequence and normalized to the gene length and library depth (reads per kilobase per million, rpkM) (26). The threshold of expression level was determined from the biological replicates and it was set for both ribosome profiling and RNA-seq data at >60 reads (24,27).
RESULTS
tRNA isoacceptors are non-uniformly charged in complex LB medium
To determine the fraction of each aminoacyl-tRNA in E. coli (strain MC4100) at various growth phases and in different media, we used tRNA-specific microarrays (17). The microarrays can be run in two modes: (i) to determine differences in tRNAs between two different organisms or cell types (28) or (ii) to quantify the fraction of aminoacyl–tRNA isoacceptor by comparing its amount to that of the total amount of each isoacceptor (17). Here, we used the latter application that takes advantage of the aminoacyl group acting as a protecting moiety of the 3′CCA ends against oxidation. Total tRNA was isolated at acidic pH, which preserves the aminoacyl moiety, and split into two even aliquots. One aliquot was deacylated and represents the total tRNAs. This was labeled with a Cy3-labeled hairpin oligonucleotide pairing to intact CCA-ends. The second aliquot was treated with periodate; only non-aminoacylated tRNAs with free 3′ CCA ends were vulnerable to periodate oxidation while the aminoacyl–tRNAs remained intact (17). Next, the intact aminoacyl–tRNAs were deacylated and Atto647-labeled oligonucleotide was ligated to unoxidized CCA-ends. Both aliquots were hybridized to the same array and the signal of Atto647-labeled aminoacyl–tRNA was calculated as a percentage of the cognate Cy3-labeled total tRNA (Figure 1A). Isoacceptors that differ by at least 8 nt can be unambiguously detected by the microarrays, thus some tDNA probes bind to more than one isoacceptor (17). In total, we were able to differentiate 40 out of 46 tRNAs and all 20 tRNA isoaceptor families of E. coli (Supplementary Table S1).
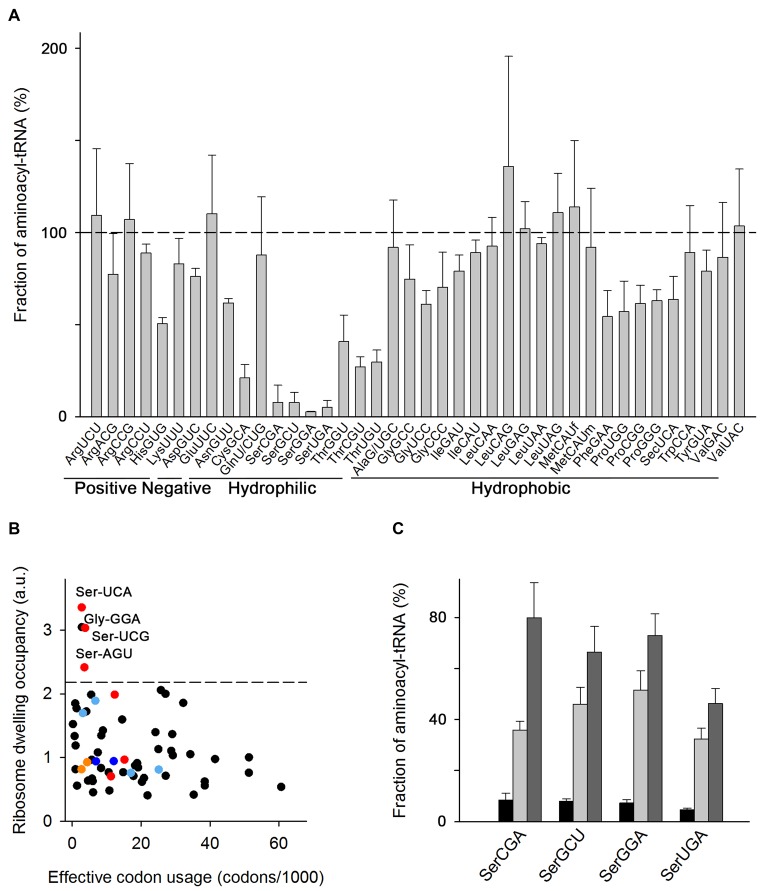
Figure 1.
Some tRNAs have suboptimal aminoacylation level in cells grown in LB medium. (A) Microarray of aminoacyl–tRNAs of E. coli MC4100 at OD600 = 0.5 grown in LB at 37°C. tRNA probes are shown with their anticodon and the corresponding amino acid and classified according to the properties of the amino acid. Data are means ± SEM of three biological replicates. The horizontal dashed line denotes the level at which all tRNAs are fully charged. MetCAUf and MetCAUm denote initiator and elongator tRNAs, respectively (see also Supplementary Table S1). (B) Average ribosome dwelling occupancy, i.e. the frequency of each codon in the ribosomal A site, as determined by ribosome profiling and plotted in correspondence to their frequency in the translatome (i.e. effective codon usage calculated from all mRNAs expressed in MC4100 cells using RNA-Seq). The codons with low levels of aminoacyl–tRNAs (from panel A) are color-coded: Ser codons in red, Cys codons in orange, His codons in blue and Thr codons in light blue. The dashed line denotes the upper bound (90% confidence interval) of the ribosome dwelling occupancies. (C) Microarray of seryl-tRNAsSer in LB medium at OD600 = 1.5 (black bars), OD600 = 2.5 (light gray bars) and OD600 = 3.5 (dark gray bars). Data are means ± S.D. For the charging pattern of the whole tRNAome see Supplementary Figure S2C.
The microarray analysis of exponentially growing E. coli MC4100 (OD600 = 0.5) revealed non-uniform charging of tRNAs. Among these, 9 isoacceptors charged between 60–75% (e.g. tRNA isoacceptors carrying Asn, Asp, Gly, Phe, Pro) and 20 nearly completely (>75%) aminoacylated (Figure 1A). Unexpectedly, we detected quite low charging levels (under 50%) for tRNA isoacceptors carrying Cys, His, Ser and Thr (Figure 1A). This finding was also corroborated by Northern blot analysis (Supplementary Figure S1A). Notably, all tRNA isoacceptors within one tRNA–isoacceptor family were charged to similar levels (Figure 1A). Similar suboptimal charging of Cys, Ser and Thr isoacceptor families was also observed in E. coli BL21(DE3) grown in LB medium, suggesting that the effect is not strain-specific (Supplementary Figure S1B).
Aminoacyl–tRNAs reach the ribosomes by diffusion and their concentration determines the translation rate of each codon (7,29). Because of the extremely low concentration of seryl–tRNAsSer, we hypothesized that Ser codons will be translated with much slower velocity than any other codon whose tRNAs are completely charged. To test this idea, we used ribosome profiling (30) to determine the average speed of translation of each codon in MC4100 cells. The frequency a codon is detected in the ribosomal A site correlates with the ribosome dwell time for that particular codon and is a measure of average codon translational speed (15,31). The ribosome profiling showed a good correlation between biological replicates (24,27). We binned the RPF by their length and calibrated them separately to the A-site codon using the 3′-assignment method that gives more precise position-specific signal for prokaryotes as discussed recently (25). Frequently used codons will statistically occupy the ribosomal A site more often, thus in parallel we performed RNA-Seq (24,27) and normalized it to the codon usage in the transcriptome (i.e. the codon usage in the genome weighted by the mRNA expression level determined by RNA-Seq). Aggregating the calibrated reads for all sequencing read lengths relative to the start and stop codon revealed a clear 3-nt phasing for the RPFs which is absent in the mRNA reads (Supplementary Figure S2A). Indeed, the occupancy of three out of six Ser codons in the ribosomal A-site was markedly high and in the 90th percentile of all codons. We attributed the increase in ribosome dwelling at these Ser codons to a decrease in translation rate, i.e. these Ser codons are among the most slowly translated codons in exponentially growing MC4100 in LB medium (Figure 1B), which correlates with the paucity of their cognate aminoacyl-tRNAs (Figure 1A), despite the fact that non-aminoacylated tRNAsSer are plentiful (2). The ribosomes dwell longer at only three (AGU, UCA and UCG) out of six Ser codons, although the four cognate seryl-tRNAsSer are low but evenly charged (Figure 1A). In searching for reason for the particular shortage of only these three codons, we realized that the transcriptome usage of AGU, UCA and UCG is an order of magnitude lower than that of the other three Ser codons (AGC, TCC and TCT); the codon usage at the whole transcriptome mirrors the abundance of their cognate tRNAs. Predictive modeling, reassuringly, identifies a potential bottleneck and effect on fitness only of rare Ser codons with rare tRNAs (10).
One of the Gly codons, the GGA codon, also exhibited higher ribosomal occupancy (Figure 1B) suggesting it is slowly translated. Its cognate tRNAGlyUCC is plentiful (2), but has the lowest charging level among the tRNAsGly (65%, Figure 1A) that may lead to a delay the ribosomes awaiting the glycyl–tRNAGlyUCC. However, tRNAGlyGCC and tRNAGlyCCC are charged to similar levels as tRNAGlyUCC (Figure 1A) for which we did not detect increased ribosome occupancy (Figure 1B). Effects beyond the concentration of the cognate charged tRNA may cause this: a transient pausing at Gly codons has also been detected in the ribosome profiling in E. coli BW25113 grown in LB and interpreted as a result of slower decoding or tRNA translocation from A- to P-site at Gly codons (32).
Strikingly, from all lowly-charged tRNA families (tRNACys, tRNAHis, tRNASer and tRNAThr) in both E. coli MC4100 and E. coli BL21(DE3) the tRNAsSer isoacceptor family exhibited the lowest aminoacylation level, at 5–10% (Figure 1A and Supplementary Figure S1B). Similarly to our observations, high ribosome occupancy at Ser codons has been reported in ribosome profiling of E. coli MG1655 in LB (33,34), however, interpreted as a result of nutrient (C-source) limitation and catabolizing Ser for energy production. Indeed, it is suggested that Ser is the first amino acid to be catabolized following shortage of a carbon source (e.g. glucose) in the medium (35). Thus, we reasoned that if nutrient limitations (e.g. limitations of the uptake of the two organic ingredients in LB, tryptone and yeast extract), are a rate-limiting step and create nutrient shortage, the addition of a favorable C-source (e.g. glucose) would be preferably used and should alleviate charging of tRNAsSer. However, we found that supplementation of the LB medium with glucose did not offset the low charging of the tRNASer isoacceptors (Supplementary Figure S1C). Also, supplementing LB with an excess of Ser did not enhance the charging level of tRNAsSer (Supplementary Figure S1C), arguing against the suggestion that the low charging level of tRNAsSer is a consequence of Ser or nutrient limitations.
Next, we asked whether the tRNA aminoacylation pattern changes with growth phase as protein synthesis declines with bacterial age (36). We analyzed the fraction of aminoacylated tRNA isoacceptors in late exponential phase (OD600 = 1.5) and early (OD600 = 2.5) and late (OD600 = 3.5) stationary phase. The tRNAsSer charging levels gradually increased at late growth phase compared to the exponential phase (Figure 1C and Supplementary Figure S2B), implying that a low charging level of tRNAsSer is not an intrinsic property of the tRNAsSer family and depends on the physiological activities of the cell. Similarly, the charging level for the most tRNA isoacceptors increased at higher OD (Supplementary Figure S2C). A likely explanation is that at higher ODs the cell ceases growth and consequently protein biosynthesis and the demand for charged tRNAs decreases. It is also possible that extracellular amino acids are completely depleted from the medium during exponential growth (35,37), thus the supply in the stationary phase is entirely maintained by the intracellular amino acid synthetic activities of the cell, a constellation which is reminiscent of growth in minimal media (see below).
tRNA aminoacylation level is more uniform by intrinsic amino acid synthesis
Growth in various media activates different physiological programs in bacteria and alters metabolic enzymes (38) to adjust the flow of carbon that is coupled to energy and intracellular nucleoside concentrations (39). rRNA and tRNA expression are physiologically connected to the metabolic state and growth rate of the cell (2,40). Thus, we next asked whether the tRNA charging levels will change when the growth rate decreases, at which time the total concentration of the tRNA isoacceptors also decreases (2). We analyzed the aminoacylation level of exponentially growing E. coli MC4100 (OD600 = 0.5) in minimal medium (MM) with glucose as a sole carbon source; the growth rate in MM is 0.016 ± 0.0023 min−1 and is about two times slower than in LB (the growth rate in LB is 0.026 ± 0.0019 min−1) (Supplementary Figure S3A). In MM all amino acids needed to serve translation activities are internally produced through the own biosynthetic activities of the cell. Notably, in MM the aminoacylation level of all tRNASer isoacceptors increased up to 50% (Figure 2A and Supplementary Figure S3B). In general, the aminoacylation level in MM is lower for all tRNA isoacceptors (Figure 2A) but far more uniform than in LB (Figure 1A). Similar aminoacylation levels were observed for another E. coli strain, CP78, when cultured in MOPS minimal medium (17).
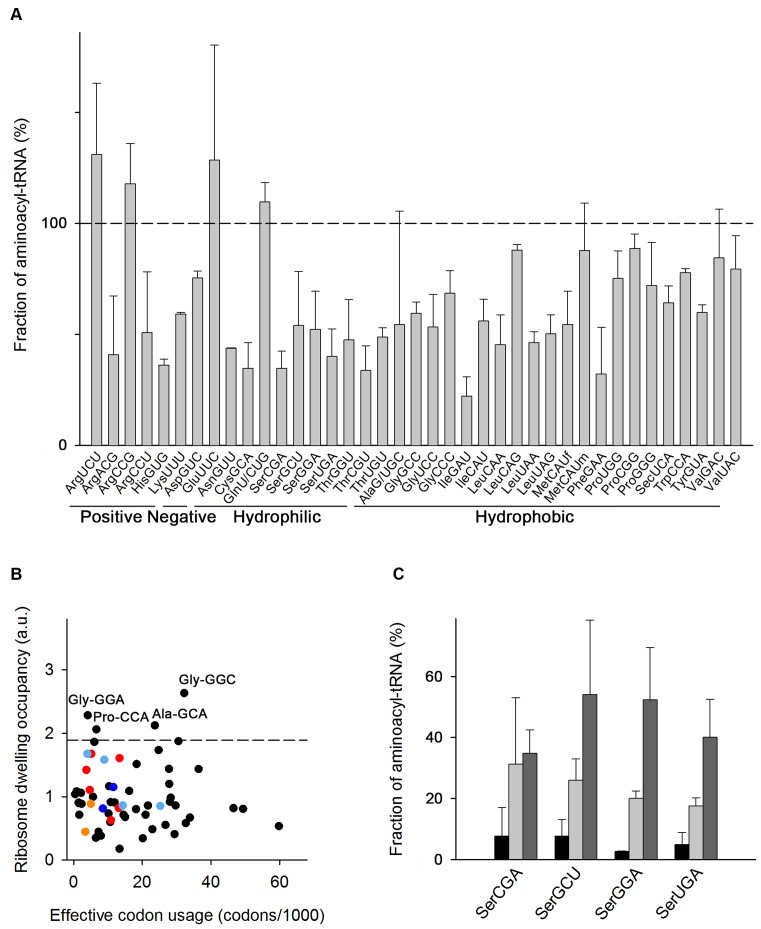
Figure 2.
Medium composition dictates the aminoacylation level of tRNA isoacceptors. (A) Microarray of aminoacyl-tRNAs of E. coli MC4100 at OD600 = 0.5 grown in MM with 0.4% glucose at 37°C.For more details see the legend to Figure 1A. (B) Average ribosome dwelling occupancy as determined by ribosome profiling and plotted in correspondence to codon frequency in the translatome (for details see the legend to Figure 1B). The following codons are color-coded: Ser codons in red, Cys codons in orange, His codons in blue and Thr codons in light blue. The dashed line denotes the upper bound (90% confidence interval) of the ribosome dwelling occupancies. (C) Microarray of seryl-tRNAsSer in LB (black), MM+AA (light gray) and MM (dark gray) at OD600 = 0.5. Data are means ± S.D. of two to three biological replicates. The microarray with all aminoacyl–tRNAs in MM+AA is included in Supplementary Figure S3C.
In MM, serine codons were not among most slowly translated and instead exhibited an average speed of elongation (Figure 2B). Two Gly codons were among the slowest elongated codons in MM although their cognate tRNAs were charged to a similar extent as the majority of the tRNAs in MM (Figure 2A). The detection of enhanced ribosome occupancy at Gly codons in MM suggests that the intimate process of decoding Gly codons, e.g. translocation, is rate-limiting (32).
tRNA aminoacylation levels mirror the degree of amino acid toxicity
What is the reason for the low charging level of some tRNAs in LB medium? LB is an undefined medium in which tryptone and yeast extract are the sources for amino acids in de novo translation. Since the composition of these complex substrates often varies and their amino acid composition may lead to a shortage of some amino acids, we next compared the tRNA aminoacylation levels in LB to those in a defined complex medium (MM+AA) containing glucose as a C-source and optimal concentrations of all 20 proteinogenic amino acids (20). The growth rate in MM+AA at 37°C of 0.023 ± 0.0015 min−1 is similar to that in LB (Supplementary Figure S3A). The charging level of the tRNA isoacceptors was slightly higher in MM+AA but more similar to that in LB than in MM (Supplementary Figure S3B). Notably, the charging level of tRNAsSer rose to ∼15–20% but remained lower than that in MM (Figure 2C). We measured an internal concentration of free Ser in MM+AA of 4.0 ± 0.3 mM (Supplementary Table S2), which is one of the highest among all amino acids and argues that Ser uptake and free Ser concentration in the cell is not a bottleneck. For comparison, the intracellular free Ser concentration in E. coli MC4100 grown in MM was two orders of magnitude lower 0.057 ± 0.005 mM (Supplementary Table S2) and of similar magnitude as measured for E. coli NCM3722 – 0.068 mM (22). Note that we were not able to quantify the amount of the intracellular free amino acids in LB because of the high background from medium remains. The similar growth rate and charging level between LB and MM+AA suggests that the intracellular free amino acids concentration will be within the same order of magnitude.
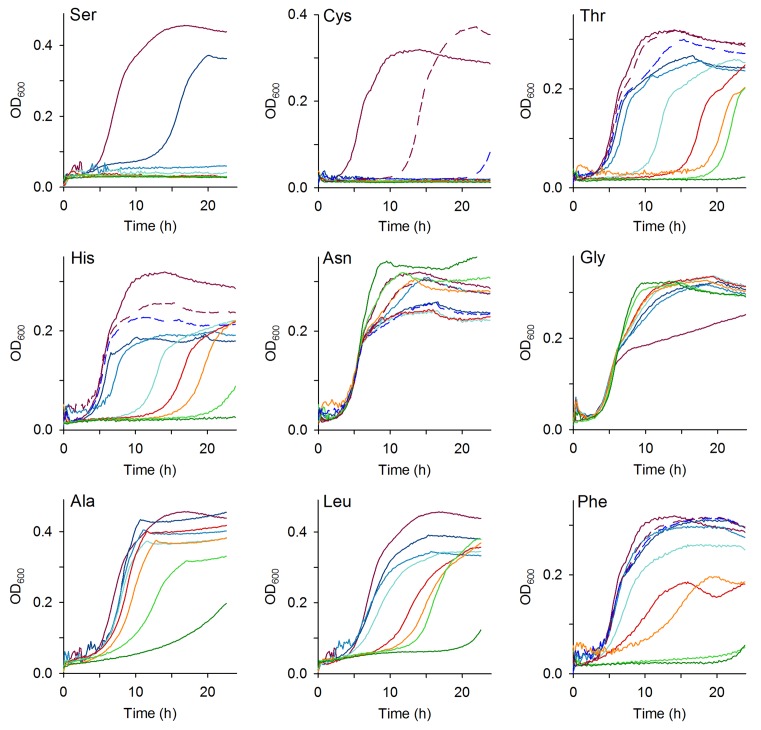
Despite the lower intracellular Ser concentration in MM as compared to MM+AA, the charging level of tRNASer isoacceptors is much higher in MM than in MM+AA (Figure 2C and Supplementary Figure S3C). Enhancing the charging of tRNAsSer by supplementation of Ser in MM was not possible, since Ser readily inhibited growth even at the lowest external concentration (Figure 3). Similarly, Cys also inhibited growth at very low concentrations when present in MM, while Thr and His showed an altered phenotype in the onset of exponential growth, but no notable effect on the growth rate (Figure 3 and Supplementary Table S3). Amino acids whose tRNA isoacceptors were charged >60% inhibited cell growth only at relatively high concentrations, (e.g. Phe, Ala, Leu) or even stimulated growth (e.g. Asn and Gly) (Figure 3). In sum, for the amino acids for which we observed suboptimal charging in LB medium (e.g. Ser, Cys, Thr and His), the degree of growth inhibition mirrored the charging level of different tRNA isoacceptors, with tRNASer and tRNACys isoacceptor families exhibiting the lowest charging level (Figure 1A) and Ser and Cys being the most toxic to E. coli MC4100 (Figure 3).
Figure 3.
Some amino acids inhibit growth of E. coli. Growth curves at 37°C in MM containing different concentrations of one single amino acid. Rainbow colors of solid lines from purple to dark green denote amino acid concentration of 0, 0.5, 1, 5, 10, 15, 30 and 50 mM, respectively. For some amino acids, additional concentrations were measured (dashed lines) to cover their concentration in MM+AA : Cys, purple dashed line—0.05 mM and dark blue dashed line—0.1 mM; His, purple dashed line—0.1 mM and dark blue dashed line—0.2 mM; Thr, Asn and Phe, purple dashed line—0.2 mM and dark blue dashed line – 0.4 mM. The charging level of tRNAs charged with Ser, Cys, Thr, His, Asn and Phe was low. For comparison, the growth in the presence of Ala, Leu and Gly is included, for which cognate tRNAs were nearly completely charged.
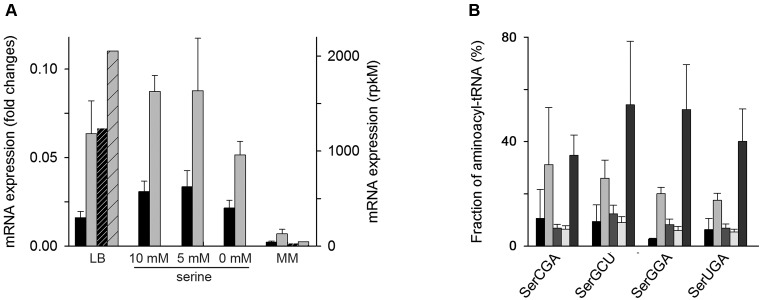
The inhibitory effect of free Ser in the growth medium has been noticed earlier (41). Ser is readily converted to pyruvate by L-Ser deaminases (42). Three genes in the E. coli genome encode L-Ser deaminases: sdaA and tdcG are expressed in anaerobic and sdaB in aerobic conditions (43,44). sdaB is within the same operon with sdaC encoding a Ser/H+ symporter. To explore the basis of the low seryl-–tRNAsSer despite the high intercellular Ser concentration in complex medium (Supplementary Table S2), we next examined the expression of sdaB and sdaC in different media using both quantitative RT-PCR and RNA-Seq data. The RNA-Seq data sets are available only for LB and MM and the sequencing reads were normalized to rpkM values which denote total reads detected per open-reading frame normalized to the depth of each library (26) (see also Materials and Methods section). Both sdaB and sdaC were highly expressed in LB medium but were barely expressed in MM (Figure 4A). Notably, in LB sdaB scored among the150 most highly expressed genes in E. coli in the RNA-Seq data set.
Figure 4.
Extracellular Ser activates the expression of sdaB and sdaC and decreases the charging of seryl-tRNAsSer. (A) mRNA expression level of sdaB (black) and sdaC (gray) as determined by qRT-PCR (right y-axis). Data are means ± SEM of three biological replicates. For LB and MM, the mRNA expression of sdaB (black striped) and sdaC (gray striped) was additionally determined from RNA-Seq (right axis). (B) Microarray of seryl-tRNAsSer from left to right: LB, MM+AA containing 10 mM, 5 mM or lacking Ser (0 mM) and MM at OD600 = 0.5. Data are means ± S.D.
Next, we analyzed whether sdaB and sdaC expression exhibits a concentration-dependent response to the amount of Ser added in the medium. Thus, to MM+AA depleted of Ser we added different concentrations of Ser (0, 5 and 10 mM; the last is the concentration in MM+AA). Both sdaB and sdaC were expressed in all conditions, even in MM+AA without Ser (Figure 4A). The expression of the Ser/H+ symporter (sdaC) in MM+AA lacking Ser is not surprising as the expression of Ser transporters is coupled to the expression of transporters for Gly and Ala (45), both amino acids were present in this growth condition. Since sdaC and sdaB are encoded within one operon and their expression is coupled, the induction of sdaC inevitably triggers the expression of sdaB encoding the L-Ser deaminase. In all of the conditions with high expression of sdaB, the charging of tRNAsSer family remained lower than in MM where the sdaB expression is nearly absent (Figure 4B), suggesting a link between low charging level of tRNAsSer and the expression of L-Ser deaminase which most likely degrades some portion of the free intracellular Ser.
Metabolic flux analysis suggests that at high intracellular concentrations Ser is redirected through L-Ser-deaminase toward pyruvate in the glycolysis pathway. However, the majority is not efficiently metabolized for energy but is instead wastefully metabolized and expelled as acetate (37). We reasoned that if excess Ser in complex media would be detoxified through L-Ser-deaminase, it would result in higher acetate production than in MM since a minimal acetate production is a common by-product of aerobic growth on glucose (46). Thus, any amount higher than MM would correlate with the metabolism of other constituents of the medium. The acetate production was higher in both complex media (LB and MM+AA) than in MM (Table 1). Our result in complex media agrees with the acetate concentration measured previously for another E. coli strain (37). Furthermore, we observed a clear increase in acetate production when adding Ser to the MM+AA (Table 1). The effect was not concentration-dependent and resembled the expression pattern of sdaB and sdaC (Figure 4A). Together, our data provide evidence that in media with low charging levels of tRNAsSer (LB and MM+AA), the expression of L-Ser-deaminase is high and acetate is excreted indicating that serine is, at least in part, degraded wastefully. In contrast, L-Ser deaminase is nearly absent in MM so that biosynthetically produced Ser is only used for tRNAsSercharging.
Table 1. Acetate concentration in the growth medium at OD600 = 0.5.
| Condition | Acetate concentration, ng/μl |
|---|---|
| LB | 403.9 ± 5.1 |
| MM+AA, 10 mM Ser | 420.1 ± 8.1 |
| MM+AA, 5 mM Ser | 439.7 ± 1.2 |
| MM+AA without Ser | 342.9 ± 11.0 |
| MM | 298.8 ± 6.0 |
DISCUSSION
In this study, we determined the aminoacylation level of tRNAs by leveraging the near single-isoacceptor resolution of tRNA-based microarray technology. Strikingly, we found that in LB and MM+AA tRNA isoacceptors are not uniformly charged with their cognate amino acids, a phenomenon which is anticodon-unspecific and relates to all tRNAs within one isoacceptor family. In MM, the charging of tRNA isoacceptors is more uniform, yet it also deviates from the assumed optimal charging level and oscillates around 50–60%. Thus, our data revisit a common assumption that E. coli tRNA isoacceptors are uniformly charged to 80–90% (15), a result that has been extrapolated from few single tRNAs (12–14).
tRNAsSer exhibit the lowest charging level in complex MM+AA and LB media followed by tRNAsCys, tRNAThr and tRNAHis. Thereby the aminoacylation level mirrors the toxicity of these amino acids when individually added to MM: the more toxic one amino acid is, the lower its aminoacylation level. Serine and cysteine are the most toxic amino acids for E. coli, as even the smallest supplementation to MM ceases cell growth completely. Cys is a potent inhibitor of growth, however, bacteria can overcome this effect (47), while the inhibition by Ser is long-lasting (41). The precise mechanism of inhibition by Ser is unclear and could branch over many different pathways as usually observed for perturbation of energy-coupled factors (42,44). Despite the high toxicity of Ser, Ser transporters are very efficient (Km = 1–10 μM (45)) and maintain high Ser uptake when this amino acid is present in the nutrient medium. The steady-state intracellular concentration of Ser is the highest among all amino acids in complex medium and is two orders of magnitude higher than that in MM (Supplementary Table S2). Thus, the free intracellular Ser should not be a limiting factor in complex (MM+AA und LB) media. However, as suggested by recent metabolic flux analysis in E. coli only 6.5% (∼0.2 mM) of the intracellular concentration of Ser is used directly for tRNASer aminoacylation, while 51% of the Ser flux is directed toward pyruvate and acetate production and 36% for glycine synthesis (37). Our measurements of the high expression of sdaB encoding L-Ser-deaminase under aerobic growth in LB and MM+AA, but not in MM and increased acetate production in complex media support the notion that intracellular Ser, at least in part, is degraded by L-Ser-deaminase, which competes with the seryl-tRNA-synthetase (Ser-RS) that charges the tRNAsSer.
Interestingly, kcat/Km values of L-Ser-deaminase of 1.6 × 105 (48) and Ser-RS of 1.5 × 104 (49) differ by an order of magnitude for their common substrate Ser. Ser-RS has much higher affinity for serine (e.g. Km of 53 μM (49) versus 2.67 mM (48) of the L-Ser-deaminase), whereas L-Ser-deaminase exhibits much higher turnover (e.g. kcat of 436 s−1 (48) versus 0.8 s−1 of Ser-RS (49)). In LB, the concentration of intracellular Ser (4.00 ± 0.30 mM, Supplementary Table S2) is higher than the Km values of both L-Ser-deaminase and Ser-RS, hence serine should undergo much higher utilization by the L-Ser-deaminase due to its high turnover number (kcat) than that of the Ser-RS. In complex medium, seryl–tRNA is also depleted in translation with turnover of 8.1 s−1 (50). Thus, two parallel pathways contribute to the low charging level of tRNAsSer: its utilization in translation and also depletion of Ser by L-Ser-deaminase, while the latter factor plays a dominant role at high Ser concentration (e.g. LB or MM+AA). Our acetate measurements and the metabolic flux analysis (15) corroborate this large role of L-Ser-deaminase in depleting free Ser in complex media.
Notwithstanding the measured low charging level of tRNAsSer, they are not correlated with fitness; the growth rate in LB and MM+AA is higher than MM (Supplementary Figure S3A). The six Ser codons are not rare on a genome scale, i.e. 5.85% of all 61 sense codons. However, Ser codons are rarer in the transcriptome, i.e. 3.9% (Figures 1B and 2B), suggesting that fast growth might be sustained even with a low-charged level of tRNAsSer. High intracellular Ser concentration at high growth rate may trigger mischarging of tRNAsThr and tRNAsAla (51,52) and consequently misincorporating Ser into proteins in lieu of Thr and Ala codons. Although it may take place, the lack of effect on bacterial fitness argues that amino acid mischarging and misincorporation is not a prevalent process.
The specificity of recognition between some tRNAs and their cognate AARS is facilitated by specific posttranscriptional modification in tRNA (53). While for some modifications (e.g. mnm5s2U34) the aminoacylation efficiency is invariant, for others (e.g. s4U34) the yield strongly depends on the growth rate (54). For example, only partial s4U modification of tRNALeu, tRNAGly, tRNAPro and tRNAGlu –is found at high growth (54). In turn, partial s4U modifications reduce tRNA charging (55). We observed reduced charging levels for tRNAGly and tRNAPro, whereas the charging of tRNALeu and tRNAGlu was nearly complete at high growth rates in MM+AA and LB. Variations in the modification pattern, particularly at high growth rate (56), may contribute to the suboptimal charging of some tRNAs, yet alone they cannot explain the very low charging of tRNAsSer, tRNAsThr and tRNAsCys.
Three of the amino acids (Ser, Thr and Cys) with suboptimal charging of the cognate tRNAs are metabolically related. For example, Ser is used to produce Cys and the byproduct of this conversion, homoserine, is the precursor for Thr biosynthesis. Thus, it cannot be ruled out that Ser indirectly influences the tRNA charging and detoxification of Thr and Cys as the metabolism of these amino acids is subordinated to Ser.
In sum, our results establish a new paradigm for biological regulatory mechanisms. To fine-tune the internal concentration of some growth-inhibiting amino acids, bacteria might trade off a high level of tRNA charging for efficient degradation and detoxification.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union [NICHE ITN] and DFG [FOR 1805 to Z.I. and N.B.]. Funding for open access charge: Deutsche Forschungsgemeinschaft [FOR 1805 to Z.I. and N.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Kirchner S., Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015;16:98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 2.Dong H., Nilsson L., Kurland C.G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 3.Dittmar K.A., Mobley E.M., Radek A.J., Pan T. Exploring the regulation of tRNA distribution on the genomic scale. J. Mol. Biol. 2004;337:31–47. doi: 10.1016/j.jmb.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 4.Bulmer M. Coevolution of codon usage and transfer RNA abundance. Nature. 1987;325:728–730. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- 5.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- 6.Novoa E.M., Ribas de Pouplana L. Speeding with control: codon usage, tRNAs and ribosomes. Trends Genet. 2012;28:574–581. doi: 10.1016/j.tig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Zhang G., Fedyunin I., Miekley O., Valleriani A., Moura A., Ignatova Z. Global and local depletion of ternary complex limits translational elongation. Nucleic Acids Res. 2010;38:4778–4787. doi: 10.1093/nar/gkq196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang G., Ignatova Z. Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr. Opin. Struct. Biol. 2011;21:25–31. doi: 10.1016/j.sbi.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Fedyunin I., Lehnhardt L., Bohmer N., Kaufmann P., Zhang G., Ignatova Z. tRNA concentration fine tunes protein solubility. FEBS Lett. 2012;586:3336–3340. doi: 10.1016/j.febslet.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Navon S., Pilpel Y. The role of codon selection in regulation of translation efficiency deduced from synthetic libraries. Genome Biol. 2011;12:R12. doi: 10.1186/gb-2011-12-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibba M., Soll D. Aminoacyl-tRNA synthesis. Ann. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 12.Sorensen M.A. Charging levels of four tRNA species in Escherichia coli Rel(+) and Rel(-) strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. J. Mol. Biol. 2001;307:785–798. doi: 10.1006/jmbi.2001.4525. [DOI] [PubMed] [Google Scholar]
- 13.Varshney U., Lee C.P., RajBhandary U.L. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 14.Yegian C.D., Stent G.S., Martin E.M. Intracellular condition of Escherichia coli transfer RNA. Proc. Natl. Acad. Sci. U.S.A. 1966;55:839–846. doi: 10.1073/pnas.55.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramaniam A.R., Zid B.M., O'Shea E.K. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell. 2014;159:1200–1211. doi: 10.1016/j.cell.2014.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elf J., Nilsson D., Tenson T., Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- 17.Dittmar K.A., Sorensen M.A., Elf J., Ehrenberg M., Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurvich O.L., Baranov P.V., Gesteland R.F., Atkins J.F. Expression levels influence ribosomal frameshifting at the tandem rare arginine codons AGG_AGG and AGA_AGA in Escherichia coli. J. Bacteriol. 2005;187:4023–4032. doi: 10.1128/JB.187.12.4023-4032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girstmair H., Saffert P., Rode S., Czech A., Holland G., Bannert N., Ignatova Z. Depletion of cognate charged transfer RNA causes translational frameshifting within the expanded CAG stretch in huntingtin. Cell Rep. 2013;3:148–159. doi: 10.1016/j.celrep.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Neidhardt F.C., Bloch P.L., Pedersen S., Reeh S. Chemical measurement of steady-state levels of ten aminoacyl-transfer ribonucleic acid synthetases in Escherichia coli. J. Bacteriol. 1977;129:378–387. doi: 10.1128/jb.129.1.378-387.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemoine A., Maya Martiotanez-Iturralde N., Spann R., Neubauer P., Junne S. Response of Corynebacterium glutamicum exposed to oscillating cultivation conditions in a two- and a novel three-compartment scale-down bioreactor. Biotechnol. Bioeng. 2015;112:1220–1231. doi: 10.1002/bit.25543. [DOI] [PubMed] [Google Scholar]
- 22.Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J., Rabinowitz J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Bartholomaus A., Fedyunin I., Feist P., Sin C., Zhang G., Valleriani A., Ignatova Z. Bacteria differently regulate mRNA abundance to specifically respond to various stresses. Philos. Trans. Maths Phys. Eng. Sci. 2016:374–380. doi: 10.1098/rsta.2015.0069. [DOI] [PubMed] [Google Scholar]
- 25.Woolstenhulme C.J., Guydosh N.R., Green R., Buskirk A.R. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 2015;11:13–21. doi: 10.1016/j.celrep.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 27.Del Campo C., Bartholomaus A., Fedyunin I., Ignatova Z. Secondary structure across the bacterial transcriptome reveals versatile roles in mRNA regulation and function. PLoS Genet. 2015;11:e1005613. doi: 10.1371/journal.pgen.1005613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dittmar K.A., Goodenbour J.M., Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang G., Hubalewska M., Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 30.Ingolia N.T. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 2010;470:119–142. doi: 10.1016/S0076-6879(10)70006-9. [DOI] [PubMed] [Google Scholar]
- 31.Ingolia N.T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 32.Balakrishnan R., Oman K., Shoji S., Bundschuh R., Fredrick K. The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res. 2014;42:13370–13383. doi: 10.1093/nar/gku1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li G.W., Oh E., Weissman J.S. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens A.T., Taylor J., Hilser V.J. Ribosome A and P sites revealed by length analysis of ribosome profiling data. Nucleic Acids Res. 2015;43:3680–3687. doi: 10.1093/nar/gkv200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruss B.M., Nelms J.M., Park C., Wolfe A.J. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 1994;176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navarro Llorens J.M., Tormo A., Martinez-Garcia E. Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 2010;34:476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 37.Selvarasu S., Ow D.S., Lee S.Y., Lee M.M., Oh S.K., Karimi I.A., Lee D.Y. Characterizing Escherichia coli DH5alpha growth and metabolism in a complex medium using genome-scale flux analysis. Biotechnol. Bioeng. 2009;102:923–934. doi: 10.1002/bit.22119. [DOI] [PubMed] [Google Scholar]
- 38.Tao H., Bausch C., Richmond C., Blattner F.R., Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saier M.H., Ramseier T.M., Reizer J. In: Escherichia coli and Salmonella: cellualr and molecular biology. 2nd edn. Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoiff WS, Riley M, Schaechter M, Umbarger HE, editors. Washington D.C.: ASM Press; 1996. pp. 1325–1343. [Google Scholar]
- 40.Gaal T., Bartlett M.S., Ross W., Turnbough C.L., Jr, Gourse R.L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 41.Hama H., Sumita Y., Kakutani Y., Tsuda M., Tsuchiya T. Target of serine inhibition in Escherichia coli. Biochem. Biophys. Res. Commun. 1990;168:1211–1216. doi: 10.1016/0006-291x(90)91157-n. [DOI] [PubMed] [Google Scholar]
- 42.Newman E.B., Malik N., Walker C. L-serine degradation in Escherichia coli K-12: directly isolated ssd mutants and their intragenic revertants. J. Bacteriol. 1982;150:710–715. doi: 10.1128/jb.150.2.710-715.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao Z., Lin R.T., Newman E.B. Sequencing and characterization of the sdaC gene and identification of the sdaCB operon in Escherichia coli K12. Eur. J. Biochem./FEBS J. 1994;222:901–907. doi: 10.1111/j.1432-1033.1994.tb18938.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X., El-Hajj Z.W., Newman E. Deficiency in L-serine deaminase interferes with one-carbon metabolism and cell wall synthesis in Escherichia coli K-12. J. Bacteriol. 2010;192:5515–5525. doi: 10.1128/JB.00748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piperno J.R., Oxender D.L. Amino acid transport systems in Escherichia coli K-12. J. Biol. Chem. 1968;243:5914–5920. [PubMed] [Google Scholar]
- 46.Leone S., Sannino F., Tutino M.L., Parrilli E., Picone D. Acetate: Friend or foe? Efficient production of a sweet protein in Escherichia coli BL21 using acetate as a carbon source. Microb. Cell Fact. 2015;14:106–116. doi: 10.1186/s12934-015-0299-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris C.L. Cysteine and growth inhibition of Escherichia coli: threonine deaminase as the target enzyme. J. Bacteriol. 1981;145:1031–1035. doi: 10.1128/jb.145.2.1031-1035.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cicchillo R.M., Baker M.A., Schnitzer E.J., Newman E.B., Krebs C., Booker S.J. Escherichia coli L-serine deaminase requires a [4Fe-4S] cluster in catalysis. J. Biol. Chem. 2004;279:32418–32425. doi: 10.1074/jbc.M404381200. [DOI] [PubMed] [Google Scholar]
- 49.Vincent C., Borel F., Willison J.C., Leberman R., Hartlein M. Seryl-tRNA synthetase from Escherichia coli: functional evidence for cross-dimer tRNA binding during aminoacylation. Nucleic Acids Res. 1995;23:1113–1118. doi: 10.1093/nar/23.7.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jakubowski H., Goldman E. Quantities of individual aminoacyl-tRNA families and their turnover in Escherichia coli. J. Bacteriol. 1984;158:769–776. doi: 10.1128/jb.158.3.769-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beebe K., Ribas De Pouplana L., Schimmel P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 2003;22:668–675. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beebe K., Mock M., Merriman E., Schimmel P. Distinct domains of tRNA synthetase recognize the same base pair. Nature. 2008;451:90–93. doi: 10.1038/nature06454. [DOI] [PubMed] [Google Scholar]
- 53.Björk G.R. In: tRNA: Structure, Biosynthesis and Function. American Society for Microbiology. Söll D., Rajbhandary U., editors. Washington D.C.: 1995. pp. 165–205. [Google Scholar]
- 54.Emilsson V., Naslund A.K., Kurland C.G. Thiolation of transfer RNA in Escherichia coli varies with growth rate. Nucleic Acids Res. 1992;20:4499–4505. doi: 10.1093/nar/20.17.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kruger M.K., Sorensen M.A. Aminoacylation of hypomodified tRNAGlu in vivo. J. Mol. Biol. 1998;284:609–620. doi: 10.1006/jmbi.1998.2197. [DOI] [PubMed] [Google Scholar]
- 56.Sakai Y., Miyauchi K., Kimura S., Suzuki T. Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons. Nucleic Acids Res. 2016;44:509–523. doi: 10.1093/nar/gkv1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.