Abstract
Chromosome stability models are usually qualitative models derived from molecular-genetic mechanisms for DNA repair, DNA synthesis, and cell division. While qualitative models are informative, they are also challenging to reformulate as precise quantitative models. In this report we explore how (A) laboratory experiments, (B) quantitative simulation, and (C) seriation algorithms can inform models of chromosome stability. Laboratory experiments were used to identify 19 genes that when over-expressed cause chromosome instability in the yeast Saccharomyces cerevisiae. To better understand the molecular mechanisms by which these genes act, we explored their genetic interactions with 18 deletion mutations known to cause chromosome instability. Quantitative simulations based on a mathematical model of the cell cycle were used to predict the consequences of several genetic interactions. These simulations lead us to suspect that the chromosome instability genes cause cell-cycle perturbations. Cell-cycle involvement was confirmed using a seriation algorithm, which was used to analyze the genetic interaction matrix to reveal an underlying cyclical pattern. The seriation algorithm searched over 1014 possible arrangements of rows and columns to find one optimal arrangement, which correctly reflects events during cell cycle phases. To conclude, we illustrate how the molecular mechanisms behind these cell cycle events are consistent with established molecular interaction maps.
INTRODUCTION
Chromosome stability mechanisms maintain the proper number of chromosomes during the cell cycle, while preventing chromosomal aberrations, such as deletion, duplication or translocation of chromosomal segments. To better understand disorders that lead to numerous inherited genetic diseases (1) and cancers (2), it is important to understand the fundamental molecular mechanisms of chromosome stability because such disorders are often associated with abnormal chromosomes.
Our understanding of chromosome stability in humans has been shaped by studies in yeast, Saccharomyces cerevisiae. Yeast has many advantages to human studies; it is genetically well characterized, easy to experiment with, and many of its genes have homologues in humans. Maintenance of chromosome stability involves the function of at least 723 genes in yeast (3). Each specific gene is implicated in chromosome stability because loss-of-function (4–8) or over-expression (9) of the specific gene causes chromosome instability. Generally, the yeast genes that affect chromosome stability function in DNA repair, replication, recombination, chromosome segregation, cell cycle control (3). Importantly, many of these genes function in processes that are similar to the processes that affect human chromosome stability in genetic disorders and/or cancers (10).
While the biology of individual genes offers insight into molecular mechanisms, the systems biology of combinations of genes is equally insightful. Consider, for example, mutations in pairs of chromosome stability genes that have been combined into a common strain to identify positive or negative genetic interactions between those genes (11–14), which often forms the basis of mechanistic models of biological processes, such as cell cycle (15). However, usually these models are either manually formulated using a limited set of pre-selected genes or automatically formulated using clustering algorithms. The phenotypes of these combinations can often reveal synergistic or suppressive effects, allowing the genes to be placed in relation to one another in the context of a molecular model, when the molecular function of at least one of the interaction partners is known (16). Formally, these genetic interactions can be represented as n x m matrices of vectors, which represent the positive or negative genetic interactions between the respective genes (17).
Automated algorithms for formulating functional mechanisms using novel sets of gene interaction data have not yet been fully developed beyond clustering functionally related interacting gene groups. Such clustering methods are mathematically arbitrary, thus have little a priori biological basis. This study uses matrix seriation (18), a method that reorders a matrix of gene–gene interaction into a cyclical arrangement, if one exists, instead of a clustered arrangement made by the currently popular clustering algorithms. The seriation method is inspired by the famous ‘Traveling Salesman Problem’ (19) to calculate an optimal cycle among the genetic interaction data, with the assumption that the optimal arrangement likely coincides with an optimum temporal biological process cycle. Therefore, seriation is particularly suited to modeling the cell cycle or chromosome maintenance during the cell cycle. In this work, we investigate the method of seriation to provide insights about the biological mechanisms of genes that affect chromosome stability.
MATERIALS AND METHODS
Strains
Chromosome stability assays and Fluorescence Activated Cell Sorting (FACS) assays were performed on cultures of the diploid strain YPH275 of S. cerevisiae (MATa/MATα ura3-52 lys2-80 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 CF [TRP1 SUP11 CEN4]), provided by Prof. Phil Hieter (20). Synthetic dosage lethality and suppression were assayed by transforming individual haploid deletion mutant strains from the gene knockout strain library (21–23) and the parental strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) as control with selected plasmids.
Plasmids
The MORF (Movable Open Reading Frame) library consists of two-micron plasmids each having one tagged gene of interest under the control of a galactose-inducible (GAL) promoter (24). Strains of E. coli containing the MORF plasmid library were obtained from Dr Elizabeth Grayhack and Dr Eric Phizicky (University of Rochester Medical School, Rochester, NY). Individual MORF clones were verified by sequencing, and were introduced into the appropriate yeast strains by transformation, and confirmed upon recovery by resequencing.
Chemicals and media
Chromosome instability reported by discrete red sectors on pink colonies was assayed on synthetic yeast growth media lacking uracil, which contained 5–6 mg/l of adenine to optimize color formation (25). Plates containing 2% galactose + 1% raffinose were used to induce gene expression regulated by the GAL promoter on MORF plasmids while plates containing 2% glucose were used to repress gene expression regulated by the GAL promoter. Enriched yeast growth medium was YP with added glucose (YPD) (25).
Transformation of MORF plasmids into yeast
To induce overexpression of genes, we transformed liquid cultures of cells obtained from uniformly pink colonies of YPH275 with plasmids selected from the MORF library. YPH275 cultures were incubated with aeration at 30°C for 2 days in 100 ml of YP media including either 2% glucose (YPD) or 1% raffinose (YPRaff) (25). Approximately 2.5 × 1011 cells were centrifuged into a pellet that was resuspended in 10 ml of transformation buffer (0.20 M LiAc, 0.10 M dithiothreitol, 40% mass per volume of polyethylene glycol, 2.6% mass per volume of dimethyl sulfoxide, and 500 μl of single-stranded DNA). Aliquots of 300 μl were mixed with 5 μl of plasmid DNA that was extracted using the Miniprep Kit from Qiagen. The mixture was heated for 30 min at 45°C and then mixed with 540 μl of YPD media. 100 μl aliquots were plated onto glucose-containing media lacking uracil and grown for two or three days at 30°C; the transformants were frozen in 8% dimethyl sulfoxide.
Prescreening MORF library plasmids for candidate chromosome instability genes
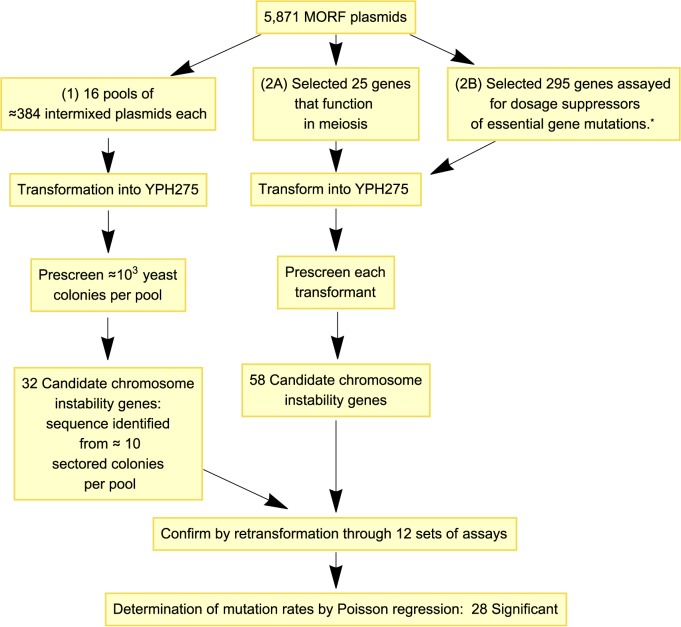
Prescreening was performed as illustrated in Figure 1. Plasmid DNA comprising the entire MORF library was partitioned into prescreening sets each including approximately 384 individual MORF plasmid DNA preparations. These DNA preparations were pooled into mixtures of 384 plasmids that were used to transform YPH275 (with the expectation that only one MORF plasmid would generally be introduced into any particular transformant cell).
Figure 1.
Flowchart illustrating the strategy for identifying chromosome instability genes. *See Patra et al. arXiv preprint arXiv:1311.2554, 2013.
Subsequently, 10 μl of transformants were spotted in a dilution series on solid synthetic media plates (containing galactose, raffinose, minimal adenine, and no uracil) using a Beckman Coulter Biomek FXP liquid handler robot; YPH275 transformed with the empty MORF vector plasmid BG1766 was the negative control and pGAL::CLB5 (26) or pGAL::YRB1 (9) MORF plasmid was the positive control.
Colonies were allowed to form, allowed to develop colony color, and dilution spots with separable colonies were examined for increased frequencies of red sectors relative to the negative control. Putative hit plasmids were extracted from the corresponding colonies, plasmid DNA confirmed by sequencing. Increased rates of plasmid instability were subsequently confirmed by retransformation into YPH275 followed by the Chromosome Stability Assay method described below.
To perform additional hypothesis-driven experiments using preselected plasmids, experiments were performed as above, but without the first step of pooling MORF plasmids and without the later step of confirming the DNA sequence of plasmids after observing frequent sectors.
Chromosome stability assay
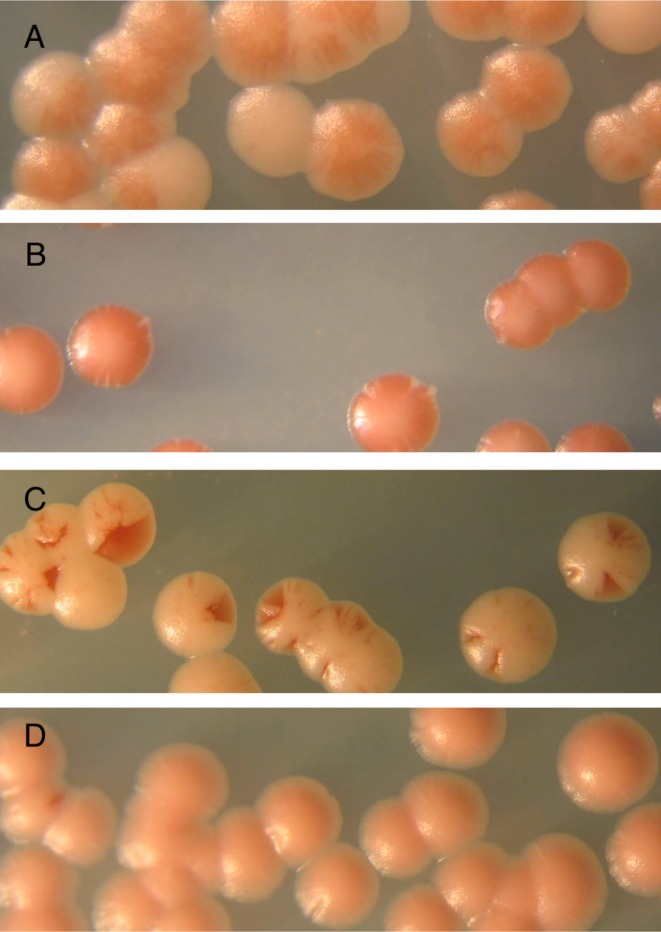
To assay for MORF plasmids that affect chromosome stability, we performed a color-based assay using colonies of the transformed strain YPH275 (Figure 2). Transformants were single colony purified on YPD plates, followed by plating on synthetic dextrose media lacking uracil to select for the plasmids. The transformants were resuspended in water, diluted, and plated for single colonies on media containing (A) glucose (synthetic dextrose containing minimal adenine and lacking uracil) and (B) galactose (galactose plus raffinose media containing minimal adenine and lacking uracil).
Figure 2.
Chromosome stability assay to confirm screened candidates. (A) Colony color of yeast strain YPH275 containing the empty vector BG1766 on media containing galactose and raffinose, or (B) glucose. (C) Example of red sectored colonies. Specifically, a MORF plasmid containing the gene NDT80 was introduced into yeast strain YPH275 and plated on media containing galactose and raffinose, or (D) glucose.
After one week of growth at 30°C, plates were shifted (in multiple replicates for each strain) to 4°C and incubated for two weeks to allow red sectors to darken. In some sets of experiments, plates remained at that temperature for an additional 2 weeks to allow any non-specific pink color to fade without compromising the darkening of the red sectors.
The number of sectors per colony was assumed to follow a Poisson (μ) distribution where 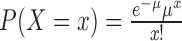 , x ≥ 0. Because the sum of n Poisson (μ) distributions is a Poisson (nμ) distribution, the total number of sectors on a plate having n colonies is Poisson (nμ), or
, x ≥ 0. Because the sum of n Poisson (μ) distributions is a Poisson (nμ) distribution, the total number of sectors on a plate having n colonies is Poisson (nμ), or 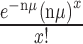 . A Poisson regression to model the mutation rate per colony μ was performed using an offset to adjust for the number of colonies on each plate; specifically, we optimized the β parameter vector in log si = log ni + β1x1 + β2x2 + ... + βnxn, where si is the number of sectors on plate i, ni is the total number of colonies on plate i, and xj equals one or zero to represent the jth gene*sugar interaction (27). (Note that the constant term β0 was omitted from the model so that each
. A Poisson regression to model the mutation rate per colony μ was performed using an offset to adjust for the number of colonies on each plate; specifically, we optimized the β parameter vector in log si = log ni + β1x1 + β2x2 + ... + βnxn, where si is the number of sectors on plate i, ni is the total number of colonies on plate i, and xj equals one or zero to represent the jth gene*sugar interaction (27). (Note that the constant term β0 was omitted from the model so that each 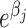 could be interpreted as the mutation rate for the jth gene*sugar.) The significance of the differences between the estimated parameters βj were computed using a Z-test using the Z values for each parameter estimate βj. Significant differences between βj for a gene on galactose versus the empty vector on galactose were determined by using a Bonferonni adjusted P value threshold of 0.01. The Poisson Regression was generated using SAS Software (University Edition). This technique builds on that of a dissertation (28), which instead reported a test of dependence of sector frequency on sugar type without using the Poisson distribution to quantify the rates of mutation per colony.
could be interpreted as the mutation rate for the jth gene*sugar.) The significance of the differences between the estimated parameters βj were computed using a Z-test using the Z values for each parameter estimate βj. Significant differences between βj for a gene on galactose versus the empty vector on galactose were determined by using a Bonferonni adjusted P value threshold of 0.01. The Poisson Regression was generated using SAS Software (University Edition). This technique builds on that of a dissertation (28), which instead reported a test of dependence of sector frequency on sugar type without using the Poisson distribution to quantify the rates of mutation per colony.
Synthetic dosage interaction
Individual MORF plasmids were introduced by transformation into specific haploid deletion mutant strains selected from the yeast knockout library. Transformants were selected on synthetic dextrose plates lacking uracil, and single colony purified. The BioRad SmartSpec 3000 optical density reader was used to establish equal titers among the transformants and to dilute each 10-, 100- and 1000-fold. Four dilutions (1×, 10×, 100× and 1000×) were spotted in duplicate onto galactose-containing and glucose-containing solid synthetic media lacking uracil (the glucose-containing media was a control to check whether all strains at all dilutions grew equally). Three replicates of each experiment were plated and incubated at 30°C.
Growth of a given transformant in a deletion mutant strain was compared with the growth of the transformant containing the same plasmid in the wild-type background strain BY4741 on the same plate by photographing and estimating the extent of growth at each dilution. The empty MORF vector plasmid was used as a control. Some genetic interactions were repeatedly validated to ensure consistency.
When the growth of the deletion mutant strain carrying the MORF plasmid was indistinguishable from that of the wild-type background strain carrying the same MORF plasmid in all dilutions, the interaction received a score of 4. Scores of 1, 2 or 3 represented poorer growth of the mutant compared to the wild-type discernible at the various dilutions; thus, score 1 represented the most affected strain, whereas score 3 represented the least affected. The interaction score of 7 represented higher levels of growth of the mutant strain carrying the MORF plasmid compared to the wild type strain carrying the same MORF plasmid (the quantity seven was chosen so that the lowest score and the highest score are numerically equidistant from the score for normal growth). See the supplementary file S1 for an example of this assay.
Seriation of matrices of genetic interaction
An unsorted matrix M was built using 19 row vectors of MORF plasmids, including the empty vector plasmid, and 18 column vectors of deletion mutants. As noted above, we used a score of 1 to represent synthetic dosage lethality, 2 or 3 to represent levels of synthetic dosage sickness, 4 to represent normal growth or that data were not available and 7 to represent suppression.
Given the matrix of genetic interactions M, it is computationally intensive (and NP hard) to find the one optimal arrangement of the 19 row vectors of M that minimizes the sum over all of the 18-dimensional Euclidean distances between the rows of data vectors (29). Nevertheless, the optimal rearrangement of the row vectors that minimizes this sum of 18-dimensional Euclidean distances can be solved using integer linear programming (30,31) because the total number of row vectors in our work is sufficiently small.
This optimization task is similar to the famous ‘Traveling Salesman Problem’, except that the two-dimensional Euclidean distance between two cities in the traditional Traveling Salesman Problem is replaced here by the 18-dimensional Euclidean distance between the data vectors of scores in rows i and j of our matrix M.
Specifically, if we denote our 18-dimensional Euclidean distances dij, our optimization task is to find a vector xij comprised of zeros and ones (where one represents the adjacency between rows i and j), while minimizing D(x) = ∑i > jdijxij subject to the constraints that 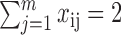 , xij ≥ 0, i = 1, 2, ..., n, i ≠ j, and xij = xji.
, xij ≥ 0, i = 1, 2, ..., n, i ≠ j, and xij = xji.
This optimization problem was solved using Mathematica Version 10.2 (a commercial software platform from Wolfram Research, Inc. 2015), by using the ‘FindShortestTour’ function, specifying the ‘Method’ ‘IntegerLinearProgramming’ and the ‘Distance Function’ of ‘EuclideanDistance’. To sort the columns in addition to the rows, the resulting matrix was transposed and ‘FindShortestTour’ was used to re-sort the transposed matrix using the same method and distance function.
Fluorescence activated cell sorting analysis
To measure the effects of MORF plasmids on the percentage of cells in G1, we prepared cultures of YPH275 for scanning by Imaging Flow Cytometry (32). Transformants were grown at 30°C in 96-well plates that contained 200 μl of synthetic dextrose media lacking uracil. After 2 days of growth, 10 μl of cells were inoculated into fresh 96-well plates and grown overnight in synthetic complete media with raffinose and lacking uracil. For 16 h at 30°C, the pGAL promoters were induced using synthetic complete media with 2% galactose and lacking uracil, and cell density was monitored to obtain cultures at the exponential growth phase.
Approximately 2 × 106 cells at the mid exponential phase were harvested by centrifugation and fixed using 70% ethanol by gently mixing at 4°C. The fixed cells were collected by centrifugation, which were then washed in water. The cells were treated with 1 mg/ml RNAse A (Sigma) for 4 h at 37°C. After centrifugation, the pellets were incubated with 1 mg/ml of Proteinase K (Invitrogen) for 1 h at 50°C. Cells were collected by centrifugation and re-suspended with 1 μM of SYTOX green (Invitrogen) in 50 mM Tris (pH 7.5).
Cells were scanned by an imaging flow cytometer, Amnis ImageStreamX (Amnis Inc., Seattle, WA, USA). 25 000 cell images were acquired and the intensity of the DNA image (green channel; 480–560 nm) was recorded along with the aspect ratio of the cell image (brightfield).
The Mathematica function ‘FindClusters’ (Wolfram Research Inc., Version 10.2 2015) was used to compute four clusters based on standardized Z-scores of the intensity and aspect ratio in each experiment (where each Z-score is 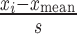 , where xi is a raw observation, xmean is the mean of those raw observations, and s is the standard deviations of those raw observations. Compared to the other clusters, the cluster for G1 was assumed to have the high aspect ratio and low DNA intensity. The percentage of cells in G1 was estimated by dividing the number of cells in the G1 cluster by the total number of cells. Note that this technique builds on that of a dissertation (28) which instead assumed predetermined thresholds of DNA intensity and aspect ratio to demarcate cells in G1.
, where xi is a raw observation, xmean is the mean of those raw observations, and s is the standard deviations of those raw observations. Compared to the other clusters, the cluster for G1 was assumed to have the high aspect ratio and low DNA intensity. The percentage of cells in G1 was estimated by dividing the number of cells in the G1 cluster by the total number of cells. Note that this technique builds on that of a dissertation (28) which instead assumed predetermined thresholds of DNA intensity and aspect ratio to demarcate cells in G1.
RESULTS
Identifying genes causing chromosome instability upon overexpression
YPH275 is a diploid strain homozygous for ade2-101 ochre mutation, and also contains the dominant ochre-suppressor gene SUP11 on a supernumerary chromosome fragment (20). The loss of the supernumerary chromosome or the SUP11 gene is signaled by the accumulation of a red pigment in the colony (or colony sector) due to defective adenine biosynthesis in the absence of SUP11. In principle, the loss of SUP11 can also result from ectopic gene conversion to sup11 present at the allelic locus, but this frequency is generally low compared to the frequency of loss of SUP11 by mitotic nondisjunction or recombination.
The increased frequency of red colonies/sectors on galactose relative to the background levels on glucose is a measure of chromosome instability induced by the MORF plasmids (26). With the expectation that many genes that affect the stability of chromosomes when over-expressed remain to be identified, we took two independent approaches for prescreening genes, which are described below. The results of confirmation assays applied to these prescreened genes are summarized in Table 1.
Table 1. Influence on chromosome loss rate of conditional over-expression of genes identified through the prescreen.
| Prescreened | μcolony | Prescreened | μcolony | Prescreened | μcolony | Prescreened | μcolony |
|---|---|---|---|---|---|---|---|
| Gene | (n colonies) | Gene | (n colonies) | Gene | (n colonies) | Gene | (n colonies) |
| Assay 1 | Assay 3 | Assay 6 | Assay 9 | ||||
| MOT2 | 0.28 * (769) | MCM3 | 0.80 * (290) | CDC4 | 0.25 * (318) | CLN1 | 0.73 * (41) |
| NRM1 | 0.26 * (828) | CDC5 | 0.47 * (104) | YDR387C | 0.07 * (2708) | GRX2 | 0.52 * (61) |
| CDC20 | 0.21 * (800) | CLB5 (+) | 0.34 * (230) | CLN1 | 0.07 * (1344) | NOP6 | 0.42 * (77) |
| CLB5 (+) | 0.09 * (1033) | IME2 | 0.19 * (148) | BMT5 | 0.06 * (455) | SPC19 | 0.34 * (96) |
| YLR053C | 0.08 * (1132) | SLD3 | 0.09 * (182) | HMLα2 | 0.05 * (3637) | YRB1 (+) | 0.32 * (275) |
| SPO7 | 0.07 * (1791) | CWC24 | 0.05 * (946) | SLM4 | 0.05 * (274) | GPG1 | 0.28 * (79) |
| CDC6 | 0.05 * (844) | YIL025C | 0.02 (284) | YGL182C | 0.05 * (1616) | YFL051C | 0.18 (60) |
| CDH1 | 0.03 * (1429) | SMC2 | 0.02 (711) | YOS1 | 0.02 (996) | GSP2 | 0.16 (95) |
| MIX23 | 0.03 (1184) | PAN6 | 0.01 (650) | YAL069W | 0.02 (1792) | DAS1 | 0.14 (132) |
| YHM2 | 0.02 (624) | BMH2 | 0.01 (564) | MOT3 | 0.02 (863) | GSH2 | 0.10 (111) |
| BFR1 | 0.02 (612) | CLB2 | 0.01 (107) | RPS24B | 0.02 (1020) | Vector (-) | 0.05 (477) |
| ARP9 | 0.02 (1108) | PRP16 | 0.01 (1242) | AIM26 | 0.02 (1640) | Assay 10 | |
| RRT6 | 0.02 (1508) | TTI2 | 0.01 (264) | PGD1 | 0.02 (299) | YRB1 (+) | 0.66 (6) |
| YDR476C | 0.02 (881) | SUA7 | 0.01 (937) | CLN2 | 0.01 (1563) | COQ10 | 0.00 (35) |
| PGA1 | 0.02 (367) | Vector (-) | 0.01 (934) | Vector (-) | 0.01 (2876) | Vector (-) | 0.00 (14) |
| TTI2 | 0.02 (1552) | YKL202W | 0.01 (394) | SYS1 | 0.01 (941) | Assay 11 | |
| Vector (-) | 0.01 (3760) | VPS9 | 0.00 (402) | CKA2 | 0.01 (288) | YRB1 (+) | 0.28 * (157) |
| YNL198C | 0.01 (1088) | WSC2 | 0.00 (813) | RPL16A | 0.01 (2898) | ERP4 | 0.05 (775) |
| CDC40 | 0.01 (1937) | PTR2 | 0.00 (567) | PCL9 | 0.00 (2100) | Q0130 | 0.04 (749) |
| MBB1 | 0.01 (2508) | MSB3 | 0.00 (609) | CLB5 (+) | 0.00 (3557) | YPL071C | 0.04 (678) |
| FAD1 | 0.01 (1286) | TRP1 | 0.00 (969) | Assay 7 | Vector (-) | 0.04 (139) | |
| SWT1 | 0.00 (2516) | Assay 4 | YRB1 (+) | 0.84 (6) | YNL226W | 0.03 (614) | |
| SRB2 | 0.00 (4916) | NDT80 | 0.79 * (498) | RPL20A | 0.08 (867) | ADY4 | 0.02 (457) |
| Assay 2 | CLB5 (+) | 0.36 * (157) | ORC1 | 0.05 (840) | Assay 12 | ||
| XRS2 | 0.54 * (66) | REC8 | 0.35 * (838) | SRP14 | 0.03 (670) | SPC19 | 0.41 * (173) |
| YRB1 (+) | 0.39 * (57) | Vector (-) | 0.07 (475) | RHO4 | 0.01 (619) | GRX2 | 0.39 * (230) |
| HFI1 | 0.06 * (842) | Assay 5 | Vector (-) | 0.00 (5) | CLN1 | 0.28 * (68) | |
| MSM1 | 0.04 (289) | YRB1 (+) | 0.60 * (58) | Assay 8 | Vector (-) | 0.00 (149) | |
| SYC1 | 0.03 (383) | ELG1 | 0.33 * (317) | YRB1 (+) | 0.78 * (82) | ||
| JID1 | 0.02 (321) | DCG1 | 0.01 (289) | POL32 | 0.08 (543) | ||
| YER152C | 0.02 (215) | Vector (-) | 0.00 (55) | Vector (-) | 0.02 (142) | ||
| Vector (-) | 0.02 (355) | YJL077W-B | 0.02 (596) | ||||
| YGL082W | 0.01 (539) | RET3 | 0.01 (450) | ||||
| TFB5 | 0.01 (375) |
*These chromosomal loss rates per colony (μcolony) are significantly different from that of the isogenic strain harboring the empty MORF vector, under identical conditions in the same experimental set (Z-test, Bonferroni-adjusted P < .01). The Z-test was used to compare the chromosomal loss rates based on the Poisson parameters estimated from a Poisson regression. See Materials and Methods.
1. In a model-independent approach, we prescreened the entire MORF library using a technique previously described (9); using this technique Ouspenski et al. discovered 30 genes that induce chromosome instability upon over-expression, but had estimated that 58 genes had remained to be discovered. That work had used a smaller cDNA library compared to the MORF library that we use in this work.
By adopting their prescreen technique followed by subsequent confirmation, we confirmed eight MORF plasmids as capable of inducing chromosome instability when over-expressed on galactose, including XRS2, HFI1, ELG1, CLN1, GRX2, NOP6, SPC19, and GPG1. These phenotypes had not previously been discovered. We also confirmed YRB1, which Ouspenski et al. had previously discovered, and we have used this plasmid as a positive control in our confirmation assays. Using the same assumptions as Ouspenski et al. (9) for sampling, we estimate to have missed at least 40 additional genes using this prescreening technique.
2A. Our second approach was a model-dependent approach. We had noted the possibility that abnormal expression of meiosis related genes in mitotic cells might cause mitotic chromosome instability. Ouspenski et al. had not explored this extensively because their cDNA library had originated from vegetatively growing cells that were presumably not sporulating. To explore this further, we chose 25 meiosis-related genes (see supplementary file S2) and prescreened these for galactose induced chromosome instability. Only two of these genes, NDT80 and REC8, induced chromosome instability in our subsequent confirmation assays (i.e. a hit-rate of 8% of the prescreened meiosis genes). Ouspenski et al. had not previously reported NDT80 and REC8.
Predominantly red sectors, characteristic of the clonal loss of the supernumerary chromosome, against a light pink colony background (characteristic of the ochre-suppressed ade2-101 phenotype) that are unaccompanied by white sectors (characteristic of cells with additional copies of the supernumerary chromosome) occurred in strains containing pGAL::NDT80 and pGAL::REC8. This observation indicated that these genes mostly induced chromosome loss without non-disjunction, or that hyperploidy and/or SUP11 dosage might be toxic to those strains, causing selective loss of aneuploid cells.
2B. In addition to prescreening meiosis-specific genes, we also prescreened various genes that function in mitotic cell cycle control or that had been previously assayed for dosage suppression based on Patra et al., arXiv preprint arXiv:1311.2554, 2013. This set also included genes that complement or suppress temperature sensitive mutations. We prescreened 295 of these plasmids (see supplementary file S2) and subsequently assayed 58 of these to confirm 19 additional genes that induce chromosome instability (about 6% of the 295 prescreened). The confirmed genes include BMT5, CDC4, CDC5, CDC6, CDC20, CDH1, CWC24, HMLα2, IME2, MCM3, MOT2, NRM1, SLD3, SLM4, SPO7, YDR387C, YGL182C and YLR053C. Note that HMLα2 is ordinarily silenced, but is regulated by an inducible pGAL promoter in the MORF library. In addition, we reconfirmed the positive control CLB5 (26).
We quantified the chromosome loss rate per colony (μcolony) induced by the prescreened MORF plasmids relative to that by the empty vector BG1766 by Poisson regression as described in Methods. Table 1 shows the results of these computations. The confirmation Assays 2, 5, 7, 8, 9, 10, 11 and 12 include genes pre-identified by the chromosome instability prescreen described above that was similar to that of Ouspenski et al. Confirmation Assay 4 includes genes pre-identified by prescreening genes that normally function during meiosis. The confirmation Assays 1, 3 and 6 include genes that were chosen because of their role in cell cycle control or because they were previously assayed for dosage suppression. In view of the results of confirmation Assay 6, we propose naming the uncharacterized ORF YDR387C to CIN10.
Note that some sector-inducing genes are not reported because red sectors were difficult to distinguish visually from pink sectors. The excluded results are those of pGAL::YML007-C, pGAL::YAR023C, pGAL::MES1, and pGAL::MHT1. In addition, one batch, confirmation Assay 7, the positive control had a small number of colonies plated and hence an insignificant mutation rate per colony.
Modeling chromosome instability, genetic interactions, and mitotic slippage
We investigated whether a formal method for discovering chromosomal instability genes could be devised by first using a mathematical model of cell cycle that is consistent with known evidence and then simulating the model to incorporate perturbations in cell mechanisms. Our expectation was that such an approach might predict chromosome instability genes and dosage suppression of cell cycle gene mutations, which indirectly should predict chromosome instability.
Specifically, we sought to model the downstream biochemical effects of the mitotic spindle checkpoint, which ensures that chromosomes are properly aligned and oriented in preparation for the metaphase-to-anaphase transition. Loss-of-function mutations of the spindle checkpoint genes mps1, mad1, mad2, mad3, bub1 or bub3, all of which affect this process, cause chromosome instability (6,33–36).
In the presence of a functional M-phase spindle checkpoint, cells will usually undergo proper cell division but do occasionally bypass checkpoint arrest and undergo premature chromosome segregation. This latter effect has been called ‘mitotic slippage’ or ‘adaptation’, and can occur after periods of cell cycle checkpoint arrest (37). The consequences of one generation of mitotic slippage may often include aneuploidy and inviability (38,39).
The spindle checkpoint genes noted above encode proteins that regulate the sequestration and subsequent release of Cdc20p to trigger the metaphase-to-anaphase transition at the appropriate time (40–42). Such mechanisms are generally thought to be important for chromosome stability and genetic interactions. We use a system of coupled linear differential equations to model cell cycle events (15), and systematically varied certain parameters for behavior of the numerical solutions that could simulate irregularities in cell cycle oscillations and thus might predict chromosome instability and dosage suppression.
In the model, protein concentration, phosphorylation, protein complex formation, etc., of the minimal components of cell cycle regulation are modeled using kinetic rate laws combined with algebraic constraints and conditional logic as described previously by Chen et al. (15). Alteration of parameters in this mathematical model can simulate gene deletion, gene dosage effects, binding affinity changes and several other perturbations (15).
For example, setting the rate constant for Cdc20p basal synthesis 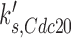 to a higher value simulates a higher level of CDC20 gene expression driven by the GAL promoter:
to a higher value simulates a higher level of CDC20 gene expression driven by the GAL promoter: 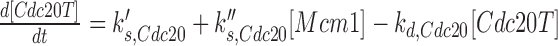 ,
,
where [Cdc20T] is the total concentration of active and inactive Cdc20p, 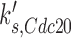 is the rate constant for basal synthesis,
is the rate constant for basal synthesis, 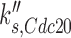 is the rate constant for Mcm1p-dependent synthesis of Cdc20p, [Mcm1] is the concentration of Mcm1p protein, and kd, Cdc20 is the rate constant for Cdc20p degradation. The reasons for using these exact variables were defined previously (15).
is the rate constant for Mcm1p-dependent synthesis of Cdc20p, [Mcm1] is the concentration of Mcm1p protein, and kd, Cdc20 is the rate constant for Cdc20p degradation. The reasons for using these exact variables were defined previously (15).
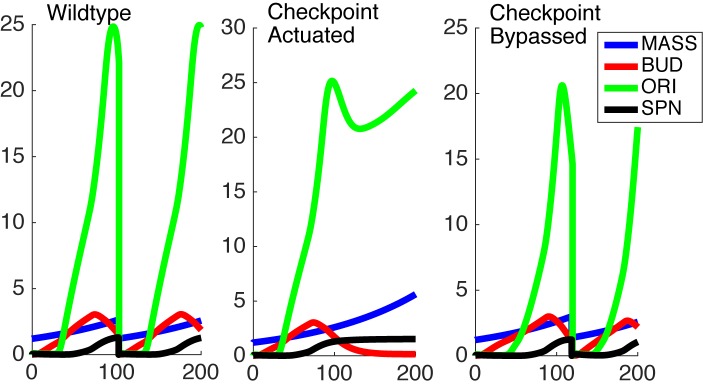
Cell cycle arrest was numerically simulated in the model in a systematic manner, resulting in loss of periodicity of protein concentrations and loss of execution of a proper sequence of cell cycle milestone events (see the ‘robustness criteria’]. Upon mathematically generating cell cycle blocks through a primary perturbation we systematically searched the parameter space in the system of equations to identify secondary perturbations that would allow resumption of the cell cycle (Figure 3). Changes of values in parameters that would cause the simulated cell cycles to resume their periodic behavior were noted.
Figure 3.
Simulations of the timecourse of in silico cellular mass (MASS), budding (BUD), development of origins of replication (ORI) and spindle formation (SPN) in a wildtype model (left), checkpoint-activated model (center) and a model with CDC20 overexpression in a checkpoint-activated background. Actuating the checkpoint signaled by Bub2p was simulated by increasing the bub2 variable to a fixed value of 1 and lowering the lte1 variable to a fixed value of 0.1, while CDC20 overexpression was simulated by increasing the basal synthesis rate of CDC20 from 0.006 to 1 dimensionless unit, with the normal period being 90 min.
For the purpose of this investigation, we focused on the simulated gain-of-function perturbations. In this manner, we identified genes that bypassed the arrest induced by loss (or ‘in silico mutation’) of genes that actuate the checkpoint signaling genes. In our simulations CDC20, CDH1, CDC15, CDC14, TEM1, CDC6, SIC1 and SWI5 bypassed the checkpoint that is actuated by the Bub2 protein (Table 2).
Table 2. Genes predicted to induce mitotic slippage, bypassing the checkpoint actuated by Bub2 protein in silico.
| Slippage predictors | Slippage parameter | |||
|---|---|---|---|---|
| Gene | Parameter | Wild-type parameter | Low | High |
| CDC20 | 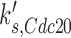 |
0.006 | 0.98 | 2.40 |
| CDH1 | 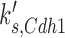 |
0.010 | 0.13 | 0.90 |
| CDC15 | [Cdc15T] | 1.000 | 5.50 | >403.00 |
| CDC14 | ks, Cdc14 | 0.200 | 0.27 | 0.40 |
| TEM1 | [Tem1p] | 1.000 | 40.40 | 403.00 |
| SIC1 | 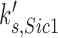 |
0.012 | 0.20 | 0.29 |
| CDC6 | 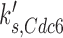 |
0.024 | 0.24 | 0.32 |
| SWI5 | 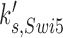 |
0.005 | 0.10 | 0.18 |
Simplistic predictions of our simulation are that CDC15 over-expression should suppress an upstream tem1Δ mutation (15,43); CDC14 over-expression should suppress an upstream cdc15Δ (44) or tem1Δ mutation (45); and either CDC20 or CDH1 overexpression should suppress a upstream tem1Δ cdc15Δ, or cdc14Δ mutation (Figure 4). While some of these are indeed true (15) a more general expectation is that over-expression of CDC20, CDC6 and CDH1 should at minimum cause checkpoint bypass in normal cell cycle, thus may cause chromosome instability. Consistently, CDC20, CDC6 and CDH1 do so (Table 1). We could not confirm the effects of CDC14, TEM1, and SIC1 over-expression because of inadequate color formation in our assays. CDC15 could not be tested because the MORF plasmid was not available.
Figure 4.
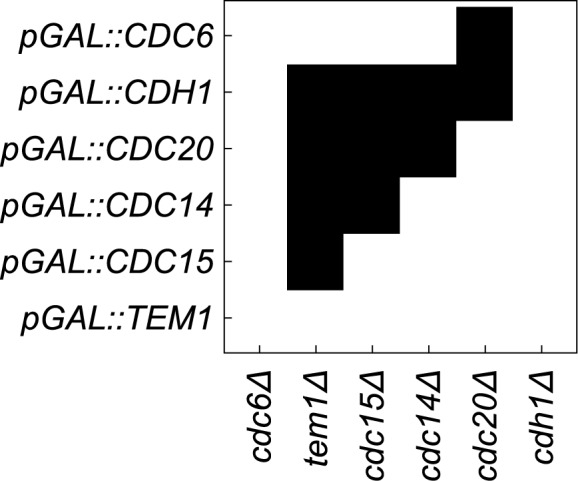
In silico predictions of genetic interactions. Cell cycle checkpoint arrest were simulated by removing the product of the genes listed along the x-axis. These blocks were overcome by simulating over-expression of the genes listed on the y-axis.
In Silico seriation of cell cycle models
The model formulated by Chen et al. (15) is a minimal model that sufficiently captures most of the landmark events of the cell cycle in budding yeast. This model however, was designed for a different intent than the one to which we subjected it; therefore, it has limited power for predicting cell cycle genes that cause chromosome instability. For example, not all genes that are known to participate in cell cycle control are engaged in the model, nor are all known functions of the genes in the model are explicitly simulated.
How does one re-engineer a complex cell cycle model to integrate additional chromosome instability genes? We propose seriating rows of novel genetic interaction data to update the underlying biochemical model that is captured by these interactions. Indeed, if the seriation approach is valid using data derived from in vivo experiments, then it should also be valid using data derived from in silico modeling.
To illustrate that the approach is valid in silico, let each of the rows of binary data in Figure 4 represent a position in hyperspace. The shortest tour is the path through the hyperspace that visits each of the data points exactly once and returns to the starting point (see Materials and Methods). As shown in Figure 4, the shortest tour determined by integer linear programming appears to re-derive the progression of gene function that qualitatively underlies this cell cycle model. Specifically, we have a cyclical series: Cdc6p→ Tem1p→ Cdc15p→ Cdc14p→ Cdc20p → Cdh1p→ Cdc6p again. This cyclical order reflects both the x-axis and the y-axis after seriation (see Materials and Methods for seriation details).
Seriation of a matrix of genetic interactions to form a genetic model for chromosome instability induction
We hypothesized that some of the chromosome-instability genes might be affecting chromosome stability due to their adverse effects on the cell cycle. If true, it is predicted that these genes should affect relative lengths of the cell cycle in a predictable manner. As demonstrated in silico, one way to numerically identify whether a set of vectors have systematic effects on a cyclically timed series of events is to conduct seriation analysis on the matrix of vectors.
Based on the μcolony estimated as described in the above section, 19 plasmids were chosen for matrix seriation analysis. The corresponding genes are CDC4, CLN1, ELG1, YGL182C, NRM1, NDT80, IME2, REC8, CDC5, YRB1, CLB5, MOT3, CDC20, XRS2, YDR387C, MOT2, HMLα2, CSE4 and HHT2 (of which CSE4 and HHT2 were included despite undetermined mutation rates). Due to resource limitations and logistical constraints, other plasmids having low μcolony estimates were not included in this set of 19 MORF plasmids.
These 19 MORF plasmids were introduced into 18 deletion mutant strains, each of which gene deletions were previously reported to cause defective chromosome stability by Yuen et al. (6) in Supplementary Table S3 of that work. Specifically we chose these deletion mutant strains because they had scored positively in all three of the assays performed by Yuen et al. (6) including the Chromosome Transmission Fidelity Assay the A-Like Faker Assay, and the Bimater Assay. Out of 33 deletion mutant strains matching these criteria, we chose those that could be transformed successfully with MORF plasmids.
The strain containing rad9Δ was included to make these genetic interaction assays comparable to previous studies which had also used rad9Δ (9). Unlike the other 17 deletion mutant strains the rad9Δ strain had scored negatively in the Chromosome Transmission Fidelity Assay but positively in the A-Like Faker Assay and the Bimater assay (6).
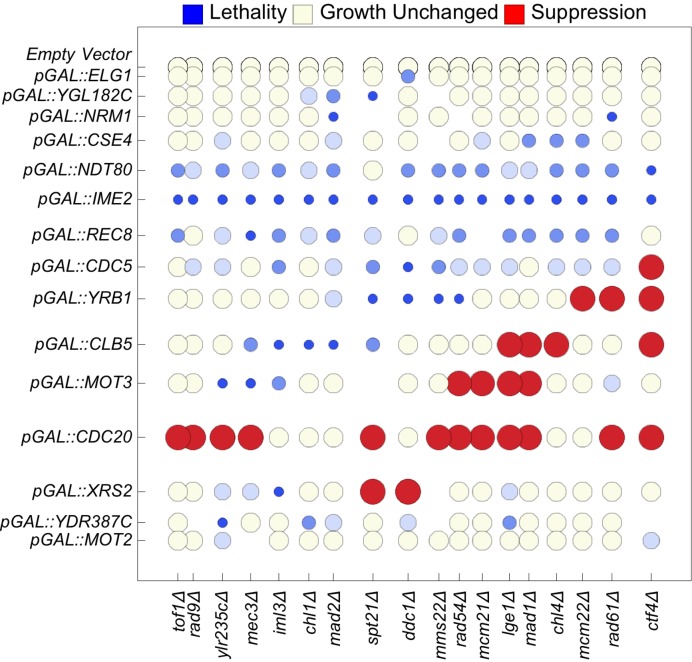
The resulting 331 combinations (11 strains could not be constructed for technical reasons) were assayed for synthetic growth defect or suppression of growth defects as described in Methods. The resulting matrix of suppression synthetic dosage lethality, and synthetic dosage sickness is illustrated in Figure 5 and supplementary file S1. Not shown are the rows for pGAL::HHT2, pGAL::CDC4, pGAL::CLN1, and pGAL::HMLα2, which are identical to the row for the empty vector. Note that this matrix has no dimension of time: the possibility that the MORF plasmids might be exhibiting the genetic interactions observed here might have to do with any temporally cyclical phenomenon (such as the cell cycle) is implicit.
Figure 5.
Seriation of genetic interactions results in a cyclical matrix. We report synthetic dosage lethality (represented as small blue circles), two levels of synthetic dosage sickness (two progressively lighter shades of blue), unchanged growth (yellow), and suppression (large red circles). To seriate the matrix, the rows (columns) of the matrix of genetic interactions are rearranged using integer linear programming to minimize the sum of the n-dimensional Euclidean distances over all pairs of adjacent n-dimensional row vectors (column vectors). The spatial distances of separation between adjacent rows and adjacent columns are proportional to the n-dimensional Euclidean distance between two separated rows or columns, respectively. The resulting order of the genes forms a model of the order of the respective functions of the genes during the cell cycle. The first row (column) is adjacent to the last row (column). Not shown are the rows for pGAL::HHT2, pGAL::CDC4, pGAL::CLN1, and pGAL::HMLα2, which are identical to the row for the empty vector.
The matrix was seriated by Integer Linear Programming (see Materials and Methods) for optimizing the total of the Euclidean-distance metrics between pairs of row vectors. The optimum sequence of the rows in the matrix had a total space of 355 687 428 095 999 permutations. The observed optimal order of the MORF genes following seriation is given in Figure 5. When we listed the genes Gene Ontology annotations (46) of the deletion mutant genes we recognized that the observed series form a kinetically coherent model approximating the temporal order of cell biological events that occur during cell cycle:
DNA damage sensing (e.g. TOF1, RAD9, MEC3);
Kinetochore-centromere cohesion (e.g. IML3, CHL1);
Helicase activity (e.g. CHL1);
Spindle checkpoint delay (e.g. MAD2);
DNA stress response (e.g. SPT21; DDC1, MMS22, RAD54);
Attachment of kinetochore to microtubules (e.g. MCM21);
Initiation of anaphase (e.g. MAD1);
The progression through the spindle assembly checkpoint (e.g. CHL4);
Chromosome segregation (e.g. MCM22);
Inhibition of cohesion between sister chromatids (e.g. RAD61);
Re-linking sister chromatid cohesion to DNA synthesis (e.g. CTF4). This function is followed cyclically by DNA damage sensing again.
We predict that one gene of unknown function, LGE1, has a similar function to the cyclically adjacent genes MCM21 (functioning in kinetochore attachment) and MAD1 (functioning in the spindle checkpoint). We predict that the dubious open reading frame YLR235C is similar to the cyclically adjacent DNA damage sensing genes RAD9 and MEC3, perhaps because the deletion of YLR235C overlaps with the deletion of TOP3 (topoisomerase III).
To arrange the over-expressed genes and the respective annotations (46) of those genes we again minimized the total of the n-dimensional Euclidean distances rather than maximizing that total. The model arrangement is:
Regulation of the G1 to S transition (e.g. CDC4, CLN1);
DNA replication and repair (e.g. ELG1);
Exit from G1 (e.g. NRM1)
Kinetochore function (e.g. CSE4)
Positive regulation of meiosis (e.g. NDT80, IME2)
Sister chromatid cohesion during meiosis (e.g. REC8)
Various functions during meiosis or mitosis (e.g. CDC5)
G1 to S transition (e.g. YRB1)
DNA synthesis during S phase (e.g. CLB5)
Regulation of stress response (e.g. MOT3)
Finish stage of Mitosis (e.g. CDC20)
Double Strand Break Repair (e.g. XRS2)
Regulation of DNA replication (e.g. MOT2). MOT2 is followed cyclically by G1 functioning genes again.
This second sequence is similar to the first sequence above. Each predicts that DNA repair is followed by kinetochore function, followed by stress response, followed by completion of mitosis, followed by DNA synthesis. This second model includes an excursion into meiosis after exiting G1 because meiosis-specific genes were used to build the matrix.
Evaluating the model using fluorescence-activated cell sorting
Does the order of the over-expressed genes in the model correctly predict the order of the respective functions of the genes during the cell cycle? To validate the model, we tested whether genes that function to transition the cell cycle out of G1 reduces the percentage of cells in G1 (47) while genes that function to transition the cell cycle out of M Phase do the opposite. The percentage of cells in G1 are inferred by clustering two-dimensional data consisting of (i) normalized DNA fluorescence and (ii) normalized aspect ratio of the brightfield image. Of importance to note here is that chromosome instability induced by plasmid overexpression was conducted in the diploid strain YPH275 whereas the temporal sequence of events were inferred from the synthetic genetic interaction matrix based on cell viability in haploid cells. Therefore the model validation experiments by FACS were conducted in the diploid YPH275 background.
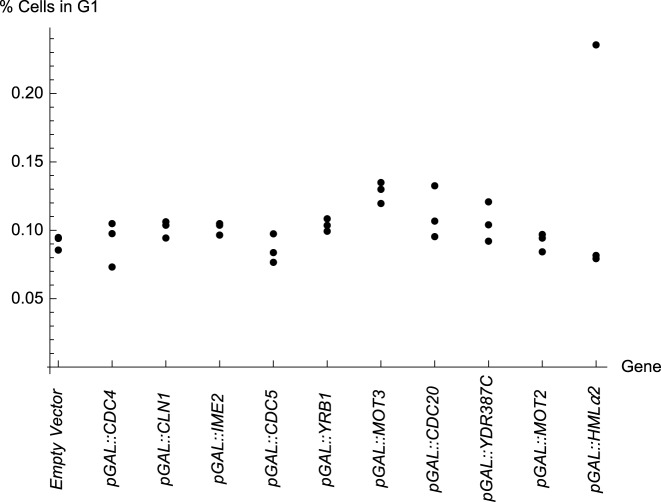
Figure 6 shows the percentage of diploid cells in the G1 phase of the cell cycle for YPH275 strains carrying the respective MORF plasmids, when grown in the presence of galactose. The order of the genes in this figure is sorted to match the order of the cycle in Figure 5. Recall that the genetic interaction data for CDC4, CLN1, and HMLα2 that are reported Figure 5 are identical to that of the empty vector. Data are not shown for AIM26, a gene of unknown function that was not tested for genetic interactions. With AIM26, we observed that the percentage of cells in G1 was 10%, 10% and 11%. Data for pGAL::NRM1 are excluded due to technical difficulties.
Figure 6.
The percentage of cells in G1 measured by Fluoresence Activated Cell Sorting (FACS). The arrangement peaks with pGAL::MOT3. The order of genes is arranged to match the order of the cycle in Figure 5. Recall from Figure 5 that pGAL::CDC4, pGAL::CLN1 and pGAL::HMLα2 had no genetic interactions and are thus similar to the empty vector. Data for pGAL::NRM1 are excluded because the results were irreprodicible among replicates.
Encouragingly we see further evidence of a cycle. The arrangement from BG1766 to pGAL::IME2 to pGAL::CDC5 to pGAL::YRB1 to pGAL::MOT3 progresses toward a peak significantly (p=0.0062, Jonckheere-Terpstra Test, a non-parametric test for ordered differences among classes). Furthermore, the arrangement from pGAL::MOT3 to pGAL::CDC20 to pGAL::YDR387C to pGAL::MOT2 to BG1766 declines from that peak significantly (P = 0.0024, Jonckheere-Terpstra Test). Hence, the arrangement supports a cyclically ordered model. Other over-expressed genes, including pGAL::CDC4, pGAL::CLN1, and pGAL::HMLα2 are not implicated in genetic interactions in this work and, therefore, it is reasonable that those percentages of cell in G1 do not appear to be trended (assuming that the data for pGAL::HMLα2 includes one outlier point).
We observed that pGAL::CDC20, induces a high percentage of cells in G1, while pGAL::CLB2 induces one of the lowest percentage of cells in G1 (data for pGAL::CLB2 that is not in Figure 6: 8%, 8.5% and 8.4%, respectively, in three replicate experiments). These results are intuitively understandable given previous observations that CDC20 functions in a manner opposite to that of CLB2 (15,48).
DISCUSSION
A healthy cell must maintain chromosome integrity during each phase of the cell cycle. Yet during the cell cycle defects in DNA repair, DNA synthesis, and chromosome transmission can cause chromosome instability. Mechanisms of chromosome instability were previously explored by others, including works that studied CDC5 over-expression (49,50) and CDC20 over-expression (42,51–54). Here, we build a cyclical model for chromosome stability by assaying the genetic interactions of pairs of genes that each normally functions to maintain chromosomes. First, we confirm that 19 genes affect chromosome stability by over-expressing the respective genes using an assay for chromosome instability. We then introduce plasmids containing these 19 genes into 18 deletion mutant strains each of which is known to cause chromosome instability. By conditionally overexpressing each of the 19 genes in each of the 18 deletion strains, we observe 35 cases of synthetic dosage lethality, 64 of synthetic dosage sickness, and 26 of suppression (out of 349 combinations tested including 18 empty-vector controls). To the best of our knowledge, these are novel findings. In addition, the genes with chromsome instability phenotypes listed in Table 1 have not been previously reported, except the positive controls YRB1 (9) and CLB5 (26).
Chromosome instability and molecular interaction maps of mitosis
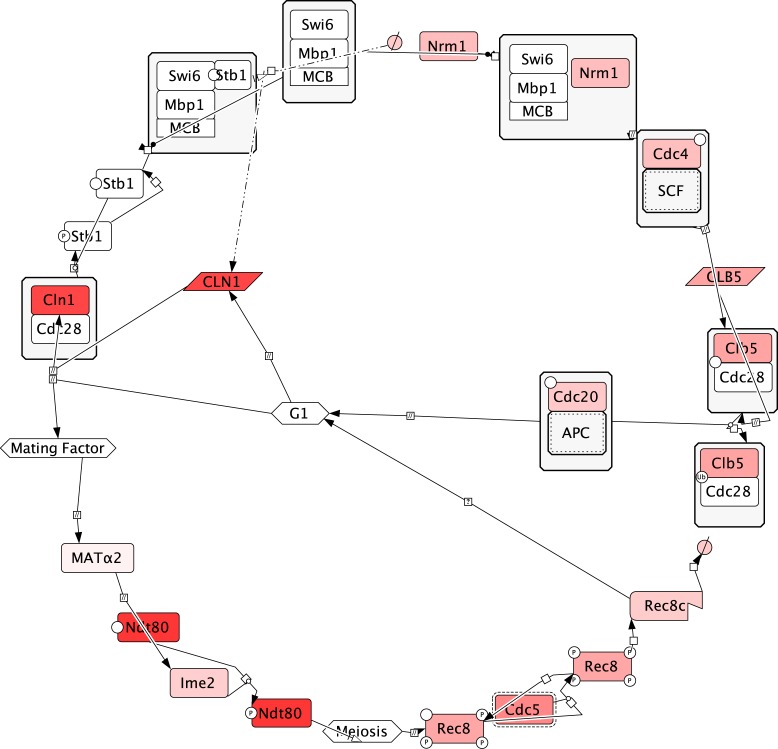
CDC20 CDC4 and CDH1 function in the ubiquitination of cell cycle control proteins including B cyclins (55–58). Therefore, it is possible that negative regulation by these genes is an indirect upstream process that causes downstream loss of function that is more directly responsible for chromosome instability. Specifically, Cdc20p ubiquitinates its target Clb5p (55); Nrm1p represses CLB5 gene (59); and Cdc4p degrades proteins that are involved in the activation of CLB5p (60). Thus clb5 loss should cause chromosome instability which has previously been reported (6,22,61). Further downstream Clb5p perturbs the Mini Chromosome Maintenance Complex by phosphorylating Mcm3p (62,63) triggering its nuclear export (64,65). Hence, loss of clb5 should cause effects similar to that of MCM3 over-expression. Moreover, we observed higher rate of chromosome instability induced by pGAL::MCM3 than by any other plasmid we tested. In conclusion, by comparing gain of function and loss of function phenotypes, we see a clearer relationship between established biochemical pathway maps and chromosome instability (Figure 7).
Figure 7.
Biochemical reactions in the merged mitotic and meiotic life cycle of yeast. Information were selected from biochemical maps of mitosis (66) and meiosis (67). Cln1p starts the mitotic cell cycle (68) Nrm1p indirectly represses expression of CLN1 (59) Cdc4p indirectly activates Clb5p (60) which is ubiquitinated by Cdc20p to restore G1 (55). Alternately the presence of mating factor should affect MATα2, indirectly causing the preexisting Ndt80p to be activated by Ime2p (69) triggering Rec8p to function in chromatid cohesion and ultimately be degraded by Cdc5p (70,71) before the cell returns to G1 again. Based on this life cycle diagram the functional arrangement of the chemical species in mitosis and meiosis, interleaved into one cycle, align to the cycle suggested by matrix seriation. Specifically, interleaving the mitotic and meiotic pathways yields the ordering CLN1/CDC4/HMLα2→ NRM1→ NDT80→ IME2→ REC8→ CDC5→ CLB5→ CDC20→ back to CLN1/CDC4/HMLα2. Omitted from the maps of mitosis and meiosis are proteins that less directly affect these processes (ELG1, YGL182C, CSE4, YRB1, MOT3, XRS2, YDR387C, and MOT2). The saturation of the red color is proportional to the highest chromosome instability rate reported for a corresponding MORF plasmid in Table 1.
While cell cycle control genes were confirmed to induce chromosome instability, the genes that function in RNA export, phospholipid biosynthesis, and regulation of nuclear envelope morphology appear to function upstream of these core cell cycle control genes. We expect that a screen having a lower false-negative rate would uncover more chromosome instability genes including upstream modulators of cell cycle control.
A paradigm for analyzing genetic interactions
Given a large matrix of genetic interactions, ordinarily it is computationally intractable (and NP hard) to compute all permutations of the arrangements of the rows and columns, which is necessary for finding an optimum arrangement of entries for a given pattern of functional significance. Here, using our small yet dense matrix we rearrange the matrix into an optimal cycle. By optimally reordering the matrix representation of these genetic interactions using integer linear programming, we find that the optimal order of the functions of the genes approximates the temporal order of processes during the yeast cell cycle. To further validate this theoretical approach, we computed in silico genetic interactions derived from a predetermined cyclical model and confirmed that the predicted cycle was consistent. These results suggest that matrices from other genetic experiments could be computationally rearranged using this method to reveal an underlying cycle that is biological meaningful.
If the seriation approach is a valid way to deduce cyclical models from genetic interactions, then we expect that seriation of the in silico genetic interaction data should reproduce the underlying qualitative assumptions about the progression of events in the budding yeast cell cycle model by Chen et al. (15). Encouragingly, we computed that seriation of the matrix of the in silico genetic interactions results in the arrangement of genes that closely matches the order of the respective function of the genes in the model.
Supplementary Material
Acknowledgments
We thank Eric Phizicky and Elizabeth Grayhack for supplying the MORF library; Phil Hieter for supplying strain YPH275; Raghavan Vasudevan, Vikashni Padmakumar, and Kankshit Bheda for transformations and plasmid purification; Arvind Kothandaraman, Rishov Chatterjee, and Hardeep Chiraya for laboratory logistics; Jose Salazar and Tanya Ferguson for Biomek FXP liquid handler support; Bryan Kraynack and Kirilynn Svay for training; Herbert Sauro, Alpan Raval, Craig Adams, Ali Nadim, and Susan Kane for discussions; David Galas for support and encouragement. We thank Dr. Babetta L. Marrone (Director, National Flow Cytometry Resource) for helpful discussions on image analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [0527023, 0523643, 0523643, 0941078 to A.R.]; National Institutes of Health [1R01GM084881-01 to A.R.]. Part of the work was supported by the Los Alamos National Laboratory National Flow Cytometry Resource funded by the National Center for Research Resources of the National Institutes of Health [P41-RR01315]. Funding for open access charge: Keck Graduate Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schinzel A. Catalogue of Unbalanced Chromosome Aberrations in Man. NY: Walter de Gruyter Berlin; 1984. [Google Scholar]
- 2.Mitelman F. Catalog of Chromosome Aberrations in Cancer. NY: Wiley-Liss; 1994. [Google Scholar]
- 3.Stirling P.C., Bloom M.S., Solanki-Patil T., Smith S., Sipahimalani P., Li Z., Kofoed M., Ben-Aroya S., Myung K., Hieter P. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 2011;7:e1002057. doi: 10.1371/journal.pgen.1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spencer F., Gerring S.L., Connelly C., Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith S., Hwang J.Y., Banerjee S., Majeed A., Gupta A., Myung K. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9039–9044. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuen K.W., Warren C.D., Chen O., Kwok T., Hieter P., Spencer F.A. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3925–3930. doi: 10.1073/pnas.0610642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanellis P., Gagliardi M., Banath J.P., Szilard R.K., Nakada S., Galicia S., Sweeney F.D., Cabelof D.C., Olive P.L., Durocher D. A screen for suppressors of gross chromosomal rearrangements identifies a conserved role for PLP in preventing DNA lesions. PLoS Genet. 2007;3:e134. doi: 10.1371/journal.pgen.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen M.P., Nelson Z.W., Hetrick E.D., Gottschling D.E. A genetic screen for increased loss of heterozygosity in Saccharomyces cerevisiae. Genetics. 2008;179:1179–1195. doi: 10.1534/genetics.108.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouspenski I.I., Elledge S.J., Brinkley B.R. New yeast genes important for chromosome integrity and segregation identified by dosage effects on genome stability. Nucleic Acids Res. 1999;27:3001–3008. doi: 10.1093/nar/27.15.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foury F. Human genetic diseases: a cross-talk between man and yeast. Gene. 1997;195:1–10. doi: 10.1016/s0378-1119(97)00140-6. [DOI] [PubMed] [Google Scholar]
- 11.Kroll E.S., Hyland K.M., Hieter P., Li J.J. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Measday V., Hieter P. Synthetic dosage lethality. Methods Enzymol. 2002;350:316–326. doi: 10.1016/s0076-6879(02)50971-x. [DOI] [PubMed] [Google Scholar]
- 13.Measday V., Baetz K., Guzzo J., Yuen K., Kwok T., Sheikh B., Ding H., Ueta R., Hoac T., Cheng B., et al. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13956–13961. doi: 10.1073/pnas.0503504102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baetz K., Measday V., Andrews B. Revealing hidden relationships among yeast genes involved in chromosome segregation using systematic synthetic lethal and synthetic dosage lethal screens. Cell Cycle. 2006;5:592–595. doi: 10.4161/cc.5.6.2583. [DOI] [PubMed] [Google Scholar]
- 15.Chen K.C., Calzone L., Csikasz-Nagy A., Cross F.R., Novak B., Tyson J.J. Integrative analysis of cell cycle control in budding yeast. Mol. Biol. Cell. 2004;15:3841–3862. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boone C., Bussey H., Andrews B.J. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 17.Raval A., Ray A. Introduction to Biological Networks. Boca Raton: CRC Press; 2013. [Google Scholar]
- 18.Kuentzer F.A., Pereira A.S., Amory A.M., Perrone G., da Silva S.R., Dinis J.M., de Almeida R. Bioinformatics and Bioengineering (BIBE), 2014 IEEE International Conference on. IEEE; 2014. pp. 231–237. [Google Scholar]
- 19.Johnson O., Liu J. A traveling salesman approach for predicting protein functions. Source Code Biol. Med. 2006;1:3. doi: 10.1186/1751-0473-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hieter P., Mann C., Snyder M., Davis R.W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- 21.Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 22.Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 23.Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 24.Gelperin D.M., White M.A., Wilkinson M.L., Kon Y., Kung L.A., Wise K.J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao W. Yeast Protocols. Totowa: Humana Press; 2006. [Google Scholar]
- 26.Sarafan-Vasseur N., Lamy A., Bourguignon J., Le Pessot F., Hieter P., Sesboue R., Bastard C., Frebourg T., Flaman J.M. Overexpression of B-type cyclins alters chromosomal segregation. Oncogene. 2002;21:2051–2057. doi: 10.1038/sj.onc.1205257. [DOI] [PubMed] [Google Scholar]
- 27.Zelterman D. Advanced Log-linear Models using SAS. Cary: SAS Institute; 2002. [Google Scholar]
- 28.Frumkin J. Claremont: The Claremont Graduate University; 2012. [Google Scholar]
- 29.Liiv I. Seriation and matrix reordering methods: an historical overview. Stat. Anal. Data Min. 2010;3:70–91. [Google Scholar]
- 30.Dantzig G., Fulkerson R., Johnson S. Solution of a large-scale traveling-salesman problem. J. Oper. Res. Soc. Am. 1954;2:393–410. [Google Scholar]
- 31.Dantzig G.B., Fulkerson D.R., Johnson S.M. On a linear-programming, combinatorial approach to the traveling-salesman problem. Oper. Res. 1959;7:58–66. [Google Scholar]
- 32.Haase S.B., Reed S.I. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle. 2002;1:132–136. [PubMed] [Google Scholar]
- 33.Winey M., Goetsch L., Baum P., Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li R., Murray A.W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 35.Dorer R.K., Zhong S., Tallarico J.A., Wong W.H., Mitchison T.J., Murray A.W. A small-molecule inhibitor of Mps1 blocks the spindle-checkpoint response to a lack of tension on mitotic chromosomes. Curr. Biol. 2005;15:1070–1076. doi: 10.1016/j.cub.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 36.Fernius J., Hardwick K.G. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 2007;3:e213. doi: 10.1371/journal.pgen.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toda K., Naito K., Mase S., Ueno M., Uritani M., Yamamoto A., Ushimaru T. APC/C-Cdh1-dependent anaphase and telophase progression during mitotic slippage. Cell Div. 2012;7:4. doi: 10.1186/1747-1028-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rieder C.L., Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell. 2004;7:637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Minn A.J., Boise L.H., Thompson C.B. Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 1996;10:2621–2631. doi: 10.1101/gad.10.20.2621. [DOI] [PubMed] [Google Scholar]
- 40.Hardwick K.G., Weiss E., Luca F.C., Winey M., Murray A.W. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 41.Fraschini R., Beretta A., Sironi L., Musacchio A., Lucchini G., Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang L.H., Lau L.F., Smith D.L., Mistrot C.A., Hardwick K.G., Hwang E.S., Amon A., Murray A.W. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 43.Shirayama M., Matsui Y., Toh E.A. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol. Cell. Biol. 1994;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visintin R., Craig K., Hwang E.S., Prinz S., Tyers M., Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 45.Jaspersen S.L., Charles J.F., Tinker-Kulberg R.L., Morgan D.O. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu W., Li Z., Zhan W., Iyer V.R., Marcotte E.M. Mechanisms of cell cycle control revealed by a systematic and quantitative overexpression screen in S. cerevisiae. PLoS Genet. 2008;4:e1000120. doi: 10.1371/journal.pgen.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cross F.R., Schroeder L., Kruse M., Chen K.C. Quantitative characterization of a mitotic cyclin threshold regulating exit from mitosis. Mol. Biol. Cell. 2005;16:2129–2138. doi: 10.1091/mbc.E04-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer R.E., Hogan E., Koshland D. Mitotic transmission of artificial chromosomes in cdc mutants of the yeast, Saccharomyces cerevisiae. Genetics. 1990;125:763–774. doi: 10.1093/genetics/125.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collins K.A., Castillo A.R., Tatsutani S.Y., Biggins S. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol. Biol. Cell. 2005;16:5649–5660. doi: 10.1091/mbc.E05-08-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan J., Chen R.H. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes Dev. 2004;18:1439–1451. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shirayama M., Zachariae W., Ciosk R., Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y., Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol. Cell. Biol. 2001;21:5190–5199. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schott E.J., Hoyt M.A. Dominant alleles of Saccharomyces cerevisiae CDC20 reveal its role in promoting anaphase. Genetics. 1998;148:599–610. doi: 10.1093/genetics/148.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shirayama M., Toth A., Galova M., Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- 56.Jackson L.P., Reed S.I., Haase S.B. Distinct mechanisms control the stability of the related S-phase cyclins Clb5 and Clb6. Mol. Cell. Biol. 2006;26:2456–2466. doi: 10.1128/MCB.26.6.2456-2466.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwab M., Lutum A.S., Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 58.Wasch R., Cross F.R. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature. 2002;418:556–562. doi: 10.1038/nature00856. [DOI] [PubMed] [Google Scholar]
- 59.de Bruin R.A., Kalashnikova T.I., Chahwan C., McDonald W.H., Wohlschlegel J., Yates J., 3rd, Russell P., Wittenberg C. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol. Cell. 2006;23:483–496. doi: 10.1016/j.molcel.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 60.Knapp D., Bhoite L., Stillman D.J., Nasmyth K. The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol. Cell. Biol. 1996;16:5701–5707. doi: 10.1128/mcb.16.10.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daniel J.A., Keyes B.E., Ng Y.P., Freeman C.O., Burke D.J. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics. 2006;172:53–65. doi: 10.1534/genetics.105.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loog M., Morgan D.O. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 63.Ubersax J.A., Woodbury E.L., Quang P.N., Paraz M., Blethrow J.D., Shah K., Shokat K.M., Morgan D.O. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 64.Liku M.E., Nguyen V.Q., Rosales A.W., Irie K., Li J.J. CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2-7 complex prevents chromosomal rereplication. Mol. Biol. Cell. 2005;16:5026–5039. doi: 10.1091/mbc.E05-05-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen V.Q., Co C., Irie K., Li J.J. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr. Biol. 2000;10:195–205. doi: 10.1016/s0960-9822(00)00337-7. [DOI] [PubMed] [Google Scholar]
- 66.Kaizu K., Ghosh S., Matsuoka Y., Moriya H., Shimizu-Yoshida Y., Kitano H. A comprehensive molecular interaction map of the budding yeast cell cycle. Mol. Syst. Biol. 2010;6:415. doi: 10.1038/msb.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hadwiger J.A., Wittenberg C., Richardson H.E., de Barros Lopes M., Reed S.I. A family of cyclin homologs that control the G1 phase in yeast. Proc. Natl. Acad. Sci. U.S.A. 1989;86:6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sopko R., Raithatha S., Stuart D. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol. Cell. Biol. 2002;22:7024–7040. doi: 10.1128/MCB.22.20.7024-7040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brar G.A., Kiburz B.M., Zhang Y., Kim J.E., White F., Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- 71.Lee B.H., Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.