Abstract
Nicotinic acetylcholine receptors (nAChRs) play a key role in carcinogenesis and progression of lung cancer; and polymorphisms in CHRNA5-A3 and CHRNB3-A6, two gene clusters encoding nAChR subunits, have been associated with lung cancer risk. In this study, we investigated whether variants in the two gene clusters were associated with prognosis of advanced non-small cell lung cancer (NSCLC). A total of 165 stage IIIB–IV NSCLC patients were enrolled in this study. Three polymorphisms (rs667282 and rs3743073 in CHRNA5-A3 and rs13280604 in CHRNB3-A6) were genotyped using the TaqMan method. Overall survival (OS) was estimated using the log-rank test and the Cox models. Our results showed that patients with CHRNA5-A3 rs667282 TT or TC genotypes had a significantly shorter OS than those carrying the CC genotype (Log-rank, P = 0.043). Furthermore, multivariate Cox regression analysis showed that rs667282 TT/TC genotypes are significantly associated with increased risk of overall deaths (adjusted hazard ratio, 1.7; 95% CI, 1.1–2.7). However, the similar results were not observed for other two polymorphisms. Furthermore, no evident association was found between these variants and clinicopathologic features of advanced NSCLC. Our present study suggested that rs667282 in CHRNA5-A3 may modify the prognosis of patients with advanced NSCLC.
Keywords: CHRNA5-A3, CHRNB3-A6, survival, nicotinic acetylcholine receptors, lung cancer
INTRODUCTION
Lung cancer is the most common cause of cancer-related deaths worldwide. Approximately 1.6 million lung cancer cases and 1.4 million lung cancer deaths occurred in 2015 globally [1]. Non-small cell lung cancer (NSCLC) accounts for about 80% of primary lung cancers, with most of the patients diagnosed at the advanced stage (stage III or IV) [2]. Despite advances in chemotherapy and radiotherapy, prognosis of advanced NSCLC patients is still unimproved with a 5-year survival rates less than 15% [3]. Historically, treatment and prediction of prognosis for these patients is usually based on clinical characteristics, such as smoking behaviors, performance status (PS) and tumor, lymph node and metastasis (TNM) staging system [4–6]. New biomarkers that allow the identification of NSCLC patients with high risk of death/recurrence will facilitate the development of specific, less toxic, more aggressive for selected, targeted treatment strategies to improve therapeutic efficacy and outcomes. Thus, identification of such genetic prognostic biomarkers is needed.
Nicotinic acetylcholine receptors (nAChRs) are ion channels that are universally expressed in the plasma membrane of all mammalian cells, including cancer cells [7]. Now nAChRs have been identified as central regulators of stimulatory and inhibitory neurotransmitters that govern the synthesis and release of growth, angiogenic and neurotrophic factors in cancer cells and cancer microenvironment [8]. Recently, several single nucleotide polymorphisms (SNPs) in two nAChR subunit-encoding gene clusters (rs1051730, rs16969968 and rs8034191 in CHRNA5-A3 and rs13280604 in CHRNB3-A6) were shown to be associated with lung cancer risk and smoking consumption in Caucasians by genome-wide association (GWA) studies [9–12]. However, the three SNPs (rs1051730, rs16969968, and rs8034191) of CHRNA5-A3 reported in GWA studies are extremely rare in Chinese population, according to the HapMap database and a study of Wu et al. [13], while two independent studies found that rs667282 and rs3743073 in CHRNA5-A3 affected lung cancer risk in Chinese population [13, 14]. On the other hand, in addition to rs13280604 there are several other SNPs, including rs6474412, rs1451240, rs10958725, rs4736835, rs10958726, rs4950, and rs6474415 in CHRNB3-A6 region, while these SNPs are either in strong linkage disequilibrium (LD) or even perfect LD (R2 > 0.8) with rs13280604. Moreover, the association of rs13280604 in CHRNB3-A6 with smoking consumption and lung cancer risk has been observed in multiple ethnic populations, including Korean from East Asia [15]. However, to date no studies have evaluated if these variants in the two gene clusters also contribute to the prognosis of lung cancer, particularly the advanced NSCLC. Consequently, we investigated the associations of these three SNPs (rs667282, rs3743073 and rs13280604) in CHRNA5-A3 and CHRNB3-A6 with survival of advanced NSCLC patients.
RESULTS
Patient characteristics
The median age of the patients was 61 years (aged 30–82). There were 134 males (81.2%), 99 ever-smokers (60%) and 140 patients (84.8%) having a performance status of 0 or 1. For the histological types, 87 (52.7%) patients were diagnosed with adenocarcinoma, 60 (36.4%) with squamous cell carcinoma and 18 (10.9%) with other types. The majority of the patients (69.1%) presented with stage IV NSCLC. A total of 106 patients received conventional, platinum-based chemotherapy, 26 patients received radiotherapy, 22 patients received a combination of radiotherapy and chemotherapy, and 11 patients received treatment by other methods. Demographic and clinical characteristics of patients are summarized in Table 1.
Table 1. Demographic and clinical characteristics of NSCLC patients.
| Characteristics | N (%) |
|---|---|
| Median age (range) | 61 (30–82) |
| ≤ 60 | 81 (49.1) |
| > 60 | 84 (50.9) |
| Gender | |
| Male | 134 (81.2) |
| Female | 31 (18.8) |
| Smoking status | |
| Ever | 99 (60) |
| Never | 66 (40) |
| ECOG PSa | |
| 0–1 | 140 (84.8) |
| 2 | 25 (15.2) |
| Histological type | |
| Squamous cell carcinoma | 60 (36.4) |
| Adenocarcinoma | 87 (52.7) |
| Others | 18 (10.9) |
| Clinical stage | |
| IIIB | 51 (30.9) |
| IV | 114 (69.1) |
| Treatment | |
| Chemotherapy | 106 (64.2) |
| Radiotherapy | 26 (15.8) |
| Chemoradiotherapy | 22 (13.3) |
| Others | 11 (6.7) |
ECOG PS, Eastern Cooperative Oncology Group performance status.
Associations of genotypes with clinicopathological characteristics of patients
All patients had the genotyping data for analysis, and the genotype distributions of patients are presented in Table 2. We found that there were no significant differences in distribution of genotypes of these polymorphisms in term of clinicopathological features, such as age, gender, smoking status, EGOG performance status, histology, stage, and treatment (P > 0.05, Table 2).
Table 2. Genotype distribution of three polymorphisms by clinicopathological characteristics.
| rs667282 | rs3743073 | rs13280604 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristics | CC n = 30 |
TT/TC n = 135 |
P* | TT n = 44 |
GG/TG n = 121 |
P* | AA n = 92 |
GG/AG n = 73 |
P* |
| Age (years) | |||||||||
| ≤ 60 | 11 | 70 | 0.13 | 23 | 58 | 0.62 | 43 | 38 | 0.50 |
| > 60 | 19 | 65 | 21 | 63 | 49 | 35 | |||
| Gender | |||||||||
| Male | 24 | 110 | 0.85 | 37 | 97 | 0.57 | 73 | 61 | 0.49 |
| Female | 6 | 25 | 7 | 24 | 19 | 12 | |||
| Smoking status | |||||||||
| Never | 12 | 54 | 1.00 | 16 | 50 | 0.56 | 35 | 31 | 0.56 |
| Ever | 18 | 81 | 28 | 71 | 57 | 42 | |||
| ECOG PSa | |||||||||
| 0–1 | 26 | 114 | 0.76 | 36 | 104 | 0.51 | 79 | 61 | 0.68 |
| 2 | 4 | 21 | 8 | 17 | 13 | 12 | |||
| Histological type | |||||||||
| Adenocarcinoma | 16 | 71 | 0.94 | 27 | 60 | 0.18 | 48 | 39 | 0.87 |
| Others | 14 | 64 | 17 | 61 | 44 | 34 | |||
| Clinical stage | |||||||||
| IIIB | 8 | 43 | 0.58 | 12 | 39 | 0.54 | 33 | 18 | 0.12 |
| IV | 22 | 92 | 32 | 82 | 59 | 55 | |||
| Treatment | |||||||||
| Chemotherapy | 25 | 103 | 0.40 | 35 | 93 | 0.72 | 67 | 61 | 0.10 |
| Others | 5 | 32 | 9 | 28 | 25 | 12 | |||
ECOG PS, Eastern Cooperative Oncology Group performance status.
P values were calculated by the χ2 tests.
Associations of genotypes with overall survival
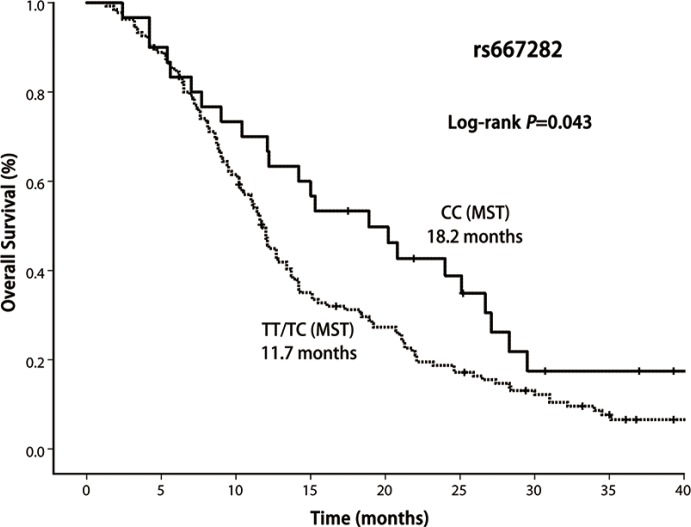
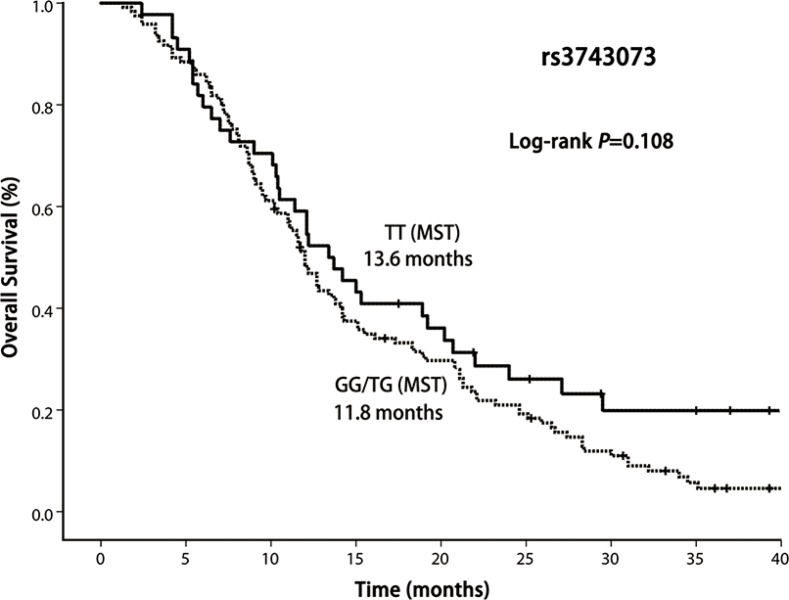
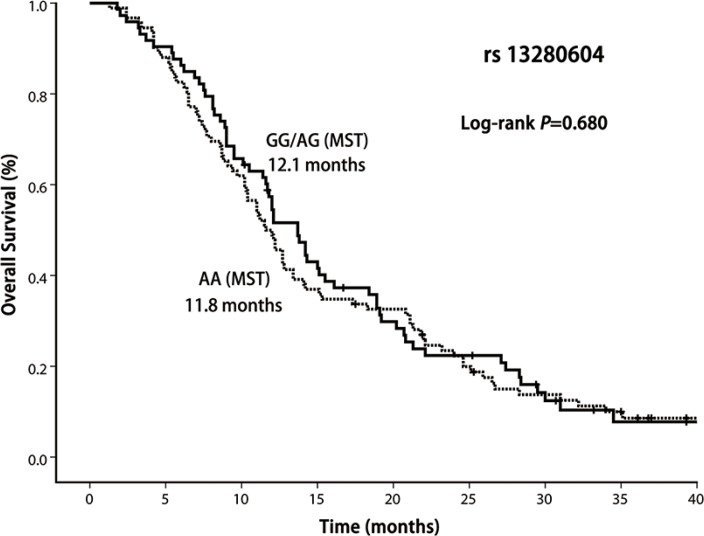
In this study, 13 patients were alive and 7 patients were lost to follow-up. In the entire group, the median survival time was 12.1 months (range 1.3–41.7 months). Our result demonstrated that OS was significantly shorter among the patients with CHRNA5-A3 rs667282 TT/TC genotypes compared with those carrying the CC genotype (Log-rank P = 0.043, Figure 1), while the similar significant differences in OS was not observed for CHRNA5-A3 rs3743073 (log-rank P = 0.108, Figure 2) and CHRNB3-A6 rs13280604 polymorphisms (log-rank, P = 0.680, Figure 3), respectively. Multivariate Cox regression analysis was used to assess the associations of three polymorphisms with full adjustment of other confounding factors including age, gender, smoking status, ECOG PS, histological type, clinical stage, and treatment, with risk of overall deaths (Table 3). The results showed that rs667282 TT/TC genotypes were significantly associated with increased risk of overall deaths among patients with advanced NSCLC (HR, 1.7, 95% CI, 1.1–2.7), independent of other prognostic factors. However, the significant associations were not found between the genotypes of other two polymorphisms and risk of overall deaths (data not shown).
Figure 1. Kaplan–Meier curve of OS according to CHRNA5-A3 rs667282 polymorphism genotypes.
Figure 2. Kaplan–Meier curve of OS according to CHRNA5-A3 rs3743073 polymorphism genotypes.
Figure 3. Kaplan–Meier curve of OS according to CHRNB3-A6 rs13280604 polymorphism genotypes.
Table 3. Multivariable analysis on association between risk of overall deaths and rs667282 genotypes in advanced NSCLC patients.
| Variables | HR* | 95% CI | P |
|---|---|---|---|
| Age (years) | |||
| ≤ 60 | |||
| > 60 | 1.1 | 0.8–1.6 | 0.53 |
| Gender | |||
| Male | |||
| Female | 0.8 | 0.5–1.2 | 0.29 |
| Smoking status | |||
| Never | |||
| Ever | 1.6 | 1.1–2.3 | 0.02 |
| ECOG PSa | |||
| 0–1 | |||
| 2 | 1.7 | 1.1–2.7 | 0.03 |
| Histological type | |||
| Adenocarcinoma | |||
| Others | 1.3 | 0.9–1.8 | 0.21 |
| Clinical stage | |||
| IIIB | |||
| IV | 1.3 | 0.9–1.9 | 0.15 |
| Treatment | |||
| Chemotherapy | |||
| Others | 1.1 | 0.7–1.6 | 0.78 |
| rs667282 | |||
| CC | |||
| TT/TC | 1.7 | 1.1–2.7 | 0.02 |
adjusted by age, gender, smoking status, ECOG PS, histological type, clinical stage, and treatment.
DISCUSSION
In the current study, we examined whether the SNPs in the two gene-clusters, which are significantly associated with lung cancer risk, are also associated with prognosis of lung cancer. According to the analysis of 165 advanced NSCLC patients, we found that rs667282 in CHRNA5-A3 was significantly associated with OS in NSCLC patients. Furthermore, multivariate Cox regression analysis showed that this SNP was an independent indicator for NSCLC prognosis after adjusting for confounding factors, including age, gender, smoking status, ECOG PS, histological type, clinical stage and treatment. No significant association was observed between the other two polymorphisms and survival.
nAChRs are the important regulators of cancer development and progression owing to their high affinity for tobacco-specific carcinogenic nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N-nitrosonornicotine (NNN) [8]. In addition, nAChR subunit-encoding gene cluster CHRNA5-A3 may have function in chemotherapy resistance to gemcitabine, cisplatin, and paclitaxel in NSCLC through the Akt-dependent proliferation and the NF-kappaB-dependent survival pathways under the stimulation of NNK [16–18]. Previous studies reported that rs667282 TT/CT genotypes in CHRNA5-A3 have been shown to be associated with increased smoking consumption which is always linked with a poor prognosis of lung cancer [13, 19], which is biologically consistent with our finding in the current study.
Although many studies have focused on the impacts of nAChR subunit-encoding genes on lung cancer risk and smoking consumption, few reports have been reported regarding the associations between the polymorphisms in these genes with survival outcomes. Recently, Jin et al. examined the association of a polymorphism rs6495309 in CHRNA5-A3, which was in strong linkage disequilibrium with rs667282 [13], with prognosis in patients with early-stage, surgically resected NSCLC [20]. They found that the polymorphism was significantly associated with survival outcome, consistent with the finding in our study in advanced NSCLC patients. In addition, there exists a risk tendency for another polymorphism rs3743073 in CHRNA5-A3, while such trend was not statistically significant. Niu's et al. found [14] that rs3743073 was associated with lung cancer risk, however, no evidence in an association with smoking consumption, while found for rs667282, was observed.
To our knowledge, this is the first study that investigated the association between polymorphisms in CHRNB3-A6 and lung cancer prognosis. The polymorphism rs13280604 was first reported to be associated with increased smoking consumption in the study of Saccone et al. [21]. They analyzed 3713 SNPs of 348 candidate genes in 1050 nicotine dependent cases and 879 controls, and observed a significant effect of the SNP on smoking behaviors in population of European descents. Subsequently, this association with smoking has been reported in multiple other ethnic populations, including African-American, European-American and Korean [15, 22–24]. Recently, a GWA study revealed that the variant in CHRNB3-A6 was significantly associated with both smoking and lung cancer risk in population of European descents; however, the effect of rs13280604 on smoking consumption was much weaker than those of the polymorphisms in CHRNA5-A3 [9]. It might implicate that the effect of rs13280604 on lung cancer prognosis may be independent of its interference with smoking consumption. In contrast, no association was found between this polymorphism, which has been found to be associated with lung cancer risk, and NSCLC survival of in the current study. Thus, further investigation in effect of this polymorphism in CHRNB3-A6 on lung cancer prognosis is warranted.
Several limitations need to be addressed in this study. Because of the limited sample sizes of NSCLC patients, results from this study may be biased. Thus future studies with a larger number of NSCLC cases are expected to further confirm our findings. Moreover, a relatively longer follow-up period is needed to investigate such associations between these variants in CHRNA5-A3 and CHRNB3-A6 with lung cancer survival outcome. In addition, in the study, we had no detailed information on smoking duration and intensity, which were required to calculate smoking pack-year. This limits our ability to assess the involvement of smoking level in the association. Finally, the majority of the study patients are diagnosed at late stage, and such analyses in clinical outcome with inclusion of early stage patients are needed in our future studies. In conclusion, this study first reveals that rs667282 in CHRNA5-A3 is significantly associated with survival outcome of NSCLC patients, indicating that the SNP may serve as a useful genetic biomarker for the prognosis of advanced NSCLC patients. However, additional large well-designed studies with different ethnic populations are needed to confirm these findings, particularly in other smoking-related cancers, such as head and neck cancers and esophageal squamous cell carcinoma in Chinese population.
MATERIALS AND METHODS
Study patients and follow-up
The subjects in this study consisted of a total of 165 patients with cytologically or histologically confirmed stage IIIB–IV NSCLC from Qilu Hospital of Shandong University between May 2009 and June 2010. We staged patients according to the 7th edition of the TNM classification of the International Union Against Cancer (UICC). Eligibility criteria included age ≥ 18 years, Eastern Cooperative Oncology Group (ECOG) performance status ≤ 2, and adequate organ function. All patients were unrelated ethnic Chinese who were all from Northern China. In this study, the primary endpoint for survival analysis was overall death. NSCLC patients were typically followed and monitored throughout their treatment and post-treatment courses with regularly scheduled clinical and radiographic examinations. Medical record review for the follow-up statuses of all patients was performed under the direct supervision of the surgeon. Overall survival (OS) was defined as the time from initial diagnosis until death from any cause or last follow-up. Participants who were alive at the end of the study period or lost to follow-up were censored. The study was performed in accordance with the Helsinki Declaration, and was approved by the Ethics Committees of Qilu Hospital of Shandong University. At recruitment, all participants were required to give written informed consent.
Genotyping
Whole-blood samples were collected from all study participants at the time of study entry. We extracted genomic DNA from the peripheral blood samples using the Tiangen Biotech kit (DP319; Tiangen Biotech (Beijing) Co., Ltd., Beijing China). The rs667282 and rs3743073 polymorphisms in CHRNA5-A3 and the rs13280604 polymorphism in CHRNB3-A6 were genotyped by TaqMan allelic discrimination technology, which uses two allele-specific TaqMan MGB probes and a PCR primer pair to detect the specific SNP target. Primers and probe information are shown in Table 4. PCR amplifications were conducted in a 96-well Applied Biosystems 7500 Real Time PCR System according to the instructions of the manufacturer and SDS 1.4 software (Applied Biosystems) was used for allelic discrimination.
Table 4. Primers and probes used for the TaqMan genotyping.
| Polymorphism | Sequence (5′-3′) |
|---|---|
| CHRNA5-A3 rs667282 | |
| Primer | F: TGGACTTTTCTACAACCTTGCTACTT |
| R: GCCTGAGACTCTGCATTTCTAACAA | |
| Probe | FAM: ACAGTATTCACGTCACCTG |
| VIC: ACAGTATTCACATCACCTG | |
| CHRNA5-A3 rs3743073 | |
| Primer | F: GGAGAAGGAGACGGTAAAAGAATCA |
| R: TGTGGTTCGGTCACATGCA | |
| Probe | FAM: TGGTTTTACTTCCCTTGGCACC |
| VIC: TTTTACTTCCCGTGGCACC | |
| CHRNB3-A6 rs13280604 | |
| Primer | F: GTGCTCCCTGTGAAGGTACA |
| R: GCTTGCTGCCCCTGGAT | |
| Probe | FAM: AATCCTGCTCTTCACCAAG |
| VIC: CCTGCTCCTCACCAAG | |
Ten percent of the samples were randomly chosen for re-testing, and the results were in 100% concordance with those of the initial assays.
Statistical analyses
Associations between genotypes of the three SNPs and individual clinicopathologic factors were assessed using the Pearson chi-square (χ2) test. OS curves were drawn with the Kaplan–Meier product limit method for each of the different genotype groups. Comparisons were tested with the log-rank test. Hazard ratios (HRs) and 95% confidence intervals (CIs) of risk genotypes were estimated by fitting the Cox model while adjusting for age, sex, smoking status, performance status, histological type, clinical stage and treatment regimen. Statistical analyses were performed with the SPSS software package (version 20). P values of less than 0.05 were taken to indicate statistically significant differences, and all of the statistical tests were two-sided.
Acknowledgments
The authors are grateful to Drs Shikun Wang and Yuanna Du for suggestions and for skillful technical assistance.
Abbreviations
- nAChRs
nicotinic acetylcholine receptors
- NSCLC
non-small cell lung cancer
- OS
overall survival
- SNP
single nucleotide polymorphisms
- HR
Hazard ratios
- CI
confidence intervals
Footnotes
CONFLICTS OF INTEREST
No potential conflicts of interest was disclosed.
GRANT SUPPORT
This work was supported by National Natural Science Foundation of China [No. 30671956, No. 81270238], and National Natural Science Foundation of Shandong Provence [No. ZR2012HM083].
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer. 1999;80:827–841. doi: 10.1002/(sici)1097-0215(19990315)80:6<827::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 3.Dillman RO, Herndon J, Seagren SL, Eaton WL, Green MR. Improved survival in stage III non-small-cell lung cancer: seven-year follow-up of cancer and leukemia group B (CALGB) 8433 trial. J Natl Cancer Inst. 1996;88:1210–1215. doi: 10.1093/jnci/88.17.1210. [DOI] [PubMed] [Google Scholar]
- 4.Nordquist LT, Simon GR, Cantor A, Alberts WM, Bepler G. Improved survival in never-smokers vs current smokers with primary adenocarcinoma of the lung. Chest. 2004;126:347–351. doi: 10.1378/chest.126.2.347. [DOI] [PubMed] [Google Scholar]
- 5.Finkelstein DM, Ettinger DS, Ruckdeschel JC. Long-term survivors in metastatic non-small-cell lung cancer: an Eastern Cooperative Oncology Group Study. J Am Societ Clin Oncol. 1986;4:702–709. doi: 10.1200/JCO.1986.4.5.702. [DOI] [PubMed] [Google Scholar]
- 6.Liu M, Jiang G, Ding J. Smoking reduces survival in young females with lung adenocarcinoma after curative resection. Med Oncol. 2012;29:570–573. doi: 10.1007/s12032-011-9826-y. [DOI] [PubMed] [Google Scholar]
- 7.Wessler I, Kirkpatrick CJ. Acetylcholine beyond neurons: the non-neuronal cholinergic system in humans. Bri J pharmacol. 2008;154:1558–1571. doi: 10.1038/bjp.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuller HM. Is cancer triggered by altered signalling of nicotinic acetylcholine receptors? Nat Rev Cancer. 2009;9:195–205. doi: 10.1038/nrc2590. [DOI] [PubMed] [Google Scholar]
- 9.Thorgeirsson TE, Gudbjartsson DF, Surakka I. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat get. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorgeirsson TE, Geller F, Sulem P. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nat. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung RJ, McKay JD, Gaborieau V. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nat. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 12.Amos CI, Wu X, Broderick P. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25. 1. Nat Get. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu C, Hu Z, Yu D. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 2009;69:5065–5072. doi: 10.1158/0008-5472.CAN-09-0081. [DOI] [PubMed] [Google Scholar]
- 14.Niu X, Chen Z, Shen S. Association of the CHRNA3 locus with lung cancer risk and prognosis in Chinese Han population. Journal of thoracic oncology. 2010;5:658–666. doi: 10.1097/JTO.0b013e3181d5e447. [DOI] [PubMed] [Google Scholar]
- 15.Cui WY, Wang S, Yang J. Significant association of CHRNB3 variants with nicotine dependence in multiple ethnic populations. Mol Psych. 2013;6:34–39. doi: 10.1038/mp.2012.190. [DOI] [PubMed] [Google Scholar]
- 16.Lam DC, Girard L, Ramirez R. Expression of nicotinic acetylcholine receptor subunit genes in non-small-cell lung cancer reveals differences between smokers and nonsmokers. Cancer Res. 2007;67:4638–4647. doi: 10.1158/0008-5472.CAN-06-4628. [DOI] [PubMed] [Google Scholar]
- 17.Dasgupta P, Kinkade R, Joshi B, Decook C, Haura E, Chellappan S. Nicotine inhibits apoptosis induced by chemotherapeutic drugs by up-regulating XIAP and survivin. Proc Natl Acad Sci USA. 2006;103:6332–6337. doi: 10.1073/pnas.0509313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsurutani J, Castillo SS, Brognard J. Tobacco components stimulate Akt-dependent proliferation and NFkappaB-dependent survival in lung cancer cells. Carcinogenesis. 2005;26:1182–1195. doi: 10.1093/carcin/bgi072. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka S, Yanagihara K, Tamaru S, Teramukai S, Kitano T, Fukushima M. Difference in survival and prognostic factors between smokers and never-smokers with advanced non-small-cell lung cancer. Int J Clin Oncol. 2013;18:17–25. doi: 10.1007/s10147-011-0334-z. [DOI] [PubMed] [Google Scholar]
- 20.Jin G, Bae EY, Yang E. A functional polymorphism on chromosome 15q25 associated with survival of early stage non-small-cell lung cancer. Journal of thoracic oncology. Int Assoc Study Lung Cancer. 2012;7:808–814. doi: 10.1097/JTO.0b013e31824c7d7c. [DOI] [PubMed] [Google Scholar]
- 21.Saccone SF, Hinrichs AL, Saccone NL. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice JP, Hartz SM, Agrawal A. CHRNB3 is more strongly associated with Fagerstrom test for cigarette dependence-based nicotine dependence than cigarettes per day: phenotype definition changes genome-wide association studies results. Addiction. 2012;107:2019–2028. doi: 10.1111/j.1360-0443.2012.03922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saccone NL, Schwantes-An TH, Wang JC. Multiple cholinergic nicotinic receptor genes affect nicotine dependence risk in African and European Americans. Genes, brain, and Behav. 2010;9:741–750. doi: 10.1111/j.1601-183X.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeiger JS, Haberstick BC, Schlaepfer I, Collins AC, Corley RP, Crowley TJ, Hewitt JK, Hopfer CJ, Lessem J, McQueen MB, Rhee SH, Ehringer MA. The neuronal nicotinic receptor subunit genes (CHRNA6 and CHRNB3) are associated with subjective responses to tobacco. Hum Mol Genet. 2008;17:724–734. doi: 10.1093/hmg/ddm344. [DOI] [PubMed] [Google Scholar]