Abstract
Designer nucleases have gained widespread attention for their ability to precisely modify genomic DNA in a programmable manner. These genome-editing nucleases make double-stranded breaks at specified loci, and desired changes can be made to modify, ablate, or excise target genes. This technology has been used widely to develop human disease models in laboratory animals and to study gene functions by silencing, activating, or modifying them. Furthermore, the recent discovery of a bacterially derived programmable nuclease termed clustered regularly interspaced palindromic repeats (CRISPR)-associated protein 9 (Cas9) has revolutionized the field because of its versatility and wide applicability. In this article, we discuss various modalities used to achieve genome editing with an emphasis on CRISPR-Cas9. We discuss genome-editing strategies to either repair or ablate target genes, with emphasis on their applications for investigating dermatological diseases. Additionally, we highlight preclinical studies showing the potential of genome editing as a therapy for congenital blistering diseases and as an antimicrobial agent, and we discuss limitations and future directions of this technology.
INTRODUCTION
Genome-editing technologies have been used widely over the last decade to develop human disease models in laboratory organisms and to study gene functions by silencing, activating, or modifying them. Furthermore, genome editing holds therapeutic potential to cure disease. A seminal example of its translational potential was its application to create HIV-resistant immune cells that were successfully transplanted into patients to control AIDS (Tebas, 2014). Although this remains the single example of its arrival to the clinic, the discovery of a prokaryotic adaptive immune system, termed clustered regularly interspaced palindromic repeats (CRISPR), and the CRISPR-associated protein 9 (Cas9), a bacterially derived programmable nuclease, has transformed the field to a point that its continued translation to the clinic appears imminent. Uses of CRISPR-Cas9 are various and include creating animal models of human disease, performing genome-wide screens to identify genes involved in complex biological processes, and genetically modifying food crops. Its ease-of-use and versatility have allowed more laboratories than ever before to work on genome editing in innovate ways. For these reasons, Science magazine named CRISPR-Cas9 the “Breakthrough of the Year” in 2015 (Travis, 2015).
PROGRAMMABLE DESIGNER NUCLEASES FOR GENOME EDITING
Designer nucleases have two components: a DNA-binding domain that guides the nuclease to the targeted genomic site and a nuclease domain that cuts the targeted locus to result in a double-stranded break (DSB). Before the advent of CRISPR, genome editing was achieved largely through the use of programmable DNA-binding proteins. These include zinc finger nucleases, transcription activator-like effector nucleases (TALENs), and meganucleases. However, these designer nucleases are of limited use because they are difficult to produce. The recent discovery of CRISPR transformed the field by allowing for the widespread use of genome-editing technology.
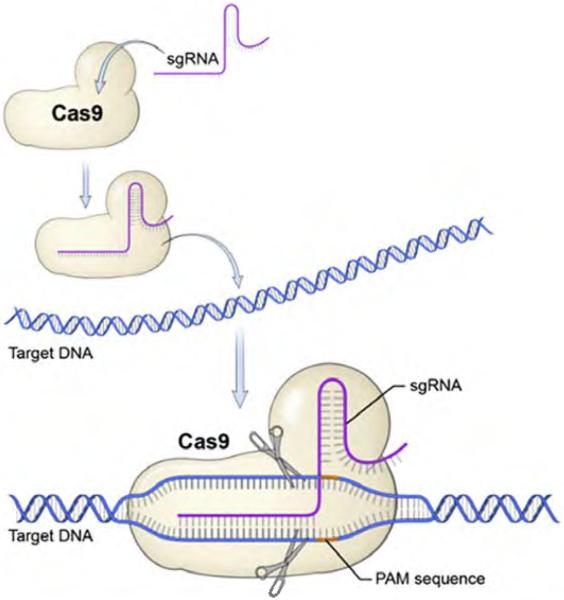
CRISPR genome editing relies on Cas9 and a single guide RNA (sgRNA). sgRNA is a custom, synthetic, single-stranded RNA that contains an 18–25-nucleotide sequence specific to the target DNA, followed by a scaffold sequence that complexes with Cas9. Hybridization of sgRNA-Cas9 complex to the targeted locus creates a conformational change that activates Cas9 nuclease activity, resulting in a DNA DSB (Figure 1). CRISPR-Cas9 is a powerful tool for genome editing because the sgRNA can be quickly designed and synthesized to target specific genomic sites. Another advantage of the sgRNA targeting mechanism is that multiple genes can be targeted simultaneously. This strategy has been used to perform genome-wide knockout screens and identify mutations involved in complex biological processes (Shalem, 2013).
Figure 1. CRISPR-Cas9–sgRNA genome targeting.
sgRNA complexes with Cas9 nuclease to hone in on the targeted genomic site containing an adjacent PAM sequence. Nucleotide hybridization of sgRNA-Cas9 complex to targeted loci creates a conformational change that activates Cas9 nuclease activity, resulting in DNA double-strand breaks. Adapted with permission from Addgene (2016). Cas9, CRISPR-associated protein 9; CRISPR, clustered regularly interspaced palindromic repeats; PAM, protospacer adjacent motif; sgRNA, single guide RNA.
STRATEGIES OF GENOME EDITING
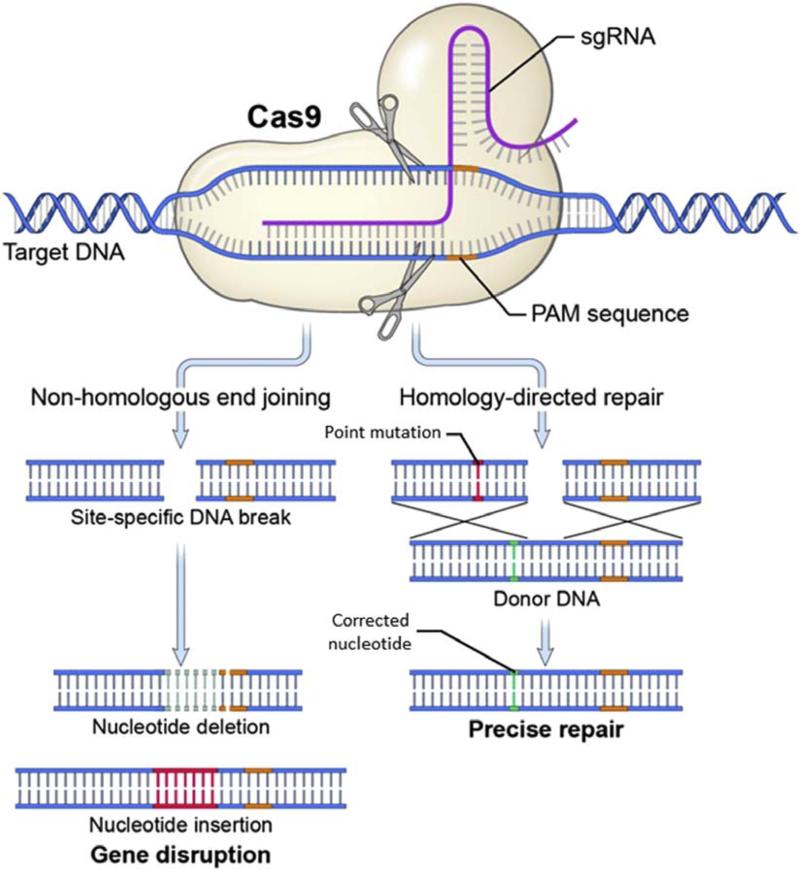
Once a DSB is made, investigators rely on two DNA DSB-repair mechanisms innate within a cell to achieve different editing outcomes—nonhomologous end joining (NHEJ) and homology-directed repair (HDR) (Figure 2). NHEJ is an inherently mutagenic process used to permanently knock out expression of a gene. During NHEJ DSB repair, the cell ligates DNA blunt ends at the DSB site. In this process, random nucleotide insertions and deletions occur, resulting in mutations and gene disruption. From a therapeutic perspective, NHEJ may be applied to monogenic diseases for which gene knockout is beneficial, such as a dominant-negative mutation, in which the mutant gene product interferes with normal cellular function.
Figure 2. CRISPR-induced NHEJ and HDR.
Upon Cas9-induced DNA DSB, the cell repairs the DSB by either NHEJ or HDR. In NHEJ, random nucleotide insertions and deletions occur as the cell ligates the DNA DSB, resulting in gene disruption. In HDR, the DSB is repaired using an externally supplied homologous DNA as a template for copying. The nucleotide sequence of the donor template is copied into the targeted site, resulting in a directed precise repair. Adapted with permission from Elsevier (Savić and Schwank, 2016). Cas9, CRISPR-associated protein 9; CRISPR, clustered regularly interspaced palindromic repeats; DSB, double-stranded break; HDR, homology-directed repair; NHEJ, nonhomologous end-joining; PAM, protospacer adjacent motif; sgRNA, single guide RNA.
Alternatively, HDR is a genome-editing strategy used to precisely correct a mutation through the use of an externally supplied homologous donor template. In a process akin to homologous recombination, the DSB is repaired using synthetic homologous DNA as the template for copying. Generally, the donor template is delivered by traditional nuclear transfection techniques or viral delivery (Osborn, 2013; Sebastiano, 2014). The nucleotide sequence of the donor template is copied at the site of the DSB. HDR allows investigators to direct the repair of a mutated gene by precisely editing the targeted nucleotide sequence. From a therapeutic perspective, HDR may be applied to monogenic diseases caused by point mutations. However, HDR is limited by its lower efficiency compared with NHEJ. In general, the genome-editing efficiency of HDR is 0.5–20.0%, whereas NHEJ can reach efficiencies of 20–60% (Maruyama, 2015). The lower efficiency is due in part to the fact that HDR relies on innate cellular proteins involved in homologous recombination expressed in the S and G2 phases of the cell cycle (Maruyama, 2015).
APPLICATION OF GENOME EDITING IN DERMATOLOGICAL RESEARCH
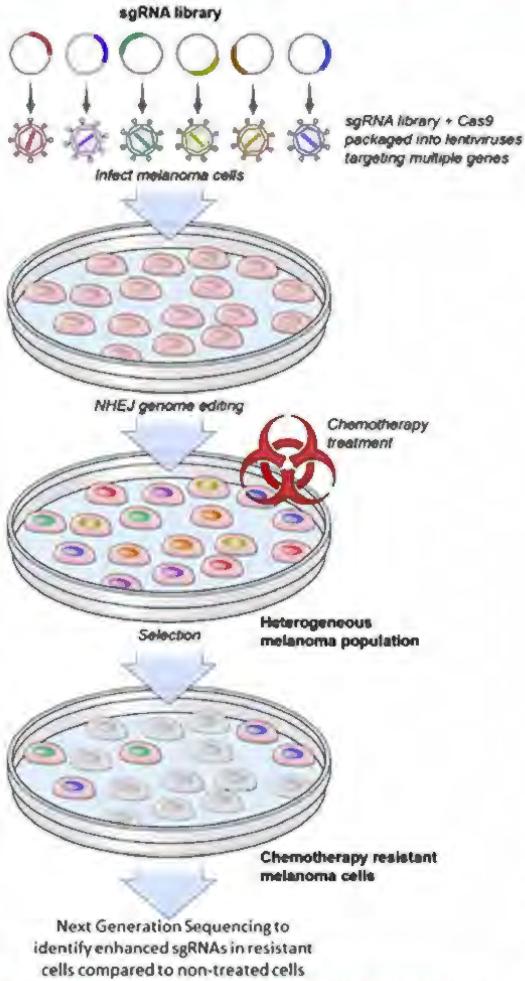
Genome editing holds great promise as a tool for understanding dermatological diseases. For example, Shalem et al. (2013) performed a genome-wide screen on a human melanoma cell line to identify mutations that confer resistance to vemurafenib, a therapeutic RAF inhibitor (Shalem, 2013). The CRISPR-mediated genome-wide screen uses a library of thousands of unique sgRNAs that target over 18,000 different genes. The sgRNA library, when delivered with a Cas9 nuclease to a melanoma cell line via lentiviruses, allowed investigators to knock out the targeted genes simultaneously via NHEJ genome editing. This experiment resulted in a heterogeneous mutant melanoma cell population that, when exposed to the RAF inhibitor PLX-4032, caused broad cell death with the exception of a few cells that had acquired mutations rendering them resistant to PLX-4032. Comparison of deep-sequencing results of the initial melanoma cell line to the PLX-4032–resistant population validated two known mutations and identified four novel mutations responsible for the acquired resistance (Figure 3).
Figure 3. CRISPR-mediated genome-wide melanoma knockout screen.
The CRISPR-mediated genome-wide screen uses a library of thousands of unique sgRNAs that target over 18,000 different genes. The sgRNA library, when delivered with a Cas9 nuclease to a melanoma cell line via lentiviruses, results in a heterogeneous mutant melanoma cell population via NHEJ genome editing. When exposed to the chemotherapy agent PLX-4032, broad cell death occurred with the exception of a small cluster of cells that had acquired mutations rendering them resistant to PLX-4032, Comparing deep-sequencing results of the initial melanoma cell line to the PLX-4032–resistant population validated two known mutations and identified four novel loss-of-function mutations responsible for the acquired resistance. Cas9, CRISPR-associated protein 9; CRISPR, clustered regularly interspaced palindromic repeats; NHEJ, nonhomologous end-joining; sgRNA, single guide RNA.
Another exciting development has been the creation of CRISPR-Cas9 derivatives for transcriptional and epigenetic modulation. By attaching transcriptional modulators onto a nuclease-defective Cas9, investigators can repress or activate transcription on a site-specific level (Thakore, 2016). Konermann et al. (2015) developed a transcription-activating Cas9 and applied it in a genome-wide screen to identify up-regulated genes involved in the acquired resistance to PLX-4032. These results helped investigators understand the mechanism of PLX-4032 action and provided leads to further understand why patients acquire resistance to melanoma therapies.
Genome editing can also be used for disease modeling. CRISPR technology allows investigators to develop transgenic mice that accurately model human diseases faster than ever before. In this regard, the technology will assist in the efforts of the International Mouse Phenotyping Consortium to create a functional catalog of the mammalian genome, which will likely elucidate genes responsible for skin abnormalities. Additionally, functions of epidermal genes may be studied by knocking out functional alleles through NHEJ or repair of gene mutations through HDR in three-dimensional skin culture models (Sebastiano, 2014).
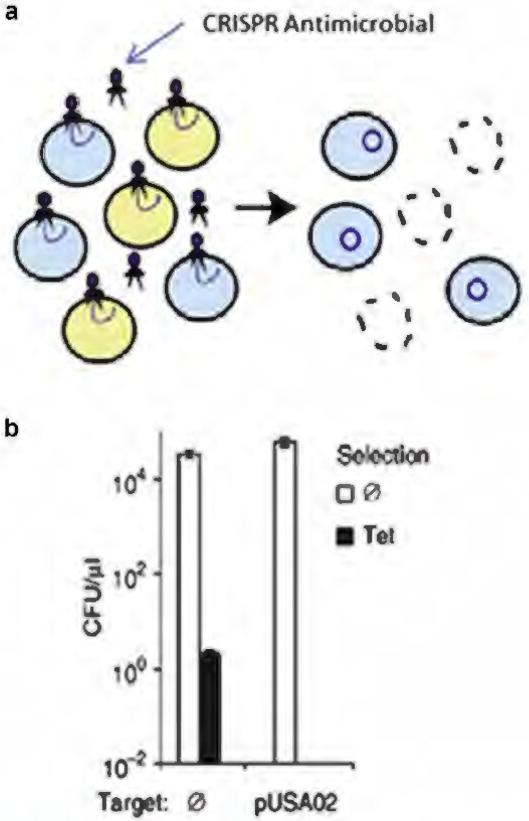
CRISPR has also been used as a selective antimicrobial agent. Acquisition of antibiotic-resistant plasmids within Staphylococcus aureus has led to an increased incidence of methicillin-resistant S. aureus infections, which are difficult to treat because of their virulence and resistance to common antibiotics. Bikard et al. (2014) developed a nonreplicative bacteriophage containing Cas9 and sgRNAs targeting antibiotic-resistant genes in staphylococci. The investigators showed that their CRISPR-Cas9 antimicrobial virus infected antibiotic-resistant staphylococci and disrupted the antibiotic-resistant genes via NHEJ genome editing, while simultaneously immunizing nonresistant staphylococci to prevent further transfer of antibiotic-resistant plasmids (Figure 4). Additionally, they showed that the CRISPR-Cas9 antimicrobial module functioned in vivo to kill antibiotic resistant S. aureus in a mouse skin colonization model. This innovative method may have clinical application as a selective antimicrobial agent to prevent methicillin-resistant S. aureus infections and the further spread of antibiotic-resistant genes.
Figure 4. CRISPR-mediated antimicrobial.
(a) CRISPR treatment of a mixed Staphylococcus aureus population of a nonresistant colony (blue) and an antibiotic-resistant colony (yellow) results in killing of the targeted strain and delivery of an immunizing phagemid to the rest of the population, (b) CRISPR-mediated immunization against tetracycline resistance. In pUSA02 condition, bacteria was treated with a CRISPR antimicrobial targeting the tetracycline resistant plasmid, pUSA02. After tetracycline-resistant plasmid transfer, bacteria subjected to CRISPR (pUSA02) were vulnerable to tetracycline and did not grow colonies, indicating NHEJ knockout of tetracycline-resistant plasmid. Adapted with permission from Macmillan Publishers Ltd: Nature Biotechnology (Bikard et al. 2014). Ø, nontreated; CFU, colony forming units; CRISPR, clustered regularly interspaced palindromic repeats; NHEJ, nonhomologous end-joining; pUSA02, plasmid conferring tetracycline resistance; Tet, tetracycline.
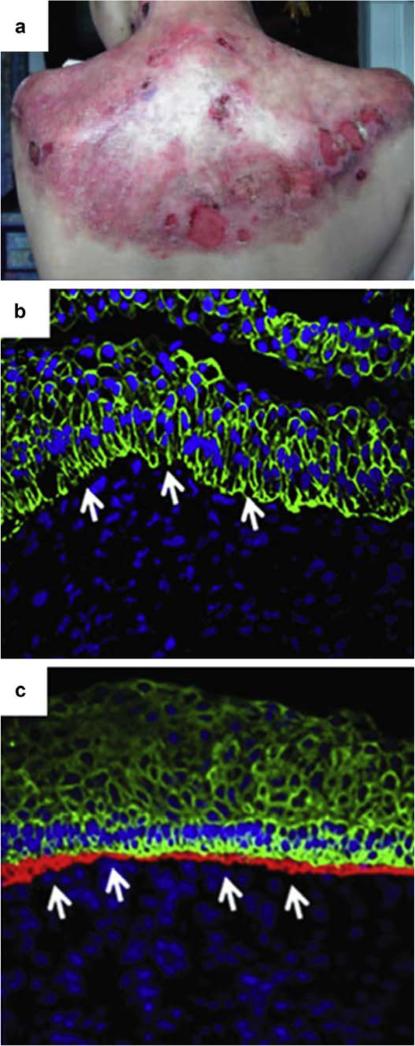
Another therapeutic application of genome editing is to produce genetically corrected epidermal cells for patients suffering from inherited skin disorders. One disease that has shown promise to be amenable to HDR genome editing is recessive dystrophic epidermolysis bullosa (RDEB). RDEB is a blistering skin disorder caused by a mutation in the COL7A1 gene, which results in a loss of wild-type VII collagen (COL7A) in the epidermal-dermal junction. RDEB is often fatal because it results in severe blistering, subsequent limb deformities, and squamous cell carcinoma (Figure 5a). Restoration of functional COL7A at the epidermal-dermal junction is probably the only therapeutic hope for a disease that is invariably lethal.
Figure 5. TALEN-mediated genome editing for correction of RDEB.
(a) RDEB patient suffering from chronic blistering. Panel a reprinted with permission from Elsevier (Petrof et al., 2015). (b) Confocal image of the epidermal-dermal junction in noncorrected RDEB cells, in which collagen VIIA is not detected. (c) Confocal image of the epidermal-dermal junction in gene-corrected RDEB cells, in which collagen VIIA is detected in the red channel. Panels b and c reprinted with permission from Macmillan Publishers Ltd: Molecular Therapy (Osborn et al., 2013). RDEB, recessive dystrophic epidermolysis bullosa; TALEN, transcription activator-like effector nuclease.
Osborn et al. (2013) developed a TALEN-based method to produce genetically corrected keratinocyte-like cells from a patient suffering from RDEB. The method involved first correcting patient-derived RDEB fibroblasts via nuclear delivery of plasmids encoding TALENs targeting the COL7A1 mutation and the HDR-inducing homologous donor template. Gene-corrected fibroblasts were then reprogrammed into induced pluripotent stem cells (iPSC) and subcutaneously injected into murine recipients to form teratomas. The genetically corrected teratomas generated a skin component with a functional epidermal-dermal junction that contained COL7A (Figure 5b and c), suggesting that genome editing could be used to treat genetic skin disorders with gene-corrected patient-derived tissue.
CHALLENGES AND FUTURE DIRECTIONS
Taken together, preclinical studies show the potential applications of genome editing for treating genetic blistering diseases and for use as an antimicrobial agent. However, the challenge remains in translating these laboratory achievements into the clinic. Ideally, a genome-editing skin therapy should not require reprogramming the primary cell target, because iPSCs can acquire mutations during reprogramming, and undifferentiated iPSCs could form tumors if transplanted into patients (Dinella, 2014). These concerns led Sebastiano et al. (2014) to develop a protocol involving whole-genome sequencing of COL7A1-corrected RDEB iPSC cells to select for clones that had a minimal mutational burden. Their strategy allowed them to generate COL7A1-corrected iPSC-derived epidermal grafts that could, in theory, be transplanted back onto an RDEB patient as an autologous skin graft (Sebastiano et al., 2014).
Limitations of CRISPR genome editing and its application in investigative dermatology include genomic target restrictions, variable sgRNA efficiencies, delivering genome-editing modules to primary keratinocytes, and off-target effects. The design of sgRNA for gene targeting is dependent on the presence of a particular protospacer adjacent motif (PAM). The PAM sequence is a Cas9-specific nucleotide motif that must be adjacent to the target site. The most frequently used Cas9 PAM site, NGG, is present in the genome every eight nucleotides (Gasiunas, 2013). However, investigators are discovering Cas9 variants that use alternate PAM sequences, which is broadening the targeting scope of CRISPR (Kleinstiver et al., 2015). Another concern is the growing number of sequence-specific features that have been shown to alter sgRNA efficiency (Peng, 2015). Delivery of genome-editing modules can be problematic, because primary keratinocytes can be difficult to transfect and have a finite lifespan. An alternative approach involves reprogramming primary epidermal cells into iPSCs, which can be genetically modified and cultured indefinitely and can be subsequently differentiated into genome-edited keratinocytes (Osborn et al., 2013; Sebastiano et al., 2014). Additionally, as is the case with all genome-editing nucleases, there are off-target effects. The genome-editing modules may cut a nontargeted site that shares sufficient sequence similarity with the targeted locus, potentially resulting in a mutation.
Because limiting off-target effects is an essential prerequisite for the application of genome editing in the clinic, strategies to overcome this challenge are being rapidly developed. For example, one method involves the use of recombinogenic adeno-associated viral vectors to mediate HDR. In this approach, the adeno-associated viral vectors contain a corrected copy of the mutated gene. Upon infection, the corrected copy replaces the mutated gene via homologous recombination. The advantage of this approach is that it does not rely on genome-editing nucleases, which can enact mutagenic DSBs at off-target sites (Mavilio, 2014; Melo, 2014; Sebastiano, 2014). Another strategy to reduce off-target effects involves a pair of Cas9-nickases that create single-stranded cuts on opposite strands, resulting in a DNA DSB. Because off-target hits result in single-stranded breaks, as opposed to a DSB, and single-stranded breaks are corrected by a fairly nonmutagenic repair pathway, off-target mutations are greatly reduced using these Cas9-nickases (Davis, 2011). Furthermore, an innovative development has been the creation of a high-fidelity Cas9 variant designed to reduce off-target effects (Kleinstiver, 2016). The high-fidelity variant retained on-target efficiencies comparable to wild-type Cas9. More importantly, most off-target mutations associated with wild-type Cas9 were not detected. These examples highlight the amazingly rapid innovation surrounding the development of CRISPR and genome editing.
In conclusion, although challenges of CRISPR remain, innovative developments will further improve this technology. This is an exciting time for the field of genome editing. We believe the technology has a bright future as a therapeutic and scientific tool to treat and study dermatologic pathologies.
Supplementary Material
CME Activity Dates: Release Date: August 16, 2016
Expiration Date: August 15, 2017
Estimated Time to Complete: 1 hour
Planning Committee/Speaker Disclosure: All speakers, planning committee members, CME committee members and staff involved with this activity as content validation reviewers have no financial relationship(s) with commercial interests to disclose relative to the content of this CME activity.
Commercial Support Acknowledgment: This CME activity is supported by an educational grant from Lilly USA, LLC.
Description: This article, designed for dermatologists, residents, fellows, and related healthcare providers, seeks to reduce the growing divide between dermatology clinical practice and the basic science/current research methodologies on which many diagnostic and therapeutic advances are built.
Objectives: At the conclusion of this activity, learners should be better able to:
Recognize the newest techniques in biomedical research.
Describe how these techniques can be utilized and their limitations.
Describe the potential impact of these techniques.
CME Accreditation and Credit Designation: This activity has been planned and implemented in accordance with the accreditation requirements and policies of the Accreditation Council for Continuing Medical Education through the joint providership of William Beaumont Hospital and the Society for Investigative Dermatology. William Beaumont Hospital is accredited by the ACCME to provide continuing medical education for physicians.
William Beaumont Hospital designates this enduring material for a maximum of 1.0 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
Method of Physician Participation in Learning Process: The content can be read from the Journal of Investigative Dermatology website: http:/www.jidonline.ong/current. Tests for CME credits may only be submitted online at https://beaumont.cloud-cme.com/RTMS-Sept16 – click ‘CME on Demand’ and locate the article to complete the test. Fax or other copies will not be accepted. To receive credits, learners must review the CME accreditation information; view the entire article, complete the post-test with a minimum performance level of 60%; and complete the online evaluation form in order to claim CME credit. The CME credit code for this activity is: 21310. For questions about CME credit cme@beaumont.edu.
ADVANTAGES.
CRISPR-Cas9 genome editing allows for precise gene repair or permanent gene knockout.
The CRISPR sgRNA honing mechanism makes genome-editing technology much easier to develop and produce compared with traditional designer nucleases.
CRISPR sgRNAs allow for multiple genes to be targeted simultaneously.
CRISPR-Cas9 genome editing can be used for genome-wide screens to identify genes and mutations responsible for complex biological processes.
Genome editing allows for the development of transgenic animal disease models.
Genome-editing technologies show promise as a therapeutic tool–for example, for treating genetic blistering diseases and for use as a selective antimicrobial agent.
LIMITATIONS.
Genome-editing modules may cut at nontargeted sites, resulting in off-target mutations with potentially adverse consequences to the host.
sgRNA design is restricted to gene targets containing a protospacer adjacent motif (PAM) sequence.
sgRNA efficiencies can be variable.
Homology-directed repair efficiency can be low.
Current genome-editing studies for treatment of recessive dystrophic epidermolysis bullosa (RDEB) rely on induced pluripotent stem cells (iPSCs), which can acquire mutations during reprogramming and form tumors if transplanted into patients.
MULTIPLE CHOICE QUESTIONS.
- A major advantage of CRISPR over traditional programmable designer nucleases is
- CRISPR's reliance on sgRNA as the targeting mechanism.
- CRISPR's higher gene-targeting efficiency.
- CRISPR's lower off-targeting efficiency.
- HDR is less efficient than NHEJ because
- HDR is inherently mutagenic and bad for the host.
- HDR relies on endogenous cellular proteins involved in homologous recombination.
- HDR relies on particular sequence-specific motifs that reduce its frequency, whereas NHEJ does not.
- Off-target mutations are a result of
- mutant nuclease variants that randomly cut and mutate the genome.
- flawed sgRNA design, which hyperactivates Cas9 nuclease activity to randomly cut and mutate nontargeted sites.
- nontargeted sites sharing sequence homology with the targeted locus.
- CRISPR-mediated genome-wide screens rely on ____ to target ____.
- sgRNA libraries; multiple genes simultaneously
- lentiviruses; gene mutations involved in the acquired resistance to melanoma chemotherapy agents
- Cas9; and kill melanoma cells
- Cas9 is guided to the targeted locus by
- endogenous genomic palindromic adjacent motifs.
- sgRNA.
- TALENs.
ACKNOWLEDGMENTS
We acknowledge Thomas Friedman, Jizhong Zou, Joan Guitart, and Jaehyuk Choi for critiques of this manuscript. This work was supported by the Intramural Research Program at the National Institute on Deafness and Other Communication Disorders (NIDCD).
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is linked to this paper. Teaching slides are available as supplementary material.
REFERENCES
- Addgene. [18 June 2016];Overview of CRISPR/Cas9. 2016 https://www.addgene.org/crispr/guide/
- Bikard D, Euler C, Jiang W, Nussenzweig P, Goldberg C, Duportet X, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–50. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Maizels N. DNA nicks promote efficient and safe targeted gene correction. PLoS One. 2011;6:e23981. doi: 10.1371/journal.pone.0023981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinella J, Koster M, Koch P. Use of induced pluripotent stem cells in dermatological research. J Invest Dermatol. 2014;134:1–5. doi: 10.1038/jid.2014.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Siksnys V. RNA-dependent DNA endonuclease Cas9 of the CRISPR system: holy grail of genome editing? Trends Microbiol. 2013;21:562–7. doi: 10.1016/j.tim.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Kleinstiver B, Pattanayak V, Prew M, Tsai S, Nguyen N, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–5. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B, Prew M, Tsai S, Topkar V, Nguyen N, Zheng Z, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–5. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham M, Trevino A, Joung J, Abudayyeh O, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–8. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Dougan S, Truttmann M, Bilate A, Ingram J, Ploegh H. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–42. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavilio F. Repairing without cutting: a safer alternative to gene correction? Mol Ther. 2014;22:690–1. doi: 10.1038/mt.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo S, Lisowski L, Bashkirova E, Zhen H, Chu K, Keene D, et al. Somatic correction of junctional epidermolysis bullosa by a highly recombino-genic AAV variant. Mol Ther. 2014;22:725–33. doi: 10.1038/mt.2013.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn M, Starker C, McElroy A, Webber B, Riddle M, Xia L, et al. TALEN-based gene correction for epidermolysis bullosa. Mol Ther. 2013;21:1151–9. doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, Lin G, Li J. Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J. 2015;283:1218–31. doi: 10.1111/febs.13586. [DOI] [PubMed] [Google Scholar]
- Petrof G, Lwin SM, Martinez-Queipo M, Abdul-Wahab A, Tso S, Mellerio JE, et al. Potential of systemic allogeneic mesenchymal stromal cell therapy for children with recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2015;135:2319–21. doi: 10.1038/jid.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savić N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl Res. 2016;168:15–21. doi: 10.1016/j.trsl.2015.09.008. [DOI] [PubMed] [Google Scholar]
- Sebastiano V, Zhen H, Haddad B, Bashkirova E, Melo SP, et al. Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci Transl Med. 2014;6:264ra163. doi: 10.1126/scitranslmed.3009540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana N, Hartenian E, Shi X, Scott D, Mikkelsen T, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2013;343:84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas P, Stein D, Tang W, Frank I, Wang S, Lee G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. New Engl J Med. 2014;370:901–10. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore P, Black J, Hilton I, Gersbach C. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods. 2016;13:127–37. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis J. Making the cut. Science. 2015;350:1456–7. doi: 10.1126/science.350.6267.1456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.