Abstract
 α-l-Threofuranosyl nucleoside triphosphates (tNTPs) are tetrafuranose nucleoside derivatives
and potential progenitors of present-day β-d-2‘-deoxyribofuranosyl nucleoside triphosphates (dNTPs).
Therminator DNA polymerase, a variant of the 9°N DNA polymerase, is an efficient DNA-directed threosyl
nucleic acid (TNA) polymerase. Here we report a detailed kinetic comparison of Therminator-catalyzed
TNA and DNA syntheses. We examined the rate of single-nucleotide incorporation for all four tNTPs and
dNTPs from a DNA primer−template complex and carried out parallel experiments with a chimeric DNA−TNA primer−DNA template containing five TNA residues at the primer 3‘-terminus. Remarkably, no drop
in the rate of TNA incorporation was observed in comparing the DNA−TNA primer to the all-DNA primer,
suggesting that few primer-enzyme contacts are lost with a TNA primer. Moreover, comparison of the
catalytic efficiency of TNA synthesis relative to DNA synthesis at the downstream positions reveals a
difference of no greater than 5-fold in favor of the natural DNA substrate. This disparity becomes negligible
when the TNA synthesis reaction mixture is supplemented with 1.25 mM MnCl2. These results indicate
that Therminator DNA polymerase can recognize both a TNA primer and tNTP substrates and is an effective
catalyst of TNA polymerization despite changes in the geometry of the reactants.
α-l-Threofuranosyl nucleoside triphosphates (tNTPs) are tetrafuranose nucleoside derivatives
and potential progenitors of present-day β-d-2‘-deoxyribofuranosyl nucleoside triphosphates (dNTPs).
Therminator DNA polymerase, a variant of the 9°N DNA polymerase, is an efficient DNA-directed threosyl
nucleic acid (TNA) polymerase. Here we report a detailed kinetic comparison of Therminator-catalyzed
TNA and DNA syntheses. We examined the rate of single-nucleotide incorporation for all four tNTPs and
dNTPs from a DNA primer−template complex and carried out parallel experiments with a chimeric DNA−TNA primer−DNA template containing five TNA residues at the primer 3‘-terminus. Remarkably, no drop
in the rate of TNA incorporation was observed in comparing the DNA−TNA primer to the all-DNA primer,
suggesting that few primer-enzyme contacts are lost with a TNA primer. Moreover, comparison of the
catalytic efficiency of TNA synthesis relative to DNA synthesis at the downstream positions reveals a
difference of no greater than 5-fold in favor of the natural DNA substrate. This disparity becomes negligible
when the TNA synthesis reaction mixture is supplemented with 1.25 mM MnCl2. These results indicate
that Therminator DNA polymerase can recognize both a TNA primer and tNTP substrates and is an effective
catalyst of TNA polymerization despite changes in the geometry of the reactants.
Introduction
Living systems have evolved to use pentofuranoses of both the ribo (RNA) and 2-deoxyribo (DNA) configurations for the backbones of polymers whose function is to faithfully store and transfer biological information necessary for normal cellular function. Although the natural selection of these two carbohydrate backbones has resulted in a successful evolutionary pathway, earlier life forms may have been based on alternative, and perhaps simpler, backbone systems before converging on pentofuranoses. One such possibility is a tetrafuranose-based backbone instead of one constructed of pentofuranoses. Of the possible tetrafuranose building blocks, the α-l-threofuranosyl nucleosides and the corresponding α-l-threofuranosyl nucleic acids (TNAs) provide a geometry most similar to the β-d-ribofuranosyl nucleosides and RNA (DNA). The geometry of a simpler progenitor need not necessarily be similar to that of RNA, but evolution might require an intermediate stage of development where both polymers are present at the same time. In that case, the ability to transfer information from the simpler polymer (e.g., one based upon α-l-threofuranose) to one at the next level of development (e.g., RNA) becomes more critical.
In addition to information transfer, primitive life forms would require these earlier polymers to also have functional characteristics including receptor−ligand binding as well as catalysis. We plan to test the idea that TNA can carry out such functional tasks by attempting to isolate functional TNA molecules in the laboratory through iterative rounds of in vitro selection and amplification.428255b00001,428255b00002 These techniques have been used to evolve functional RNA and DNA molecules capable of binding (aptamers) and/or catalysis (ribozymes). To extend this approach to structurally diverse nucleic acids such as TNA,428255b00003 it is necessary to show that the TNAs can be prepared effectively using DNA or RNA templates and wild-type or mutant (if necessary) DNA (or RNA) polymerases. TNA is capable of Watson−Crick base pairing with complementary DNA, RNA, and TNA oligonucleotides, despite having a backbone unit one atom shorter than that of DNA or RNA. To examine the functional characteristics of TNAs, we need to determine whether (i) TNA aptamers and catalysts can be selected from random sequence libraries and (ii) how well the functional properties of TNA compare with RNA.428255b00004 Results from these experiments will provide insight into the fitness of TNA as a potential genetic polymer.
The ability to enzymatically synthesize TNA is a prerequisite for in vitro selection and directed evolution experiments. Several studies have examined the effect of phosphate substitutions on polymerase-mediated nucleic acid replication. The replacement of nonbridging oxygen atoms with sulfur,428255b00005 methyl,428255b00006 or BH3 groups428255b00007 is compatible with limited polymerase-mediated primer-extension synthesis, while acyclic and 4‘-modified sugar residues generally act as chain terminators.428255b00008,428255b00009 The poor enzymatic recognition of such modified nucleoside triphosphates highlights the difficulties involved in using natural enzyme polymerases to synthesize unnatural polynucleotides.428255b00010
To evaluate the potential of tNTPs as enzyme substrates for the preparation of TNAs, we screened a wide variety of naturally occurring DNA polymerases for the ability to recognize TNA either in the template or as a nucleoside triphosphate. From those studies, we identified several DNA polymerases capable of faithfully synthesizing short sequences of DNA on a TNA template,428255b00011 as well as DNA polymerases able to catalyze the synthesis of limited stretches of TNA on a DNA template.428255b00012 Unfortunately, these DNA polymerases are far too inefficient to be used for the iterative in vitro selection and amplification experiments. For example, DNA synthesis on a TNA template by Bst Pol I and MMLV reverse transcriptase required at least 60 min to obtain full-length (n + 9) extension. By comparison, TNA synthesis on a DNA template is even more challenging, requiring a 24-h incubation with Deep Vent (exo-) before the insertion of three contiguous tTTP residues occurred. Herdewijn has reported a similar finding for the incorporation of tTTP by Vent (exo-), suggesting that tNTPs are not easily accepted by many DNA polymerases.428255b00013 In an effort to locate effective polymerization catalysts, we have continued to screen DNA polymerases for improved TNA polymerase activity. We have recently identified a mutant archeal family B DNA polymerase commonly referred to as Therminator DNA polymerase428255b00014 as a reasonably efficient DNA-dependent TNA polymerase. This polymerase was previously shown to exhibit decreased sugar specificity and more efficient incorporation of acyclic chain terminating nucleotides.428255b00014 Here we report a detailed kinetic analysis of Therminator-catalyzed TNA synthesis using a DNA template.
Materials and Methods
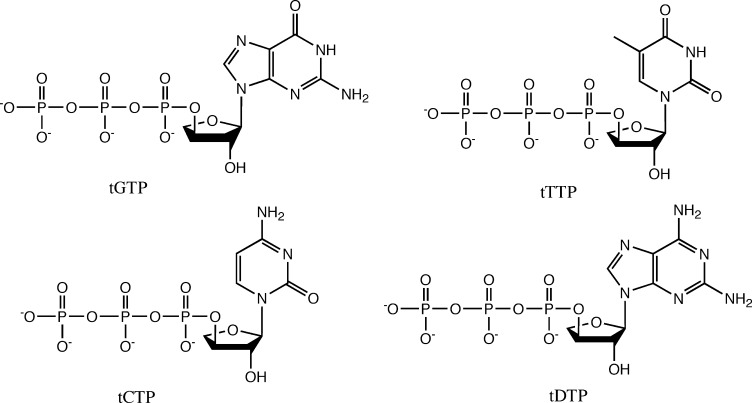
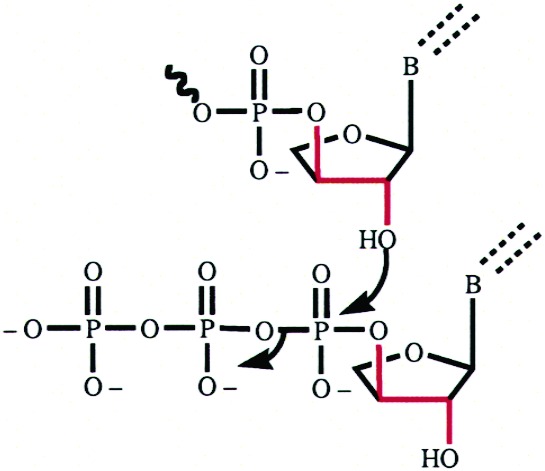
Oligonucleotides and Triphosphate Derivatives. Solid-phase oligonucleotide syntheses of DNA primers and templates were performed on an automated ABI 394 DNA synthesizer (Applied Biosystems, Foster City, CA) using standard β-cyanoethyl phosphoramidite chemistry. DNA synthesis reagents and phosphoramidites were purchased from Glen Research (Sterling, VA). Oligonucleotides were deprotected in concentrated NH4OH (55 °C, 1 h), lyophilized to dryness, and purified by preparative denaturing polyacrylamide gel electrophoresis. Oligonucleotides were quantified by absorbance at 260 nm using standard molar extinction coefficients. TNA triphosphate derivatives (Figure 1) were synthesized in ≥95% purity as described previously by Zou et al.428255b00015
Figure 1.
Structures of the four tNTP residues used in kinetic assays.
Enzyme Screen. DNA primers were labeled at the 5‘-terminus by incubating in the presence of [γ-32P]-ATP (Amersham, Billireca, MA) and T4 polynucleotide kinase (New England Biolabs, Beverly, MA) for 1 h at 37 °C. Labeled primer was annealed to the DNA template in ThermoPol buffer [20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, and 20 mM MgSO4] by heating to 95 °C for 3 min and cooling to room temperature for 15 min. Polymerase reactions were started by adding DNA polymerase to a premixed solution containing DNA primer−template complex, tNTP substrates (tTTP, tGTP, and tDTP), BSA, and DTT in thermopol buffer at 55 °C. Reaction progress was monitored over time by 20% denaturing gel electrophoresis. At designated time points, aliquots (1 μL) were removed and combined with 19 μL of stop buffer [8 M urea/ 1X-TBE buffer (89 mM Tris/89 mM boric acid/2 mM EDTA)/20 mM EDTA (pH = 8.0), 0.05% xylene cyanol and bromophenol blue]. Polymerase reactions contained primer−template complex (750 nM), tNTPs (20 μM each), BSA (1 μg), DTT (1 mM), and 0.25 μL of DNA polymerase in a final reaction volume of 10 μL. For each DNA polymerase, a volume of 0.25 μL is equivalent to 0.5 units for DNA polymerases 9°N, Therminator, Deep Vent (exo-), and Vent (exo-); 0.75 units for Y409V/A485L mutant 9°N DNA polymerase; and 3.0 units for Y409V mutant 9°N polymerase. All DNA polymerases reported in this study are commercially available with the exception of mutant 9°N DNA polymerase Y409V/A485L and Y409V, which were generously provided by W. Jack and A. Gardner at New England Biolabs and mutant versions of Taq DNA polymerase recently identified and described by Holliger.428255b00019 Taq DNA polymerase mutants were not used beyond the initial screen due to poor tNTP substrate recognition. Unit definition: one unit catalyzes the incorporation of 10 nmol of dNTP into an acid precipitable material in 30 min at 75 °C.
MALDI-TOF Mass Spectrometry. The products of each single-nucleotide addition reaction were analyzed by MALDI-TOF mass spectrometry analysis. For each reaction, 10 μM primer−template were annealed in 1.1X Thermopol buffer by heating to 95 °C for 3 min followed by incubation at room temperature for 5 min. We added 1 mM MnCl2 (tNTP reactions only), 20 μM appropriate NTP (except tTTP reaction, which contained 250 μM tTTP), and 0.25 units of Therminator to a final volume of 10 μL. Reactions were incubated at 55 °C. dNTP reactions were stopped after 5 min, and tNTP reactions were stopped after 10−120 min by addition of 20 mM EDTA. Reactions were precipitated in ethanol/2 M NH4AC/0.25 μL glycogen solution and purified by C18 ZipTip (Millipore). Samples were eluted with 1 μL of matrix solution consisting of 3-hydroxypicolinic acid in 50% acetonitrile/diammonium citrate in water in a ratio of 9:1. Eluates were directly spotted onto a gold-coated MALDI-TOF plate and analyzed in negative mode on a Voyager MALDI-TOF mass spectrometer (Applied Biosystems). Spectra are an average of >150 scans.
Steady-State Kinetics. Kinetic measurements were carried out as described for standing-start single-nucleotide insertions.428255b00016 The final DNA duplex concentration was 250 nM in all cases. The chimeric DNA−TNA primer used to measure the kinetics of tNTP extension from the TNA terminus of a DNA−TNA primer was constructed by template-directed synthesis using Therminator DNA polymerase to extend a DNA primer with five tNTP residues (see above). Polymerization reactions were initiated by adding 10 μL of 2 × dNTP or tNTP solution (0.01−10 μM) to an equal volume of the reaction mixture containing primer−template complex, 20 mM Tris-HCl, 10 mM KCl, 10 mM (NH4)2SO4, 20 mM MgSO4, 0.1% Triton X-100, 0.25 μg/μL BSA, 100 μM DTT, and either 0.05 units of Therminator DNA polymerase (final concentration 9.3 nM) (New England Biolabs, 2 u/μL) or 0.1 units of Deep Vent (exo-) DNA polymerase (final concentration 1.82 nM) (New England Biolabs, 2 u/μL). Some reactions were also supplemented with 1.25 mM MnCl2 (freshly prepared). The amount of dNTP or tNTP used and reaction time (1−3 min) were adjusted to limit polymerization to 20% or less. Polymerization reactions were incubated at 55 °C and quenched with 10 μL of stop buffer [8 M urea/TBE/20 mM EDTA (pH = 8.0), 0.05% xylene cyanol and bromophenol blue]. Extents of reaction were determined by 20% denaturing polyacrylamide gel electrophoresis and quantifying the resulting bands by phosphorimaging (Bio-Rad Molecular Imager FX, Hercules, CA). Reaction velocities were calculated as the extent of reaction divided by reaction time. Kinetic parameters for KM and Vmax were determined by linear regression analysis of a Hanes−Woolf plot with reported values being the average of three or more independent experiments (see below).
Kinetic Measurements. We determined the kinetic parameters (Vmax and Km) for each enzyme using the “steady-state method” for assessing DNA polymerase single-nucleotide incorporation efficiency.428255b00016 This is a kinetic assay in which the polymerase concentration is far below the primer−template concentration and the reaction time is short. By varying the concentrations of dNTPs, kinetic parameters for Km and Vmax can be obtained from a Hanes−Woolf plot of [dNTP]/v versus [dNTP] through a linear least-squares fit of the data. The intercept and slope of the resulting plot correspond to the values for Km/Vmax and 1/Vmax of the reaction process, respectively. In this case, Km is defined as the [dNTP] when the reaction velocity is half of the maximum velocity under saturating substrate conditions and Vmax is the maximum velocity as determined by a hyperbolic function of relative velocity verses [dNTP]. Thus, this assay provides a straightforward method for determining the enzymatic efficiency (Vmax/Km) for a given enzyme−substrate pair. The ratio of the Vmax/Km values for different substrates provides a quantitative measurement describing the selectivity of a given polymerase for dNTP versus tNTP nucleotides. These selectivity values are independent of enzyme activity and thus allow the comparison of different enzymes.
Results and Discussion
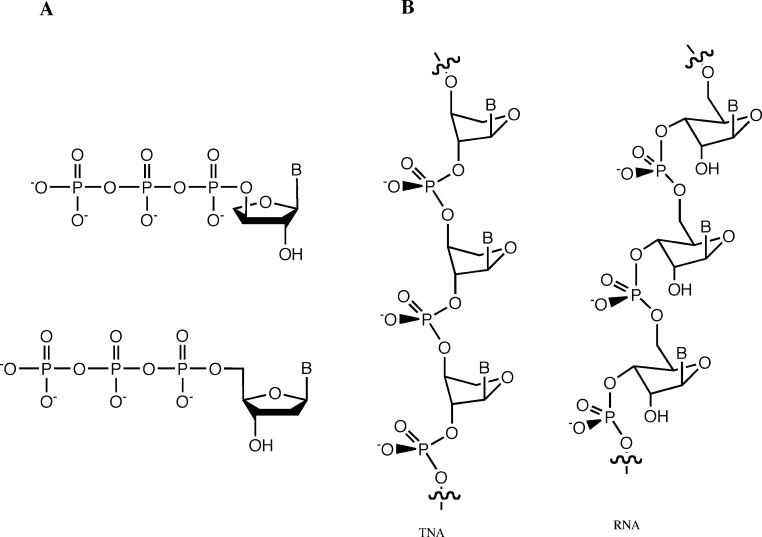
The α-l-threofuranosyl nucleoside triphosphates (tNTPs), considered as potential substrates for DNA polymerase, suffer from the absence of one carbon atom in the helical repeat unit, relative to the native dNTPs (Figure 2A). The absence of this methylene group from the carbohydrate backbone (relative to ribofuranosyl) means that with the triphosphate portion of the tNTP bound correctly within the active site, the nucleobase/sugar residue will necessarily be pulled toward the triphosphate by the absence of the 5‘-carbon and may not correctly interact with the template residue. Such effects could alter enzyme substrate affinity and result in higher apparent KM values characterizing the polymerization reaction. In the absence of tNTP−template pairing, the catalytic step necessary to incorporate the tNTP residue into the primer strand may be correspondingly reduced (with lowered Vmax values). Alternatively, with the nucleobase/sugar residue correctly bound in the active site, and paired with the template residue, the absence of the 5‘ carbon may result in a distortion in the position of the triphosphate, also reducing overall binding affinity (with increasing KM values) and placing the α-phosphate residue in a less optimal position for the requisite chemistry to take place (reducing Vmax).
Figure 2.
Structures of (A) tNTP and dNTP building blocks and (B) 3‘ → 2‘ linked α-l-threofuranosyl nucleotides (TNA) and 5‘ → 3‘ linked β-d-ribofuranosyl nucleotides (RNA).
On the other hand, the hydroxyl that functions as the nucleophile (3‘OH in RNA, 2‘OH in TNA) by virtue of the regioisomeric shift by one carbon unit should still be located more or less in the catalytically active site. Despite the shorter backbone length, it appears that TNA and RNA can both form polymers in which the nucleobases are spaced regularly at the same distances (Figure 2B). Crystallographic studies indicate that the TNA backbone can reach far enough to link two stacked base pairs and result in an essentially native DNA−TNA duplex.428255b00017
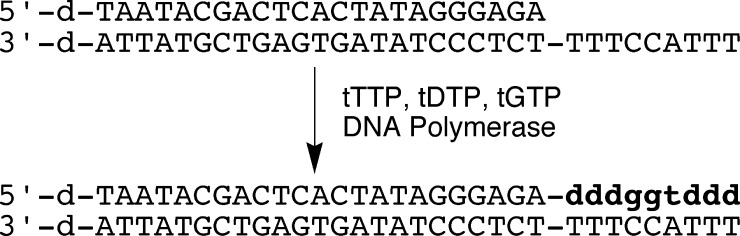
Polymerase Screens. On the basis of our previous demonstration that Deep Vent (exo-) DNA polymerase (DVexo-) was capable of extending a DNA primer with three residues of α-l-threofuranosyl thymidine using tTTP as the substrate,428255b00012 we decided to examine a number of other thermophilic DNA polymerases for improved TNA polymerase activity. A synthetic primer−template complex was constructed that consisted of a DNA primer annealed to a longer DNA template such that nine DNA residues were available to function as a template for the synthesis of TNA (Figure 3). Polymerases were challenged to extend the DNA primer using a mixture of tTTP, tGTP, and tDTP (see Figure 1). The diaminopurine analogue (tDTP) of adenine was used in place of threosyl adenosine 3‘-triphosphate, because this substitution slightly raises the thermodynamic stability of the DNA−TNA heteroduplex. It has additionally been shown to increase the efficiency of nonenzymatic TNA-directed ligation reactions.428255b00018 We have also reported significant rate enhancements as a result of replacing A residues in the template with D residues in previous primer-extension reactions using tTTP.428255b00012
Figure 3.
TNA polymerase assay. Sequence of primer−template complex. TNA elongation product of DNA template is shown as lowercase bold letters.
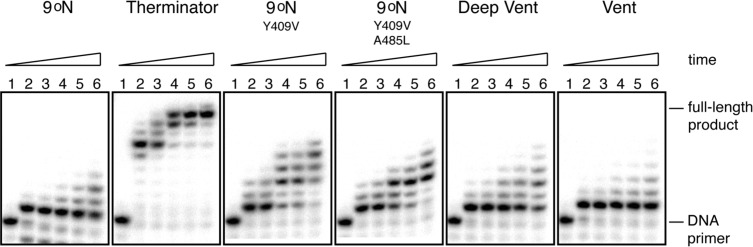
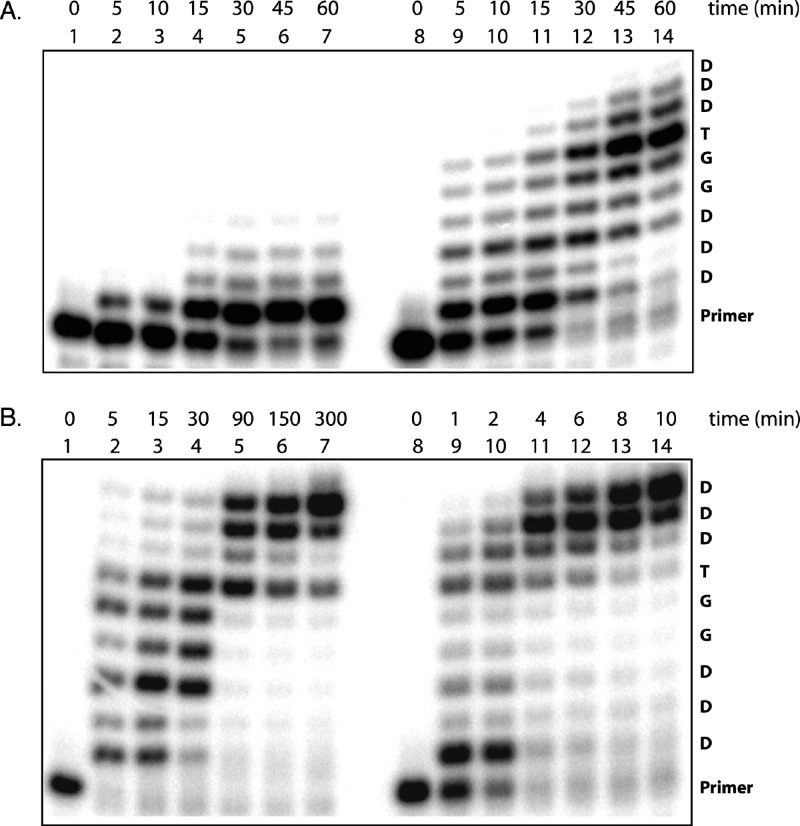
To compare qualitatively the ability of various DNA polymerases to extend a DNA primer with sequential tNTP residues, we screened a series of mutant versions of Taq polymerase (data not shown)19 and several exonuclease-deficient archeal family B DNA polymerases, including 9°N, 9°N variants A485L (Therminator), Y409V, and the Y409V/A485L double mutant, as well as deep vent and vent polymerases (Figure 4). This screen revealed that the Therminator DNA polymerase is a much more efficient TNA polymerase than any other family B polymerase. For instance, in the Therminator-catalyzed TNA synthesis reaction, full-length product is visible by 90 min, and the reaction appears largely complete by 300 min. The remaining polymerases are only able to insert a limited number of tNTP residues within that same time period. To compare the activity of DVexo- and Therminator DNA polymerases in greater detail, additional reaction time courses were performed (Figure 5A,B, lanes 1−7). In these reactions, DVexo- rapidly catalyzed primer extension by one nucleotide followed by very slow and incomplete addition of a second and third nucleotide. Under the same conditions, the Therminator-catalyzed reaction is characterized by the continuous incorporation of consecutive residues, proceeding almost to completion after 300 min. Since Therminator is able to continue synthesis even after all primer positions expected to contact the enzyme are composed of TNA, it seems likely that DVexo- loses essential contacts with the primer as additional TNA nucleotides are added.
Figure 4.
TNA polymerase screen. Products of tNTP elongation on a 5‘[-P32]-labeled 23-mer primer annealed to a DNA template. Reaction progress over time was analyzed by denaturing polyacrylamide gel electrophoresis for experiments using exonuclease deficient family B DNA polymerases: 9°N, Therminator, 9°N single mutant Y409V, 9°N double mutant Y409V and A485L, Deep Vent, and Vent. Time points were taken for each polymerase reaction at 0 (no enzyme), 15, 30, 90, 150, and 300 min, lanes 1−6, respectively.
Figure 5.
TNA primer extension reactions. Reaction progress over time was analyzed by denaturing polyacrylamide gel electrophoresis for (A) Deep Vent exo- (DVexo-) and (B) Therminator-catalyzed DNA polymerase reactions. Primer-extension reactions were performed in the absence (lanes 1−7) and presence (lanes 8−14) of 1.25 mM MnCl2.
Encouraged by this result, we attempted to optimize the reaction conditions for TNA polymerase activity. Variations in salt concentration or pH had little effect, but the presence of Mn2+ ions dramatically increased the reaction efficiency. Mn2+ ions are known to relax the substrate specificity of many DNA polymerases, possibly by allowing enhanced binding to the β- and γ-phosphates of the dNTPs.428255b00020 We have previously used this strategy to improve the rate of DNA synthesis on a TNA template.428255b00011 As illustrated in lanes 8−14 of Figure 5, supplementing the reaction mixture with 1.25 mM MnCl2 enhances the efficiency of TNA synthesis for both DVexo- and Therminator polymerases. With MnCl2 we observed the presence of trace amounts of fully elongated product, visible after 1 h, for reactions catalyzed by DVexo-. Therminator-catalyzed reactions appear essentially complete in just 10 min with Mn2+ versus 150−300 min in the absence of Mn2+.
To verify the addition of tNTPs to the 3‘-terminus of the DNA primer, Therminator-catalyzed single-nucleotide addition reactions were analyzed by MALDI-TOF mass spectrometry. As shown in Table 1, the observed mass for each of the four tNTP and dNTP primer extension reactions was found to be within 1 atomic mass unit of the calculated value. Thus, the extension of a DNA primer with tNTPs does not appear to be the result of unwanted dNTP contamination as TNA and DNA differ by one methylene group or 14 atomic mass units.
Table 1.
MALDI-TOF Mass Spectrometry Analysis of Therminator-Mediated Single-Nucleotide Extension of dNTPs and tNTPs from a DNA Primer
| 5‘-d-TAATACGACTCACTATAGGGAGA 3‘-d-ATTATGCTGAGTGATATCCCTCTXYZCTTT | ||||
| XYZ (DNA) | NTP | expected mass (m/z) | observed mass (m/z) | |
| 1 | TGG | dDTP | 7385.8 | 7386.7 |
| 2 | CGG | dGTP | 7386.8 | 7386.9 |
| 3 | DGG | dTTP | 7361.8 | 7362.6 |
| 4 | GAA | dCTP | 7346.8 | 7345.9 |
| 5 | TGG | tDTP | 7371.8 | 7372.7 |
| 6 | CGG | tGTP | 7372.8 | 7372.3 |
| 7 | DGG | tTTP | 7347.8 | 7346.8 |
| 8 | GAA | tCTP | 7332.8 | 7333.5 |
| 9 | CGG | none | 7058.6 | 7058.9 |
To quantify these initial observations, we decided to perform two types of experiments: (i) the extension of an all-DNA primer by one TNA versus one DNA nucleotide and (ii) the extension of a DNA primer containing five 3‘-terminal TNA residues by one TNA or DNA nucleotide. We determined the catalytic efficiency of TNA and DNA synthesis at each of these positions by measuring single-nucleotide insertion kinetics using the steady-state method.428255b00016 Values for Vmax/KM were determined for DVexo- and Therminator-catalyzed insertion of all four α-l-threofuranosyl nucleoside 3‘-triphosphates (Figure 1) opposite their cognate Watson−Crick complement. This approach provides an accurate method for evaluating the catalytic efficiency for the overall reaction process, but does not identify the rate-limiting step of the reaction. For native DNA nucleotides, the rate-limiting step typically involves a conformational change prior to chemical bond formation.428255b00021 Conversely, for mismatched and many non-natural DNA substrates, phosphodiester bond formation tends to become rate-limiting.428255b00022 By comparing the catalytic efficiency of DV (exo-) and Therminator DNA polymerases for tNTP relative to that of dNTP substrates, we are able to determine the enzymatic selectivity (e.g., the ability for each polymerase to distinguish threose substrates from deoxyribose substrates) for each enzyme substrate pair. For a DNA polymerase to be an efficient TNA polymerase it must exhibit little to no selectivity between correctly paired dNTP and tNTP substrates.
Single-Nucleotide TNA Elongation of a DNA Primer. Primer-extension experiments with tNTP substrates and an all-DNA primer and template specifically examine the effects of changes in the nucleotide substrate, since enzyme binding to the primer−template complex should be normal, and the reactive groups (3‘-OH of the primer and the α-phosphate of the triphosphate) are unchanged. Thus, any observed effects should reflect changes in substrate binding, changes in the orientation of the bound substrate, or altered conformational changes in the enzyme as a result of alterations in the substrate.
We measured single nucleotide incorporation kinetics for DVexo--catalyzed synthesis of TNA from a DNA primer. The catalytic efficiency (Vmax/KM) for DVexo--catalyzed extension of a DNA primer was 20−100 fold lower for tNTP incorporation compared to that of dNTP incorporation (Table 2, entries 5−8 versus 1−4). The observed kinetic selectivity is greatest between tCTP and dCTP (∼90-fold), with the least discrimination occurring between tTTP and TTP (∼20-fold). Comparison of the individual KM and Vmax values for TNA and DNA shows that the loss in catalytic efficiency is dominated by increases in KM. The KM for tNTPs is 5−45-fold higher than that for dNTPs, while the Vmax drops by only 2−5-fold. While the large increases in KM may reflect loss of binding interactions with the tNTP substrate, this interpretation is complicated by the possibility of changes in the rate-limiting step of the reaction. The changes in Vmax suggest that the TNA triphosphate may not be properly positioned in the active site of the enzyme for inline attack by the 3‘-hydroxyl of the DNA primer; alternatively, the conformational changes known to follow substrate binding may be slower with tNTP substrates.
Table 2.
Steady-State Kinetic Parameters for Deep Ventexo--Mediated Single-Nucleotide Extension of dNTPs versus tNTPs from a DNA Primer
| 5‘-d-TAATACGACTCACTATAGGGAGA 3‘-d-ATTATGCTGAGTGATATCCCTCTYZCGTTT | |||||
| YZ (DNA) | NTP | KM (μM) | Vmax (%/min) | efficiency (Vmax/KM) | |
| 1 | TC | dATP | 1.4 ± 0.09 | 46 ± 13 | 3.3 × 107 |
| 2 | CA | dGTP | 0.66 ± 0.18 | 33 ± 9.0 | 5.0 × 107 |
| 3 | AC | dTTP | 0.65 ± 0.33 | 15 ± 1.5 | 2.3 × 107 |
| 4 | GC | dCTP | 0.18 ± 0.12 | 14 ± 3.5 | 7.8 × 107 |
| 5 | TC | tDTP | 7.5 ± 5.0 | 8.3 ± 2.4 | 1.1 × 106 |
| 6 | CA | tGTP | 9.3 ± 2.5 | 7.5 ± 1.0 | 8.6 × 105 |
| 7 | AC | tTTP | 6.2 ± 1.5 | 7.0 ± 1.0 | 1.1 × 106 |
| 8 | GC | tCTP | 8.2 ± 0.3 | 7.4 ± 1.0 | 8.5 × 105 |
The catalytic efficiency for Therminator-catalyzed single-nucleotide addition of tNTPs and dNTPs to a DNA primer is significantly better than that for DVexo--catalyzed synthesis (Table 3). Comparison of the Vmax/KM values for the two enzymes reveals a 17−200-fold improvement in the incorporation of tNTP substrates for Therminator versus DVexo-. This large value does not solely reflect better incorporation of tNTPs by Therminator as this enzyme also shows 4−10-fold increases in catalytic efficiency with dNTPs when compared to DVexo-. Thus, under these conditions Therminator polymerase is simply a more active polymerase than DVexo-.
Table 3.
Steady-State Kinetic Parameters for Therminator-Mediated Single-Nucleotide Extension from DNA
| 5‘-d-TAATACGACTCACTATAGGGAGA 3‘-d-ATTATGCTGAGTGATATCCCTCTYZCGTTT | ||||||
| YZ (DNA) | NTP | KM (μM) | Vmax (%/min) | efficiency (Vmax/KM) | MnCl2a | |
| 1 | TC | dATP | 0.12 ± 0.14 | 21 ± 7.5 | 1.8 × 108 | − |
| 2 | CA | dGTP | 0.21 ± 0.97 | 49 ± 2.4 | 2.3 × 108 | − |
| 3 | AC | dTTP | 0.21 ± 0.12 | 40 ± 2.0 | 1.9 × 108 | − |
| 4 | GC | dCTP | 0.12 ± 0.07 | 36 ± 8.6 | 3.0 × 108 | − |
| 5 | TC | tDTP | 0.73 ± 0.2 | 48 ± 33 | 6.6 × 107 | − |
| 6 | CA | tGTP | 0.46 ± 0.12 | 25 ± 2.9 | 5.4 × 107 | − |
| 7 | AC | tTTP | 1.5 ± 0.17 | 28 ± 1.0 | 1.9 × 107 | − |
| 8 | GC | tCTP | 0.32 ± 0.02 | 55 ± 1.4 | 1.7 × 108 | − |
| 9 | TC | tDTP | 0.19 ± 0.01 | 25 ± 21 | 1.3 × 108 | + |
| 10 | CA | tGTP | 0.39 ± 0.01 | 33 ± 1.0 | 8.5 × 107 | + |
| 11 | AC | tTTP | 0.19 ± 0.11 | 30 ± 7.0 | 1.6 × 108 | + |
| 12 | GC | tCTP | 0.13 ± 0.04 | 29 ± 2.0 | 2.2 × 108 | + |
| 13 | CA | ddGTP | 0.13 ± 0.03 | 19 ± 2.0 | 1.5 × 108 | − |
| 14 | CA | acycGTP | 0.05 ± 0.03 | 14 ± 2.0 | 2.8 × 108 | − |
| 15 | DC | dTTP | 0.05 ± 0.03 | 14 ± 3.3 | 2.9 × 108 | − |
| 16 | DC | tTTP | 0.06 ± 0.05 | 15 ± 5.7 | 2.3 × 108 | − |
| 17 | DC | tTTP | 0.04 ± 0.03 | 6.0 ± 0.9 | 1.6 × 108 | + |
a Reaction mixture supplemented with 1.25 mM MnCl2.
Inspection of the individual KM and Vmax values for the Therminator-catalyzed reactions suggests the slight drop in catalytic efficiency with tNTPs is primarily due to a modest increase in KM; there is no significant difference between Vmax values with tNTP and dNTP substrates. Examination of Therminator-catalyzed TNA synthesis relative to DNA synthesis (entries 5−8 versus 1−4) indicates that tNTPs are only slightly less efficient (2−10-fold) than dNTP substrates. Because a small degree of kinetic selectivity was observed between tTTP and TTP (∼10-fold), we tested the idea that tTTP incorporation could be improved by replacing dA with dD in the DNA template. While this substitution provided a modest increase in the catalytic efficiency for DNA (1.5-fold, entry 3 versus 15), its effect on TNA synthesis was much more pronounced (12-fold, entry 7 versus 16). Interestingly, the enhanced efficiency of both TTP and tTTP incorporation opposite D in the template is due to a decrease in KM, consistent with enhanced binding due to the increased strength of the T−D base pair.
Because of the increased rate of TNA synthesis observed in the presence of Mn2+, we decided to measure Therminator-catalyzed single-nucleotide tNTP insertion efficiencies in the presence of 1.25 mM MnCl2. As expected, supplementing the reaction mixture with Mn2+ ions improves the enzymatic efficiency of the tNTP incorporation to values closely approximating those observed with dNTPs in the absence of Mn2+ (0.8−0.3-fold as efficient, entries 9−12 versus 1−4). Evaluating the catalytic efficiency of Therminator-catalyzed TNA synthesis in the presence (entries 9−12) and absence (entries 5−8) of Mn2+ ions reveals that the presence of manganese lowers the KM of tNTPs, with an especially significant decrease in the KM for tTTP. This change might be due to Mn2+ ions relaxing some conformational constraints within the enzyme active site, thus restoring weakened contacts to the substrate. Mn2+ ions have a similar but smaller effect on the KM for tTTP incorporation opposite dD (entry 17 versus 16).
Therminator-Catalyzed TNA Synthesis from a TNA Primer. To examine the effect of TNA residues in the primer on continued TNA synthesis, we constructed a chimeric DNA−TNA primer containing five TNA residues at the 3‘-terminus of the DNA primer. The length of the TNA segment was chosen to ensure that only the TNA region of the primer contacted the polymerase.428255b00023,428255b00024 Using this new primer, we then measured Therminator-catalyzed single-nucleotide incorporation kinetics (Table 4) for all four tNTP and dNTP substrates in the absence (entries 1−8) and presence (entries 9−16) of Mn2+ ions. The extension of the TNA terminus of the DNA−TNA primer remains slightly faster with dNTPs than with tNTPs (approximately 2−4-fold in nearly every case). However, the rate of TNA synthesis in the presence of Mn2+ ions (entries 13−16) is nearly identical to the rate of DNA synthesis in the absence of Mn2+ (entries 1−4). This result suggests that it should be possible to synthesize long stretches of TNA in a polymerase-mediated reaction with catalytic rates that rival natural DNA synthesis. Surprisingly, no drop in the rate of incorporation was observed for any of the four tNTP substrates at the downstream positions (compare Table 3 with Table 4). On the contrary, a slight increase in the catalytic efficiency (2−6-fold) was noted for most substrates. By comparison, similar experiments previously performed using DVexo- DNA polymerase to catalyze the incorporation of tTTP from a DNA−TNA chimeric primer ending in either one or two tT residues resulted in a 2600-fold drop in catalytic efficiency.428255b00012 This comparison suggests that Therminator polymerase may form more direct contacts to the TNA primer, thus permitting continued TNA synthesis.
Table 4.
Steady-State Kinetic Parameters for Therminator-Mediated Single-Nucleotide Extension from the TNA Terminus of a DNA−TNA Primer
| 5‘-d-TAATACGACTCACTATAGGGAGAatagg 3‘-d-ATTATGCTGAGTGATATCCCTCTTATCCYZCGTTT | ||||||
| YZ (DNA) | NTP | KM (μM) | Vmax (%/min) | efficiency Vmax/KM | MnCl2a | |
| 1 | TC | dATP | 0.040 ± 0.045 | 22 ± 11 | 5.5 × 108 | − |
| 2 | CA | dGTP | 0.019 ± 0.01 | 16 ± 7.7 | 8.4 × 108 | − |
| 3 | AC | dTTP | 0.054 ± 0.001 | 31 ± 0.8 | 5.7 × 108 | − |
| 4 | GC | dCTP | 0.066 ± 0.02 | 32 ± 14 | 4.8 × 108 | − |
| 5 | TC | tDTP | 0.18 ± 0.1 | 21 ± 9.4 | 1.2 × 108 | − |
| 6 | CA | tGTP | 0.09 ± 0.02 | 29 ± 1.6 | 3.2 × 108 | − |
| 7 | AC | tTTP | 0.12 ± 0.03 | 27 ± 2.7 | 2.3 × 108 | − |
| 8 | GC | tCTP | 0.21 ± 0.04 | 36 ± 2.6 | 1.7 × 108 | − |
| 9 | TC | dATP | 0.025 ± 0.009 | 43 ± 7.8 | 1.7 × 109 | + |
| 10 | CA | dGTP | 0.031 ± 0.007 | 31 ± 5.4 | 1.0 × 109 | + |
| 11 | AC | dTTP | 0.007 ± 0.001 | 35 ± 0.7 | 5.0 × 109 | + |
| 12 | GC | dCTP | 0.019 ± 0.007 | 39 ± 8.3 | 2.1 × 109 | + |
| 13 | TC | tDTP | 0.04 ± 0.01 | 31 ± 5.9 | 8.7 × 108 | + |
| 14 | CA | tGTP | 0.08 ± 0.03 | 39 ± 3.1 | 4.8 × 108 | + |
| 15 | AC | tTTP | 0.09 ± 0.03 | 38 ± 4.7 | 4.4 × 108 | + |
| 16 | GC | tCTP | 0.11 ± 0.08 | 49 ± 1.1 | 4.8 × 108 | + |
a Reaction mixture supplemented with 1.25 mM MnCl2.
Rationalizing the superior catalytic efficiency of Therminator-mediated TNA polymerization relative to other family B DNA polymerases (including its parental 9°N polymerase) requires understanding the role of its A485L mutation. Previous work by Gardner and Jack has indicated that family B archaeal DNA polymerases bearing this mutation exhibit increased efficiency of incorporation of chain-terminating dideoxy and acyclic nucleoside triphosphates.428255b00014 To quantitatively compare these analogues with tNTPs, we measured the insertion kinetics for ddGTP and the acyclic derivative, 2-hydroxyethoxymethyl-GTP, using Therminator DNA polymerase and an all-DNA primer (see entries 13 and 14 of Table 3). Comparison of the catalytic efficiency of incorporation of all four nucleotides (Vmax/KM) results in the following substrate order: acGTP ≈ dGTP > ddGTP > tGTP. Therminator-mediated incorporation of acycloGTP is approximately 2-fold more efficient than ddGTP and 5-fold more efficient than tGTP. In light of this result, it appears that the mutation A485L allows efficient nucleotide incorporation largely independent of sugar recognition. The crystal structures of the apo 9°N DNA polymerase and family B RB69 DNA polymerase replication complex offer some insight into the structural influence of the A485L substitution.428255b00025,428255b00026 The A485 residue faces away from, and does not interact directly with, the incoming nucleoside triphosphate. Instead, the increased bulk of the leucine residue appears to favor the rotation of the fingers domain, which is known to follow nucleotide binding and precede phosphodiester bond formation. If this conformational change is normally slow in nucleotides with sugar analogues, the A485L mutation would be expected to facilitate their incorporation by speeding up this rate-limiting transition.
We have not undertaken a detailed analysis of the fidelity of tNTP incorporation by the Therminator polymerase using the DNA template. However, under the conditions of the kinetic characterization of tNTP single-nucleotide incorporation studies, we never observe the appearance of the n + 2 product (specific incorporation followed by nonspecific incorporation). We observe exclusively the n + 1 primer product. If TNA synthesis by the Therminator polymerase occurred with low levels of sequence fidelity, we would expect some nonspecific elongation of the n + 1 primer. Although qualitative in nature, this observation suggests that under the concentrations and conditions employed for the described elongations, sequence fidelity is unlikely to be a major concern. A detailed kinetic analysis of the fidelity of TNA synthesis is currently in progress.
In summary, we have identified Therminator DNA polymerase as an enzyme capable of significant TNA synthesis activity. Using this polymerase, we have found that despite considerable differences in the sugar−phosphate backbone, the enzymatic efficiency of TNA substrates compares closely with natural DNA substrates. The fact that present-day polymerases can incorporate simpler versions of the nucleoside triphosphate building blocks under conditions where rates of polymerization rival those of the dNTPs suggests that simpler progenitors could form the basis for an alternative genetic system. The observed polymerization rates for tNTPs also suggest that the iterative process of in vitro selection and amplification of TNA aptamers and enzymes may now be possible through a DNA display approach similar to mRNA display,428255b00027 in which random sequence TNA oligonucleotides are displayed on their encoding DNA message.428255b00002
Acknowledgments
We wish to thank R. Krishnamurthy and A. Eschenmoser for generously providing the advanced intermediate nucleoside derivatives for the synthesis of tTTP, tDTP, and tGTP. We would also like to thank P. Holliger and colleagues for providing several mutant versions of Taq DNA polymerase for us to test as potential DNA-dependent TNA polymerases and W. Jack and A. Gardner at New England Biolabs for providing samples of mutant 9°N DNA polymerase Y409V/A485L and Y409V. This work was supported by grants from the NIH (RO1 GM053936) and the NASA Astrobiology Institute (NNA04CC12A) to J.W.S. and the NSF (MCB0451488) to LWM. J.W.S. is an Investigator and J.C.C. was a Research Associate of the Howard Hughes Medical Institute. J.K.I. was supported in part by a Ford Foundation predoctoral fellowship.
Supporting Information Available
Supporting Information Available
Detailed experimental methods for steady-state kinetic studies, enzyme saturation plots, Hanes−Woolf plots of kinetic data for nucleotide insertions, and MALDI-TOF mass spectra for Therminator-catalyzed single-nucleotide addition of tNTPs and dNTPs. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- For recent reviews, see: (a) Famulok M. Curr. Opin. Struct. Biol.1999, 9, 324−329. (b) Wilson D. S.; Szostak J. W. Annu. Rev. Biochem. 1999, 68, 611−647. (c) Breaker R. R. Chem. Rev.1997, 97, 371−390. (d) Lin H.; Cornish V. W. Angew. Chem., Int. Ed.2002, 41, 4402−4425. 10.1016/S0959-440X(99)80043-8 [DOI]
- Ichida J. K.; Zou K.; Horhota A.; Yu B.; McLaughlin L. W.; Szostak J. W. J. Am. Chem. Soc. 2005, 127, 2802–2803. 10.1021/ja045364w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Schoning K.-U.; Scholz P.; Guntha S.; Wu X.; Krishnamurthy R.; Eschenmoser A. Science 2000, 290, 1347–1351. 10.1126/science.290.5495.1347. [DOI] [PubMed] [Google Scholar]; b Eschenmoser A. Science 1999, 284, 2118–2124. 10.1126/science.284.5423.2118. [DOI] [PubMed] [Google Scholar]
- a Ellington A. D.; Szostak J. W. Nature 1990, 346, 818–822. 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]; b Tuerk C.; Gold L. Science 1990, 249, 505–510. 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Brody R. S.; Frey P. A. Biochemistry 1981, 20, 1245–1252. 10.1021/bi00508a030. [DOI] [PubMed] [Google Scholar]
- Dineva M. A.; Chakurov S.; Bratovanova E. K.; Devedjiev I.; Petkov D. D. Bioorg. Med. Chem. 1993, 1, 411–414. 10.1016/S0968-0896(00)82151-3. [DOI] [PubMed] [Google Scholar]
- He K. Z.; Porter K. W.; Hasan A.; Briley J. D.; Shaw B. R. Nucleic Acids Res. 1999, 27, 1788–1794. 10.1093/nar/27.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez C. I.; Ansari M. A.; Gibbs R.; Burgess K. Bioorg. Med. Chem. Lett. 1997, 7, 3013–3016. 10.1016/S0960-894X(97)10135-4. [DOI] [Google Scholar]
- a Hess M. T.; Schwitter U.; Petretta M.; Giese B.; Naegeli H. Biochemistry 1997, 36, 2332–2337. 10.1021/bi961689g. [DOI] [PubMed] [Google Scholar]; b Marx A.; MacWilliams M. P.; Bickle T. A.; Schwitter U.; Giese B. J. Am. Chem. Soc. 1997, 119, 1131–1132. 10.1021/ja9627813. [DOI] [Google Scholar]
- a Suzuki M.; Avicola A. K.; Hood L.; Loeb L. A. J. Biol. Chem. 1997, 272, 11228–11235. 10.1074/jbc.272.17.11228. [DOI] [PubMed] [Google Scholar]; b Astatke M.; Ng K.; Grindley N. D.; Joyce C. M. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 3402–3407. 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Fa M.; Radeghieri A.; Henry A. A.; Romesberg F. E. J. Am. Chem. Soc. 2004, 126, 1748–1754. 10.1021/ja038525p. [DOI] [PubMed] [Google Scholar]
- Chaput J. C; Ichida J. K.; Szostak J. W. J. Am. Chem. Soc. 2003, 125, 856–857. 10.1021/ja028589k. [DOI] [PubMed] [Google Scholar]
- Chaput J. C.; Szostak J. W. J. Am. Chem. Soc. 2003, 125, 9274–9275. 10.1021/ja035917n. [DOI] [PubMed] [Google Scholar]
- Kempeneers V.; Vastmans K.; Rozenski J.; Herdewijn P. Nucleic Acids Res. 2003, 31, 6221–6226. 10.1093/nar/gkg833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Gardner A. F.; Jack W. E. Nucleic Acids Res. 2002, 30, 605–613. 10.1093/nar/30.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Gardner A. F.; Jack W. E. Nucleic Acids Res. 1999, 27, 2545–2553. 10.1093/nar/27.12.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou K.; Horhota A.; Yu B.; Szostak J. W.; McLaughlin L. W. Org. Lett. 2005, 7, 1485–1487. 10.1021/ol050081+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Boosalis M. S.; Petruska J.; Goodman M. F. J. Biol. Chem. 1987, 262, 14689–14696. [PubMed] [Google Scholar]; b Goodman M. F.; Creighton S.; Bloom L. B.; Petruska J. Crit. Rev. Biochem. Mol. Biol. 1993, 28, 83–126. 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- a Wilds C. J.; Wawrzak Z.; Krishnamurthy R.; Eschenmoser A.; Egli M. J. Am. Chem. Soc. 2002, 124, 13716–13721. 10.1021/ja0207807. [DOI] [PubMed] [Google Scholar]; b Pallan P. S.; Wilds C. J.; Wawrzak Z.; Krishnamurthy R.; Eschenmoser A.; Egli M. Angew. Chem., Int. Ed. 2003, 42, 5893–5895. 10.1002/anie.200352553. [DOI] [PubMed] [Google Scholar]
- Wu X.; Delgado G.; Krishnamurthy R.; Eschenmoser A. Org. Lett. 2002, 4, 1283–1286. 10.1021/ol020016p. [DOI] [PubMed] [Google Scholar]
- Ghadessy F. J.; Ramsay N.; Boudsocq F.; Loakes D.; Brown A.; Iwai S.; Vaisman A.; Woodgate R.; Holliger P. Nat. Biotechnol. 2004, 22, 755–759. 10.1038/nbt974. [DOI] [PubMed] [Google Scholar]
- a Cadwell R. C.; Joyce G. F. PCR Methods Appl. 1992, 2, 28–33. 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]; b Tabor S.; Richardson C. C. Proc. Natl. Acad. Sci. U.S.A. 1989, 86, 4076–4080. 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astatke M.; Grindley N. D. F.; Joyce C. M. J. Mol. Biol. 1988, 278, 147–165. 10.1006/jmbi.1998.1672. [DOI] [PubMed] [Google Scholar]
- Kool E. T. Curr. Opin. Chem. Biol. 2000, 4, 602–608. 10.1016/S1367-5931(00)00141-1. [DOI] [PubMed] [Google Scholar]
- a Ollis D. L.; Brick P.; Hamlin R.; Xuong N. G.; Steitz T. A. Nature 1985, 313, 762–766. 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]; b Doublie S.; Tabor S.; Long A. M.; Richardson C. C.; Ellenberger T. Nature 1998, 391, 251–258. 10.1038/34593. [DOI] [PubMed] [Google Scholar]; c Kiefer J. R.; Mao C.; Braman J. C.; Beese L. S. Nature 1998, 391, 304–307. 10.1038/34693. [DOI] [PubMed] [Google Scholar]; d Li Y.; Korolev S.; Waksman G. EMBO J. 1998, 17, 7514–7525. 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Franklin M. C.; Wang J.; Steitz T. A. Cell 2001, 105, 657–667. 10.1016/S0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]; f Silvian L. F.; Toth E. A.; Pham P.; Goodman M. F.; Ellenberger T. Nat. Struct. Biol. 2001, 8, 984–989. 10.1038/nsb1101-984. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A.; Bebenek K. Annu. Rev. Biochem. 2000, 69, 497–529. 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- Rodriguez A. C.; Park H.-W.; Mao C.; Besse L. A. J. Mol. Biol. 2000, 299, 447–462. 10.1006/jmbi.2000.3728. [DOI] [PubMed] [Google Scholar]
- Franklin M. C.; Wang J.; Steitz T. A. Cell 2001, 105, 657–667. 10.1016/S0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Roberts R. W.; Szostak J. W. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 12297–12302. 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.