A combination of experimental data obtained in somatic cells and Xenopus egg extracts and modeling suggests a novel function for centrosome maturation that balances the activity of the mitotic microtubule assembly pathways favoring bipolar spindle formation.
Abstract
Centrosome maturation is the process by which the duplicated centrosomes recruit pericentriolar components and increase their microtubule nucleation activity before mitosis. The role of this process in cells entering mitosis has been mostly related to the separation of the duplicated centrosomes and thereby to the assembly of a bipolar spindle. However, spindles can form without centrosomes. In fact, all cells, whether they have centrosomes or not, rely on chromatin-driven microtubule assembly to form a spindle. To test whether the sequential activation of these microtubule assembly pathways, defined by centrosome maturation and nuclear envelope breakdown, plays any role in spindle assembly, we combined experiments in tissue culture cells and Xenopus laevis egg extracts with a mathematical model. We found that interfering with the sequential activation of the microtubule assembly pathways compromises bipolar spindle assembly in tissue culture cells but not in X. laevis egg extracts. Our data suggest a novel function for centrosome maturation that determines the contribution of the chromosomal microtubule assembly pathway and favors bipolar spindle formation in most animal cells in which tubulin is in limiting amounts.
INTRODUCTION
In most animal cells, the mitotic spindle assembles in the presence of two centrosomes. Before mitosis, during G2, a complex network of kinases and feedback loops drives centrosome maturation (Lindqvist et al., 2009). This process promotes the recruitment of various proteins around the centrosome, increasing the amount of pericentriolar material (PCM) and centrosome microtubule (MT) nucleation activity (Palazzo et al., 2000; Wiese and Zheng, 2006). Centrosome maturation has been proposed to be essential for centrosome separation (Wang et al., 2014) and, thus, bipolar spindle formation and for spindle orientation by defining the position of the spindle poles (Lu and Johnston, 2013).
Although centrosomes were once believed to be essential for spindle assembly, it is now established that acentrosomal MTs assembled through a chromatin-dependent pathway are necessary and sufficient for spindle formation in all cell types (Karsenti et al., 1984; Heald et al., 1996; Khodjakov et al., 2000). This pathway becomes active only after nuclear envelope breakdown (NEBD), when nuclear and cytoplasmic contents mix and the centrosomes are already actively assembling MTs. Thus direct visualization of MTs generated by the chromatin-dependent pathway is difficult in centrosome-containing cells. To overcome this limitation, we and others have extensively used MT regrowth upon nocodazole washout (Tulu et al., 2006; Meunier and Vernos, 2011). This simple technique is sufficient to detect MT assembly at the centrosomes and in the chromatin region. However, MT regrowth experiments can also be used to follow spindle formation (Ferenz et al., 2009) and determine how the interplay between the MT assembly pathways leads to spindle formation.
Here we combine MT regrowth experiments and mathematical modeling to investigate the interplay between MT assembly pathways during spindle assembly. Our data suggest that, in most animal cells, centrosome maturation defines the level of activity of the chromosome-dependent MT assembly pathway, thus establishing a balance that favors spindle bipolarity.
RESULTS
Bipolar spindle assembly is compromised in cells undergoing MT regrowth
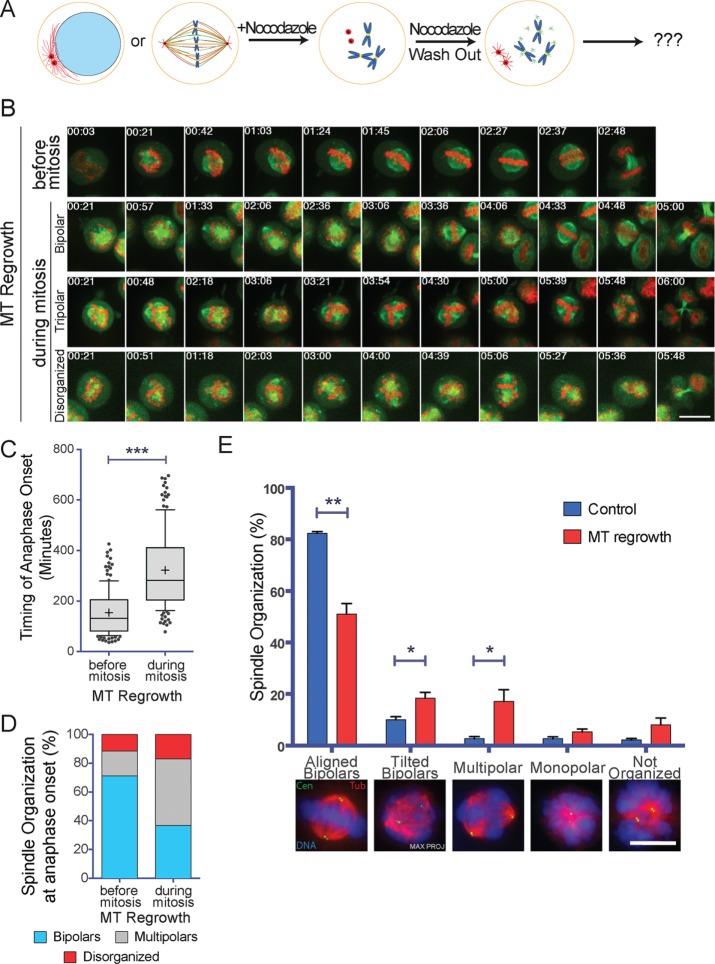
Using time-lapse fluorescence microscopy, we followed mitotic HeLa H2B-eGFP/α-tubulin–monomeric red fluorescent protein (mRFP) cells undergoing MT regrowth after nocodazole washout (Figure 1A). Cells that entered mitosis within the 12-h imaging period after nocodazole washout were considered as controls (Figure 1B and Supplemental Video S1), as in these cells, MT regrowth occurred during interphase. The quantification of the time required for the cells to enter anaphase showed that mitotic cells undergoing MT regrowth required a significantly longer time (322 ± 152 min) than control cells (154 ± 88 min; Figure 1C). Moreover, the proportion of cells entering anaphase with a multipolar spindle (46.2 ± 4.8%) or disorganized MTs (17.1 ± 3.1%) was significantly higher than in control cells (17.3 ± 2.9 and 11.6 ± 1.4%, respectively, for multipolar spindles and disorganized MT organization; Figure 1D). Most of the multipolar spindles were tripolars (97% of multipolars). The delay in prometaphase for the cells undergoing MT regrowth was also associated with the assembly of unstable spindles that collapsed several times before eventually progressing into anaphase (Figure 1B and Supplemental Videos S2–S4).
FIGURE 1:
Spindle assembly is compromised in mitotic cells that undergo MT regrowth. (A) Experimental design. Centrosome MTs are in red, chromatin dependent MTs are in green. (B) Images from time-lapse movies of H2B-eGFP/α-tubulin-mRFP HeLa cells undergoing mitosis after nocodazole washout. Top, control cell (MT Regrowth before mitosis). The three bottom rows show arrested mitotic cells going through mitosis after nocodazole washout (MT Regrowth during mitosis). In the two bottom rows, the cells enter anaphase after >5 h with a tripolar and a disorganized spindle. Tubulin is in green and H2B in red. Time is in hours:minutes. Time 0 corresponds to NEBD for the control cell and to nocodazole washout for MT Regrowth cells. Scale bar, 10 μm. (C) Time required for H2B-eGFP/α-tubulin-mRFP HeLa cells to enter anaphase after nocodazole washout. For control cells (MT Regrowth before mitosis), the values correspond to the time from NEBD to anaphase entry. For mitotic cells undergoing MT regrowth (MT Regrowth during mitosis), the values correspond to the time from nocodazole washout to anaphase entry. Box-and-whiskers plot. A total of 185 control cells and 149 MT regrowth cells were analyzed in four independent experiments. ***p < 0.001. (D) Spindle organization at anaphase onset. The proportion of bipolar (light blue), multipolar (gray), and disorganized (red) spindles was quantified for the cells analyzed in C. Mitotic cells undergoing MT regrowth (MT Regrowth) assemble significantly more multipolar and disorganized spindles than control cells (Control; p < 0.05). (E) Spindle organization in mitotic HeLa cells (Control, blue) or HeLa cells undergoing MT regrowth (MT Regrowth, red). Proportion of different spindle organizations 30 min after nocodazole washout or in control cells. MT regrowth cells have significantly fewer bipolar spindles properly oriented and a higher number of tilted and multipolar spindles than control cells. Representative IF images of different spindle organizations are shown below the graph. Data from 847 controls cells and 864 MT regrowth cells from four independent experiments, counting in each at least 200 cells/condition. DNA is in blue, centrin in green, and tubulin in red. Scale bar, 10 μm. Error bars, SEM. *p < 0.05; **p < 0.01.
To rule out that these phenotypes could be cell-line specific, we performed live imaging under similar conditions in two different cell lines: U2OS γ-tubulin–photoactivatable green fluorescent protein (paGFP)/α-tubulin-mCherry and LLC-PK1α α-tubulin-GFP (Supplemental Videos S5 and S6). The analysis of the movies showed that these cells also exhibited spindle organization defects similar to those observed in HeLa cells under the same conditions (Supplemental Figure S1, A and B).
A closer examination of the time-lapse recordings from the mitotic HeLa cells undergoing MT regrowth suggested that spindles rotated frequently (Figure 1B and Supplemental Videos S2 and S3). To characterize this phenotype (nonfluorescent) HeLa cells were fixed 30 min after nocodazole washout and processed for immunofluorescence to visualize the centrosomes (anti-centrin signal), the MTs, and the chromosomes (Figure 1E). Spindle organization and the relative position of the centrosomes in the z-axis were then analyzed by selecting cells having only two centrosomes. Bipolar spindles having the centrosomes separated by >5 μm in the z-axis were classified as tilted. Consistent with the live-imaging data, cells undergoing MT regrowth had spindle assembly and orientation defects. Indeed, the proportion of tilted bipolar spindles and of multipolar spindles increased significantly from 10.0 ± 1.2 and 2.7 ± 0.7%, respectively, in controls cells to 18.4 ± 2.3 and 17.1 ± 4.6%, respectively, in cells undergoing MT regrowth (Figure 1E). These results confirmed that mitotic cells undergoing MT regrowth have spindle organization and orientation defects.
To rule out that the observed phenotypes were associated to the use of nocodazole, we performed MT regrowth experiments after cold treatment and quantified the mitotic phenotypes in fixed HeLa cells containing only two centrosomes as described earlier (Supplemental Figure S2). These cells showed similar spindle assembly and orientation defects to those treated with nocodazole. These results rule out the possibility that these defects are caused by the presence of low concentrations of nocodazole potentially remaining after washout during spindle assembly.
Taken together, these data show that mitotic cells undergoing MT regrowth have problems in assembling and orienting the bipolar spindle, leading to an extended prometaphase that results in mitotic arrest or catastrophic anaphase.
Centrosome positioning at the time of MT regrowth does not influence spindle assembly
To test whether some of the phenotypes observed upon MT regrowth were due to centrosome separation defects, we incubated HeLa cells with the Eg5 inhibitor monastrol before placing them in nocodazole (Figure 2A). As expected, a large proportion of mitotic cells (64.7 ± 1.5%) had closely juxtaposed centrosomes (distance <3 μm) under these conditions, compared with 35.8 ± 3.8% in cells incubated only in nocodazole (Supplemental Figure S3A). Consistently, upon nocodazole washout, 64.1 ± 6.4% of the cells kept in monastrol assembled a monopolar spindle, whereas only 3.7 ± 1.0% of the control dimethyl sulfoxide (DMSO)–treated cells did (Figure 2B).
FIGURE 2:
The spindle assembly defects observed upon nocodazole washout are not directly related to centrosome positioning, whereas they correlate with lower levels of PCM components at the centrosomes. (A) Experimental design of the experiments shown in B. (B) Spindle organization in cells preincubated in DMSO (MT Regrowth Control, blue) or monastrol followed by nocodazole incubation and washout in the absence (MT Regrowth after Monastrol, red) or presence of monastrol (MT Regrowth in Monastrol, cyan). Proportion of different spindle organizations 30 min after nocodazole washout. Representative IF images of different spindle organizations are shown below the graph. Data from 694 control cells, 672 MT regrowth after monastrol cells, and 630 MT regrowth in monastrol cells from three independent experiments, counting in each at least 200 cells/condition. DNA is in blue, centrin in green, and tubulin in red. Scale bar, 10 μm. Error bars, SEM. *p < 0.05; **p < 0.01. (C) Cells undergoing MT regrowth form astral MTs less efficiently. Quantification of the proportion of control cells (dark gray) and MT regrowth–treated cells (light gray) having long astral MTs (Long MTs), short astral MTs (Short MTs), or no detectable astral MT (None). Representative IF images of different spindle organizations are shown below the graph. Data from 854 control cells and 802 MT regrowth cells from four independent experiments, counting in each at least 200 cells/condition. DNA is in blue, centrin in green, and tubulin in red. Scale bar, 10 μm. Error bars, SEM. ***p < 0.001. (D) On MT regrowth, centrosomes recruit less pericentrin. Representative IF images of two spindles assembled in control cells or in MT regrowth–treated cells. DNA is in blue, pericentrin in green, and tubulin in red. The pericentrin staining is reported also in gray scale. Scale bar, 10 μm. (E) On MT regrowth, centrosomes recruit less γ-tubulin. Representative IF images of two spindles assembled in control cells or in MT regrowth–treated cells. DNA is in blue, γ-tubulin in green, and tubulin in red. The γ-tubulin staining is reported also in gray scale. Scale bar, 10 μm. (F) Quantification of the pericentrin and γ-tubulin signal to the mitotic centrosomes of control cells (dark gray) and MT regrowth–treated cells (light gray). The signals of both proteins are significantly reduced in MT regrowth–treated cells. Data from 370 cells for pericentrin control, 371 cells for pericentrin MT regrowth, 371 cells for γ-tubulin control, and 363 cells for γ-tubulin MT regrowth from six independent experiments, counting in each at least 50 cells/condition. Error bars, SEM. *p < 0.05; ****p < 0.0001.
Of interest, however, after simultaneous washout of monastrol and nocodazole, a similar proportion of multipolar (11.4 ± 0.9%) and tilted spindles (25.4 ± 3.3%) assembled in these cells compared with the control DMSO-treated cells (12.1 ± 1.7% multipolar spindles and 24.3 ± 2.3% tilted spindles; Figure 2B). These data indicate that the respective position of the two centrosomes at the time of MT regrowth has no major effect on spindle assembly.
Centrosome MT assembly capacity is reduced upon MT regrowth
The spindle orientation phenotype (Figure 1E) suggested defects at the level of the astral MTs that would be consistent with a defective centrosome capacity for assembling MTs (Gergely et al., 2000; Lu and Johnston, 2013). To determine whether this was the case, we fixed HeLa cells 30 min after nocodazole washout and categorized the bipolar spindles according to their astral MTs (long, short, or no astral MTs; Figure 2C). Control, untreated cells had a significantly higher proportion of bipolar spindles with long astral MTs than mitotic cells undergoing MT regrowth (77.1 ± 4.2 and 60.0 ± 4.0%, respectively). Consistently, control, untreated cells had a lower proportion of spindles with short astral MTs than mitotic cells undergoing MT regrowth (19.4 ± 1.9 and 34.4 ± 1.8%, respectively; Figure 2C). These data provide further support to the idea that the capacity of mitotic centrosomes for MT assembly is reduced in mitotic cells undergoing MT regrowth. However, we cannot rule out that other defects in these cells may promote spindle misorientation (Collins et al., 2012; Kern et al., 2016).
To address more directly whether the centrosome capacity for assembling MTs may be altered upon MT regrowth, we quantified the levels of pericentrin and γ-tubulin at the centrosomes in bipolar spindles assembled after nocodazole washout and in control, untreated cells (Figure 2, D and E). We found that after nocodazole washout, the levels of pericentrin were reduced to 72.0 ± 3.5% and those of γ-tubulin to 87.1 ± 3.3% of the corresponding levels in control, untreated cells (Figure 2F). These data suggest that after nocodazole washout, the mitotic centrosomes indeed have reduced capacity for MT assembly.
The centrosomal and the chromosomal pathways compete for a limiting component
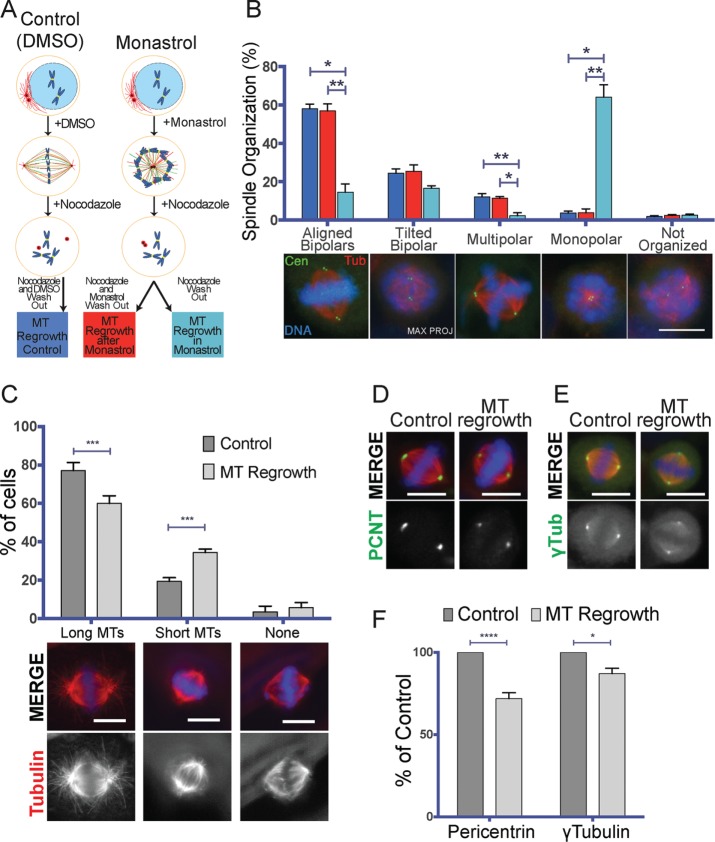
We next tried to understand why bipolar spindle assembly was compromised in mitotic cells undergoing MT regrowth. We reasoned that the reduction of MT assembly at the centrosomes in these cells could potentially change the contribution of the chromosomal MT assembly pathway if there is a competition between the two pathways for a common limiting component. A similar mechanism was proposed to be at play between actin nucleators in fission yeast. By suppressing formin or the Arp2/3 pathways, it was found that the unperturbed pathway was favored (Burke et al., 2014). To use an analogous approach, we tested whether reducing the activity of one pathway during MT regrowth would increase the activity of the other. We performed a MT regrowth experiment in cells that cannot nucleate chromosome-dependent MTs because TPX2 is silenced (Gruss et al., 2001; Supplemental Figure S3B). We then quantified the tubulin signal around the mitotic centrosomes at the early stages of MT regrowth (Lüders et al., 2006; Figure 3A) and found that the tubulin signal associated with the mitotic centrosomes was significantly higher in TPX2-silenced cells than in control cells (respectively 164.4 ± 87.3 and 100.0 ± 29.3%; Figure 3A). This result indicates that preventing chromosome-dependent MT nucleation by TPX2 silencing promotes an increase of centrosomal MT nucleation.
FIGURE 3:
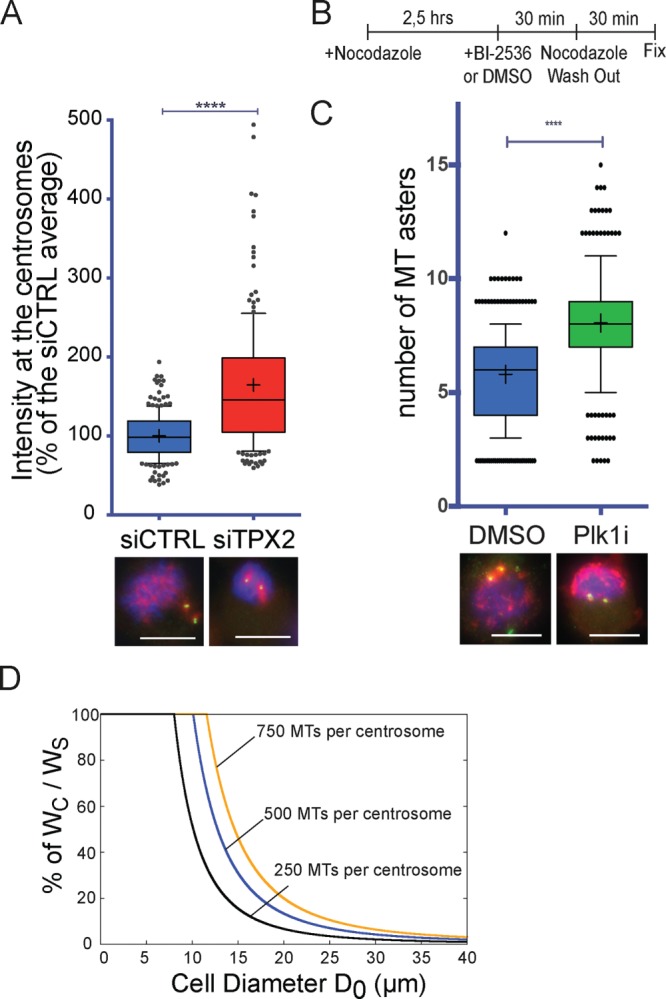
The MT assembly pathway activities are interdependent. (A) Tubulin signal on the mitotic centrosomes 1 min, 30 s after nocodazole washout in mitotic cells silenced using a scrambled siRNA (siCTRL, blue) or a TPX2 siRNA (siTPX2, red). Box-and-whiskers plot showing the intensity of the tubulin signal associated with the mitotic centrosome expressed as percentage of the average of the control. Representative IF images of siCTRL and siTPX2 are shown below the graph. DNA is in blue, centrin in green, and tubulin in red. Data from 205 siCTRL cells and 196 siTPX2 cells from four independent experiments, counting in each at least 40 cells/condition. To pool experiments, in each experiment, the mean tubulin signal of the control condition was considered as 100%. ****p < 0.0001. Scale bar, 10 μm. (B) Experimental design of the MT regrowth performed in Plk1-inhibited cells. (C) Number of MT asters generated 3 min after nocodazole washout in cells treated with DMSO (blue) or BI-2536 (Plk1i, green) as in B. Box-and-whiskers plot showing number of MT asters counted in each condition. Representative IF images of DMSO and Plk1i cells are shown below the graph. DNA is in blue, pericentrin in green, and tubulin in red. Data from 339 DMSO cells and 346 Plk1i cells from three independent experiments counting in each at least 100 cells/condition. ****p < 0.0001. Scale bar, 10 μm. (D) Percentage of the theoretical ratio of tubulin incorporated into centrosomal MTs (WC) to the total tubulin that will constitute the spindle MTs (WS), depending on cell diameter. Three theoretical levels of centrosome maturation (defined as number of MTs nucleated by the centrosomes) were modeled: 250, 500, and 750 MTs per centrosome with average MT length of 3 μm (respectively, black, blue, and orange curves). As the cell size increases, centrosomes use proportionally less of the total tubulin available for the spindle MTs. For cell diameters within 10–30 μm, altering the level of centrosome maturation has a dramatic consequence on the availability of tubulin for the chromosomal pathway. Most dividing animal cells are in this range (Milo and Phillips, 2015), and thus the model suggests that centrosome maturation at NEBD may be a tightly controlled process.
We then tested whether reducing the MT assembly capacity of the centrosome has an effect on the assembly of chromosome-dependent MTs under regrowth conditions. We applied BI-2536, the specific inhibitor of the centrosomal kinase Plk1 (lane and Nigg, 1996), to the cells 30 min before nocodazole washout (Lenart et al., 2007; Steegmaier et al., 2007), using DMSO as control (Figure 3B). We then quantified the number of MT asters present in mitotic cells fixed after 1.5 min of nocodazole washout as previously described (Meunier and Vernos, 2011). Plk1 inhibition resulted in the assembly of significantly more MT asters than in the control conditions (respectively, 8.1 ± 2.2 and 5.8 ± 2.1 MT asters; Figure 3C). This result indicates that impairing centrosome maturation through Plk1 inhibition promotes an increase of chromosome-dependent MT nucleation.
Taken together, our results, in agreement with other reports (Torosantucci et al., 2008; Hayward et al., 2013), suggest that the centrosomal and chromosomal pathways compete for some limiting component(s), hypothesized to be tubulin (Torosantucci et al., 2008).
A limiting-component model for tubulin describes the balance between the mitotic MT assembly pathways
It was recently proposed that most animal cells have limiting amounts of tubulin during mitosis due to their small size (Good et al., 2013). This could be important because in most animal cells, two sequential waves of MT nucleation and assembly activation occur: first, through centrosome maturation starting in G2, and, later, through the chromatin-driven pathway after NEBD. Thus the level of centrosome maturation that occurs before NEBD may have a strong effect on the chromatin-dependent MT assembly pathway by restricting the amount of available tubulin (Supplemental Figure S4, A and B). Centrosome maturation might therefore have a novel function for bipolar spindle formation in defining the balance between the two MT assembly pathways in small cells. Because large cells have tubulin in nonlimiting amounts (Good et al., 2013), this putative novel function may be specific to small cells.
We extended a mathematical model using numerical solutions adopted previously to describe the contribution of the MT assembly pathways to the spindle based on their competition for available tubulin (Table 1; Ferenz et al., 2009; Good et al., 2013). We considered that the origin of the total tubulin incorporated into the spindle (WS) corresponds to centrosomal MTs (WC) and to chromosome-dependent MTs (WR). We assumed that WC is a controlled parameter defined by centrosome maturation occurring before NEBD (Verde et al., 1992) and independent of the cell size (Table 1). The model provides a quantitative description of the competition between WC and WR in small cells, whereas in big cells, it describes no competition (Supplemental Figure S4C). Moreover, it defines quantitatively that the degree of competition is set by the level of centrosome maturation (WC) at NEBD, depending on the cell size (Figure 3D). According to this model, in most dividing animal cells whose diameter is in the range of 10–30 μm (Milo and Phillips, 2015), small changes in centrosome maturation will alter significantly the strength of the chromosome-dependent MT assembly pathway and therefore the relative contribution of centrosomal and chromosomal MTs to the spindle (Figure 3D). In contrast, in large cells (diameter >30 μm), the level of centrosome maturation is irrelevant because it will not limit tubulin for the chromosomal pathway (Figure 3D).
TABLE 1:
Variables and parameters used for the mathematical model.
| Notation | Meaning | Value |
|---|---|---|
| WS | Tubulin incorporated by spindle MTs | WR + WC |
| WR | Tubulin incorporated by chromatin-dependent MTs | 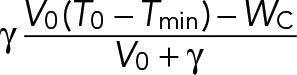 |
| WC | Tubulin incorporated by centrosomal MTs | 2NLδ/NA |
| V0 | Volume of the cell | 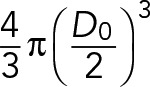 |
| D0 | Diameter of the cell | Variable |
| T0 | Concentration of tubulin in the cell | 25 μM (Gard and Kirschner, 1987; Good et al., 2013) |
| Tmin | Minimum free tubulin concentration | 10 μM (Good et al., 2013) |
| γ | MT dynamics parameter | 36π103 μm3 derived from Good et al. (2013) |
| δ | Density of tubulin dimers per µm of MT | 1625 dimers/μm (McIntosh et al., 2009) |
| N | Number of MTs per centrosome | 250–750 (Ferenz et al., 2009) |
| L | Average centrosomal MT length | 2.5–6 μm (Peset et al., 2005; Ferenz et al., 2009) |
| NA | Avogadro constant | 6.022 × 1023 mol−1 |
The model therefore suggests that competition for a limited pool of tubulin can change substantially the relative contribution of the centrosomal and chromosomal MTs in spindle assembly. Furthermore, our experimental results suggest that interfering with the way in which this competition occurs by forcing the centrosomal and chromosomal pathways to start simultaneously is deleterious for spindle assembly in mammalian cells.
So far, our data indicate that correct spindle assembly depends on the balance between the MT assembly pathways. The establishment of the correct balance requires centrosome maturation and the sequential activation of the MT assembly pathways.
Sequential activation of centrosomal and chromosomal MT assembly pathways is not required for bipolar spindle assembly in Xenopus egg extracts
To test whether the competition model may be relevant for spindle assembly, we first decided to explore whether spindle assembly is compromised under MT regrowth conditions in a system in which tubulin is not limiting, like Xenopus laevis egg extracts (Good et al., 2013).
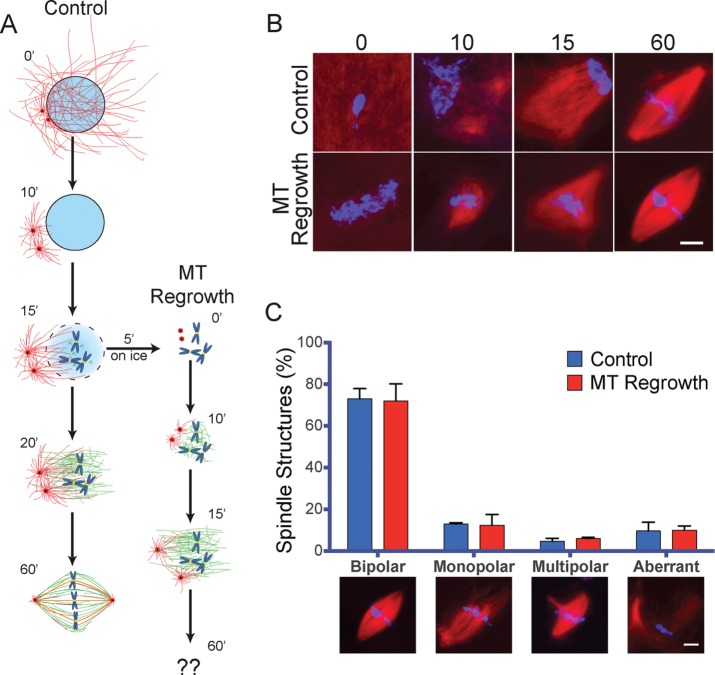
Cytostatic factor (CSF)–arrested extract containing sperm nuclei was released into interphase by calcium addition and cycled back into M phase. Samples were taken at different time points of incubation to monitor MT organization and spindle assembly by fluorescence microscopy. As previously described, 10 min after entry into M phase, we visualized two prominent MT asters emanating from the centrosomes but no chromatin-associated MTs (Figure 4B). This suggested that in this system, the centrosomal and the chromosomal pathways are sequentially activated as in somatic cells. Indeed, at later time points, an array of MTs appeared on the chromosomes.
FIGURE 4:
MT regrowth does not impair spindle assembly in X. laevis egg extracts. (A) Schematic representation of spindle assembly in cycled Xenopus egg extract (Control) and upon MT regrowth after cold treatment (MT Regrowth). Time is in minutes. (B) Representative images of mitotic structures assembled in Xenopus egg extracts in control conditions and after cold-induced MT depolymerization (MT Regrowth) at the indicated times (in minutes) as shown in A. At 60 min, bipolar spindles assemble in both conditions. DNA is in blue and tubulin in red. Scale bar, 10 μm. (C) Control and MT regrowth extracts assemble spindles with similar efficiency. Spindle organization in control egg extracts (blue) or cold-treated egg extracts (MT Regrowth, red). Proportion of spindle organizations after 60 min of incubation. Representative images are shown below the graph. Data from 321 control structures and 312 MT regrowth structures from three independent experiments, counting in each at least 100 mitotic structures/condition. Error bars, SEM. DNA is in blue and tubulin in red. Scale bar, 10 μm. No statistically significant difference was detected between conditions.
To perform a MT regrowth experiment, 15 min after cycling into M phase, the extract was placed on ice for 5 min to promote MT depolymerization and then returned to 20ºC to allow MT regrowth and spindle assembly (Figure 4A). Samples were taken at several time points of incubation to monitor MT organization by fluorescence microscopy (Figure 4B). In contrast to control conditions, 10 min after MT regrowth, a dense MT network covered the chromosomes, suggesting that the centrosomal and chromosomal pathways nucleated MTs concomitantly (Figure 4B). Of interest, after 60 min of regrowth, bipolar spindles formed with similar efficiency as in the control conditions (73.0 ± 4.9 and 72.0 ± 8.2% of bipolar spindles, respectively, for control and MT regrowth conditions; Figure 4C).
Similar results were obtained when we looked at spindle formation during MT regrowth in noncycled egg extracts (CSF extracts) in the presence of only one centrosome. In this case, most spindles are monopolar spindles (Supplemental Figure S5A) because the centrosome acts as a dominant site for pole formation (Heald et al., 1997). After 15 min of incubation, a single MT aster in the proximity of the DNA was observed in control reactions corresponding to the centrosomally nucleated MTs. Under MT regrowth conditions, MTs were also visualized on the chromosomal mass (Supplemental Figure S5B). After 60 min of incubation at 20°C, the proportion of monopolar and bipolar spindles was similar in control and MT regrowth conditions (Supplemental Figure S5C). Therefore, also upon MT regrowth, the individual centrosomes retain the ability to constrain the spindles into a monopolar organization.
These results indicate that altering the sequential activation of the centrosomal and the chromosomal MT assembly pathways has no consequences for spindle assembly in Xenopus egg extracts. However, they do not provide any evidence supporting the idea that this is due to the nonlimiting amounts of tubulin in this system.
Sequential activation of centrosomal and chromosomal MT assembly pathways is necessary for bipolar spindle assembly in Xenopus egg extracts with decreased tubulin levels
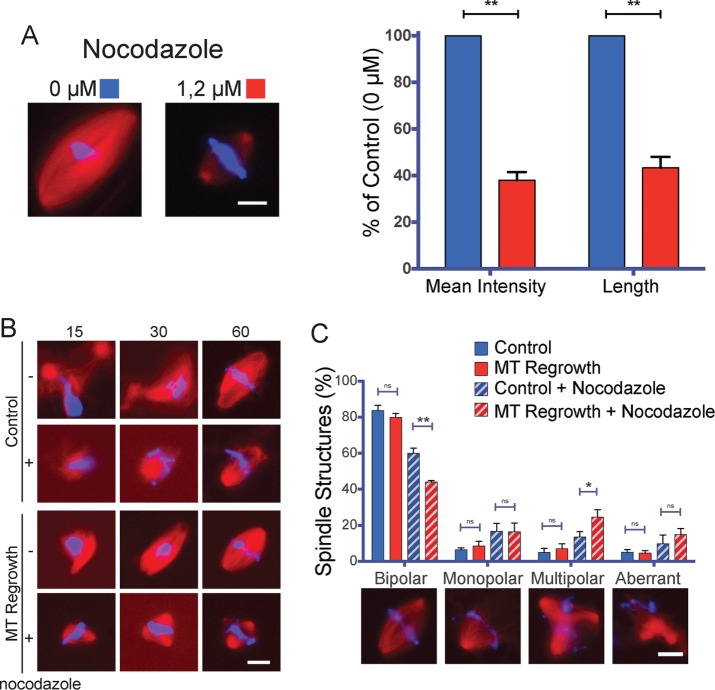
To test directly whether tubulin availability is a critical parameter during spindle assembly in Xenopus egg extracts, we sought to decrease the pool of available tubulin by supplementing the egg extracts with nocodazole (Head et al., 1985). We extrapolated that a concentration of nocodazole that reduces the length and the tubulin density of the spindles would also significantly reduce the availability of free tubulin. Addition of 1.2 μM nocodazole to the egg extract resulted in some reduction in bipolar spindle formation (see later discussion), and these spindles were 56.7 ± 4.7% shorter than controls, and their tubulin density was reduced to 62.0 ± 3.5% (Figure 5A).
FIGURE 5:
Reduced tubulin amounts impair spindle assembly in X. laevis egg extracts undergoing MT regrowth. (A) Spindles assembled in the presence of 1.2 μM nocodazole are shorter and have reduced tubulin density. Left, representative images of spindles assemble in control extract (0 μM, blue) and nocodazole-treated extract (1.2 μM, red). Right, mean tubulin density and mean spindle length as percentage of control (0 μM). Data from 172 control and 158 nocodazole-treated spindles from three independent experiments, quantifying in each at least 50 spindles/condition. DNA is in blue and tubulin in red. Scale bar, 10 μm. **p < 0.01. (B) Representative images of mitotic structures assembled in Xenopus egg extracts treated or not with 1.2 μM nocodazole in control (Control) conditions and after cold-induced MT depolymerization (MT Regrowth) at the indicated times (in minutes), treated as shown in Figure 5A. At 60 min, bipolar spindles assemble in all conditions but with different efficiencies. DNA is in blue and tubulin in red. Scale bar, 10 μm. (C) MT regrowth compromises spindle assembly only in extracts treated with nocodazole. Spindle organization in control egg extracts (blue) or cold-treated egg extracts (MT Regrowth red), treated (diagonal lines pattern) or not (no pattern) with 1.2 μM nocodazole as indicated. Proportion of spindle organizations after 60 min of incubation. Representative images are shown below the graph. Data from 479 control structures, 648 MT regrowth structures, 706 control in the presence of nocodazole, and 778 MT regrowth in the presence of nocodazole structures from four independent experiments, counting in each at least 100 mitotic structures/condition. Error bars: SEM. DNA is in blue and tubulin in red. Scale bar, 10 μm. *p < 0.05 and **p < 0.01.
We next performed a MT regrowth experiment as described earlier (Figure 4A) but in the presence of 1.2 μM nocodazole. Consistent with our previous results, the proportion of bipolar spindles assembled in control or regrowth conditions was similar (Figure 5C). In contrast, MT regrowth in extracts containing 1.2 μM nocodazole resulted in a significantly reduced proportion of bipolar spindles (44.0 ± 0.7% instead of 60.0 ± 2.9% in control conditions) and a significantly increased proportion of multipolar spindles (24.6 ± 4.1% instead of 13.6 ± 2.9% in control conditions; Figure 5C). These data indicate that bipolar spindle assembly is compromised in egg extracts by interference with the sequential activation of the centrosomal and chromosomal MT assembly pathways when tubulin is limiting, similar to what happens in small somatic cells.
Taken together, these results suggest that the sequential activation of the centrosomal and the chromosome-dependent MT assembly pathways is important for bipolar spindle assembly when tubulin is a limiting factor.
DISCUSSION
A novel role for centrosome maturation in regulating the initial contribution of the chromatin-dependent MT assembly pathway in spindle formation
We presented data suggesting that in most animal cells entering mitosis, the sequential activation of MT nucleation, first at the centrosomes and later around the chromatin, plays an important role to ensure bipolar spindle formation and faithful chromosome segregation. Indeed, we show that if in mitotic cells the centrosomal and chromosomal pathways start MT nucleation concomitantly, then bipolar spindle assembly is compromised, leading to mitotic delays and abnormal cell division. This suggests a novel and important function for centrosome maturation that goes beyond centrosome separation. We propose that the level of centrosome maturation sets the initial MT nucleation of the chromosomal pathway because the two MT assembly pathways compete for the limiting pool of tubulin. The resulting balance favors bipolar spindle assembly, correct chromosome attachment, and therefore faithful cell division. This idea is consistent with previously published data (Cimini et al., 2001; Torosantucci et al., 2008; Hasegawa et al., 2013; Hayward et al., 2013). More direct experimental evidence could be gained only by manipulating each of the two pathways at different times as the cell enters mitosis and forms the bipolar spindle. Unfortunately, this is currently technically not possible. Spindle assembly involves another MT-dependent MT assembly pathway that we did not consider specifically in our study. This pathway, involving the augmin complex (Goshima et al., 2008; Uehara et al., 2009), promotes MT amplification by using both centrosomal and acentrosomal MTs as substrates (Lawo et al., 2009; Hayward et al., 2013). In principle, it may not have a strong influence on our conclusions. However, it will be of interest to investigate further the onset timing of the augmin pathway and its effects on spindle assembly.
Our data showing spindle assembly defects in cells undergoing MT regrowth are robust, although we found small differences when MT depolymerization was induced by nocodazole or cold. However, upon these treatments, cells are not in identical conditions. Cells placed in the cold do not progress in the cell cycle, and therefore only cells that were already in mitosis, having some level of chromosome organization and centrosome positioning, are monitored for spindle assembly after being placed them back at 37°C (Supplemental Figure S2A). Instead, cells incubated in nocodazole enter mitosis without MTs, and their chromosomes and centrosomes are more randomly organized and positioned. This may explain the slightly stronger phenotypes we observed after nocodazole washout.
The importance for cell division of the sequential activation of the centrosomal and chromosome-dependent MT assembly pathways may be a general principle in most somatic animal cells (Cimini et al., 2001). We found some differences in the penetrance of the phenotypes in different cell lines, but this may be due to specific external factors, such as cell shape (Kwon et al., 2008). For instance, the reduced sensitivity of the LLC-PK1a cells to MT regrowth treatment may be related to their shape, as they remain relatively flat in mitosis compared with other cells.
The spindle assembly defects observed upon MT regrowth may result from errors in spindle pole organization problems and kinetochore–MT attachments
The mechanism favoring multipolar spindle formation and the anaphase onset delay after mitotic MT regrowth is unclear. It is possible that the increased activity of the chromosome-dependent pathway in these conditions favors the formation of self-organized spindle poles that are not easily clustered with the two centrosomes. An alternative and complementary mechanism may be that the increased MT assembly in the proximity of the chromosomes favors erroneous MT–kinetochore attachments and thereby delays anaphase entry due to the activation of the spindle assembly checkpoint. Consistent with these ideas, PtK1 cells that were used in another study also showed spindle assembly defects during MT regrowth and a higher frequency of lagging chromosomes during anaphase due to an increase number of merotelic attachments (Cimini et al., 2001). Thus the sequential activation of the MT assembly pathways may favor bipolar spindle assembly, as well as the establishment of correct kinetochore–MT attachments.
Bipolar spindle assembly and correct kinetochore–MT attachment are both critical events for faithful mitosis and may be interconnected. However, bipolar spindles can form in the absence of k-fibers (Heald et al., 1996; DeLuca et al., 2002; Cai et al., 2009). MT regrowth impairs both spindle assembly (this study) and kinetochore–MT attachments (Cimini et al., 2001). Although it is therefore difficult to determine the specific mechanism(s) inducing the mitotic delay after MT regrowth, it is likely to depend on an active spindle assembly checkpoint preventing cells from entering anaphase.
Tubulin is a limiting component that may define the balance between the centrosomal and chromosomal MT assembly pathways in mitosis
Our data indicate that the sequential activation of the MT assembly pathways is necessary because they are interdependent (Figure 3). Using a mathematical model, we describe this phenomenon as based on the competition for a limiting component. A prediction of the limiting-component model is that it should be possible to rescue spindle assembly after MT regrowth by tuning the concentration of tubulin or the levels of the proteins involved in MT nucleation. However, these experiments are not trivial. We did seek to interfere with each of the pathways either by silencing TPX2, which is essential for the chromosome-dependent MT nucleation pathway, or inhibiting Plk1, which is essential for centrosome maturation. However, our analysis had to be restricted to the initial phases of MT regrowth (Figure 3) because both proteins have pleiotropic functions in spindle assembly (Eckerdt et al., 2008; Li et al., 2010; Ma et al., 2011).
As previously proposed (Good et al., 2013), tubulin may be an important limiting component because its intracellular concentration is tightly regulated at several levels, including mRNA transcription and stability, tubulin dimer cofolding, and tubulin dimer sequestration (Andersen, 2000; Lopez-Fanarraga et al., 2001; Saussede-Aim and Dumontet, 2009). Nevertheless, we cannot rule out that other factors may also be limiting for spindle assembly. For instance, there are substantial differences between cell lines concerning cellular concentrations of PCM components and microtubule-associated proteins, cell size (Milo and Phillips, 2015), levels of centrosome maturation (unpublished data), and the steepness of the RanGTP gradient governing the chromatin MT pathway (Hasegawa et al., 2013). Thus we propose that optimal equilibrium between the level and the timing of centrosome maturation (Kress et al., 2013), the chromatin MT assembly pathway activity, and the timing of NEBD may be adapted to determine specific spindle assembly dynamics in every cell type (Hasegawa et al., 2013).
Conclusion
In summary, we propose that a novel important mechanism determines the correct balance between the mitotic MT assembly pathways, ensuring correct bipolar spindle formation and cell division fidelity. This mechanism relies on the sequential activation of the MT assembly pathways defined by centrosome maturation and NEBD.
MATERIALS AND METHODS
Cell culture and small interfering RNA transfection
All human cells were grown at 37ºC in a 5% CO2 humid atmosphere in DMEM, 4.5 g/l glucose, supplemented Ultraglutamine (Lonza, Basel, Switzerland), 10% fetal bovine serum (FBS; Invitrogen, Waltham, MA), 100 U/ml penicillin, and 100 μg/ml streptomycin. The stable HeLa cells expressing H2B–eGFP/α-tubulin–mRFP were a gift from P. Meraldi (University of Geneva, Geneva, Switzerland; McAinsh et al., 2006) and were grown in the presence of 400 μg/ml G418 and 20 μg/ml puromycin. The stable U2OS cells expressing γ-tubulin-paGFP/α-tubulin-mCherry were a gift from J. Lüders (Institute for Research in Biomedicine, Barcelona, Spain; Lecland and Lüders, 2014) and were grown in the presence of 400 μg/ml G418 and 20 μg/ml puromycin.
The stable porcine LLC-PK1α cells expressing α-tubulin-GFP were a gift from P. Wadsworth (University of Massachusetts, Amherst, MA) (Tulu et al., 2006). Cells were grown in a 1:1 mixture of Opti-MEM (Invitrogen) and F-10 (GIBCO, Waltham, MA) supplemented with 7.5% FBS, l-glutamine (Invitrogen), penicillin, and streptomycin.
Small interfering RNAs (siRNAs) were transfected with Lipofectamine RNAiMAX (Invitrogen) using 100 pmol per well in six-well plates according to the manufacturer’s protocol. Cells were analyzed 48 h after transfection. The TPX2 siRNA was previously described (Gruss et al., 2002). The control siRNA is a scrambled siRNA from Dharmacon (5′-CGUACGCGGAAUACUUCGAUU-3′).
Plk1 inhibitor BI-2536 (ref 1129; Axon Medchem, The Netherlands) was diluted to 10 mM using DMSO and supplemented to cells at 100 nM.
Live-cell imaging
For fluorescence time-lapse imaging, cells were imaged every 3 min for a total duration of 12 h with a 40× objective on a Zeiss Cell Observer microscope (Zeiss, Jena, Germany). Each image corresponds to the maximal projection of five sections at 2.5-μm intervals. Images were processed using ImageJ (National Institutes of Health, Bethesda, MD) and Photoshop CS5.1 (Adobe, San Jose, CA).
Microtubule regrowth assays
Briefly, cells were plated 2 d before the experiment, and nocodazole (2 μM for the human cell lines and 3.3 μM nocodazole for the LLC-PK1α porcine cells) was added to the medium for 3 h and then washed out four times with phosphate-buffered saline (PBS) and twice with medium at 37°C. For quantitative analysis, cells were fixed in methanol immediately before nocodazole washout (time 0) or 30 min later, and samples were processed for immunofluorescence (IF). Results were quantified counting 200 mitotic cells per condition. For live-imaging observations, cells were mounted on stage immediately after nocodazole washout; thus, depending on the experiment, the first image was obtained between 12 and 21 min after the washout.
For MT regrowth in monastrol (Sigma-Aldrich, St. Louis, MO)-preincubated cells, HeLa cells were placed in medium containing 100 μM monastrol. At 15 h later, 2 μM nocodazole was added to the medium. After 2.5 h, monastrol was washed out four times with medium supplemented with nocodazole. At 30 min later, MT regrowth was performed as described earlier.
For MT regrowth after cold treatment, cells were supplemented with cold medium and incubated on ice for 1½ h. For regrowth, warm medium was supplemented (time 0), and 30 min later, samples were processed for IF.
Xenopus egg extract preparation
Preparation of fresh CF-arrested Xenopus egg extract (CSF extract) and cycled spindle assembly reactions were performed as previously described (Desai et al., 1999). For MT regrowth, 15 min after cycling the extract back into mitosis by addition of CSF extract, the reaction was incubated on ice for 5 min and placed back at 20ºC. Squashes were taken at the indicated times to analyze the state of the MT assembly and organization. MT regrowth using CSF extracts was performed similarly to that fir cycled extracts, incubating the sample on ice for 5 min after 15 min of incubation at 20ºC. Nocodazole was added to the final concentration of 1.2 μM. Intermediate dilutions were done using CSF-XB buffer from the 20 mM stock.
Immunofluorescence
Cells were grown on coverslips and fixed in methanol at −20°C for 10 min. For the visualization of astral MTs, cells were fixed using paraformaldehyde and 4%-PHEM (60 mM 1,4-piperazinediethanesulfonic acid, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10 mM ethylene glycol tetraacetic acid, 8 mM MgSO4, pH 7.00). Fixative was added to cells directly after aspiration of the medium. Cells were incubated for 10 min at room temperature and later washed twice with PBS. The blocking and antibody dilution buffer was 2% BSA (Sigma-Aldrich) and 0.1% Triton X-100 (Sigma-Aldrich). Coverslips were mounted in 10% Mowiol (Calbiochem, Kenilworth, NJ) in 0.1 M Tris-HCl, pH 8.2, and 25% glycerol (Merck, Kenilworth, NJ). Fixed cells were visualized using 63 or 100× objectives on an inverted DMI-6000 Leica wide-field fluorescence microscope.
Spindles assembled in egg extracts were diluted in spindle dilution buffer (30% glycerol, 1% Triton X-100, BRB80) and spun down on a coverslip through a 4-ml spindle cushion (40% glycerol, BRB80). Coverslips were processed for IF as described . Samples were visualized with a 40× objective on an inverted DMI-6000 Leica wide-field fluorescence microscope.
Antibodies
The homemade rabbit anti-hTPX2 was previously described (Gruss et al., 2002) and was used at 10 μg/ml for IF and at 1 μg/ml for Western blot analysis.
The following antibodies were used: mouse anti–α-tubulin (DM1A; Sigma-Aldrich) at 1:1000 for IF; rabbit anti–β-tubulin (ab6046; Abcam, Cambridge, UK) at 1:700 for IF; mouse anti-centrin (20H5; Millipore, Kenilworth, NJ) at 1:2000 for IF; rabbit anti-pericentrin (ab4448; Abcam) at 1:1000 for IF; and mouse anti–γ-tubulin GTU-488 (T6556; Sigma-Aldrich) 1:2000 for IF. Secondary antibodies anti-rabbit and anti-mouse conjugated to Alexa 488, 568, or 680 (Invitrogen) or IRdye 800 CW (Li-Cor, Lincoln, NE) were used at 1:1000 for IF and 1:10,000 for Western blot. Samples were visualized with a 40× objective on an inverted DMI-6000 Leica wide-field fluorescence microscope. Pictures were acquired with the Leica Application Suite software (Leica Microsystems, Wetzlar, Germany). Images were processed and mounted using Photoshop CS5.1.
Western blot
Blots were developed using Alexa Fluor 680– and IRdye 800CW–labeled antibodies and analyzed with the Odyssey Infrared imaging system (Li-Cor).
Quantifications
For the quantification of the recruitment of PCM components to the mitotic centrosomes, pictures were obtained using the 100× objective and identical camera settings. The quantification was performed in ImageJ as previously described (Lüders et al., 2006). Briefly, a circle of defined size (2 μm in diameter) was drawn on the focused mitotic centrosome, and the intensity of the fluorescence signal was measured. To compare experiments, in each experiment, the values were normalized on the average intensity of the control cells.
For the quantification of spindle organization in cells undergoing MT regrowth, we considered only cells containing two centrosomes.
For the measurement of spindle length and MT density in spindles assembled in egg extracts, the pictures were taken using the 40× objective and identical camera settings. Images were opened using ImageJ, and spindle areas were drawn manually using the freehand selection tool. Spindle length was measured using the straight-line tool.
Statistics
Statistical analysis of the data was done by performing an independent two-sample t test in Prism 6 (GraphPad Software, La Jolla, CA) or Excel (Microsoft, Redmond, WA).
In box-and-whiskers plots, boxes show values between the 25th and the 75th percentiles, with a line at the median and a cross (+) at the mean, whiskers extend from the 10th to the 90th percentiles, and dots correspond to outlier values.
Mathematical model
In our model, we considered that the total amount of tubulin in the cell is constant, deriving the following conservation law:
|
|
in which the total amount of tubulin V0T0 (where T0 is the concentration of free tubulin before MT polymerization and V0 is the cell volume) is the sum of the tubulin incorporated by centrosomal MTs (WC), chromatin-driven MTs (WR), and the free tubulin (V0T1).
For WC, we assumed that centrosomal MT dynamics reaches a steady state before the chromatin-driven MT assembly pathway is activated (Verde et al., 1992). This assumption is based on data showing that centrosomal MTs need <5 min to reach a steady state, whereas chromatin-driven MTs need >10 min to start polymerizing MTs (Verde et al., 1992). Therefore WC is determined solely by centrosome MT dynamics because for simplicity we assumed that the chromatin-driven MT assembly pathway does not affect WC. This is certainly true before NEBD and WR onset. After NEBD, we cannot exclude feedback between WC and WR, but they are likely very complex, and we do not account for them in the model. However, our conclusions concern mostly the WR onset.
Because WC is a controlled defined parameter, we extended the assumption made by Good et al. (2013) that WS = γ(T1 − Tmin) to the tubulin available for the chromatin MTs: WR = γ(T1 − Tmin).
Based on this assumption, the amount of tubulin available for the chromatin MTs, WR, is proportional to the amount of free tubulin present at the initiation of the chromatin pathway. Here Tmin is the minimum concentration of tubulin necessary for the chromatin pathway to start, and γ is a proportionality constant that can be estimated from experimental data in the limit of infinite tubulin (Good et al., 2013). (Here γ is an effective parameter accounting for all of the MT dynamics parameters.)
We can therefore define the amount of tubulin available for the chromatin MTs as
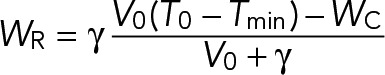 (2) (2)
|
This equation predicts that the amount of tubulin incorporated by the centrosomal MTs (WC) determines the level of tubulin available for the chromatin MTs. In cells having nonlimiting amounts of tubulin (V0T0 → ∞), we can disregard the competition between the two pathways and also the presence of a minimum amount of tubulin required (controlled by Tmin). On the contrary, when the total amount of tubulin is limiting, centrosomal MTs strongly affect the outcome of the chromatin pathway, eventually preventing MT assembly through this pathway for WC > V0(T0 − Tmin) (i.e., when centrosomal MTs incorporate a large proportion of the available tubulin). If the remaining available tubulin is >Tmin, the chromatin MT pathway cannot generate MTs.
Supplementary Material
Acknowledgments
We thank N. Mallol and J. Cela (Centre for Genomic Regulation, Barcelona Institute of Science and Technology) for excellent technical help and the rest of the Vernos lab for comments. We thank S. Meunier, J. Solon, and M. Rodriguez (Centre for Genomic Regulation) for comments during project development and manuscript preparation. We thank R. Heald (University of California, Berkeley, CA) and T. Stearns (Stanford University, Stanford, CA) for useful advice on centrin immunofluorescence staining in Xenopus egg extracts. We thank D. Kern (Cheesman Laboratory, Whitehead Institute, Cambridge, MA) for advice on MT aster visualization. We thank J. Lüders (Institute for Research in Biomedicine, Barcelona, Spain), P. Meraldi (University of Geneva, Geneva, Switzerland), and P. Wadsworth (University of Massachusetts, Amherst, MA) for the gift of stable cell lines. T.C. was supported by Formación de Personal Investigador (FPI) Fellowship BES-2010-031355. This work was supported by Spanish ministry grants BFU2009-10202 and BFU2012-37163. We acknowledge support of the Spanish Ministry of Economy and Competitiveness, Centro de Excelencia Severo Ochoa 2013-2017, SEV-2012-0208.
Abbreviations used:
- MT
microtubule
- NEBD
nuclear envelope breakdown
- PCM
pericentriolar material.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-05-0322) on August 3, 2016.
REFERENCES
- Andersen SS. Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol. 2000;10:261–267. doi: 10.1016/s0962-8924(00)01786-4. [DOI] [PubMed] [Google Scholar]
- Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol. 2014;24:579-585. doi: 10.1016/j.cub.2014.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, O’Connell C, Khodjakov AL, Walczak CE. Chromosome congression in the absence of kinetochore fibres. Nat Cell Biol. 2009;11:832–838. doi: 10.1038/ncb1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins ES, Balchand SK, Faraci JL, Wadsworth P, Lee WL. Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol Biol Cell. 2012;23:3380–3390. doi: 10.1091/mbc.E12-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Murray AW, Mitchison TJ, Walczak CE. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- Eckerdt F, Eyers PA, Lewellyn AL, Prigent C, Maller JL. Spindle pole regulation by a discrete Eg5-interacting domain in TPX2. Curr Biol. 2008;18:519–525. doi: 10.1016/j.cub.2008.02.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz NP, Paul R, Fagerstrom C, Mogilner A, Wadsworth P. Dynein antagonizes Eg5 by crosslinking and sliding antiparallel microtubules. Curr Biol. 2009;19:1833–1838. doi: 10.1016/j.cub.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Kirschner MW. Microtubule assembly in cytoplasmic extracts of Xenopus oocytes and eggs. J Cell Biol. 1987;105:2191–2201. doi: 10.1083/jcb.105.5.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Kidd D, Jeffers K, Wakefield JG, Raff JW. D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 2000;19:241–252. doi: 10.1093/emboj/19.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. Cytoplasmic volume modulates spindle size during embryogenesis. Science. 2013;342:856–860. doi: 10.1126/science.1243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Wittmann M, Yokoyama H, Pepperkok R, Kufer T, Silljé H. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat Cell Biol. 2002;4:871–879. doi: 10.1038/ncb870. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Ryu SJ, Kalab P. Chromosomal gain promotes formation of a steep RanGTP gradient that drives mitosis in aneuploid cells. J Cell Biol. 2013;200:151–161. doi: 10.1083/jcb.201206142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward D, Metz J, Pellacani C, Wakefield JG. Synergy between multiple microtubule-generating pathways confers robustness to centrosome-driven mitotic spindle formation. Dev Cell. 2013;28:81–93. doi: 10.1016/j.devcel.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head J, Lee LL, Field DJ, Lee JC. Equilibrium and rapid kinetic studies on nocodazole-tubulin interaction. J Biol Chem. 1985;260:11060–11066. [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J Cell Biol. 1997;138:615–628. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E, Newport J, Kirschner MW. Respective roles of centrosomes and chromatin in the conversion of microtubule arrays from interphase to metaphase. J Cell Biol. 1984;99:47s–54s. doi: 10.1083/jcb.99.1.47s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern DM, Nicholls PK, Page DC, Cheeseman IM. A mitotic SKAP isoform regulates spindle positioning at astral microtubule plus ends. J Cell Biol. 2016;213:315–328. doi: 10.1083/jcb.201510117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov AL, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Kress E, Schwager F, Holtackers R, Seiler J, Prodon F, Zanin E, Eiteneuer A, Toya M, Sugimoto A, Meyer H, et al. The UBXN-2/p37/p47 adaptors of CDC-48/p97 regulate mitosis by limiting the centrosomal recruitment of Aurora A. J Cell Biol. 2013;201:559–575. doi: 10.1083/jcb.201209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, Habermann B, Tagliaferro A, Poser I, Hutchins JRA, Hegemann B, et al. HAUS, the 8-subunit human augmin complex, regulates centrosome and spindle integrity. Curr Biol. 2009;19:816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- Lecland N, Lüders J. The dynamics of microtubule minus ends in the human mitotic spindle. Nat Cell Biol. 2014;16:770–778. doi: 10.1038/ncb2996. [DOI] [PubMed] [Google Scholar]
- Lenart P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304-315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- Li H, Liu XS, Yang X, Wang Y, Wang Y, Turner JR, Liu X. Phosphorylation of CLIP-170 by Plk1 and CK2 promotes timely formation of kinetochore–microtubule attachments. EMBO J. 2010;29:2953–2965. doi: 10.1038/emboj.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A, Rodriguez-Bravo V, Medema RH. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J Cell Biol. 2009;185:193–202. doi: 10.1083/jcb.200812045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Fanarraga M, Avila J, Guasch A, Coll M, Zabala JC. Review: postchaperonin tubulin folding cofactors and their role in microtubule dynamics. J Struct Biol. 2001;135:219–229. doi: 10.1006/jsbi.2001.4386. [DOI] [PubMed] [Google Scholar]
- Lu MS, Johnston CA. Molecular pathways regulating mitotic spindle orientation in animal cells. Development. 2013;140:1843–1856. doi: 10.1242/dev.087627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J, Patel UK, Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol. 2006;8:137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- Ma N, Titus J, Gable A, Ross JL, Wadsworth P. TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J Cell Biol. 2011;195:87–98. doi: 10.1083/jcb.201106149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh AD, Meraldi P, Draviam VM, Toso A, Sorger PK. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J. 2006;25:4033–4049. doi: 10.1038/sj.emboj.7601293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Morphew MK, Grissom PM, Gilbert SP, Hoenger A. Lattice structure of cytoplasmic microtubules in a cultured mammalian cell. J Mol Biol. 2009;394:177–182. doi: 10.1016/j.jmb.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier S, Vernos I. K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat Cell Biol. 2011;13:1406–1414. doi: 10.1038/ncb2372. [DOI] [PubMed] [Google Scholar]
- Milo R, Phillips R. Cell Biology by the Numbers. New York: Garland Science; 2015. p. 426. [Google Scholar]
- Palazzo RE, Vogel JM, Schnackenberg BJ, Hull DR, Wu X. Centrosome maturation. Curr Top Dev Biol. 2000;49:449–470. doi: 10.1016/s0070-2153(99)49021-0. [DOI] [PubMed] [Google Scholar]
- Peset I, Seiler J, Sardon T, Bejarano LA, Rybina S, Vernos I. Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J Cell Biol. 2005;170:1057–1066. doi: 10.1083/jcb.200504037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saussede-Aim J, Dumontet C. Regulation of tubulin expression: multiple overlapping mechanisms. Int J Med Med Sci. 2009:290–296. [Google Scholar]
- Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, Gurtler U, Garin-Chesa P, Lieb S, Quant J, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Torosantucci L, De Luca M, Guarguaglini G, Lavia P, Degrassi F. Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol Biol Cell. 2008;19:1873–1882. doi: 10.1091/mbc.E07-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulu US, Fagerstrom C, Ferenz NP, Wadsworth P. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr Biol. 2006;16:536–541. doi: 10.1016/j.cub.2006.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Nozawa R-S, Tomioka A, Petry S, Vale RD, Obuse C, Goshima G. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc Natl Acad Sci USA. 2009;106:6998–7003. doi: 10.1073/pnas.0901587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Dogterom M, Stelzer E, Karsenti E, Leibler S. Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J Cell Biol. 1992;118:1097–1108. doi: 10.1083/jcb.118.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Jiang Q, Zhang C. The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. J Cell Sci. 2014;127:4111–4122. doi: 10.1242/jcs.151753. [DOI] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. Microtubule nucleation: gamma-tubulin and beyond. J Cell Sci. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.