Abstract
Objective
The disease process in rheumatoid arthritis (RA) starts years before clinical diagnosis, and elevated disease-specific autoantibodies can be detected in this period. Early responses to known or novel autoantigens likely drive the eventual production of pathogenic autoimmunity. Importantly, the presence of disease-specific autoantibodies can identify individuals who are at high-risk for future RA onset but are currently without arthritis. The goal of the current studies is to characterize plasmablasts in these individuals.
Methods
We investigated the antibody-secreting plasmablasts of a well characterized cohort of individuals at-risk for RA based on serum RA-related autoantibody positivity (Ab+) in comparison to patients with early (<1 yr) seropositive RA and healthy controls. The plasmablast antibody repertoires of at-risk subjects were analyzed using DNA barcode-based methods with paired heavy- and light-chain gene sequencing. Cells were single-cell sorted prior to sequentially adding cell- and plate-specific DNA barcodes, followed by next-generation sequencing.
Results
Total plasmablast levels were similar in Ab+ individuals (1%) and controls (0.4–1.6%). However, increased frequencies of IgA+ vs. IgG+ plasmablasts were observed in Ab+ individuals (39% IgA+, 37% IgG+ plasmablasts) as compared to other groups (1–9% IgA+, 71–87% IgG+ plasmablasts). Paired antibody sequences from Ab+ subjects revealed cross-isotype clonal families and similar sequence characteristics between the IgA and IgG plasmablast repertoires. Ab+ individuals also demonstrated elevated serum levels of IgA isotype anti-CCP3 antibodies.
Conclusion
The IgA plasmablast dominance in these Ab+ individuals suggests that a subset of RA-related autoantibodies may arise from mucosal immune responses and be involved in early disease pathogenesis in individuals who are at-risk for developing RA.
Introduction
Rheumatoid arthritis (RA) is an autoimmune disease affecting about 1% of the population (1), and is associated with disease-specific autoantibodies, including anti-citrullinated protein antibodies (ACPAs) and rheumatoid factors (RF). The disease process begins many years before the clinical diagnosis of RA, with RA-specific autoantibodies present in individuals for up to 5–10 years (2–8). This phase is followed by the development of increased systemic levels of pro-inflammatory cytokines and chemokines as well as epitope spreading of individual ACPAs, culminating in the onset of clinically apparent arthritis and classifiable disease (7). RA-related autoantibodies play a key role in the pathogenesis of RA. For example, murine models of RA show that anti-citrullinated protein/peptide responses can lead to and exacerbate arthritis (9–13). T cell responses to citrullinated proteins can also enhance disease (14).
First degree relatives (FDRs) of RA patients and other subjects identified during screening programs may be classified as being at “high risk” for developing RA in the future when they exhibit one or more of these RA-related autoantibodies, despite not exhibiting any current signs or symptoms of arthritis (15, 16). Importantly, these “high-risk” subjects demonstrate features consistent with a preclinical disease state, including an expanded ACPA repertoire (17) and elevated cytokine/chemokine levels (18). They also demonstrate pulmonary inflammation (19) and expression of RA-related autoantibodies in the lung (20) suggesting the presence of an ongoing mucosal immune response. Individuals who are within one year after their RA diagnosis (Early RA) are also of interest due to their recent transition from preclinical to clinically apparent RA, which is a poorly understood process (21).
Several groups have sequenced single B cells from the synovium (22) and circulating plasmablasts in patients with established RA (23). However, determining the earliest citrullinated and other potentially novel epitopes to which B cells respond is important for the efforts to understand the origins of RA.
During an immune response, peripheral blood plasmablasts arise from both naïve and memory B cells activated by inciting antigens (23–28). In order to study an ongoing antibody response, it is highly informative to study the antibody repertoires of circulating plasmablasts. Herein we have identified and characterized plasmablasts derived from 3 groups: 1) RA-related antibody positive (Ab+) subjects without a history of or current inflammatory arthritis (IA) (Ab+ At-Risk); 2) individuals with Early RA; and 3) normal controls. Utilizing a recently developed method of barcode-enabled single cell antibody sequencing (29), plasmablasts sorted from Ab+ At-Risk individuals were sequenced and characterized.
Patients and Methods
Human samples
Subjects were selected under the aegis of the Studies of the Etiology of RA (SERA) project (30), a natural history study of individuals who are at elevated risk for developing RA because of a first-degree relative (FDR) with RA, a genetic risk factor for RA (shared epitope), or serum RA-related autoantibody positivity identified through community screening efforts. Subjects were selected as follows from the Colorado site of the SERA cohort: 1) Ab+ At-Risk subjects were defined as being positive for 1 or more commercial ACPA tests (CCP2 or CCP3.1) and/or 2 or more RF isotypes in the absence of a history of or presence of IA on examination at the time of the study. These autoantibody criteria are sensitive (~56%) and highly specific (>96%) for future RA and therefore serve as appropriate criteria for identifying subjects for evaluation of early mechanisms of disease initiation and evolution (7). 2) Early RA individuals were all seropositive (CCP and/or RF), met 1987 ACR classification criteria (31), and were within 1 year of the onset of symptoms attributed to clinically apparent RA. 3) Normal controls were recruited from the Denver community through advertising efforts and had no personal history of autoimmune rheumatic disease, no examination evidence of IA at the time of the study, and were ACPA and RF negative.
Autoantibody testing
Anti-CCP3.1 (IgG/IgA Inova Diagnostics, San Diego, CA, USA) and anti-CCP2 (IgG Diastat; Axis-Shield, Dundee, UK) ELISAs were performed and analyzed according to the manufacturers’ instructions. RF was measured using nephelometry (Siemens, Munich GE) with a positivity cutoff of ≥15 IU/ml, and RF isotypes. (IgG/A/M) were measured by ELISA (QUANTA Lite) following manufacturer recommendations (Inova Diagnositics, San Diego, CA, USA). Cutoffs for positivity for RF nephelometry and isotypes were defined using a threshold that was higher than that observed in 95% of 490 randomly selected blood donor controls from the Denver area. All subjects were tested for ACPA IgG/A/M using ELISAs coated with the CCP3 antigen (research use only; donated by Inova Diagnostics, Inc., San Diego, CA, USA) by a technician blinded to sample status. A standard curve was established by performing serial dilutions on a pooled serum sample from patients with RA with known high serum levels of anti-CCP.
Single-cell sorting of plasmablasts
Sorting was performed as previously described (29). Peripheral blood mononuclear cells (PBMCs) were stained with fluorochrome-conjugated antibodies against CD19 (HIB19; Biolegend), CD3 (UCHT1; eBioscience), CD33 (WM53; Biolegend), CD14 (M5E2; Fisher), CD20 (L27; Fisher Scientific), CD27 (M-T271; Fisher Scientific), CD38 (HB7; Fisher Scientific), IgA (IS11-8E10; Miltenyi Biotec), and IgM (G20-127; Fisher Scientific). Analyses were performed on a doublet cell excluded, live, and lymphoid cell gate. IgG+ and IgA+ plasmablasts were single-cell sorted into 96-well plates using a MoFlo XDP100 Cell Sorter (Beckman Coulter), with plasmablasts defined as CD19+CD3-CD33-CD14-CD20-CD27+CD38hi. As IgG-producing plasmablasts have low surface BCR expression, ‘IgG+’ cells were identified by the absence of IgA and IgM staining. The isotype of sorted cells was subsequently confirmed by gene-specific PCR and by identification of isotype-specific sequences in the final data (Supplemental Table S1).
Barcode-enabled antibody sequencing
Antibody repertoire sequencing was performed as previously described (23, 29). Briefly, plasmablasts were sorted into 96-well plates containing lysis buffer (10mM Tris with 1U/μL RiboLock RNAse inhibitor) and stored at −80C. One of 96 unique well-specific DNA barcodes was added to the cDNA of each cell by template switching during reverse transcription with Maxima Reverse Transcriptase (Thermo Fisher Scientific). Well ID-tagged cDNA was pooled from each plate, and plate-specific indices were added during PCR with primers specific for the HC and LC constant regions. Gamma, Kappa, and Lambda primers were as described in (23), and the nested Alpha gene-specific primer sequences were 1: ATT CGT GTA GTG CTT CAC GTG; 2: CTA TGC GCC TTG CCA GCC CGC GGG AAG ACC TTG GGG CTG GT. Barcoded amplicons from multiple plates were pooled prior to the addition of sequencing adaptors and final purification with AMPure XP beads (Beckman Coulter). Samples sequenced prior to Fall 2014 underwent Roche 454 sequencing using Lib-L adaptors and Titanium chemistry. Following the development of 600 bp read lengths on the Illumina MiSeq platform, samples underwent 2 × 300 MiSeq analysis.
Compound barcode assignment and assembly of sequences
Compound barcode assignment and assembly of amplicon sequences was performed as previously described (23). Sequence data was demultiplexed using a custom software pipeline to separate reads from each single cell according to its unique compound (plate + well) barcode ID. Sff output files from 454 sequencing were read into Python by using the Biopython package, and sequences were grouped and parsed into separate sff files on the basis of their compound ID. To correct for sequencing errors, Newbler 2.6 was used to assemble reads from each sff file into consensus sequences by using the “-cdna”, “-ud” and “-urt” options. For MiSeq, poor quality reads and bases were trimmed (Trimmomatic-0.32) and remaining paired reads stitched together (FLASH-1.2.10). The stitched sequences were then read into Python, grouped by compound ID, and parsed into separate fastq files. Consensus sequences were identified by clustering reads within each fastq file into operational taxonomic units (OTUs) (32–34).
V(D)J assignment and production of phylogenetic trees
HC V(D)J and LC VJ sequences were analyzed with version 1.3.1 of the IMGT HighV-QUEST database (35). IMGT HighV-Quest outputs including gene segment usage and the numbers of silent and non-silent mutations from germline were further analyzed and plotted using GraphPad Prism software and the ggplot2 package in R. Sequences were binned according to their HC V-gene usage, and HC and LC were then concatenated and aligned with Muscle to produce phylogenetic trees as previously described (23, 36). Tree images were drawn using ETE (37).
Analysis of clonality
IMGT HighV-Quest data were read into R and B cells with shared HC VJ and LC VJ gene segments were grouped together. Within these groups, CDR3 amino acid sequences were compared using the stringdist package to calculate Levenshtein distance. Clonal families were defined as sharing HC and LC VJ genes and having a CDR3 AA Levenshtein distance of ≤2 for both HC and LC. Clonal Families were numbered and counted in R prior to statistical analysis with GraphPad Prism.
Confirmation of antibody isotypes
In addition to FACS analysis of plasmablast antibody isotypes, the production of IgG or IgA by cells from a given patient was confirmed in two ways: i) gene-specific PCR during library preparation for sequencing, in which sorted IgA plates were not amplified with IgG primers and vice-versa; ii) the presence of isotype-specific sequences (“ASTKG” for IgG versus “ASPTS” for IgA).
Multiplex autoantigen arrays
Levels of IgG and IgA antibodies targeting 51 putative RA-associated autoantigens were measured using a custom Bio-Plex assay as previously described (38), and analyzed on a Luminex 200 instrument running Bio-Plex Manager software version 6.1. Three subjects, including 2 normal controls and 1 early RA patient, were excluded from analysis due broad, non-citrulline specific IgG reactivity (12 or more native antigens elevated 3 fold or greater over the cohort average).
Statistical Analysis
Data are presented as the mean ± SD. Two-tailed unpaired Student’s t tests were used to assess differences between 2 groups, substituting the nonparametric Mann-Whitney test when data did not follow a normal Gaussian distribution, or Welch’s correction when SDs were not equal. For multiple comparisons, 1-way ANOVA was used, substituting the nonparametric Kruskal-Wallis test when data did not follow Gaussian distribution, followed by Geisser-Greenhouse method to correct for violations of the sphericity assumption and either Dunn’s multiple comparisons test or Tukey’s multiple comparisons test. All tests were performed using Prism software (GraphPad Software). P < 0.05 was considered indicative of statistical significance. Fisher’s exact test was used to determine significance for comparisons of prevalence and ANOVA was used for comparisons of continuous variables in tables. Significance Analysis of Microarrays (SAM) version 4.0 was used to run a multiclass comparison between subject groups in the multiplex autoantigen arrays. SAM hits with a q-value of less than 0.1% were chosen for display.
Study Approval
Human samples were collected after obtaining written informed consent from participants prior to their inclusion in the study, and according to the human subject protocols approved by the Investigational Review Board at the University of Colorado Denver Anschutz Medical Campus.
Results
Subject characteristics and autoantibody results
Characteristics of the study subjects are described in Table 1. Ab+ and Early RA subjects are similar in age, with normal controls significantly younger. There are no significant differences in gender or race. Subjects with Early RA reported significantly higher rates of ever and current smoking compared to other groups.
Table 1.
Characteristics of the study subjects.
| Demographic | A Normal ¥ | B Ab+ ‡ | C Early RA § |
|---|---|---|---|
| Age at visit (Mean ± SD) | 38.9 ± 13.4 ‡§ | 52.9 ± 11.2 | 52.6 ± 9.6 |
| Sex (% Female) | 86.36 | 76.47 | 69.23 |
| Race (% NHW) | 59.09 | 85.29 | 69.23 |
| Ever Smoke (% Yes) | 22.73 | 35.29 | 84.6 ¥‡ |
| Current Smoker (% Yes) | 4.55 | 5.88 | 38.5 ¥‡ |
| Pack years (Mean ± SD) | 1.0 ± 0.6 ‡§ | 8.5 ± 7.3 | 12.0 ± 11.1 |
| CCP2 (% Positive) | 0 | 14.71 | 76.9 ¥‡ |
| CCP2 (Mean ± SD) | NA | 163.7 ± 171.8 | 250.1 ± 242.6 |
| CCP3.1 (% Positive) | 0 | 61.8 ¥§ | 92.3 ¥‡ |
| CCP3.1 (Mean ± SD) | NA | 241.5 ± 557.2 | 700.0 ± 978.1 |
| RF (% Positive) | 0 | 32.4 | 53.8 ¥‡ |
| RF (Mean ± SD) | NA | 128.0 ± 193.0 | 272.7 ± 319.3 |
| n=22 | n=33 | n=13 |
Recruited through community screening efforts. Individuals without personal history of autoimmune rheumatic disease, had no examination evidence of IA at the time of the plasmablast study, and who were ACPA and RF negative.
Recruited from the Studies of the Etiologies of Rheumatoid Arthritis project. Subjects were first-degree relatives of probands with RA or were seropositive for RA-related autoantibodies identified through community screening efforts. Subjects were designated seropositive based on serum testing for RA-related autoantibodies at time of original sample collection and were defined as being positive for 1 or more commercial ACPA tests (CCP2 or CCP3.1) and/or 2 or more RF isotypes in absence of a history of or presence of inflammatory arthritis (IA) on examination at the time of the plasmablast study.
<12 months’ duration since onset of symptoms attributed to clinically apparent RA.
Bolded values indicate significance (p value ≤ 0.05, nonparametric testing) with specified groups. Groups are identified with the following symbols: Normal - ¥, Ab+ FDR - ‡, Early RA - §. Samples were compared and significance was determined using chi-squared testing for comparisons of prevalence and ANOVA for continuous variables.
Participants with Early RA exhibited the highest prevalence of seropositivity for CCP2, CCP3.1, and RF of any isotype. Normal controls were negative for all antibodies tested. Supplemental Figure S1 displays the results of CCP and RF testing for all groups.
IgA+ peripheral blood plasmablasts are elevated in Ab+ At-Risk subjects
Plasmablasts, defined as CD19+CD20−CD27+CD38hi (Supplemental Figure S2), were stained and single-cell sorted from peripheral blood mononuclear cells (PBMCs) into 96-well plates as previously described (29). Plasmablasts were sorted into separate plates based on isotype (IgA, IgM, or IgG). A difference in the relative frequency of total plasmablasts between Ab+ At-Risk subjects and normal controls, as well as normal controls compared to those with Early RA, was noted (Figure 1B).
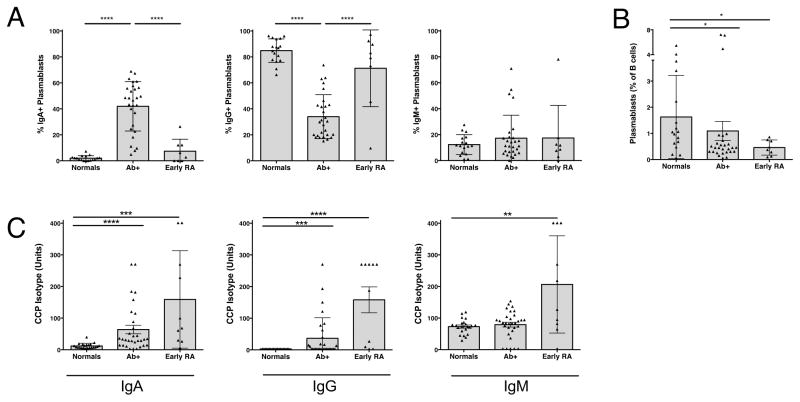
Figure 1. Early RA and Ab+ At-Risk individuals display elevated IgA+ peripheral blood plasmablasts and serum ACPA reactivity.
Plots depict autoantibody and plasmablast levels in the serum from healthy controls (Normals, n = 17), at-risk seropositive subjects (Ab+, n = 28), and subjects with early rheumatoid arthritis (Early RA, n = 8). (A) Antibody isotype distribution of plasmablasts. Individual isotypes were determined by their expression of IgA or IgM. IgG plasmablasts were defined as IgA−IgM− plasmablasts. Each data point represents a single subject; individuals were excluded from reported plasmablast levels if it was determined there was insufficient data to accurately define frequencies of plasmablast isotypes. (B) Total peripheral blood plasmablasts (CD19+CD20−CD27+CD38hi cells) as a percent of total B cells. (C) Serum levels of CCP3-reactive autoantibodies were measured with isotype-specific detection antibodies. Samples were compared using ANOVA utilizing the Geisser-Greenhouse correction, followed by Tukey’s multiple comparisons test (A), or Dunn’s multiple comparisons test (B, C). Data represent mean ± SD. * = P < 0.05; ** = P < 0.01; *** = P < 0.001; **** = P < 0.0001.
Although total plasmablast numbers were not elevated in the Ab+ At-Risk group compared to other groups, the Ab+ group demonstrated a significant increase in IgA+ plasmablasts compared to all other groups (Figure 1A). Conversely, those with Early RA have significantly lower relative levels of IgA+ plasmablasts and instead demonstrate much higher relative levels of IgG+ plasmablasts (Figure 1A). No difference in the relative levels of IgM+ plasmablasts between any of the groups was observed. The pattern of plasmablasts found in individual subjects within the different groups is displayed in Supplemental Figure S3.
We have follow-up data on 17 of these individuals, of which 2 (~12%) developed classifiable RA at 4 and 13 months, respectively, after the plasmablast visit. Overall follow-up of these individuals was of a median of 24 months (range 4–35 months). Additional follow-up of these cases is underway.
When serum samples from these subjects were tested for isotype-specific reactivity to CCP3 plates, both anti-CCP3 IgA and IgG levels were elevated above normal controls in Ab+ At-Risk subjects as well as Early RA subjects (Figure 1C). These results are consistent with our plasmablast isotype findings, as well as our total ACPA level findings, and may suggest that circulating plasmablasts contribute to IgA ACPA production in Ab+ At-Risk individuals.
Sequencing of paired heavy- and light-chain antibody sequences expressed by Ab+ At-Risk subjects demonstrates shared clonal families between IgA and IgG isotypes
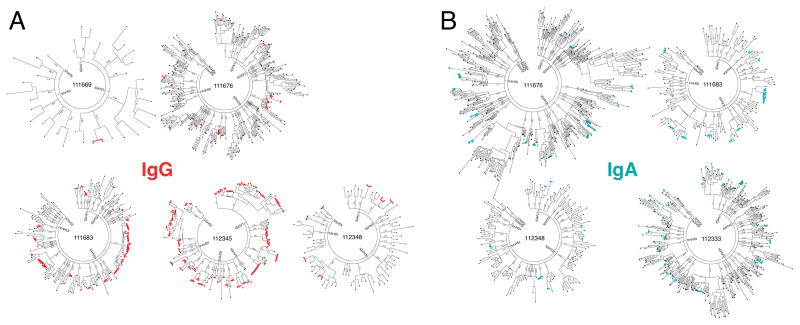
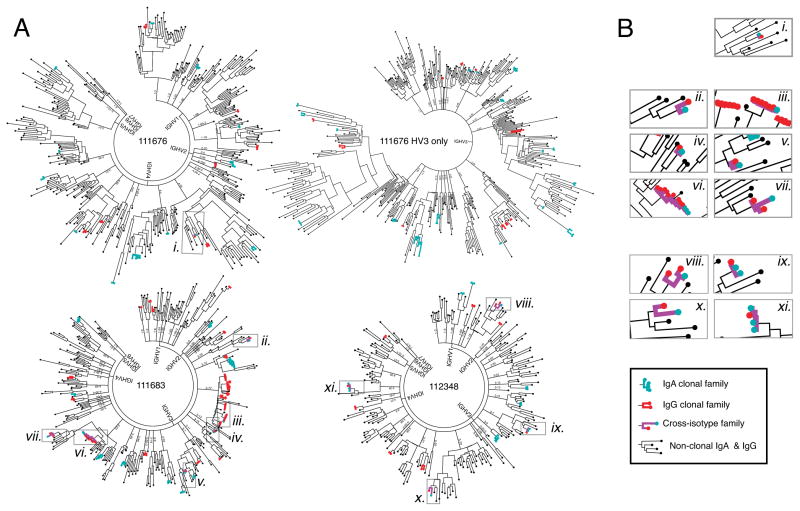
Plasmablasts from five Ab+ At-Risk subjects were used for paired HC + LC repertoire sequencing (Figure 2). Phylogenetic trees of paired IgG (Figure 2A) and IgA (Figure 2B) sequences were generated as previously described (29). In addition to isotype-specific flow cytometric staining, the isotype of IgA+ and IgG+ plasmablasts was confirmed by gene-specific PCR performed during the sequencing process, and by identification of isotype-specific constant region sequences in the final data. These methods confirmed the high fidelity of the method for sequencing the desired isotype (Table S1). From these data, we were able to identify clonal families of antibodies that use the same heavy-chain V and J gene segments, same light-chain V and J gene segments, and have a Levenshtein distance ≤2 in the CDR3 amino acid sequences. Combined IgG + IgA phylogenetic trees were generated to demonstrate the relationship between sequences across these isotypes (Figure 3A). IgG/IgA cross-isotype clonal families of varying sizes were observed in all three Ab+ At-Risk subjects for whom both isotypes had been sequenced (Figure 3B).
Figure 2. Sequencing of plasmablast antibody repertoires in Ab+ At-Risk individuals.
(A) Phylogenetic trees of paired IgG antibody sequences from the peripheral blood plasmablasts of five Ab+ At-Risk subjects. (B) Phylogenetic trees of paired IgA antibody sequences from the peripheral blood plasmablasts of four Ab+ At-Risk subjects. Clonally related sequences (defined as sharing HC and LC V and J genes and HC and LC CDR3 AA sequence Levenshtein distance ≤ 2) are marked in red for IgG and teal for IgA. Phylogenetic trees were generated by concatenating and clustering (MUSCLE alignment) the heavy- and light-chain sequences and arranging them by heavy-chain V-gene family. Each peripheral node represents a single paired antibody.
Figure 3. Shared clonality among IgG and IgA repertoires of Ab+ At-Risk subjects.
(A) Combined trees of IgA and IgG sequences from three antibody positive subjects. Subject 111676 was divided into two trees (IgHV3 family and IgHV1,2,4–7) for better viewing. (B) Magnified views of cross-isotype clonal families showing IgG-IgA shared clonality. Clonal family sequences are marked in red for IgG and teal for IgA; cross-isotype families are indicated by purple lines. Non-clonal sequences of either isotype are marked in black.
Repertoire sequence characteristics are similar between IgG and IgA repertoires
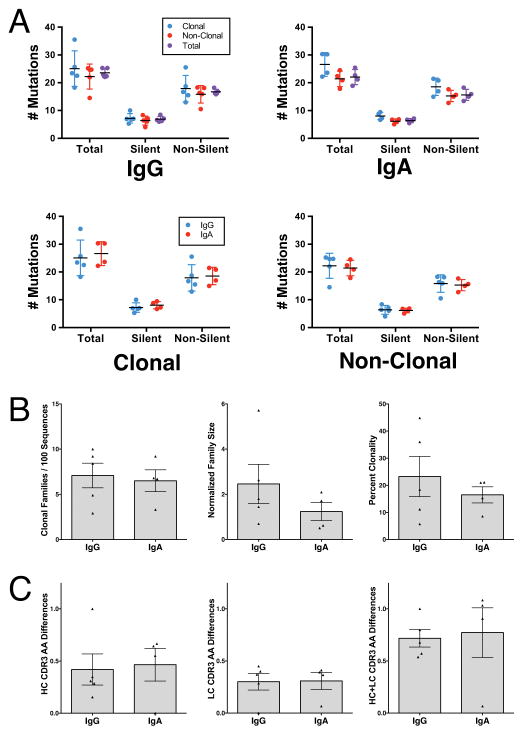
Plasmablast antibodies produced by Ab+ At-Risk subjects contained mutations from the germline gene segments consistent with affinity matured plasmablasts (Figure 4A). Mutation rates did not differ between clonal and non-clonal sequences, nor did they differ between IgG and IgA repertoires. However, within cross-isotype clonal families the IgA family members were more mutated from the germline sequence than their closest IgG relatives (Figure 4A). This is consistent with the fact that IgG-producing cells can class switch to IgA production, but IgA-producing cells cannot switch back to IgG. The number of clonal families per tree sequence did not differ between IgG and IgA repertoires (Figure 4B), nor did the degree of similarity within the CDR3 sequences of each family (Figure 4C).
Figure 4. Antibody repertoire characteristics.
(A) Average numbers of mutations from germline compared between IgA and IgG, as well as between clonal and non-clonal antibody sequences. Each dot represents the mean number of mutations for a single subject (IgG: n = 5; IgA: n = 4). (B) Number, size, and sequence percent of clonal families for each Ab+ At-Risk subject’s antibody repertoire. Each dot represents a single subject (IgG: n = 5; IgA: n = 4). (C) Amount of variation (Levenshtein distance) within the CDR3 amino acid sequences of clonal families from each Ab+ At-Risk subject. Each dot represents the average level of difference for all clonal families within a single subject. Samples were compared using 2-tailed Student’s t test with Welch’s correction. Samples were compared using ANOVA (A) or 2-tailed Student’s t test with Welch’s correction (B, C). Data represent mean ± SD.
Serum IgA and IgG are reactive to known and putative RA-related antigens
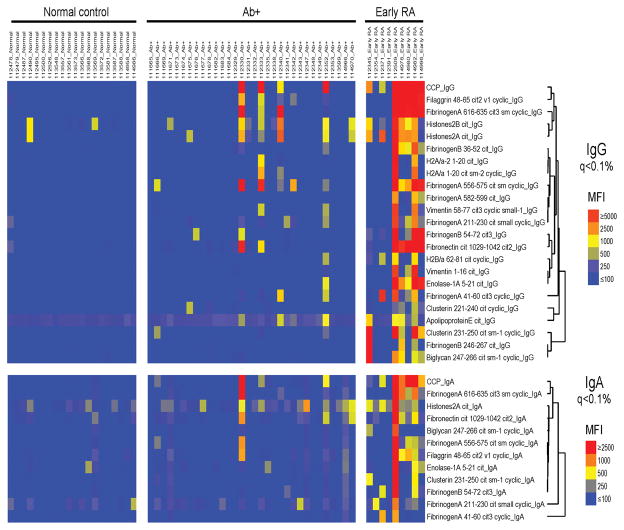
To identify possible IgA autoantigens associated with Ab+ At-Risk status, we utilized a multiplex antigen array to compare IgA and IgG ACPA specificities present in the serum. Figure 5 demonstrates that levels of IgG antibodies to 21 of 51 known and putative RA-related autoantigens are differentially observed in normal controls, Ab+ At-Risk, and Early RA subjects. Levels of IgA antibodies to 13 of 51 antigens were also differentially observed in these groups. Antibodies detected in Ab+ At-Risk individuals targeted many of the same antigens that Early RA subject antibodies.
Figure 5. Serum IgG and IgA reactivity to ACPA.
Heatmap showing levels of anti-citrullinated protein antibodies in the serum of Normals (n = 20), Ab+ At-Risk subjects (n = 32), and Early RA patients (n = 9). Levels of autoantibodies against 51 putative targets of the RA immune response were compared between the described groups using a bead-based multiplex antigen microarray. Significance Analysis of Microarrays (SAM) software was used to sort output based on false discovery rates in order to identify antigens with the greatest differences in autoantibody reactivity between groups. SAM hits with a q-value of less than 0.1% were chosen for display. Labels on the color key are fluorescence intensity relative to the average values in the evaluated cohort.
Discussion
Herein we report the sequence characterization of paired heavy- and light-chain immunoglobulin genes expressed by single B cells in subjects at a high risk for future development of clinically apparent RA. Identification of clonally related antibodies enables analysis of the evolution of responses to potential known and unknown RA-related autoantigens that could characterize the initial phases of clinical autoimmunity and eventually lead to clinical disease.
RA-related autoantibodies can be elevated in the serum years prior to the onset of clinical disease during a period of autoimmunity that has come to be classified as ‘preclinical RA’ (4, 5). Previous studies of preclinical RA have demonstrated that the progression of autoimmunity is characterized by initial reactivity to a limited number of known citrullinated antigens, with epitope spreading over time to the point where clinically-apparent articular RA appears (4, 5, 39). However, what is not known are the mechanisms by which autoimmunity develops, a process that may be brought into better understanding by identifying the potential inciting antigen(s) and ultimately targeting these for disease prevention.
Examining plasmablasts may prove useful in elucidating the initiation and propagation of autoimmunity during preclinical RA. Prior studies in patients with established RA have revealed multiple clonal families of antibodies, many of which bind to known RA-related autoantigens. Thus, this method is capable of isolating monoclonal autoantibodies and identifying their cognate antigens (23, 40–43).
Similar approaches can be used in preclinical RA to identify plasmablast abnormalities with relevance to the pathogenesis of RA. While we did not identify specific antigenic targets of these plasmablasts, it is particularly intriguing that the IgA+ plasmablasts are increased in Ab+ At-Risk individuals without IA. The predominant antibody of mucosal immunity is IgA (44), and IgA ACPAs have been shown to be highly specific for RA when present in both preclinical and recently diagnosed RA patients (45, 46). Furthermore, our findings correlate with previous data in that we observed increased levels of IgA ACPAs using a CCP3 assay within our At-Risk population (46).
This observation suggests several possibilities as to the origin of autoantibodies seen years prior to joint disease onset. In particular, the associations between smoking and RA, as well as emerging data regarding the potential role of oral, lung and gut inflammation in the development of RA all point to a mucosal site of origination of disease (19, 20, 47–50). In particular, the occurrence of severe periodontitis is increased in patients with RA, and these patients also exhibit higher joint disease activity scores (51). Moreover, several oral pathogens are expanded in patients with untreated Early RA, and antibodies to these pathogens are increased in these patients (48, 52, 53). Evidence that the lung may play a role in the mucosal origin of ACPA is also quite strong. Specifically, cigarette smoke exposure is highly associated with future RA development (54, 55). Individuals with RA also often present with pulmonary abnormalities (56), and RA-related autoantibody positive individuals without RA demonstrate airways abnormalities on imaging (19). Studies of sputum from at-risk individuals have also demonstrated lung production of RA-related antibodies, which again supports a mucosal origin (20). Finally, gut-residing bacteria help drive autoimmune arthritis in mice (57), and changes in gut microbiota have been associated with early untreated RA in humans (47, 58).
Steady state levels of IgA+ plasmablasts exist in normal individuals, and these levels are not affected by systemic vaccination against non-mucosal pathogens, which may indicate that systemic and mucosal humoral immune responses are regulated independently of each other (40). Our results demonstrating relatively low IgA plasmablasts in normal individuals are not in alignment with all prior studies (40); however, we have utilized three methods (flow cytometry, isotype-specific primers and DNA sequence analysis) to assure the correct isotype identification in our cohort.
Plasmablast repertoires in Ab+ At-Risk subjects show equivalent mutation rates to those that have been reported for established RA (23), suggesting that these cells are part of an ongoing chronic immune response and not an unrelated gastrointestinal infection. Similarly, the size and frequency of clonal families in Ab+ At-Risk subjects was lower than what we have observed for acute infection and vaccination responses (29, 59), and was more in line with chronic autoimmune activation ((23) and unpublished observations). Whether plasmablasts in at-risk subjects are induced by a sudden trigger (such as viral or bacterial infection) or progress steadily towards autoreactivity over time due to low-level stimulation of the gingival and pulmonary mucosa is not yet clear. However, sequence characteristics consistent with an acute triggering infection were not observed in this study.
Unexpectedly, individuals with Early RA within our study did not have elevated frequencies of IgA+ plasmablasts; it may be that once clinically-apparent RA is present, subjects have already progressed past the early mucosal inflammatory stage. In addition, we speculate that DMARDs used to treat RA could affect total plasmablast levels as well as the different isotypes. It is also not clear why anti-CCP3 IgA levels were elevated in the At-Risk subjects while RF IgA levels were not. One possible explanation is that these two autoantibody pathways are generated at different sites and/or times in the evolution of RA. These issues will need to be addressed in future studies.
We determined that there was no correlation between shared epitope positivity and IgA/IgG predominance of a subject’s plasmablasts. Additionally, there was not a specific pattern of elevated ACPA or RF levels in the individuals with elevated IgA+ plasmablasts, nor were there differences in total serum IgG, IgA, or IgM (data not shown). At-risk individuals with high levels of IgA+ plasmablasts were not more likely to have ever been smokers (data not shown). Our conclusions may be limited by small sample size; however, these findings lead us to speculate that there is a causal relationship between elevated IgA+ plasmablasts and an as yet unknown factor. Our results demonstrate elevated serum IgA anti-CCP antibodies and show peripheral blood IgA+ plasmablasts of as yet unknown specificity, so we cannot directly link the source of IgA anti-CCP to the IgA+ plasmablasts. In addition, whether IgA+ plasmablasts are in fact producing the RF antibodies that are detected serologically will be answered in future studies by expressing and testing monoclonal antibodies derived from IgA+ plasmablast sequences.
Although IgA+ cells were increased and IgG+ cells decreased in Ab+ At-Risk subjects relative to other groups, no significant differences were observed in the antibody sequence characteristics of IgA+ vs. IgG+ plasmablasts. Given that IgG-expressing cells represent an earlier class switching state than IgA+ cells, the equivalent sequence characteristics of these isotypes implies that nascent IgG responses are forming at the same time that other IgG+ cells are class switching to IgA. This is consistent with the transition towards IgG predominance in Early RA subject plasmablasts. The presence of cross-isotype clonal families supports the idea that we have captured a snapshot of the incipient immune response as it transitions from IgG to IgA. Although average mutation rates did not differ between IgG and IgA repertoires of Ab+ At-Risk subjects overall, IgA members of cross-isotype families were more mutated than their IgG counterparts. This is to be expected, given that class-switch recombination can only move in one direction, in this case from IgG to IgA.
We have previously observed large clonal expansions following acute infection or vaccination and smaller clonally expanded families in subjects with chronic infection or autoimmunity (23, 29, 59). Cells of the small clonal families retained during a chronic response may be better able to bind their cognate antigens and compete for resources than their non-surviving clonal relatives. The clonality of both IgG and IgA+ plasmablast repertoires from Ab+ At-Risk individuals is similar to what has been previously reported in established RA (23). Although we have measured an actively developing immune response in the process of switching from IgG to IgA, we have not captured the absolute earliest time point (within weeks of initiation), which would likely be required to observe acute expansion of large clonal families.
There are several caveats to our findings. Because individuals in our control groups did not exhibit increased relative levels of IgA+ plasmablasts, we were not able to sort enough of these cells to generate a meaningful population with which to sequence and compare IgG/IgA antibody repertoires with Ab+ At-Risk subjects. Thus we have not compared cross-isotype clonality between controls and Ab+ At-Risk subjects. We have previously observed cross-isotype families in Lyme disease, pulmonary arterial hypertension, and following Influenza vaccination (Blum & Robinson, unpublished observations); the ability to capture developing clones in the process of class switching makes sense in the context of sequencing plasmablasts, cells that represent nascent immune responses, rather than established memory.
It is also possible that vaccinations or illnesses influenced the presence of circulating plasmablasts in our subjects. In particular, the timing of standard influenza vaccinations (October to March) coincided with the collection of samples from cases who had increased plasmablast levels. We did not collect vaccination information, nor did we exclude individuals who had recently had any infection. However, a critical finding was that there was a significant increase in the relative frequency of IgA+ plasmablasts in Ab+ At-Risk subjects. This increase was not limited to individuals in whom we observed increased plasmablast levels, and was thus likely to be independent of any potential vaccination or illness that might have caused a general increase in plasmablasts.
Another caveat is that the antibodies of interest may be responding to as of yet undetermined pathogens, though immune response to these pathogens may be the initial response that leads down the path of autoimmunity and eventual clinical disease.
These studies enhance our understanding of potentially pathogenic immune responses in the preclinical stages of RA. By utilizing barcode-enabled antibody sequencing to study this unique population of at-risk individuals, we are able to find potentially pathogenic autoantibodies that could be implicated in the development of clinical disease. The discovery of these antibodies and their cognate antigens may lead to the development of tolerizing therapies for at-risk individuals. Study of synovial fluids and tissues from individuals with early and long standing RA may allow us to determine if cells from mucosal tissues drive local joint inflammation. Future studies will focus on longitudinal analyses of plasmablast evolution in preclinical RA to the development of classifiable RA, sequencing plasmablasts that stain positive with citrullinated antigen multimers in order to identify specific antigen reactivity, characterization of plasmablasts at mucosal sites, recombinant expression and characterization of sequence-derived antibodies, and further bioinformatic analyses of antibody sequence characteristics that may be shared across multiple subjects.
Supplementary Material
Serum from each group (Normals: n = 22; Ab+: n = 33; Early RA: n = 13) was collected at the time of the study visit and subsequently tested for levels of CCP2, CCP3.1 and RF by nephelometry, respectively. Samples were compared using ANOVA followed by Dunn’s multiple comparisons test. Data represent mean ± SD. **** P < 0.0001.
IgG+ and IgA+ plasmablasts were single-cell sorted into 96-well plates with plasmablasts defined as CD19+CD3-CD33-CD14-CD20-CD27+CD38hi. Cells were gated on lymphocytes determined by FSC and SSC. Doublets were excluded using SSC-Height and SSC-Width. CD3, CD33, and CD14 positive cells were excluded with a dump gate. Dead cells were excluded with Sytox Blue, and CD19+ cells were selected. CD20 low/negative cells were selected. CD38 high/CD27+ cells were then defined as plasmablasts. As IgG-producing plasmablasts have low surface BCR expression, ‘IgG+’ cells were identified by the absence of IgA and IgM staining, while IgA+ plasmablasts were identified by IgM-IgA+ surface staining.
The pattern of plasmablast isotypes found in individual subjects in the various groups. Normals (n = 17) and individuals in the Early RA group (n = 8) exhibited high levels of IgG plasmablasts, and low levels of both IgA and IgM plasmablasts. Many subjects in the Ab+ At-Risk group (n = 28) displayed higher levels of IgA plasmablasts, and lower levels of both IgG and IgM plasmablasts.
Fidelity of the sequencing method to the desired isotype was determined by identifying isotype-specific sequences in the IgH constant region (i.e., ASTKG for gamma and ASPTS for alpha). ‘Sequencing method’ refers to the isotype we intended to sequence, while ‘sequences yielded’ refers to the actual isotype of each sequence in the resulting data.
Acknowledgments
We are indebted to Marie Feser for her coordination of the SERA projects, Chelsea Fleisher and Courtney Anderson for patient recruitment and sample collection, Laurie Moss for expertly managed databases, and Ryan Gan for his input and assistance. We thank INOVA Diagnostics for donating the CCP isotype kits.
These studies were supported by NIH U01 AI101981 (VMH, WHR) and NIH T32 AR07534 (JDK).
Footnotes
Conflict of Interest Statement: W.H.R. is a consultant to and owns equity in Atreca, Inc. All other authors have declared that no conflict of interest exists.
References
- 1.Neovius M, Simard JF, Askling J group As. Nationwide prevalence of rheumatoid arthritis and penetration of disease-modifying drugs in Sweden. Ann Rheum Dis. 2011 Apr;70(4):624–9. doi: 10.1136/ard.2010.133371. [DOI] [PubMed] [Google Scholar]
- 2.del Puente A, Knowler WC, Pettitt DJ, Bennett PH. The incidence of rheumatoid arthritis is predicted by rheumatoid factor titer in a longitudinal population study. Arthritis Rheum. 1988 Oct;31(10):1239–44. doi: 10.1002/art.1780311004. [DOI] [PubMed] [Google Scholar]
- 3.Aho K, Palosuo T, Heliovaara M, Knekt P, Alha P, von Essen R. Antifilaggrin antibodies within “normal” range predict rheumatoid arthritis in a linear fashion. J Rheumatol. 2000 Dec;27(12):2743–6. [PubMed] [Google Scholar]
- 4.Rantapaa-Dahlqvist S, de Jong BA, Berglin E, et al. Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum. 2003 Oct;48(10):2741–9. doi: 10.1002/art.11223. [DOI] [PubMed] [Google Scholar]
- 5.Nielen MM, van Schaardenburg D, Reesink HW, et al. Specific autoantibodies precede the symptoms of rheumatoid arthritis: a study of serial measurements in blood donors. Arthritis Rheum. 2004 Feb;50(2):380–6. doi: 10.1002/art.20018. [DOI] [PubMed] [Google Scholar]
- 6.Karlson EW, Chibnik LB, Tworoger SS, et al. Biomarkers of inflammation and development of rheumatoid arthritis in women from two prospective cohort studies. Arthritis Rheum. 2009 Mar;60(3):641–52. doi: 10.1002/art.24350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deane KD, O’Donnell CI, Hueber W, et al. The number of elevated cytokines and chemokines in preclinical seropositive rheumatoid arthritis predicts time to diagnosis in an age-dependent manner. Arthritis Rheum. 2010 Nov;62(11):3161–72. doi: 10.1002/art.27638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van de Stadt LA, de Koning MH, van de Stadt RJ, et al. Development of the anti-citrullinated protein antibody repertoire prior to the onset of rheumatoid arthritis. Arthritis Rheum. 2011 Nov;63(11):3226–33. doi: 10.1002/art.30537. [DOI] [PubMed] [Google Scholar]
- 9.Lundberg K, Nijenhuis S, Vossenaar ER, et al. Citrullinated proteins have increased immunogenicity and arthritogenicity and their presence in arthritic joints correlates with disease severity. Arthritis Res Ther. 2005;7(3):R458–67. doi: 10.1186/ar1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn KA, Kulik L, Tomooka B, et al. Antibodies against citrullinated proteins enhance tissue injury in experimental autoimmune arthritis. J Clin Invest. 2006 Apr;116(4):961–73. doi: 10.1172/JCI25422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill JA, Bell DA, Brintnell W, et al. Arthritis induced by posttranslationally modified (citrullinated) fibrinogen in DR4-IE transgenic mice. J Exp Med. 2008 Apr 14;205(4):967–79. doi: 10.1084/jem.20072051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uysal H, Bockermann R, Nandakumar KS, et al. Structure and pathogenicity of antibodies specific for citrullinated collagen type II in experimental arthritis. J Exp Med. 2009 Feb 16;206(2):449–62. doi: 10.1084/jem.20081862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho PP, Lee LY, Zhao X, et al. Autoimmunity against fibrinogen mediates inflammatory arthritis in mice. J Immunol. 2010 Jan 1;184(1):379–90. doi: 10.4049/jimmunol.0901639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cordova KN, Willis VC, Haskins K, Holers VM. A citrullinated fibrinogen-specific T cell line enhances autoimmune arthritis in a mouse model of rheumatoid arthritis. J Immunol. 2013 Feb 15;190(4):1457–65. doi: 10.4049/jimmunol.1201517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deane KD, Norris JM, Holers VM. Preclinical rheumatoid arthritis: identification, evaluation, and future directions for investigation. Rheum Dis Clin North Am. 2010 May;36(2):213–41. doi: 10.1016/j.rdc.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramos-Remus C, Castillo-Ortiz JD, Aguilar-Lozano L, et al. Autoantibodies in predicting rheumatoid arthritis in healthy relatives of rheumatoid arthritis patients. Arthritis Rheumatol. 2015 Aug 5; doi: 10.1002/art.39297. [DOI] [PubMed] [Google Scholar]
- 17.Young KA, Deane KD, Derber LA, et al. Relatives without rheumatoid arthritis show reactivity to anti-citrullinated protein/peptide antibodies that are associated with arthritis-related traits: studies of the etiology of rheumatoid arthritis. Arthritis Rheum. 2013 Aug;65(8):1995–2004. doi: 10.1002/art.38022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes-Austin JM, Deane KD, Derber LA, et al. Multiple cytokines and chemokines are associated with rheumatoid arthritis-related autoimmunity in first-degree relatives without rheumatoid arthritis: Studies of the Aetiology of Rheumatoid Arthritis (SERA) Ann Rheum Dis. 2013 Jun;72(6):901–7. doi: 10.1136/annrheumdis-2012-201505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demoruelle MK, Weisman MH, Simonian PL, et al. Brief report: airways abnormalities and rheumatoid arthritis-related autoantibodies in subjects without arthritis: early injury or initiating site of autoimmunity? Arthritis Rheum. 2012 Jun;64(6):1756–61. doi: 10.1002/art.34344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willis VC, Demoruelle MK, Derber LA, et al. Sputum autoantibodies in patients with established rheumatoid arthritis and subjects at risk of future clinically apparent disease. Arthritis Rheum. 2013 Oct;65(10):2545–54. doi: 10.1002/art.38066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arend WP, Firestein GS. Pre-rheumatoid arthritis: predisposition and transition to clinical synovitis. Nat Rev Rheumatol. 2012 Oct;8(10):573–86. doi: 10.1038/nrrheum.2012.134. [DOI] [PubMed] [Google Scholar]
- 22.Amara K, Steen J, Murray F, et al. Monoclonal IgG antibodies generated from joint-derived B cells of RA patients have a strong bias toward citrullinated autoantigen recognition. J Exp Med. 2013 Mar 11;210(3):445–55. doi: 10.1084/jem.20121486. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Tan YC, Kongpachith S, Blum LK, et al. Barcode-enabled sequencing of plasmablast antibody repertoires in rheumatoid arthritis. Arthritis Rheumatol. 2014 Oct;66(10):2706–15. doi: 10.1002/art.38754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odendahl M, Mei H, Hoyer BF, et al. Generation of migratory antigen-specific plasma blasts and mobilization of resident plasma cells in a secondary immune response. Blood. 2005 Feb 15;105(4):1614–21. doi: 10.1182/blood-2004-07-2507. [DOI] [PubMed] [Google Scholar]
- 25.Radbruch A, Muehlinghaus G, Luger EO, et al. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006 Oct;6(10):741–50. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- 26.Wrammert J, Smith K, Miller J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008 May 29;453(7195):667–71. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fink A, Lemmermann NA, Gillert-Marien D, et al. Antigen presentation under the influence of ‘immune evasion’ proteins and its modulation by interferon-gamma: implications for immunotherapy of cytomegalovirus infection with antiviral CD8 T cells. Med Microbiol Immunol. 2012 Nov;201(4):513–25. doi: 10.1007/s00430-012-0256-z. [DOI] [PubMed] [Google Scholar]
- 28.Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunol Rev. 2012 May;247(1):52–63. doi: 10.1111/j.1600-065X.2012.01124.x. [DOI] [PubMed] [Google Scholar]
- 29.Tan YC, Blum LK, Kongpachith S, et al. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clin Immunol. 2014 Mar;151(1):55–65. doi: 10.1016/j.clim.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolfenbach JR, Deane KD, Derber LA, et al. A prospective approach to investigating the natural history of preclinical rheumatoid arthritis (RA) using first-degree relatives of probands with RA. Arthritis Rheum. 2009 Dec 15;61(12):1735–42. doi: 10.1002/art.24833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988 Mar;31(3):315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 32.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011 Nov 1;27(21):2957–63. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013 Oct;10(10):996–8. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 34.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014 Aug 1;30(15):2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alamyar E, Duroux P, Lefranc MP, Giudicelli V. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- 36.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004 Aug 19;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huerta-Cepas J, Dopazo J, Gabaldon T. ETE: a python Environment for Tree Exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sokolove J, Lindstrom TM, Robinson WH. Development and deployment of antigen arrays for investigation of B-cell fine specificity in autoimmune disease. Front Biosci (Elite Ed) 2012;4:320–30. doi: 10.2741/379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holers VM. Autoimmunity to citrullinated proteins and the initiation of rheumatoid arthritis. Curr Opin Immunol. 2013 Dec;25(6):728–35. doi: 10.1016/j.coi.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mei HE, Yoshida T, Sime W, et al. Blood-borne human plasma cells in steady state are derived from mucosal immune responses. Blood. 2009 Mar 12;113(11):2461–9. doi: 10.1182/blood-2008-04-153544. [DOI] [PubMed] [Google Scholar]
- 41.van Beers JJ, Schwarte CM, Stammen-Vogelzangs J, Oosterink E, Bozic B, Pruijn GJ. The rheumatoid arthritis synovial fluid citrullinome reveals novel citrullinated epitopes in apolipoprotein E, myeloid nuclear differentiation antigen, and beta-actin. Arthritis Rheum. 2013 Jan;65(1):69–80. doi: 10.1002/art.37720. [DOI] [PubMed] [Google Scholar]
- 42.Ossipova E, Cerqueira CF, Reed E, et al. Affinity purified anti-citrullinated protein/peptide antibodies target antigens expressed in the rheumatoid joint. Arthritis Res Ther. 2014 Aug 12;16(4):R167. doi: 10.1186/ar4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ytterberg AJ, Joshua V, Reynisdottir G, et al. Shared immunological targets in the lungs and joints of patients with rheumatoid arthritis: identification and validation. Ann Rheum Dis. 2014 May 9; doi: 10.1136/annrheumdis-2013-204912. [DOI] [PubMed] [Google Scholar]
- 44.Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. The immune geography of IgA induction and function. Mucosal Immunol. 2008 Jan;1(1):11–22. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- 45.Svard A, Kastbom A, Soderlin MK, Reckner-Olsson A, Skogh T. A comparison between IgG- and IgA-class antibodies to cyclic citrullinated peptides and to modified citrullinated vimentin in early rheumatoid arthritis and very early arthritis. J Rheumatol. 2011 Jul;38(7):1265–72. doi: 10.3899/jrheum.101086. [DOI] [PubMed] [Google Scholar]
- 46.Kokkonen H, Mullazehi M, Berglin E, et al. Antibodies of IgG, IgA and IgM isotypes against cyclic citrullinated peptide precede the development of rheumatoid arthritis. Arthritis Res Ther. 2011;13(1):R13. doi: 10.1186/ar3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol. 2008 Aug;35(8):1500–5. [PubMed] [Google Scholar]
- 48.Mikuls TR, Thiele GM, Deane KD, et al. Porphyromonas gingivalis and disease-related autoantibodies in individuals at increased risk of rheumatoid arthritis. Arthritis Rheum. 2012 Nov;64(11):3522–30. doi: 10.1002/art.34595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harvey GP, Fitzsimmons TR, Dhamarpatni AA, Marchant C, Haynes DR, Bartold PM. Expression of peptidylarginine deiminase-2 and -4, citrullinated proteins and anti-citrullinated protein antibodies in human gingiva. J Periodontal Res. 2013 Apr;48(2):252–61. doi: 10.1111/jre.12002. [DOI] [PubMed] [Google Scholar]
- 50.Liu X, Zou Q, Zeng B, Fang Y, Wei H. Analysis of fecal Lactobacillus community structure in patients with early rheumatoid arthritis. Curr Microbiol. 2013 Aug;67(2):170–6. doi: 10.1007/s00284-013-0338-1. [DOI] [PubMed] [Google Scholar]
- 51.de Smit M, Westra J, Vissink A, Doornbos-van der Meer B, Brouwer E, van Winkelhoff AJ. Periodontitis in established rheumatoid arthritis patients: a cross-sectional clinical, microbiological and serological study. Arthritis Res Ther. 2012;14(5):R222. doi: 10.1186/ar4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wegner N, Wait R, Sroka A, et al. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and alpha-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 2010 Sep;62(9):2662–72. doi: 10.1002/art.27552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scher JU, Ubeda C, Equinda M, et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 2012 Oct;64(10):3083–94. doi: 10.1002/art.34539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klareskog L, Stolt P, Lundberg K, et al. A new model for an etiology of rheumatoid arthritis: smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 2006 Jan;54(1):38–46. doi: 10.1002/art.21575. [DOI] [PubMed] [Google Scholar]
- 55.Klareskog L, Malmstrom V, Lundberg K, Padyukov L, Alfredsson L. Smoking, citrullination and genetic variability in the immunopathogenesis of rheumatoid arthritis. Semin Immunol. 2011 Apr;23(2):92–8. doi: 10.1016/j.smim.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 56.Metafratzi ZM, Georgiadis AN, Ioannidou CV, et al. Pulmonary involvement in patients with early rheumatoid arthritis. Scand J Rheumatol. 2007 Sep-Oct;36(5):338–44. doi: 10.1080/03009740701393957. [DOI] [PubMed] [Google Scholar]
- 57.Wu HJ, Ivanov II, Darce J, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010 Jun 25;32(6):815–27. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scher JU, Ubeda C, Artacho A, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015 Jan;67(1):128–39. doi: 10.1002/art.38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu DR, Tan YC, Kongpachith S, et al. Identifying functional anti-Staphylococcus aureus antibodies by sequencing antibody repertoires of patient plasmablasts. Clin Immunol. 2014 May-Jun;152(1–2):77–89. doi: 10.1016/j.clim.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Serum from each group (Normals: n = 22; Ab+: n = 33; Early RA: n = 13) was collected at the time of the study visit and subsequently tested for levels of CCP2, CCP3.1 and RF by nephelometry, respectively. Samples were compared using ANOVA followed by Dunn’s multiple comparisons test. Data represent mean ± SD. **** P < 0.0001.
IgG+ and IgA+ plasmablasts were single-cell sorted into 96-well plates with plasmablasts defined as CD19+CD3-CD33-CD14-CD20-CD27+CD38hi. Cells were gated on lymphocytes determined by FSC and SSC. Doublets were excluded using SSC-Height and SSC-Width. CD3, CD33, and CD14 positive cells were excluded with a dump gate. Dead cells were excluded with Sytox Blue, and CD19+ cells were selected. CD20 low/negative cells were selected. CD38 high/CD27+ cells were then defined as plasmablasts. As IgG-producing plasmablasts have low surface BCR expression, ‘IgG+’ cells were identified by the absence of IgA and IgM staining, while IgA+ plasmablasts were identified by IgM-IgA+ surface staining.
The pattern of plasmablast isotypes found in individual subjects in the various groups. Normals (n = 17) and individuals in the Early RA group (n = 8) exhibited high levels of IgG plasmablasts, and low levels of both IgA and IgM plasmablasts. Many subjects in the Ab+ At-Risk group (n = 28) displayed higher levels of IgA plasmablasts, and lower levels of both IgG and IgM plasmablasts.
Fidelity of the sequencing method to the desired isotype was determined by identifying isotype-specific sequences in the IgH constant region (i.e., ASTKG for gamma and ASPTS for alpha). ‘Sequencing method’ refers to the isotype we intended to sequence, while ‘sequences yielded’ refers to the actual isotype of each sequence in the resulting data.