Abstract
Microtubule-associated protein 7 (MAP7) plays an important role in cancer cells. In this study, we identified the prognostic significance of MAP7 expression in cytogenetically normal acute myeloid leukemia (CN-AML) patients (aged <60 years) based on several microarray datasets. In the first group (n = 129), high MAP7 expression (MAP7high) was associated with adverse overall survival (OS; P = 0.0441) and event-free survival (EFS; P = 0.0114) compared with low MAP7 expression (MAP7low). In addition, the prognostic significance of MAP7 was confirmed by European Leukemia Net (ELN) intermediate-I genetic categories and multivariable analysis. In the second independent group of CN-AML patients (aged <60 years), MAP7high was also associated with adverse OS (n = 88, OS; P = 0.00811). To understand the inherent mechanisms of MAP7’s prognosis, we investigated genome-wide gene/microRNA expression signatures associated with MAP7 expression. Several known oncogenic genes/microRNAs and anti-oncogenic genes/microRNAs were disordered in MAP7high CN-AML patients. In conclusion, MAP7high is an adverse prognostic biomarker for CN-AML, which may be attributed to the distinctive genome-wide gene/microRNA expression and related cell signaling pathways.
Cytogenetically normal acute myeloid leukemia is the largest cytogenetic subset in AML patients1. CN-AML is defined by the lack of detectable chromosome abnormalities and other sensitive prognostic biomarkers of chromosome abnormalities. However, they may contain insidious mutations, aberrantly expressed genes, or microRNAs that are potential prognosticators. Studies have already indicated several factors associated with favorable or poor outcomes. The former group includes NPM1 and double CEBPA mutations. The latter group includes FLT3-ITD and high expression of ERG2, BAALC2, WT1, DNMT3B3, ITPR24, MAPKBP15, and ATP1B16, as well as low expression of LEF17. Such biomarkers can be useful indicators for risk stratification as well as provide insight into the pathogenesis of CN-AML and thus inspire novel targeted therapies.
In this study, we used multiple types of Gene Expression Omnibus (GEO) microarray datasets and different bioinformatic approaches to systematically screen for possible prognostic biomarkers. Gene signatures with both aberrant expression and significant prognostic values were filtered out (5 genes), including MAP7. Overlapping our results with previous reports included in a 24-gene AML prognostic signature8, MAP7 was the only gene that was identified in common across all reports (Supplementary Figure 1).
MAP7 is one of the microtubule-associated proteins (MAPs) and is predominantly expressed in cells of epithelial origin. MAPs are involved in microtubule dynamics and are indispensable in cell division, motility, differentiation and other important cellular and intracellular activities9. MAP7 has shown important prognostic value in several types of malignancy. Recently, MAP7high was shown to be associated with tumor recurrence and poor prognosis in stage II colon cancer patients10. However, the possible impact of MAP7 expression on the prognosis of CN-AML has yet to be examined. Therefore, we aimed to explore the prognostic significance and mechanism of action for MAP7 in CN-AML.
Results
Expression of MAP7 in AML patients and normal controls
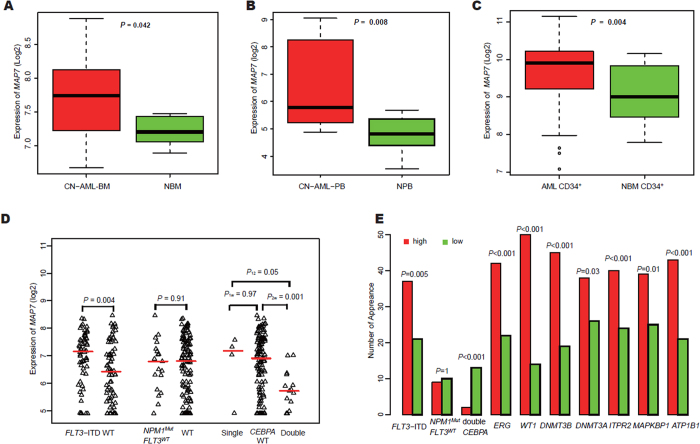
A microarray dataset of BM samples was used for expression analysis, including 116 CN-AML BM and 5 normal bone marrow (NBM) samples (GEO accession number GSE1159). Remarkably higher expression of MAP7 was evident in CN-AML BM than NBM (P = 0.042) (Fig. 1A). Overexpression of MAP7 was further validated by another microarray dataset of peripheral blood (PB) samples [7 CN-AML PB samples (PB samples contained 70–97% blast cells) vs. 10 normal PB (NPB) samples (P = 0.008), GEO accession number GSE9476] (Fig. 1B). Moreover, a third database that included AML CD34+ cells (n = 46) and NBM CD34+ cells (n = 31) derived from potential donors for allogeneic BM transplantation was used for the MAP7 expression analysis (GEO accession number GSE30029). The results showed a significant increase in expression of MAP7 in AML CD34+ cells compared with NBM CD34+ cells (P = 0.004) (Fig. 1C). Furthermore, MAP7high was found in CN-AML (n = 116) among the different subgroups of AML patients as follows: 19 CBFβ-MYH11 patients, 10 patients with complex karyotypes, 17 patients with MLL-translocation, 86 patients with other findings, 18 PML-RARA patients, and 22 AML-ETO patients. Additionally, MAP7high was found in 5 NBM patients (Supplementary Figure 2). These findings showed that MAP7 was widely expressed at high levels in CN-AML patients.
Figure 1. Differential expression of MAP7.
(A) CN-AML-BM cases (n = 116) compared with NBM samples (n = 5); (B) CN-AML-PB cases (n = 7) compared with NPB samples (n = 10); (C) AML CD34+ cells (n = 46) compared with NBM CD34+ cells (n = 31); (D) Expression of MAP7 in CN-AML patients with FLT3-ITD and the mutation of NPM1 and CEBPA; (E) Associations between MAP7 expression and known prognostic biomarkers in CN-AML patients.
Associations between MAP7 expression and other prognostic biomarkers in CN-AML
The 129 CN-AML patients were further divided into subgroups by the presence of FLT3-ITD and mutation status of NPM1 and CEBPA. Levels of MAP7 expression were compared among different subgroups (GEO accession number GSE6891) (Fig. 1C). MAP7 was significantly more highly expressed in samples with FLT3-ITD and single and wild-type CEBPA compared with samples without FLT3-ITD and samples with double CEBPA mutations (P = 0.004, P = 0.05, and P = 0.001, respectively, Fig. 1D). No significant differences were detected between NPM1-mutated (no FLT3) and wild-type samples (P = 0.91) or between single CEBPA-mutated and wild-type samples (P = 0.97, Fig. 1E).
Differences in gene mutation and expression profiles between MAP7high and MAP7low groups
In the 129-patient cohort, patients in the MAP7high group were more likely to carry an FLT3-ITD mutation and less likely to carry a double CEBPA mutation (P = 0.005, P = 0.005) compared with the MAP7low group. No additional links between MAP7 expression and other mutations were found. MAP7high patients with CN-AML were more likely to have a higher expression of ERG, WT1, DNMT3B, DNMT3A, MAPKBP1, ITPR2 and ATP1B1 mutations than MAP7low patients (P = 0.0004, P < 0.0001, P < 0.0001, P = 0.03, P = 0.01, P = 0.005, and P < 0.001, respectively). See Table 1 and Fig. 1D.
Table 1. Patients’ characteristics in the first cohort of 129 CN-AML patients according to MAP7 expression levels.
| Variable | MAP7high, n = 64 | MAP7low, n = 65 | P |
|---|---|---|---|
| Median age, y (range) | 47.5 (18–59) | 45 (16–59) | 0.55 |
| FAB subtype, no (%) | |||
| M0 | 2 (3.1) | 0 (0.0) | 0.24 |
| M1 | 21 (32.8) | 20 (30.8) | 0.85 |
| M2 | 15 (23.4) | 7 (10.8) | 0.06 |
| M4 | 8 (12.5) | 13 (20.0) | 0.34 |
| M5 | 14 (21.9) | 21 (32.3) | 0.24 |
| M6 | 1 (1.6) | 0 (0.0) | 0.5 |
| Other | 3 (4.7) | 4 (6.2) | 1 |
| FLT3-ITD, no (%) | 37 (57.8) | 21 (32.3) | 0.005 |
| FLT3-TKD, no (%) | 9 (14.0) | 9 (13.8) | 1 |
| NPM1Mut/FLT3WT, no (%) | 9 (14.0) | 10 (15.4) | 1 |
| CEBPA, mutated, no (%) | |||
| Single | 3 (4.7) | 1 (1.5) | 0.37 |
| Double | 2 (3.1) | 13 (20.0) | 0.005 |
| IDH1 mutated, no (%) | 8 (12.5) | 10 (15.4) | 0.8 |
| IDH2 mutated, no (%) | 8 (12.5) | 4 (6.15) | 0.24 |
| High ERG, no (%) | 42 (65.6) | 22 (33.8) | 0.0004 |
| High BAALC, no (%) | 34 (53.1) | 30 (46.2) | 0.48 |
| High LEF1, no (%) | 27 (42.1) | 37 (57.0) | 0.11 |
| High WT1, no (%) | 50 (78.1) | 14 (21.5) | <0.0001 |
| High DNMT3B, no (%) | 45 (70.3) | 19 (29.2) | <0.0001 |
| High DNMT3A, no (%) | 38 (59.4) | 26 (40.0) | 0.03 |
| High MAPKBP1, no (%) | 39 (61.0) | 25 (38.5) | 0.01 |
| High ITPR2, no (%) | 40 (62.5) | 24 (36.9) | 0.005 |
| High ATP1B1, no (%) | 43 (67.2) | 21 (32.3) | <0.001 |
FAB, French-American-British classification; ITD, internal tandem duplication; TKD, tyrosine kinase domain; WT: wild type.
High ERG, BAALC, LEF1, WT1, DNMT3B, DNMT3A, MAPKP1, ITPR2 and ATP1B1 expression were defined as an expression level above the median of all samples. NPM1Mut/FLT3WT was defined as CN-AML patients with a mutation of NPM1 (NPM1Mut) and without FLT3-ITD/TKD (FLT3WT).
MAP7high was associated with adverse outcomes
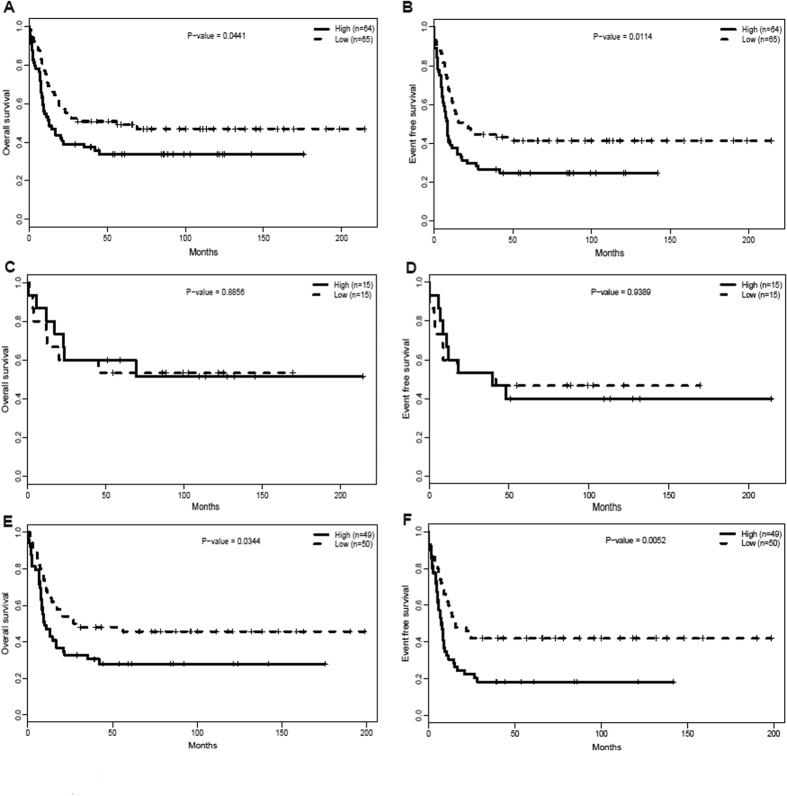
In the 129-CN-AML patient cohort, the MAP7high group had markedly shorter OS (Fig. 2A, P = 0.0441) and EFS (Fig. 2B, P = 0.0114) compared with the MAP7low group. The associations between MAP7 expression and prognostic significance within the European Leukemia Net (ELN) favorable group and intermediate-I genetic group were also separately analyzed. Within the ELN favorable group (n = 30), no significant difference was observed in OS (Fig. 2C, P = 0.8856) or EFS (Fig. 2D, P = 0.9389) between the MAP7high and MAP7low patients. In the ELN Intermediate-I group (n = 99), however, MAP7high patients had a significantly shorter OS (Fig. 2E, P = 0.0344) and EFS (Fig. 2F, P = 0.0052) than MAP7low patients.
Figure 2. Prognostic value of MAP7 expression.
(A) OS and (B) EFS in the 129 CN-AML patients; (C) OS and (D) EFS in the ELN Favorable category; (E) OS and (F) EFS in the ELN Intermediate-I category.
MAP7 expression was associated with shorter OS and EFS in multivariable analyses
After adjusting for the impact of known risk factors, we performed multivariable analyses to confirm the prognostic significance of MAP7 expression. In the multivariable models for OS and EFS, MAP7high had adverse impacts on OS (P = 0.03, Table 2) as well as EFS (P = 0.01, Table 2). The other factors negatively correlated with EFS were NPM1 wild-type and FLT3-ITD mutations (P = 0.01 and P = 0.04, respectively, Table 2).
Table 2. Multivariable analysis of OS and EFS in the first cohort of 129 CN-AML patients.
| Variables in Final Model by End Point | HR | 95% CI | P |
|---|---|---|---|
| OS | |||
| MAP7 expression | 1.7 | 1.05–2.74 | 0.03 |
| NPM1, mutated VS wild type | 0.63 | 0.38–1.05 | 0.07 |
| CEBPA, mutated VS wild type | 0.57 | 0.27–1.21 | 0.14 |
| IDH1, mutated VS wild type | 0.95 | 0.48–1.85 | 0.87 |
| IDH2, mutated VS wild type | 0.58 | 0.25–1.36 | 0.21 |
| EFS | |||
| MAP7 expression | 1.79 | 1.13–2.84 | 0.01 |
| NPM1, mutated VS wild type | 0.52 | 0.31–0.86 | 0.01 |
| CEBPA, mutated VS wild type | 0.69 | 0.35–1.37 | 0.29 |
| FLT3-ITD, presented VS others | 1.6 | 1.0–2.55 | 0.04 |
| IDH1, mutated VS wild type | 1.31 | 0.71–2.4 | 0.39 |
| IDH2, mutated VS wild type | 0.65 | 0.28–1.53 | 0.32 |
HR, hazard ratio; CI, confidence interval.
MAP7high was associated with adverse outcomes in the second independent CN-AML group
We studied an independent group of 88 previously untreated CN-AML patients (aged <60 years). Patients with the FAB-M5 mutation were found to have more MAP7low among all FAB subtypes (P = 0.033). In this cohort, MAP7high patients with CN-AML were more likely to have higher expressions of ERG, WT1, DNMT3B, DNMT3A, ITPR2, MAPKBP1 and ATP1B1 (P < 0.001, P < 0.001, P < 0.001, P = 0.02, P < 0.001, P < 0.001, and P < 0.001, respectively) and lower expression of LEF1 (P = 0.001) compared with MAP7low patients (Supplementary Table 1). In addition, MAP7high patients had significantly shorter OS rates than MAP7low patients (n = 44 vs. n = 44, P = 0.00811; Supplementary Figure 3).
Associations between genome-wide gene-expression profiles and MAP7 expression
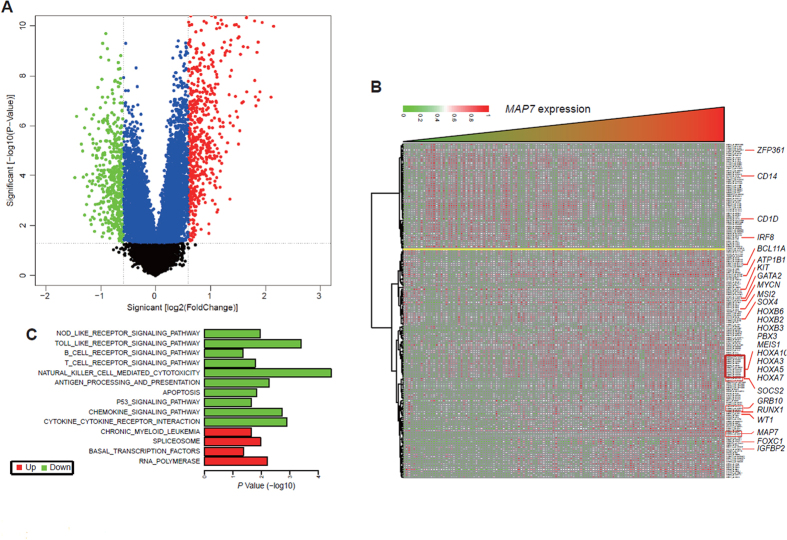
To further assess the role of MAP7 in CN-AML, we derived MAP7-associated gene expression profiles by microarray analysis. We first identified 586 up-regulated and 482 down-regulated genes that were significantly associated with MAP7 expression (P < 0.05, Fold Change = 1.5, Fig. 3A). With a more rigorous analysis (Fold Change = 2, and profiles with NA values were all deleted), 180 genes were filtered and presented in an aberrant expression heat map (Fig. 3B). The up-regulated genes included the following: 1) genes involved in leukemogenesis (MYCN, Sox411), tumorigenesis promoters (HOXA2, 3, 5, 7, 10 and HOXB2, 3, 612,BCL11A13, FOXC114, RUNX115, Pbx316 and Meis116), and tyrosine kinase genes (c-KIT, GRB10); 2) independent adverse prognostic factors in AML including WT1, RUNX117, SOCS218, GATA219, ATP1B16 and MSI220; and 3) genes correlating with chemotherapy resistance in adult AML patients (IGFBP221). The down-regulated genes included the following: 1) immune system activators such as CD14 and CD1d; 2) hematopoietic tumor suppressor IRF822; and 3) ZFP36L1, a promotor of monocyte/macrophage differentiation that represses CDK623. These dysregulated genes might explain the correlation between MAP7 and the prognosis of CN-AML.
Figure 3. Genome-wide gene expression profile and cell signaling pathways associated with MAP7 expression is shown.
(A) Volcano plot of differential gene expression; MAP7high and MAP7low were marked by red and green circles, respectively; (B) Expression heat map of associated genes; (C) Cell signaling pathways.
Tumorigenesis is closely associated with different cell signaling pathways, each composed of many genes. To assess the biological features associated with MAP7, cell signaling pathways in MSigDB were evaluated, and the mean expression of all genes in a pathway was used to quantize its expression level (Fig. 3C). Cell signaling pathways involved in “RNA polymerase”, “basal transcription factors”, “spliceosome”, and “chronic myeloid leukemia” were significantly up-regulated, whereas immune response pathways such as “cytokine-cytokine receptor interaction”, “chemokine signaling”, “antigen processing and presentation”, “natural killer-cell-mediated cytotoxicity”, “T-cell receptor signaling”, “B-cell receptor signaling”, “toll-like receptor signaling”, and “NOD-like receptor signaling” were down-regulated. Apoptotic pathways including “p53 signaling” and “apoptosis” were also down-regulated. These findings were consistent with the dysregulated gene expressions, collectively, these genes and pathways might be involved in the development of CN-AML.
Associations between genome-wide microRNA profiles and MAP7 expression
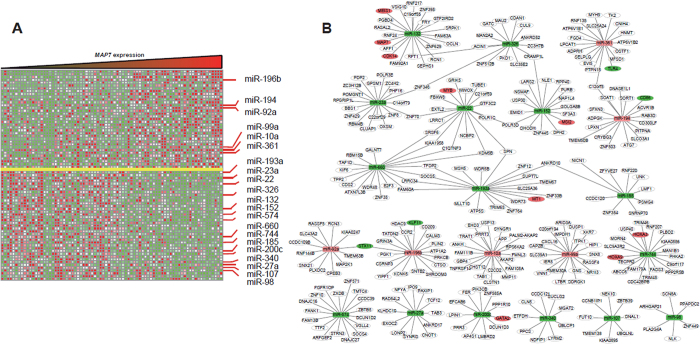
An analysis of microRNA genome-wide profiles revealed 145 microRNAs that were strongly associated with MAP7 expression (P < 0.05) (Fig. 4A). First, MAP7high was positively correlated with levels of miR-196b, miR-92a, miR-99a, miR-10a, miR-361 and miR-194. In previous reports, these microRNAs were shown to have important tumor-promoting properties. miR-196b targets tumor-suppressor genes such as Fas24, and overexpression of miR-196b is associated with aggressive leukemia in mice and a poor prognosis in AML. miR-92a promotes cell proliferation in acute promyelocytic leukemia and induces erythroleukemia through p53 down-regulation25. miR-99a serves as a potential oncogene in pediatric myeloid leukemia26. Overexpression of miR-10a is associated with poor OS in AML patients27. The level of miR-361 was found to be decreased after chemotherapy28. An increased expression of miR-194 is associated with an increased risk of a poor prognosis in CN-AML patients29. Second, miR-193a, miR-23a, miR-22, miR-326, miR-132, miR-152, miR-574, miR-660, miR-744, miR-185, miR-200c, miR-340, miR-27a, miR-107 and miR-98 were down-regulated. In our previous study, we showed that miR-193a targeted c-kit30. The down-regulation of miR-193a could lead to a higher expression of c-kit, consistent with the above-stated gene-expression profiles. Other microRNAs in this group have important tumor-suppression roles. miR-23a targets the BCR/ABL oncogene in CML31. miR-22 regulates the expression of oncogenic NET1 in CML32. miR-326 represses the oncogenic Hedgehog pathway in CML by targeting the signal transducer Smo33. miR-132 targets the p53-down-regulator SIRT1, which in turn promotes p53 expression34. miR-152 is crucial for the anti-tumor effect of natural killer cells by up-regulating HLA-G35. miR-574 is a tumor suppressor in imatinib-resistant CML36. miR-660 is down-regulated in lung cancer patients, and it inhibits lung tumorigenesis by targeting the MDM2-p53 interaction37. Proto-oncogene eEF1A2 is a target of miR-74438. miR-185 suppresses tumor proliferation in breast cancer by directly targeting E2F6 and DNMT1 and indirectly up-regulating BRCA139. miR-200c inhibits breast cancer proliferation by targeting KRAS40. miR-340 inhibits glioblastoma cell proliferation by suppressing CDK6, cyclin-D1 and cyclin-D241. miR-27a functions as a tumor suppressor in acute leukemia by regulating 14-3-3θ42. Epigenetic silencing of miR-107 regulates CDK6 expression in pancreatic cancer43. miR-98 inhibits tumor angiogenesis and invasion by targeting ALK4 and MMP1144.
Figure 4. Genome-wide microRNA expression profile associated with MAP7 expression is shown.
(A) Expression heat map of associated microRNAs; (B) Candidate miRNAs with their target genes.
To explore the dynamic microRNA-mRNA regulatory relationship associated with MAP7 expression in CN-AML, we used microRNA-target prediction algorithms to analyze the target genes of the microRNAs mentioned above45 (Fig. 4B). In the group of up-regulated microRNAs, TLR4, CD86 and KLF11 were the target genes of miR-361, miR-194 and miR-196b, respectively. STX11 was a common target of miR-92a and miR-196b. These target genes have been proven to exert important anti-tumor effects as follows: TLR4 and CD86 can activate the anti-tumor effect of NK-cells, KLF11 is a tumor suppressor gene in myelodysplastic syndrome (MDS)46, and STX11 defects may be associated with MDS and AML47. In the group of down-regulated microRNAs, MEIS1, MAP7 and CDK14 were the target genes of miR-132. MSI2 and MYB were the target genes of miR-152 and miR-22, respectively. WT1 was the target gene of miR-193a. HOXA3 and HOXA9 were the target genes of miR-744. GATA2 was the target gene of miR-200c. Collectively, alterations of the microRNA profiles might contribute to the prognosis of MAP7 through their regulation of target genes.
Discussion
CN-AMLs constitute the largest proportion of all primary AMLs. The leukemic blasts of CN-AML patients contain none of the detectable chromosome abnormalities that are traditionally sensitive prognosticators. Identification of universal prognostic biomarkers has been a key focus in CN-AML research. MAP7, a gene involved in microtubule stabilization and epithelial cell differentiation, had shown promising prognostic values in our serial experiments. First, we found that MAP7 expression was up-regulated in two independent CN-AML cohorts. This finding indicated that MAP7 might play an active role in leukemogenesis. In the first cohort of 129 patients (aged <60 years), our study demonstrated that MAP7high was associated with shorter OS and EFS. In this cohort, we found that MAP7high was associated with the presence of other adverse prognosticators, such as FLT3-ITD, and high ERG, WT1, DNMT3B, ITPR2, MAPKBP1 and ATP1B1 expression. After adjusting for known prognosticators by multivariable analyses, the association of MAP7high with adverse OS and EFS still existed. These results indicated that MAP7high might be an adverse prognostic biomarker and could substitute other adverse factors. Using ELN genetic categories, our results suggested that the prognostic impact of MAP7 expression was most pronounced in the ELN intermediate-I genetic group, and MAP7 expression could therefore be used to refine the risk stratification for these patients.
Given that the first CN-AML group (n = 129, aged <60 years) included a small number of patients who had received Allogeneic Hematopoietic Stem Cell Transplantation (AlloSCT), the prognostic value of MAP7 was further confirmed in the second independent group of CN-AML patients without AlloSCT who received intensive double induction and consolidation chemotherapy (n = 88, aged <60 years). Thus, it seemed that MAP7high could be used as an adverse prognostic biomarker for CN-AML patients.
CN-AML patients show a degree of genetic uniformity, which facilitates the identification of new biomarkers but limits the scope of their application. However, MAP7 was the only gene that was identified by overlapping our results with previous reports included in a 24-gene AML prognostic signature8, indicating that MAP7high may be an adverse biomarker in both CN-AML and AML.
Genome-wide expression profile analysis showed that in the MAP7high CN-AML patients, genes related to cell proliferation regulation were up-regulated, in particular, c-KIT and MYCN. Genes that were independent adverse prognostic factors were also overexpressed. By contrast, genes related to tumor suppression and immune activation were down-regulated in MAP7high patients. Several important cell signaling pathways that promote cell proliferation or contribute to leukemogenesis, such as “RNA polymerase”, “basal transcription factors” and “chronic myeloid leukemia” were up-regulated, whereas some immune activation signaling pathways, such as “toll-like receptor signaling” and “NOD-like receptor signaling” were down-regulated. “P53 signaling” and “apoptosis” pathways were also down-regulated. The down-regulation of these pathways might explain the immune escape and apoptosis blockage in MAP7high CN-AML patients.
Recently, it has been found that microRNAs play an important role in regulating the lineage differentiation of hematopoietic cells. MicroRNAs modulate the expression of oncogenes or tumor suppressors. In our study, the MAP7-associated microRNA profile revealed that in MAP7high CN-AML patients, some oncogenic microRNAs such as miR-196b and miR-99a were up-regulated, whereas anti-tumor microRNAs such as miR-193a and miR-27a were down-regulated.
These dysregulated genes and/or microRNAs could potentially interact, contributing to leukemogenesis. For example, among the dysregulated genes, FOXC1, a gene almost exclusively associated with expression of the HOXA/B locus, could block monocyte/macrophage differentiation and enhance clonogenecity14. Another dangerous liaison observed among Pbx3, Meis1 and Hoxa9 contributed to leukemogenesis16. In the network of dysregulated microRNA-mRNA/pathways, the following associations were made: 1) miR-132 could target MEIS1 and MAP7; 2) miR-193a could target WT1 and c-kit; 3) miR-744 could target HOXA3; 4) miR-152 could target MSI2; 5) miR-200c could target GATA2; 6) miR-92a and miR-660 could cause p53 down-regulation directly or indirectly in AML and lung cancer; and 7) miR-132 could promote p53 down-regulation by targeting SIRT1. These microRNA expression profiles were consistent with the observed gene-expression profiles in the network of dysregulated-mRNA/pathways. The above findings of MAP7-associated gene/microRNA profiles and cell signaling pathways helped explain the leukemogenesis process and the adverse outcomes in MAP7high patients with CN-AML.
With respect to recent findings that promoter methylation of certain genes played an important role in tumorigenesis48 and MAP7 expression and was significantly associated with the increased expression of DNMT3A and DNMT3B in both cohorts, we explored the association between MAP7 expression levels and the methylation levels of genome-wide gene promoters. Although genome-wide deregulated gene promoter methylation profiles were found in CN-AML patients when high vs low MAP7 expressers were compared, none of the genes that contributed to leukemogenesis in AML were found (data not shown). Overall, these data suggest that although MAP7high is a potentially predictive marker in CN-AML, genome-wide deregulated gene promoter methylation profiles do not provide insights into the pathogenesis of CN-AML.
In summary, our study is the first to provide evidence that MAP7high is associated with adverse outcomes in CN-AML patients, even after adjusting for other known molecular risk factors. Previous findings demonstrated the consistency and validity of microarray expression data from quantitative real-time PCR (qPCR). For example, in the second cohort, the microarray expression data and the qPCR data of LEF1 expression were in good agreement. Compared with NBM, MAP7 is widely expressed at a higher level in CN-AML patients, and its expression can be more readily measured in clinical settings. Its overexpression may be a valuable new marker for risk stratification of CN-AML patients. Moreover, distinctive gene/microRNA expression patterns in CN-AML patients provide insights into the pathogenesis processes associated with varying MAP7 expression levels. Our results also indicated that MAP7 may be a promising therapeutic target for CN-AML.
Methods
Patients and treatment
The first group included 129 untreated primary CN-AML patients diagnosed between 1990 and 2008 (median age, 46 years; range, 16–59 years). Per diagnosis, all were uniformly treated based on study protocols of the Dutch-Belgian Cooperative Trial Group for Hematology Oncology (HOVON) (details of the therapeutic protocol available at http://www.hovon.nl). Normal karyotype was established with a conventional cytogenetic examination of at least 20 metaphases from bone marrow (BM). All samples contained 80–100% blast cells after thawing. NPM1, CEBPA, IDH1, and IDH2 mutations, FLT3-ITD and tyrosine kinase domain mutations (FLT3-TKD [D835]) were examined by RT-PCR assays. All clinical, cytogenetic, molecular information, as well as gene expression profiles of the 129 patients, can be publicly downloaded (www.ncbi.nlm.nih.gov/geo, accession number GSE6891). This research was approved by the institutional review boards at Weill Cornell Medical College and Erasmus University Medical Center, and written donor informed consent was obtained in accordance with the Declaration of Helsinki. The methods were carried out in accordance with the approved guidelines.
The second independent group of 88 CN-AML patients (median age: 47 years; range: 17–59 years) also received uniform treatment provided by the multicenter AMLCG-1999 trial. These patients received intensive double induction and consolidation chemotherapy. Gene expression data are publicly available (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE12417). The AMLCG-1999 clinical trials were approved by the local institutional review boards, and informed consent from all patients was obtained in accordance with the Declaration of Helsinki. The methods were carried out in accordance with the approved guidelines.
Microarray analyses
Gene/microRNA expression and methylation data were previously published (accession number GSE1159,GSE9476, GSE30029, GSE6891 and GSE12417 for gene expression). The Cancer Genome Atlas (TCGA) database was used for mRNA/microRNA expression and genome-wide promoter methylation. Briefly, gene expression data were obtained by Affymetrix Human Genome 133 plus 2.0 and U133A Gene Chips. All designs and quality control of the microarray experiment and data normalization were in line with the standard Affymetrix protocols. RNA-Seq data of mRNA/microRNA and genome-wide promoter methylation levels were derived from TCGA obtained by whole-genome high-throughput sequencing and Illumina 450 K chips, respectively, which provided 73 CN-AML patients with all data for mRNA, microRNA and methylation. Patients with MAP7 expression values (whether microarray in GEO or RNA-Seq in TCGA) above the median for all patients were classified as having MAP7high, and the others were considered to have MAP7low. ERG, BAALC, LEF1, WT1, DNMT3A, DNMT3B, MAPKBP1, ITPR2, and ATP1B1 expression levels were also determined from the microarray data.
Statistical analyses
OS was defined as the time from date of diagnosis to death due to any causes. EFS was defined as the time from date of diagnosis to removal from the study because of the absence of complete remission, relapse or death. First, because MAP7 expression was found to be normally distributed in 129 CN-AML patients (Supplementary Figure 4A), we subdivided 129 CN-AML patients into four quartiles (Q1: <25%, Q2: 25~50%, Q3: 50~75%, Q4: >75%) based on MAP7 expression values to determine the best classification method for this group. Second, we found that no significant difference was observed between Q1 and Q2 (OS: Q12, P = 0.852; EFS: Q12, P = 0.777), and the same result was also observed between Q3 and Q4 (OS: Q34, P = 0.342; EFS: Q34, P = 0.524). Although this result was also observed between Q2 and Q3 (OS: Q23, P = 0.283; EFS: Q23, P = 0.151), it had the smallest P value compared with Q12 and Q34 (Supplementary Figure 4B and C). Thus, the cohort was divided into MAP7high and MAP7low groups according to the median of all samples for further analysis. The Kaplan-Meier method and log-rank test were used to estimate the association between MAP7 expression and OS/EFS. Fisher’s exact test and Wilcoxon rank-sum test were used to investigate the associations between MAP7 expression levels and clinical and molecular characteristics for categorical and continuous variables, respectively. Multivariable Cox proportional hazards models were used to study the association between MAP7 expression levels and OS/EFS in the presence of other known risk factors. Between MAP7high and MAP7low groups, Student’s t-test and multiple hypothesis corrections (False Discovery Rate, FDR) were used to identify differences in gene/microRNA expression and genome-wide promoter methylation profiles. The statistical cutoff values were a fold-change of 1.5 and an adjusted P-value of ≤ 0.05. All analyses were performed using the R 3.1.1 software packages.
Additional Information
How to cite this article: Fu, L. et al. High expression of MAP7 predicts adverse prognosis in young patients with cytogenetically normal acute myeloid leukemia. Sci. Rep. 6, 34546; doi: 10.1038/srep34546 (2016).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81500118, 61501519, 81270611).
Footnotes
Author Contributions L.F., H.F. and L.Z. designed the study and wrote the manuscript. K.X., Y.P., Jing W., K.H. and L.T. performed statistical analyses. Y.L., Jijun W. and H.J. analyzed the data. W.H., X.K. and J.S. coordinated the study for the entire time. All authors approved the final manuscript.
References
- Mrozek K., Heerema N. A. & Bloomfield C. D. Cytogenetics in acute leukemia. Blood Rev 18, 115–136 (2004). [DOI] [PubMed] [Google Scholar]
- Schwind S. et al. BAALC and ERG expression levels are associated with outcome and distinct gene and microRNA expression profiles in older patients with de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood 116, 5660–5669 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederwieser C. et al. Prognostic and biologic significance of DNMT3B expression in older patients with cytogenetically normal primary acute myeloid leukemia. Leukemia 29, 567–575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. L., Fu L. & Wang W. D. High expression of inositol 1,4,5-trisphosphate receptor, type 2 (ITPR2) as a novel biomarker for worse prognosis in cytogenetically normal acute myeloid leukemia. Oncotarget 6, 5299–5309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L. et al. Mitogen-activated protein kinase binding protein 1 (MAPKBP1) is an unfavorable prognostic biomarker in cytogenetically normal acute myeloid leukemia. Oncotarget 6, 8144–8154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. L. et al. Overexpression of ATP1B1 predicts an adverse prognosis in cytogenetically normal acute myeloid leukemia. Oncotarget (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzeler K. H. et al. High expression of lymphoid enhancer-binding factor-1 (LEF1) is a novel favorable prognostic factor in cytogenetically normal acute myeloid leukemia. Blood 120, 2118–2126 (2012). [DOI] [PubMed] [Google Scholar]
- Li Z. et al. Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J Clin Oncol 31, 1172–1181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K. M. & Setaluri V. Microtubule-associated proteins as targets in cancer chemotherapy. Clin Cancer Res 13, 2849–2854 (2007). [DOI] [PubMed] [Google Scholar]
- Blum C. et al. The expression ratio of Map7/B2M is prognostic for survival in patients with stage II colon cancer. Int J Oncol 33, 579–584 (2008). [PMC free article] [PubMed] [Google Scholar]
- Omidvar N. et al. PML-RARalpha co-operates with Sox4 in acute myeloid leukemia development in mice. Haematologica 98, 424–427 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N. & Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer 10, 361–371 (2010). [DOI] [PubMed] [Google Scholar]
- Satterwhite E. et al. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood 98, 3413–3420 (2001). [DOI] [PubMed] [Google Scholar]
- Somerville T. D. et al. Frequent Derepression of the Mesenchymal Transcription Factor Gene FOXC1 in Acute Myeloid Leukemia. Cancer Cell 28, 329–342 (2015). [DOI] [PubMed] [Google Scholar]
- Goyama S. et al. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J Clin Invest 123, 3876–3888 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cuellar M. P., Steger J., Fuller E., Hetzner K. & Slany R. K. Pbx3 and Meis1 cooperate through multiple mechanisms to support Hox-induced murine leukemia. Haematologica 100, 905–913 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L. et al. High expression of RUNX1 is associated with poorer outcomes in cytogenetically normal acute myeloid leukemia. Oncotarget (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo G. S. et al. High expression of suppressor of cytokine signaling-2 predicts poor outcome in pediatric acute myeloid leukemia: a report from the Children’s Oncology Group. Leuk Lymphoma 55, 2817–2821 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesink M. et al. High GATA2 expression is a poor prognostic marker in pediatric acute myeloid leukemia. Blood 120, 2064–2075 (2012). [DOI] [PubMed] [Google Scholar]
- Byers R. J., Currie T., Tholouli E., Rodig S. J. & Kutok J. L. MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood 118, 2857–2867 (2011). [DOI] [PubMed] [Google Scholar]
- Kuhnl A. et al. High expression of IGFBP2 is associated with chemoresistance in adult acute myeloid leukemia. Leuk Res 35, 1585–1590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano G. et al. The hematopoietic tumor suppressor interferon regulatory factor 8 (IRF8) is upregulated by the antimetabolite cytarabine in leukemic cells involving the zinc finger protein ZNF224, acting as a cofactor of the Wilms’ tumor gene 1 (WT1) protein. Leuk Res 40, 60–67 (2016). [DOI] [PubMed] [Google Scholar]
- Chen M. T. et al. ZFP36L1 promotes monocyte/macrophage differentiation by repressing CDK6. Sci Rep 5, 16229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun 3, 688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi M., Salehi R., Gheisari Y. & Kazemi M. Inhibition of MicroRNA miR-92a Inhibits Cell Proliferation in Human Acute Promyelocytic Leukemia. Turk J Haematol 30, 157–162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. MiR-99a may serve as a potential oncogene in pediatric myeloid leukemia. Cancer Cell Int 13, 110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi Y. et al. Serum level of miR-10-5p as a prognostic biomarker for acute myeloid leukemia. Int J Hematol 102, 296–303 (2015). [DOI] [PubMed] [Google Scholar]
- Koutova L. et al. The impact of standard chemotherapy on miRNA signature in plasma in AML patients. Leuk Res 39, 1389–1395 (2015). [DOI] [PubMed] [Google Scholar]
- Marcucci G. et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med 358, 1919–1928 (2008). [DOI] [PubMed] [Google Scholar]
- Gao X. N. et al. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene 30, 3416–3428 (2011). [DOI] [PubMed] [Google Scholar]
- Xishan Z., Xianjun L., Ziying L., Guangxin C. & Gang L. The malignancy suppression role of miR-23a by targeting the BCR/ABL oncogene in chromic myeloid leukemia. Cancer Gene Ther 21, 397–404 (2014). [DOI] [PubMed] [Google Scholar]
- Ahmad H. M. et al. miR-22 regulates expression of oncogenic neuro-epithelial transforming gene 1, NET1. FEBS J 281, 3904–3919 (2014). [DOI] [PubMed] [Google Scholar]
- Babashah S. et al. Targeting of the signal transducer Smo links microRNA-326 to the oncogenic Hedgehog pathway in CD34+ CML stem/progenitor cells. Int J Cancer 133, 579–589 (2013). [DOI] [PubMed] [Google Scholar]
- Dal Bo M. et al. The SIRT1/TP53 axis is activated upon B-cell receptor triggering via miR-132 up-regulation in chronic lymphocytic leukemia cells. Oncotarget 6, 19102–19117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X. et al. Down-expression of miR-152 lead to impaired anti-tumor effect of NK via upregulation of HLA-G. Tumour Biol (2015). [DOI] [PubMed] [Google Scholar]
- Jurkovicova D. et al. microRNA expression profiling as supportive diagnostic and therapy prediction tool in chronic myeloid leukemia. Neoplasma 62, 949–958 (2015). [DOI] [PubMed] [Google Scholar]
- Fortunato O. et al. Mir-660 is downregulated in lung cancer patients and its replacement inhibits lung tumorigenesis by targeting MDM2-p53 interaction. Cell Death Dis 5, e1564 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vislovukh A. et al. Proto-oncogenic isoform A2 of eukaryotic translation elongation factor eEF1 is a target of miR-663 and miR-744. Br J Cancer 108, 2304–2311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. et al. miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol Cancer Ther 13, 3185–3197 (2014). [DOI] [PubMed] [Google Scholar]
- Song C. et al. miR-200c inhibits breast cancer proliferation by targeting KRAS. Oncotarget 6, 34968–34978 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. et al. miR-340 inhibits glioblastoma cell proliferation by suppressing CDK6, cyclin-D1 and cyclin-D2. Biochem Biophys Res Commun 460, 670–677 (2015). [DOI] [PubMed] [Google Scholar]
- Scheibner K. A. et al. MiR-27a functions as a tumor suppressor in acute leukemia by regulating 14-3-3theta. PLoS One 7, e50895 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H. et al. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology 9, 293–301 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siragam V. et al. MicroRNA miR-98 inhibits tumor angiogenesis and invasion by targeting activin receptor-like kinase-4 and matrix metalloproteinase-11. Oncotarget 3, 1370–1385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y. C. et al. Prognostic significance of NPM1 mutation-modulated microRNA-mRNA regulation in acute myeloid leukemia. Leukemia (2015). [DOI] [PubMed] [Google Scholar]
- Potapova A. et al. Epigenetic inactivation of tumour suppressor gene KLF11 in myelodysplastic syndromes*. Eur J Haematol 84, 298–303 (2010). [DOI] [PubMed] [Google Scholar]
- Rudd E. et al. Spectrum and clinical implications of syntaxin 11 gene mutations in familial haemophagocytic lymphohistiocytosis: association with disease-free remissions and haematopoietic malignancies. J Med Genet 43, e14 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. et al. Cancer epigenetics and methylation. Science 297, 1807–1808; discussion 1807-1808 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.