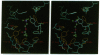Abstract
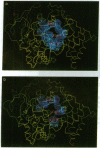
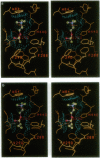
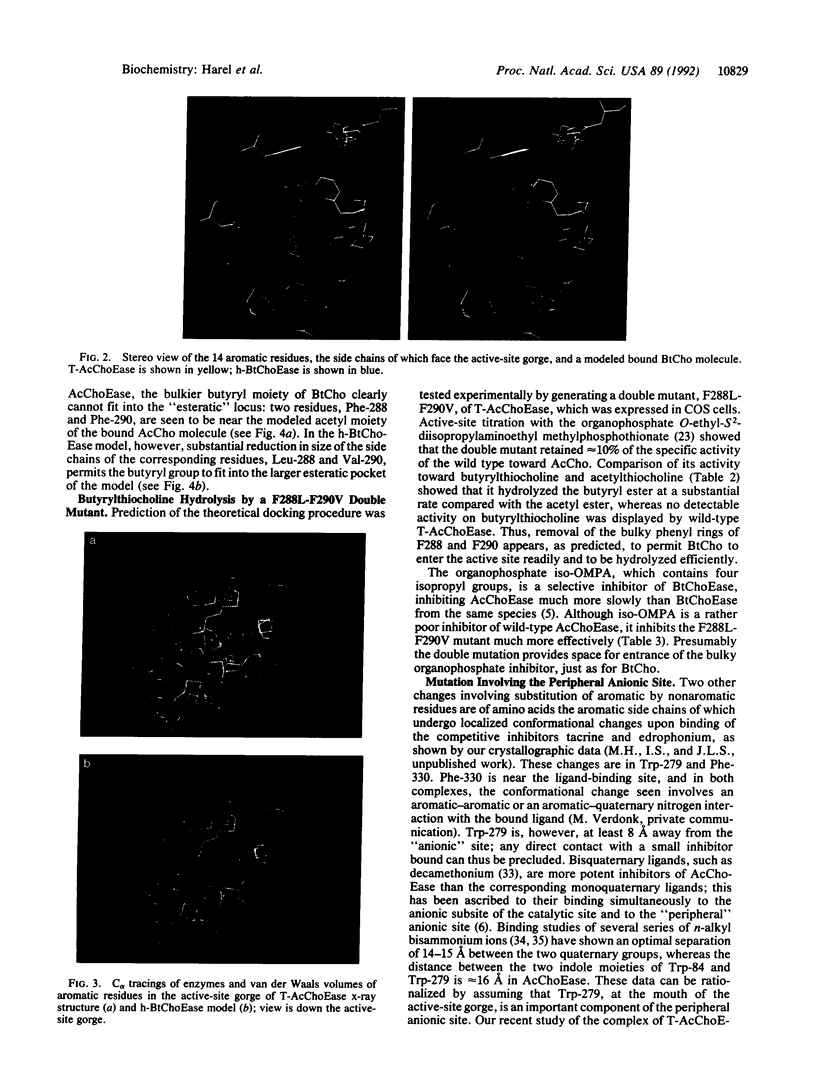
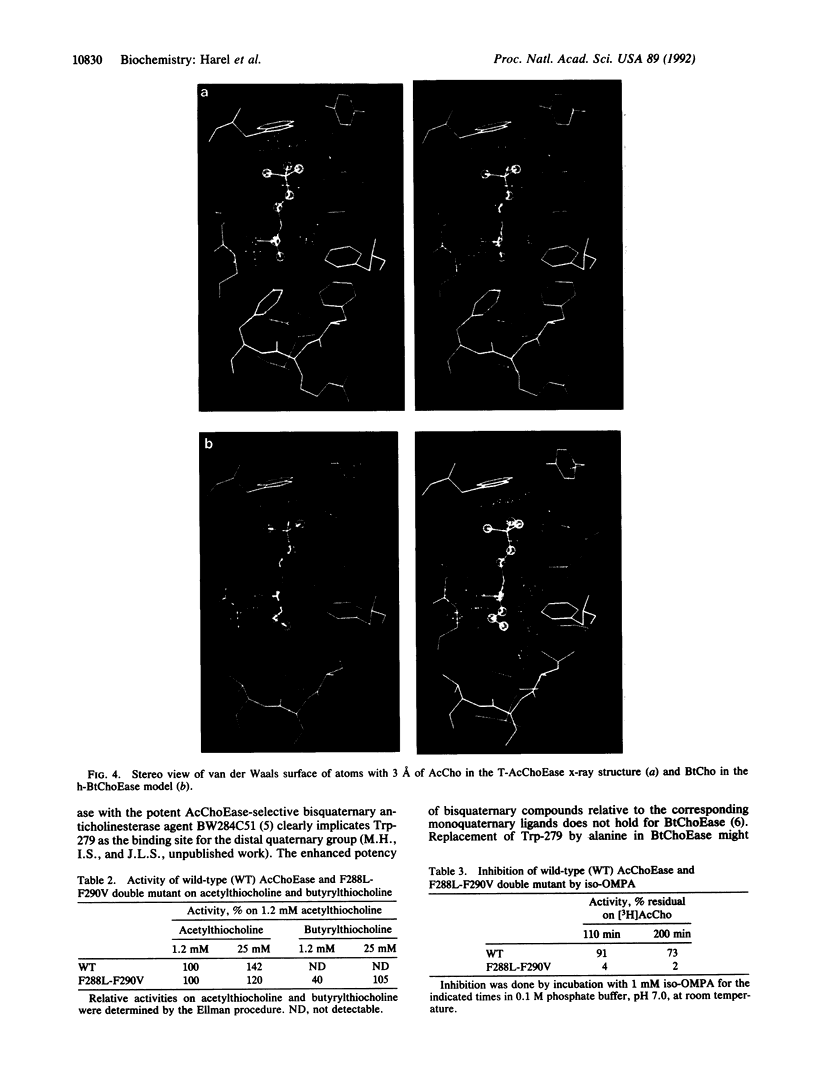
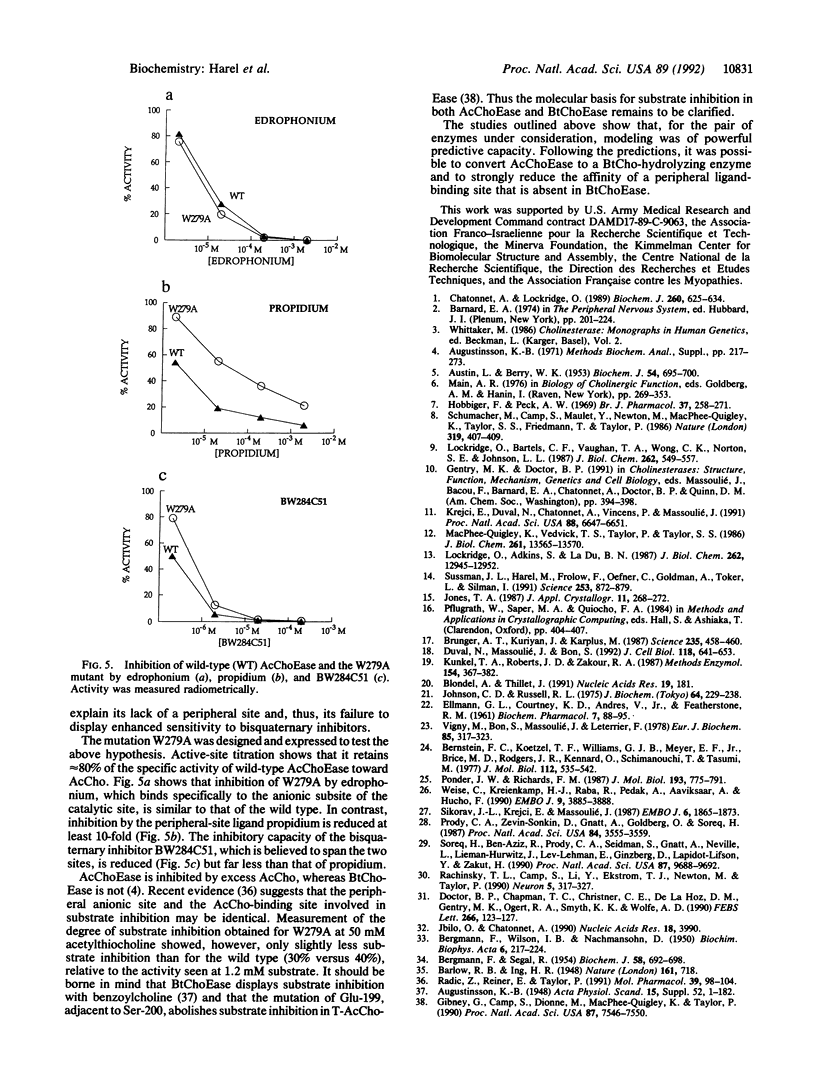
Torpedo acetylcholinesterase (AcChoEase, EC 3.1.1.7) and human butyrylcholinesterase (BtChoEase, EC 3.1.1.8), while clearly differing in substrate specificity and sensitivity to inhibitors, possess 53% sequence homology; this permitted modeling human BtChoEase on the basis of the three-dimensional structure of Torpedo AcChoEase. The modeled BtChoEase structure closely resembled that of AcChoEase in overall features. However, six conserved aromatic residues that line the active-site gorge, which is a prominent feature of the AcChoEase structure, are absent in BtChoEase. Modeling showed that two such residues, Phe-288 and Phe-290, replaced by leucine and valine, respectively, in BtChoEase, may prevent entrance of butyrylcholine into the acyl-binding pocket. Their mutation to leucine and valine in AcChoEase, by site-directed mutagenesis, produced a double mutant that hydrolyzed butyrylthiocholine almost as well as acetylthiocholine. The mutated enzyme was also inhibited well by the bulky, BtChoEase-selective organophosphate inhibitor (tetraisopropylpyrophosphoramide, iso-OMPA). Trp-279, at the entrance of the active-site gorge in AcChoEase, is absent in BtChoEase. Modeling designated it as part of the "peripheral" anionic site, which is lacking in BtChoEase. The mutant W279A displayed strongly reduced inhibition by the peripheral site-specific ligand propidium relative to wild-type Torpedo AcChoEase, whereas inhibition by the catalytic-site inhibitor edrophonium was unaffected.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AUSTIN L., BERRY W. K. Two selective inhibitors of cholinesterase. Biochem J. 1953 Jul;54(4):695–700. [PMC free article] [PubMed] [Google Scholar]
- Augustinsson K. B. Determination of activity of cholinesterases. Methods Biochem Anal. 1971;(Suppl):217–273. doi: 10.1002/9780470110409.ch8. [DOI] [PubMed] [Google Scholar]
- BERGMANN F., SEGAL R. The relationship of quaternary ammonium salts to the anionic sites of true and pseudo cholinesterase. Biochem J. 1954 Dec;58(4):692–698. doi: 10.1042/bj0580692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERGMANN F., WILSON I. B., NACHMANSOHN D. The inhibitory effect of stilbamidine, curare and related compounds and its relationship to the active groups of acetylcholine esterase; action of stilbamidine upon nerve impulse conduction. Biochim Biophys Acta. 1950 Sep;6(1):217–224. doi: 10.1016/0006-3002(50)90094-1. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Blondel A., Thillet J. A fast and convenient way to produce single stranded DNA from a phagemid. Nucleic Acids Res. 1991 Jan 11;19(1):181–181. doi: 10.1093/nar/19.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Chatonnet A., Lockridge O. Comparison of butyrylcholinesterase and acetylcholinesterase. Biochem J. 1989 Jun 15;260(3):625–634. doi: 10.1042/bj2600625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doctor B. P., Chapman T. C., Christner C. E., Deal C. D., De La Hoz D. M., Gentry M. K., Ogert R. A., Rush R. S., Smyth K. K., Wolfe A. D. Complete amino acid sequence of fetal bovine serum acetylcholinesterase and its comparison in various regions with other cholinesterases. FEBS Lett. 1990 Jun 18;266(1-2):123–127. doi: 10.1016/0014-5793(90)81522-p. [DOI] [PubMed] [Google Scholar]
- Duval N., Massoulié J., Bon S. H and T subunits of acetylcholinesterase from Torpedo, expressed in COS cells, generate all types of globular forms. J Cell Biol. 1992 Aug;118(3):641–653. doi: 10.1083/jcb.118.3.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELLMAN G. L., COURTNEY K. D., ANDRES V., Jr, FEATHER-STONE R. M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol. 1961 Jul;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- Gibney G., Camp S., Dionne M., MacPhee-Quigley K., Taylor P. Mutagenesis of essential functional residues in acetylcholinesterase. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7546–7550. doi: 10.1073/pnas.87.19.7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbiger F., Peck A. W. Hydrolysis of suxamethonium by different types of plasma. Br J Pharmacol. 1969 Sep;37(1):258–271. doi: 10.1111/j.1476-5381.1969.tb09543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jbilo O., Chatonnet A. Complete sequence of rabbit butyrylcholinesterase. Nucleic Acids Res. 1990 Jul 11;18(13):3990–3990. doi: 10.1093/nar/18.13.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. D., Russell R. L. A rapid, simple radiometric assay for cholinesterase, suitable for multiple determinations. Anal Biochem. 1975 Mar;64(1):229–238. doi: 10.1016/0003-2697(75)90423-6. [DOI] [PubMed] [Google Scholar]
- Krejci E., Duval N., Chatonnet A., Vincens P., Massoulié J. Cholinesterase-like domains in enzymes and structural proteins: functional and evolutionary relationships and identification of a catalytically essential aspartic acid. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6647–6651. doi: 10.1073/pnas.88.15.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lockridge O., Adkins S., La Du B. N. Location of disulfide bonds within the sequence of human serum cholinesterase. J Biol Chem. 1987 Sep 25;262(27):12945–12952. [PubMed] [Google Scholar]
- Lockridge O., Bartels C. F., Vaughan T. A., Wong C. K., Norton S. E., Johnson L. L. Complete amino acid sequence of human serum cholinesterase. J Biol Chem. 1987 Jan 15;262(2):549–557. [PubMed] [Google Scholar]
- MacPhee-Quigley K., Vedvick T. S., Taylor P., Taylor S. S. Profile of the disulfide bonds in acetylcholinesterase. J Biol Chem. 1986 Oct 15;261(29):13565–13570. [PubMed] [Google Scholar]
- Ponder J. W., Richards F. M. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987 Feb 20;193(4):775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Prody C. A., Zevin-Sonkin D., Gnatt A., Goldberg O., Soreq H. Isolation and characterization of full-length cDNA clones coding for cholinesterase from fetal human tissues. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3555–3559. doi: 10.1073/pnas.84.11.3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachinsky T. L., Camp S., Li Y., Ekström T. J., Newton M., Taylor P. Molecular cloning of mouse acetylcholinesterase: tissue distribution of alternatively spliced mRNA species. Neuron. 1990 Sep;5(3):317–327. doi: 10.1016/0896-6273(90)90168-f. [DOI] [PubMed] [Google Scholar]
- Radić Z., Reiner E., Taylor P. Role of the peripheral anionic site on acetylcholinesterase: inhibition by substrates and coumarin derivatives. Mol Pharmacol. 1991 Jan;39(1):98–104. [PubMed] [Google Scholar]
- Schumacher M., Camp S., Maulet Y., Newton M., MacPhee-Quigley K., Taylor S. S., Friedmann T., Taylor P. Primary structure of Torpedo californica acetylcholinesterase deduced from its cDNA sequence. 1986 Jan 30-Feb 5Nature. 319(6052):407–409. doi: 10.1038/319407a0. [DOI] [PubMed] [Google Scholar]
- Sikorav J. L., Krejci E., Massoulié J. cDNA sequences of Torpedo marmorata acetylcholinesterase: primary structure of the precursor of a catalytic subunit; existence of multiple 5'-untranslated regions. EMBO J. 1987 Jul;6(7):1865–1873. doi: 10.1002/j.1460-2075.1987.tb02445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq H., Ben-Aziz R., Prody C. A., Seidman S., Gnatt A., Neville L., Lieman-Hurwitz J., Lev-Lehman E., Ginzberg D., Lipidot-Lifson Y. Molecular cloning and construction of the coding region for human acetylcholinesterase reveals a G + C-rich attenuating structure. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9688–9692. doi: 10.1073/pnas.87.24.9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Harel M., Frolow F., Oefner C., Goldman A., Toker L., Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- Vigny M., Bon S., Massoulié J., Leterrier F. Active-site catalytic efficiency of acetylcholinesterase molecular forms in Electrophorus, torpedo, rat and chicken. Eur J Biochem. 1978 Apr 17;85(2):317–323. doi: 10.1111/j.1432-1033.1978.tb12241.x. [DOI] [PubMed] [Google Scholar]
- Weise C., Kreienkamp H. J., Raba R., Pedak A., Aaviksaar A., Hucho F. Anionic subsites of the acetylcholinesterase from Torpedo californica: affinity labelling with the cationic reagent N,N-dimethyl-2-phenyl-aziridinium. EMBO J. 1990 Dec;9(12):3885–3888. doi: 10.1002/j.1460-2075.1990.tb07607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]