Abstract
Phosphoramide mustard (PM), the ovotoxic metabolite of the anti-cancer agent cyclophosphamide (CPA), destroys rapidly dividing cells by forming NOR-G-OH, NOR-G and G-NOR-G adducts with DNA, potentially leading to DNA damage. A previous study demonstrated that PM induces ovarian DNA damage in rat ovaries. To investigate whether PM induces DNA adduct formation, DNA damage and induction of the DNA repair response, rat spontaneously immortalized granulosa cells (SIGCs) were treated with vehicle control (1% DMSO) or PM (3 or 6 μM) for 24 or 48 h. Cell viability was reduced (P < 0.05) after 48 h of exposure to 3 or 6 μM PM. The NOR-G-OH DNA adduct was detected after 24 h of 6 μM PM exposure, while the more cytotoxic G-NOR-G DNA adduct was formed after 48 h by exposure to both PM concentrations. Phosphorylated H2AX (γH2AX), a marker of DNA double stranded break occurrence, was also increased by PM exposure, coincident with DNA adduct formation. Additionally, induction of genes (Atm, Parp1, Prkdc, Xrcc6, and Brca1) and proteins (ATM, γH2AX, PARP-1, PRKDC, XRCC6, and BRCA1) involved in DNA repair were observed in both a time- and dose-dependent manner. These data support that PM induces DNA adduct formation in ovarian granulosa cells, induces DNA damage and elicits the ovarian DNA repair response.
Keywords: Cyclophosphamide, Phosphoramide mustard, DNA adduct, DNA damage, DNA repair, Granulosa cells
Introduction
Infertility is a side effect of female cancer treatment. Female cancer treatment survivors have reduced fecundity, relative to their non-treated counterparts (Hudson, 2010). In addition, cancer treatment can reduce the ovarian follicular reserve, resulting in premature ovarian failure (POF) (Hudson, 2010). Fewer primordial follicles have been demonstrated in autopsied ovarian samples (Himelstein-Braw et al., 1977) and fewer antral follicles detected by ultrasound (Larsen et al., 2003) in females that received anti-neoplastic treatment compared to their age-matched control subjects.
Cyclophosphamide (CPA) is an alkylating agent used to treat both cancer and autoimmune disorders. CPA causes acute ovarian failure in childhood cancer survivors (Chemaitilly et al., 2006) and rapid amenorrhea in women undergoing adjuvant treatment for breast cancer (Minton and Munster, 2002). CPA is a pro-drug requiring hepatic biotransformation by cytochrome P-450 enzymes, to generate an active cytotoxic metabolite, phosphoramide mustard (PM) (Madden et al., 2014; Plowchalk and Mattison, 1991). PM is the anti-neoplastic as well as the ovotoxic metabolite of CPA (Desmeules and Devine, 2006; Petrillo et al., 2011). PM causes primordial and small primary follicle loss (Petrillo et al., 2011), and a volatile metabolite of PM, chloroethylaziridine (CEZ) is also likely involved in PM-induced ovotoxicity (Madden et al., 2014).
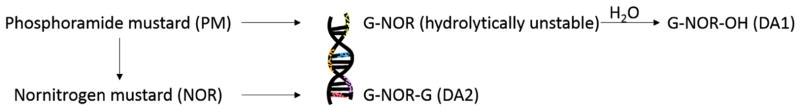
PM induces DNA or chromosomal damage in mammalian cells, which could be considered as genotoxicity markers (Anderson et al., 1995). PM is also an alkylating agent, known to cause cytotoxicity through forming cross-linked DNA adducts which inhibit DNA strand separation during replication (Phillips et al., 2000). DNA alkylation by PM occurs primarily at the N-7 position of guanine (Mehta et al., 1980) giving rise to the first product, a guanosine-PM adduct, which is unstable with a half-life of ~2–3 h (Mehta and Ludlum, 1982). PM forms a mono-functional DNA adduct; N-(2-chloroethyl)-N-[2-(7-guaninyl)ethyl]amine (NOR-G) and two bi-functional DNA adducts; N-(2-hydroxyethyl)-N-[2-(N7-guaninyl)ethyl]-amine (NOR-G-OH) and N,N-bis[2-(N7-guaninyl)ethyl]amine (G-NOR-G), both in vitro (Cushnir et al., 1990) and in vivo (Malayappan et al., 2010). The cross-linked adduct G-NOR-G is responsible for the cytotoxicity and teratological effects that contribute to the therapeutic uses of CPA/PM (Little and Mirkes, 1987).
Upon DSB induction, cells activate DNA damage responses (DDR) that comprise of cell cycle arrest, DNA damage repair, and subsequent cell cycle resumption or cell death (Giunta et al., 2010). One of the most immediate DDR events is phosphorylation of histone H2AX (γH2AX), considered the gold standard for localizing DSBs since it recruits and maintains DNA repair molecules at damage sites until repair is completed (Svetlova et al., 2010). Some DDR proteins activated due to DSBs include ataxia–telangiectasia mutated (ATM), ATM related (ATR) and DNA-dependent protein kinases (DNA-PKs) (Svetlova et al., 2010). DNA DSBs can be repaired by both the non-homologous end joining (NHEJ; Chiruvella et al., 2012) and homologous recombination (HR; Scully et al., 1997) pathways.
The granulosa cell is the somatic cell component of the oocyte-containing follicle, and close association between the granulosa cell and oocyte is required for follicular development. Some functions of granulosa cells include the production of sex steroids (Bjersing and Carstensen, 1967) and a myriad of growth factors that interact with the oocyte during development (Forde et al., 2008). Loss of granulosa cells during preantral and antral stages of follicular development leads to a premature reduction in female fecundity through reduced follicle health and oocyte viability (Walters et al., 2012). An in vivo study demonstrated that the destruction of granulosa cells by the PM parent metabolite, CPA, potentially occurs through oxidative stress-induced DNA damage leading to apoptosis in rats (Lopez and Luderer, 2004). Additionally, PM has been shown to induce DNA damage in exposed ovaries of mice and rats (Petrillo et al., 2011). In this current study, we hypothesized that PM causes DNA adduct formation which precipitates the granulosa cell towards demise due to DNA damage induction. In addition, we proposed that the DDR would be induced to counteract PM-induced ovarian DNA damage.
Methods and materials
Reagents
Phosphoramide mustard (PM) was obtained from the National Cancer Institute (Bethesda, MD). 2-β-Mercaptoethanol, 30% acrylamide/0.8% bisacrylamide, ammonium persulfate, glycerol, N′N′N′N′-tetramethylethylenediamine (TEMED), Tris base, Tris HCL, sodium chloride, Tween-20, bis-2-chloroethylamine hydrochloride, 2′-deoxyguanosine, dimethyl sulfoxide (DMSO) and sodium acetate were purchased from Sigma-Aldrich Inc. (St. Louis, MO). Dulbecco’s Modified Eagle Medium (D-MEM)/F12 (1×), 0.25% trypsin-ethylenediaminetetraacetic acid (EDTA), Pen Strep and fetal bovine serum (FBS) were from Gibco by Life Technologies (Grand Island, NY). Millicell-EZ slides were from Millipore (Bedford, MA). A Corning vacuum filter/storage system and cell culture flasks were purchased from Corning Inc. (Corning, NY). RNeasy Mini Kit, QIAshredder Kit, RNeasy MinElute Kit, Quantitect™ SYBR Green PCR Kit and Blood and Cell Culture DNA Mini Kit were purchased from Qiagen Inc. (Valencia, CA). All primers were purchased from the Iowa State University DNA facility. All primary antibodies were purchased from Abcam (Cambridge, MA). RNAlater was obtained from Ambion Inc. (Austin, TX). The polyclonal goat anti-rabbit secondary was obtained from Pierce Biotechnology (Rockford, IL). Ponceau S was from Fisher Scientific. An ECL Plus chemical luminescence detection kit was obtained from GE Healthcare, Amersham (Buckinghamshire, UK).
Rat spontaneously immortalized granulosa cell culture
A spontaneously immortalized clonal granulosa cell (SIGC) line derived from primary rat ovarian granulosa cell cultures were obtained as a gift from Dr. Burghardt at Texas A&M University. SIGC (2.5 × 104 cells) were cultured in 25-cm2 flasks in media (DMEM/F12 plus 5% FBS and 50 mg/ml of Pen Strep) at 37 °C and 5% CO2 until 80% confluent.
Cell viability
SIGCs were treated with DMSO and/or PM (0.5 μM, 1 μM, 3 μM or 6 μM) for 48 h to perform cell viability measurements (n = 3 per treatment). Cells were harvested by trypsinization and 100 μl of the cell suspension was stained with Trypan blue (1:1). Trypan blue selectively penetrates cell membranes of dead cells, coloring them blue, whereas it is not absorbed by membranes of live cells, thus excluding live cells from staining. Cell counting was performed using a hemocytometer and the percentage of viable cells calculated. Since it is difficult to ascertain the level of PM reaching the ovary in CPA-treated patients, we used a bioassay endpoint approach in which ~20% depletion of cell viability was achieved and DNA adduct formation as well as gene and protein assays were performed at preceding time points.
DNA isolation and mass spectrometry
Cells were harvested by trypsinization and pelleted by centrifugation at 1200 rpm for 5 min, washed twice in phosphate buffer saline (PBS) and resuspended in cold PBS (4 °C). DNA was isolated from cells using a Qiagen blood and cell culture DNA kit according to the manufacturer’s protocol. DNA concentration was determined using NanoDrop (λ = 260/280 nm; ND 1000; Nanodrop Technologies Inc., Wilmington, DE). Internal standards of NOR-G-OH (DNA adduct 1; DA1) and G-NOR-G (DNA adduct 2; DA2) were prepared according to a modified procedure (Hemminki, 1985; Malayappan et al., 2010). DNA samples from cell lines (10–50 μg in 200 μl of buffer) were used to perform Agilent QTOF6540 LC/MS analysis at the Iowa State University Chemical Instrumentation Facility. Each sample (1 μl) was injected into the JetStream ESI ion source and water/MeOH (20/80) was used as effluent solvent. The LC column used in this experiment was C18, 4.6 * 150 mm, 1.8 μm. Accurate mass measurement was achieved by constantly infusing a calibrant (masses: 121.0508 and 922.0098). Detection of PM-DNA adducts were performed once.
Immunofluorescence
Cells (1 × 105 cells/well) were transferred to Millicell-EZ slide wells (Millipore, Bedford, MA) and treated with DMSO ± PM (3 or 6 μM) for 24 or 48 h (n = 3 per timepoint/treatment). Cells were washed with PBS three times and fixed with 95% ethanol for 5 min on ice, followed by incubation in 0.25–0.5% Triton X-100 containing PBS for 10 min to permeabilize the membranes. After washing, slides were blocked with 5% normal serum in PBS for 1 h and washed again with PBS. Slides were incubated with a primary antibody directed against γH2AX (1:50) overnight at 4 °C. After washing in PBS, slides were incubated with a donkey anti-rabbit IgG-FITC secondary antibody (1:3000) for 1 h. Slides were then counterstained with 4–6-diamidino-2-phenylindole (DAPI). Images were taken using a Leica fluorescent microscope and the percentage of cells with foci for γH2AX was calculated (n = 3 slides/treatment). Total cell number per slide was approximately 150.
RNA isolation and qRT-PCR
SIGCs were treated with DMSO ± PM (3 or 6 μM) for 24 or 48 h (n = 3 per timepoint/treatment). RNA was isolated using an RNeasy Mini Kit and the concentration was determined using NanoDrop (λ = 260/280 nm; ND 1000; Nanodrop Technologies Inc., Wilmington, DE). Total RNA (1 μg) was reverse transcribed to cDNA utilizing the Superscript III One-Step qRT-PCR. Diluted cDNA (2 μl; 1:20) was amplified on an Eppendorf PCR Master cycler using a Quantitect™ SYBR Green PCR Kit (n = 3 samples per treatment/timepoint; n = 3 repetitions per sample). Primers for Atm, Prkdc, Xrcc6, Parp1, Rad51, Brca1 and Gapdh were designed by Primer 3 Input Version (0.4.0) (Table 1). The regular cycling program consisted of a 15 min hold at 95 °C and 45 cycles of denaturing at 95 °C for 15 s, annealing at 58 °C for 15 s, and extension at 72 °C for 20 s at which point data were acquired. Each sample was normalized to Gapdh before quantification using the 2−ΔΔCt method (Livak and Schmittgen, 2001; Pfaffl, 2001)
Table 1.
Primer sequences used for qPCR.
| Genes | Forward primer | Reverse primer |
|---|---|---|
| Atm | TCAGCAGCACCTCTGATTCTT | AGACAGACATGCTGCCTCCT |
| Brca1 | CCCTCTTAGTCTGCTGAGCT | CCTTTGGGTGGCTGTACTGA |
| Parp1 | AAGTGCCAGTGTCAAGGAGA | ACAGGGAGCAAAAGGGAAGA |
| Prkdc | GCCACAGACCCCAATATCCT | TATCTGACCATCTCGCCAGC |
| Rad51 | ATCCCTGCATGCTTGTTCTC | CTGCAGCTGACCATAACGAA |
| Xrcc6 | GATCTGACACTGCCCAAGGT | TGCTTCTTCGGTCCACTCTT |
| Gapdh | GTGGACCTCATGGCCTACAT | GGATGGAATTGTGAGGGAGA |
Protein isolation and Western blot
SIGCs were treated with DMSO ± PM (3 or 6 μM) for 24 or 48 h (n = 3 per timepoint/treatment) and protein isolated using lysis buffer containing protease and phosphatase inhibitors as previously described (Ganesan et al., 2013). Briefly, cells were placed on ice for 10 min, followed by two rounds of centrifugation at 10,000 rpm for 15 min and protein concentration was measured using a BCA protocol. Protein was stored at −80 °C until further use. SDS-PAGE was used to separate the proteins and then transferred to a nitrocellulose membrane. Membranes were blocked for 1 h in 5% milk in Tris-buffered saline containing Tween-20. Membranes were incubated in one of: anti-PARP-1 antibody (1:200), anti-ATM antibody (1:100), anti-rabbit phosphorylated H2AX antibody (γH2AX; 1:100), anti-rabbit BRCA1 antibody (1:500), anti-rabbit PRKDC antibody (1:100), anti-RAD 51 antibody (1:500) and anti-mouse XRCC6 antibody (1:100) for 36 h at 4 °C. Following three washes in TTBS (1×), membranes were incubated with a species specific secondary antibody (1:2000) for 1 h at room temperature. Membranes were washed three times in TTBS and one time in TBS and then incubated in an enhanced chemiluminescence (ECL) detection substrate for 5 min and exposed to X-ray film. Densitometry of the appropriate bands was performed using ImageJ software (NCBI). Background densitometry values were subtracted and protein of interest densitometry values were normalized to those of Ponceau S.
Statistical analysis
Data were analyzed by unpaired t-tests comparing treatment with control raw data at each individual time-point using Prism 5.04 software (Graph Pad Software). Where there are more than two treatments (mRNA and protein), ANOVA was utilized (Graph Pad Software). Statistical significance was defined as P < 0.05. For graphical purposes, protein expression is presented as a mean ± SE of the respective control.
Results
Cell viability was impacted by PM exposure
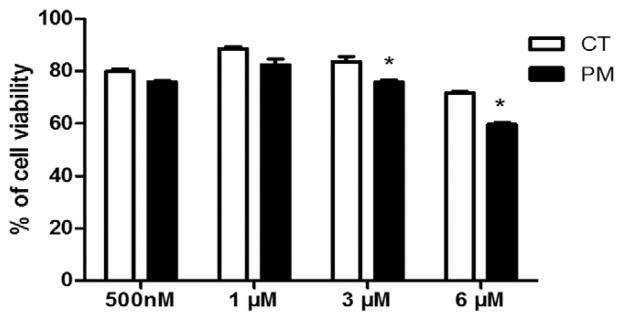
SIGCs were treated with DMSO or PM (0.5, 1, 3 or 6 μM) for 48 h and cell viability was quantified using Trypan blue staining. PM reduced (P < 0.05) cell viability at concentrations of 3 μM and higher. Relative to control, the percentage of viable cells was 91% and 83.6% after exposure to 3 μM and 6 μM PM, respectively (Fig. 2). (See Fig. 1.)
Fig. 2.
Cell viability is impacted by PM exposure. SIGCs were treated with DMSO or PM (0.5, 1, 3 or 6 μM) for 48 h. Trypan blue staining was used to determine the cell viability. Values represent % of viable cells ± SE. *P < 0.05; different from control (n = 3 per treatment/timepoint). Loss of cell viability was observed at the 3 and 6 μM PM exposures.
Fig. 1.
Formation of phosphoramide mustard-induced DNA adducts. PM undergoes nonenzymatic degradation to produce nornitrogen mustard (NOR). Both PM and NOR alkylate guanine bases within DNA form guanine monoadducts, N-[2-(N7-guaninyl)ethyl]-N-[2-hydroxyethyl]-amine (G-NOR-OH), and DNA–DNA cross-links, N,N-bis[2-(N7-guaninyl) ethyl] amine (G-NOR-G), although PM is considered the major DNA reactive metabolite.
Adapted from Malayappan et al. (2010).
PM exposure induces DNA adduct formation
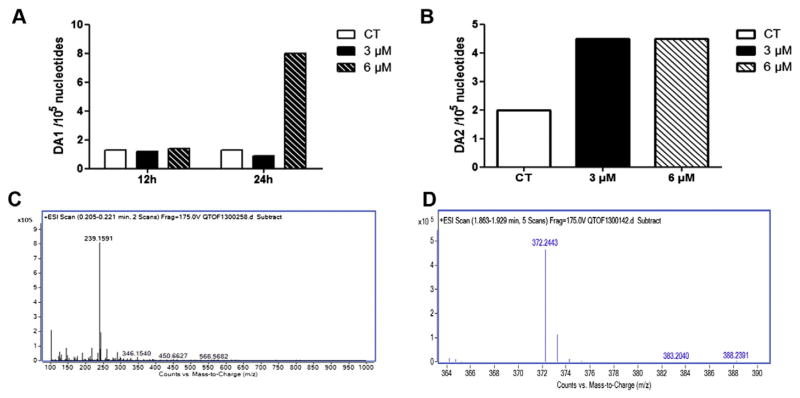
SIGCs were treated with DMSO or PM (3 or 6 μM) for 12, 24 or 48 h and LC/MS analysis was performed. DA1 (Fig. 3A, B; mol. weight = 239) was observed after 24 h of exposure to 6 μM PM, while DA2 (Fig. 3C, D; mol. weight = 372) was formed after 48 h with both the 3 and 6 μM PM exposures.
Fig. 3.
PM exposure induces DNA adduct formation. SIGCs were treated with DMSO or PM (3 or 6 μM) for 12, 24 or 48 h for DNA isolation. Agilent QTOF6540 liquid chromatography (LC) mass spectrometry (MS) analysis was performed to detect DA1 and DA2 formation. Internal standards of DA1 and DA2 were prepared according to a modified procedure (Hemminki, 1985; Malayappan et al., 2010). Values represent the relative abundance of DNA adducts (1 × 105 nucleotides). (A) Quantification of DA1 appearance: DA1 was observed after 24 h of exposure to 6 μM PM. (B) Quantification of DA2 appearance: DA2 was formed after 48 h at both the 3 and 6 μM concentrations. (C) LC/MS spectrum indicating the DA1 product of 239 m. wt. (D) LC/MS spectrum indicating the DA2 product of 372 m. wt.
DNA damage was induced by PM exposure as determined by localization of γH2AX
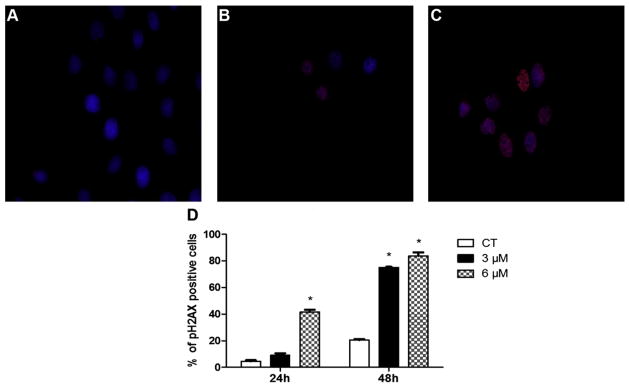
SIGCs were treated with DMSO or PM (3 or 6 μM) for 24 or 48 h to perform γH2AX protein localization (Fig. 4A–C) using immunofluorescence. The percentage of cells with positive γH2AX foci were increased (P < 0.05) by the 6 μM PM exposure after 24 h, relative to control treated cells. After 48 h of PM exposure (both concentrations) increased levels of γH2AX foci were observed (Fig. 4D).
Fig. 4.
DNA damage was induced by PM exposure as determined by localization of γH2AX. SIGCs were treated with (A) DMSO, (B) 3 μM PM or (C) 6 μM PM for 24 or 48 h and γH2AX protein localized using immunofluorescence staining (A–C). Blue represents DAPI nuclear stain; red staining indicates a FITC labeled primary antibody against γH2AX protein. (D) Values represent % of γH2AX positive cells (out of a total of ~150 cells per slide) ± SE; n = 3 per treatment/timepoint. Different letters indicate difference from CT; P < 0.05. γH2AX protein appearance indicates DNA DSB formation, which was subsequent to DNA adduct detection.
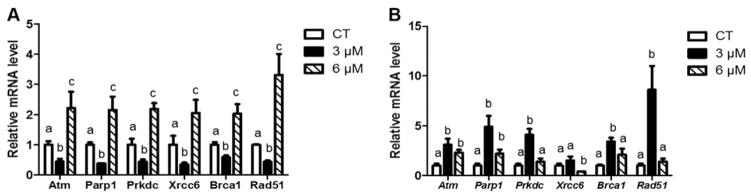
PM exposure increases DDR gene mRNA expression level
Following 24 h of PM exposure, the DDR genes Atm, Parp1, Prkdc, Xrcc6, and Brca1 and Rad51 mRNA expression were increased (P < 0.05) after 6 μM concentration compared to both control and cells exposed to 3 μM PM. Atm, Parp1, Prkdc, Xrcc6, Brca1 and Rad51 mRNA expression were decreased (P < 0.05) by 3 μM PM exposure after 24 h compared to both control and 6 μM PM treated cells (Fig. 5A).
Fig. 5.
PM exposure increases DDR gene mRNA expression levels. RNA was isolated from SIGCs treated with DMSO or PM (3 or 6 μM) for (A) 24 h or (B) 48 h, followed by qRT-PCR (n = 3 samples per treatment/timepoint; n = 3 repetitions per sample). Genes were normalized to Gapdh and values reported as fold change due to PM exposure relative to the control value of 1 ± SEM. Different letters represent differences between treatments; P < 0.05. Increased mRNA levels were observed in cells exposed to the 6 μM PM concentration at 24 h before the increase was evident in cells exposed to 3 μM PM at 48 h, indicating a temporal pattern of DDR gene induction in response to PM.
After PM exposure for 48 h, mRNA abundance of the DNA repair response genes Atm, Parp1, Prkdc, and Brca1 and Rad51 mRNA expression were increased (P < 0.05) at 3 μM concentration while no changes in Xrcc6 mRNA expression were observed compared to control and 6 μM PM. At the same time Atm, Parp1, and Brca1 were increased at the 6 μM PM concentration compared to control and 3 μM PM. No changes were detected in Prkdc, Xrcc6, or Rad51 mRNA expression at 3 μM compared to control and 6 μM PM exposure (Fig. 5B).
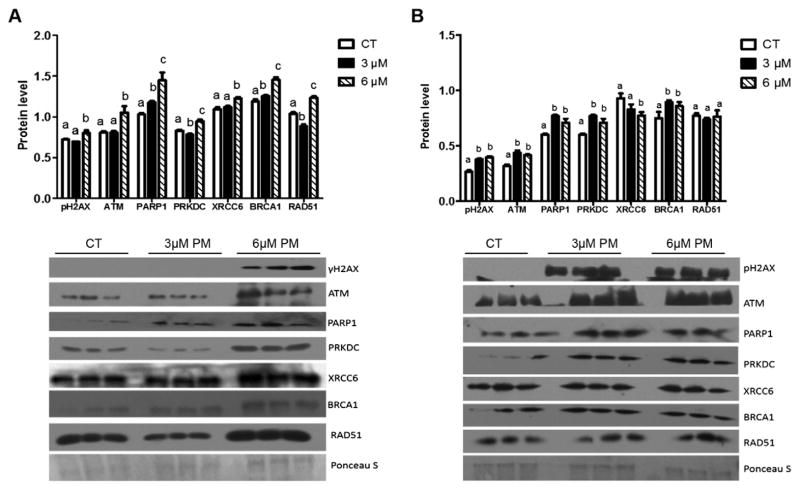
DDR proteins were increased in response to PM exposure
Following 24 h of PM exposure, the DDR proteins γH2AX, ATM, PRKDC, XRCC6 and RAD51 were increased (P < 0.05) by exposure to 6 μM PM. BRCA1 and PARP-1 were increased (P < 0.05) by both concentrations of PM compared to control. In contrast, PRKDC and RAD51 were decreased (P < 0.05) by 3 μM PM compared to both control and 6 μM PM treatments (Fig. 6A).
Fig. 6.
DDR proteins are generally increased in response to PM exposure. Total protein was isolated from SIGCs treated with DMSO or PM (3 or 6 μM) for (A) 24 h or (B) 48 h, followed by Western blotting (n = 3 per treatment/timepoint). All treatments were run on the same gel per timepoint (n = 3/treatment) as depicted in the blots presented. Ponceau S was used for normalization with protein of interest and values were reported as raw mean value ± SEM. Different letters indicate differences between treatments; P < 0.05. PM induced increased γH2AX, ATM, PARP-1, PRKDC, XRCC6, BRCA1, and RAD51 after 24 h of the 6 μM PM exposure, while the 3 μM PM exposure increased γH2AX, ATM, PARP-1, PRKDC, and BRCA1 after 48 h.
After 48 h of PM exposure, the DDR proteins γH2AX, ATM, PARP-1, PRKDC and BRCA1 were increased (P < 0.05) at both PM concentrations compared to control while XRCC6 protein was decreased (P < 0.05) by 6 μM, but not 3 μM, PM concentration. RAD51 protein was not altered by either concentration of PM compared to control (Fig. 6B).
Discussion
In the current study, we describe DNA adduct formation followed by the appearance of DNA DSBs which occurs after PM exposure. In addition, we illustrate the subsequent induction of the DDR in a rat ovarian SIGC line. Our rationale for these studies was that CPA has been reported to destroy ovarian follicles by targeting granulosa cells in mice and rats (Desmeules and Devine, 2006). Biotransformation of CPA to PM, the anti-neoplastic and ovotoxic form, has been described (Plowchalk and Mattison, 1991) and it is recognized that PM can be detected in the urine of CPA-treated patients, approximately 3 h following exposure at levels of 50–100 nmol/ml (Jardine et al., 1978). Using an in vitro ovarian culture system, we have previously reported that PM induces follicle loss after 4 days of exposure (Madden et al., 2014). Also, evidence supporting that PM induces DNA DSBs was shown in oocyte and granulosa cell nuclei (Petrillo et al., 2011). In addition, CPA-induced oxidative stress-induced DNA damage has been described (Tsai-Turton et al., 2007). Additionally, we have reported that PM is metabolized to a volatile ovotoxicant, 2-chloroethylaziridine (CEZ; Madden et al., 2014), with a longer half-life than PM (Lu and Chan, 1996), therefore, PM exposed ovaries can be considered to also be exposed to CEZ.
It is known that the cytotoxic properties of PM occur by the formation of DNA adducts, thereby preventing cell division by inhibiting DNA strand separation (Cushnir et al., 1990), required for cancer cell destruction. In addition, the cross-linked adduct DA2 is responsible for the cytotoxicity and teratological effects of CPA (Little and Mirkes, 1987) and these events are required for tumor destruction. PM formed the DA2 adduct in exposed rat embryos (Mirkes et al., 1992) as well as in white blood cells of cancer patients (Malayappan et al., 2010). PM-induced cell death was observed in granulosa cells after 48 h. PM binds to DNA to first form DA1 at a higher PM concentration after 24 h of exposure followed rapidly by the formation of DA2, observed in both PM exposures after 48 h. Thus, these results lend support to our proposal that DNA adduct formation is involved in the ovotoxic, in addition to the anti-neoplastic properties of PM.
To next evaluate whether DNA damage occurs in granulosa cells after PM-induced DNA adduction, we determined any alterations to ATM, a protein kinase that is recruited and activated upon DNA DSB. Increased Atm mRNA and protein were observed after 24 h of exposure to 6 μM PM, and by both PM concentrations after 48 h, indicating a dose-and time-dependent impact of PM on Atm induction. A number of chemicals including CPA activate ATM signaling eventually resulting in apoptosis in extra-ovarian mammalian cells (Ganesan et al., 2013; Toulany et al., 2014). ATM phosphorylates histone H2AX in response to DNA DSBs (Burma et al., 2001) and γH2AX protein was previously localized within the oocyte and granulosa cell of PM exposed mouse ovaries (Petrillo et al., 2011). Both ATM activation and γH2AX appearance serve as proxy markers of cellular DNA damage (Tanaka et al., 2006). Thus, we determined if γH2AX was increased in granulosa cells post-PM exposure and show that PM increased the percentage of γH2AX-positive cells and also increased γH2AX protein in total ovarian protein homogenates in both a dose- and time-dependent manner. These data indicate that DNA damage is induced by PM exposure after DNA adduct formation in rat ovarian granulosa cells.
In order to next sequentially follow whether DNA DSB induction results in a protective response in the form of the DDR within granulosa cells, we quantified changes in poly(ADP-ribose) polymerase-1 (PARP-1) which is rapidly activated by DNA DSBs. Additionally, PARP-1 is involved in posttranslational modification of nuclear proteins during DNA damage and activates cellular processes such as DNA replication, DNA repair, apoptosis and genome stability (Yu et al., 2002). We demonstrate that Parp1 mRNA and protein were increased in a similar manner to that observed in Atm — by 6 μM PM after 24 h of PM exposure and by both PM concentrations after 48 h. We also found that ATM protein level was coincident with PARP-1 protein level after PM exposure. A link between ATM and PARP-1 has been previously found; PARP-1 deficient mice were extremely sensitive to low doses of γ-radiation, and this phenotype could be ascribed to a deficient ATM-kinase activation in tissues such as the intestine epithelium (de Murcia et al., 1997).
PARP-1 competes with XRCC6 to repair a DSB as part of the NHEJ pathway but, in contrast, PARP-1 can also be recruited to a DSB in the absence of XRCC6 when the classical pathway of NHEJ is absent (Wang et al., 2006). XRCC6 is a KU protein which forms a heterodimer with XRCC5 (Calsou et al., 2003). A deficiency of XRCC6 protein causes genomic instability and malignant formation (Wang et al., 2013). We found that Xrcc6 mRNA was reduced by the lower PM exposure after 24 h, but increased by the 6 μM PM concentration at the same time point. Subsequently, Xrcc6 mRNA in 3 μM PM-treated cells returned to control levels, while it was reduced at the higher PM concentration. XRCC6 protein levels were initially increased by the 6 μM PM exposure, but subsequently decreased. These results indicate that there is a dynamic response of Xrcc6 mRNA to PM exposure, and may suggest that XRCC6 is involved in the earlier DDR via the NHEJ repair pathway, but at later times of exposure, and that PARP-1 could potentially compete with XRCC6 to repair DNA damage through an XRCC6-independent DNA repair pathway; however, these possibilities cannot be confirmed by the current study.
We found a dose-dependent temporal pattern of Prkdc mRNA and protein increased expression in response to PM exposure. Our study also indicates that PM could induce PRKDC as part of the mechanism to repair DNA damage and possibly result in improved survivability of SIGCs. PRKDC is a DNA-dependent protein kinase recruited and activated at the site of DNA damage by KU proteins to repair DSBs repaired by NHEJ pathways. Interestingly, capsaicin-induced autophagy of MCF-7 cells was shown to occur via activation of the PRKDC, ATM and PARP-1 DDR proteins (Yoon et al., 2012). Further, increased ionizing radiation-induced Prkdc mRNA was observed in mouse leukemic monocyte macrophage cell line RAW264.7 and was thought to be involved in the prevention of cell death (Dhariwala et al., 2012).
We also observed a temporal pattern of increased Brca1 mRNA and protein due to PM exposure, supporting that the HR DDR pathway is induced and that ovarian Brca1 is responsive to PM exposure. BRCA1 is a breast cancer protein required for repair of DSBs via the HR pathway. Previously, we have found that Brca1 mRNA and protein were increased in response to a chemical exposure (7,12-dimethylbenz[a]anthracene) that induced DSBs in rat ovaries (Ganesan et al., 2013). In cells that were resistant to 5-azacytidine activation of ATM and BRCA1 inhibited 5-azacytidine-induced apoptosis by repairing DNA damage but this activation of ATM and BRCA1 was absent in the parental 5-azacytidine-sensitive leukemia cell line (Imanishi et al., 2014). Thus, these data support that Brca1 is involved in the ovarian response to PM exposure, likely as an attempt to prevent ovotoxicity.
We found that PM increased Rad51 mRNA and protein at the 6 μM PM concentration and that a similar response occurred with the 3 μM PM exposure, but this lagged from that of the higher concentration, again indicating a dynamic, temporal pattern of the DDR to PM exposure. BRCA1 can interact with RAD 51 during HR to regulate the function of RAD51 in response to DSBs and block sister chromatid replication slippage (Cousineau et al., 2005). Downregulation of RAD51 previously indicated failure of the HR pathway in hypoxic cancer cells (Bindra et al., 2004). Also, Rad51 mRNA was elevated in tumor cells, suggesting that it plays a role in repair of DNA damage (Raderschall et al., 2002).
Taken together, these data demonstrate that PM exposure to an ovarian granulosa cell line caused formation of two DNA adducts. In addition, loss of cell viability and induction of the DDR, in both a time- and dose-dependent manner was observed in all of the genes and their protein products that were examined. There was apparent activation of both the NHEJ as well as the HR pathways of DNA repair, thus, one was not favored over another. Interestingly, the lower concentration of PM resulted in decreased mRNA levels for all genes investigated, despite there being no effect on the internal housekeeping gene used. Potentially this decline in mRNA levels may indicate a mechanism of ovotoxicity that is occurring at low level PM exposure, or may represent a time lag in mRNA induction that may have also occurred at an earlier time point at the 6 μM PM concentration. It is probable and likely that there are posttranslational modifications and regulatory pathways being induced that were outside the scope of this study. Activation of the DDR appears to be temporal, eventually returning to that of the control treated cells, which could represent that the DNA DSB has either been repaired, or that the cell has been shunted towards an atretic fate. However, the data reported herein may provide initial targets for amelioration of CPA/PM-induced infertility. Taken together, PM, the active metabolite of CPA, may induce ovarian follicle death via the formation of DNA adducts and the ovary induces mechanisms to respond to this genotoxic insult likely in order to protect the precious oocyte-containing follicle pool.
Acknowledgments
The project described was supported the National Institutes of Environmental Health Sciences [R00ES016818].
Abbreviations
- CPA
cyclophosphamide
- PM
phosphoramide mustard
- NOR-G-OH
N-(2-hydroxyethyl)-N-[2-(N7-guaninyl)ethyl]-amine
- NOR-G
N-(2-chloroethyl)-N-[2-(7-guaninyl)ethyl]amine
- G-NOR-G
N-bis[2-(N7-guaninyl)ethyl]amine
- SIGCs
spontaneously immortalized granulosa cells
- CEZ
chloroethylaziridine
- DNA-PKs
DNA-dependent protein kinases
- DDR
DNA damage response
- DSBs
double-strand breaks
- NHEJ
non-homologous end joining
- HR
homologous recombination
- EDTA
ethylenediaminetetraacetic acid
- FBS
fetal bovine serum
- PBS
phosphate buffered saline
- ECL
enhanced chemiluminescence
- DMSO
dimethyl sulfoxide
- ATM
ataxia telangiectasia mutated
- ATR
ATM-related
- XRCC6
X-ray repair complementing defective repair in Chinese hamster cells 6
- BRCA1
breast cancer type 1
- RAD 51
Rad 51 homolog
- PARP-1
poly(ADP-ribose) polymerase-1
- PRKDC
protein kinase, DNA-activated, catalytic polypeptide
- γH2AX
phosphorylated histone H2AX
- TEMED
N′N′N′N′-tetramethylethylenediamine
Footnotes
Conflict of interest statement
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Environmental Health Sciences or the National Institutes of Health.
References
- Anderson D, Bishop JB, Garner RC, Ostrosky-Wegman P, Selby PB. Cyclophosphamide: review of its mutagenicity for an assessment of potential germ cell risks. Mutat Res. 1995;330:115–181. doi: 10.1016/0027-5107(95)00039-l. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Schaffer PJ, Meng A, Woo J, Måseide K, Roth ME, Lizardi P, Hedley DW, Bristow RG, Glazer PM. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24:8504–8518. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjersing L, Carstensen H. Biosynthesis of steroids by granulosa cells of the porcine ovary in vitro. J Reprod Fertil. 1967;14:101–111. doi: 10.1530/jrf.0.0140101. [DOI] [PubMed] [Google Scholar]
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- Calsou P, Delteil C, Frit P, Drouet J, Salles B. Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4-ligase IV recruitment. J Mol Biol. 2003;326:93–103. doi: 10.1016/s0022-2836(02)01328-1. [DOI] [PubMed] [Google Scholar]
- Chemaitilly W, Mertens AC, Mitby P, Whitton J, Stovall M, Yasui Y, Robison LL, Sklar CA. Acute ovarian failure in the Childhood Cancer Survivor Study. J Clin Endocrinol Metab. 2006;91:1723–1728. doi: 10.1210/jc.2006-0020. [DOI] [PubMed] [Google Scholar]
- Chiruvella KK, Sebastian R, Sharma S, Karande AA, Choudhary B, Raghavan SC. Time-dependent predominance of nonhomologous DNA end-joining pathways during embryonic development in mice. J Mol Biol. 2012;417:197–211. doi: 10.1016/j.jmb.2012.01.029. [DOI] [PubMed] [Google Scholar]
- Cousineau I, Abaji C, Belmaaza A. BRCA1 regulates RAD51 function in response to DNA damage and suppresses spontaneous sister chromatid replication slippage: implications for sister chromatid cohesion, genome stability, and carcinogenesis. Cancer Res. 2005;65:11384–11391. doi: 10.1158/0008-5472.CAN-05-2156. [DOI] [PubMed] [Google Scholar]
- Cushnir JR, Naylor S, Lamb JH, Farmer PB, Brown NA, Mirkes PE. Identification of phosphoramide mustard/DNA adducts using tandem mass spectrometry. Rapid Commun Mass Spectrom. 1990;4:410–414. doi: 10.1002/rcm.1290041014. [DOI] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. PNAS. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmeules P, Devine PJ. Characterizing the ovotoxicity of cyclophosphamide metabolites on cultured mouse ovaries. Toxicol Sci. 2006;90:500–509. doi: 10.1093/toxsci/kfj086. [DOI] [PubMed] [Google Scholar]
- Dhariwala F, Narang H, Krishna M. Ionizing radiation induced signaling of DNA damage response molecules in RAW 264.7 and CD4+ T cells. Mol Cell Biochem. 2012;363:43–51. doi: 10.1007/s11010-011-1156-z. [DOI] [PubMed] [Google Scholar]
- Forde N, Mihm M, Canty MJ, Zielak AE, Baker PJ, Park S, Lonergan P, Smith GW, Coussens PM, Ireland JJ, Evans ACO. Differential expression of signal transduction factors in ovarian follicle development: a functional role for betaglycan and FIBP in granulosa cells in cattle. Physiol Genomics. 2008;33:193–204. doi: 10.1152/physiolgenomics.00274.2007. [DOI] [PubMed] [Google Scholar]
- Ganesan S, Bhattacharya P, Keating AF. 7,12-Dimethylbenz[a]anthracene exposure induces the DNA repair response in neonatal rat ovaries. Toxicol Appl Pharmacol. 2013;272:690–696. doi: 10.1016/j.taap.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta S, Belotserkovskaya R, Jackson SP. DNA damage signaling in response to double-strand breaks during mitosis. J Cell Biol. 2010;190:197–207. doi: 10.1083/jcb.200911156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki K. Binding of metabolites of cyclophosphamide to DNA in a rat liver microsomal system and in vivo in mice. Cancer Res. 1985;45:4237–4243. [PubMed] [Google Scholar]
- Himelstein-Braw R, Peters H, Faber M. Influence of irradiation and chemotherapy on the ovaries of children with abdominal tumours. Br J Cancer. 1977;36:269–275. doi: 10.1038/bjc.1977.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson MM. Reproductive outcomes for survivors of childhood cancer. Obstet Gynecol. 2010;116:1171–1183. doi: 10.1097/AOG.0b013e3181f87c4b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi S, Umezu T, Ohtsuki K, Kobayashi C, Ohyashiki K, Ohyashiki JH. Constitutive activation of the ATM/BRCA1 pathway prevents DNA damage-induced apoptosis in 5-azacytidine-resistant cell lines. Biochem Pharmacol. 2014;89:361–369. doi: 10.1016/j.bcp.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Jardine I, Fenselau C, Appler M, Kan MN, Brundrett RB, Colvin M. Quantitation by gas chromatography–chemical ionization mass spectrometry of cyclophosphamide, phosphoramide mustard, and nornitrogen mustard in the plasma and urine of patients receiving cyclophosphamide therapy. Cancer Res. 1978;38:408–415. [PubMed] [Google Scholar]
- Larsen EC, Müller J, Schmiegelow K, Rechnitzer C, Andersen AN. Reduced ovarian function in long-term survivors of radiation- and chemotherapy-treated childhood cancer. J Clin Endocrinol Metab. 2003;88:5307–5314. doi: 10.1210/jc.2003-030352. [DOI] [PubMed] [Google Scholar]
- Little SA, Mirkes PE. DNA cross-linking and single-strand breaks induced by teratogenic concentrations of 4-hydroperoxycyclophosphamide and phosphoramide mustard in postimplantation rat embryos. Cancer Res. 1987;47:5421–5426. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopez SG, Luderer U. Effects of cyclophosphamide and buthionine sulfoximine on ovarian glutathione and apoptosis. Free Radic Biol Med. 2004;36:1366–1377. doi: 10.1016/j.freeradbiomed.2004.02.067. [DOI] [PubMed] [Google Scholar]
- Lu H, Chan KK. Gas chromatographic–mass spectrometric assay for N-2-chloroethylaziridine, a volatile cytotoxic metabolite of cyclophosphamide, in rat plasma. J Chromatogr B Biomed Appl. 1996;678:219–225. doi: 10.1016/0378-4347(95)00528-5. [DOI] [PubMed] [Google Scholar]
- Madden JA, Hoyer PB, Devine PJ, Keating AF. Involvement of a volatile metabolite during phosphoramide mustard-induced ovotoxicity. Toxicol Appl Pharmacol. 2014;277:1–7. doi: 10.1016/j.taap.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malayappan B, Johnson LA, Nie B, Panchal D, Matter B, Jacobson P, Tretyakova N. Quantitative high-performance liquid chromatography–electrospray ionization tandem mass spectrometry analysis of bis-N7-guanine DNA–DNA cross-links in white blood cells of cancer patients receiving cyclophosphamide therapy. Anal Chem. 2010;82:3650–3658. doi: 10.1021/ac902923s. [DOI] [PubMed] [Google Scholar]
- Mehta JR, Ludlum DB. Trapping of DNA-reactive metabolites of therapeutic or carcinogenic agents by carbon-14-labeled synthetic polynucleotides. Cancer Res. 1982;42:2996–2999. [PubMed] [Google Scholar]
- Mehta JR, Przybylski M, Ludlum DB. Alkylation of guanosine and deoxyguanosine by phosphoramide mustard. Cancer Res. 1980;40:4183–4186. [PubMed] [Google Scholar]
- Minton SE, Munster PN. Chemotherapy-induced amenorrhea and fertility in women undergoing adjuvant treatment for breast cancer. Cancer Control. 2002;9:466–472. doi: 10.1177/107327480200900603. [DOI] [PubMed] [Google Scholar]
- Mirkes PE, Brown NA, Kajbaf M, Lamb JH, Farmer PB, Naylor S. Identification of cyclophosphamide-DNA adducts in rat embryos exposed in vitro to 4-hydroperoxycyclophosphamide. Chem Res Toxicol. 1992;5:382–385. doi: 10.1021/tx00027a010. [DOI] [PubMed] [Google Scholar]
- Petrillo SK, Desmeules P, Truong TQ, Devine PJ. Detection of DNA damage in oocytes of small ovarian follicles following phosphoramide mustard exposures of cultured rodent ovaries in vitro. Toxicol Appl Pharmacol. 2011;253:94–102. doi: 10.1016/j.taap.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DH, Farmer PB, Beland FA, Nath RG, Poirier MC, Reddy MV, Turteltaub KW. Methods of DNA adduct determination and their application to testing compounds for genotoxicity. Environ Mol Mutagen. 2000;35:222–233. doi: 10.1002/(sici)1098-2280(2000)35:3<222::aid-em9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Plowchalk DR, Mattison DR. Phosphoramide mustard is responsible for the ovarian toxicity of cyclophosphamide. Toxicol Appl Pharmacol. 1991;107:472–481. doi: 10.1016/0041-008x(91)90310-b. [DOI] [PubMed] [Google Scholar]
- Raderschall E, Stout K, Freier S, Suckow V, Schweiger S, Haaf T. Elevated levels of Rad51 recombination protein in tumor cells. Cancer Res. 2002;62:219–225. [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- Svetlova MP, Solovjeva LV, Tomilin NV. Mechanism of elimination of phosphorylated histone H2AX from chromatin after repair of DNA double-strand breaks. Mutat Res. 2010;685:54–60. doi: 10.1016/j.mrfmmm.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Halicka HD, Huang X, Traganos F, Darzynkiewicz Z. Constitutive histone H2AX phosphorylation and ATM activation, the reporters of DNA damage by endogenous oxidants. Cell Cycle. 2006;5:1940–1945. doi: 10.4161/cc.5.17.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai-Turton M, Luong BT, Tan Y, Luderer U. Cyclophosphamide-induced apoptosis in COV434 human granulosa cells involves oxidative stress and glutathione depletion. Toxicol Sci. 2007;98(1):216–230. doi: 10.1093/toxsci/kfm087. [DOI] [PubMed] [Google Scholar]
- Toulany M, Mihatsch J, Holler M, Chaachouay H, Rodemann HP. Cisplatin-mediated radiosensitization of non-small cell lung cancer cells is stimulated by ATM inhibition. Radiother Oncol. 2014;111:228–236. doi: 10.1016/j.radonc.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Walters KA, Middleton LJ, Joseph SR, Hazra R, Jimenez M, Simanainen U, Allan CM, Handelsman DJ. Targeted loss of androgen receptor signaling in murine granulosa cells of preantral and antral follicles causes female subfertility. Biol Reprod. 2012;87(151):1–11. doi: 10.1095/biolreprod.112.102012. [DOI] [PubMed] [Google Scholar]
- Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Lin H, Hua F, Hu ZW. Repairing DNA damage by XRCC6/KU70 reverses TLR4-deficiency-worsened HCC development via restoring senescence and autophagic flux. Autophagy. 2013;9:925–927. doi: 10.4161/auto.24229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Ahn SG, Lee BH, Jung SH, Oh SH. Role of autophagy in chemoresistance: regulation of the ATM-mediated DNA-damage signaling pathway through activation of DNA–PKcs and PARP-1. Biochem Pharmacol. 2012;83:747–757. doi: 10.1016/j.bcp.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]