ABSTRACT
Chicken anemia virus (CAV) is a single-stranded circular DNA virus that carries 3 genes, the most studied of which is the gene encoding VP3, also known as apoptin. This protein has been demonstrated to specifically kill transformed cells while leaving normal cells unharmed in a manner that is independent of p53 status. Although the mechanistic basis for this differential activity is unclear, it is evident that the subcellular localization of the protein is important for the difference. In normal cells, apoptin exists in filamentous networks in the cytoplasm, whereas in transformed cells, apoptin is present in the nucleus and appears as distinct foci. We have previously demonstrated that DNA damage signaling through the ataxia telangiectasia mutated (ATM) pathway induces the translocation of apoptin from the cytoplasm to the nucleus, where it induces apoptosis. We found that apoptin contains four checkpoint kinase consensus sites and that mutation of either threonine 56 or 61 to alanine restricts apoptin to the cytoplasm. Furthermore, treatment of tumor cells expressing apoptin with inhibitors of checkpoint kinase 1 (Chk1) and Chk2 causes apoptin to localize to the cytoplasm. Importantly, silencing of Chk2 rescues cancer cells from the cytotoxic effects of apoptin. Finally, treatment of virus-producing cells with Chk inhibitor protects them from virus-mediated toxicity and reduces the titer of progeny virus. Taken together, our results indicate that apoptin is a sensor of DNA damage signaling through the ATM-Chk2 pathway, which induces it to migrate to the nucleus during viral replication.
IMPORTANCE The chicken anemia virus (CAV) protein apoptin is known to induce tumor cell-specific death when expressed. Therefore, understanding its regulation and mechanism of action could provide new insights into tumor cell biology. We have determined that checkpoint kinase 1 and 2 signaling is important for apoptin regulation and is a likely feature of both tumor cells and host cells producing virus progeny. Inhibition of checkpoint signaling prevents apoptin toxicity in tumor cells and attenuates CAV replication, suggesting it may be a future target for antiviral therapy.
INTRODUCTION
Circoviruses are a diverse group of nonenveloped, icosahedral viruses containing circular, single-stranded DNA genomes (1, 2). These viruses have been shown to infect a wide range of hosts, and they are the causative agents of several serious diseases in animals. In particular, chicken anemia virus (CAV), a member of the genus Gyrovirus, is the etiological agent of chicken infectious anemia. CAV infects several bone marrow-derived cells, resulting in severe anemia and immunosuppression in young chickens and compromised immune response in older birds (3, 4). CAV can lead to considerable economic loss during intensive chicken farming, and control of the virus through vaccination is currently standard practice. Recently, a novel circovirus with partial homology to CAV isolated from a skin swab was designated human gyrovirus (HGyV) (5). The identification of HGyV indicates a distinct potential for circovirus human pathogenesis, warranting further investigation.
The negative-sense CAV genome consists of 2,319 nucleotides and is replicated by a rolling-circle mechanism, although the packaging and egress of viral particles are poorly characterized (2, 6). It encodes multiple overlapping open reading frames (7) that are translated into three distinct polypeptides. CAV viral protein 1 (VP1) is the major structural protein, while VP2 is a replicase and has dual-specificity phosphatase activity (8). VP3, also called apoptin, is a nonstructural protein implicated in the induction of apoptosis and viral cytotoxicity in host cells. It is currently believed that the induction of apoptosis via apoptin by host cell factors represents a late step in the viral replication cycle whereby the resultant cell death facilitates the egress of novel viral particles, perhaps via association with apoptotic bodies.
Apoptin has attracted considerable interest due to its ability to mediate cell death selectively in cells that have undergone an oncogenic transformation (9–11). Preclinical in vitro and in vivo proof-of-concept studies performed with apoptin have demonstrated some success, with several small-animal models showing effective tumor regression while displaying minimal systemic toxicity (reviewed in reference 12).
Apoptin is a compact, 14-kDa polypeptide consisting of 121 amino acids enriched in basic residues (13). An N-terminal leucine-rich sequence (LRS) mediates nuclear export and multimerization of apoptin into globular complexes that interact with multiple cellular partners, notably the promyelocytic leukemia (PML) protein (9, 14). The C-terminal domain of apoptin contains both a bipartite nuclear localization sequence (NLS) and a secondary nuclear export sequence (NES), and together, these motifs confer the nucleocytoplasmic shuttling activity of the protein (9, 15). The extreme C-terminal residues of apoptin constitute a functional protein interaction motif mediating interaction with the anaphase-promoting complex or cyclosome (APC/C) ubiquitin E3 ligase. The APC/C is a 1.5-MDa protein complex that is essential for mitotic progression by ubiquitinating securin, cyclins, and other proteins that prevent mitotic exit (16). Interestingly, this inhibitory interaction has been shown to mediate p53-independent G2/M cell cycle arrest and apoptosis in cancer cells by means of APC/C dissociation and degradation within PML nuclear bodies (PML-NB) (17, 18).
The differential subcellular localization of apoptin has been implicated as the major mechanism mediating tumor-specific cytotoxicity (9, 19). In transformed cells, apoptin predominantly localizes to the nucleus, whereas this accumulation is impaired in normal cells. In the host setting of viral replication, CAV induces marked toxicity with tropism for cells of the hematopoietic lineage (20). These cell types undergo rapid proliferation and hence are prime candidates for exhibiting replication-associated stress responses. Stress responses result from the generation of aberrant DNA replication structures and exposure of the viral single-stranded genome. Similarly, activation of oncogenes in human cancer promotes deregulated firing of origins of replication, further contributing to replication stress and facilitating genomic instability (21–24). Collectively, these lesions induce a DNA damage response (DDR) mediated by the phosphatidylinositol 3-kinase-like (PIKK) ataxia telangiectasia mutated (ATM)-checkpoint kinase 2 (Chk2) and ATM-Rad3 related (ATR)-Chk1 axes (25–27).
Our previous studies have shown that DDR signaling regulates the subcellular localization of apoptin, requiring both ATM and DNA-dependent protein kinase (DNA-PK) (28). However, the primary structure of apoptin lacks consensus motifs for ATM or DNA-PK [p(Ser/Thr)-Gln], suggesting that a downstream component activated in response to ATM or DNA-PK could act as a functional apoptin kinase in human and avian host cells. In support of this notion, we identified four Chk1 and Chk2 phosphorylation consensus motifs [Arg-X-X-p(Ser/Thr)] within apoptin.
In this study, we observed that functional inhibition of Chk1/2 activity was accompanied by impaired apoptin nuclear localization and concomitant apoptosis. Moreover, we demonstrate that apoptin constitutes a veritable checkpoint kinase substrate in cancer cells. Two phosphorylation site mutants in the N-terminal domain of apoptin (T56A and T61A) showed significant impairment in nuclear accumulation, suggesting that these residues are the principal sites of Chk-mediated phosphorylation. Importantly, inhibition of the Chk proteins resulted in reduced cytopathic effect (CPE) in virus-producing cells, as well as reduced production of progeny virus. In summary, the regulation of apoptin described in this study provides mechanistic insight necessary for the establishment of a cohesive model of apoptin function and contributes to a broader understanding of dysregulated DNA damage response signaling in human cancer.
MATERIALS AND METHODS
Cell lines, viruses, and drugs.
H1299 non-small-cell lung adenocarcinoma cells were obtained from the ATCC and were maintained in Dulbecco's modified Eagle medium (Wisent Inc., QC, Canada) supplemented with 10% fetal bovine serum (FBS) (Sigma) and 0.1% gentamicin (Wisent Inc., QC, Canada) at 37°C with 5% CO2. MDCC MSB-1 cells (CLS Cell Lines Services, Eppelheim, Germany), a chicken lymphoblast line immortalized with Marek's disease virus, were cultured in 1× RPMI 1640 (Wisent Inc., QC, Canada) supplemented with 10% FBS and 50 μg/ml gentamicin at 41°C under 5% CO2. The adenoviruses encoding apoptin (Ad-Apwt) or LacZ (Ad-LacZ) were described previously (17) and used to infect H1299 cells at a multiplicity of infection (MOI) of 35. The Chk1/2 inhibitors AZD7762 and CHIR-124 were purchased from Selleck (Houston, TX) and used at the indicated concentrations. Paclitaxel (originally named taxol) was used at 100 nM. Cells were irradiated using a Radsource RS-2000 irradiator.
Site-directed mutagenesis and plasmid construction.
Flag-apoptin point mutants were generated by the QuikChange (Stratagene) protocol using synthetic primers (Sigma) and PCR amplification of a p3×Flag-myc-CMV-26 expression vector encoding wild-type (WT) apoptin. The N-terminal green fluorescent protein (GFP)-apoptin expression vector was subcloned from Flag vectors as described previously (17). To generate N-terminal hemagglutinin (HA)-tagged apoptin fusion proteins, wild-type and mutant apoptin open reading frames were amplified using PCR, digested by restriction endonucleases, and ligated into the pcDNA3-HA vector. Mutant clones were confirmed by sequencing.
Transfection.
H1299 cells were seeded and transfected with the indicated plasmids the following day at 70% confluence using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Silencing of Chk1 and Chk2 was carried out by overnight transfection of H1299 cells seeded at low confluence in 6-well plates or 60-mm dishes using 50 nmol of scrambled small interfering RNA (siRNA) (5′-AAU UCU CCG AAC GUG UCA CGU dTdT-3′), Chk1-1 (5′-GCG UGC CGU AGA CUG UCC AdTdT-3′), Chk1-2 (5′-GCA ACA GUA UUU CGG UAU A dTdT-3′), Chk2-1 (5′-GAA CCU GAG GAC CAA GAA C dTdT-3′), or Chk2-2 (5′-AAC GCC GUC CUU UGA AUA ACA dTdT-3′) siRNA duplexes (Sigma) and 5 μl Lipofectamine 2000 (Invitrogen). The cells were allowed to proliferate for 18 h prior to infection with Ad-LacZ or Ad-Apwt.
Western blotting.
SDS-PAGE and Western blotting were performed with standard protocols using the following antibodies at the indicated dilutions: 1:1,000 mouse monoclonal anti-Flag M2 (Sigma), 1:1,000 rabbit monoclonal anti-HA (BioLegend), 1:2,000 mouse monoclonal anti-GFP (Clontech), 1:2,000 monoclonal rat anti-pSer10 histone H3 (Sigma), 1:500 monoclonal anti-pSer1981 ATM (Santa Cruz), 1:1,000 monoclonal rabbit anti-ATM (Cell Signaling), 1:1,000 monoclonal rabbit anti-Chk1 (Cell Signaling), 1:50 rabbit polyclonal anti-APC1 (described in reference 17), 1:1,000 mouse monoclonal anti-Cdc20 (Santa Cruz), 1:500 mouse monoclonal anti-Cdh1 (NeoMarkers), 1:1,000 mouse monoclonal anti-53BP1 (BD), 1:500 polyclonal rabbit anti-pSer25 53BP1 (Bethyl), 1:1,000 rabbit monoclonal anti-pThr68 Chk2 (Cell Signaling), 1:500 mouse monoclonal anti-Chk2 (Santa Cruz), 1:1,000 rabbit polyclonal anti-APC2 (BioLegend), 1:500 mouse monoclonal anti-Cdc27 (BD), 1:1,000 mouse polyclonal anti-APC7 (a gift of A. S. Turnell), 1:1,000 rabbit polyclonal anti-APC8 (Santa Cruz), 1:1,000 rabbit polyclonal anti-APC5 (a gift of A. S. Turnell), and 1:1,000 rabbit polyclonal antibodies against apoptin and phosphoapoptin (T108), which have been previously described (29), and 1:1,000 rabbit polyclonal anti-actin (Sigma). The secondary antibodies used were 1:5,000 goat polyclonal anti-mouse and mouse anti-rabbit horseradish peroxidase (HRP) (Jackson ImmunoResearch, Inc.). The blots were developed with Western Lightning Plus ECL (PerkinElmer) or SuperSignal West Femto (Thermo Scientific) chemiluminescent substrates and exposed on film. The secondary antibodies used for immunofluorescence assays were Alexa Fluor 488- and 594-conjugated goat polyclonal anti-mouse, -rat, and -rabbit (ThermoFisher Scientific).
Immunofluorescence.
Cells grown on glass coverslips or 6-well plates were fixed in ice-cold methanol at −20°C for 20 min. The cells were washed twice with phosphate-buffered saline (PBS) and blocked in 10% antibody dilution buffer (ADB) (10% FBS, 0.05% Triton X-100 in PBS) for 15 min. Staining was carried out by incubation with the primary antibody in 10% ADB for 1 h at room temperature. The cells were washed 3 times for 5 min each time with PBS, followed by incubation with Alexa Fluor-conjugated secondary antibodies diluted 1:500 in 10% ADB for 1 h at room temperature. The cells were washed again, counterstained with DAPI (4′-6-diamidino-2-phenylindole; 1 μg/ml), and mounted on slides using ImmuMount mounting medium (Thermo Scientific Shanton). The cells were analyzed using a Zeiss AxioVert fluorescence microscope with AxioVision and cropped for presentation with ImageJ software.
Cell extract preparation.
H1299 cells were seeded on 10-cm dishes prior to transfection with the indicated Flag-apoptin constructs or infection with adenoviral vectors and treatment with dimethyl sulfoxide (DMSO) or AZD7762. Immunoprecipitation experiments were performed using 1,000 μg of whole-cell extract. For whole-cell extract preparation, cells were harvested as described above and resuspended in 100 μl of NETN buffer (50 mM Tris-Cl, pH 7.5, 2 mM MgCl2, 150 mM NaCl, 1% NP-40) per 1 × 106 cells, vortexed briefly, and lysed on ice for 30 min. The lysates were then centrifuged at maximum speed for 15 min at 4°C, and 4× Laemmli buffer was added to 60 μl of lysate (∼200 μg) and set aside as input for immunoblot analysis. The supernatants were incubated with 10 μl of packed EZview red anti-Flag M2 affinity gel (Sigma) for 2 h at 4°C on an end-over-end rotator. Following incubation, the supernatant was discarded and the affinity gel was washed five times with NETN, eluted in 60 μl 1× Laemmli buffer, and boiled for 5 min prior to use. Subcellular fractionation was performed as described previously (28). All the buffers were supplemented with one Complete Mini protease inhibitor cocktail tablet (Roche) per 10 ml. 32P metabolic labeling was performed on apoptin as previously described (30). Following detection of the 32P signal on film, the same gel was rehydrated and then stained with Coomassie to demonstrate equal expression of apoptin.
In vitro kinase assay.
H1299 cells were seeded on 10-cm dishes prior to transfection with 5 μg empty vector or apoptin constructs and 12.5 μl Lipofectamine 2000 in 5 ml Opti-MEM for 4 h. At 48 h posttransfection, the cells were harvested and then lysed in 1 ml modified RIPA buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitors. Flag-apoptin was then immunoprecipitated as described above. Following five washes with RIPA buffer, the resin was washed three times with wash buffer (25 mM HEPES, pH 7.5, 50 mM NaCl, 0.1% Triton X-100) and then finally washed twice with kinase assay buffer (5 mM MOPS [morpholinepropanesulfonic acid], pH 7.2, 2.5 mM glycerol 2-phosphate, 5 mM MgCl2,1 mM EGTA, 0.4 mM EDTA, 0.25 mM dithiothreitol [DTT]). The resin was then incubated with 100 ng Chk2 (Biovision) in kinase assay buffer and 5 μl ATP cocktail {575 μl kinase buffer, 15 μl 10 mM cold ATP, and 10 μl of 32P-labeled 10-mCi/ml (3,000-Ci/mmol) γ-ATP ([γ-32P]ATP)} for 15 min at 30°C. The total reaction volume was 25 μl. Residual ATP was then washed out, and 4× sample buffer was added to the resin slurry. The samples were then resolved by SDS-PAGE. The gel was stained with Coomassie to determine apoptin expression levels and finally dried and exposed to film to capture apoptin phosphorylation.
Two-dimensional gel electrophoresis.
H1299 cells were seeded on 15-cm dishes prior to infection with Ad-Apwt and treatment with DMSO or AZD7762 (6.3 μM). Cells were harvested as described above, and whole-cell extracts were prepared using 100 μl of modified RIPA buffer. Flag-apoptin was purified by immunoprecipitation as described above, with washing performed using RIPA buffer. Proteins were eluted with 60 μl DeStreak rehydration solution (GE) and boiled for 5 min prior to loading on an Ettan IPGPhor I isoelectric-focusing system (Amersham Biosciences) using Immobiline DryStrip gels, pH 6 to 11 (GE). The gels were rehydrated with DeStreak rehydration solution in ceramic strip holders overnight prior to isoelectric focusing following the manufacturer's recommendation. The gel strips were then incubated sequentially in two aliquots of equilibration buffer (50 mM Tris-Cl, pH 8.8, 6 M urea, 2% SDS, 30% glycerol) supplemented with 2% DTT and 2.5% iodoacetamide, respectively, on a rotating platform for 15 min each time at room temperature. The equilibrated strips were installed into standard SDS-PAGE cassettes and sealed with 0.5% agarose. The proteins were then resolved in the second dimension, transferred to nitrocellulose membranes, and probed by Flag immunoblotting.
Flow cytometry.
H1299 cells were seeded at low confluence in 6-well plates or 60-mm dishes and transfected overnight with siRNA duplexes. The cells were then infected with Ad-LacZ or Ad-Apwt as described above. For cell cycle analysis, cells were trypsinized at 24 h postinfection, counted, and resuspended in ice-cold PBS prior to overnight fixation of 1 × 106 cells in 70% ethanol at −20°C. The cells were subsequently washed and resuspended in 500 μl of propidium iodide (PI) staining buffer (200 μg/ml PI, 0.5 mg/ml RNase A, 0.1% Triton X-100 in PBS) and incubated for 15 min at 37°C in the dark. For apoptosis quantification, cells were harvested at 24, 48, or 72 h postinfection; washed twice; and resuspended at 1 × 106 cells/ml in ice-cold 1× annexin V binding buffer (10 mM HEPES, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2). One hundred microliters of cell suspension (1 × 105 cells) was incubated with 5 μl phycoerythrin (PE)-conjugated annexin V (BD BioSciences) and 5 μg 7-aminoactinomycin D (7-AAD) (AG Scientific) for 30 min at room temperature in the dark. Samples were diluted with 400 μl 1× binding buffer prior to data acquisition. The stained cell preparations were analyzed immediately on a FACSCalibur flow cytometer. Appropriate unstained and single-stained controls were prepared for determination of compensation using CellQuest Pro software (BD). Detection of mitotic cells was performed by staining for pSer10 histone H3 as previously described (31). The data were subsequently processed with FlowJo software.
Transfections and preparation of replicative CAV DNA.
The full-length CAV replicon used in these studies has been previously described (7). Viral genomes were recircularized with T4 DNA ligase to generate replicative-form (RF) viral genomes. MSB-1 cells were electroporated using the Amaxa Nucleofector system kit T (Lonza) according to the manufacturer's instructions. MSB-1 cells (2.0 × 106) and 400 ng of RF viral DNA were used in each transfection. Following electroporation, cells were allowed to recover for 1 h before being washed three times in PBS to eliminate residual RF viral DNA. The cells were then suspended at 2.5 × 105 cells/ml in growth medium.
Quantification of viral DNA.
Viral DNA was harvested from MSB-1 cells using the QiaAmp Viral RNA minikit (Qiagen) according to the manufacturer's instructions. The purified DNA was then digested with DpnI to minimize background generated by the RF viral DNA, derived from bacteria, used in the initial electroporation. Quantitative PCR (qPCR) amplification was carried out using the QuantiFast SYBR green qPCR kit. Primers targeting the Vp1 gene were designed so that the product spanned a DpnI restriction site. The cycling parameters were as follows: 95°C for 5 min, 95°C for 15 s, 60°C for 30 s, 95°C for 15 s, and 60°C for 20 min; melting curve, 95°C. The primers used to detect CAV were as follows: FW, ATGACCCTGCAAGACATGGG; RV, CTTTTTGCCACCGGTTCTGG.
Statistical analysis.
Quantitative data are presented as means and standard errors of the mean. Where indicated, statistical analyses were performed using the Student two-tailed t test with data collected from 3 or more biological repeats.
RESULTS
Chemical inhibition of Chk1/2 induces cytoplasmic accumulation of apoptin in tumor cells.
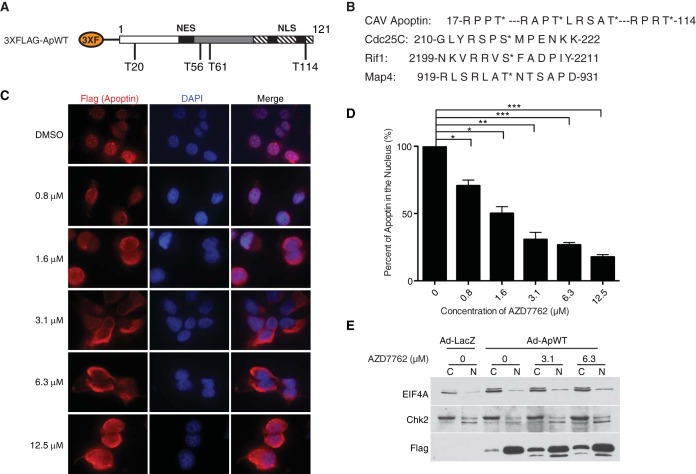
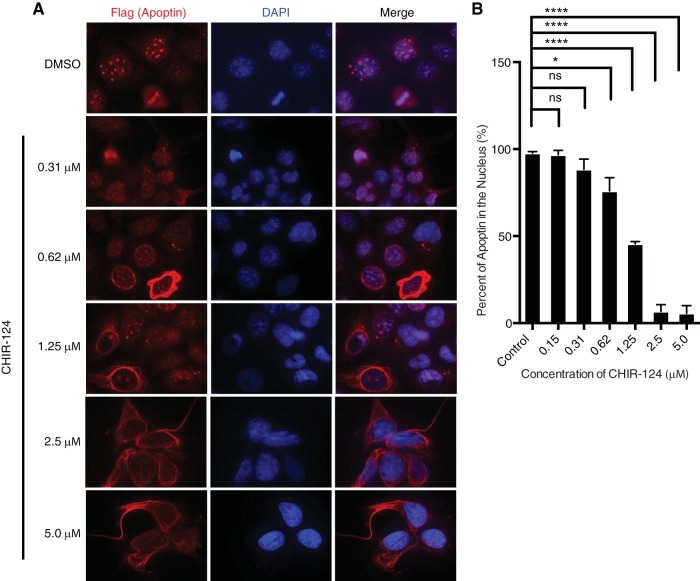
Regulation of apoptin nucleocytoplasmic shuttling by posttranslational modifications, principally phosphorylation, has been proposed to regulate the balance between NES and NLS activities, maintaining apoptin in the nuclei of transformed cells. We previously demonstrated that inhibition of ATM and DNA-PK in H1299 non-small-cell lung adenocarcinoma cells efficiently impaired the nuclear localization of apoptin (28). Given that the primary structure of apoptin lacks ATM or DNA-PK consensus motifs, we hypothesized that downstream checkpoint kinases, namely, Chk1 and Chk2, could phosphorylate apoptin and regulate its subcellular localization. In support of this notion, we identified four Chk consensus motifs within the primary structure of apoptin [Arg-X-X-p(Ser/Thr)] at threonine residues 20, 56, 61, and 114 (Fig. 1A and B). These motifs are very similar to those in known Chk substrates (32, 33) (Fig. 1B). In order to assess the involvement of checkpoint kinases in apoptin localization within transformed cells, we employed the small-molecule ATP-competitive kinase inhibitor AZD7762 in H1299 cells expressing Flag-tagged apoptin. To examine the effects of AZD7762 on apoptin localization, cells were infected and treated with various concentrations of AZD7762 or dimethyl sulfoxide (DMSO) prior to processing for Flag immunofluorescence. Examination of the cells revealed mostly cytoplasmic localization of apoptin in treated cells (Fig. 1C). Quantification of the cells revealed a significant dose-dependent increase in cytoplasmic apoptin, consistent with what is shown in representative micrographs (Fig. 1D). We then confirmed these observations by biochemical fractionation of apoptin-expressing cells treated with AZD7762. We found that apoptin accumulated in the cytoplasm in response to treatment with the inhibitor (Fig. 1E). We also tested a second inhibitor, CHIR-124, that primarily inhibits Chk1 and found that it efficiently inhibited nuclear accumulation of apoptin, further confirming the role of these kinases in regulating apoptin localization (Fig. 2A and B).
FIG 1.
Treatment with the pan-Chk inhibitor AZD7762 impairs nuclear localization of apoptin in a dose-dependent manner. (A) Schematic of 3×Flag-apoptin showing possible Chk1 and Chk2 phosphorylation sites (not drawn to scale). (B) Comparison of the amino acid sequence of apoptin with those of known checkpoint kinase substrates. Asterisks indicate that the preceding amino acid is the one that is phosphorylated (i.e., the threonine); hyphens indicate a stretch of irrelevant amino acids between the sequences that are shown. (C) H1299 cells were infected with Ad-Apwt and treated with DMSO or the indicated concentrations of AZD7762. At 24 h postinfection, the cells were processed for Flag immunofluorescence. (D) Quantification of cells displaying nuclear apoptin was determined by scoring at least 100 Flag-positive cells per condition. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.0005. The data are presented as means and standard errors of the mean (SEM). (E) Cells were prepared as for panel C but fractionated into nuclear (N) and cytoplasmic (C) fractions and then analyzed by SDS-PAGE and Western blotting. Numbers indicate the concentrations of AZD7762 with which the cells were treated.
FIG 2.
Treatment with a selective Chk1 inhibitor impairs nuclear localization of apoptin in a dose-dependent manner. (A) H1299 cells were infected with Ad-Apwt and treated with DMSO or the indicated concentrations of CHIR-124 (Chk1-i). At 24 h postinfection, the cells were processed for Flag immunofluorescence. (B) Quantification of cells displaying nuclear apoptin was determined by scoring at least 100 Flag-positive cells per condition. The data are presented as means and SEM (n = 3). Statistical significance was tested for treatment against the DMSO control (Student's t test; *, P < 0.05; ****, P < 0.0001).
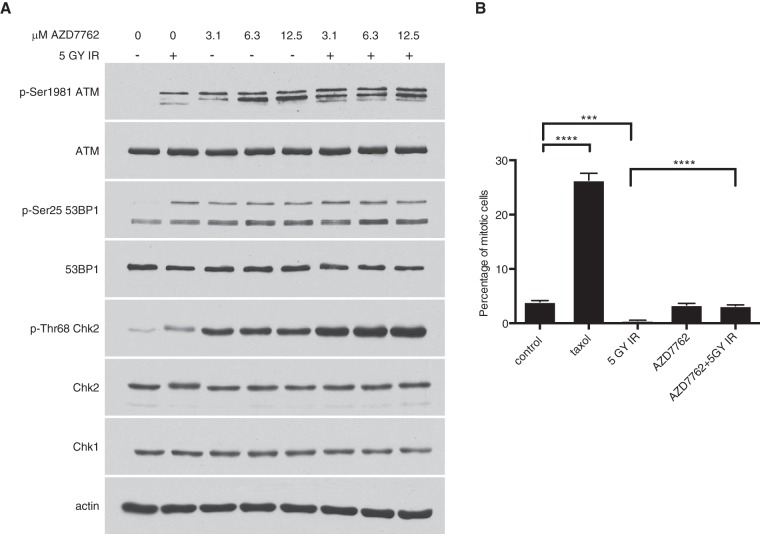
We next checked the effects of AZD7762 treatment on DNA damage signaling in H1299 cells and found that treatment induced DNA damage signaling through ATM, likely due to aberrant DNA replication during S phase (34). Treatment of irradiated H1299 cells with AZD7762 did not markedly affect ATM signaling (Fig. 3A), confirming the specificity of the inhibitor. Since a function of the ATM-Chk2 pathway is to induce G2 arrest and block mitotic entry in response to DNA damage, we tested the efficacy of the inhibitor by measuring the percentage of mitotic cells in irradiated cells treated with the inhibitor. We found that treatment with AZD7762 completely abolished the cell cycle arrest induced by Chk2 in response to irradiation (Fig. 3B). The accumulation of mitotic cells in response to treatment with the microtubule-stabilizing drug paclitaxel served as a positive control for the assay.
FIG 3.
Treatment with the pan-Chk inhibitor AZD7762 impairs checkpoint kinase activity, but not ATM kinase activity. ATM S1981, 53BP1 S25, and Chk2 T68 are characterized ATM consensus sites. (A) H1299 cells were pretreated for 30 min with the indicated concentrations of AZD7762 (Chk-i) and then irradiated with 5 Gy ionizing radiation (IR). At 6 h posttreatment, the cells were harvested and processed for SDS-PAGE. (B) Quantification of mitotic cells by flow cytometry measurement of pSer10 histone 3-positive cells following the same treatment as for panel A. The data are presented as means and SEM (n = 3). Statistical significance was tested for treatment against the control (Student's t test; ****, P < 0.0001; ***, P = 0.0001).
Silencing of Chk1/2 expression via siRNA induces cytoplasmic accumulation of apoptin in tumor cells.
In order to determine if Chk1 or Chk2 affects localization of apoptin, we designed siRNAs to specifically knock down expression of the kinases. Immunofluorescence staining of apoptin showed that specific targeting of Chk1 and Chk2 alone or in combination resulted in an increase in cytoplasmic accumulation of apoptin comparable to that with a nontargeting control sequence (Fig. 4A and C). Protein levels of Chk1 and Chk2 were analyzed by immunoblotting to verify the knockdown efficiency (Fig. 4B). Furthermore, combined knockdown using both targeting sequences did not yield further impairment of nuclear localization. Also, knockdown of Chk1 and Chk2 with a second set of siRNA duplexes yielded nearly identical results (data not shown). Thus, apoptin phosphorylation by individual checkpoint kinases may represent interdependent events, raising the possibility that Chk1 and Chk2 may regulate activation of distinct apoptin functions in a context-dependent manner.
FIG 4.
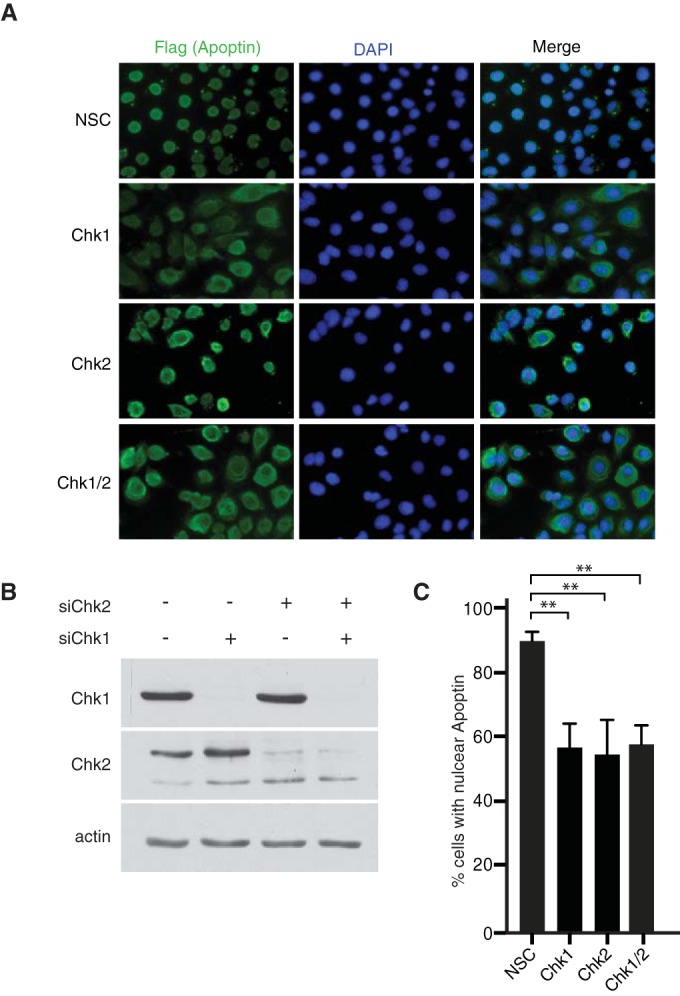
RNAi knockdown of Chk1/2 induces cytoplasmic localization of wild-type apoptin. (A) H1299 cells were transfected at low confluence with the indicated siRNA duplexes and allowed to proliferate for 16 h. The cells were then infected with Ad-Apwt and processed for Flag immunofluorescence at 24 h postinfection. (B) Immunoblot analysis of the experiment shown in panel A verifying knockdown efficiency and specificity. (C) Quantification of cells displaying nuclear apoptin was determined by scoring at least 100 Flag-positive cells per condition. **, P ≤ 0.01. NSC indicates a nonsilencing control.
Apoptin is a checkpoint kinase substrate.
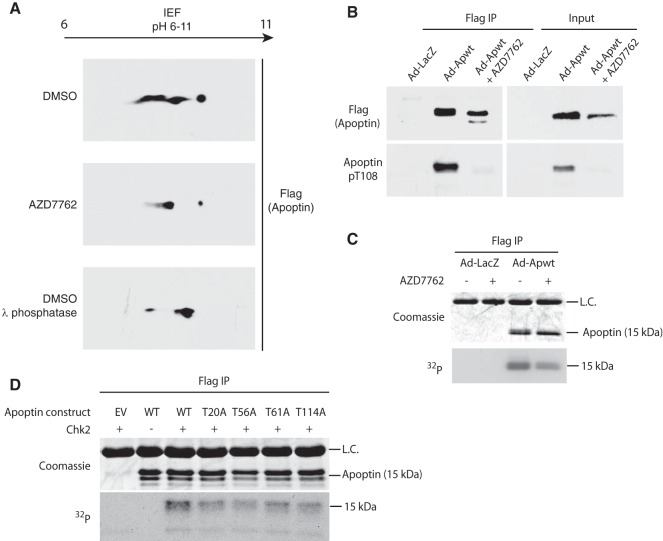
To confirm that the regulation of apoptin subcellular localization by the checkpoint kinases is a result of phosphorylation, we performed two-dimensional gel electrophoresis to analyze Flag-tagged apoptin peptides in the presence or absence of inhibitor. Flag-apoptin was introduced by adenoviral infection into H1299 cells with concurrent treatment with the Chk1/2 inhibitor AZD7762 or DMSO (vehicle control). Flag-apoptin was immunoprecipitated under stringent conditions and analyzed in the first dimension by isoelectric focusing and then by standard SDS-PAGE. The migration of apoptin peptides at a higher isoelectric point can be observed in the presence of AZD7762 relative to control treatment (Fig. 5A). Since phosphorylated peptides migrate at a lower isoelectric point, owing to the negative charge of phosphoryl groups at physiological pH, the compaction of peptides is consistent with a loss of phosphorylation. The migration of apoptin in the presence of AZD7762 was comparable to that of Flag-apoptin incubated with lambda phosphatase prior to isoelectric focusing.
FIG 5.
Apoptin is a checkpoint kinase substrate. (A) H1299 cells were infected with Ad-Apwt and treated with DMSO or 6.3 μM AZD7762 (Chk-i). At 24 h postinfection, Flag-apoptin was purified by immunoprecipitation under stringent conditions and resolved in the first dimension with a pH 6 to 11 gradient and then analyzed by SDS-PAGE in the second dimension. Immunoblotting for Flag was performed to visualize Flag-apoptin peptides. Lambda-phosphatase treatment of purified Flag-apoptin served as a positive control. IEF, isoelectric focusing. (B) H1299 cells were treated with DMSO or 6.3 μM Chk-i and infected with Ad-Apwt or Ad-LacZ for 24 h. Whole-cell extracts were prepared for immunoprecipitation (IP) of Flag complexes and immunoblot analysis with the indicated antibodies (pT108, phospho-specific antibody [Ab] for phosphorylated Thr108). (C) H1299 cells were infected with Ad-Apwt or Ad-LacZ and treated with DMSO or 6.3 μM AZD7762. At 23 h postinfection, the medium was changed to include 250 mCi/ml [32P]orthophosphate. Following 1 h of labeling, Flag-apoptin was purified by immunoprecipitation under stringent conditions and analyzed by SDS-PAGE. (D) H1299 cells were transfected with the indicated constructs. At 48 h posttransfection, Flag-apoptin was immunoprecipitated with anti-Flag resin. The resin was then incubated with 100 ng Chk2 and [γ-32P]ATP for 15 min at 30°C. Residual γ-ATP was washed out, and 4× sample buffer was added to the samples, which were then analyzed by SDS-PAGE. L.C. indicates IgG light chain.
Previous studies have implicated the phosphorylation of T108 as a critical mediator of apoptin localization and function (35). To address whether impairment of Chk1 and Chk2 alter the phosphorylation status of T108, we isolated Flag-apoptin complexes from H1299 cells by immunoprecipitation and performed immunoblot analysis with a phosphospecific antibody for apoptin T108. Interestingly, there was a marked reduction in T108 phosphorylation following treatment with AZD7762 (Fig. 5B). Moreover, chemical inhibition of Chk1 and Chk2 affected the migration of Flag-apoptin on SDS-PAGE gels, so that a faster-migrating, lower-molecular-weight band is clearly visible (Fig. 5B). Our data suggest that inhibition of Chk1 and Chk2 results in cytoplasmic relocalization of apoptin and loss of phosphorylation at the T108 site by nuclear cyclin-Cdk2 complexes, although we cannot dismiss the possibility that Chk1 or Chk2 acts directly on the nonconsensus T108 residue (36).
To confirm that inhibition of Chk1 and Chk2 directly alters apoptin phosphorylation, we performed [32P]orthophosphate metabolic labeling in the presence and absence of AZD7762, followed by immunoprecipitation of apoptin under stringent conditions. We found that 32P labeling of apoptin was reduced in the presence of AZD7762, indicating that apoptin is less phosphorylated in treated cells. Moreover, the apoptin immunoprecipitated from the treated cells migrated faster in the gel, consistent with a loss of phosphorylation (Fig. 5C).
Finally, to determine if Chk2 directly phosphorylates apoptin, recombinant apoptin was immunoprecipitated from H1299 cells and added to purified Chk2 in the presence of [γ-32P]ATP. Figure 5D shows that Chk2 was indeed able to directly phosphorylate apoptin. In vitro kinase reactions were also performed using apoptin mutants in which each of the four predicted Chk1/2 phosphorylation sites was changed to alanines (T20A, T56A, T61A, and T114A). Each of the four mutants displayed less 32P incorporation than wild-type apoptin, suggesting that at least in vitro, all four sites are phosphorylated by Chk2 (Fig. 5D). Taken together, our observations support the hypothesis that Chk-mediated phosphorylation is a regulatory mechanism of apoptin activation.
Mutation of N-terminal threonine residues impairs nuclear localization of apoptin in tumor cells.
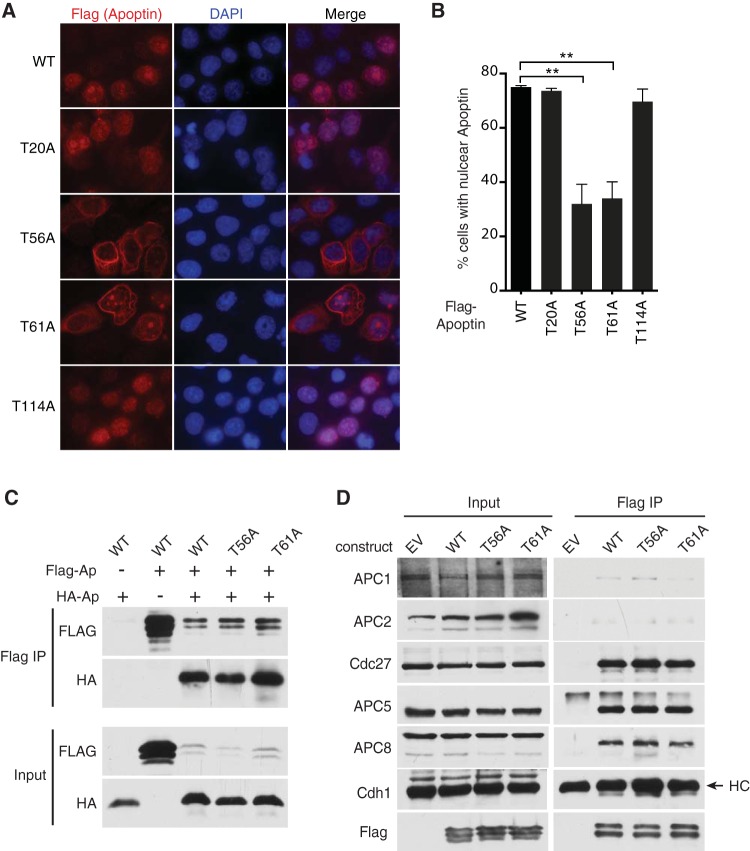
To determine which of the possible phosphorylation sites on apoptin are regulated by the Chk proteins in vivo, we transiently transfected the Chk1/2 phosphorylation site mutants into H1299 cells and compared their localization patterns with that of wild-type apoptin. The T56A and T61A mutants displayed the greatest defect in nuclear accumulation, with cells displaying filamentous apoptin located in the cytoplasm (Fig. 6A and B). Therefore, our data suggest that phosphorylation at N-terminal threonine residues facilitates nuclear localization of apoptin. Given that the T56A and T61A mutants failed to localize to the nucleus, we determined if these mutants were otherwise functional. We therefore tested whether the apoptin mutants had defects in multimerization activity, which was previously shown to be essential for induction of apoptosis (9). The aggregation of monomers and interaction of apoptin with PML bodies has been shown to require the N-terminal leucine-rich motif (14, 18). We cotransfected H1299 cells with HA-tagged apoptin and mutant Flag-apoptin and tested the interaction of differentially tagged apoptin. Immunoprecipitation of Flag-apoptin captured both wild-type and mutant apoptin, indicating that interaction between apoptin monomers was unaffected (Fig. 6C).
FIG 6.
Mutation of Thr56 and Thr61 impairs nuclear localization of apoptin in H1299 cells. (A) H1299 cells were transfected with Flag-tagged WT or mutant apoptin. The cells were allowed to proliferate for 24 h prior to processing for Flag immunofluorescence with nuclear staining (DAPI). (B) Quantification of cells displaying nuclear apoptin was determined by scoring at least 100 Flag-positive cells per condition. **, P ≤ 0.01. The error bars indicate SEM. (C and D) H1299 cells were transfected with the indicated constructs for 24 h. Whole-cell extracts were prepared for immunoprecipitation of Flag complexes and immunoblot analysis with the indicated antibodies. E.V. indicates empty vector.
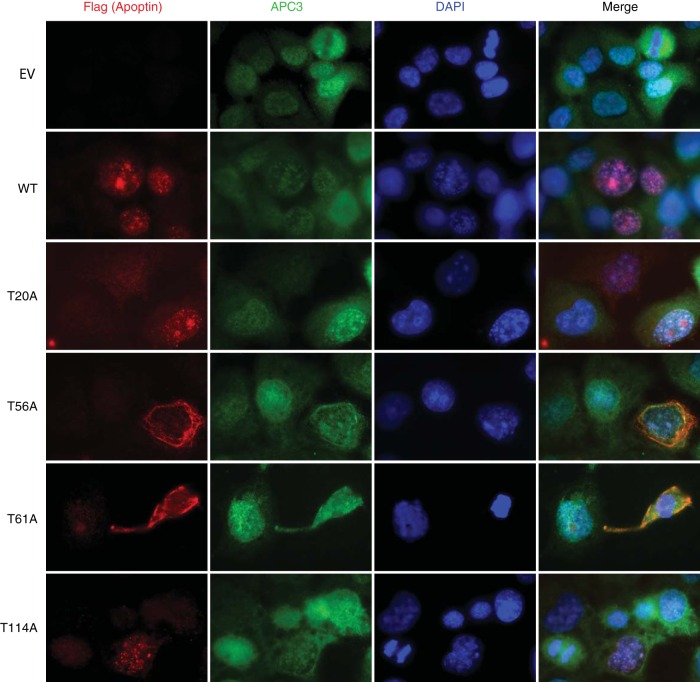
Similarly, we determined if the phosphorylation mutants were still able to interact with the anaphase-promoting complex. Consistent with previous studies (17), wild-type apoptin was able to pull down both core (APC1, APC2, APC3 or Cdc27, APC5, and APC8) and coactivator (Cdc20 and Cdh1) components of the complex under the conditions employed in our assay (Fig. 6D). T56A and T61A mutants immunoprecipitated APC/C subunits with efficiency similar to that of wild-type apoptin, indicating that these mutants are otherwise functional. We confirmed these results by staining cells for both APC3 and WT or mutated apoptin. We found that the apoptin T56A and T61A mutants continued to colocalize with APC3 despite being cytoplasmic (Fig. 7). Together, these results indicate that residues T56 and T61 are important for regulating the localization of apoptin but do not affect other activities of the protein, including multimerization and interaction with the APC/C.
FIG 7.
Mutation of Thr56 and Thr61 does not affect colocalization of the APC/C with apoptin. H1299 cells were transfected with Flag-tagged WT or mutant apoptin. The cells were allowed to proliferate for 24 h prior to processing for Flag and APC3 immunofluorescence with nuclear staining (DAPI). E.V. indicates empty vector.
Chk1/2 activity regulates apoptin-induced G2/M arrest and apoptosis.
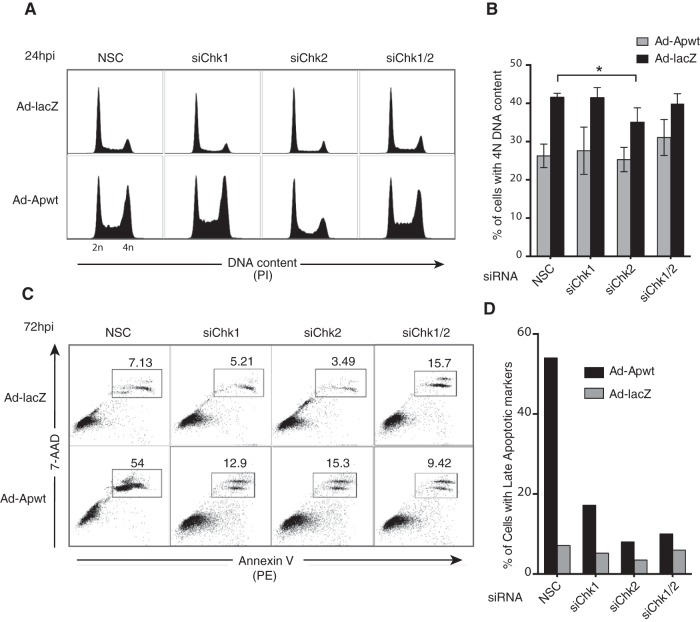
If checkpoint kinases regulate the subcellular localization of apoptin, chemical inhibition or knockdown of Chk1 and Chk2 would be predicted to impair apoptin functions in the nucleus. The interaction of apoptin with the APC/C has been shown to result in the sequestration of the APC/C to PML bodies, resulting in G2/M arrest (17). To determine whether Chk1 and Chk2 action is required for apoptin-induced cell cycle arrest, we performed cell cycle analysis of H1299 cells transfected with siRNAs targeting Chk1 and/or Chk2 and transduced with an apoptin-expressing vector. Individual or combined knockdown of Chk1 and Chk2 alone did not alter the cell cycle, as evaluated by control LacZ-expressing-cell profiles (Fig. 8A). Analysis of cell cycle profiles from apoptin-expressing cells showed a significant decrease in cells with 4 N DNA content upon transfection with siRNA targeting Chk2 (Fig. 8B).
FIG 8.
RNAi knockdown of Chk2 impairs apoptin-induced G2/M arrest and apoptosis. (A) H1299 cells were transfected at low confluence with the indicated siRNA duplexes prior to infection with Ad-Apwt or Ad-LacZ. At 24 h postinfection (hpi), the cells were fixed and stained with PI for flow cytometry. (B) Quantification of cells containing 4 N (G2/M phase) DNA content derived from replicate experiments, as shown in panel A. *, P ≤ 0.05. The error bars indicate SEM. (C) Representative flow cytometric profiles of cells treated as for panel A and harvested and stained at 72 hpi with 7-AAD and PE annexin V. Percentages of PE- and 7-AAD-double-positive cells corresponding to the late apoptotic population are boxed. (D) Quantification of late apoptotic cells at 72 hpi performed by manual gating analysis.
We then investigated the kinetics of apoptin-induced cell death. H1299 cells were treated as described above for cell cycle analysis, and cell death was assessed at various time points by annexin V/7-AAD staining (Fig. 8C and D). In accordance with previous studies (17), at 72 h postinfection, approximately 54% of apoptin cells were apoptotic in the nonsilencing controls. Notably, we observed a significant decrease in late apoptotic cells in cells treated with siRNA targeting either Chk1 or Chk2 (Fig. 8C and D). These data show that silencing of Chk1 and Chk2 functionally delays apoptin-induced cell death, presumably by impaired accumulation of apoptin in the nucleus so that it cannot execute its proapoptotic functions.
Inhibition of Chk activity in CAV-infected cells attenuates virus production and cytopathic effect.
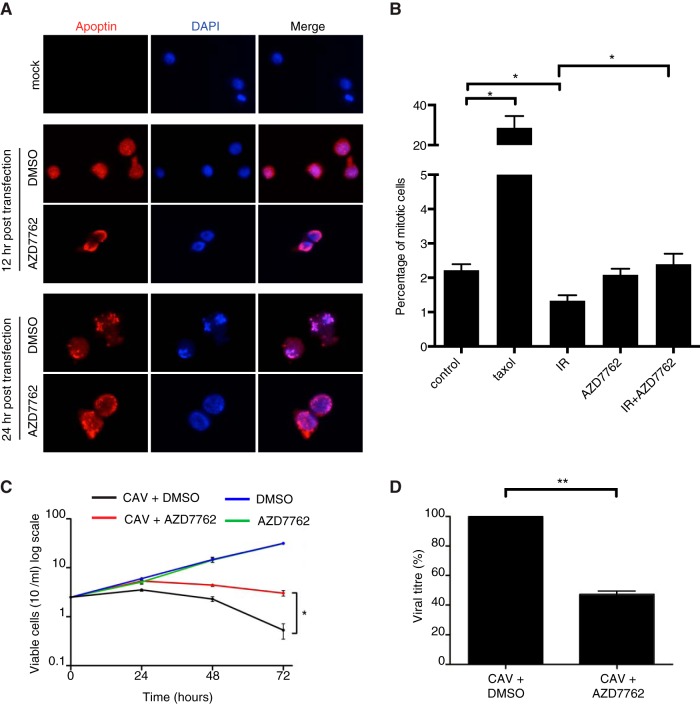
Given the importance of the Chk proteins in regulating apoptin, we asked whether this regulation plays a role during CAV replication. To accomplish this, we used a chicken lymphoid cell line transformed with Marek's disease virus (MSB-1) that permits CAV replication (37). We first determined if AZD7762 was able to affect the localization of apoptin in the context of viral replication. MSB-1 cells were infected with wild-type CAV, and the localization of apoptin was examined by immunofluorescence using an anti-apoptin antibody (29). Figure 9A shows that apoptin localized to the nuclei of CAV-infected cells but that addition of AZD7762 resulted in cytoplasmic retention of the protein. We then checked if AZD7762 was able to abolish the cell cycle arrest normally induced by the Chk proteins in response to ionizing radiation, as we did in H1299 cells. We found that while the irradiation caused cell cycle arrest in untreated cells, treatment with AZD7762 abolished this arrest in MSB-1 cells (Fig. 9B). These experiments confirmed that AZD7762 is able to inhibit chicken Chk1/2 and that inhibition of the Chk proteins can affect localization of apoptin during viral replication. CAV is known to induce a potent CPE (7), and we therefore determined if AZD7762 had an effect on CPE mediated by viral replication. Figure 9C shows that MSB-1 cells infected with CAV rapidly decreased in viability while mock-infected cells continued to grow normally. Interestingly, inhibition of Chk proteins with AZD7762 was able to significantly protect CAV-infected cells from death. We next asked whether treatment of CAV-producing cells with the inhibitor would have an effect on viral replication. When fresh cells were infected with CAV and treated with AZD7762, we observed an almost 60% decrease in virus production in the treated population (Fig. 9D). Taken together, these data show that Chk signaling affects apoptin activity during viral infection and is required for effective CAV replication.
FIG 9.
Inhibition of Chk activity in CAV-producing cells attenuates virus production and cytopathic effect. (A) MSB-1 cells were transfected with CAV and treated with AZD7762 (350 nM) or DMSO. The cells were then fixed and stained for immunofluorescence as indicated. (B) MSB-1 cells were pretreated with AZD7762 (350 nm) for 30 min and then irradiated with 3 Gy ionizing radiation. Mitotic cells were quantified by flow cytometry measurement of pSer10 histone 3-positive cells. (C) MSB-1 cells were transfected with CAV and treated with AZD7762 (350 nm) or DMSO. Cellular viability was monitored over time by trypan blue staining. (D) MSB-1 cells were transfected with CAV and treated with AZD7762 (350 nm) or DMSO. Newly synthesized viral genomes were extracted 72 h posttransfection and quantified by quantitative PCR. Statistical significance was tested for treatment against the control (Student's t test; *, P < 0.05; **, P < 0.01).
DISCUSSION
Subcellular localization has been implicated as the main characteristic contributing to the selective mechanism of action of apoptin (9, 19). Although the importance of apoptin localization in apoptosis has been well characterized, the mechanism of regulation and how it relates to viral replication has yet to be addressed. In the current study, we show that chemical inhibition or RNA interference (RNAi)-mediated silencing of Chk1 and Chk2 abrogates nuclear accumulation of apoptin in transformed cells. Consistent with the localization data, we demonstrated that cell cycle arrest and apoptosis induced by apoptin were impaired in cells with Chk1 and Chk2 knockdown. Our data show that two threonine residues (T56 and T61) adjacent to the N-terminal NES motif are sites of Chk phosphorylation, providing a mechanistic basis for regulation of apoptin localization. Importantly, we show that inhibition of Chk1/2-mediated apoptin phosphorylation and nuclear localization impairs viral replication.
Although the subcellular localization of apoptin appears to be a major mechanism of regulation, there is evidence that other factors may also be required. For example, it has been demonstrated that forced localization of apoptin to the nucleus of primary cells by fusing a canonical nuclear localization signal is insufficient to induce apoptosis (9, 15). Previous studies have shown that threonines 107 and 108 on apoptin are phosphorylated in transformed cells and that this modification is important in regulating apoptotic activity (35, 38, 39). Neither of these sites has the consensus sequence of a Chk1/2 target, and others have suggested that they may be targeted by a nuclear kinase, such as HIPK2 (40). Full activation of apoptin may therefore require a 2-step process whereby DDR signaling promotes nuclear localization and a nuclear kinase, such as HIPK2, then phosphorylates T-108. Consistent with this model, we observed that T-108 phosphorylation was reduced in cells treated with Chk inhibitors (Fig. 3B). Once apoptin becomes localized to the nucleus, additional modifications, such as sumoylation, may also contribute to full functional activation of the protein (18).
The notion that Chk1 and Chk2 phosphorylation may regulate apoptin function by altering localization may explain the transformation-specific localization of the protein. Cancer cells have been shown to have elevated basal levels of DDR signaling and therefore to also have elevated Chk activity (24, 41, 42). In the context of viral infection, the insights into selective activation of apoptin presented in our study may aid in the clarification of apoptin function in viral replication and pathogenesis. It is likely that during early stages of viral infection in normal (nontransformed) host cells, apoptin remains cytoplasmic and inactive. During rolling-circle replication of the CAV genome, single-stranded DNA is generated and serves as a trigger for DNA damage sensors, including RPA and other components involved in ATR activation (3). Abnormal replication fork structures subsequently generate double-strand breaks (DSB) and induce recruitment and activation of ATM (25, 26). In response to signaling events emanating from these foci, Chk1 and Chk2 subsequently initiate activity on downstream substrates, mediating appropriate repair responses. The abundance of apoptin and activated Chk1 and Chk2 would steadily increase upon the onset of viral DNA replication, eventually reaching a threshold analogous to that of transformed cells, whereby apoptin-induced cell death is initiated. This model is consistent with evidence indicating that nuclear apoptin is associated with productive CAV replication and cytopathogenicity (43).
Our group previously characterized an inhibitory consequence of nuclear apoptin for DDR focus formation, principally mediated by proteasomal degradation of the essential DSB mediator MDC1 (28). It is possible that apoptin senses ATM-Chk2 and ATR-Chk1 signaling by direct phosphorylation events, promoting subsequent impairment of the host response and thus preventing it from interfering with viral replication. The inhibitory role of apoptin is similar in principle to reported functions of several viral effectors, including the herpes simplex virus (HSV) immediate-early protein ICP0. HSV ICP0 is a functional RING ubiquitin E3 ligase inducing the degradation of RNF8 and RNF168, which are host factors downstream of MDC1 that facilitate DDR mediator recruitment at DSB-induced foci (27, 44, 45). Additionally, other DNA tumor viruses, including adenovirus, polyomaviruses, and human papillomavirus, encode multiple proteins that regulate the DDR (46–48).
Despite our identification of the mechanistic basis for apoptin activation by DDR signaling, the mechanism by which nuclear apoptin induces apoptosis remains incompletely defined. Previous work by our group and others has suggested that interaction with the APC/C results in mitotic catastrophe and cell death (9, 17, 49). Inhibition of APC/C function is a common mechanism exploited by several metazoan viruses, presumably to maintain favorable cellular conditions for viral replication (50, 51). Interestingly, in light of essential functions of DDR kinases and mediators in the coordination of cell cycle checkpoints, the described interplay of apoptin with the DDR may represent a strategy common to many viruses. Indeed, understanding the mechanism of apoptin may contribute to the elucidation of novel links between the DNA damage response, the spindle assembly checkpoint, and virus replication.
ACKNOWLEDGMENTS
We thank Mathieu Noteborn for the CAV replicon.
This work was supported by grants from the Canadian Institute of Health Research (CIHR), MOP-179122, and the Natural Science and Engineering Research Council (NSERC) of Canada to J.G.T. T.J.K. was supported by studentships from the Fonds de la Recherche en Santé du Québec (FRSQ) and CIHR. T.F.N. was supported by an NSERC undergraduate studentship research award.
REFERENCES
- 1.Fauquet CM, Fargette D. 2005. International Committee on Taxonomy of Viruses and the 3,142 unassigned species. Virol J 2:64. doi: 10.1186/1743-422X-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung AK. 2012. Porcine circovirus: transcription and DNA replication. Virus Res 164:46–53. doi: 10.1016/j.virusres.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Schat KA. 2009. Chicken anemia virus. Curr Top Microbiol Immunol 331:151–183. [DOI] [PubMed] [Google Scholar]
- 4.Todd D. 2000. Circoviruses: immunosuppressive threats to avian species: a review. Avian Pathol 29:373–394. doi: 10.1080/030794500750047126. [DOI] [PubMed] [Google Scholar]
- 5.Sauvage V, Cheval J, Foulongne V, Gouilh MA, Pariente K, Manuguerra JC, Richardson J, Dereure O, Lecuit M, Burguiere A, Caro V, Eloit M. 2011. Identification of the first human gyrovirus, a virus related to chicken anemia virus. J Virol 85:7948–7950. doi: 10.1128/JVI.00639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prasetyo AA, Kamahora T, Kuroishi A, Murakami K, Hino S. 2009. Replication of chicken anemia virus (CAV) requires apoptin and is complemented by VP3 of human torque teno virus (TTV). Virology 385:85–92. doi: 10.1016/j.virol.2008.10.043. [DOI] [PubMed] [Google Scholar]
- 7.Noteborn MH, de Boer GF, van Roozelaar DJ, Karreman C, Kranenburg O, Vos JG, Jeurissen SH, Hoeben RC, Zantema A, Koch G, van Ormondt H, van der Eb AJ. 1991. Characterization of cloned chicken anemia virus DNA that contains all elements for the infectious replication cycle. J Virol 65:3131–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters MA, Jackson DC, Crabb BS, Browning GF. 2002. Chicken anemia virus VP2 is a novel dual specificity protein phosphatase. J Biol Chem 277:39566–39573. doi: 10.1074/jbc.M201752200. [DOI] [PubMed] [Google Scholar]
- 9.Heilman DW, Teodoro JG, Green MR. 2006. Apoptin nucleocytoplasmic shuttling is required for cell type-specific localization, apoptosis, and recruitment of the anaphase-promoting complex/cyclosome to PML bodies. J Virol 80:7535–7545. doi: 10.1128/JVI.02741-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeurissen SH, Wagenaar F, Pol JM, van der Eb AJ, Noteborn MH. 1992. Chicken anemia virus causes apoptosis of thymocytes after in vivo infection and of cell lines after in vitro infection. J Virol 66:7383–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Los M, Panigrahi S, Rashedi I, Mandal S, Stetefeld J, Essmann F, Schulze-Osthoff K. 2009. Apoptin, a tumor-selective killer. Biochim Biophys Acta 1793:1335–1342. doi: 10.1016/j.bbamcr.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Backendorf C, Visser AE, de Boer AG, Zimmerman R, Visser M, Voskamp P, Zhang YH, Noteborn M. 2008. Apoptin: therapeutic potential of an early sensor of carcinogenic transformation. Annu Rev Pharmacol Toxicol 48:143–169. doi: 10.1146/annurev.pharmtox.48.121806.154910. [DOI] [PubMed] [Google Scholar]
- 13.Leliveld SR, Dame RT, Rohn JL, Noteborn MH, Abrahams JP. 2004. Apoptin's functional N- and C-termini independently bind DNA. FEBS Lett 557:155–158. doi: 10.1016/S0014-5793(03)01465-0. [DOI] [PubMed] [Google Scholar]
- 14.Leliveld SR, Zhang YH, Rohn JL, Noteborn MH, Abrahams JP. 2003. Apoptin induces tumor-specific apoptosis as a globular multimer. J Biol Chem 278:9042–9051. doi: 10.1074/jbc.M210803200. [DOI] [PubMed] [Google Scholar]
- 15.Danen-Van Oorschot AA, Zhang YH, Leliveld SR, Rohn JL, Seelen MC, Bolk MW, Van Zon A, Erkeland SJ, Abrahams JP, Mumberg D, Noteborn MH. 2003. Importance of nuclear localization of apoptin for tumor-specific induction of apoptosis. J Biol Chem 278:27729–27736. doi: 10.1074/jbc.M303114200. [DOI] [PubMed] [Google Scholar]
- 16.Peters J-M. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 7:644–656. [DOI] [PubMed] [Google Scholar]
- 17.Teodoro JG, Heilman DW, Parker AE, Green MR. 2004. The viral protein Apoptin associates with the anaphase-promoting complex to induce G2/M arrest and apoptosis in the absence of p53. Genes Dev 18:1952–1957. doi: 10.1101/gad.1198404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janssen K, Hofmann TG, Jans DA, Hay RT, Schulze-Osthoff K, Fischer U. 2007. Apoptin is modified by SUMO conjugation and targeted to promyelocytic leukemia protein nuclear bodies. Oncogene 26:1557–1566. doi: 10.1038/sj.onc.1209923. [DOI] [PubMed] [Google Scholar]
- 19.Danen-Van Oorschot AA, Fischer DF, Grimbergen JM, Klein B, Zhuang S, Falkenburg JH, Backendorf C, Quax PH, Van der Eb AJ, Noteborn MH. 1997. Apoptin induces apoptosis in human transformed and malignant cells but not in normal cells. Proc Natl Acad Sci U S A 94:5843–5847. doi: 10.1073/pnas.94.11.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuscu B, Gurel A. 2008. Lesions in the thymus and bone marrow in chicks with experimentally induced chicken infectious anemia disease. J Vet Sci 9:15–23. doi: 10.4142/jvs.2008.9.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hills SA, Diffley JF. 2014. DNA replication and oncogene-induced replicative stress. Curr Biol 24:R435–R444. doi: 10.1016/j.cub.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Negrini S, Gorgoulis VG, Halazonetis TD. 2010. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 23.Halazonetis TD, Gorgoulis VG, Bartek J. 2008. An oncogene-induced DNA damage model for cancer development. Science 319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 24.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 25.Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat Cell Biol 16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol Cell 40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukas J, Lukas C, Bartek J. 2011. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 28.Kucharski TJ, Gamache I, Gjoerup O, Teodoro JG. 2011. DNA damage response signaling triggers nuclear localization of the chicken anemia virus protein Apoptin. J Virol 85:12638–12649. doi: 10.1128/JVI.05009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang J, Cole D, Westwood N, Macpherson L, Farzaneh F, Mufti G, Tavassoli M, Gaken J. 2010. Crucial roles for protein kinase C isoforms in tumor-specific killing by apoptin. Cancer Res 70:7242–7252. doi: 10.1158/0008-5472.CAN-10-1204. [DOI] [PubMed] [Google Scholar]
- 30.Lu Y, Kucharski TJ, Gamache I, Blanchette P, Branton PE, Teodoro JG. 2014. Interaction of adenovirus type 5 E4orf4 with the nuclear pore subunit Nup205 is required for proper viral gene expression. J Virol 88:13249–13259. doi: 10.1128/JVI.00933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juan G, Darzynkiewicz Z. 2004. Detection of mitotic cells. Curr Protoc Cytom Chapter 7:Unit 7.24. doi: 10.1002/0471142956.cy0724s28. [DOI] [PubMed] [Google Scholar]
- 32.O'Neill T, Giarratani L, Chen P, Iyer L, Lee CH, Bobiak M, Kanai F, Zhou BB, Chung JH, Rathbun GA. 2002. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J Biol Chem 277:16102–16115. doi: 10.1074/jbc.M111705200. [DOI] [PubMed] [Google Scholar]
- 33.Blasius M, Forment JV, Thakkar N, Wagner SA, Choudhary C, Jackson SP. 2011. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol 12:R78. doi: 10.1186/gb-2011-12-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buisson R, Boisvert JL, Benes CH, Zou L. 2015. Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress during S phase. Mol Cell 59:1011–1024. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohn JL, Zhang YH, Aalbers RI, Otto N, Den Hertog J, Henriquez NV, Van De Velde CJ, Kuppen PJ, Mumberg D, Donner P, Noteborn MH. 2002. A tumor-specific kinase activity regulates the viral death protein Apoptin. J Biol Chem 277:50820–50827. doi: 10.1074/jbc.M208557200. [DOI] [PubMed] [Google Scholar]
- 36.Maddika S, Panigrahi S, Wiechec E, Wesselborg S, Fischer U, Schulze-Osthoff K, Los M. 2009. Unscheduled Akt-triggered activation of cyclin-dependent kinase 2 as a key effector mechanism of apoptin's anticancer toxicity. Mol Cell Biol 29:1235–1248. doi: 10.1128/MCB.00668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goryo M, Suwa T, Matsumoto S, Umemura T, Itakura C. 1987. Serial propagation and purification of chicken anaemia agent in MDCC-MSB1 cell line. Avian Pathol 16:149–163. doi: 10.1080/03079458708436360. [DOI] [PubMed] [Google Scholar]
- 38.Lee YH, Cheng CM, Chang YF, Wang TY, Yuo CY. 2007. Apoptin T108 phosphorylation is not required for its tumor-specific nuclear localization but partially affects its apoptotic activity. Biochem Biophys Res Commun 354:391–395. doi: 10.1016/j.bbrc.2006.12.201. [DOI] [PubMed] [Google Scholar]
- 39.Lanz HL, Florea BI, Noteborn MH, Backendorf C. 2012. Development and application of an in vitro apoptin kinase assay. Anal Biochem 421:68–74. doi: 10.1016/j.ab.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 40.Poon IK, Oro C, Dias MM, Zhang J, Jans DA. 2005. Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res 65:7059–7064. doi: 10.1158/0008-5472.CAN-05-1370. [DOI] [PubMed] [Google Scholar]
- 41.DiTullio RA Jr, Mochan TA, Venere M, Bartkova J, Sehested M, Bartek J, Halazonetis TD. 2002. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol 4:998–1002. doi: 10.1038/ncb892. [DOI] [PubMed] [Google Scholar]
- 42.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 43.Renshaw RW, Soine C, Weinkle T, O'Connell PH, Ohashi K, Watson S, Lucio B, Harrington S, Schat KA. 1996. A hypervariable region in VP1 of chicken infectious anemia virus mediates rate of spread and cell tropism in tissue culture. J Virol 70:8872–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaurushiya MS, Lilley CE, Aslanian A, Meisenhelder J, Scott DC, Landry S, Ticau S, Boutell C, Yates JR III, Schulman BA, Hunter T, Weitzman MD. 2012. Viral E3 ubiquitin ligase-mediated degradation of a cellular E3: viral mimicry of a cellular phosphorylation mark targets the RNF8 FHA domain. Mol Cell 46:79–90. doi: 10.1016/j.molcel.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panier S, Durocher D. 2013. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat Rev Mol Cell Biol 14:661–672. doi: 10.1038/nrm3659. [DOI] [PubMed] [Google Scholar]
- 46.Carson CT, Orazio NI, Lee DV, Suh J, Bekker-Jensen S, Araujo FD, Lakdawala SS, Lilley CE, Bartek J, Lukas J, Weitzman MD. 2009. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J 28:652–662. doi: 10.1038/emboj.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moody CA, Laimins LA. 2009. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog 5:e1000605. doi: 10.1371/journal.ppat.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turnell AS, Grand RJ. 2012. DNA viruses and the cellular DNA-damage response. J Gen Virol 93:2076–2097. doi: 10.1099/vir.0.044412-0. [DOI] [PubMed] [Google Scholar]
- 49.Lanz HL, Zimmerman RME, Brouwer J, Noteborn MHM, Backendorf C. 2013. Mitotic catastrophe triggered in human cancer cells by the viral protein apoptin. Cell Death Dis 4:e487. doi: 10.1038/cddis.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mo M, Shahar S, Fleming SB, Mercer AA. 2012. How viruses affect the cell cycle through manipulation of the APC/C. Trends Microbiol 20:440–448. doi: 10.1016/j.tim.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Smolders L, Teodoro JG. 2011. Targeting the anaphase promoting complex: common pathways for viral infection and cancer therapy. Expert Opin Ther Targets 15:767–780. doi: 10.1517/14728222.2011.558008. [DOI] [PubMed] [Google Scholar]