Abstract
An expanded polyglutamine (polyQ) tract at the amino-terminus of the androgen receptor (AR) confers toxic properties responsible for neuronal and non-neuronal degeneration in spinal and bulbar muscular atrophy (SBMA), one of nine polyQ expansion diseases. Both lower motor neurons and peripheral tissues, including skeletal muscle, are affected, supporting the notion that SBMA is not a pure motor neuron disease but a degenerative disorder of the neuromuscular system. Here, we review experimental evidence demonstrating both nerve and muscle degeneration in SBMA model systems and patients. We propose that polyQ AR toxicity targets these components in a time-dependent fashion, with muscle pathology predominating early and motor neuron loss becoming more significant at late stages. This model of pathogenesis has important therapeutic implications, suggesting that symptoms arising from degeneration of nerve or muscle predominate at different points and that directed interventions targeting these components will be variably effective depending upon disease progression.
Keywords: Spinal and bulbar muscular atrophy, CAG/polyglutamine disorder, Androgen receptor, Skeletal muscle, Motor neuron
Introduction
Over the last two and a half decades, neurogeneticists have identified nine degenerative disorders caused by abnormally long CAG/polyglutamine (polyQ) tracts in the coding sequence of disease-causing genes [1]. This type of mutation was first identified in the androgen receptor (AR) gene as the cause of spinal and bulbar muscular atrophy (SBMA; Kennedy disease) [2]. Since then, similar coding region microsatellite expansions have been identified as the causative mutation in otherwise unrelated genes encoding huntingtin protein, responsible for Huntington’s disease, in atrophin-1, causing dentatorubral-pallidoluysian atrophy, and in ataxin-1, -2, and -3, in the Cav2.1 P/Q voltage-dependent calcium channel, in ataxin-7, and in TATA-binding protein, causing spinocerebellar ataxias type 1, 2, 3, 6, 7, and 17, respectively.
As a group, these disorders share both a common mutational mechanism and several important clinical features that suggest the existence of shared underlying pathogenic mechanisms. All are chronic, progressive neurodegenerative diseases with symptom onset most frequently in mid-life and phenotypes that are slowly progressive. In each case, the length of the glutamine tract is negatively correlated with the age of onset and positively correlated with the disease severity. As the expanded repeat is unstable and shifts in length as it is passed from one generation to the next, intergenerational expansions underlie genetic anticipation [1]. Importantly, in each disease, the mutation leads to toxicity as a result of misfolding of the mutant protein. While these mutations impact normal protein function to yield disease-specific manifestations, proteotoxicity unrelated to normal function is also common to all of these disorders and is a significant mediator of cell dysfunction and death. Notably, while eight of the polyQ disorders are inherited as autosomal dominant disorders, the penetrance of the SBMA phenotype is dependent upon male levels of circulating androgens. This feature leads to the occurrence of disease only in men.
Spinal and bulbar muscular atrophy
The initial clinical description of SBMA appeared in the Japanese literature over a century ago and is attributed to Hiroshi Kawahara [3]. Nearly three quarters of a century later, William Kennedy documented details of the clinical and pathological features of this disorder and described its X-linked pattern of inheritance [4]. Anita Harding provided a subsequent comprehensive clinical description, noting that symptoms are frequently late in onset and do not occur in heterozygous female carriers [5], features that make the family history of the disease less obvious and the diagnosis more difficult. Additional analyses have confirmed that female carriers show only subclinical disease manifestations [6, 7]. SBMA patients often present with symptoms of muscle weakness between 30 and 60 years of age. These features are typically preceded by tremors, muscle cramps, and elevated serum levels of creatine kinase. Bulbar, facial, and proximal muscles of the arms and legs are involved as the disease progresses. Dysphagia and aspiration can occur in advanced cases. In addition to neuromuscular symptoms, patients may develop sensory neuropathy, particularly affecting vibration in the distal legs, as well as signs of partial androgen insensitivity, including gynecomastia, reduced fertility, and testicular atrophy [8, 9]. The prevalence of the disease is estimated at 1–2 per 100,000 individuals, although precise numbers are difficult to ascertain. SBMA patients may be misdiagnosed with other neuromuscular diseases, such as amyotrophic lateral sclerosis, from which it is distinguished clinically by its striking sex bias, slow progression, and absence of upper motor neuron signs. The sex bias of disease reflects the importance of male levels of circulating androgens in triggering toxicity of the polyQ AR protein. This feature of pathogenesis was first established in mouse models of disease by Gen Sobue and colleagues, where surgical or chemical castration of transgenic male mice produced marked improvement of symptoms, whereas testosterone administration to female transgenic mice markedly exacerbated symptoms and pathologic features; subsequent studies in additional mouse models have confirmed these observations [10–12].
Androgen receptor
Linkage studies initially localized the genetic defect responsible for SBMA to the proximal long arm of the X chromosome (Xq11–12) in a region containing the AR gene [13]. Analysis of this candidate gene by Kenneth Fischbeck and colleagues identified an expansion of a CAG trinucleotide repeat encoding a polyglutamine tract in the coding sequence of exon 1 [2]. This repeat encodes 9–34 glutamines in normal individuals and is expanded from 38 up to the mid-60s in SBMA patients. The AR protein is a 110-kDa nuclear receptor that belongs to the steroid/thyroid hormone receptor family. After binding ligand, either testosterone (T) or dihydrotestosterone (DHT), AR translocates to the nucleus and regulates the expression of target genes. Notably, DHT is much more potent than T as an AR ligand because it has a fourfold higher affinity and a fivefold slower dissociation rate from the receptor [14]. DHT is synthetized from T by the enzyme 5α-reductase, especially in the prostate. In other tissues where 5α-reductase activity is absent, such as skeletal muscle, T is the natural ligand of the AR. Variation in the length of the CAG repeat, even within normal alleles, influences AR function as a ligand activated transcription factor, such that shorter repeats are associated with a more transcriptionally active receptor. This variation in AR function has been associated with a variety of phenotypic outcomes in men [15–17]. AR is expressed in reproductive tissues as well as in skeletal muscle, liver, kidney, adrenal gland, skin, and the central nervous system, indicating that it supports the development and/or the maintenance of many cell types [18].
AR structure
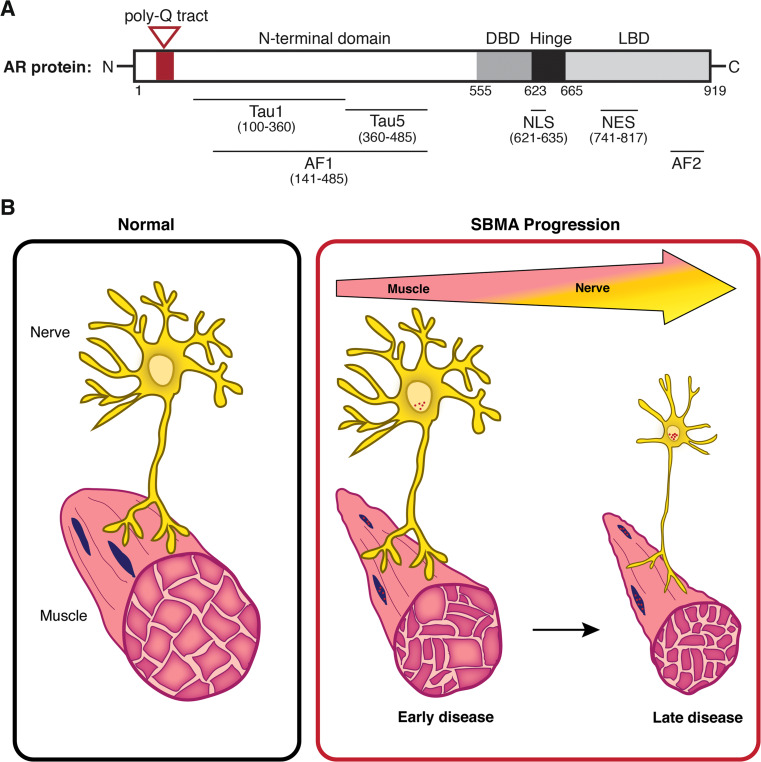
Of the eight exons composing the AR gene, exon 1 encodes an amino-terminal transactivation domain (NTD) that contains the CAG trinucleotide repeat (Fig. 1a). The domain encoded by exon 1 possesses the major transcriptional regulatory activity of the receptor, including the strong activation function 1 (AF-1, residues 142–485) region that contains the two transcription activation units, Tau-1 (residues 100–360) and Tau-5 (residues 360–485) [19, 20]. These regions are required for AR’s interaction with transcriptional coregulators. Notably, AF-1 mediates interaction with members of the p160 family of nuclear receptor coactivators. Through this mechanism, steroid receptor coactivator-1 (SRC-1) and c-AMP responsive element binding protein (CBP) are recruited to the AR [21–23]; these interactions are altered in disease models [24, 25] (see below). In addition to the polyQ tract, the amino-terminal domain of the AR protein contains polyglycine and polyproline stretches, but these polyamino acid tracts are not implicated in disease, and their functional roles are poorly understood.
Fig. 1.
Expansion of the androgen receptor’s polyglutamine tract triggers neuromuscular degeneration in SBMA. a Linear diagram of the AR, indicating the presence of the glutamine tract in the N-terminal domain (highlighted in red). This track is expanded in SBMA patients to ≥38 glutamines. Also shown are the central DBD and hinge regions, C-terminal LBD, and several additional functional domains that are detailed in the text. Numbers indicate amino acid residues. b Model of time-dependent pathology in SBMA resulting in skeletal muscle and motor neuron degeneration. Skeletal muscle alterations occur early in disease progression and are reflected by myopathic symptoms, including elevated serum creatine kinase levels and muscle cramps. Symptoms arising from motor neuron degeneration predominate at later stages and progress to motor neuron loss at end stage. Nuclear accumulation and intranuclear inclusions of the polyQ AR (in red) occur throughout the disease in both nerve and muscle cells. Data from model systems suggest that expression of the polyQ AR in skeletal muscle plays an important role in disease symptoms and progression
The central DNA-binding domain (DBD, residues 556–623) and the hinge region (residues 624–665) are encoded by exons 2 and 3. The DBD is composed of two zinc fingers that allow specificity in DNA binding and stabilization of DNA–protein interactions, whereas the hinge region contains a PEST sequence (P proline, E glutamic acid, S serine, T threonine) that has been suggested to target proteins for degradation through the proteasome [26]. In addition, the junction between the DBD and hinge region contains a nuclear localization signal (NLS, residues 621–635) that interacts with α-importin and drives AR into the nucleus following ligand binding [27]. In addition to influencing nuclear localization and proteasomal degradation, these regions have complex affects on DNA binding, on the interaction of amino- and carboxy-terminal domains (so-called N/C interaction), and on the recruitment of coregulators [28, 29]. These regions are also the site of functionally important post-translational modifications, including acetylation, ubiquitylation, and methylation [30]. For example, acetylation at Lys residues 630, 632, and 633 affects subcellular localization, interaction with coregulators, and transcriptional activity [31].
The carboxy-terminal ligand-binding domain (LBD, residues 666–919) is encoded by exons 4–8, and undergoes ligand-dependent conformational changes that lead to the assembly of the less potent activation function 2 (AF-2) region [32]. This region has weak interactions with transcriptional coregulators, such as SRC-1. The hydrophobic surface of AF-2 is essential for the interaction of the carboxy-terminus of the receptor with the NTD and, therefore, for the full activity of the AR [33]. The LBD is also important for the regulation of AR nuclear export. It contains a nuclear export signal (NES, residues 742–817) that helps to complete AR nuclear/cytoplasmic shuttling after ligand withdrawal [34].
AR in SBMA
Expansion of the AR’s polyQ tract results in ligand-dependent protein unfolding/misfolding as well as a partial loss of transactivation function. Proteotoxicity caused by the polyQ tract expansion occurs through mechanisms that are distinct from normal AR function (so-called toxic gain of function) [35] and is manifest in several cell types, including both lower motor neurons and skeletal muscle cells [36–38]. In addition, the polyQ tract expansion in AR is also responsible for the partial loss of normal function that is characteristic of SBMA. The mutation seems to disrupt the interaction between the amino-terminal transactivation domain and transcriptional coactivators [14]. Reduced AR activity likely contributes to the endocrine symptoms observed in many SBMA patients. Notably, endocrine target tissues also express high levels of the polyQ AR protein and show effects of proteotoxicity [39]. This complex mixture of effects mediated by protein misfolding and loss of normal function is a feature shared by SBMA and several other degenerative proteinopathies.
Underlying proteotoxicity is the expanded polyQ tract in the amino-terminus of the AR protein that alters the conformation of this domain from a random coil to a β-sheet. This change is believed to favor the formation of soluble oligomeric species that are considered to be rate-limiting in aggregation [40]. These oligomers are thought to be a toxic species that initiates a complex downstream series of events that lead to cell degeneration [41–43]. Oligomers are also capable of coalescing into insoluble fibrils, which are the building blocks of histopathologically visible nuclear inclusions that are present in SBMA tissue [44, 45]. While these AR immunoreactive intranuclear inclusions are indicative of protein unfolding/misfolding as a result of the CAG repeat expansion, several studies demonstrated that they may play a protective role in pathogenesis by sequestering the mutant protein and preventing its toxicity [42]. In fact, diffuse accumulation of misfolded polyQ proteins, rather than protein aggregates, correlates more closely with the initiation of neurodegeneration in several polyglutamine diseases [46]. In support of this notion, Adachi et al. [38] demonstrated that diffuse nuclear accumulation of the polyQ AR is more frequent than nuclear inclusions in the anterior horn of the spinal cord and that it is strictly correlated with the number of glutamines in the AR polyQ tract. It has been suggested that the polyQ AR principally accumulates within the nuclei of motor neurons in a diffusible form, leading to neural dysfunction and eventual cell death (reviewed in [32]). Reports have also suggested that the polyQ AR accumulates in the cytoplasm of certain types of cells, including sensory neurons [38]. The importance of these cytoplasmic aggregates is uncertain as studies in model systems have established that nuclear localization of the polyQ AR is essential for toxicity. Indeed, mutation of the nuclear localization signal or addition of a nuclear export signal reduces toxicity in cellular and Drosophila models of SBMA [47, 48].
Regulation by the Hsp90/Hsp70-based chaperone machinery
Both the normal and expanded polyQ ARs are client proteins of the heat shock protein 90 (Hsp90) and Hsp70-based chaperone machinery [49, 50]. Unlike the classical notion of individual chaperones binding to newly synthesized or unfolded/misfolded proteins to help them attain their folded conformation, the Hsp90/Hsp70-based chaperone machinery interacts with pre-folded proteins in their native or near-native conformations. Through this interaction, the chaperone machinery regulates essential aspects of client protein proteostasis, including ligand binding, cytoplasmic to nuclear shuttling, and degradation [49, 51]. In the protein quality control function of the chaperone machinery, Hsp90 and Hsp70 have opposing effects on client protein stability [52]. For instance, when cycling with Hsp90 is blocked by specific inhibitors, client proteins undergo rapid ubiquitination and degradation through the proteasome, thereby revealing an essential role of Hsp90 in maintaining protein stability. In contrast, Hsp70, along with its co-chaperone Hsp40, mediates turnover of unfolded/misfolded client proteins by facilitating the recruitment of chaperone-dependent E3 ubiquitin ligases, such as CHIP (carboxyl terminus of Hsc70-interacting protein) [53]. These mechanisms tightly regulate degradation of the polyQ AR. In fact, Hsp90 inhibition or Hsp70 or CHIP overexpression ameliorates disease in a transgenic mouse model of SBMA [54, 55]. Notably, both Hsp90 and Hsp70 have ATP binding sites and intrinsic ATPase activity. Both of these chaperones have low affinity for their substrates in their ATP-bound states, but the hydrolysis of ATP to ADP changes their conformation and increases their binding affinity [51]. This allosteric regulation of Hsp70 binding affinity to unfolded/misfolded clients has enabled the identification of genetic and pharmacological strategies to favor the ADP-bound form of Hsp70 and, thereby, promote ubiquitination and clearance of the polyQ AR [56].
When T or the more potent ligand DHT binds to the polyQ AR, its interaction with the chaperone machinery becomes much more dynamic. This permits the receptor to undergo conformational changes resulting in the disassociation of Hsp90 and the rapid initiation of intramolecular interactions between the amino- and carboxy-terminus of the AR (N/C interaction), followed by nuclear translocation. After nuclear translocation, the AR dimer stably binds to androgen response elements in promoter or enhancer regions of target genes [57]. As a result, AR mediates the effects of the androgens by modulating the expression of target genes. These steps are also critical in the pathogenesis of SBMA: nuclear translocation of the polyQ AR is necessary, but not sufficient for its toxicity [38]. Moreover, ligand-dependent interactions of the carboxy-terminal AF-2 domain with coregulators [58] and N/C interaction [59, 60] are both required for polyQ AR toxicity in model systems.
Altered transcriptional activity
The AR NTD mediates the majority of the AR’s transcriptional activity and serves as a surface for the recruitment and assembly of transcriptional coregulators. The polyQ tract expansion in the NTD alters interaction with these coregulators and impacts gene expressions. The mutant receptor undergoes a partial loss of function, and, thereby, fails to regulate a subset of genes that are normally androgen-responsive [61]. This likely contributes to a loss of trophic support for target cells, including both skeletal muscle and lower motor neurons. The coactivator SRC-1, a member of the p160 family, interacts directly with the NTD domain of AR, where Tau-5 has high affinity for a glutamine-rich domain in the p160 proteins [19]. This coactivator also interacts with AF-2 in the LBD of AR, but this particular interaction is not considered essential for AR transcriptional function [23]. The interaction between AR and SRC-1 potentiates receptor activity by recruitment of additional cofactors, including CREB-binding protein (CBP)/p300 and coactivator-associated arginine methyltransferase 1 (CARM1) that modify chromatin structure by histone acetyltransferase and histone methyltransferase activity [21, 62]. The presence of the polyQ tract expansion in SBMA reduces p160-mediated coactivation and, thereby, affects AR regulation of gene expression. It has been proposed that a shorter polyQ tract gives the AR a more accessible or stable surface for AR-interacting proteins like SRC-1 [22]. Conversely, there is evidence that polyQ AR toxicity is mediated by abnormal interactions with components of the transcriptional regulatory apparatus. For example, the expanded polyQ tract may favor aberrant association with coregulators, such as NF-Y and p300/CBP-associated factor to alter the expression of critical genes [63]. These data are consistent with the observation that toxicity of the polyQ AR in a Drosophila model requires DNA binding followed by association with coregulators [58]. In addition, nuclear accumulation of the polyQ AR is suggested to cause broad effects on gene expression by sequestering and interfering with the function of essential transcriptional coregulators [24]. For example, sequestration of Sp1 and CBP by the polyQ AR has been shown to reduce histone acetyltransferase activity in cellular models of disease [24]. The decrease in histone acetylation may alter chromatin structure and diminish expression of genes (e.g., vascular endothelial factor and type II TGF-beta receptor) that promote neuronal survival [63, 64]. In support of this view, increased acetylation by treatment with histone deacetylase inhibitors mitigates neuronal dysfunction and ameliorates motor phenotype in cellular and animal models of polyglutamine repeat diseases [65–68].
Post-translational modifications
A large number of post-translational modifications have been identified that influence AR function and stability, including phosphorylation, acetylation, and sumoylation. PolyQ AR phosphorylation on Ser 215 and Ser 792, mediated by the serine-threonine protein kinase Akt, inhibits ligand binding and mitigates toxicity in cultured motor neurons [69]. Insulin-like growth factor 1 (IGF-1) increases this effect in cell culture. Muscle-specific IGF-1 overexpression attenuates both muscle and spinal cord pathology in SBMA transgenic mice, and its beneficial effects may occur through phosphorylation and inactivation of the polyQ AR by Akt [70]. Acetylation at Lys 630, 632, and 633 has been shown to increase polyQ AR cytotoxicity. PolyQ AR nuclear hyperacetylation is suppressed by sirtuin 1 (SIRT1, a nicotinamide adenine dinucleotide-dependent histone deacetylase), an effect that reduces polyQ AR nuclear accumulation and toxicity in primary motor neurons [71]. Sumoylation inhibits the transcriptional regulatory activity of the polyglutamine protein [72]. Disruption of polyQ AR sumoylation at Lys 385 and 518 enhances its activity as a hormone-dependent transcriptional regulator and increases its trophic support. In a knock-in mouse model, disruption of polyQ AR sumoylation rescues exercise endurance, type I muscle fiber atrophy, and early death [73], thereby revealing ligand-induced trophic effects that are beneficial in disease.
Motor neuron degeneration
Lower motor neuron degeneration has long been considered to be a primary pathological event in SBMA [4, 5, 74]. In subjects evaluated at autopsy, loss of lower motor neurons and, to a lesser extent, primary sensory neurons of the dorsal root ganglia is observed [74]. Motor neuron degeneration occurs in the anterior horn of the spinal cord and in brainstem motor nuclei, and is associated with decreased axon numbers in the ventral spinal nerve roots. In contrast, neurons in Onuf’s nucleus, the intermediolateral columns, and Clarke’s columns of the spinal cord are generally well preserved [74]. Electromyography of SBMA subjects shows significantly decreased motor conduction velocities, compound muscle action potentials, sensory conduction velocities, and sensory nerve action potentials. These features are indicative of axonal degeneration that involves both motor and sensory nerves [75, 76]. Similarly, motor unit number estimation revealed that the number of functioning motor units is reduced in SBMA patients compared with controls, a defect that correlates with diminished ipsilateral grip strength and disease duration [77].
Pathological analyses provide evidence of polyQ AR unfolding/misfolding within lower motor neurons. Nuclear inclusions of the mutant AR protein are present in motor neurons of the spinal cord and brainstem of SBMA patients. Similar inclusions are also found in non-neuronal tissues, including scrotal skin, dermis, kidney, heart, and testis [44]. These inclusions show immunoreactivity with antibodies specific for epitopes at the amino-terminus of the androgen receptor and with antibodies against ubiquitin; antibodies recognizing epitopes at the carboxy-terminus of the protein are unable to detect nuclear inclusions, thus suggesting that the carboxy-terminal domain is cleaved and not present in the inclusions or is masked within the inclusion itself [44, 45].
Skeletal muscle degeneration
Clinical reports have established that SBMA patients frequently present with initial myopathic symptoms, including both muscle cramping and serum creatine kinase levels that exceed those which are typically observed in denervating diseases [78, 79]. Moreover, muscle biopsies from SBMA patients show myopathic features, indicating that expression of the polyQ AR in skeletal muscle exerts cell autonomous toxic effects [78–80].
These clinical reports are buttressed by findings in a variety of model systems that support the occurrence of skeletal muscle degeneration in SBMA. Analysis of a knock-in mouse model shows that skeletal muscle pathology characterized by both myopathic and neurogenic features occurs early in the disease and precedes the occurrence of spinal cord pathology by many months [10]. Interestingly, muscle atrophy in diseased mice is promoted by robust activation of macroautophagy [81–83], raising the possibility that overactivity of this pathway may be deleterious in SBMA. Supporting the concept that skeletal muscle is an important component of disease pathogenesis are data from transgenic mice that overexpress the wild type AR only in skeletal muscle and show hormone-dependent myopathy and motor axon loss [84]. Moreover, muscle-specific overexpression of IGF-1 or peripheral IGF-1 administration mitigates SBMA symptoms in transgenic mice [70, 85]. In addition, skeletal muscle satellite cells isolated from SBMA patients show impaired myogenic capacity in cell culture [86]. The notion that skeletal muscle is an important and therapeutically attractive target was recently tested in SBMA mouse models. Subcutaneous delivery of antisense oligonucleotides to knock-in and BAC transgenic models of SBMA diminished polyQ AR expression in the periphery but not spinal cord, and rescued muscle atrophy, neuromuscular function and survival [36]. These findings were corroborated by genetic studies in which deletion of a floxed polyQ AR allele in muscles of BAC transgenic mice produced similar robust effects [37].
Neuromuscular degeneration
We have come to view SBMA as a degenerative disorder of the neuromuscular system, with contributions to pathogenesis by both skeletal muscle cells and lower motor neurons. This model is distinct from the idea that degeneration in SBMA exclusively targets the central nervous system (CNS), leading to secondary effects in skeletal muscle that are wholly attributable to motor neuron loss. We propose that the relative contributions of each of these cell types changes as the disease progresses (Fig. 1b). Early in the disease course, when patients exhibit prominent myopathic features (muscle cramping, elevated serum creatine kinase), contributions from skeletal muscle may be most significant. It is at this stage that impaired trophic support by affected muscle may expedite the occurrence of motor neuron dysfunction. Diminished expression of several neurotrophic factors by diseased muscle has been demonstrated in SBMA mouse models, including neurotrophin-4, glial-cell derived neurotrophic factor, and brain derived neurotrophic factor; the loss of these factors or other signals may eventually promote motor neuron degeneration [10, 87]. Experimental evidence also suggests that polyQ AR expression in motor neurons contributes to disease manifestations. Indeed, transgenic expression of the polyQ AR only in motor neurons [88] or intrathecal gene knockdown by antisense oligonucleotide administration to SBMA transgenic mice [89] both support a contribution of motor neurons to pathogenesis. By end-stage, when subjects come to autopsy, frank motor neuron degeneration is present, indicating that late in the disease symptoms attributable to motor neuron loss likely predominate. This model of pathogenesis has important therapeutic implications. It suggests that interventions that occur early, prior to neuron loss, are most likely to be beneficial. Moreover, this model implies that strategies to target peripherally expressed polyQ AR in muscle may be effective, particularly early in the disease course. It is our hope that as new and promising therapies move into the clinic, these concepts will help to guide trial design to enable the discovery and validation of disease modifying therapies.
Acknowledgments
We thank Kayla Capper for help creating the illustration. Supported by the National Institutes of Health (R01 NS055746, R21 NS089516 to A.P.L.) and by the University of Michigan Protein Folding Disease Initiative.
Abbreviations
- SBMA
Spinal and bulbar muscular atrophy
- CAG
Cytosine adenine guanine
- PolyQ
Polyglutamine
- AR
Androgen receptor
- T
Testosterone
- DHT
Dihydrotestosterone
- NTD
Amino-terminal domain
- DBD
DNA-binding domain
- LBD
Ligand-binding domain
- AF-1/AF-2
Activation function
- Tau-1/Tau-5
Transcription activation unit
- SRC-1
Steroid receptor coactivator-1
- NLS
Nuclear localization signal
- PEST
Proline, glutamic acid, serine, threonine
- CBP
c-AMP responsive element binding protein
- Hsp
Heat shock proteins
- CHIP
Carboxyl terminus of Hsc70-interacting protein
- N/C interaction
Amino- and carboxy-terminus interaction
- Lys
Lysine
- Ser
Serine
- SIRT1
Sirtuin 1
- IGF-1
Insulin-like growth factor-1
- CNS
Central nervous system
References
- 1.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.La Spada AR, et al. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352(6330):77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 3.Kawahara H. A family of progressive bulbar palsy. Aichi Med School J. 1897;16:3–4. [Google Scholar]
- 4.Kennedy W, Alter M, Sung J. Progressive proximal spinal and bulbar muscular atrophy of late onset. Neurology. 1968;18(7):671–680. doi: 10.1212/WNL.18.7.671. [DOI] [PubMed] [Google Scholar]
- 5.Harding AE, et al. X-linked recessive bulbospinal neuronopathy: a report of ten cases. J Neurol Neurosurg Psychiatry. 1982;45(11):1012–1019. doi: 10.1136/jnnp.45.11.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt BJ, et al. Expression of X-linked bulbospinal muscular atrophy (Kennedy disease) in two homozygous women. Neurology. 2002;59(5):770–772. doi: 10.1212/WNL.59.5.770. [DOI] [PubMed] [Google Scholar]
- 7.Sobue G, et al. Subclinical phenotypic expressions in heterozygous females of X-linked recessive bulbospinal neuronopathy. J Neurol Sci. 1993;117(1–2):74–78. doi: 10.1016/0022-510X(93)90157-T. [DOI] [PubMed] [Google Scholar]
- 8.Nagashima T, et al. Familial bulbo-spinal muscular atrophy associated with testicular atrophy and sensory neuropathy (Kennedy–Alter–Sung syndrome). Autopsy case report of two brothers. J Neurol Sci. 1988;87(2–3):141–152. doi: 10.1016/0022-510X(88)90240-7. [DOI] [PubMed] [Google Scholar]
- 9.Battaglia F, et al. Kennedy’s disease initially manifesting as an endocrine disorder. J Clin Neuromuscul Dis. 2003;4(4):165–167. doi: 10.1097/00131402-200306000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Yu Z, et al. Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. J Clin Invest. 2006;116(10):2663–2672. doi: 10.1172/JCI28773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chevalier-Larsen ES, et al. Castration restores function and neurofilament alterations of aged symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. J Neurosci. 2004;24(20):4778–4786. doi: 10.1523/JNEUROSCI.0808-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katsuno M, et al. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35(5):843–854. doi: 10.1016/S0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 13.Fischbeck K, et al. Localization of the gene for X-linked spinal muscular atrophy. Neurology. 1986;36(12):1595–1598. doi: 10.1212/WNL.36.12.1595. [DOI] [PubMed] [Google Scholar]
- 14.Poletti A. The polyglutamine tract of androgen receptor: from functions to dysfunctions in motor neurons. Front Neuroendocrinol. 2004;25:1–26. doi: 10.1016/j.yfrne.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Clark P, Irvine R, Coetzee G. The androgen receptor CAG repeat and prostate cancer risk. Methods Mol Med. 2003;81:255–266. doi: 10.1385/1-59259-372-0:255. [DOI] [PubMed] [Google Scholar]
- 16.Davis-Dao C, et al. Shorter androgen receptor CAG repeat lengths associated with cryptorchidism risk among Hispanic white boys. J Clin Endocrinol Metab. 2012;97(3):E393–E399. doi: 10.1210/jc.2011-2439. [DOI] [PubMed] [Google Scholar]
- 17.Palazzolo I, et al. The role of the polyglutamine tract in androgen receptor. J Steroid Biochem Mol Biol. 2008;108(3–5):245–253. doi: 10.1016/j.jsbmb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Chang C, et al. Androgen receptor: an overview. Crit Rev Eukaryot Gene Expr. 1995;5(2):97–125. doi: 10.1615/CritRevEukarGeneExpr.v5.i2.10. [DOI] [PubMed] [Google Scholar]
- 19.Callewaert L, Van Tilborgh N, Claessens F. Interplay between two hormone-independent activation domains in the androgen receptor. Cancer Res. 2006;66(1):543–553. doi: 10.1158/0008-5472.CAN-05-2389. [DOI] [PubMed] [Google Scholar]
- 20.Tan EM, et al. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin. 2015;36(1):3–23. doi: 10.1038/aps.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christiaens V, et al. Characterization of the two coactivator-interacting surfaces of the androgen receptor and their relative role in transcriptional control. J Biol Chem. 2002;277(51):49230–49237. doi: 10.1074/jbc.M209322200. [DOI] [PubMed] [Google Scholar]
- 22.Callewaert L, et al. Implications of a polyglutamine tract in the function of the human androgen receptor. Biochem Biophys Res Commun. 2003;306(1):46–52. doi: 10.1016/S0006-291X(03)00902-1. [DOI] [PubMed] [Google Scholar]
- 23.Bevan CL, et al. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol Cell Biol. 1999;19(12):8383–8392. doi: 10.1128/MCB.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCampbell A, et al. CREB-binding protein sequestration by expanded polyglutamine. Hum Mol Genet. 2000;9(4):2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- 25.Stenoien DL, et al. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum Mol Genet. 1999;8(5):731–741. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- 26.Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21(6):267–271. doi: 10.1016/S0968-0004(96)10031-1. [DOI] [PubMed] [Google Scholar]
- 27.Ni L, et al. Androgen induces a switch from cytoplasmic retention to nuclear import of the androgen receptor. Mol Cell Biol. 2013;33(24):4766–4778. doi: 10.1128/MCB.00647-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanner TM, et al. A 629RKLKK633 motif in the hinge region controls the androgen receptor at multiple levels. Cell Mol Life Sci. 2010;67(11):1919–1927. doi: 10.1007/s00018-010-0302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haelens A, et al. The hinge region regulates DNA binding, nuclear translocation, and transactivation of the androgen receptor. Cancer Res. 2007;67(9):4514–4523. doi: 10.1158/0008-5472.CAN-06-1701. [DOI] [PubMed] [Google Scholar]
- 30.Clinckemalie L, et al. The hinge region in androgen receptor control. Mol Cell Endocrinol. 2012;358(1):1–8. doi: 10.1016/j.mce.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Fu M, et al. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem. 2000;275(27):20853–20860. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- 32.Katsuno M, et al. Pathogenesis and therapy of spinal and bulbar muscular atrophy (SBMA) Prog Neurobiol. 2012;99(3):246–256. doi: 10.1016/j.pneurobio.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 33.He B, et al. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH(2)-terminal domain. J Biol Chem. 1999;274(52):37219–37225. doi: 10.1074/jbc.274.52.37219. [DOI] [PubMed] [Google Scholar]
- 34.Saporita AJ, et al. Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J Biol Chem. 2003;278(43):41998–42005. doi: 10.1074/jbc.M302460200. [DOI] [PubMed] [Google Scholar]
- 35.Kratter IH, Finkbeiner S. PolyQ disease: too many Qs, too much function? Neuron. 2010;67(6):897–899. doi: 10.1016/j.neuron.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieberman AP, et al. Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell Rep. 2014;7(3):774–784. doi: 10.1016/j.celrep.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cortes CJ, et al. Muscle expression of mutant androgen receptor accounts for systemic and motor neuron disease phenotypes in spinal and bulbar muscular atrophy. Neuron. 2014;82(2):295–307. doi: 10.1016/j.neuron.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adachi H, et al. Widespread nuclear and cytoplasmic accumulation of mutant androgen receptor in SBMA patients. Brain. 2005;128(3):659–670. doi: 10.1093/brain/awh381. [DOI] [PubMed] [Google Scholar]
- 39.Yu Z, et al. Abnormalities of germ cell maturation and sertoli cell cytoskeleton in androgen receptor 113 CAG knock-in mice reveal toxic effects of the mutant protein. Am J Pathol. 2006;168(1):195–204. doi: 10.2353/ajpath.2006.050619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyttenbach A. Role of heat shock proteins during polyglutamine neurodegeneration. J Mol Neurosci. 2004;23(1–2):69–95. doi: 10.1385/JMN:23:1-2:069. [DOI] [PubMed] [Google Scholar]
- 41.Jochum T, et al. Toxic and non-toxic aggregates from the SBMA and normal forms of androgen receptor have distinct oligomeric structures. Biochim Biophys Acta. 2012;1822(6):1070–1078. doi: 10.1016/j.bbadis.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 42.Taylor JP, et al. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum Mol Genet. 2003;12(7):749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- 43.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6(1):11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 44.Li M, et al. Nonneural nuclear inclusions of androgen receptor protein in spinal and bulbar muscular atrophy. Am J Pathol. 1998;153(3):695–701. doi: 10.1016/S0002-9440(10)65612-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, et al. Nuclear inclusions of the androgen receptor protein in spinal and bulbar muscular atrophy. Ann Neurol. 1998;44(2):249–254. doi: 10.1002/ana.410440216. [DOI] [PubMed] [Google Scholar]
- 46.Miller J, et al. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat Chem Biol. 2011;7(12):925–934. doi: 10.1038/nchembio.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montie HL et al (2009) Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet 18(11) [DOI] [PMC free article] [PubMed]
- 48.Takeyama K-I, et al. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron. 2002;35(5):855–864. doi: 10.1016/S0896-6273(02)00875-9. [DOI] [PubMed] [Google Scholar]
- 49.Pratt WB, Toft DO (2003) Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) [DOI] [PubMed]
- 50.Pratt WB et al (2014) A model in which heat shock protein 90 targets protein-folding clefts: rationale for a new approach to neuroprotective treatment of protein folding diseases. Exp Biol Med 1–9 [DOI] [PMC free article] [PubMed]
- 51.Pratt WB, et al. Proposal for a role of the Hsp90/Hsp70-based chaperone machinery in making triage decisions when proteins undergo oxidative and toxic damage. Exp Biol Med (Maywood) 2010;235(3):278–288. doi: 10.1258/ebm.2009.009250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pratt WB, et al. Targeting Hsp90/Hsp70-based protein quality control for treatment of adult onset neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2014;55:353–371. doi: 10.1146/annurev-pharmtox-010814-124332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pratt WB, Morishima Y, Osawa Y. The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J Biol Chem. 2008;283(34):22885–22889. doi: 10.1074/jbc.R800023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adachi H, et al. CHIP overexpression reduces mutant androgen receptor protein and ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model. Neurobiol Dis. 2007;27(19):5115–5126. doi: 10.1523/JNEUROSCI.1242-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waza M, et al. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat Med. 2005;11:1088–1095. doi: 10.1038/nm1298. [DOI] [PubMed] [Google Scholar]
- 56.Wang AM, et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat Chem Biol. 2013;9:112–118. doi: 10.1038/nchembio.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Royen ME, et al. Stepwise androgen receptor dimerization. J Cell Sci. 2012;125(8):1970–1979. doi: 10.1242/jcs.096792. [DOI] [PubMed] [Google Scholar]
- 58.Nedelsky NB, et al. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 2010;67(6):936–952. doi: 10.1016/j.neuron.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orr CR, et al. An interdomain interaction of the androgen receptor is required for its aggregation and toxicity in spinal and bulbar muscular atrophy. J Biol Chem. 2010;285(46):35567–35577. doi: 10.1074/jbc.M110.146845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zboray L, et al. Preventing the androgen receptor N/C interaction delays disease onset in a mouse model of SBMA. Cell Rep. 2015;13(10):2312–2323. doi: 10.1016/j.celrep.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lieberman AP, et al. Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum Mol Genet. 2002;11(17):1967–1976. doi: 10.1093/hmg/11.17.1967. [DOI] [PubMed] [Google Scholar]
- 62.Powell SM, et al. Mechanisms of androgen receptor signalling via steroid receptor coactivator-1 in prostate. Endocr Relat Cancer. 2004;11(1):117–130. doi: 10.1677/erc.0.0110117. [DOI] [PubMed] [Google Scholar]
- 63.Katsuno M, et al. Disrupted transforming growth factor-beta signaling in spinal and bulbar muscular atrophy. J Neurosci. 2010;30(16):5702–5712. doi: 10.1523/JNEUROSCI.0388-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sopher B, et al. Androgen receptor YAC transgenic mice recapitulate SBMA motor neuronopathy and implicate VEGF164 in the motor neuron degeneration. Neuron. 2004;41(5):687–699. doi: 10.1016/S0896-6273(04)00082-0. [DOI] [PubMed] [Google Scholar]
- 65.Minamiyama M, et al. Sodium butyrate ameliorates phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2004;13(11):1183–1192. doi: 10.1093/hmg/ddh131. [DOI] [PubMed] [Google Scholar]
- 66.Butler R, Bates GP. Histone deacetylase inhibitors as therapeutics for polyglutamine disorders. Nat Rev Neurosci. 2006;7(10):784–796. doi: 10.1038/nrn1989. [DOI] [PubMed] [Google Scholar]
- 67.McCampbell A, et al. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc Natl Acad Sci USA. 2001;98(26):15179–15184. doi: 10.1073/pnas.261400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413(6857):739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 69.Palazzolo I, et al. Akt blocks ligand binding and protects against expanded polyglutamine androgen receptor toxicity. Hum Mol Genet. 2007;16(13):1593–1603. doi: 10.1093/hmg/ddm109. [DOI] [PubMed] [Google Scholar]
- 70.Palazzolo I, et al. Overexpression of IGF-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron. 2009;63(3):316–328. doi: 10.1016/j.neuron.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Montie HL, Pestell RG, Merry DE. SIRT1 modulates aggregation and toxicity through deacetylation of the androgen receptor in cell models of SBMA. Neurobiol Dis. 2011;31(48):17425–17436. doi: 10.1523/JNEUROSCI.3958-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mukherjee S, et al. Small ubiquitin-like modifier (SUMO) modification of the androgen receptor attenuates polyglutamine-mediated aggregation. J Biol Chem. 2009;284(32):21296–21306. doi: 10.1074/jbc.M109.011494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chua JP et al (2014) Disrupting SUMOylation potentiates transactivation function and ameliorates polyglutamine AR-mediated disease. J Clin Invest [DOI] [PMC free article] [PubMed]
- 74.Sobue G, et al. X-linked recessive bulbospinal neuronopathy. A clinicopathological study. Brain. 1989;112:209–232. doi: 10.1093/brain/112.1.209. [DOI] [PubMed] [Google Scholar]
- 75.Suzuki K, et al. CAG repeat size correlates to electrophysiological motor and sensory phenotypes in SBMA. Brain. 2008;131(1):229–239. doi: 10.1093/brain/awm289. [DOI] [PubMed] [Google Scholar]
- 76.Katsuno M, et al. Pathogenesis, animal models and therapeutics in spinal and bulbar muscular atrophy (SBMA) Exp Neurol. 2006;200(1):8–18. doi: 10.1016/j.expneurol.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 77.Suzuki K, et al. The profile of motor unit number estimation (MUNE) in spinal and bulbar muscular atrophy. J Neurol Neurosurg Psychiatry. 2010;81(5):567–571. doi: 10.1136/jnnp.2009.190462. [DOI] [PubMed] [Google Scholar]
- 78.Atsuta N, et al. Natural history of spinal and bulbar muscular atrophy (SBMA): a study of 223 Japanese patients. Brain. 2006;129(6):1446–1455. doi: 10.1093/brain/awl096. [DOI] [PubMed] [Google Scholar]
- 79.Rhodes LE, et al. Clinical features of spinal and bulbar muscular atrophy. Brain. 2009;132(12):3242–3251. doi: 10.1093/brain/awp258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sorarù G, et al. Spinal and bulbar muscular atrophy: skeletal muscle pathology in male patients and heterozygous females. J Neurol Sci. 2008;264(1–2):100–105. doi: 10.1016/j.jns.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 81.Chua JP, et al. Transcriptional activation of TFEB/ZKSCAN3 target genes underlies enhanced autophagy in spinobulbar muscular atrophy. Hum Mol Genet. 2014;23(5):1376–1386. doi: 10.1093/hmg/ddt527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu Z et al (2011) Macroautophagy is regulated by the UPR-mediator CHOP and accentuates the phenotype of SBMA mice. PLoS Genet 7(10) [DOI] [PMC free article] [PubMed]
- 83.Rusmini P et al (2015) Aberrant autophagic response in the muscle of a knock-in mouse model of spinal and bulbar muscular atrophy. Sci Rep 5(15174) [DOI] [PMC free article] [PubMed]
- 84.Monks DA, et al. Overexpression of wild-type androgen receptor in muscle recapitulates polyglutamine disease. Proc Natl Acad Sci USA. 2007;104(46):18259–18264. doi: 10.1073/pnas.0705501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rinaldi C, et al. Insulin like growth factor (IGF)-1 administration ameliorates disease manifestations in a mouse model of spinal and bulbar muscular atrophy. Mol Med. 2012;18(1):1261–1268. doi: 10.2119/molmed.2012.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Malena A, et al. Androgen-dependent impairment of myogenesis in spinal and bulbar muscular atrophy. Acta Neuropathol. 2013;126:109–121. doi: 10.1007/s00401-013-1122-9. [DOI] [PubMed] [Google Scholar]
- 87.Funakoshi H, et al. Differential expression of mRNAs for neurotrophins and their receptors after axotomy of the sciatic nerve. J Cell Biol. 1993;123(2):455–465. doi: 10.1083/jcb.123.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramzan F, et al. Distinct etiological roles for myocytes and motor neurons in a mouse model of Kennedy’s disease/spinobulbar muscular atrophy. J Neurosci. 2015;35(16):6444–6451. doi: 10.1523/JNEUROSCI.3599-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sahashi K, et al. Silencing neuronal mutant androgen receptor in a mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2015;24(21):5985–5994. doi: 10.1093/hmg/ddv300. [DOI] [PubMed] [Google Scholar]