Abstract
TOXCAT is a widely used genetic assay to study interactions of transmembrane helices within the inner membrane of the bacterium Escherichia coli. TOXCAT is based on a fusion construct that links a transmembrane domain of interest with a cytoplasmic DNA-binding domain from the Vibrio cholerae ToxR protein. Interaction driven by the transmembrane domain results in dimerization of the ToxR domain, which, in turn, activates the expression of the reporter gene chloramphenicol acetyl transferase (CAT). Quantification of CAT is used as a measure of the ability of the transmembrane domain to self-associate. Because the quantification of CAT is relatively laborious, we developed a high-throughput variant of the assay, TOXGREEN, based on the expression of super-folded GFP and detection of fluorescence directly in unprocessed cell cultures. Careful side-by-side comparison of TOXCAT and TOXGREEN demonstrates that the methods have comparable response, dynamic range, sensitivity and intrinsic variability both in LB and minimal media. The greatly enhanced workflow makes TOXGREEN much more scalable and ideal for screening, since hundreds of constructs can be rapidly assessed in 96 well plates. Even for small scale investigations, TOXGREEN significantly reduces time, labor and cost associated with the procedure. We demonstrate applicability with a large screening for self-association among the transmembrane domains of bitopic proteins of the divisome (FtsL, FtsB, FtsQ, FtsI, FtsN, ZipA and EzrA) belonging to 11 bacterial species. The analysis confirms a previously reported tendency for FtsB to self-associate, and suggests that the transmembrane domains of ZipA, EzrA and FtsN may also possibly oligomerize.
Graphical abstract
1. Introduction
Among the helical membrane proteins, the class that spans the bilayer with a single transmembrane (TM) domain is the most prevalent, accounting for 20% or more of all membrane proteins in most organisms [1]. The TM domains of these “single-pass” or “bitopic” membrane proteins are sometimes referred as “membrane anchors”. However, it is becoming increasingly evident that these segments – which bridge the two universes across membrane – often play active roles in biological function [2]. These roles are generally established through the formation of oligomeric complexes, where modulation of association or conformational changes can be part of mechanisms that regulate the biological activity of these proteins [3]. For this reason, there is great interest for methods suitable for investigating whether TM helices associate, for measuring the strength of their interactions, and for identifying which amino acids are involved at their interaction interfaces.
Quantitative measurements of TM helix oligomerization in vitro can be obtained with a variety of biophysical methods. For example, Förster Resonance Energy Transfer (FRET) [4–9] and sedimentation equilibrium analytical ultracentrifugation (SE-AUC) [10,11] are widely used as complementary methods. SE-AUC is directly sensitive to the oligomeric mass of a complex and can provide accurate energetics for association in detergent micelles. FRET has been particularly important for investigating the energetics of TM helix association in lipid bilayers. Disulfide exchange equilibrium [12,13] and, most recently, steric trapping [14,15] can also be applied to determine TM helix equilibria both in detergents and in lipid bilayers. Finally, SDS-PAGE [16,17] is applicable to the subset of TM complexes that are sufficiently stable to remain oligomeric in the harsh detergent SDS, and has also been widely applied to screening TM helix association.
Another common approach for studying TM helix association is to utilize a number of genetic reporter systems, which are applied in vivo in the membrane of Escherichia coli. These systems complement the above biophysical methods in a number of ways. The genetic systems do not suffer from the many of the issues that can arise when working with membrane proteins in vitro, where many steps (sample expression or synthesis, reconstitution, labeling, data acquisition and analysis) can be laborious or technically challenging. The biological methods do not provide a measure of the specific stoichiometry of a complex nor quantitative energetics data, but they enable valuable relative comparison. Therefore, they are useful for assessing whether a TM helix has a tendency to form oligomers in membranes, and they are ideal for the identification of the interaction interface of a complex, which can be explored by exhaustive mutagenesis. The genetic systems are also suitable for larger scale screening or selection, which are generally unattainable in vitro.
A distinctive feature of the genetic reporter systems is that the measurements are performed within biological membranes, as opposed to membrane mimics such as synthetic bilayers or detergent micelles in solution. A FRET (QI-FRET) method exists for the quantitative measurement of association of TM complexes directly within eukaryotic membranes [18,19]. QI-FRET is powerful and sophisticated but requires specialized knowledge and instrumentation. In comparison, the genetic reporter assays represent a less quantitative but more approachable way to assess TM helix oligomerization in the complexity of a living membrane. For these reasons, the various genetic reporter assays have been widely adopted. In their history, they have contributed immensely to our understanding of association in the membrane, as well as to the functional characterization of many important biological complexes.
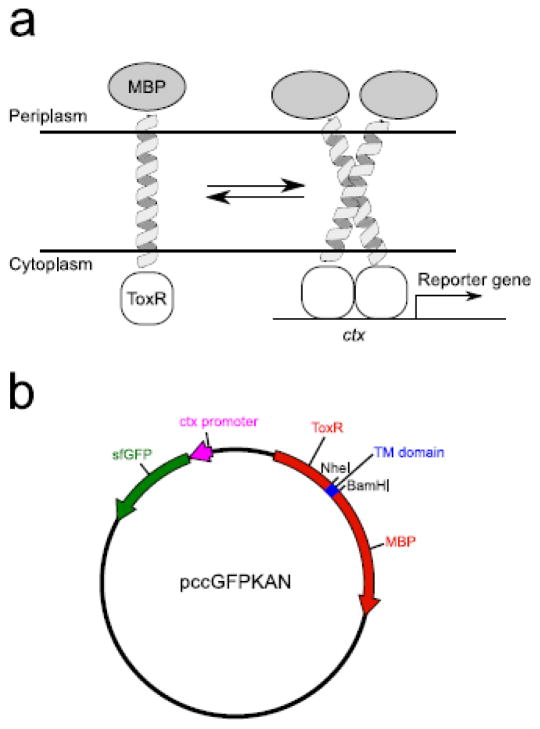
The first genetic reporter assays were developed following the discovery of the V. cholera transcription factor ToxR [20]. The ToxR system was used by Langosch to develop a reporter assay in which the N-terminal ToxR domain was fused to a TM domain of interest and a C-terminal Maltose Binding Protein (MBP) [21]. As illustrated in Fig. 1a, dimerization induced by the TM domain causes the ToxR transcriptional activator to bind the ctx promoter and this initiates the transcription of the lacZ reporter gene. The TOXCAT assay was later developed by Engelman and coworkers [22]. In TOXCAT, a similar ToxR-TM-MBP fusion protein regulates the expression of chloramphenicol acetyltransferase (CAT). The adoption of an antibiotic resistance gene enabled the application of assay to selecting strongly oligomerizing sequences out of randomized libraries [23,24].
Figure 1. The TOXGREEN Assay.
(a) An overview of the ToxR-based assays. TM domain of interest is expressed as a ToxR-TM-MBP fusion protein which is biologically inserted into the inner membrane of E. coli. Upon dimerization of the TM region, ToxR will bind the ctx promoter and activate transcription of a reporter gene (lacZ, CAT, sfGFP). (b) TOXGREEN expression vector. The gene of the TOXCAT fusion is represented in red. The TM domain inserted at NheI and BamHI cut sites is highlighted in blue. The ctx promoter (magenta) and sfGFP reporter gene (green) are also shown.
Since these initial systems, a large variety of other ToxR based methods have been developed. The Langosch group developed POSSYCCAT, which integrated a CAT reporter gene directly in the chromosome of E. coli, thus creating a system suitable for selection [25]. POSSYCCAT was further refined exploiting the ability of ToxR to act as activator or repressor when it binds to alternative DNA sequences, allowing for the selection of TM domains that had an intermediate oligomerization propensity [26]. TOXCAT has been adapted to oligomerizing multi-pass membrane proteins [27], and a version of TOXCAT was created in which the CAT reporter gene was replaced by luciferase (ToxLux) for improved detection [28]. Dominant-negative ToxR systems have also been developed, i.e. systems that rely on disruption of a homo-oligomer by a competing helix fused to an inactivated ToxR domain. In these dominant negative systems hetero-oligomerization causes a reduction in reporter gene expression [29,30]. DN-ToxRed, in particular, was the first system that introduced the use of a fluorescent protein (mCherry) as the reporter gene [29].
Transcription regulators other than ToxR have also been used for genetic reporter assays. GALLEX utilizes the LexA transcriptional repressor and the lacZ reporter gene [31]. A major innovation of GALLEX was the use of two LexA DNA binding domains with different DNA sequence specificity, enabling the measurement of hetero-oligomeric association. The recently introduced AraTM assay allows for exploration of the role of both transmembrane and juxtamembrane regions in dimerization of transmembrane proteins [32]. A dominant-negative version of AraTM has also been produced [33]. In addition, unlike the ToxR- and LexA-based assays, AraTM uses the signal sequence of MBP to target the complex to the membrane, thus decoupling membrane trafficking from the specific sequence of the TM domain. This promotes Type I orientation, which is advantageous for the study of a large number of important mammalian receptors in their native cellular orientation [32].
Since its development in 1999, TOXCAT has been used by over thirty distinct research groups, resulting in more than 70 publications in which the assay has been applied to a broad variety of membrane proteins systems. In most of these studies, TOXCAT contributed to defining the biological role of TM homo-oligomers, in integration with data obtained by other biophysical or biological experiments. The rich spectrum of subjects that have been studied with TOXCAT is apparent in Table 1, which lists them categorized by their biological functions. In the majority of the cases, TOXCAT has been applied to a plasma membrane single-pass protein of human or mammalian origin. However, the list also includes studies involving plant, yeast, bacterial and viral proteins, as well as proteins of intracellular compartments such as the mitochondrion and the endoplasmic reticulum. The biological functions of the proteins examined with TOXCAT are just as diverse. They include many receptors and proteins involved in cellular adhesion, but also amyloid forming proteins, chaperones, toxins, enzymes, photosynthetic proteins and more. Interestingly, on multiple occasions TOXCAT has been applied to study the self-association of individual helices of polytopic membrane proteins such GPCRs and channels.
Table 1.
membrane protein systems investigated with TOXCAT
| Receptor Tyrosine Kinases: | ErbB family [28,70,71], insulin receptor [28], RET [72,73], PDGFβR [74], DDR1 [75], FGFR3, INSR, LKT, TIE2 [76] and others [77] |
| Receptor-like protein tyrosyne phosphatases: | 19 RPTPs [78] |
| Plant receptor-like kinases: | CR4 and ACR4 [79,80] |
| Cytokine receptors: | thrombopoietin receptor [81], p75 neurotrophin receptor [82] |
| Cellular adhesion: | integrin αIIbβ3 [83–85], syndecan family [86], E-cadherin [87], Na,K-ATPase subunit β [88], L-selectin [89], GP Ib-IX complex [90,91] |
| Neurological proteins: | M6a [92], sigma-1 receptor [67], myelin proteolipid protein [93], myelin protein zero [94], p75 neurotrophin receptor [82], synaptobrevin [95], drosophila Wnt receptor [96] |
| Mitochondrial proteins: | UbiB protein kinase-like ADCK3 [69], rat carnitine palmitoyltransferase 1A [97,98], Bcl-2 family apoptosis regulator BNIP3 [35,99] |
| Amyloid forming proteins: | pigment cell-specific protein PMEL [100], amyloid precursor protein [101] |
| Immunological: | major histocompatibility complex (MHC) 1 class II-associated invariant chain [102] |
| Transporters: | ABC transporter ABCG2 (TM helix 1) [103] |
| GPCRs: | Mam2 (TM helix 1) [104] |
| Chaperones: | yeast ER chaperone Rot1 [105] |
| Channels: | gap junction protein connexin 26 (TM helix 1) [106], voltage-dependent potassium channel BK (TM helix 1) [107], Epithelial Na+ channel EnaC subunit α (TM helix 1) [108] |
| Hematopoietic proteins: | mediator of receptor activation DAP12 [109], spleen focus forming virus gp55-A and gp55-P [110], thrombopoietin receptor [81] |
| Bacterial proteins: | cell division proteins FtsB [41], twin-arginine translocase protein component TatA [111], reaction center-light-harvesting 1 complex protein PufX [68,112] and assembly factor PuhB (individual TM helices) [113] |
| Viral Proteins: | spleen focus forming virus gp55-A and gp55-P [110], HIV-1 gp41 [114], SARS ScoV [115], HPV minor capsid protein L2 [116], BPV E5 [117,118], bacteriophage M13 major coat protein [119–122] |
| Toxin related: | Helicobacter pylori toxin VacA [123–125], Human Anthrax Toxin Receptor ANTXR1 [126] |
| Designed, randomized or model TM helices: | designed [127–132], randomized [23,24,133], glycophorin A as model sequence [134,135], δ-helices [136] |
In addition to the study of biological systems, TOXCAT has also been often used for motif analysis by the groups of Deber, Engelman, Mingarro, MacKenzie and others, to investigate the determinant of helix-helix association in membranes. These studies have involved a variety of constructs, from designed sequences to randomized libraries, as well as several studies that use models systems such as glycophorin A. Remarkably, such studies led to the discovery of the GxxxG motif as a major driver for TM-helix association [24,34].
The practical nature of TOXCAT makes it suitable for measuring numerous samples, such as for large scale mutagenesis. However, when scaled up to tens of samples the analysis becomes laborious. The quantification of CAT expression, performed either enzymatically [22,35] or via ELISA [36], requires a large number of steps, limiting the number of samples or the number of biological replicas of the same sequence that can be analyzed in a single session. Even the ToxLux variant, which enhances the work-flow by using a luciferase reporter gene, still requires manipulation of each individual sample (cell lysis and addition of reagents) [28].
Here we address these limitations by reporting the conversion of TOXCAT into a high-throughput variant called TOXGREEN. TOXGREEN is based on a Green Fluorescent Protein (GFP) reporter gene, which can be rapidly quantified directly in untreated cell culture samples in a fluorescence plate reader. We show with careful side-by-side testing that the responses of the two assays are indistinguishable and that the characteristics of the original TOXCAT, such as sensitivity and response, are maintained in TOXGREEN. With a much more simplified workflow, TOXGREEN saves time and cost for small scale analysis and enables large scale screening of TM helix association. We demonstrate TOXGREEN’s applicability by screening the TM domains of seven bitopic proteins of the bacterial divisome complex for self-association.
2. Materials and Methods
2.1 Subcloning of the TOXGREEN plasmid
The CAT reporter gene was replaced with the gene for sfGFP [37,38] in the pccKAN plasmid, using the Restriction Free Quikchange method [39], to generate the plasmid pccGFPKAN (Fig. 1b).
Genes encoding for the TM domains of interest were digested with NheI and DpnII and ligated into the compatible NheI-BamHI restriction sites of the pccKAN and pccGFPKAN plasmids (Fig. 1b) using Quick Ligase (NEB), resulting in the protein sequences reported in the supplementary Table S1 and Table S2. All constructs were confirmed by DNA sequencing (QuintaraBio). The plasmids have been deposited on the AddGene repository with the following accession numbers: TOXGREEN empty plasmid pccGFPKAN: # 73649; glycophorin A, G83I mutant, pccGFPG83I: #73650; glycophorin A wild type, pccGFPGpA: #73651.
2.2 TOXGREEN assay growth conditions
TOXGREEN constructs were transformed into E. coli MM39 cells. sfGFP expression was quantified in two conditions, log and stationary phase, and two different types of media, LB or M9 minimal media (230 mM Na2HPO4, 110 mM KH2PO4, 43 mM NaCl, 93 mM NH4Cl, 1 mM MgSO4, 0.4% glucose, 18.8 μM thiamine). For log phase conditions, a freshly streaked colony was inoculated into 3 mL of either LB broth or M9 media, containing 100 μg/mL ampicillin and grown overnight at 37 °C. 30 μL of the overnight culture were inoculated into fresh 3mL LB Broth containing 100 μg/mL ampicillin and incubated at 37 °C until an optical density of approximately 0.6 at 600 nm was reached. Fluorescent scans were performed on M9 samples directly in the undiluted cultures. To reduce background for samples grown in LB, 1.5 mL of cells were collected by centrifugation at 17,000 g and concentrated three-fold by re-suspending them in 0.5 mL in PBS solution (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), prior to fluorescence measurements.
For stationary phase conditions, individual colonies were inoculated into 3 mL of LB broth or M9 minimal media containing 100 μg/mL ampicillin and incubated for 16 hours at 37 °C. Fluorescent scans were performed on these cells directly in the undiluted LB or M9 cultures. For both log and stationary phase samples, aliquots were removed and stored in SDS-PAGE loading buffer for immunoblotting.
2.3 Fluorescence measurements of sfGFP expression
300 μL of each cell sample was transferred to a 96-well black walled, clear bottom plate (Fisher Scientific). Fluorescence measurements were performed using an Infinite M1000 Pro plate reader (Tecan), using an excitation wavelength of 485 nm and recording emission from 500 to 600 nm. The relative sfGFP expression (TOXGREEN signal) was calculated by normalizing the fluorescence emission at 512 nm to the optical density of the sample at 600 nm. The normalized fluorescence of each sample was then subtracted of the normalized fluorescence of cells that contained the no-TM control plasmid pccGFPKAN to remove non-specific background (Fig. 2b and d).
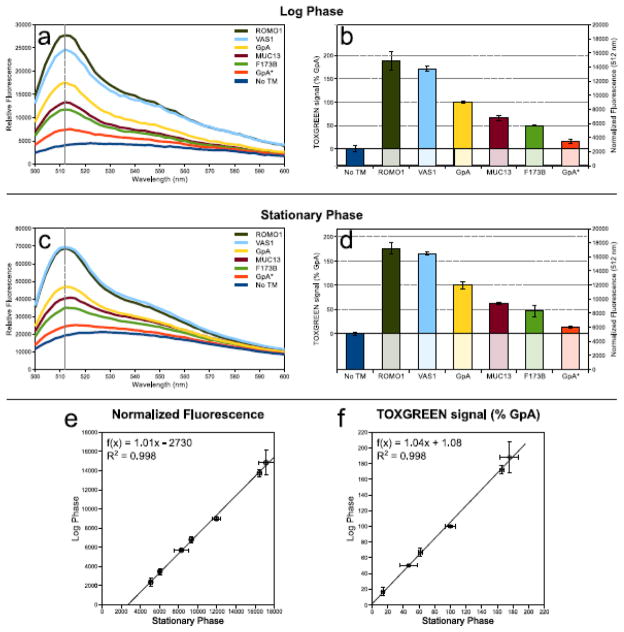
Figure 2. Log and stationary phase cell cultures performs equivalently in TOXGREEN.
Fluorescence measurements of seven TOXGREEN constructs, including the “no-TM” control. The dashed vertical line indicates the readout wavelength used (512 nm). a) Fluorescence spectra of cells in log phase concentrated 3x and resuspended in PBS buffer. b) Conversion of log phase cell’s fluorescence at 512 nm to TOXGREEN signal. The fluorescence is normalized to cell density and the background fluorescence of the “no TM” construct is subtracted. The signal is here normalized to the GpA sample. c) Spectra of stationary phase cells, measured directly in LB media. d) Conversion of stationary phase cell’s fluorescence to TOXGREEN signal. e) Comparison of fluorescence of log and stationary phase cells after normalization to cell density. f) Comparison of relative TOXGREEN signal for log and stationary phase cells. Wester blots of the relative to these experiment are shown in supplementary Figure S4.
2.4 TOXCAT assay
The TOXCAT constructs were transformed into MM39 cells. A freshly streaked colony was inoculated into 3 mL of LB broth containing 100 μg/mL ampicillin and grown overnight at 37°C. 30 μL of overnight cultures were inoculated into 3 mL of LB broth and grown to an OD600 of approximately 0.6 at 37 °C. After recording the optical density, 1.5 mL of cells were harvested by centrifugation for 10 min at 17000 g and resuspended in 0.5 mL of lysis buffer (25 mM Tris-HCl, 2 mM EDTA, pH 8.0). Cells were lysed by probe sonication at medium power for 15 seconds over ice. An aliquot was removed from each sample and stored in SDS-PAGE loading buffer for immunoblotting. The lysates were then cleared by centrifugation at 17000 g and the supernatant was kept on ice for CAT activity assay.
CAT activity was measured as described [40,41]. Briefly, 1 mL of buffer containing 0.1 mM acetyl CoA, 0.4 mg/mL 5,5′-dithiobis-(2-nitrobenzoic acid) or Ellman’s reagent, and 0.1 M Tris-HCl pH 7.8, were mixed with 40 μL of cleared cell lysates and the absorbance at 412 nm was measured for two minutes to establish basal enzyme activity rate. After addition of 40 μL of 2.5 mM chloramphenicol in 10% ethanol, the absorbance was measured for an additional two minutes to determine CAT activity. The basal CAT activity was subtracted and the value was normalized by the cell density measured as OD600.
2.5 Maltose test and immunoblotting
To confirm proper membrane insertion and orientation of the TOXCAT and TOXGREEN constructs, overnight cultures were plated on M9 minimal medium plates containing 0.4% maltose as the only carbon source and grown at 37 °C for 72 hours.
For immunoblotting, the equivalent of 200 μL of culture media at a cell density of 1 OD600 were pelleted and chemically lysed with SoluLyse (Genlantis). 3 μL of cell lysates were mixed with 2× loading buffer and loaded onto a NuPAGE 4–12% Bis-Tris SDS-PAGE gel (Invitrogen), each construct was run in duplicate. Proteins were transferred to PVDF membranes (VWR) for 1 hour at 100 millivolts. Blots were blocked using 5% bovine serum albumin (US Biologicals) in TBS-Tween buffer (50 mM Tris, 150 mM NaCl, 0.05% Tween 20) overnight at 4 °C, incubated with goat biotinylated anti-Maltose Binding Protein antibodies (Vector labs) at 25 °C for 2 hours, followed by peroxidase-conjugated streptavidin anti-goat secondary antibodies (Jackson ImmunoResearch) at 4 °C for 2 hours. Blots were developed with the Pierce ECL Western Blotting Substrate Kit, 1 mL of ECL solution was added to the blot and incubated for 90 seconds. Chemiluminescence was measured using an ImageQuant LAS 4000 (GE Healthsciences). Immunoblots of samples used for direct comparison (Figs. 5, S3 and S4) were processed and developed in parallel.
3. Results
The fluorescent protein chosen to replace CAT was superfolded GFP (sfGFP), an enhanced version of GFP that has improved folding and maturation kinetics and greater resistance to denaturation. There are precedents for the use of a fluorescent protein in a genetic assay for membrane protein interaction. Berger and coworker chose the eGFP variant as the fluorescent reporter gene of the AraC-based assay [32], whereas the dominant-negative DN-ToxRed assay is based on mCherry [29].
3.1 TOXGREEN response in log phase cultures
Fig. 2a shows the emission spectra recorded for bacterial cultures harvested in log phase condition to a cell density of 0.6 OD600. The samples consist of cells expressing six different ToxR-TM-MBP chimeras and a no-TM control (cells transformed with the pccGFPKAN plasmid). The constructs include the wild-type sequence of the glycophorin A (GpA) and the monomeric G83I variant (GpA*), which are typically included in TOXCAT as positive and negative controls. The other four constructs were selected to cover a range of high, medium and low associating sequences.
We found that direct quantification of the log phase cultures in the LB culturing media was not possible because of the high background produced by the media (supplementary Fig. S1a). Harvesting the cells by centrifugation and resuspending them in the same volume of PBS solution solved the background problem (supplementary Fig. S1b). However, to obtain satisfactory signal-to-noise, we found that it was necessary to concentrate the cells by resuspending them in PBS in one third of the original volume (Fig. 2a).
The fluorescence profiles have an emission maximum around 512 nm. The monomeric GpA* variant displayed an emission at 512 nm that was approximately 43% of the dimeric GpA construct. The no-TM control showed a broad baseline with a reading at 512 nm that was approximately 23% of the GpA construct. The basal signal of the no-TM control may be due to autofluorescence or scattering, whereas it is unlikely that this fluorescence is due to background expression of sfGFP because no peak is apparent around the characteristic wavelength of sfGFP.
Fig. 2b demonstrates how the raw fluorescence at 512 nm can converted to a quantity that reflects the relative expression of the reporter gene. First, the fluorescence is normalized by dividing its value by cell density (OD600). Then the basal reading of the no-TM construct is subtracted to account for background. In the figure, the signal is expressed as a percentage of the wild-type GpA construct. The corrected signal of the GpA* mutant corresponded to 16% of the GpA wild-type, which is in line with the range of values normally reported in the literature.
3.2 TOXGREEN response in stationary phase cultures
Stationary phase conditions were also tested in which the cells were grown for 16 hours. These cultures have a high cell density (approximately 3.8 OD600), which results in better fluorescence readings and improved signal to noise. This can indeed be observed in Fig. 2c, which shows the fluorescence scans of the stationary phase samples. Because of the strong fluorescence signal, these samples can be measured directly in the LB cell culture media (for comparison, measurements for same cells after centrifugation and resuspension in PBS is illustrated in supplementary Fig. S1b).
We compared the response observed in stationary phase to that of log phase, which is the typical growth regime of the original TOXCAT [22]. When the raw fluorescence values are normalized to cell density, the stationary and log phase values become very close (Fig. 2c vs Fig. 2a). When the background fluorescence of the no-TM control is subtracted, the TOXGREEN signals are very similar in both conditions (Fig. 2d vs Fig. 2b). This is further confirmed by the direct comparison in the XY plots of Fig. 2e and 2f, in which the data is reported as normalized fluorescence and percent of GpA signal, respectively. The results are essentially identical, indicating that TOXGREEN can be carried out in either condition. The advantage of stationary phase conditions is that the cultures are measured directly without the additional centrifugation and resuspension steps.
3.3 TOXGREEN response in minimal media cultures
To further address the autofluorescence background issue experienced in LB media, we tested culturing the cells in a chemically defined media, such as M9 (supplementary Fig. S2). M9 cultures were measured directly in the growth media, both in log phase (Fig. S2a,b) and stationary phase (Fig. S2c,d) conditions. As expected, the switch to M9 media reduces background fluorescence, enabling the measurements to be taken directly in the culture media even at the lower cell density of log phase cultures. The reporter gene expression pattern remained similar to LB cultures. When cells grown to log phase in M9 (measured directly in media) are compared to cells grown to log phase in LB (resuspended in PBS), the correlation coefficient of the linear regression is good (R2=0.95, Fig. S2e,f). The same comparison between cells grown to stationary phase in either M9 or LB (in both cases, measured directly in the culture media) is excellent (R2=0.99, Fig. S2g,h). Therefore M9 media is indeed a feasible alternative for performing the TOXGREEN assay.
3.4 Comparison with TOXCAT
To directly compare TOXGREEN to the original assay, we used a library of known TOXCAT constructs that contain predicted helix-helix interfaces from human single-span transmembrane proteins [42]. We choose 18 constructs (listed in Table S1) that covered wide a range of CAT expression levels, from approximately 25% to 175% relative to the CAT expression of the GpA standard.
Fig. 3 shows a direct comparison of the constructs measured with TOXCAT and TOXGREEN in stationary phase. CAT expression was quantified based on its enzymatic activity, whereas sfGFP was quantified using fluorescence. In the figure, both sets are normalized to the expression level observed for the respective GpA standard. Regression analysis shows an excellent linear relationship between TOXCAT and TOXGREEN (R2 = 0.910). Neither the value of the slope (1.10 ± 0.08, standard error) nor the value of the intercept (3.7 ± 8.4) are statistically different (p ≫ 0.05) from the expected relationship of equivalent responses (slope = 1, intercept = 0). Therefore this analysis indicates that TOXGREEN produces outcomes that are indistinguishable from the original TOXCAT.
Figure 3. TOXGREEN and TOXCAT are in excellent agreement.
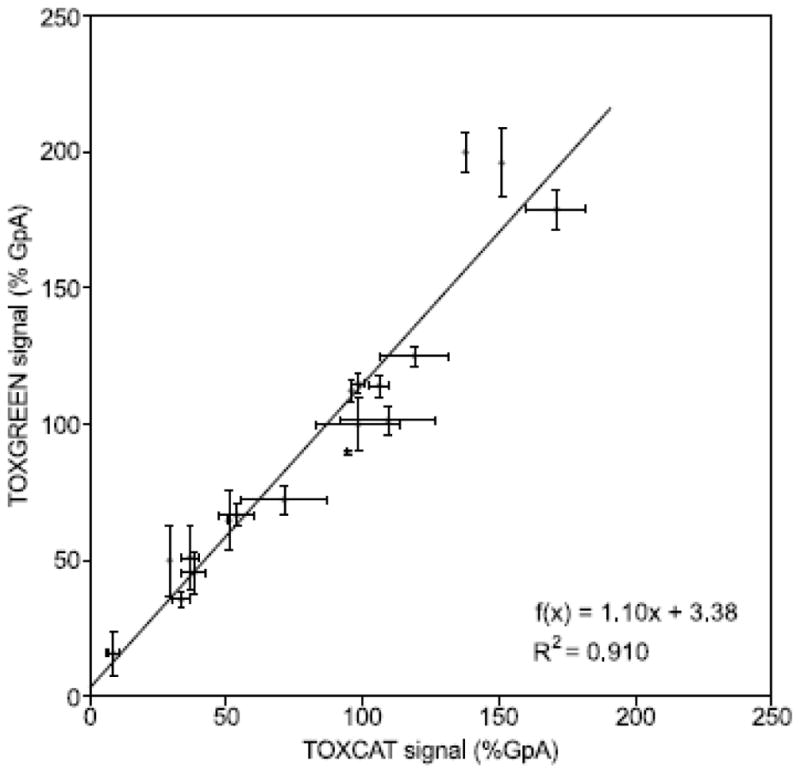
Comparison of reporter gene expression between TOXCAT (measured as CAT enzymatic activity in lysates) and TOXGREEN (measured as fluorescence intensity whole cells in stationary phase). The values have been normalized to their respective value of the GpA sample (100%). The linear regression fit is also shown (blue line). The values of the slope and intercept are not statistically significant from the values expected if the two assays had identical response (i.e. slope = 1, intercept = 0).
The expression level of the ToxR-TM-MBP chimeric constructs in TOXCAT and TOXGREEN was also compared by immunoblotting. As shown in supplementary Fig. S3, the expression level of the various chimeras have similar patterns, which is consistent with very similar levels of expression in the two assays. This is expected since the chimeras and their promoter sequence are identical in both assays.
3.5 Analysis of variability and reproducibility
Given the biological nature of the assay, variability can be an issue. Comparison of the variation within sets of biological replicas of the same construct in TOXCAT and TOXGREEN shows that the two assays perform similarly (Fig. 3). Among the 18 samples tested, the average standard deviation expressed relative to the GpA signal was 5.2% and 6.5% for TOXCAT and TOXGREEN, respectively. When the standard deviation was normalized to the signal of each respective sample, the average variation was also similar (8.9% for TOXCAT and 8.6% for TOXGREEN).
The long term reproducibility of TOXGREEN was also tested by repeating the assay on the same set of five constructs over multiple days (eight biological replicas per construct per day, Fig. 4). The results demonstrates that the day-by-day variability of TOXGREEN is generally comparable to the variability observed within a single-day, and that there is also relatively comparable expression of the chimera across multiple days (supplementary Fig. S4c).
Figure 4. Multi-day variability.
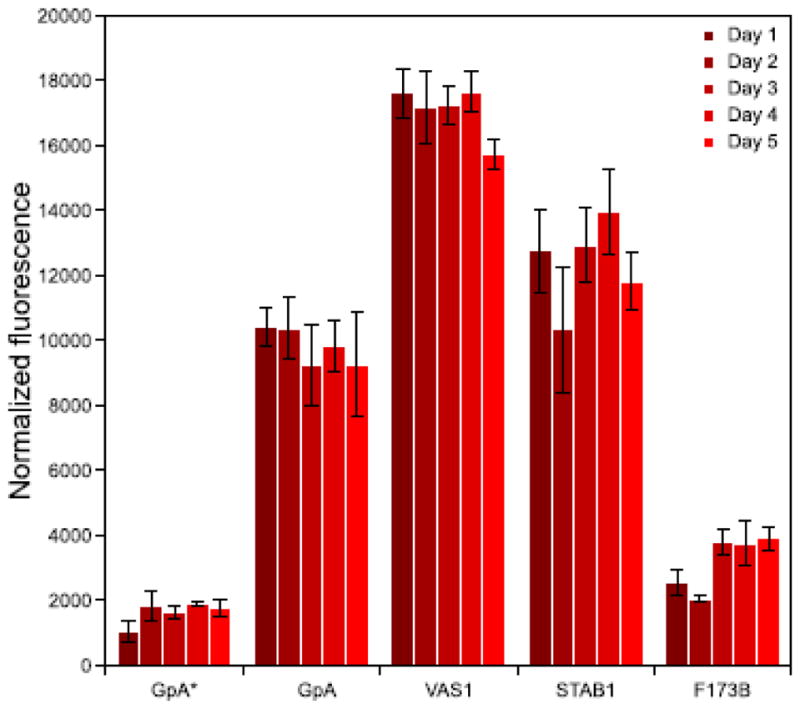
To test the reproducibility of TOXGREEN over multiple days, the same five constructs were assayed over multiple days. The bars represent the standard deviation of eight independent biological replica per day (i.e. cultures inoculated from different colonies). The per day variability is in line with the variability observed over multiple days. Wester blots relative to these experiment are shown in supplementary Figure S4.
3.6 High-throughput screening of TM helix self-association in bacterial divisome proteins
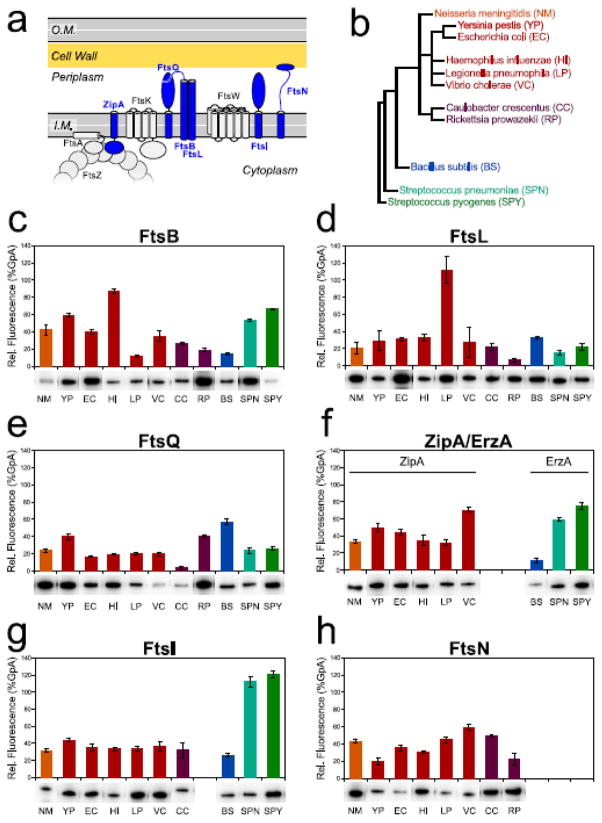
To test the high-throughput capabilities of TOXGREEN, we performed a large-scale screening for TM helix self-association in membrane proteins of the bacterial divisome. The divisome is the large and still poorly understood multi-protein complex that operates bacterial cell division [43,44]. The divisome of E. coli comprises many essential integral membrane proteins (Fig. 5a), six of which are bitopic (ZipA, FtsQ, FtsB, FtsL, FtsI and FtsN). We have analyzed the propensity of the TM region of these bitopic proteins to self-associate, using the sequences from eleven diverse bacterial species (Fig. 5b). In total, we have analyzed 60 individual TM sequences (supplementary Table S2), each measured with at least 4 independent biological replica, for a total of more than 240 individual measurements.
Figure 5. TOXGREEN analysis of bacterial divisome proteins.
a) Schematic representation of the essential membrane proteins of the bacterial divisome of E. coli. The six bitopic proteins are highlighted in blue. ZipA contributes to tethering to the membrane the polymeric FtsZ, which forms the scaffold of the divisome. FtsB and FtsL form a hetero-tetramer mediated by their TM helices and the periplasmic coiled coiled domains. They are recruited to the divisome by FtsQ, which forms with them a ternary complex. FtsI is a Pennicilline Binding Protein important for the synthesis of septal cell wall. FtsN plays an important roled in the regulation of cell divison. It contains a SPOR domain that recognizes the septal peptidoglycan. b) Evolutionary tree of the 11 bacterial species selected for the analysis. These include the gram negative alpha- (C. crescentus, R. prowazekii), beta- (N. meningitidis) and gamma-proteobacteria species (Y. pestis, E. coli, H. influenzae, L. pneumophila, V. cholera), as well as gram positive bacilli (B. subtilis) and cocci (S. pneumonia, S. pyogenes) species. c–h) TOXGREEN analysis of FtsB, FtsL, FtsQ, ZipA/EzrA, FtsI and FtsN sequences from the 11 species. Not all proteins are present in all the species. In particular, ZipA (f) is present only some classes of proteobacteria; for the gram positive species we analyzed EzrA, which is an FtsZ modulator topologically similar to ZipA. TOXGREEN chimera expression levels were verified by Western blot using anti-MBP antibodies (bands displayed under the histograms). It is notable how the chimeras expression levels vary among the samples. In general, it is not possible to draw a precise relationship between the physical strength of association and reporter gene expression in TOXCAT/TOXGREEN, even when the chimera’s expression levels are considered. Empirically, 40% GpA can be taken as a reasonable limit under which the confidence in discriminating specific association from background expression is low.
FtsB and FtsL are the only membrane proteins of the divisome whose oligomeric state has been biophysically characterized. FtsB and FtsL form a higher-oligomeric complex (likely a 2:2 hetero-tetramer) that is mediated by the TM domains and juxta-membrane coiled coil domains of the two proteins [6]. Their complex is essential for the recruitment of the late components of the divisome [45,46]. Using FRET in vitro [6] and TOXCAT [41], we reported previously that the TM domain of FtsB self-associates, albeit weakly. Here (Fig. 5c), we found that among the beta and gamma proteobacteria (N. meningitis, Y. pestis, E. coli, H. influenzae and V. cholera) FtsB retains a moderate to strong tendency to self associate (40–90% of GpA). Within this group the only exception is the FtsB-TM sequence of L. pneumophila. Interestingly, L. pneumophila is also the species that displays strong self-association for FtsL (Fig. 5d), which is low in all other sequences. The gram-positive bacteria S. pneumoniae and S. pyogenes also display significant self-association for FtsB. Overall, the data confirm that FtsB and FtsL retain some propensity to self-associate, although it is unknown whether their homo-oligomerization has a physiological role in vivo.
FtsQ is the protein responsible for recruiting the FtsBL complex to the division site, forming a ternary complex [47,48]. The main interactions between FtsQ and the FtsBL complex are believed to occur in C-terminal region of the periplasmic soluble domain of the proteins [47,49,50]. The TM domain of FtsQ is not essential for its function because it can be swapped with the TM domain of an unrelated protein [51]. The TOXGREEN analysis shows that in the majority of cases the FtsQ sequences produced near basal GFP expression (around 20% GpA, Fig. 5e). The main exception is the TM domain of the FtsQ of B. subtilis, whose GFP expression level is near 60% of the GpA standard.
ZipA is a protein unique to gamma, and perhaps beta, proteobacteria. It is essential for tethering the tubulin homolog FtsZ to the membrane. Working in concert with the peripheral membrane protein FtsA, ZipA contributes to the formation of the filamentous scaffold that supports the assembly of the divisome (the Z-ring) [52,53]. ZipA is type I bitopic protein with intracellular globuler domains, unlike FtsB, FtsL, FtsQ, FtsI and FtsN, which are all type II proteins with periplasmic soluble domains (Fig. 5a). Several of the TOXGREEN ZipA constructs yielded GFP expression levels above 40% of the GpA standard, including 70% expression for the V. cholera sequence (Fig. 5f). In general, it is not possible to draw a precise relationship between physical strength of association and reporter gene expression (TOXCAT/TOXGREEN response is sensitive to the specific nature and length of sequence used in the construct) but 40% GpA can be empirically taken as a reasonable limit under which the confidence in discriminating specific association from background expression is low. Both FtsZ and FtsA (an actin homolog) are homo-polymeric proteins. Therefore the notion that ZipA may self-associate forming dimers or higher-oligomers would not be surprising. This finding highlights the need for further investigation into the self-association ability of the TM region of ZipA.
EzrA is a type I bitopic protein, similar to ZipA, that is found only in gram positive bacteria. Though it shares structural similarities and possibly homology with ZipA [54], EzrA appears to be a negative regulator of FtsZ assembly [55]. It has been well characterized in B. subtilis, where it inhibits FtsZ polymerization and bundling by reducing FtsZ GTP hydrolysis [55,56]. Our TOXGREEN analysis found only basal level of reporter gene expression for the B. subtilis TM sequence, but rather significant (60–80% GpA) GFP expression levels for the two Streptococcus species (Fig. 5f).
FtsI is a Penicilling Binding Protein (PBP), which are transpeptidases and transglycosilases involved in the final stages of the synthesis of the cell-wall peptidoglycan during bacterial cell division [57]. FtsI interacts with its TM domain and works in association with FtsW [58,59], a large multispan lipid flippase that is responsible for the export of peptidoglycan precursor to the periplasm [60]. All species of FtsI-TM promoted relatively low expression of GFP (30–40%), with the exception of the homologs annotated for S. pneumoniae and S. pyogenes, which appear to have a strong tendency to self-associate (>100% GpA) (Fig. 5g). Interestingly, FtsI represents the third case presenting a strong propensity for TM self-association among the Streptococcus proteins (FtsB and ErzA being the other two cases, Fig. 5c and 5f).
The last protein examined is FtsN. FtsN consists of a small cytoplasmic region, a transmembrane domain and a long periplasmic region that includes a long linker peptide and a C-terminal globular SPOR domain, which binds specifically to septal wall peptidoglycan (Fig. 5a) [61]. FtsN is the last of the essential proteins to accumulate at the division site [62,63]. Various biological evidence suggests that FtsN interacts with many other division proteins, including FtsZ and FtsA, the FtsBLQ subcomplex and the peptidoglycan synthase subcomplex (FtsW, FtsI) [64–66]. We found that FtsN-TM sequences of a number of proteobacterial species promote levels of GFP expression that are in the 40–60% range of the GpA standard (N. meningitis, L. pneumophila, V. cholera and C. crescentus). As in the case of ZipA, these findings represents an interesting lead for further investigations into the potential role of TM self-association for FtsN function.
4. Discussion
We have demonstrated that the replacement of CAT for sfGFP greatly simplifies the operations of TOXCAT and significantly enhances its throughput. We have shown that fluorescence can be measured directly in live cell culture without the need of a lysis step. TOXGREEN can be performed on log phase cell cultures, the same growth conditions of the original TOXCAT. In these conditions, the cells need to be harvested and resuspended in buffer for improved detection. We found that TOXGREEN can be performed on cultures in stationary phase, producing indistinguishable results, as well as in log or stationary phase in M9 media. In these conditions, the workflow of TOXGREEN becomes extremely simple, since fluorescence is measured directly in the original cultures without any further processing.
In comparison, working with TOXCAT is significantly more laborious. CAT can be quantified either enzymatically [22,35] or immunochemically (ELISA) [36]. Both method requires lysis of the bacteria and significant processing. For example, enzymatic quantification with the Ellman’s reagent [35], which is the method that our group has adopted [41,67–69], requires approximately 90 minutes of preparatory work and four minutes per sample at the spectrophotometer, which translates into a day of continuous operator work for measuring forty individual samples. Similarly, ELISA requires several incubations and washes. The protocol is more rapid, but still laborious and time consuming, and requires expensive reagents.
The data show that the change in reporter gene does not alter the nature of the assay, its response, its dynamic range, its sensitivity or the level of its intrinsic variability. TOXGREEN loses a distinctive feature of TOXCAT, the ability to select strongly self-associating sequences in large combinatorial libraries, which exploits the resistance against the antibiotic chloramphenicol conferred by CAT. For this particular feature a user should revert to the original TOXCAT. The selection is, however, a specialized feature that is seldom used, since TOXCAT is almost always applied in the literature to assessing a specific set of constructs.
The new protocol has enabled the rapid screen for self-association a large set of TM domains of bitopic proteins of the bacterial divisome, which identified several cases of TM domains that display an apparent tendency to self-associate. The divisome is a biologically important and still poorly understood multi-protein complex, as well as a potential target for novel antibiotics. Although TOXGREEN measurement are not sufficient on their own to determine whether these domains are oligomeric in their physiological form, our results provide several interesting leads for further biophysical and biological investigation that may reveal further insights on the structural organization of the divisome. TOXGREEN could be readily used to determine the helix-helix interface of these potential oligomers using exhaustive mutagenesis, an approach that we have successfully applied to the E. coli FtsB dimer [41].
Given the ease in testing multiple replica of multiple construct at once, we conclude that the TOXGREEN variant of TOXCAT represents a simpler, less expensive, and high-throughput version of the assay. Plasmids, cells and a detailed protocol are available upon request.
Supplementary Material
Highlights.
TOXCAT is a widely used genetic assay for transmembrane helix association
We have developed a high-throughput version of TOXCAT called TOXGREEN
TOXGREEN be assayed directly in cell culture media without any manipulation
We present carefully tested protocols for alternative conditions for the assay
We applied TOXGREEN to the analysis of bitopic proteins of the bacterial divisome
Acknowledgments
We thank Dr. Ben Mueller for providing the TOXCAT constructs used in this study. We are grateful to Samantha Anderson and Elizabeth Caselle for helpful discussion and critical reading of the manuscript and to Dan Stevens and the Biophysics Instrumentation Facility of the Dept. of Biochemistry at the University of Wisconsin-Madison for experimental support. The work was supported by National Institutes of Health Grant R01GM0997522 and National Science Foundation Grant CHE-1415910. CRA was also supported in part by a Biochemistry Undergraduate Summer Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 2.Hubert P, Sawma P, Duneau JP, Khao J, Hénin J, Bagnard D, Sturgis J. Single-spanning transmembrane domains in cell growth and cell-cell interactions: More than meets the eye? Cell Adh Migr. 2010;4:313–324. doi: 10.4161/cam.4.2.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore DT, Berger BW, DeGrado WF. Protein-Protein Interactions in the Membrane: Sequence, Structural, and Biological Motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher LE, Engelman DM, Sturgis JN. Detergents modulate dimerization, but not helicity, of the glycophorin A transmembrane domain. Journal of Molecular Biology. 1999;293:639–651. doi: 10.1006/jmbi.1999.3126. [DOI] [PubMed] [Google Scholar]
- 5.Khadria A, Senes A. Measurement of transmembrane peptide interactions in liposomes using Förster resonance energy transfer (FRET) Methods Mol Biol. 2013;1063:19–36. doi: 10.1007/978-1-62703-583-5_2. [DOI] [PubMed] [Google Scholar]
- 6.Khadria AS, Senes A. The Transmembrane Domains of the Bacterial Cell Division Proteins FtsB and FtsL Form a Stable High-Order Oligomer. Biochemistry. 2013;52:7542–7550. doi: 10.1021/bi4009837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khadria AS, Senes A. Fluorophores, environments, and quantification techniques in the analysis of transmembrane helix interaction using FRET. Biopolymers. 2015;104:247–264. doi: 10.1002/bip.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merzlyakov M, You M, Li E, Hristova K. Transmembrane Helix Heterodimerization in Lipid Bilayers: Probing the Energetics behind Autosomal Dominant Growth Disorders. Journal of Molecular Biology. 2006;358:1–7. doi: 10.1016/j.jmb.2006.01.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.You M, Li E, Wimley WC, Hristova K. Förster resonance energy transfer in liposomes: Measurements of transmembrane helix dimerization in the native bilayer environment. Analytical Biochemistry. 2005;340:154–164. doi: 10.1016/j.ab.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleming KG. Standardizing the Free Energy Change of Transmembrane Helix-Helix Interactions. Journal of Molecular Biology. 2002;323:563–571. doi: 10.1016/s0022-2836(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 11.Choma C, Gratkowski H, Lear JD, DeGrado WF. Asparagine-mediated self-association of a model transmembrane helix. Nat Struct Mol Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- 12.Cristian L, Lear JD, DeGrado WF. Determination of membrane protein stability via thermodynamic coupling of folding to thiol-disulfide interchange. Protein Sci. 2003;12:1732–1740. doi: 10.1110/ps.0378603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cristian L, Lear JD, DeGrado WF. Use of thiol-disulfide equilibria to measure the energetics of assembly of transmembrane helices in phospholipid bilayers. Proc Natl Acad Sci USA. 2003;100:14772–14777. doi: 10.1073/pnas.2536751100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong H, Blois TM, Cao Z, Bowie JU. Method to measure strong protein – protein interactions in lipid bilayers using a steric trap. PNAS. 2010;107:19802–19807. doi: 10.1073/pnas.1010348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong H, Chang YC, Bowie JU. Measuring transmembrane helix interaction strengths in lipid bilayers using steric trapping. Methods Mol Biol. 2013;1063:37–56. doi: 10.1007/978-1-62703-583-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemmon MA, Flanagan JM, Treutlein HR, Zhang J, Engelman DM. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 17.Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. PNAS. 2009;106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Novicky L, Merzlyakov M, Hristov T, Hristova K. Measuring the energetics of membrane protein dimerization in mammalian membranes. J Am Chem Soc. 2010;132:3628–3635. doi: 10.1021/ja910692u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li E, Placone J, Merzlyakov M, Hristova K. Quantitative measurements of protein interactions in a crowded cellular environment. Anal Chem. 2008;80:5976–5985. doi: 10.1021/ac800616u. [DOI] [PubMed] [Google Scholar]
- 20.Kolmar H, Hennecke F, Götze K, Janzer B, Vogt B, Mayer F, Fritz HJ. Membrane insertion of the bacterial signal transduction protein ToxR and requirements of transcription activation studied by modular replacement of different protein substructures. EMBO J. 1995;14:3895–3904. doi: 10.1002/j.1460-2075.1995.tb00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langosch D, Brosig B, Kolmar H, Fritz HJ. Dimerisation of the glycophorin A transmembrane segment in membranes probed with the ToxR transcription activator. J Mol Biol. 1996;263:525–530. doi: 10.1006/jmbi.1996.0595. [DOI] [PubMed] [Google Scholar]
- 22.Russ WP, Engelman DM. TOXCAT: a measure of transmembrane helix association in a biological membrane. Proc Natl Acad Sci USA. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawson JP, Weinger JS, Engelman DM. Motifs of serine and threonine can drive association of transmembrane helices. J Mol Biol. 2002;316:799–805. doi: 10.1006/jmbi.2001.5353. [DOI] [PubMed] [Google Scholar]
- 24.Russ WP, Engelman DM. The GxxxG motif: A framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 25.Gurezka R, Langosch D. In vitro selection of membrane-spanning leucine zipper protein-protein interaction motifs using POSSYCCAT. J Biol Chem. 2001;276:45580–45587. doi: 10.1074/jbc.M105362200. [DOI] [PubMed] [Google Scholar]
- 26.Lindner E, Unterreitmeier S, Ridder ANJA, Langosch D. An extended ToxR POSSYCCAT system for positive and negative selection of self-interacting transmembrane domains. J Microbiol Methods. 2007;69:298–305. doi: 10.1016/j.mimet.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 27.Joce C, Wiener AA, Yin H. Multi-Tox: application of the ToxR-transcriptional reporter assay to the study of multipass protein transmembrane domain oligomerization. Biochim Biophys Acta. 2011;1808:2948–2953. doi: 10.1016/j.bbamem.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennasroune A, Gardin A, Auzan C, Clauser E, Dirrig-Grosch S, Meira M, Appert-Collin A, Aunis D, Crémel G, Hubert P. Inhibition by transmembrane peptides of chimeric insulin receptors. Cell Mol Life Sci. 2005;62:2124–2131. doi: 10.1007/s00018-005-5226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger BW, Kulp DW, Span LM, DeGrado JL, Billings PC, Senes A, Bennett JS, DeGrado WF. Consensus motif for integrin transmembrane helix association. Proc Natl Acad Sci USA. 2010;107:703–708. doi: 10.1073/pnas.0910873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindner E, Langosch D. A ToxR-based dominant-negative system to investigate heterotypic transmembrane domain interactions. Proteins. 2006;65:803–807. doi: 10.1002/prot.21226. [DOI] [PubMed] [Google Scholar]
- 31.Schneider D, Engelman DM. GALLEX, a measurement of heterologous association of transmembrane helices in a biological membrane. J Biol Chem. 2003;278:3105–3111. doi: 10.1074/jbc.M206287200. [DOI] [PubMed] [Google Scholar]
- 32.Su PC, Berger BW. Identifying key juxtamembrane interactions in cell membranes using AraC-based transcriptional reporter assay (AraTM) J Biol Chem. 2012;287:31515–31526. doi: 10.1074/jbc.M112.396895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su PC, Berger BW. A novel assay for assessing juxtamembrane and transmembrane domain interactions important for receptor heterodimerization. J Mol Biol. 2013;425:4652–4658. doi: 10.1016/j.jmb.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 34.Brosig B, Langosch D. The dimerization motif of the glycophorin A transmembrane segment in membranes: importance of glycine residues. Protein Sci. 1998;7:1052–1056. doi: 10.1002/pro.5560070423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sulistijo ES, Jaszewski TM, MacKenzie KR. Sequence-specific dimerization of the transmembrane domain of the “BH3-only” protein BNIP3 in membranes and detergent. J Biol Chem. 2003;278:51950–51956. doi: 10.1074/jbc.M308429200. [DOI] [PubMed] [Google Scholar]
- 36.Li R, Gorelik R, Nanda V, Law PB, Lear JD, DeGrado WF, Bennett JS. Dimerization of the transmembrane domain of Integrin alphaIIb subunit in cell membranes. J Biol Chem. 2004;279:26666–26673. doi: 10.1074/jbc.M314168200. [DOI] [PubMed] [Google Scholar]
- 37.Cotlet M, Goodwin PM, Waldo GS, Werner JH. A comparison of the fluorescence dynamics of single molecules of a green fluorescent protein: one- versus two-photon excitation. Chemphyschem. 2006;7:250–260. doi: 10.1002/cphc.200500247. [DOI] [PubMed] [Google Scholar]
- 38.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 39.van den Ent F, Löwe J. RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods. 2006;67:67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Shaw WV. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Meth Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 41.LaPointe LM, Taylor KC, Subramaniam S, Khadria A, Rayment I, Senes A. Structural organization of FtsB, a transmembrane protein of the bacterial divisome. Biochemistry. 2013;52:2574–2585. doi: 10.1021/bi400222r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mueller BK, Anderson SM, Subramaniam S, Lange E, Senes A. The strength of GxxxG-mediated association in transmembrane dimers correlates with optimization of Cα — H hydrogen bonding and van der Walls. 2016 in prepartion. [Google Scholar]
- 43.Rowlett VW, Margolin W. The bacterial divisome: ready for its close-up. Philos Trans R Soc Lond, B, Biol Sci. 2015;370 doi: 10.1098/rstb.2015.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsang MJ, Bernhardt TG. Guiding divisome assembly and controlling its activity. Curr Opin Microbiol. 2015;24:60–65. doi: 10.1016/j.mib.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghigo JM, Beckwith J. Cell Division in Escherichia coli: Role of FtsL Domains in Septal Localization, Function, and Oligomerization. J Bacteriol. 2000;182:116–129. doi: 10.1128/jb.182.1.116-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen JC, Weiss DS, Ghigo JM, Beckwith J. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J Bacteriol. 1999;181:521–530. doi: 10.1128/jb.181.2.521-530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalez MD, Beckwith J. Divisome under construction: distinct domains of the small membrane protein FtsB are necessary for interaction with multiple cell division proteins. J Bacteriol. 2009;191:2815–2825. doi: 10.1128/JB.01597-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez MD, Akbay EA, Boyd D, Beckwith J. Multiple interaction domains in FtsL, a protein component of the widely conserved bacterial FtsLBQ cell division complex. J Bacteriol. 2010;192:2757–2768. doi: 10.1128/JB.01609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van den Berg van Saparoea HB, Glas M, Vernooij IGWH, Bitter W, den Blaauwen T, Luirink J. Fine-mapping the Contact Sites of the Escherichia coli Cell Division Proteins FtsB and FtsL on the FtsQ Protein. J Biol Chem. 2013;288:24340–24350. doi: 10.1074/jbc.M113.485888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masson S, Kern T, Le Gouëllec A, Giustini C, Simorre JP, Callow P, Vernet T, Gabel F, Zapun A. Central domain of DivIB caps the C-terminal regions of the FtsL/DivIC coiled-coil rod. J Biol Chem. 2009;284:27687–27700. doi: 10.1074/jbc.M109.019471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guzman LM, Weiss DS, Beckwith J. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J Bacteriol. 1997;179:5094–5103. doi: 10.1128/jb.179.16.5094-5103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ortiz C, Natale P, Cueto L, Vicente M. The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol Rev. 2016;40:57–67. doi: 10.1093/femsre/fuv040. [DOI] [PubMed] [Google Scholar]
- 53.Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung KM, Hsu HH, Govindan S, Chang BY. Transcription regulation of ezrA and its effect on cell division of Bacillus subtilis. J Bacteriol. 2004;186:5926–5932. doi: 10.1128/JB.186.17.5926-5932.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh JK, Makde RD, Kumar V, Panda D. A membrane protein, EzrA, regulates assembly dynamics of FtsZ by interacting with the C-terminal tail of FtsZ. Biochemistry. 2007;46:11013–11022. doi: 10.1021/bi700710j. [DOI] [PubMed] [Google Scholar]
- 56.Haeusser DP, Schwartz RL, Smith AM, Oates ME, Levin PA. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol Microbiol. 2004;52:801–814. doi: 10.1111/j.1365-2958.2004.04016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wissel MC, Weiss DS. Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J Bacteriol. 2004;186:490–502. doi: 10.1128/JB.186.2.490-502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mercer KLN, Weiss DS. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J Bacteriol. 2002;184:904–912. doi: 10.1128/jb.184.4.904-912.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohammadi T, van Dam V, Sijbrandi R, Vernet T, Zapun A, Bouhss A, Diepeveen-de Bruin M, Nguyen-Distèche M, de Kruijff B, Breukink E. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang JC, Van Den Ent F, Neuhaus D, Brevier J, Löwe J. Solution structure and domain architecture of the divisome protein FtsN. Mol Microbiol. 2004;52:651–660. doi: 10.1111/j.1365-2958.2004.03991.x. [DOI] [PubMed] [Google Scholar]
- 62.Addinall SG, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 63.Chen JC, Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol Microbiol. 2001;42:395–413. doi: 10.1046/j.1365-2958.2001.02640.x. [DOI] [PubMed] [Google Scholar]
- 64.Di Lallo G, Fagioli M, Barionovi D, Ghelardini P, Paolozzi L. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation, Microbiology (Reading. Engl) 2003;149:3353–3359. doi: 10.1099/mic.0.26580-0. [DOI] [PubMed] [Google Scholar]
- 65.Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol. 2005;187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexeeva S, Gadella TWJ, Jr, Verheul J, Verhoeven GS, den Blaauwen T. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol Microbiol. 2010;77:384–398. doi: 10.1111/j.1365-2958.2010.07211.x. [DOI] [PubMed] [Google Scholar]
- 67.Gromek KA, Suchy FP, Meddaugh HR, Wrobel RL, LaPointe LM, Chu UB, Primm JG, Ruoho AE, Senes A, Fox BG. The oligomeric States of the purified sigma-1 receptor are stabilized by ligands. J Biol Chem. 2014;289:20333–20344. doi: 10.1074/jbc.M113.537993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsin J, Lapointe LM, Kazy A, Chipot C, Senes A, Schulten K. Oligomerization state of photosynthetic core complexes is correlated with the dimerization affinity of a transmembrane helix. J Am Chem Soc. 2011;133:14071–14081. doi: 10.1021/ja204869h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khadria AS, Mueller BK, Stefely JA, Tan CH, Pagliarini DJ, Senes A. A Gly-zipper motif mediates homodimerization of the transmembrane domain of the mitochondrial kinase ADCK3. J Am Chem Soc. 2014;136:14068–14077. doi: 10.1021/ja505017f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beevers AJ, Damianoglou A, Oates J, Rodger A, Dixon AM. Sequence-dependent oligomerization of the Neu transmembrane domain suggests inhibition of “conformational switching” by an oncogenic mutant. Biochemistry. 2010;49:2811–2820. doi: 10.1021/bi902087v. [DOI] [PubMed] [Google Scholar]
- 71.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J Biol Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 72.Benej M, Fekecsova S, Poturnajova M. Assessing the effect of RET transmembrane domain mutations in receptor self-association capability using the in vivo TOXCAT system. Neoplasma. 2013;60:111–120. doi: 10.4149/neo_2013_015. [DOI] [PubMed] [Google Scholar]
- 73.Kjaer S, Kurokawa K, Perrinjaquet M, Abrescia C, Ibáñez CF. Self-association of the transmembrane domain of RET underlies oncogenic activation by MEN2A mutations. Oncogene. 2006;25:7086–7095. doi: 10.1038/sj.onc.1209698. [DOI] [PubMed] [Google Scholar]
- 74.Oates J, King G, Dixon AM. Strong oligomerization behavior of PDGFbeta receptor transmembrane domain and its regulation by the juxtamembrane regions. Biochim Biophys Acta. 2010;1798:605–615. doi: 10.1016/j.bbamem.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 75.Noordeen NA, Carafoli F, Hohenester E, Horton MA, Leitinger B. A transmembrane leucine zipper is required for activation of the dimeric receptor tyrosine kinase DDR1. J Biol Chem. 2006;281:22744–22751. doi: 10.1074/jbc.M603233200. [DOI] [PubMed] [Google Scholar]
- 76.Anbazhagan V, Munz C, Tome L, Schneider D. Fluidizing the membrane by a local anesthetic: phenylethanol affects membrane protein oligomerization. J Mol Biol. 2010;404:773–777. doi: 10.1016/j.jmb.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 77.Finger C, Escher C, Schneider D. The single transmembrane domains of human receptor tyrosine kinases encode self-interactions. Sci Signal. 2009;2:ra56. doi: 10.1126/scisignal.2000547. [DOI] [PubMed] [Google Scholar]
- 78.Chin CN, Sachs JN, Engelman DM. Transmembrane homodimerization of receptor-like protein tyrosine phosphatases. FEBS Lett. 2005;579:3855–3858. doi: 10.1016/j.febslet.2005.05.071. [DOI] [PubMed] [Google Scholar]
- 79.Stokes KD, Gururaj Rao A. Dimerization properties of the transmembrane domains of Arabidopsis CRINKLY4 receptor-like kinase and homologs. Arch Biochem Biophys. 2008;477:219–226. doi: 10.1016/j.abb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 80.Stokes KD, Rao AG. The role of individual amino acids in the dimerization of CR4 and ACR4 transmembrane domains. Arch Biochem Biophys. 2010;502:104–111. doi: 10.1016/j.abb.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 81.Matthews EE, Thévenin D, Rogers JM, Gotow L, Lira PD, Reiter LA, Brissette WH, Engelman DM. Thrombopoietin receptor activation: transmembrane helix dimerization, rotation, and allosteric modulation. FASEB J. 2011;25:2234–2244. doi: 10.1096/fj.10-178673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vilar M, Charalampopoulos I, Kenchappa RS, Simi A, Karaca E, Reversi A, Choi S, Bothwell M, Mingarro I, Friedman WJ, Schiavo G, Bastiaens PIH, Verveer PJ, Carter BD, Ibáñez CF. Activation of the p75 neurotrophin receptor through conformational rearrangement of disulphide-linked receptor dimers. Neuron. 2009;62:72–83. doi: 10.1016/j.neuron.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li W, Metcalf DG, Gorelik R, Li R, Mitra N, Nanda V, Law PB, Lear JD, Degrado WF, Bennett JS. A push-pull mechanism for regulating integrin function. Proc Natl Acad Sci USA. 2005;102:1424–1429. doi: 10.1073/pnas.0409334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yin H, Litvinov RI, Vilaire G, Zhu H, Li W, Caputo GA, Moore DT, Lear JD, Weisel JW, Degrado WF, Bennett JS. Activation of platelet alphaIIbbeta3 by an exogenous peptide corresponding to the transmembrane domain of alphaIIb. J Biol Chem. 2006;281:36732–36741. doi: 10.1074/jbc.M605877200. [DOI] [PubMed] [Google Scholar]
- 85.Zhu H, Metcalf DG, Streu CN, Billings PC, Degrado WF, Bennett JS. Specificity for homooligomer versus heterooligomer formation in integrin transmembrane helices. J Mol Biol. 2010;401:882–891. doi: 10.1016/j.jmb.2010.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dews IC, Mackenzie KR. Transmembrane domains of the syndecan family of growth factor coreceptors display a hierarchy of homotypic and heterotypic interactions. Proc Natl Acad Sci USA. 2007;104:20782–20787. doi: 10.1073/pnas.0708909105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu L, Hu TT, Luo SZ. Leucine Zipper Motif Drives the Transmembrane Domain Dimerization of E-cadherin. Int J Pept Res Ther. 2013;20:95–102. [Google Scholar]
- 88.Barwe SP, Kim S, Rajasekaran SA, Bowie JU, Rajasekaran AK. Janus model of the Na,K-ATPase beta-subunit transmembrane domain: distinct faces mediate alpha/beta assembly and beta-beta homo-oligomerization. J Mol Biol. 2007;365:706–714. doi: 10.1016/j.jmb.2006.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Srinivasan S, Deng W, Li R. L-selectin transmembrane and cytoplasmic domains are monomeric in membranes. Biochim Biophys Acta. 2011;1808:1709–1715. doi: 10.1016/j.bbamem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Luo SZ, Li R. Specific heteromeric association of four transmembrane peptides derived from platelet glycoprotein Ib-IX complex. J Mol Biol. 2008;382:448–457. doi: 10.1016/j.jmb.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wei P, Liu X, Hu MH, Zuo LM, Kai M, Wang R, Luo SZ. The dimerization interface of the glycoprotein Ibβ transmembrane domain corresponds to polar residues within a leucine zipper motif. Protein Sci. 2011;20:1814–1823. doi: 10.1002/pro.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Formoso K, García MD, Frasch AC, Scorticati C. Filopodia formation driven by membrane glycoprotein M6a depends on the interaction of its transmembrane domains. J Neurochem. 2015;134:499–512. doi: 10.1111/jnc.13153. [DOI] [PubMed] [Google Scholar]
- 93.Ng DP, Deber CM. Modulation of the oligomerization of myelin proteolipid protein by transmembrane helix interaction motifs. Biochemistry. 2010;49:6896–6902. doi: 10.1021/bi100739r. [DOI] [PubMed] [Google Scholar]
- 94.Plotkowski ML, Kim S, Phillips ML, Partridge AW, Deber CM, Bowie JU. Transmembrane domain of myelin protein zero can form dimers: possible implications for myelin construction. Biochemistry. 2007;46:12164–12173. doi: 10.1021/bi701066h. [DOI] [PubMed] [Google Scholar]
- 95.Bowen ME, Engelman DM, Brunger AT. Mutational Analysis of Synaptobrevin Transmembrane Domain Oligomerization†. Biochemistry. 2002;41:15861–15866. doi: 10.1021/bi0269411. [DOI] [PubMed] [Google Scholar]
- 96.Petrova IM, Lahaye LL, Martiáñez T, de Jong AWM, Malessy MJ, Verhaagen J, Noordermeer JN, Fradkin LG. Homodimerization of the Wnt receptor DERAILED recruits the Src family kinase SRC64B. Mol Cell Biol. 2013;33:4116–4127. doi: 10.1128/MCB.00169-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jenei ZA, Borthwick K, Zammit VA, Dixon AM. Self-association of transmembrane domain 2 (TM2), but not TM1, in carnitine palmitoyltransferase 1A: role of GXXXG(A) motifs. J Biol Chem. 2009;284:6988–6997. doi: 10.1074/jbc.M808487200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jenei ZA, Warren GZL, Hasan M, Zammit VA, Dixon AM. Packing of transmembrane domain 2 of carnitine palmitoyltransferase-1A affects oligomerization and malonyl-CoA sensitivity of the mitochondrial outer membrane protein. FASEB J. 2011;25:4522–4530. doi: 10.1096/fj.11-192005. [DOI] [PubMed] [Google Scholar]
- 99.Lawrie CM, Sulistijo ES, MacKenzie KR. Intermonomer hydrogen bonds enhance GxxxG-driven dimerization of the BNIP3 transmembrane domain: roles for sequence context in helix-helix association in membranes. J Mol Biol. 2010;396:924–936. doi: 10.1016/j.jmb.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Watt B, Tenza D, Lemmon MA, Kerje S, Raposo G, Andersson L, Marks MS. Mutations in or near the transmembrane domain alter PMEL amyloid formation from functional to pathogenic. PLoS Genet. 2011;7:e1002286. doi: 10.1371/journal.pgen.1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang H, Barreyro L, Provasi D, Djemil I, Torres-Arancivia C, Filizola M, Ubarretxena-Belandia I. Molecular determinants and thermodynamics of the amyloid precursor protein transmembrane domain implicated in Alzheimer’s disease. J Mol Biol. 2011;408:879–895. doi: 10.1016/j.jmb.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dixon AM, Stanley BJ, Matthews EE, Dawson JP, Engelman DM. Invariant chain transmembrane domain trimerization: a step in MHC class II assembly. Biochemistry. 2006;45:5228–5234. doi: 10.1021/bi052112e. [DOI] [PubMed] [Google Scholar]
- 103.Polgar O, Ierano C, Tamaki A, Stanley B, Ward Y, Xia D, Tarasova N, Robey RW, Bates SE. Mutational analysis of threonine 402 adjacent to the GXXXG dimerization motif in transmembrane segment 1 of ABCG2. Biochemistry. 2010;49:2235–2245. doi: 10.1021/bi902085q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lock A, Forfar R, Weston C, Bowsher L, Upton GJG, Reynolds CA, Ladds G, Dixon AM. One motif to bind them: A small-XXX-small motif affects transmembrane domain 1 oligomerization, function, localization, and cross-talk between two yeast GPCRs. Biochim Biophys Acta. 2014;1838:3036–3051. doi: 10.1016/j.bbamem.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 105.Martínez-Garay CA, Juanes MA, Igual JC, Mingarro I, Bañó MC. A transmembrane serine residue in the Rot1 protein is essential for yeast cell viability. Biochemical Journal. 2014;458:239–249. doi: 10.1042/BJ20131306. [DOI] [PubMed] [Google Scholar]
- 106.Jara O, Acuña R, García IE, Maripillán J, Figueroa V, Sáez JC, Araya-Secchi R, Lagos CF, Pérez-Acle T, Berthoud VM, Beyer EC, Martínez AD. Critical role of the first transmembrane domain of Cx26 in regulating oligomerization and function. Mol Biol Cell. 2012;23:3299–3311. doi: 10.1091/mbc.E11-12-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Morera FJ, Alioua A, Kundu P, Salazar M, Gonzalez C, Martinez AD, Stefani E, Toro L, Latorre R. The first transmembrane domain (TM1) of β2-subunit binds to the transmembrane domain S1 of α-subunit in BK potassium channels. FEBS Lett. 2012;586:2287–2293. doi: 10.1016/j.febslet.2012.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kashlan OB, Maarouf AB, Kussius C, Denshaw RM, Blumenthal KM, Kleyman TR. Distinct structural elements in the first membrane-spanning segment of the epithelial sodium channel. J Biol Chem. 2006;281:30455–30462. doi: 10.1074/jbc.M604615200. [DOI] [PubMed] [Google Scholar]
- 109.Wei P, Zheng BK, Guo PR, Kawakami T, Luo SZ. The association of polar residues in the DAP12 homodimer: TOXCAT and molecular dynamics simulation studies. Biophys J. 2013;104:1435–1444. doi: 10.1016/j.bpj.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Constantinescu SN, Keren T, Russ WP, Ubarretxena-Belandia I, Malka Y, Kubatzky KF, Engelman DM, Lodish HF, Henis YI. The erythropoietin receptor transmembrane domain mediates complex formation with viral anemic and polycythemic gp55 proteins. J Biol Chem. 2003;278:43755–43763. doi: 10.1074/jbc.M302974200. [DOI] [PubMed] [Google Scholar]
- 111.Warren G, Oates J, Robinson C, Dixon AM. Contributions of the transmembrane domain and a key acidic motif to assembly and function of the TatA complex. J Mol Biol. 2009;388:122–132. doi: 10.1016/j.jmb.2009.02.060. [DOI] [PubMed] [Google Scholar]
- 112.Aklujkar M, Beatty JT. Investigation of Rhodobacter capsulatus PufX interactions in the core complex of the photosynthetic apparatus. Photosyn Res. 2006;88:159–171. doi: 10.1007/s11120-006-9047-y. [DOI] [PubMed] [Google Scholar]
- 113.Aklujkar M, Prince RC, Beatty JT. The PuhB protein of Rhodobacter capsulatus functions in photosynthetic reaction center assembly with a secondary effect on light-harvesting complex 1. J Bacteriol. 2005;187:1334–1343. doi: 10.1128/JB.187.4.1334-1343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim JH, Hartley TL, Curran AR, Engelman DM. Molecular dynamics studies of the transmembrane domain of gp41 from HIV-1. Biochim Biophys Acta. 2009;1788:1804–1812. doi: 10.1016/j.bbamem.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Arbely E, Granot Z, Kass I, Orly J, Arkin IT. A trimerizing GxxxG motif is uniquely inserted in the severe acute respiratory syndrome (SARS) coronavirus spike protein transmembrane domain. Biochemistry. 2006;45:11349–11356. doi: 10.1021/bi060953v. [DOI] [PubMed] [Google Scholar]
- 116.Bronnimann MP, Chapman JA, Park CK, Campos SK. A transmembrane domain and GxxxG motifs within L2 are essential for papillomavirus infection. J Virol. 2013;87:464–473. doi: 10.1128/JVI.01539-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Freeman-Cook LL, Dixon AM, Frank JB, Xia Y, Ely L, Gerstein M, Engelman DM, DiMaio D. Selection and characterization of small random transmembrane proteins that bind and activate the platelet-derived growth factor beta receptor. J Mol Biol. 2004;338:907–920. doi: 10.1016/j.jmb.2004.03.044. [DOI] [PubMed] [Google Scholar]
- 118.Talbert-Slagle K, Marlatt S, Barrera FN, Khurana E, Oates J, Gerstein M, Engelman DM, Dixon AM, Dimaio D. Artificial transmembrane oncoproteins smaller than the bovine papillomavirus E5 protein redefine sequence requirements for activation of the platelet-derived growth factor beta receptor. J Virol. 2009;83:9773–9785. doi: 10.1128/JVI.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dawson JP, Melnyk RA, Deber CM, Engelman DM. Sequence context strongly modulates association of polar residues in transmembrane helices. J Mol Biol. 2003;331:255–262. doi: 10.1016/s0022-2836(03)00714-9. [DOI] [PubMed] [Google Scholar]
- 120.Johnson RM, Rath A, Deber CM. The position of the Gly-xxx-Gly motif in transmembrane segments modulates dimer affinity. Biochem Cell Biol. 2006;84:1006–1012. doi: 10.1139/o06-192. [DOI] [PubMed] [Google Scholar]
- 121.Johnson RM, Rath A, Melnyk RA, Deber CM. Lipid solvation effects contribute to the affinity of Gly-xxx-Gly motif-mediated helix-helix interactions. Biochemistry. 2006;45:8507–8515. doi: 10.1021/bi060063f. [DOI] [PubMed] [Google Scholar]
- 122.Melnyk RA, Kim S, Curran AR, Engelman DM, Bowie JU, Deber CM. The affinity of GXXXG motifs in transmembrane helix-helix interactions is modulated by long-range communication. J Biol Chem. 2004;279:16591–16597. doi: 10.1074/jbc.M313936200. [DOI] [PubMed] [Google Scholar]
- 123.McClain MS, Cao P, Cover TL. Amino-terminal hydrophobic region of Helicobacter pylori vacuolating cytotoxin (VacA) mediates transmembrane protein dimerization. Infect Immun. 2001;69:1181–1184. doi: 10.1128/IAI.69.2.1181-1184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McClain MS, Iwamoto H, Cao P, Vinion-Dubiel AD, Li Y, Szabo G, Shao Z, Cover TL. Essential role of a GXXXG motif for membrane channel formation by Helicobacter pylori vacuolating toxin. J Biol Chem. 2003;278:12101–12108. doi: 10.1074/jbc.M212595200. [DOI] [PubMed] [Google Scholar]
- 125.McClain MS, Czajkowsky DM, Torres VJ, Szabo G, Shao Z, Cover TL. Random mutagenesis of Helicobacter pylori vacA to identify amino acids essential for vacuolating cytotoxic activity. Infect Immun. 2006;74:6188–6195. doi: 10.1128/IAI.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Go MY, Kim S, Partridge AW, Melnyk RA, Rath A, Deber CM, Mogridge J. Self-association of the transmembrane domain of an anthrax toxin receptor. J Mol Biol. 2006;360:145–156. doi: 10.1016/j.jmb.2006.04.072. [DOI] [PubMed] [Google Scholar]
- 127.Cunningham F, Poulsen BE, Ip W, Deber CM. Beta-branched residues adjacent to GG4 motifs promote the efficient association of glycophorin A transmembrane helices. Biopolymers. 2011;96:340–347. doi: 10.1002/bip.21565. [DOI] [PubMed] [Google Scholar]
- 128.Johnson RM, Heslop CL, Deber CM. Hydrophobic helical hairpins: design and packing interactions in membrane environments. Biochemistry. 2004;43:14361–14369. doi: 10.1021/bi0492760. [DOI] [PubMed] [Google Scholar]
- 129.Johnson RM, Hecht K, Deber CM. Aromatic and cation-pi interactions enhance helix-helix association in a membrane environment. Biochemistry. 2007;46:9208–9214. doi: 10.1021/bi7008773. [DOI] [PubMed] [Google Scholar]
- 130.Tulumello DV, Deber CM. SDS micelles as a membrane-mimetic environment for transmembrane segments. Biochemistry. 2009;48:12096–12103. doi: 10.1021/bi9013819. [DOI] [PubMed] [Google Scholar]
- 131.Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat Struct Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- 132.Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proc Natl Acad Sci USA. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cohen EB, Jun SJ, Bears Z, Barrera FN, Alonso M, Engelman DM, DiMaio D. Mapping the homodimer interface of an optimized, artificial, transmembrane protein activator of the human erythropoietin receptor. PLoS ONE. 2014;9:e95593. doi: 10.1371/journal.pone.0095593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bañó-Polo M, Baeza-Delgado C, Orzáez M, Marti-Renom MA, Abad C, Mingarro I. Polar/Ionizable residues in transmembrane segments: effects on helix-helix packing. PLoS ONE. 2012;7:e44263. doi: 10.1371/journal.pone.0044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Duong MT, Jaszewski TM, Fleming KG, MacKenzie KR. Changes in apparent free energy of helix-helix dimerization in a biological membrane due to point mutations. J Mol Biol. 2007;371:422–434. doi: 10.1016/j.jmb.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cunningham F, Rath A, Johnson RM, Deber CM. Distinctions between hydrophobic helices in globular proteins and transmembrane segments as factors in protein sorting. J Biol Chem. 2009;284:5395–5402. doi: 10.1074/jbc.M809017200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.