Summary
CD4+ T cells, or T helper cells, are critical mediators and coordinators of adaptive immunity. Unique effector T helper cell populations have been identified that perform distinct functions in response to pathogenic infection. The T follicular helper (Tfh) cells are one such subset, which has been identified as the primary T‐cell population responsible for interacting with B cells to promote effective antibody‐mediated immune responses. Since their initial description at the turn of the century, and subsequent classification as a distinct T helper cell subset, there has been substantial interest in elucidating the regulatory mechanisms that govern Tfh cell formation. The collective insight from this body of work has demonstrated that Tfh cell differentiation is a complex and multistage process regulated by a litany of cell‐intrinsic and cell‐extrinsic factors. As with the development of the other recognized T helper cell subsets, specific cytokines exercise prominent roles in both the positive and negative regulation of Tfh cell development. However, the exact composition of, and stage‐specific requirements for, these environmental factors in the governance of Tfh cell differentiation remain incompletely understood. In this review, we summarize what is known regarding the role of cytokines in both the promotion and inhibition of Tfh cell differentiation and function.
Keywords: cytokines, T follicular helper cells, T helper cell differentiation, transcription factors
Abbreviations
- Bcl‐6
B‐cell lymphoma‐6
- Blimp‐1
B lymphocyte induced maturation protein 1
- Cxcr5
C‐X‐C chemokine receptor type 5
- EBI3
Epstein–Barr‐virus‐induced gene 3
- GC
germinal centre
- ICOS
inducible T‐cell co‐stimulator
- IL‐4
interleukin‐4
- Jak
Janus kinase
- NK
natural killer cell
- NKT
natural killer T cell
- STAT
signal transducer and activator of transcription
- Tcm
central memory T cell
- Tfh
T follicular helper
- TGF‐β
transforming growth factor β
- Th1
T helper 1
Introduction
The generation of pathogen‐neutralizing antibodies by B lymphocytes is a hallmark feature of the adaptive immune system and a necessary step in mounting an effective immune response to viral, bacterial and parasitic infections. While B cells are responsible for the production of secreted antibodies, it has been known for over 50 years that T cells are also a critical component of functional humoral immunity.1, 2, 3 CD4+ T helper cells were identified as logical candidates for fulfilling this functional requirement, given their demonstrated roles in coordinating immune responses through their influences on other immune cells. With the identification of T helper 1 (Th1) and Th2 cells in 1986 by Mosmann and Coffman, it was thought that one or both of these T‐cell populations may contribute to B‐cell help.4 Initially, Th2 cells were thought to be the ‘B helper’ T‐cell population because of their secretion of cytokines that favourably influenced B‐cell function and antibody production, such as interleukin 4 (IL‐4). However, almost 15 years later, an additional CD4+ T helper cell subset was described that expressed high levels of the chemokine receptor C‐X‐C chemokine receptor type 5 (Cxcr5).5, 6, 7 Subsequently, this novel T helper cell subset was defined as the population that contributed most prominently to B‐cell help. These CD4+ T cells were termed B follicular helper T cells, and are now commonly referred to as T follicular helper (Tfh) cells.
Tfh cells function as critical mediators of the humoral immune response through direct interactions with B lymphocytes. Mechanistically, Tfh cells first engage B cells in cognate interactions at the T‐cell–B‐cell border in secondary lymphoid tissues such as the lymph nodes and spleen.8, 9, 10 Upon initial engagement, CD40–CD40 ligand interactions result in B‐cell proliferation, differentiation and antibody isotype switching.11 Consequently, germinal centres (GC) form, wherein antibody diversification and affinity maturation occur, resulting in the production of the neutralizing antibodies that are a critical component in the adaptive immune response to pathogenic infection.
The multistage process of Tfh cell differentiation
While Tfh cells have a well‐defined role in the immune response, the molecular mechanisms that underlie their formation are less clear. Following their initial description, there was much debate regarding whether Tfh cells, as with the previously defined Th1 and Th2 cell subsets, were a unique T helper cell population. Historically, distinct T helper cell populations have been defined based on their ability to perform non‐redundant effector functions, and their expression of unique gene programmes dictated by cell‐specific ‘lineage‐defining’ transcription factors. For example, T‐bet and Gata3 have been identified as the lineage‐defining transcription factors for the Th1 and Th2 cell fates, respectively.12, 13 Indeed, numerous studies have shown that Tfh cells possess a distinct gene expression profile and that the transcriptional repressor B‐cell lymphoma‐6 (Bcl‐6) is required for Tfh cell development.14, 15, 16, 17, 18 Hence, it is now readily accepted that Tfh cells comprise a specialized subset of T helper cells that are critical to mounting effective humoral immune responses.
The generation of Tfh cell populations is a multistep process comprised of both pre‐GC Tfh and mature GC Tfh stages.19, 20 Tfh cell differentiation is initiated when naive T cells undergo antigen‐dependent activation in the presence of specific environmental signals including those derived from cytokines (Fig. 1). This results in an initial increase in Bcl‐6 expression and the concomitant up‐regulation of genes associated with the Tfh cell programme, including the cell surface receptors Cxcr5, inducible T‐cell co‐stimulator (ICOS), and programmed cell death protein 1.14, 19, 20, 21, 22, 23 The importance of Bcl‐6 to Tfh cell development is demonstrated by the striking lack of GCs in mice with T‐cell‐specific deletion of Bcl‐6 expression.14, 15, 16 This prominent role for Bcl‐6 in the initiation of Tfh differentiation is due at least in part, to its ability to repress the expression of B‐lymphocyte‐induced maturation protein 1 (Blimp‐1). Blimp‐1 has been identified as an antagonistic factor of the Tfh gene programme.14, 24 Following early Bcl‐6 up‐regulation and Blimp‐1 repression, Cxcr5‐directed homing of pre‐GC Tfh cells results in trafficking to the follicles of secondary lymphoid tissues where cognate interactions with B cells – notably those between ICOS and ICOS ligand (ICOSL) – take place.19, 20 This initial interaction results in a further increase in Tfh‐associated gene expression patterns, commitment to the functional GC Tfh cell programme, and subsequent GC formation (Fig. 1).22
Figure 1.
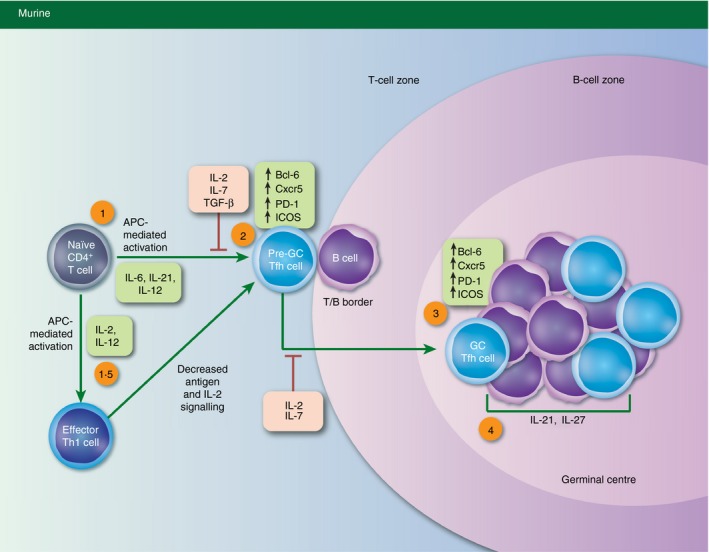
T follicular helper (Tfh) cell development and the impact of cytokine signals. Schematic depicting the multistage process of Tfh cell differentiation. The reported impacts of cytokine signals on murine Tfh cell development have been highlighted in red (negative) and green (positive). Tfh cell development is initiated upon antigen‐presenting‐cell‐mediated activation of naive CD4+ T cells in the presence of the indicated cytokines (1). This results in the up‐regulation of the Tfh lineage‐defining transcription factor B‐cell lymphoma 6 (Bcl‐6), as well as the expression of additional canonical Tfh genes including Cxcr5, programmed cell death protein 1 (PD‐1), and inducible T‐cell co‐stimulator (ICOS). Pre‐germinal centre (‘pre‐GC’) Tfh cells traffic to the T–B border, where they participate in cognate interactions with B cells (2). This results in the further up‐regulation of Tfh gene expression patterns including increased expression of Bcl‐6, Cxcr5, PD‐1 and ICOS (3). Subsequent cognate interactions between T and B lymphocytes result in the formation of the germinal centre and maintenance of the GC Tfh cell phenotype (4).
Interestingly, despite this well‐established sequence of events, many questions remain regarding the maturation of Tfh cell populations. For example, although Tfh cells develop during the initial response to antigen through the events described above, it has also been shown that additional effector T helper cell subsets are capable of up‐regulating a Tfh‐like gene expression profile, making alternative Tfh developmental pathways plausible as well (Fig. 1).24, 25, 26, 27 Similarly, studies have provided compelling evidence to suggest that differentiated Tfh cells are capable of up‐regulating gene expression programmes associated with alternative T helper cell types.28, 29, 30 Furthermore, it has been demonstrated that these multidirectional plasticity events in T helper cell populations are due in large part to an ‘accessible’ chromatin structure present at gene loci encoding key lineage‐defining transcription factors, including Bcl‐6.28, 31, 32 These previous studies are intriguing, because they support the possibility that, in addition to effector Tfh cells, other T helper populations may assist in antibody‐mediated immunity by co‐opting certain aspects of the Tfh cell gene programme. Hence, the concept of T helper cell flexibility between Tfh and other T helper cell populations is an important consideration in the complex process of Tfh cell differentiation.
Factors that regulate Tfh cell development
The regulatory mechanisms that govern the differentiation of Tfh cells, like those of other T helper cell subsets, are guided by the coordinated interplay between cell‐extrinsic signals (i.e. cytokines or ligand–receptor interactions) and cell‐intrinsic transcriptional networks. In addition to the required role for Bcl‐6 mentioned previously, a number of other transcription factors have been identified that play prominent roles in establishing the Tfh gene expression profile, including Irf4, Batf, Ascl2, Maf, Lef‐1, Tcf‐1, and a number of signal transducer and activator of transcription (STAT) factors.27, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43 Receptor–ligand interactions, notably those that induce ICOS signalling, are known to promote Bcl‐6 expression and play additional, critical roles in Tfh cell development.22, 40, 44, 45, 46 These mechanisms of regulation have been comprehensively reviewed elsewhere.19, 20, 47 Herein, we will focus primarily on the roles of cytokines in the regulation of Tfh cell development and function. In the past decade, novel scientific tools and elegant experimental design have combined to result in an emerging body of scientific literature that has provided important, and sometimes surprising, insights into the molecular mechanisms by which these environmental mediators regulate multiple aspects of Tfh cell biology. Specifically, we will discuss the impact of individual cytokines, and the downstream signalling pathways and transcription factors that they modulate, on the promotion and inhibition of Tfh cell development and the corresponding immune functions that Tfh cells perform (Table 1).
Table 1.
Cytokines involved in the positive and negative regulation of human and murine T follicular helper (Tfh) cell development
| Species | Cytokine | Downstream factor(s) | Role in Tfh development |
|---|---|---|---|
| Mouse | IL‐6 | STAT1, STAT3 | + |
| Mouse | IL‐21 | STAT1, STAT3 | + |
| Mouse | IL‐12 | STAT4 | + |
| Mouse | IL‐27 | STAT1, STAT3 | + |
| Mouse | TGF‐β | STAT3, STAT4 | − |
| Mouse | IL‐2 | STAT5 | − |
| Mouse | IL‐7 | STAT5 | − |
| Human | IL‐6 | STAT1, STAT3 | + |
| Human | IL‐21 | STAT1, STAT3 | + |
| Human | IL‐12 | STAT4 | + |
| Human | IL‐23 | STAT4, STAT3 | + |
| Human | TGF‐β | STAT3, STAT4 | + |
| Human | Activin A | SMAD2, SMAD3 | + |
| Human | IL‐2 | STAT5 | − |
The shading is to differentiate cytokines that negatively regulate TFH cell development from those that positively regulate Tfh development.
Cytokine signalling pathways that promote Tfh cell differentiation
Interleukin‐6
Interleukin‐6 was originally characterized as a factor that could both influence the activation of T cells and stimulate some aspects of B‐cell differentiation.48 It is produced by many of the professional antigen‐presenting cells of the immune system, with follicular dendritic cells being a primary contributor. Interleukin‐6 signals through a receptor composed of IL‐6‐specific (IL‐6Rα) and gp130 receptor subunits.48 Initially, IL‐6 binds to IL‐6Rα and subsequently interacts with gp130 to form the IL‐6 receptor (IL‐6R) signalling complex. Downstream intracellular signalling is mediated through the intracellular domain of gp130, resulting in the activation of the Janus kinase/STAT (Jak/STAT) pathway and the phosphorylation of STAT3 and STAT1 via Janus kinase 1 (Jak1). Following phosphorylation, STAT transcription factors dimerize and translocate to the nucleus where they regulate target gene expression. Interestingly, both STAT3 and STAT1 have been implicated in the direct regulation of Bcl‐6 expression (Fig. 2). Hence, given the critical role for Bcl‐6 in Tfh cell development, it is perhaps not surprising that mice lacking IL‐6 or a functional IL‐6R signalling complex have deficiencies in Tfh cell formation.34, 44, 49, 50 However, this effect on the Tfh population is only partial, suggesting that there are redundant, IL‐6‐independent pathways that can result in Tfh cell generation. Much of the data surrounding IL‐6 suggest that it functions early in Tfh cell formation. However, there are reports that IL‐6 produced late in chronic viral infection is required for optimal Tfh cell responses and antibody production.51, 52 In humans, IL‐6 has also been implicated in the promotion of Tfh cell responses, suggesting that the role of IL‐6 in Tfh development may be conserved across species.53 Hence, the collective data suggest that IL‐6 probably plays an important role in the promotion of the Tfh cell fate, but perhaps not an essential one.
Figure 2.
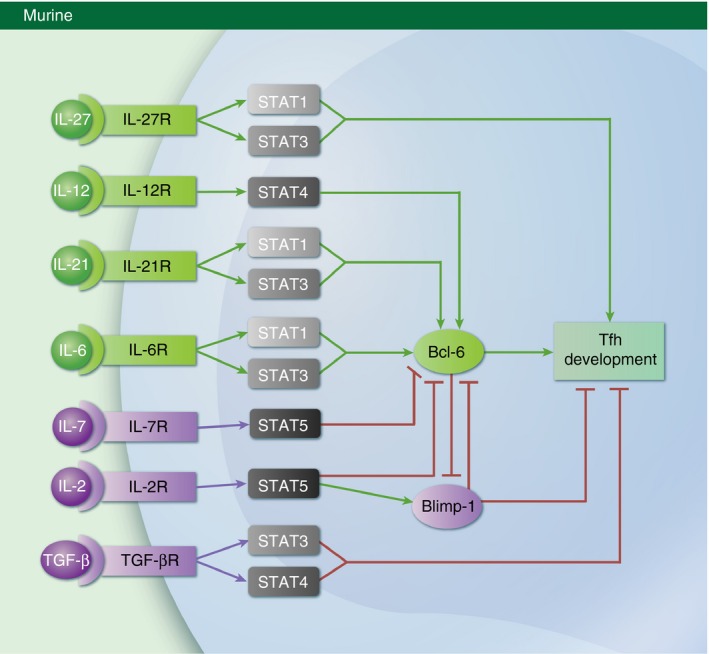
Cytokines that promote or inhibit murine follicular helper T (Tfh) cell differentiation. An illustrated diagram of the mechanisms by which cytokines regulate the development of Tfh cells in mice. Individual cytokines and the signal transducer and activator of transcription (STAT) transcription factors they activate are shown. Additionally, the impact of each cytokine on B‐cell lymphoma 6 (Bcl‐6) expression and Tfh cell development is indicated. It is important to note that the function of the depicted cytokines is not conserved across species, as transforming growth factor‐β (TGF‐β) signalling has been shown to negatively regulate Tfh development in mice, yet induces the expression of the Tfh gene programme in human cells.
Interleukin‐21
Interleukin‐21 is a cytokine of the common γ (γ c)‐chain family that is produced by natural killer T (NKT) and select CD4+ T cells, including Th17 and Tfh populations.54 Interleukin‐21 signals are received through a cell surface cytokine receptor composed of IL‐21Rα and γ c subunits.54 As with IL‐6, IL‐21 signalling results in activation of STAT3 and STAT1 transcription factors (Fig. 2). Additionally, similar to mice with deficiencies in IL‐6 signalling, IL‐21‐deficient mice display lower percentages of Tfh cells upon infection and mice lacking functional IL‐21 cytokine receptors display impaired Tfh formation.44, 55 As Tfh cells produce IL‐21, it is likely that IL‐21‐mediated autocrine effects play a significant role in the maintenance and augmentation of Tfh cell gene programming. Hence, whereas IL‐6 may be important in the initial priming of Tfh cells through up‐regulation of STAT1‐ or STAT3‐dependent Bcl‐6 expression, IL‐21 may play a more prominent role in sustaining Tfh cell identity and function.38 Still, the fact that mice with impaired IL‐6 or IL‐21 signalling have only partially compromised Tfh cell populations, combined with the shared activation of STAT3 downstream of IL‐6 and IL‐21 signalling, led to the hypothesis that these cytokines may play redundant roles in Tfh cell formation. Indeed, mice deficient in both IL‐6 and IL‐21 displayed a further reduction in Tfh numbers compared with IL‐6 or IL‐21 deficiency alone.49, 50 Interestingly, the continued, though significantly diminished, formation of Tfh cells in the absence of these cytokines, suggests that there are still as yet undiscovered IL‐6‐ and IL‐21‐independent regulatory mechanisms that contribute to Tfh cell generation.56
Interleukin‐12
Interleukin‐12 was originally identified as a factor that stimulated natural killer (NK) cell populations to produce IFN‐γ.57 More recently, IL‐12 has been shown to be a cytokine required for Th1 cell development.58, 59, 60 Mechanistically, IL‐12 signals through a heterodimer composed of IL‐12Rβ1 and IL‐12Rβ2 subunits, with signalling that ultimately results in the phosphorylation of STAT4.61, 62 Activated STAT4 is a critical driver of Th1 differentiation as it directly induces the expression of the hallmark Th1 genes IFN‐γ and T‐bet.63, 64, 65, 66 As such, it was somewhat surprising when IL‐12‐dependent activation of STAT4 was demonstrated to be an early inducer of Bcl‐6 expression in murine naive CD4+ T cells (Fig. 2).27 Interestingly, in humans, IL‐12 also appears to play a prominent role in the positive regulation of Tfh cell development, where it has been implicated in the activation of STAT3.67, 68 In vitro, the combination of IL‐12 and transforming growth factor‐β (TGF‐β) drives the expression of Bcl‐6, Cxcr5 and several other canonical Tfh genes.39 In further support of a role for IL‐12 in Tfh cell development, humans with mutations disrupting the function of IL‐12Rβ1 have diminished Tfh cell numbers.38, 69 Hence, though it seems somewhat counterintuitive given its required role in Th1 development, IL‐12 may also be an important contributor, especially in humans, to the promotion of Tfh cell differentiation.
Interleukin‐23
A second member of the IL‐12 cytokine family, IL‐23, has also been implicated in the promotion of human Tfh cell development. Interestingly, it has been shown that the combination of either IL‐12 or IL‐23 and TGF‐β is sufficient to drive in vitro development of human Tfh cells.39 Indeed, IL‐23 and IL‐12 share overlapping features such as the requirement for IL‐12Rβ1 as part of their receptor complex, and the downstream activation of STAT4.69 Similar to the previously discussed roles of IL‐6 and IL‐21 in murine Tfh cell development, this is yet another example of the redundancy in the environmental cues and downstream signalling events that promotes Tfh cell formation.
Interleukin‐27
Interleukin‐27 is a heterodimeric cytokine composed of p28 (IL‐27) and Epstein–Barr virus‐induced gene 3 (EBI3) subunits.70 Interleukin‐27 signals through a receptor composed of the IL‐27‐specific subunit IL‐27Rα and the gp130 subunit, which is shared with other cytokines including IL‐6.70 Similar to IL‐6, IL‐27 signalling primarily activates STAT1 and STAT3 through their phosphorylation by Jak1. Given the similarities between IL‐6 and IL‐27 signalling, it is logical that IL‐27 might also play a role in Tfh cell development. Interestingly, whereas IL‐27 does not appear to influence early murine Tfh cell development, it has been shown to contribute to Tfh cell maintenance, due to IL‐27‐mediated activation of IL‐21 expression (Fig. 2).71, 72 As discussed previously, IL‐21 is an important promoter of Tfh cell homeostasis and function. Indeed, it has been shown that in the absence of IL‐27 signalling, there is a reduction in IL‐21 expression and antibody production in mice, highlighting the role of this cytokine in the humoral immune response.71 Alternatively, it has also been suggested that IL‐27 may play an important role in Tfh development by antagonizing IL‐2 signalling, a known negative regulator of the Tfh cell fate.24, 70, 73, 74, 75
Activin A
Recently, an exciting finding has shed light on a novel cytokine involved in the differentiation of human Tfh cells. Crotty and colleagues used an in vitro screen of a collection of recombinant human proteins to identify Activin A as a novel inducer of the Tfh gene programme.76 Specifically, when combined with IL‐12, Activin A stimulation resulted in the significant up‐regulation of many Tfh‐associated proteins including BCL‐6, CXCR5 and programmed cell death protein 1 in human and non‐human primate cells. Importantly, treatment with Activin A also resulted in the repression of the Tfh antagonist Blimp‐1. Subsequently, the authors demonstrated that Sma and mothers against decapentaplegic (SMAD) SMAD2/SMAD3 signalling downstream of Activin A was responsible for the promotion of the Tfh‐like phenotype. Hence, these findings help to establish Activin A as yet another cytokine implicated in Tfh cell development in humans.76
Transforming growth factor‐β
The role of TGF‐β with regards to Tfh cell differentiation is more nuanced, as it has been implicated in both the positive and negative regulation of Tfh development. In mice, TGF‐β inhibits Tfh gene expression patterns including the expression of Bcl‐6 (Fig. 2).44, 77 In contrast, as previously mentioned, TGF‐β in conjunction with either IL‐12 or IL‐23 results in the STAT3‐ and STAT4‐dependent generation of in vitro‐derived human cells with Tfh‐like gene profiles and functions.39 These studies highlight that despite some similarities, the combination of cytokines that influence murine and human Tfh cell development and their function is not entirely conserved.
Type I interferons (IFN‐α/β)
Early after infection, type 1 interferons are produced by innate immune cells to initiate a cascade of immune responses. Like TGF‐β, the role for type 1 interferons in the promotion of Tfh development is complicated. Early in vitro work described a role for IFN‐α/β‐induced STAT1 in the activation of many Tfh genes including Bcl6.78 However, in a subsequent in vivo study using lymphocytic choriomeningitis virus infection, an antagonistic relationship between IFN‐activated STAT1 and STAT3 was described.79 Additional factors present in vivo (versus in vitro) may explain the discrepancies between the two studies. Ultimately, the authors of the second study concluded that exposure to type 1 IFN signalling repressed Tfh cell development in favour of Th1 differentiation.79 As with many of the cytokines described above, future work will be necessary to determine the exact role for type 1 IFN signalling in the regulation of Tfh cell differentiation.
Cytokine signalling pathways that inhibit Tfh cell development and function
Interleukin‐2
Interleukin‐2 is a member of the γ c cytokine family that signals through a heterotrimeric receptor comprised of IL‐2Rα, IL‐2Rβ and γ c.80, 81 High‐affinity IL‐2 signalling via this heterotrimeric form of the IL‐2 receptor results in the robust activation of the transcription factor STAT5.80, 81 It is now well established that the IL‐2/STAT5 axis is a negative regulator of Tfh cell development.24, 73, 74, 75, 82 In a study using an in vivo mouse model of influenza infection, it was demonstrated that exogenous administration of IL‐2 resulted in the inhibition of antigen‐specific Tfh cell generation, a lack of GC formation, and a reduction in neutralizing antibody production.73 Furthermore, it was shown that T‐cell‐specific deletion of IL‐2Rα resulted in an increase in the number of Tfh cells generated in response to infection.73
Mechanistically, many regulatory pathways have been implicated in the IL‐2‐dependent repression of Tfh cell development. First, IL‐2 signalling has been shown to promote the expression of the transcriptional repressor Blimp‐1, a known antagonist of Bcl‐6 expression and the Tfh gene programme.14, 83 Additionally, IL‐2‐activated STAT5 has been shown to directly bind to the Bcl6 locus and repress its expression (Fig. 2).24 Interestingly, the association of STAT5 with the Bcl6 promoter correlates with a reduction in bound STAT3, suggesting that these two STAT factors compete for identical DNA binding sites.24 A similar mechanism has been proposed in the regulation of Th17 cell formation, which also relies on STAT3 signalling.84 Finally, IL‐2 signalling is known to regulate the expression of a number of cytokine receptors. It has been shown that increased IL‐2 signalling results in the repression of IL‐6Rα and to a lesser extent, gp130.24, 85 Given the demonstrated role for IL‐6 in promoting the initiation of Tfh cell development, this is yet another mechanism by which IL‐2 signalling may limit Tfh cell differentiation. Hence, collectively, there is a large body of evidence establishing IL‐2 as a potent negative regulator of the Tfh cell fate.
Interleukin‐7
Like IL‐2, IL‐7 is a γ c cytokine family member with downstream effects propagated through the activation of STAT5.86 Interleukin‐7 signalling is mediated through a heterodimeric receptor composed of IL‐7Rα and γ c subunits. It is well established that IL‐2Rα and IL‐7Rα expression patterns inversely correlate in a T‐cell stage‐specific manner.86, 87 For example, IL‐2Rα expression is limited to effector T helper cell populations, whereas IL‐7Rα expression predominates during naive and memory cell homeostasis. Interestingly, two recent reports suggest that the IL‐7/STAT5 signalling axis may function to negatively regulate Bcl‐6 expression – and that of the Tfh gene programme – during naive and memory T‐cell stages (Fig. 2).25, 88 In one study, it was demonstrated that Bcl‐6 directly represses IL‐7R expression during early effector Tfh cell development. The authors then went on to show that this was a critical regulatory event in the initiation of Tfh cell generation, as IL‐7 signalling negatively regulated the generation of effector Tfh cells.88
In the second recent study, our laboratory identified IL‐7 signalling as a negative regulator of Bcl‐6 expression and the expression of many additional Tfh genes in post‐effector Th1 cells.25 First, we demonstrated that in response to decreased IL‐2 and T‐cell receptor signalling, Th1 cells were capable of up‐regulating the Bcl‐6‐dependent expression of both Tfh and central memory T cell gene expression patterns, including the expression of IL‐7R. Intriguingly, when these cells were exposed to IL‐7, the expression of several Tfh‐associated genes, including Bcl‐6, was repressed, whereas the expression of central memory T genes remained relatively unaltered. As with IL‐2 signalling, our data suggested that STAT5 association with the Bcl6 promoter was responsible for the IL‐7‐dependent repression of Bcl‐6 expression. Hence, the combined data from these studies indicate that IL‐2 and IL‐7 may direct conserved STAT5‐dependent regulatory mechanisms that govern Bcl‐6 expression through multiple stages of T‐cell differentiation.24, 25, 74, 75, 88
The above findings were somewhat surprising, as increased Bcl‐6 expression and IL‐7 signalling are important factors in both the differentiation and homeostasis of memory cell populations, including central memory T cells.82, 83, 89, 90 We postulated that stage‐specific IL‐7‐mediated repression of Bcl‐6 may be important in directing specific memory cell functions by differential regulation of the Tfh and central memory T gene programmes, which include the expression of cellular trafficking receptors such as Cxcr5, CD62L and Ccr7.25 However, it is important to point out that, in addition to its required role in regulating the expression of the Tfh gene programme, Bcl‐6 is also a demonstrated regulator of metabolic and cell cycle pathways.91, 92, 93 Hence, it is possible that the IL‐7/STAT5 regulatory axis may play key roles in governing aspects of memory cell survival and metabolism by tightly regulating Bcl‐6 expression. Indeed, the expression of Bcl‐6 appears to be regulated in a dynamic manner, as levels of Bcl‐6 have been observed to decrease in memory cell populations post‐infection.82 Future work will be required to comprehensively assess the differential contributions of Bcl‐6 and IL‐7 signalling to the establishment and maintenance of Tfh and memory cell populations.
Concluding remarks
Cytokines and cytokine signalling pathways have been the targets of therapeutic strategies since pioneering immunotherapeutic treatments using exogenous IL‐2 to promote immune‐mediated destruction of tumours.80, 94, 95 As this review has attempted to highlight, the contribution of cytokine‐dependent environmental cues to developing Tfh cells is a central, yet complicated, component of the differentiation process. Continued research in this area will be instrumental in unlocking the complexity of the molecular mechanisms underlying Tfh cell formation, which in turn has the potential to greatly impact human health. Acquiring a comprehensive understanding of Tfh cell biology will lead to the design of more efficacious vaccine strategies, and also allow for the targeted treatment of the aberrant Tfh activities that are the hallmarks of numerous autoimmune diseases.96 Hence, although significant insight into the contributions of cytokines to the governance of Tfh cell differentiation has been achieved in the past decade, there is still much to discover about the integrated mechanisms by which these environmental factors regulate Tfh‐mediated immune responses.
Disclosures
The authors declare no conflict of interest with regards to the contents of this manuscript.
Acknowledgements
The authors would like to sincerely thank members of the Oestreich Laboratory for thoughtful discussions and critical reading of the manuscript. The authors would also like to apologize to those whose work could not be adequately cited and discussed due to length restrictions. This work was supported by funds from the Virginia Tech Carilion Research Institute.
References
- 1. Claman HN, Chaperon EA, Triplett RF. Thymus‐marrow cell combinations. Synergism in antibody production. Proc Soc Exp Biol Med 1966; 122:1167–71. [DOI] [PubMed] [Google Scholar]
- 2. Miller JF. Effect of thymectomy in adult mice on immunological responsiveness. Nature 1965; 208:1337–8. [DOI] [PubMed] [Google Scholar]
- 3. Mitchell GF, Miller JF. Cell to cell interaction in the immune response. II. The source of hemolysin‐forming cells in irradiated mice given bone marrow and thymus or thoracic duct lymphocytes. J Exp Med 1968; 128:821–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol 1986; 136:2348–57. [PubMed] [Google Scholar]
- 5. Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M et al Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med 2000; 192:1545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim CH, Rott LS, Clark‐Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center‐localized subset of CXCR5+ T cells. J Exp Med 2001; 193:1373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J Exp Med 2000; 192:1553–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garside P, Ingulli E, Merica RR, Johnson JG, Noelle RJ, Jenkins MK. Visualization of specific B and T lymphocyte interactions in the lymph node. Science 1998; 281:96–9. [DOI] [PubMed] [Google Scholar]
- 9. Toellner KM, Gulbranson‐Judge A, Taylor DR, Sze DM, MacLennan IC. Immunoglobulin switch transcript production in vivo related to the site and time of antigen‐specific B cell activation. J Exp Med 1996; 183:2303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van den Eertwegh AJ, Noelle RJ, Roy M, Shepherd DM, Aruffo A, Ledbetter JA et al In vivo CD40‐gp39 interactions are essential for thymus‐dependent humoral immunity. I. In vivo expression of CD40 ligand, cytokines, and antibody production delineates sites of cognate T–B cell interactions. J Exp Med 1993; 178:1555–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Noelle RJ, Roy M, Shepherd DM, Stamenkovic I, Ledbetter JA, Aruffo A. A 39‐kDa protein on activated helper T cells binds CD40 and transduces the signal for cognate activation of B cells. Proc Natl Acad Sci U S A 1992; 89:6550–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T‐bet, directs Th1 lineage commitment. Cell 2000; 100:655–69. [DOI] [PubMed] [Google Scholar]
- 13. Zheng W, Flavell RA. The transcription factor GATA‐3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 1997; 89:587–96. [DOI] [PubMed] [Google Scholar]
- 14. Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, et al Bcl6 and Blimp‐1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 2009; 325:1006–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD et al Bcl6 mediates the development of T follicular helper cells. Science 2009; 325:1001–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL et al The transcriptional repressor Bcl‐6 directs T follicular helper cell lineage commitment. Immunity 2009; 31:457–68. [DOI] [PubMed] [Google Scholar]
- 17. Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS et al T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non‐Th1/Th2 effector cells that provide help for B cells. J Immunol 2004; 173:68–78. [DOI] [PubMed] [Google Scholar]
- 18. Kim CH, Lim HW, Kim JR, Rott L, Hillsamer P, Butcher EC. Unique gene expression program of human germinal center T helper cells. Blood 2004; 104:1952–60. [DOI] [PubMed] [Google Scholar]
- 19. Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity 2014; 41:529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular helper T cells. Annu Rev Immunol 2016; 34:335–68. [DOI] [PubMed] [Google Scholar]
- 21. Baumjohann D, Okada T, Ansel KM. Cutting edge: distinct waves of BCL6 expression during T follicular helper cell development. J Immunol 2011; 187:2089–92. [DOI] [PubMed] [Google Scholar]
- 22. Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, et al ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 2011; 34:932–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choi YS, Yang JA, Crotty S. Dynamic regulation of Bcl6 in follicular helper CD4 T (Tfh) cells. Curr Opin Immunol 2013; 25:366–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl‐6 in TH1 cells to regulate flexibility with a TFH‐like gene profile. Nat Immunol 2012; 13:405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McDonald PW, Read KA, Baker CE, Anderson AE, Powell MD, Ballesteros‐Tato A et al IL‐7 signalling represses Bcl‐6 and the TFH gene program. Nat Commun 2016; 7:10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moon H, Park C, Lee JG, Shin SH, Lee JH, Kho I et al Early development in the peritoneal cavity of CD49dhigh Th1 memory phenotype CD4+ T cells with enhanced B cell helper activity. J Immunol 2015; 195:564–75. [DOI] [PubMed] [Google Scholar]
- 27. Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA et al Early Th1 cell differentiation is marked by a Tfh cell‐like transition. Immunity 2011; 35:919–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu KT, Kanno Y, Cannons JL, Handon R, Bible P, Elkahloun AG et al Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro‐generated and in vivo‐derived follicular T helper cells. Immunity 2011; 35:622–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luthje K, Kallies A, Shimohakamada Y, Belz GT, Light A, Tarlinton DM et al The development and fate of follicular helper T cells defined by an IL‐21 reporter mouse. Nat Immunol 2012; 13:491–8. [DOI] [PubMed] [Google Scholar]
- 30. Ballesteros‐Tato A, Randall TD, Lund FE, Spolski R, Leonard WJ, Leon B. T follicular helper cell plasticity shapes pathogenic T helper 2 cell‐mediated immunity to inhaled house dust mite. Immunity 2016; 44:259–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei G, Wei L, Zhu J, Zang C, Hu‐Li J, Yao Z et al Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 2009; 30:155–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oestreich KJ, Weinmann AS. Encoding stability versus flexibility: lessons learned from examining epigenetics in T helper cell differentiation. Curr Top Microbiol Immunol 2011; 356:145–64. [DOI] [PubMed] [Google Scholar]
- 33. Bollig N, Brustle A, Kellner K, Ackermann W, Abass E, Raifer H et al Transcription factor IRF4 determines germinal center formation through follicular T‐helper cell differentiation. Proc Natl Acad Sci U S A 2012; 109:8664–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choi YS, Eto D, Yang JA, Lao C, Crotty S. Cutting edge: STAT1 is required for IL‐6‐mediated Bcl6 induction for early follicular helper cell differentiation. J Immunol 2013; 190:3049–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U et al The transcription factor BATF controls the global regulators of class‐switch recombination in both B cells and T cells. Nat Immunol 2011; 12:536–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, et al Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol 2012; 188:3734–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F et al Transcription factor achaete‐scute homologue 2 initiates follicular T‐helper‐cell development. Nature 2014; 507:513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma CS, Avery DT, Chan A, Batten M, Bustamante J, Boisson‐Dupuis S et al Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood 2012; 119:3997–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmitt N, Liu Y, Bentebibel SE, Munagala I, Bourdery L, Venuprasad K et al The cytokine TGF‐β co‐opts signaling via STAT3‐STAT4 to promote the differentiation of human TFH cells. Nat Immunol 2014; 15:856–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH et al The costimulatory molecule ICOS regulates the expression of c‐Maf and IL‐21 in the development of follicular T helper cells and TH‐17 cells. Nat Immunol 2009; 10:167–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F et al LEF‐1 and TCF‐1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol 2015; 16:980–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu L, Cao Y, Xie Z, Huang Q, Bai Q, Yang X et al The transcription factor TCF‐1 initiates the differentiation of T(FH) cells during acute viral infection. Nat Immunol 2015; 16:991–9. [DOI] [PubMed] [Google Scholar]
- 43. Wu T, Shin HM, Moseman EA, Ji Y, Huang B, Harly C et al TCF1 is required for the T follicular helper cell response to viral infection. Cell Rep 2015; 12:2099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L et al Generation of T follicular helper cells is mediated by interleukin‐21 but independent of T helper 1, 2, or 17 cell lineages. Immunity 2008; 29:138–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stone EL, Pepper M, Katayama CD, Kerdiles YM, Lai CY, Emslie E et al ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity 2015; 42:239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weber JP, Fuhrmann F, Feist RK, Lahmann A, Al Baz MS, Gentz LJ et al ICOS maintains the T follicular helper cell phenotype by down‐regulating Kruppel‐like factor 2. J Exp Med 2015; 212:217–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Crotty S. Follicular helper CD4 T cells [T(FH)]. Annu Rev Immunol 2011; 29:621–63. [DOI] [PubMed] [Google Scholar]
- 48. Hunter CA, Jones SA. IL‐6 as a keystone cytokine in health and disease. Nat Immunol 2015; 16:448–57. [DOI] [PubMed] [Google Scholar]
- 49. Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R et al IL‐21 and IL‐6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS One 2011; 6:e17739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karnowski A, Chevrier S, Belz GT, Mount A, Emslie D, D'Costa K et al B and T cells collaborate in antiviral responses via IL‐6, IL‐21, and transcriptional activator and coactivator, Oct2 and OBF‐1. J Exp Med 2012; 209:2049–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harker JA, Lewis GM, Mack L, Zuniga EI. Late interleukin‐6 escalates T follicular helper cell responses and controls a chronic viral infection. Science 2011; 334:825–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC et al CD4 T follicular helper cell dynamics during SIV infection. J Clin Invest 2012; 122:3281–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chavele KM, Merry E, Ehrenstein MR. Cutting edge: circulating plasmablasts induce the differentiation of human T follicular helper cells via IL‐6 production. J Immunol 2015; 194:2482–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Leonard WJ, Wan CK. IL‐21 signaling in immunity. F1000Res 2016; 5:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vogelzang A, McGuire HM, Yu D, Sprent J, Mackay CR, King C. A fundamental role for interleukin‐21 in the generation of T follicular helper cells. Immunity 2008; 29:127–37. [DOI] [PubMed] [Google Scholar]
- 56. Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, et al In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol 2010; 185:313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kobayashi M, Fitz L, Ryan M, Hewick RM, Clark SC, Chan S et al Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J Exp Med 1989; 170:827–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O'Garra A, Murphy KM. Development of TH1 CD4+ T cells through IL‐12 produced by Listeria‐induced macrophages. Science 1993; 260:547–9. [DOI] [PubMed] [Google Scholar]
- 59. Manetti R, Parronchi P, Giudizi MG, Piccinni MP, Maggi E, Trinchieri G et al Natural killer cell stimulatory factor (interleukin 12 [IL‐12]) induces T helper type 1 (Th1)‐specific immune responses and inhibits the development of IL‐4‐producing Th cells. J Exp Med 1993; 177:1199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Seder RA, Gazzinelli R, Sher A, Paul WE. Interleukin 12 acts directly on CD4+ T cells to enhance priming for interferon‐γ production and diminishes interleukin 4 inhibition of such priming. Proc Natl Acad Sci U S A 1993; 90:10188–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL‐12 responses and enhanced development of Th2 cells in Stat4‐deficient mice. Nature 1996; 382:174–7. [DOI] [PubMed] [Google Scholar]
- 62. Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT et al Requirement for Stat4 in interleukin‐12‐mediated responses of natural killer and T cells. Nature 1996; 382:171–4. [DOI] [PubMed] [Google Scholar]
- 63. Nishikomori R, Usui T, Wu CY, Morinobu A, O'Shea JJ, Strober W. Activated STAT4 has an essential role in Th1 differentiation and proliferation that is independent of its role in the maintenance of IL‐12Rβ2 chain expression and signaling. J Immunol 2002; 169:4388–98. [DOI] [PubMed] [Google Scholar]
- 64. Park WR, Nakahira M, Sugimoto N, Bian Y, Yashiro‐Ohtani Y, Zhou XY et al A mechanism underlying STAT4‐mediated up‐regulation of IFN‐γ induction inTCR‐triggered T cells. Int Immunol 2004; 16:295–302. [DOI] [PubMed] [Google Scholar]
- 65. Yang Y, Ochando JC, Bromberg JS, Ding Y. Identification of a distant T‐bet enhancer responsive to IL‐12/Stat4 and IFNγ/Stat1 signals. Blood 2007; 110:2494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu X, Sun YL, Hoey T. Cooperative DNA binding and sequence‐selective recognition conferred by the STAT amino‐terminal domain. Science 1996; 273:794–7. [DOI] [PubMed] [Google Scholar]
- 67. Ma CS, Suryani S, Avery DT, Chan A, Nanan R, Santner‐Nanan B et al Early commitment of naive human CD4+ T cells to the T follicular helper [T(FH)] cell lineage is induced by IL‐12. Immunol Cell Biol 2009; 87:590–600. [DOI] [PubMed] [Google Scholar]
- 68. Schmitt N, Morita R, Bourdery L, Bentebibel SE, Zurawski SM, Banchereau J et al Human dendritic cells induce the differentiation of interleukin‐21‐producing T follicular helper‐like cells through interleukin‐12. Immunity 2009; 31:158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schmitt N, Bustamante J, Bourdery L, Bentebibel SE, Boisson‐Dupuis S, Hamlin F et al IL‐12 receptor β1 deficiency alters in vivo T follicular helper cell response in humans. Blood 2013; 121:3375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yoshida H, Hunter CA. The immunobiology of interleukin‐27. Annu Rev Immunol 2015; 33:417–43. [DOI] [PubMed] [Google Scholar]
- 71. Batten M, Ramamoorthi N, Kljavin NM, Ma CS, Cox JH, Dengler HS et al IL‐27 supports germinal center function by enhancing IL‐21 production and the function of T follicular helper cells. J Exp Med 2010; 207:2895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Harker JA, Dolgoter A, Zuniga EI. Cell‐intrinsic IL‐27 and gp130 cytokine receptor signaling regulates virus‐specific CD4+ T cell responses and viral control during chronic infection. Immunity 2013; 39:548–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ballesteros‐Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE et al Interleukin‐2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity 2012; 36:847–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med 2012; 209:243–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nurieva RI, Podd A, Chen Y, Alekseev AM, Yu M, Qi X et al STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J Biol Chem 2012; 287:11234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Locci M, Wu JE, Arumemi F, Mikulski Z, Dahlberg C, Miller AT et al Activin A programs the differentiation of human TFH cells. Nat Immunol 2016; 17:976–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. McCarron MJ, Marie JC. TGF‐β prevents T follicular helper cell accumulation and B cell autoreactivity. J Clin Invest 2014; 124:4375–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nakayamada S, Poholek AC, Lu KT, Takahashi H, Kato M, Iwata S et al Type I IFN induces binding of STAT1 to Bcl6: divergent roles of STAT family transcription factors in the T follicular helper cell genetic program. J Immunol 2014; 192:2156–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ray JP, Marshall HD, Laidlaw BJ, Staron MM, Kaech SM, Craft J. Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity 2014; 40:367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liao W, Lin JX, Leonard WJ. Interleukin‐2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 2013; 38:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Malek TR, Castro I. Interleukin‐2 receptor signaling: at the interface between tolerance and immunity. Immunity 2010; 33:153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the Bcl6 transcription factor and the interleukin‐2 receptor generate T helper 1 central and effector memory cells. Immunity 2011; 35:583–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl‐6 and Blimp‐1 in T and B lymphocyte differentiation. Nat Immunol 2010; 11:114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z et al Interleukin‐2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 2007; 26:371–81. [DOI] [PubMed] [Google Scholar]
- 85. Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL‐2 broadly regulates differentiation into helper T cell lineages. Nat Immunol 2011; 12:551–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Carrette F, Surh CD. IL‐7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin Immunol 2012; 24:209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Liao W, Lin JX, Leonard WJ. IL‐2 family cytokines: new insights into the complex roles of IL‐2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol 2011; 23:598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liu X, Lu H, Chen T, Nallaparaju KC, Yan X, Tanaka S et al Genome‐wide analysis identifies Bcl6‐controlled regulatory networks during T follicular helper cell differentiation. Cell Rep 2016; 14:1735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ichii H, Sakamoto A, Arima M, Hatano M, Kuroda Y, Tokuhisa T. Bcl6 is essential for the generation of long‐term memory CD4+ T cells. Int Immunol 2007; 19:427–33. [DOI] [PubMed] [Google Scholar]
- 90. Ichii H, Sakamoto A, Hatano M, Okada S, Toyama H, Taki S et al Role for Bcl‐6 in the generation and maintenance of memory CD8+ T cells. Nat Immunol 2002; 3:558–63. [DOI] [PubMed] [Google Scholar]
- 91. Basso K, Dalla‐Favera R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv Immunol 2010; 105:193–210. [DOI] [PubMed] [Google Scholar]
- 92. Oestreich KJ, Read KA, Gilbertson SE, Hough KP, McDonald PW, Krishnamoorthy V et al Bcl‐6 directly represses the gene program of the glycolysis pathway. Nat Immunol 2014; 15:957–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Phan RT, Saito M, Basso K, Niu H, Dalla‐Favera R. BCL6 interacts with the transcription factor Miz‐1 to suppress the cyclin‐dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol 2005; 6:1054–60. [DOI] [PubMed] [Google Scholar]
- 94. Rosenberg SA, Yang JC, Topalian SL, Schwartzentruber DJ, Weber JS, Parkinson DR et al Treatment of 283 consecutive patients with metastatic melanoma or renal cell cancer using high‐dose bolus interleukin 2. JAMA 1994; 271:907–13. [PubMed] [Google Scholar]
- 95. Rosenberg SA. Raising the bar: the curative potential of human cancer immunotherapy. Sci Transl Med 2012; 4:127ps8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Tangye SG, Ma CS, Brink R, Deenick EK. The good, the bad and the ugly – TFH cells in human health and disease. Nat Rev Immunol 2013; 13:412–26. [DOI] [PubMed] [Google Scholar]


