Abstract
Antibiotic resistance among pathogenic bacteria is an ever-increasing issue worldwide. Unfortunately, very little has been achieved in the pharmaceutical industry to combat this problem. This has led researchers and the medical field to revisit past drugs that were deemed too toxic for clinical use. In particular, the cyclic cationic peptides polymyxin B and colistin, which are specific for Gram-negative bacteria, have been used as “last resort” antimicrobials. Before the 1980s, these drugs were known for their renal and neural toxicities; however, new clinical practices and possibly improved manufacturing have made them safer to use. Previously suggested to primarily attack the membranes of Gram-negative bacteria and to not easily select for resistant mutants, recent research exploring resistance and mechanisms of action has provided new perspectives. This review focuses primarily on the proposed alternative mechanisms of action, known resistance mechanisms, and how these support the alternative mechanisms of action.
Polymyxins are typically reserved for treating serious Gram-negative bacterial infections. New insights into their mechanisms of action and resistance may lead to more widespread and informed use of this group of antibiotics.
Public news outlets, academic articles (primary literature and reviews), government documents, and countless other sources echo the alarm regarding multiple antibiotic resistance development in nosocomial pathogens (Fernández et al. 2011; CDC 2013; Taylor et al. 2014; Berendonk et al. 2015). Unfortunately, recent antibiotic research has encountered numerous obstacles, notably limited discovery of new antibiotic compounds. This relates to the poor economic incentives for pharmaceutical companies to invest in antibiotic development given pharmacoeconomic considerations (a combination of poor success rates, limited markets because of the large number of different antibiotics on the market, and the fact that antibiotics are generally used acutely and, thus, do not have “repeat customers”) and limited funding for academic laboratories working in the field. Even though occasional breakthroughs are made, for example, the antibiotic teixobactin (Ling et al. 2015), it is noteworthy that while chemically novel, this compound addresses a known target and has yet to be examined clinically. The medical community has recognized the critical importance of combating bacterial resistance, and this has led to the revival of disfavored antibacterials such as the cyclic peptide polymyxin B and its relatives (Falagas and Kasiakou 2006; Landman et al. 2008; Falagas et al. 2010).
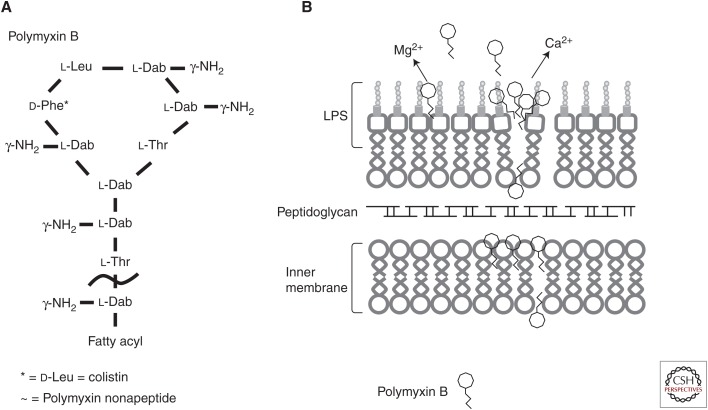
Polymyxin was first isolated in 1947 from the Gram-positive soil bacterium Bacillus polymyxa, which was reclassified as Paenibacillus polymyxa in 1993 (Storm et al. 1977; Ash et al. 1994). Fifteen different molecular variations are known and are produced by P. polymyxa subspecies, including polymyxin E (colistin) from ssp. Colistinus and polymyxin M (mattacin) from Paenibacillus kobensis (Storm et al. 1977; Martin et al. 2003; Choi et al. 2009b; Tambadou et al. 2015). The polymyxins are small lipopeptide molecules of ∼1200 Da in mass and are characterized by a polycationic peptide ring with a short protruding peptide attached to a hydrophobic fatty acid tail (Fig. 1A) (Newton 1956; Evans 1999; Nation et al. 2014). The clinically used polymyxins, polymyxin B (a mixture of composed of polymyxins B3, B6, and minor components B1, B1-I, B2) and colistin (containing two major components colistin A and B and ∼30 minor components) (Orwa et al. 2001), differ primarily by a single d-phenylalanine replaced by a d-leucine within the peptide ring (Landman et al. 2008; Yu et al. 2015). The cationic ring makes these drugs soluble in aqueous environments, whereas the hydrophobic acyl chain facilitates insertion into bacterial membranes (Evans 1999; Nation et al. 2014).
Figure 1.
The structure of polymyxin B and likely mode of membrane interaction. (A) The general structure of the cyclic cationic peptide polymyxin B. Colistin (polymyxin E) replaces the phenylalanine (d-Phe*) in polymyxin B with a leucine. Attenuated polymyxin nonapeptide has the fatty acyl tail removed adjacent to the threonine outside the ring structure (∼). (B) Polymyxin B interacts with the lipid A portion of the lipopolysaccharide (LPS) outer membrane. The peptides cross the outer membrane through a “self-promoted uptake” mechanism and then interact with the cytoplasmic membrane to inhibit cellular energization, and possibly cause inhibition of cell division and/or cytoplasmic membrane permeabilization and subsequent cell death.
All polymyxins are nonribosomally produced (i.e., produced by large enzymes called nonribosomal peptide synthetases) (Finking and Marahiel 2004), and although their structures have been known for years, the mechanisms for their in vivo synthesis are still being elucidated. The peptides are formed in steps by enzyme modules with specific domains that govern adenylation, thiolation (peptidyl carrier protein [PCP]), and condensation (Finking and Marahiel 2004; Choi et al. 2009b). To date, the nonribosomal peptide synthetases have been identified for polymyxin M (Martin et al. 2003), polymyxin A (Choi et al. 2009b), variants of polymyxin B (Shaheen et al. 2011), and polymyxin P (Niu et al. 2013).
All of the polymyxin peptides have similar bactericidal activities with efficacy against Gram-negative organisms and a few Gram-positive species (see section ExPortal) (Newton 1956; Storm et al. 1977; Vega and Caparon 2012). Polymyxin and its relatives were commonly used in clinical treatment until the 1980s, when their nephrotoxicity caused a decline in usage (Falagas and Kasiakou 2006; Landman et al. 2008; Falagas et al. 2010). The nephrotoxicity is drug-related acute tubular necrosis leading to acute renal failure and stems from accumulation and persistence in the body, with preferences for kidney and brain tissues (Kunin and Bugg 1971; Evans 1999). Similar to the drug’s interaction with the bacterial outer membrane, it permeabilizes eukaryotic membranes leading to swelling and lysis (Berg et al. 1996, 1998; Lewis and Lewis 2004; Falagas and Kasiakou 2006). However, recent studies have indicated that the incidence of nephrotoxicity is less common and severe compared with older studies (Berg et al. 1996, 1998; Lewis and Lewis 2004; Falagas and Kasiakou 2006) and, intriguingly, polymyxin B was found to be less nephrotoxic than the prodrug colistimethate in which the positive charges on colistin are neutralized (Phe et al. 2014). In addition, polymyxins can bind to neurons and block the release of acetylcholine, preventing neurotransmission and affecting muscular activity, although neurotoxic effects are usually mild and resolve on discontinuation of therapy (Falagas and Kasiakou 2006).
After usage was reduced because of toxicity concerns, polymyxins were reserved for treatment of persistent infections in cystic fibrosis patients, ophthalmic conjunctivitis infections, and in over-the-counter topical antibiotic ointments (Hancock and Chapple 1999; Hancock 2000; Falagas et al. 2010). The usage of polymyxins has seen a recent upsurge despite issues with toxicity because of increasing resistance to other drugs. Currently, they are the go-to drug for serious multidrug-resistant (MDR) Gram-negative bacterial infections, particularly those caused by MDR Pseudomonas aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae, in which they have become the last-line treatment for infections that are resistant to other available antibiotics (Nation et al. 2015). Polymyxin B is administered directly as an antibiotic, whereas colistin is administered as the prodrug colistin methanesulfonate (in which diaminobenzoate residues are derivatized as methane sulphonates), which is hydrolyzed into colistin in vivo (Zavascki et al. 2007; Landman et al. 2008; Nation et al. 2014). As mentioned above, recent examinations into the reported renal and neural toxicities indicate that the severity and incidences of these effects remain high but not nearly as high as previously suggested (Falagas and Kasiakou 2006; Kwa et al. 2008; Landman et al. 2008). Thus, although renal and neural toxicity remain an adverse side effect, our current handling of the drugs has more positive outcomes (Nation et al. 2015). Correct formulations, dosage (lower than past usage), discontinuation on adverse symptoms, avoidance of coadministration with other potential nephro/neurotoxic dugs, and overall better critical care and, possibly, better manufacturing have led to lower incidences of nephro and neural toxicities (Falagas and Kasiakou 2006). Consensus recommendations for their clinical usage were recently published (Nation et al. 2015).
Disruption and/or permeabilization of Gram-negative bacterial cytoplasmic membrane has been suggested as the main mechanism of action by polymyxin B and colistin (Teuber 1974). However, doubt regarding the singular nature of this mechanism has been raised (Zhang et al. 2000). Because of resurgence of their usage, research into the mechanisms of action and resistance of polymyxin B and colistin has intensified. The purpose of this review is to highlight some of the alternative and less characterized mechanisms of action of polymyxin B and colistin as well as the bacterial resistance to these drugs.
MECHANISMS OF ACTION
Membrane Target and Evidence for Alternative Mechanisms of Action
Since their discovery many years ago, much has been learned regarding the mechanism of action of polymyxins. Newton (1956) originally suggested that the mechanism of polymyxin B involved its “ability to combine with and disorganize structures of the bacterial cell which are responsible for the maintenance of the osmotic equilibrium of the cell.” It is generally accepted that the Gram-negative selectivity is mediated by initial interaction with the outer membrane of Gram-negative bacteria. Early studies showed that Gram-negative bacteria pretreated with polymyxin B were more susceptible to lysozyme treatment, indicating that polymyxin B disrupted the outer membrane, forming visible protrusions, thereby exposing the underlying peptidoglycan layer to lysozyme (Warren et al. 1957; Koike et al. 1969). Additionally, treatment of Escherichia coli with polymyxin B increases susceptibility to β-lactam antibiotics, which target the peptidoglycan synthesis machinery (Rosenthal and Storm 1977). It was subsequently proposed and shown that the cationic charges in the polypeptide portion of polymyxin electrostatically bind to the negatively charged lipopolysaccharide (LPS) that is the predominant (or only) surface lipid of the outer membrane in Gram-negative bacteria, assisted by the interaction of the lipid tail with the fatty acids of the lipid A moiety of the LPS molecule (Fig. 1B). When it binds to phosphate residues of lipid A, polymyxin displaces the membrane-stabilizing magnesium and calcium ions that cross-bridge adjacent lipid A molecules and stabilize the outer membrane (Moore et al. 1986; Evans 1999; Falagas and Kasiakou 2005; Landman et al. 2008; Fernández et al. 2013). By displacing these divalent cations with the bulkier polycationic polymyxin, the membrane becomes weakened and the permeability barrier is disrupted allowing for uptake of previously nonpermeating or weakly permeating molecules and leakage of periplasmic proteins (Hancock 1984). The displacement of divalent cations and permeabilization allows for self-promoted uptake of the polymyxin molecule itself enabling it to penetrate the periplasm and approach the cytoplasmic membrane (Hancock and Bell 1988; Hancock 1997). Polymyxin B nonapeptide (PMBN), lacking the fatty acyl tail, had virtually no antibiotic activity but was still able to compromise the outer membrane (Vaara and Vaara 1983; Daugelavicius et al. 2000; Zhang et al. 2000; Lu et al. 2014), indicating that outer membrane permeabilization could occur because of charge:charge interactions alone, although larger concentrations of this attenuated polymyxin were required for permeabilization, suggesting that the hydrophobic acyl tail promotes outer membrane permeabilization. The lack of activity of PMBN, however, indicates that the outer membrane is a site of interaction, but it is not the killing target. In addition to binding to LPS, a polymyxin B photoprobe was recently shown to bind to unspecified outer membrane proteins (van der Meijden and Robinson 2015). However, for killing to occur, according to the membrane target model, the cytoplasmic membrane must be compromised, whereas other studies have suggested alternative membrane-associated or cytoplasmic targets (Zhang et al. 2000).
The membrane-active mechanism of action for polymyxins has been well studied, but it is unclear how polymyxin interacts with and disrupts the cytoplasmic membrane (Landman et al. 2008; Falagas et al. 2010; Yu et al. 2015). Early experiments showed that high concentrations of polymyxin caused the release of cytoplasmic material (Cerny and Teuber 1971; Schindler and Teuber 1975; Dixon and Chopra 1986; Landman et al. 2008). It was shown that polymyxin B caused lipid exchange between outer and inner membrane leaflet binding in experiments with simulated Gram-negative membrane (Clausell et al. 2007). Similarly, Berglund et al. (2015) used computer modeling to propose that polymyxin B initially aggregates at the LPS surface by burying their acyl tails into micelle/pore-like structures. Conversely, polymyxin B did not form aggregates in the inner membrane model, suggesting that the membrane interactions are dependent on the membrane architecture of the inner and outer membranes in Gram-negative bacteria.
Despite this evidence and overall assumption for the membrane as the sole target for the polymyxins, there is evidence for alternative mechanisms of action. An early study investigating how different environmental ions affected polymyxin killing of P. aeruginosa (Klemperer et al. 1979) showed that lysis is not required for cell death. In P. aeruginosa, at concentrations greater than the minimum inhibitory concentration (MIC), polymyxin led to substantial cell death while only marginally increasing cytoplasmic membrane permeability, as assessed by reduction in the transmembrane potential gradient (Zhang et al. 2000). Similarly, the cytoplasmic membrane of polymyxin-treated E. coli was permeabilized, as determined by the measured release of ions, only at concentrations well above the minimal bactericidal range (Daugelavicius et al. 2000). This high cell death at concentrations showing little increase in cytoplasmic membrane permeability indicates that alternative or additional mechanisms of action are likely to contribute to the antibacterial activity of polymyxins. Studies with a modified, fluorescently labeled polymyxin B that retained activity showed that it coalesced at the outer membrane before penetration into the periplasm, cytoplasmic membrane, and, finally, the cytoplasm in K. pneumoniae (Deris et al. 2014), indicating a plethora of potential sites of action. Some of the alternative mechanisms of activity that have been proposed are described below.
Ribosome Binding
In early investigations of the mechanism of action of polymyxins and colistin, it was discovered that polymyxin could precipitate E. coli ribosomes (Nakajima and Kawamata 1966; Teuber 1967), similar to other polycationic antibiotics. Despite these seminal papers, there was no follow-up for ∼50 years. Recently, McCoy et al. (2013) reestablished this link by examining the relationship of polymyxin to aminoglycosides, which are cationic saccharide bactericidal antibiotics that also bind to ribosomes. They posited that the shared cationic nature, nephrotoxicity, and self-promoted uptake (Hancock 1997) needed further exploration. Specifically, they examined the ability of polymyxin to bind to bacterial 16S ribosomal RNA using a modified fluorescence resonance energy transfer assay. Polymyxin showed moderate binding to the 16S A-site of E. coli ribosomes, 10 times weaker than kanamycin yet 10 times stronger than the negative control, an inactive colistin derivative with sulfomethylated (charge-neutralized) amino groups. Intriguingly, an analog of polymyxin, with a biphenyl acyl tail and a d-octylglycine at the phenylalanine position, showed similar binding affinity to kanamycin. Also, PMBN showed two times better binding than intact polymyxin B. This is interesting because, in vivo, the nonapeptide apparently cannot pass the outer membrane (Vaara and Vaara 1983). Despite these abilities to bind to the ribosome, bacterial translation was not affected; however, in contrast, eukaryotic translation was negatively affected. Nevertheless, we deduce that ribosome binding is not an alternative or additional mechanism because there were apparently no obvious consequences of ribosome binding on prokaryotic translation nor evidence that polymyxin penetrates sufficiently into the cytoplasm to inhibit the large number of ribosomes in bacterial cells.
Bacterial Respiration
Strictly speaking, membrane permeabilization has many cellular consequences beyond a compromised cell structure or lysis. Indeed, disruption of cytoplasmic membrane integrity can impact cellular energetics, cell division, and cell wall biosynthesis. Bacterial respiration, for instance, requires an intact membrane to function properly (Storm et al. 1977). Consistent with this concept, sublethal concentrations of polymyxin B inhibited oxygen consumption in A. baumannii without penetrating the cytoplasmic membrane (Saugar et al. 2002).
Despite intrinsic resistance of Gram-positive bacteria to polymyxin, Tochikubo et al. (1986) examined nicotinamide adenine dinucleotide (NADH) oxidase activity following germination of Bacillus subtilis spores. They determined that polymyxin B inhibited NADH oxidase activity as well as NADH cytochrome c reductase activity, but not NADH dehydrogenase activity. It was proposed that the spore pericortex prevents direct interaction with the inner membrane, but somehow the activity of polymyxin still inhibited respiration (Tochikubo et al. 1986). Another group has shown similar effects on bacterial respiration machinery as polymyxin B inhibited the activity of the alternative NADH dehydrogenase and the malate: quinone oxidoreductase in Mycobacterium smegmatis (Mogi et al. 2009).
A screen for polymyxin B–resistant mutants in the susceptible Gram-positive Streptococcus pyogenes, identified the fatty acid biosynthesis master regulator, fabT (Dalebroux and Swanson 2012; Port et al. 2014). Fatty acid biosynthesis is known to activate the stringent response. Similarly, polymyxin treated cells show an increase in ppGpp, but not pppGpp (Cortay and Cozzone 1983b; de la Fuente-Núñez et al. 2014). This increase was independent of RelA, which is the normal producer of (p)ppGpp (Cortay and Cozzone 1983a). It was suspected that the increase in ppGpp was because of a lack of cellular ATP powering the enzymatic degradation by SpoT (Cortay and Cozzone 1983b). Indeed, inhibition of respiration by polymyxin leads to a decrease in the ATP pool of the cell (Storm et al. 1977). These reports provide compelling evidence that respiration inhibition is a target of polymyxin beyond membrane disruption. Similarly, other bactericidal cyclic cationic peptides are known to permeabilize the outer membrane and cause inhibition of respiration at their minimal effective doses (Skerlavaj et al. 1990; Wu and Hancock 1999; Spindler et al. 2011). Nevertheless, inhibition of respiration has not been shown to be a bactericidal target so it is likely that it contributes to, rather than causes, cell killing.
Cell Division
Bacterial cell division is an essential process that depends on cytoplasmic membrane–associated machinery and the membrane potential (Strahl and Hamoen 2010). For example, it is well established that many third-generation cephalosporin antibiotics act directly on penicillin binding protein 3 to inhibit correct cell division leading the formation of long filamentous bodies in which individual cells have grown but not separated. A recent paper using atomic force microscopy (AFM) on colistin-treated P. aeruginosa provided evidence that colistin also affects cells division (Mortensen et al. 2009). A significant decrease in dividing cells was observed when cells were treated with sublethal concentrations of colistin in addition to a slight decrease in colony-forming units, but no significant killing (Mortensen et al. 2009). The investigators also observed an increase in cellular rigidity and speculated that colistin binding to peptidoglycan stiffens the bacterial cell wall and speculated that this prevented proper cell division (Mortensen et al. 2009). This conclusion, however, contrasts to another AFM study that showed reduced membrane integrity (Lu et al. 2014). Thus, an alternative possibility might be that polymyxin inserts into the cytoplasmic membrane and interferes with the cell division machinery. A cell division defect was also reported in Mycobacterium aurum in which sub-MIC doses of colistin reversibly arrested cell division without killing cells (David and Rastogi 1985). It was proposed that growth arrest arose because of membrane perturbation that disrupted genome attachment and inhibited DNA replication (David and Rastogi 1985), a process that has also been postulated to explain the bactericidal activity of the cationic aminoglycosides (Hancock 1981).
Other cationic amphipathic peptides have also been proposed to affect cell division. The cationic antimicrobial peptide indolicidin has been shown to induce filamentation in E. coli (Subbalakshmi and Sitaram 1998), although the investigators proposed that filamentation is a secondary effect because of inhibition of DNA synthesis by indolicidin. Conversely, it was shown that the mouse cathelicidin antimicrobial peptide (CRAMP) caused filamentation in B. subtilis by inhibiting FtsZ-mediated ring formation, which is a critical step in cell division (Handler et al. 2008).
At this stage, we cannot conclude that cell division is a direct target of the polymyxins. However, cell division is an extremely complex process, with many moving parts, and much of the cell division machinery lies within the inner membrane (Margolin 2005; Egan and Vollmer 2015; Gray et al. 2015). Therefore, it is important to further examine whether polymyxin may directly (e.g., through FtsZ) or indirectly (through antagonism of energetics) affect cell division as part of its overall mechanism of action.
ExPortal
The precise mechanism for Gram-positive bacteria resistance to polymyxin and colistin is largely unknown (Storm et al. 1977). It is believed that the large peptidoglycan layer prevents interaction with the cytoplasmic membrane, because B. subtilis and Staphylococcus aureus protoplasts are susceptible to polymyxin (LaPorte et al. 1977; Xiong et al. 2005). Interestingly, there are a few noted instances of Gram-positives showing polymyxin susceptibility. For example, S. pyogenes is susceptible to polymyxin B, likely because of the absence of the mprF gene, which encodes an enzyme that modifies the negative charge of phosphatidylglycerol on its capsular polysaccharide (Vega and Caparon 2012). It was observed that polymyxin B targeted the ExPortal structure of S. pyogenes (Vega and Caparon 2012). The ExPortal is a microdomain for the Sec secretion system (Rosch and Caparon 2005). Each cell has only one ExPortal found adjacent to the future division site (Rosch and Caparon 2005). Sublethal concentrations of polymyxin B disrupted the lipid structure around the ExPortal and impeded the secretion of the cysteine protease, SpeB, and the streptolysin O cytolysin (Rosch et al. 2007; Vega and Caparon 2012). Interestingly, despite the disruption in the lipids by sublethal doses, membrane integrity was not compromised (Vega and Caparon 2012), supporting the idea of an alternative mechanism of action different from direct membrane disruption. It was hypothesized that the defect in the secretion mechanism and/or another unknown internal target leads to the demise of the cells (Vega and Caparon 2012).
Reactive Oxygen Species
Recently, a highly controversial mode of bacterial cell death by bactericidal antibiotics was proposed. Some groups suggested that these antibiotics, including polymyxin B, colistin, and kanamycin kill through oxidative stress and reactive oxygen species (ROS) generation (Dwyer et al. 2007, 2009; Brochmann et al. 2014; Dong et al. 2015). However, these overall conclusions regarding this proposed common mechanism were disputed by several investigators, in particular, by showing that antibiotic killing was identical under aerobic and anaerobic conditions indicating independence of oxygen and hydroxyl radicals (Kindrachuk et al. 2011; Keren et al. 2013; Liu and Imlay 2013; Paulander et al. 2014). Similarly, ROS independent killing by colistin has been shown (Brochmann et al. 2014). Conversely, Dong et al. (2015) showed that polymyxin B induced transcription in E. coli of the oxidative stress response gene soxS, and caused an increase in ROS, although they did not imply that this explained the mechanism of killing. A variety of reports have discussed potential mechanisms of resistance mediated by oxidative enzymes but alternative explanations should be sought (Antonic et al. 2013; Heindorf et al. 2014; Pournaras et al. 2014). Despite all the studies and evidence for mechanisms of killing, clearly the exact mechanism of polymyxin action remains to be determined.
POLYMYXIN RESISTANCE
It has been observed that, compared with many other antibiotics, resistance to polymyxins is difficult to attain in the laboratory. Because polymyxins are being used as a last line of defense in treating MDR bacterial infections, it is of considerable interest to examine mechanisms of resistance that have arisen in the clinic or laboratory (Falagas et al. 2010; Fernández et al. 2011; Olaitan et al. 2014). Known resistance mechanisms include intrinsic, mutational, and adaptive, although there are no known horizontally acquired resistance mechanisms. The majority of known mechanisms address the first point of attack by polymyxin, namely, the outer membrane LPS. Overall resistance mechanisms include alterations to reduce the net negative charge or fluidity of LPS, increase in drug efflux, reduced porin pathway, capsule formation, and hypervesiculation. For brevity, this section will focus on the most studied mechanisms of resistance and ones related to alternative mechanisms of action.
LPS–Lipid A Modifications
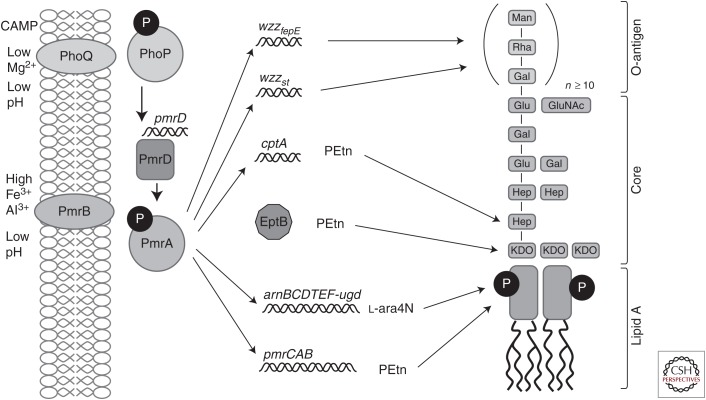
In most Gram-negative bacteria, the two-component PhoP/Q and PmrA/B (response regulator/sensor kinase) regulatory systems mediate polymyxin resistance by governing the mechanisms that modify LPS (Fig. 2; Table 1). These systems regulate cationic antimicrobial resistance in response to low environmental Mg2+ and Ca2+ as well as cationic antimicrobial peptides (CAMPs) and other inducers, such as low pH, excess Fe3+, excess Al3+, and macrophage phagosomes (Brown and Melling 1969; McPhee et al. 2003; Nishino et al. 2006; Prost et al. 2007; Gunn 2008; Gooderham et al. 2009). In Salmonella enterica serovar Typhimurium, PhoP activates the expression of pmrD whose product was shown to activate the PmrA/B system (Kox et al. 2000; Kato et al. 2003), although PmrD is absent in P. aeruginosa and instead PhoP directly interacts with the pmrAB promoter while the two response regulators PhoP and PmrA bind to and maybe compete for binding to the same promoter site(s) on the LPS modification operon. Other two-component systems like ParR/S, CprR/S, and ColR/S in P. aeruginosa have been also implicated in the regulation of lipid A modification enzymes (McPhee et al. 2003; Moon and Gottesman 2009; Fernández et al. 2010; Muller et al. 2011; Fernández et al. 2012; Gutu et al. 2013). In addition, P. aeruginosa has a two-component sensor kinase cbrA that responds to carbon source/catabolite repression and adaptively affects polymyxin B resistance by altering the transcriptional expression of the oprH-phoPQ operon, the pmrAB operon, and the LPS modification arn operon (Yeung et al. 2011). The most likely physiological adaptive resistance mechanism is because of the direct response of the sensor kinases of two component regulators to the presence of peptides, both natural host defense (antimicrobial) peptides and polymyxins, which interact directly to activate these sensor kinases (Bader et al. 2005). In Salmonella, the peptide-specific sensor kinase is PhoQ, whereas in Pseudomonas, a pair of sensor kinases, ParS and CprS, respond to polymyxin and/or peptides (Fernández et al. 2010, 2012). Thus, adaptive resistance can be a result of both self-induced resistance during polymyxin therapy and the antimicrobial peptide component of host defense responses to infections.
Figure 2.
General mechanisms of resistance to polymyxin by lipopolysaccharide (LPS) modifications in Salmonella (mechanisms differ in Pseudomonas). The two-component system PhoQP, activates expression of pmrD. PmrD activates PmrA, which activates cptA and the pmr and arn operons. Along with EptB, CptA modifies the LPS core. The products pmr and arn substitute the phosphates the lipid A portion of the LPS for PEtn and l-Ara4N, respectively. Collectively, the net charge of the outer membrane changes, resulting in repulsion of polymyxin.
Table 1.
Lipopolysaccharide (LPS) modifications leading to polymyxin resistance or supersusceptibility
| Modification | Function | Genesa | Bacterial spp. | References |
|---|---|---|---|---|
| l-Ara4N or PEtn modification of lipid A | Two-component system (TCS) kinase/response regulator (K/RR) | phoP/phoQ | E. coli | Moon and Gottesman 2009 Gunn et al. 1998 Macfarlane et al. 2000 |
| S. Typhimurium | ||||
| P. aeruginosa | ||||
| pmrA/pmrB |
P. aeruginosa S. Typhimurium A. baumannii K. pneumoniae E. coli |
McPhee et al. 2003 Gunn et al. 1998 Arroyo et al. 2011 Choi and Ko 2014 Trent et al. 2001 |
||
| parR/parS | P. aeruginosa | Fernández et al. 2010 | ||
| cprR/cprS | P. aeruginosa | Fernández et al. 2012 | ||
| colR/colS | P. aeruginosa | Gutu et al. 2013 | ||
| Rcs system | S. Typhimurium | Mouslim and Groisman 2003 | ||
| cbrAB | P. aeruginosa | Yeung et al. 2011 | ||
| Proposed response regulator | rppA | P. mirabilis | Wang et al. 2008 | |
| Activates pmrAB | pmrD | S. Typhimurium | Kato et al. 2003 | |
| UDP-glucose dehydrogenase | pmrE (pagA or ugd) |
P. mirabilis S. Typhimurium |
Jiang et al. 2010a Mouslim and Groisman 2003 Gunn et al. 1998 |
|
|
arnBCADTEF (pmrHFIJKLMpbgP) |
P. aeruginosa E. coli S. Typhimurium P. mirabilis Y. pseudotuberculosis K. pneumoniae |
Muller et al. 2011 Yan et al. 2007 Gunn et al. 2000 Lee et al. 2004 Jiang et al. 2010b Marceau et al. 2004 Cheng et al. 2010 |
||
| l-Ara4N transferase |
arnT (pmrK) (pqaB) |
E. coli S. Typhimurium S. typhi |
Subashchandrabose et al. 2013 Trent et al. 2001 Baker et al. 1999 |
|
| Mannosyltransferase | pbgE1 | P. luminescens | Bennett and Clarke 2005 | |
| UDP-glucose dehydrogenase | ugdBCAL2946 | B. cenocepacia | Loutet et al. 2009 | |
| Ugd phosphorylation | etk | E. coli | Lacour et al. 2008 | |
| Oxidoreductase | DsbA3, DsbA2, and DsbA3 | N. meningitidis | Piek et al. 2014 | |
| PEtn modification of LOS/LPS core | TCS (kinase/response regulator) | misS/misR | N. meningitidis | Tzeng et al. 2004 |
| Transferase | eptB |
E. coli S. Typhimurium |
Reynolds et al. 2005 Gibbons et al. 2008 |
|
| PEtn transferase | pmrC (eptA and lptA) |
S. Typhimurium A. baumannii E. coli |
Lee et al. 2004 Arroyo et al. 2011 Herrera et al. 2010 |
|
| Phosphotransferase | cptA | S. Typhimurium | Tamayo et al. 2005a | |
| sRNA | mgrR | E. coli | Moon and Gottesman 2009 | |
| Transferase (also affects PEtn modification of flagella and glycans | eptC | C. jejuni | Cullen et al. 2012, 2013; Scott et al. 2012 | |
| Deacylation of lipid A | Deacetylase | naxD | F. tularensis | Llewellyn et al. 2012 |
| Deacetylase | pagL | S. enterica | Kawasaki et al. 2007 | |
| Deacetylase | yfbH (pmrJ) | E. coli | Subashchandrabose et al. 2013 | |
| sRNA | lpxR | S. Typhimurium | Reynolds et al. 2006 | |
| Repressor of phoPQ expression | Transmembrane regulator | mgrB (yobG) | K. pneumoniae | Lopez-Camacho et al. 2014 |
| MerR-like regulator | brlR | P. aeruginosa | Chambers and Sauer 2013 | |
| sRNA | micA | E. coli | Coornaert et al. 2010 | |
| l-Lys modification of cell membrane phosphatidylglycerol | Transferase | mprF (lpiA) | S. aureus, E. faecalis, and R. tropici | Yang et al. 2012 Bao et al. 2012 Sohlenkamp et al. 2007 |
| Lysinylation | lysX | M. tuberculosis | Maloney et al. 2009 | |
| Phosphorylation of lipid A | Phosphotransferase | lpxT |
S. Typhimurium E. coli |
Herrera et al. 2010 Touze et al. 2008 |
| waaP (rfaP) |
E. coli S. Typhimurium P. aeruginosa |
Yethon et al. 2000 Tsai et al. 2012 Delucia et al. 2011 |
||
| Dephosphorylation of lipid A | Phosphatase | lpxE and lpxF | H. pylori, F. novicida, and R. etli | Tran et al. 2006 Wang et al. 2006 Ingram et al. 2010 |
| Phosphatase | PG1587 and PG1773 | P. gingivalis | Coats et al. 2009 | |
| Transcriptional activator | slyA |
D. dadantii S. Typhimurium |
Haque et al. 2009 Shi et al. 2004b |
|
| ugtL | S. Typhimurium | Shi et al. 2004a,b | ||
| Glycylation of lipid A | TCS (K/RR) | vprA/vprB | V. cholerae | Herrera et al. 2014 |
| TCS (K/RR) | carR/carS | V. cholerae | Bilecen et al. 2015 | |
| almEFG | V. cholerae | Hankins et al. 2012 | ||
| Addition of amide-linked acyl chains in lipid A | Dehydrogenase and transaminase | gnnA and gnnB | C. jejuni | van Mourik et al. 2010 |
| Glucosamine modification | lgmABC | B. pertussis | Shah et al. 2014 | |
| pgaABCD |
E. coli A. baumannii |
Amini et al. 2009 Choi et al. 2009a |
aFor brevity, not all genes are mentioned in the text. Gene contained within parentheses are alternative names or homologous genes.
The output targets of the two component systems are diverse, but commonly make modifications to the phosphate groups of lipid A in LPS to amine substituents, such as 4-amino-4-deoxy-l-arabinose (l-Ara4N), or phosphoethanolamine (PEtn). The addition of l-Ara4N or PEtn reduces the net negative charge of the bacterial surface and decreases the electrostatic interaction with polycationic peptides, such as the polymyxins, thus inhibiting self-promoted uptake. The arnBCADTEF operon, activated by ParR/S and PmrA/B in P. aeruginosa and Salmonella, etc. (Subashchandrabose et al. 2013) adds an l-Ara4N modification to lipid A (Aguirre et al. 2000; Gunn et al. 2000; Tamayo et al. 2005b; Raetz et al. 2007; Barrow and Kwon 2009; Gooderham et al. 2009; Fernández et al. 2010).
Conversely, in Salmonella pmrC (also named eptA and lptA), which is also activated by PmrA, is involved in PEtn modification of lipid A (Lee et al. 2004). Similarly, the pmrC gene encodes a phosphoethanolamine transferase and modifies LPS with PEtn in A. baumannii (Arroyo et al. 2011). The LPS core may also be modified with PEtn by the eptB (analogous to pmrC) and cptA gene products. EptB regulates PEtn addition to 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) in E. coli (Reynolds et al. 2005) and CptA in Salmonella modifies phosphorylated heptose-I residue of the LPS core (Tamayo et al. 2005a). Paradoxically, activation of PhoP/Q system activates the sRNA mgrR, which is a repressor of eptB (Moon and Gottesman 2009). Various oxidoreductases, such as DsbA1, DsbA2, and DsbA3, contribute to PEtn modifications of lipid A in Neisseria meningitidis. Specifically, DsbA3 was shown to be important for the activity of LptA, a PEtn transferase (Piek et al. 2014).
Independent of the PhoP/Q and PmrA/B, the Rcs system (RcsC–YojN–RcsB) in S. enterica has been shown to regulate the expression of ugd, which encodes a UDP-glucose dehydrogenase, an enzyme important in the synthesis of l-Ara4N (Mouslim and Groisman 2003). Also the tyrosine-kinase etk in E. coli phosphorylates Ugd independently of the two-component systems (Lacour et al. 2008).
In addition to l-Ara4N or PEtN modifications, deacylation of lipid A occurs to decrease the fluidity of LPS and, thus, the ability of cationic peptides to partition into the outer membrane leading to self-promoted uptake (Olaitan et al. 2014). In Salmonella, the activation of PhoP/Q system activates pagL, the product of which deacylates lipid A (Kawasaki et al. 2007). Conversely, the naxD gene in Francisella tularensis has also been shown to be responsible for deacylation of lipid A (Llewellyn et al. 2012). Further, PmrA-activated genes include lpxR, which is associated with deacylation of lipid A and is repressed by the sRNA micF (Papenfort et al. 2006; Reynolds et al. 2006; Corcoran et al. 2012). Additional modifications to lipid A include phosphorylation, dephosphorylation, glycylation, and glucosylation. It has also been shown that misR mutants in N. meningitidis lose all of their lipooligosaccharide (LOS) core HepII PEtn decorations leading to an increase in polymyxin B sensitivity (Tzeng et al. 2004). Similar to LPS, LOS is essential for maintaining the integrity of the outer membrane and provides protection from antimicrobial peptides. The waaP (rfaP) gene product is also involved in LPS core modification by phosphorylation of lipid A and was required for polymyxin resistance in S. enterica and P. aeruginosa (Yethon et al. 2000; Delucia et al. 2011; Tsai et al. 2012).
Although it is more difficult to obtain polymyxin resistance than resistance to other antibiotics, the increased use of polymyxin has started to lead to clinical resistance. Mutations in phoQ genes have been identified in colistin-resistant clinical isolates of P. aeruginosa and K. pneumoniae (Choi and Ko 2014; Olaitan et al. 2014). Further mutations in the pmrA or pmrB genes resulted in activation of arnB (pmrH) or other PmrA-activated loci that are required for polymyxin resistance (Sun et al. 2009). Mutations in arnT (also known as pmrK and pqaB) have been shown to affect resistance to polymyxin B in E. coli and S. enterica (Trent et al. 2001). Conversely, S. enterica and Yersinia pseudotuberculosis mutants in the pmrF gene, which is necessary for l-Ara4N substitution on bactoprenol phosphate, displayed an increased susceptibility to polymyxin B (Gunn et al. 2000; Marceau et al. 2004). The most exotic polymyxin resistance mutation is the complete loss of LPS by clinical isolates of A. baumannii (Moffatt et al. 2010, 2011), although many polymyxin resistant isolates in this bacterium have pmrB mutations influencing expression of the upstream pmrC gene (Arroyo et al. 2011). Another interesting clinical mutant was in the parR gene (one of the polymyxin receptors of Pseudomonas), which influenced both inducible and constitutive multidrug resistance to four different classes of antibiotics through the activation of three distinct mechanisms (efflux, porin loss, and LPS modification) (Muller et al. 2011).
The tolA gene is involved in LPS production and this gene has been shown to be necessary for polymyxin B resistance in S. typhimurium (Paterson et al. 2009). Further, LPS modification loci—wzzst and wzzfep activated by PmrA/P, wbaP (rfbP), waaG (rfaG), waaI (rfaI), rfaH, waaJ (rfaJ), waaL (rfaL), or wzy (rfc)—are involved in synthesis of the LPS rough core in S. enterica and appear to be required for polymyxin B resistance, likely because of effects on phosphorylation and/or acylation of LPS (Delgado et al. 2006; Holzer et al. 2009; Ilg et al. 2009; Kong et al. 2011; Pescaretti et al. 2011). Similar mutations in waaL and rfbA (rffH) contribute to resistance to colistin in K. pneumoniae (Sassera et al. 2014). Similarly, in E. coli, five lipid A–modifying genes, ais (b2252), b2253 (yfbE, orf1), b2254 (yfbF, orf2), b2256 (yfbH, orf4), and crcA were associated with antimicrobial peptide resistance (Kruse et al. 2009). Overexpression of K. pneumoniae RamA, which regulates the lpxC, lpxL-2, and lpxO genes associated with lipid A biosynthesis, resulted in increased polymyxin B resistance because of LPS alterations (De Majumdar et al. 2015). An LPS synthesis gene encoding the glycosyltransferase, lpsB, has also been implicated in colistin resistance in A. baumannii (Hood et al. 2013). In addition, the S. typhimurium PhoP-activated gene, mgtA, is also required for resistance to the cationic antibiotic polymyxin B. MgtA controls modification of phosphate residues in the lipid A portion of the LPS (Park and Groisman 2014).
Stress Responses
Biofilm formation is thought to be a response to stress (de la Fuente-Núñez et al. 2014). The P. aeruginosa genes psrA and cbrA, and V. cholerae carR, regulate biofilm formation and polymyxin B resistance (Gooderham et al. 2008; Bilecen et al. 2015). Interestingly, psrA responds to cationic peptides, selected antibiotics (Gooderham et al. 2008) as well as an increase in fatty acids and is also an activator of the stationary phase σ factor, RpoS (Kang et al. 2009). As mentioned above, mutations in FabT, the fatty acid biosynthesis master regulator of S. pyogenes, led to an increase in fatty acids and polymyxin resistance (Port et al. 2014). The increase in fatty acids may activate RpoS through PsrA, which in turn activates the stringent response, biofilm formation, and resistance. Additionally, PhoP/Q has been shown to stabilize RpoS in S. enterica (Tu et al. 2006). Moreover, biofilm genes of P. aeruginosa have been correlated with increased resistance toward colistin because of the complex architecture of biofilms (Folkesson et al. 2008). Interestingly, it was shown that P. aeruginosa biofilm cells with high metabolic activity are more resistant to colistin through up-regulation of the arn operon (Pamp et al. 2008); in principle, this could be mediated through either PsrA or the carbon stress triggered regulator CbrA.
σ Factor RpoE in Burkholderia cenocepacia and other pathogens has been shown to contribute to polymyxin B resistance, likely through the regulation of genes controlling envelope and oxidative stress responses (Sikora et al. 2009; Loutet and Valvano 2011). Moreover, stationary phase σ factor RpoS regulon genes (katE, osmY, wrbA, yciF) are required for polymyxin resistance in E. coli and S. enterica (Oh et al. 1998, 2000; Bader et al. 2003). In S. enterica, an rpoN (nitrogen responsive σ factor σ54) mutant increased polymyxin resistance. It was suggested that rpoN induced resistance to polymyxin via regulation of PTS transporters (Barchiesi et al. 2009).
Polymyxin Efflux
Multidrug efflux pumps play an important role in antibiotic and polymyxin resistance in bacteria but have only been shown to influence polymyxin resistance in a small number of instances. In different organisms, the MtrC–MtrD–MtrE, RosAB, AcrAB–TolC, NorM, KpnEF, and VexAB efflux pumps have been described to confer tolerance toward polymyxin B (Bengoechea and Skurnik 2000; Fehlner-Gardiner and Valvano 2002; Tzeng et al. 2005; Bina et al. 2008; Padilla et al. 2010; Warner and Levy 2010; Tsai et al. 2012; Srinivasan et al. 2014). Further, sap has been implicated in polymyxin B resistance in E. coli because of its homology with ABC effluxers that function against antibiotics (Subashchandrabose et al. 2013). The marRAB operon (activator of efflux) was associated with polymyxin B resistance in E. coli through interactions with Rob and up-regulation of the AcrAB–TolC efflux pump (Warner and Levy 2010). Similarly, overexpression of RamA in K. pneumoniae resulted in increased polymyxin B resistance by mechanisms, including modulation of efflux pump genes such as acrAB, oqxAB, and yrbB-F (De Majumdar et al. 2015). Mutation of the outer membrane porins, PorB and OmpU, conferred resistance to polymyxin B in N. meningitidis and V. cholerae, suggesting a potential role in uptake, perhaps by influencing an active efflux process (Mathur and Waldor 2004; Tzeng et al. 2005).
Plasmid-Mediated Resistance
Until now, polymyxin and colistin resistance was thought to be strictly limited to chromosomal mutations. Very recently, Liu et al. (2016) noted an increase of colistin resistance among food-borne commensal E. coli in China. They identified the resistance gene and defined it as mcr-1. The gene product, MCR-1, was found to share sequence similarities to phosphoethanolamine transferases suggesting resistance by modifying the phosphate groups of lipid A in LPS. An expanded search identified the mcr-1 gene in E. coli and K. pneumoniae from both animal and human isolates. The initial publication of this work online immediately drew the attention of research groups worldwide initiating searches for the mcr-1 gene in public and local databases. By the time this initial report was published, other instances of mcr-1 in Gram-negative bacteria had been found in North America, Europe, South America, and Africa (Arcilla et al. 2016; Hu et al. 2016; Olaitan et al. 2016; Tse and Yuen 2016; Webb et al. 2016). The potential for widespread uncontrolled transmission is undeniable, and every precaution should be made to prevent further dissemination of resistance.
CONCLUDING REMARKS
The cyclic cationic peptides polymyxin B and colistin are potent bactericidal agents against Gram-negative bacteria. Despite their traditional record of neural and renal toxicities, they have performed well as the last line of defense against persistent MDR infections. Although neural and renal toxicities still occur, physicians are more knowledgeable and better equipped to manage toxicity (Falagas and Kasiakou 2006), which has led to a resurgence in the interest and use of this class of antibiotics to combat serious infections.
As discussed in this review, the widely accepted mode of action of polymyxins involves targeting the membranes of Gram-negative bacteria, leading to permeabilized outer and inner membranes and resulting in lysis and cell death. However, there are substantial reasons to doubt this mechanism of action as outlined here. Conversely, groups have explored the ability of polymyxins to bind ribosomes, prevent cell division, and inhibit bacterial respiration. It is hoped that a more profound understanding of the mechanisms of action and resistance will improve our ability to design and develop more potent and less toxic derivatives of polymyxin and colistin. Indeed, some groups are exploring these novel polymyxins (Katsuma et al. 2009; Velkov et al. 2014) and leading the charge in combating bacterial resistance.
ACKNOWLEDGMENTS
We thank Dr. Evan Haney for critical reading of the manuscript. We also recognize funding support No. CZ.1.07/2.3.00/30.0004 to P.M. and funding from the Canadian Institutes for Health Research and Cystic Fibrosis Canada to R.E.W.H., who was also supported by a Canada Research Chair in Health and Genomics.
Footnotes
Editors: Lynn L. Silver and Karen Bush
Additional Perspectives on Antibiotics and Antibiotic Resistance available at www.perspectivesinmedicine.org
REFERENCES
- Aguirre A, Lejona S, Vescovi EG, Soncini FC. 2000. Phosphorylated PmrA interacts with the promoter region of ugd in Salmonella enterica serovar Typhimurium. J Bacteriol 182: 3874–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amini S, Goodarzi H, Tavazoie S. 2009. Genetic dissection of an exogenously induced biofilm in laboratory and clinical isolates of E. coli. PLoS Pathog 5: e1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonic V, Stojadinovic A, Zhang B, Izadjoo MJ, Alavi M. 2013. Pseudomonas aeruginosa induces pigment production and enhances virulence in a white phenotypic variant of Staphylococcus aureus. Infect Drug Resist 6: 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcilla MS, van Hattem JM, Matamoros S, Melles DC, Penders J, de Jong MD, Schultsz C, consortium C. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16: 147–149. [DOI] [PubMed] [Google Scholar]
- Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, Hancock RE. 2011. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother 55: 3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash C, Priest FG, Collins MD. 1994. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie van Leeuwenhoek 64: 253–260. [DOI] [PubMed] [Google Scholar]
- Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol 50: 219–230. [DOI] [PubMed] [Google Scholar]
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122: 461–472. [DOI] [PubMed] [Google Scholar]
- Baker SJ, Gunn JS, Morona R. 1999. The Salmonella typhi melittin resistance gene pqaB affects intracellular growth in PMA-differentiated U937 cells, polymyxin B resistance and lipopolysaccharide. Microbiology 145: 367–378. [DOI] [PubMed] [Google Scholar]
- Bao Y, Sakinc T, Laverde D, Wobser D, Benachour A, Theilacker C, Hartke A, Huebner J. 2012. Role of mprF1 and mprF2 in the pathogenicity of Enterococcus faecalis. PLoS ONE 7: e38458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchiesi J, Espariz M, Checa SK, Soncini FC. 2009. Downregulation of RpoN-controlled genes protects Salmonella cells from killing by the cationic antimicrobial peptide polymyxin B. FEMS Microbiol Lett 291: 73–79. [DOI] [PubMed] [Google Scholar]
- Barrow K, Kwon DH. 2009. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 53: 5150–5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengoechea JA, Skurnik M. 2000. Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol Microbiol 37: 67–80. [DOI] [PubMed] [Google Scholar]
- Bennett HPJ, Clarke DJ. 2005. The pbgPE operon in Photorhabdus luminescens is required for pathogenicity and symbiosis. J Bacteriol 187: 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, Bürgmann H, Sørum H, Norström M, Pons MN, et al. 2015. Tackling antibiotic resistance: The environmental framework. Nat Rev Microbiol 13: 310–317. [DOI] [PubMed] [Google Scholar]
- Berg JR, Spilker CM, Lewis SA. 1996. Effects of polymyxin B on mammalian urinary bladder. J Membr Biol 154: 119–130. [DOI] [PubMed] [Google Scholar]
- Berg JR, Spilker CM, Lewis SA. 1998. Modulation of polymyxin B effects on mammalian urinary bladder. Am J Physiol Renal Physiol 275: F204–F215. [DOI] [PubMed] [Google Scholar]
- Berglund NA, Piggot TJ, Jefferies D, Sessions RB, Bond PJ, Khalid S. 2015. Interaction of the antimicrobial peptide polymyxin B1 with both membranes of E. coli: A molecular dynamics study. PLoS Comput Biol 11: e1004180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilecen K, Fong JC, Cheng A, Jones CJ, Zamorano-Sanchez D, Yildiz FH. 2015. Polymyxin B resistance and biofilm formation in Vibrio cholerae are controlled by the response regulator CarR. Infect Immun 83: 1199–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina XR, Provenzano D, Nguyen N, Bina JE. 2008. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect Immun 76: 3595–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochmann RP, Toft A, Ciofu O, Briales A, Kolpen M, Hempel C, Bjarnsholt T, Høiby N, Jensen PØ. 2014. Bactericidal effect of colistin on planktonic Pseudomonas aeruginosa is independent of hydroxyl radical formation. Int J Antimicrob Agents 43: 140–147. [DOI] [PubMed] [Google Scholar]
- Brown MR, Melling J. 1969. Role of divalent cations in the action of polymyxin B and EDTA on Pseudomonas aeruginosa. J Gen Microbiol 59: 263–274. [DOI] [PubMed] [Google Scholar]
- CDC. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta. [Google Scholar]
- Cerny G, Teuber M. 1971. Differential release of periplasmic versus cytoplasmic enzymes from Escherichia coli B by polymyxin B. Arch Mikrobiol 78: 166–179. [DOI] [PubMed] [Google Scholar]
- Chambers JR, Sauer K. 2013. The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ. J Bacteriol 195: 4678–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Chen YF, Peng HL. 2010. Molecular characterization of the PhoPQ–PmrD–PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J Biomed Sci 17: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi MJ, Ko KS. 2014. Mutant prevention concentrations of colistin for Acinetobacter baumannii, Pseudomonas aeruginosa and Klebsiella pneumoniae clinical isolates. J Antimicrob Chemother 69: 275–277. [DOI] [PubMed] [Google Scholar]
- Choi AHK, Slamti L, Avci FY, Pier GB, Maira-Litran T. 2009a. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol 191: 5953–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SK, Park SY, Kim R, Kim SB, Lee CH, Kim JF, Park SH. 2009b. Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis. J Bacteriol 191: 3350–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausell A, Garcia-Subirats M, Pujol M, Busquets MA, Rabanal F, Cajal Y. 2007. Gram-negative outer and inner membrane models: Insertion of cyclic cationic lipopeptides. J Phys Chem B 111: 551–563. [DOI] [PubMed] [Google Scholar]
- Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, Goodlett DR, Ernst RK, Darveau RP. 2009. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell Microbiol 11: 1587–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert A, Lu A, Mandin P, Springer M, Gottesman S, Guillier M. 2010. MicA sRNA links the PhoP regulon to cell envelope stress. Mol Microbiol 76: 467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran CP, Podkaminski D, Papenfort K, Urban JH, Hinton JC, Vogel J. 2012. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol Microbiol 84: 428–445. [DOI] [PubMed] [Google Scholar]
- Cortay JC, Cozzone AJ. 1983a. A study of bacterial response to polypeptide antibiotics. FEBS Lett 157: 307–310. [DOI] [PubMed] [Google Scholar]
- Cortay JC, Cozzone AJ. 1983b. Accumulation of guanosine tetraphosphate induced by polymixin and gramicidin in Escherichia coli. Biochim Biophys Acta 755: 467–473. [DOI] [PubMed] [Google Scholar]
- Cullen TW, Madsen JA, Ivanov PL, Brodbelt JS, Trent MS. 2012. Characterization of unique modification of flagellar rod protein FlgG by Campylobacter jejuni lipid A phosphoethanolamine transferase, linking bacterial locomotion and antimicrobial peptide resistance. J Biol Chem 287: 3326–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, O’Brien JP, Hendrixson DR, Giles DK, Hobb RI, Thompson SA, Brodbelt JS, Trent MS. 2013. EptC of Campylobacter jejuni mediates phenotypes involved in host interactions and virulence. Infect Immun 81: 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Swanson MS. 2012. ppGpp: Magic beyond RNA polymerase. Nat Rev Microbiol 10: 203–212. [DOI] [PubMed] [Google Scholar]
- Daugelavicius R, Bakiene E, Bamford DH. 2000. Stages of polymyxin B interaction with the Escherichia coli cell envelope. Antimicrob Agents Chemother 44: 2969–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David HL, Rastogi N. 1985. Antibacterial action of colistin (polymyxin E) against Mycobacterium aurum. Antimicrob Agents Chemother 27: 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente-Núñez C, Reffuveille F, Haney EF, Straus SK, Hancock REW. 2014. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Path 10: e1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado MA, Mouslim C, Groisman EA. 2006. The PmrA/PmrB and RcsC/YojN/RcsB systems control expression of the Salmonella O-antigen chain length determinant. Mol Microbiol 60: 39–50. [DOI] [PubMed] [Google Scholar]
- Delucia AM, Six DA, Caughlan RE, Gee P, Hunt I, Lam JS, Dean CR. 2011. Lipopolysaccharide (LPS) inner-core phosphates are required for complete LPS synthesis and transport to the outer membrane in Pseudomonas aeruginosa PAO1. MBio 2: e00142–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Majumdar S, Yu J, Fookes M, McAteer SP, Llobet E, Finn S, Spence S, Monahan A, Monaghan A, Kissenpfennig A, et al. 2015. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog 11: e1004627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deris ZZ, Swarbrick JD, Roberts KD, Azad MAK, Akter J, Horne AS, Nation RL, Rogers KL, Thompson PE, Velkov T, et al. 2014. Probing the penetration of antimicrobial polymyxin lipopeptides into Gram-negative bacteria. Bioconjug Chem 25: 750–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Chopra I. 1986. Polymyxin B and polymyxin B nonapeptide alter cytoplasmic membrane permeability in Escherichia coli. J Antimicrob Chemother 18: 557–563. [DOI] [PubMed] [Google Scholar]
- Dong TG, Dong S, Catalano C, Moore R, Liang X, Mekalanos JJ. 2015. Generation of reactive oxygen species by lethal attacks from competing microbes. Proc Natl Acad Sci 112: 2181–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. 2007. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol 3: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol 12: 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AJF, Vollmer W. 2015. The stoichiometric divisome: A hypothesis. Front Microbiol 6: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. 1999. Polymyxin B sulfate and colistin: Old antibiotics for emerging multiresistant Gram-negative bacteria. Ann Pharmacother 33: 960–967. [DOI] [PubMed] [Google Scholar]
- Falagas ME, Kasiakou SK. 2005. Colistin: The revival of polymyxins for the management of multidrug-resistant Gram-negative bacterial infections. Clin Infect Dis 40: 1333–1341. [DOI] [PubMed] [Google Scholar]
- Falagas ME, Kasiakou SK. 2006. Toxicity of polymyxins: A systematic review of the evidence from old and recent studies. Crit Care 10: R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas ME, Rafailidis PI, Matthaiou DK. 2010. Resistance to polymyxins: Mechanisms, frequency and treatment options. Drug Resist Updat 13: 132–138. [DOI] [PubMed] [Google Scholar]
- Fehlner-Gardiner CC, Valvano MA. 2002. Cloning and characterization of the Burkholderia vietnamiensis norM gene encoding a multi-drug efflux protein. FEMS Microbiol Lett 215: 279–283. [DOI] [PubMed] [Google Scholar]
- Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock REW. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR–ParS. Antimicrob Agents Chemother 54: 3372–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L, Breidenstein EBM, Hancock REW. 2011. Creeping baselines and adaptive resistance to antibiotics. Drug Resist Updat 14: 1–21. [DOI] [PubMed] [Google Scholar]
- Fernández L, Jenssen H, Bains M, Wiegand I, Gooderham WJ, Hancock REW. 2012. The two-component system CprRS senses cationic peptides and triggers adaptive resistance in Pseudomonas aeruginosa independently of ParRS. Antimicrob Agents Chemother 56: 6212–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L, Alvarez-Ortega C, Wiegand I, Olivares J, Kocíncová D, Lam JS, Martínez JL, Hancock REW. 2013. Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob Agents Chemother 57: 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finking R, Marahiel MA. 2004. Biosynthesis of nonribosomal peptides1. Annu Rev Microbiol 58: 453–488. [DOI] [PubMed] [Google Scholar]
- Folkesson A, Haagensen JA, Zampaloni C, Sternberg C, Molin S. 2008. Biofilm induced tolerance towards antimicrobial peptides. PLoS ONE 3: e1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons HS, Reynolds CM, Guan Z, Raetz CR. 2008. An inner membrane dioxygenase that generates the 2-hydroxymyristate moiety of Salmonella lipid A. Biochemistry 47: 2814–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock REW. 2008. Induction by cationic antimicrobial peptides and involvement in intrinsic polymyxin and antimicrobial peptide resistance, biofilm formation, and swarming motility of PsrA in Pseudomonas aeruginosa. J Bacteriol 190: 5624–5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooderham WJ, Gellatly SL, Sanschagrin F, McPhee JB, Bains M, Cosseau C, Levesque RC, Hancock REW. 2009. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology 155: 699–711. [DOI] [PubMed] [Google Scholar]
- Gray AN, Egan AJ, Van’t Veer IL, Verheul J, Colavin A, Koumoutsi A, Biboy J, Altelaar AFM, Damen MJ, Huang KC, et al. 2015. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. eLife 4: e07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn JS. 2008. The Salmonella PmrAB regulon: Lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol 16: 284–290. [DOI] [PubMed] [Google Scholar]
- Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. 1998. PmrA–PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol 27: 1171–1182. [DOI] [PubMed] [Google Scholar]
- Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. 2000. Genetic and functional analysis of a PmrA–PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect Immun 68: 6139–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutu AD, Sgambati N, Strasbourger P, Brannon MK, Jacobs MA, Haugen E, Kaul RK, Johansen HK, Høiby N, Moskowitz SM. 2013. Polymyxin resistance of Pseudomonas aeruginosa phoQ mutants is dependent on additional two-component regulatory systems. Antimicrob Agents Chemother 57: 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RE. 1981. Aminoglycoside uptake and mode of action—With special reference to streptomycin and gentamicin. II: Effects of aminoglycosides on cells. J Antimicrob Chemother 8: 429–445. [DOI] [PubMed] [Google Scholar]
- Hancock RE. 1984. Alterations in outer membrane permeability. Annu Rev Microbiol 38: 237–264. [DOI] [PubMed] [Google Scholar]
- Hancock RE. 1997. Peptide antibiotics. Lancet 349: 418–422. [DOI] [PubMed] [Google Scholar]
- Hancock RE. 2000. Cationic antimicrobial peptides: Towards clinical applications. Expert Opin Investig Drugs 9: 1723–1729. [DOI] [PubMed] [Google Scholar]
- Hancock REW, Bell A. 1988. Antibiotic uptake into Gram-negative bacteria. Eur J Clin Microbiol Infect Dis 7: 713–720. [DOI] [PubMed] [Google Scholar]
- Hancock RE, Chapple DS. 1999. Peptide antibiotics. Antimicrob Agents Chemother 43: 1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler AA, Lim JE, Losick R. 2008. Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol Microbiol 68: 588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankins JV, Madsen JA, Giles DK, Brodbelt JS, Trent MS. 2012. Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in Gram-positive and Gram-negative bacteria. Proc Natl Acad Sci 109: 8722–8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque MM, Kabir MS, Aini LQ, Hirata H, Tsuyumu S. 2009. SlyA, a MarR family transcriptional regulator, is essential for virulence in Dickeya dadantii 3937. J Bacteriol 191: 5409–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindorf M, Kadari M, Heider C, Skiebe E, Wilharm G. 2014. Impact of Acinetobacter baumannii superoxide dismutase on motility, virulence, oxidative stress resistance and susceptibility to antibiotics. PLoS ONE 9: e101033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CM, Hankins JV, Trent MS. 2010. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol Microbiol 76: 1444–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CM, Crofts AA, Henderson JC, Pingali SC, Davies BW, Trent MS. 2014. The Vibrio cholerae VprA–VprB two-component system controls virulence through endotoxin modification. MBio 5: e02283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer SU, Schlumberger MC, Jackel D, Hensel M. 2009. Effect of the O-antigen length of lipopolysaccharide on the functions of type III secretion systems in Salmonella enterica. Infect Immun 77: 5458–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood MI, Becker KW, Roux CM, Dunman PM, Skaar EP. 2013. Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect Immun 81: 542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Liu F, Lin IY, Gao GF, Zhu B. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16: 146–147. [DOI] [PubMed] [Google Scholar]
- Ilg K, Endt K, Misselwitz B, Stecher B, Aebi M, Hardt WD. 2009. O-antigen-negative Salmonella enterica serovar Typhimurium is attenuated in intestinal colonization but elicits colitis in streptomycin-treated mice. Infect Immun 77: 2568–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram BO, Sohlenkamp C, Geiger O, Raetz CR. 2010. Altered lipid A structures and polymyxin hypersensitivity of Rhizobium etli mutants lacking the LpxE and LpxF phosphatases. Biochim Biophys Acta 1801: 593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SS, Lin TY, Wang WB, Liu MC, Hsueh PR, Liaw SJ. 2010a. Characterization of UDP-glucose dehydrogenase and UDP-glucose pyrophosphorylase mutants of Proteus mirabilis: Defectiveness in polymyxin B resistance, swarming, and virulence. Antimicrob Agents Chemother 54: 2000–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SS, Liu MC, Teng LJ, Wang WB, Hsueh PR, Liaw SJ. 2010b. Proteus mirabilis pmrI, an RppA-regulated gene necessary for polymyxin B resistance, biofilm formation, and urothelial cell invasion. Antimicrob Agents Chemother 54: 1564–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Lunin VV, Skarina T, Savchenko A, Schurr MJ, Hoang TT. 2009. The long-chain fatty acid sensor, PsrA, modulates the expression of rpoS and the type III secretion exsCEBA operon in Pseudomonas aeruginosa. Mol Microbiol 73: 120–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Latifi T, Groisman EA. 2003. Closing the loop: The PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc Natl Acad Sci 100: 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuma N, Sato Y, Ohki K, Okimura K, Ohnishi K, Sakura N. 2009. Development of des-fatty acyl-polymyxin B decapeptide analogs with Pseudomonas aeruginosa-specific antimicrobial activity. Chem Pharm Bull (Tokyo) 57: 332–336. [DOI] [PubMed] [Google Scholar]
- Kawasaki K, China K, Nishijima M. 2007. Release of the lipopolysaccharide deacylase PagL from latency compensates for a lack of lipopolysaccharide aminoarabinose modification-dependent resistance to the antimicrobial peptide polymyxin B in Salmonella enterica. J Bacteriol 189: 4911–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339: 1213–1216. [DOI] [PubMed] [Google Scholar]
- Kindrachuk KN, Fernandez L, Bains M, Hancock RE. 2011. Involvement of an ATP-dependent protease, PA0779/AsrA, in inducing heat shock in response to tobramycin in Pseudomonas aeruginosa. Antimicrob Agents Chemother 55: 1874–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemperer RM, Gilbert P, Meier AM, Cozens RM, Brown MR. 1979. Influence of suspending media upon the susceptibility of Pseudomonas aeruginosa NCTC 6750 and its spheroplasts to polymyxin B. Antimicrob Agents Chemother 15: 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M, Iida K, Matsuo T. 1969. Electron microscopic studies on mode of action of polymyxin. J Bacteriol 97: 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Yang J, Liu Q, Alamuri P, Roland KL, Curtiss R III. 2011. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar Typhimurium. Infect Immun 79: 4227–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kox LF, Wösten MM, Groisman EA. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J 19: 1861–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Christensen B, Raventos D, Nielsen AK, Nielsen JD, Vukmirovic N, Kristensen HH. 2009. Transcriptional profile of Escherichia coli in response to novispirin G10. Int J Pept Res Ther 15: 17–24. [Google Scholar]
- Kunin CM, Bugg A. 1971. Binding of polymyxin antibiotics to tissues: The major determinant of distribution and persistence in the body. J Infect Dis 124: 394–400. [DOI] [PubMed] [Google Scholar]
- Kwa AL, Tam VH, Falagas ME. 2008. Polymyxins: A review of the current status including recent developments. Ann Acad Med Singapore 37: 870–883. [PubMed] [Google Scholar]
- Lacour S, Bechet E, Cozzone AJ, Mijakovic I, Grangeasse C. 2008. Tyrosine phosphorylation of the UDP-glucose dehydrogenase of Escherichia coli is at the crossroads of colanic acid synthesis and polymyxin resistance. PLoS ONE 3: e3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman D, Georgescu C, Martin DA, Quale J. 2008. Polymyxins revisited. Clin Microbiol Rev 21: 449–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPorte DC, Rosenthal KS, Storm DR. 1977. Inhibition of Escherichia coli growth and respiration by polymyxin B covalently attached to agarose beads. Biochemistry 16: 1642–1648. [DOI] [PubMed] [Google Scholar]
- Lee H, Hsu FF, Turk J, Groisman EA. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J Bacteriol 186: 4124–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JR, Lewis SA. 2004. Colistin interactions with the mammalian urothelium. Am J Physiol Cell Physiol 286: C913–C922. [DOI] [PubMed] [Google Scholar]
- Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, et al. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517: 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339: 1210–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, et al. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect Dis 16: 161–168. [DOI] [PubMed] [Google Scholar]
- Llewellyn AC, Zhao J, Song F, Parvathareddy J, Xu Q, Napier BA, Laroui H, Merlin D, Bina JE, Cotter PA, et al. 2012. NaxD is a deacetylase required for lipid A modification and Francisella pathogenesis. Mol Microbiol 86: 611–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Camacho E, Gomez-Gil R, Tobes R, Manrique M, Lorenzo M, Galvan B, Salvarelli E, Moatassim Y, Salanueva IJ, Pareja E, et al. 2014. Genomic analysis of the emergence and evolution of multidrug resistance during a Klebsiella pneumoniae outbreak including carbapenem and colistin resistance. J Antimicrob Chemother 69: 632–636. [DOI] [PubMed] [Google Scholar]
- Loutet SA, Valvano MA. 2011. Extreme antimicrobial peptide and polymyxin B resistance in the genus Burkholderia. Front Microbiol 2: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutet SA, Bartholdson SJ, Govan JRW, Campopiano DJ, Valvano MA. 2009. Contributions of two UDP-glucose dehydrogenases to viability and polymyxin B resistance of Burkholderia cenocepacia. Microbiology 155: 2029–2039. [DOI] [PubMed] [Google Scholar]
- Lu S, Walters G, Parg R, Dutcher JR. 2014. Nanomechanical response of bacterial cells to cationic antimicrobial peptides. Soft Matter 10: 1806–1815. [DOI] [PubMed] [Google Scholar]
- Macfarlane EL, Kwasnicka A, Hancock RE. 2000. Role of Pseudomonas aeruginosa PhoP–PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146: 2543–2554. [DOI] [PubMed] [Google Scholar]
- Maloney E, Stankowska D, Zhang J, Fol M, Cheng QJ, Lun S, Bishai WR, Rajagopalan M, Chatterjee D, Madiraju MV. 2009. The two-domain LysX protein of Mycobacterium tuberculosis is required for production of lysinylated phosphatidylglycerol and resistance to cationic antimicrobial peptides. PLoS Pathog 5: e1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau M, Sebbane F, Ewann F, Collyn F, Lindner B, Campos MA, Bengoechea JA, Simonet M. 2004. The pmrF polymyxin-resistance operon of Yersinia pseudotuberculosis is upregulated by the PhoP–PhoQ two-component system but not by PmrA–PmrB, and is not required for virulence. Microbiology 150: 3947–3957. [DOI] [PubMed] [Google Scholar]
- Margolin W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol 6: 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NI, Hu H, Moake MM, Churey JJ, Whittal R, Worobo RW, Vederas JC. 2003. Isolation, structural characterization, and properties of mattacin (polymyxin M), a cyclic peptide antibiotic produced by Paenibacillus kobensis M. J Biol Chem 278: 13124–13132. [DOI] [PubMed] [Google Scholar]
- Mathur J, Waldor MK. 2004. The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect Immun 72: 3577–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy LS, Roberts KD, Nation RL, Thompson PE, Velkov T, Li J, Tor Y. 2013. Polymyxins and analogues bind to ribosomal RNA and interfere with eukaryotic translation in vitro. ChemBioChem 14: 2083–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee JB, Lewenza S, Hancock REW. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA–PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol 50: 205–217. [DOI] [PubMed] [Google Scholar]
- Moffatt JH, Harper M, Harrison P, Hale JDF, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, et al. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54: 4971–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt JH, Harper M, Adler B, Nation RL, Li J, Boyce JD. 2011. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob Agents Chemother 55: 3022–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi T, Murase Y, Mori M, Shiomi K, Omura S, Paranagama MP, Kita K. 2009. Polymyxin B identified as an inhibitor of alternative NADH dehydrogenase and malate: Quinone oxidoreductase from the Gram-positive bacterium Mycobacterium smegmatis. J Biochem 146: 491–499. [DOI] [PubMed] [Google Scholar]
- Moon K, Gottesman S. 2009. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol Microbiol 74: 1314–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RA, Bates NC, Hancock RE. 1986. Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob Agents Chemother 29: 496–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen NP, Fowlkes JD, Sullivan CJ, Allison DP, Larsen NB, Molin S, Doktycz MJ. 2009. Effects of colistin on surface ultrastructure and nanomechanics of Pseudomonas aeruginosa cells. Langmuir 25: 3728–3733. [DOI] [PubMed] [Google Scholar]
- Mouslim C, Groisman EA. 2003. Control of the Salmonella ugd gene by three two-component regulatory systems. Mol Microbiol 47: 335–344. [DOI] [PubMed] [Google Scholar]
- Muller C, Plésiat P, Jeannot K. 2011. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother 55: 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Kawamata J. 1966. Studies on the mechanism of action of colistin. IV: Activation of “latent” ribonuclease in Escherichia coli by colistin. Biken J 9: 115–123. [PubMed] [Google Scholar]
- Nation RL, Velkov T, Li J. 2014. Colistin and polymyxin B: Peas in a pod, or chalk and cheese? Clin Infect Dis 59: 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nation RL, Li J, Cars O, Couet W, Dudley MN, Kaye KS, Mouton JW, Paterson DL, Tam VH, Theuretzbacher U, et al. 2015. Framework for optimisation of the clinical use of colistin and polymyxin B: The Prato polymyxin consensus. Lancet Infect Dis 15: 225–234. [DOI] [PubMed] [Google Scholar]
- Newton BA. 1956. The properties and mode of action of the polymyxins. Bacteriol Rev 20: 14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K, Hsu FF, Turk J, Cromie MJ, Wösten MMSM, Groisman EA. 2006. Identification of the lipopolysaccharide modifications controlled by the Salmonella PmrA/PmrB system mediating resistance to Fe(III) and Al(III). Mol Microbiol 61: 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B, Vater J, Rueckert C, Blom J, Lehmann M, Ru JJ, Chen XH, Wang Q, Borriss R. 2013. Polymyxin P is the active principle in suppressing phytopathogenic Erwinia spp. by the biocontrol rhizobacterium Paenibacillus polymyxa M-1. BMC Microbiol 13: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JT, Van Dyk TK, Cajal Y, Dhurjati PS, Sasser M, Jain MK. 1998. Osmotic stress in viable Escherichia coli as the basis for the antibiotic response by polymyxin B. Biochem Biophys Res Commun 246: 619–623. [DOI] [PubMed] [Google Scholar]
- Oh JT, Cajal Y, Skowronska EM, Belkin S, Chen J, Van Dyk TK, Sasser M, Jain MK. 2000. Cationic peptide antimicrobials induce selective transcription of micF and osmY in Escherichia coli. Biochim Biophys Acta 1463: 43–54. [DOI] [PubMed] [Google Scholar]
- Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front Microbiol 5: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaitan AO, Chabou S, Okdah L, Morand S, Rolain JM. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16: 147. [DOI] [PubMed] [Google Scholar]
- Orwa JA, Govaerts C, Busson R, Roets E, Van Schepdael A, Hoogmartens J. 2001. Isolation and structural characterization of colistin components. J Antibiot (Tokyo) 54: 595–599. [DOI] [PubMed] [Google Scholar]
- Padilla E, Llobet E, Domenech-Sanchez A, Martinez-Martinez L, Bengoechea JA, Alberti S. 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 54: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB–oprM genes. Mol Microbiol 68: 223–240. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. 2006. σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol 62: 1674–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Groisman EA. 2014. Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system. Mol Microbiol 91: 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson GK, Northen H, Cone DB, Willers C, Peters SE, Maskell DJ. 2009. Deletion of tolA in Salmonella typhimurium generates an attenuated strain with vaccine potential. Microbiology 155: 220–228. [DOI] [PubMed] [Google Scholar]
- Paulander W, Wang Y, Folkesson A, Charbon G, Lobner-Olesen A, Ingmer H. 2014. Bactericidal antibiotics increase hydroxyphenyl fluorescein signal by altering cell morphology. PLoS ONE 9: e92231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescaretti MdLM, Lopez FE, Morero RD, Delgado MA. 2011. The PmrA/PmrB regulatory system controls the expression of the wzzfepE gene involved in the O-antigen synthesis of Salmonella enterica serovar Typhimurium. Microbiology 157: 2515–2521. [DOI] [PubMed] [Google Scholar]
- Phe K, Lee Y, McDaneld PM, Prasad N, Yin T, Figueroa DA, Musick WL, Cottreau JM, Hu M, Tam VH. 2014. In vitro assessment and multicenter cohort study of comparative nephrotoxicity rates associated with colistimethate versus polymyxin B therapy. Antimicrob Agents Chemother 58: 2740–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piek S, Wang Z, Ganguly J, Lakey AM, Bartley SN, Mowlaboccus S, Anandan A, Stubbs KA, Scanlon MJ, Vrielink A, et al. 2014. The role of oxidoreductases in determining the function of the neisserial lipid a phosphoethanolamine transferase required for resistance to polymyxin. PLoS ONE 9: e106513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port GC, Vega LA, Nylander AB, Caparon MG. 2014. Streptococcus pyogenes polymyxin B–resistant mutants display enhanced ExPortal integrity. J Bacteriol 196: 2563–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pournaras S, Poulou A, Dafopoulou K, Chabane YN, Kristo I, Makris D, Hardouin J, Cosette P, Tsakris A, De E. 2014. Growth retardation, reduced invasiveness, and impaired colistin-mediated cell death associated with colistin resistance development in Acinetobacter baumannii. Antimicrob Agents Chemother 58: 828–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit RE, Miller SI. 2007. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell 26: 165–174. [DOI] [PubMed] [Google Scholar]
- Raetz CRH, Reynolds CM, Trent MS, Bishop RE. 2007. Lipid A modification systems in Gram-negative bacteria. Annu Rev Biochem 76: 295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CM, Kalb SR, Cotter RJ, Raetz CRH. 2005. A phosphoethanolamine transferase specific for the outer 3-deoxy-d-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J Biol Chem 280: 21202–21211. [DOI] [PubMed] [Google Scholar]
- Reynolds CM, Ribeiro AA, McGrath SC, Cotter RJ, Raetz CRH, Trent MS. 2006. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J Biol Chem 281: 21974–21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch JW, Caparon MG. 2005. The ExPortal: An organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol 58: 959–968. [DOI] [PubMed] [Google Scholar]
- Rosch JW, Hsu FF, Caparon MG. 2007. Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J Bacteriol 189: 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal KS, Storm DR. 1977. Disruption of the Escherichia coli outer membrane permeability barrier by immobilized polymyxin B. J Antibiot (Tokyo) 30: 1087–1092. [DOI] [PubMed] [Google Scholar]
- Sassera D, Comandatore F, Gaibani P, D’Auria G, Mariconti M, Landini MP, Sambri V, Marone P. 2014. Comparative genomics of closely related strains of Klebsiella pneumoniae reveals genes possibly involved in colistin resistance. Ann Microbiol 64: 887–890. [Google Scholar]
- Saugar JM, Alarcon T, Lopez-Hernandez S, Lopez-Brea M, Andreu D, Rivas L. 2002. Activities of polymyxin B and cecropin A-melittin peptide CA(1-8)M(1-18) against a multiresistant strain of Acinetobacter baumannii. Antimicrob Agents Chemother 46: 875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler PRG, Teuber M. 1975. Action of polymyxin B on bacterial membranes: Morphological changes in the cytoplasm and in the outer membrane of Salmonella typhimurium and Escherichia coli B. Antimicrob Agents Chemother 8: 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott NE, Nothaft H, Edwards AVG, Labbate M, Djordjevic SP, Larsen MR, Szymanski CM, Cordwell SJ. 2012. Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine. J Biol Chem 287: 29384–29396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NR, Hancock RE, Fernandez RC. 2014. Bordetella pertussis lipid A glucosamine modification confers resistance to cationic antimicrobial peptides and increases resistance to outer membrane perturbation. Antimicrob Agents Chemother 58: 4931–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen M, Li J, Ross AC, Vederas JC, Jensen SE. 2011. Paenibacillus polymyxa PKB1 produces variants of polymyxin B–type antibiotics. Chem Biol 18: 1640–1648. [DOI] [PubMed] [Google Scholar]
- Shi Y, Cromie MJ, Hsu FF, Turk J, Groisman EA. 2004a. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol Microbiol 53: 229–241. [DOI] [PubMed] [Google Scholar]
- Shi Y, Latifi T, Cromie MJ, Groisman EA. 2004b. Transcriptional control of the antimicrobial peptide resistance ugtL gene by the Salmonella PhoP and SlyA regulatory proteins. J Biol Chem 279: 38618–38625. [DOI] [PubMed] [Google Scholar]
- Sikora AE, Beyhan S, Bagdasarian M, Yildiz FH, Sandkvist M. 2009. Cell envelope perturbation induces oxidative stress and changes in iron homeostasis in Vibrio cholerae. J Bacteriol 191: 5398–5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerlavaj B, Romeo D, Gennaro R. 1990. Rapid membrane permeabilization and inhibition of vital functions of Gram-negative bacteria by bactenecins. Infect Immun 58: 3724–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlenkamp C, Galindo-Lagunas KA, Guan Z, Vinuesa P, Robinson S, Thomas-Oates J, Raetz CRH, Geiger O. 2007. The lipid lysyl-phosphatidylglycerol is present in membranes of Rhizobium tropici CIAT899 and confers increased resistance to polymyxin B under acidic growth conditions. Mol Plant Microbe Interact 20: 1421–1430. [DOI] [PubMed] [Google Scholar]
- Spindler EC, Hale JDF, Giddings TH, Hancock REW, Gill RT. 2011. Deciphering the mode of action of the synthetic antimicrobial peptide Bac8c. Antimicrob Agents Chemother 55: 1706–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan VB, Singh BB, Priyadarshi N, Chauhan NK, Rajamohan G. 2014. Role of novel multidrug efflux pump involved in drug resistance in Klebsiella pneumoniae. PLoS ONE 9: e96288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm DR, Rosenthal KS, Swanson PE. 1977. Polymyxin and related peptide antibiotics. Annu Rev Biochem 46: 723–763. [DOI] [PubMed] [Google Scholar]
- Strahl H, Hamoen LW. 2010. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci 107: 12281–12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subashchandrabose S, Smith SN, Spurbeck RR, Kole MM, Mobley HLT. 2013. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog 9: e1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbalakshmi C, Sitaram N. 1998. Mechanism of antimicrobial action of indolicidin. FEMS Microbiol Lett 160: 91–96. [DOI] [PubMed] [Google Scholar]
- Sun S, Negrea A, Rhen M, Andersson DI. 2009. Genetic analysis of colistin resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother 53: 2298–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Choudhury B, Septer A, Merighi M, Carlson R, Gunn JS. 2005a. Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar Typhimurium lipopolysaccharide core. J Bacteriol 187: 3391–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Prouty AM, Gunn JS. 2005b. Identification and functional analysis of Salmonella enterica serovar Typhimurium PmrA-regulated genes. FEMS Immunol Med Microbiol 43: 249–258. [DOI] [PubMed] [Google Scholar]
- Tambadou F, Caradec T, Gagez AL, Bonnet A, Sopéna V, Bridiau N, Thiéry V, Didelot S, Barthélémy C, Chevrot R. 2015. Characterization of the colistin (polymyxin E1 and E2) biosynthetic gene cluster. Arch Microbiol 197: 521–532. [DOI] [PubMed] [Google Scholar]
- Taylor PK, Yeung ATY, Hancock REW. 2014. Antibiotic resistance in Pseudomonas aeruginosa biofilms: Towards the development of novel anti-biofilm therapies. J Biotechnol 191: 121–130. [DOI] [PubMed] [Google Scholar]
- Teuber M. 1967. Precipitation of ribosomes from E. coli B by polymyxin B. Die Naturwissenschaften 54: 71. [DOI] [PubMed] [Google Scholar]
- Teuber M. 1974. Action of polymyxin B on bacterial membranes. III: Differential inhibition of cellular functions in Salmonella typhimurium. Arch Microbiol 100: 131–144. [Google Scholar]
- Tochikubo K, Yasuda Y, Kozuka S. 1986. Decreased particulate NADH oxidase activity in Bacillus subtilis spores after polymyxin B treatment. Microbiology 132: 277–287. [DOI] [PubMed] [Google Scholar]
- Touze T, Tran AX, Hankins JV, Mengin-Lecreulx D, Trent MS. 2008. Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Mol Microbiol 67: 264–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran AX, Whittimore JD, Wyrick PB, McGrath SC, Cotter RJ, Trent MS. 2006. The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin. J Bacteriol 188: 4531–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: Induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J Biol Chem 276: 43122–43131. [DOI] [PubMed] [Google Scholar]
- Tsai MH, Wu SR, Lee HY, Chen CL, Lin TY, Huang YC, Chiu CH. 2012. Recognition of mechanisms involved in bile resistance important to halting antimicrobial resistance in nontyphoidal Salmonella. Int J Antimicrob Agents 40: 151–157. [DOI] [PubMed] [Google Scholar]
- Tse H, Yuen KY. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16: 145–146. [DOI] [PubMed] [Google Scholar]
- Tu X, Latifi T, Bougdour A, Gottesman S, Groisman EA. 2006. The PhoP/PhoQ two-component system stabilizes the alternative σ factor RpoS in Salmonella enterica. Proc Natl Acad Sci 103: 13503–13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng YL, Datta A, Ambrose K, Lo M, Davies JK, Carlson RW, Stephens DS, Kahler CM. 2004. The MisR/MisS two-component regulatory system influences inner core structure and immunotype of lipooligosaccharide in Neisseria meningitidis. J Biol Chem 279: 35053–35062. [DOI] [PubMed] [Google Scholar]
- Tzeng YL, Ambrose KD, Zughaier S, Zhou X, Miller YK, Shafer WM, Stephens DS. 2005. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J Bacteriol 187: 5387–5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M, Vaara T. 1983. Polycations sensitize enteric bacteria to antibiotics. Antimicrob Agents Chemother 24: 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meijden B, Robinson JA. 2015. Synthesis of a polymyxin derivative for photolabeling studies in the Gram-negative bacterium Escherichia coli. J Pept Sci 21: 231–235. [DOI] [PubMed] [Google Scholar]
- van Mourik A, Steeghs L, van Laar J, Meiring HD, Hamstra HJ, van Putten JPM, Wosten MMSM. 2010. Altered linkage of hydroxyacyl chains in lipid A of Campylobacter jejuni reduces TLR4 activation and antimicrobial resistance. J Biol Chem 285: 15828–15836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega LA, Caparon MG. 2012. Cationic antimicrobial peptides disrupt the Streptococcus pyogenes ExPortal. Mol Microbiol 85: 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velkov T, Roberts KD, Nation RL, Wang J, Thompson PE, Li J. 2014. Teaching “old” polymyxins new tricks: New-generation lipopeptides targeting Gram-negative “superbugs.” ACS Chem Biol 9: 1172–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McGrath SC, Cotter RJ, Raetz CR. 2006. Expression cloning and periplasmic orientation of the Francisella novicida lipid A 4′-phosphatase LpxF. J Biol Chem 281: 9321–9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WB, Chen IC, Jiang SS, Chen HR, Hsu CY, Hsueh PR, Hsu WB, Liaw SJ. 2008. Role of RppA in the regulation of polymyxin b susceptibility, swarming, and virulence factor expression in Proteus mirabilis. Infect Immun 76: 2051–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner DM, Levy SB. 2010. Different effects of transcriptional regulators MarA, SoxS and Rob on susceptibility of Escherichia coli to cationic antimicrobial peptides (CAMPs): Rob-dependent CAMP induction of the marRAB operon. Microbiology 156: 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren GH, Gray J, Yurchenco JA. 1957. Effect of polymyxin on the lysis of Neisseria catarrhalis by lysozyme. J Bacteriol 74: 788–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb HE, Granier SA, Marault M, Millemann Y, den Bakker HC, Nightingale KK, Bugarel M, Ison SA, Scott HM, Loneragan GH. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16: 144–145. [DOI] [PubMed] [Google Scholar]
- Wu M, Hancock REW. 1999. Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J Biol Chem 274: 29–35. [DOI] [PubMed] [Google Scholar]
- Xiong YQ, Mukhopadhyay K, Yeaman MR, Adler-Moore J, Bayer AS. 2005. Functional interrelationships between cell membrane and cell wall in antimicrobial peptide-mediated killing of Staphylococcus aureus. Antimicrob Agents Chemother 49: 3114–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan A, Guan Z, Raetz CR. 2007. An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J Biol Chem 282: 36077–36089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-J, Bayer AS, Mishra NN, Meehl M, Ledala N, Yeaman MR, Xiong YQ, Cheung AL. 2012. The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect Immun 80: 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yethon JA, Gunn JS, Ernst RK, Miller SI, Laroche L, Malo D, Whitfield C. 2000. Salmonella enterica serovar Typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence in vivo. Infect Immun 68: 4485–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung ATY, Bains M, Hancock REW. 2011. The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J Bacteriol 193: 918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Qin W, Lin J, Fang S, Qiu J. 2015. Antibacterial mechanisms of polymyxin and bacterial resistance. Biomed Res Int 2015: 679109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: A critical review. J Antimicrob Chemother 60: 1206–1215. [DOI] [PubMed] [Google Scholar]
- Zhang L, Dhillon P, Yan H, Farmer S, Hancock REW. 2000. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob Agents Chemother 44: 3317–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]