Abstract
Prion diseases are disorders that share several characteristics that are typical of many neurodegenerative diseases. Recently, several studies have extended the prion concept to pathological aggregation in malignant tumors involving misfolded p53, a tumor-suppressor protein. The aggregation of p53 and its coaggregation with p53 family members, p63 and p73, have been shown. Certain p53 mutants exert a dominant-negative regulatory effect on wild-type (WT) p53. The basis for this dominant-negative effect is that amyloid-like mutant p53 converts WT p53 into an aggregated species, leading to a gain-of-function (GoF) phenotype and the loss of its tumor-suppressor function. Recently, it was shown that p53 aggregates can be internalized by cells and can coaggregate with endogenous p53, corroborating the prion-like properties of p53 aggregates. The prion-like behavior of oncogenic p53 mutants provides an explanation for its dominant-negative and GoF properties, including the high metastatic potential of cancer cells carrying p53 mutations. The inhibition of p53 aggregation appears to represent a promising target for therapeutic intervention in patients with malignant tumors.
Amyloid-like mutant p53 can convert wild-type p53 into an aggregated form, and p53 aggregates can be transmitted from cell to cell. These properties may contribute to p53’s role in tumor formation and malignancy.
Prion diseases are disorders that share several characteristics that are typical of many neurodegenerative diseases (Prusiner 1998, 2013, 2014). Recently, studies have provided evidence that the prion concept may be extended to several neurodegenerative diseases (Silva et al. 2008; Polymenidou and Cleveland 2011; Prusiner 2012; Soto 2012; Irwin et al. 2013). Key proteins involved in these diseases, such as Aβ, tau, α-synuclein, SOD1, and TDP43, may act as prions, as shown by their transmissibility in animals and in cultured mammalian cells (Polymenidou and Cleveland 2011; Prusiner 2012; Soto 2012; Irwin et al. 2013; Prusiner et al. 2015). These degenerative diseases are based on the prion-like conversion of a correctly folded protein into a misfolded version, resulting in a toxic gain-of-function (GoF) that leads to cell death. New findings have extended the concept of pathological aggregation to malignant tumors involving misfolded p53, a tumor-suppressor protein (Ishimaru et al. 2003a; Silva et al. 2010, 2014; Ano Bom et al. 2012; Wilcken et al. 2012). Numerous studies have established the aggregation of p53 and its coaggregation with the paralogous proteins p63 and p73 into different types of aggregates, including amyloid oligomers and fibrils (Bullock et al. 1997; Ishimaru et al. 2003a; Lee et al. 2003; Rigacci et al. 2008; Silva et al. 2010; Xu et al. 2011; Ano Bom et al. 2012; Wilcken et al. 2012).
p53 is a tetrameric protein that is encoded by the TP53 gene. It is a master regulator of cell homeostasis and DNA stability. p53 acts as a transcription factor by binding to specific DNA sequences and inducing the transcription of genes involved in the regulation of cell-cycle arrest for DNA repair, senescence, and apoptosis, as well as other processes (Vousden and Lane 2007). Because of its importance in cell homeostasis, its function is finely regulated by several mechanisms. The half-life of p53 in the cell is very short, and p53 is the target of cellular proteins such as MDM2 and MDM4 (also referred to as MDMX) (Schon et al. 2002; Linares et al. 2003). Both of these proteins bind to and inactivate p53, but only MDM2 exhibits ubiquitin E3-ligase activity that directs p53 to proteasomal degradation (Fang et al. 2000).
p53 mutations are the most common genetic alterations found in cancers and are observed in >50% of all tumors. From the approximately 200 different single mutations already described in p53, several residues are considered as hotspots, including R248, R175, G245, R273, R249, and R282 (Petitjean et al. 2007). All of these residues are found in p53's core domain, which is responsible for its interactions with DNA. For this reason, most of these mutants are incapable of exerting the wild-type (WT) level of transcriptional activity (Bullock et al. 1997). The most frequent effect of p53 mutations is loss-of-function (LoF); however, GoF effects, such as increased migration, invasion, and metastasis, have also been observed (Muller and Vousden 2013). Another important characteristic related to p53 mutations is a dominant-negative effect (Freed-Pastor and Prives 2012) that is exerted by mutant p53 on WT p53, owing to the presence of different mutant alleles in the same cell. In these cases, p53 is typically inactivated and loses its function in the cell. The classical explanation for this effect is that heterotetramerization of mutant and WT p53 produces an inactive conformation that cannot bind to gene targets. Alternatively, our group has suggested that p53 inactivation is related to a prion-like effect of mutant p53 on WT p53 (Ano Bom et al. 2012). This concept explains additional mutant p53 GoF effects and mutant p53-specific protein–protein interactions, which are further discussed below.
In this review, we describe the properties of p53 aggregation and comment on the cellular fates and consequences of these aggregates. The prion-like properties and cell-to-cell transmissibility of these proteins and the role of p53 oligomers in this process are also considered.
THE ROLE OF ORDERED AND INTRINSICALLY DISORDERED DOMAINS IN TUMOR-SUPPRESSOR PROTEIN FUNCTION AND CANCER DEVELOPMENT
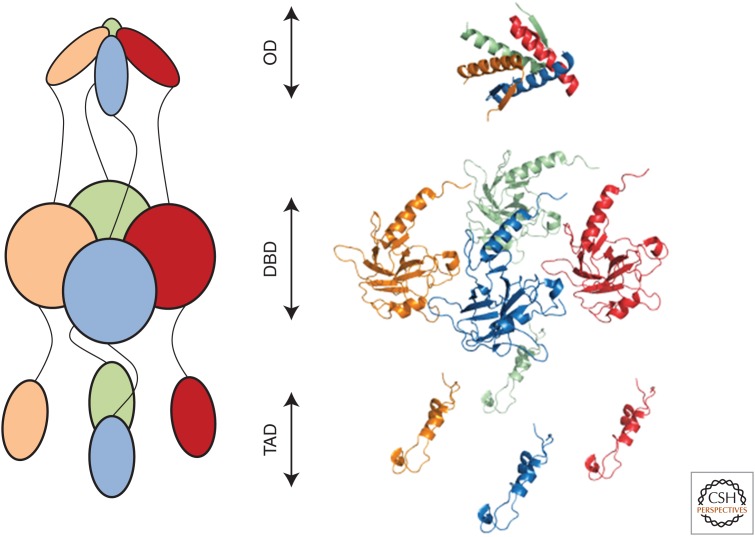
Tumor suppressors are typically multidomain proteins flanked by disordered segments. p53 includes three well-organized regions: an N-terminal transactivation domain spanning residues 1-70 (Dawson et al. 2003), a sequence-specific DNA-binding domain within residues 94-293, and an oligomerization domain consisting of residues 324-355 (Fig. 1). Flanking these regions are two disordered polyproline regions: one polyproline region composed of residues 71-93, which links the transactivation domain to the DNA-binding domain, and a second polyproline region consisting of residues 294-323, which links the DNA-binding domain to the oligomerization domain. The C-terminus (residues 356-393) contains an unstructured basic region.
Figure 1.
The three-dimensional structure of p53. Active p53 is a tetrameric protein consisting of an oligomerization domain (OD), a DNA-binding domain (DBD), and a transactivation domain (TAD). Schematic representation (left) and crystal structures (right; PDB IDs: OD, 2J0Z; DBD, 2XWR; and TAD, 2L14) of p53 monomers (green, red, blue, and gold) in the predicted active conformation based on electron microscopy and small-angle X-ray-scattering reconstructions (Tidow et al. 2007; Melero et al. 2011).
p53 activity and stability are regulated by the phosphorylation of specific serine and threonine regions of its transactivation domain (Botuyan et al. 1997; Lee et al. 2000). The transcription factors p300/CBP and the ubiquitin protein ligase MDM2 (HDM2 for the human ortholog) bind to overlapping binding sites within this N-terminal region. Following DNA damage, phosphorylation of p53 at Ser15, Thr18, and Ser20 dissociates MDM2 from the transactivation domain and increases its affinity for p300/CBP, facilitating p53 transcriptional activity (Kussie et al. 1996; Ferreon et al. 2009; Lee et al. 2010). The direct effect of the binding of MDM2 to the transactivation domain is the inhibition of p53 transcriptional activity, although the major effect of MDM2 on p53 occurs via its E3 ubiquitin-ligase activity (Michael and Oren 2003). The p53–MDM2 interaction was shown to be critical because the lethality of MDM2-null mice was rescued by the simultaneous deletion of the TP53 gene (Jones et al. 1995; Montes de Oca Luna et al. 1995). In addition, a second MDM2 binding site in the p53 core domain has been speculated to stabilize the MDM2–p53 interaction during degradation (Yu et al. 2006).
MDMX (HDMX for the human ortholog) is another key regulator of p53 that binds to the transactivation domain of p53 and inhibits its transcriptional activity (Shvarts et al. 1996, 1997; Böttger et al. 1999). MDMX is also a partner of MDM2 (Sharp et al. 1999; Tanimura et al. 1999) and contributes to tumor formation, as shown by the retrovirus-mediated MDMX overexpression in primary mouse embryonic fibroblasts, which induced immortalization and neoplastic transformation in combination with HRasV12 (Danovi et al. 2004). A significant proportion of tumor cell lines express increased levels of HDMX compared with normal cell lines (Ramos et al. 2001). Systematic screening of HDMX expression or amplification in more than 500 human tumors of different origins revealed HDMX overexpression in many of them (Danovi et al. 2004), suggesting that HDMX is an oncogene. In addition, MDMX was shown to block the p300/CBP-mediated acetylation of p53 (Sabbatini and McCormick 2002), a modification that is critical for the tumor-suppressor functions of p53 (Brooks and Gu 2011).
The first disordered polyproline region of p53 contains five partially conserved PxxP motifs, which regulate its activity. The abrogation of this polyproline region does not influence p53-mediated transcriptional transactivation but severely affects its suppression of tumor growth (Walker and Levine 1996). The study by Walker and Levine (1996) provided the first evidence that the transcriptional activity and tumor growth suppression of p53 are uncoupled events and that the first polyproline region mediates a critical activity involved in p53-dependent tumor suppression. Further investigation has shown that this region is crucial for p53-mediated apoptosis but is not necessary for p53-mediated cell growth arrest or suppression of cell transformation (Sakamuro et al. 1997). The first polyproline region is also important for p53 regulation. The absence of this domain increases the affinity of MDM2 for p53, rendering p53 more susceptible to negative regulation and facilitating the ubiquitination and nuclear export of this protein (Berger et al. 2011). Further studies clarified the mechanism by which the polyproline region renders p53 as sensitive to MDM2 inhibition. Pro82 in the first polyproline region is required for the p53–Chk2 interaction in response to DNA damage and the subsequent phosphorylation of p53 at Ser20 (Berger et al. 2005). The presence of germline Pro82 (P82L) mutations in cancer patients with Li–Fraumeni syndrome and ovarian carcinoma (Sun et al. 1996) and somatic mutations (P85S and P89S) in bladder tumors (Taylor et al. 1996) supports the importance of the first polyproline region of p53 in regulating the activity of this protein.
The disordered C-terminal basic region of p53 is another site of multiple posttranslational modifications, especially acetylation. The basic region contains multiple acetylated lysines that function similarly to histone tails. The binding of p300/CBP acetyltransferase to the transactivation domain of p53 acetylates not only histones but also p53 itself (Gu and Roeder 1997). The basic C-terminal region of p53 binds to nonspecific sequences in DNA (Wang et al. 1993; Wu et al. 1995; Jayaraman and Prives 1999) and regulates the sequence-specific binding of its core domain (Hupp et al. 1992; Ahn and Prives 2001).
CONSEQUENCES OF MUTATIONS AND MISFOLDING OF p53 IN CANCER DEVELOPMENT
From 3281 samples clustered into 12 tumor types, 127 mutated genes involved in diverse signaling and enzymatic processes were identified. In these samples, TP53 was the most frequently mutated gene (42% of the samples) (Kandoth et al. 2013). The current database of TP53 mutants (p53.iarc.fr) includes 45,000 somatic mutations, most of which provide a selective advantage to a specific cell clone in its microenvironment, increasing its survival or reproduction. These variants, also known as driver mutations, are commonly involved in clonal expansion and tumorigenesis (Stratton et al. 2009).
At least three primary factors influence the mutational frequency of TP53 in tumors: (1) the tumors are highly heterogeneous and comprise different subtypes; (2) the stage of development; and (3) exogenous factors, such as viral or bacterial infection. Depending on the type of cancer, the rate of TP53 mutations range from <5% (as in cervical carcinoma) to 90% (as in ovarian carcinoma). In breast carcinoma, for example, molecular profiling revealed four major subtypes displaying variable frequencies of TP53 mutations: 12%, 30%, 72%, and 80% for the luminal A, luminal B, HER2-E, and basal-like subtypes, respectively (Weigelt et al. 2010; Curtis et al. 2012). Considering the stage of development, a lower frequency of TP53 mutations was reported in primary prostate tumors (between 10% and 20%) than in metastatic tumors (up to 50%) (Schlomm et al. 2008). In biphasic chronic myeloid leukemia, TP53 mutations most frequently occur during the blastic phase (Calabretta and Perrotti 2004; Malcikova et al. 2014). Among exogenous factors, several human viruses impair p53 activity. In cervical cancer, the human papillomavirus E6 protein targets p53 for degradation (Scheffner et al. 1990). Additionally, the p53 R249S variant is commonly observed in liver cancer associated with aflatoxin-B1 food contamination (Aguilar et al. 1993).
Nonsynonymous single-nucleotide variants or missense mutations (i.e., those affecting amino-acid sequences) are the most common TP53 alterations. Biallelic substitutions are very common in tumor-suppressor genes, but TP53 is most commonly affected by monoallelic alterations, including at six hotspots within the DNA-binding domain (R175, G245, R248, R249, R273, and R282). Despite the dominance of nonsynonymous single-nucleotide variants, synonymous single-nucleotide variants, frameshift mutations, silent mutations, splice mutations, CpG dinucleotide transitions, and mutations affecting posttranslational regions of TP53 have also been reported in different tumor types (Leroy et al. 2014). In osteosarcoma, for example, a high frequency of TP53 gene deletion was reported (Masuda et al. 1987; Barretina et al. 2010). Although noninherited variants of TP53 are associated with several types of cancer, germline mutations cause a rare autosomal-dominant predisposition to cancer, termed Li–Fraumeni syndrome (Li 1969a,b). Most Li–Fraumeni patients do not present with a single site-specific tumor, but rather present at an early age with a variety of tumor types, most of which carry a specific p53 germline mutation displaying approximately 90%–95% penetrance (Malkin et al. 1990). In sporadic glioblastomas, dominant-negative p53 mutants were shown to accelerate tumor growth and development, because the average age at diagnosis was younger for patients carrying these mutations than for those carrying recessive mutations (Marutani et al. 1999).
Mutations in the p53 gene occur in more than half of human cancers and often result in altered transcriptional activities. In contrast, mutations in the p63 and p73 genes are not commonly observed in cancer (Levrero et al. 2000). In most cases, defective p53 is caused by a single amino-acid mutation in the DNA-binding domain. Such mutations are divided into two classes: class I (conformation mutants), which involve amino acids important for maintaining p53 structure (i.e., R175H, G245S, R249S, and R273H); and class II (contact mutants), which involve amino acids that directly interact with DNA (i.e., R248 and R273) (p53.free.fr). It has also been shown that contact mutants, such as R248Q, can have structural consequences (Wong et al. 1999; Ishimaru et al. 2003a,b).
At present, no consensus explanation for the circumstances under which p53 acts as a tumor suppressor or oncogene is available. Yeast model analysis of more than 2000 p53 variants revealed that the transcriptional activity of p53 variants ranges from complete inactivation to enhanced activation compared with WT p53 (Kato et al. 2003). The high frequency of p53 mutations in several tumors and the observation that p53−/− mice show a dramatic predisposition to early-onset cancer confirm that p53 acts as an important tumor suppressor (Vogelstein et al. 2000; Kenzelmann and Attardi 2010). Controversially, mouse models have shown that as a tumor suppressor, p53 responds not to acute DNA damage but to oncogene-induced expression of the tumor suppressor p19ARF, which activates p53 via the sequestration and inhibition of MDM2 (Christophorou et al. 2006; Efeyan et al. 2007). Additionally, mice deficient in p21, Puma, and Noxa were unable to undergo p53-mediated apoptosis, G1/S cell-cycle arrest, or senescence but remained free of tumor development for at least 500 days (Velente et al. 2013). These findings suggest that the induction of apoptosis, cell-cycle arrest, and senescence are not required for the p53-mediated suppression of tumor development (Li et al. 2012).
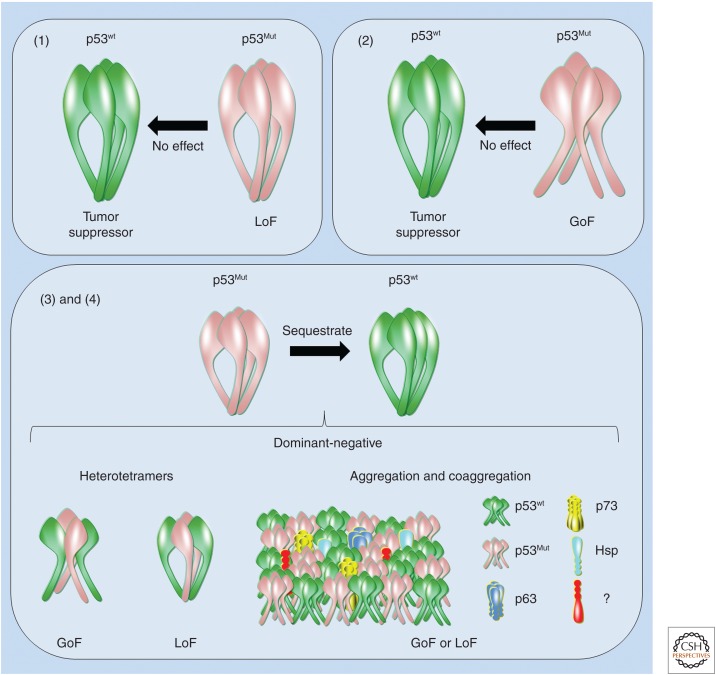
At the molecular level, and from the most simplistic and combinatorial perspective, monoallelic mutations in p53 result in the following effects: (1) certain p53 mutants lack the activity of WT p53; (2) certain p53 mutants acquire oncogenic activity without disturbing the activity of WT p53; (3) certain p53 mutants inhibit the WT protein via a dominant-negative effect and exhibit oncogenic activity; and (4) certain p53 mutants inhibit the WT protein via a dominant-negative effect but exert no other activity (Fig. 2). The gain of oncogenic activity by mutant p53 was first evidenced by the transfection of mutant p53 into TP53-null cells, conferring these cells with the ability to generate tumors in mice (Wolf et al. 1984; Halevy et al. 1990; Shaulsky et al. 1991). In addition to this GoF effect, certain p53 mutants act via a dominant-negative mechanism in which hetero-oligomerization/aggregation may occur between the mutant and WT p53 proteins (Milner and Medcalf 1991; Milner et al. 1991). In this model, a dominant-negative effect might also be observed for mutant p53 and its ancestral p63 and p73 paralogs. Using nanoflow electrospray ionization mass spectrometry, p63 and p73 homotetramers were shown to form after 30 min of incubation, along with mixed tetramers at 3:1, 2:2, and 1:3 ratios. In contrast to these results, neither incubation of p53 homotetramers in p73 nor p63 homotetramers resulted in exchange after 24 h, indicating the divergent evolution of the oligomerization domain within the p53 family (Joerger et al. 2009). Furthermore, a GoF phenotype associated with certain p53 mutants was shown based on their coaggregation with p63 and p73 (Xu et al. 2011). A dominant-negative effect also likely occurs via high-level oligomeric states in which aggregated mutant p53 sequesters WT p53 into mixed oligomers (Ano Bom et al. 2012). Several oncogenic functions of mutant p53 have been characterized (Muller and Vousden 2013; Bisio et al. 2014), but these functions are beyond the scope of this review.
Figure 2.
Effects of p53 mutation on its activity. Certain p53 mutants (1) lose wild-type (WT) activity (loss-of-function [LoF]); (2) acquire oncogenic activity without disturbing the activity of WT p53 (gain-of-function [GoF]); (3) inhibit the WT p53 protein via a dominant-negative effect and display oncogenic activity (GoF); and (4) inhibit the WT p53 protein via a dominant-negative effect but display no other activity (LoF). Dominant-negative mechanisms are exemplified (i.e., heterotetramers, aggregation, or coaggregation). Cellular partners (e.g., WT p53, p63, p73, HSPs, and proteins that have yet to be discovered [?]) coaggregate with mutant p53.
In the late 1990s, Sir Alan Fersht’s group used differential scanning calorimetry and spectroscopy to evaluate the thermodynamics of WT and mutant (R175H, C242S, R248Q, R249S, and R273H) p53, and they observed irreversible denaturation and aggregation of these forms under certain conditions (Bullock et al. 1997). In 2003, a study by our group provided the basis for understanding p53 core domain aggregation (Ishimaru et al. 2003a,b). Recently, we examined whether WT and a p53 hotspot mutant aggregate as an amyloid fibril under physiological conditions and whether the mutant seeds the aggregation of WT p53 (Ano Bom et al. 2012). Using several structural and cellular approaches, we showed the amyloid nature of the WT and mutant p53 aggregates. Furthermore, our study revealed that a seed of amyloid aggregates formed from the R248Q p53 hotspot mutant accelerated the aggregation of WT p53 (Ano Bom et al. 2012). We established that prion-like behavior by p53 may explain the dominant-negative and GoF effects of certain p53 mutations.
Supporting the role of aggregated p53 and its prion-like behavior in cancer, we observed greater co-localization of mutant p53 with amyloid oligomers in breast cancer MDA-MB-231 cells than in WT p53-expressing (MCF7) cells (Ano Bom et al. 2012). Suggesting direct implications for cancer pathogenesis, similar results were observed in biopsies from breast cancer patients carrying specific p53 mutations (R175H, H193L, I195L, Y234C, G245S, or R248Q) (Levy et al. 2011) and in biopsies from the skin of six patients with basal-cell carcinoma (Lasagna-Reeves et al. 2013). Furthermore, a high level of p53 immunostaining was observed within aggregates containing mutant and WT p53 in prostate cancer samples (Kluth et al. 2014). Finally, it was shown that in a population of high-grade serous ovarian carcinoma (HGSOC) cancer cells exhibiting cancer stem cell properties, p53 aggregation is associated with p53 LoF and platinum resistance; however, when HGSOC cells differentiated into their chemosensitive progeny, they lost their capacity for both tumor initiation and p53 aggregation (Yang-Hartwich et al. 2015). Furthermore, the overexpression of p14ARF, a positive regulator of p53, inhibited MDM2-mediated p53 degradation and led to an imbalance in p53 turnover that promoted the formation of p53 aggregates. When p14ARF was inhibited, p53 aggregation was suppressed, and the cancer cells became sensitized to platinum treatment. Moreover, using two-dimensional gel electrophoresis and mass spectrometry, Yang-Hartwich et al. (2015) discovered that aggregated p53 acts uniquely by interacting with proteins involved in cancer cell survival and tumor progression. This correlation between p53 aggregation and platinum resistance is relevant to the poor prognosis of patients with HGSOC (Yang-Hartwich et al. 2015). Together, these recent ex vivo experiments on p53 confirm the involvement of p53 aggregation in cancer. Although a prion-like mechanism would explain the dominant-negative and GoF effects observed during p53 aggregation, several questions remain to be answered before this mechanism can be defined as an etiologic factor for cancer pathogenesis, invasiveness, and metastasis (Silva et al. 2014).
HOW WOULD THE AMYLOID AGGREGATION OF p53 CONTRIBUTE TO ONCOGENESIS?
The term “amyloid” is generically used for all proteins capable of forming large, insoluble fibrils (Fig. 3A). At the atomic level, amyloid fibrils are initiated from small aggregates, or seeds, of the nucleating amino-acid sequence in a β-sheet conformation (Sawaya et al. 2007). The general structure consists of a few β-strands that associate in parallel or antiparallel orientations via backbone hydrogen bonding to form a β-sheet. Two protofilaments then associate via side-chain interactions. The close interdigitation of the side chains into a complimentary structure results in a steric zipper conformation that excludes water from the core of the interface (Foguel et al. 2003; Sawaya et al. 2007; Reddy et al. 2010). Most frequently, the interface is composed of hydrophobic residues, although polar sequences can also form steric zippers (Balbirnie et al. 2001). The addition of protofilaments along the fiber axis results in the elongation of the fibril structures. Dyes such as thioflavin T and Congo red tend to bind to amyloid structures, causing changes in their spectrophotometric properties. Although these dyes are generally specific enough to be used as diagnostic tools, varying binding modes are possible depending on the fibril type, which may cause false positive or negative results (Groenning 2010). Evidence suggests that, although large amyloid fibrils physically disrupt organelles and tissues, smaller aggregates are more toxic (Buxbaum 2004; Bitan et al. 2005). There are several hypotheses as to why this is the case, but, currently, the exact mechanisms of amyloid oligomer toxicity are not well established (Kayed and Lasagna-Reeves 2013). Although amyloid fibrils are typically associated with pathology, some amyloid fibrils perform biological functions. For instance, amyloid fibril formation by a domain of Pmel17 plays a role in melanin polymerization in animal cells, and some spider silk fibers consist of amyloid fibrils formed from spidroin (Chiti and Dobson 2006; Hammer et al. 2008; Shewmaker et al. 2011). Amyloid fibril formation has been shown for numerous proteins under the appropriate conditions; however, not all of these proteins are considered prion-like (Bucciantini et al. 2002). A prion is a type of amyloid fibril that is capable of converting a normal endogenous cellular protein to an amyloid conformation, spreading these fibrils between cells (Prusiner 1982, 1998; Prusiner et al. 1998). p53 and other amyloid fibrils have been referred to as prion-like, owing to the ability of these aggregates to propagate to other cells by penetrating cell membranes (Brundin et al. 2010; Ano Bom et al. 2012; Forget et al. 2013; Rangel et al. 2014).
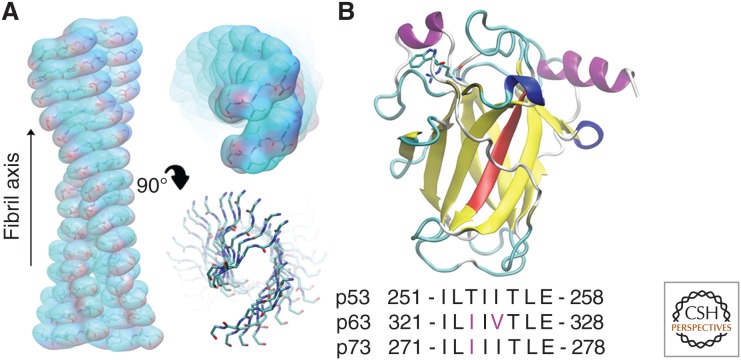
Figure 3.
Amyloid formation by the p53 DNA-binding domain (DBD). (A) Model of the p53 252-256 amyloid fibril generated based on molecular dynamics (MDs) simulations of the aggregation-prone sequence. (B) The DBD structure of p53 (PDB ID: 2XWR) (Natan et al. 2011) showing the amyloidogenic 251-258 region (red), Trp91/Arg174 (atomic detail), and equivalent sequence comparison.
The 1997 Nobel Prize in physiology or medicine was awarded to Stanley Prusiner for his discovery of prions, transmissible polypeptide particles that undergo a conformational change from their cellular form (PrPC) to a β-sheet-rich pathogenic form (PrPSc). This conformational modification of the prion protein is the basis of its transmissibility and the pathogenesis of several diseases, including bovine spongiform encephalopathy in cows and Creutzfeldt–Jakob disease in humans (Prusiner 1998; Prusiner et al. 1998, 2015). Thus, to be classified as a prion, a protein must be transmissible in vitro and in vivo. The first characteristic shared between p53 and prions is the conformational conversion of p53 during tumorigenesis (WT p53 to mutant p53), and the second characteristic is the ability of mutant p53 to sequester WT p53 into amyloid species. Despite these findings, it may be premature to classify p53 as a prion because the mechanisms underlying its transmission have yet to be definitively shown. However, recent studies have shown independent mechanisms of p53 secretion and uptake by cells (Lee et al. 2009a, 2013; Forget et al. 2013), suggesting that p53 may act as a transmissible agent. In initial experiments, the oncogenic protein Kristen-Ras (K-Ras) was shown to participate in p53 suppression by inducing Snail. The depletion of Snail induced p53 expression in K-Ras mutant cancer cells but not in WT K-Ras cancer cells (Halaschek-Wiener et al. 2004; Lee et al. 2009b). Thus, a direct correlation between K-Ras-mediated p53 suppression and tumorigenesis was established for lung and pancreatic cancers, which display a higher frequency of K-Ras mutations than other cancer types (Karnoub and Weinberg 2008). Snail suppresses p53 in response to the oncogenic functions of K-Ras, including the secretion of p53 from cells and its subsequent uptake by K-Ras mutant cells via caveolin-1-mediated endocytosis (Lee et al. 2013). Furthermore, aggregates of full-length p53 were shown to penetrate HeLa and NIH-3T3 cells via macropinocytosis and to induce the aggregation of intracellular p53 (Forget et al. 2013). Although mouse models remain inadequate, these surprising mechanisms of between-cell p53 transmissibility have prompted new discussions about its proposed prion-like mechanism and have indicated the need for ongoing experiments by our group and others to demonstrate the involvement of p53 aggregates in cancer pathogenesis and progression.
THE p53 FAMILY AND THE INTERACTIONS BETWEEN ITS MEMBERS
Almost 20 years after the discovery of p53, two related genes, TP63 and TP73, were identified. These genes encode proteins with structures and functions that are similar to but distinguishable from those of p53 (Kaghad et al. 1997; Yang et al. 1998; Levrero et al. 2000). Similar to p53, family members include transcription factors involved in the regulation of the cell cycle, proliferation, differentiation, DNA damage response, and apoptosis (Levrero et al. 2000; Collavin et al. 2010). p63 and p73 not only bind to and activate consensus p53 response elements, such as p21 and Puma, but also have unique gene targets (Fontemaggi et al. 2002; Osada et al. 2005). Despite their partially redundant or unknown functions, p63, p73, and their various isoforms have been reported to regulate genes involved in embryonic development, including loricrin and involucrin (De Laurenzi et al. 2000). Mouse models have provided insight into the distinct biological roles of p53 and its paralogs (Yang et al. 2002; Lu et al. 2009). The crucial role of p53 in averting the formation of spontaneous tumors was shown by p53-null mouse studies, which showed that these mice die of cancer at a young age (Donehower et al. 1992; Yamamoto et al. 2000). Although a subset of p53-null mice show developmental and fertility defects, they are generally viable and undergo normal embryonic development (Rotter et al. 1993; Sah et al. 1995). Conversely, p73-null mice are born viable but show nervous-system abnormalities, hydrocephalus, and immunological disorders such as chronic inflammation (Yang et al. 2000). These mice also show reproductive and behavioral defects and generally die within the first 2 months. p63-null mice are born alive but die immediately after birth (Mills et al. 1999; Yang et al. 1999). They show a severe phenotype, lacking limbs and a wide range of epithelial structures, including the skin, prostate, breasts, and urothelia, indicating that p63 is required to maintain the pool of proliferating stem cells during epithelial development (Senoo et al. 2007; Su et al. 2009).
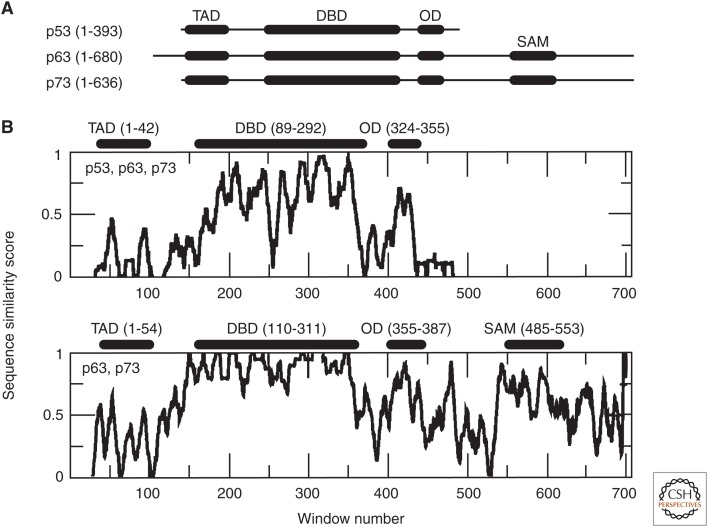
The p53 family of proteins displays sequence similarity in its transactivation domains, DNA-binding domains, and oligomerization domains (Fig. 4). p63 and p73 are more homologous than p53 and contain a sterile α-motif domain, which is present in a variety of proteins and plays a role in the regulation of developmental processes (Schultz et al. 1997). Each protein is expressed as numerous isoforms, which typically lack the N- or C-terminal region. Their DNA-binding domains display the highest homology, sharing ∼60% identity; the DNA-binding domains of p63 and p73 share ∼86% identity. The p53 DNA-binding domain sequence most strongly differs from that of p63 and p73 in the L2 loop (surrounding window 250; residues 176-192). p63 is slightly longer than p73, primarily because it contains additional N-terminal residues. Structurally, the DNA-binding domain is highly similar among these three proteins, maintaining the same β-sandwich fold (Joerger and Fersht 2008). The oligomerization domains of p63 and p73 can form weak heterotetramers with each other, but not with the oligomerization domain of p53 because of their high sequence similarity, which is less similar to the sequence of p53 (Li and Prives 2007; Joerger et al. 2009).
Figure 4.
Domain structure and sequence comparisons of p53 family proteins. (A) The predominant domain structures of p53, p63, and p73, depicting the transactivation domain (TAD), DNA-binding domain (DBD), oligomerization domain (OD), and sterile α-motif (SAM) domains. (B) Sequence similarity scores for the comparison between p53, p63, and p73 (top), and between p63 and p73 (bottom). The similarity scores were generated based on the Clustal Omega (Sievers et al. 2011) gapped alignment of the full-length protein sequences using a 10-residue moving window average and considering the fully conserved, highly conserved, moderately conserved, nonconserved, and gapped positions.
In vivo and in vitro studies revealed that the DNA-binding domain of certain p53 mutants directly associate with p63 and p73 (Di Como et al. 1999; Strano et al. 2000, 2002; Gaiddon et al. 2001). The association of mutant p53 with WT p53 or p63/p73 results in a dominant-negative effect in which tumor-suppressor functions are impaired and oncogenicity is increased (Li and Prives 2007; Xu et al. 2011; Muller and Vousden 2014).
MECHANISMS UNDERLYING THE PRION-LIKE AGGREGATION OF TUMOR SUPPRESSORS: DOMINANT-NEGATIVE AND GAIN-OF-FUNCTION EFFECTS
Over the last 20 years, many researchers have detected abnormal p53 accumulation in cells. In the early 1990s, studies showed the large, insoluble buildup of p53 in cancer cells carrying or lacking p53 mutations (Moll et al. 1992, 1995, 1996). Subsequently, p53 aggregates induced by low concentrations of guanidine hydrochloride were found to dissociate under pressure, indicating that these aggregates contain water-excluded cavities, a property of amyloid fibrils (Ishimaru et al. 2004). In 2003, it was shown that the DNA-binding domains of WT and R248Q mutant p53 form fibrillar aggregates displaying a high β-sheet content (Ishimaru et al. 2003a,b). It was also shown that class I mutations increased the rate of aggregation relative to the WT sequence (Levy et al. 2011; Xu et al. 2011; Ano Bom et al. 2012). Several class I mutants also co-localized with p63/p73 (Strano et al. 2000, 2002; Gaiddon et al. 2001; Puca et al. 2011). In vitro and in vivo studies of p53 deletion mutants confirmed direct interactions between the DNA-binding domains of mutant p53 and p63/p73. Harshly prepared or repeatedly frozen/thawed p53 was also shown to bind to p73 (Bensaad et al. 2003). Furthermore, the sensitivity of p53 mutants to an antibody that binds to a normally inaccessible region of the p53 DNA-binding domain was associated with p63/p73-binding ability (Gannon et al. 1990). Change in the conformation of the DNA-binding domain of p53 is the only key element for interacting and interfering with p73 (Bensaad et al. 2003). The loss of a structural Zn2+ ion within the DNA-binding domain also promotes p53 aggregation (Butler and Loh 2003). Under physiological conditions, a considerable proportion of total p53 exists in a Zn2+-free state, and this population of Apo DNA-binding domain is able to promote the aggregation of Zn2+-bound p53 DNA-binding domains. The R175H p53 mutant shows significantly accelerated Zn2+ loss compared with WT p53, potentially explaining its high propensity for aggregation (Butler and Loh 2003; Xu et al. 2011). Interestingly, R175H and other structural mutants, such as G245S, R249S, and R248Q, are located near the Zn2+-binding loops of p53.
A link between conformational changes in the p53 DNA-binding domain and an increased propensity for aggregation has been recognized for some time, but the molecular details of these aggregates have only recently emerged. Bioinformatic scanning of the p53 sequence suggested that a conserved aggregation-prone peptide (residues 251-258) within the DNA-binding domain of WT p53 might represent the nucleation site for aggregate formation (Fig. 3) (Xu et al. 2011; Ghosh et al. 2014; Rangel et al. 2014; Soragni et al. 2016). Further studies of this short segment using synthetic peptides revealed that it formed amyloid-like aggregates with WT p53 and coaggregated with p63/p73, leading to the inhibition of their functions (Xu et al. 2011; Ghosh et al. 2014). Furthermore, p53 aggregation and coaggregation was suppressed by inserting arginine mutations (p53 I254R, p63 I324R, and p73 I274R) or by scrambling the p53 peptide sequence. Arginine, lysine, and proline often flank sequences displaying a high propensity to aggregate and are considered as gatekeeper amino acids because of their ability to oppose aggregation (Rousseau et al. 2006). Together, the inability of these arginine mutants and the scrambled p53 peptide to aggregate demonstrate that fibril construction based on the p53 sequence is stringent and requires defined amino-acid composition and order. This finding is consistent with the high complementarity required for steric zipper formation. Molecular dynamics (MDs) simulations have also shown that residues 251-258 are aggregation prone. Simulations performed by Ghosh et al. (2014) on multiple copies of the 251-258 sequence revealed that it was inclined to self-associate, forming clusters enriched in β-sheet content. Our tests showed similar results, in which the 252-256 LTIIT region formed stable structures that tended to propagate, enabling the modeling of a potential fibril structure based on this sequence (Fig. 3). These and other investigations have shown the utility of MD simulations for assessing the conformational biases of specific amino-acid sequences and for providing insight into the self-assembly of amyloid fibrils (Jang and Shin 2006; Cino et al. 2013).
Although the DNA-binding domains of p63 and p73 contain similar amino-acid sequences in the regions corresponding to the aggregation-prone sequence of p53 (Fig. 3), they appear to have a much lower tendency to aggregate. Nonetheless, the coaggregation of p63/p73 with p53 does appear to occur via their similar motifs (p63 321-328 and p73 271-278) because the p63 I324R and p73 I274R mutants failed to interact with mutant p53 (Xu et al. 2011). Notably, p63 and p73 contain an isoleucine instead of a threonine in the i + 2 position, corresponding to T253 of p53, located near the center of the p53 aggregation region. The presence of this bulky isoleucine in this position rather than threonine may impair the steric complementarity required for self-aggregation, although this mechanism has yet to be shown.
A recent in vitro study further investigated the mechanism of how a destabilized p53 mutant may coaggregate with WT p53 and its paralogs p63 and p73 (Wang and Fersht 2015). According to Wang and Fersht (2015), coaggregation would occur mostly by trapping rather than seeding and inducing propagation.
The currently available evidence supports the hypothesis that intrinsic instability, mutations, and Zn2+ loss can lead to the exposure of the aggregation-prone region in the p53 DNA-binding domain, leading to self-association and the formation of amyloid fibrils; however, the mechanisms underlying the coaggregation of p53 with p63/p73 are uncertain (Muller and Vousden 2013; de Oliveira et al. 2015; Wang and Fersht 2015). The interaction between Trp91 and Arg174 in the p53 DNA-binding domain increases the melting temperature by several degrees and decreases the rate of aggregation compared with constructs that begin just a few residues away, such as at Ser94 (Natan et al. 2011). In the DNA-binding domain structure, this interaction may hold the N-terminal region in an orientation that counteracts the exposure of the aggregation-prone region (Fig. 3). The p63 and p73 DNA-binding domains do not contain a corresponding W91/R174 pair, which may facilitate the exposure of their segments that coaggregate with p53. Further investigation of the structure and dynamics of the p63 and p73 DNA-binding domains is necessary to test this hypothesis and to provide deeper insight into the molecular mechanisms underlying the coaggregation of p53 with p63 and p73.
NEW APPROACHES FOR CANCER THERAPY INVOLVING p53 AGGREGATION AND PRION-LIKE BEHAVIOR
The modulation of p53 aggregation and the prion-like behavior of oncogenic p53 mutants are attractive for therapeutic applications to cancer and other protein-misfolding diseases (Silva et al. 2014; de Oliveira et al. 2015). The prevention of aggregation is a widely studied strategy focused on identifying new compounds that interfere with the formation of aggregates and with the accumulation of misfolded proteins or prefibrillar aggregates. These compounds include natural or synthetic small molecules, peptides, and nucleic acid aptamers that stabilize proteins and inhibit oligomerization and/or fibrillization (Chiti and Dobson 2006; Silva et al. 2010; Soto 2012). Other promising targets for the development of new anticancer therapies include templating, multiplication, and spreading to other cells (Rangel et al. 2014).
p53-induced gene expression and its function can be affected by incorporating mutant p53 into the WT p53 tetramers or by aggregating WT p53 via its DNA-binding domain (Ishimaru et al. 2003a; Li and Prives 2007; Magzoub and Miranker 2011; Xu et al. 2011; Muller and Vousden 2013; Wang and Fersht 2015). Alternatively, the coaggregation of p53 with p63/p73 occurs via interactions between similar sequences within their DNA-binding domains. Ultimately, the sequestration of these proteins into inactive aggregates is detrimental to their tumor-suppressor functions, and various approaches are being tested to restore normal p53 functionality.
The large number of protein–protein interactions in which p53 is involved can be exploited for therapeutic purposes. For instance, many potential treatments aim to inhibit the negative regulation of p53 by MDM2. In cases in which WT p53 is expressed, small molecules or peptides that interfere with the p53–MDM2 interaction either directly or via secondary pathways can prevent p53 degradation, restoring its functions (Selivanova 2014; Stindt et al. 2014; Yu et al. 2014). Although this approach has yielded promising results, the p53 interactome is highly complex, and any therapy that targets a specific protein or interaction may produce unexpected side effects. Alternatively, for cases in which WT p53 is not expressed, therapies using viruses to deliver functional p53 genes are being pursued (Lane et al. 2010). Although this may appear to be a more straightforward approach, resulting in fewer off-target effects, the methodology for gene therapy using viruses remains to be perfected.
Because even the WT p53 DNA-binding domain is only marginally stable, rendering it prone to aggregation, there have been efforts to improve its stability. As described previously, the DNA-binding properties of p53 are frequently lost on p53 aggregation. Our group showed that small cognate double-stranded DNA stabilizes both the p53 DNA-binding domain and full-length p53, preventing amyloid formation. Therefore, such DNA sequences might be useful as part of a new approach to cancer therapy (Ishimaru et al. 2009).
Additional efforts are aiming to restore the normal functionality of mutant p53. Compensating mutations within the p53 DNA-binding domain have been shown to increase stability and to diminish the high aggregation propensity of certain common p53 mutants, but the use of such information to generate a therapy is not straightforward (Bullock and Fersht 2001). Most efforts have focused on identifying small molecules that can reactivate mutant p53. There are currently several candidate compounds under various stages of development (Wassman et al. 2013; Yu et al. 2014). Drugs such as CDB3 are able to rescue the conformation of unstable p53 mutants (Friedler et al. 2003), allowing these proteins that show an increased half-life to reach the nucleus and act as tumor suppressors. For example, PRIMA-1 is converted to compounds that form adducts with thiols in mutant p53, inducing the apoptosis of tumor cells (Lambert et al. 2009). Furthermore, CP31398 has been shown to increase the stability of several class I mutant p53 DNA-binding domains, restoring their tumor-suppressor functions (Foster et al. 1999; Tang et al. 2007); however, the mechanism of action of CP31398 is unclear because it appears to bind to target p53 DNA sequences rather than the p53 DNA-binding domain (Rippin et al. 2002). Alternatively, PRIMA-1, which has shown positive results in stage I/II clinical trials, covalently binds to Cys124 in the p53 DNA-binding domain and restores the functionality of several class I mutants (Lambert et al. 2009; Lehmann et al. 2012; Wassman et al. 2013; Bykov and Wiman 2014). Docking and MD simulations were used to identify a partially accessible pocket surrounding C124 that, in addition to serving as the binding site of PRIMA-1, may bind to other restorative molecules such as stictic acid. Treatment of cells carrying the R175H or G245S class I p53 mutant with PRIMA-1 or stictic acid considerably increased the transcription of the p53 gene targets p21 and Puma relative to the control treatment (Wassman et al. 2013). Some p53-reactivating compounds, such as NSC319725 and PhiKan083, have been shown to induce the transcription of p53 gene targets; however, these compounds are specific for certain DNA-binding domain mutations (R175H and Y220C, respectively) (Boeckler et al. 2008; Yu et al. 2012). Allele-specific drugs represent more targeted approaches, but compounds displaying the ability to bind to a diverse set of p53 mutants may be more practical.
Nucleic acid aptamers and glycosaminoglycans may also prevent the aggregation and the prion-like conversion of p53 in cancers related to mutant p53 (Ishimaru et al. 2009). Similar approaches have shown positive results in other protein misfolding–related diseases, such as transmissible spongiform encephalopathies (Vieira et al. 2011, 2014; Silva et al. 2013). The restoration of mutant p53 to an active conformation via ZnCl2 supplementation has also been shown. Zinc treatment reduced the interaction of p53 with p73 and restored the binding of both p53 and p73 to their target gene promoters (Puca et al. 2011; D’Orazi and Givol 2012; Garufi et al. 2013). As discussed above, Zn2+ is a major factor that modulates the stability and aggregation rate of p53. The established crucial role of Zn2+ in stabilizing and inhibiting DNA-binding domain aggregation is highly consistent with the positive results observed for small molecules that rescue p53 function via the stabilization of the DNA-binding domain. In addition to serving as one of the simplest potential therapeutics, Zn2+ easily crosses the blood–brain barrier, which is often a barrier for small molecules. Regardless of which treatments emerge, the abundance of methodologies and agents under development highlights the frequency and importance of p53 mutations.
Although compounds that stabilize the p53 DNA-binding domain would likely inhibit p53 aggregation, thereby slowing disease progression, it is unclear whether p53 aggregates and coaggregates can be dissolved. It is unlikely that the therapeutics discussed above would be able to reverse existing damage. Studies have shown that resveratrol may clear Aβ plaques in mouse models of Alzheimer's disease (Marambaud et al. 2005; Karuppagounder et al. 2009) and may inhibit carcinogenesis via the induction of p53-dependent cell death. Indeed, the transient transfection of H1299 lung-cancer cells with WT p53 sensitized these cells to the pro-apoptotic effects of resveratrol (Ferraz da Costa et al. 2012). Additionally, the curry spice curcumin has been shown to bind to and disaggregate Aβ fibrils in vivo (Yang et al. 2005). Further investigation is necessary to assess the efficacy of these compounds and to better understand their mechanisms of action and their effects on p53 and other amyloid fibrils.
In a recent study, a protein assembly modulator (CLR01) showed an intriguing effect on p53 DNA-binding domain aggregation (Herzog et al. 2015). Whereas CLR01 induced rapid formation of p53 aggregates of intermediate sizes, it inhibited additional p53 aggregation and reduced the cytotoxicity of the amyloid aggregates (Herzog et al. 2015). To some extent, this behavior is similar to that found for the PrP protein, in which some compounds, such as polyanions, stimulate or inhibit aggregation depending on the condition (Gomes et al. 2008; Silva et al. 2008; Vieira et al. 2014).
In an elegant recent study, Soragni et al. (2016) designed a cell-penetrating peptide (named RecACp53) that is able to inhibit mutant p53 amyloid aggregation. RecACp53 works by binding to the amyloidogenic segment (252-258) of p53, preventing aggregation. RecACp53 was able to rescue p53 function in cell lines and organoids derived from HGSOCs with p53 mutations, particularly in residues R175 and R248 (the most commonly mutated residues). R248Q mutant p53 has been previously shown to aggregate in breast cancer biopsies and cell lines (Levy et al. 2011; Ano Bom et al. 2012). RecACp53 was also able to reduce in vivo xenograft growth and metastasis (Soragni et al. 2016). RecACp53 had no effects when the cells had WT p53.
Using a different strategy, Yang-Hartwich et al. (2015) found that WT p53 in stem cells of HGSOC also had the property to aggregate. These cells lost the capacity for p53 aggregation and tumor development when they differentiated into chemosensitive progenies. Because p53 aggregation was dependent on the overexpression of p14ARF in the stem cell lines, its inhibition resulted in suppression of p53 aggregation (Yang-Hartwich et al. 2015). It seems that WT p53 behaves as a mutant conformation in stem cells of HGSOC. In fact, our group showed that interconversion of WT p53 into a conformation that mimics the R248Q mutant occurred in vitro after reversal of mild denaturation conditions (pressure and low temperature) (Ishimaru et al. 2003b). In tumors harboring WT p53, mild denaturing conditions, such as acidic pH in cellular microenvironments, may favor the conversion into a mutant-like conformation with prion-like and amyloidogenic properties (Ano Bom et al. 2010).
CONCLUDING REMARKS
Recently discovered characteristics of the tumor suppressor p53 include its prion-like properties and cellular uptake mechanisms, which are related to its GoF and are associated with tumor formation and malignancy. Thus, small molecules that bind to p53 and prevent its aggregation may represent a suitable strategy to prevent cancer. Currently, efforts to improve health are based on prevention, diagnosis, and treatment. Cancer has been selected as a target for precision medicine (Collins and Varmus 2015), especially because of the highly individual characteristics of malignant tumors. Each mutation of p53 exerts a different effect, many of which are likely related to the formation of homo- or heteroaggregates. Although there appears to be a direct correlation between the prion-like effect of aggregated p53 and tumorigenesis, much more research is required before these findings yield medical benefits from therapeutic intervention, early diagnosis, and prevention of cancer.
ACKNOWLEDGMENTS
Our laboratory is supported by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq awards and the Institutos Nacionais de Ciência e Tecnologia [INCT] Program), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Biology available at www.cshperspectives.org
REFERENCES
- Aguilar F, Hussain SP, Cerutti P. 1993. Aflatoxin B1 induces the transversion of G→T in codon 249 of the p53 tumor suppressor gene in human hepatocytes. Proc Natl Acad Sci 90: 8586–8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Prives C. 2001. The C-terminus of p53: The more you learn the less you know. Nat Struct Biol 8: 730–732. [DOI] [PubMed] [Google Scholar]
- Ano Bom AP, Freitas MS, Moreira FS, Ferraz D, Sanches D, Gomes AM, Valente AP, Cordeiro Y, Silva JL. 2010. The p53 core domain is a molten globule at low pH: Functional implications of a partially unfolded structure. J Biol Chem 285: 2857–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ano Bom AP, Rangel LP, Costa DC, de Oliveira GA, Sanches D, Braga CA, Gava LM, Ramos CH, Cepeda AO, Stumbo AC, et al. 2012. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: Implications for cancer. J Biol Chem 287: 28152–28162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbirnie M, Grothe R, Eisenberg DS. 2001. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated β-sheet structure for amyloid. Proc Natl Acad Sci 98: 2375–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho A, et al. 2010. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet 42: 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Le Bras M, Unsal K, Strano S, Blandino G, Tominaga O, Rouillard D, Soussi T. 2003. Change of conformation of the DNA-binding domain of p53 is the only key element for binding of and interference with p73. J Biol Chem 278: 10546–10555. [DOI] [PubMed] [Google Scholar]
- Berger M, Stahl N, Del Sal G, Haupt Y. 2005. Mutations in proline 82 of p53 impair its activation by Pin1 and Chk2 in response to DNA damage. Mol Cell Biol 25: 5380–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M, Vogt Sionov R, Levine AJ, Haupt Y. 2011. A role for the polyproline domain of p53 in its regulation by Mdm2. J Biol Chem 276: 3785–3790. [DOI] [PubMed] [Google Scholar]
- Bisio A, Ciribilli Y, Fronza G, Inga A, Monti P. 2014. TP53 mutants in the Tower of Babel of cancer progression. Hum Mutat 35: 689–701. [DOI] [PubMed] [Google Scholar]
- Bitan G, Fradinger EA, Spring SM, Teplow DB. 2005. Neurotoxic protein oligomers—What you see is not always what you get. Amyloid 12: 88–95. [DOI] [PubMed] [Google Scholar]
- Boeckler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR. 2008. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci 105: 10360–10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttger V, Böttger A, Garcia-Echeverria C, Ramos YF, van der Eb AJ, Jochemsen AG, Lane DP. 1999. Comparative study of the p53-mdm2 and p53-MDMX interfaces. Oncogene 18: 189–199. [DOI] [PubMed] [Google Scholar]
- Botuyan MV, Momand J, Chen Y. 1997. Solution conformation of an essential region of the p53 transactivation domain. Fold Des 2: 331–342. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. 2011. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell 2: 456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundin P, Melki R, Kopito R. 2010. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol 11: 301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. 2002. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416: 507–511. [DOI] [PubMed] [Google Scholar]
- Bullock AN, Fersht AR. 2001. Rescuing the function of mutant p53. Nat Rev Cancer 1: 68–76. [DOI] [PubMed] [Google Scholar]
- Bullock AN, Henckel J, De Decker BS, Johnson CM, Nikolova PV, Proctor MR, Lane DP, Fersht AR. 1997. Thermodynamic stability of wild-type and mutant p53 core domain. Proc Natl Acad Sci 94: 14338–14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JS, Loh SN. 2003. Structure, function, and aggregation of the Zinc-free form of the p53 DNA binding domain. Biochemistry 42: 2396–2403. [DOI] [PubMed] [Google Scholar]
- Buxbaum JN. 2004. The systemic amyloidoses. Curr Opin Rheumatol 16: 67–75. [DOI] [PubMed] [Google Scholar]
- Bykov VJ, Wiman KG. 2014. Mutant p53 reactivation by small molecules makes its way to the clinic. FEBS Lett 588: 2622–2627. [DOI] [PubMed] [Google Scholar]
- Calabretta B, Perrotti D. 2004. The biology of CML blast crisis. Blood 103: 4010–4022. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. 2006. Protein misfolding, functional amyloid and human disease. Annu Rev Biochem 75: 333–366. [DOI] [PubMed] [Google Scholar]
- Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. 2006. The pathological response to DNA damage does not contribute to p53-mediated tumor suppression. Nature 443: 214–217. [DOI] [PubMed] [Google Scholar]
- Cino EA, Choy WY, Karttunen M. 2013. Conformational biases of linear motifs. J Phys Chem B 117: 15943–15957. [DOI] [PubMed] [Google Scholar]
- Collavin L, Lunardi A, Del Sal G. 2010. p53-family proteins and their regulators: Hubs and spokes in tumor suppression. Cell Death Differ 17: 901–911. [DOI] [PubMed] [Google Scholar]
- Collins FS, Varmus H. 2015. A new initiative on precision medicine. N Engl J Med 372: 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. 2012. The genomic and transcriptomic architecture of 2,000 breast tumors reveals novel subgroups. Nature 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, et al. 2004. Amplification of MdmX (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor. Mol Cell Biol 24: 5835–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson R, Müller L, Dehner A, Klein C, Kessler H, Buchner J. 2003. The N-terminal domain of p53 is natively unfolded. J Mol Biol 332: 1131–1141. [DOI] [PubMed] [Google Scholar]
- De Laurenzi V, Rossi A, Terrinoni A, Barcaroli D, Levrero M, Costanzo A, Knight RA, Guerrieri P, Melino G. 2000. p63 and p73 transactivate differentiation gene promoters in human keratinocytes. Biochem Biophys Res Commun 273: 342–346. [DOI] [PubMed] [Google Scholar]
- de Oliveira GA, Rangel LP, Costa DC, Silva JL. 2015. Misfolding, aggregation, and disordered segments in c-Abl and p53 in human cancer. Front Oncol 5: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como CJ, Gaiddon C, Prives C. 1999. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol 19: 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Butel JS, Bradley A. 1992. Deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215–221. [DOI] [PubMed] [Google Scholar]
- D’Orazi G, Givol D. 2012. p53 reactivation: The link to zinc. Cell Cycle 11: 2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Collado M, Velasco-Miguel S, Serrano M. 2007. Genetic dissection of the role of p21Cip1/Waf1 in p53-mediated tumor suppression. Oncogene 26: 1645–1649. [DOI] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. 2000. Mdm2 is a RING finger–dependent ubiquitin protein ligase for itself and p53. J Biol Chem 275: 8945–8951. [DOI] [PubMed] [Google Scholar]
- Ferraz da Costa DC, Casanova FA, Quarti J, Malheiros MS, Sanches D, Dos Santos PS, Fialho E, Silva JL. 2012. Transient transfection of a wild-type p53 gene triggers resveratrol-induced apoptosis in cancer cells. PLoS ONE 7: e48746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreon JC, Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE. 2009. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci 106: 6591–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foguel D, Suarez MC, Ferrão-Gonzales AD, Porto TC, Palmieri LC, Einsiedler CM, Andrade LR, Lashuel HA, Lansbury PT, Kelly JW, et al. 2003. Dissociation of amyloid fibrils of α-synuclein and transthyretin by pressure reveals their reversible nature and the formation of water-excluded cavities. Proc Natl Acad Sci 100: 9831–9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontemaggi G, Kela I, Amariglio N, Rechavi G, Krishnamurthy J, Strano S, Sacchi A, Givol D, Blandino G. 2002. Identification of direct p73 target genes combining DNA microarray and chromatin immunoprecipitation analyses. J Biol Chem 277: 43359–43368. [DOI] [PubMed] [Google Scholar]
- Forget KJ, Tremblay G, Roucou X. 2013. p53 aggregates penetrate cells and induce the co-aggregation of the intracellular p53. PLoS ONE 8: e69242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster BA, Coffey HA, Morin MJ, Rastinejad F. 1999. Pharmacological rescue of mutant p53 conformation and function. Science 286: 2507–2510. [DOI] [PubMed] [Google Scholar]
- Freed-Pastor WA, Prives C. 2012. Mutant p53: One name, many proteins. Genes Dev 26: 1268–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedler A, Veprintsev DB, Hansson LO, Fersht AR. 2003. Kinetic instability of p53 core domain mutants: Implications for rescue by small molecules. J Biol Chem 278: 24108–24112. [DOI] [PubMed] [Google Scholar]
- Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. 2001. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol 21: 1874–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon JV, Greaves R, Iggo R, Lane DP. 1990. Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J 9: 1595–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garufi A, Trisciuoglio D, Porru M, Leonetti C, Stoppacciaro A, D’Orazi V, Avantaggiati M, Crispini A, Pucci D, D’Orazi G. 2013. Fluorescent curcumin-based Zn(II)-complex reactivates mutant (R175H and R273H) p53 in cancer cells. J Exp Clin Cancer Res 32: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Ghosh D, Ranganathan S, Anoop A, P SK, Jha NN, Padinhateeri R, Maji SK. 2014. Investigating the intrinsic aggregation potential of evolutionarily conserved segments in p53. Biochemistry 53: 5995–6010. [DOI] [PubMed] [Google Scholar]
- Gomes MP, Millen TA, Ferreira PS, Cunha e Silva NL, Vieira TC, Almeida MS, Silva JL, Cordeiro Y. 2008. Prion protein complexed to N2a cellular RNAs through its N-terminal domain forms aggregates and is toxic to murine neuroblastoma cells. J Biol Chem 283: 19616–19625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenning M. 2010. Binding mode of thioflavin T and other molecular probes in the context of amyloid fibrils–Current status. J Chem Biol 3: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal region. Cell 90: 595–606. [DOI] [PubMed] [Google Scholar]
- Halaschek-Wiener J, Wacheck V, Kloog Y, Jansen B. 2004. Ras inhibition leads to transcriptional activation of p53 and down-regulation of Mdm2: Two mechanisms that cooperatively increase p53 function in colon cancer cells. Cell Signal 16: 1319–1327. [DOI] [PubMed] [Google Scholar]
- Halevy O, Michalovitz D, Oren M. 1990. Different tumor-derived p53 mutants exhibit distinct biological activities. Science 250: 113–116. [DOI] [PubMed] [Google Scholar]
- Hammer ND, Wang X, McGuffie BA, Chapman MR. 2008. Amyloids: Friend or foe? J Alzheimers Dis 13: 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog G, Shmueli MD, Levy L, Engel L, Gazit E, Klärner FG, Schrader T, Bitan G, Segal D. 2015. The Lys-specific molecular tweezer, CLR01, modulates aggregation of the mutant p53 DNA binding domain and inhibits its toxicity. Biochemistry 54: 3729–3738. [DOI] [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgley CA, Lane DP. 1992. Regulation of the specific DNA binding function of p53. Cell 71: 875–886. [DOI] [PubMed] [Google Scholar]
- Irwin DJ, Abrams JY, Schonberger LB, Leschek EW, Mills JL, Lee VM, Trojanowski JQ. 2013. Evaluation of potential infectivity of Alzheimer and Parkinson disease proteins in recipients of cadaver-derived human growth hormone. JAMA Neurol 70: 462–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru D, Andrade LR, Teixeira LS, Quesado PA, Maiolino LM, Lopez PM, Cordeiro Y, Costa LT, Heckl WM, Weissmüller G, et al. 2003a. Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry 42: 9022–9027. [DOI] [PubMed] [Google Scholar]
- Ishimaru D, Maia LF, Maiolino LM, Quesado PA, Lopez PC, Almeida FC, Valente AP, Silva JL. 2003b. Conversion of wild-type p53 core domain into a conformation that mimics a hot-spot mutant. J Mol Biol 333: 443–451. [DOI] [PubMed] [Google Scholar]
- Ishimaru D, Lima LM, Maia LF, Lopez PM, Ano Bom AP, Valente AP, Silva JL. 2004. Reversible aggregation plays a crucial role on the folding landscape of p53 core domain. Biophys J 87: 2691–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru D, Ano Bom AP, Lima LM, Quesado PA, Oyama MF, De Moura Gallo CV, Cordeiro Y, Silva JL. 2009. Cognate DNA stabilizes the tumor suppressor p53 and prevents misfolding and aggregation. Biochemistry 48: 6126–6135. [DOI] [PubMed] [Google Scholar]
- Jang S, Shin S. 2006. Amyloid β-peptide oligomerization in silico: Dimer and trimer. J Phys Chem B 110: 1955–1958. [DOI] [PubMed] [Google Scholar]
- Jayaraman L, Prives C. 1999. Covalent and noncovalent modifiers of the p53 protein. Cell Mol Life Sci 55: 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR. 2008. Structural biology of the tumor suppressor p53. Annu Rev Biochem 77: 557–582. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Rajagopalan S, Natan E, Veprintsev DB, Robinson CV, Fersht AR. 2009. Structural evolution of p53, p63 and p73: Implication for heterotetramer formation. Proc Natl Acad Sci 106: 17705–17710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378: 206–208. [DOI] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, et al. 1997. Monoallelically expressed gene related to p53 at 1P36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90: 809–819. [DOI] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkkowski MA, et al. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub AE, Weinberg RA. 2008. Ras oncogenes: Split personalities. Nat Rev Mol Cell Biol 9: 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuppagounder SS, Pinto JT, Xu H, Chen HL, Beal MF, Gibson GE. 2009. Dietary supplementation with resveratrol reduces plaque pathology in a transgenic model of Alzheimer’s disease. Neurochem Int 54: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C. 2003. Understanding the function–structure and function–mutation relationships of the p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci 100: 8424–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R, Lasagna-Reeves CA. 2013. Molecular mechanisms of amyloid oligomers toxicity. J Alzheimers Dis 33: S67–S78. [DOI] [PubMed] [Google Scholar]
- Kenzelmann Broz D, Attardi LD. 2010. In vivo analysis of p53 tumor suppressor function using genetically engineered mouse models. Carcinogenesis 31: 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluth M, Harasimowicz S, Burkhardt L, Grupp K, Krohn A, Prien K, Gjoni J, Haß T, Galal R, Graefen M, et al. 2014. Clinical significance of different types of p53 gene alteration in surgically treated prostate cancer. Int J Cancer 135: 1369–1380. [DOI] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau L, Levine AJ, Pavletich NP. 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274: 948–953. [DOI] [PubMed] [Google Scholar]
- Lambert JM, Gorzov P, Veprintsev DB, Söderqvist M, Segerbäck D, Bergman J, Fersht AR, Hainaut P, Wiman KG, Bykov VJ. 2009. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 15: 376–388. [DOI] [PubMed] [Google Scholar]
- Lane DP, Cheok CF, Lain S. 2010. p53-based cancer therapy. Cold Spring Harb Perspect Biol 2: a001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasagna-Reeves CA, Clos AL, Castillo-Carranza D, Sengupta U, Guerrero-Muñoz M, Kelly B, Wagner R, Kayed R. 2013. Dual role of p53 amyloid formation in cancer; loss of function and gain of toxicity. Biochem Biophys Res Commun 430: 963–968. [DOI] [PubMed] [Google Scholar]
- Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, Chang J, Sung YC, Choi KY, Han KH. 2000. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem 275: 29426–29432. [DOI] [PubMed] [Google Scholar]
- Lee AS, Galea C, DiGiammarino EL, Jun B, Murti G, Ribeiro RC, Zambetti G, Schultz CP, Kriwacki RW. 2003. Reversible amyloid formation by the p53 tetramerization domain and a cancer-associated mutant. J Mol Biol 327: 699–709. [DOI] [PubMed] [Google Scholar]
- Lee SH, Lee SJ, Chung JY, Jung YS, Choi SY, Hwang SH, Choi D, Ha NC, Park BJ. 2009a. p53 secreted by K-Ras-Snail pathway, is endocytosed by K-Ras-mutated cells; implication of target-specific drug delivery and early diagnostic marker. Oncogene 28: 2005–2014. [DOI] [PubMed] [Google Scholar]
- Lee SH, Lee SJ, Jung YS, Xu Y, Kang HS, Ha NC, Park BJ. 2009b. Blocking of p53–Snail binding, promoted by oncogenic K-Ras, recovers p53 expression and function. Neoplasia 11: 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Martinez-Yamout MA, Dyson HJ, Wright PE. 2010. Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding domain. Biochemistry 49: 9964–9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Woo TG, Lee SJ, Kim JS, Ha NC, Park BJ. 2013. Extracellular p53 fragment re-enters K-Ras mutated cells through the caveolin-1 dependent early endosomal system. Oncotarget 4: 2523–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S, Bykov VJ, Ali D, Andrén O, Cherif H, Tidefelt U, Uggla B, Yachnin J, Juliusson G, Moshfegh A, et al. 2012. Targeting p53 in vivo: A first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol 30: 3633–3639. [DOI] [PubMed] [Google Scholar]
- Leroy B, Girard L, Hollestelle A, Minna JD, Gazdar AF, Soussi T. 2014. Analysis of TP53 mutation status in human cancer cell lines: A reassessment. Hum Mutat 35: 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levrero M, De Laurenzi V, Costanzo A, Gong J, Wang JY, Melino G. 2000. The p53/p63/p73 family of transcription factors: Overlapping and distinct functions. J Cell Sci 113: 1661–1670. [DOI] [PubMed] [Google Scholar]
- Levy CB, Stumbo AC, Ano Bom AP, Portari EA, Cordeiro Y, Silva JL, De Moura-Gallo CV. 2011. Co-localization of mutant p53 and amyloid-like protein aggregates in breast tumors. Int J Biochem Cell Biol 43: 60–64. [DOI] [PubMed] [Google Scholar]
- Li FP, Fraumeni JF Jr. 1969a. Rhabdomyosarcoma in children: Epidemiologic study and identification of a familial cancer syndrome. J Natl Cancer Inst 43: 1365–1373. [PubMed] [Google Scholar]
- Li FP, Fraumeni F Jr. 1969b. Soft-tissue sarcomas, breast cancer, and other neoplasms. A familiar syndrome? Ann Intern Med 71: 747–752. [DOI] [PubMed] [Google Scholar]
- Li Y, Prives C. 2007. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene 26: 2220–2225. [DOI] [PubMed] [Google Scholar]
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W. 2012. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis and senescence. Cell 149: 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. 2003. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci 100: 12009–12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu WJ, Amatruda JF, Abrams JM. 2009. p53 ancestry: Gazing through an evolutionary lens. Nat Rev Cancer 9: 758–762. [DOI] [PubMed] [Google Scholar]
- Magzoub M, Miranker AD. 2011. Protein aggregation: p53 succumbs to peer pressure. Nat Chem Biol 7: 248–249. [DOI] [PubMed] [Google Scholar]
- Malcikova J, Pavlova S, Kozubik KS, Pospisilova S. 2014. TP53 mutation analysis in clinical practice: Lessons from chronic lymphocytic leukemia. Hum Mutat 35: 663–671. [DOI] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JF Jr, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. 1990. Germ line p53 mutations in a familiar syndrome of breast cancer, sarcomas, and other neoplasms. Science 250: 1233–1238. [DOI] [PubMed] [Google Scholar]
- Marambaud P, Zhao H, Davies P. 2005. Resveratrol promotes clearance of Alzheimer’s disease amyloid-β peptides. J Biol Chem 280: 37377–37382. [DOI] [PubMed] [Google Scholar]
- Marutani M, Tonoki H, Tada M, Takahashi M, Kashiwazaki H, Hida Y. 1999. Dominant-negative mutations of the tumor suppressor p53 relating to early onset of glioblastoma multiforme. Cancer Res 59: 4765–4769. [PubMed] [Google Scholar]
- Masuda H, Miller C, Koeffler HP, Battifora H, Cline MJ. 1987. Rearrangement of the p53 gene in human osteogenic sarcomas. Proc Natl Acad Sci 84: 7716–7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melero R, Rajagopalan S, Lázaro M, Joerger AC, Brandt T, Veprintsev DB, Lasso G, Gil D, Scheres SHW, Carazo JM, et al. 2011. Electron microscopy studies on the quaternary structure of p53 reveal different binding modes for p53 tetramers in complex with DNA. Proc Natl Acad Sci 108: 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael D, Oren M. 2003. The p53–Mdm2 module and the ubiquitin system. Semin Cancer Biol 13: 49–58. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398: 708–713. [DOI] [PubMed] [Google Scholar]
- Milner J, Medcalf EA. 1991. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell 65: 765–774. [DOI] [PubMed] [Google Scholar]
- Milner J, Medcalf EA, Cook AC. 1991. Tumor suppressor p53: Analysis of wild-type and mutant p53 complexes. Mol Cell Biol 11: 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Riou G, Levine AJ. 1992. Two distinct mechanisms alter p53 in breast cancer: Mutation and nuclear exclusion. Proc Natl Acad Sci 89: 7262–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, LaQuaglia M, Bénard J, Riou G. 1995. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc Natl Acad Sci 92: 4407–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Ostermeyer AG, Haladay R, Winkfield B, Frazier M, Zambetti G. 1996. Cytoplasmic sequestration of wild-type p53 protein impairs the G1 checkpoint after DNA damage. Mol Cell Biol 16: 1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206. [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH. 2013. p53 mutations and cancer. Nat Cell Biol 15: 2–8. [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH. 2014. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 25: 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natan E, Baloglu C, Pagel K, Freund SM, Morgner N, Robinson CV, Fersht AR, Joerger AC. 2011. Interaction of the p53 DNA-binding domain with its N-terminal extension modulates the stability of the p53 tetramer. J Mol Biol 409: 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada M, Park HL, Nagakawa Y, Yamashita K, Fomenkov A, Kim MS, Wu G, Nomoto S, Trink B, Sidransky D. 2005. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol Cell Biol 25: 6077–6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. 2007. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat 28: 622–629. [DOI] [PubMed] [Google Scholar]
- Polymenidou M, Cleveland DW. 2011. The seeds of neurodegeneration: Prion-like spreading in ALS. Cell 147: 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216: 136–144. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. 1998. Prions. Proc Natl Acad Sci 95: 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. 2012. Cell biology. A unifying role for prions in neurodegenerative diseases. Science 336: 1511–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. 2013. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet 47: 601–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. 2014. Madness and memory. Yale University Press, New Haven, CT. [Google Scholar]
- Prusiner SB, Scott MR, De Armond SJ, Cohen FE. 1998. Prion protein biology. Cell 93: 337–348. [DOI] [PubMed] [Google Scholar]
- Prusiner SB, Woerman AL, Mordes DA, Watts JC, Rampersaud R, Berry DB, Patel S, Oehler A, Lowe JK, Kravitz SN, et al. 2015. Evidence for α-synuclein prions causing multiple system atrophy in humans with Parkinsonism. Proc Natl Acad Sci 112: E5308–E5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca R, Nardinocchi L, Porru M, Simon AJ, Rechavi G, Leonetti C, Givol D, D’Orazi G. 2011. Restoring p53 active conformation by zinc increases the response of mutant p53 tumor cells to anticancer drugs. Cell Cycle 10: 1679–1689. [DOI] [PubMed] [Google Scholar]
- Ramos YF, Stad R, Attema J, Peltenburg LT, van der Eb AJ, Jochemsen AG. 2001. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res 61: 1839–1842. [PubMed] [Google Scholar]
- Rangel LP, Costa DC, Vieira TC, Silva JL. 2014. The aggregation of mutant p53 produces prion-like properties in cancer. Prion 8: 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy G, Straub JE, Thirumalai D. 2010. Dry amyloid fibril assembly in a yeast prion peptide is mediated by long-lived structures containing water wires. Proc Natl Acad Sci 107: 21459–21464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigacci S, Bucciantini M, Relini A, Pesce A, Gliozzi A, Berti A, Stefani M. 2008. The (1–63) region of the p53 transactivation domain aggregates in vitro into cytotoxic amyloid assemblies. Biophys J 94: 3635–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippin TM, Bykov VJ, Freund SM, Selivanova G, Wiman KG, Fersht AR. 2002. Characterization of the p53-rescue drug CP-31398 in vitro and in living cells. Oncogene 21: 2119–2129. [DOI] [PubMed] [Google Scholar]
- Rotter V, Schwartz D, Almon E, Goldfinger N, Kapon A, Meshorer A, Donehower LA, Levine AJ. 1993. Mice with reduced levels of p53 protein exhibit the testicular giant-cell degenerative syndrome. Proc Natl Acad Sci 90: 9075–9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F, Serrano L, Schymkowitz JW. 2006. How evolutionary pressure against protein aggregation shaped chaperone specificity. J Mol Biol 355: 1037–1047. [DOI] [PubMed] [Google Scholar]
- Sabbatini P, McCormick F. 2002. MDMX inhibits the p300/CBP-mediated acetylation of p53. DNA Cell Biol 21: 519–525. [DOI] [PubMed] [Google Scholar]
- Sah VP, Attardi LD, Mulligan GJ, Williams BO, Bronson RT, Jacks T. 1995. A subset of p53-deficient embryos exhibit exencephaly. Nat Genet 10: 175–180. [DOI] [PubMed] [Google Scholar]
- Sakamuro D, Sabbatini P, White E, Prendergast GC. 1997. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene 15: 887–898. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. 2007. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447: 453–457. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63: 1129–1136. [DOI] [PubMed] [Google Scholar]
- Schlomm T, Iwers L, Kirstein P, Jessen B, Köllermann J, Minner S, Passow-Drolet A, Mirlacher M, Milde-Langosch K, Graefen M, et al. 2008. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod Pathol 21: 1371–1378. [DOI] [PubMed] [Google Scholar]
- Schon O, Friedler A, Bycroft M, Freund SMV, Fersht AR. 2002. Molecular mechanism of the interaction between MDM2 and p53. J Mol Biol 323: 491–501. [DOI] [PubMed] [Google Scholar]
- Schultz J, Ponting CP, Hofmann K, Bork P. 1997. SAM as a protein interaction domain involved in developmental regulation. Protein Sci 6: 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selivanova G. 2014. Wild type p53 reactivation: From lab bench to clinic. FEBS Lett 588: 2628–2638. [DOI] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. 2007. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129: 523–536. [DOI] [PubMed] [Google Scholar]
- Sharp DA, Kratowicz SA, Sank MJ, George DL. 1999. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem 274: 38189–38196. [DOI] [PubMed] [Google Scholar]
- Shaulsky G, Goldfinger N, Rotter V. 1991. Alterations in tumor development in vivo mediated by expression of wild type or mutant p53 proteins. Cancer Res 51: 5232–5237. [PubMed] [Google Scholar]
- Shewmaker F, McGlinchey RP, Wickner RB. 2011. Structural insights into functional and pathological amyloid. J Biol Chem 286: 16533–16540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvarts A, Steegenga WT, Riteco N, Van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, et al. 1996. MDMX: A novel p53-binding protein with some functional properties of MDM2. EMBO J 15: 5349–5357. [PMC free article] [PubMed] [Google Scholar]
- Shvarts A, Bazuine M, Dekker P, Ramos YF, Steegenga WT, Merckx G, van Ham RC, van der Houven van Oordt W, van der Eb AJ, et al. 1997. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics 43: 34–42. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Mol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JL, Lima LM, Foguel D, Cordeiro Y. 2008. Intriguing nucleic-acid-binding features of mammalian prion protein. Trends Biochem Sci 33: 132–140. [DOI] [PubMed] [Google Scholar]
- Silva JL, Vieira TC, Gomes MP, Bom AP, Lima LM, Freitas MS, Ishimaru D, Cordeiro Y, Foguel D. 2010. Ligand binding and hydration in protein misfolding: Insights from studies of prion and p53 tumor suppressor proteins. Acc Chem Res 43: 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JL, Rangel LP, Costa DC, Cordeiro Y, De Moura Gallo CV. 2013. Expanding the prion concept to cancer biology: Dominant-negative effect of aggregates of mutant p53 tumour suppressor. Biosci Rep 33: 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JL, De Moura Gallo CV, Costa DC, Rangel LP. 2014. Prion-like aggregation of mutant p53 in cancer. Trends Biochem Sci 39: 260–267. [DOI] [PubMed] [Google Scholar]
- Soragni A, Janzen DM, Johnson LM, Lindgren AG, Thai-Quynh Nguyen A, Tiourin E, Soriaga AB, Lu J, Jiang L, Faull KF, et al. 2016. A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell 29: 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C. 2012. Transmissible proteins: Expanding the prion heresy. Cell 149: 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stindt MH, Muller PA, Ludwig RL, Kehrloesser S, Dötsch V, Vousden KH. 2014. Functional interplay between MDM2, p63/p73 and mutant p53. Oncogene 34: 4300–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Cristofanelli B, Shaul Y, Castagnoli L, Levine AJ, Sacchi A, Cesareni G, Oren M, et al. 2000. Physical and functional interaction between p53 mutants and different isoroforms of p73. J Biol Chem 275: 29503–29512. [DOI] [PubMed] [Google Scholar]
- Strano S, Fontemaggi G, Costanzo A, Rizzo MG, Monti O, Baccarini A, Del Sal G, Levrero M, Sacchi A, Oren M, et al. 2002. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J Biol Chem 277: 18817–18826. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. 2009. The cancer genome. Nature 458: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, Biernaskie JA, Sinha S, Prives C, Pevny LH, et al. 2009. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell 5: 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XF, Johannsson O, Håkansson S, Sellberg G, Nordenskjöld B, Olsson H, Borg A. 1996. A novel p53 germline alteration identified in a late onset breast cancer kindred. Oncogene 13: 407–411. [PubMed] [Google Scholar]
- Tang X, Zhu Y, Han L, Kim AL, Kopelovich L, Bickers DR, Athar M. 2007. CP-31398 restores mutant p53 tumor suppressor function and inhibits UVB-induced skin carcinogenesis in mice. J Clin Invest 117: 3753–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. 1999. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett 447: 5–9. [DOI] [PubMed] [Google Scholar]
- Taylor JA, Li Y, He M, Mason T, Mettlin C, Vogler WJ, Maygarden S, Liu E. 1996. p53 mutations in bladder tumors from arylamine-exposed workers. Cancer Res 56: 294–298. [PubMed] [Google Scholar]
- Tidow H, Melero R, Mylonas E, Freund SM, Grossmann JG, Carazo JM, Svergun DI, Valle M, Fersht AR. 2007. Quaternary structures of tumor suppressor p53 and a specific p53-DNA complex. Proc Natl Acad Sci 104: 12324–12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velente LJ, Gray DH, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL, Janic A, Strasser A. 2013. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma and Noxa. Cell Rep 3: 1339–1345. [DOI] [PubMed] [Google Scholar]
- Vieira TC, Reynaldo DP, Gomes MPB, Almeida MS, Cordeiro Y, Silva JL. 2011. Heparin binding by murine recombinant prion protein leads to transient aggregation and formation of RNA-resistant species. J Am Chem Soc 133: 334–344. [DOI] [PubMed] [Google Scholar]
- Vieira TC, Cordeiro Y, Caughey B, Silva JL. 2014. Heparin binding confers prion stability and impairs its aggregation. FASEB J 28: 2667–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. 2000. Surfing the p53 network. Nature 408: 307–310. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. 2007. p53 in health and disease. Nat Rev Mol Cell Biol 8: 275–283. [DOI] [PubMed] [Google Scholar]
- Walker KK, Levine AJ. 1996. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci 93: 15335–15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Fersht AR. 2015. Propagation of aggregated p53: Cross-reaction and coaggregation vs. seeding. Proc Natl Acad Sci 112: 2443–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Reed M, Wang P, Stenger JE, Mayr G, Anderson ME, Schwedes JF, Tegtmeyer P. 1993. P53 domains: Identification and characterization of two autonomous DNA-binding regions. Genes Dev 7: 2575–2586. [DOI] [PubMed] [Google Scholar]
- Wassman CD, Baronio R, Demir Ö, Wallentine BD, Chen CK, Hall LV, Salehi F, Lin DW, Chung BP, Hatfield GW, et al. 2013. Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53. Nat Commun 4: 1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Geyer FC, Reis-Filho JS. 2010. Histological types of breast cancer: How special are they? Mol Oncol 4: 192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcken R, Wang G, Boeckler FM, Fersht AR. 2012. Kinetic mechanism of p53 oncogenic mutant aggregation and its inhibition. Proc Natl Acad Sci 109: 13584–13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Harris N, Rotter V. 1984. Reconstitution of p53 expression in a nonproducer Ab-MuLV-transformed cell line by transfection of a functional p53 gene. Cell 38: 119–126. [DOI] [PubMed] [Google Scholar]
- Wong KB, DeDecker BS, Freund SM, Proctor MR, Bycroft M, Fersht AR. 1999. Hot-spot mutants of p53 core domain evidence characteristic local structural changes. Proc Natl Acad Sci 96: 8438–8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Bayle JH, Elenbaas B, Pavletich NP, Levine AJ. 1995. Alternatively spliced forms in the carboxy-terminal domain of the p53 protein regulate its ability to promote annealing of complementary single strands of nucleic acid. Mol Cell Biol 15: 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, Cornelis A, Rozenski J, Zwolinska A, Marine JC, et al. 2011. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol 7: 285–295. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Tsukamoto T, Sakai H, Shirai N, Ohgaki H, Furihata C, Donehower LA, Yoshida K, Tatematsu M. 2000. p53 knockout mice (−/−) are more susceptible than (+/−) or (+/+) mice to N-methyl-N-nitrosourea stomach carcinogenesis. Carcinogenesis 21: 1891–1897. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dötsch V, Andrews NC, Caput D, McKeon F. 1998. p63, a p53 homolog at 3Q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 2: 305–316. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398: 714–718. [DOI] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lacks spontaneous tumours. Nature 404: 99–103. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Caput D, Mckeon F. 2002. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet 18: 90–95. [DOI] [PubMed] [Google Scholar]
- Yang F, Lim GP, Begum AN, Ubeda OJ, Simmons MR, Ambegaokar SS, Chen PP, Kayed R, Glabe CG, Frautschy SA, et al. 2005. Curcumin inhibits formation of amyloid β oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J Biol Chem 280: 5892–5901. [DOI] [PubMed] [Google Scholar]
- Yang-Hartwich Y, Soteras MG, Lin ZP, Holmberg J, Sumi N, Craveiro V, Liang M, Romanoff E, Bingham J, Garofalo F, et al. 2015. p53 protein aggregation promotes platinum resistance in ovarian cancer. Oncogene 34: 3605–3616. [DOI] [PubMed] [Google Scholar]
- Yu GW, Rudiger S, Veprintsev D, Freund S, Fernandez-Fernandez MR, Fersht AR. 2006. The central region of HDM2 provides a second binding site for p53. Proc Natl Acad Sci 103: 1227–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Vazquez A, Levine AJ, Carpizo DR. 2012. Allele-specific p53 mutant reactivation. Cancer Cell 21: 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Narayanan S, Vazquez A, Carpizo DR. 2014. Small molecule compounds targeting the p53 pathway: Are we finally making progress? Apoptosis 19: 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]