Abstract
Host-associated microbiomes perform many beneficial functions including resisting pathogens and training the immune system. Here, we show that amphibians developing in captivity lose substantial skin bacterial diversity, primarily due to reduced ongoing input from environmental sources. We combined studies of wild and captive amphibians with a database of over 1 000 strains that allows us to examine antifungal function of the skin microbiome. We tracked skin bacterial communities of 62 endangered boreal toads, Anaxyrus boreas, across 18 time points, four probiotic treatments, and two exposures to the lethal fungal pathogen Batrachochytrium dendrobatidis (Bd) in captivity, and compared these to 33 samples collected from wild populations at the same life stage. As the amphibians in captivity lost the Bd-inhibitory bacteria through time, the proportion of individuals exposed to Bd that became infected rose from 33% to 100% in subsequent exposures. Inoculations of the Bd-inhibitory probiotic Janthinobacterium lividum resulted in a 40% increase in survival during the second Bd challenge, indicating that the effect of microbiome depletion was reversible by restoring Bd-inhibitory bacteria. Taken together, this study highlights the functional role of ongoing environmental inputs of skin-associated bacteria in mitigating a devastating amphibian pathogen, and that long-term captivity decreases this defensive function.
Keywords: captivity, probiotics, amphibian, Anaxyrus boreas, Batrachochytrium dendrobatidis, microbiome
1. Introduction
The symbiotic microbiome is increasingly recognized as important to host health. Host-associated microbiomes confer diverse benefits ranging from digestive to immunological functions, including mediation of pathogens [1–3]. Whether loss of symbiont diversity in turn leads to a reduced functional capacity of the microbiome is not well understood. The skin microbiome of amphibians is increasingly recognized for influencing the disease outcome when amphibians are infected with Batrachochytrium dendrobatidis (Bd) [4–7]. In susceptible amphibians, Bd infects the skin, reproduces and ruptures cells, causing electrolyte imbalance and organ failure [8]. Because Bd attacks the external skin surface, the presence of Bd-inhibitory bacteria in the skin mucosome can mitigate the establishment and proliferation of Bd [9]. Thus, the community composition of bacteria is predicted to have a large influence on Bd establishment. Previous studies have focused on bacterial taxa that are negatively or positively correlated with Bd [6,7], but did not directly examine the Bd-inhibitory capacity of those bacteria. We have developed a database of 1 127 Bd-inhibitory bacterial isolates that have been cultured from amphibians, tested in co-culture with Bd, Sanger-sequenced (16S rRNA, 1 600 bp), and identified. This provides a unique resource to examine which members of a given bacterial community probably have Bd-inhibitory function based on matches to this culture-tested database [10].
We compared the skin microbiome of the highly Bd-susceptible [11], Colorado endangered boreal toad (Anaxyrus boreas) in the wild to individuals collected from the same population and reared in captive enclosures from the egg stage onwards without environmental substrates (aquatic sediments and soil). We also sampled microbes from the natural environment (wetland water, aquatic sediment) of the wild toads. With individually housed captive toads, we conducted experimental manipulations aimed at understanding how captivity affects the skin microbiome through time, during exposure to Bd-inhibitory bacterial isolates (‘probiotics’), and during challenges with Bd. Specifically, we examined shifts in Bd-inhibitory bacteria through time and across treatments by filtering the entire bacterial community data (16S rRNA marker gene sequencing) through our Bd-inhibitory isolates database, which provides the basis for our functional assay of the boreal toad microbiome. Our confidence in using this database was bolstered by an independent study to culture bacteria from boreal toads, where we found that out of 48 different isolates tested in co-culture with Bd, eight (17%) were inhibitory (see the electronic supplementary material), which is on par with the results of matches to the database presented below.
2. Material and methods
(a). Animal husbandry and sample collection
Boreal toads were obtained as juveniles from the Native Aquatic Restoration Facility (NASRF), in Alamosa, Colorado. At NASRF, individual sibling toads were reared from one egg mass that was collected from the wild, from the Southern Rocky Mountains in Colorado. Juveniles that were reared at NASRF were transported to the University of Colorado at Boulder and housed in an experimental chamber from 22 November 2012 to 20 June 2013. All amphibians were housed individually in Sterilite containers (5.7 l) on wire racks within a walk-in experimental chamber. Temperature was held constant at 19°C to benefit both the alpine toads and growth for Bd. An automatic twelve-hour light cycle was used. Amphibian containers were pre-rinsed with sterile Holtfreter's solution. Containers were propped up one inch to allow each amphibian a wet and dry portion within the enclosure. Crickets (pinhead size, Fluker Farms) and fresh Holfreter's solution were given to all individuals every third day. Water was removed from each container and 10 crickets were given to each individual. Any remaining crickets were removed after 2 h. Containers were given a rinse with Holtfreter's solution and a new 450 ml was added to each container.
Toads grew during the experiment, averaging 9.52 grams at the beginning of the experiment and left averaging 17.64 grams. Manipulation 1 (probiotic and Bd exposure 1) ran from 29 November 2012 to 23 December 2012. Before sampling, all individuals were allowed one week to acclimate to their new environment. Bacterial swab samples were collected every 3 days. Manipulation 2 (probiotic and Bd exposure 2) ran from 18 February 2013 to 20 June 2013. Bacterial swab samples were collected once per week until March 22, and then once per two weeks thereafter. Before sample collection, amphibians were rinsed twice with 50 ml of sterile nuclease-free water (Berdick-Jackson Water). A sterile cotton-tipped swab (BBL, Culture Swab; Becton, Dickinson and Company) was used to brush along the amphibian's skin thereby obtaining a microbial sample. From these microbial swab samples, DNA was extracted and processed as in [12]. A separate swab was collected for Bd quantitative real-time polymerase chain reaction (qPCR) analysis (see below). Samples were tested for Bd, following standard protocols [13].
(b). Probiotic treatments and Bd exposures
Probiotic selection occurred by identification of Bd-inhibitory strains using in vitro co-culture assays and 96 well assays [9]. Bacterial isolates were cultured from the skin of wild boreal toads, captive boreal toads, and wild American bullfrogs, Lithobates catesbeiana, from Colorado. The strongest Bd-inhibitory bacterial strains (strains that demonstrated visually clear zones of inhibition in co-culture and also significantly reduced optical density in 96 well assays) were processed for Sanger sequencing of the 16S rRNA gene (1 500 bp). Three of these strains were chosen for use in experiments, to diversify the source of the strain (e.g. captive, wild, and amphibian host species) (electronic supplementary material, table S1).
During probiotic and Bd treatments, all individuals of A. boreas were held in individual plastic containers (4 oz) with 50 ml of Holtfreter's solution, enough to cover a juvenile toad's body. Probiotic-treated individuals were given bacterial cells (1 × 106) suspended in sterile water [4]. We used a Bd strain recently isolated from bullfrogs in Colorado that was confirmed as the global pandemic lineage (Bd-GPL) [14], and we used qPCR for measuring Bd loads [13]. In manipulation 1, Bd exposures (1 × 105 Bd zoospores) occurred 2 days after the probiotic treatment. In manipulation 2, Bd exposures occurred one week after two sequential Janthinobacterium lividum inoculations that were 8 days apart. Control individuals were treated with sterile water only. Their containers were agitated to ensure toadlet exposure to treatments. After treatments, toads were then returned to their respective larger enclosures. Subsequent doses of J. lividum were added starting at week 4 after Bd exposure to ensure persistence of the probiotic treatment (electronic supplementary material, figure S7).
(c). Bacterial sample processing and bioinformatics
Microbial swab samples were collected, DNA extracted and processed as in [12]. Amplicons were sequenced on one Illumina MiSeq run at the University of Colorado, Boulder, yielding 150 bp reads of the V4 region of the 16S rRNA gene, using 515f primers. Sequence data from captive and wild toad samples were joined together before operational taxonomic unit (OTU) picking and subsequent filtering. Using QIIME, sequences were filtered for quality and assigned to their respective sample using default settings. OTUs were picked with open reference and clustered into OTUs at 97% similarity according to the subsampling open reference protocol [15], using the August 2013 version of the Greengenes reference database [16]. Taxonomy was assigned to de novo picked OTUs using the Ribosomal Database Project (RDP) classifier with an 80% confidence threshold. Sequences were aligned to the Greengenes reference alignment using PyNAST and a tree was constructed with FastTree2 according to standard procedures within QIIME. OTUs with less than 0.005 per cent total abundance were filtered out of our analysis according to recommendations from [17]. Remaining samples were rarefied to 12 700 sequences per sample, to maximize sequence depth and conserve sample abundance in both captive and wild toad samples. For subsequent analyses, samples with fewer than 12 700 sequences per sample, including experimental and sequencing controls were removed from the analysis. The resulting 12 026 900 million sequences yielded 726 unique OTUs. Analyses were performed using QIIME v. 1.9.0 unless otherwise specified. Alpha diversity visualization and statistics were conducted in R (R Core Team (2013)). Survival curve statistics and visualization were performed using MedCalc v. 16.2.1.
(d). Bd-inhibitory bacterial database
We used a Bd-inhibitory database that was constructed from greater than 2 000 isolates of bacteria cultured from the skin of amphibians from around the globe, including boreal toads and tested against Bd in co-culture [10]. The bacterial isolates that exhibited inhibition of Bd in co-culture in vitro were Sanger sequenced (16S rRNA gene, 1 500 bp), then trimmed and used to pick OTUs. This was achieved by trimming the Sanger sequence to the first 150 bp beyond primer 515f, so that the inhibitory sequence reads would be the same length as the Illumina MiSeq reads. The sequenced Bd-inhibitory bacteria underwent closed reference OTU picking with the Greengenes reference database (August 2013 version), and this resulted in 304 unique inhibitory OTUs, forming the basis of the Bd-inhibitory database used in this study. To examine how many matches were in our toad dataset (MiSeq 16S rRNA marker gene sequence results for wild/captive toads), we used the closed reference picks from the MiSeq processed samples to check for exact 150 bp matches to the 304 Bd-inhibitory OTU dataset. From the 642 unique OTUs that we observed in our explicitly wild and captive juvenile boreal toad dataset, 31 (5%) were exact matches to the Bd-inhibitory set, and these are shown in electronic supplementary material, figure S3.
3. Results
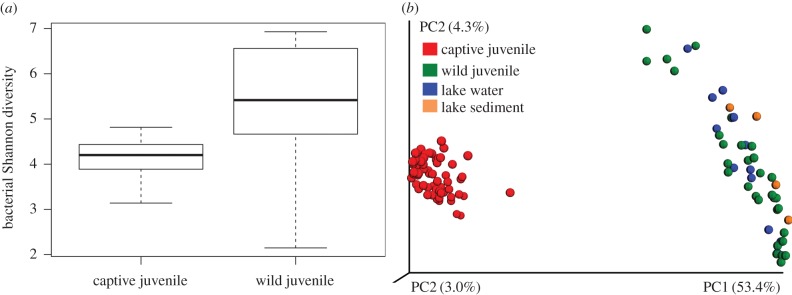
The effect of captivity on the boreal toad skin microbiome was large. Both the overall bacterial diversity and the community variability were far reduced on the captive toads compared with wild toads (figure 1). Only one bacteria in the genus Cellulosimicrobium was an abundant member (more than 13%) on both captive and wild juveniles; otherwise, there was little overlap at finer taxonomic resolution (electronic supplementary material, figures S1 and S2). In comparing wild toads of various life stages with their environment (wetland water and sediment), it is clear that wild toads acquire their diverse microbiome as a subset of the microbes present in their environment [19,20] (figure 1b).
Figure 1.
(a) Shannon diversity of bacterial OTUs on captive and wild juvenile boreal toads. Data shown for captive toads (n = 62) represent the communities sampled on day 4 in captivity and wild toads (n = 30) were sampled in a single high elevation wetland habitat in Colorado. Wild toads had a significantly more diverse skin microbiome compared with captive toads (ANOVA F = 41.47, p < 0.0001). (b) Beta-diversity of captive juveniles toads, wild juveniles, and environmental samples (water and sediment) from the wild. Bacterial communities on captive toads differ dramatically from wild toads and their environment (ANOSIM R2 = 0.8554, p < 0.001). Wild toads were rinsed with sterile water to remove transient environmental microbes [18]. Each point represents the bacterial community, by sample type: red = captive juvenile skin (n = 42), green = wild juvenile skin (n = 42), blue = lake water (n = 13), orange = lake sediment (n = 4). Diversity patterns are visualized using principal coordinate plots of unweighted UniFrac distances.
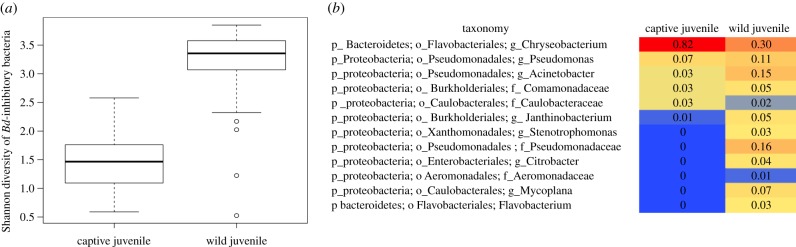
Wild toads have a significantly higher diversity of Bd-inhibitory bacteria on their skin than captives (figure 2a; electronic supplementary material, figure S3). However, captive toads share some of the same abundant Bd inhibitors with wild toads, in particular bacteria in the genus Chryseobacterium (figure 2b). The key difference is that Chryseobacterium comprises less than 5% of the community on wild toads and up to 26% of the total skin microbiome on captive toads (early in captivity, figure 3). The most abundant Chryseobacterium in this study matches sequences that have demonstrated Bd inhibition in co-culture from independent studies on unrelated amphibians [21,22]. Considering the broader diversity of Bd-inhibitory bacteria beyond this dominant group, wild juvenile toads have only 1.6% of the skin community composed of Bd-inhibitory bacteria, while up to 29.1% of the skin community on captive juvenile toads was Bd inhibitory (figure 2b; electronic supplementary material, figure S4). We did not determine whether captive toads were more resistant to Bd than wild toads, given the obvious difficulty of infecting wild, endangered boreal toads. Instead, we conducted a time series observation of the bacterial skin communities on captive toads and challenged them with Bd, as described below. Furthermore, the marker gene sequencing approach does not provide information about the absolute abundance of any of these bacteria, which precludes a comparison of whether captive or wild toads have higher protective bacterial cell counts.
Figure 2.
(a) Shannon diversity of Bd-inhibitory taxa found on captive and wild juveniles rarefied to 12 700 sequences per sample. Captive juveniles (n = 62) sampled at day 4 and wild juveniles (n = 30) sampled in the field; ANOVA F = 157.7, p < 0.001. (b) The heatmap depicts the proportional abundance (mean number of sequences per taxon divided by total sequences per host group) of only Bd-inhibitory bacterial OTUs across captive boreal toads and wild boreal toads. Heatmap includes OTUs found with 0.3% or greater of the proportional abundance. Electronic supplementary material, figure S3 includes all inhibitory OTUs found on wild (n = 30 OTUs) and captive (n = 12) juvenile boreal toads. Heatmap is ordered by decreasing taxonomic abundance in captive juvenile individuals. (Online version in colour.)
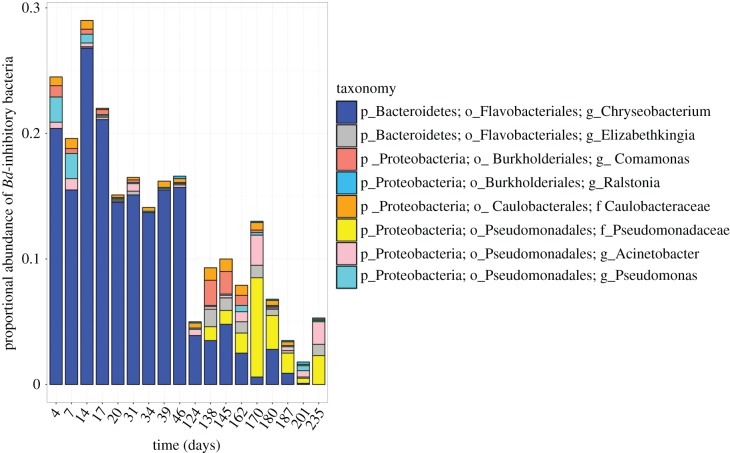
Figure 3.
The proportional abundance (as a mean per cent of total sequences per sample) of bacterial taxa on 62 captive boreal toads through time. Bd-inhibitory bacterial taxa are determined as sequence matches to the independent Bd-inhibitory isolates database [10]. Other bacterial taxa that do not match Bd inhibitors are not shown. The proportional abundance of Chryseobacterium decreases through time, ranging from 26% to 0.1% of the total community. (Online version in colour.)
Although the proportional abundance of Bd-inhibitory bacteria on captive toads increased briefly to an exceedingly high level early in captivity, it then decreased considerably (figure 3; electronic supplementary material, figure S3). The dominant Bd-inhibitory bacterium Chryseobacterium decreased from a maximum of 26% of the total skin bacterial community on day 17, to a minimum of 0.1% on day 235 (figure 3). Consequently, the toads had a higher proportion of Bd-inhibitory bacteria at the time of first Bd exposure (day 16) than at the time of the second Bd exposure (day 127, figure 4a and electronic supplementary material, figure S7). During the first Bd exposure (when the Bd inhibitors were high), 13 out of 39 (30%) of the exposed toads became infected. During the second Bd exposure (when the Bd inhibitors were low), all 31 (100%) of the exposed toads became infected (figure 4b; electronic supplementary material, figure S8). Additionally, Bd loads reached fatal levels 15 days faster in the second exposure, when there were fewer Bd-inhibitory bacteria (figure 4c).
Figure 4.
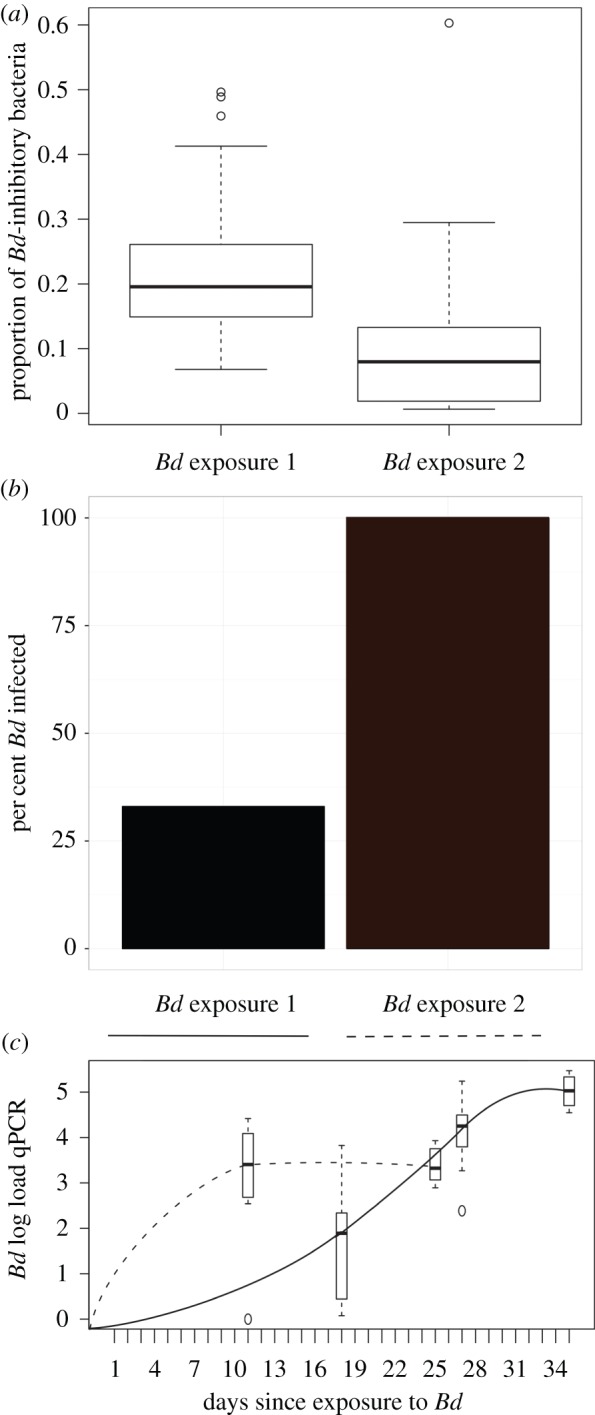
The proportion of Bd-inhibitory bacteria on captive toads, the per cent infected, and the pathogen load during the first and second exposures to Bd. (a) The proportion of sequences that match the Bd-inhibitory database found on individuals at the time of Bd exposure 1 (n = 39 toads on day 17) and Bd exposure 2 (n = 31 toads on day 138). There is a higher proportion of the skin community that is Bd inhibitory at the time of first Bd exposure than there was at the time of the second Bd exposure (ANOVA F = 16.22, p < 0.00016). (b) Of 39 toads exposed to Bd in the first trial, 13 (33%) became infected. During the second Bd exposure, 31 out of 31 (100%) toads became infected. (c) Bd load is presented as the number of log-transformed DNA copies (qPCR) through time during Bd exposure 1 (solid line) and Bd exposure 2 (dotted line). Bd load means are calculated from 13 infected individuals in exposure 1 and from 10 infected individuals in exposure 2 (these include the Bd-only infected group and excludes the J. lividum treated group which differed significantly in the second trial, see figure 5; electronic supplementary material, figure S4). (Online version in colour.)
To test whether the addition of further Bd-inhibitory bacteria could alter the disease outcome when toads were exposed to Bd, we included ‘probiotic’ treatment groups. We inoculated toads with cultured strains of Bd-inhibitory bacteria prior to each of the two sequential pathogen exposures (electronic supplementary material, table S1). Treatment categories included controls, probiotics only, probiotics and Bd, and Bd only. Between Bd exposures, probiotic treated toads (excluding the controls) were randomly divided and re-assigned to treatment groups in the second probiotic trial to control for an effect of the previous treatment. All control toads and probiotic-only treated toads survived and maintained body weight, indicating adequate husbandry and that the probiotic treatments had no adverse effects. During the first Bd exposure, when all captive toads had a high proportion of Bd-inhibitory bacteria already present, none of the three different strains of probiotic-treated groups (Chryseobacterium indolgenes and two Pseudomonas isolates) demonstrated an increased survival compared with the Bd-only group, presumably because Chryseobacterium was already dominant on all captive toads (figure 3; electronic supplementary material, figure S5). For the second Bd challenge, we used J. lividum because it demonstrated a protective effect on other amphibians [4,23]. More than 100 days later, when the previously abundant Bd-inhibitory bacteria on toads had diminished (as in figure 3), all toads exposed to Bd became infected (figure 4b). Toads exposed to only Bd experienced 100% mortality, while toads treated with J. lividum prior to Bd experienced 60% mortality (figure 5), thus the group treated with J. lividum and Bd had a significantly higher survival rate compared with the Bd-only group (Mantel-Cox, χ2 = 6.021, p = 0.0141; figure 5). Overall, the J. lividum probiotic-treated group experienced a trend toward lower Bd loads in addition to reduced or delayed mortality (figure 5; electronic supplementary material, figure S8).
Figure 5.
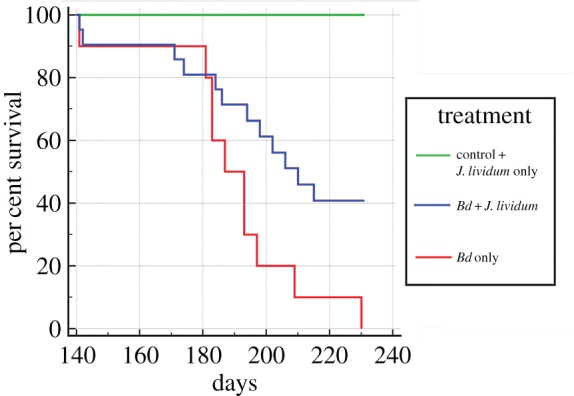
Survival of toads treated with Janthinobacterium lividum (J. lividum) prior to exposure to Bd (J. lividum plus Bd, n = 21, blue), toads exposed to Bd only (n = 10, red), toads exposed to J. liv only (n = 10), and controls exposed to a sterile sham treatment (n = 5), combined as green. J. lividum treated toads plus Bd experienced a 40 per cent increase in survival compared to the Bd-only treated toads (Mantel-Cox, χ2 6.021, p = 0.0141). No mortality was observed for control toads or toads treated with J. lividum only. (Online version in colour.)
4. Discussion
Captivity resulted in a massive loss of diversity of the amphibian skin microbiome (also see [24,25]) and increased dominance of a few Bd-inhibiting bacteria. While our study demonstrates that these dominant Bd-inhibitory bacteria provide a protective effect against the fungal pathogen, it is likely that the larger loss of microbiome diversity impacts the broader health of the host. Furthermore, the long-term persistence of only a few Bd-inhibitory bacteria is unlikely, and if they decrease in abundance, as they did in our study, the protective effect also diminishes. The J. lividum probiotic-treated group experienced higher survival and a trend toward decreased Bd loads. This finding is important because it demonstrates that after a long time in captivity, when the skin communities of the toads had lost a large proportion of the Bd-inhibitory microbiome, treatment with J. lividum significantly reversed the disease outcome. Given that we conducted serial Bd exposures on individual boreal toads (equally re-distributed across treatment groups), there is some concern that the toads could have developed increased tolerance to Bd (as has been observed in Cuban treefrogs [26]), and that the role of the skin microbiome could be confounded by the adaptive immune response. However, the second Bd exposure yielded 100% infection, after only 30% infection in the first exposure (the opposite of what would be expected if an effective immune response had occurred). Taken together, these findings support a very strong role of the skin microbiome in mediating the fungal pathogen.
Our study demonstrates that when hosts are removed from native habitats and reared in captive environments without environmental substrates (aquatic sediments and soil), there is a substantial effect on the microbiome. However, gaining insight into the resulting consequences of the loss of microbiome diversity has not been examined in previous studies. Apparent in this study is the essential role of the natural environmental substrates in replenishing the diversity of the amphibian skin microbiome (figure 1; electronic supplementary material, figure S4). Only one taxon, an OTU in the genus Cellulosimicrobium, was an abundant member on both captive and wild juveniles, when considering the entire microbiome. Overall, the captive toads retained 30% of the OTUs found on wild toads (electronic supplementary material, figure S2), but the large majority of these are very rare on the captive toads in terms of proportional abundance (electronic supplementary material, figure S1). Loudon et al. demonstrated that moving wild salamanders (Plethodon cinereus) into captivity had a strong effect on the skin microbiome, but that they maintained a closer to ‘wild-type’ skin microbiome when housed with soil substrate taken from their site of capture [25]. Becker et al. demonstrated that Panamanian golden frogs (Atelopus zeteki) reared in captivity (up to 8 years) shared 70% of their skin bacteria with wild counterparts [26]. A key difference between these two studies and this study is that the others examined amphibians in captivity with natural substrates (e.g. soil, rocks, water, plant material, etc.), whereas the boreal toads were housed without such substrates. Taken together, these studies indicate that environmental substrates do substantially increase the maintenance of a ‘wild-type’ skin microbiome, but whether the protective function of those skin communities is retained has not previously been addressed and further study is needed to tease apart how the type and source of environmental substrates can maximize the similarity to conditions in the wild.
Evidence suggests that wild boreal toads acquire associations with antifungal bacteria very early in development (larval stage) and these communities re-assemble on the skin of A. boreas upon metamorphosis [19]. While some of the larval microbiota are retained, metamorphs gain a new community of skin microbes from terrestrial inocula [19]. Here, we observed a crash in the abundance of an antifungal taxon, Chryseobacterium, once dominant on juveniles in captivity, and other antifungal bacteria typically available to wild toads from soil or other natural inocula did not replace this deficit. Thus, in captivity, sources of protective microbiota may be needed throughout development in order to maintain disease defences [6]. We underscore the importance of preserving the natural habitat of such species being raised in captivity as a unique source of the environmental microbes that species ultimately require for their health and development.
For many species of conservation concern, captive assurance colonies are needed to preserve biological diversity until natural environments become more suitable [27], or new conservation tools are developed that can mitigate the effects of pathogens [28,29,30]. The results of this study underscore the importance of the microbiome in the health of amphibians and inform how we can improve captive rearing programmes [26]. We demonstrate that relatively few Bd-inhibitory bacterial taxa can have a large effect at reducing disease caused by Bd—as long as they represent a large proportion of the community. If animals are removed from the environment and the potential inocula contained therein, the genetic diversity of the host microbiome can be lost. Our study suggests that a microbiome-inclusive strategy to manage animal health in captivity should include maintaining contact with natural substrates that contain protective microbial inocula sources.
Supplementary Material
Acknowledgements
We thank the Colorado Parks and Wildlife and the NASRF for providing boreal toads for this work. Thank you to Graham Goodman and Julia Moy who helped with collection of the data and care of the amphibians.
Ethics
Permits and authorization were granted by Colorado Parks and Wildlife (CPW) and the University of Colorado Institutional Animal Care and Use Committee (IACUC).
Data accessibility
Bacterial data, including raw bacterial sequence reads and processed OTU tables with mapping files, are available through Dryad at: http://dx.doi.org/10.5061/dryad.ck1qs.
Authors' contributions
V.J.M., J.G.K., and D.C.W. designed the study. J.G.K., D.C.W., and H.M.A. collected the data. H.M.A. and J.G.K. conducted the molecular laboratory work. J.G.K., V.J.M., and D.C.W. analysed the data, and all authors contributed to writing the manuscript.
Competing interests
We have no competing interests.
Funding
Funding support was provided by research grants from the NSF (DEB 1146284 to V.J.M. and R.K., and 1136602 to R.H.) and the Keck Foundation and John S. Templeton Foundation to V.J.M. and R.K.
References
- 1.McFall-Ngai M, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236. ( 10.1073/pnas.1218525110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caballero S, Pamer EG. 2015. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu. Rev. Immunol. 33, 227–256. ( 10.1146/annurev-immunol-032713-120238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser M, Bork P, Fraser C, Knight R, Wang J. 2013. The microbiome explored: recent insights and future challenges. Nat. Rev. Microbiol. 11, 213–217. ( 10.1038/nrmicro2973) [DOI] [PubMed] [Google Scholar]
- 4.Harris RN, et al. 2009. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 3, 818–824. ( 10.1038/ismej.2009.27) [DOI] [PubMed] [Google Scholar]
- 5.Lam BA, Walke JB, Vredenburg VT, Harris RN. 2010. Proportion of individuals with anti-Batrachochytrium dendrobatidis skin bacteria is associated with population persistence in the frog Rana muscosa. Biol. Conserv. 143, 529–531. ( 10.1016/j.biocon.2009.11.015) [DOI] [Google Scholar]
- 6.Becker MH, et al. 2015. Composition of symbiotic bacteria predicts survival in Panamanian golden frogs infected with a lethal fungus. Proc. R. Soc. B 282, 20142881 ( 10.1098/rspb.2014.2881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jani AJ, Briggs CJ. 2014. The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. Proc. Natl Acad. Sci. USA 111, E5049–E5058. ( 10.1073/pnas.1412752111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voyles J, et al. 2009. Pathogenesis of chytridiomycosis, a cause of catastrophic amphibian declines. Science 326, 582–585. ( 10.1126/science.1176765) [DOI] [PubMed] [Google Scholar]
- 9.Woodhams DC, et al. 2014. Interacting symbionts and immunity in the amphibian skin mucosome predict disease risk and probiotic effectiveness. PLoS ONE 9, e96375 ( 10.1371/journal.pone.0096375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodhams DC, et al. 2015. Antifungal isolates database of amphibian skin-associated bacteria and function against emerging fungal pathogens. Ecol. Ecol. Arch. 96, 595–595. ( 10.1890/14-1837.1) [DOI] [Google Scholar]
- 11.Carey C, Bruzgul JE, Livo LJ, Walling ML, Kuehl KA, Dixon BF, Pessier AP, Alford RA, Rogers KB. 2006. Experimental exposures of boreal toads (Bufo boreas) to a pathogenic chytrid fungus (Batrachochytrium dendrobatidis). EcoHealth, 3, 5–21 ( 10.1007/s10393-005-0006-4) [DOI] [Google Scholar]
- 12.Kueneman JG, Parfrey LW, Woodhams DC, Archer HM, Knight R, McKenzie VJ. 2014. The amphibian skin-associated microbiome across species, space and life history stages. Mol. Ecol. 23, 1238–1250. ( 10.1111/mec.12510) [DOI] [PubMed] [Google Scholar]
- 13.Boyle DG, Boyle DB, Olsen V, Morgan JAT, Hyatt AD. 2004. Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis. Aquat. Organ. 60, 141–148. ( 10.3354/dao060141) [DOI] [PubMed] [Google Scholar]
- 14.Peterson AC, McKenzie VJ. 2014. Investigating differences across host species and scales to explain the distribution of the amphibian pathogen Batrachochytrium dendrobatidis. PLoS ONE, 9, e107441 ( 10.1371/journal.pone.0107441) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rideout JR, et al. 2014. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2, e545 ( 10.7717/peerj.545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. ( 10.1038/ismej.2011.139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. ( 10.1038/nmeth.2276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenzie VJ, Bowers RM, Fierer N, Knight R, Lauber CL. 2012. Co-habiting amphibian species harbor unique skin bacterial communities in wild populations. ISME J. 6, 588–596. ( 10.1038/ismej.2011.129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kueneman JG, Woodhams DC, Van Treuren W, Archer HM, Knight R, McKenzie VJ. 2015. Inhibitory bacteria reduce fungi on early life stages of endangered Colorado boreal toads (Anaxyrus boreas). ISME J. 10, 934–944. ( 10.1038/ismej.2015.168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walke JB, Becker MH, Loftus SC, House LL, Cormier G, Jensen RV, Belden LK. 2014. Amphibian skin may select for rare environmental microbes. ISME J. 8, 2207–2217. ( 10.1038/ismej.2014.77) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker MH, et al. 2015. Phylogenetic distribution of symbiotic bacteria from Panamanian amphibians that inhibit growth of the lethal fungal pathogen Batrachochytrium dendrobatidis. Mol. Ecol. 24, 1628–1641. ( 10.1111/mec.13135) [DOI] [PubMed] [Google Scholar]
- 22.Walke JB, Becker MH, Loftus SC, House LL, Teotonio TL, Minbiole KP, Belden LK. 2015. Community structure and function of amphibian skin microbes: an experiment with bullfrogs exposed to a Chytrid fungus. PLoS ONE 10, e0139848 ( 10.1371/journal.pone.0139848) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker MH, Brucker RM, Schwantes CR, Harris RN, Minbiole KP. 2009. The bacterially produced metabolite violacein is associated with survival of amphibians infected with a lethal fungus. Appl. Environ. Microbiol. 75, 6635–6638. ( 10.1128/AEM.01294-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loudon AH, Woodhams DC, Parfrey LW, Archer H, Knight R, McKenzie VJ, Harris RN. 2014. Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus). ISME J. 8, 830–840. ( 10.1038/ismej.2013.200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker MH, Richards-Zawacki CL, Gratwicke B, Belden LK. 2014. The effect of captivity on the cutaneous bacterial community of the critically endangered Panamanian golden frog (Atelopus zeteki). Biol. Conserv. 176, 199–206. ( 10.1016/j.biocon.2014.05.029) [DOI] [Google Scholar]
- 26.McMahon TA, et al. 2014. Amphibians acquire resistance to live and dead fungus overcoming fungal immunosuppression. Nature 511, 224–227. ( 10.1038/nature13491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendelson JR, et al. 2006. Confronting amphibian declines and extinctions. Science 313, 5783 ( 10.1126/science.1128396) [DOI] [PubMed] [Google Scholar]
- 28.Bletz MC, Loudon AH, Becker MH, Bell SC, Woodhams DC, Minbiole KP, Harris RN. 2013. Mitigating amphibian chytridiomycosis with bioaugmentation: characteristics of effective probiotics and strategies for their selection and use. Ecol. Lett. 16, 807–820. ( 10.1111/ele.12099) [DOI] [PubMed] [Google Scholar]
- 29.Woodhams DC, Bletz MC, Kueneman JG, McKenzie VJ. 2016. Managing amphibian disease with skin microbiota. Trends Microbiol. 24, 161–164. ( 10.1016/j.tim.2015.12.010) [DOI] [PubMed] [Google Scholar]
- 30.Rebollar EA, et al. 2016. Using ‘omics’ and integrated multi-omics approaches as guides to probiotic selection to mitigate chytridiomycosis. Front. Microbiol. 7, 68 ( 10.3389/fmicb.2016.00068) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bacterial data, including raw bacterial sequence reads and processed OTU tables with mapping files, are available through Dryad at: http://dx.doi.org/10.5061/dryad.ck1qs.