SHORT HYPOCOTYL1 (SH1), a chromatin remodeler, regulates hypocotyl elongation in cucumber through modulating the UVR8-mediated UVB signaling pathway.
Abstract
In Arabidopsis (Arabidopsis thaliana), the UVR8-mediated signaling pathway is employed to attain UVB protection and acclimation to deal with low-dosage UVB (LDUVB)-induced stresses. Here, we identified SHORT HYPOCOTYL1 (SH1) in cucumber (Cucumis sativus), which regulates LDUVB-dependent hypocotyl elongation by modulating the UVR8 signaling pathway. We showed that hypocotyl elongation in cucumbers carrying the recessive sh1 allele was LDUVB insensitive and that Sh1 encoded a human SMARCA3-like chromatin remodeling factor. The allele frequency and distribution pattern at this locus among natural populations supported the wild cucumber origin of sh1 for local adaptation, which was under selection during domestication. The cultivated cucumber carries predominantly the Sh1 allele; the sh1 allele is nearly fixed in the semiwild Xishuangbanna cucumber, and the wild cucumber population is largely at Hardy-Weinberg equilibrium for the two alleles. The SH1 protein sequence was highly conserved among eukaryotic organisms, but its regulation of hypocotyl elongation in cucumber seems to be a novel function. While Sh1 expression was inhibited by LDUVB, its transcript abundance was highly correlated with hypocotyl elongation rate and the expression level of cell-elongation-related genes. Expression profiling of key regulators in the UVR8 signaling pathway revealed significant differential expression of CsHY5 between two near isogenic lines of Sh1. Sh1 and CsHY5 acted antagonistically at transcriptional level. A working model was proposed in which Sh1 regulates LDUVB-dependent hypocotyl elongation in cucumber through changing the chromatin states and thus the accessibility of CsHY5 in the UVR8 signaling pathway to promoters of LDUVB-responsive genes for hypocotyl elongation.
The hypocotyl of dicotyledonous plants is the embryonic stem connecting the two embryonic leaves (cotyledons) and the primary root (radicle). The hypocotyl, which has a relatively simple architecture (Scheres et al., 1994), is a very plastic organ, strongly influenced by both external and internal cues known to regulate cell elongation, such as light, gravity, temperature, and hormones. In Arabidopsis (Arabidopsis thaliana), after germination, hypocotyl growth relies mainly on longitudinal cell elongation rather than cell divisions; its length can increase by more than 10-fold (Gendreau et al., 1997). The morphological simplicity, growth behavior, availability of mutants, and ease of phenotyping of mutants of this organ makes it a model for understanding the mechanism of cell elongation and various biological events that participate in its control (Gendreau et al., 1997; Vandenbussche et al., 2005; Boron and Vissenberg, 2014).
In nature, the development of germinated seeds starts under soil cover; the seedling undergoing skotomorphogenic growth has a very long hypocotyl, which allows quick attaining of light and de-etiolation after underground germination. The dark growth of seedlings is also characterized by closed cotyledons, reduced root growth, and development of apical hook on the hypocotyl. Once out of the soil, the seedlings undergo light-induced photomorphogenesis, which is associated with inhibition of hypocotyl elongation, opening of apical hook, expansion of cotyledons, and accumulation of chlorophyll and anthocyanin (Nemhauser and Chory, 2002).
Plants have evolved complex and sophisticated transcriptional networks regulating seedling photomorphogenesis, hence hypocotyl development, which has been well characterized in Arabidopsis (for review, see Jiao et al., 2007; Boron and Vissenberg, 2014; Galvao and Fankhauser, 2015). Plants are able to sense multiple parameters of ambient light signals, including light quantity (fluence), quality (wavelengths), direction, and duration. Light signals at different wave lengths are perceived through at least four distinct families of photoreceptors which, in Arabidopsis, include phytochromes (PHYA–PHYE, for red and far red light), cryptochromes and phototropins (CRY1 and CRY2, blue/UVA light), and UVR8 (UV RESISTANCE LOCUS8, for UVB; for review, see Jiao et al., 2007; Liu et al., 2011; Heijde and Ulm, 2012).
During the shift from skotomorphogenesis to photomorphogenesis, approximately one-third of genes in the Arabidopsis genome are altered (Ma et al., 2001; Tepperman et al., 2001). The coordinated activation or repression of downstream genes during this transition is realized mainly through transcriptional regulatory networks. Several important players connecting light perception by photoreceptors and downstream signaling cascades have been extensively studied including CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), ELONGATED HYPOCOTYL5 (HY5), phytochrome-interacting factors, and PICKLE. COP1 acts as an E3 ubiquitin ligase and is a repressor of photomorphogenesis in the dark, which targets photomorphogenesis promoting factors, such as HY5, for degradation in the dark (Osterlund et al., 2000; Holm et al., 2002; Saijo et al., 2003; Seo et al., 2003).
HY5 is a bZIP transcription factor and a positive regulator of photomorphogenesis through physical interaction with COP1 (Koornneef et al., 1980; Oyama et al., 1997; Ang et al., 1998; Chattopadhyay et al., 1998; Li et al., 2010). In the dark, HY5 is ubiquitinated by COP1 and degraded by the proteasome; in light, HY5 is stabilized and acts as a promoter of seedling de-etiolation. HY5 binds to the promoters of a large number of light-responsive genes (for example, cell-elongation-related genes for hypocotyl elongation; Lee et al., 2007; Zhang et al., 2011; Abbas et al., 2014). HY5 also plays key roles in other plant growth and development processes such as integrating multiple hormonal signaling pathways (Cluis et al., 2004; Vandenbussche et al., 2007; Alabadí et al., 2008), as well as mediating plant responses to environmental cues like cold (Lau and Deng 2010; Catalá et al., 2011) and UVB (see below).
UVB light (280–320 nm) is a small but important component of sunlight. Plants use UV as an environmental cue to regulate a wide range of physiological processes. Under high-fluence UVB light, plants exhibit more generic stress responses, which may include DNA damage, generation of reactive oxygen species, and inhibition of photosynthesis (Brosché et al., 2002; Frohnmeyer and Staiger, 2003; Robson et al., 2015). In contrast, low fluence UVB acts as a positive signal that promotes plant photomorphogenic development that is characterized by hypocotyl growth inhibition, accumulation of phenolic “sunscreen” metabolites, and acclimation to UVB stress (Christie and Jenkins, 1996; Kim et al., 1998; Kliebenstein et al., 2002; Tilbrook et al., 2016). In Arabidopsis, UVR8 is a photomorphogenic UVB-specific signaling component that orchestrates the expression of a range of genes with vital UVB-protective or acclimation functions (Christie et al., 2012). However, few molecular players involved in UVR8-mediated UVB signal transduction are currently known, including COP1 and HY5 (Ulm et al., 2004; Brown et al., 2005; Oravecz et al., 2006; Favory et al., 2009; Huang et al., 2012; Binkert et al., 2014; Ulm and Jenkins 2015). Under this model, the monomerized active UVR8 interacts with COP1, leading to the expression and stabilization of HY5, which directly bind to the promoter of several UVB responsive genes (Rizzini et al., 2011; Cloix et al., 2012; Binkert et al., 2014; Jenkins, 2014; Yin et al., 2015, 2016).
While the details on the UVR8-signaling pathways are largely unknown, Gardner et al. (2009) found that a mutant of UVR8 had similar hypocotyl growth inhibition to the wild type after UVB irradiation, suggesting a UVR8-independent UVB-responsive pathway. Biever et al. (2014) suggested that UVB inhibition of hypocotyl growth in etiolated Arabidopsis seedlings (that had already undergone photomorphogenesis) is a consequence of cell cycle arrest initiated by the accumulation of photodimers (UVB-induced covalent linkage of adjacent pyrimidine bases). Morales et al. (2013) studied UVR8 pathway functions under outdoor natural sunlight, which includes UVA, UVB, and high irradiance of photosynthetically active radiation. They found that UVR8 is required for transcript accumulation of genes involved in UV protection, oxidative stress, hormone signal transduction, and defense against herbivores under solar UV. Morales et al. (2013) suggest that, under natural UVA irradiance, UVR8 is likely to interact with the UVA/blue light signaling pathways to moderate UVB-driven transcript accumulation; thus, UVR8-dependent UVB acclimation during the early stages of plant development may enhance normal growth under long-term exposure to solar UV.
Our knowledge of photoreceptor-mediated light signaling in plants has been obtained mainly from Arabidopsis under controlled environments. Recent studies have revealed that at least some orthologous genes in crop plants may have additional or altered functions that are not detectable in Arabidopsis and vice versa (Liu et al., 2004; Giliberto et al., 2005; Chatterjee et al., 2006). For example, the HP2 gene encodes the tomato (Solanum lycopersicum) homolog of DET1, and dramatic variations between Arabidopsis det1 and tomato hp2 phenotypes have been observed (Mustilli et al., 1999). Bhatia et al. (2008) identified and characterized a regulatory gene SHORT HYPOCOTYL IN WHITE LIGHT1 (SHW1) from chickpea (Cicer arietinum), which encodes a unique Ser-Arg-Asp-rich protein. In Arabidopsis, SHW1 interacts with HY5 and COP1, promoting COP1-mediated degradation of HY5 during seedling development (Srivastava et al., 2015). These studies illustrate how findings in nonmodel species may be important to provide novel insights into the mechanisms of light-regulated plant growth and development.
Cucumber, Cucumis sativus (2n = 2x = 14), is an important vegetable crop worldwide and is a model system for studying several important biological processes. Cucumber is native to the Southern Asia continent (Candolle, 1959; Sebastian et al., 2010). Three botanical varieties of C. sativus have been recognized, including cultivated cucumber C. sativus L. var sativus (CSS hereinafter), the wild cucumber C. sativus L. var hardwickii (Royle) Alef. (CSH hereinafter; Royle, 1835; Duthie, 1903; Yang et al., 2012), and the semiwild Xishuangbanna cucumber C. sativus L. var xishuangbannanesis Qi et Yuan (XIS hereinafter; Qi, 1983). The XIS originated in tropical southwest China, and the surrounding regions exhibit a number of characteristic traits useful for cucumber improvement (Bo et al., 2015). In the XIS accession SWCC8, we found that hypocotyl length remained very short under a variety of different light conditions. In this study, we characterized hypocotyl elongation in SWCC8 with great details. We conducted map-based cloning of the SHORT HYPOCOTYL1 (Sh1) gene and identified a candidate gene encoding a homolog of chromatin remodeling factor SMARCA3. The link between a SMARCA3 chromatin remodeling factor and UVB-dependent hypocotyl elongation has not been found in any plant species; thus, it may represent a new player in the UVB signaling pathway. The objectives of this study were to clone and characterize the structure, function, and evolution of Sh1 gene in cucumber with the aim of understanding the molecular mechanisms of Sh1-mediated hypocotyl elongation. We first confirmed the low-dosage UVB-insensitive hypocotyl elongation in SWCC8. Then, we cloned the Sh1 locus with a map-based cloning strategy and validated its function with gene expression and phylogenetic analysis. We also examined allelic diversity of this locus in natural cucumber populations, which revealed the origin and evolution of this gene. Finally, we proposed a model to explain the possible roles of SH1 protein in regulating hypocotyl elongation in cucumber.
RESULTS
Hypocotyl Elongation in Semiwild XIS Cucumber Is LDUVB Insensitive
We previously observed differential hypocotyl growth of CSS and XIS lines in the greenhouses and growth chambers. We reasoned that this may be due to influence of light quality or quantity in the environments. We examined the wavelength spectra of different light sources and found that UVB could be effectively filtered out with glasses (Supplemental Fig. S1, A–D). Thus, we defined that the glass-roofed greenhouses and growth chambers with a glass plate shield we used in this study were UVB-free. The growth chamber with fluorescent lamps still had low dosage of UVB (LDUVB) and was defined as a LDUVB environment.
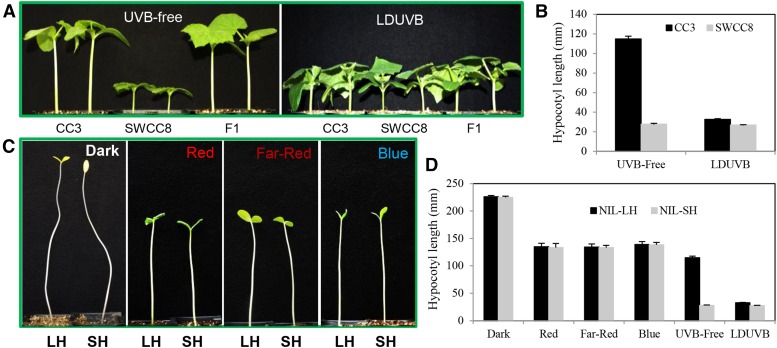
Significant differences in seedling hypocotyl length were observed between SWCC8 and CC3 under UVB-free but not under LDUVB conditions (Fig. 1, A and B). Under UVB-free white light, at 10 d after germination, the mean hypocotyl length of CC3 was ∼120 mm, whereas that of SWCC8 was around 27 mm; under LDUVB, the hypocotyl length of both lines was similar (28–33 mm). The much longer hypocotyl in CC3 under the UVB-free condition was due to a much faster daily elongation rate in the first week after germination than SWCC8, which was similar to that under LDUVB (Fig. 2). These results suggested that hypocotyl elongation of SWCC8 is LDUVB insensitive and that of CC3 is inhibited under LDUVB.
Figure 1.
LDUVB-dependent hypocotyl elongation in CC3 and SWCC8 cucumbers. Hypocotyl elongation is LDUVB insensitive in SWCC8 and is inhibited by LDUVB in CC3 (A and B). There is no significant difference in hypocotyl elongation between SWCC8 and CC3 under dark, red, far-red, or blue light conditions (C and D), but there seems to be photomorphogenic hypocotyl elongation inhibition under monochromic light or UVB-free white light (D). Hypocotyl length data in B and D were collected at 10 d after germination.
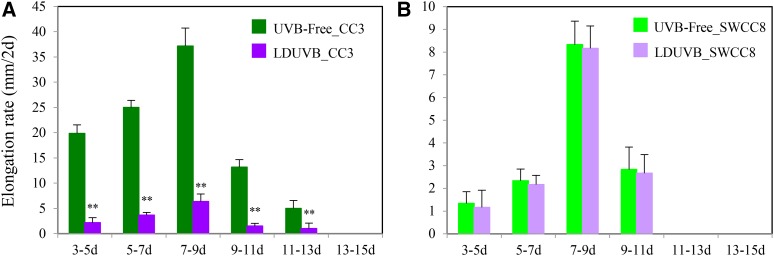
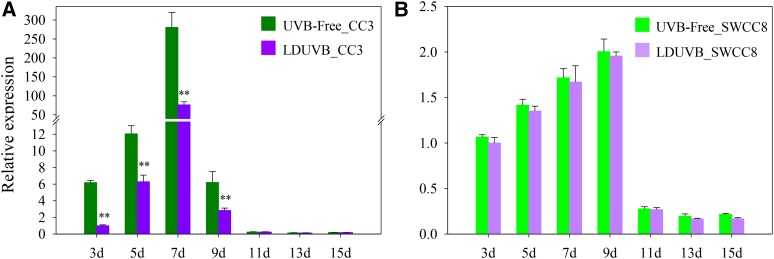
Figure 2.
Hypocotyl elongation rate in CC3 and SWCC8 under UVB-free white light and LDUVB conditions. Under UVB-free light, CC3 hypocotyl elongation rate (mm/2 d) reached its peak at 7 d after germination, which is much faster than that under LDUVB (A). SWCC8 has a much slow hypocotyl elongation rate (B). **Statistically significant differences of expression level based on t test (P < 0.01).
To investigate if SWCC8 had different responses to low and high level of UV irradiation, we measured hypocotyl length of SWCC8 and CC3 at different UV fluence rates (Supplemental Fig. S2). The results indicated that high dosage of UV irradiation resulted in seedling damage and stunted growth, confirming SWCC8 was insensitive to LDUVB.
In addition to UVB, the light spectrum in the greenhouses and growth chambers also differed in the fluence of lights at other wavelengths (Supplemental Fig. S1). To examine possible effects of light quantity on hypocotyl elongation, we measured hypocotyl length of two near isogenic lines of the Sh1 gene, NIL-SH (NIL carrying homologous short hypocotyl allele sh1) and NIL-LH (NIL carrying homologous long hypocotyl allele Sh1) under four light conditions: dark, blue, red, and far-red LED lights (see Supplemental Fig. S1E for wavelength of each monochromic light source). Three days after germination, no difference in hypocotyl length was observed between the two NILs when they were grown in the dark (Fig. 1C). Similar results were found under monochromic or multispectrum white lights (Fig. 1D). These results further confirmed that hypocotyl elongation in SWCC8 or WI7167 (another accession of the semiwild XIS cucumber) was LDUVB insensitive. However, despite the similar hypocotyl length between the two lines under each light condition, the absolute length in the dark (skotomorphogenesis) was clearly longer than that under the red, far-red, and blue light, with that under UVB-free white light being the shortest (Fig. 1, C and D). This was likely due to the photoreceptor-induced photomorphogenic effects of hypocotyl elongation inhibition. The data also suggested that the inhibition on hypocotyl elongation was stronger under multispectrum UVB-free white light than that under monochromic blue, red, or far-red light.
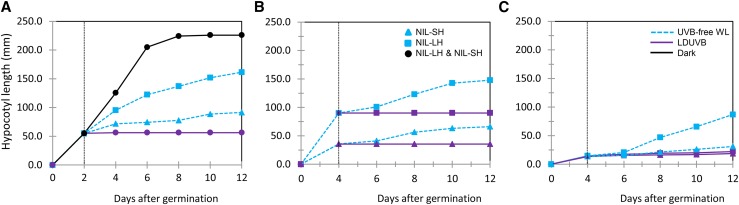
To separate the effects of non-UVB photoreceptor-induced photomorphogenic and LDUVB-induced inhibition on hypocotyl elongation, we performed three additional experiments. In the first experiment, the two NILs were first grown in the dark for 2 d after germination and then moved to LDUVB or UVB-free light or kept in dark for 10 d. In the second and third experiments, the two NILs were grown in UVB-free or LDUVB condition for 4 d and then moved to LDUVB and UVB-free lights, respectively. Hypocotyl length of each seedling plant in each treatment was measured every other day for 12 d. The results from the three pilot studies are shown in Figure 3, which provided more insights into LDUVB-dependent hypocotyl elongation in the two NILs. As shown in Figure 3A, under dark, hypocotyl length of both NILs was ∼50 mm by day 2, which reached the peak of ∼226 mm at day 12. After day 2, when exposed under LDUVB, hypocotyl elongation of both NILs was almost completely inhibited throughout the duration of the experiment. When exposed to UVB-free white light, the hypocotyl length of both NILs showed continuing increase, which was, at day 12, ∼161 mm in NIL-LH and ∼91 mm in NIL-SH. Thus, the reduced hypocotyl length of NIL-LH under UVB-free light as compared with that in the dark may represent photomorphogenic inhibition and that between the two NILs under UVB-free light may be due to the effect of the sh1 allele (defective function in hypocotyl elongation). Results from the second and third experiments (Fig. 3, B and C) further supported this conclusion. In addition, it was clear that the action of the Sh1 locus on hypocotyl elongation in response to LDUVB and UVB-free lights was very fast and effective.
Figure 3.
Hypocotyl elongation of near isogenic lines of the Sh1 locus under different light treatments. In A, the two NILs were grown under dark for 2 d, then moved to LDUVB or UVB-free lights or continued in the dark for 10 d. In B and C, the NILs were grown under either UVB-free WL (white light) or LDUVB light for 4 d, then moved to LDUVB and UVB-free WL, respectively. Hypocotyl length was measured every other day after switch the lights for 12 d. Each data point was the mean from three plants. The solid square, triangle, or circle represents NIL-LH, NIL-SH or both, respectively. Solid dark, purples, and dashed blue lines indicate dark, LDUVB, and UVB-free WL, respectively.
Recent work in Arabidopsis indicated that hypocotyl elongation during photomorphogenesis was also affected by temperature (Delker et al., 2014; Johansson et al., 2014). We investigated the effect of temperature on hypocotyl elongation in two NILs under UVB-free and LDUVB conditions at 17°C, 27°C, and 36°C (Supplemental Fig. S3). We found that hypocotyl elongation of NIL-SH remained insensitive to LDUVB under all three temperatures, whereas NIL-LH exhibited longer hypocotyl under UVB-free than under LDUVB. The differences were the largest at optimal growth temperature (27°C; Supplemental Fig. S3). This was probably due to the pleiotropic effects of the sh1 allele on slower hypocotyl elongation.
To summarize, the above studies all supported the hypothesis that hypocotyl elongation in XIS was LDUVB insensitive, whereas LDUVB inhibited hypocotyl elongation in sensitive CSS genotypes. This LDUVB-dependent hypocotyl elongation in cucumber was regulated through the Sh1 gene.
A Recessive Locus, sh1, Controls LDUVB-Insensitive Hypocotyl Elongation
Under UVB-free light condition, CC3 × SWCC8 F1 plants exhibited long hypocotyl comparable to the female parent, CC3 (Fig. 1A). Of the 124 recombinant inbred lines (RILs) derived from this cross, 67 and 57 had long and short hypocotyls, respectively. Among 1183 F2 plants, 878 and 305 had long and short hypocotyls, respectively. Goodness-of-fit χ2 tests indicated that the data in the RIL (P = 0.4190) and F2 (P = 0.5569) populations were consistent with the expected 1:1 and 3:1 segregation, respectively, suggesting a single recessive gene underlying the short hypocotyl phenotype in SWCC8. In the WI7200 × WI7167 F2:3 population, hypocotyl length was treated as a quantitative trait. The hypocotyl length of each plant was measured in each F3 family. The frequency distribution of mean hypocotyl length among 165 F3 was largely normal with the two extreme values corresponding to the two parental lines and the F1 close to WI7200 (Supplemental Fig. S4A). If the cutoff of mean hypocotyl length was set to 2.1 cm, the segregation of long (128) and short hypocotyl (37) families in this population was also consistent to 3:1 expected ratio (P = 0.4448 in χ2 test). Linkage analysis (below) indicated the short hypocotyl in both SWCC8 and WI7167 was controlled by the same recessive gene, which is designated as sh1 hereinafter.
From our multiple-year observations, the short hypocotyl length in CSH and XIS was often associated with slow growth at the seedling stage. This was probably the result of local adaptation to the high UVB fluence (see “Discussion”). To ascertain the linkage between the two traits, we conducted QTL mapping of seedling stage plant height (vine length at 5-true leaf stage) (Bo et al., 2011) and hypocotyl length in the 124-RIL population; the LOD profiles are shown in Supplemental Figure S5. The results further confirmed the pleiotropic effect of sh1 allele on slow seedling growth as measured with vine length.
SH1 Encodes a Homolog of Human SMARCA3-Like Chromatin Remodeling Factor
Initial screening with 480 simple sequence repeat (SSR) markers identified nine polymorphic ones between CC3 and SWCC8, as well as between the short hypocotyl bulk (S-bulk) and the long hypocotyl bulk (L-bulk) of RILs, all of which were located in cucumber chromosome 3. Linkage analysis in 124 RILs revealed that the Sh1 locus was flanked with markers SSR03409 and SSR31430 that were 0.8 and 1.3 cM away from the Sh1 locus, respectively. Physically, both markers were located in an interval of approximately 812 kb in the Gy14 scaffold02229.
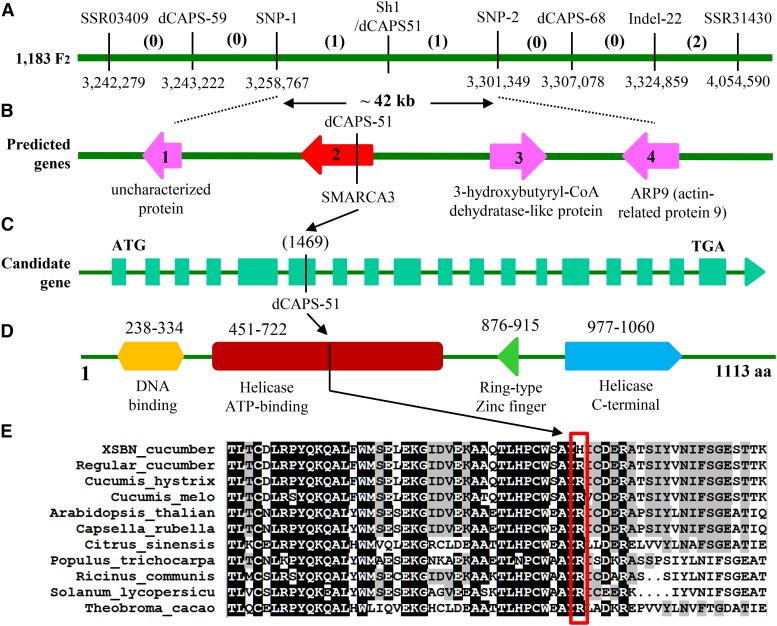
Among 1183 F2 plants of CC3 × SWCC8 screened with the two flanking markers, four recombinants were identified. Bioinformatic analysis of the resequencing data of the two parental lines identified new markers in this interval, six of which were mapped, including Indel-22, dCAPS-51, dCAPS-59, dCAPS-68, SNP-1, and SNP-2. The resulting genetic and physical maps for the Sh1 locus with the eight markers are illustrated in Figure 4A. Among the eight markers, dCAPS-51 was cosegregating with the Sh1 locus, whereas SNP-1 and SNP-2 flanked the sh1 locus and were ∼42 kb apart in scaffold02229. Four genes (gene 1 to gene 4) were annotated in this 42 kb genomic region (Fig. 4B). Alignment of genomic DNA sequences of this region between the two parental lines identified four indels and 23 SNPs, but all of them were silent mutations occurring in either intergenic regions or introns of these candidate genes, except for the SNP (C in CC3 versus T in SWCC8) in the exon of gene 2, from which the cosegregating marker dCAPS-51 was derived (Fig. 4C).
Figure 4.
Map-based cloning of Sh1 locus. Fine mapping with 1183 F2 plants delimits the Sh1 locus into a 42-kb region (A) that contains four predicted genes (B). Of the four genes, only the second one is the candidate gene for Sh1, which encodes a SMARCA3 homolog. There are 18 exons of this gene, and the cosegregating marker dCAPS-51 was developed from a SNP within the sixth exon of the candidate gene (C). The SMARCA3-like protein contains all four functional domains, which are characteristic of the helicase-like transcription factor subfamily chromatin remodelers (D). The SNP in the sixth exon results in an amino acid substitution from R (Arg) to H (His) in the helicase ATP-binding domain (D). The amino acid sequence of this domain is highly conserved in plants. Of the nine species compared, only SWCC8 cucumber has H, and all others have R at this SNP location (E).
We examined expression dynamics within 15 d after germination at 2 d intervals with quantitative PCR (qPCR) of the four genes in SWCC8 and CC3 under UVB-free and LDUVB conditions. There were no differences in expression of genes 1, 3, and 4 between LDUVB and UVB-free treatments (Supplemental Fig. S6), excluding them as possible candidates for Sh1. We further investigated the association of gene 2 with LDUVB-dependent hypocotyl elongation in cucumber natural populations. Hypocotyl lengths of 211 cucumber lines from various sources were measured under both UVB-free and LDUVB conditions (data presented in Supplemental Table S1). Among the 211 lines, 197 showed LDUVB-sensitive hypocotyl elongation (shorter hypocotyl under LDUVB) and 14 insensitive. Representative images of selected cucumber lines in each group and corresponding hypocotyl data are shown in Supplemental Figure S7, A to D. All 211 cucumber lines were genotyped with the dCAPS-51 (examples in Supplemental Fig. S7E). We found that 192, 14, and five lines carried the Sh1 (from CC3), sh1 (from SWCC8), or both (heterozygotes) alleles, respectively (complete data in Supplemental Table S1). All 14 LDUVB-insensitive lines carried the SWCC8 allele, further confirming that gene 2 was indeed the candidate gene for Sh1.
Gene 2 was predicted to encode a SMARCA3-like protein. The genomic DNA sequence of this Sh1 candidate gene was 5686 bp in length for both CC3 and SWCC8. Annotation predicted 18 exons in this gene (Fig. 4C), which was confirmed with cloning of full-length cDNA from both parental lines. Genomic DNA and cDNA sequences for the two lines are provided in Supplemental File S1 (GenBank accession no. KX639507). BLAST searches in the Gy14 and 9930 draft genomes indicated only one copy of this gene in the cucumber genome. The SMARCA3-like protein is a helicase-like transcription factor (HLTF) of the snf2 family and Rad5/16 subfamily (Debauve et al., 2008). The SMARCA3 protein contains multiple functional domains, including DNA-binding, helicase ATP-binding, RING-type Zinc finger, and helicase C-terminal domains, which all existed in the deduced SH1 protein (Fig. 4D). The SNP within the sixth exon of the Sh1 candidate gene resulted in an amino acid substitution from R (Arg) in CC3 to H (His) in SWCC8, which was located in the helicase ATP-binding domain of the predicted protein (Fig. 4D). Many plant genomes contain sequences that have been annotated to encode SMARCA3-like proteins. We aligned the amino acid sequences of the SMARCA3 helicase ATP-binding domain of CC3 and SWCC8 cucumbers and that from nine other plant species including two cucumber relatives, Cucumis hystrix and melon (Cucumis melo; Fig. 4E). We found that the amino acid sequence in this region was highly conserved. All compared sequences had the amino acid R at the SNP position except for that of SWCC8, which was H, confirming the critical SNP mutation associated with the short hypocotyl phenotype in SWCC8 (Fig. 4E).
SH1 Protein Is Highly Conserved across Different Eukaryotic Kingdoms of the Life Tree
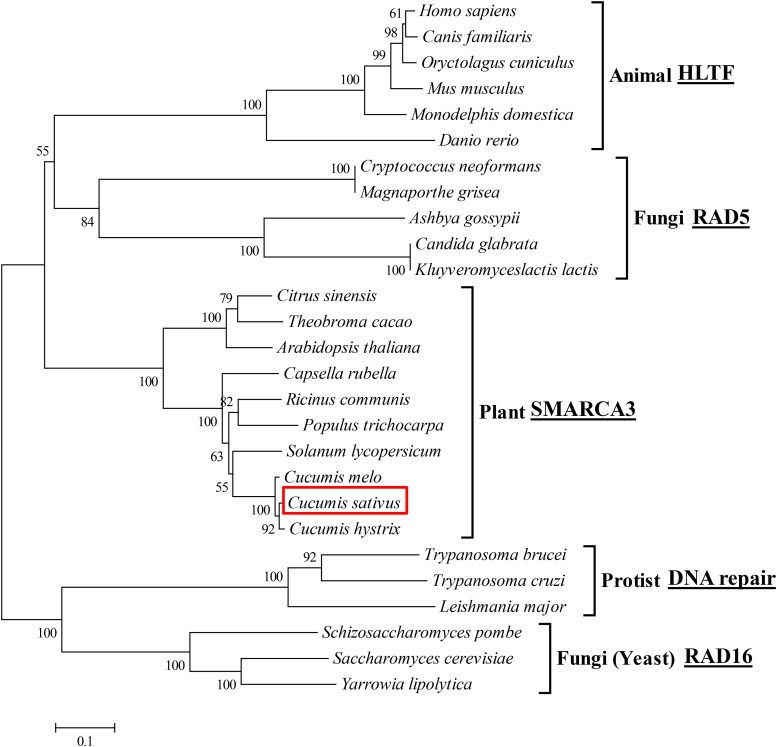
To understand the structural and functional relationship between SH1 in cucumber and its homologs in other species, we constructed a phylogenic tree using amino sequences from cucumber SH1 and 19 other Rad5/16 subfamily proteins, including eight from fungi, six from animals, nine from plants, and three from protists. The resulting neighbor-joining tree is shown in Figure 5. It was clear that the HLTF-type SMARCA3 protein was highly conserved in eukaryotes during evolution. The clustering of these sequences was largely consistent with known evolutionary relationships of these organisms (Ciccarelli et al., 2006), suggesting possible conservation of functions of this gene as a chromatin remodeler across the very diverse organisms.
Figure 5.
Phylogenetic tree of SH1 protein in cucumber and its homologs in 26 other species. The neighbor-joining phylogenetic tree was constructed with RAD5/RAD16 subfamily protein sequences from 27 species. The clustering is consistent with known phylogenetic relationships of different eukaryotic kingdoms of the life tree. The number at each node is the probability that this node is supported (in percentages).
Expression of SH1 Is Development Stage Dependent and Is Regulated by LDUVB
We examined the dynamics of hypocotyl elongation in CC3 and SWCC8 seedlings at different time points after germination under UVB-free and LDUVB light conditions. In both lines, under both conditions hypocotyl elongation stopped at ∼15 d after germination, and the fastest elongation occurred ∼7 to 9 d after germination (Fig. 2; Supplemental Fig. S8). In SWCC8, the absolute length or elongation rate at any time point under UVB-free white light was slightly higher than that under LDUVB; in contrast, under UVB-free condition, CC3 showed much faster hypocotyl elongation and longer hypocotyl than that under LDUVB (Fig. 2A; Supplemental Fig. S8B), indicating hypocotyl growth was inhibited by LDUVB. In addition, under LDUVB, the hypocotyl length of SWCC8 at each time point was shorter than that of CC3.
Under both light conditions, the expression level of the Sh1 gene in the two lines was highly consistent with the hypocotyl elongation rate. In CC3, mirroring the fastest elongation rate around 7 to 9 d after germination (Fig. 2A), Sh1 also had the highest expression level at this time, which was a 286- (UVB-free) and 77-fold (LDUVB) increase, respectively, as compared with that at 3 d after germination (Fig. 6A). This result also suggested that Sh1 expression is inhibited by LDUVB. Meanwhile, in SWCC8, the expression of Sh1 gene under UVB-free light was only slightly higher than that under LDUVB at any time point, and the peak expression of Sh1 occurred at 9 d after germination, which showed an approximate 2-fold increase under both UVB-free and LDUVB conditions (Fig. 6B). Clearly, the low expression in SWCC8 was primarily due to the defective function of the sh1 allele. Nevertheless, similar to CC3, LDUVB also seemed to inhibit Sh1 expression in the SWCC8 background: transcript level of Sh1 under LDUVB was ∼90% of that under UVB-free condition in the first 9 d after germination (but not statistically significant; Fig. 6B).
Figure 6.
Expression level of the Sh1 gene CC3 and SWCC8 under UVB-free white light and LDUVB conditions. In CC3, Sh1 expression is inhibited significantly by LDUVB (A). In SWCC8, there is no difference in the level of Sh1 expression under UVB-free white light and LDUVB conditions, which leaked at 9 d after germination (B). **Statistically significant differences of expression level based on t test (P < 0.01).
We calculated Pearson’s correlation coefficient (r) using hypocotyl length and Sh1 fold changes in the days before Sh1 expression reached its peak in both CC3 and SWCC8 under both light conditions. We found that there was a high correlation between hypocotyl length and Sh1 transcript abundance (r > 0.9 in all cases). Thus, qPCR data suggested that Sh1 expression was inhibited by LDUVB and the sh1 allele was defective in transcript accumulation, which was much lower than Sh1 even under UVB-free condition. The expression of both Sh1 and sh1 alleles was inhibited by LDUVB. The abundance of Sh1 transcript was highly correlated with the absolute length of hypocotyl and elongation rate in both lines under both conditions.
SH1 Regulates LDUVB-Dependent Hypocotyl Elongation through Activation of UVR8-Mediated Signaling Pathway
In Arabidopsis, hypocotyl growth relies mainly on longitudinal cell elongation (Gendreau et al., 1997). We examined the microscopic cell structure of the hypocotyls of both SWCC8 and CC3 growing under UVB-free and LDUVB conditions. We found that the cells of the hypocotyl in CC3 grown under the UVB-free condition were five to eight times as long as those in SWCC8 or CC3 under LDUVB; however, the widths of these cells were largely similar (Fig. 7, A1 and A2), suggesting that the long hypocotyl in CC3 under the UVB-free condition was due primarily to longitudinal cell elongation.
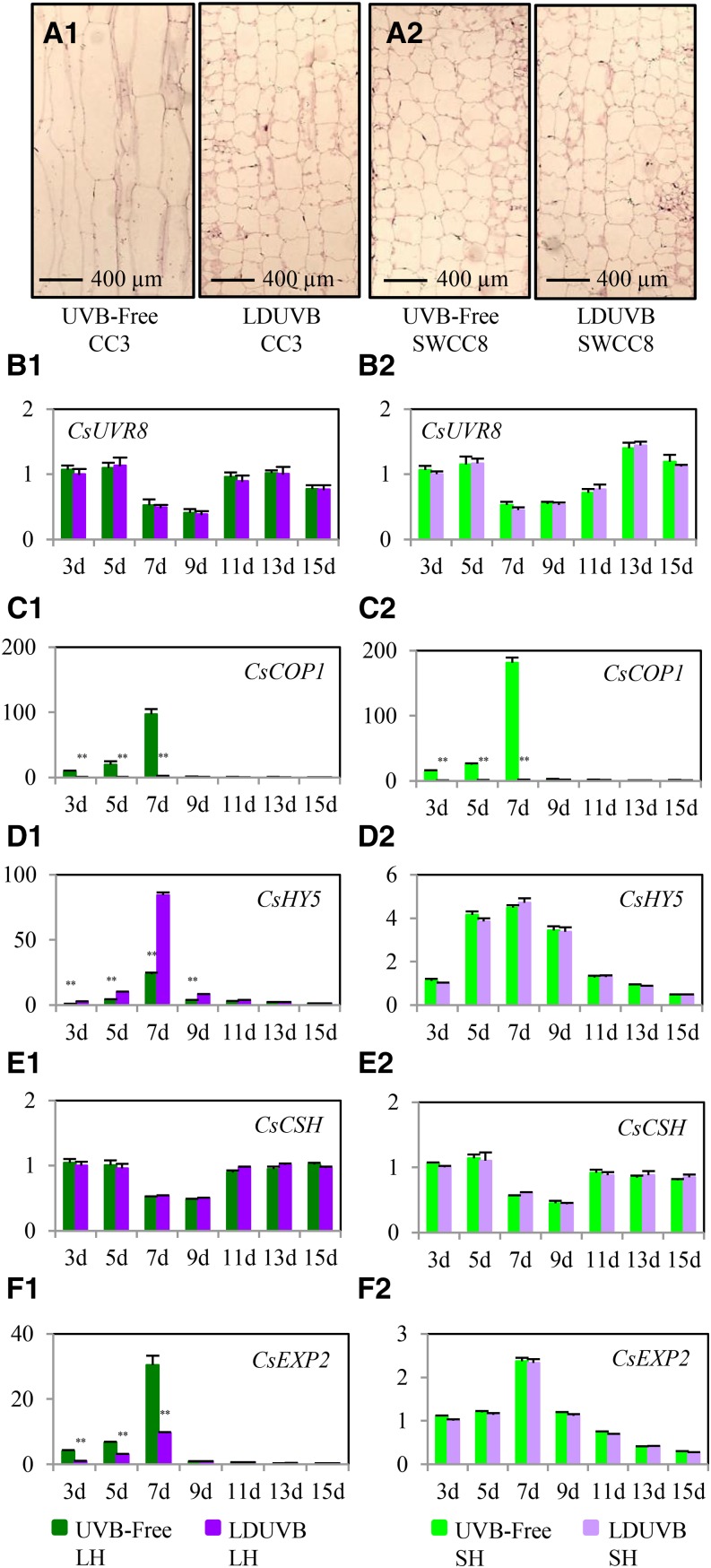
Figure 7.
Expression profiles of genes in cucumber UVR8 signaling pathway in two near isogenic lines under LDUVB and UVB-free light conditions. Microscopic observation indicates primarily longitudinal cell growth during hypocotyl elongation in CC3 under UVB-free light (A1). Hypocotyl growth is inhibited by LDUVB in CC3 or SWCC8 under either condition due to lack of cell elongation (A2). The hypocotyl elongation in two near isogenic lines (NIL-LH and NIL-SH) under UVB-free light or its inhibition under LDUVB is not regulated by CsUVR8 (B1 and B2), CsCOP1 (C1 and C2), or CsCSH (E1 and E2) at the transcription level but is consistent with the alternated expression of CsHY5 and cell-elongated-related genes (F1 and F2). From B to F, the y axis is relative expression level of each gene. The left and right panels are for NIL-LH and NIL-SH, respectively.
In the UVR8-mediated UVB responsive signaling pathway of Arabidopsis, HY5 and COP1 interact to regulate the expression of many other genes downstream of the pathway, including cell-elongation-related genes for hypocotyl elongation. HY5 also plays important roles in cross talks with multiple hormonal signaling pathways (see “Introduction”). To understand the molecular mechanism by which Sh1 regulates hypocotyl cell elongation, we investigated the expression pattern of cucumber CsUVR8, CsCOP1, CsHY5, CsCHS, and six cell-elongation-related genes (CsEXT3, CsEXP2, CsXTH17, CsXTR6, CsDWF4, and CsIAA19) in the two NILs of Sh1 (NIL-LH and NIL-SH) under UVB-free and LDUVB light conditions. The expression patterns of CsUVR8, CsCOP1, CsHY5, CsCHS, and CsEXP2 are illustrated in Figure7, B to F, respectively. Those for the remaining five genes are presented in Supplemental Figure S9.
The UVR8 pathway starts with its perception of UVB light resulting in monomizeration of the UVR8 dimer. The UVR8 monomer then physically interacts with COP1 to regulate expression of HY5 (Ulm and Jenkins, 2015). No difference in CsUVR8 expression was found between UVB-free and LDUVB conditions in both NIL lines; this was consistent with its constitutive expression found in Arabidopsis. It’s interesting to see the fluctuation of UVR8 transcript abundance by time. The expression level was reduced by ∼50% 7 to 9 d after germination and then returned to normal level (Fig. 7B).
At day 7 after germination, under UVB-free light, there was nearly a 150- and 180-fold increase of CsCOP1 expression in NIL-LH and NIL-SH (as compared to day 1), respectively; but under LDUVB, its expression showed only a 2- to 3-fold increase (Fig. 7C) indicating CsCOP1 expression was downregulated by LDUVB regardless of the Sh1/sh1 genetic backgrounds. Therefore, both CsUVR8 and CsCOP1 did not seem to be related with Sh1 gene expression and LDUVB-dependent hypocotyl elongation.
The expression pattern of CsHY5 in NIL-LH under the two light conditions was the opposite of that of CsCOP1 (Fig. 7D). At 7 d after germination, CsHY5 reached its peak level of expression, an 85-fold increase under LDUVB and approximately a 24-fold increase under UVB-free light; in NIL-SH, no difference in the expression of CsHY5 was observed, and the peak expression also occurred at 7 d with approximately a 5-fold increase under both UVB-free and LDUVB conditions. These results indicated that CsCOP1 and CsHY5 had opposite actions in regulating SH1-directed, LDUVB-dependent hypocotyl elongation.
The expression pattern of the cucumber chalcone synthase gene CsCHS was largely similar to that of CsUVR8; no significant difference was found between the two NIL lines at any time point under either light condition, although it was a bit higher in NIL-SH at 11 to 13 d under LDUVB (Fig. 7E). The expression patterns of the six cell-elongation-related genes (Fig. 7F; Supplemenatl Fig. S9) were largely consistent with that of the Sh1 gene (Fig. 6). That is, in NIL-LH, the expression of all six genes was drastically increased in UVB-free condition compared with that under LDUVB, with the biggest difference of expression level being around 7 to 9 d after germination, whereas in NIL-SH there was no significant difference in expression of these genes between UVB-free and LDUVB conditions. These results suggest that expression of all these genes was regulated by the Sh1 locus.
Yeast Two-Hybrid Growth Assay Reveals Interaction between SH1 and CsHY5
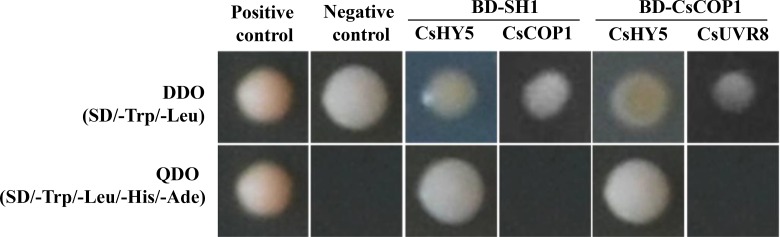
To further confirm the relationship of Sh1 gene expression and the UVR8 signaling pathway, we conducted the GAL4-based yeast two-hybrid (Y2H) assay to explore possible protein level interactions of SH1 with CsHY5, CsCOP1, and CsUVR8. The following four bait-prey mating pairs were examined: BD-SH1+AD-CsHY5, BD-SH1+AD-CsCOP1, BD-CsCOP1+AD-CsUVR8, and BD-CsCOP1+AD-CsHY5. Interactions were found between SH1 and CsHY5, CsCOP1, and CsHY5, but not between SH1 and CsCOP1 or between CsCOP1 and CsUVR8 (Fig. 8). In Arabidopsis, the COP1-HY5 interaction has been well established (e.g. Ang et al., 1998); the interaction between COP1 and HY5 is UVB dependent: there is no interaction in the absence of photomorphogenic UVB (Favory et al., 2009; Yin et al., 2016). Therefore, our Y2H experimental data among CsUVR8, CsCOP1, and CsHY5 were consistent with findings in Arabidopsis, suggesting that cucumber may take a similar UVR8 signaling pathway for UVB stress responses. These data provided additional evidence for the involvement of the UVR8 signaling pathway in Sh1-regulated hypocotyl regulation through CsHY5 in cucumber.
Figure 8.
Protein-protein interactions revealed from year two-hybrid assay. Y2HGold yeast strain with SH1 or CsCOP1 in the bait vector and CsHY5 and CsCOP1 in the prey vector were assayed for growth on double dropout (DDO; SD/-Trp/-Leu) or quadruple dropout (QDO; SD/-Leu-Trp-His-Ade) growth media. The positive control (pGBKT7-53 + pGADT7-T) and negative control (empty vectors pGBKT7-Lam + pGADT7-T) were included in the assay. Y2H assay revealed interactions between SH1 and CsHY5 as well as between CsCOP1 and CsHY5.
The sh1 Allele Was of Wild Cucumber Origin, Which Was under Selection during Domestication
We measured hypocotyl elongation under UVB-free and LDUVB conditions of 211 C. sativus accessions, of which 204, one, and six belonged to CSS, XIS, and CSH, respectively (Supplemental Table S1). Genotyping of these materials with the SNP-derived dCAPS-51 marker indicated that 192 (90%), 14 (8%), and five (2%) accessions carried Sh1, sh1, and both (heterozygotes) alleles, respectively. The 14 accessions carrying the sh1 allele included one XIS, three CSH, and 10 CSS, all of which showed LDUVB insensitive hypocotyl elongation (Supplemental Table S1; Supplemental Fig. S7).
To gain a more complete picture on allele distribution at this locus, we genotyped 291 additional cucumber lines with dCAPS-51. Information about all 502 lines is listed in Supplemental Table S1. Genotypic and allele frequencies of this locus in each population are presented in Table I. Among the 502 lines, 48, 35, and 419 were CSH, XIS, and CSS, respectively. Consistent with results in the 211 accessions with hypocotyl length data, the majority of CSS (380/502 = 75.7%) carried the Sh1 allele, whereas 33 XIS accessions carried the recessive sh1 allele, and two (XIS3-4 and XIS26) were heterozygous at this locus. Almost half (44%) of the 48 CSH accessions were homozygous recessive (sh1sh1); 38% and 19% of them were homozygous dominant (Sh1Sh1) and heterozygous (Sh1sh1), respectively. The approximate geographic coordinates (altitude and latitude) of the origin of each line (Supplemental Table S1) were used to plot these 502 lines on a world map, and the result is shown in Supplemental Figure S10. It was clear that accessions from India and the XIS from southwest China were highly enriched with the recessive sh1 allele: 54% of the CSS and CSH accessions from India (49 out of 91) and 94.3% of XIS accessions carried either sh1 or both alleles. These results suggested that the sh1 allele originated in CSH and was inherited by the XIS lineage. The allele frequency of Sh1 and sh1 in the CSH population was 0.469 and 0.531, respectively (Table I), which was largely at Hardy-Weinberg equilibrium, suggesting free gene flow between CSS and CSH cucumbers. In CSS populations, ∼7% accessions carried the sh1 (allele frequency 0.080), which may be an unintended consequence of diversifying selection in cucumber breeding (see “Discussion”).
Table I. Genotypic and allele frequencies at the Sh1 locus among 502 accessions from three C. sativus botanical varieties.
Note: The Sh1 (LDUVB-sensitive) and sh1 (LDUVB-insensitive) alleles are from CC3 and SWCC8, respectively.
| Populations | Sh1Sh1 (%) | sh1sh1 (%) | Sh1sh1 (%) | f(Sh1) | f(sh1) | Total |
|---|---|---|---|---|---|---|
| Wild cucumber (CSH) | 18 (38%) | 21 (44%) | 9 (18%) | 0.469 | 0.531 | 48 |
| XIS cucumber (XIS) | 0 | 33 (95%) | 2 (5%) | 0.029 | 0.971 | 35 |
| Cultivated cucumber (CSS) | 380 (90%) | 28 (7%) | 11 (3%) | 0.920 | 0.080 | 419 |
| Sum | 398 | 82 | 22 | 502 |
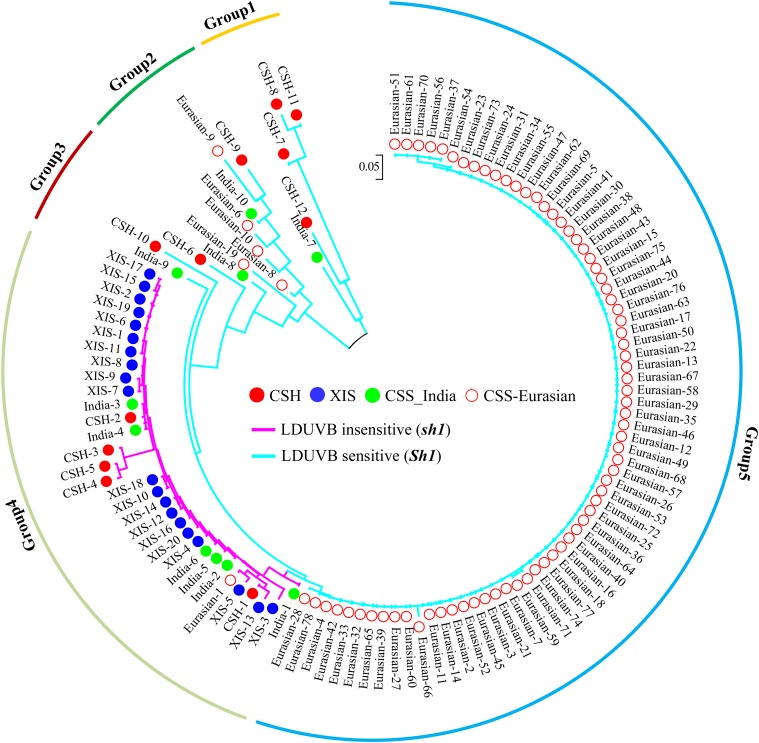
To ascertain the CSH origin of the sh1, we examined the linkage disequilibrium at the Sh1 locus among 120 cucumber accessions with resequencing data including 12 CSH, 20 XIS, and 88 CSS accessions (Supplemental Table S2). Of the 88 CSS lines, 10 were from India (India-1 to India-10), and 78 were from various geographic regions in Asia, Europe, and the Americas, which were assigned Eurasian-1 to Eurasia-78, respectively. Geographic distribution of these lines is highlighted in Supplemental Figure S10B. One hundred and ninety-three SNPs in a 15 kb vicinity of the Sh1 locus were used for linkage disequilibrium analysis and construction of a phylogenetic tree for the 120 accessions, which is illustrated in Figure 9. The grouping result with principal component analysis based on 67 of the 193 SNPs is illustrated in Supplemental Figure S10C. From a simply visual inspection of the nucleotide variations in this region, it was easy to see limited nucleotide polymorphism among the 20 XIS or 78 Eurasia lines. In the resulting NJ tree, the 120 lines were placed in five groups, with group 5 including 72 of 78 lines in the Eurasia group that carried only the Sh1 allele (LDUVB-sensitive hypocotyl elongation). Groups 1, 2, and 3 were composed of 16 accessions, most of which were from India (CSS and CSH), with high genetic diversity at these SNP loci. All of the 16 accessions carried the Sh1 allele. The last group, group 4, contained 32 accessions, including 20 XIS, five CSH, six Indian CSS, and one Eurasian. A common feature of these lines in group 4 was that all lines carried the sh1 allele. These results further confirmed the early notion that the LDUVB-insensitive sh1 allele had its origin in CSH, and XIS inherited this allele in its whole lineage.
Figure 9.
A dendrogram of 120 cucumber accessions from CSS, XIS, and CSH cucumbers based on 193 SNPs within 15-kb region of the Sh1 locus. All XIS accessions (blue solid dots) were from the Xishuangbanna region of Southwest China and carry the LDUVB-insensitive sh1 allele (all in group 4). All CSH accessions were from India, and approximately half of them carry the LDUVB-sensitive Sh1 allele and the other half the sh1 allele (in groups 1, 2, 3, and 4). The majority of CSS cucumber accessions carry the Sh1 allele (group 5, the Eurasian group).
DISCUSSION
The Origin and Evolution of sh1 Allele in Cucumber
Three botanical varieties have been recognized in cucumber: the CSS, CSH, and XIS cucumbers. We found that the LDUVB-insensitive hypocotyl elongation in the XIS cucumber was caused by a SNP within the Sh1 locus (Fig. 4). Among the 502 cucumber lines examined, 398, 82, and 22 carried the Sh1, sh1, and both alleles, respectively (Table I). All XIS accessions carried the LDUVB-insensitive sh1 allele (two were heterozygous), and the majority of CSS carried LDUVB-sensitive Sh1 allele. Of the 48 CSH accessions from India, 18 (38%), 21 (44%), and nine (18%) carried Sh1, sh1, and both alleles, respectively (Table I). To further trace the origin of the sh1 allele, we also genotyped nine C. hystrix (2n = 2x = 24) accessions (Supplemental Table S1) with the dCAPS-51 marker. C. hystrix is the closest, and the only sexually compatible, relative of cucumber. All C. hystrix accessions carried the dominant Sh1 allele, suggesting that the sh1 allele might have arisen de novo in CSH. All XIS accessions carried the sh1 allele, which is probably due to a LDUVB-insensitive initial founder. Of the 35 XIS accessions examined, two were heterozygous at the Sh1 locus (Supplemental Table S1), which was probably due to unintentional crosses of XIS with CSS by local people or insects. The relatively low frequency of the sh1 allele in CSS was likely the result of gene flow for LDUVB-insensitive lines in India or other regions overlapping with CSH, or it may have been an unintended introduction during the use of Indian germplasm in cucumber breeding. Meanwhile, the prevalence of the Sh1 allele in CSS is likely due to selection during domestication.
The rise of the sh1 allele in CSH is easy to understand in the evolutionary context in its natural habitats. The wild cucumber is distributed mainly in the foothills of the Himalaya mountains, where the elevation and UVB radiation is high. Although UVB is a minor component of sunlight reaching the Earth’s surface, it has major biological effects on plants. Plants have developed certain mechanisms to adapt to solar UVB radiation, such as leaf morphological features to improve epidermal reflectance of UVB radiation (e.g. Holmes and Keiller, 2002) and UV screening mechanism by accumulation of UV-absorbing compounds (i.e. flavonoids; e.g. Filella and Penuelas, 1999). It is conceivable that CSH employed LDUVB-insensitive short hypocotyl as a tactic to cope with possible damage. In CSH and XIS, seedlings with short hypocotyl often also grow very slowly in the early stage, which might be an additional general adaptation to avoid UVB damage to the seedlings during the plants’ most vulnerable stage. This association can be seen in the colocalization of hypocotyl length and vine length at the seedling stage in the RIL population (Supplemental Fig. S5). On the other hand, the slow seedling growth associated with the short hypocotyl may be the very reason why it was selected against during domestication, which favored the quick establishing seedlings desirable for agricultural production. Since LDUVB is always present in nature, it may be argued that selection of the Sh1 allele during domestication may not be efficient since Sh1 expression was also inhibited by LDUVB (Fig. 6). This can be explained by the fact that there was less inhibition of Sh1 expression and hypocotyl elongation by LDUVB in LDUVB-sensitive lines than that in LDUVB-insensitive one (Fig. 3; Supplemental Figs. S7 and S8).
This short hypocotyl allele may have practical use in cucumber breeding. The last decade has seen significant increase of large-scale vegetable production in protected environments. Efficient control of hypocotyl elongation is important for vegetable production through transplanting or grafting. Often, seedlings are prepared in plugs in the greenhouses in which solar UV exclusion may cause excessive hypocotyl elongation (e.g. Shinkle et al., 2004; Zhang et al., 2014b; Hernández and Kubota, 2016) that are more likely to suffer mechanical damage when using automated transplanting (Styler and Koranski, 1997). At present, plant hormone spray is used to control hypocotyl elongation in greenhouse seedling production. Use of the UVB-insensitive short hypocotyl gene such as sh1 may help modulate hypocotyl growth for high-quality plugs for transplanting in cucumber production. This could be an interesting example of dedomestication due to the changing practice of agricultural production.
SMARCA3, a Chromatin Remodeler as the Candidate Gene of Sh1
We show that Sh1 encoded a homolog of the human SMARCA3 (Fig. 4D) of the helicase-like transcription factor (HLTF) family. The HLTF sequences are highly conserved during evolution and can be identified in the genomes of all eukaryotes (Fig. 5). However, little is known about the normal functions of HLTF. The gene encoding HLTF is frequently subjected to hypermethylation in human cancers, suggesting that it may normally act as a tumor suppressor (Debauve et al., 2008; MacKay et al., 2009). The human HLTF is more closely related to the yeast (Saccharomyces cerevisiae) RAD5 and RAD16 proteins (Flaus et al., 2006; Fig. 5). RAD5 is the key component in the RAD5-dependent error-free branch of postreplication repair in yeast (for review, see Friedberg et al., 2006). In Arabidopsis, the yeast RAD5 homolog AtRAD5a is involved in DNA repair and homologous recombination (Chen et al., 2008).
UVB irradiation may cause DNA damage, which, if not repaired, may result in cell arrest and hypocotyl elongation inhibition (Biever et al., 2014). Since RAD5 is involved in DNA repair, is it possible that SH1 plays a similar role in DNA repair in cucumber? This is unlikely for the following reasons. First, the hypocotyl elongation in CC3 was sensitive to LDUVB. This may be similar to photomorphogenic UVB fluence (Supplemental Fig. S1), which does not seem to cause DNA damage. From an evolutionary perspective, it is unlikely that the short hypocotyl in SWCC8 or other CSH or XIS cucumbers was due to DNA damage-induced cell arrest under the long domestication process in natural environments. Second, although HLTF is structurally similar to yeast RAD5, the DNA repair function of RAD5 has not been found in HLTF (MacKay et al., 2009). In addition, in Arabidopsis, expression of AtRAD5 showed a moderate increase under gamma-ray irradiation or mutagen bleomycin treatment; also, its expression occurred in whole seedlings and in various adult tissues (Chen et al., 2008). If SH1 performs functions similar to AtRAD5, we would expect increased expression of Sh1 under LDUVB, but we saw the opposite (Fig. 6).
ATPase-dependent chromatin remodelers can serve as spatial and temporal regulators of gene expression in growth and development by altering the accessibility of genes using energy derived from the hydrolysis of ATP to change the positioning, occupancy, and composition of nucleosomes rendering genomic regions accessible to the transcriptional machinery or transcription factors (Clapier and Cairns, 2009; Narlikar et al., 2013). In plants, accumulating evidence suggests that light-mediated gene expression and responses also require chromatin reorganization (Chua et al., 2003; Bertrand et al., 2005; Benhamed et al., 2006; Fisher and Franklin, 2011; Han et al., 2015; Bourbousse et al., 2015). For example, in Arabidopsis, the transcription factor PICKLE encodes an ATP-dependent Chromodomain Helicase-DNA Binding3 (CHD3)-type chromatin remodeling factor of the SWI/SNF family (Ogas et al., 1999; Kwon and Wagner, 2007). Jing et al. (2013) and Zhang et al. (2014a) found that PICKLE interacts with HY5 and negatively regulates its activity by repressing the trimethylation of histone H3 Lys-27 (H3K27me3) at loci involved in cell elongation during photomorphogenesis. Based on the expression pattern of relevant genes in the UVB signaling pathways, it is possible that the SH1 may use a similar mechanism as PICKLE in regulating hypocotyl elongation in cucumber.
Sh1 Regulates LDUVB-Dependent Hypocotyl Elongation via the UVR8 Signaling Pathway
It is well known that UVB irradiation inhibits hypocotyl elongation. We found that under UVB-free light, the hypocotyl length of LDUVB-sensitive CC3 at 10 d after germination was more than 6-fold longer than that under LDUVB, whereas the nonsensitive SWCC8 did not show any significant changes under both light conditions (Fig. 1, A and B). No difference in hypocotyl length was found between the two lines under dark, red, far-red, or blue lights (Fig. 1, C and D). However, at 3 d after germination, the hypocotyl length of the two lines under monochromic lights was less than half of that under the dark; the difference was even bigger under UVB-free white light, where the hypocotyl length was roughly one-eighth of that in the dark (Fig. 1D). The inhibition of hypocotyl elongation under red, far-red, or blue lights as compared with the dark condition seems to be a photomorphogenic response mediated by photoreceptors (Jiao et al., 2007). Since the UVB-free white light in the greenhouse (Supplemental Fig. S1B) covers a major part of the solar irradiance spectrum, the very short hypocotyl of CC3 under UVB-free light may be the result of the interaction of multiple photomorphogenic light signaling pathways (e.g. Morales et al., 2013), which seems to be more effective in hypocotyl elongation inhibition (Figs. 1 and 3). Thus, the hypocotyl elongation inhibition in CC3 under LDUVB was clearly not the result of UVB-free white light-induced photomorphogenic effects.
On the ninth day after germination under UVB-free light, the hypocotyl elongation rate of CC3 reached its maximum, and the hypocotyl length of CC3 was nearly five times that of SWCC8 (Fig. 2; Supplemental Fig. S8B). Consistent with this, the expression level of Sh1 showed a nearly 280-fold increase in CC3 at this time point (Fig. 6A); the expression of CsCOP1, CsHY5, and cell-elongation-related genes all had 30- to 85-fold increases (Fig. 7; Supplemental Fig. S9). In Arabidopsis, hypocotyl inhibition under the UVB treatment was generally less than 2-fold (e.g. Oravecz et al., 2006; Zhang et al., 2014a; Yin et al., 2015). While it was evident that the UVR8-mediated signaling pathway was involved in regulating hypocotyl elongation in the two lines, hypocotyl inhibition by UVB was much stronger in magnitude and time duration than what observed in Arabidopsis. Thus, the dramatic difference in LDUVB-dependent, Sh1-regulated hypocotyl elongation in cucumber could be attributed to the UVR8 photomorphogenic UVB signaling pathway that has been recruited by Sh1 during evolution with much more intensified and amplified effects on regulating hypocotyl elongation.
We investigated expression dynamics of selected genes in the UVR8 signaling pathway including CsUVR8, CSCOP1, CsHY5, CsCHS, and six cell-elongation-related genes. We found no difference in the expression level of CsUVR8 between the two NILs of Sh1 at any time point (Fig. 6), which was consistent with early findings in Arabidopsis that UVR8 is constitutively expressed (Kaiserli and Jenkins, 2007; Favory et al., 2009; Heilmann and Jenkins, 2013). Similarly, no difference in CsCOP1 expression pattern was found between NIL-LH and NIL-SH (Fig. 7) suggesting that CsCOP1 transcript abundance or CsUVR8-CsCOP1 interaction is not linked with the differential responses of hypocotyl elongation to LDUVB in CC3. On the other hand, the expression of CsHY5 in NIL-LH increased by 84-fold under LDUVB and 23-fold under UVB-free white light in the first 7 d. However, no-differential, low-level expression of CsHY5 was observed in LDUVB-insensitive NIL-SH line under both light conditions (Fig. 7, D1 and D2). This CsHY5 expression pattern was consistent with the increased expression of HY5-responsive genes (Fig. 7; Supplemental Fig. S9). Thus, the expression of CsCOP1, CsHY5, and cell-elongation-related genes were in agreement with CsUVR8-mediated photomorphogenic responses. That is, CsCOP1 and CsHY5 act antagonistically in regulating hypocotyl elongation, but the magnitude of effects in terms of expression level of relevant genes and hypocotyl length was intensified to a much higher level in the SH1-regulated pathway.
Role of SH1 in LDUVB-Dependent Hypocotyl Elongation: A Model
In Arabidopsis, it is well established that, under UVB, interaction of UVR8 with COP1 stabilizes HY5 (Favory et al., 2009; Huang et al., 2012; Cloix et al., 2012; Yin et al., 2015, 2016) and enhances the association of HY5 with target promoters, including its own (Oravecz et al., 2006; Abbas et al., 2014; Binkert et al., 2014). HY5 then activates transcription of many UVR8-regulated genes, which are associated with hypocotyl elongation inhibition, UVB protection, or acclamation (Brown et al., 2005; Oravecz et al., 2006; Huang et al., 2012; Binkert et al., 2014). How UVR8 regulates gene expression through HY5 is still unknown. Although HY5 plays critical roles in UVB signaling, it lacks a transcriptional activation domain and cannot alone activate transcription (Ang et al., 1998; Stracke et al., 2010). In addition, UVR8 does not seem to interact directly with HY5 (Heijde et al., 2013; Binkert et al., 2016). Binkert et al. (2016) suggested that there may be HY5 partner protein(s) that provide a transcriptional activation domain and enable UVB responsive gene expression. In this study, significant differences in expression for CsHY5, but not for CsCOP1 and CsUVR8, were observed between UVB-sensitive and insensitive NILs. Y2H assays revealed interactions between SH1 and CsHY5, as well as between CsCOP1 and CsHY5, but not between CsCOP1 and CsUVR8 (due to lack of UVB stress; Fig. 9). These data strongly suggest that the UVR8 signaling pathway exists in cucumber, and Sh1 plays an important role in linking CsHY5 and UVB responsive genes.
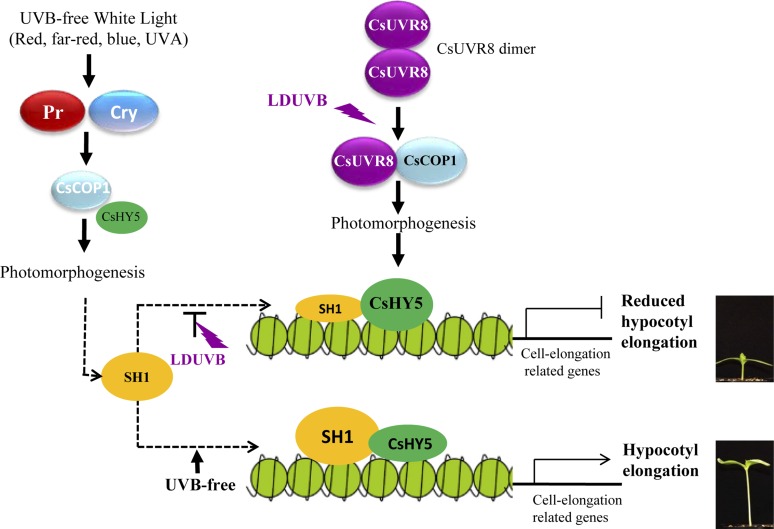
Chromatin remodelers have been shown to play important roles in plant light signaling (reviewed Barneche et al., 2014; Han et al., 2015; Bourbousse et al., 2015). For example, the CHD3 subfamily chromatin remodeling factor PICKLE represses photomorphogenesis (Jing et al., 2013). PICKLE physically interacts with key components of the light and brassinosteroid signaling pathways by repressing the trimethylation of histone H3 Lys-27 (H3K27me3) on target promoters, which promotes association of PICKLE with cell-elongation-related genes (Zhang et al., 2014a). Thus, we proposed a working model (Fig. 10) for SH1, a chromatin remodeling factor, which modulates the CsUVR8 signaling pathway by changing the chromatin states to control the accessibility of CsHY5 to the promoter of cell-elongation-related genes. Under UVB-free white light or monochromic lights, plants carrying either Sh1 or sh1 allele will undergo photomorphogenesis mediated by phytochromes or cryptochromes via the COP1/HY5 pathway. In LDUVB-sensitive plants, the Sh1 gene has high expression. The SH1 alters the accessibility of the CsHY5 to the promoters of cell-elongation-related genes to activate those genes for hypocotyl elongation. LDUVB inhibits Sh1 expression, reducing its abundance and thus SH1 protein levels, which allows increased CsHY5 expression and more binding to the promoter of cell-elongation-related genes to inactivate their expression, therefore hypocotyl elongation. In plants carrying the sh1 allele, the expression of sh1 was always low regardless of the light conditions resulting in similar outcome as Sh1 under LDUVB. In this rather simplified model, the interaction between SH1 and CsHY5 is largely quantitative depending on the transcript abundance of Sh1. Clearly, further investigations are needed to understand the exact roles of the novel player SH1 in LDUVB-dependent hypocotyl elongation.
Figure 10.
A working model on SH1-regulated hypocotyl elongation through modulating of the CsUVR8-mediated signaling pathway. Under UVB-free white light or monochromic lights, the plants will undergo photomorphogenesis mediated by phytochromes (Pr) or cryptochromes (Cry) via the CSCOP1/CsHY5 pathway. In LDUVB-sensitive plants, the Sh1 gene has high level of expression. The abundant SH1 chromatin remodeler protein alters the accessibility of CsHY5 transcription factor to activate cell-elongation-related genes for hypocotyl elongation. LDUVB inhibits Sh1 expression, reduces its abundance, and thus SH1 protein level, which allows increased CsHY5 expression and more binding to the promoter of cell-elongation-related genes to inactivate their expression, therefore hypocotyl elongation. In UVB-insensitive plants, the expression of sh1 is always low regardless of the light conditions resulting in a similar outcome as Sh1 under LDUVB.
CONCLUSION
1. The short hypocotyl in XIS cucumber is controlled by a simply inherited recessive allele, sh1. Cucumber lines carrying homozygous sh1 allele were LDUVB insensitive, and those carrying the dominant allele Sh1 are LDUVB sensitive.
2. Sh1 encoded a human SMARCA3-like chromatin remodeling factor. The SH1 protein sequence was highly conserved among eukaryotic organisms. Regulation of hypocotyl elongation in cucumber was a novel function of this family of proteins.
3. The sh1 allele was originated from wild cucumber for local adaptation, and was under selection during domestication. The cultivated cucumber carries predominantly the Sh1 allele; the sh1 allele is nearly fixed in the semiwild Xishuangbanna cucumber, and the wild cucumber population is largely at Hardy-Weinberg equilibrium for the alleles.
4. LDUVB inhibits Sh1 expression; Sh1 transcript abundance was highly correlated with hypocotyl elongation rate and the expression level of cell-elongation-related genes.
5. SH1 regulates LDUVB-dependent hypocotyl elongation in cucumber by modulating the UVR8 signaling pathway. SH1 interacts with CsHY5 during this process. SH1 seems to be able to change the chromatin states thus the accessibility of CsHY5 to promoters of LDUVB-responsive genes for hypocotyl elongation.
6. More work is needed to investigate the details of SH1-CsHY5 interaction to elucidate its roles in regulating hypocotyl elongation.
MATERIALS AND METHODS
Plant Materials
Three segregating populations were employed for genetic mapping and cloning of the sh1 locus including 124 RILs (Bo et al., 2011, 2015), 1183 F2 individuals from SWCC8 × CC3, and 164 F3 families derived from WI7200 × WI7167 (Qu et al., 2014). CC3, SWCC8, and derived RILs were originally developed by Nanjing Agricultural University, China. Both CC3 (aka, Beijingjietou, north China fresh market type) and WI7200 (derived from PI 249561; Li et al., 2011) were cultivated cucumbers. SWCC8 and WI7167 are semiwild XIS cucumbers with short hypocotyls conferred by the sh1 allele. To reduce genetic background noises, two NILs, NIL-LH (long hypocotyl, Sh1Sh1) and NIL-SH (short hypocotyl, sh1sh1) were developed with marker-assisted backcrossing of the progeny of WI7167 × WI7200 with WI7200 as the recurrent parent. The two NILs had similar flowering times and growth vigor, and the same responses to LDUVB as the respective parent and were used in various investigations on gene expression and effects of light quality and temperature on hypocotyl elongation in this study.
To study allelic diversity of the sh1 locus in natural cucumber populations and the evolution of hypocotyl length during domestication, 502 cucumber lines from various sources were employed, including 419 CSS, 48 CSH, and 35 XIS accessions. Among them, 99 with whole-genome resequencing data were described in Qi et al. (2013). Moreover, nine Cucumis hystrix lines were also employed for the study of the sh1 allele evolution. The names, origin, seed, or DNA source, and taxonomic status of all 511 entries are listed in Supplemental Table S1.
Phenotypic Characterization of Low-Dosage UVB Sensitivity of Cucumber Lines
Define the LDUVB Environment
Phenotypic data on the hypocotyl length of various materials was collected from three environments, including the University of Wisconsin-Madison Walnut Street Greenhouses (with glass roof), the Biotron facility (climate control room), and growth chambers with fluorescent lamps (model GSE-41L2; Geneva Scientific). Photometric and colorimetric measurements of different light sources were performed with a spectrometer (model USB2000+) and associated SpectraSuite software from Ocean Optics. The spectra of different light sources are presented in Supplemental Fig. S1, A to D. It is easy to see that the UVB part of various light sources could be efficiently filtered out with glass. Thus, the greenhouses and growth chamber with glass covers between the plants and the fluorescent lamps (UVB-free growth chamber) used in this study were considered UVB-free, and the growth chamber with uncovered fluorescent lamps (LDUVB growth chamber) was used for the LDUVB treatment.
Phenotyping Hypocotyl Length in Segregating Populations
Measurements of the hypocotyl length of the plants in all biparental segregating or natural populations were conducted at the seedling stage (10–15 d after germination) in the UVB-free greenhouses. Conditions in the UVB greenhouses were 22°C to 30°C day and 20°C to 25°C night with a 14 h light/10 h dark photoperiod. For each RIL or F3 family, data from at least 15 seedlings was collected, and the RIL or family means were used in linkage analysis or QTL mapping with molecular markers.
Effects of Light Quality, UV Fluence, and Temperature on Hypocotyl Elongation
Comparative analysis of hypocotyl elongation was conducted in both LDUVB and UVB-free conditions. The effect of light quality on hypocotyl elongation was investigated in the Biotron with blue, red, and far-red LED light sources with a peak wavelength of 440, 620, and 740 nm, respectively (Supplemental Fig. S1E). The photon flux density was adjusted to approximately 10 µmol with a photosynthetically active radiation quantum light meter. Seedlings were grown under 28°C day and 17°C night with a 14 h photoperiod in the growth chambers or in the Biotron. A parallel dark treatment was also applied with these experiments. Hypocotyl length was measured at 3 d after germination for a total of 10 d.
To evaluate the effects of light intensity of UV light on hypocotyl elongation, SWCC8 and CC3 seedlings (five seedlings each) were exposed to UV light from UV bulbs (model GEG30T8) at three intensity levels: 0 (control), low (∼0.15 Jm−2 min−1), and high (∼3.0 Jm−2 min−1). UV irradiation treatment started at the third day after germination with 5 hour’s exposure per day. Hypocotyl length was measured at 7 d after germination. The effect of temperature on hypocotyl elongation was evaluated in a growth chamber in UVB-free (with glass plate cover) and LDUVB (no glass plate cover) conditions at three temperature settings: 17°C, 27°C, and 36°C. Hypocotyl length of NIL-SH and NIL-LH (five seedlings per entry) was measured from 10-d-old-seedlings.
Microscopic measurement of hypocotyl cell length was performed under a dissecting microscope with 10-d-old seedlings of SWCC8 and CC3.
Fine Genetic Mapping of Sh1 Locus
Bulked segregation analysis was used for the quick identification of markers linked with the Sh1 locus. Two DNA bulks, the S-bulk and L-bulk, were constructed each consisting of 10 short and 10 long hypocotyl RIL plants of the CC3 × SWCC8 cross, respectively (one plant per RIL). A total of 480 microsatellite or SSR markers evenly distributed in seven cucumber chromosomes (Yang et al., 2012, 2013, 2014) were selected to screen for polymorphisms between the two bulks. After initial placement of Sh1 in chromosome 3, a scaffold-based chromosome walking strategy (Li et al., 2011) was taken to identify more closely linked markers. At the fine mapping stage, the genomes of all parental lines were resequenced with Illumina Hi-SEquation 2000 at >15× coverage each (100 bp paired end) at the University of Wisconsin Biotech Center. Bioinformatic analysis of resequencing data for marker discovery and SNP genotyping all followed Pan et al. (2015) and Li et al. (2016). Very closely linked or cosegregating markers in the RIL population were further applied to 1183 F2 plants. For each recombinant F2 plant defined in an interval with two flanking markers, at least 30 F2-derived F3 plants in each family were scored to infer the genotype at the Sh1 locus in the F2 plants.
DNA extraction, PCR amplification, marker analysis, and gel electrophoresis followed Li et al. (2011). Linkage analysis of the sh1 locus with molecular markers was performed with the Kosambi mapping function using JoinMap 4.0 with the threshold LOD score of 4.0. In our previous study, we developed a linkage map with 225 SSR loci using the WI7200 × WI7167 F2:3 population (Qu et al., 2014). This population was also segregating for hypocotyl length and was used in this study to confirm results from the CC3 × SWCC8 population. However, we took a QTL mapping approach in this population. QTL analysis was conducted using the Multiple QTL (MQM) model in R/qtl following Weng et al. (2015).
DNA Annotation, Gene Predication, and Full-Length cDNA Cloning
Fine genetic mapping delimited the sh1 locus in a 42 kb region. Gene prediction in the genomic DNA region was performed with the computer program FGENESH (http://sunl.softberry.com/). Gene annotation was based on BLASTx at NCBI (http://blast.ncbi.nlm.nih.gov/) and checked manually. To validate the intron-exon structure predicted by the computer software, full-length cDNA was cloned for the candidate gene. Information about all of the primers used in this study is presented in Supplemental Table S3.
Gene Expression Analysis
We investigated the time-course expression of the Sh1 candidate gene with real-time quantitative PCR (qPCR). The hypocotyl samples from NIL-SH and NIL-LH were collected at seven different time points (third, fifth, seventh, ninth, eleventh, thirteenth, and fifteenth day after germination) growing under UVB-free and LDUVB conditions. Total RNA extraction, qPCR procedure followed Li et al. (2016) with the cucumber ubiquitin extension protein gene as the reference (Wan et al., 2010). Each sample was run with three biological and technical replicates and significance tests among replications were performed with t tests.
To further explore the molecular mechanism by which SH1 regulates hypocotyl elongation, we examined the expression dynamics of selected cucumber homologous genes that may play important roles in the UVR8 signaling pathways including CsUVR8, CsCOP1, CsHY5, and CsCSH. The CsCHS gene encodes chalcone synthase, which catalyzes the first committed step in the flavonoid biosynthesis pathway, and its expression is often used as a marker in UVB irradiation-induced photomorphogenic protection (e.g. Fuglevand et al., 1996). We conducted qPCR on six cell-elongation-related genes, which are involved in encoding cell wall loosening or hydrolytic enzymes or encoding components in the biosynthesis or signaling of brassinosteroids or auxin (Jing et al., 2013). Homologous genomic DNA sequences of the following Arabidopsis (Arabidopsis thaliana) genes were extracted from the Gy14 cucumber draft gene assembly: EXTENSIN3 (EXT3, GenBank accession no. At1g21310), EXPANSIN2 (EXP2, At5g05290), DWARF4 (DWF4, At3g50660), INDOLE-3-ACETIC ACID INDUCIBLE19 (IAA19, At3g15540), XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE17 (XTH17, At1g65310), and XYLOGLUCAN ENDOTRANSGLYCOSYLASE6 (XTR6, At4g25810; Jing et al., 2013). These cucumber homologs were assigned gene symbols CsEXT3, CsEXP2, CsDWF4, CsXTH17, CsXTR6, and CsIAA19, respectively. Sequence information from all primers used in qPCR in this study is presented in Supplemental Table S3.
Phylogenetic Analysis of SH1 Protein Sequences
We showed that the candidate gene of Sh1 was a homolog of the human SMARCA3 chromatin remodeling factor, which belongs to the HLTF family and Rad5/16 subfamily. We investigated the phylogenetic relationships of cucumber SH1 protein and other Rad5/16 subfamily protein members. HLTF/SMARCA3 or RAD5/RAD16 protein sequences of eight fungal, six animal, three protist, and nine plant species were downloaded from the SNF2 family protein database (http://www.snf2.net/snf2/subfamilies/rad516.shtml). The eight fungal species included Cryptococcus neoformans (yeast, accession no. P0CQ67.1), Magnaporthe grisea (rice blast fungus, accession no. XP_778272.1), Ashbya gossypii (mold, accession no. Q753V5.2), Candida glabrata (haploid yeast, accession no. Q6FY76.1), Kluyveromyces lactis (yeast, accession no. Q6CJM4.1), Schizosaccharomyces pombe (fission yeast, accession no. CAA21065.1), Saccharomyces cerevisiae (baker’s yeast, accession no. P31244.1), and Yarrowia lipolytica (yeast, accession no. XP_504855.1). The six animals were Canis familiaris (dog, accession no. 477106), Homo sapiens (human, accession no. 6596), Oryctolagus cuniculus (European rabbit, accession no. 100009232), Monodelphis domestica (short-tailed opossum, accession no. 100617443), Mus musculus (house mouse, accession no. Q6PCN7.1), and Danio rerio (zebrafish, accession no. 564651). The three protists included Trypanosoma brucei (accession no. EAN79056.1), Trypanosoma cruzi (accession no. XP_821558.1), and Leishmania major (accession no. CAJ04924.1).
The sequences with predicted SMARCA3 or RAD5 proteins were downloaded from NCBI database for the following nine species: Cucumis melo (melon, accession no. XP_008438555.1), C. hystrix (the senior author’s lab), Arabidopsis (Q9FNI6.1), Capsella rubella (the pink shepherd’s-purse, XP_006279560.1), Citrus sinensis (orange, XP_006484966.1), Populus trichocarpa (black cottonwood, XP_006385564.1), Ricinus communis (castorbean, XP_002529311.1), Solanum lycopersicum (tomato, XP_004234234.1), and Theobroma cacao (cacao, EOY33587.1). These sequences were used to build a neighbor-joining tree (Saitou and Nei, 1987) that was constructed by the MEGA 5.0 software (http://www.megasoftware.net/) with 1000 bootstrap replications.
Allelic Diversity of Sh1 Locus in Cucumber Natural Populations
The LDUVB insensitivity in short hypocotyl cucumber lines (SWCC8 and WI7167) was due to an SNP within the candidate gene. We examined allelic diversities at the Sh1 locus among 511 Cucumis accessions, of which 48, 35, 419, and nine belonged to the CSH, XIS, CSS, and C. hystrix, respectively (Supplemental Table S1). The genotypes of 120 Cucumis sativus lines at this SNP site were inferred from resequenced data; 367 were genotyped with a dCAPS marker (dCAPS-51) derived from the causal SNP within the Sh1 locus. For the remaining 24 C. hystrix or XIS accessions (with XIS or HYS in accession names, Supplemental Table S1), SNP genotyping was performed using the KBiosciences Competitive Allele-Specific PCR SNP genotyping system (KASPar).
To validate the association between the allelic diversity at the Sh1 SNP locus with hypocotyl elongation, we measured hypocotyl length under both UVB-free (greenhouse) and LDUVB (University of Wisconsin Biotron facility) conditions for 211 of 502 cucumber lines.
Whole-genome resequencing data were available for 120 of the 502 lines; 99 were downloaded from the NCBI database (Qi et al., 2013), and 21 were resequenced with the Illumina Hi-SEquation 2000 platform by the senior author’s lab. In order to further understand the origin and evolution of the LDUVB-insensitive sh1 allele, we explored the nucleotide variations in the Sh1 region in 120 resequenced cucumber lines including 12 CSH, 20 XIS, and 88 CSS lines (listed in Supplemental Table S1). Illumina sequence reads of these 120 lines were aligned with GS Reference Mapper (V2.8) software using Gy14 as the reference. We selected 193 SNPs in the upstream and downstream of sh1 SNP position, respectively, and used them to construct a phylogenetic tree. Multiple sequence alignment was performed with the Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/).
We further performed principal component analysis (Mardia et al., 1979) for the 120 resequenced cucumber lines with 67 of 193 SNPs in the 15 kb region whose minor allele frequency was > 0.05 and missing data were <5%.
Y2H Assay
Seedlings of the cucumber inbred line 9930 were grown in a UVB-free environment. Leaf and hypocotyl samples were collected for total RNA with the BioFast BIOZOL Total RNA Extraction Reagent (Bioer). The RNA qualities and concentrations were determined with agarose gel electrophoresis and NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). First strand cDNA was synthesized using AffinityScript Multiple Temperature cDNA Synthesis Kit (Agilent). The cDNA sequences for four cucumber genes, Sh1, CsHY5, CsCOP1, and CsUVR8 were amplified and cloned. The sequences of primer pairs for each gene are presented in Supplemental Table S3. Full-length cDNAs of CsHY5, CsUVR8, and CsCOP1 were inserted into vector pGADT7 (AD) as prey, and the coding sequence of SH1 and CsCOP1 were fused to vector pGBKT7 (BD) as bait using One Step Seamless Cloning kit (Novoprotein). The insert sequence in each vector was verified by PCR amplification and sequencing. Full-length cDNA sequences of these genes are shown in Supplemental File S2.
The Matchmaker Gold Yeast Two-Hybrid System (Clontech) was used to perform the Y2H assay. The following bait-prey pairs were used in cotransformation into BD-SH1+AD-CsHY5, BD-SH1+AD-CsCOP1, BD-CsCOP1+AD-CsHY5, and BD-CsCOP1+AD-CsUVR8. Setting up positive and negative controls and testing bait for autoactivation all followed manufacturer’s instructions. Three days after transformation, colonies growing on DDO double dropout synthetically defined (SD/-Trp/-Leu) growth media were transferred to higher stringency QDO agar plates (SD/-His/-Ade/-Trp/-Leu) to continue to grow at 30°C for 3 to 5 d for observation of the results. For each bait-prey pair, at least three independent experiments were performed.
Accession Numbers
The cucumber SH1 genomic DNA sequence has been deposited to GenBank under the accession number KX639507.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Wavelength spectra of different light sources that define UVB-free and LDUVB experimental conditions in this study.
Supplemental Figure S2. Hypocotyl elongation of SWCC8 and CC3 under LDUVB, high dosage UVB, and UVB-free conditions.
Supplemental Figure S3. Effect of temperature on hypocotyl elongation in near isogenic lines of Sh1 gene under LDUVB and UVB-free conditions.
Supplemental Figure S4. QTL analysis in the WI7167 × WI7200 F2:3 population identifies a major-effect QTL for hypocotyl length that colocalizes with the Sh1 locus.
Supplemental Figure S5. QTL analysis in SWCC8 × CC3 RIL population suggests that the slow vine growth in seedling stage in SWCC8 is a pleiotropic effect of the sh1 locus.
Supplemental Figure S6. Expression patterns of gene 1, gene 3, and gene 4 in NILs of Sh1 under UVB-free and LDUVB light conditions exclude the three genes as the candidate of the Sh1 locus.
Supplemental Figure S7. Association of LDUVB-dependent hypocotyl elongation with the Sh1 locus in cucumber natural populations.
Supplemental Figure S8. Hypocotyl elongation dynamics of SWCC8 and CC3 under LDUVB and UVB-free conditions within 15 d after germination.
Supplemental Figure S9. Expression dynamics of five cell-elongation-related genes in two NILs of the Sh1 gene under UVB-free and LDUVB light conditions in 15 d after germination.
Supplemental Figure S10. Worldwide geographic distribution of the Sh1 and sh1 alleles among natural populations of the CSS, CSH, and XIS cucumbers.
Supplemental Table S1. Cucumber materials used in this study for genome-wide association and phylogenetic analysis at Sh1 locus.
Supplemental Table S2. 193 SNPs in a 15-kb region of the Sh1 locus among 120 accessions of CSS, CSH, and XIS cucumbers used for phylogenetic analysis.
Supplemental Table S3. Information about the primers used in genetic mapping, gene expression, and Y2H studies.
Supplemental File 1. Genomic DNA sequences of Sh1 candidate gene of SWCC8 and CC3. Introns are highlighted in yellow.
Supplemental File 2. Complementary DNA (cDNA) sequences of cucumber genes SH1, CsHY5, CsCOP1, and CsUVR8 used in yeast two-hybrid assay.
Supplementary Material
Acknowledgments
We thank Kristin Haider for technical help and English editing of the manuscript. Names are necessary to report factually on available data; however, the United States Department of Agriculture (USDA) neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval of the product to the exclusion of others that may also be suitable. USDA is an equal opportunity provider and employer.
Glossary
- LDUVB
low-dosage UVB
- HLTF
helicase-like transcription factor
- RIL
recombinant inbred line
Footnotes
This work was supported by a United States Department of Agriculture Specialty Crop Research Initiative grant (SCRI, project no. 2011-51181-30661) to Y.We. The work in Y.L.’s lab was supported by the National Natural Science Foundation of China (31171955 and 31471891). J.C.'s work was supported by the National Natural Science Foundation of China (31430075).
Articles can be viewed without a subscription.
References
- Abbas N, Maurya JP, Senapati D, Gangappa SN, Chattopadhyay S (2014) Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 26: 1036–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D, Gallego-Bartolomé J, Orlando L, García-Cárcel L, Rubio V, Martínez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, et al. (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53: 324–335 [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Barneche F, Malapeira J, Mas P (2014) The impact of chromatin dynamics on plant light responses and circadian clock function. J Exp Bot 65: 2895–2913 [DOI] [PubMed] [Google Scholar]
- Benhamed M, Bertrand C, Servet C, Zhou DX (2006) Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18: 2893–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand C, Benhamed M, Li YF, Ayadi M, Lemonnier G, Renou J-P, Delarue M, Zhou DX (2005) Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1, is required to integrate light signals to regulate gene expression and growth. J Biol Chem 280: 1465–1473 [DOI] [PubMed] [Google Scholar]
- Bhatia S, Gangappa SN, Kushwaha R, Kundu S, Chattopadhyay S (2008) SHORT HYPOCOTYL IN WHITE LIGHT1, a serine-arginine-aspartate-rich protein in Arabidopsis, acts as a negative regulator of photomorphogenic growth. Plant Physiol 147: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biever JJ, Brinkman D, Gardner G (2014) UV-B inhibition of hypocotyl growth in etiolated Arabidopsis thaliana seedlings is a consequence of cell cycle arrest initiated by photodimer accumulation. J Exp Bot 65: 2949–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkert M, Crocco CD, Ekundayo B, Lau K, Raffelberg S, Tilbrook K, Yin R, Chappuis R, Schalch T, Ulm R (2016) Revisiting chromatin binding of the Arabidopsis UV-B photoreceptor UVR8. BMC Plant Biol 16: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkert M, Kozma-Bognár L, Terecskei K, De Veylder L, Nagy F, Ulm R (2014) UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26: 4200–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo K, Ma Z, Chen J, Weng Y (2015) Molecular mapping reveals structural rearrangements and quantitative trait loci underlying traits with local adaptation in semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis Qi et Yuan). Theor Appl Genet 128: 25–39 [DOI] [PubMed] [Google Scholar]
- Bo KL, Shen J, Qian CT, Song H, Chen JF (2011) Genetic analysis of the important agronomic traits on Beijingjietou × Xishuangbanna cucumber recombinant inbred lines. J Nanjing Agri Univ 34: 20–24 [Google Scholar]
- Boron AK, Vissenberg K (2014) The Arabidopsis thaliana hypocotyl, a model to identify and study control mechanisms of cellular expansion. Plant Cell Rep 33: 697–706 [DOI] [PubMed] [Google Scholar]
- Bourbousse C, Mestiri I, Zabulon G, Bourge M, Formiggini F, Koini MA, Brown SC, Fransz P, Bowler C, Barneche F (2015) Light signaling controls nuclear architecture reorganization during seedling establishment. Proc Natl Acad Sci USA 112: E2836–E2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosché M, Schuler MA, Kalbina I, Connor L, Strid A (2002) Gene regulation by low level UV-B radiation: identification by DNA array analysis. Photochem Photobiol Sci 1: 656–664 [DOI] [PubMed] [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102: 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candolle AD. (1959) Origin of Cultivated Plants. Hafner Publishing, New York [Google Scholar]
- Catalá R, Medina J, Salinas J (2011) Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci USA 108: 16475–16480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Sharma P, Khurana JP (2006) Cryptochrome 1 from Brassica napus is up-regulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation. Plant Physiol 141: 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I-P, Mannuss A, Orel N, Heitzeberg F, Puchta H (2008) A homolog of ScRAD5 is involved in DNA repair and homologous recombination in Arabidopsis. Plant Physiol 146: 1786–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, et al. (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Jenkins GI (1996) Distinct UV-B and UV-A/blue light signal transduction pathways induce chalcone synthase gene expression in Arabidopsis cells. Plant Cell 8: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua YL, Watson LA, Gray JC (2003) The transcriptional enhancer of the pea plastocyanin gene associates with the nuclear matrix and regulates gene expression through histone acetylation. Plant Cell 15: 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Cloix C, Kaiserli E, Heilmann M, Baxter KJ, Brown BA, O’Hara A, Smith BO, Christie JM, Jenkins GI (2012) C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc Natl Acad Sci USA 109: 16366–16370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS (2004) The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J 38: 332–347 [DOI] [PubMed] [Google Scholar]
- Debauve G, Capouillez A, Belayew A, Saussez S (2008) The helicase-like transcription factor and its implication in cancer progression. Cell Mol Life Sci 65: 591–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, Sonntag L, James GV, Janitza P, Ibañez C, Ziermann H, Peterson T, Denk K, Mull S, Ziegler J, et al. (2014) The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Reports 9: 1983–1989 [DOI] [PubMed] [Google Scholar]
- Duthie JF. (1903) Flora of the Upper Gangetic Plain, and of the Adjacent Siwalik and Sub-Himalayan Tracts. Superintendent of Government Printing, Calcutta, India [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filella I, Penuelas J (1999) Altitudinal differences in UV absorbance, UV reflectance and related morphological traits of Quercus ilex and Rhododendron ferrugineum in the Mediterranean region. Plant Ecol 145: 157–165 [Google Scholar]
- Fisher AJ, Franklin KA (2011) Chromatin remodelling in plant light signalling. Physiol Plant 142: 305–313 [DOI] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T (2006) Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34: 2887–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T (2006) DNA Repair and Mutagenesis. ASM Press, Washington, DC [Google Scholar]
- Frohnmeyer H, Staiger D (2003) Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol 133: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglevand G, Jackson JA, Jenkins GI (1996) UV-B, UV-A, and blue light signal transduction pathways interact synergistically to regulate chalcone synthase gene expression in Arabidopsis. Plant Cell 8: 2347–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC, Fankhauser C (2015) Sensing the light environment in plants: photoreceptors and early signaling steps. Curr Opin Neurobiol 34: 46–53 [DOI] [PubMed] [Google Scholar]
- Gardner G, Lin C, Tobin EM, Loehrer H, Brinkman D (2009) Photobiological properties of the inhibition of etiolated Arabidopsis seedling growth by ultraviolet-B irradiation. Plant Cell Environ 32: 1573–1583 [DOI] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giliberto L, Perrotta G, Pallara P, Weller JL, Fraser PD, Bramley PM, Fiore A, Tavazza M, Giuliano G (2005) Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol 137: 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S-K, Wu M-F, Cui S, Wagner D (2015) Roles and activities of chromatin remodeling ATPases in plants. Plant J 83: 62–77 [DOI] [PubMed] [Google Scholar]
- Heijde M, Binkert M, Yin R, Ares-Orpel F, Rizzini L, Van De Slijke E, Persiau G, Nolf J, Gevaert K, De Jaeger G, et al. (2013) Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc Natl Acad Sci USA 110: 20326–20331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M, Ulm R (2012) UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci 17: 230–237 [DOI] [PubMed] [Google Scholar]
- Heilmann M, Jenkins GI (2013) Rapid reversion from monomer to dimer regenerates the ultraviolet-B photoreceptor UV RESISTANCE LOCUS8 in intact Arabidopsis plants. Plant Physiol 161: 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández R, Kubota C (2016) Physiological responses of cucumber seedlings under different blue and red photon flux ratios using LEDs. Environ Exp Bot 121: 66–74 [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes MG, Keiller DR (2002) Effects of pubescence and waxes on the reflectance of leaves in the ultraviolet and photosynthetic wavebands: a comparison of a range of species. Plant Cell Environ 25: 85–93 [Google Scholar]
- Huang X, Ouyang X, Yang P, Lau OS, Li G, Li J, Chen H, Deng XW (2012) Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24: 4590–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI. (2014) The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26: 21–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jing Y, Zhang D, Wang X, Tang W, Wang W, Huai J, Xu G, Chen D, Li Y, Lin R (2013) Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson H, Jones HJ, Foreman J, Hemsted JR, Stewart K, Grima R, Halliday KJ (2014) Arabidopsis cell expansion is controlled by a photothermal switch. Nat Commun 5: 4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiserli E, Jenkins GI (2007) UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Tennessen DJ, Last RL (1998) UV-B-induced photomorphogenesis in Arabidopsis thaliana. Plant J 15: 667–674 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lim JE, Landry LG, Last RL (2002) Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol 130: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP (1980) Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana L. heynh. Z Pflanzenphysiol 100: 147–160 [Google Scholar]
- Kwon CS, Wagner D (2007) Unwinding chromatin for development and growth: a few genes at a time. Trends Genet 23: 403–412 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee JH, Zhang H, Shen Y, et al. (2010) Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Pan Y, Wen C, Li Y, Liu X, Zhang X, Behera TK, Xing G, Weng Y (2016) Integrated analysis in bi-parental and natural populations reveals CsCLAVATA3 (CsCLV3) underlying carpel number variations in cucumber. Theor Appl Genet 129: 1007–1022 [DOI] [PubMed] [Google Scholar]
- Li Y, Yang L, Pathak M, Li D, He X, Weng Y (2011) Fine genetic mapping of cp: a recessive gene for compact (dwarf) plant architecture in cucumber, Cucumis sativus L. Theor Appl Genet 123: 973–983 [DOI] [PubMed] [Google Scholar]
- Liu H, Liu B, Zhao C, Pepper M, Lin C (2011) The action mechanisms of plant cryptochromes. Trends Plant Sci 16: 684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Roof S, Ye Z, Barry C, van Tuinen A, Vrebalov J, Bowler C, Giovannoni J (2004) Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc Natl Acad Sci USA 101: 9897–9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay C, Toth R, Rouse J (2009) Biochemical characterisation of the SWI/SNF family member HLTF. Biochem Biophys Res Commun 390: 187–191 [DOI] [PubMed] [Google Scholar]
- Mardia KV, Kent JT, Bibby JM (1979) Multivariate Analysis. Academic Press, London [Google Scholar]
- Morales LO, Brosché M, Vainonen J, Jenkins GI, Wargent JJ, Sipari N, Strid Å, Lindfors AV, Tegelberg R, Aphalo PJ (2013) Multiple roles for UV RESISTANCE LOCUS8 in regulating gene expression and metabolite accumulation in Arabidopsis under solar ultraviolet radiation. Plant Physiol 161: 744–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Fenzi F, Ciliento R, Alfano F, Bowler C (1999) Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Sundaramoorthy R, Owen-Hughes T (2013) Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154: 490–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J, Chory J (2002) Photomorphogenesis. In Meyerowitz E, Somerville C, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–12 [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adám E, Schäfer E, Nagy F, Ulm R (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Bo K, Cheng Z, Weng Y (2015) The loss-of-function GLABROUS 3 mutation in cucumber is due to LTR-retrotransposon insertion in a class IV HD-ZIP transcription factor gene CsGL3 that is epistatic over CsGL1. BMC Plant Biol 15: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi CZ. (1983) A new type of cucumber, Cucumis sativus L. var. xishuangbannanesis Qi et Yuan. Acta Hort Sinica 10: 259–263 (in Chinese) [Google Scholar]
- Qi J, Liu X, Shen D, Miao H, Xie B, Li X, Zeng P, Wang S, Shang Y, Gu X, et al. (2013) A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat Genet 45: 1510–1515 [DOI] [PubMed] [Google Scholar]
- Qu SP, Pan YP, Weng Y (2014) QTL Mapping of flowering time and fruit shape in Xishuangbana cucumber (Cucumis sativus L. var. xishuangbannanesis Qi et Yuan). Proc Cucurbitaceae 2014: 54–56 [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Robson TM, Klem K, Urban O, Jansen MAK (2015) Re-interpreting plant morphological responses to UV-B radiation. Plant Cell Environ 38: 856–866 [DOI] [PubMed] [Google Scholar]
- Royle JF. (1835) Illustrations of the Botany of the Himalayan Mountains. Wm. H. Alland & Co, London, England. [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P (1994) Embryonic origin of the Arabidopsis root and root meristem initials. Development 120: 2475–2487 [Google Scholar]
- Sebastian P, Schaefer H, Telford IRH, Renner SS (2010) Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc Natl Acad Sci USA 107: 14269–14273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Shinkle JR, Atkins AK, Humphrey EE, Rodgers CW, Wheeler SL, Barnes PW (2004) Growth and morphological responses to different UV wavebands in cucumber (Cucumis sativum) and other dicotyledonous seedlings. Physiol Plant 120: 240–248 [DOI] [PubMed] [Google Scholar]
- Srivastava AK, Senapati D, Srivastava A, Chakraborty M, Gangappa SN, Chattopadhyay S (2015) Short Hypocotyl in White Light1 interacts with Elongated Hypocotyl5 (HY5) and Constitutive Photomorphogenic1 (COP1) and promotes COP1-mediated degradation of HY5 during Arabidopsis seedling development. Plant Physiol 169: 2922–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R (2010) The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ 33: 88–103 [DOI] [PubMed] [Google Scholar]
- Styler RC, Koranski DS (1997) Plug and Transplant Production: A Grower’s Guide. Ball Publishing, Batavia, IL [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook K, Dubois M, Crocco CD, Yin R, Chappuis R, Allorent G, Schmid-Siegert E, Goldschmidt-Clermont M, Ulm R (2016) UV-B perception and acclimation in Chlamydomonas reinhardtii. Plant Cell 28: 966–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Máté Z, Adám E, Oakeley EJ, Schäfer E, Nagy F (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Jenkins GI (2015) Q&A: How do plants sense and respond to UV-B radiation? BMC Biol 13: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Habricot Y, Condiff AS, Maldiney R, Van der Straeten D, Ahmad M (2007) HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J 49: 428–441 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Verbelen J-P, Van Der Straeten D (2005) Of light and length: regulation of hypocotyl growth in Arabidopsis. BioEssays 27: 275–284 [DOI] [PubMed] [Google Scholar]
- Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399: 257–261 [DOI] [PubMed] [Google Scholar]
- Weng Y, Colle M, Wang Y, Yang L, Rubinstein M, Sherman A, Ophir R, Grumet R (2015) QTL mapping in multiple populations and development stages reveals dynamic quantitative trait loci for fruit size in cucumbers of different market classes. Theor Appl Genet 128: 1747–1763 [DOI] [PubMed] [Google Scholar]
- Yang L, Li D, Li Y, Gu X, Huang S, Garcia-Mas J, Weng Y (2013) A 1,681-locus consensus genetic map of cultivated cucumber including 67 NB-LRR resistance gene homolog and ten gene loci. BMC Plant Biol 13: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LM, Koo D-H, Li DW, Zhang T, Jiang JM, Luan FS, Renner SS, Hénaff E, Sanseverino W, Garcia-Mas J, et al. (2014) Next-generation sequencing, FISH mapping, and synteny-based modeling reveal mechanisms of decreasing dysploidy in Cucumis. Plant J 77: 16–30 [DOI] [PubMed] [Google Scholar]
- Yang LM, Koo DH, Li Y, Zhang X, Luan F, Havey MJ, Jiang J, Weng Y (2012) Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J 71: 895–906 [DOI] [PubMed] [Google Scholar]
- Yin R, Arongaus AB, Binkert M, Ulm R (2015) Two distinct domains of the UVR8 photoreceptor interact with COP1 to initiate UV-B signaling in Arabidopsis. Plant Cell 27: 202–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Skvortsova MY, Loubéry S, Ulm R (2016) COP1 is required for UV-B-induced nuclear accumulation of the UVR8 photoreceptor. Proc Natl Acad Sci USA 113: E4415–E4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Jing Y, Jiang Z, Lin R (2014a) The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 26: 2472–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65: 346–358 [DOI] [PubMed] [Google Scholar]
- Zhang LX, Allen LH Jr, Vaughan MM, Hauser BA, Boote KJ (2014b) Solar ultraviolet radiation exclusion increases soybean internode lengths and plant height. Agric For Meteorol 184: 170–178 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.