The 9-1-1 complex member RAD1 directly interacts with HUS1 and RAD9 and facilitates accurate meiotic recombination by suppressing nonhomologous end joining in rice.
Abstract
During meiosis, programmed double-strand breaks (DSBs) are generated to initiate homologous recombination, which is crucial for faithful chromosome segregation. In yeast, Radiation sensitive1 (RAD1) acts together with Radiation sensitive9 (RAD9) and Hydroxyurea sensitive1 (HUS1) to facilitate meiotic recombination via cell-cycle checkpoint control. However, little is known about the meiotic functions of these proteins in higher eukaryotes. Here, we characterized a RAD1 homolog in rice (Oryza sativa) and obtained evidence that O. sativa RAD1 (OsRAD1) is important for meiotic DSB repair. Loss of OsRAD1 led to abnormal chromosome association and fragmentation upon completion of homologous pairing and synapsis. These aberrant chromosome associations were independent of OsDMC1. We found that classical nonhomologous end-joining mediated by Ku70 accounted for most of the ectopic associations in Osrad1. In addition, OsRAD1 interacts directly with OsHUS1 and OsRAD9, suggesting that these proteins act as a complex to promote DSB repair during rice meiosis. Together, these findings suggest that the 9-1-1 complex facilitates accurate meiotic recombination by suppressing nonhomologous end-joining during meiosis in rice.
Meiosis comprises two successive cell divisions after a single S phase, generating four haploid products. To ensure proper chromosome segregation at the first meiotic division, crossovers (COs) are formed between homologous chromosomes (Kleckner, 2006). CO formation requires faithful repair of programmed DNA double-strand breaks (DSBs) introduced by the protein SPO11 (Keeney et al., 1997; Shinohara et al., 1997).
Mitotic cells employ two basic strategies for DSB repair: homologous recombination (HR) and classical nonhomologous end-joining (C-NHEJ; Deriano and Roth, 2013). HR requires an undamaged template sequence for repair, while the C-NHEJ pathway involves direct ligation of the broken ends in a Ku-dependent manner. Both HR and C-NHEJ safeguard genome integrity during mitosis (Ceccaldi et al., 2016; Symington and Gautier, 2011). However, DSBs are preferentially repaired by HR during meiosis, because only this pathway generates COs. C-NHEJ competes with HR and creates de novo mutations in the gametes, indicating that this activity should be restricted during meiotic DSB repair. Previous studies have identified several factors essential for preventing C-NHEJ in meiosis (Goedecke et al., 1999; Martin et al., 2005; Adamo et al., 2010; Lemmens et al., 2013).
Although the mechanism inhibiting C-NHEJ during meiosis is still elusive, regulators guaranteeing the success of the HR pathway have been extensively studied. Radiation sensitive1 (RAD1) is an evolutionarily conserved protein whose best-known function is checkpoint signaling. RAD1, a member of the ring-shaped RAD9-RAD1-HUS1 (9-1-1) complex, plays a crucial role in activating the pachytene checkpoint, a surveillance mechanism for monitoring the progression of meiotic HR in many organisms (Lydall et al., 1996; Hong and Roeder, 2002; Eichinger and Jentsch, 2010).
In addition to their well-known roles in checkpoint signaling, members of 9-1-1 complex may also play a direct role in facilitating DSB repair and HR during meiosis. RAD1 is associated with both synapsed and unsynapsed chromosomes during prophase I in mouse (Freire et al., 1998). The homolog of RAD1 in Saccharomyces cerevisiae is Rad17, and rad17 mutant exhibits persistent Rad51 foci (Shinohara et al., 2003). Moreover, mutations in Rad17 lead to a reduced frequency of interhomolog recombination, aberrant synapsis, increased rates of ectopic recombination events, and illegitimate repair from the sister chromatids during meiosis (Grushcow et al., 1999). Recently, Rad17 was shown to be necessary for the efficient assembly of ZMM proteins (Shinohara et al., 2015). Apart from RAD1, the other partners of 9-1-1 were also shown to be involved in DSB repair. HUS1 is proved essential for meiotic DSB repair in Drosophila (Peretz et al., 2009). Moreover, Hus1 inactivation in mouse testicular germ cells results in persistent meiotic DNA damage, chromosomal defects, and germ cell depletion (Lyndaker et al., 2013). Nevertheless, little is known about the role of 9-1-1 proteins in higher plants. In Arabidopsis (Arabidopsis thaliana), mutants of RAD9 show increased sensitivity to genotoxic agents and delayed general repair of mitotic DSBs (Heitzeberg et al., 2004). A recent study indicates that HUS1 is involved in DSB repair of both mitotic and meiotic cells in rice (Che et al., 2014).
In this study, we showed that OsRAD1 was required for the accurate repair of DSBs in rice during meiosis. Importantly, we demonstrated that the defective meiotic DSB repair in the Osrad1 mutants could be partially suppressed by blocking the C-NHEJ pathway. We also investigated the relationship between OsRAD1 and other key recombination proteins. Together, our findings indicated that the 9-1-1 complex plays a crucial role in the meiotic DSB repair mechanism.
RESULTS
Characterization of OsRAD1
In a screen for meiotic mutants, we obtained two mutant rice lines. Through map-based cloning and sequencing (see “Materials and Methods”), we found that the mutant lines were allelic for a disruption in LOC_Os06g04190. This gene was designated Oryza sativa RAD1 (OsRAD1) based on the homology of the protein sequence (see below), and the corresponding mutants were designated Osrad1-1 and Osrad1-2.
We isolated the full-length cDNA sequence of OsRAD1 by performing RT-PCR and RACE on young panicles. OsRAD1 has seven exons and six introns, with a 915-nucleotide open reading frame encoding a protein of 304 amino acids. Osrad1-1 has a single base-pair deletion in the first exon, which causes a frame-shift and a premature stop codon. In Osrad1-2, a 17-bp deletion in the fifth intron was detected, resulting in incorrect mRNA splicing (Fig. 1A).
Figure 1.
OsRAD1 is required for fertility in rice. A, Gene structure and mutation sites of OsRAD1. Exons are shown as black boxes. Untranslated regions are shown in white. B and C, Comparison of the wild-type (B) and Osrad1 (C) plant. D, Comparison of the wild-type (left) and Osrad1 (right) panicle. E and F, I2-KI staining of the wild-type (E) and Osrad1 pollen grains (F). Bars = 50 µm.
We carried out BLAST searches in public databases using the deduced protein sequence of OsRAD1, finding that OsRAD1 shares significant similarity with RAD1 proteins from other species (Supplemental Fig. S1). As revealed by qRT-PCR, OsRAD1 was highly expressed in young panicles and seedlings, although it was also expressed in leaves, roots, and sheaths (Supplemental Fig. S2).
Nonhomologous Chromosome Associations Are Observed in Osrad1 Meiocytes
Both Osrad1 mutant lines exhibited normal vegetative growth but complete sterility (Fig. 1, B–D). I2-KI staining showed that the pollen grains were completely nonviable in the mutants (Fig. 1, E and F). In addition, pollinating the mutant flowers with wild-type pollen did not result in seed production, suggesting that the mutants were both male- and female-sterile.
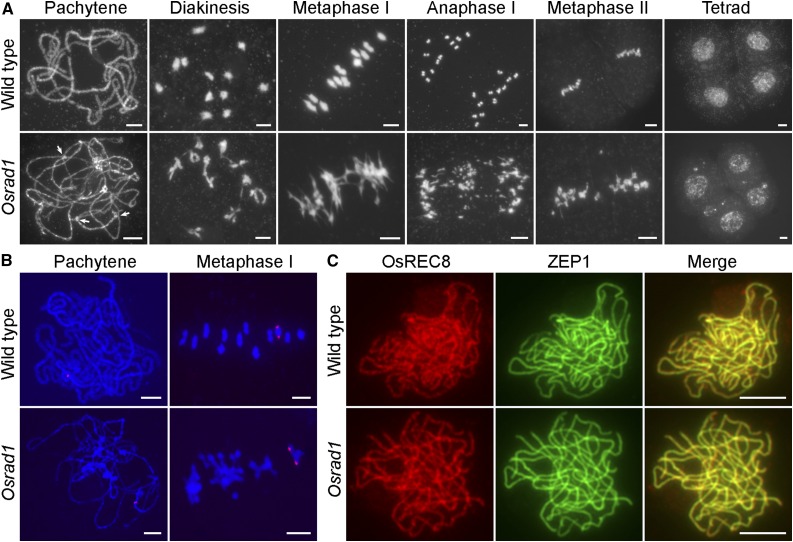
We examined meiotic chromosomes in meiocytes from both the wild-type and Osrad1 lines to clarify the cause of sterility in these mutants. In the wild type, meiotic chromosomes started to condense and appeared as thin threads at leptotene. At zygotene, pairing and synapsis of homologous chromosomes began. Because synapsis was completed at pachytene, fully synapsed chromosome pairs could be detected at this stage. After disassembly of the synaptonemal complex (SC) at diplotene, the 12 bivalents further condensed, revealing the presence of chiasmata at diakinesis. At metaphase I, all 12 highly condensed bivalents aligned in the center of the cell. During anaphase I and telophase I, homologous chromosomes separated and migrated toward opposite poles of the cell, leading to the formation of dyads. The sister chromatids then segregated equationally at the second meiotic division, which gave rise to tetrad spores (Fig. 2A).
Figure 2.
OsRAD1 is essential for meiosis. A, Chromosome spreads of male meiocytes in wild type (top row) and Osrad1 (bottom row). The abnormal chromosome associations in Osrad1 are pointed out with arrows. B, Homologous bivalents are formed in Osrad1. FISH analysis of the chromosome interaction in wild type and Osrad1. C, Immunostaining for ZEP1 (green) in wild type and Osrad1 at pachytene. OsREC8 signals (red) were used to indicate the meiocytes. Bars = 5 µm.
In the Osrad1-1 mutant, the chromosome behavior was similar to that of wild type from leptotene to zygotene. Anomalies, however, were observed in the Osrad1-1 mutant thereafter. Chromosomes alignment was roughly normal during pachytene, but associations between parallel chromosome pairs were detected. At diakinesis, this type of associations became more evident, although bivalent-like structures existed in most cells. And univalents were detected in some cells (12.9%, n = 85). At metaphase I, the putative bivalents aligned on the equatorial plate due to the drag force of spindle fibers. During anaphase I, the bivalents separated due to the pulling of microtubules, leading to unequal segregation of the chromosomes to the two daughter cells. During this process, extensive chromosome bridges and fragments were always observed. At telophase I, two masses of chromosomes arrived at opposite poles of the cell, and several acentric chromosome fragments remained on the equatorial plate. Subsequently, chromosome bridges between sister chromatids as well as fragments were detected in meiosis II, and tetrads with unequal numbers of chromosomes and several micronuclei eventually formed (Fig. 2A). The chromosome behavior of Osrad1-2 resembled that of Osrad1-1 (Supplemental Fig. S3). Taken together, cytological analysis of the Osrad1 mutants indicated that these plants had serious defects in HR.
Homolog Pairing and Synapsis Are Independent of OsRAD1
To explore the nature of these bivalent-like structures in the mutants, we performed fluorescent in situ hybridization (FISH) experiments using 5S rDNA as a probe to determine whether the structures were composed of homologous chromosomes. The 5S rDNA was located on the centromere-proximal region of chromosome 11. At pachytene, wild-type meiocytes exhibited one 5S rDNA signal, representing completely synapsed chromosome 11 (Fig. 2B). The staining pattern of the probes in the mutant resembled that in the wild type, indicating that the loss of OsRAD1 did not disturb homologous pairing. When metaphase I meiocytes were probed, we found that the bivalent-like structures were composed of associated homologous chromosomes (Fig. 2B).
To further verify SC assembly in the absence of OsRAD1, we performed immunolocalization analysis using an antibody against ZEP1 on spread preparations of meiocytes. ZEP1, a central element of the SC, is a perfect marker for detecting the status of rice chromosome synapsis (Wang et al., 2010). The localization of ZEP1 in Osrad1 was indistinguishable from that in wild type at pachytene, when ZEP1 patterns appeared as linear signals extending along the entire chromosomes (Fig. 2C). This result suggests that SC assembly is unaffected by the mutation of OsRAD1.
Early Homologous Recombination Is Not Disturbed in Osrad1
Homologous pairing in mammals and higher plants relies on interhomolog recombination. The normal SC assembly observed in the Osrad1 mutant is reminiscent of the location of important factors participating in early HR.
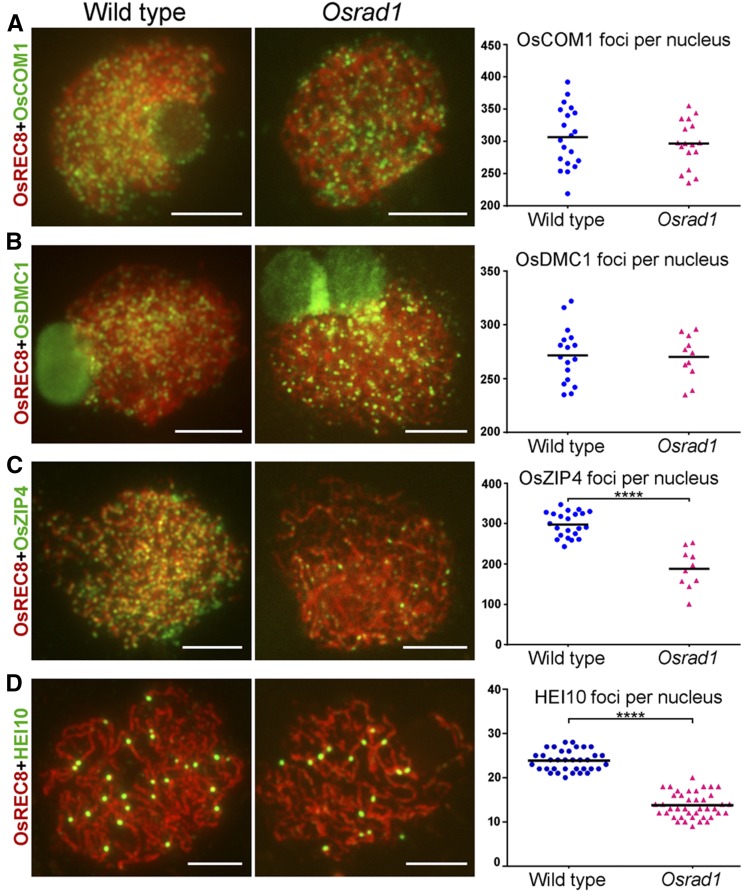
OsCOM1 participates in meiotic DSB end resection, while OsDMC1 mediates the single-strand invasion in recombination (Ji et al., 2012; Wang et al., 2016). Both of these proteins are required for homologous recombination during meiosis in rice. In immunostaining assays, these proteins were observed as punctuate foci on wild-type chromosomes during early meiosis. Similar signals were observed in the Osrad1 mutants (Fig. 3, A and B). The number of OsCOM1 foci was comparable between the wild type (306.7 ± 10.5, n = 20) and the Osrad1 mutants (296.5 ± 8.8, n = 17). The number of OsDMC1 foci in the Osrad1 mutants (270.1 ± 6.3, n = 11) was also similar to that of the wild type (271.5 ± 6.3, n = 17). In addition, the localization of OsMRE11 and OsRAD51C was not affected by mutation of OsRAD1 (Supplemental Fig. S4). These results suggest that early HR could be implemented in Osrad1.
Figure 3.
OsRAD1 contributes to recruitment of ZMM proteins. A, Immunostaining and scatter plot of OsCOM1 in wild type and Osrad1. P = 0.4709, two-tailed Student’s t test. B, Immunostaining and scatter plot of OsDMC1 in wild type and Osrad1. P = 0.8781, two-tailed Student’s t test. C, Immunostaining and scatter plot of OsZIP4 in wild type and Osrad1. ****P < 0.0001, two-tailed Student’s t test. D, Immunostaining and scatter plot of HEI10 in wild type and Osrad1. ****P < 0.0001, two-tailed Student’s t test. Bars = 5 µm.
Osrad1 Mutants Exhibit Defects in Crossover Formation
ZMM proteins such as OsZIP4 and HEI10 are thought to be necessary for the interference-sensitive CO formation pathway, which accounts for most of the COs in rice (Shen et al., 2012; Wang et al., 2012). Our data demonstrated the normal localization of early HR factors in the Osrad1 mutants. We wanted to explore whether the formation of interference-sensitive COs was affected by the mutation of OsRAD1. The number of OsZIP4 foci (188.3 ± 15.4, n = 10) in Osrad1 meiocytes was reduced to 63% that of the wild type (298.1 ± 6.5, n = 22) at zygotene (Fig. 3C). We compared the number of HEI10 foci (13.76 ± 0.44, n = 41) in Osrad1 with that in wild type (23.83 ± 0.39, n = 35), finding that there were significantly fewer bright HEI10 foci in Osrad1 than in wild type (Fig. 3D). Therefore, the number of interference-sensitive COs was reduced in this mutant due to the loss of OsRAD1.
Ectopic Interactions in Osrad1 Are Dependent on Meiotic DSBs
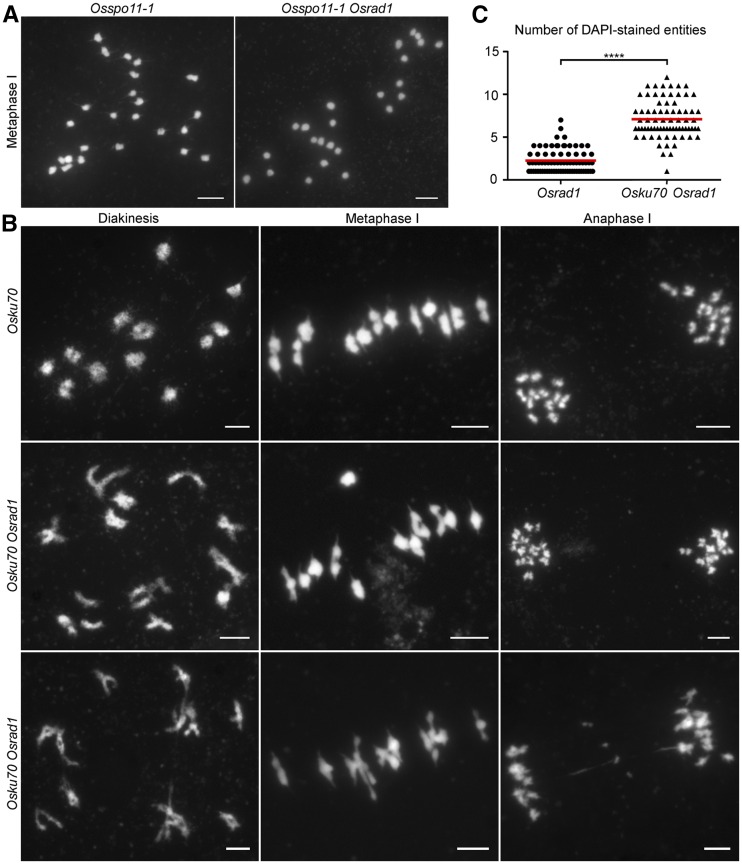
To investigate whether the abnormal chromosome behavior in Osrad1 meiocytes was due to deficient meiotic DSB repair, we carried out genetic analysis. Meiotic recombination is initiated by SPO11-generated DSBs. We recently isolated a spo11-1 rice mutant in our laboratory and used it to generate an Osspo11–1 Osrad1 double mutant. The double mutant exhibited the typical Osspo11–1 phenotype: 24 asynaptic univalents scattered throughout the entire nucleus at metaphase I and a lack of chromosome fragments at anaphase I (Fig. 4A). Therefore, the formation of ectopic associations and chromosome fragmentations in Osrad1 is dependent on the presence of meiotic DSBs.
Figure 4.
Abnormal meiotic DSBs repair leads to ectopic chromosome interactions in Osrad1 meiocytes. A, The Osrad1 meiotic phenotype depends on OsSPO11-1. Bars = 5 µm. B, Osku70 mutation relieves the meiotic defects in Osrad1. Bars = 5 μm. C, Number of DAPI-stained entities per nucleus in metaphase I meiocytes. Each symbol (filled circle for Osrad1 and triangle for Osku70 Osrad1) represents a single nucleus. Red lines indicate the average numbers of DAPI-stained entities. ****P < 0.0001, two-tailed Student’s t test.
Loss of Ku Partially Reduces Meiotic Defects in the Osrad1 Mutants
To help verify that the chromosomal aggregations in Osrad1 mutant meiocytes were due to inappropriate DSB repair, we created an Osku70 Osrad1 double mutant. OsKU70 is a rice homolog of Ku70, a key factor in the C-NHEJ repair pathway. We carefully observed the meiotic chromosomal behaviors of Osku70 Osrad1. Strikingly, disruption of OsKu70 led to a reduced frequency of abnormal chromosome associations in the Osrad1 mutants (Fig. 4B). Similar results were observed in Osku70 Oshus1 (Supplemental Fig. S5). We analyzed the extent of chromosome aggregations in Osrad1 and Osku70 Osrad1 by quantifying the 4,6-diamidinophenylindole (DAPI)-stained entities. There are 12 DAPI-stained entities (isolated bivalents) that could be detected in wild type at metaphase I. Although bivalent-like structures existed in Osrad1 and Osku70 Osrad1, they are constantly held together by aberrant associations. The connected chromosomes are deemed to be one DAPI-stained entity. Statistically, the number of DAPI-stained entities per cell in Osrad1 was 2.25 ± 0.16 (n = 75), while that in the Osku70 Osrad1 double mutant was 7.11 ± 0.26 (n = 73; Fig. 4C), implying a significant alleviation of chromosome aggregation. As mentioned above, the mutation of Osrad1 obstructed CO formation. To determine whether Osku70 deficiency alleviated this defect, we carried out statistical analysis of HEI10 signals in Osku70 Osrad1 meiocytes. The number of HEI10 foci was 17.75 ± 0.75 (n = 24). Thus, lack of OsKU70 restored a portion of CO formation. These data suggest that removal of OsKU70 partially suppresses the chromosome aggregation and CO reduction in OsRAD1-deficient meiocytes, implying that the formation of some univalents and ectopic interactions in Osrad1 is due to the repair of meiotic DSBs through the C-NHEJ pathway.
OsRAD1 Acts in a COM1-Dependent Manner and Independently of ZMM Proteins
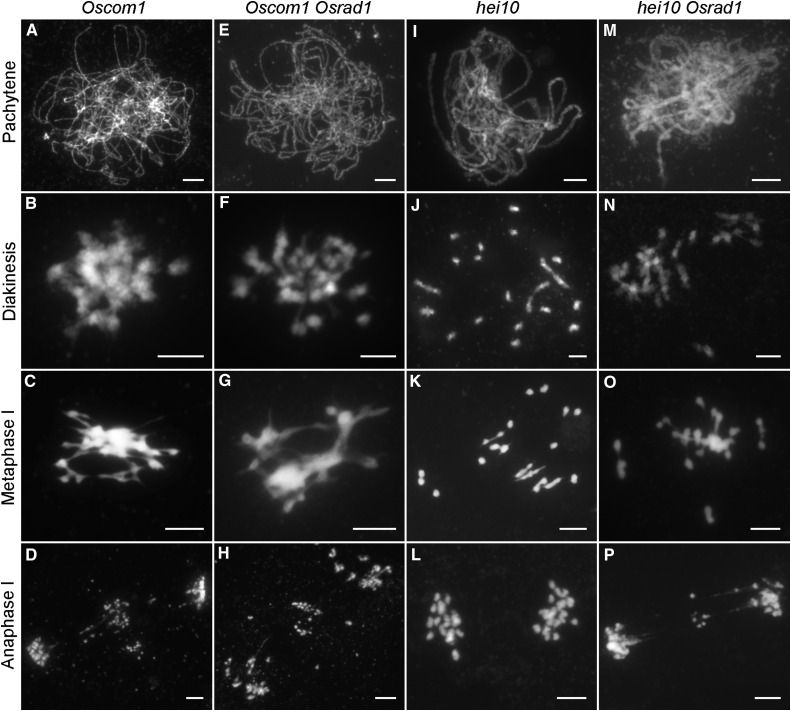
To obtain more precise information about the function and position of OsRAD1 in the DSB repair pathway, we generated Oscom1 Osrad1 double mutants. OsCOM1 participates in meiotic DSB end processing and is required for HR during rice meiosis. Both homologous pairing and synapsis were suppressed in the Oscom1 mutants (Fig. 5A). Aberrant chromosome interactions and fragments were also detected (Fig. 5, B–D). The phenotype of the Oscom1 Osrad1 double mutant could not be distinguished from that of the Oscom1 single mutant (Fig. 5, E–H), suggesting that OsRAD1 acts after OsCOM1 in the DSB repair cascade.
Figure 5.
Genetic analysis of OsRAD1 with OsCOM1 and HEI10. A to D, Oscom1 shows abolished homologous pairing and ectopic associations. E to H, The Oscom1 Osrad1 double mutant displays similar chromosome behaviors to those of the Oscom1 single mutant. I to L, Crossovers formation is disturbed in hei10. M to P, The hei10 Osrad1 double mutant exhibits nonhomologous chromosome associations. Bars = 5 µm.
Since COs are associations that are generated deliberately between homologous chromosomes, we wanted to know whether the chromosome phenotype of the Osrad1 mutant is related to impaired CO regulation. We decided to investigate the relationship between ectopic associations and interference-sensitive COs, because most COs in rice are derived from the interference-sensitive pathway. We therefore inquire into the involvement of two ZMM proteins, OsMER3 and HEI10, in this process. ZMM proteins were previously shown to be involved in the maturation of interference-sensitive COs. Osmer3 and hei10 mutants exhibit reduced chiasmata frequency in rice (Wang et al., 2009, 2012). HEI10 mutation has no effect on early meiotic events but results in a highly pronounced decrease in CO formation, leading to a large number of univalents at metaphase I (Fig. 5, I –L). We examined the chromosome behaviors in the hei10 Osrad1 double mutant, finding that this mutant exhibited nonhomologous chromosome associations as the Osrad1 mutant (Fig. 5, M–P). The number of univalents in hei10 Osrad1 (3.5 ± 0.39, n = 22) decreased compared to the hei10 mutant (15.77 ± 0.75, n = 26). The same results were obtained by analyzing the chromosome behaviors of the Osmer3 Osrad1 double mutant (Supplemental Fig. S6). The number of univalents (17.85 ± 0.67, n = 27) in Osmer3 was decreased to 3.96 ± 0.51 (n = 26) in the Osmer3 Osrad1 double mutant. While aberrant chromosome associations were also detected, this data implied that chromosomal aggregates in Osrad1 arise independently of the ZMM-mediated interference-sensitive CO pathway.
OsRAD1 Functions in Parallel with OsRAD51C and OsDMC1 during Meiotic DSB Repair
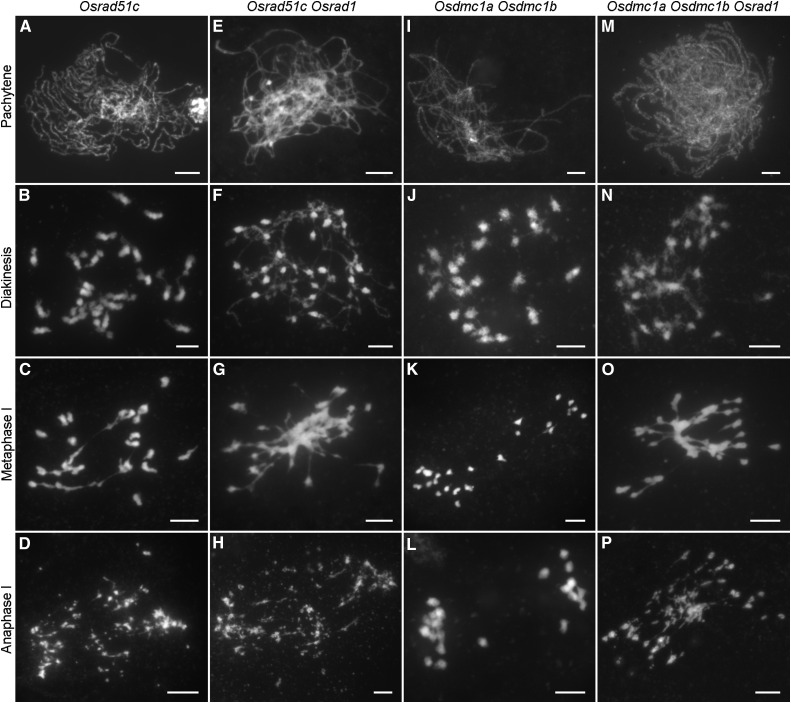
OsRAD51C is essential for DSB repair during meiosis (Tang et al., 2014). In the Osrad51c mutant, homologous pairing and synapsis were abolished at pachytene (Fig. 6A), and univalents were observed at diakinesis (Fig. 6B). After anaphase I, all of the univalents broke into fragments (Fig. 6D). Homologous pairing was abolished, and extensive chromosome fragmentations, as well as ectopic chromosome associations, were detected in the Osrad51c Osrad1 double mutant (Fig. 6, E–H).
Figure 6.
Genetic analysis of OsRAD1 with OsRAD51C and OsDMC1. A to D, Osrad51c displays asynapsis, univalents, and chromosome fragmentations. I to L, Univalents are observed in the Osdmc1a Osdmc1b double mutant. M to P, The Osdmc1a Osdmc1b Osrad1 triple mutant shows a cumulative effect of the three mutations. Bars = 5 µm.
Next we analyzed the genetic relationship between OsDMC1 and OsRAD1. Previous studies have shown that DMC1 mediates single-strand invasion and double Holliday junction formation during HR. Our previous study showed that OsDMC1A and DMC1B were functionally redundant in HR during rice meiosis (Wang et al., 2016). The situation of the Osdmc1a Osdmc1b double mutant was investigated. A typical asynaptic phenotype was observed in Osdmc1a Osdmc1b, with nearly 24 univalents distributed in the nuclei at metaphase I (Fig. 6, I–L). A cumulative effect of the mutations was observed in the Osdmc1a Osdmc1b Osrad1 triple mutant. Synapsis was disrupted in this background, as was the case in Osdmc1a Osdmc1b double mutant (Fig. 6M). At metaphase I, ectopic chromosome associations were detected in all meiocytes observed (Fig. 6O). Finally, chromosome bridges and fragmentation were observed in anaphase I (Fig. 6P).
The cumulative phenotype indicates that the occurrence of ectopic interactions in Osrad1 does not require OsRad51C or OsDMC1. However, DSB repair in Osrad1 partially relies on these proteins. Taken together, these results suggest that OsRAD1 may function in parallel with OsDMC1 and OsRAD51C during meiotic DSB repair. This notion is also supported by the observation that OsRAD51C and OsDMC1 were distributed normally in Osrad1 (Fig. 3B and Supplemental Fig. S4).
The 9-1-1 Complex May Be Involved in DSB Repair during Rice Meiosis
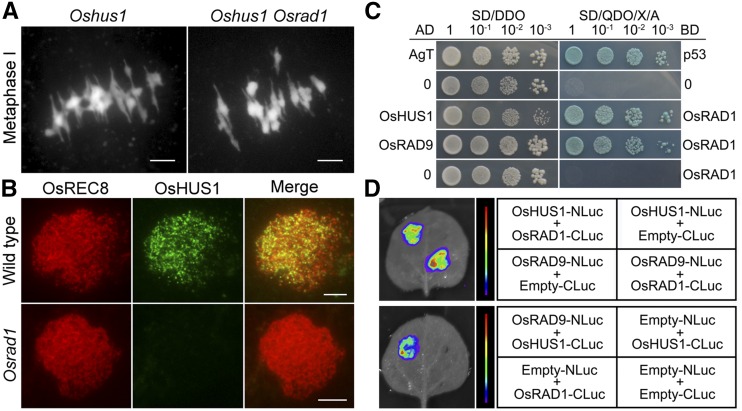
In yeast and mammalian cells, RAD1 interacts with RAD9 and HUS1, forming a heterotrimeric complex (the 9-1-1 complex). This complex is required for the activation of checkpoint signaling pathways involved in DNA damage responses. The existence of this complex has not yet been verified in plants. OsHUS1 was required for HR during meiosis. Oshus1 mutant showed similar meiotic defects to Osrad1, with aberrant chromosome associations at metaphase I (Fig. 7A). The phenotype of the Oshus1 Osrad1 double mutant is indistinguishable from that of each single mutant (Fig. 7A), implying that these proteins function in the same pathway. To further assess whether OsRAD1 functions as a component of the 9-1-1 complex during meiosis, we conducted immunostaining for OsHUS1 in Osrad1. In the wild type, OsHUS1 localizes to meiotic chromosomes as intensive foci. OsHUS1 signal was not detected in Osrad1 (Fig. 7B), indicating that OsHUS1 is loaded onto meiotic chromosomes in an OsRAD1-dependent fashion. We therefore investigated whether these three proteins interacted using multiple assays. In yeast two-hybrid analysis, ORFs of OsHUS1 and OsRAD9 were cloned into the related yeast AD vector pGADT7, while the ORF of OsRAD1 was cloned into the BD vector pGBKT7. After detecting coexpression on the double dropout medium, the interactions were verified by the growth of yeast on selective medium. Two-hybrid assays revealed that OsRAD1 was capable of interacting with OsHUS1 and OsRAD9 (Fig. 7C). Therefore, it is likely that a 9-1-1 complex also exists in plants, despite the lack of an interaction between OsHUS1 and OsRAD9 detected in this system. We also verified these interactions in Nicotiana benthamiana using split-luciferase complementation assays. The ORFs of OsRAD1, OsHUS1 and OsRAD9 were cloned into the pCAMBIA-NLuc and pCAMBIA-CLuc. The constructs were transiently expressed in N. benthamiana, and a luciferase complementation imaging assay was conducted. The results confirmed the OsRAD1–OsHUS1 and OsRAD1–OsRAD9 interactions (Fig. 7D). An OsHUS1–OsRAD9 interaction was also detected in this system. Therefore, the 9-1-1 complex may function in meiotic DSB repair in rice.
Figure 7.
The 9-1-1 complex is involved in DSB repair during rice meiosis. A, The Oshus1 Osrad1 double mutant showing similar chromosome behaviors to those of the single mutant. Bars = 5 μm. B, OsHUS1 localization relies on OsRAD1. Bars = 5 µm. C, OsRAD1 interacts with OsHUS1 and OsRAD9 in yeast two-hybrid assays. SD/DDO was used to test the cotransformation efficiency. The interactions were verified by the growth of yeast on selective medium SD/QDO/X/A. AgT and murine p53 were used as positive controls. D, OsRAD1, OsHUS1, and OsRAD9 interact with each other in N. benthamiana. The indicated constructs were transiently expressed in N. benthamiana, and a luciferase complementation imaging assay was conducted. AD: prey vector pGADT7; AgT: SV40 large-T antigen; BD: bait vector pGBKT7; CLuc: C-terminal fragment of firefly luciferase; NLuc: N-terminal fragment of firefly luciferase; SD/DDO: synthetic drop-out medium lacking Leu and Trp; SD/QDO/X/A: synthetic drop-out medium lacking Leu, Trp, His, and Ade, with X-α-Gal and aureobasidin A.
DISCUSSION
The 9-1-1 complex has been intensively investigated in yeast and mammals, with studies primarily focusing on checkpoint signaling (Parrilla-Castellar et al., 2004; Longhese et al., 2009). However, little is known about the meiotic function of 9-1-1 in higher eukaryotes, because mutation of these genes in mammals is lethal (Weiss et al., 2000; Hopkins et al., 2004; Han et al., 2010). OsHUS1 was recently identified as a key factor facilitating accurate meiotic recombination in rice (Che et al., 2014). In this study, we characterized the Osrad1 mutant, which has normal vegetative development, but has defects in gametogenesis. According to our data, OsRAD1 is required for suppression of the aberrant chromosome associations during meiosis. We showed that the Osrad1 meiotic phenotype depends on DSB formation. Thus, it is likely that the chromosome associations and fragmentation observed in the Osrad1 mutant reflect defects in meiotic DSB repair. Our results suggested that although OsRAD1 is dispensable for homologous pairing and synapsis, it is necessary for CO formation. Interestingly, only some of the COs appeared to be affected by OsRAD1 mutation. In support of this notion, fluorescent signals from key recombination factors, such as OsDMC1 and OsZIP4, were detected on meiotic chromosomes of Osrad1. Our results also suggest that OsRAD1 may function in meiotic DSB repair by forming the 9-1-1 complex, as determined by genetic analysis as well as various protein interaction assays. All of these findings suggested that the 9-1-1 complex plays a key role in the repair of meiotic DSBs.
Importantly, we demonstrated that the 9-1-1 checkpoint complex is an important inhibitory factor for NHEJ during meiotic DSB repair. C-NHEJ and HR are two major pathways for DSB repair in mitotic cells. During meiosis, programmed DSBs are channeled into HR to guarantee accurate chromosome segregation. MRN/X-dependent DSB end resection is a critical determinant of pathway choice (Takeda et al., 2007; Zhang et al., 2009; Yin and Smolikove, 2013). COM1 promotes meiotic recombination by counteracting the NHEJ complex Ku during Caenorhabditis elegans gametogenesis (Lemmens et al., 2013). Studies in plants have also demonstrated a pivotal role for MRE11 and COM1 in preventing nonhomologous chromosome associations (Uanschou et al., 2007; Ji et al., 2012). We therefore blocked the C-NHEJ pathway in OsRAD1-deficient meiotic cells by removing OsKU70, an essential protein for C-NHEJ-mediated DSB repair, through mutant analysis. The results indicate that the ectopic associations in the Osrad1 as well as Oshus1 mutants were partially suppressed by the removal of C-NHEJ.
However, the mechanism by which the 9-1-1 complex affects DSB repair pathway choice appears to differ from that of MRE11 and COM1, since both homologous pairing and synapsis were completed in Osrad1. By contrast, the Osmre11 and Oscom1 mutants were incapable of homologous recognition. Thus, a clear absence of homologous pairing was observed in these mutants at pachytene stage (Ji et al., 2012, 2013). Our results provide evidence that the function of OsCOM1 is epistatic to that of OsRAD1 in rice. Moreover, we observed normal localization of several meiotic factors (OsMRE11, OsCOM1, OsRAD51C, and OsDMC1) in the Osrad1 mutant, demonstrating that early HR events were not disturbed in this mutant. Thus, we propose that most DSBs are repaired by interhomolog recombination in the Osrad1 mutants and that these HR events are adequate for ensuring homologous pairing and synapsis, although a portion of DSBs are repaired through a Ku-dependent C-NHEJ pathway.
Notably, Ku deficiency did not fully suppress the ectopic chromosome interactions in the Osrad1 mutants. Moreover, the loss of OsKU70 could not fully restore the number of HEI10 foci in Osrad1 meiocytes, indicating that the reduction of CO in Osrad1 mutant was not entirely ascribed to C-NHEJ. Other mutagenic DSB repair pathways may account for the remaining ectopic chromosome associations in the Osku70 Osrad1 double mutant. Therefore, the aberrant chromosome associations in Osrad1 could be divided into two kinds. Some associations are derived from C-NHEJ, and the rest of the associations are not.
One hypothesis to explain the existence of residual aberrant associations involves the alternative end-joining (Alt-EJ) pathway. Alt-EJ is independent of Ku70/Ku80 and Xrcc4 and is highly error-prone (Guirouilh-Barbat et al., 2004; Boboila et al., 2010; Grabarz et al., 2012). Recently, the 9-1-1 complex was shown to be required for efficient HR, as Rad9 knockdown resulted in an increased frequency of Alt-EJ (Tsai and Kai, 2014). An alternative interpretation for the presence of residual ectopic interactions involves a disorder in HR—specifically, an ectopic recombination. Ectopic recombination, also known as “nonallelic HR”, refers to recombination between nonallelic DNA segments that share high sequence similarity. This type of recombination is usually suppressed to maintain genome stability (Stankiewicz and Lupski, 2002; Chen et al., 2010; Sasaki et al., 2010). The role of RAD1 in suppressing ectopic recombination during meiosis was first uncovered in budding yeast (Grushcow et al., 1999), but this function has not been reported in higher organisms. Here we confirmed the notion that OsRAD1 is necessary for suppressing nonhomologous associations during meiotic recombination. It is tempting to speculate that 9-1-1 is an important component of the surveillance mechanism that specifically eliminates nonallelic HR intermediates during meiosis.
Collectively, the 9-1-1 complex may function to ensure HR at multiple ways. We proposed a schematic model depicting the function of 9-1-1 complex in meiotic recombination (Fig. 8). How the 9-1-1 complex prevents nonhomologous associations at the molecular level is currently unknown. Studies in yeast have indicated that 9-1-1 serves as a stimulatory factor for both Dna2-Sgs1 and Exo1 during DNA resection (Blaikley et al., 2014; Ngo et al., 2014). Moreover, the human RAD1 was proved to have an exonuclease activity in vitro (Parker et al., 1998). Based on the current models of DNA end resection at meiotic DSBs, we propose that 9-1-1 stimulates DNA resection to inhibit mutagenic DSB repair pathways and thereby reinforce HR.
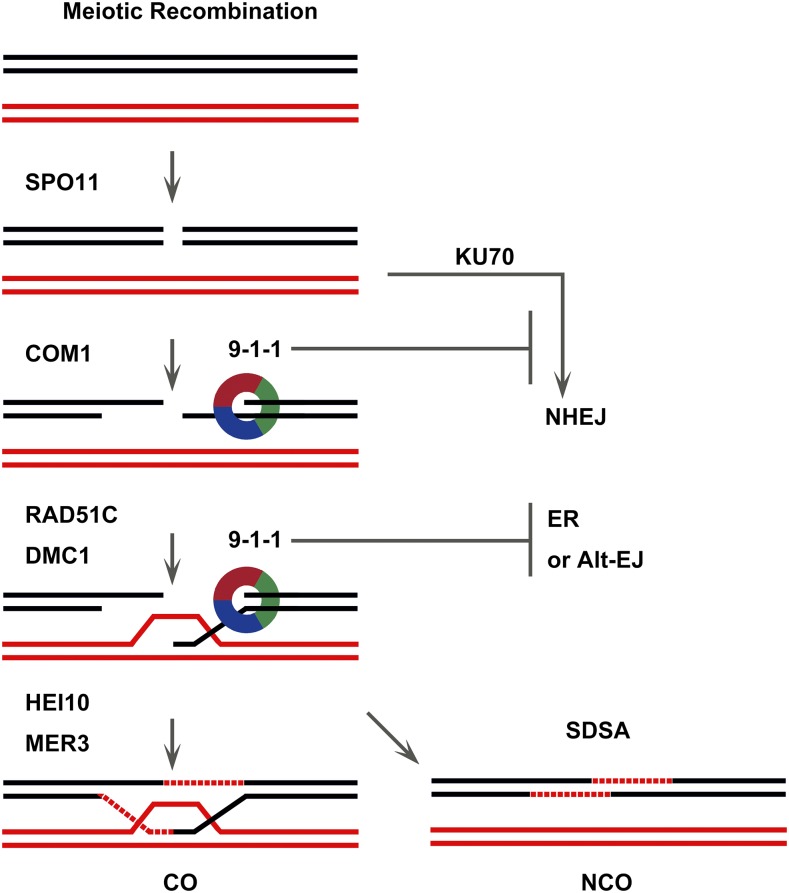
Figure 8.
A model for 9-1-1 complex in meiotic recombination. Black and red lines indicate dsDNA molecules of two nonsister chromatids. Only key steps and proteins mentioned in this article are labeled. Meiotic recombination starts with the formation of DSBs mediated by SPO11. These DSBs are processed by proteins including COM1 to yield the 3′ single-stranded tails, which can suppress DSB repair by C-NHEJ. Under the catalyzation of RAD51C and DMC1, the 3′ tails invade the homolog. Then ZMMs (such as HEI10 and MER3) promote the generation of COs. We propose that the 9-1-1 complex may reinforce meiotic recombination in two processes: (1) stimulating DNA resection and thereby inhibiting C-NHEJ; and (2) assisting end invasion to restrain ectopic recombination or Alt-EJ.
MATERIALS AND METHODS
Plant Materials and Culture Conditions
The Osrad1 rice (Oryza sativa) mutant lines were selected from a collection of mutants induced by 60CoƳ-ray irradiation. Mutant alleles involved in this study were: Osspo11-1, Oscom1, Osrad51c, Osmer3, hei10, Oshus1, Osku70, and Osdmc1a Osdmc1b. The Osku70 mutant line (3A-01546) was obtained from the Rice T-DNA Insertion Sequence Database (http://signal.salk.edu/cgi-bin/RiceGE) and the other mutant lines were previously isolated in our laboratory. Nipponbare was used as the wild type in all experiments. All plants were grown in paddy fields in Beijing (China) or Sanya (Hainan Province, China) during the natural growing season.
Map-Based Cloning of OsRAD1
In a screen for rice meiotic mutants, two mutant lines that segregated 3:1 (indicating a single recessive mutation) for sterility and meiotic defects were identified. Osrad1-1 was obtained from indica rice variety Guangluai 4, and Osrad1-2 was from japonica rice variety Yandao 8.
A map-based cloning approach was adopted to isolate the target gene. The heterozygous line Osrad1-1+/− was crossed with the japonica cultivar Nipponbare, and Osrad1-2+/− was crossed with indica rice variety 3037, to produce the mapping populations. Using sterile plants that segregated in the F2 population (28 plants for Osrad1-1 and 22 for Osrad1-2), the marker M1 was found to be the most closely linked marker in the two mutants. Fine gene mapping was then carried out with additional sterile F2 and F3 plants to pinpoint the target gene within a 280-kilobase region between markers P1 and P2. Based on the MSU Rice Genome Annotation Project Database and Resource (http://rice.plantbiology.msu.edu/), a candidate OsRAD1 gene (LOC_Os06g04190) was identified, which was annotated as encoding the putative cell cycle checkpoint protein RAD1. Sequencing of OsRAD1 in the two mutant lines showed that both mutants were disrupted in this genetic region.
All of the makers mentioned above were insertion-deletion markers, which were developed based on sequence differences between indica variety 9311 and japonica variety Nipponbare according to the data published at http://www.ncbi.nlm.nih.gov. Primer sequences are listed in Supplemental Table S1.
Cloning of Full-Length cDNA of OsRAD1
Total RNA extraction was conducted using the TRIzol reagent (Invitrogen). Reverse transcription was performed with primer Adaptor-T (18) using Superscript III RNaseH reverse transcriptase (Invitrogen). For 3′ RACE, gene-specific primers R3-1 and R3-2 were used, along with universal primers. PCR using primers RO-F and RO-R was performed to amplify the open reading frame. The PCR products were cloned into the PMD18-T vector (TaKaRa) and sequenced. The sequences were then spliced together to obtain the full-length cDNA sequence.
Real-Time RT-PCR for Transcript Expression Analysis
Total RNA was extracted individually from the roots, leaf sheaths, leaves, young panicles (5 cm to 7 cm), and seedlings (7 d) of Nipponbare, and reverse-transcribed into cDNA. Real-time RT-PCR analysis was performed using the Bio-Rad CFX96 real-time PCR instrument and EvaGreen (Biotium) with gene-specific primers (RAD1RT-F, RAD1RT-R) and standard control primers (Actin-F, Actin-R; Supplemental Table S2).
Cytology
Young panicles were fixed in Carnoy’s solution for at least 24 h at room temperature. Anthers at the proper developmental stage were then squashed in acetocarmine solution. After washing with 45% acetic acid, the chromosome preparations were frozen in liquid nitrogen for 2 min. The slides were dehydrated through an ethanol series (70, 90, and 100%) as soon as the coverslips were removed. Chromosomes on the slides were then counterstained with 4,6-diamidinophenylindole (DAPI) in antifade solution (Vector Laboratories). FISH analysis was conducted as described in Zhang et al. (2005). Immunofluorescence analysis was performed according to Shen et al. (2012). The antibodies against OsREC8, ZEP1, OsCOM1, OsMRE11, OsDMC1, OsZIP4, HEI10, and OsHUS1 were previously described in the literature (Wang et al., 2010; Shao et al., 2011; Ji et al., 2012; Shen et al., 2012; Wang et al., 2012; Ji et al., 2013; Miao et al., 2013; Che et al., 2014). Images were captured under a Zeiss A2 fluorescence microscope with a micro CCD camera.
Yeast Two-Hybrid Assay
The yeast two-hybrid (Y2H) assays were performed using the Matchmaker Gold Yeast Two-Hybrid System (Clontech). The open reading frames of OsRAD1, OsRAD9, and OsHUS1 were amplified from rice panicle cDNA clones (japonica) using specific primers and cloned into Clontech vectors (pGADT7 and pGBKT7). Yeast strain Y2HGold was used in the assays. Serial 1:10 dilutions were prepared in water and 5 µL of each dilution was used per spot.
Split-Luciferase Complementation Assay
The split-luciferase complementation assay was performed as previously described in Chen et al. (2008). Agrobacterium tumefaciens (GV3101) harboring the empty or recombinant vectors was infiltrated into expanded Nicotiana benthamiana leaves, followed by incubation in a growth room for 72 h For LUC activity measurements; 1 mm luciferin was sprayed onto the leaves. After incubation in the dark for 5 min, a cooled CCD imaging apparatus was used to capture the LUC images.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers OsRAD1, BAB19377; OsHUS1, BAS90186; OsRAD9, XP_015632035; BdRAD1(Brachypodium distachyon), XP_003561137; SbRAD1(Sorghum bicolor), XP_002436412; ZmRAD1(Zea mays), NP_001149448; AtRAD1(Arabidopsis thaliana), NP_193511; GmRAD1(Glycine max), XP_003555794; HsRAD1(Homo sapiens), NP_002844; MmRAD1(Mus musculus), NP_035362; DmRAD1(Drosophila melanogaster), NP_477440; CeRAD1(Caenorhabditis elegans), NP_499521; SpRad1(Schizosaccharomyces pombe), NP_594809; ScRad17(Saccharomyces cerevisiae), EHN04542.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Multiple sequence alignment of OsRAD1 homologs.
Supplemental Figure S2. Expression analysis of OsRAD1 in leaf, young panicle, sheath, root, and seeding by real-time q-PCR.
Supplemental Figure S3. Meiotic defects in Osrad1-2.
Supplemental Figure S4. Localization of OsMRE11 and OsRAD51C in Osrad1.
Supplemental Figure S5. Genetic analysis of OsHUS1 with OsKU70.
Supplemental Figure S6. Chromosome behaviors in Osmer3 and Osmer3 Osrad1.
Supplemental Table S1. Markers used in map-based cloning.
Supplemental Table S2. Primers for RACE, RT-PCR, and plasmid construction.
Supplementary Material
Footnotes
Articles can be viewed without a subscription.
References
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A (2010) Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell 39: 25–35 [DOI] [PubMed] [Google Scholar]
- Blaikley EJ, Tinline-Purvis H, Kasparek TR, Marguerat S, Sarkar S, Hulme L, Hussey S, Wee BY, Deegan RS, Walker CA, Pai CC, Bähler J, et al. (2014) The DNA damage checkpoint pathway promotes extensive resection and nucleotide synthesis to facilitate homologous recombination repair and genome stability in fission yeast. Nucleic Acids Res 42: 5644–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, Fazeli A, Feldman L, Nussenzweig A, Nussenzweig M, Alt FW (2010) Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci USA 107: 3034–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Rondinelli B, D’Andrea AD (2016) Repair pathway choices and consequences at the double-strand break. Trends Cell Biol 26: 52–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che L, Wang K, Tang D, Liu Q, Chen X, Li Y, Hu Q, Shen Y, Yu H, Gu M, Cheng Z (2014) OsHUS1 facilitates accurate meiotic recombination in rice. PLoS Genet 10: e1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-M, Cooper DN, Férec C, Kehrer-Sawatzki H, Patrinos GP (2010) Genomic rearrangements in inherited disease and cancer. Semin Cancer Biol 20: 222–233 [DOI] [PubMed] [Google Scholar]
- Deriano L, Roth DB (2013) Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet 47: 433–455 [DOI] [PubMed] [Google Scholar]
- Eichinger CS, Jentsch S (2010) Synaptonemal complex formation and meiotic checkpoint signaling are linked to the lateral element protein Red1. Proc Natl Acad Sci USA 107: 11370–11375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire R, Murguía JR, Tarsounas M, Lowndes NF, Moens PB, Jackson SP (1998) Human and mouse homologs of Schizosaccharomyces pombe rad1+ and Saccharomyces cerevisiae RAD17: linkage to checkpoint control and mammalian meiosis. Genes Dev 12: 2560–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedecke W, Eijpe M, Offenberg HH, van Aalderen M, Heyting C (1999) Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet 23: 194–198 [DOI] [PubMed] [Google Scholar]
- Grabarz A, Barascu A, Guirouilh-Barbat J, Lopez BS (2012) Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am J Cancer Res 2: 249–268 [PMC free article] [PubMed] [Google Scholar]
- Grushcow JM, Holzen TM, Park KJ, Weinert T, Lichten M, Bishop DK (1999) Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics 153: 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS (2004) Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell 14: 611–623 [DOI] [PubMed] [Google Scholar]
- Han L, Hu Z, Liu Y, Wang X, Hopkins KM, Lieberman HB, Hang H (2010) Mouse Rad1 deletion enhances susceptibility for skin tumor development. Mol Cancer 9: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzeberg F, Chen IP, Hartung F, Orel N, Angelis KJ, Puchta H (2004) The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J 38: 954–968 [DOI] [PubMed] [Google Scholar]
- Hong EJE, Roeder GS (2002) A role for Ddc1 in signaling meiotic double-strand breaks at the pachytene checkpoint. Genes Dev 16: 363–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins KM, Auerbach W, Wang XY, Hande MP, Hang H, Wolgemuth DJ, Joyner AL, Lieberman HB (2004) Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol Cell Biol 24: 7235–7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Tang D, Wang K, Wang M, Che L, Li M, Cheng Z (2012) The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis. Plant J 72: 18–30 [DOI] [PubMed] [Google Scholar]
- Ji J, Tang D, Wang M, Li Y, Zhang L, Wang K, Li M, Cheng Z (2013) MRE11 is required for homologous synapsis and DSB processing in rice meiosis. Chromosoma 122: 363–376 [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Kleckner N. (2006) Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115: 175–194 [DOI] [PubMed] [Google Scholar]
- Lemmens BBLG, Johnson NM, Tijsterman M (2013) COM-1 promotes homologous recombination during Caenorhabditis elegans meiosis by antagonizing Ku-mediated non-homologous end joining. PLoS Genet 9: e1003276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Bonetti D, Guerini I, Manfrini N, Clerici M (2009) DNA double-strand breaks in meiosis: checking their formation, processing and repair. DNA Repair (Amst) 8: 1127–1138 [DOI] [PubMed] [Google Scholar]
- Lydall D, Nikolsky Y, Bishop DK, Weinert T (1996) A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383: 840–843 [DOI] [PubMed] [Google Scholar]
- Lyndaker AM, Lim PX, Mleczko JM, Diggins CE, Holloway JK, Holmes RJ, Kan R, Schlafer DH, Freire R, Cohen PE, Weiss RS (2013) Conditional inactivation of the DNA damage response gene Hus1 in mouse testis reveals separable roles for components of the RAD9-RAD1-HUS1 complex in meiotic chromosome maintenance. PLoS Genet 9: e1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JS, Winkelmann N, Petalcorin MI, McIlwraith MJ, Boulton SJ (2005) RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol Cell Biol 25: 3127–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C, Tang D, Zhang H, Wang M, Li Y, Tang S, Yu H, Gu M, Cheng Z (2013) Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25: 2998–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo GH, Balakrishnan L, Dubarry M, Campbell JL, Lydall D (2014) The 9-1-1 checkpoint clamp stimulates DNA resection by Dna2-Sgs1 and Exo1. Nucleic Acids Res 42: 10516–10528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker AE, Van de Weyer I, Laus MC, Oostveen I, Yon J, Verhasselt P, Luyten WH (1998) A human homologue of the Schizosaccharomyces pombe rad1+ checkpoint gene encodes an exonuclease. J Biol Chem 273: 18332–18339 [DOI] [PubMed] [Google Scholar]
- Parrilla-Castellar ER, Arlander SJ, Karnitz L (2004) Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 3: 1009–1014 [DOI] [PubMed] [Google Scholar]
- Peretz G, Arie LG, Bakhrat A, Abdu U (2009) The Drosophila hus1 gene is required for homologous recombination repair during meiosis. Mech Dev 126: 677–686 [DOI] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S (2010) Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol 11: 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao T, Tang D, Wang K, Wang M, Che L, Qin B, Yu H, Li M, Gu M, Cheng Z (2011) OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol 156: 1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Tang D, Wang K, Wang M, Huang J, Luo W, Luo Q, Hong L, Li M, Cheng Z (2012) ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J Cell Sci 125: 2581–2591 [DOI] [PubMed] [Google Scholar]
- Shinohara A, Gasior S, Ogawa T, Kleckner N, Bishop DK (1997) Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2: 615–629 [DOI] [PubMed] [Google Scholar]
- Shinohara M, Hayashihara K, Grubb JT, Bishop DK, Shinohara A (2015) DNA damage response clamp 9-1-1 promotes assembly of ZMM proteins for formation of crossovers and synaptonemal complex. J Cell Sci 128: 1494–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Sakai K, Ogawa T, Shinohara A (2003) The mitotic DNA damage checkpoint proteins Rad17 and Rad24 are required for repair of double-strand breaks during meiosis in yeast. Genetics 164: 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18: 74–82 [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J (2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271 [DOI] [PubMed] [Google Scholar]
- Takeda S, Nakamura K, Taniguchi Y, Paull TT (2007) Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell 28: 351–352 [DOI] [PubMed] [Google Scholar]
- Tang D, Miao C, Li Y, Wang H, Liu X, Yu H, Cheng Z (2014) OsRAD51C is essential for double-strand break repair in rice meiosis. Front Plant Sci 5: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FL, Kai M (2014) The checkpoint clamp protein Rad9 facilitates DNA-end resection and prevents alternative non-homologous end joining. Cell Cycle 13: 3460–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uanschou C, Siwiec T, Pedrosa-Harand A, Kerzendorfer C, Sanchez-Moran E, Novatchkova M, Akimcheva S, Woglar A, Klein F, Schlögelhofer P (2007) A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J 26: 5061–5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu Q, Tang D, Liu X, Du G, Shen Y, Li Y, Cheng Z (2016) OsDMC1 is not required for homologous pairing in rice meiosis. Plant Physiol 171: 230–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Tang D, Wang M, Lu J, Yu H, Liu J, Qian B, Gong Z, Wang X, Chen J, Gu M, Cheng Z (2009) MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J Cell Sci 122: 2055–2063 [DOI] [PubMed] [Google Scholar]
- Wang K, Wang M, Tang D, Shen Y, Miao C, Hu Q, Lu T, Cheng Z (2012) The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet 8: e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang K, Tang D, Wei C, Li M, Shen Y, Chi Z, Gu M, Cheng Z (2010) The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RS, Enoch T, Leder P (2000) Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev 14: 1886–1898 [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Smolikove S (2013) Impaired resection of meiotic double-strand breaks channels repair to nonhomologous end joining in Caenorhabditis elegans. Mol Cell Biol 33: 2732–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Yang Q, Bao W, Zhang Y, Han B, Xue Y, Cheng Z (2005) Molecular cytogenetic characterization of the Antirrhinum majus genome. Genetics 169: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shim EY, Davis M, Lee SE (2009) Regulation of repair choice: Cdk1 suppresses recruitment of end joining factors at DNA breaks. DNA Repair (Amst) 8: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.