Significance
Pyruvate dehydrogenase (PDH) deficiency is the cause of several human metabolic diseases. In mammals, the transcriptional control of PDH complex components and its impact on pathophysiology remain poorly understood. We show that E4 transcription factor 1 (E4F1) controls a transcriptional program essential for PDH activity that involves genes linked to human metabolic syndromes. Genetic inactivation of murine E4f1 results in a strong decrease of PDH activity and severe perturbations of pyruvate metabolism. In concordance with the work of Legati et al., we show that striated muscle-specific E4f1 KO animals display phenotypes that recapitulate these clinical symptoms, providing an exciting clinical perspective to the present work.
Keywords: E4F1, PDH, pyruvate, muscle, endurance
Abstract
The mitochondrial pyruvate dehydrogenase (PDH) complex (PDC) acts as a central metabolic node that mediates pyruvate oxidation and fuels the tricarboxylic acid cycle to meet energy demand. Here, we reveal another level of regulation of the pyruvate oxidation pathway in mammals implicating the E4 transcription factor 1 (E4F1). E4F1 controls a set of four genes [dihydrolipoamide acetlytransferase (Dlat), dihydrolipoyl dehydrogenase (Dld), mitochondrial pyruvate carrier 1 (Mpc1), and solute carrier family 25 member 19 (Slc25a19)] involved in pyruvate oxidation and reported to be individually mutated in human metabolic syndromes. E4F1 dysfunction results in 80% decrease of PDH activity and alterations of pyruvate metabolism. Genetic inactivation of murine E4f1 in striated muscles results in viable animals that show low muscle PDH activity, severe endurance defects, and chronic lactic acidemia, recapitulating some clinical symptoms described in PDC-deficient patients. These phenotypes were attenuated by pharmacological stimulation of PDH or by a ketogenic diet, two treatments used for PDH deficiencies. Taken together, these data identify E4F1 as a master regulator of the PDC.
The pyruvate dehydrogenase (PDH) complex (PDC) is a mitochondrial multimeric complex that catalyzes the oxidative decarboxylation of pyruvate into acetyl-CoA (AcCoA), thus linking pyruvate metabolism to the tricarboxylic acid (TCA) cycle. Localized in the mitochondrial matrix, the core PDC is composed of multiple copies of three catalytic enzymes: PDHA1/E1, dihydrolipoamide transacetylase (DLAT)/E2, and dihydrolipoamide dehydrogenase (DLD)/E3 (1). To fuel the PDC, pyruvate translocates across the inner mitochondrial membrane through the heterodimeric pyruvate transporter mitochondrial pyruvate carrier 1 (MPC1)/MPC2 (2, 3). The activity of the PDC depends on several cofactors, including lipoate, CoEnzymeA (CoA), FAD+, NAD+, and thiamine pyrophosphate, the latter being imported in the mitochondria by the SLC25A19 transporter (4). So far, fine-tuning of PDC activity has been mainly attributed to posttranslational modifications of its subunits (5, 6), including the extensively studied phosphorylation of PDHA1/E1 modulated by PDH kinases (PDK1–4) and phosphatases (PDP1–2). However, in lower organisms, such as Escherichia coli and Candida albicans, PDC is also controlled at the transcriptional level by the coordinated regulation of genes encoding its components and regulators (7, 8). The importance of such transcriptional regulation of the PDC in mammals remains elusive. Physiological regulation of PDC plays a pivotal role in metabolic flexibility to adjust energetic metabolism and biosynthesis to nutrient availability and energy demand (9), such as in skeletal muscles during exercise (10). PDH activity is altered in several human metabolic syndromes associated with chronic lactate acidosis, progressive neurological degeneration, and muscular atonia (11). Genetic mitochondrial disorders associated with PDH deficiency mainly result from hypomorphic mutations in genes encoding subunits or regulators of the PDC, including in PDHA1, DLAT, DLD, PDP1, MCP1, and SLC25A19 (3, 11–13). The diverse clinical manifestations of PDC-deficient patients are significantly, but only partly, improved by ketogenic diets that provide alternative energetic substrates or by treatment with PDK inhibitors, such as dicholoroacetate (DCA). Thiamine/lipoic acid supplementations that favor optimal PDH activity, or bicarbonate treatment that buffers lactate acidosis, have also been tested, although with moderate efficiency (14, 15). The design of new and more efficient therapeutic approaches will require a better understanding of PDH regulation and the development of clinically relevant animal models.
Here we reveal another level of regulation of the pyruvate oxidation pathway in mammals that implicates the E4 transcription factor 1 (E4F1). Initially identified as a cellular target of the E1A viral oncoprotein (16), E4F1 was then described as a physical interactor of several tumor suppressors that gate cell division and survival in proliferating cells, including pRB, RASSF1A, p14ARF, and p53 (17–21). E4F1 is essential for early embryonic mouse development (22), and for either proliferation or survival of actively dividing mammalian cells (23–25). In proliferating cells, we have recently shown that E4F1 controls genes implicated in cell-cycle checkpoints and genome surveillance, but also unexpectedly, a transcriptional program involved in mitochondria functions (24, 26). Here we further characterized this mitochondria-associated program and found that E4F1 coordinates the transcription of a set of genes involved in PDH-mediated pyruvate oxidation. Accordingly, tissue-specific inactivation of murine E4f1 in the postmitotic and differentiated compartment of striated muscles resulted in a strong reduction of muscular PDH activity. Surprisingly, this constitutively low PDH activity did not compromise animal viability, although these animals displayed chronic lactic acidemia and endurance defects that recapitulate some clinical symptoms described in PDC-deficient patients.
Results
E4F1 Controls Genes Involved in PDH Activity.
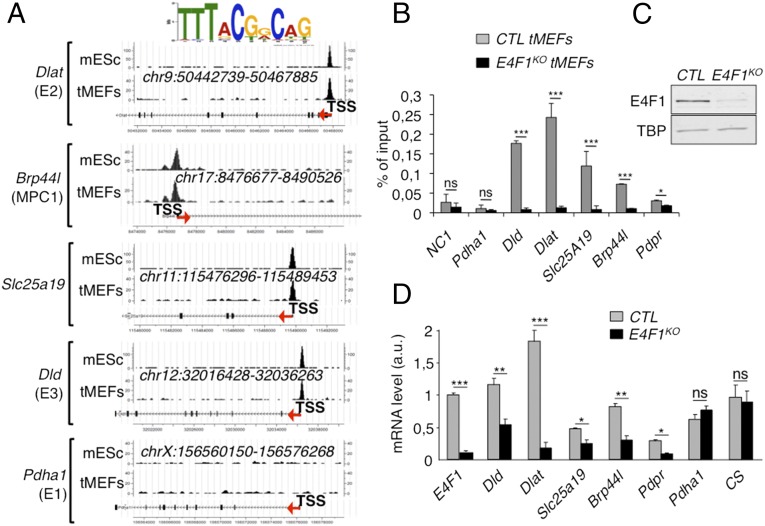
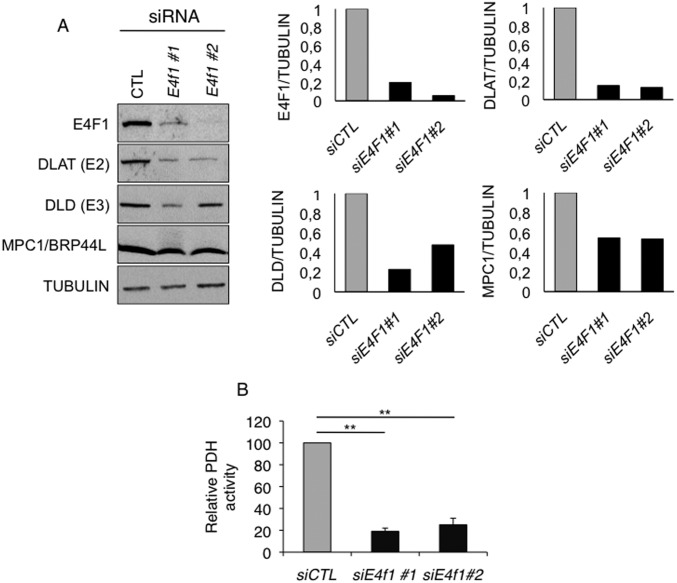
We recently identified by ChIP, combined with deep sequencing (ChIP-seq), the repertoire of endogenous target DNA sites bound by E4F1 in primary and Ha-RasV12 transformed mouse embryonic fibroblasts (tMEFs) (24, 26). We completed this gene list by performing additional E4F1 ChIP-seq analyses in murine embryonic stem (ES) cells and defined a common set of promoter regions that were bound by E4F1 in these two cell types (Fig. 1A) (GSE57221 and GSE57228). Gene ontology analysis of E4F1 target genes revealed an unexpected enrichment for nuclear genes encoding mitochondrial proteins (24). Surprisingly, a closer analysis of this subprogram identified four genes, located on distinct chromosomes, which are directly involved in PDH function. These genes encode the E2 and E3 subunits of the PDH core enzyme (Dlat, Dld), the mitochondrial pyruvate transporter MPC1 (Brp44l), and the mitochondrial transporter of the PDH cofactor thiamine pyrophosphate (Slc25a19/DNC). A fifth gene, encoding the negative regulator of the PDH phosphatases (Pdpr) (27) was also identified as an E4F1 target gene by ChIP-seq in transformed fibroblasts, but not in ES cells (data not shown). Sequence analyses revealed that these genes contained one or two bona fide E4F1 binding sites nearby their transcription start site (TSS) (Fig. 1A). These E4F1 direct target genes were further validated by ChIP-quantitative PCR (qPCR) experiments performed upon Cre-mediated inactivation of E4f1 in E4f1−/flox tMEFs (hereafter referred to as E4f1KO) (Fig. 1 B and C). Consistent with a role for E4F1 as a bona fide transcriptional activator for these PDH-related genes, the mRNA levels of Dlat, Brp44l/Mpc1, Dld, Scl25A19, and Pdpr decreased in E4f1KO cells, although to various extents (Fig. 1D). In contrast, the transcript levels of another PDH core component, Pdha1, and of the mitochondrial enzyme citrate synthase (CS), which were not identified as E4F1 direct target genes, did not vary upon acute E4f1 inactivation (Fig. 1D). At the protein level, a strong down-regulation of DLAT and BRP44L/MPC1, and a moderate decrease of DLD were observed in E4f1KO cells (Fig. 2A). Of note, siRNA-mediated depletion of E4f1 in fibroblasts also resulted in the down-regulation of DLAT, BRP44L/MPC1 and DLD proteins (Fig. S1A), confirming the role of E4F1 in the control of these genes. Taken together, our data highlight a previously undescribed function of E4F1 in the transcriptional control of genes involved in PDH activity in mammals.
Fig. 1.
E4F1 directly controls a transcriptional program involved in PDH activity in mammals. (A) E4F1 ChIP-Seq read densities in tMEFs and mouse ES cells (mES) at the Dlat, Dld, Mpc1/Brp44l, Slc25a19/DNC (deoxynucleotide carrier) genes, and at the Pdha1 gene as a representative control locus to which E4F1 does not bind. The E4F1 consensus motif determined by MEME is shown and arrows indicate the genes orientation. (B) Validation of these E4F1 target genes by ChIP-qPCR assays performed in E4f1KO and control (CTR) tMEFs, 3 d after transduction with a self-excising Cre-retrovirus. A gene-poor noncoding region of chromosome 8 (NC1) and the Pdha1 promoter region (TSS) were used as controls. Enrichments are represented as percentages of input (mean value ± SEM, n = 3). (C) Protein levels of E4F1, and of the loading control Tata binding protein (TBP), determined by immunoblotting of total protein extracts prepared from E4f1KO, and control tMEFs. (D) mRNA levels of Dlat, Dld, Slc25a19, Brp44l/Mpc1, Pdpr, and of two control genes (CS and Pdha1) determined by RT-qPCR analysis in E4f1KO and control tMEFs. Histobars represent the mean value ± SEM (n = 6). ***P < 0.001; **P < 0.01; *P < 0.05; ns, not significant.
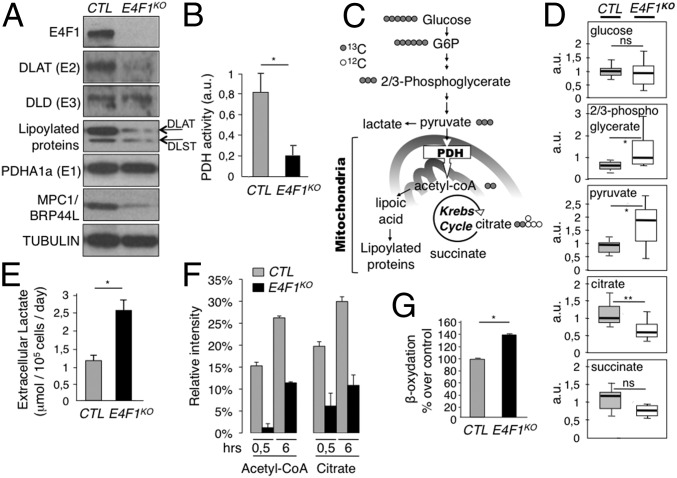
Fig. 2.
Impaired PDH activity and deregulation of the pyruvate pathway in E4f1KO cells. (A) Protein levels of E4F1, DLAT, DLD, lipoylated proteins (DLAT and DLST), PDHE1a, MPC1/BRP44L, and Tubulin (loading control) determined by immunoblotting of total cell extracts prepared from E4f1KO and control (CTR) tMEFs. (B) PDH enzymatic activity measured in E4f1KO and CTR tMEFs. (C) Schematic representation of the pyruvate–AcCoA pathway. (D) Relative levels of several metabolites linked to the pyruvate pathway in E4f1KO and CTR tMEFs, measured by LC/MS (n = 8). (E) Extracellular lactate level in the medium of E4f1KO and CTR tMEFs. Histobars represent the mean value ± SEM (n = 5). (F) Relative abundance of M+2 isotopomers of AcCoA and citrate that derive from pyruvate oxidation as determined by LC-MS in E4f1KO and match CTR tMEFs cultured in d-[U-13C]glucose for 30 min or 6 h (mean ± SD, experiment performed in triplicate). (G) Relative levels of FAO measured upon incubation of E4f1KO and CTR tMefs with 3H-palmitate (mean value ± SEM, n = 3). **P < 0.01; *P < 0.05; ns, not significant.
Fig. S1.
Impact of E4f1 depletion on PDC. (A) Protein levels of E4F1, DLAT, DLD, MPC1, and of the loading control Tubulin, determined by immunoblotting of total protein extracts prepared from tMEFs, 3 d after transfection of control or E4f1-siRNAs. (Right) Quantification of this representative experiment using imageJ. Data were normalized according to Tubulin levels. (B) PDH activity measured by DipStick assay in protein extracts prepared from tMEFs, 3 d after transfection of control or E4f1-siRNAs. Histobars represent the mean value ± SEM (n = 3). **P < 0.01.
E4f1 Inactivation Results in Reduced PDH Activity and Metabolic Reprogramming.
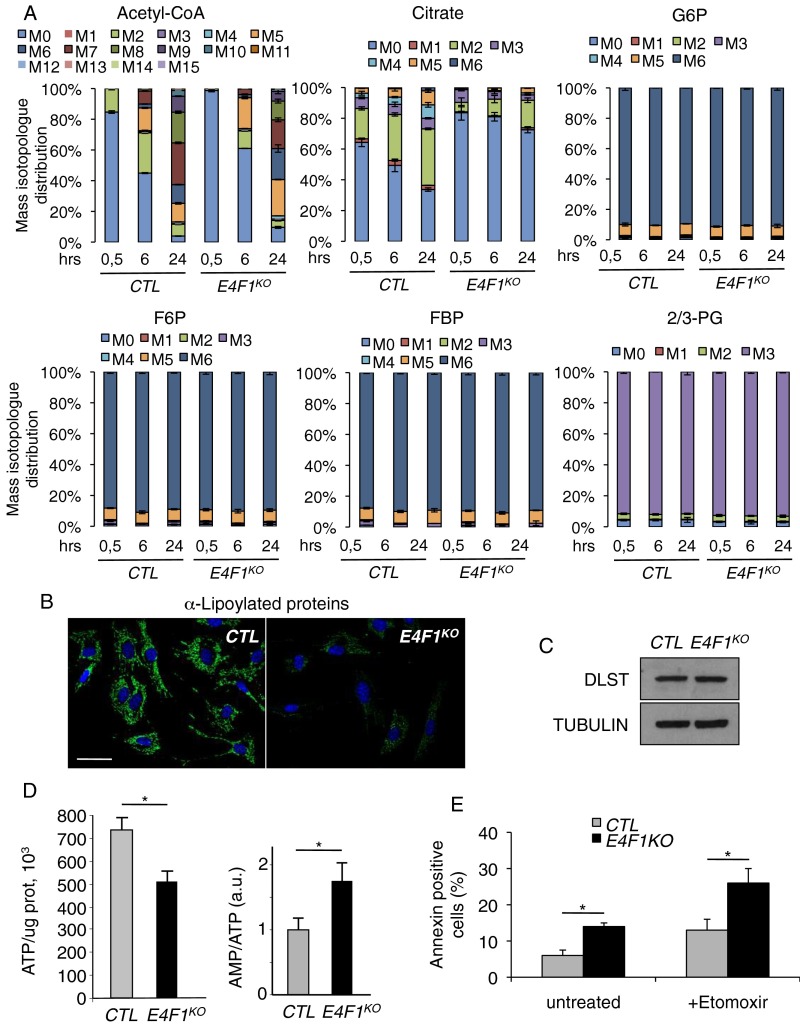
As a direct consequence of decreased expression of PDC subunits, E4f1KO fibroblasts and E4f1 siRNA-treated cells exhibited a marked decrease of PDH enzymatic activity (Fig. 2B and Fig. S1B). In E4F1-deficient cells, this reduced PDH activity should impact on pyruvate-derived mitochondrial AcCoA production, lead to accumulation of glycolytic intermediates, and induce the redirection of the glycolytic flux. We addressed this notion by performing comparative nontargeted gas chromatography/liquid chromatography-mass spectrometry (GC/LC-MS) metabolomic analyses in control and E4f1KO fibroblasts. As predicted, these analyses showed an accumulation of intracellular pyruvate and of its upstream precursor 2/3-phosphoglycerate (2/3PG) in E4f1KO cells, as well as lower levels of citrate and succinate, two intermediates of the TCA cycle (Fig. 2 C and D). E4F1-deficient cells also exhibited increased level of extracellular lactate in their culture medium (Fig. 2E). To further assess the PDH-dependent pyruvate oxidation pathway, we next performed stable isotope tracing experiments in control and E4f1KO fibroblasts cultured in presence of uniformly labeled [U-13C]glucose. Comparative LC-MS analyses of intracellular metabolites clearly showed a strong decrease of 13C incorporation into AcCoA (M+2 isotopomer) and in its downstream metabolite, citrate (M+2 isotopomer), in E4f1KO cells (Fig. 2F and Fig. S2A). Of note, the relative 13C enrichment in the first glycolytic intermediates (glucose-6-phosphate, fructose-6-phosphate, fructose-1,6-biphosphate, 2/3-PG) was unaffected, suggesting that both control and E4f1KO fibroblasts display comparable glycolytic fluxes. Taken together, these analyses indicate that E4F1-deficiency impairs PDH activity with impacts on the pyruvate oxidation pathway (Fig. S2A).
Fig. S2.
Metabolic consequences of PDH deficiency in E4f1KO cells. (A) The mass isotopomer distribution (without correction for natural isotope abundance) for AcCoA, citrate, glucose-6 phosphate (G6P), fructose-6 phosphate (F6P), fructose-1,6 biphosphate (FBP), 2/3 phosphoglycerate (2/3PG) was determined by LC-MS in E4f1KO tMefs and match control cells cultured with d-[U-13C]glucose for 30 min, 6 or 24 h, as indicated (mean ± SD). (B) Immunofluorescence staining of lipoylated proteins in E4f1KO and CTR tMEFs. (Scale bar, 50 μm.) (C) Protein levels of DLST and of the loading control Tubulin, determined by immunoblotting of total protein extracts prepared from E4f1KO tMEFs and match control cells. (D) Total ATP level (Left) and the AMP/ATP ratio (Right) were determined in E4f1KO and control (CTR) tMEFs. Histobars represent the mean value ± SEM of n = 3 independent experiments. (E) FACS analysis of apoptotic cells in E4f1KO and control (CTR) tMEFs, 24 h after incubation with the FAO-inhibitor etomoxir. Histobars represent the mean value of annexin-V+ cells ± SEM (n = 5 independent experiments). All analyses were performed in E4f1KO tMEFs and match control cells, 5 d after transduction with a self-excising Cre-encoding retrovirus. *P < 0.05.
Of note, we also assessed mitochondrial protein lipoylation as both a direct readout of DLAT expression and of an indirect readout of defective AcCoA production by PDH in E4f1KO cells (2, 28). Indeed, the precursor of lipoic acid, octanoic acid, is synthesized from mitochondrial AcCoA through fatty acid biosynthesis. Lipoic acid is then covalently attached to few proteins, for which it serves as a cofactor. At first glance, total protein lipoylation seemed to be strongly reduced in E4f1KO cells, as revealed by immunofluorescence (Fig. S2B) using an antibody that recognizes all lipoylated proteins. Although this strong decrease of total protein lipoylation likely reflects mainly DLAT protein down-regulation (the most abundant lipoylated protein), we also observed a moderate down-regulation of the lipoylation of dihydrolipoamide-S-succinyl tranferase (DLST) by immunoblotting (Fig. 2A and Fig. S2C), suggesting that the AcCoA-dependent lipoylation pathway is also partly affected in E4f1KO cells, and could also contribute to the phenotype of E4f1KO. Finally, despite their low PDH activity, E4f1KO cells showed a moderate, but significant, decrease of intracellular ATP, as described in our previous report (see also ref. 24) (Fig. S2D). This moderate alteration of ATP levels suggests that alternative energetic pathways were activated in E4f1KO cells. Indeed, E4f1KO cells show signs of adaptive metabolic responses, as illustrated by increased fatty acid oxidation (FAO) (Fig. 2G). Accordingly, these cells were highly sensitive to the FAO inhibitor Etomoxir (Fig. S2E). Taken together, these data show that PDH activity and the mitochondrial pyruvate pathway are impaired in E4f1KO cells.
E4f1 KO in Striated Skeletal Muscles Results in PDH Dysfunction.
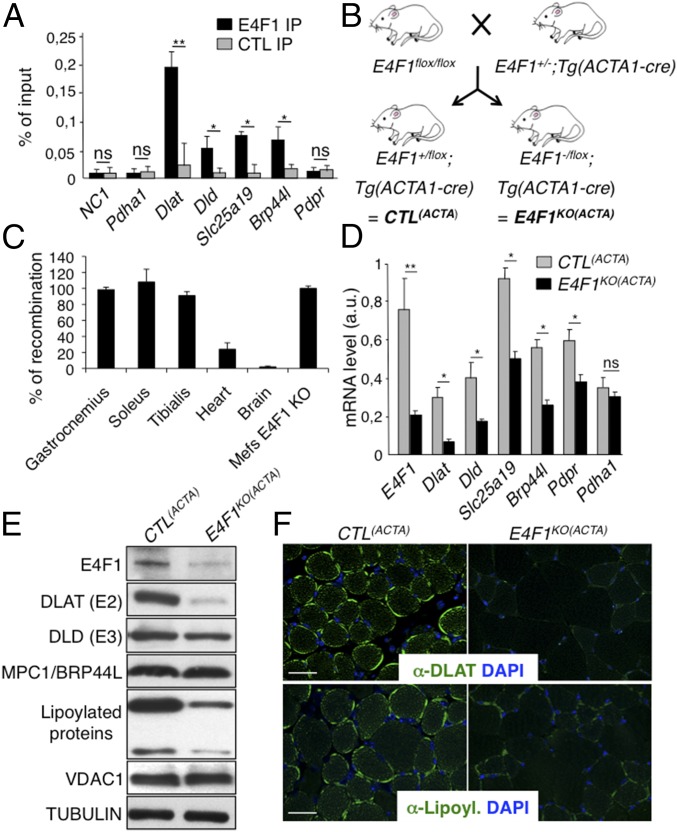
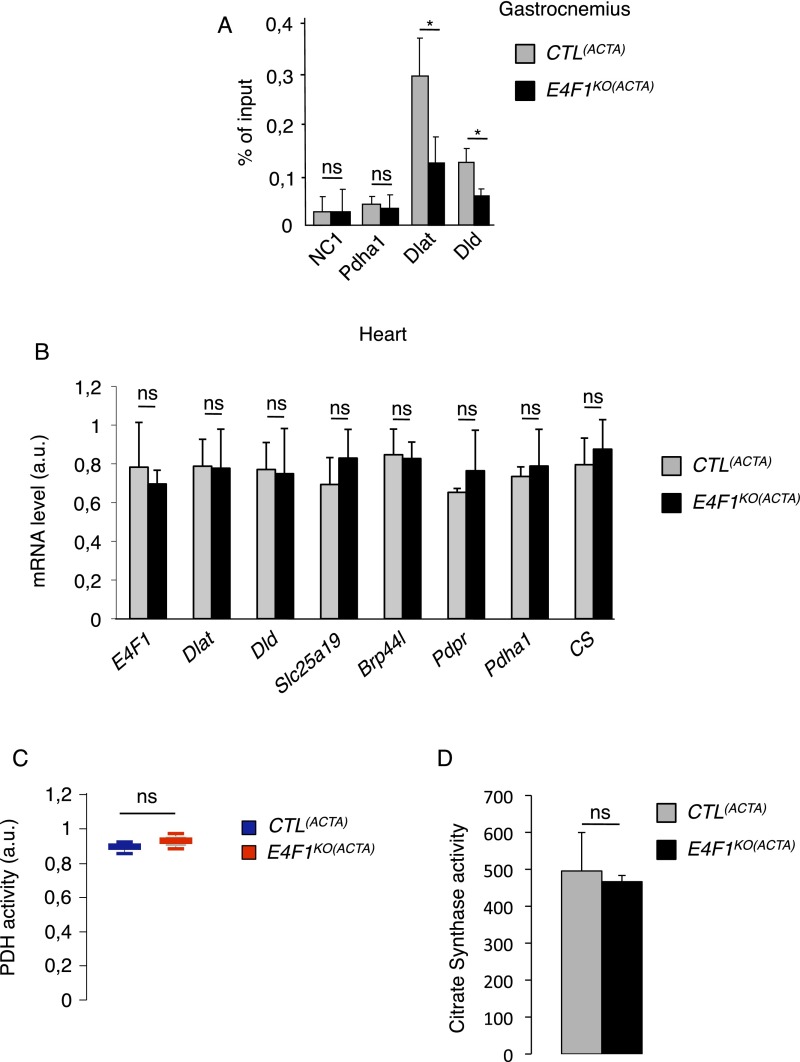
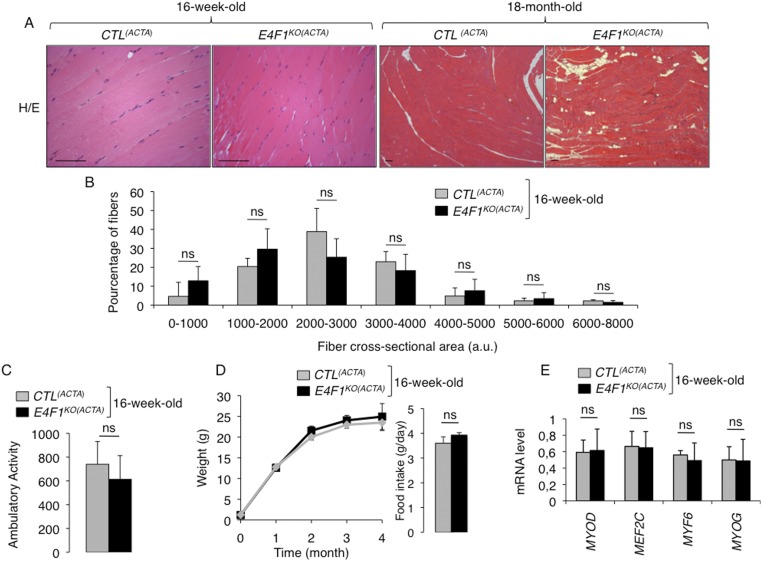
We next assessed the in vivo relevance of E4F1-mediated control of this PDC transcriptional program in striated muscle, a tissue which physiological function relies on high PDH activity during exercise (10). First, we confirmed by ChIP-qPCR the recruitment of E4F1 on Dlat, Dld, Slc25a19, and Brp44l/Mpc1 promoters in adult tibialis and gastrocnemius muscles of resting mice, indicating that E4F1–PDH program also exists in adult muscle cells (Fig. 3A and Fig. S3A). Then, we inactivated E4f1 in vivo in striated muscles by crossing E4f1−/flox mice with Acta1-Cre transgenic (Tg) mice that express the Cre recombinase under the control of the skeletal α-actin promoter [hereafter referred to as Tg(Acta1-Cre)] (Fig. 3B) (29). We verified the efficiency and the tissular specificity of Cre-driven recombination of the E4f1flox allele in E4f1−/flox; Tg(Acta1-Cre) and E4f1+/flox; Tg(Acta1-Cre) control littermates [hereafter referred to as CTL(ACTA) and E4f1KO(ACTA), respectively]. Cre-driven inactivation of the E4f1flox allele was largely restricted to striated skeletal muscles, as shown by the strong reduction of E4f1 mRNA and protein levels in gastrocnemius of adult E4f1KO(ACTA) mice (Fig. 3 D and E). Although limited Cre-mediated recombination (20% efficiency) was also detected in heart (Fig. 3C), this did not impair significantly the cardiac mRNA level of E4f1 when assessed at the whole tissue level (Fig. S3B). E4f1KO(ACTA) mice were healthy and viable, and detailed anatomo-pathological analyses of skeletal muscles at 16 wk of age revealed neither major histological alterations nor significant differences in the number and size of muscle fibers compared with control littermates (Fig. S4 A, B, and D). Accordingly, mRNA levels of muscular differentiation markers and inducers such as Mif6, Mef2c, MyoD, and Myogenin were similar in adult striated muscles of 16-wk-old E4f1KO(ACTA) and control mice (Fig. S4E). However, alterations resembling degenerative to necrotizing and diffuse myopathy were gradually detected in older animals. Thus, H&E staining of striated muscle sections prepared from 18-mo-old animals showed that E4f1 KO led to myophagocytosis, hypercontracted fibers, centralized regenerative fibers, immune cell infiltration, and the presence of adipocytes (Fig. S4A). These data suggest that in the long term, E4F1 deficiency results in skeletal muscle disorganization and histological alterations.
Fig. 3.
The PDH transcriptional program controlled by E4F1 is essential to sustain PDH activity in skeletal muscles. (A) ChIP-qPCR experiments performed with anti-E4F1 antibody or an irrelevant control antibody on the promoter of Dlat, Dld, Brp44l/Mpc, Slc25a19, and Pdpr in murine striated skeletal muscles. A gene-poor noncoding region of chromosome 8 (NC1) and the Pdha1 promoter region (TSS) were used as controls. Enrichments are represented as percentages of input (mean value ± SEM, n = 4). (B) Generation of striated skeletal muscle-specific E4f1KO mice. Mice harboring the E4f1 null and flox alleles were intercrossed with Tg(Acta1-Cre) transgenic mice to generate E4f1−/flox; Tg(Acta1-Cre) and E4f1+/flox; Tg(Acta1-Cre) control littermates [E4f1KO(ACTA) and CTL(ACTA), respectively]. (C) Cre-mediated recombination efficiency of the E4f1 flox allele in vivo was assessed by qPCR analysis on genomic DNA prepared from striated muscles, heart, and brain. Cre-mediated recombination in E4f1flox tMEFs was used to normalize (100% efficiency). Histobars represent the mean value ± SEM (n = 3). (D) mRNA levels of Dlat, Dld, Slc25a19, Brp44l/Mpc1, Pdpr and of a control gene (Pdha1) in striated muscles prepared from 16-wk-old E4f1KO(ACTA) and CTL(ACTA) animals. Histobars represent the mean value ± SEM (n = 5) measured by RT-qPCR. (E) Protein levels of E4F1, DLAT, DLD, MPC1/BRP44L, lipoylated proteins, VDAC and Tubulin (loading control) determined by immunoblotting of total cell extracts prepared from the same animals than in D. (F) Immunofluorescence analysis of DLAT (Upper) and lipoylated proteins (Lower) in muscle sections prepared from E4f1KO(ACTA) and CTL(ACTA) animals. (Scale bars, 200 μm.) **P < 0.01; *P < 0.05; ns, not significant.
Fig. S3.
E4F1 regulates the PDC in skeletal muscles. (A) ChIP-qPCR experiments performed with an anti-E4F1 antibody on the promoter region of Dlat and Dld in murine striated skeletal muscles of E4f1KO(ACTA) mice and CTL(ACTA) littermates. A gene-poor noncoding region of chromosome 8 (NC1) and the Pdha1 promoter region (TSS) were used as controls. Enrichments are represented as percentages of input (mean value ± SEM of n = 3 independent experiments). (B) mRNA levels of E4f1, Dlat, Dld, Slc25A19, Brp44l/Mpc1, Pdpr, and of two control genes (CS and Pdha1) determined by RT-qPCR analysis in the heart of E4f1KO(ACTA) mice and CTL(ACTA) littermates. Histobars represent the mean value ± SEM of n = 6 animals per experimental group. (C) PDH activity (arbitrary unit) measured in protein extracts prepared from heart of resting 16 wk-old E4f1KO(ACTA) and CTL(ACTA) animals. Histobars represent the mean value ± SEM of n = 6 animals per experimental group. (D) CS activity (arbitrary unit) measured in protein extracts prepared from gastrocnemius of resting 16-wk-old E4f1KO(ACTA) and CTL(ACTA) animals. Histobars represent the mean value ± SEM of n = 2 animals. *P < 0.05; ns, not significant.
Fig. S4.
E4F1 deficiency does not induce morphological alterations of skeletal muscles in young E4f1KO(ACTA) mice but results in progressive skeletal muscle disorganization and histological alterations during aging. (A) H&E staining of striated muscle sagittal sections prepared from 16-wk-old (Left) or 18-mo-old (Right) E4f1KO(ACTA) males and control littermates. Note that old E4f1KO(ACTA) animals display myophagocytosis, hypercontracted fibers, centralized regenerative fibers, and evidence of replacement of muscular cells by adipocytes. (Scale bars, 200 μm.) (B) Fiber size distribution (cross-sectional area) in skeletal muscles of 16-wk-old E4f1KO(ACTA) and control animals. Histobars represent the mean value ± SEM of cross sectional area of muscular fibers (arbitrary unit) (n = 5 males per experimental group). (C) Spontaneous locomotor activity of 16-wk-old E4f1KO(ACTA) males and control littermates was determined using an automated system measuring infrared beam break. Histobars represent the number of times the animals broke the beam by spontaneous locomotor activity in the cage during 24 h (mean value ± SEM, n = 5 animals per group). (D, Left) Body weight of E4f1KO(ACTA) males and control littermates was measured over a period of 4 mo. (Right) Histobars represent the food intake of 16-wk-old E4f1KO(ACTA) males and control littermates, measured over 96h (mean ± SEM, n = 6 animals per group). (E) mRNA levels of the muscular differentiation markers and inducers Mif6, Mef2c, MyoD, and Myogenin determined by RT-qPCR analysis in adult striated muscles of 16-wk-old E4f1KO(ACTA) and control mice. Histobars represent the mean value ± SEM (n = 5 animals per experimental group). ns, not significant.
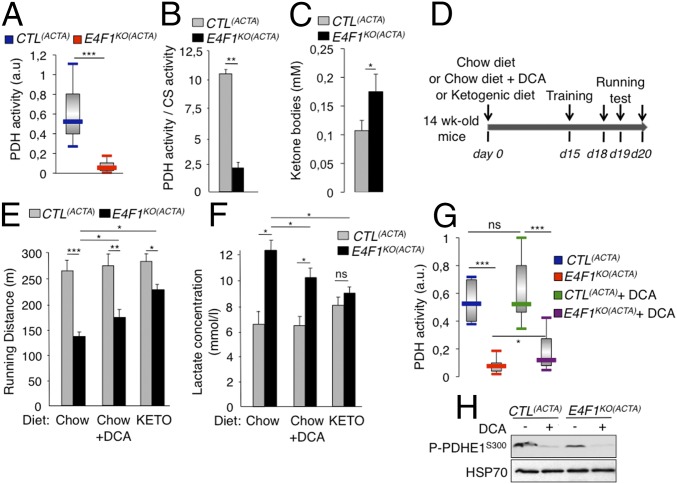
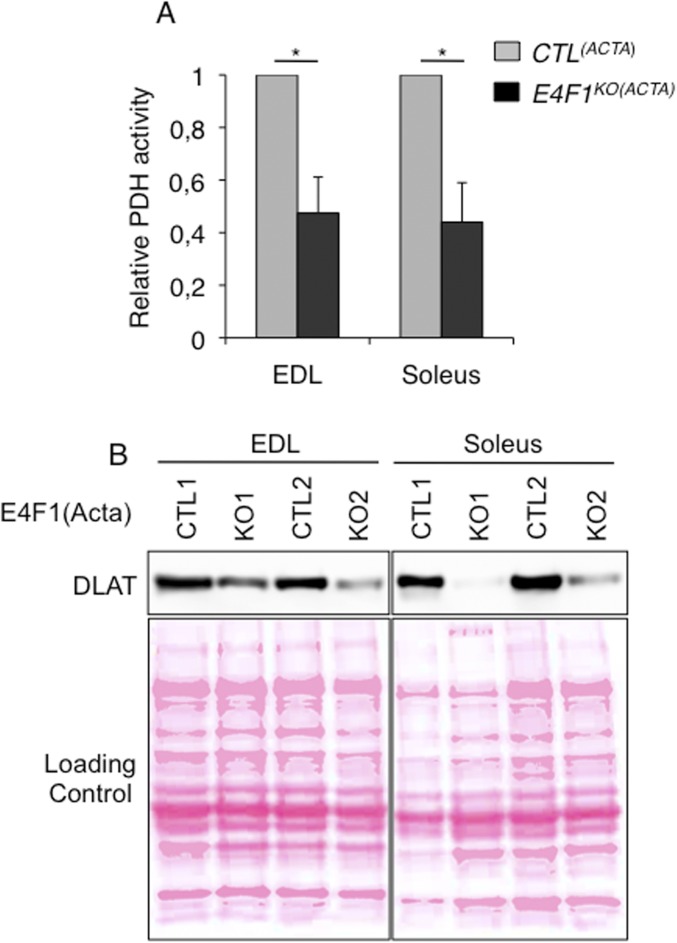
Next, we evaluated the consequences of E4f1 inactivation in vivo on this PDH transcriptional program. In skeletal, but not in cardiac E4f1KO muscles, mRNA, and protein levels of Dlat were strongly altered, whereas the mRNA level of Pdha1, used as control, remained unchanged (Fig. 3 D–F and Fig. S3B). Expression of Brp44l, Slc25a19, and Dld was also slightly decreased at the mRNA level (Fig. 3D), although to a lesser extent than Dlat. As in E4f1KO fibroblasts (tMEFs) in culture, protein lipoylation was also markedly decreased in E4f1KO muscles, as shown by immunoblotting on proteins extracts and immunostaining of tissue sections (Fig. 3 E and F). Impaired expression of these PDC components in E4f1KO muscles resulted in 80–90% reduction of PDH enzymatic activity in gastrocnemius, as measured by two independent methods (Fig. 4 A and B and Fig. S3D). E4f1KO(ACTA) mice also exhibited increased level of circulating ketone bodies, suggesting that E4f1KO muscles activated FAO as in E4f1KO tMEFs (Fig. 4C). Of note, DLAT expression and PDH activity were also strongly down-regulated in extensor digitorum longus and soleus striated muscles isolated from E4f1KO(ACTA) mice, indicating that both red and white muscle fibers are equally affected by E4F1-deficiency (Fig. S5 A and B). Consistent with the absence of depletion of E4f1 mRNA in cardiac tissue in this animal model (Fig. S3B), no significant difference in PDH activity was detected in the heart of E4f1KO(ACTA) mice (Fig. S3C). These data show that the E4F1–PDH connection is critical for the pyruvate-AcCoA metabolic pathway in adult striated skeletal muscles, confirming its biological relevance in vivo.
Fig. 4.
E4f1 inactivation in skeletal muscles results in reduced PDH activity, chronic lactate acidosis and reduced muscular endurance. (A and B) PDH activity measured in protein extracts prepared from gastrocnemius of resting 16-wk-old E4f1KO(ACTA) animals and CTL(ACTA) littermates using two different methods, DipStick Assay (A) or [14C]-Pyruvate oxidation assay (B). CS enzymatic activity that does not vary (Fig. S3D) was used to normalize PDH activity in B. Histobars represent the mean value ± SEM of n = 5 (A) and n = 2 (B) independent experiments. (C) Ketone bodies level in serum of E4f1KO(ACTA) and CTL(ACTA) males. (D) Schematic representation of the experimental design to measure the impact of E4f1 inactivation on physical endurance. A shift from a chow to a ketogenic (KETO) diet or addition of DCA in drinking water was done 2 wk before the first training session. (E) Locomotor performance (running distance before exhaustion) of 16-wk-old E4f1KO(ACTA) and CTL(ACTA) animals under chow or KETO diets, or after administration of DCA, was evaluated using forced treadmill running. Histobars represent the mean value ± SEM of three independent measurements for each animal (n = 8 males per group). (F) Lactate level (serum) of E4f1KO(ACTA) and CTL(ACTA) males under chow or ketogenic diets, or after administration of DCA, measured after running. Histobars represent the mean value ± SEM (n = 8 males per group). (G) PDH activity was measured after running in protein extracts from striated muscles of E4f1KO(ACTA) and CTL(ACTA) males, in the presence or absence of DCA. Histobars represent the mean value ± SEM (n = 5). (H) Phosphorylation level of serine 300 of PDHE1 assessed by immunoblotting of total protein extracts from gastrocnemius of E4F1KO(ACTA) and CTL(ACTA) animals, in the absence or presence of DCA. HSP70 protein was used as loading control. ***P < 0.001; **P < 0.01; *P < 0.05; ns, not significant.
Fig. S5.
E4F1 controls PDC in red and white muscle fibers. (A) PDH activity (arbitrary unit) measured in protein extracts prepared from extensor digitorum longus (EDL) and soleus striated muscles of resting 16 wk-old E4f1KO(ACTA) and CTL(ACTA) animals, as indicated. Histobars represent the mean value ± SEM of n = 5 animals per experimental group. (B) Protein level of DLAT in EDL and soleus muscles was determined by immunoblotting of total protein extracts prepared from two independent E4f1KO(ACTA) and CTL(ACTA) animals. Red ponceau staining of the same membranes was performed to ensure equal loading (Lower). *P < 0.05.
E4f1 Inactivation in Skeletal Muscles Results in Lactate Acidosis and Muscular Endurance Defects.
Although E4f1-deficient muscles display a strong reduction of basal PDH activity, 16-wk-old E4f1KO(ACTA) mice did not show spontaneous locomotor deficiency in normal housing conditions, as quantified by infrared light beam interruption in cages (Fig. S4C). This surprising result indicates that a low muscular PDH activity (10–20% of normal levels) (Fig. 4 A and B) is sufficient to sustain basal locomotor activity and viability. PDH activity has been documented to increase in skeletal muscles during high-intensity exercise and to contribute to muscular endurance (10). Therefore, we hypothesized that the residual PDH activity in muscles of E4f1KO(ACTA) mice might not be sufficient to support the energetic demand that occurs during an acute and high exercise workload. Locomotor activity of control and E4f1KO(ACTA) adult mice was assessed upon forced treadmill running (Fig. 4D). Although PDH activity increased in all animals in this experimental setting, it remained much lower in E4f1KO(ACTA) mice relative to control littermates (Fig. 4G). Accordingly, E4f1KO(ACTA) animals displayed a marked decrease of their physical endurance, as documented by a twofold reduction of their running performance [total running distance (Fig. 4E) and time to exhaustion (Fig. S6A)].
Fig. S6.
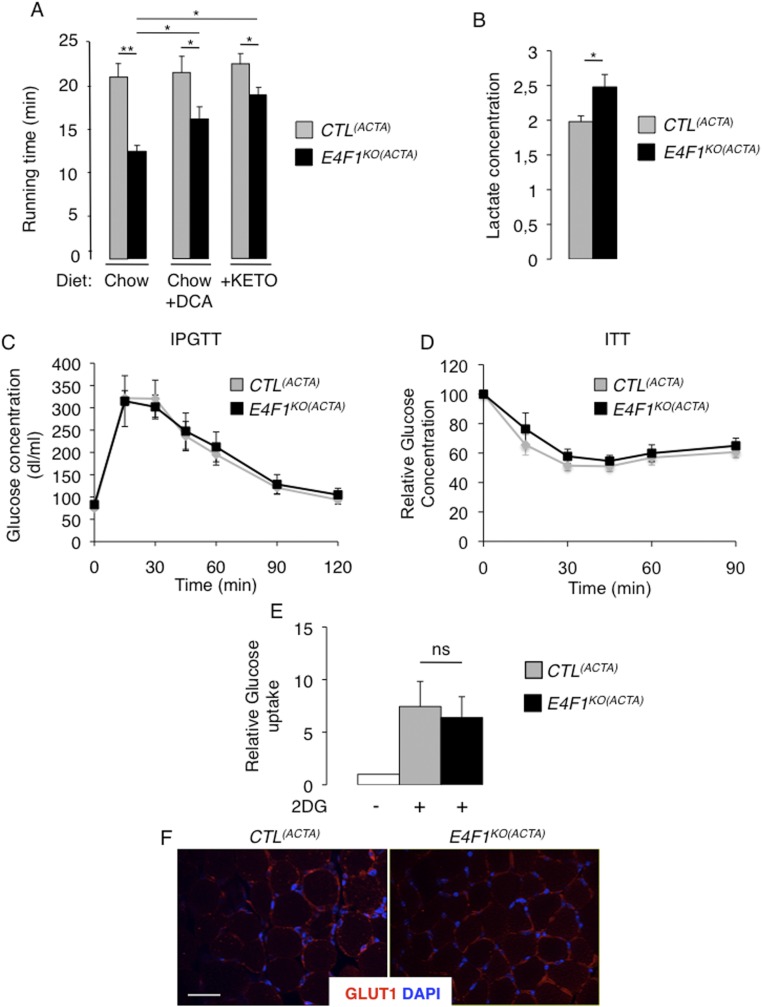
Phenotypic characterization of E4f1KO(ACTA) animals. (A) Locomotor performances (running time before exhaustion) of E4f1KO(ACTA) and CTL(ACTA) males under chow or ketogenic (KETO) diets, or under chow diet in presence of DCA, were evaluated using forced treadmill running. Three independent measurements were performed 1 d apart for each animal. Histobars represent the mean value ± SEM of n = 8 males for each experimental group. (B) Lactate level in the serum of resting 16 wk-old E4f1KO(ACTA) and CTL(ACTA) animals. Histobars represent the mean value ± SEM of n = 6 animals per experimental group. (C and D) Intraperitoneal glucose tolerance test (IPGTT) (C) and ITT (D), performed on E4f1KO(ACTA) and control littermates (mean ± SEM, n = 5 males per group). (E) Relative glucose uptake was measured in gastrocnemius of E4f1KO(ACTA) and CTL(ACTA) males after an acute exercise workload. Histobars represent the mean value ± SEM of n = 4 males for each experimental group. (F) GLUT1 immunofluorescence performed on striated muscle (gastrocnemius) tissue sections prepared from E4f1KO(ACTA) and CTL(ACTA) animals. Sections were counterstained with DAPI. (Scale bar, 200 μm.) **P < 0.01; *P < 0.05; ns, not significant.
Because PDH deficiency results in chronic lactic acidemia in patients, we measured lactate levels in the serum of E4f1KO(ACTA) and control mice. E4f1KO(ACTA) animals exhibited increased lactate levels relative to controls under normal housing conditions and regular chow diet (Fig. S6B). Lactic acidemia was further exacerbated upon acute exercise (Fig. 4F). Importantly, 16-wk-old E4f1KO(ACTA) mice showed no apparent alterations of glucose homeostasis, as assessed by insulin- and glucose-tolerance tests (Fig. S6 C and D), glucose uptake, and expression of the glucose transporter GLUT1 (Fig. S6 E and F). Collectively, our data show that E4f1KO(ACTA) mice display phenotypes that recapitulate some clinical symptoms observed in PDC-deficient patients, including lactic acidemia and exercise intolerance (11, 30).
Muscular defects of E4f1KO(ACTA) Mice Are Rescued upon Pharmacological Reactivation of PDH or Under a Ketogenic Diet.
We next evaluated in E4f1KO(ACTA) mice the impact of DCA treatment and of a ketogenic diet, two therapies that are included in the standard care of many PDC-deficient patients. E4f1KO(ACTA) and control animals were treated for 2 wk with the PDK-inhibitor DCA before evaluating their endurance capacity (Fig. 4D). This treatment reduced the inhibitory phosphorylation of the PDHE1 subunit on serine 300 (Fig. 4H), resulting in a moderate but significant increase of muscular PDH activity in E4f1KO(ACTA) mice (Fig. 4G). This finding suggested that the residual pool of PDC in E4f1KO muscles was amenable to DCA treatment. Remarkably, as in PDC-deficient patients, this treatment improved the physical endurance and reduced the lactic acidemia of E4f1KO(ACTA) mice exposed to intense treadmill exercise (Fig. 4 E and F and Fig. S6A).
Ketogenic diet provides an alternative source of energy and AcCoA through increased FAO that partly improve physical fitness and lactate acidemia in PDC-deficient patients. Importantly, its efficacy was confirmed in zebrafish models with mutations in Dlat or Pdhb (31, 32). Accordingly, feeding E4f1KO(ACTA) mice with a ketogenic diet (Fig. 4D) significantly improved their running capacity and normalized their blood lactate level (Figs. 4 E and F and Fig. S6A). Of note, beneficial effects of these two treatments on E4f1KO(ACTA) mice were clear but remained partial, a situation also observed in PDC-deficient patients (14). Collectively, these rescue experiments confirm that the muscular phenotypes observed in E4F1-deficient animals result from PDH deficiency (Fig. 5).
Fig. 5.
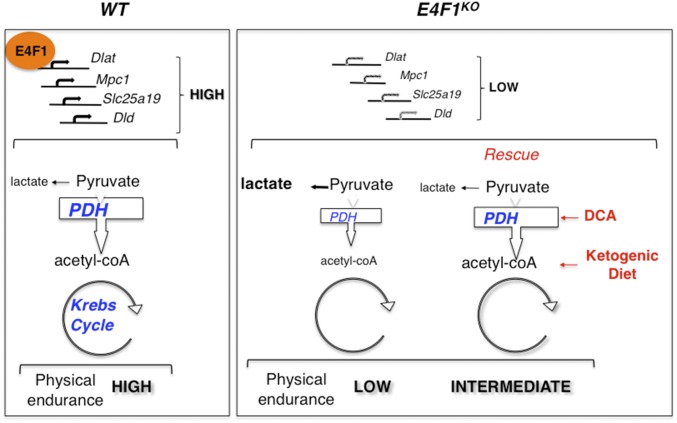
Schematic representation of E4F1-mediated effects on pyruvate metabolism. E4F1 protein controls transcription of a set of genes encoding core components or regulators of the PDC. E4F1 deficiency results in impaired PDH activity and redirection of the glycolytic flux toward lactate production. Mice with E4f1-deficient skeletal muscles exhibit muscular endurance defects and chronic lactate acidemia. Like in PDC-deficient patients, these defects are partly improved by ketogenic diet (KETO) or by treatment with a PDH activator, DCA.
Discussion
Here, we describe a level of regulation of PDH in mammals implicating the E4F1 transcription factor. To characterize its role in PDH-regulation, we developed a skeletal muscle-specific E4f1 KO mouse model, the phenotypes of which recapitulate some clinical symptoms observed in PDC-deficient patients.
In mammals, the control of PDC activity has been mainly attributed to posttranslational modifications of its subunits by PDKs and PDPs. Its control at the transcriptional level has been far less investigated and mainly concerns the regulation of individual Pdk genes by multiple transcription factors, including Foxo1/3a (33), Hif1a (34), or p53 (35). Nevertheless, in more primitive organisms, it was demonstrated that genes coding for other regulators, but also for core PDC components, are coregulated and organized in regulons or operons. Thus, in E. coli, the aceE/E1, the aceF/E2, and IpdA/E3 genes form a single operon that also includes the transcriptional regulator of this operon PdpR (7). In C. albicans, the Gal4p transcriptional regulator controls the expression of the five main components of the PDC complex, including the Pda1/E1, Pdb1/E1, Dlat1/E2, Lpd1/E3, and Pdx1 subunits (8). Our data reveal that such a coordinated transcriptional program, important for PDH-mediated pyruvate oxidation, also exists in mammals. Composed of at least four genes—Dlat/E2, Dld/E3, Brp44l/MPC1, and Slc25a19—this program is controlled by E4F1, a sequence-specific transcription factor bound nearby the TSS of these genes.
This E4F1-controlled transcription program is a main contributor of the total PDH activity, as demonstrated by the impact of conditional gene targeting of E4f1 in proliferating cells and in postmitotic differentiated muscular cells that resulted in a 80–90% reduction of the basal PDH activity. Surprisingly, we show that, despite their very weak muscular PDH activity, animals lacking E4F1 in their striated muscles were viable and displayed normal basal locomotor activity, at least in normal housing conditions. Nevertheless, these animals exhibited lactic acidemia and severe exercise intolerance that were partly rescued by the pharmacological reactivation of the remaining pool of PDC by DCA, or by shunting the need for PDH activity by promoting FAO using a ketogenic diet. Our histological analyses indicate that although E4f1 inactivation did not result in major disorganization of this tissue in young animals, long-term PDH-deficiency led to a degenerative muscular myopathy in older animals. So far, such clinical symptoms have not been described in PDC patients, likely because most of these patients do not live long enough to develop myopathies. On the other hand, it is commonly described that these patients often exhibit epileptic seizures and microcephalies. These symptoms were not observed in our muscle-specific E4f1KO mice, despite these animals displayed chronic lactic acidemia. This finding questions the origin of the neurological manifestations observed in PDC patients and suggests that the latter symptoms do not result solely from chronic systemic lactic acidemia, but could also arise from multiple brain-specific metabolic alterations. Tissue-specific inactivation of E4f1 in the central nervous system may provide a definitive answer to this clinically relevant question.
Strikingly, recent genetic studies designed to identify new mutated genes involved in unsolved cases of primary mitochondrial human disorders, led to the identification of a homozygous nonsynonymous mutation in the E4F1 gene of a patient showing reduced PDH complex activity, muscular defects, and lactate acidemia (36). This first indication that the E4F1-controlled program could be deregulated in a pathological situation provides an exciting clinical perspective to the present work. Indeed, our E4f1KO(ACTA) animal models display phenotypes that recapitulate some clinical symptoms observed in this PDC-deficient patient. Thus, these animals could represent potential models for preclinical studies aiming at testing new therapeutic strategies to improve the consequences of PDH deficiency.
Furthermore, it should be noted that missense mutations in the E4F1-target genes Dlat, Dld, Brp44l, and Slc25a19 have been identified in several congenital metabolic disorders associated with reduced PDH activity and alteration of the pyruvate oxidation pathway. These disorders include PDC deficiency, lipoamide dehydrogenase deficiency, or Amish lethal microcephaly syndromes (3, 11, 13). It is also worth noting that complete KO mouse models for Pdha1, Dld, Slc25a19, as well as for E4f1, all show severe developmental defects and lethality during early embryonic development (4, 22, 37–39). This finding raises interesting questions about the importance of the E4F1-controlled PDH-program during embryogenesis and beyond, about the poorly characterized metabolic rewiring of the pyruvate pathway that may occur during development.
Altogether, our data highlight the role of E4F1 in PDH-dependent metabolic homeostasis and pave the way for new studies on the physiological rewiring of the pyruvate pathway. This work should also stimulate new research aiming at exploring the role of nuclear transcription factors in unsolved cases of mitochondrial diseases.
Experimental Procedures
Accession Numbers.
The full series of data, including expression arrays and ChIP-Seq data reported in this paper were deposited on the Gene Expression Omnibus (GEO) dataset repository (see GEO SuperSeries GSE57242 and GSE57221) (24, 26). E4F1 binding regions were defined by combining bioinformatic toolboxes provided by CisGenome and Qeseq software systems. Detailed protocols, bioinformatic tools and primers used for ChIp-seq and ChIP-qPCR validations were as previously described (24, 26) and detailed in SI Experimental Procedures.
Mouse Models and Experimental Treatment.
E4f1−/+, E4f1+/flox, and E4f1−/flox mice (22, 40) were intercrossed with ActaCre mice to obtain E4f1+/flox; ActaCre and E4f1−/flox; ActaCre compound mice on a mixed 129Sv/J/DBA/C57BL/6 background. Phenotypic characterization of compound E4f1KO(ACTA) and CTL(ACTA) mice was performed on 16-wk-old or 18-mo-old littermates. Mice were housed in a pathogen-free barrier facility in accordance with the ethic Committee for Animal Welfare (Comité d'Ethique en Experimenttion Animal - Languedoc Rossillon, CEEA-LR-12116). Mice were maintained under chow diet (A03, Safe; i.e., 22 kcal% protein, 65 kcal% carbohydrate, and 13 kcal% fat) in the presence or absence of DCA in the drinking water (added to the drinking water for 2 wk at a final concentration of 2 g/L), or were fed with a ketogenic diet (F3666, Bio-Serv; 5kcal% protein, 2kcal% carbohydrate, and 93kcal% fat) for 2 wk before assessing their physical performances, as detailed in SI Experimental Procedures.
PDH Activity and Lactate Measurements, Metabolomics.
Two different protocols were used in parallel to measure PDH activity in protein extracts prepared from cells or muscles (gastrocnemius and heart). PDH activity DipStick Assay Kit (ab109882, Abcam) was used on cell (25 μg) or muscle (5 µg) extracts prepared according to the manufacturer’s protocol, and quantified by ImageJ. PDH enzymatic activity was also assessed by measuring the release of 14CO2 after incubation of protein extracts (1 mg of protein per milliliter) with [1-14C] pyruvate, as previously described and detailed in SI Experimental Procedures (41). To determine the concentration of glucose, 2/3PG, pyruvate, citrate, and succinate in E4f1 WT and KO tMEFs, extracts were prepared from 2 × 107 cells and analyzed by GC/MS and LC/MS/MS platforms (Metabolon). Eight independent samples were analyzed for each cell lines. Lactate production by cells was measured in culture medium using a l-Lactate Assay Kit (Eton Bioscience). Lactate and ketone bodies concentration in blood were measured using a lactometer (EKF Diagnostics) and β-ketone strips (Optium, Abbott), respectively.
Statistical Analysis.
Unpaired Student’s t test was used in all analyses. Statistical significance was expressed as follows: *P < 0.05, **P < 0.01, ***P < 0.001.
Supplemental Materials and Methods.
Experimental procedures relative to genotyping of animal models, generation of cells, RT-qPCR analyses, siRNA-mediated E4f1-depletion, immunoblotting, immunohistochemistry, and immunofluorescence assays, measurement of FAO, and stable isotope tracing, insulin- and glucose-tolerance tests, and in vivo glucose uptake, are described in SI Experimental Procedures.
SI Experimental Procedures
Genotyping of Mouse Models.
E4f1KO(ACTA) and CTL(ACTA) mice were genotyped by PCR on tail genomic DNA using Red-N extract kit (Sigma), with the following primers:
E4f1 WT (E4f1+) and conditional KO flox alleles (E4f1flox): Fwd, 5′-CCTTGAGCACGGAGGAGAGC-3′ and Rev, 5′-GCCCTAGCCTGCTCTGCCATC-3′.
E4f1 null allele (E4f1−): Fwd, 5′-CACTGCCTTGGAGGACTTTG-3′ and Rev, 5′-CCTCTGTTCCACATACACTTCATTC-3′. Acta1-Cre allele: Fwd, 5′-GCGGTCTGGCAGTAAAAACTATC-3′ and Rev, 5′-GTGAAACAGCATTGCTGTCACTT-3′.
Endurance Test.
Normal activity (average daily distance) was gauged for 24 h in cages equipped with infrared beams in the x and y dimensions. For the running test, mice were first trained to run on a treadmill (Colombus Instruments) for 10 min at slow speed (10 m/min). Seventy-two hours later, their running capacity was measured by gradually increasing speed (every 5 min) from 5 m/min to 25 m/min until exhaustion.
Insulin and Glucose Tolerance Tests.
For insulin resistance (ITT) and glucose tolerance (GTT) tests, 16-wk-old males were fasted 4 h or overnight, and injected intraperitoneally with 0.75 U/kg of insulin or 2 g/kg of glucose, respectively. Glucose concentration was measured with a blood glucometer (Accu-Chek, Roche).
Generation of E4f1KO Cells and siRNA-Mediated Depletion.
HaRasV12-transformed E4f1−/flox and E4f1+/flox tMEFs were generated and grown as previously described (24). Conditional inactivation of the E4f1 flox allele was achieved by transduction of tMEFS with retroviral particles encoding a self-excising Cre recombinase (42) produced in Platinum-E retroviral packaging cell lines (Cell Biolabs). For siRNA-mediated depletion of E4f1, tMEFs were transfected with 50 nM of siRNA (Sigma Mission siRNA 00060727 and 00060728) using Lipofectamine RNAimax according to the manufacturer’s recommendations.
Quantitative ChIP Assays.
Detailed protocols used for ChIP, as well as Bioinformatic tools and parameters used to treat ChIP-seq data and annotate E4F1 bound regions were previously described (24, 26). Briefly, trypsinized cells (3 × 107) or gastrocnemius muscle (100 mg) were cross-linked (1% formaldehyde), nuclei were isolated, and chromatin was extracted and sonicated (vibralcell, bioblock). E4F1 ChIPs were carried out with affinity-purified rabbit anti-E4F1 polyclonal antibody (21) and pulled down by Dynabeads coupled to protein G. Input and immunoprecipitated DNA were decross-linked, treated with RNAseA and proteinase K, and purified by phenol-chloroform-isomalylic-alcohol extraction/precipitation and then chromatography (nucleospin extract II columns, Macherey-Nagel). Immunoprecipitated DNAs were analyzed by qPCR (MXPro stratagene and SYBRGreen mix) with promoter-specific primers (see below) or processed for deep-sequencing (Hi-SEq. 2000, Illumina) analyzed using the mm9 mouse genome sequence. ChIP-qPCR were performed as previously described (24) using a validated anti-E4F1 rabbit polyclonal antibody (21) and the following primers:
Dlat: Fwd, 5′-ACAGACGCGCCACATTACTGC and Rev, 5′-GCTGCTCTTGGAGAGGTCACT;
Dld: Fwd, 5′TACACACGACTCCAGCTCTGCAT and Rev, 5′-ATAAGTCTTACCAGGCGTTCAGCG;
Brp44l: Fwd, 5′ ACACTTCTGGAGACTGAGGCTCTT and Rev, 5′- ACAGAGACGGTGAGATCCTGCAAA;
Pdpr: Fwd, 5′ TCTGAGGCTCCAGTGAACAATGCT and Rev, 5′- TAAGGCCTTTCAGTGCTTGGCTTG;
Slc25a19: Fwd, 5′ TGAAGTCTGCGCGGCTATGGAATA and Rev, 5′- TAATACCAGGCTTCCCGCCATCTT;
NC1 (gene-poor non coding region of chromosome 8): Fwd, 5′- AAGGGGCCTCTGCTTAAAAA and Rev, 5′- AGAGCTCCATGGCAGGTAGA;
Pdha1: Fwd, 5′- AGGAACATGTGGCCGTCCATTA and Rev, 5′- TTCACCACTTCTTCGCTGGTCTGT.
14C-Pyruvate Assay to Assess PDH Activity in Protein Extract.
Briefly, we measured the release of 14CO2 after incubation of protein extracts [1 mg of protein per milliliter, prepared by sonication in ice-cold homogenizing buffer (100 mM phosphate buffer, 2 mM EDTA, 1 mM DTT)] with 0.2 mM [1-14C] pyruvate in the presence of 20 mM MgCl2, 0.5 mM CaCl2, 0.03 mM cytochrome C, 480 U/L cytochrome C reductase, 5 mM l-carnitine. CS activity was measured in parallel in the same extracts and used as a reference to normalize PDH activities (41).
RNA Extraction and RT-qPCR.
mRNA expression was evaluated in tMEFs and muscles by RT-qPCR. Cells or muscles were lysed in TRIZOL reagent (Invitrogen), and total RNAs were isolated according to the manufacturer’s recommendations and loaded on eukaryote total RNA 6000 nano chips (Agilent) to verify quality. cDNAs were synthesized from 1 µg of total RNA using random hexamers and SuperScript III Reverse transcription (Invitrogen). Real-time qPCR was performed on a LightCycler 480SW 1.5 apparatus (Roche) with Platinium Taq DNA polymerase (Invitrogen) and an SYBR Green mix containing 3 mM MgCl2, and dNTPs 30 µM each; 45 cycles of 95 °C for 4 s, 65 °C for 10 s, and 72 °C for 30 s. Rpl13a transcripts were used for normalization. Primers sequences were as follows:
E4f1: Fwd, 5′-CTCAAGGCCCACATGGTAA and Rev, 5′-CACACTTGGCACATTTGTAGG;
Dlat: Fwd, 5′- TTGGCCTGTCTGAAAGTTCC and Rev, 5′-TTACCCTCTCTCGCTTTGGA;
Dld: Fwd, 5′-TGGAGTCGTGTGTACCGCTCCTT and Rev, 5′-GGAACTGAAGAAACTCCTTGTAGGCCA;
Slc25a19: Fwd, 5′-TCAGTGTCAGGATTTGTCACCCGT and Rev, 5′- AGAATGCTCTTGGGCCTTCCTCTT;
Brp44L: Fwd, 5′- TCCAGAGATTATCAGTGGGCGGAT and Rev, 5′-GCCAGTTTCGAGGTTGTACCTTGT;
Pdpr: Fwd, 5′- GAACAAGAAACAGGGATCCAAAC and Rev, 5′-CTTGAGTTGATACGCTTCAGAGA;
Pdha1: Fwd, 5′- GCTGGTTGCTTCCCGTAAT and Rev, 5′-TAGTACTTGAGCCCATCCTCTC
CS: Fwd, 5′- TCTACTCACTGCAGCAACC and Rev, 5′-TGGAAGAAGCACTGGCAT.
Protein Extraction, Immunoprecipitation, and Western Blotting.
Total protein extracts were prepared by lysing cells or muscles extracts in Triton X-100 lysis buffer (50 mM Tris⋅HCl, pH 7.4, 100 mM NaCl, 50 mM NaF, 5 mM EDTA, 40 mM β-glycerophosphate, 1 mM Na orthovanadate, 10−4 M PMSF, 10−6 M leupeptin, 10−6 M pepstatin A, and 1% Triton X-100). Protein extracts were separated by SDS/PAGE and transferred to nitrocellulose membranes, blocked in TBS containing 5% (wt/vol) nonfat milk for 1 h at room temperature and incubated overnight at 4 °C with primary antibodies. The following antibodies were used: anti-E4F1 (2), DLAT (sc32925, Santa Cruz), DLD (sc135027, Santa Cruz), BRP44L/MPC1 (HPA045119, Sigma), lipoic acid (ab58724, Abcam and 437695, Calbiochem), VDAC1 (ab14734, Abcam), Phospho (Ser300) PDHE1-A type I (ABS 204, Millipore), HSP70 (MA3-028, Thermo Fisher Scientific), DLST (ab95946, Abcam), and Tubulin (T9026, Sigma).
Immunostainings and Muscle Fiber Size Distribution.
Muscle biopsies were fixed in 4% (vol/vol) neutral-buffered formalin (VWR Chemicals) for 24 h and paraffin-embedded tissues were sectioned (4 μm) and processed for immunostainings or for H&E staining. Immunostainings were performed on muscle sections and tMEFs using anti-lipoic acid (437695, Calbiochem), anti-DLAT (H-160, Santa Cruz), and glucose transporter GLUT1 (ab15309, Abcam) antibodies in PBS supplemented with 1.5% (wt/vol) BSA (Roche). Alexa488-conjugated secondary anti-rabbit IgG antibody (Thermofischer) was incubated for 2 h at room temperature. Cover glasses were mounted with Mowiol (Biovalley). To measure the size of muscle fibers, gastrocnemius muscles were frozen in Tissue-Tek OCT (Sakura), cryosections (10 μm) were stained with Azarubin dye, and fiber size and banding were analyzed and quantified (ImageJ software) by high-resolution light microscopy, as previously described (43). For immunofluorescence analyses, tMEFs were fixed with methanol (Sigma) at −20 °C for 5 min.
FAO.
FAO was measured in triplicates by quantifying the production of 3H2O from [9,10-3H]palmitate. Briefly, the cells were trypsinized, counted, plated at 5 × 104 cells per well in 12-well culture plates, and allowed to grow for 2 d. Cultured cells were washed three times with Dulbecco’s PBS. Then, 200 μL of [9,10(n)-3H]palmitic acid (60 Ci/mmol; NEN) bound to fatty-acid–free albumin (final concentration 125 μM) containing 1 mM carnitine was added per well. After 2-h incubation at 37 °C, the mixture was removed and added to a tube containing 200 μL of cold 10% (vol/vol) TCA. The tubes were then centrifuged for 10 min at 2,200 × g at 4 °C, and aliquots of supernatants (350 μL) were removed, mixed with 55 μL of 6 M NaOH, and applied to ion-exchange resin (Dowex). The columns were washed twice with 750 μL of water, and the eluates were counted. The results of FAO were normalized according to the total protein content that was determined by the BCA method (Pierce).
Annexin Staining.
tMEFs were incubated with the FAO-inhibitor etomoxir (100 μM final, 24 h) before staining with Annexin-V-FITC (Invitrogen) and propidium iodide and then analyzed by flow-cytometry (FACSCalibur; BD).
In Vivo Glucose Uptake.
Glucose uptake was assessed using the Glucose uptake-Glo assay kit (Promega). Briefly, E4f1KO(ACTA) and control littermates were injected intraperitoneally with 100 mg/kg of unlabeled 2-deoxy-glucose (2DG) and then subjected to an acute exercise (10 min). Skeletal muscles (gastrocnemius) of these animals were harvested as fast as possible and snap frozen in liquid nitrogen. Tissues were lysed in PBS/0.2 N NaOH/0.5% DTAB and 2DGG6P was measured according to the manufacturer’s instructions.
Stable Isotope Tracing Experiments.
E4f1KO tMEFs and match control cells were incubated in DMEM without glucose (01-056-01, Clinisciences) complemented with 4.5 g/L 13C-d-glucose (CLM-1396–1, Euriso-Top) for 30 min, 6 or 24 h. Intracellular metabolites were sampled according to Martano et al. (44). Central metabolites were extracted at −20 °C with 8 mL of acetonitrile/methanol/water+0.1% of formic acid (2:2:1), except for AcCoA that was extracted at −20 °C with 12 mL of a solution containing 125 mM formic acid in 80% (vol/vol) methanol at pH 2.9. Samples were evaporated in a Rotavap RII (Buchi), and resuspended in 100 µL of water for analysis of central metabolites, or in 100 µL of a solution of 25 mM ammonium formate with 2% (vol/vol) of methanol for analysis of AcCoA. Debris were removed by centrifugation at 10,000 × g for 5 min and samples were then analyzed by LC-MS. Carbon isotopolog distributions were expressed relative to the sum of all analyzed isotopologs.
Acknowledgments
This work was supported by grants from the Ligue Nationale Contre le Cancer (LNCC, équipes labelisées 2011 (to C.S.) and 2016 (to L.L.C.); Agence Nationale pour la Recherche Grants SVSE2-YinE4F1Yang2 and MetaboCycle2 (to C.S. and L.L.C.); grants from the Fondation ARC and from the Canceropole Grand Sud Ouest (to G.R.); institutional support from INSERM (L.L.C. and C.S.), the Site de Recherche Intégrée sur le Cancer Montpellier Cancer Grant INCa-DGOS-Inserm 6045 (to C.S.) and CNRS (C.S.); fellowships from LNCC (to T.H. and M.L.); fellowships financed on the Agence Nationale pour la Recherche Grants JCJC-0014-01 and SVSE2-YinE4F1Yang2 (to O.K. and H.D.); LabEx EpiGenMed ANR-10-LABX-12-01 (to B.S.); and the technical support of Montpellier Rio Imaging (MRI), Genomics (MGX), Metamus and Animal (RAM) facilities.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602754113/-/DCSupplemental.
References
- 1.Patel MS, Nemeria NS, Furey W, Jordan F. The pyruvate dehydrogenase complexes: Structure-based function and regulation. J Biol Chem. 2014;289(24):16615–16623. doi: 10.1074/jbc.R114.563148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herzig S, et al. Identification and functional expression of the mitochondrial pyruvate carrier. Science. 2012;337(6090):93–96. doi: 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- 3.Bricker DK, et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science. 2012;337(6090):96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindhurst MJ, et al. Knockout of Slc25a19 causes mitochondrial thiamine pyrophosphate depletion, embryonic lethality, CNS malformations, and anemia. Proc Natl Acad Sci USA. 2006;103(43):15927–15932. doi: 10.1073/pnas.0607661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel MS, Korotchkina LG. Regulation of the pyruvate dehydrogenase complex. Biochem Soc Trans. 2006;34(Pt 2):217–222. doi: 10.1042/BST20060217. [DOI] [PubMed] [Google Scholar]
- 6.Mathias RA, et al. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159(7):1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogasawara H, Ishida Y, Yamada K, Yamamoto K, Ishihama A. PdhR (pyruvate dehydrogenase complex regulator) controls the respiratory electron transport system in Escherichia coli. J Bacteriol. 2007;189(15):5534–5541. doi: 10.1128/JB.00229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Askew C, et al. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog. 2009;5(10):e1000612. doi: 10.1371/journal.ppat.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajagopalan KN, et al. Metabolic plasticity maintains proliferation in pyruvate dehydrogenase deficient cells. Cancer Metab. 2015;3:7. doi: 10.1186/s40170-015-0134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters SJ. Regulation of PDH activity and isoform expression: Diet and exercise. Biochem Soc Trans. 2003;31(Pt 6):1274–1280. doi: 10.1042/bst0311274. [DOI] [PubMed] [Google Scholar]
- 11.Patel KP, O’Brien TW, Subramony SH, Shuster J, Stacpoole PW. The spectrum of pyruvate dehydrogenase complex deficiency: Clinical, biochemical and genetic features in 371 patients. Mol Genet Metab. 2012;106(3):385–394. doi: 10.1016/j.ymgme.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maj MC, et al. Pyruvate dehydrogenase phosphatase deficiency: identification of the first mutation in two brothers and restoration of activity by protein complementation. J Clin Endocrinol Metab. 2005;90(7):4101–4107. doi: 10.1210/jc.2005-0123. [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg MJ, et al. Mutant deoxynucleotide carrier is associated with congenital microcephaly. Nat Genet. 2002;32(1):175–179. doi: 10.1038/ng948. [DOI] [PubMed] [Google Scholar]
- 14.Abdelmalak M, et al. Long-term safety of dichloroacetate in congenital lactic acidosis. Mol Genet Metab. 2013;109(2):139–143. doi: 10.1016/j.ymgme.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferriero R, et al. Phenylbutyrate therapy for pyruvate dehydrogenase complex deficiency and lactic acidosis. Sci Transl Med. 2013;5(175):175ra31. doi: 10.1126/scitranslmed.3004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raychaudhuri P, Rooney R, Nevins JR. Identification of an E1A-inducible cellular factor that interacts with regulatory sequences within the adenovirus E4 promoter. EMBO J. 1987;6(13):4073–4081. doi: 10.1002/j.1460-2075.1987.tb02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rizos H, et al. Association of p14ARF with the p120E4F transcriptional repressor enhances cell cycle inhibition. J Biol Chem. 2003;278(7):4981–4989. doi: 10.1074/jbc.M210978200. [DOI] [PubMed] [Google Scholar]
- 18.Le Cam L, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127(4):775–788. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 19.Fenton SL, et al. Identification of the E1A-regulated transcription factor p120 E4F as an interacting partner of the RASSF1A candidate tumor suppressor gene. Cancer Res. 2004;64(1):102–107. doi: 10.1158/0008-5472.can-03-2622. [DOI] [PubMed] [Google Scholar]
- 20.Sandy P, et al. p53 is involved in the p120E4F-mediated growth arrest. Oncogene. 2000;19(2):188–199. doi: 10.1038/sj.onc.1203250. [DOI] [PubMed] [Google Scholar]
- 21.Fajas L, et al. pRB binds to and modulates the transrepressing activity of the E1A-regulated transcription factor p120E4F. Proc Natl Acad Sci USA. 2000;97(14):7738–7743. doi: 10.1073/pnas.130198397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Cam L, Lacroix M, Ciemerych MA, Sardet C, Sicinski P. The E4F protein is required for mitotic progression during embryonic cell cycles. Mol Cell Biol. 2004;24(14):6467–6475. doi: 10.1128/MCB.24.14.6467-6475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatchi E, et al. E4F1 deficiency results in oxidative stress-mediated cell death of leukemic cells. J Exp Med. 2011;208(7):1403–1417. doi: 10.1084/jem.20101995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodier G, et al. The transcription factor E4F1 coordinates CHK1-dependent checkpoint and mitochondrial functions. Cell Reports. 2015;11(2):220–233. doi: 10.1016/j.celrep.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 25.Grote D, et al. E4F1 is a master regulator of CHK1-mediated functions. Cell Reports. 2015;11(2):210–219. doi: 10.1016/j.celrep.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 26.Houlès T, Rodier G, Le Cam L, Sardet C, Kirsh O. Description of an optimized ChIP-seq analysis pipeline dedicated to genome wide identification of E4F1 binding sites in primary and transformed MEFs. Genom Data. 2015;5:368–370. doi: 10.1016/j.gdata.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan J, Lawson JE, Reed LJ. Role of the regulatory subunit of bovine pyruvate dehydrogenase phosphatase. Proc Natl Acad Sci USA. 1996;93(10):4953–4956. doi: 10.1073/pnas.93.10.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawson JE, Niu XD, Reed LJ. Functional analysis of the domains of dihydrolipoamide acetyltransferase from Saccharomyces cerevisiae. Biochemistry. 2001;30(47):11249–11254. doi: 10.1021/bi00111a009. [DOI] [PubMed] [Google Scholar]
- 29.Miniou P, et al. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res. 1999;27(19):e27. doi: 10.1093/nar/27.19.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cameron JM, et al. Identification of a canine model of pyruvate dehydrogenase phosphatase 1 deficiency. Mol Genet Metab. 2007;90(1):15–23. doi: 10.1016/j.ymgme.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Taylor MR, Hurley JB, Van Epps HA, Brockerhoff SE. A zebrafish model for pyruvate dehydrogenase deficiency: Rescue of neurological dysfunction and embryonic lethality using a ketogenic diet. Proc Natl Acad Sci USA. 2004;101(13):4584–4589. doi: 10.1073/pnas.0307074101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurer CM, Schönthaler HB, Mueller KP, Neuhauss SCF. Distinct retinal deficits in a zebrafish pyruvate dehydrogenase-deficient mutant. J Neurosci. 2010;30(36):11962–11972. doi: 10.1523/JNEUROSCI.2848-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon H-S, Huang B, Unterman TG, Harris RA. Protein kinase B-alpha inhibits human pyruvate dehydrogenase kinase-4 gene induction by dexamethasone through inactivation of FOXO transcription factors. Diabetes. 2004;53(4):899–910. doi: 10.2337/diabetes.53.4.899. [DOI] [PubMed] [Google Scholar]
- 34.Kim J-W, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3(3):177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Contractor T, Harris CR. p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res. 2012;72(2):560–567. doi: 10.1158/0008-5472.CAN-11-1215. [DOI] [PubMed] [Google Scholar]
- 36.Legati A, et al. New genes and pathomechanisms in mitochondrial disorders unraveled by NGS technologies. Biochim Biophys Acta. 2016;1857(8):1326–1335. doi: 10.1016/j.bbabio.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 37.Johnson MT, et al. Inactivation of the murine pyruvate dehydrogenase (Pdha1) gene and its effect on early embryonic development. Mol Genet Metab. 2001;74(3):293–302. doi: 10.1006/mgme.2001.3249. [DOI] [PubMed] [Google Scholar]
- 38.Johnson MT, Yang HS, Magnuson T, Patel MS. Targeted disruption of the murine dihydrolipoamide dehydrogenase gene (Dld) results in perigastrulation lethality. Proc Natl Acad Sci USA. 1997;94(26):14512–14517. doi: 10.1073/pnas.94.26.14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pliss L, et al. Cerebral developmental abnormalities in a mouse with systemic pyruvate dehydrogenase deficiency. PLoS One. 2013;8(6):e67473. doi: 10.1371/journal.pone.0067473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lacroix M, et al. Transcription factor E4F1 is essential for epidermal stem cell maintenance and skin homeostasis. Proc Natl Acad Sci USA. 2010;107(49):21076–21081. doi: 10.1073/pnas.1010167107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brivet M, et al. Impaired mitochondrial pyruvate importation in a patient and a fetus at risk. Mol Genet Metab. 2003;78(3):186–192. doi: 10.1016/s1096-7192(03)00016-7. [DOI] [PubMed] [Google Scholar]
- 42.Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8(1):233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 43.Pessemesse L, et al. Depletion of the p43 mitochondrial T3 receptor in mice affects skeletal muscle development and activity. FASEB J. 2012;26(2):748–756. doi: 10.1096/fj.11-195933. [DOI] [PubMed] [Google Scholar]
- 44.Martano G, et al. Fast sampling method for mammalian cell metabolic analyses using liquid chromatography-mass spectrometry. Nat Protoc. 2015;10(1):1–11. doi: 10.1038/nprot.2014.198. [DOI] [PubMed] [Google Scholar]