Abstract
Mitochondrial gene expression uses a non‐universal genetic code in mammals. Besides reading the conventional AUG codon, mitochondrial (mt‐)tRNAM et mediates incorporation of methionine on AUA and AUU codons during translation initiation and on AUA codons during elongation. We show that the RNA methyltransferase NSUN3 localises to mitochondria and interacts with mt‐tRNAM et to methylate cytosine 34 (C34) at the wobble position. NSUN3 specifically recognises the anticodon stem loop (ASL) of the tRNA, explaining why a mutation that compromises ASL basepairing leads to disease. We further identify ALKBH1/ABH1 as the dioxygenase responsible for oxidising m5C34 of mt‐tRNAM et to generate an f5C34 modification. In vitro codon recognition studies with mitochondrial translation factors reveal preferential utilisation of m5C34 mt‐tRNA Met in initiation. Depletion of either NSUN3 or ABH1 strongly affects mitochondrial translation in human cells, implying that modifications generated by both enzymes are necessary for mt‐tRNAM et function. Together, our data reveal how modifications in mt‐tRNAM et are generated by the sequential action of NSUN3 and ABH1, allowing the single mitochondrial tRNAM et to recognise the different codons encoding methionine.
Keywords: ABH1, mitochondria, NSUN3, RNA modification, translation
Subject Categories: Protein Biosynthesis & Quality Control, RNA Biology
Introduction
More than a hundred different chemical modifications of ribonucleosides have been identified in cellular RNAs (Czerwoniec et al, 2009; Motorin & Helm, 2011). Modifications regulate the biogenesis, structure and function of the corresponding RNAs and RNA–protein complexes (RNPs). Many modifications occur in RNAs involved in translation and are therefore likely to affect protein synthesis. Several modified ribonucleosides including 6‐methyladenosine (m6A), 5‐methylcytidine (m5C), 1‐methyladenosine (m1A) and pseudouridine have recently been shown to occur in messenger (m)RNAs and to affect their biogenesis, translation and stability (see e.g. Carlile et al, 2014; Liu & Jia, 2014; Dominissini et al, 2016). Methylated nucleosides can undergo further modification and proteins of the AlkB family of alpha‐ketoglutarate and Fe(II)‐dependent dioxygenases (ALKBH1‐8 and FTO in human cells) can oxidise or even remove modifications in DNA and RNA (Fedeles et al, 2015; Ougland et al, 2015), increasing the dynamics and regulation of RNA modifications and their roles in RNA metabolism. Compared to mRNAs and other cellular RNAs, transfer (t)RNAs and ribosomal (r)RNAs contain the highest proportion of modified nucleosides. The large majority of rRNA modifications are already installed co‐transcriptionally by small nucleolar (sno)RNPs, and only few base modifications require the action of lone‐standing enzymes (Watkins & Bohnsack, 2012; Sharma & Lafontaine, 2015). tRNAs contain the largest variety of nucleoside modifications, and many of them are suggested to affect tRNA biogenesis and nuclear export, tRNA structure, interaction with aminoacyl‐tRNA‐sythetases or codon recognition during translation (Agris et al, 2007; Leisegang et al, 2012; Hori, 2014; Duechler et al, 2016; Ranjan & Rodnina, 2016). Many tRNAs contain base modifications of the nucleoside at position 34 of the tRNA anticodon (the “wobble position”). These modifications modulate codon–anticodon basepairing, often allowing one tRNA to recognise several different nucleosides in the third position of the codon. Mutations in enzymes responsible for introducing these “wobble base” modifications or genetic alterations in tRNA sequences that affect such modifications are often associated with disease, especially in mitochondrial tRNAs (Lott et al, 2013; Powell et al, 2015).
One ribonucleoside modification that has been identified in several tRNAs, in both cytoplasmic and mitochondrial rRNA, in other non‐coding RNAs and in mRNAs is 5‐methylcytosine (m5C). m5C modifications can be installed by any of the seven proteins of the Nol1/Nop2/SUN domain (NSUN) family and by an enzyme named DNA methyltransferase 2 (DNMT2). DNMT2 mainly catalyses the m5C modification in position 38 of tRNAAsp in human cells (Goll et al, 2006), while the so far characterised NSUN proteins show specificity for tRNAs (NSUN2, NSUN6; Schaefer et al, 2010; Tuorto et al, 2012; Blanco et al, 2014; Haag et al, 2015a) or rRNA (NSUN1/NOP2, NSUN5; Sloan et al, 2013; Tafforeau et al, 2013; Schosserer et al, 2015). NSUN2 can also modify vault RNAs and mRNAs (Hussain et al, 2013), and NSUN4 was described to localise to mitochondria where it was shown to methylate the mitochondrial 12S rRNA in mice (Cámara et al, 2011; Metodiev et al, 2014).
NSUN3 was one of the last uncharacterised members of the family, and we show here that this RNA methyltransferase localises to the mitochondrial matrix in human cells. Using in vivo UV cross‐linking and analysis of cDNA (CRAC) and 5‐azacytidine (5‐AzaC) CRAC, we show that NSUN3 specifically interacts with the mitochondrial tRNAMet where it is responsible for introducing a 5‐methylcytosine (m5C) modification at the “wobble position”. In addition, we find that the m5C modification can be further oxidised by the alpha‐ketoglutarate and Fe(II)‐dependent dioxygenase ALKBH1/ABH1, generating a 5‐formylcytidine (f5C) at this position. Analysis of mt‐tRNAMet synthesised with the different cytosine modifications in the wobble position revealed that codon recognition in an in vitro translation system utilising mitochondrial initiation and elongation factors depends on the modification state of C34 in mt‐tRNAMet. In vivo, knock‐down of ABH1 abolishes f5C34 formation, while depletion of NSUN3 leads to a decrease in mt‐tRNAMet modification. Furthermore, reducing the levels of either NSUN3 or ABH1 leads to a significant decrease in mitochondrial translation in vivo, suggesting important roles for the modifications installed by the two enzymes in mt‐tRNAMet function. Interestingly, our data also show that NSUN3 requires the anticodon stem loop for substrate recognition and a pathogenic mutation in the ASL abolishes C34 methylation, implying that lack of this modification can lead to disease.
Results
NSUN3 localises to the mitochondrial matrix
More than 10 years ago computational analysis identified NSUN3 as a member of the Nol1/Nop2/Sun domain (NSUN) family of putative m5C RNA methyltransferases (Bujnicki et al, 2004). NSUN3 was suggested to localise to mitochondria (Rhee et al, 2013); however, the target spectrum and biological function of the protein have remained unknown. To confirm the mitochondrial localisation of NSUN3, we generated a HEK293 cell line stably expressing NSUN3‐GFP from a tetracycline‐inducible promoter. Confocal fluorescence microscopy revealed that NSUN3‐GFP localises to distinct cytoplasmic foci that showed co‐localisation with a mitotracker (Fig 1A), indicating a mitochondrial localisation of NSUN3. To determine whether NSUN3 is imported into mitochondria, we performed protease protection assays using a tetracycline‐inducible NSUN3‐HisPrcFLAG (Hexahistidine‐PreScission protease cleavage site‐2×FLAG tagged NSUN3) cell line. We isolated mitochondria that were then either left intact, subjected to swelling to rupture the outer mitochondrial membrane and generate mitoplasts or were disrupted using sonication before treatment with different concentrations of proteinase K. While treatment of intact mitochondria led to the degradation of the outer membrane protein TOM70, the intermembrane space domain of TIM23 was digested in mitoplasts. Similar to the matrix‐localised domain of TIM44, NSUN3 only became susceptible to proteinase K digestion upon rupture of mitochondria by sonication (Fig 1B), indicating that NSUN3 is localised in the mitochondrial matrix in human cells.
Figure 1. NSUN3 localises to the mitochondrial matrix.
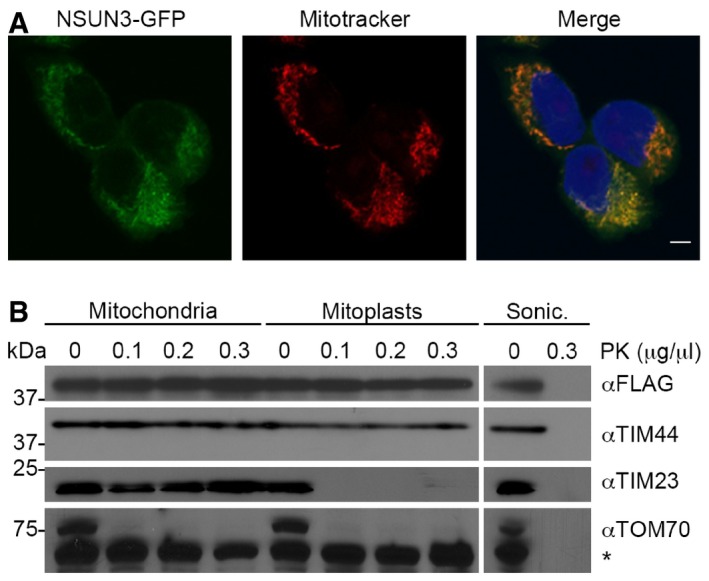
- The localisation of NSUN3 was analysed in HEK293 cells stably expressing NSUN3‐GFP. NSUN3‐GFP (green) and staining with Mitotracker (red) are shown separately and in an overlay with DAPI to indicate nuclei. The scale bar represents 5 μm.
- To analyse submitochondrial localisation of NSUN3, human mitochondria were isolated and either left untreated, swollen in hypotonic buffer (Mitoplasts) or disrupted by sonication (Sonic.) before treatment with different amounts of proteinase K (PK) where indicated, followed by SDS–PAGE and Western blotting using antibodies against human TIM44, TIM23, TOM70 or FLAG‐tagged NSUN3. Note that TIM44 extends into the matrix, while the N‐terminus of TIM23 localises to the intermembrane space and TOM70 is largely exposed on the mitochondrial surface. The asterisk indicates a cross‐reaction of the TOM70 antibody.
NSUN3 associates with mitochondrial tRNAMet
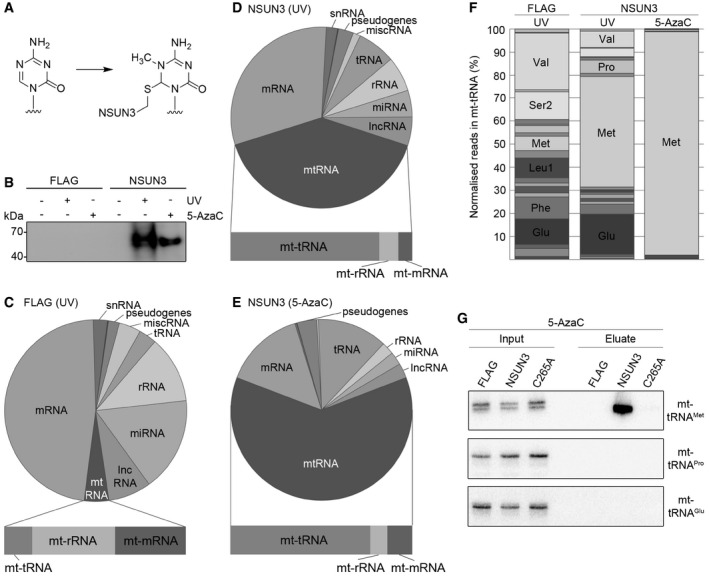
To identify NSUN3 target RNAs, we performed UV cross‐linking and analysis of cDNA (CRAC; Bohnsack et al, 2012; Sloan et al, 2015) experiments using the NSUN3‐HisPrcFLAG cell line; a HEK293 cell line expressing only the HisPrcFLAG tag was used as a control. In addition, cells expressing NSUN3‐HisPrcFLAG were treated with the cytidine derivative 5‐azacytidine (5‐AzaC) as a cross‐linking reagent, which is incorporated into nascent RNA and specifically traps m5C RNA methyltransferases on their target nucleotides in a covalent protein–RNA intermediate during the methylation reaction (Fig 2A; Khoddami & Cairns, 2013). Without cross‐linking or after UV or 5‐AzaC cross‐linking in vivo, protein–RNA complexes were purified followed by RNA trimming, radiolabelling and ligation of adaptors to the bound RNA. Protein–RNA complexes were separated by SDS–PAGE, transferred to a membrane and exposed to an X‐ray film. Both UV and 5‐AzaC cross‐linking of NSUN3‐HisPrcFLAG resulted in a strong specific signal not observed for the controls (Fig 2B), indicating association of NSUN3 with cellular RNAs. Interacting RNAs were then extracted from the membrane and subjected to RT–PCR to generate a cDNA library for Illumina deep sequencing. Mapping of the obtained sequence reads on the human genome resulted in a strong over‐representation of mitochondrial‐encoded RNA (mt‐RNA). mt‐RNA represented 40% and 62% of total reads obtained upon UV or 5‐AzaC cross‐linking of NSUN3, respectively, compared to only 4% of sequence reads from the HisPrcFLAG control (Figs 2C–E and EV1A), suggesting a specific association of NSUN3 with mitochondrial RNA. As sequences from mitochondrial tRNAs were strongly enriched in the NSUN3‐cross‐linked fractions (Fig 2D and E, lower panels) compared to the control (Fig 2C, lower panel), we analysed the distribution of reads between the 22 mitochondrial tRNAs. Strikingly, 50 and 95% of the reads mapped to mt‐tRNAMet in the NSUN3 UV and 5‐AzaC cross‐linking experiments, respectively (Fig 2F). In contrast, the data obtained for the HisPrcFLAG control contained only 5% sequencing reads that mapped to mt‐tRNAMet, indicating that NSUN3 specifically interacts with this tRNA (Fig 2F). To confirm the specificity of this interaction, we performed 5‐AzaC cross‐linking using cells expressing the HisPrcFLAG control, NSUN3‐HisPrcFLAG and the catalytically inactive NSUN3(C265A)‐HisPrcFLAG mutant, in which the catalytic cysteine of the TCT tripeptide that is conserved in motif IV in m5C methyltransferases of the NSUN family is replaced by alanine (C265A). After cross‐linking and isolation of complexes via the FLAG‐tagged proteins, interacting RNAs were analysed by Northern blotting using probes for the detection of the mitochondrial tRNAs mt‐tRNAPro, mt‐tRNAGlu and mt‐tRNAMet (Fig 2G). While mt‐tRNAPro and mt‐tRNAGlu could not be detected in any of the eluates, mt‐tRNAMet was strongly enriched in the eluate from the NSUN3 wild‐type sample, but was not detected in any of the controls, further supporting that mt‐tRNAMet specifically interacts with NSUN3. The specific requirement for the conserved catalytic cysteine and the efficient cross‐linking of NSUN3 to 5‐AzaC containing mt‐tRNAMet strongly suggest that NSUN3 is an active m5C RNA methyltransferase that uses the conserved mechanism of the NSUN family to mediate m5C methylation of its substrate mt‐tRNAMet in human mitochondria.
Figure 2. NSUN3 cross‐links to the mitochondrial tRNAM et in vivo .
-
AStructure of 5‐azacytidine and formation of a covalent RNA methyltransferase adduct.
-
BHEK293 cells expressing NSUN3‐HisPrcFLAG (NSUN3) or the HisPrcFLAG tag alone (FLAG) were either not cross‐linked (−), UV cross‐linked (UV) or treated with 5‐azacytidine (5‐AzaC). The protein–RNA complexes were affinity purified and the bound RNA was trimmed, end‐labelled with 32P phosphate and ligated to linkers. Protein–RNA complexes were separated by SDS–PAGE, transferred to nitrocellulose and exposed to an X‐ray film.
-
C–EThe UV or 5‐AzaC cross‐linking and analysis of cDNA (CRAC) experiments with NSUN3‐HisPrcFLAG (D, E) or the FLAG control (C) samples were treated as described in (B). The RNA was isolated from the nitrocellulose membrane‐bound protein–RNA complexes and converted into cDNA for sequence library production and Illumina deep sequencing. Pie charts present different RNA classes and the relative distribution of Illumina sequence reads that were obtained after mapping of the reads on the human genome. Bar graphs below indicate the distribution of mitochondrial (mt‐)tRNA, mt‐rRNA and mt‐mRNA sequence reads among the reads mapped to the mitochondrial genome. Abbreviations: tRNA, transfer RNA; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; rRNA, ribosomal RNA; mtRNA, mitochondrial‐encoded RNA; miscRNA, miscellaneous RNA; miRNA, microRNA; lncRNA, long non‐coding RNA.
-
FRelative distribution of mitochondrial tRNA sequence reads obtained from the CRAC experiments using UV or 5‐AzaC cross‐linking with cells expressing the NSUN3‐HisPrcFLAG (NSUN3) protein or control cells (FLAG). Only mt‐tRNAs that were represented by more than 5% of all mt‐tRNAs reads are labelled.
-
G5‐AzaC cross‐linking was performed and RNA associated with wild‐type NSUN3, the catalytically inactive NSUN3 mutant (C265A) or the FLAG tag alone was isolated as described in (B). The RNA was analysed by Northern blot using probes against the mt‐tRNAMet, mt‐tRNAPro and mt‐tRNAGlu. Inputs (0.1%) are shown on the left and eluates (50%) on the right.
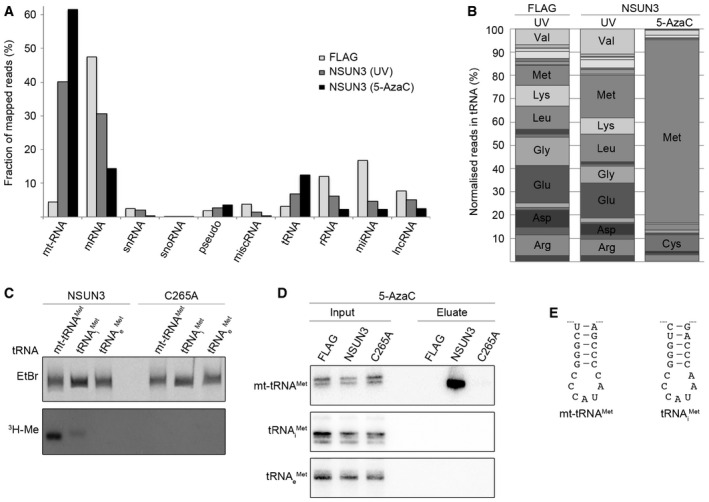
Figure EV1. The cytoplasmic tRNA Met‐i and Met‐e do not represent substrates of NSUN3 in vivo .
-
A, BThe UV or 5‐AzaC cross‐linking and analysis of cDNA (CRAC) experiments with NSUN3‐HisPrcFLAG or FLAG control cells were performed as described for Fig 2. (A) The percentages of the Illumina sequence reads mapped to individual classes of RNA are given graphically for each sample. Abbreviations: tRNA, transfer RNA; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; rRNA, ribosomal RNA; mtRNA, mitochondrial‐encoded RNA; miscRNA, miscellaneous RNA; miRNA, microRNA; lncRNA, long non‐coding RNA. (B) The relative distribution of cytoplasmic tRNA sequence reads obtained from the CRAC experiments is shown. Only tRNAs that were represented by more than 5% of all cytoplasmic tRNA reads are labelled.
-
CIn vitro methylation reactions were performed using recombinant His14‐MBP‐NSUN3 (NSUN3) or the catalytically inactive mutant His14‐MBP‐NSUN3‐C265A (C265A), [3H‐methyl]‐labelled S‐adenosylmethionine as a methyl group donor and in vitro‐transcribed mitochondrial mt‐tRNAMet, cytoplasmic tRNAi Met and tRNAe Met. The RNA was then separated on a denaturing polyacrylamide gel, stained with ethidium bromide (EtBr) to indicate inputs and exposed to an X‐ray film to analyse methylation (3H‐Me).
-
D5‐AzaC cross‐linking was performed and RNA‐associated with wild‐type NSUN3, the catalytic NSUN3 mutant (C265A) or the FLAG tag alone was isolated as described in (A). The RNA was isolated from the purified protein–RNA complexes and analysis by Northern blot using probes against the mt‐tRNAMet, mt‐tRNAi Met and mt‐tRNAe Met. Inputs are shown on the left and eluates on the right. The mt‐tRNAMet panel is identical to that shown in Fig 2G.
-
EThe nucleotide sequences of the anticodon stem loops of mt‐tRNAMet (left) and tRNAi Met (right) are shown.
NSUN3 specifically methylates cytosine 34 in mt‐tRNAMet
To gain further insight into the catalytic activity of NSUN3, we prepared recombinant NSUN3 protein and the catalytically inactive mutant (NSUN3‐C265A) and performed in vitro methylation experiments using in vitro T7 RNA‐polymerase transcripts of mt‐tRNAMet, mt‐tRNAPro and mt‐tRNAGlu in the presence of S‐[3H‐methyl] adenosylmethionine (SAM) as a methyl group donor. NSUN3 efficiently methylated mt‐tRNAMet, but not the other transcripts, and the catalytic activity of NSUN3 was abolished by mutation of the catalytic cysteine (Fig 3A).
Figure 3. NSUN3 modifies the wobble position of mt‐tRNAM et .
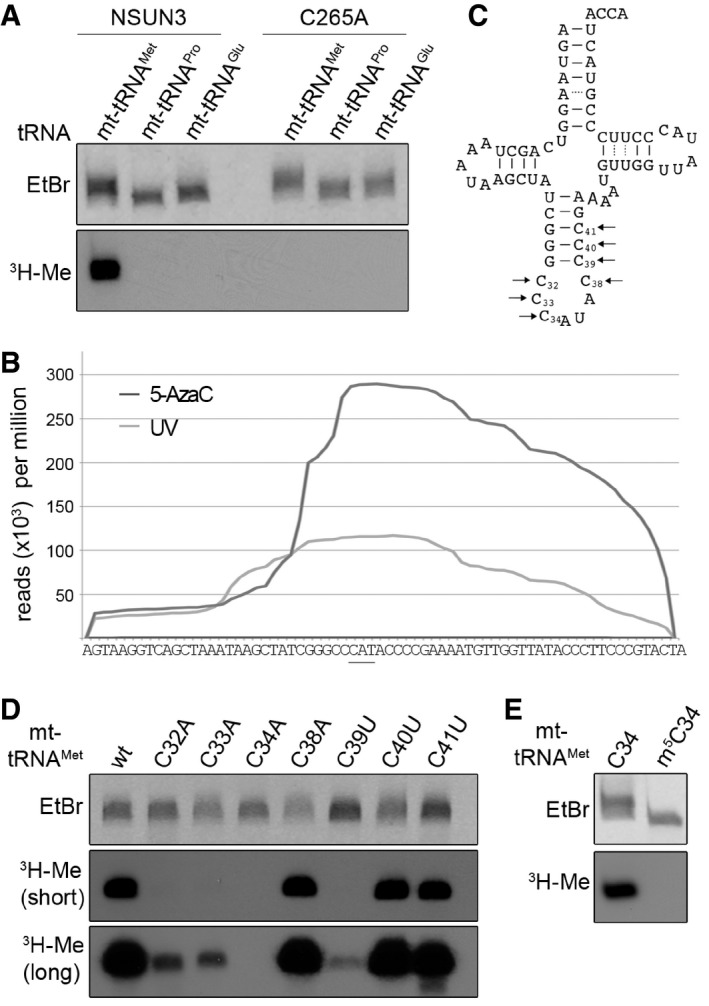
- In vitro methylation reactions were performed using recombinant His14‐MBP‐NSUN3 (NSUN3) or the catalytically inactive mutant His14‐MBP‐NSUN3‐C265A (C265A), 3H‐labelled S‐adenosylmethionine as a methyl group donor and in vitro‐transcribed mt‐tRNAMet, mt‐tRNAPro and mt‐tRNAGlu. The RNA was then separated on a denaturing polyacrylamide gel, stained with ethidium bromide (EtBr) to indicate inputs and exposed to an X‐ray film to analyse methylation (3H‐Me).
- The distribution of Illumina sequence reads along the mt‐tRNAMet sequence obtained from CRAC experiments with NSUN3 after UV (light grey) or 5‐AzaC cross‐linking (dark grey) is given as reads per million mapped reads. The position of the anticodon is indicated by a bar.
- Cloverleaf scheme of the mt‐tRNAMet sequence. Nucleosides that were exchanged in the mutational analysis shown in the following panels are marked with arrows, and the nucleotide positions in the tRNA are given.
- In vitro methylation assays were performed as described in (A) with His14‐MBP‐NSUN3 and in vitro‐transcribed wild‐type mt‐tRNAMet and cytidine mutants of the anticodon stem and loop region indicated in (C). Two exposure times of X‐ray films are shown 16 h (short) and 3 days (long).
- In vitro methylation assay of in vitro‐transcribed mt‐tRNAMet and chemically synthesised mt‐tRNAMet containing an m5C modification at the wobble position. The experiment and analysis were performed as described in (A).
Besides the strong enrichment of reads from mt‐tRNAMet in the CRAC data sets, we had observed that reads mapping to the cytoplasmic tRNAs that mediate incorporation of methionine during translation initiation (tRNAi Met) and elongation (tRNAe Met) were over‐represented in the NSUN3 cross‐linking data (8% of reads mapped to cytoplasmic tRNA were tRNAMet reads in FLAG control; 18% after UV and 79% after 5‐AzaC cross‐linking; Fig EV1B). We therefore tested whether NSUN3 could methylate transcripts of tRNAi Met and tRNAe Met in in vitro methyltransferase assays. While mt‐tRNAMet was methylated very efficiently by NSUN3, only very weak or no methylation was observed for the tRNAi Met and tRNAe Met transcripts, respectively (Fig EV1C). To analyse possible interactions between NSUN3 and tRNAi Met or tRNAe Met in vivo, we performed 5‐AzaC cross‐linking and immunoprecipitation experiments using HEK293 cells expressing the HisPrcFLAG tag alone, wild‐type or mutant (C265A) NSUN3‐HisPrcFLAG and analysed the co‐precipitation of tRNAs by Northern blotting. While mt‐tRNAMet was strongly enriched with wild‐type NSUN3, no association of the cytoplasmic tRNAi Met or tRNAe Met could be detected (Fig EV1D), indicating that NSUN3 does not specifically bind cytoplasmic tRNAs in vivo and that the interactions observed in the 5‐AzaC CRAC likely occurred after cell lysis due to similar sequences of the anticodon stem loop of tRNAi Met and mt‐tRNAMet (Fig EV1E). Together with the mitochondrial localisation of NSUN3 (Fig 1), these data indicate that NSUN3 can weakly recognise the tRNAi Met as a substrate in vitro, but that mt‐tRNAMet, rather than tRNAi Met, represents its genuine methylation substrate in vivo.
In order to identify which region of mt‐tRNAMet interacts with NSUN3, we analysed the distribution of reads obtained by NSUN3 cross‐linking to mt‐tRNAMet. Analysis of both UV and 5‐AzaC cross‐linking experiments showed that the highest read density was obtained with sequences corresponding to the anticodon stem loop (ASL) of mt‐tRNAMet (Fig 3B) suggesting that the NSUN3 target residue lies within this region. As NSUN3 is a member of the cytosine methyltransferase family of NSUN proteins, we generated in vitro transcripts of mt‐tRNAMet in which each cytosine present in the ASL was individually mutated to an adenosine (ASL loop cytosines) or uracil (cytosines in the stem of the ASL; Fig 3C). Although mutation of several cytosines affected NSUN3‐mediated methylation in in vitro methylation assays, only mutation of cytosine 34 abolished the modification (Fig 3D), suggesting that the C34 wobble nucleotide is the NSUN3 target in mt‐tRNAMet. This conclusion was confirmed by a lack of methylation when chemically synthesised mt‐tRNAMet containing an m5C34 was treated with NSUN3 in methylation assays (Fig 3E), supporting the finding that NSUN3 generates an m5C moiety at position 34 in mt‐tRNAMet.
Among the mt‐tRNAMet mutants (Fig 3D), the C39U mutant, which has previously been identified in patients with mitochondrial dysfunction (Lott et al, 2013; Tang et al, 2013), was a particularly poor substrate for NSUN3, suggesting that this residue might be critical for methylation or that a stable stem in the ASL could be required for NSUN3 recognition. To distinguish between these possibilities, we generated a series of ASL mutants where individual cytosines in the stem were either replaced by uracil allowing for less stable G:U basepairing or mutants in which guanosine and cytosine in G:C basepairs were swapped between the strands of the stem, resulting in identical stability of the stem but a change in the sequence (Fig 4A). While no reduction in NSUN3 methylation was observed for the mutants generated by swapping the G:C basepairs in the stem, mutations to G:U basepairs reduced methylation efficiency and again almost abolished it for the C39U mutant (Fig 4B). These data indicate that a stable stem in the ASL is required for NSUN3 substrate recognition and methylation of C34 in mt‐tRNAMet. Incubation of synthesised mt‐tRNAMet ASL with NSUN3 in a methylation assay further revealed that the ASL is sufficient for recognition and methylation (Fig 4C).
Figure 4. NSUN3 requires a stable anticodon stem loop of mt‐tRNAM et for methylation of cytosine 34.
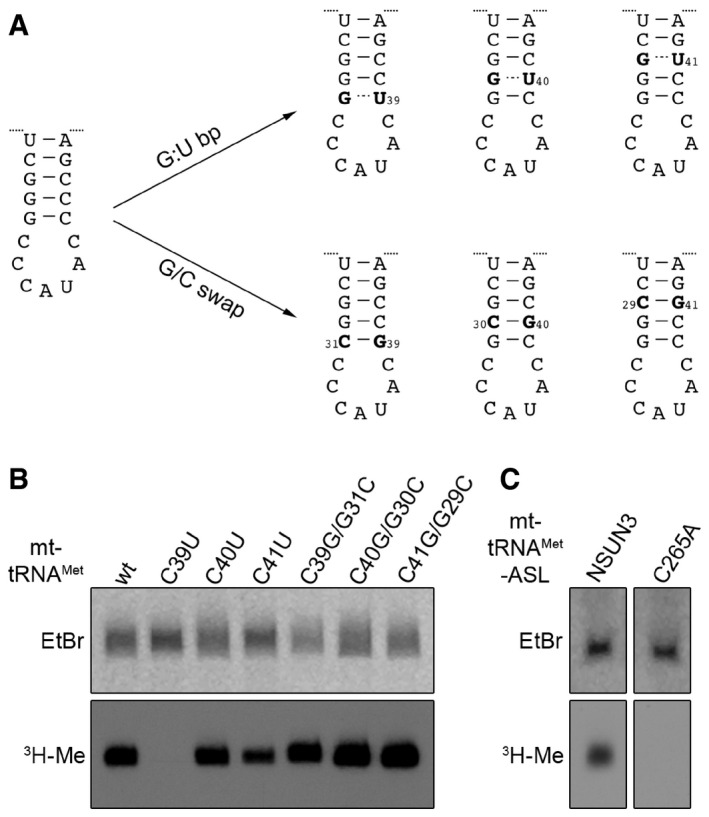
- Scheme showing the mutations introduced in the anticodon stem loop (ASL) of mt‐tRNAMet for analysing tRNA substrate recognition by NSUN3. ASL mutants included G:U basepairs (G:U bp) to affect the stability of basepairing and sequence of the stem or mutants were generated by swapping G:C basepairs (G/C swap), leading to changes in sequence without affecting basepairing stability.
- In vitro methylation assays were performed using [3H‐methyl]‐labelled S‐adenosylmethionine, the in vitro transcripts of the mt‐tRNAMet mutants described in (A) and recombinant His14‐MBP‐NSUN3. RNA was then separated on a denaturing polyacrylamide gel, stained with ethidium bromide (EtBr), dried and exposed to an X‐ray film to detect methylated transcripts (3H‐Me).
- In vitro methylation assay using chemically synthesised ASL. The experiment and analysis were performed as described in (B).
Taken together, we have identified the mitochondrial tRNAMet as the methylation substrate of the RNA methyltransferase NSUN3. NSUN3 recognises the ASL of mt‐tRNAMet and requires a stable stem structure for substrate recognition and generation of m5C34. Furthermore, a pathogenic mutation in the stem loop abolishes NSUN3‐mediated modification, indicating that lack of modification of C34 can lead to disease.
ALKBH1/ABH1 localises in mitochondria and specifically interacts with mt‐tRNAMet
Previous reports suggested that mt‐tRNAMet can be modified at position 34 to contain a 5‐formylcytosine (f5C; Moriya et al, 1994; Suzuki & Suzuki, 2014). We hypothesised that a specific oxygenase might oxidise the m5C34 moiety established by NSUN3 to generate an f5C34 modification in mt‐tRNAMet. While the Ten‐Eleven Translocation (TET) protein family of dioxygenases primarily mediates oxidation of m5C in nuclear DNA and has also been implicated in histone modification, most of the members of the AlkB‐like Fe(II)/alpha‐ketoglutarate‐dependent dioxygenases (ALKBH) have been shown to act on RNA (reviewed in Shen et al, 2014; Fedeles et al, 2015; Li et al, 2015; Ougland et al, 2015). These include FTO (ALKBH9) that is implicated together with ALKBH5 in the oxidative removal of several modifications including 6‐methyladenosine (m6A) from RNA and ALKBH8 that is involved in the generation of 5‐methoxycarbonylmethyluridine (mcm5U) in cytoplasmic tRNAs (Fu et al, 2010a,b; Songe‐Møller et al, 2010; Jia et al, 2011; Thalhammer et al, 2011; Berulava et al, 2013; Zheng et al, 2013). So far, only ALKBH7, which was suggested to act on protein substrates during necrosis (Fu et al, 2013; Solberg et al, 2013; Wang et al, 2014), and ALKBH1/ABH1 have been reported to localise to mitochondria; however, the cellular localisation of ABH1 has been a matter of debate (Pan et al, 2008; Westbye et al, 2008; Ougland et al, 2012). We therefore analysed the cellular localisation of ABH1 in HEK293 cells by immunofluorescence analysis and co‐staining with a mitotracker (Fig 5A). The ABH1 antibody showed a cytoplasmic localisation with enrichment in foci that were also stained by the mitotracker, indicating that ABH1 is largely present in mitochondria in HEK293 cells, which could allow it to act on mt‐tRNAMet. Partial localisation of ABH1 to the mitochondrial matrix was further supported by proteinase K protection assays, in which ABH1 remained intact in mitoplasts and was only degraded upon mitochondrial lysis that allowed access of the protease to the matrix (Fig EV2). To test whether ABH1 specifically interacts with mt‐tRNAMet, we generated a HEK293 cell line expressing ABH1‐HisPrcFLAG and performed UV cross‐linking and pull‐down experiments followed by Northern blotting to analyse for ABH1‐associated RNAs. mt‐tRNAMet (and not mt‐tRNAGlu) was retrieved with ABH1‐HisPrcFlag, but not with the HisPrcFLAG control (Fig 5B), indicating that ABH1 interacts specifically and directly with mt‐tRNAMet in mitochondria.
Figure 5. ABH1 localises to mitochondria in HEK293 cells and specifically interacts with mt‐tRNAM et .
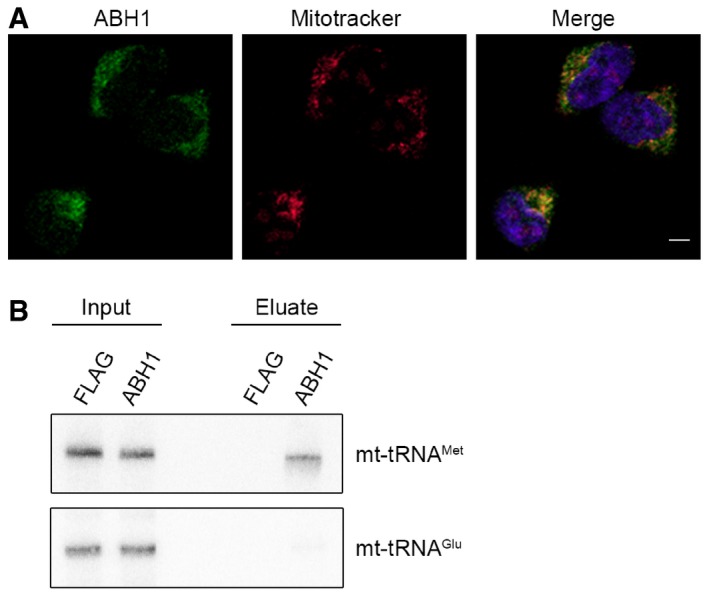
- The localisation of ABH1 was analysed by immunofluorescence in HEK293 cells. ABH1 (green) localisation and mitochondria stained with Mitotracker (red) are shown separately and in an overlay with DAPI to indicate nuclei. The scale bar represents 5 μm.
- HEK293 cells expressing ABH1‐HisPrcFLAG (ABH1) or the HisPrcFLAG tag alone (FLAG) were UV cross‐linked (UV), and protein–RNA complexes were affinity purified. Co‐precipitated RNA was isolated and analysed by Northern blot using probes against mt‐tRNAMet and mt‐tRNAGlu. Inputs (0.1%) are shown on the left and eluates (50%) on the right.
Figure EV2. The mitochondrial pool of ABH1 is localised in the matrix.
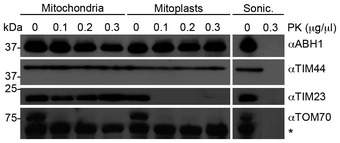
To analyse submitochondrial localisation of ABH1, human mitochondria were isolated and either left untreated, swollen in hypotonic buffer (Mitoplasts) or disrupted by sonication (Sonic.) before treatment with different amounts of proteinase K (PK) where indicated, followed by SDS–PAGE and Western blotting using antibodies against human TIM44, TIM23, TOM70 or FLAG‐tagged NSUN3. Note that TIM44 extrudes into the matrix, while a major domain of TIM23 localises to the intermembrane space and TOM70 is largely exposed on the mitochondrial surface. The asterisk indicates a cross‐reaction of the TOM70 antibody. The panels of TIM44, TIM23 and TOM70 are identical with those shown in Fig 1B.
ABH1 mediates oxidation of m5C34 in mt‐tRNAMet
The interaction of ABH1 with mt‐tRNAMet suggests that it might mediate oxidation of m5C34 in mt‐tRNAMet. We therefore radioactively methylated mt‐tRNAMet using [3H‐methyl]‐labelled S‐adenosylmethionine and recombinant NSUN3 and generated recombinant ABH1 and FTO for in vitro oxidation assays. The oxidation assays were performed in the presence of alpha‐ketoglutarate and Fe2+ either without enzyme, with maltose binding protein (MBP), wild‐type ABH1, the ABH1 alpha‐ketoglutarate/Fe2+‐binding mutants R338A or D233A (Westbye et al, 2008), or FTO, and oxidation was monitored by measuring tritium release from the methyl group. Only wild‐type ABH1 could oxidise m5C34 in mt‐tRNAMet and the reaction required the presence of alpha‐ketoglutarate and Fe2+ (Fig 6A), which further supports the notion that mt‐tRNAMet is a genuine substrate of ABH1 and that ABH1 utilises the conserved mechanism of the ALKBH family.
Figure 6. ABH1 can oxidise m5C34 in mt‐tRNAM et in vitro .
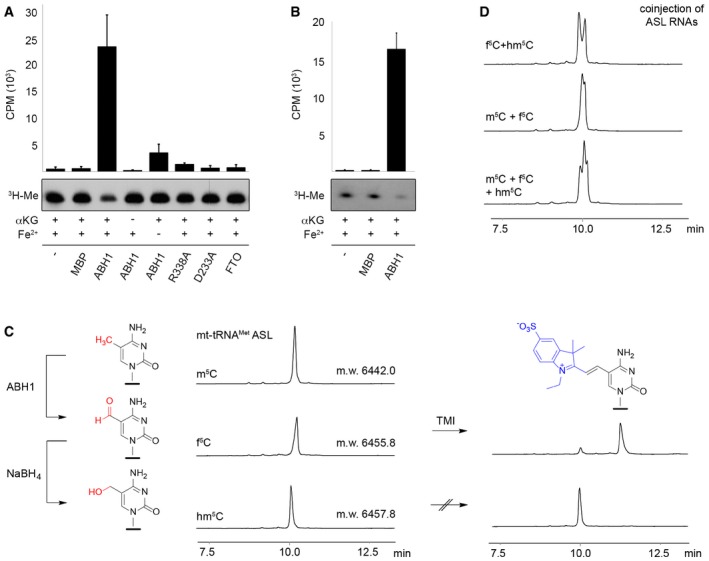
- In vitro‐transcribed mt‐tRNAMet was methylated at C34 using recombinant NSUN3 and 3H‐labelled S‐adenosylmethionine as a methyl group donor. Radiolabelled mt‐tRNAMet was re‐extracted and then subjected to oxidation assays without protein (−), with maltose binding protein (MBP), with the dioxygenase FTO or using wild‐type (ABH1) or mutant (R338A, D233A) His14‐MBP‐ABH1. Besides ABH1 controls lacking α‐ketoglutarate (αKG) or Fe2+ ions, all samples contained α‐ketoglutarate and Fe2+ ions. After oxidation, RNA was precipitated and the tritium released upon oxidation of radiolabelled mt‐tRNAMet was quantified in the supernatant. Counts per minute (CPM) are shown for experiments performed in triplicate with error bars indicating ± SD (upper panel). Pelleted RNA was separated on a denaturing polyacrylamide gel and exposed to an X‐ray film to analyse the tritium retained (3H‐Me).
- Synthetic anticodon stem loop (ASL) was radioactively labelled and subjected to oxidation assays that were performed and analysed as described in (A) using no protein (−), MBP or wild‐type His14‐MBP‐ABH1 (ABH1). Experiments were performed in triplicate with error bars indicating ± SD.
- Anion exchange HPLC analysis was performed on synthetic m5C‐containing ASL (20 nt) before and after oxidation by ABH1. The small shift in retention time indicates formation of f5C‐modified RNA. The ABH1 oxidation product was then treated with NaBH4 to generate hm5C‐modified RNA. All three samples were analysed by ESI‐MS, and the molecular weight (m.w.) is indicated on the HPLC trace. Only the f5C‐containing RNA was labelled efficiently with 1‐ethyl‐2,3,3‐trimethylindoleninium‐5‐sulphonate (TMI).
- The different retention times of m5C‐, hm5C‐ and f5C‐modified ASL RNA were confirmed by co‐injection of samples shown in (C). HPLC was performed as in (C).
We next analysed whether the mt‐tRNAMet ASL alone is sufficient for in vitro recognition by ABH1 and oxidation of m5C34. Indeed, m5C34‐containing mt‐tRNAMet ASL was efficiently oxidised by ABH1 (Fig 6B), allowing further characterisation of the oxidation product by HPLC. Treatment of chemically synthesised m5C34‐containing ASL with ABH1 resulted in almost quantitative oxidation of m5C to 5‐formylcytosine (f5C). The presence of f5C was confirmed by mass spectrometry and by the efficient conversion in a 5‐formylpyrimidine‐specific reaction with the trimethylindol derivative TMI (Fig 6C; Samanta et al, 2016). Upon treatment of the oxidation product with NaBH4, f5C was chemically reduced to 5‐hydroxymethyl‐cytosine (hm5C), which did not react with TMI (Fig 6C). Co‐injection of mt‐tRNAMet ASL derivatives further confirmed that the different oxidation states can be distinguished by HPLC (Fig 6D).
Together, these data show that ABH1 is present in mitochondria of HEK293 cells where the enzyme can mediate oxidation of the m5C34‐containing mt‐tRNAMet generated by NSUN3 to provide f5C‐containing mt‐tRNAMet for mitochondrial translation.
Modifications of C34 modulate codon recognition by mt‐tRNAMet
To understand how the modification state of C34 in mt‐tRNAMet affects its function in translation, we studied codon recognition by mt‐tRNAMet variants on the ribosome. As a reconstituted mitochondrial in vitro translation system is not readily available, we tested binding of different modification states of mt‐tRNAMet in the presence of purified recombinant human mitochondrial translation factors on ribosomes from Escherichia coli. Even though the structure of bacterial and mitochondrial ribosomes is significantly different, the structure of the decoding centre of the ribosome is highly conserved (reviewed in Greber & Ban, 2016), allowing mitochondrial translation factors to bind at the conserved sites of bacterial ribosomes. To mimic codon recognition during translation initiation, we used recombinant human mitochondrial initiation factor 2 (MTIF2), which recruits mt‐tRNAMet to the P site of the ribosome. Codon recognition during the elongation phase was studied with the human mitochondrial translation elongation factor TUFM, which delivers the tRNA to the A site. We used chemically synthesised mt‐tRNAMet containing either unmodified C34, m5C34, hm5C34 or f5C34 aminoacylated with [14C]Met. Ribosomes programmed with mRNAs presenting AUG, AUA or AUU codons in the P or A site were mixed with MTIF2–GTP or TUFM–GTP and one of the mt‐tRNAMet variants. mt‐tRNAMet–ribosome complexes were retrieved by nitrocellulose filtration and quantified by scintillation counting. The universal AUG codon or the AUA codon in the P site was preferentially recognised by the m5C34‐modified mt‐tRNAMet (Fig 7A). Binding to the ribosomes containing an AUU codon in the P site was generally lower and less specific with respect to mt‐tRNAMet modification. Controls showed only weak binding of mt‐tRNAMet in the absence of ribosomes or mRNA independent of the modification status of the tRNA, indicating that the observed differences in the P site binding are due to specific recognition of these codons by mt‐tRNAMet in complex with MTIF2. Also the TUFM‐dependent decoding in the A site was generally more efficient on AUG than on AUA codons (Fig 7B). The modified and unmodified mt‐tRNAMet variants were capable of reading the AUG codon. Notably the m5C‐modified mt‐tRNAMet was less efficient than other variants in AUG decoding, while AUA was read with similar efficiencies by all variants of mt‐tRNAMet. Together these data indicate that the modification state of C34 in mt‐tRNAMet influences codon recognition by the tRNA in the P and A site, with m5C acting as a predominant decoder during initiation at AUG and all tRNAs capable of decoding during elongation. We note that the kinetics of decoding may be different depending on the modification and thus some mt‐tRNAMet variants may be kinetically preferred over the others. However, a kinetic analysis of decoding upon initiation and elongation is beyond the scope of the present work.
Figure 7. Modification of cytosine 34 modulates codon recognition by mt‐tRNAM et in vitro .
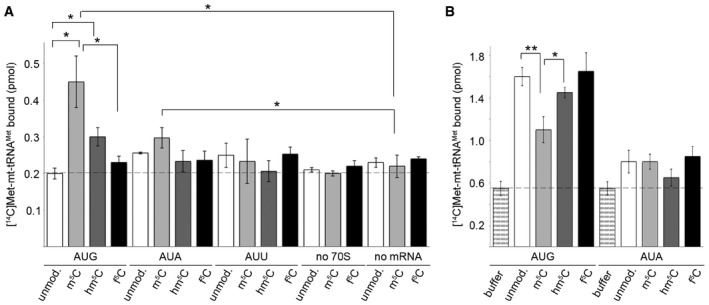
- MTIF2‐dependent reading of initiation codons AUG, AUA or AUU in the P site of the ribosome by unmodified (unmod.) or C34‐modified [14C]Met‐tRNAMet. Binding was determined by nitrocellulose filtration, and [14C]Met‐tRNAMet retrieved on the membrane was quantified by scintillation counting. Binding in the absence of ribosomes (no 70S) or mRNA (no mRNA) served as controls. Data from three independent experiments are presented with error bars indicating ± SEM. The statistical significance of the results was analysed by t‐test and is indicated by the asterisks in the graph (*P < 0.05).
- TUFM‐dependent recognition of A site codons during elongation. Data from three independent experiments are presented with error bars indicating ± SEM and statistical analysis as in (A) (*P < 0.05, **P < 0.01).
Different modification states of cytosine 34 occur in mt‐tRNAMet in vivo
The cross‐linking and in vitro modification data show that cytosine 34 in mt‐tRNAMet can be methylated by NSUN3 to generate m5C and then further oxidised by the dioxygenase ABH1 to f5C. In addition, these different modifications in mt‐tRNAMet may influence codon recognition. To gain insight into the occurrence of the mt‐tRNAMet modification states in vivo, we first established RNAi‐mediated depletion of NSUN3 and ABH1 (Fig 8A). After siRNA treatment, analysis of mRNA levels showed an ≈80% decrease in NSUN3 or ABH1 mRNA levels. Equal amounts of RNA extracted from knock‐down cells were then treated with the 5‐formylpyrimidine‐specific TMI to convert f5C into a hemicyanine derivative, which blocks primer extension by reverse transcriptase at the site of modification, thereby allowing to analyse the presence of f5C in the RNA (Samanta et al, 2016). Primer extension analysis revealed that the fraction of f5C34‐containing mt‐tRNAMet decreased by more than three‐fold (NSUN3) and more than four‐fold (ABH1) when NSUN3 and ABH1 were depleted (Fig 8B and C), confirming the roles of these enzymes in establishing the modification in vivo. To identify the presence of other modification states at C34, RNA from wild‐type cells or cells transfected with non‐target siRNAs or those targeting NSUN3 or ABH1 was first subjected to DNase digest and then treated with bisulphite. Alternatively, the DNase digest was followed by chemical reduction of the RNA with NaBH4 to convert f5C to hm5C and bisulphite treatment. In both cases, after deamination and desulphonation, mt‐tRNAMet was specifically amplified by reverse transcription and PCR and then cloned. Analysis of sequences derived from wild‐type RNA after reduction indicated that mt‐tRNAMet is fully modified at position C34. Comparison to the non‐reduced sample suggested that although the majority of these modifications are f5C, a portion of cytosines at this position are not converted by the bisulphite treatment, indicating that they carry the m5C34 modification installed by NSUN3 (Fig 8D). Consistent with these data, depletion of NSUN3 resulted in a decrease in the mt‐tRNAMet fraction carrying a modification on C34 and a decrease in the portion of m5C, while upon depletion of ABH1, position 34 was almost exclusively read as cytosine independent of whether the RNA had been reduced. These results confirm methylation of C34 by NSUN3 and further show that the ABH1 knock‐down abolishes the formation of f5C34 in mt‐tRNAMet in vivo. We note that the bisulphite data do not rule out the presence of hm5C, which is also resistant to bisulphite conversion. Oxidative bisulphite sequencing, which can distinguish m5C and hm5C in DNA (Booth et al, 2013), resulted in degradation of the RNA. However, hm5C was not observed upon ABH1 oxidation in vitro and had no beneficial effect in ribosome binding assays, suggesting that this modification might not play a major role for mt‐tRNAMet.
Figure 8. Knock‐down of NSUN3 or ABH1 leads to a reduction in the modification of cytosine 34 in mt‐tRNAM et in vivo .
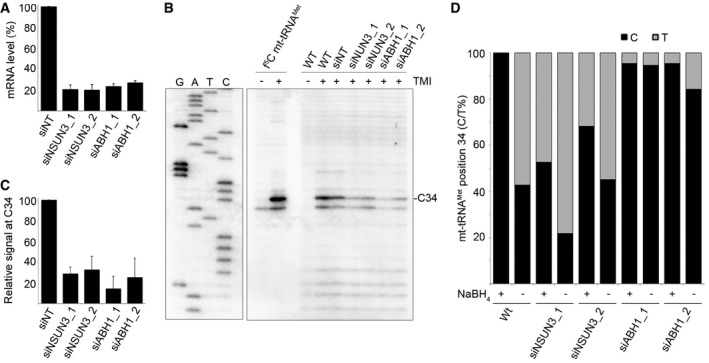
- HeLa cells were transfected with two different siRNAs against NSUN3 (siNSUN3_1, siNSUN3_2), ABH1 (siABH1_1, siABH1_2) or with non‐target (siNT) siRNA, and the knock‐down efficiency was analysed by quantitative PCR. The relative abundance of the NSUN3 or ABH1 mRNA was normalised to GAPDH levels. Data are presented as mean ± SD.
- Chemically synthesised f5C modified mt‐tRNAMet and total RNA from wild‐type (WT) cells or those transfected with siRNAs as in (A) were treated with TMI to specifically label f5C residues. Primer extension, using a radiolabelled antisense primer, was performed under limited dNTP conditions. Products were separated on a denaturing polyacrylamide gel alongside a sequencing ladder, and RNAs were detected using a phosphorimager.
- Primer extension reactions were performed on total RNA from cells transfected with siRNAs as described in (B). Stops corresponding to position C34 in mt‐tRNAMet were quantified in three independent experiments, and results are shown graphically as mean ± SD.
- RNA from wild‐type HeLa cells and cells treated with siRNAs against NSUN3 or ABH1 (as in A) was either first reduced with NaBH4 or directly treated with bisulphite. After deamination and desulphonation, mt‐tRNAMet RNAs were reverse transcribed, amplified, cloned and sequenced. The proportions of thymine (grey) generated by bisulphite conversion or non‐converted cytosine (black) at position 34 of mt‐tRNAMet are shown. Note that for sequences from non‐reduced samples, thymine can also originate from unmodified or f5C‐containing mt‐tRNAMet, while in reduced samples, it originates from unmodified cytosine.
Modifications of cytosine 34 in mt‐tRNAMet are required for mitochondrial translation in vivo
Our finding that the mt‐tRNAMet C39U mutation, which has previously been identified in patients with mitochondrial dysfunction (Lott et al, 2013; Tang et al, 2013), largely abolishes m5C34 formation by NSUN3, suggests that the C34 modification is required for mt‐tRNAMet function in vivo and that mt‐tRNAMet malfunction might cause the disease in these patients. To analyse the requirement for the modifications installed by NSUN3 and ABH1 for translation in mitochondria, we measured the amount of 35S‐methionine incorporated into proteins during mitochondrial translation in vivo after depletion of NSUN3 or ABH1. Indeed, depletion of either NSUN3 or ABH1 resulted in reduced 35S‐incorporation, suggesting that the modifications installed by these proteins are required for mt‐tRNAMet function. Close inspection of the individual synthesis rates of the mitochondrial proteins revealed that the translation of all mitochondrial proteins was affected by NSUN3 or ABH1 depletion, which is in line with the presence of both AUG and non‐canonical codons encoding methionine (AUA, AUU) in all of these mRNAs. Moreover, we observed that cell growth was affected by knock‐down of either NSUN3 or ABH1 (Fig EV3), further supporting the important roles of the modifications installed by these enzymes for mitochondrial function and the cellular metabolism.
Figure EV3. RNAi‐mediated depletion of NSUN3 or ABH1 leads to reduced cell growth.
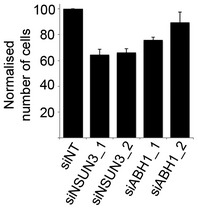
HeLa cells that had been transfected with siRNAs against NSUN3 (siNSUN3_1 or siNSUN3_2), ABH1 (siABH1_1 or siABH1_2) or non‐target siRNAs (siNTs) were harvested and counted. Cell numbers from three experiments were normalised to the non‐target control, and results are given graphically as mean ± SD.
Discussion
Expression of the mitochondrial genome is fundamental in eukaryotes for maintaining the cellular energy metabolism and various metabolic pathways. The human mitochondrial DNA encodes 13 mRNAs that are translated on mitochondrial ribosomes to generate proteins of the respiratory chain complexes, which are essential for oxidative phosphorylation. The mitochondrial protein synthesis machinery employs a minimalistic set of 22 mitochondrial tRNAs and, even though they contain a reduced number of modified residues compared to their cytoplasmic counterparts, mitochondrial tRNAs possess multiple RNA modifications that require import of the corresponding modification enzymes from the cytoplasm (Watanabe & Yokobori, 2011; Suzuki & Suzuki, 2014; Powell et al, 2015). The largest diversity of modifications in these tRNAs occurs in and around the anticodon, especially at the wobble position. This coincides with the extreme reduction in isoacceptors, requiring most tRNAs to act in decoding of several different codons, and with specific mitochondrial changes in the universal genetic code. Despite the importance of the tRNA modifications for mitochondrial translation and physiology, many of the modification pathways, the enzymes involved and the roles of these modifications in mitochondrial translation have remained unknown so far.
Here, we describe the biosynthetic pathway that introduces modifications at the wobble position of the mitochondrial tRNAMet. We show that the RNA methyltransferase NSUN3 efficiently methylates C34 of mt‐tRNAMet to produce m5C, which can then be oxidised by the alpha‐ketoglutarate and Fe(II)‐dependent dioxygenase ALKBH1/ABH1. Mammalian mt‐tRNAMet can be modified to f5C at the wobble position, and our data demonstrate that this modification is introduced in vivo by the consecutive action of NSUN3 and ABH1 (Fig 9C). Interestingly, the bisulphite sequencing data further suggest that after methylation by NSUN3 only a part of the mitochondrial pool of mt‐tRNAMet is oxidised by ABH1, indicating the presence of m5C34‐containing mt‐tRNAMet in vivo.
Figure 9. NSUN3 and ABH1 are both required for efficient mitochondrial translation in vivo .
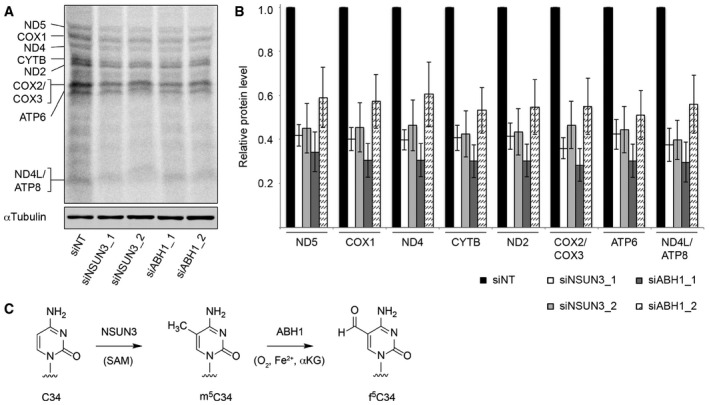
- HeLa cells were treated with non‐target siRNAs (siNT) or those targeting NSUN3 (siNSUN3_1 or siNSUN3_2) or ABH1 (siABH1_1 or siABH1_2) for 72 h before labelling of mitochondrial translation products with [35S]methionine. Protein samples were separated by SDS–PAGE then transferred to a membrane. Labelled proteins were detected using a phosphorimager, and the levels of tubulin were determined by Western blotting using an antibody against the endogenous protein for normalisation.
- Mitochondrially translated proteins that could be clearly detected were quantified in three independent experiments, and the results are shown graphically as mean ± SD.
- Overview of the modification pathway of C34 in mt‐tRNAMet. NSUN3 introduces an m5C methylation on C34 using S‐adenosylmethionine (SAM) as methyl group donor, and this can be further oxidised by ABH1 in the presence of O2, Fe(II) (Fe2+) and alpha‐ketoglutarate (αKG) to produce f5C34.
The ability of ABH1 to oxidise m5C to f5C is striking with respect to the previously described substrate specificity of this oxygenase enzyme. ABH1 can demethylate single‐stranded DNA and RNA in vitro with low efficiency, with a preference for oxidation of N3‐methylcytosine (m3C) (Westbye et al, 2008), and has been suggested to act as histone demethylase and abasic site lyase (Müller et al, 2010; Ougland et al, 2012). While the homologous E. coli AlkB cannot oxidise m5C in vitro (Li et al, 2010) and human ALKBH2 and ALKBH3 preferentially repair alkylation at nucleobase heteroatoms such as m3C and 1‐methyladenosine (m1A) (Aas et al, 2003; Falnes et al, 2004), the oxidation of m5C involves transformation of a pseudobenzylic methyl group. In DNA, this reaction is catalysed by related Fe(II)/α‐ketoglutarate‐dependent oxygenases of the TET enzyme family, and the oxidation products play a significant role in epigenetic regulation in mammals (Tahiliani et al, 2009; Breiling & Lyko, 2015; Li et al, 2015). The TET enzymes produce hm5C as primary stable oxidation product, which can be further oxidised to f5C and 5‐carboxycytosine (ca5C), although these higher oxidation products are 10‐ to 100‐fold less abundant than hm5C in DNA and are mainly linked to active demethylation (Ito et al, 2011; Pfaffeneder et al, 2011; Wagner et al, 2015). In RNA, the analogous oxidation of m5C to hm5C has been reported by catalytic domains of mammalian TET enzymes (Fu et al, 2014) and the homologous Drosophila protein dTET (Delatte et al, 2016). f5C was detected as minor oxidation product in total cellular RNA by mass spectrometry‐based isotope tracing (Huber et al, 2015), but the enzymes generating this modification have remained unknown. The observation that oxidation products of m5C have been detected in RNA from all domains of life, including organisms that do not contain homologous TET enzymes, suggests that m5C can be metabolically oxidised by enzymes other than those of the TET family. We have identified ABH1 as the first such enzyme that produces f5C in human mitochondria. Under the conditions tested, f5C was the only oxidation product detected in vitro; hm5C did not accumulate as intermediate and no further oxidation to ca5C was detected. In the absence of a three‐dimensional structure of ABH1, the molecular reasons for the apparent specificity of ABH1 for the biosynthesis of f5C remain unknown. With the broad target spectrum reported for ABH1, it will also be interesting to understand on the structural level how this enzyme can accommodate interactions with diverse protein and RNA substrates and modulate their modification state.
In mitochondria, NSUN3 and ABH1 act on mt‐tRNAMet, which represents the only tRNAMet that acts both in translation initiation and elongation, in contrast to bacterial and eukaryotic cytoplasmic translation systems. Besides reading the universal AUG codons, mt‐tRNAMet is employed for decoding of AUA codons during initiation and elongation, as well as an AUU initiation codon in the case of the NADH dehydrogenase 2 (ND2) mRNA. Our data obtained with synthetic aminoacylated mt‐tRNAMet containing unmodified C34, m5C34, hm5C34 or f5C34 and the human mitochondrial translation initiation factor MTIF2 reveal that the presence of the m5C modification in the wobble position enhances codon reading of the AUG and, to a lesser extent, AUA initiation codons in the P site of the ribosome, suggesting a specific role of m5C34 modification during translation initiation. The AUU initiation codon, which is only present in the ND2 mRNA, is recognised, albeit poorly, by non‐modified or f5C‐modified mt‐tRNAMet. The recognition efficiency of the AUA and AUU initiation codons is low, consistent with the previous results obtained with mt‐tRNAMet anticodon stem loop (Bilbille et al, 2011). However, given that translation in mitochondria is generally slow and the mRNA recruitment for translation often relies on protein factors specific for each mRNA (Kuzmenko et al, 2014), it is conceivable that even weak codon–anticodon interaction with mt‐tRNAMet may be sufficient to start translation. While m5C‐modified mt‐tRNAMet preferentially acts in translation initiation, results from A site binding studies in the presence of the mitochondrial elongation factor TUFM suggest that mt‐tRNAMet variants other than m5C34 are more efficient in decoding of the internal AUG codons during translation elongation. In combination with the generally lower efficiency of the alternative codons in the in vitro binding assays, only small differences between the binding of the unmodified, m5C‐ or f5C‐modified mt‐tRNAMet to AUA codons in the ribosomal A site were observed, while the binding of hm5C‐containing mt‐tRNAMet was less efficient. Previous reports with the unmodified or f5C34‐containing ASL of mt‐tRNAMet suggested that the formyl group might stabilise the non‐conventional basepairing of f5C34 with an adenosine in the third position of an AUA codon (Bilbille et al, 2011; Cantara et al, 2013). These studies also observed that binding of mt‐tRNAMet to alternative codons was weaker than to AUG and it is likely that ribosome interactions with mt‐tRNAMet outside of the ASL, that is with the tRNA body, further influence mt‐tRNAMet binding. Together, our data indicate that the different modification states of cytosine 34 in mt‐tRNAMet can expand the ability of the single tRNAMet to read the different codons encoding methionine in mitochondrial translation initiation and elongation.
The modification state of C34 in mt‐tRNAMet is controversially discussed in the literature and two reports that were published while this manuscript was under consideration find different levels of NSUN3‐dependent m5C34 and f5C34 in human mt‐tRNAMet (Nakano et al, 2016; Van Haute et al, 2016). Our findings imply that in vivo a large fraction of the m5C34‐containing mt‐tRNAMet is oxidised by ABH1, which is in line with previous reports that found the f5C34 modification in mt‐tRNAMet (Moriya et al, 1994; Takemoto et al, 2009; Suzuki & Suzuki, 2014). However, we also observed that mt‐tRNAMet carrying m5C34 is present in vivo, which is supported by findings of Van Haute et al (2016), and that this modification state of mt‐tRNAMet is efficiently recruited to the P site of the ribosome in vitro. Importantly, mutations in mt‐tRNAMet itself have been shown to cause severe mitochondrial disorders (Lott et al, 2013; Tang et al, 2013) and we found that one such mutation (C39U), which leads to destabilisation of the anticodon stem structure, largely abolishes mt‐tRNAMet methylation by NSUN3. These results indicate that NSUN3 malfunction and a lack in mt‐tRNAMet modification might represent the molecular cause of such diseases. An important role of the modifications installed in mt‐tRNAMet by NSUN3 and ABH1 is further supported by our findings that knock‐down of either NSUN3 or ABH1 affects mitochondrial translation and leads to reduced cell survival. While mt‐tRNAMet likely represents the only substrate of NSUN3, ABH1 has a broader target spectrum and its depletion might also influence other molecules affecting mitochondrial translation. Interestingly, previous reports have suggested a differential localisation of ABH1 in different cell types. While the dioxygenase is mainly localised in mitochondria and the cytoplasm in HEK293 and HeLa cells (Fig 5; Westbye et al, 2008), it has been reported to be nuclear in embryonic stem cells (Ougland et al, 2012, 2016). Together, these findings suggest that the methylation of cytosine 34 in mt‐tRNAMet by NSUN3 represents an important modification present in many, if not all cell types, while the different localisation of ABH1 might result in differential modification of mt‐tRNAMet on cytosine 34 in different cell types, tissues and developmental stages and might thereby fine tune mitochondrial translation in vivo.
Materials and Methods
Human cell culture, stable cell lines and in vivo cross‐linking
HeLa CCL2 and HEK293 Flp‐In T‐Rex cells (Life Technologies) were cultured with 5% CO2 at 37°C in DMEM supplemented with 10% FCS and 2 mM glutamine. For generation of tetracycline‐inducible stable cell lines the NSUN3 or ABH1 CDS were cloned into the pcDNA5 vector with C‐terminal GFP or His‐PreScission protease cleavage site‐2×FLAG (HisPrcFlag) tag. The catalytically inactive NSUN3 C265A mutant was generated by site‐directed mutagenesis (Haag et al, 2015b). The constructs were transfected into HEK293 Flp‐In T‐Rex cells according to the manufacturer's instructions and as described (Sloan et al, 2015). UV and 5‐AzaC cross‐linking and analysis of cDNA (CRAC) experiments were carried out as previously described (Bohnsack et al, 2012; Haag et al, 2015a; see also Appendix Supplementary Methods). Detection of co‐immunoprecipitated tRNA by Northern blot was performed as previously described (Haag et al, 2015a). In brief, after cross‐linking and immunoprecipitation of protein–RNA complexes the RNA was eluted by proteinase K digestion for 16 h, precipitated and resuspended in loading dye (95% formamide, 5 mM EDTA, bromophenol blue). The RNA was separated on a denaturing 12% polyacrylamide gel (7M urea), transferred to a nylon membrane and selected tRNAs were detected by Northern blotting using specific 32P‐5′ end‐labelled probes (anti‐mt‐tRNAMet, anti‐mt‐tRNAGlu, anti‐mt‐tRNAPro, anti‐tRNAi Met, anti‐tRNAe Met; Appendix Table S1) on a phosphorimager.
Microscopy, isolation of mitochondria and protease protection assays
HEK293 cells expressing NSUN3‐GFP under the control of a tetracycline‐inducible promoter were selected and NSUN3‐GFP expression was induced by 1 μg/mL doxycycline treatment for 24 h. Cells were treated with MitoTracker® Orange CMTMRos (Life Technologies) in PBS for 20 min at 37°C, washed in PBS and fixed with 4% formaldehyde in PBS for 10 min at room temperature. After washing with PBS, cells were mounted on coverslips using Vectashield® (Vector labs) for confocal microscopy and localisation analysis. Alternatively, immunofluorescence using an antibody against ABH1 (see Appendix Table S2) was performed as previously described (Haag et al, 2015a). Isolation of mitochondria, analysis of submitochondrial localisation and protease protection assays were performed as described using the antibodies listed in Appendix Table S2 (Dennerlein et al, 2015).
RNA interference, RNA isolation and qRT–PCR
HeLa CCL2 cells were transfected with siRNAs (40 nM) targeting NSUN3 (siNSUN3_1, siNSUN3_2) or ABH1 (siABH1_1, siABH1_2) or a non‐target siRNA (siNT) using Lipofectamine RNAiMax (Life Technologies) according to the manufacturer's instructions. Cells were harvested 96 h after siRNA transfection and total RNA was isolated using TRI reagent (Sigma‐Aldrich). The knock‐down efficiency was determined by qRT–PCR and relative quantification was performed using primers for NSUN3 (NSUN3_qPCR_fwd, NSUN3_qPCR_rev), ABH1 (ABH1_qPCR_fwd, ABH1_qPCR_rev) and GAPDH (GAPDH_qPCR_fwd, GAPDH_qPCR_rev; for siRNA and primer sequences see Appendix Table S1 and S3).
NaBH4 treatment and bisulphite reaction
To analyse the cytosine modification status of mt‐tRNAMet, DNase I treated total RNA from wild‐type, NSUN3 or ABH1 knock‐down cells was either directly subjected to bisulphite sequencing (Schaefer et al, 2009) or treated with 0.25 M NaBH4 in 200 mM Tris–HCl pH 7.5, 20 mM MgCl2, 200 mM KCl for 30 min on ice and precipitated prior to the bisulphite reaction. Reduced or untreated RNA was bisulphite treated using the Qiagen bisulphite kit according to the manufacturer's instructions. The deamination reaction was carried out in a thermocycler with 5 min at 70°C, 60 min at 60°C (3 times). Samples were desalted using 6×SSC Micro bio spin chromatography columns and subsequently desulphonated by incubation in Tris pH 9 for 30 min at 37°C. The RNA was precipitated and reverse transcribed using the mt‐tRNAMet_RT primer and Superscript III reverse transcriptase (Thermo) according to the manufacturer's instructions. PCR products were then cloned using a TOPO‐TA kit (Thermo) and sequenced. At least 50 sequences were analysed per sample and only sequences in which all other cytosines besides C34 in mt‐tRNAMet were converted were used for the analysis presented.
Cloning and recombinant expression of proteins and in vitro transcription of tRNAs
The coding sequences of human NSUN3, ABH1 or FTO were cloned into a pQE80 derivative encoding an N‐terminal His14‐MBP‐tag (Weis et al, 2014) and the CDS of MTIF2 or TUFM into a pQE80 derivative encoding a C‐terminal His10‐tag (Mingot et al, 2004). The ABH1 D233A and R233A, and NSUN3 C265A mutants were generated by site‐directed mutagenesis (Haag et al, 2015b). Recombinant proteins were expressed in Escherichia coli (DE3) Rosetta pLysS (NSUN3, ABH1) or (BL21) Codon Plus (MTIF2, TUFM) cells and details of protein purification are given in the Appendix Supplementary Methods. Mt‐tRNAMet, mt‐tRNAGlu, mt‐tRNAPro, tRNAi Met and tRNAe Met sequences were generated by recursive PCR as described (Müller et al, 2013) using four overlapping oligonucleotides each and cloned into a pQE vector derivative lacking an internal T7 promoter. The CCA tail and a BsaI restriction site were added at the 3′ end of the tRNA gene and the forward primer contained the sequence of the T7 promoter. Point mutations were introduced by site‐directed mutagenesis. For in vitro transcription, 500 ng of BsaI‐linearised plasmid were incubated with 1 mM NTPs, T7‐RNA polymerase, 1× transcription buffer (Thermo) and RiboLock (Thermo) for 1 h at 37°C. After transcription, samples were treated with DNase I for 15 min and purified over a Sephadex G‐25 spin column (Roche).
Preparation of synthetic tRNAs and ribosome binding assays
RNA oligonucleotides were prepared by solid‐phase synthesis using 2′‐O‐TOM‐protected ribonucleotide phosphoramidites, chemically phosphorylated on solid support, deprotected in two steps with methylamine in water/ethanol, followed by 1 M tetrabutylammonium fluoride in tetrahydrofuran, purified by denaturing PAGE, and analysed by analytical anion exchange chromatography under denaturing conditions (6M urea, 80°C) and ESI‐MS. Synthetic tRNAs were prepared by enzymatic ligation of chemically synthesised RNA fragments using T4 DNA ligase and DNA splint oligonucleotides (2–5 nmol scale, incubation at 30°C for 12 h), analogous to previously reported procedures (Rieder et al, 2009). The full‐length tRNAs were isolated by denaturing PAGE, extracted into Tris–NaCl buffer, precipitated with ethanol and re‐dissolved in water. To generate f5C34 or hm5C‐containing mt‐tRNAMet, ligation was performed with m5C34‐ASL RNA oligonucleotides that were treated with recombinant ABH1 on preparative scale (5–10 nmol) (see oxidation assays for conditions), or treated with ABH1 and then reduced with NaBH4 (see NaBH4 treatment). The modified ASLs were PAGE purified and their homogeneity and identity were confirmed by anion exchange HPLC and ESI‐MS. Labelling of f5C‐RNA with 1‐ethyl‐2,3,3‐trimethylindoleninium‐5‐sulphonate (TMI) and analysis of primer extension stops on sequencing gels were performed as described (van Nues et al, 2011; Samanta et al, 2016). Ribosome binding assays were performed as described (Rezgui et al, 2013; see also Appendix Supplementary Methods).
In vitro methylation and oxidation assays
Methylation of RNAs was carried out essentially as described (Jurkowski et al, 2008; Müller et al, 2013). Reactions containing 1 μM recombinant NSUN3 and 1 μM of tRNA or 10 μg of total RNA in 1× methylation buffer (50 mM Tris–HCl pH 7.0, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT) and 1.7 μM [3H]‐SAM (Hartmann), 1 unit/ml RiboLock (Thermo) were incubated at 22°C for 2 h. After addition of proteinase K for 30 min to stop the reaction, RNAs were separated on a 12% denaturing (7 M urea) polyacrylamide gel, stained with ethidium bromide, fixed and immersed in amplify solution (Amersham) for 1 h. After drying, the gel was exposed to a X‐ray film for 16 h to 2 weeks at −80°C. For in vitro oxidation reactions mt‐tRNAMet or mt‐tRNAMet ASL were labelled with a [3H]‐containing methyl group by in vitro methylation with NSUN3. The methylated RNA was precipitated and incubated with 1 μM recombinant wild‐type or mutant His14‐MBP‐ABH1, MBP or His14‐MBP‐FTO in the presence of 50 mM HEPES pH 6.9, 5 mM MgCl2, 4 mM ascorbic acid and 100 μM Fe(NH4)2(SO4)2 and 100 μM α‐ketoglutarate for 1 h at 22°C. The reaction was stopped by addition of proteinase K and the RNA was precipitated. The supernatant containing released [3H] was analysed by scintillation counting, and the corresponding RNA pellets were separated by denaturing gel electrophoresis and analysed as described for the methylation assay. Preparative scale oxidation of synthetic m5C34 ASL for preparation of mt‐tRNAMet by ligation was performed under analogous conditions, followed by PCI extraction and PAGE purification.
In vivo analysis of mitochondrial translation
In vivo labelling was performed as previously described (Chomyn, 1996). HeLa cells were transfected with siRNAs (non‐target, NSUN3 or ABH1) and cultivated for 72 h. Before labelling, cells were starved in medium lacking serum and methionine. Cytosolic translation was inhibited by treating cells with 100 μg/ml emetine (Sigma‐Aldrich) for 10 min. Translation of mitochondrial proteins was pulsed with 0.2 mCi/ml 35S methionine for 30 min. Cells were harvested, and proteins were separated on a 10–18% Tricin–SDS–PAGE followed by transfer onto a PVDF membrane and exposure to a phosphor screen. Autoradiography signals were measured by a phosphorimager (Typhoon FLA 9500) and quantified by Imagequant TL software (GE Healthcare). Equivalent amounts of samples were run on SDS–PAGE for fluorescent Western blot analysis for normalisation.
Data availability
The primary high‐throughput sequencing data of the UV and 5‐azacytidine cross‐linking and analysis of cDNA (CRAC) experiments have been submitted to the GEO SRA database and assigned the identifier GSE84664.
Author contributions
SH, KES, CB, ASW, BH and MTB purified proteins and performed methyltransferase and oxidation assays; SH and KES performed and analysed cross‐linking experiments; SH, KES and CB did bisulphite treatment and analysis; KES, JK and MTB performed bioinformatics analysis; ASW, SD and PR analysed NSUN3 and ABH1 localisation; CH, SH, JS and KES synthesised and analysed RNAs; NR and MVR designed and performed ribosome binding assays; ASW, SD, KES, SH and PR performed RNAi and mitochondrial translation assays; PR, MVR, CH and MTB designed the study and analysed data; MTB, CH and KES wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank Philipp Hackert for technical assistance and Sebastian Leidel for helpful discussions. This work was supported by the Deutsche Forschungsgemeinschaft (SPP 1784: BO3442/2‐1 to M.T.B., HO4436/2‐1 to C.H.; SFB1190 to M.T.B., M.V.R. and P.R.), the Alexander von Humboldt Foundation (postdoctoral fellowship to K.E.S.), European Research Council (AdG No. 339580) to P.R., the Faculty of Medicine, Georg‐August‐University Göttingen (M.T.B., P.R. and “Startförderung” to S.H.) and the Max Planck Society (M.V.R., C.H and P.R.).
The EMBO Journal (2016) 35: 2104–2119
See also: F Boos et al (October 2016)
Contributor Information
Claudia Höbartner, Email: claudia.hoebartner@chemie.uni-goettingen.de.
Markus T Bohnsack, Email: Markus.Bohnsack@med.uni-goettingen.de.
References
- Aas PA, Otterlei M, Falnes PO, Vågbø CB, Skorpen F, Akbari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, Krokan HE (2003) Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421: 859–863 [DOI] [PubMed] [Google Scholar]
- Agris PF, Vendeix FA, Graham WD (2007) tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol 366: 1–13 [DOI] [PubMed] [Google Scholar]
- Berulava T, Ziehe M, Klein‐Hitpass L, Mladenov E, Thomale J, Rüther U, Horsthemke B (2013) FTO levels affect RNA modification and the transcriptome. Eur J Hum Genet 21: 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbille Y, Gustilo EM, Harris KA, Jones CN, Lusic H, Kaiser RJ, Delaney MO, Spremulli LL, Deiters A, Agris PF (2011) The human mitochondrial tRNAMet: structure/function relationship of a unique modification in the decoding of unconventional codons. J Mol Biol 406: 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, Kellner S, Hölter SM, Garrett L, Wurst W, Becker L, Klopstock T, Fuchs H, Gailus‐Durner V, Hrabĕ de Angelis M, Káradóttir RT et al (2014) Aberrant methylation of tRNAs links cellular stress to neuro‐developmental disorders. EMBO J 33: 2020–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Tollervey D, Granneman S (2012) Identification of RNA helicase target sites by UV cross‐linking and analysis of cDNA. Methods Enzymol 511: 275–288 [DOI] [PubMed] [Google Scholar]
- Booth MJ, Ost TW, Beraldi D, Bell NM, Branco MR, Reik W, Balasubramanian S (2013) Oxidative bisulfite sequencing of 5‐methylcytosine and 5‐hydroxymethylcytosine. Nat Protoc 8: 1841–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A, Lyko F (2015) Epigenetic regulatory functions of DNA modifications: 5‐methylcytosine and beyond. Epigenetics Chromatin 8: 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki JM, Feder M, Ayres CL, Redman KL (2004) Sequence‐structure‐function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res 32: 2453–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cámara Y, Asin‐Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, Franz T, Erdjument‐Bromage H, Tempst P, Hallberg BM, Gustafsson CM, Larsson NG (2011) MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab 13: 527–539 [DOI] [PubMed] [Google Scholar]
- Cantara WA, Murphy FV 4th, Demirci H, Agris PF (2013) Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc Natl Acad Sci USA 110: 10964–10969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas‐Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV (2014) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn A (1996) In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol 264: 197–211 [DOI] [PubMed] [Google Scholar]
- Czerwoniec A, Dunin‐Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K (2009) MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res 37: D118–D121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, Wetzel C, Kreher J, Soin R, Creppe C, Limbach PA, Gueydan C, Kruys V, Brehm A, Minakhina S, Defrance M et al (2016) Transcriptome‐wide distribution and function of RNA hydroxymethylcytosine. Science 351: 282–285 [DOI] [PubMed] [Google Scholar]
- Dennerlein S, Oeljeklaus S, Jans D, Hellwig C, Bareth B, Jakobs S, Deckers M, Warscheid B, Rehling P (2015) MITRAC7 Acts as a COX1‐Specific Chaperone and Reveals a Checkpoint during Cytochrome c Oxidase Assembly. Cell Rep 12: 1644–1655 [DOI] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch‐Moshkovitz S, Peer E, Kol N, Ben‐Haim MS, Dai Q, Di Segni A, Salmon‐Divon M, Clark WC, Zheng G, Pan T, Solomon O, Eyal E, Hershkovitz V, Han D, Doré LC, Amariglio N, Rechavi G, He C (2016) The dynamic N(1)‐methyladenosine methylome in eukaryotic messenger RNA. Nature 530: 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duechler M, Leszczyńska G, Sochacka E, Nawrot B (2016) Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell Mol Life Sci 73: 3075–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PØ, Bjørås M, Aas PA, Sundheim O, Seeberg E (2004) Substrate specificities of bacterial and human AlkB proteins. Nucleic Acids Res 32: 3456–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM (2015) The AlkB Family of Fe(II)/α‐Ketoglutarate‐dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. J Biol Chem 290: 20734–20742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD (2010a) Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol Cell Biol 30: 2449–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dai Q, Zhang W, Ren J, Pan T, He C (2010b) The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5‐methoxycarbonylmethyluridine at the wobble position of tRNA. Angew Chem Int Ed Engl 49: 8885–8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Jordan JJ, Samson LD (2013) Human ALKBH7 is required for alkylation and oxidation‐induced programmed necrosis. Genes Dev 27: 1089–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, Cai Q, Ji D, Jin SG, Niedernhofer LJ, Pfeifer GP, Xu GL, Wang Y (2014) Tet‐mediated formation of 5‐hydroxymethylcytosine in RNA. J Am Chem Soc 136: 11582–11585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH (2006) Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311: 395–398 [DOI] [PubMed] [Google Scholar]
- Greber BJ, Ban N (2016) Structure and Function of the Mitochondrial Ribosome. Annu Rev Biochem 85: 103–132 [DOI] [PubMed] [Google Scholar]
- Haag S, Warda AS, Kretschmer J, Günnigmann MA, Höbartner C, Bohnsack MT (2015a) NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA 21: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag S, Kretschmer J, Bohnsack MT (2015b) WBSCR22/Merm1 is required for late nuclear pre‐ribosomal RNA processing and mediates N7‐methylation of G1639 in human 18S rRNA. RNA 21: 180–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H (2014) Methylated nucleosides in tRNA and tRNA methyltransferases. Front Genet 5: 144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SM, van Delft P, Mendil L, Bachman M, Smollett K, Werner F, Miska EA, Balasubramanian S (2015) Formation and abundance of 5‐hydroxymethylcytosine in RNA. ChemBioChem 16: 752–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, Frye M (2013) NSun2‐mediated cytosine‐5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 4: 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y (2011) Tet proteins can convert 5‐methylcytosine to 5‐formylcytosine and 5‐carboxylcytosine. Science 333: 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C (2011) N6‐methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nat Chem Biol 7: 885–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A (2008) Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase‐like catalytic mechanism. RNA 14: 1663–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, Cairns BR (2013) Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol 31: 458–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmenko A, Atkinson GC, Levitskii S, Zenkin N, Tenson T, Hauryliuk V, Kamenski P (2014) Mitochondrial translation initiation machinery: conservation and diversification. Biochimie 100: 1321–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisegang MS, Martin R, Ramírez AS, Bohnsack MT (2012) Exportin t and Exportin 5: tRNA and miRNA biogenesis ‐ and beyond. Biol Chem 393: 599–604 [DOI] [PubMed] [Google Scholar]
- Li D, Delaney JC, Page CM, Chen AS, Wong C, Drennan CL, Essigmann JM (2010) Repair of DNA Alkylation Damage by the Escherichia coli Adaptive Response Protein AlkB as Studied by ESI‐TOF Mass Spectrometry. J Nucleic Acids 2010: 369434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Guo B, Wu H, Tan L, Lu Q (2015) TET Family of Dioxygenases: Crucial Roles and Underlying Mechanisms. Cytogenet Genome Res 146: 171–180 [DOI] [PubMed] [Google Scholar]
- Liu J, Jia G (2014) Methylation modifications in eukaryotic messenger RNA. J Genet Genomics 41: 21–33 [DOI] [PubMed] [Google Scholar]
- Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, Procaccio V, Wallace DC (2013) mtDNA Variation and Analysis Using Mitomap and Mitomaster. Curr Protoc Bioinformatics 44: 1.23.1–1.23.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metodiev MD, Spåhr H, Loguercio Polosa P, Meharg C, Becker C, Altmueller J, Habermann B, Larsson NG, Ruzzenente B (2014) NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet 10: e1004110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingot JM, Bohnsack MT, Jäkle U, Görlich D (2004) Exportin 7 defines a novel general nuclear export pathway. EMBO J 23: 3227–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya J, Yokogawa T, Wakita K, Ueda T, Nishikawa K, Crain PF, Hashizume T, Pomerantz SC, McCloskey JA, Kawai G, Hayashi N, Yokoyama S, Watanabe K (1994) A novel modified nucleoside found at the first position of the anticodon of methionine tRNA from bovine liver mitochondria. Biochemistry 33: 2234–2239 [DOI] [PubMed] [Google Scholar]
- Motorin Y, Helm M (2011) RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2: 611–631 [DOI] [PubMed] [Google Scholar]
- Müller TA, Meek K, Hausinger RP (2010) Human AlkB homologue 1 (ABH1) exhibits DNA lyase activity at abasic sites. DNA Repair (Amst) 9: 58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Windhof IM, Maximov V, Jurkowski T, Jeltsch A, Förstner KU, Sharma CM, Gräf R, Nellen W (2013) Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2‐homologue (DnmA). Nucleic Acids Res 4: 8615–8627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T (2016) NSUN3 methylase initiates 5‐formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat Chem Biol 12: 546–551 [DOI] [PubMed] [Google Scholar]
- van Nues RW, Granneman S, Kudla G, Sloan KE, Chicken M, Tollervey D, Watkins NJ (2011) Box C/D snoRNP catalysed methylation is aided by additional pre‐rRNA base‐pairing. EMBO J 30: 2420–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ougland R, Lando D, Jonson I, Dahl JA, Moen MN, Nordstrand LM, Rognes T, Lee JT, Klungland A, Kouzarides T, Larsen E (2012) ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem Cells 30: 2672–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ougland R, Rognes T, Klungland A, Larsen E (2015) Non‐homologous functions of the AlkB homologs. J Mol Cell Biol 7: 494–504 [DOI] [PubMed] [Google Scholar]
- Ougland R, Jonson I, Moen MN, Nesse G, Asker G, Klungland A, Larsen E (2016) Role of ALKBH1 in the Core Transcriptional Network of Embryonic Stem Cells. Cell Physiol Biochem 38: 173–184 [DOI] [PubMed] [Google Scholar]
- Pan Z, Sikandar S, Witherspoon M, Dizon D, Nguyen T, Benirschke K, Wiley C, Vrana P, Lipkin SM (2008) Impaired placental trophoblast lineage differentiation in Alkbh1(‐/‐) mice. Dev Dyn 237: 316–327 [DOI] [PubMed] [Google Scholar]
- Pfaffeneder T, Hackner B, Truss M, Münzel M, Müller M, Deiml CA, Hagemeier C, Carell T (2011) The discovery of 5‐formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl 50: 7008–7012 [DOI] [PubMed] [Google Scholar]
- Powell CA, Nicholls TJ, Minczuk M (2015) Nuclear‐encoded factors involved in post‐transcriptional processing and modification of mitochondrial tRNAs in human disease. Front Genet 6: 79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan N, Rodnina MV (2016) tRNA wobble modifications and protein homeostasis. Translation 4: e1143076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezgui VA, Tyagi K, Ranjan N, Konevega AL, Mittelstaet J, Rodnina MV, Peter M, Pedrioli PG (2013) tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine‐tune protein translation by promoting ribosome A‐site binding. Proc Natl Acad Sci USA 110: 12289–12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY (2013) Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339: 1328–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder R, Höbartner C, Micura R (2009) Enzymatic ligation strategies for the preparation of purine riboswitches with site‐specific chemical modifications. Methods Mol Biol 540: 15–24 [DOI] [PubMed] [Google Scholar]
- Samanta B, Seikowski J, Höbartner C (2016) Fluorogenic Labeling of 5‐Formylpyrimidine Nucleotides in DNA and RNA. Angew Chem Int Ed Engl 55: 1912–1916 [DOI] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Lyko F (2009) RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 37: e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F (2010) RNA methylation by Dnmt2 protects transfer RNAs against stress‐induced cleavage. Genes Dev 24: 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, Calle‐Perez A, Pircher A, Gerstl MP, Pfeifenberger S, Brandl C, Sonntagbauer M, Kriegner A, Linder A, Weinhäusel A, Mohr T, Steiger M, Mattanovich D, Rinnerthaler M, Karl T et al (2015) Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun 6: 6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Lafontaine DL (2015) ‘View From A Bridge’: A New Perspective on Eukaryotic rRNA Base Modification. Trends Biochem Sci 40: 560–575 [DOI] [PubMed] [Google Scholar]
- Shen L, Song CX, He C, Zhang Y (2014) Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem 83: 585–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Bohnsack MT, Watkins NJ (2013) The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep 5: 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Leisegang MS, Doebele C, Ramírez AS, Simm S, Safferthal C, Kretschmer J, Schorge T, Markoutsa S, Haag S, Karas M, Ebersberger I, Schleiff E, Watkins NJ, Bohnsack MT (2015) The association of late‐acting snoRNPs with human pre‐ribosomal complexes requires the RNA helicase DDX21. Nucleic Acids Res 43: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg A, Robertson AB, Aronsen JM, Rognmo Ø, Sjaastad I, Wisløff U, Klungland A (2013) Deletion of mouse Alkbh7 leads to obesity. J Mol Cell Biol 5: 194–203 [DOI] [PubMed] [Google Scholar]
- Songe‐Møller L, van den Born E, Leihne V, Vågbø CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PØ, Klungland A (2010) Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol Cell Biol 30: 1814–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Suzuki T (2014) A complete landscape of post‐transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 42: 7346–7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL (2013) The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre‐rRNA processing factors. Mol Cell 51: 539–551 [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A (2009) Conversion of 5‐methylcytosine to 5‐hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto C, Spremulli LL, Benkowski LA, Ueda T, Yokogawa T, Watanabe K (2009) Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res 37: 1616–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Wang J, Zhang VW, Li FY, Landsverk M, Cui H, Truong CK, Wang G, Chen LC, Graham B, Scaglia F, Schmitt ES, Craigen WJ, Wong LJ (2013) Transition to next generation analysis of the whole mitochondrial genome: a summary of molecular defects. Hum Mutat 34: 882–893 [DOI] [PubMed] [Google Scholar]
- Thalhammer A, Bencokova Z, Poole R, Loenarz C, Adam J, O'Flaherty L, Schödel J, Mole D, Giaslakiotis K, Schofield CJ, Hammond EM, Ratcliffe PJ, Pollard PJ (2011) Human AlkB homologue 5 is a nuclear 2‐oxoglutarate dependent oxygenase and a direct target of hypoxia‐inducible factor 1α (HIF‐1α). PLoS ONE 6: e16210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F (2012) RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19: 900–905 [DOI] [PubMed] [Google Scholar]
- Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, Rorbach J, Lantaff R, Blanco S, Sauer S, Kotzaeridou U, Hoffmann GF, Memari Y, Kolb‐Kokocinski A, Durbin R, Mayr JA, Frye M, Prokisch H, Minczuk M (2016) Deficient methylation and formylation of mt‐tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun 7: 12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Steinbacher J, Kraus TF, Michalakis S, Hackner B, Pfaffeneder T, Perera A, Müller M, Giese A, Kretzschmar HA, Carell T (2015) Age‐dependent levels of 5‐methyl‐, 5‐hydroxymethyl‐, and 5‐formylcytosine in human and mouse brain tissues. Angew Chem Int Ed Engl 54: 12511–12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, He Q, Feng C, Liu Y, Deng Z, Qi X, Wu W, Mei P, Chen Z (2014) The atomic resolution structure of human AlkB homolog 7 (ALKBH7), a key protein for programmed necrosis and fat metabolism. J Biol Chem 289: 27924–27936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Yokobori S (2011) tRNA Modification and Genetic Code Variations in Animal Mitochondria. J Nucleic Acids 2011: 623095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT (2012) The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 3: 397–414 [DOI] [PubMed] [Google Scholar]
- Weis BL, Missbach S, Marzi J, Bohnsack MT, Schleiff E (2014) The 60S associated ribosome biogenesis factor LSG1‐2 is required for 40S maturation in Arabidopsis thaliana. Plant J 80: 1043–10456 [DOI] [PubMed] [Google Scholar]
- Westbye MP, Feyzi E, Aas PA, Vågbø CB, Talstad VA, Kavli B, Hagen L, Sundheim O, Akbari M, Liabakk NB, Slupphaug G, Otterlei M, Krokan HE (2008) Human AlkB homolog 1 is a mitochondrial protein that demethylates 3‐methylcytosine in DNA and RNA. J Biol Chem 283: 25046–25056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K et al (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File
Data Availability Statement
The primary high‐throughput sequencing data of the UV and 5‐azacytidine cross‐linking and analysis of cDNA (CRAC) experiments have been submitted to the GEO SRA database and assigned the identifier GSE84664.


