Abstract
Endogenous retroviruses (ERVs) are transcriptionally active in cleavage stage embryos, yet their functions are unknown. ERV sequences are present in the majority of long intergenic noncoding RNAs (lincRNAs) in mouse and humans, playing key roles in many cellular processes and diseases. Here, we identify LincGET as a nuclear lincRNA that is GLN‐, MERVL‐, and ERVK‐associated and essential for mouse embryonic development beyond the two‐cell stage. LincGET is expressed in late two‐ to four‐cell mouse embryos. Its depletion leads to developmental arrest at the late G2 phase of the two‐cell stage and to MAPK signaling pathway inhibition. LincGET forms an RNA–protein complex with hnRNP U, FUBP1, and ILF2, promoting the cis‐regulatory activity of long terminal repeats (LTRs) in GLN, MERVL, and ERVK (GLKLTRs), and inhibiting RNA alternative splicing, partially by downregulating hnRNP U, FUBP1, and ILF2 protein levels. Hnrnpu or Ilf2 mRNA injection at the pronuclear stage also decreases the preimplantation developmental rate, and Fubp1 mRNA injection at the pronuclear stage causes a block at the two‐cell stage. Thus, as the first functional ERV‐associated lincRNA, LincGET provides clues for ERV functions in cleavage stage embryonic development.
Keywords: ERV, exon skipping, lincRNA, transcription regulation, two‐cell block
Subject Categories: Development & Differentiation, RNA Biology
Introduction
Many events are crucial for the whole embryonic development during the cleavage stage, including nuclear programming 1, zygote genome activation (ZGA) 2, and the first cell fate specification 3. The active transcription of endogenous retroviruses (ERVs) has been described during cleavage stage, yet little is known about their function 4. In mouse and humans, more than two‐thirds of long intergenic noncoding RNAs (lincRNAs) are ERV‐associated 5, indicating the possibility that ERVs regulate cleavage stage embryonic development via their associated lincRNAs 6, 7. However, ERV‐associated lincRNAs have been almost exclusively treated as an aggregate class of repetitive elements. Recent studies revealed that families of ERV‐associated lincRNAs are closely related to pluripotency—the transcription of MERVL 4 and HERVH 8, 9, 10 or HERVK 11 can act as a hallmark of two‐cell embryo‐like totipotent mouse embryonic stem cells (ESCs) and naïve‐like human ESCs, respectively. Functional data on individual ERV‐associated lincRNA are limited due to the highly repetitive nature of its sequence and its low expression level. It was recently reported that some human inner cell mass (ICM) expressing ERV‐associated lincRNAs such as Hpat2, Hpat3, and Hpat5 modulate the acquisition of pluripotency and the formation of the ICM 12. However, whether active ERVs in cleavage stage embryos function through ERV‐associated lincRNAs remains unknown. If any, functional studies of cleavage stage‐specific individual ERV‐associated lincRNA remain challenging.
Although a few have been mechanistically characterized, lincRNAs have emerged as a new field in biology, playing significant roles in many biological functions, including X chromosome inactivation 13, imprinting 14, Hox‐associated pattern formation 15, 16, neuronal fate specification 16, 17, pluripotency and differentiation control 18, 19, 20, cell apoptosis and cell cycle control 21, 22, immune response 23, 24, and mitochondria regulation 25.
The underlying molecular mechanisms related to lincRNAs remain unclear. Overall, lincRNAs and/or their transcripts program various biological functions via epigenetic and nonepigenetic mechanisms at the pre‐transcriptional, transcriptional, and post‐transcriptional levels. At the pre‐transcriptional level, lincRNAs are known as epigenetic modifiers through recruiting chromatin‐modifying complexes in cis or in trans and modeling histone lysine methylation 15, 16, 26, 27, histone acetylation 28, 29, or DNA methylation 30. At the transcriptional level, lincRNAs act as enhancer 31, 32, insulator releaser 33, transcription interferer 34, or transcription factor controller 35. At the post‐transcriptional level, lncRNAs regulate the splicing 36, localization 22, and stability 37 of the target RNAs by base pairing with them or work as miRNA sponge 38, weakening RNAi‐mediated negative regulation.
Here, we identified LincGET as a novel nuclear lincRNA, which is late two‐ to four‐cell mouse embryo‐specific and associated with mouse ERVs. LincGET depletion leads to the developmental arrest at late G2 phase of two‐cell stage with normal initiation of major ZGA and reorganization of pericentric rings into chromocenters. Further, we determined that LincGET acts as a transcription factor and RNA alternative splicing factor via forming a RNA–protein complex with hnRNP U, FUBP1, and ILF2. LincGET mediates the cis‐regulatory activity of GLKLTRs and protects some exons from exon skipping splicing, partially through downregulation of hnRNP U, FUBP1, and ILF2 at the post‐transcriptional level. Hnrnpu or Ilf2 mRNA injection at the pronuclear stage decreased the preimplantation developmental rate and Fubp1 mRNA injection at the pronuclear stage caused two‐cell block. Overall, our results show that LincGET is essential for correct major ZGA processes and furthers the cleavage of two‐cell embryos via regulating the transcription and RNA alternative splicing.
Results
LincGET and Dyei are ERV‐associated two‐ to four‐cell‐specific nuclear lincRNAs
To identify functional ERV‐associated lincRNAs in mouse preimplantation embryos, specific primers toward the mouse ERV sequence (Table EV1) were designed for directional random amplification. Fifty two‐cell, four‐cell, eight‐cell, 16‐cell, 32‐cell, and blastocyst stage embryos were collected and mixed, and total RNA was extracted. Thirty‐six novel transcripts (named by neighbor genes, except for LincGET, named according to its function) were identified using reverse‐transcription polymerase chain reaction (RT–PCR), and the sequencing results were analyzed through UCSC blat tool (http://genome.ucsc.edu/cgi-bin/hgBlat). As expected, most of these transcripts (28/36) were ERV‐associated. Twenty‐three are GLN‐associated, 5 are MERVL‐associated, 4 are LINE‐associated, 1 is in the intergenic region between Gm7627 and Nr2f2, 1 is located in the intron of Ak045672, 1 is 5′‐UTR‐associated, and 1 is an antisense transcript of Serpinc1 (Fig EV1A and Table EV2).
Figure EV1. LincGET and Dyei are ERV‐associated transcripts.
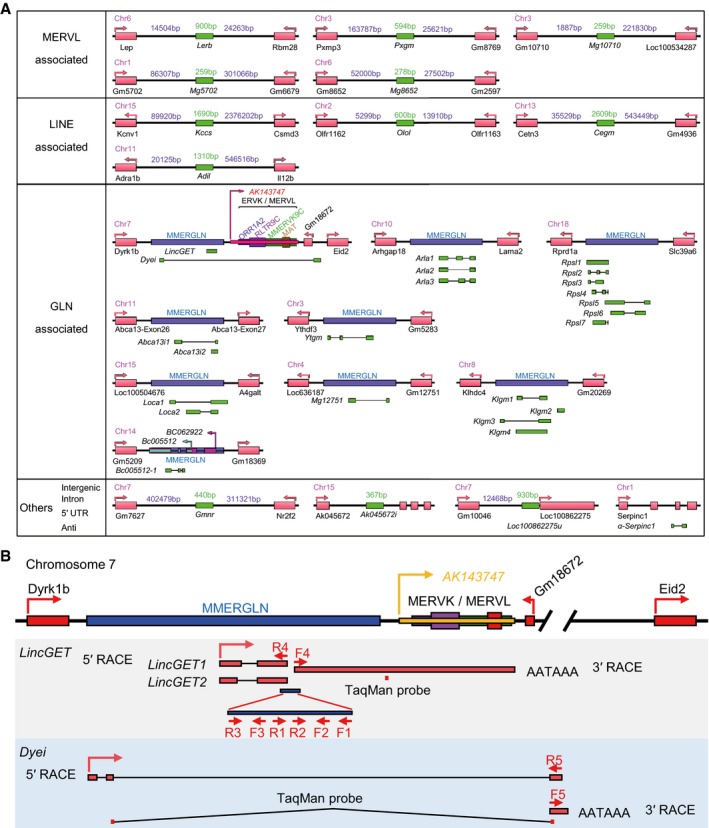
- Gene loci of 36 novel transcripts in mouse preimplantation embryos (GenBank accession numbers are shown in Table EV2). Twenty‐three are GLN‐associated, 5 are MERVL‐associated, 4 are LINE‐associated, 1 is in the intergenic region between Gm7627 and NR2F2, 1 is located in the intron of Ak045672, 1 is 5′ UTR‐associated, and 1 is an antisense transcript of Serpinc1.
- Gene locus of LincGET and Dyei. Primers and TaqMan probes are shown. RACE, rapid amplification of cDNA end. AATAAA is the polyadenylated signal site.
The expression pattern of these 28 ERV‐associated novel transcripts was analyzed by SYBR Green real‐time quantitative PCR (SG‐qPCR) in order to screen functional individual ERV‐associated lincRNAs (Appendix Figs S1 and S2A). The result showed that LincGET and Dyei are the most two‐cell and four‐cell specific, indicating a potential role of LincGET and (or) Dyei in the cleavage stage during mouse development. We choose them for further studies.
In order to obtain the full‐length LincGET, we first performed strand‐specific RT–PCR (SSRT–PCR) analysis because the random amplification fragment has no intron. The results showed that LincGET is only transcribed from one DNA strand that is active for Dyrk1b (Figs 1A and B, and EV1B). The reliability of SSRT–PCR results was confirmed by sequencing (data not shown). For its ERV association, the 3′‐rapid amplification of cDNA ends (RACE) showed multiple bands, but only one band (the marked band in Fig 1C) was confirmed as being LincGET‐specific by sequencing. 3′‐RACE showed that the end of LincGET is similar to that of Ak143743 and that LincGET is a GLN, MERVL, and ERVK‐associated polyadenylated transcript (Figs 1A and EV1B). 5′‐RACE result showed that LincGET presents 2 variants, LincGET1 (6,285 nucleotide (nt), GenBank accession number, KU245560) and LincGET2 (6,107 nt, GenBank accession number, KU245561), resulting from different splice donor of its only intron (Figs 1A, C and D, and EV1B, and Table EV2). The Dyei fragment from random amplification has one intron; thus, its transcription direction is known. Molecular cloning by RACE showed that Dyei (GenBank accession number, KU258809) is a 665‐nt GLN‐associated polyadenylated transcript that possesses 3 exons (Figs 1A–C and EV1B, and Table EV2).
Figure 1. LincGET and Dyei are two‐ to four‐cell embryo‐specific, ERV‐associated nuclear lincRNAs.
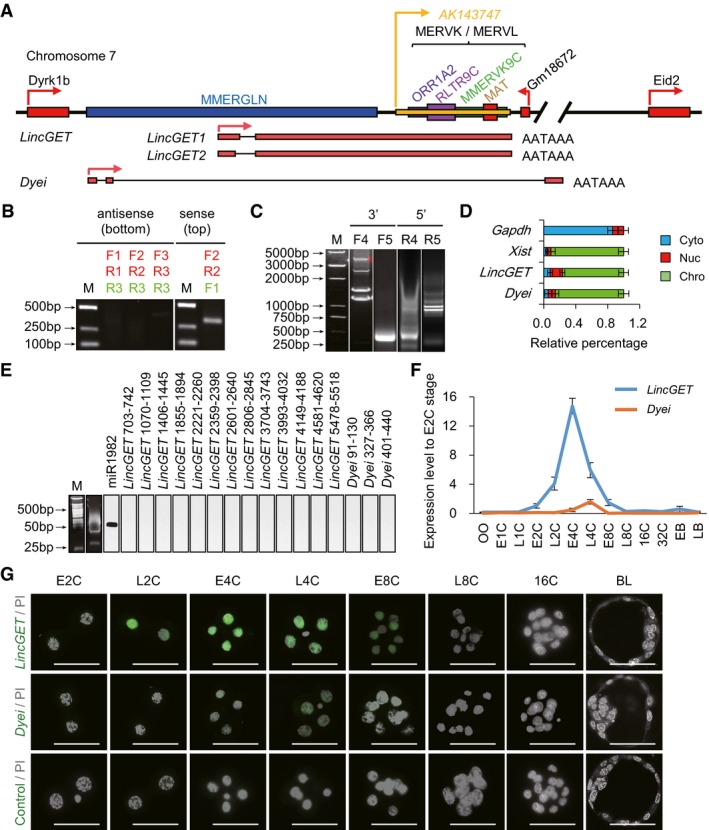
- Gene locus of LincGET and Dyei. LincGET is between Dyrk1b and Gm18672, and Dyei is between Dyrk1b and Eid2. There are one GLN sequence and some ERV fragments of MERVK or MERVL between Dyrk1b and Gm18672. AATAAA is the polyadenylated signal site.
- Single‐strand RT–PCR (SSRT–PCR) results for LincGET. LincGET is transcribed from the top strand. Primers used in strand‐specific reverse transcription are shown in green, while primers used in PCR are shown in red. About 50 early four‐cell embryos were used for each experiment, and three experimental replicates were performed.
- 3′ RACE and 5′ RACE results for LincGET and Dyei. Gene‐specific primers (F4, F5, R4, and R5) are shown in Fig EV1B. *Indicates the bands corresponding to the correct band of 3′ RACE for LincGET. About 200 early four‐cell embryos were used for each RACE experiment, and three experimental replicates were performed.
- Subcellular localization analysis of LincGET and Dyei by RNA fractionation and TM‐qPCR analysis. The results show that LincGET and Dyei locate in the nucleus and are mainly associated with chromatin. The error bars represent s.e.m. Chro, chromosome; Nuc, nucleoplasm; Cyto, cytoplasm. Gapdh and Xist act as cyto and chro control, respectively. About 500 early four‐cell embryos were used for each experiment, and three experimental replicates were performed.
- miRNA reverse Northern blot for LincGET and Dyei. miRNAs were isolated and amplified by RT–PCR after adding double adaptors; then Southern blot (reverse Northern) was performed. It shows no evidence of small RNA products, indicating that LincGET and Dyei do not function as pre‐miRNA. The miR1982 was used as a miRNA positive control. About 400 early four‐cell embryos were used for each experiment, and three experimental replicates were performed.
- Expression pattern of LincGET and Dyei at different stages of preimplantation mouse embryos by TM‐qPCR. The error bars represent s.e.m. About 50 embryos of each stage were used, and three experimental replicates were performed.
- RNA‐FISH in early two‐ to 16‐cell embryos for LincGET and Dyei. The results show that LincGET is present in the nucleus of late two‐ to four‐cell embryos and weakly expressed in early eight‐cell embryos, while Dyei is present in the nucleus of two‐ to four‐cell embryos. E2C, early two‐cell stage (n = 6 for each probe); L2C, late two‐cell stage (n = 7 for each probe); E4C, early four‐cell stage (n = 6 for each probe); L4C, late four‐cell stage (n = 5 for each probe); E8C, early eight‐cell stage (n = 4 for each probe); L8C, late eight‐cell stage (n = 5 for each probe); 16C, 16‐cell stage (n = 4 for each probe); BL, blastocyst (n = 5 for each probe). Scale bar, 50 μm. Three experimental replicates were performed.
We next evaluated whether LincGET and Dyei are lincRNAs. We first analyzed their open‐reading frames (ORFs) by NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/) to test the coding potential of LincGET and Dyei. The test only recognized several mini‐ORFs in LincGET and Dyei (Appendix Fig S2B). We analyzed the subcellular localization of LincGET and Dyei through RNA fractionation and following TaqMan real‐time quantitative PCR (TM‐qPCR) assay. Mouse four‐cell embryos were separated into cytoplasmic (Cyto), nuclear‐soluble (Nuc), and chromatin‐bound (Chro) fractions. Gapdh and Xist were analyzed as control. The results showed that LincGET and Dyei are located in the nucleus and mainly associated with chromatin (Fig 1D). In order to ensure the reliability of TM‐qPCR results, which might be challenged by the presence of many ERV sequences on the mouse genome, we designed probes in the unique regions of LincGET and Dyei (Fig EV1B) and sequenced the TM‐qPCR products. There was no LincGET or Dyei‐like sequence, suggesting the reliability of TM‐qPCR. The nuclear localization strongly suggests that LincGET and Dyei are noncoding. Moreover, computational secondary structure analysis (Mfold web server version 3.2, http://mfold.rna.albany.edu/?q=mfold) did not reveal obvious stem loops—a key characteristic of pre‐miRNAs—in LincGET and Dyei (Appendix Fig S3A). Consistently, miRNA reverse Northern blot analysis of total miRNA of two‐ to four‐cell embryos showed no evidence of LincGET or Dyei‐derived small RNA products. As a positive control, we detected miR1982, which is present in two‐ to four‐cell mouse embryos according to the published RNA‐seq data 39 (Fig 1E). Thus, we excluded the possibility that LincGET and Dyei are primary miRNAs. These results collectively confirmed that LincGET and Dyei are lincRNAs.
Furthermore, we analyzed LincGET and Dyei expression pattern in different stages of mouse preimplantation embryos with TM‐qPCR. LincGET is relatively constant at an about‐zero level during MII oocyte to one‐cell embryo, rises in two‐ to four‐cell embryo, and peaks in early four‐cell stage. Its expression then dramatically decreases to the zero level in blastocysts, while Dyei is only detected in late four‐cell stage (Fig 1F). We also analyzed the expression of LincGET and Dyei in ESCs, induced pluripotent stem cells (iPSCs), and various tissues, but no expression was detected for both (Appendix Fig S3B). Consistently, RNA‐FISH revealed that LincGET level is high in the nuclei of late two‐ to four‐cell embryos and decreases dramatically after early eight‐cell embryos, while Dyei is only detected in the nuclei of four‐cell embryos (Fig 1G). Collectively, these results suggest that LincGET and Dyei are late two‐ to four‐cell embryo‐specific, ERV‐associated nuclear lincRNAs.
LincGET depletion results in developmental arrest at late G2 phase of two‐cell stage
To explore the role of LincGET and Dyei in mouse embryonic development, RNA interference (RNAi) assay was performed. Since small‐interfering RNAs (siRNAs) and double‐strand RNAs (dsRNAs) failed to effectively knock down the nuclear LincGET and Dyei (Fig EV2A), we chose locked nucleic acid (LNA), which has been proven to successfully interfere with nuclear transcripts 40. To do so, zygotes were microinjected with control‐LNA (control‐LNA), the LNA targeting LincGET (LincGET‐LNA), or the LNA targeting Dyei (Dyei‐LNA) at the pronuclear stage (Fig 2A). With a near 100% interference efficiency (Figs 2B and EV2B), LincGET depletion caused the developmental arrest at two‐cell stage in mouse embryos (Figs 2C and EV2C–E, and Table 1). However, Dyei depletion had no effect on embryonic development (Fig EV2D–F). The co‐injection of the full‐length LincGET1 with mutations at the LNA target sites, but not partial sequences, partially rescued the embryonic development to our‐ to eight‐cell stage, and some embryos developed to the blastocyst stage (Fig EV2C–E and Table 1).
Figure EV2. LincGET depletion results in developmental arrest at the late G2 phase of two‐cell stage, while Dyei depletion has no effect on the preimplantation development.
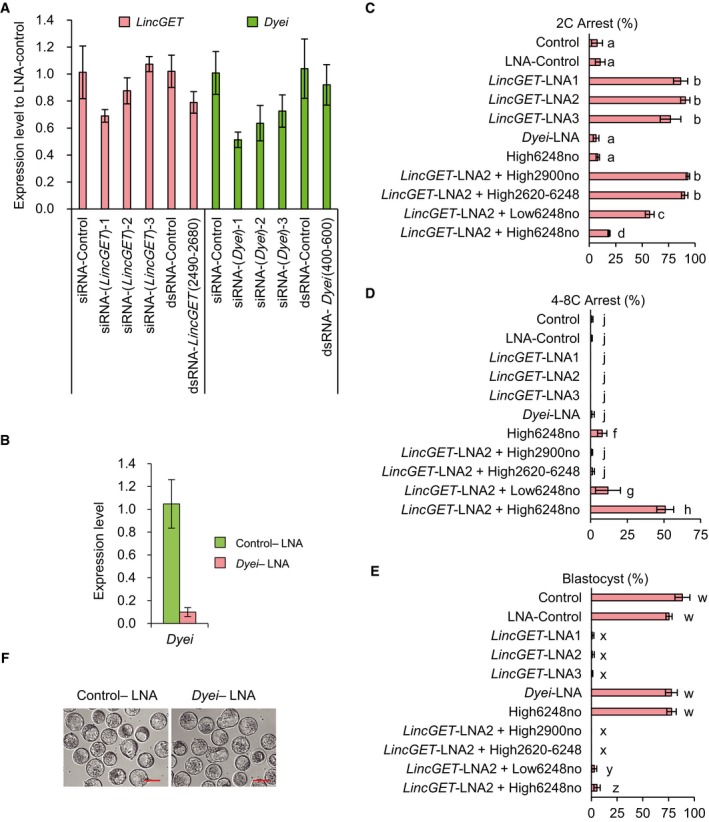
-
AsiRNA or dsRNA do not efficiently interfere with nuclear LincGET and Dyei. siRNA or dsRNA was injected at phCG 25 h, and embryos were collected at phCG 48 h at the late two‐cell stage for TM‐qPCR analysis. The error bars represent s.e.m. Three experimental replicates were performed, and about 50 embryos were used for each time.
-
BDyei‐LNA can efficiently deplete nuclear Dyei. LNA was injected at phCG 25 h, and embryos were collected at phCG 48 h at the late two‐cell stage for TM‐qPCR analysis. The error bars represent s.e.m. Three experimental replicates were performed, and about 50 embryos were used for each time.
-
C–ELincGET depletion leads to developmental arrest at the late G2 phase of two‐cell stage and the co‐injection of full‐length of LincGET1 lacking LincGET‐LNA2 target site, but not that of partial sequences can partially rescue the embryonic development. Dyei depletion has no effect on the preimplantation development. 2C, two‐cell stage; 4–8C, four‐ to eight‐cell stage; BL, blastocyst stage; 6,248no, full‐length LincGET1 with LNA target sites mutation; 2,900no, 1–2,900 nt of LincGET1 with LNA target sites mutation; 2,620–6,248, 2,620–6,248 nt of LincGET1. High, 400 ng/μl; low, 150 ng/μl. Different letters in same panel indicate significant difference (P < 0.001).
-
FInjection of Dyei‐LNA has no significant impact on preimplantation development. LNA was injected at phCG 25 h, and photographs were taken at phCG 114 h at the late blastocyst stage. Scale bar, 50 μm.
Figure 2. LincGET depletion results in developmental arrest at the late G2 phase of two‐cell stage with no effect on ZGA initiation and pericentric rings reorganization.
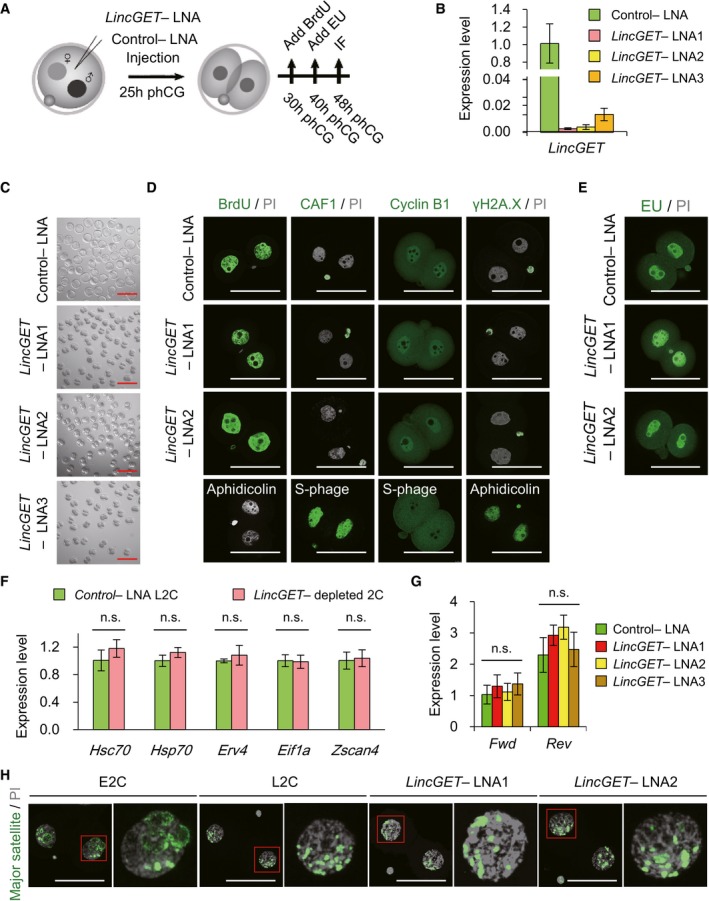
- Experimental scheme to analyze the effects of LincGET depletion on embryonic development. phCG, post‐human chorionic gonadotropin injection; LNA, locked nucleic acid; IF, immunofluorescence. LNA was injected at phCG 25 h. For BrdU staining, BrdU was added at phCG 30 h. For EU staining, EU was added at phCG 40 h. IF, including detection of BrdU and EU, was performed at phCG 48 h.
- LNA efficiently mediated LincGET knockdown. LNA was injected at phCG 25 h, and embryos were collected at phCG 48 h at late two‐cell stage for TM‐qPCR analysis. The error bars represent s.e.m. About 50 embryos of each stage were used, and three experimental replicates were performed.
- LincGET‐depleted embryos arrest at the two‐cell stage. LNA was injected at phCG 25 h, and photographs were taken at phCG 114 h at the late blastocyst stage. Embryos injected with control‐LNA can develop to the late blastocyst stage, while LincGET‐depleted embryos arrest at the two‐cell stage. Scale bar, 100 μm. At least three experimental replicates were performed for each LNA injection (Table 1).
- LincGET depletion results in developmental arrest at the G2 phase of two‐cell stage without affecting DNA integrity and replication. We used BrdU to visualize S and G2 phases, CAF‐1 to visualize S phase, and PI to visualize the M phase. Cyclin B1 is a marker of G2 stage, and H2A.X is a marker of DNA damage. Aphidicolin‐treated embryos arrest at the S phase without DNA replication. LNA was injected at phCG 25 h, and embryos were collected at phCG 48 h at the late two‐cell stage for IF analysis. Scale bar, 50 μm. Three experimental replicates were performed, and about 15 embryos were used in each group.
- EU staining indicates the normal major ZGA process after LincGET depletion. EU was added to the culture medium at phCG 40 h, and EU signals were detected at phCG 48 h. Scale bar, 50 μm. Three experimental replicates were performed, and about 15 embryos were used in each group.
- Genes related to major ZGA initiation, like Hsc70, Hsp70, Erv4, Eif1a, and Zscan4, are expressed normally in LincGET‐depleted L2C embryos compared to that in control embryos. Embryos injected with LNA were collected at phCG 48 h at the late two‐cell stage for TM‐qPCR analysis. The error bars represent s.e.m. About 100 embryos were used for each group, and three experimental replicates were performed. n.s., P > 0.05.
- The transcription of pericentric satellites is normal after LincGET depletion. Embryos injected with LNA were collected at phCG 50 h at the late two‐cell stage for TM‐qPCR analysis. The error bars represent s.e.m. About 50 embryos were used for each group, and three experimental replicates were performed. n.s., P > 0.05.
- DNA‐FISH analysis of major transcripts shows that the pericentric domain reorganization toward chromocenters is not affected by LincGET depletion. Scale bar, 50 μm. Three experimental replicates were performed, and about 15 embryos were used in each group.
Table 1.
Embryonic development after LNA microinjection
| Experiments (n) | Embryos (n) | 2C arrest (%) | 4–8C arrest (%) | BL (%) | |
|---|---|---|---|---|---|
| Control | 3 | 218 | 7.38 ± 5.17a | 0.56 ± 0.96j | 88.30 ± 7.23w |
| Control‐LNA | 5 | 227 | 10.31 ± 4.59a | 0.32 ± 0.72j | 75.36 ± 2.95w |
| LincGET‐LNA1 | 6 | 367 | 86.39 ± 6.72b | 0.00 ± 0.00j | 0.89 ± 1.56x |
| LincGET‐LNA2 | 10 | 860 | 90.76 ± 4.28b | 0.00 ± 0.00j | 0.87 ± 1.94x |
| LincGET‐LNA3 | 4 | 189 | 76.80 ± 9.72b | 0.00 ± 0.00j | 0.49 ± 0.98x |
| Dyei‐LNA | 3 | 149 | 6.39 ± 2.70a | 0.88 ± 1.52j | 77.67 ± 5.81w |
| High6248no | 4 | 255 | 8.01 ± 1.55a | 7.78 ± 3.17f | 77.80 ± 4.55w |
| LincGET‐LNA2 + High2900no | 3 | 186 | 93.21 ± 1.77b | 0.49 ± 0.84j | 0.00 ± 0.00x |
| LincGET‐LNA2 + High2620‐6248 | 3 | 106 | 90.17 ± 3.08b | 1.22 ± 1.28j | 0.00 ± 0.00x |
| LincGET‐LNA2 + Low6248no | 3 | 310 | 57.08 ± 4.17c | 11.78 ± 8.49 g | 3.41 ± 1.89y |
| LincGET‐LNA2 + High6248no | 4 | 421 | 18.57 ± 1.03d | 50.82 ± 5.76 h | 5.52 ± 3.07z |
| Control‐LNA + si‐Egfp | 3 | 74 | 4.06 ± 0.26a | 1.45 ± 2.51j | 79.51 ± 5.37w |
| Control‐LNA + si‐Hnrnpu | 3 | 84 | 3.67 ± 0.78a | 2.30 ± 3.98j | 81.21 ± 7.65w |
| Control‐LNA + si‐Fubp1 | 3 | 83 | 3.74 ± 0.89a | 0.00 ± 0.00j | 79.69 ± 1.44w |
| Control‐LNA + si‐Ilf2 | 3 | 79 | 3.93 ± 0.94a | 2.86 ± 2.58j | 77.37 ± 8.79w |
| LincGET‐LNA2 + si‐Egfp | 3 | 106 | 82.29 ± 4.90b | 0.00 ± 0.00j | 0.00 ± 0.00x |
| LincGET‐LNA2 + si‐Hnrnpu | 3 | 118 | 85.78 ± 5.25b | 0.76 ± 1.31j | 0.00 ± 0.00x |
| LincGET‐LNA2 + si‐Fubp1 | 3 | 108 | 54.04 ± 3.51c | 21.24 ± 2.40i | 12.85 ± 1.30v |
| LincGET‐LNA2 + si‐Ilf2 | 3 | 118 | 81.44 ± 3.13b | 5.90 ± 1.23f | 0.00 ± 0.00x |
| Control‐LNA + OE‐Egfp | 3 | 79 | 3.04 ± 2.64a | 2.54 ± 2.40j | 81.64 ± 3.77w |
| Control‐LNA + OE‐Hnrnpu | 3 | 96 | 6.16 ± 2.88e | 17.88 ± 7.68i | 62.42 ± 6.41u |
| Control‐LNA + OE‐Fubp1 | 3 | 84 | 89.05 ± 1.91b | 0.00 ± 0.00j | 0.00 ± 0.00x |
| Control‐LNA + OE‐Ilf2 | 3 | 108 | 8.06 ± 4.90e | 18.31 ± 7.27i | 63.16 ± 6.53u |
2C, two‐cell stage; 4–8C, four‐ to eight‐cell stage; BL, blastocyst stage; 6,248no, full‐length LincGET1 lacking LincGET‐LNA2 target site; 2,900no, 1–2,900 nt of LincGET1 lacking LincGET‐LNA2 target site; 2,620–6,248, 2,620–6,248 nt of LincGET1. High, 400 ng/μl; low, 150 ng/μl. si‐, siRNA. OE‐, overexpression. Different letters in same column indicate significant difference (P < 0.001).
To narrow down the cell cycle phase of developmental arrest by LincGET depletion, we used immunofluorescence staining (IF) for BrdU [added at post‐human chorionic gonadotropin injection (phCG) 30 h (Fig 2A)] to visualize the S and G2 phases, CAF‐1 to visualize the S phase, and PI to visualize the M phase. Similar to embryos injected with control‐LNA at phCG 48 h (control‐LNA L2C), LincGET‐depleted two‐cell embryos (LincGET‐depleted 2C) had an interphase chromatin status and were strongly stained with BrdU, but not with CAF1 (Figs 2D and EV3A), indicating that they were arrested at the G2 phase. Detection of cyclin B1 (Fig 2D) and histone H3 serine 10 phosphorylation (H3S10ph), which are detected at the G2 to M transition stage, further indicated that LincGET‐depleted 2C was arrested at the late G2 phase. In addition, we added BrdU at the late two‐cell stage (phCG 48 h) and tested the BrdU signal at the late four‐cell stage (phCG 62 h) to determine whether the arrested embryos presented an S phase. The results showed that embryos injected with control‐LNA were BrdU positive and reached the four‐cell stage, while embryos injected with LincGET‐LNA were BrdU negative and still arrested at the two‐cell stage (Fig EV3B), indicating that the embryos injected with LincGET‐LNA were arrested at the G2 phase and no DNA replication could happen. Furthermore, we utilized γH2AX, a marker of DNA damage in eukaryotes 41, to exclude that the arrest resulted from DNA damage or replication stress rather than LincGET depletion. We barely observed any γH2AX signals in LincGET‐depleted 2C, while embryos treated with aphidicolin showed strong γH2AX signals (Fig 2D). The arrest at late G2 phase of two‐cell stage caused by LincGET depletion and the booming of LincGET transcription level in late two‐cell to early four‐cell stage indicate that LincGET may be essential for the second cleavage of mouse embryos.
Figure EV3. LincGET depletion results in developmental arrest at the late G2 phase of two‐cell stage with no effect on ZGA initiation and pericentric rings reorganization.
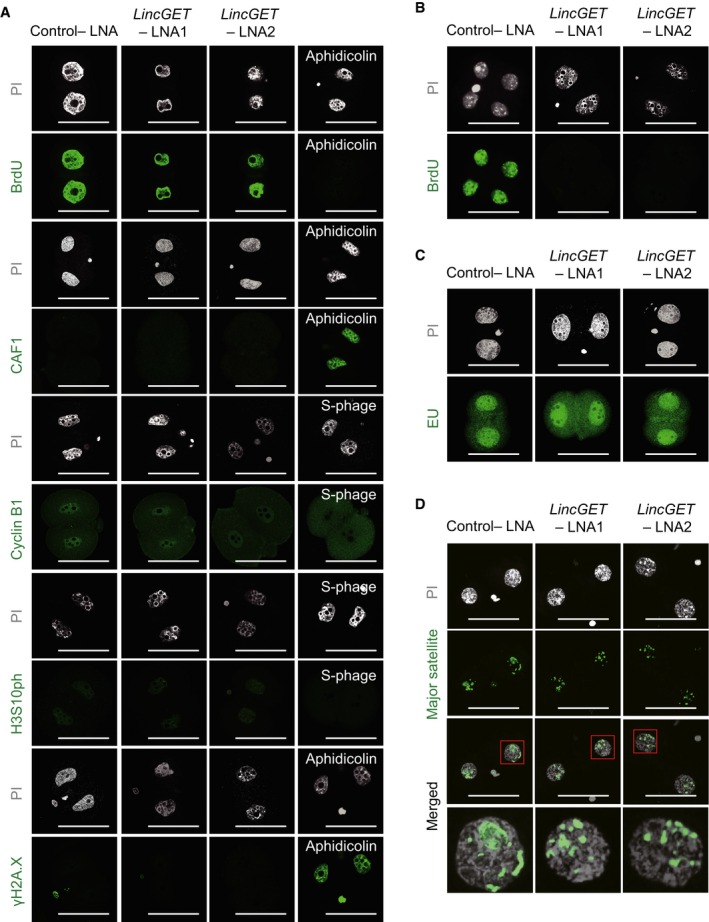
- LincGET depletion results in developmental arrest at the G2 phase of two‐cell stage without affecting DNA integrity and replication. We used BrdU to visualize S and G2 phases, CAF‐1 to visualize S phase, and PI to visualize the M phase. Cyclin B1 and histone H3 serine 10 phosphorylation (H3S10ph) are markers of the G2 stage, and H2A.X is a marker of DNA damage. Aphidicolin‐treated embryos arrested at S phase without DNA replication. LNA was injected at phCG 25 h, and embryos were collected at phCG 48 h at late two‐cell stage for IF analysis. Scale bar, 50 μm. Three experimental replicates were performed, and about 15 embryos were used in each group.
- Embryos injected with LincGET‐LNA were arrested at the G2 phase, and no DNA replication was observed. Results showed that embryos injected with control‐LNA were BrdU positive and reached the four‐cell stage, while embryos injected with LincGET‐LNA were BrdU negative and arrested at the two‐cell stage. BrdU was added at the late two‐cell stage (phCG 48 h) and measured at the late four‐cell stage (phCG 62 h). Scale bar, 50 μm. Three experimental replicates were performed, and about 15 embryos were used in each group.
- EU staining indicated the normal major ZGA process after LincGET depletion. EU was added to the culture medium at phCG 40 h, and EU signals were detected at phCG 48 h. Scale bar, 50 μm. Three experimental replicates were performed, and about 15 embryos were used in each group.
- DNA‐FISH analysis of major transcripts shows that the pericentric domain reorganization toward chromocenters is not affected by LincGET depletion. Scale bar, 50 μm. Three experimental replicates were performed, and about 15 embryos were used in each group.
LincGET depletion did not affect the initiation of major ZGA or the reorganization of pericentric rings
Mouse ZGA contains the minor wave (minor ZGA) at late one‐cell stage and the major wave (major ZGA) at early two‐cell stage 2. ZGA, especially the major ZGA, is necessary for maternal‐to‐embryonic transition and results in of the establishment of the totipotent state of each blastomere in early two‐cell stage embryos, required for developing to four‐cell stage 42. Several genes such as Hsc70, Hsp70, Erv4, Eif1a, and Zscan4 have been identified as being actively transcribed at the onset of major ZGA 43. The delay of major ZGA initiation results in the two‐cell block of mouse embryos 44. Thus, we hypothesized that the two‐cell block caused by LincGET depletion may be associated with the delay of major ZGA initiation. To evaluate the initiation of major ZGA in LincGET‐depleted 2C, we first used 5′‐ethynyluridine (EU) staining (added at phCG 40 h) for total de novo transcripts. The results showed that there was no significant difference in EU signals between control‐LNA L2C and LincGET‐depleted 2C (Figs 2E and EV3C). Secondly, we analyzed the expression level of ZGA initiation genes, Hsc70, Hsp70, Erv4, Eif1a, and Zscan4, by SG‐qPCR. These genes were expressed normally in LincGET‐depleted 2C (Fig 2F). Thus, LincGET depletion has no significant effect on initiation of ZGA.
Depletion of transcripts from major satellites also caused embryonic development arrest at the G2 phase of two‐cell stage, and the pericentric rings would not reorganized into chromocenters 40. Thus, we question whether the embryonic development arrest by LincGET depletion is related with a decrease in major satellite transcripts. We examined the expression level of forward and reverse transcripts of major satellites in control‐LNA L2C and LincGET‐depleted 2C by SSRT followed by SG‐qPCR. No significant change was observed in the expression levels of forward and reverse transcripts of major satellites have (Fig 2G). Additionally, DNA‐FISH showed that the reorganization into chromocenters of the pericentric satellites was normally processed (Figs 2H and EV3D). Thus, LincGET depletion has no effect on the transcription or the reorganization of pericentric satellites.
LincGET depletion leads to MAPK signaling pathway inhibition
To clarify the mechanism underlying the arrest at late two‐cell stage induced by LincGET depletion, we performed low initial amount RNA‐seq to compare gene expression during major ZGA of control‐LNA L2C (2,225 embryos) and LincGET‐depleted 2C (2,042 embryos). From a general view, compared with the control‐LNA L2C, 1,244 genes were deregulated [723 genes were upregulated and 521 genes were downregulated, called differentially expressed genes (DEGs) (FDR ≤ 0.0001, RPKM ≥ 1, and fold change > 2)] (Fig 3A). KEGG pathway analysis of DEGs suggested that LincGET depletion disturbed the MAPK signaling pathway (Fig 3B and Appendix Fig S4A), by inhibiting key factors in the ERK1/2‐MAPK and JNK/P38‐MAPK signaling pathways. The RNA‐sequencing results were confirmed by SG‐qPCR (Fig 3C). Furthermore, both the total and phosphorylated protein levels of P38 and ERK1/2 dramatically decreased as shown by Western blot (Fig 3D) and IF (Fig 3E and F, and Appendix Fig S4B). Additionally, downregulating target genes of the ERK1/2‐MAPK and JNK/P38‐MAPK signaling pathways were upregulated, while upregulating target genes were downregulated upon LincGET depletion (Fig 3G). These results validate the inhibition of ERK1/2‐MAPK and JNK/P38‐MAPK signaling pathways by LincGET depletion. The inhibition of MAPK signaling pathway in early two‐cell stage can induce developmental arrest at G2 phase of two‐cell stage in mouse embryos 45. However, adding anisomycin, a MAPK signaling pathway agonists, failed to rescue the developmental arrest caused by LincGET depletion (data not shown), indicating that the inhibition of the MAPK signaling pathway is not essential or that the effects of LincGET depletion are irreversible.
Figure 3. LincGET depletion results in the inhibition of the MAPK signaling pathway.
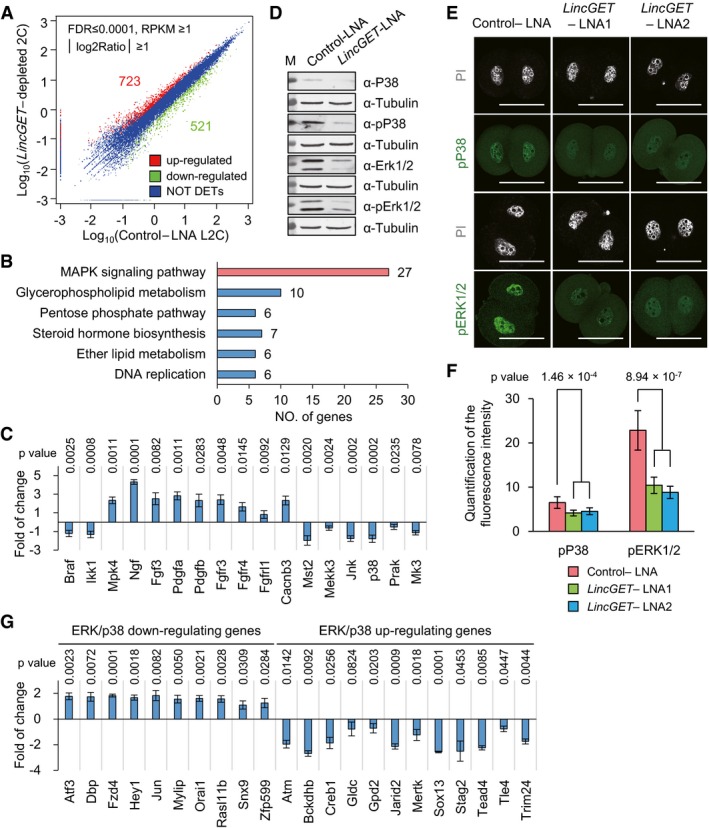
- Differentially expressed genes (DEGs) analysis based on RNA‐seq data. Compared to the control‐LNA L2C, 723 genes were upregulated, and 521 genes were downregulated in LincGET‐depleted embryos.
- KEGG pathway analysis of DEGs showed that the MAPK signaling pathway is mainly affected by LincGET depletion.
- The expression of key factors in the ERK1/2‐MAPK or LNK/P38‐MAPK signaling pathways was significantly affected by LincGET depletion. Embryos injected with LNA were collected at phCG 48 h at late two‐cell stage for SG‐qPCR analysis. The error bars represent s.e.m. About 150 embryos were used for each group, and three experimental replicates were performed. Two‐tailed Student's t‐test was used for statistical analysis.
- Western blot analysis indicates that the protein and phosphorylation level of p38 and ERK1/2, key kinases in the MAPK signaling pathway, decreased in LincGET‐depleted 2C. Embryos injected with LNA were collected at phCG 48 h at late two‐cell stage for Western blot analysis, and about 200 embryos were used for each lane. Three experimental replicates were performed.
- Immunofluorescence indicates that the phosphorylation level of P38 and ERK1/2 decreased dramatically in LincGET‐depleted 2C. Embryos injected with LNA were collected at phCG 48 h at late two‐cell stage for IF analysis. Scale bar, 50 μm. Three experimental replicates were performed, and about 15 embryos were used in each group.
- Quantification of the fluorescence intensity indicates that the phosphorylation level of P38 and ERK1/2 decreased dramatically in LincGET‐depleted 2C. Two‐tailed Student's t‐test was used for statistical analysis. The error bars represent s.e.m. Three experimental replicates were performed, and about 15 embryos were used in each group. P‐value between Control LNA group and LincGET‐LNA group was shown. There was no difference between LincGET‐LNA1 and LincGET‐LNA2. Related to panel (E) and Appendix Fig S4B.
- The expression of some target genes of the ERK1/2‐MAPK or LNK/P38‐MAPK signaling pathways was significantly affected by LincGET depletion. Embryos injected with LNA were collected at phCG 48 h at late two‐cell stage for SG‐qPCR analysis. The error bars represent s.e.m. About 150 embryos were used for each group, and three experimental replicates were performed. Two‐tailed Student's t‐test was used for statistical analysis.
LincGET binds to hnRNP U, FUBP1, and ILF2
As we know that lincRNAs always function through interaction with proteins. Thus, we performed RNA pull‐down followed by mass spectrometry (pd‐MS) with biotin‐labeled LincGET and early four‐cell embryos (6,367 embryos) lysates to identify the proteins to which LincGET binds. Three specific bands in LincGET lane compared to the anti‐LincGET control lane were identified as hnRNP U (also known as SAF‐A), FUBP1, and ILF2 (also known as NF45) (Fig 4A). Due to resource constraints, pd‐MS was only performed once. However, the results were confirmed by using Western blot analysis, which was repeated thrice (Fig 4B).
Figure 4. LincGET binds to hnRNP U, FUBP1, and ILF2.
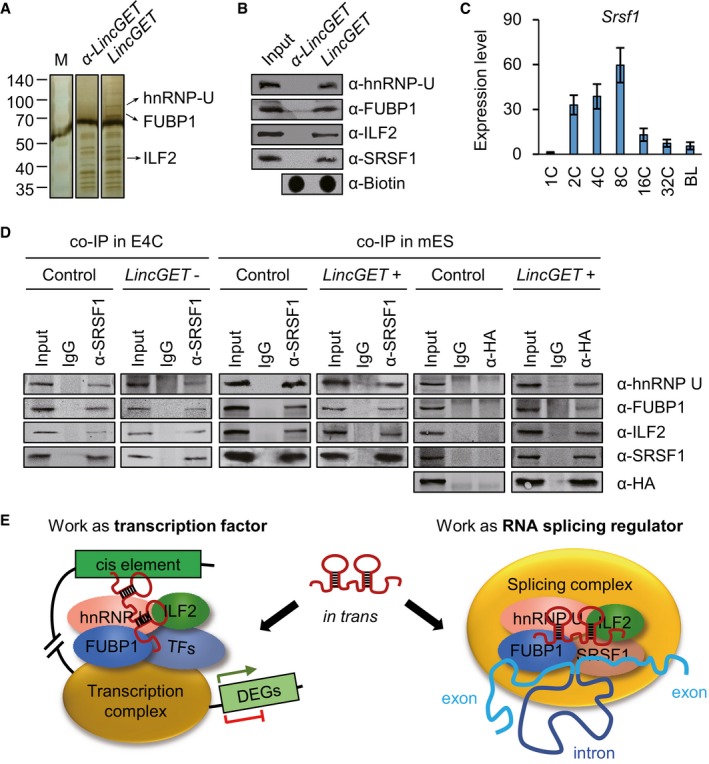
- LincGET interacts with hnRNP U, FUBP1, and ILF2 in vitro. Silver staining of SDS–PAGE gel following RNA pull‐down assay shows the proteins bound to LincGET (right lane) and antisense LincGET (α‐LincGET, middle lane). Only one pull‐down assay for mass spectrometry analysis was performed with 6,367 early four‐cell embryos. Three specific bands in the right lane (arrow) were analyzed through mass spectrometry and confirmed as hnRNP U, FUBP1, and ILF2.
- Mass spectrometry results of hnRNP U and ILF2 were confirmed by Western blot following RNA pull‐down assay (pull‐down WB); α‐, anti‐. For each pull‐down WB assay, about 1,500 early four‐cell embryos were used and three experimental replicates were performed.
- Expression pattern of Srsf1 in preimplantation mouse embryos measured by SG‐qPCR. 1C, zygote stage; 2C, two‐cell stage; 4C, four‐cell stage; 8C, eight‐cell stage; 16C, 16‐cell stage; 32C, 32‐cell stage; BL, blastocyst stage. The error bars represent s.e.m. About 50 embryos of each stage were used, and three experimental replicates were performed.
- Co‐IP results in early four‐cell embryos using anti‐SRSF1 and in mouse ESCs using anti‐SRSF1 or anti‐HA (for HA‐labeled MS2 coat protein). The results show that LincGET forms an RNA–protein complex with hnRNP U, FUBP1, ILF2, and SRSF1. Moreover, hnRNP U, FUBP1, ILF2, and SRSF1 can form protein complex without LincGET. For each co‐IP assay, about 2,500 early four‐cell embryos or 1 × 106 mouse ESCs were used, and three experimental replicates were performed. E4C, early four‐cell embryos; mES, mouse ESCs; LincGET‐, LincGET elimination; LincGET+, LincGET overexpression.
- Model showing that LincGET acts as a transcription factor and RNA alternative splicing factor through binding to hnRNP U, FUBP1, and ILF2.
hnRNP U, FUBP1, and ILF2 are not only transcription factors 46, 47, 48, 49, 50, 51, 52, 53, but also alternative splicing regulators 54, 55, 56, 57, 58. Thus, we tested whether SRSF1, which is a well‐known alternative splicing regulator 59 and is highly expressed in two‐ to eight‐cell stage mouse embryos (Fig 4C), is involved in the LincGET‐protein complex. SRSF1 was detected in the LincGET‐pull‐down results by Western blot (Fig 4B). In order to verify the LincGET‐protein complex, co‐immunoprecipitation assays (co‐IP) were performed in both early four‐cell embryos and mouse ESCs with HA‐tagged MS2‐labeled LincGET overexpression. The co‐IP results using anti‐SRSF1 and anti‐HA antibodies (with IgG as control) showed that LincGET truly formed an RNA–protein complex with hnRNP U, FUBP1, ILF2, and SRSF1 (Fig 4D). In addition, hnRNP U, FUBP1, ILF2, and SRSF1 can form a protein complex without LincGET.
Taken together, these findings suggest a dual function of LincGET that work as a transcription factor and RNA alternative splicing regulator during the major ZGA stage (Fig 4E).
LincGET acts as a transcription factor
Enhancer‐like lincRNAs mediate the enhancer activity by binding and tethering transcription factors to the target genes in cis 31, 32, 60, 61. In addition, ERV LTRs can act as enhancers 62, 63. Thus, we speculated that LincGET may bind to its associated GLKLTRs and mediate their cis‐regulatory activity in trans. In other word, LincGET acts as a transcription factor. To verify this hypothesis, we compared the median distance of DEGs to neighboring GLKLTR and 10,000 times that of equal amount of random genes by Wilcoxon rank single test. The results showed that the rank of DEGs is the smallest compared to the distribution of 10,000‐time random controls (P < 2.2 × 10−16) (Fig 5A), which means DEGs prefer to locate close to GLKLTR (Fig EV4A). Furthermore, using dox‐induced LincGET expression assay and dual‐luciferase reporter system, we determined that LincGET increased the enhancer activities of GLKLTRs in 293T cells in a dose‐dependent manner (Fig 5B). Moreover, overexpression of hnRNP U and ILF2 further reinforced, while overexpression of FUBP1 weakened the enhancer‐like activity of LincGET toward GLKLTRs without influencing the level of LincGET (Fig 5B). These results suggested that LincGET can act as a transcription factor to bind GLKLTRs and mediate their cis‐regulatory activities during major ZGA.
Figure 5. LincGET acts as both a transcription factor and an exon skipping splicing inhibitor.
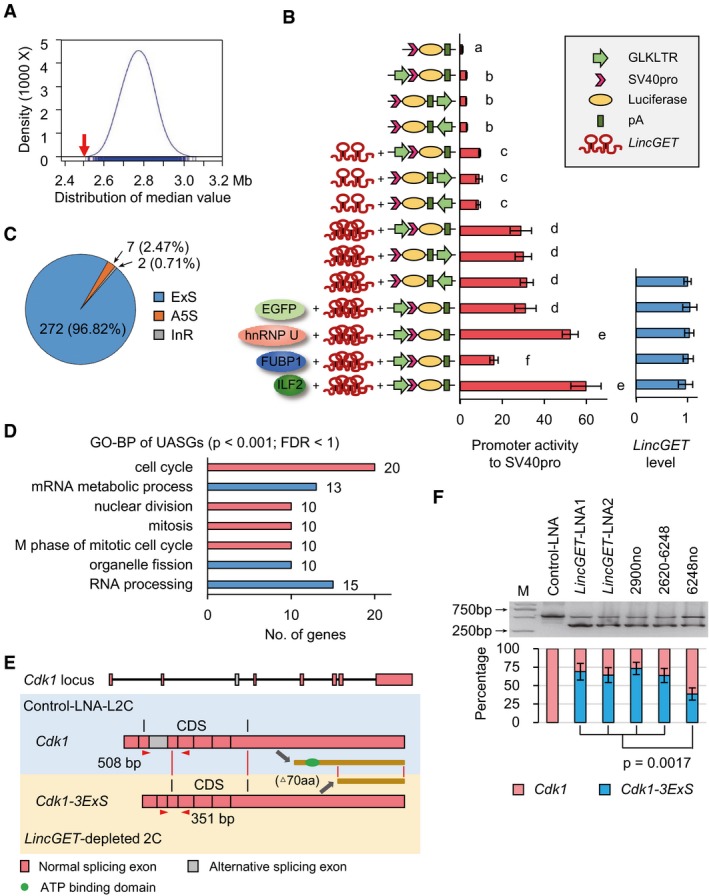
- The median distance of DEGs to neighbor GLKLTRs (red arrow) and the distribution of that of random genes (blue) (P < 2.2 × 10−16) measured by Wilcoxon rank single test. GLKLTRs are gene loci that contain whole LTR of GLN, MERVL, or ERVK. The results show that DEGs prefer neighboring to GLKLTRs.
- Dox‐induced LincGET expression assay and dual‐luciferase reporter system show that LincGET increased the enhancer activity of GLKLTRs in 293T cells in a dose‐dependent manner. The enhancer activity of GLKLTRs is also increased by overexpression of hnRNP U or ILF2 and decreased by FUBP1 overexpression, which have no effect on LincGET levels. The y‐axis shows the construction of luciferase reporter plasmids and overexpressed genes. Three experimental replicates were performed. Two‐tailed Student's t‐test was used for the statistical analysis. Different letters indicate significant difference (P < 0.01).
- Number and percentage of unusual alternative splicing genes (UASGs, FDR < 0.05) in LincGET‐depleted 2C compared to control‐LNA L2C. ExS, exon skipping; A5S, alternative 5′ splicing; InR, intron retention.
- GO‐BP analysis of UASGs showed that “cell cycle” especially the “M phase of the mitotic cell cycle” is primarily affected. The terms with P < 0.001 and FDR < 1 are shown.
- Cdk1 was exon skipping spliced in LincGET‐depleted 2C, resulting in Cdk1‐3ExS where the third exon was deleted. The abnormal spliced Cdk1‐3ExS variant translated into a truncated CDK1 that lacks the N‐terminal 70 amino acids (aa), which affects the kinase activity of CDK1 lacking a conservative ATP binding domain predicted by InterPro. PCR primers for panel (F) (Cdk1‐Ex3‐F and Cdk1‐Ex3‐R, sequences are shown in Table EV1) are shown in red triangles.
- Exon skipping event of Cdk1‐3ExS in LincGET‐depleted 2C is confirmed by RT–PCR. Injection of LincGET fragments at the pronuclear stage was unable to inhibit the exon 3 skipping of Cdk1, while injection of full‐length LincGET1 lacking LincGET‐LNA2 target site partially inhibited the exon 3 skipping of Cdk1. For each lane, about 50 embryos were used. The error bars represent s.e.m. Three experimental replicates were performed. Two‐tailed Student's t‐test was used for the statistical analysis.
Figure EV4. Some features of DEGs and splicing events in LincGET‐depleted 2C.
- Loci of DEGs and GLKLTRs. Red lines represent upregulated genes; blue lines represent downregulated genes; green blots represent GLKLTRs.
- Three types of alternative splicing events were observed and different between control‐LNA L2C and LincGET‐depleted 2C.
- CDK1 protein domain prediction by InterPro (http://www.ebi.ac.uk/interpro/). An ATP binding domain is located at the N‐terminus.
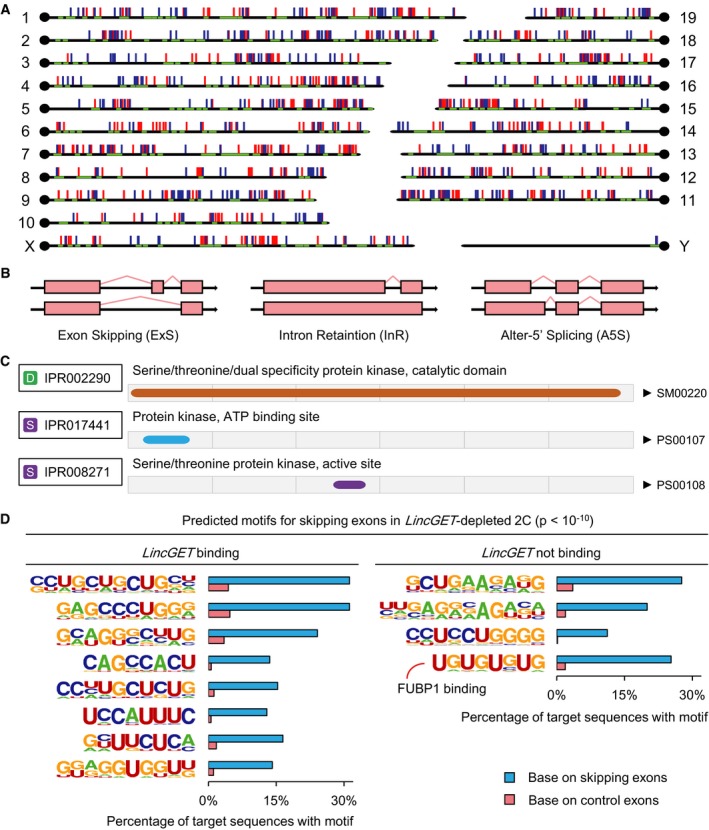
- Predicted motifs for skipping exons in LincGET‐depleted 2C. We analyzed the sequences of skipping exons in LincGET‐depleted 2C with flanked 200‐bp intron sequences, and 12 motifs that were enriched in these skipping exons relative to random control exons (P < 10−10) were detected. Eight of these 12 motifs (left) are affiliated to LincGET. One of the other 4 LincGET, which are not binding motifs (right, the UGUGUGUG motif), is the FUBP1 targeting sequence in Dmd intron 38 (UUGUGUGUGU) required for exon 39 skipping splicing.
LincGET acts as an RNA exon skipping splicing inhibitor
Alternative splicing is one way of gene regulation at the RNA level in a developmental stage‐specific manner. Abnormal alternative splicing is adverse to development 64. We analyzed the alternative splicing events 65 in control‐LNA L2C and LincGET‐depleted 2C from the RNA‐seq data. Two hundred and eighty‐one genes presented unusual alternative splicing events (FDR < 0.05) (Fig 5C) in LincGET‐depleted 2C, involving three different types of alternative splicing types (Fig EV4B). Surprisingly, 272/281 (96.82%) of unusual alternative splicing genes (UASGs) present with exon skipping (Fig 5C), implying that LincGET mainly participates in the inhibition of exon skipping.
To explore the role played by genes protected from exon skipping by LincGET, we analyzed these 272 UASGs by Gene Ontology Biological Process (GO‐BP). Interestingly, the GO‐BP suggested that main “cell cycle”, especially the “M phase of mitotic cell cycle” was severely affected (Fig 5D). Thus, unusual alternative splicing during major ZGA, including the M‐phase key regulators, may be one of the mechanism underlying the late two‐cell arrest of LincGET‐depleted mouse embryos. Moreover, in LincGET‐depleted 2C, the key G2 to M transition‐associated gene, cyclin‐dependent kinase 1 (Cdk1), was abnormally spliced, its third exon was moved from the mature Cdk1 mRNA (Cdk1‐3ExS, Fig 5E). The abnormal spliced Cdk1‐3ExS variant translated into a truncated CDK1 that lacks the N‐terminal 70 amino acids (aa) predicted by GENSCAN (http://genes.mit.edu/GENSCAN.html) (Fig 5E), which would affect the kinase activity of CDK1, lacking a conservative ATP binding domain predicted by InterPro (http://www.ebi.ac.uk/interpro/) (Fig EV4C).
We confirmed the exon 3 skipping splicing event of Cdk1 in LincGET‐depleted 2C by RT–PCR and determined that about 70% Cdk1 was spliced into Cdk1‐3ExS in LincGET‐depleted 2C (Fig 5F). Injection of LincGET fragments at the pronuclear stage could not inhibit the exon 3 skipping of Cdk1 and full‐length LincGET1 lacking LincGET‐LNA2 target site injection partially inhibited the exon 3 skipping of Cdk1 (Fig 5F), which was consistent with the development rescue experiment (Table 1). Thus, the full‐length and, at least, the LincGET‐LNA2 target site are needed for the exon skipping inhibitor function of LincGET. Furthermore, we analyzed the sequences of skipping exons in LincGET‐depleted 2C with flanked 200‐bp intron sequence, and 12 motifs that were enriched in these skipping exons relative to random control exons (P < 10−10) were increased (Fig EV4D). Eight of these 12 motifs are affiliated to LincGET, which raises the possibility that LincGET binds to the target exon or its neighboring intron to protect it from exon skipping splicing.
LincGET‐induced downregulation of hnRNP U, FUBP1, and ILF2 protein levels underlies exon skipping splicing inhibition
hnRNP U (for Smn2 exon 7) 66, FUBP1 (for Dmd exon 39) 67, ILF2 (for Smn2 exon 7) 57, and SRSF1 (for Smn2 exon 7) 66 promote exon skipping. Additionally, one of these 4 LincGET not binding motifs (Fig EV4D, the UGUGUGUG motif) is the FUBP1 targeting sequence in Dmd intron 38 (UUGUGUGUGU) required for exon 39 skipping splicing 67. Thus, we questioned whether LincGET inhibits exon skipping partially through downregulation of hnRNP U, FUBP1, and ILF2, which can form a RNA–protein complex with LincGET.
Immunostaining for hnRNP U, FUBP1, ILF2, and SRSF1 was performed in normal late two‐cell, normal early four‐cell embryos, control‐LNA L2C, and LincGET‐depleted 2C to evaluate the effect of LincGET on their expression. The results showed that all of them are located in nuclei of normal or arrested late two‐cell and early four‐cell embryos, and except SRSF1, hnRNP U, FUBP1, and ILF2 are dramatically enriched after LincGET depletion, even at levels higher than those detected in normal four‐cell embryos (Fig 6A and B, and Appendix Fig S5). SG‐qPCR indicated that LincGET depletion had no effect on Hnrnpu, Fubp1, Ilf2, and Srsf1 expression at the RNA level (Fig 6C).
Figure 6. LincGET decreases hnRNP U, FUBP1, and ILF2 protein levels.
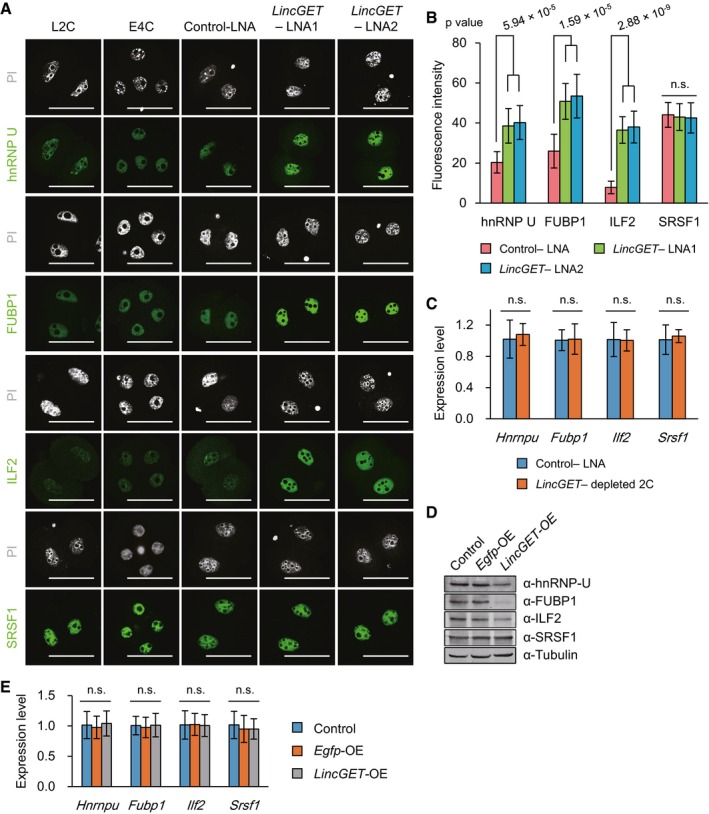
- IF staining of hnRNP U, FUBP1, ILF2, and SRSF1 in normal L2C, E4C, and embryos injected with control‐LNA or LincGET‐LNA. The results show that hnRNP U, FUBP1, ILF2, and SRSF1 are present in the nuclei of two‐ to four‐cell embryos, and the expression of hnRNP U, FUBP1, and ILF2 increased significantly after LincGET depletion. Normal L2C and four‐cell embryos were collected at phCG 48 h and 54 h, respectively. Embryos injected with LNA were collected at phCG 48 h. Scale bar, 50 μm. Three experimental replicates were performed, and about 15 embryos were used in each group.
- Quantification of the fluorescence intensity shows that the expression of hnRNP U, FUBP1, and ILF2 increased significantly after LincGET depletion. Two‐tailed Student's t‐test was used for the statistical analysis. The error bars represent s.e.m. Three experimental replicates were performed, and about 15 embryos were used in each group. Related to panel (A) and Appendix Fig S5. n.s., P > 0.05.
- LincGET depletion in embryos had no effect on RNA levels of Hnrnpu, Fubp1, Ilf2, and Srsf1 measured by SG‐qPCR. Three experimental replicates were performed, and about 50 embryos were used for each time. Two‐tailed Student's t‐test was used for the statistical analysis. n.s., P > 0.05. The error bars represent s.e.m.
- LincGET overexpression in mouse ESCs decreases the protein level of hnRNP U, FUBP1, and ILF2 by Western blot. Three experimental replicates were performed, and about 1 × 106 cells were used each time.
- LincGET overexpression in mouse ESCs had no effect on RNA levels of Hnrnpu, Fubp1, Ilf2, and Srsf1 measured by SG‐qPCR. Three experimental replicates were performed, and about 1 × 106 cells were used each time. Two‐tailed Student's t‐test was used for the statistical analysis; n.s., P > 0.05. The error bars represent s.e.m.
Furthermore, we overexpressed LincGET in mouse ESCs where there is no LincGET expression (Appendix Fig S3B) and evaluated hnRNP U, FUBP1, ILF2, and SRSF1 protein and RNA levels by Western blot and SG‐qPCR, respectively. LincGET overexpression decreased the protein level of hnRNP U, FUBP1, and ILF2, especially that of FUBP1 (Fig 6D), while it had no effect on SRSF1 protein level (Fig 6D) and RNA levels of Hnrnpu, Fubp1, Ilf2, and Srsf1 (Fig 6D and E). Thus, LincGET decreases the protein level of hnRNP U, FUBP1, and ILF2.
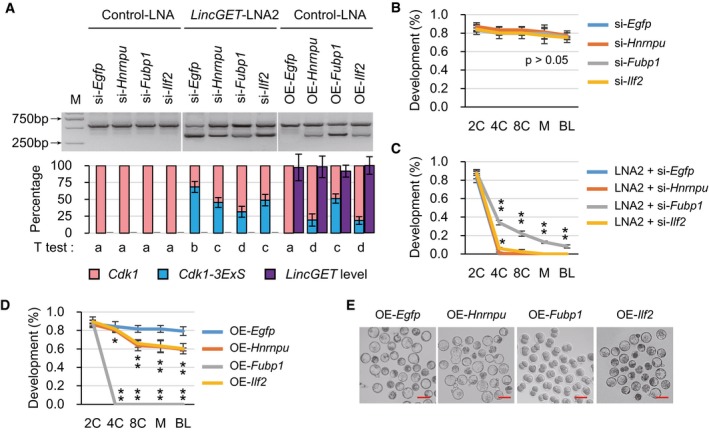
Furthermore, to evaluate the effect of hnRNP U, FUBP1, and ILF2 on exon skipping and preimplantation development with or without LincGET, we injected control‐LNA together with siRNAs targeting for one of these genes, LincGET‐LNA2 together with siRNA, or control‐LNA together with mRNA were injected at the pronuclear stage (12 groups), then tested the Cdk1 and Cdk1‐3ExS levels at late two‐cell stage (phCG 48 h) and tracked the preimplantation development. The results showed that knockdown of Hnrnpu, Fubp1, or Ilf2 in the presence of LincGET had no effect on exon 3 skipping of Cdk1 or preimplantation development (Fig 7A and B). Knockdown of Hnrnpu, Fubp1, or Ilf2, in absence of LincGET, lowers the Cdk1‐3ExS level (Fig 7A), especially si‐Fubp1 which partially rescued the arrested preimplantation development caused by LincGET depletion (Fig 7C). Overexpression of Hnrnpu, Fubp1, or Ilf2, in the presence of LincGET, promoted exon 3 skipping of Cdk1 (Fig 7A), especially Fubp1 which caused two‐cell block, while Hnrnpu and Ilf2 only lower the developmental rate (Fig 7D and E) without affecting LincGET levels (Fig 7A).
Figure 7. hnRNP U, FUBP1, and ILF2 promote exon 3 skipping of Cdk1 .
-
ART–PCR results showed that hnRNP U, FUBP1, and ILF2 promote exon 3 skipping of Cdk1. We injected control‐LNA or LincGET‐LNA2 together with siRNA for either Egfp, Hnrnpu, Fubp1, or Ilf2 or control‐LNA together with mRNA for either one of them at the pronuclear stage (12 groups) and assessed Cdk1 and Cdk1‐3ExS levels at the late two‐cell stage (phCG 48 h). For each lane, about 50 embryos were used. Three experimental replicates were performed. The results show that knockdown of either of Hnrnpu, Fubp1, or Ilf2, but not Egfp in the presence of LincGET had no effect on exon 3 skipping of Cdk1 (left 4 lanes). Knockdown of either Hnrnpu, Fubp1, or Ilf2, but not Egfp in absence of LincGET decreased the Cdk1‐3ExS level (middle 4 lanes), especially si‐Fubp1. Overexpression of Hnrnpu, Fubp1, or Ilf2, but not Egfp in the presence of LincGET promoted exon 3 skipping of Cdk1 (right 4 lanes), especially Fubp1. The band intensities were quantitated and are shown under the gel image. Two‐tailed Student's t‐test was used for the statistical analysis and different letters indicate significant difference (P < 0.05). si‐, siRNA; OE‐, overexpression. The error bars represent s.e.m.
-
BKnockdown of either Hnrnpu, Fubp1, or Ilf2 in the presence of LincGET had no effect on preimplantation development. We injected control‐LNA together with siRNA for Egfp, Hnrnpu, Fubp1, or Ilf2 at the pronuclear stage, and development was assessed at the two‐cell (2C), four‐cell (4C), eight‐cell (8C), morula (M), and blastocyst (BL) stage. Three experimental replicates were performed (Table 1). Two‐tailed Student's t‐test was used for the statistical analysis. si‐, siRNA. The error bars represent s.e.m.
-
CFubp1 knockdown can partially rescue the 2C arrest caused by LincGET depletion to the blastocyst stage, and Ilf2 knockdown can improve the development rate of 4C, but Hnrnpu knockdown cannot improve the development. We injected LincGET‐LNA2 together with siRNA against Egfp, Hnrnpu, Fubp1, or Ilf2 at the pronuclear stage, and development was assessed at the 2C, 4C, 8C, M, and BL stage. Three experimental replicates were performed (Table 1). Two‐tailed Student's t‐test was used for the statistical analysis. *P < 0.05, and **P < 0.01. LNA2, LincGET‐LNA2. si‐, siRNA. The error bars represent s.e.m.
-
D, EFubp1 overexpression in the presence of LincGET causes 2C arrest, and Hnrnpu or Ilf2 overexpression can lower the preimplantation development rate. We injected control‐LNA together with mRNA for Egfp, Hnrnpu, Fubp1, or Ilf2 at the pronuclear stage, and development was assessed at the 2C, 4C, 8C, M, and BL stage. Images were acquired at the BL stage (E). Three experimental replicates were performed (Table 1). Two‐tailed Student's t‐test was used for the statistical analysis. *P < 0.05, and **P < 0.01. OE‐, overexpression. The error bars represent s.e.m. (D). Scale bar, 100 μm (E).
Therefore, our data showed that LincGET can act both as a transcription factor and as exon skipping splicing inhibitor at the transcriptional and post‐transcriptional levels, respectively, to guarantee a correct major ZGA in mouse embryos and a smooth transition from two‐cell to four‐cell stage (Fig EV5).
Figure EV5. Model representing LincGET function.
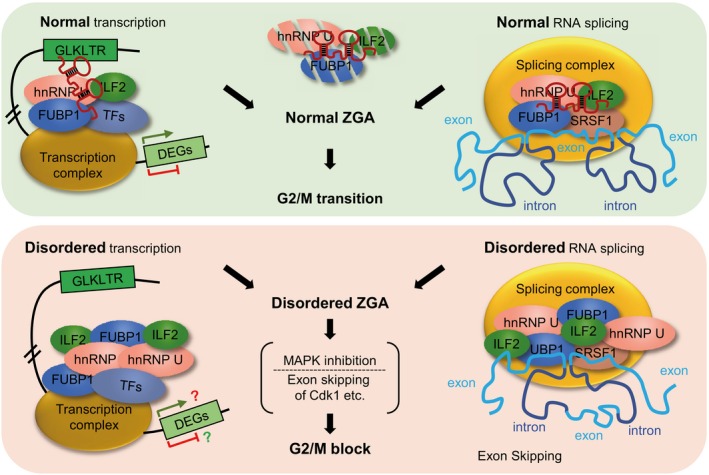
In normal two‐cell mouse embryos, LincGET works as a transcription factor, mediating the cis‐regulatory activity of GLKLTRs, and an inhibitor of exon skipping, partially through decreasing hnRNP U, FUBP1, and ILF2 protein levels (up). However, when LincGET is depleted, disordered transcription and disordered RNA splicing would result in disordered ZGA, MAPK signaling inhibition, and exon skipping of some M‐phase‐associated genes, including Cdk1, leading to G2/M block (down).
Discussion
In the present study, we identified a late two‐ to four‐cell mouse embryo‐specific, GLN‐, MERVL‐, and ERVK‐associated nuclear lincRNA, termed LincGET. LincGET depletion led to two‐cell block at late G2 phase with normal major ZGA initiation and pericentric rings reorganization into chromocenters, but inhibition of MAPK signaling pathway. Moreover, through interaction with hnRNP U, FUBP1, and ILF2, LincGET plays key roles in transcription and RNA splicing during major ZGA stage. On the one hand, LincGET works as a transcription factor mediating the cis‐regulatory activity of GLKLTRs. On the other hand, LincGET works as an inhibitor of exon skipping of some major ZGA transcripts, especially some M‐phase‐associated genes, including Cdk1. Hence, LincGET is essential for correct major ZGA process and further cleavage of two‐cell embryos (Fig EV5).
The discovery of Xist in 1992 68, 69 showed for the first time that lincRNAs can act as key regulators of biological processes. However, few lincRNAs are well studied, especially in cleavage stage mammalian embryos. This is the first study to discover and report a cleavage stage embryo‐specific functional lincRNA, LincGET. Depletion of LincGET resulted in that almost no embryo developed beyond the two‐cell stage, indicating that LincGET is essential for the second cleavage of mouse embryos. Studies on preimplantation development, especially the cleavage stage development, are important for both reproductive biology and regenerative medicine. Besides, understanding the nature of reprogramming and totipotency of cleavage stage, embryos will enlighten the research on and utilization of ESCs and iPSCs. However, cleavage stage embryos are special cells, which carry out a series of important distinctive developmental events such as genomewide reprogramming with protection of imprinting regions 1, 70, ZGA 2, and segregation of ICM and trophectoderm 71. Therefore, identification of cleavage stage embryo‐specific lincRNAs is very important. Many two‐cell mouse embryo‐specific transcripts (coding or noncoding) derived from MERVL can work as unique markers of two‐cell mouse embryos 4 and a large number of lincRNAs have been identified in preimplantation embryos 6, 7. However, the function of these lincRNAs was unclear. Recently, bidirectional promoter‐associated noncoding RNAs (pancRNAs) have been shown to be important for mouse preimplantation development 72. Nevertheless, apoptosis was induced in both blastomere and ESCs after pancIl17 depletion, indicating that the function of pancIl17d was not embryo‐specific. In contrast, LincGET is late two‐ to four‐cell embryo‐specific, which is essential for major ZGA and further cleavage of two‐cell embryos. Thus, the identification of LincGET has a profound significance for understanding the cleavage stage development.
There are many active ERVs in cleavage stage embryos, yet little is known about their functions 6, 7. Enhancer‐like lincRNAs mediate the enhancer activity by binding to its own locus and tethering transcription regulators to the target genes in cis 31, 32, 60, 61 and ERV LTRs can act as enhancers 62, 63. Here, we show that LincGET is a GLN, MERVL, and ERVK‐associated lincRNA, acting as a transcription factor, binding to its associated GLKLTRs and mediating their cis‐regulatory activity in trans. Thus, LincGET is similar to enhancer‐like lincRNAs, but it works in trans. Dual‐luciferase reporter assays confirmed the transcription activator activity of LincGET, which can be increased by hnRNP U or FUBP1 and lowered by FUBP1. LincGET, acting as a transcription factor, sheds lights on new functioning patterns of ERV sequences and repeat sequences.
Here, we determined that LincGET is also an important alternative splicing component together with hnRNP U, FUBP1, ILF2, and SRSF1, where LincGET may mainly function through decreasing the protein level of hnRNP U, FUBP1, and ILF2. Alternative splicing is one type of gene regulatory mechanisms at the RNA level. Through alternative splicing, the first transcript can either become different RNA transcripts encoding different proteins or even become noncoding RNAs. Alternative splicing is carried out in a developmental stage‐specific manner 64. Abnormal alternative splicing is adverse to development. Many lincRNAs are regulators of alternative splicing, like Malat1 59, 73, 51A‐ncRNA 74, and Zeb2 antisense RNA 36. Although we showed that LincGET acts as an exon skipping inhibitor partially through decreasing protein levels of hnRNP U, FUBP1, and ILF2, whose overexpression improves exon 3 skipping of Cdk1, the functioning mechanism of LincGET in RNA splicing is still worth exploring and future studies are warranted.
FUBP1, single strand far upstream element (FUSE)‐binding protein 1, is known as a transcription activator of c‐Myc 75 and is considered a proto‐oncogene due to its role in the etiology of several types of cancer where it is overexpressed 76. Additionally, FUBP1 is associated with exon 39 skipping splicing of Dmd 67. Recently, FUBP1 was shown to be an essential factor of hematopoietic stem cell self‐renewal. FUBP1 functional inactivation resulted in embryonic lethal anemia at around E15.5 caused by diminished severe decrease in the number of hematopoietic stem cells 53. Here, we showed that FUBP1 overexpression by mRNA injection at the pronuclear stage leads to two‐cell block, similar to LincGET depletion, both of which are valuable to study two‐cell specific transcription and RNA alternative splicing.
Our current study identified a novel GLN‐, MERVL‐, and ERVK‐associated lincRNA, LincGET, which is essential for mouse embryonic development beyond the two‐cell stage via regulating the transcription and RNA alternative splicing at major ZGA stage. As the first functional ERV‐associated lincRNA revealed in two‐ to two‐cell stage mammalian embryos, LincGET provides clues for ERV functions in cleavage stage development, even the nature of totipotency.
Materials and Methods
Primer and probe design
All primers (except major transcripts primers 40) and probes were designed using PrimerPremier5 (Table EV1) and synthesized from Invitrogen.
Antibodies
The following antibodies were used for immunoblotting and (or) immunostaining: anti‐hnRNP U 49, anti‐FUBP1, (Abcam, #ab181111), anti‐ILF2 77, anti‐BrdU (Abcam, #ab1893), anti‐CAF1 p150 (Abcam, #ab126625), anti‐γH2A.X (Abcam, #ab2893), anti‐H3S10ph (Abcam, #ab5176), anti‐cyclin B1 (Abcam, #ab32053), anti‐DIG (Roche, #11333062910), anti‐ERK1/2 (Abcam, #ab17942), anti‐ERK1/2 (phospho‐T185/Y187) (Abcam, #ab200807), anti‐P38 (Abcam, #ab7952), anti‐P38 (phospho‐T180) (Abcam, #ab178867), and anti‐SRSF1 73.
Embryo collection
All experiments were performed in accordance with ARRIVE guidelines and regulations. Embryos were collected from 6‐week‐old ICR superovulated female mice crossed with ICR males, at the following times post‐human chorionic gonadotropin (phCG) injection: early one‐cell stage (phCG 19 h), late one‐cell stage (phCG 30 h), early two‐cell stage (phCG 39 h), late two‐cell stage (phCG 48 h), early four‐cell stage (phCG 54 h), late four‐cell stage (phCG 62 h), early eight‐cell stage (phCG 68 h), late eight‐cell stage (phCG 74 h), 16‐cell stage (phCG 80 h), 32‐cell stage (phCG 90 h), early blastocyst stage (phCG 98 h), and late blastocyst stage (phCG 114 h). Additionally, oocytes were collected from 6‐week‐old ICR superovulated females at phCG 12 h.
RNA extraction, reverse transcription, PCR, TM‐qPCR, and RACE
RNA was extracted by RNeasy Mini Kit (QIAGEN, #74104), and the RNase‐Free DNase Set (QIAGEN, #79254) was used to ensure no DNA contamination. Reverse transcription was performed by High Capacity cDNA Reverse Transcription Kit (ABI, #4368814). PCR was performed by LongAmpTM Taq DNA polymerase (NEB, #M0534L). TM‐qPCR was performed by TaqMan Universal Master Mix II (Life, #4440048). 5′‐RACE was performed by SMARTer™ RACE cDNA Amplification Kit (Clontech, #634923). 3′‐RACE was performed by 3′‐Full RACE Core Set ver.2.0 (TaKaRa, #D314). All of the above experiments were performed according to manufacturers' protocols.
Plasmid vectors construction
To obtain the DIG‐labeled RNA probes, the LincGET‐specific region (2574‐2763), Dyei‐specific region (384‐642), and a part of Egfp sequence (362‐668) as control were amplified using LongAmpTM Taq DNA polymerase (NEB, #M0534L) and were subcloned into the plasmid pEASY‐T3 cloning vector (TransGen, #CT301‐02), which contains the T7 promoter. Full‐length LincGET1 lacking LincGET‐LNA2 target site and partial sequences of LincGET were also subcloned into the plasmid pEASY‐T3 cloning vector. For dox‐induced expression of MS2‐labeled LincGET, 7× MS2‐LincGET1 was cloned into EcoRI digested TetO‐FUW‐OSKM (Addgene, #20321). For co‐IP experiment of MS2 coat protein (MS2P), HA‐labeled MS2P was cloned into EcoRI digested TetO‐FUW‐OSKM (Addgene, #20321).
miRNA reverse Northern blotting
Total miRNAs in 500 early four‐cell embryos were extracted by mirVana miRNA isolation kit (Ambion, AM1560). Then adaptors were added to 5′ and 3′ end of miRNAs followed by reverse transcription and PCR, using Small RNA Cloning Kit (TaKaRa, #DRR065), resulting in microDNAs. We run the microDNAs on 15% urea‐PAGE gel (15 ml; 7.2 g urea, 1.5 ml 10× TBE, 5.6 ml 40% acrylamide (acryl:bis acryl = 19:1), 75 μl 10% ammonium persulfate, and 15 μl TEMED) at 45 mA for 1 h, soaked the gel for 5 min in a 0.5–1 μg/ml solution of ethidium bromide in 1× TBE, and visualized using a UV transilluminator. After staining, we transferred the microDNAs to a nylon membrane (Life) by electroblotting at 200 mA for at least 1 h. After blotting, we cross‐linked the microDNAs to the membranes using a commercial UV cross‐linking device (120 mJ burst over 30 s). We then pre‐hybridized the membrane in 10 ml pre‐hybridization solution (6× SSC (Sigma), 10× Denhardt's solution (Invitrogen), 0.2% SDS) for at least 1 h at 65°C. Next, we hybridized membrane in 10 ml of hybridization solution (6× SSC, 5× Denhardt's solution, 0.2% SDS) containing 0.1 μM 3′‐end‐DIG‐labeled single‐strand DNA oligonucleotide for 8–24 h with gentle agitation at room temperature (RT). We then washed the blot with 10 ml wash solution (6× SSC, 0.2% SDS) with gentle agitation at RT for 5 min for three times and once at 42°C for 10 min. After the final wash, signals were detected using the DIG detection kit (Roche, #11093657910) according to the manufacturer's protocol.
RNA‐FISH
The probes were labeled by in vitro transcription using DIG RNA Labeling Kit (SP6/T7) (Roche, #11175025910). After removal of the zona pellucida with acidic Tyrode's solution, mouse embryos were incubated in 1× PBS containing 6 mg/ml BSA for 15 min. Then, embryos were transferred on coverslips coated with Denhardt's solution (Sigma, #30915‐5ML) and dried for 30 min at RT 40. Embryos were fixed in 3% paraformaldehyde (PFA) for 12 min followed by permeabilization in RNA‐FISH permeabilizing solution (0.5% Triton X‐100, 10 mM Vanadyl ribonucleoside complex (Sigma, #94742‐1ML), in 1× PBS) for 6 min on ice. After two washes in 70% EtOH for 5 min each, dehydration was performed in 80%, 95%, twice 100% EtOH, each for 5 min at RT, and the slides were dried for 5 min. The embryos were hybridized in hybridization solution (50% formamide (Sigma), 2× SSC, 10% dextran sulfate (Sigma), 10 mM VRC, 2 mg/ml BSA (Sigma)) containing 0.1 nM DIG‐labeled RNA probes at 37°C overnight. After three washes for 5 min each in hybridization washing solution (50% formamide, 2× SSC) at 42°C and four washes for 5 min each in PBT (1% Tween‐20, in 1× PBS), we blocked the embryos in blocking solution (10% sheep serum, 0.05% BSA, in 1× PBS) for 1 h at RT followed by incubation in antibody hybridization solution (2% sheep serum, 0.05% BSA, anti‐DIG antibody (1:200), in 1× PBS) for 2–3 h at RT. After four washes for 5 min each in PBT, embryos were stained with PI (10 μg/ml in PBS) for 7 min. Then, embryos were mounted on glass slides after three washes.
DNA‐FISH
The oligonucleotide probes were labeled with DIG by Invitrogen. After removal of the zona pellucida with acidic Tyrode's solution, mouse embryos were incubated in 1× PBS containing 6 mg/ml BSA for 15 min. Embryos were then transferred on coverslips coated with Denhardt's solution (Sigma) and dried for 30 min at RT 40. Embryos were fixed in 3% PFA for 15 min followed by permeabilization in DNA‐FISH permeabilizing solution I (0.2% Triton X‐100 in 1× PBS) for 30 min at RT and then in DNA‐FISH permeabilizing solution II (0.7% Triton X‐100, 0.1 m HCl, in 1× PBS) for 15 min on ice. After two washes in 70% EtOH for 5 min each, dehydration was performed in 80%, 95%, twice 100% EtOH, each for 5 min at RT. The embryos were then dried for 5 min. Embryos were then denatured in hybridization washing solution (50% formamide, 2× SSC) at 80°C for 30–45 min, washed two times in 100% cold EtOH for 5 min each, and dry for 5 min, and hybridized in hybridization solution (50% formamide (Sigma), 2× SSC, 10% dextran sulfate (Sigma), 10 mM VRC, 2 mg/ml BSA (Sigma)) containing 0.1 μM each DIG‐labeled RNA probes at 37°C overnight. After three washes for 5 min each in hybridization washing solution (50% formamide, 2× SSC) at 42°C and four washes for 5 min each in PBT (1% Tween‐20, in 1× PBS), we blocked embryos in blocking solution (10% sheep serum, 0.05% BSA, in 1× PBS) for 1 h at RT followed by incubating in antibody hybridization solution (2% sheep serum, 0.05% BSA, anti‐DIG antibody (1:200), in 1× PBS) for 2–3 h at RT. After four washes for 5 min each in PBT, embryos were stained with PI (10 μg/ml in PBS) for 7 min. Then, embryos were mounted on glass slides after three washes.
RNA interference
We isolated zygotes from superovulated mated ICR females at post‐human chorionic gonadotropin injection (phCG) 20 h and microinjected about 10 pl (10 μM) LNA (Exiqon) into the cytoplasm between 24 and 27 h after hCG injection, using an Eppendorf micromanipulator on a Nikon inverted microscope.
BrdU staining
To identify the phase at which LincGET‐depleted 2C embryos arrest, we added 20 μg/ml (final concentration) BrdU to the cultured mouse embryos injected with control‐LNA or LNA targeting LincGET at phCG 30 h. The immunofluorescence staining was performed at phCG 48 h. Additionally, to determine whether DNA replication would happen in arrested LincGET‐depleted 2C, we added BrdU at late two‐cell stage at phCG 48 h and assessed the BrdU signal at phCG 62 h. After removal of the zona pellucida with acidic Tyrode's solution, mouse two‐cell embryos were washed two times for 5 min each in washing solution (0.1% Tween‐20, 0.01% Triton X‐100 in 1× PBS). Embryos were fixed in 3% PFA for 30 min followed by incubation in 1.5 M HCl diluted in 1× PBS. After three washes for 5 min each in washing solution, embryos were permeabilized in normal permeabilizing solution (1% Triton X‐100 in 1× PBS) overnight at 4°C. Embryos were then blocked in blocking solution (1% BSA in 1× PBS) for 1 h at RT after three washes for 5 min each in washing solution (0.1% Tween‐20, 0.01% Triton X‐100 in 1× PBS), followed by incubation with primary antibody diluted with blocking solution overnight at 4°C. After three washes for 5 min each in washing solution, embryos were incubated with Alexa series fluorescent tag‐conjugated secondary antibody diluted with washing solution for 1 h at RT. After three washes in washing solution, nuclei were stained with PI (10 μg/ml in 1× PBS) for 7 min. Embryos were then mounted on glass slides after three washes.
EU staining
EU (10 μM, final concentration) was added to the cultured mouse embryos injected with control‐LNA or LNA targeting LincGET at phCG 40 h. After removal of the zona pellucida with acidic Tyrode's solution, mouse two‐cell embryos were washed two times for 5 min each in washing solution at phCG 48 h. Embryos were then fixed with 3% PFA in 1× PBS for 30 min, followed by permeabilization with normal permeabilizing solution. Incorporated EU was detected using the click‐iT® RNA Alexa Fluor 488 Imaging Kit (Invitrogen, #C10329).
Immunofluorescence staining
After removal of the zona pellucida with acidic Tyrode's solution, mouse embryos were fixed in 3% PFA for 40 min at RT, followed by permeabilization in normal permeabilizing solution overnight at 4°C. The following steps are as for BrdU staining.
Microscope analysis and image processing
We acquired bright field images of embryos under a Nikon inverted microscope eclipse TS100 equipped with a Digital Sight camera system (Nikon), and fluorescence staining was imaged using the inverted microscope (Leica DMI3000B) or laser‐scanning confocal microscope (LSM 780). We used Adobe Photoshop CS4 for further processing.
RNA pull‐down assay
RNAs were in vitro‐transcribed with mMESSAGEmMACHINE® T7 ULTRA Kit (Ambion, #AMB1345‐5) and biotinylated with Pierce RNA 3′‐End Desthiobiotinylation Kit (Pierce, #20163) following the manufacturer's manual. Slot blot was performed to demonstrate that RNAs were efficiently biotinylated. Fifty picomoles of biotinylated RNA were heated to 85°C for 2 min, put immediately on ice for at least 2 min, and an equal volume of RNA structure buffer (10 mM Tris pH 7.0, 0.1 M KCl, 10 mM MgCl2) was added. The samples were then shifted to RT for at least 20 min to allow proper secondary structure formation. Early four‐cell stage embryos (for pull‐down mass spectrum, 6,367 embryos were used, and for pull‐down Western blot, about 1,500 embryos were used for each time) were digested with Pierce IP Lysis Buffer (Pierce, #87787) supplied with protease inhibitor cocktail (Pierce, #78441) according to the manufacturer's protocol. RNA pull‐down was performed by Pierce Magnetic RNA‐Protein Pull‐Down Kit (Pierce, #20164) according to the manufacturer's protocol. The retrieved protein was detected by mass spectrometry or Western blot.
Mass spectrometry
We run the protein retrieved by RNA pull‐down on 10% SDS–PAGE gel (separation gel 10 ml: 3.3 ml 30% acrylamide (acryl:bis acryl = 29:1), 2.5 ml 1.5 M‐pH 8.8‐Tris–HCl, 100 μl 10% SDS, 50 μl 10% ammonium persulfate, and 5 μl TEMED; spacer gel 10 ml: 1.7 ml 30% acrylamide (acryl:bis acryl = 29:1), 2.5 ml 0.5M‐pH 6.8‐Tris–HCl, 100 μl 10% SDS, 50 μl 10% ammonium persulfate, and 10 μl TEMED) at 150 V for 1 h. After electrophoresis, we removed the gel from the cassette and stain the gel using SilverQuest™ Silver Staining kit (Invitrogen, #LC6100) according to manufacturer's protocol. The specific bands were cut out and subjected to mass spectrometry after destaining. The mass spectrometry analysis was performed by BGI Company.
Western blot
The protein retrieved by RNA pull‐down assay or from 200 embryos digested with Pierce IP lysis buffer (10 μl/lane) was mixed with 30 μl sample buffer (10 ml; 1.25 ml 0.5 M‐pH 6.8‐Tris–HCl, 2.5 ml glycerin, 2 ml 10% SDS, 200 μl 0.5% bromophenol blue, 3.55 ml H2O, and 0.5 ml β‐mercaptoethanol) and incubated for 5 min in boiling water. The samples were separated on SDS–PAGE with a 5% stacking gel (10 ml; 5.7 ml ddH2O, 2.5 ml 1.5 M pH 6.8 Tris–HCl, 1.7 ml 30% acrylamide (acryl:bis acryl = 29:1), 100 μl 10% SDS, 50 μl 10% ammonium persulfate, and 10 μl TEMED) and a 10% separating gel (10 ml; 4.1 ml ddH2O, 2.5 ml 1.5 M pH 8.8 Tris–HCl, 3.3 ml 30% acrylamide (acryl:bis acryl = 29:1), 100 μl 10% SDS, 50 μl 10% ammonium persulfate, and 5 μl TEMED) at 120 V for 1.5 h and then electrophoretically transferred onto a nitrocellulose membrane at 300 mA for 1 h. Membranes were blocked in TBST buffer (10 mM Tris, 150 mM NaCl, 0.1% Tween‐20, pH 7.4) containing 3% BSA, for 1 h at RT and then incubated with primary antibody, diluted in TBST containing 1% BSA, overnight at 4°C. After three washes for 10 min each in TBST, the membrane was incubated for 1 h at RT with the secondary antibody diluted in TBST. After three washes for 10 min each, the signals were detected using ECL and films.
Co‐immunoprecipitation
We collected about 2,500 early four‐cell stage embryos (phCG 50–52 h) and added 100 μl RIPA buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.4, 1 mM EDTA, 0.1% SDS, 1% NP‐40, 0.5% sodium deoxycholate, 0.5 mM DTT, 1 mM PMSF/cocktail) and 5 μl RNase inhibitor (Ambion, #AM2694) followed by incubation on ice for 10 min. We then harvested 10 μl of lysates used as inputs. The remaining lysates were divided into two parts and incubated with anti‐SRSF1 or anti‐HA antibody overnight at 4°C. Next, 10 μl of protein A agarose beads (Novex, #15918014) was added and the mixture was rotated for 4 h at 4°C, followed by microcentrifugation at 900 g for 5 min at 4°C. The beads were washed three times with RIPA buffer for 5 min each. The results were mixed with 15 μl of Western blot sample buffer and incubated for 5 min in boiling water. Western blot was performed to detect hnRNP U, FUBP1, ILF2, and SRSF1 (HA‐MS2P was detected in the anti‐HA co‐IP group).
Low initial amount RNA‐seq and data analysis
We microinjected LNA at phCG 25 h and collected control‐LNA L2C (2,225 embryos) and LincGET‐depleted 2C (2,042 embryos) embryos strictly at phCG 48 h. Low initial amount RNA‐seq and alternative splicing events analysis were performed by BGI Company. The gene expression level was calculated by using the reads per kilobase transcriptome per million mapped reads (RPKM) method 78. For gene expression difference analysis, we treated genes with FDR ≤ 0.0001, RPKM ≥ 1, and changed > twofold as DEGs. To select the software for alternative splicing event detection, we evaluated four pieces of software (SOAPsplice 79, TopHat 80, SpliceMap 81, and MapSplice 82) based on 50‐nt reads simulated by Maq 83. Our evaluation indicated that TopHat was the best. Therefore, it was used in our pipeline to perform this analysis for genes with RPKM ≥ 1. We only detected four types of alternative splicing events, exon skipping, intron retention, alternative 5′ splice site, and alternative 3′ splice site. The other three types were not included in our report due to high false‐positive results with the present program. We performed Wilcoxon rank single test (P < 2.2 × 10−16) of median distance of DEGs to neighbor NLK‐LTRs and that of 10,000 times of random control to test the relationship between DEGs and GLKLTRs. GLKLTRs correspond to the gene loci that contain whole LTR of GLN, MERVL, or ERVK. For motif analysis in Fig EV4D, we first obtained sequences of skipping exons or total exons in genome with extending 100 nt in two directions; Secondly, we used Homer to enrich motifs in sequences from special exons and sequences from total exons as background under default parameters. Finally, we searched reverse complementary pairing regions for these motifs in LincGET.
Statistical analysis
Differences of data [mean ± standard error of the mean (s.e.m.)] were analyzed by using two‐tailed Student's t‐test.
Author contributions
JW, QZ, and ZL conceived and designed the study. JW, XL, LW, YZ, YL, GJ, XS, RW, SL, and BX performed the experiments; JW, XL, JL, GB, LL, and WL analyzed the data. QZ and ZL supervised the project. JW, QZ, and ZL designed and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Acknowledgements
The authors thank all members of the Laboratory of Embryo Biotechnology in Northeast agricultural university; Ming Ge, Xiaolong Cui, Guihai Feng, and Shiwen Li for technical assistance. This study was supported by the National Natural Science Foundation of China (31371457, J1210069, and 31471395), China National Basic Research Program (2012CBA01300, 2016YFA0100200), and Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01020100). We thank Eppendorf, Leica, and Beckman for supplying the devices.
EMBO Reports (2016) 17: 1452–1470
Contributor Information
Qi Zhou, Email: qzhou@ioz.ac.cn.
Zhonghua Liu, Email: liu086@yahoo.com.
References
- 1. Probst AV, Almouzni G (2011) Heterochromatin establishment in the context of genome‐wide epigenetic reprogramming. Trends Genet 27: 177–185 [DOI] [PubMed] [Google Scholar]
- 2. Schultz RM (2002) The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update 8: 323–331 [DOI] [PubMed] [Google Scholar]
- 3. Bedzhov I, Graham SJ, Leung CY, Zernicka‐Goetz M (2014) Developmental plasticity, cell fate specification and morphogenesis in the early mouse embryo. Philos Trans R Soc Lond B Biol Sci 369: 20130538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL (2012) Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C (2013) Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB (2004) Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606 [DOI] [PubMed] [Google Scholar]
- 7. Evsikov AV, de Vries WN, Peaston AE, Radford EE, Fancher KS, Chen FH, Blake JA, Bult CJ, Latham KE, Solter D et al (2004) Systems biology of the 2‐cell mouse embryo. Cytogenet Genome Res 105: 240–250 [DOI] [PubMed] [Google Scholar]
- 8. Wang J, Xie G, Singh M, Ghanbarian AT, Rasko T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV et al (2014) Primate‐specific endogenous retrovirus‐driven transcription defines naive‐like stem cells. Nature 516: 405–409 [DOI] [PubMed] [Google Scholar]
- 9. Lu X, Sachs F, Ramsay L, Jacques PE, Goke J, Bourque G, Ng HH (2014) The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol 21: 423–425 [DOI] [PubMed] [Google Scholar]
- 10. Ohnuki M, Tanabe K, Sutou K, Teramoto I, Sawamura Y, Narita M, Nakamura M, Tokunaga Y, Nakamura M, Watanabe A et al (2014) Dynamic regulation of human endogenous retroviruses mediates factor‐induced reprogramming and differentiation potential. Proc Natl Acad Sci USA 111: 12426–12431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY et al (2015) Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522: 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Durruthy‐Durruthy J, Sebastiano V, Wossidlo M, Cepeda D, Cui J, Grow EJ, Davila J, Mall M, Wong WH, Wysocka J et al (2015) The primate‐specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat Genet 48: 44–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gendrel AV, Heard E (2014) Noncoding RNAs and epigenetic mechanisms during X‐chromosome inactivation. Annu Rev Cell Dev Biol 30: 561–580 [DOI] [PubMed] [Google Scholar]
- 14. Koerner MV, Pauler FM, Huang R, Barlow DP (2009) The function of non‐coding RNAs in genomic imprinting. Development 136: 1771–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bertani S, Sauer S, Bolotin E, Sauer F (2011) The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell 43: 1040–1046 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hawkins PG, Morris KV (2010) Transcriptional regulation of Oct4 by a long non‐coding RNA antisense to Oct4‐pseudogene 5. Transcription 1: 165–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rosa A, Ballarino M (2016) Long noncoding RNA regulation of pluripotency. Stem Cells Int 2016: 1797692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Williams GT, Mourtada‐Maarabouni M, Farzaneh F (2011) A critical role for non‐coding RNA GAS5 in growth arrest and rapamycin inhibition in human T‐lymphocytes. Biochem Soc Trans 39: 482–486 [DOI] [PubMed] [Google Scholar]
- 22. Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grander D, Morris KV (2013) A pseudogene long‐noncoding‐RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol 20: 440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K (2013) The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon‐gamma locus. Cell 152: 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X (2014) The STAT3‐binding long noncoding RNA lnc‐DC controls human dendritic cell differentiation. Science 344: 310–313 [DOI] [PubMed] [Google Scholar]
- 25. Wang K, Sun T, Li N, Wang Y, Wang JX, Zhou LY, Long B, Liu CY, Liu F, Li PF (2014) MDRL lncRNA regulates the processing of miR‐484 primary transcript by targeting miR‐361. PLoS Genet 10: e1004467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD (2009) Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci 12: 1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R (2008) Induced ncRNAs allosterically modify RNA‐binding proteins in cis to inhibit transcription. Nature 454: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Imamura T, Yamamoto S, Ohgane J, Hattori N, Tanaka S, Shiota K (2004) Non‐coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem Biophys Res Commun 322: 593–600 [DOI] [PubMed] [Google Scholar]
- 31. Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q (2010) Long noncoding RNAs with enhancer‐like function in human cells. Cell 143: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Orom UA, Shiekhattar R (2011) Noncoding RNAs and enhancers: complications of a long‐distance relationship. Trends Genet 27: 433–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C (2008) The LPS‐induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell 32: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR (2009) Toggle involving cis‐interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci USA 106: 18321–18326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP (2010) Noncoding RNA gas5 is a growth arrest‐ and starvation‐associated repressor of the glucocorticoid receptor. Sci Signal 3: ra8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG (2008) A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1‐induced epithelial‐mesenchymal transition. Genes Dev 22: 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ, Wahlestedt C (2008) Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed‐forward regulation of beta‐secretase. Nat Med 14: 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H (2013) Endogenous miRNA sponge lincRNA‐RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self‐renewal. Dev Cell 25: 69–80 [DOI] [PubMed] [Google Scholar]
- 39. Cao S, Han J, Wu J, Li Q, Liu S, Zhang W, Pei Y, Ruan X, Liu Z, Wang X (2014) Specific gene‐regulation networks during the pre‐implantation development of the pig embryo as revealed by deep sequencing. BMC Genom 15: 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G (2010) A strand‐specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell 19: 625–638 [DOI] [PubMed] [Google Scholar]
- 41. van Attikum H, Gasser SM (2009) Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 19: 207–217 [DOI] [PubMed] [Google Scholar]
- 42. Minami N, Suzuki T, Tsukamoto S (2007) Zygotic gene activation and maternal factors in mammals. J Reprod Dev 53: 707–715 [DOI] [PubMed] [Google Scholar]
- 43. Chu DP, Tian S, Qi L, Hao CJ, Xia HF, Ma X (2013) Abnormality of maternal‐to‐embryonic transition contributes to MEHP‐induced mouse 2‐cell block. J Cell Physiol 228: 753–763 [DOI] [PubMed] [Google Scholar]
- 44. Qiu JJ, Zhang WW, Wu ZL, Wang YH, Qian M, Li YP (2003) Delay of ZGA initiation occurred in 2‐cell blocked mouse embryos. Cell Res 13: 179–185 [DOI] [PubMed] [Google Scholar]
- 45. Maekawa M, Yamamoto T, Kohno M, Takeichi M, Nishida E (2007) Requirement for ERK MAP kinase in mouse preimplantation development. Development 134: 2751–2759 [DOI] [PubMed] [Google Scholar]
- 46. Ranpura SA, Deshmukh U, Reddi PP (2008) NF45 and NF90 in murine seminiferous epithelium: potential role in SP‐10 gene transcription. J Androl 29: 186–197 [DOI] [PubMed] [Google Scholar]
- 47. Kiesler P, Haynes PA, Shi L, Kao PN, Wysocki VH, Vercelli D (2010) NF45 and NF90 regulate HS4‐dependent interleukin‐13 transcription in T cells. J Biol Chem 285: 8256–8267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao G, Shi L, Qiu D, Hu H, Kao PN (2005) NF45/ILF2 tissue expression, promoter analysis, and interleukin‐2 transactivating function. Exp Cell Res 305: 312–323 [DOI] [PubMed] [Google Scholar]
- 49. Bi HS, Yang XY, Yuan JH, Yang F, Xu D, Guo YJ, Zhang L, Zhou CC, Wang F, Sun SH (2013) H19 inhibits RNA polymerase II‐mediated transcription by disrupting the hnRNP U‐actin complex. Biochim Biophys Acta 1830: 4899–4906 [DOI] [PubMed] [Google Scholar]
- 50. Vizlin‐Hodzic D, Johansson H, Ryme J, Simonsson T, Simonsson S (2011) SAF‐A has a role in transcriptional regulation of Oct4 in ES cells through promoter binding. Cell Reprogram 13: 13–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vizlin‐Hodzic D, Runnberg R, Ryme J, Simonsson S, Simonsson T (2011) SAF‐A forms a complex with BRG1 and both components are required for RNA polymerase II mediated transcription. PLoS ONE 6: e28049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao J, Ding J, Li Y, Ren K, Sha J, Zhu M, Gao X (2009) HnRNP U mediates the long‐range regulation of Shh expression during limb development. Hum Mol Genet 18: 3090–3097 [DOI] [PubMed] [Google Scholar]
- 53. Rabenhorst U, Thalheimer FB, Gerlach K, Kijonka M, Bohm S, Krause DS, Vauti F, Arnold HH, Schroeder T, Schnutgen F (2015) Single‐stranded DNA‐binding transcriptional regulator FUBP1 Is essential for fetal and adult hematopoietic stem cell self‐renewal. Cell Rep 11: 1847–1855 [DOI] [PubMed] [Google Scholar]
- 54. Vu NT, Park MA, Shultz JC, Goehe RW, Hoeferlin LA, Shultz MD, Smith SA, Lynch KW, Chalfant CE (2013) hnRNP U enhances caspase‐9 splicing and is modulated by AKT‐dependent phosphorylation of hnRNP L. J Biol Chem 288: 8575–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xiao R, Tang P, Yang B, Huang J, Zhou Y, Shao C, Li H, Sun H, Zhang Y, Fu XD (2012) Nuclear matrix factor hnRNP U/SAF‐A exerts a global control of alternative splicing by regulating U2 snRNP maturation. Mol Cell 45: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Obrdlik A, Kukalev A, Louvet E, Ostlund Farrants AK, Caputo L, Percipalle P (2008) The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol Cell Biol 28: 6342–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rigo F, Hua Y, Chun SJ, Prakash TP, Krainer AR, Bennett CF (2012) Synthetic oligonucleotides recruit ILF2/3 to RNA transcripts to modulate splicing. Nat Chem Biol 8: 555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jacob AG, Singh RK, Mohammad F, Bebee TW, Chandler DS (2014) The splicing factor FUBP1 is required for the efficient splicing of oncogene MDM2 pre‐mRNA. J Biol Chem 289: 17350–17364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA (2010) The nuclear‐retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R (2013) Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494: 497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G (2010) A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pi W, Zhu X, Wu M, Wang Y, Fulzele S, Eroglu A, Ling J, Tuan D (2010) Long‐range function of an intergenic retrotransposon. Proc Natl Acad Sci USA 107: 12992–12997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li J, Akagi K, Hu Y, Trivett AL, Hlynialuk CJ, Swing DA, Volfovsky N, Morgan TC, Golubeva Y, Stephens RM (2012) Mouse endogenous retroviruses can trigger premature transcriptional termination at a distance. Genome Res 22: 870–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J (2013) Single‐cell RNA‐Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20: 1131–1139 [DOI] [PubMed] [Google Scholar]
- 65. Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y (2014) rMATS: robust and flexible detection of differential alternative splicing from replicate RNA‐Seq data. Proc Natl Acad Sci USA 111: E5593–E5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wee CD, Havens MA, Jodelka FM, Hastings ML (2014) Targeting SR proteins improves SMN expression in spinal muscular atrophy cells. PLoS ONE 9: e115205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Miro J, Laaref AM, Rofidal V, Lagrafeuille R, Hem S, Thorel D, Mechin D, Mamchaoui K, Mouly V, Claustres M (2015) FUBP1: a new protagonist in splicing regulation of the DMD gene. Nucleic Acids Res 43: 2378–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S (1992) The product of the mouse Xist gene is a 15 kb inactive X‐specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526 [DOI] [PubMed] [Google Scholar]
- 69. Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF (1992) The human xist gene – analysis of a 17 Kb inactive X‐specific Rna that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542 [DOI] [PubMed] [Google Scholar]
- 70. Wu SC, Zhang Y (2010) Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 11: 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Papaioannou VE (1982) Lineage analysis of inner cell mass and trophectoderm using microsurgically reconstituted mouse blastocysts. J Embryol Exp Morphol 68: 199–209 [PubMed] [Google Scholar]
- 72. Hamazaki N, Uesaka M, Nakashima K, Agata K, Imamura T (2015) Gene activation‐associated long noncoding RNAs function in mouse preimplantation development. Development 142: 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F (2010) A long nuclear‐retained non‐coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29: 3082–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ciarlo E, Massone S, Penna I, Nizzari M, Gigoni A, Dieci G, Russo C, Florio T, Cancedda R, Pagano A (2013) An intronic ncRNA‐dependent regulation of SORL1 expression affecting Abeta formation is upregulated in post‐mortem Alzheimer's disease brain samples. Dis Model Mech 6: 424–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, Avigan M, Levens D (1994) Sequence‐specific, single‐strand binding‐protein activates the far upstream element of C‐Myc and defines a new DNA‐binding motif. Genes Dev 8: 465–480 [DOI] [PubMed] [Google Scholar]
- 76. Zhang F, Tian Q, Wang Y (2013) Far upstream element‐binding protein 1 (FUBP1) is overexpressed in human gastric cancer tissue compared to non‐cancerous tissue. Onkologie 36: 650–655 [DOI] [PubMed] [Google Scholar]
- 77. Shamanna RA, Hoque M, Pe'ery T, Mathews MB (2013) Induction of p53, p21 and apoptosis by silencing the NF90/NF45 complex in human papilloma virus‐transformed cervical carcinoma cells. Oncogene 32: 5176–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA‐Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- 79. Huang S, Zhang J, Li R, Zhang W, He Z, Lam TW, Peng Z, Yiu SM (2011) SOAPsplice: genome‐Wide ab initio detection of splice junctions from RNA‐Seq data. Front Genet 2: 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA‐Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Au KF, Jiang H, Lin L, Xing Y, Wong WH (2010) Detection of splice junctions from paired‐end RNA‐seq data by SpliceMap. Nucleic Acids Res 38: 4570–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM (2010) MapSplice: accurate mapping of RNA‐seq reads for splice junction discovery. Nucleic Acids Res 38: e178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li H, Ruan J, Durbin R (2008) Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File


