The silencing of StNAC103, a phellem-induced gene, reveals its repressor role in suberin and wax deposition and provides evidence of fine control over the formation of the apoplastic barrier.
Keywords: Apoplastic barriers, NAC transcription factor, periderm, phellem, regulation, StNAC103, suberin, suberin-associated wax, tuber
Abstract
Suberin and wax deposited in the cork (phellem) layer of the periderm form the lipophilic barrier that protects mature plant organs. Periderm lipids have been widely studied for their protective function with regards to dehydration and for how they respond to environmental stresses and wounding. However, despite advances in the biosynthetic pathways of suberin and associated wax, little is known about the regulation of their deposition. Here, we report on a potato NAC transcription factor gene, StNAC103, induced in the tuber phellem (skin). The StNAC103 promoter is active in cells undergoing suberization such as in the basal layer of the phellem, but also in the root apical meristem. Gene silencing in potato periderm correlates with an increase in the suberin and wax load, and specifically in alkanes, ω-hydroxyacids, diacids, ferulic acid, and primary alcohols. Concomitantly, silenced lines also showed up-regulation of key genes related to the biosynthesis and transport of suberin and wax in the tuber periderm. Taken together, our results suggest that StNAC103 has a role in the tight regulation of the formation of apoplastic barriers and is, to the best of our knowledge, the first candidate gene to be identified as being involved in the repression of suberin and wax deposition.
Introduction
The phellem or cork is a physical barrier at the interface between (mature) secondary growth tissues and the environment. It prevents dehydration and facilitates resistance to pathogens and mechanical wounds. The phellem is located within the outer face of the periderm, the dermal structure that replaces the epidermis in secondary stems, secondary roots, tubers, and tissues healing from wounds. The phellem (cork) cells undergo suberization, a process in which suberin and wax are deposited onto the inner face of primary cell walls during a differentiation process that ends with cell death (Pollard et al., 2008). Suberin, like cutin, is a polyester consisting of fatty acids and glycerol forming a matrix in which wax is bound. However, in contrast to cutin, suberin contains longer chain length (>20 carbons) fatty acids and derivatives, a greater proportion of dicarboxylic acid monomers, and a covalently linked polyaromatic moiety (Bernards, 2002). The suberin-associated wax mostly consists of linear, very long chain aliphatic compounds of up to C32, including alkanes, primary alcohols, fatty acids, and feruloyl esters of primary alcohols. It has recently been shown in Arabidopsis roots that suberin-associated wax mainly consists of primary alcohols, predominantly in the form of alkyl hydroxycinnamates (Delude et al., 2016). The role of both cuticular and suberin-associated wax in a reduction of water loss has been clearly demonstrated (Schreiber et al., 2005; Zhang et al., 2005; Kosma et al., 2009; Lee et al., 2014; Zhu et al., 2014). Moreover, it has been shown that an increased production of cuticular wax leads to improved drought tolerance (Zhang et al., 2005; Kosma et al., 2009; Lee et al., 2014; Zhu et al., 2014). Analyses of Arabidopsis mutants with altered suberin accumulation demonstrated that increased suberin content resulted in better tolerance to salt stress and wilting (Beisson et al., 2007; Baxter et al., 2009; Gou et al., 2009). The study of Arabidopsis suberin and wax mutants has facilitated the characterization of many enzymes involved in their biosynthetic pathways (for reviews, see Beisson et al., 2012; Yeats and Rose, 2013). However, potato tuber skin has played an important part in the study of the influence of suberin and wax chemical composition on periderm physiology as sufficient amounts of periderm can be easily obtained. The main advantage of the potato periderm model is that it makes it possible to quantify water permeability to study the contribution of down-regulation of a gene in the water barrier properties of the periderm (Schreiber et al., 2005; Serra et al., 2009a, b, 2010; Landgraf et al., 2014). For example, in potato tuber phellem, the silencing of CYP86A33 greatly decreased suberin C18:1 diacids and ω-hydroxyacids, altered suberin lamellation, and resulted in a 3.5-fold higher permeability to water (Serra et al., 2009b). In contrast, the silencing of FHT (fatty ω-hydroxyacid/fatty alcohol hydroxycinnamoyl transferase) resulted in a lack of feruloyl esters of primary alcohols in both suberin and associated wax, yielding potato tubers that were 15 times more permeable to water, although the suberin ultrastructure remained unchanged (Serra et al., 2010).
Land plants commonly encounter adverse conditions that result in temperature, water, light, or salt stress, and have evolved regulatory mechanisms to adapt lipid barriers to changing environmental conditions. Plants respond to water deficit treatment by increasing the deposition of both leaf cuticular wax and cutin monomers (Kosma et al., 2009). Moreover, it has been reported that Arabidopsis plants respond to salt stress by increasing the amount of alkyl coumarates in root wax (Kosma et al., 2012). For cuticle regulation, a complex regulatory network responsive to developmental and environmental cues and mediated by hormones, transcription factors, and epigenetic and post-transcriptional mechanisms has been suggested (Yeats and Rose, 2013). In this regulatory network, abscisic acid (ABA) is thought to stimulate cuticle deposition together with a suite of AP2/EREBP, MYB, MADS-box, and HD-Zip IV transcription factors (Samuels et al., 2008; Yeats and Rose, 2013; Hen-Avivi et al., 2014). Little is currently known about the regulatory network controlling suberin and the associated wax. Wounding and abiotic stresses activate suberin and wax synthesis through the induction of genes such as KCS (3-ketoacyl-CoA synthase: Franke et al., 2009; Lee et al., 2009), FAR (fatty acid reductase: Domergue et al., 2010), and FHT (Boher et al., 2013). In healing tissues, suberin is deposited in suberizing cells of the wound-closing layer and the wound periderm. ABA induces suberin biosynthetic genes in Arabidopsis (Lee et al., 2009; Barberon et al., 2016), potato (Lulai et al., 2008; Boher et al., 2013), and tomato (Leide et al., 2012), triggers suberin accumulation in potato tuber wound-healing tissue (Lulai et al., 2008), and gives rise to ectopic suberin deposition in root (Barberon et al., 2016). It has been shown that the application of ethylene leads to the disappearance of pre-formed suberin lamellae as well as to the finding that the plant’s nutritional status is key to the regulation of suberization, revealing an unexpected flexibility in the regulation (Barberon et al., 2016). However, the transcription factors that regulate suberization are relatively unknown. Until recently, information was limited to the expression profile of specific regulatory genes induced in suberized tissues: an Arabidopsis AP2/ERF transcription factor (Lasserre et al., 2008) and the cork oak (Quercus suber) and apple (Malus×domestica) transcription factors WRKY, NAM, and R2R3-MYB classes (Soler et al., 2007, 2008; Almeida et al., 2013; Legay et al., 2015). The first and only evidence of a regulatory effect on suberin is AtMYB41, which ectopically activates the biosynthesis and assembly of suberin concurrently with the induction of suberin and lignin genes in Arabidopsis (Kosma et al., 2014).
NAC transcription factors belong to one of the largest plant-specific transcription factor families and are represented by 110 genes in potato (Singh et al., 2013). Typically, NAC proteins have a well conserved N-terminal region, known as the NAC domain, and a diversified C-terminal region known as a TAR (transcription activation region) domain (Olsen et al., 2005). The NAC domain is involved in DNA binding whereas the C-terminal region acts in transcription regulation and protein–protein interactions. NAC transcription factors are involved in a diverse array of functions including development, lateral root formation, auxin signaling, secondary cell wall biosynthesis, and stress regulation (Olsen et al., 2005; Zhong et al., 2010; Puranik et al., 2012). In potato, a large proportion of NAC genes are sensitive to abiotic stress (Gong et al., 2015). In this study, we investigate the potato NAC transcription factor gene, StNAC103 (Singh et al., 2013), in tuber periderm. StNAC103 is the putative ortholog to a cork oak NAM transcription factor gene highlighted as a phellem regulation candidate based on its up-regulation in cork oak phellem (Soler et al., 2007) and its co-expression throughout the growing season with well-known suberin genes (Soler et al., 2008). More recently, the putative ortholog of StNAC103 in apple (Malus×domestica) was also identified in the skin of apple fruits showing russeting, a skin defect due to the accumulation of suberin on the fruit exocarp (Legay et al., 2015). StNAC103 belongs to the potato NAC subgroup C, which contains genes related to the stress response (StNAC017 and StNAC030: Singh et al., 2013) and organ development (ATNAC4 and CUC3: Hibara et al., 2006; Vidal et al., 2013). The Arabidopsis putative ortholog of StNAC103 (ANAC058) confers hypersensitivity to ABA and it has been suggested that it may be a modulator of ABA-mediated germination potential (Coego et al., 2014). Here, we show that StNAC103 is up-regulated in cells undergoing suberization and induced by wounding and ABA treatment. The silencing of StNAC103 in potato periderm correlates with an increase in suberin and wax compounds concomitant with the up-regulation of suberin and wax genes, hence suggesting a negative regulatory effect on the deposition of apoplastic lipids. Taken as a whole, this work suggests that the periderm apoplastic barrier is finely regulated and that StNAC103 is involved in this regulatory network.
Materials and methods
Plant material
Potato plants (Solanum tuberosum Group Tuberosum) cvs Desirée and Andigena were propagated as described previously by Serra et al. (2010). Solanum tuberosum Group Tuberosum cv. Desirée was used for silencing. For Desirée tubers, in vitro plants grown in MS (Murashige and Skoog) medium supplemented with 2% sucrose were transferred to soil and grown for 3 months in a walk-in chamber before harvesting. Andigena plants were used for detection of the promoter activity given that in this cultivar tuber onset is induced by photoperiod and tuber maturation can be more finely regulated than for Desirée plants. For Andigena plants, tuberization was induced in vitro by growing two-node stem explants in MS medium supplemented with 8% sucrose in short-day conditions (8h light/16h dark) at 22 ºC for 1 week and then transferred to the dark. We also used S. tuberosum Group Tuberosum cv. Monalisa potatoes, purchased at a local supermarket, for the wound healing experiment. For this experiment, potato tuber discs (3mm thick and 13mm in diameter) were obtained by cutting tuber flesh (parenchyma) excised with a cork borer and left to heal at room temperature in saturated humidity and dark conditions.
ABA treatment of roots in hydroponic cultures
Desirée plants were propagated first in vitro and after 3 weeks plants were transferred to hydroponic culture and grown for 15 d in half-strength Hoagland’s solution (Arnon and Hoagland, 1939) in a 10 liter container in a growth chamber (light/dark photoperiod cycle of 12/12h at 22 ºC and 67 µmol m−2 s−1). Subsequently, a solution of ABA (Sigma, A-1049) suspended in ethanol was added to the cultures at a final concentration of 50 µM and roots were incubated for 5h and 26h. Control plants were treated with a mock solution of ethanol. Each time point had 4–6 biological replicates, each consisting of a pool of three roots from different plants.
RNA extraction of potato tissues
Periderm tissue was manually dissected using sterile scalpels. Once harvested, tissue samples were immediately frozen in liquid nitrogen. Total RNA was isolated using the guanidine hydrochloride method (Logemann et al., 1987) and genomic DNA was removed by on-column DNase digestion using the RNeasy MinElute (Qiagen) in accordance with the manufacturer’s instructions. RNA concentration and purity were determined by formamide–formaldehyde denaturing agarose gel electrophoresis and a Nanodrop spectrophotometer. First-strand cDNA was synthesized from 2 µg of DNase-digested RNA. We used the Superscript III reverse transcriptase (Invitrogen) with oligo(dT)18 primers for the synthesis of full-length cDNA before cloning, or the high capacity cDNA reverse transcription kit (Life Technologies) with random primers to synthesize cDNA for real-time reverse transcription–PCR (RT–PCR) analysis.
Recombinant vector construction and plant transformation
Since information about the potato genome was not available at that time, primers to amplify and clone the StNAC103 coding sequence and the region used for silencing were designed using the information available from the potato Expressed Sequence Tag assembly (TC143904) with homology to the cork oak sequence (EE743827) encoding a NAC transcription factor isolated from a SSH phellem library by Soler et al. (2007). We also used the most similar sequences from Arabidopsis (At3g18840) and Petunia×hybrid (GI:21389167) to design primers when potato sequence information was not available. For StNAC103 silencing, a specific fragment of 253bp located primarily in the TAR domain was used. The amplification product was first cloned into the pENTR/D TOPO vector (Life Technologies) and then transferred into the binary destination vector pBIN19RNAi (Serra et al., 2009b) using LR clonase II (Invitrogen). Using the potato genome sequence available, specific primers were designed to get a 2291 bp sequence upstream of the start codon (primers are shown in Supplementary Table S1 at JXB online). The PCR product was then inserted into the pDONR207 vector using the BP clonase II (Life Technologies) and subsequently transferred to the vector pKGWFS7 (Karimi et al., 2002) using LR clonase II.
Recombinant plasmids were inserted into Agrobacterium tumefaciens (GV2260) following the protocol of Höfgen and Willmitzer (1988), and leaves were infected with transformed Agrobacterium tumefaciens cells. Kanamycin-resistant plants were regenerated in vitro by organogenesis according to Banerjee et al. (2006).
Histochemical GUS staining of ProStNAC103:GUS-GFP transgenic plants
β-Glucuronidase (GUS) histochemical staining was performed using Andigena transgenic potato plants containing the ProStNAC103:GUS-GFP construct. Fixation was done by immersion of the transformed tissue in an ice-chilled 90% acetone (v/v) bath, followed by incubation for 20min on ice and rinsing twice with water before infiltration for 20min under vacuum with the staining solution [1mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic sodium salt 3·H2O (X-GlcA, Duchefa), 50mM sodium phosphate buffer (pH 7), 1.25mM potassium ferrocyanide, 1.25mM potassium ferricyanide, 10mM EDTA, and 0.05% (v/v) Triton X-100]. Incubation in staining solution was extended for a maximum of 48h at 37 ºC and then cleared with 70% (v/v) ethanol. Transformed GUS-stained and control tissues were examined with a bright-field microscope (AH2 Vanox-T microscope Olympus) and a stereomicroscope (Wild M420 Makroskop), and photographed with a digital camera (Olympus Camedic C-4040ZOOM).
Real-time PCR analysis
Real-time PCRs were performed in an optical 96-well plate with an ABI PRISM 7300 Sequence Detector System (Applied Biosystems) and a Fluidigm (Biomark). Gene-specific PCR primers were designed with Primer 3 (Untergasser et al., 2012) and checked with Primer Express 3.0 (Applied Biosystems). Reactions contained 1× Power SYBR Green Master Mix reagent (Applied Biosystems), 300nM of the respective primers, and 5 µl of a 25-fold dilution of the corresponding cDNA in a final volume of 20 µl. The following standard thermal profile was used for all PCRs: 95 °C for 10min; 40 cycles of 95 °C for 15s and 60 °C for 1min. A dissociation step was performed after amplification to confirm the presence of a single amplicon. All data were processed and analyzed with 7300 SDS 1.3.1 software (Applied Biosystems). For microfluidics quantitative PCR (qPCR; Fluidigm), 1.25 μl of the synthesized cDNA was pre-amplified and then purified with exonuclease treatment. Pre-amplified and purified cDNAs were diluted to 1:5 and used for real-time qPCR amplification the BioMark™ system (Fluidigm). A dissociation curve was obtained to check primer specificity for each amplification reaction. Data collection and analysis was performed using Fluidigm Real-Time PCR analysis software 3.0.2 (Fluidigm). For each primer pair, standard curves with a 5-fold dilution series (1/1, 1/5, 1/25, 1/125, 1/625) of template were obtained to determine the amplification efficiency.
mRNA abundance was calculated as relative transcript abundance=(Etarget)ΔCt target (control–sample)/(Ereference)ΔCt reference (control–sample) (Pfaffl, 2001). The controls used to standardize data were as follows: for the StNAC103 transcript accumulation pattern in native tissues, a mix with equal amounts of all samples; for transcript accumulation in tuber wound healing time course, a pool of each biological replicate was obtained at 144h post-wounding; for wound-healing leaves, a pool of each biological replicate was obtained at 72h post-wounding; for transcript accumulation in silenced transgenic lines, a pool of biological replicates of the control line (Desirée) was used. The housekeeping gene APRT encoding an adenine phosphoribosyl transferase was used to normalize the results (Nicot et al., 2005; Soler et al., 2011), except for the time course experiment of the tuber discs in which the constitutive gene EF1α was employed as it has less variation in these conditions. To check the absence of genomic DNA contamination, reverse transcriptase controls were used, whereas to check the absence of environmental contamination we used non-template controls (NTCs). The sequence of primers used for real-time PCR is shown in Supplementary Table S1.
Isolation of periderm membranes
Periderm membranes used for chemical and water permeance analyses consist of the phellem (cork) layer after removal of the non-suberized tissue by enzymatic digestion as described by Schreiber et al. (2005). In short, fragments of tuber skin were separated from the inner flesh with a cork borer and incubated in a 2% (v/v) cellulase and a 2% (v/v) pectinase solution to remove all unsuberized (parenchymal) cells. After digestion, the remaining phellem membranes were washed, dried, and stored at room temperature until used. All membranes were obtained from tubers after 1 month storage at room temperature to allow post-harvest periderm maturation.
Suberin and wax chemical analyses
Suberin and wax analyses were performed following the procedure described by Schreiber et al. (2005). Briefly, the wax fraction was extracted by a treatment with a mixture of chloroform and methanol (1:1 v/v) for 18h. For depolymerization of the aliphatic suberin, wax-free membranes were then trans-esterified by incubation at 70 ºC for 18h with methanol/boron trifluoride (~10% BF3 in methanol; Fluka). All compounds were quantified and analyzed as trimethylsilyl (TMS) derivatives, obtained by N,O-bis (trimethylsilyl) trifluoroacetamide (BSTFA; Macherey-Nagel) derivatization. Derivative products were identified by GC with a selective mass detector with ion trap (Trace GC 2 000 series coupled to a Thermo Scientific Polaris Q a mass spectrometer). For each peak, the mass spectrum was compared with data reported in the literature (Kolattukudy and Agrawal, 1974; Zeier and Schreiber, 1997; Zeier and Schreiber, 1998). Quantification was performed using a GC-FID (flame ionization detector; Shimadzu GC- 2010 Plus, Kyoto, Japan) by comparison of peak areas with an internal standard [for wax compounds, tetracosane (Fluka); for suberin monomers, dotrioacontane (Fluka)]. Results were statistically analyzed using Student’s t-test.
Measurement of peridermal permeance
Peridermal permeance to water was measured using transpiration chambers by a gravimetric method (Schreiber et al., 2005; Serra et al., 2009b). Weight loss was measured at regular time periods during 30 d in an analytical balance. Water permeance (P, m s−1) was obtained using only the slope values from chambers in which water loss (g) across the periderm against time (s) fit a linear regression of R2 ≥0.99.
Accession numbers
Accession numbers of the StNAC103 promoter region (2291bp upstream of the translation initiation codon) and the StNAC103 full-length coding sequence of the S. tuberosum Group Tuberosum are KT582103 and KT598221, respectively.
Results
StNAC103 is expressed in suberized potato tissues
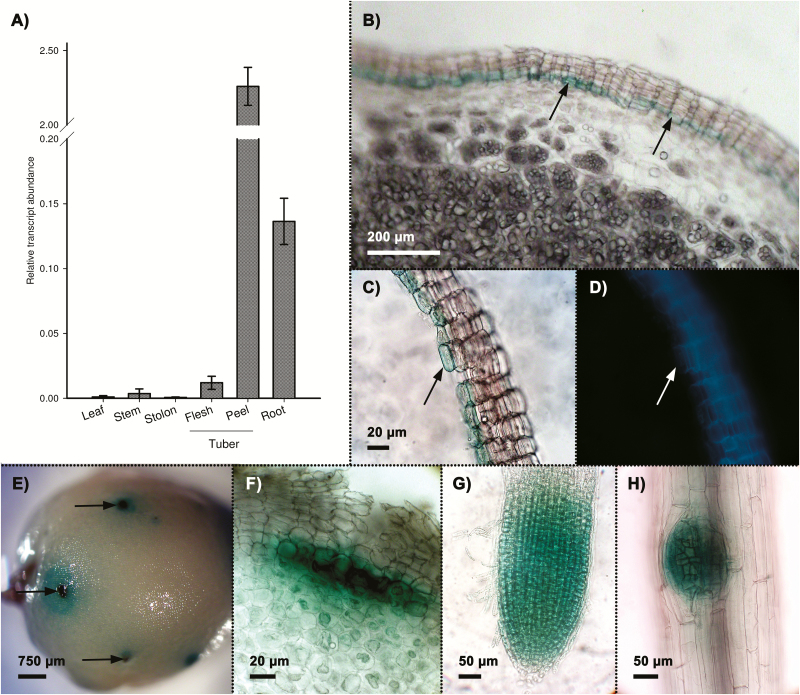
The protein sequence of the S. tuberosum Group Tuberosum StNAC103 (KT598221) shares 98.41% identity with StNAC103 from S. tuberosum Group Phureja (Supplementary Fig. S1A). Both sequences are identical in the conserved NAC domain, with the differences restricted to the TAR domain. The expression profile of StNAC103 Group Tuberosum in vegetative tissues was analyzed by quantitative RT–PCR; transcripts were mainly detected in tissues in which suberin is constitutively present, specifically tuber skin (phellem) and root (Fig. 1A). To analyze StNAC103 expression further, potato plants were stably transformed with a construct containing the StNAC103 promoter region fused to the coding sequence of the double transcriptional reporter GUS–green fluorescent protein (GFP). Tubers bearing the ProStNAC103:GUS-GFP were analyzed for GUS activity. The blue color, which is indicative of promoter activity, was localized in the periderm and restricted to the internal (basal) cell layers of the phellem (Fig. 1B). This localization was confirmed in skin sheets containing only the phellem peeled off from the tubers (Fig. 1C). Phellem cells in which the promoter was active corresponded to living cells undergoing differentiation but in which suberin autofluorescence is very faint or absent (Fig. 1D, arrow). GUS staining of whole tubers revealed large areas of the tuber surface with weak GUS expression, but also high GUS expression corresponding to the lenticels (Fig. 1E, arrows). The deep blue labeling of lenticels matched with the lenticular phellogen at the base of the lenticel (Fig. 1F). StNAC103 promoter activity was also analyzed in primary roots obtained from hydroponic cultures in which promoter activity was not localized in the suberized root barriers, exodermis, or endodermis, but in lateral root primordia and root apical meristems (Fig. 1G, H). Such a pattern contrasts with that of FHT, a well-studied gene involved in suberin biosynthesis (Serra et al., 2010) which is active only in suberized cell layers (Boher et al., 2013). Both patterns can be compared in Supplementary Fig. S2, showing potato roots from plants bearing, respectively, the ProStNAC103:GUS-GFP and the ProFHT:GUS-GFP constructs, grown and stained in parallel.
Fig. 1.
StNAC103 transcript accumulation and promoter activity in potato native tissues. (A) Transcript abundance in different potato organs and tissues. Relative mRNA expression levels were expressed as the mean ±SD (the biological replicates were four for stem, leaf, and root, three for stolon and parenchyma, and two for phellem). (B–F) StNAC103 promoter activity in ProStNAC103:GUS-GFP tuber. (B) Section of a tuber showing GUS staining specific to the inner phellem cell layers (arrows). (C) Sheet of mechanically isolated phellem showing the GUS blue marker and the (D) suberin autofluorescence. (E) The GUS signal observed through the tuber surface showing lenticels as dark blue dots (arrows). (F) Detail of a lenticel seen in cross-section showing the GUS blue marker located to the lenticular phellogen. (G, H) Root whole mounts showing the GUS signal localized in (G) apical and (H) lateral root meristems.
StNAC103 expression is triggered by wounding stress and ABA treatment
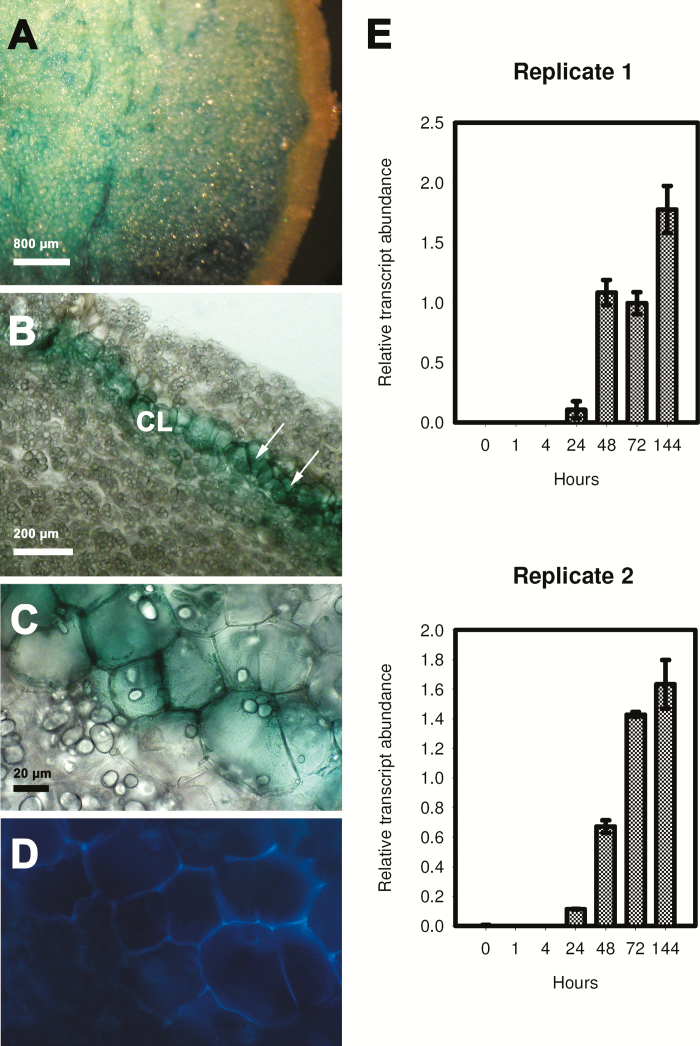
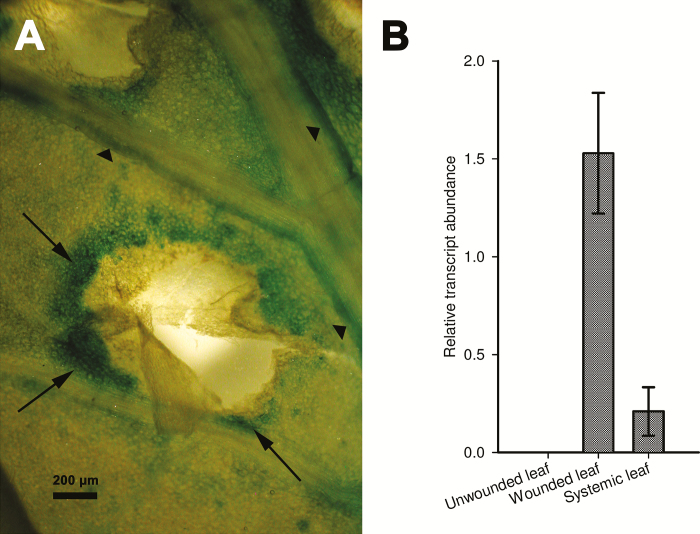
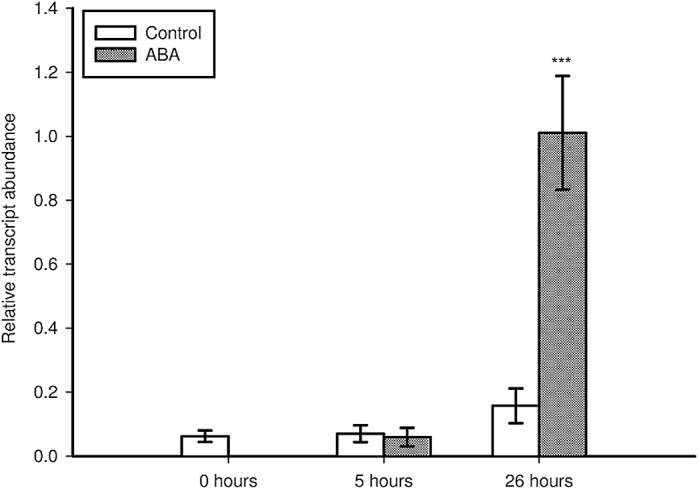
To examine the activation of StNAC103 in healing tissues, we injured tubers and leaves in ProStNAC103:GUS-GFP plants. After 48h of wounding by a single cut, tubers showed reporter expression at the severed surface (Fig. 2A). Microscopic examination revealed that it was confined to the cell layers in which suberization occurs (Fig. 2B). In healing tissues, GUS-stained cells are living cells depleted of starch grains (Fig. 2C) whose cell walls display the typical suberin autofluorescence under UV excitation (Fig. 2D). A time course quantitative RT–PCR analysis of StNAC103 expression throughout the healing process was performed in healing tuber flesh (parenchyma) discs obtained from commercial potatoes. StNAC103 transcripts were detected beginning at 24h after injury, and levels increased gradually during the wound healing (Fig. 2E). Expanded leaves were also injured by pinching the leaflets with tweezers and then left to heal. In healing leaves (72h after wounding), the GUS activity was mainly restricted to the wound margins (Fig. 3A, arrows) but was also detected in some surrounding tissues (Fig. 3A, arrowheads). To evaluate a systemic activation of the promoter as a consequence of a local wounding, we compared StNAC103 transcript levels in damaged leaves with those of undamaged leaves on the node immediately below (systemic leaves) and also with the expression in leaves of unwounded plants. Both damaged and systemic leaves showed transcript accumulation, although levels were lower in systemic leaves (Fig. 3B). We further investigated StNAC103 induction in response to ABA in potato roots grown in hydroponic cultures. As can be seen in Fig. 4, after 26h of ABA treatment (50 µM) there was a strong accumulation of StNAC103 transcript in roots. Hence, this gene is also inducible by abiotic stress signals such as wounding and ABA, and this responsiveness occurs in roots but also in tissues where StNAC103 is not preferentially expressed, such as tuber flesh and leaf.
Fig. 2.
StNAC103 expression in wound-healing tubers. (A–D) StNAC103 promoter activity in ProStNAC103:GUS-GFP plants. (A) GUS staining of wound-healing tubers 48h after wounding observed through the cut surface. (B) In a section through the cut surface, the blue color localized to the wound-healing tissue (arrows). (C) Detail of (B) under bright field and (D) under UV excitation to indicate suberin autofluorescence. (E) Time course accumulation of StNAC103 transcript in potato tuber healing discs during 144h presented separately for two biological replicates. The mean ±SD of three technical replicates is shown. CL, closing layer.
Fig. 3.
StNAC103 in wound-healing leaves. (A) GUS staining of a leaf 72h after wounding showing the blue color in the wound margins (arrows) and in a nearby minor vein (arrowheads). (B) StNAC103 transcript accumulation in wounded, unwounded, and systemic leaf is reported. Relative mRNA expression levels were expressed as the mean ±SD of two biological replicates.
Fig. 4.
StNAC103 induction by ABA in roots grown in hydroponic culture. The transcript abundance was analyzed in roots of plants treated with 50 µM ABA during 5h and 26h. Relative mRNA expression levels were expressed as the mean ±SD of 4–6 biological replicates. Pronounced statistically significant differences are shown with three asterisks (P<0.001). Results were statistically analyzed using Student’s t-test.
StNAC103 silencing
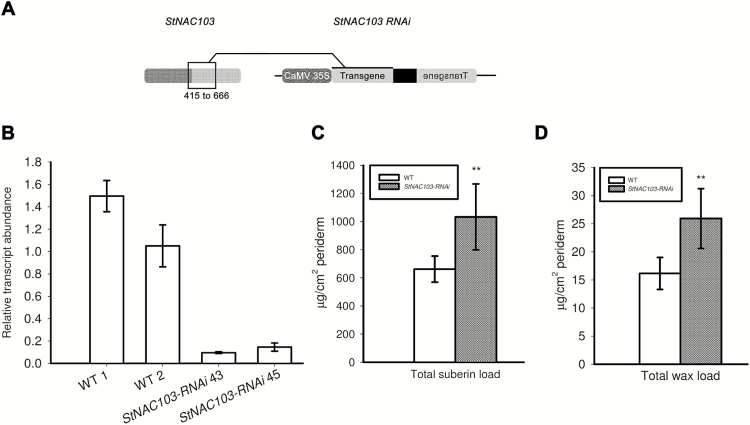
To evaluate the role of StNAC103 in the phellem, we generated transgenic potato plants that silence StNAC103. The silencing construct was targeted primarily to the variable TAR domain, the most specific region of the StNAC103 gene, and was designated as StNAC103-RNAi (Supplementary Fig. S1B; Fig. 5A). In order to check the specificity of the silencing, putative targets of the StNAC103-RNAi construct were investigated by a blastN performed in the potato genome database (Potato Genome Sequencing Consortium et al., 2011; PGSC S. tuberosum group Phureja, DM1-3 Transcripts v3.4, expected threshold of 1) using the silencing RNAi sequence as a query. The blastN identified the StNAC103 transcript with 0 mismatches along all the query sequence and a partial match with StNAC057 in one region with two mismatches in 21 consecutive nucleotides. To check if despite these two mismatches StNAC057 could be an off-target of the sequence used for RNAi silencing, the relative transcript abundance of StNAC057 was analyzed in StNAC103-RNAi and wild-type periderms (Supplementary Fig. S3). No decrease was observed in StNAC057 transcript levels in the StNAC103-RNAi periderms, so ruling out cross-silencing.
Fig. 5.
Effects of StNAC103 silencing in the apoplastic lipid load of the tuber periderm. (A) Scheme of the StNAC103-RNAi construct designed for gene silencing targeting the variable TAR doamin. The NAC domain is shown in dark gray and the TAR domain in light gray. (B) StNAC103 relative transcript abundance in the periderm of silencing and wild-type lines. Relative mRNA expression levels were expressed as the mean ±SD of three technical replicates. (C) Total suberin and (D) total wax load in the wild-type and StNAC103-RNAi periderm from tubers stored for 39 d. Significant differences are shown with two asterisks (P<0.01).
StNAC103 gene silencing in tuber periderm was confirmed by RT–PCR in a number of kanamycin-resistant lines, and those lines showing a substantial decrease in transcript levels were selected (Fig. 5B). All potato plants deficient in StNAC103 developed normally either in vitro or in pots. Likewise, transgenic tubers displayed no visual differences compared with the wild type during tuber development, at harvest, or during storage. Two StNAC103-RNAi lines (43 and 45) corresponding to different transformation events were propagated for tuber production and used for phenotypic analyses.
StNAC103 impacts the suberin and wax composition of the potato tuber periderm
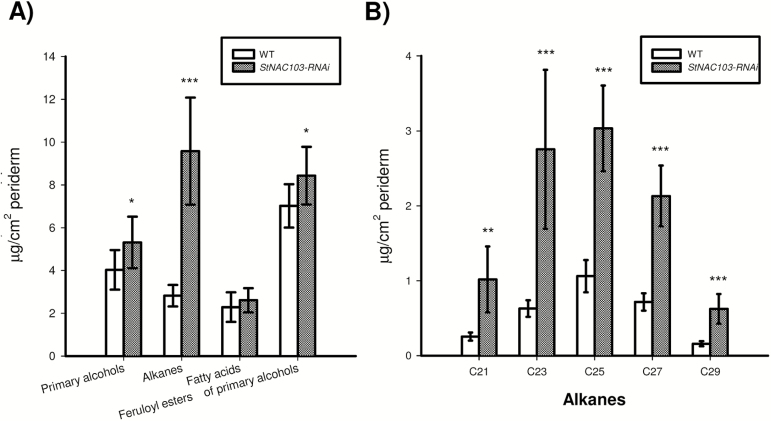
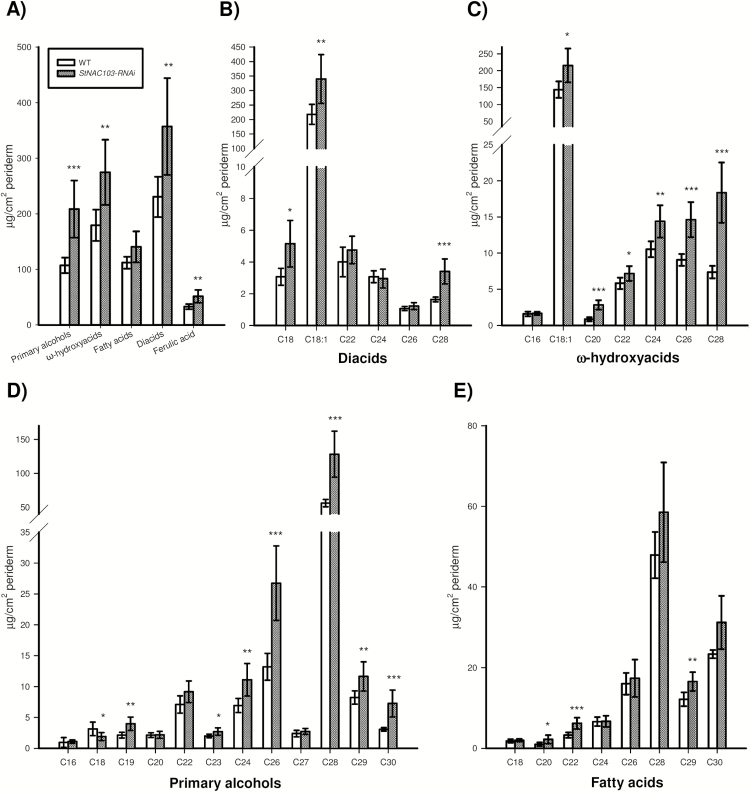
The effect of StNAC103 on the chemical composition of the suberin polymer and solvent-extractable wax was analyzed by GC-MS and quantified by GC-FID from enzymatically isolated periderm membranes obtained as described in the Materials and methods. Tubers deficient in StNAC103 showed a significant increase in wax and suberin load and important changes in their composition (Fig. 5C, D). The total amount of each wax substance class showed that the most affected substance class is that of alkanes (Fig. 6A), which significantly increased in the periderm of silenced lines. Primary alcohols and feruloyl esters of primary alcohols were also affected. The chemical composition analysis of the wax fraction showed that individual alkanes doubled their amounts in StNAC103-RNAi periderm, compared with the wild type (Fig. 6B), while feruloyl esters of primary alcohols and primary alcohols showed smaller differences between the wild type and silenced periderms (Supplementary Fig. S4). As can be seen in Supplementary Fig. S4, only C22 and C28 primary alcohols increased. Regarding feruloyl esters of primary alcohols: C16, C18, and C20 decreased and C23, C24, C26, and C30 increased. For the suberin fraction (Fig. 7A), StNAC103 silencing resulted in an increase in the primary alcohols, ω-hydroxyacids, diacids, and ferulic acid. Amongst single monomers, the levels of the two main monomers in the suberin from potato periderm, C18:1 diacid and C18:1 ω-hydroxyacid, were increased (Fig. 7B, C), but the C18 and C28 diacid were also augmented. In addition, all the ω-hydroxyacids longer than C18 were also increased in the silenced lines (C20, C22, C24, C26, and C28 ω-hydroxyacids). Among primary alcohols, the amounts of C19, C23, C24, C26, C28, C29, and C30 alcohol were enhanced, but the C18 alcohol decreased (Fig. 7D). Results for the separate lines are plotted in Supplementary Figs S5 and S6. The significant role of StNAC103 in periderm lipids led us to investigate its contribution to the water barrier function. To this end, we analyzed the water permeance in enzymatically isolated periderm membranes using a gravimetric method. As shown in Supplementary Fig. S7, the permeance to water was not statistically different between StNAC103-RNAi and wild-type periderms, and thus the increase in suberin and wax load was not found to play a significant role in the physiology of the periderm.
Fig. 6.
Effects of StNAC103 silencing on the wax chemical composition shown as the amount of compounds per surface area from tuber periderm stored for 39 d. (A) Composition by substance classes of the wild-type and StNAC103-RNAi periderm. (B) Profile of alkanes identified in the periderm of wild-type and silencing lines. Values are the the mean ±SD of the wild type (five biological replicates) and two independent transformation events for StNAC103-RNAi, lines 43 and 45 (four and three biological replicates, respectively). Significant differences (P<0.05) are denoted with one asterisk, whereas pronounced significant differences (P<0.01; P<0.001) are shown with two and three asterisks, respectively.
Fig. 7.
Effect of StNAC103 silencing on suberin composition shown as the amount of compounds per surface area from tuber periderm stored for 39 d. (A) Composition by substance classes, (B) profile of diacids, (C) profile of ω-hydroxyacids, (D) profile of primary alcohols, and (E) profile of fatty acids represented by their carbon chain length of the wild type and StNAC103-RNAi lines. Values are the mean ±SD of the wild type (five biological replicates) and two independent transformation events for StNAC103-RNAi, lines 43 and 45 (four and three biological replicates, respectively). Significant differences (P<0.05) are denoted with one asterisk, whereas particularly pronounced significant differences (P<0.01; P<0.001) are shown with two and three asterisks, respectively.
StNAC103 silencing results in the induction of some genes key for suberin and wax accumulation
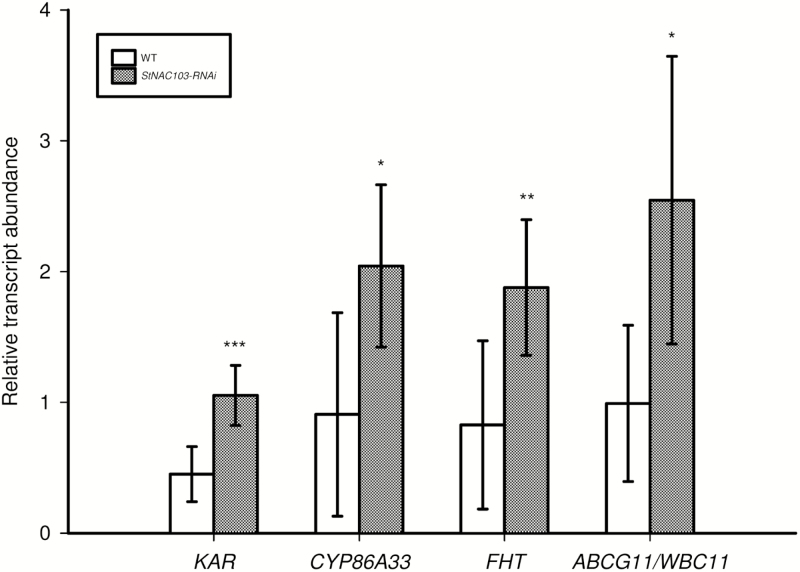
To understand further the molecular mechanisms of StNAC103 function, the relative expression of a selected set of genes was analyzed by microfluidics qPCR in silenced and wild-type periderms. The genes analyzed were representative of de novo synthesis of fatty acids (KAR, ketoacyl-ACP reductase), ω-hydroxylation of fatty acids (CYP86A33, cytochrome P450), cross-linking of the fatty acid polyester to the ferulic acid (FHT, fatty ω-hydroxyacid/fatty alcohol hydroxycinnamoyl), and export of suberin and wax compounds (ABCG11/WBC11). Relative transcript abundance shows that all these genes are up-regulated in the periderm of StNAC103-RNAi lines in comparison with the wild type (Fig. 8), and hence they act downstream of StNAC103. As such they are either direct or indirect targets of this transcription factor.
Fig. 8.
Microfluidics qPCR in StNAC103-RNAi and wild-type periderm of some key genes involved in suberin and wax accumulation. The relative transcript abundances of KAR, CYP86A33, FHT, and ABCG11/WBC11 are shown. Values are the mean ±SD of the wild type (four biological replicates) and two independent transformation events for StNAC103-RNAi, lines 43 and 45 (five and four biological replicates, respectively). Significant differences (P<0.05) are denoted with one asterisk, whereas pronounced significant differences (P<0.01; P<0.001) are shown with two and three asterisks, respectively.
Discussion
The StNAC103-silenced potato plants show an increase of the total load of suberin and wax in the periderm. In the wax fraction, the silencing mainly produces an increase in the alkane accumulation whereas in the suberin fraction there is an increase in the ω-hydroxyacids, diacids, primary alcohols, and ferulic acid. In agreement with this phenotype, some key genes that are representative of the biosynthesis and transport of suberin and wax display up-regulation in the periderm of StNAC103-RNAi lines. All of the above suggest a repressor role for StNAC103 in the suberin and suberin-associated wax. Together with AtMYB41, StNAC103 is the second transcription factor characterized with a clear effect in apoplastic lipids (Kosma et al., 2014). However, AtMYB41 works as an activator showing a correlation between gene overexpression, up-regulation of suberin-related genes, and ectopic deposition of suberin and associated wax in leaf epidermis (Kosma et al., 2014).
The NAC family of transcription factors comprises both transcriptional activators and repressors. A transcription repression function has been specifically demonstrated for calmodulin-binding NAC protein (Kim et al., 2007), the vascular-related NAC-domain-interacting 2 protein (Yamaguchi et al., 2010), and the NAC050 and NAC052 (Ning et al., 2015). Moreover, some NAC-type transcription factors, such as GmNAC20, contain an active repression domain of 35 amino acids (NARD) with a highly conserved sequence among repressor proteins, which includes the hydrophobic LVFY motif also thought to contribute to the repression function (Hao et al., 2010). The alignment of the NARD-like sequence from StNAC103 and proteins showing suppression of transcriptional activity (Hao et al., 2010) shows conservation of the LVFY motif (Supplementary Fig. S8) and 16 identical residues from the 17 highlighted previously by Hao et al. (2010) in StNAC103. This is compatible with a repression activity of StNAC103 and is in agreement with the increase of suberin and wax observed in StNAC103-silenced lines.
Phellem cells are formed with the activity of a lateral meristem called phellogen, and phellem initials should be elongated and produce thick suberized cell walls before suffering from programmed cell death and becoming mature cork cells, in a similar way to as is described for xylem formation (Hertzberg et al., 2001). StNAC103 is highly expressed in tissues under the suberization process, but this gene induction is counter-intuitive with the suggested repressor role of this gene, whose silencing yields an increase in suberin and associated wax in StNAC103-RNAi periderm. It is probable that the differentiation of phellem cells, as is the case for xylem cells (Schuetz et al., 2012), is a process that is tightly regulated transcriptionally. Hence, it can be surmised that StNAC103 functions by preventing a premature or inappropriate suberization of phellem initials, probably by competing with other regulatory proteins that promote suberization such as MYB41. The competition of positive and negative stimuli would provide a fine-tuning mechanism to control the suberin deposition process spatially and temporally. There are several cases in the literature in which certain biological processes must be rigorously regulated and the repressor is expressed concurrently with onset of the process. For instance, EgMYB1, a key repressor of lignin biosynthesis in Eucalyptus, is preferentially expressed during xylem differentiation (Legay et al., 2010). Moreover, in Arabidopsis, the ABA repressor 1 (ABR1), which is a negative regulator of ABA response, is highly responsive to ABA treatment (Pandey et al., 2005), and RAP2.1, a DREB (dehydration-responsive element-binding protein) transcriptional repressor, is induced by drought (Dong and Liu 2010). Apart from its role in phellem cells undergoing suberization, StNAC103 also turns on in cells involved in wound healing, similarly to the suberin biosynthetic gene FHT (Boher et al., 2013). It is widely known that a wound, which is a potential focus of infection due to the loss of the protective barrier, induces biotic and abiotic stress responses in plants. Healing of the wound tuber requires rapid suberization of existing parenchyma cells bordering the wound surface and later development of a deeper wound phellogen layer. The induction of genes involved in suberin deposition, cell wall, and cell proliferation during the first days after wounding in potato tuber has been reported (Neubauer et al., 2012; Lulai and Neubauer, 2014). The induction of StNAC103 probably mediates the amount of suberin that is synthesized during the formation of this protective healing tissue.
The induction of StNAC103 in the proliferative cells of lenticular phellogen is consistent with a repression function in cells that should remain undifferentiated without suberin in their cell walls. Lenticels are responsible for gas exchange in the periderm and undergo long-term structural changes by means of the production of non-suberized and suberized filling tissues depending on the physiological and environmental requirements (Hayward, 1974; Groh et al., 2002). The regulation of this gas exchange involves cell proliferation to increase gas exchange and suberin biosynthesis to restrict gas exchange (Lendzian, 2006), and StNAC103 can participate in maintaining this balance. In the same line of evidence, the promoter activity in young roots also reinforces the involvement of StNAC103 in the suggested repressor role. In roots undergoing primary growth, cell wall suberization is confined to the root water barriers: the exodermis and endodermis. Accordingly, the typical suberin genes such as GPAT5, ASFT, and FHT show their promoter activity localized in these suberized cell layers (Beisson et al., 2007; Molina et al., 2009; Boher et al., 2013). In contrast, the StNAC103 promoter is very active in the apical meristem of potato primary and lateral roots, suggesting a role for this gene in the root meristem, probably preventing a premature suberization in meristematic cells. Remarkably, the lipid transporter ABCG11/WBC11/Desperado, whose putative ortholog is up-regulated in the periderm of StNAC103-RNAi lines, is also expressed in the apical and lateral meristems of roots (Bird et al., 2007; Luo et al., 2007; Panikashvili et al., 2007, 2010).
Our results in potato roots showed induction of StNAC103 by ABA treatment. This is in agreement with the prediction of one ABA-responsive element (ABRE) in the StNAC103 promoter according to the PLACE and PlantCare databases (Higo et al., 1999; Lescot et al., 2002) but also with the ABA-responsive induction of StNAC103 in the RNA sequencing (RNA-seq) data from different potato cultivars (Massa et al., 2011; Singh et al., 2013). ABA is known to be involved in suberin deposition. In wound-healing potato tubers, the ABA treatment enhances an increase in the suberin poly(aliphatic) domain (Lulai et al., 2008), a fact that is also supported by the ability of ABA to induce suberin-associated genes such as GPAT5, FHT, AtMYB41, and ABCG6 (Boher et al., 2013; Kosma et al., 2014; Yadav et al., 2014; Barberon et al., 2016).
Despite the notable increase in the amount of suberin and associated wax, the periderm from silenced StNAC103 lines did not show any reduction in water permeability. A correlation has been found in several studies between a decrease in suberin or the associated wax load and the impairment of the barrier function in potato tuber periderm and Arabidopsis seeds (i.e. Beisson et al., 2007; Serra et al., 2009a). However, the accumulation of suberin monomers has not always been associated with an improvement in the barrier function. For instance, ectopic accumulation of suberin monomers in the leaf cuticle has been reported as even resulting in an impairment of the water barrier function (Li et al., 2007; Cominelli et al., 2008; Kosma et al., 2014). These mixed findings have led to the suggestion that in addition to the monomer load, the structural organization may also be crucial for the water permeability of the apoplastic barrier.
We do not know the direct target genes of StNAC103, but four genes involved in several key steps of wax and suberin accumulation (KAR, FHT, CYP86A33, and WBC11) have been transcriptionally up-regulated in the periderm of StNAC103-RNAi lines. The induction of these genes is in agreement with the modified lipid profile observed in the periderm of StNAC103-silenced lines. For example, KAR is involved in the de novo synthesis of fatty acids (Li-Beisson et al., 2013) and its activation meets the need of a higher demand for fatty acids. Taking into consideration the phenotype of the mutants (Gou et al., 2009; Molina et al., 2009; Serra et al., 2010), the up-regulation of FHT supports the increase in wax ferulate esters, ω-hydroxyacids, primary alcohols, and ferulic acid observed. The up-regulation of CYP86A33, based on the chemical composition of CYP86A1 and CYP86A33 mutants (Li et al., 2007; Höfer et al., 2008; Serra et al., 2009b), is in agreement with the increased presence of the two most abundant suberin monomers, C18:1 ω-hydroxyacid and diacid. The induction of ABCG11/WBC11, a key component of the export pathway for cutin, wax, and suberin, agrees with the accumulation of alkanes, primary alcohols, and C18:1 ω-hydroxyacid suberin monomers (Bird et al., 2007; Luo et al., 2007; Panikashvili et al., 2007, 2010; Ukitsu et al., 2007).
In conclusion we report StNAC103 as a candidate gene for suberin and associated wax repression that probably acts by means of transcriptional regulation of genes related to suberin and wax accumulation. StNAC103 is induced not only in cells undergoing suberization, but also in cells where suberization does not take place but which are physically close to suberized cells, such as lenticular phellogen and root apical meristem. Taking into account the findings presented, it is tempting to speculate that the activation of StNAC103 has a common goal: to prevent suberin deposition. This control can occur either in cells where premature deposition of suberin can be detrimental to the proper functioning of the tissue or as a suberin deposition brake in cells where suberin accumulation takes place, suggesting that the deposition of apoplastic lipids must be under tight control. Further research is needed to elucidate the direct targets of StNAC103 transcription factor. To the best of our knowledge, this is the first regulatory gene characterized as having a role in the repression of suberin and associated wax.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. StNAC103 protein and nucleotide sequences showing the NAC and TAR domains and the region used for silencing.
Figure S2. StNAC103 and FHT promoter activity in potato primary root.
Figure S3. Relative transcript abundance of StNAC057 in StNAC103-RNAi and wild-type periderm.
Figure S4. Wax fatty acids, primary alcohols, and feruloyl esters of primary alcohols in StNAC103-RNAi and the wild type.
Figure S5. Wax composition plotted for the different lines of StNAC103-RNAi analyzed.
Figure S6. Suberin composition for the different lines of StNAC103-RNAi analyzed.
Figure S7. Water permeance of StNAC103-silenced periderm from tubers stored for 39 d.
Figure S8. Alignment of the NARD-like sequences from StNAC103 (99–134) and proteins showing suppression of transcriptional activity.
Table S1. List of genes, loci, and primers used.
Supplementary Material
Acknowledgements
This work was supported by the Ministerio de Educación y Ciencia (AGL2006-07342, FPI grant to OS), the Ministerio de Innovación y Ciencia (AGL2009-13745), the Ministerio de Economía y Competitividad (AGL2012-36725, AGL2015-67495-C2-1-R), Departament d’Universitats, Investigació i Societat de la Informació of Catalonia (PhD grants to RV and MS), and the University of Girona (PhD grant to DCA and SF, and grant SING11/1). The authors are grateful to Professor C. Robin Buell for helpful suggestions on the manuscript (Michigan State University) and to Professor S. Prat (CNB, Madrid) for providing the silencing vector and helpful advice during this study. The authors thank S. Gómez (Departament de Biologia, UdG, Girona) for her valuable assistance in carrying out the laboratory work and taking care of plants, and N. Salvatella, A. Quintana, and I. Armendáriz who assisted with some of the experiments while they were doing their end-of-degree final projects. We would also like to thank Professor V. Salvadó (Departament de Química, UdG, Girona) and Professor C. Pla (Departament de Biologia, UdG, Girona) for kindly lending us the GC-MS and the Thermocycler, respectively.
Glossary
Abbreviations:
- ABA
abscisic acid
- ABCG11/WBC11
ATP-binding cassette/white–brown complex
- AtMYB41
myeloblastosis transcription factor
- CYP86A33
cytochrome P450
- FHT
fatty ω-hydroxyacid/fatty alcohol hydroxycinnamoyl transferase
- KAR
ketoacyl-ACP reductase
- NAC
NAM, ATAF, and CUC
- NARD
NAC repression domain
- qPCR
quantitative PCR
- RT–PCR
reverse transcription–PCR.
References
- Almeida T, Menéndez E, Capote T, Ribeiro T, Santos C, Gonçalves S. 2013. Molecular characterization of Quercus suber MYB1, a transcription factor up-regulated in cork tissues. Journal of Plant Physiology 170, 172–178. [DOI] [PubMed] [Google Scholar]
- Arnon D, Hoagland D. 1939. A comparison of water culture and soil as media for crop production. Science 2, 512–514. [DOI] [PubMed] [Google Scholar]
- Banerjee AK, Prat S, Hannapel DJ. 2006. Efficient production of transgenic potato (S. tuberosum L. ssp. andigena) plants via Agrobacterium tumefaciens-mediated transformation. Plant Science 170, 732–738. [Google Scholar]
- Barberon M, Vermeer JEM, De Bellis D, et al. 2016. Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell 164, 1–13. [DOI] [PubMed] [Google Scholar]
- Baxter I, Hosmani PS, Rus A, Lahner B, Borevitz JO, Muthukumar B, Mickelbart MV, Schreiber L, Franke RB, Salt DE. 2009. Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genetics 5, e1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Li-Beisson Y, Pollard M. 2012. Solving the puzzles of cutin and suberin polymer biosynthesis. Current Opinion in Plant Biology 15, 329–337. [DOI] [PubMed] [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. 2007. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. The Plant Cell 19, 351–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards MA. 2002. Demystifying suberin. Canadian Journal of Botany 80, 227–240. [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu X, Yephremov A, Samuels L. 2007. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. The Plant Journal 52, 485–498. [DOI] [PubMed] [Google Scholar]
- Boher P, Serra O, Soler M, Molinas M, Figueras M. 2013. The potato suberin feruloyl transferase FHT which accumulates in the phellogen is induced by wounding and regulated by abscisic and salicylic acids. Journal of Experimental Botany 64, 3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coego A, Brizuela E, Castillejo P, et al. 2014. The TRANSPLANTA collection of Arabidopsis lines: a resource for functional analysis of transcription factors based on their conditional overexpression. The Plant Journal 77, 944–953. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Sala T, Calvi D, Gusmaroli G, Tonelli C. 2008. Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. The Plant Journal 53, 53–64. [DOI] [PubMed] [Google Scholar]
- Delude C, Fouillen L, Bhar P, Cardinal M-J, Pascal S, Santos P, Kosma DK, Joubès J, Rowland O, Domergue F. 2016. Primary fatty alcohols are major components of suberized root tissues of Arabidopsis in the form of alkyl hydroxycinnamates. Plant Physiology 171, 1934–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domergue F, Vishwanath SJ, Joubès J, Ono J, Lee JA, Bourdon M, Alhattab R, Lowe C, Pascal S, Lessire R, Rowland O. 2010. Three Arabidopsis fatty acyl-Coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiology 153, 1539–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C-J, Liu J-Y. 2010. The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biology 10, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Höfer R, Briesen I, Emsermann M, Efremova N, Yephremov A, Schreiber L. 2009. The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. The Plant Journal 57, 80–95. [DOI] [PubMed] [Google Scholar]
- Gong L, Zhang H, Gan X, et al. 2015. Transcriptome profiling of the potato (Solanum tuberosum L.) plant under drought stress and water-stimulus conditions. PLoS One 10, e0128041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J-Y, Yu X-H, Liu C-J. 2009. A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 18855–18860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh B, Hübner C, Lendzian KJ. 2002. Water and oxygen permeance of phellems isolated from trees: the role of waxes and lenticels. Planta 215, 794–801. [DOI] [PubMed] [Google Scholar]
- Hao Y-J, Song Q-X, Chen H-W, Zou H-F, Wei W, Kang X-S, Ma B, Zhang W-K, Zhang J-S, Chen S-Y. 2010. Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232, 1033–1043. [DOI] [PubMed] [Google Scholar]
- Hayward P. 1974. Waxy structures in the lenticels of potato tubers and their possible effects on gas exchange. Planta 120, 273–277. [DOI] [PubMed] [Google Scholar]
- Hen-Avivi S, Lashbrooke J, Costa F, Aharoni A. 2014. Scratching the surface: genetic regulation of cuticle assembly in fleshy fruit. Journal of Experimental Botany 65, 4653–4664. [DOI] [PubMed] [Google Scholar]
- Hertzberg M, Aspeborg H, Schrader J, et al. 2001. A transcriptional roadmap to wood formation. Proceedings of the National Academy of Sciences, USA 98, 14732–14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara K, Karim MR, Takada S, Taoka K, Furutani M, Aida M, Tasaka M. 2006. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. The Plant Cell 18, 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. 1999. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfer R, Briesen I, Beck M, Pinot F, Schreiber L, Franke R. 2008. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid ω-hydroxylase involved in suberin monomer biosynthesis. Journal of Experimental Botany 59, 2347–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L. 1988. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Research 16, 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY(TM) vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kim HS, Park BO, Yoo JH, et al. 2007. Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. Journal of Biological Chemistry, 282, 36292–36302. [DOI] [PubMed] [Google Scholar]
- Kolattukudy P, Agrawal V. 1974. Structure and composition of aliphatic constituents of potato tuber skin (suberin). Lipids 9, 682–691. [Google Scholar]
- Kosma DK, Bourdenx B, Bernard A, Parsons EP, Lu S, Joubès J, Jenks MA. 2009. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiology 151, 1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Molina I, Ohlrogge JB, Pollard M. 2012. Identification of an Arabidopsis fatty alcohol: caffeoyl-Coenzyme A acyltransferase required for the synthesis of alkyl hydroxycinnamates in root waxes. Plant Physiology 160, 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Murmu J, Razeq FM, Santos P, Bourgault R, Molina I, Rowland O. 2014. AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. The Plant Journal 80, 216–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf R, Smolka U, Altmann S, et al. 2014. The ABC transporter ABCG1 is required for suberin formation in potato tuber periderm. The Plant Cell 26, 3403–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasserre E, Jobet E, Llauro C, Delseny M. 2008. AtERF38 (At2g35700), an AP2/ERF family transcription factor gene from Arabidopsis thaliana, is expressed in specific cell types of roots, stems and seeds that undergo suberization. Plant Physiology and Biochemistry 46, 1051–1061. [DOI] [PubMed] [Google Scholar]
- Lee S-B, Jung S-J, Go Y-S, Kim H-U, Kim J-K, Cho H-J, Park O-K, Suh M-C. 2009. Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. The Plant Journal 60, 462–475. [DOI] [PubMed] [Google Scholar]
- Lee SB, Kim H, Kim RJ, Suh MC. 2014. Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Reports 33, 1535–1546. [DOI] [PubMed] [Google Scholar]
- Legay S, Guerriero G, Deleruelle A, Lateur M, Evers D, André C, Hausman J-F. 2015. Apple russeting as seen through the RNA-seq lens: strong alterations in the exocarp cell wall. Plant Molecular Biology 88, 21–40. [DOI] [PubMed] [Google Scholar]
- Legay S, Sivadon P, Blervacq AS, et al. 2010. EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytologist 188, 774–786. [DOI] [PubMed] [Google Scholar]
- Leide J, Hildebrandt U, Hartung W, Riederer M, Vogg G. 2012. Abscisic acid mediates the formation of a suberized stem scar tissue in tomato fruits. New Phytologist 194, 402–415. [DOI] [PubMed] [Google Scholar]
- Lendzian KJ. 2006. Survival strategies of plants during secondary growth: barrier properties of phellems and lenticels towards water, oxygen, and carbon dioxide. Journal of Experimental Botany 57, 2535–2546. [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJK, Molina I, Pollard M, Ohlrogge J. 2007. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proceedings of the National Academy of Sciences, USA 104, 18339–18344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, et al. 2013Acyl-lipid metabolism. The Arabidopsis Book. 11, e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. 1987. Improved method for the isolation of RNA from plant tissues. Analytical Biochemistry 163, 16–20. [DOI] [PubMed] [Google Scholar]
- Lulai EC, Neubauer JD. 2014. Wound-induced suberization genes are differentially expressed, spatially and temporally, during closing layer and wound periderm formation. Postharvest Biology and Technology 90, 24–33. [Google Scholar]
- Lulai EC, Suttle JC, Pederson SM. 2008. Regulatory involvement of abscisic acid in potato tuber wound-healing. Journal of Experimental Botany 59, 1175–1186. [DOI] [PubMed] [Google Scholar]
- Luo B, Xue X-Y, Hu W-L, Wang L-J, Chen X-Y. 2007. An ABC transporter gene of Arabidopsis thaliana, AtWBC11, is involved in cuticle development and prevention of organ fusion. Plant and Cell Physiology 48, 1790–1802. [DOI] [PubMed] [Google Scholar]
- Massa AN, Childs KL, Lin H, Bryan GJ, Giuliano G, Buell CR. 2011. The transcriptome of the reference potato genome Solanum tuberosum group Phureja clone DM1-3 516R44. PLoS One 6, e26801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I, Li-Beisson Y, Beisson F, Ohlrogge JB, Pollard M. 2009. Identification of an Arabidopsis feruloyl-coenzyme A transferase required for suberin synthesis. Plant Physiology 151, 1317–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer JD, Lulai EC, Thompson AL, Suttle JC, Bolton MD. 2012. Wounding coordinately induces cell wall protein, cell cycle and pectin methyl esterase genes involved in tuber closing layer and wound periderm development. Journal of Plant Physiology 169, 586–595. [DOI] [PubMed] [Google Scholar]
- Nicot N, Hausman J-F, Hoffmann L, Evers D. 2005. Housekeeping gene selection for real-time RT–PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany 56, 2907–2914. [DOI] [PubMed] [Google Scholar]
- Ning Y-Q, Ma Z-Y, Huang H-W, Mo H, Zhao T-T, Li L, Cai T, Chen S, Ma L, He X-J. 2015. Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Research 43, 1469–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K. 2005. DNA-binding specificity and molecular functions of NAC transcription factors. Plant Science 169, 785–797. [DOI] [PubMed] [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S. 2005. ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiology 139, 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D, Savaldi-Goldstein S, Mandel T, Yifhar T, Franke RB, Höfer R, Schreiber L, Chory J, Aharoni A. 2007. The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiology 145, 1345–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D, Shi JX, Bocobza S, Franke RB, Schreiber L, Aharoni A. 2010. The Arabidopsis DSO/ABCG11 transporter affects cutin metabolism in reproductive organs and suberin in roots. Molecular Plant 3, 563–575. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Research 29, 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. 2008. Building lipid barriers: biosynthesis of cutin and suberin. Trends in Plant Science 13, 236–246. [DOI] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium , Xu X, Pan S, et al. 2011. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. [DOI] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M. 2012. NAC proteins: regulation and role in stress tolerance. Trends in Plant Science 17, 369–381. [DOI] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R. 2008. Sealing plant surfaces: cuticular wax formation by epidermal cells. Annual Review of Plant Biology 59, 683–707. [DOI] [PubMed] [Google Scholar]
- Schreiber L, Franke R, Hartmann K. 2005. Wax and suberin development of native and wound periderm of potato (Solanum tuberosum L.) and its relation to peridermal transpiration. Planta 220, 520–530. [DOI] [PubMed] [Google Scholar]
- Schuetz M, Smith R, Ellis B. 2012. Xylem tissue specification, patterning, and differentiation mechanisms. Journal of Experimental Botany 64, 11–31. [DOI] [PubMed] [Google Scholar]
- Serra O, Hohn C, Franke R, Prat S, Molinas M, Figueras M. 2010. A feruloyl transferase involved in the biosynthesis of suberin and suberin-associated wax is required for maturation and sealing properties of potato periderm. The Plant Journal 62, 277–290. [DOI] [PubMed] [Google Scholar]
- Serra O, Soler M, Hohn C, Franke R, Schreiber L, Prat S, Molinas M, Figueras M. 2009a Silencing of StKCS6 in potato periderm leads to reduced chain lengths of suberin and wax compounds and increased peridermal transpiration. Journal of Experimental Botany 60, 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra O, Soler M, Hohn C, Sauveplane V, Pinot F, Franke R, Schreiber L, Prat S, Molinas M, Figueras M. 2009b CYP86A33-targeted gene silencing in potato tuber alters suberin composition, distorts suberin lamellae, and impairs the periderm’s water barrier function. Plant Physiology 149, 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Sharma V, Pal AK, Acharya V, Ahuja PS. 2013. Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Research 20, 403–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler M, Serra O, Fluch S, Molinas M, Figueras M. 2011. A potato skin SSH library yields new candidate genes for suberin biosynthesis and periderm formation. Planta 233, 933–945. [DOI] [PubMed] [Google Scholar]
- Soler M, Serra O, Molinas M, García-Berthou E, Caritat A, Figueras M. 2008. Seasonal variation in transcript abundance in cork tissue analyzed by real time RT–PCR. Tree Physiology 28, 743–751. [DOI] [PubMed] [Google Scholar]
- Soler M, Serra O, Molinas M, Huguet G, Fluch S, Figueras M. 2007. A genomic approach to suberin biosynthesis and cork differentiation. Plant Physiology 144, 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukitsu H, Kuromori T, Toyooka K, et al. 2007. Cytological and biochemical analysis of COF1, an Arabidopsis mutant of an ABC transporter gene. Plant and Cell Physiology 48, 1524–1533. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3-new capabilities and interfaces. Nucleic Acids Research 40, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Riveras E, Contreras-López O, Gutiérrez RA. 2013. Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proceedings of the National Academy of Sciences, USA 110, 12840–12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Molina I, Ranathunge K, Castillo IQ, Steven J, Rothstein SJ, Reeda JW. 2014. ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. The Plant Cell 26, 3569–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H, Demura T. 2010. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. The Plant Cell 22, 1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JKC. 2013. The formation and function of plant cuticles. Plant Physiology 163, 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeier J, Schreiber L. 1997. Chemical composition of hypodermal and endodermal cell walls and xylem vessels isolated from Clivia miniata (identification of the biopolymers lignin and suberin). Plant Physiology 113, 1223–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeier J, Schreiber L. 1998. Comparative investigation of primary and tertiary endodermal cell walls isolated from the roots of five monocotyledoneous species: chemical composition in relation to fine structure. Planta 206, 349–361. [Google Scholar]
- Zhang J-Y, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LlW2, Wang Z-Y. 2005. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). The Plant Journal 42, 689–707. [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye Z-H. 2010. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends in Plant Science 15, 625–632. [DOI] [PubMed] [Google Scholar]
- Zhu L, Guo J, Zhu J, Zhou C. 2014. Enhanced expression of EsWAX1 improves drought tolerance with increased accumulation of cuticular wax and ascorbic acid in transgenic Arabidopsis. Plant Physiology and Biochemistry 75, 24–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.