Abstract
Sumoylation is a powerful regulatory system that controls many of the critical processes in the cell, including DNA repair, transcriptional regulation, nuclear transport, and DNA replication. Recently, new functions for SUMO have begun to emerge. SUMO is covalently attached to components of each of the four major cytoskeletal networks, including microtubule‐associated proteins, septins, and intermediate filaments, in addition to nuclear actin and actin‐regulatory proteins. However, knowledge of the mechanisms by which this signal transduction system controls the cytoskeleton is still in its infancy. One story that is beginning to unfold is that SUMO may regulate the microtubule motor protein dynein by modification of its adaptor Lis1. In other instances, cytoskeletal elements can both bind to SUMO non‐covalently and also be conjugated by it. The molecular mechanisms for many of these new functions are not yet clear, but are under active investigation. One emerging model links the function of MAP sumoylation to protein degradation through SUMO‐targeted ubiquitin ligases, also known as STUbL enzymes. Other possible functions for cytoskeletal sumoylation are also discussed. © 2015 The Authors. Cytoskeleton Published by Wiley Periodicals, Inc.
Keywords: microfilaments, septins, MT, IF, SUMO, microtubule‐associated proteins, MAPs
Abbreviations used
- MAP
microtubule‐associated protein
- SIMs
SUMO interacting motifs;
- SPB
spindle pole body
- STUbL
SUMO‐targeted ubiquitin ligase
Introduction
Sumoylation is a fascinating regulatory system that displays several similarities to the more well‐known process of ubiquitination, but is nevertheless a distinct signal transduction system. Like ubiquitin, the SUMO moiety is conjugated to target proteins on lysine residues and this can drastically alter that protein's localization, protein‐protein interactions, and even its stability. Sumoylation regulates many of the basic cellular processes, including DNA replication, translation, ribosomal maturation [Finkbeiner et al., 2011], DNA repair [Prudden et al., 2011], PML (promyelocytic leukemia) nuclear body formation [Nagai et al., 2011], nucleo‐cytoplasmic trafficking [Wang et al., 2012], kinetochore function and chromosome segregation [Stead et al., 2003; Yong‐Gonzales et al., 2012; Pinder et al., 2013], transcription and transcriptional repression [Garcia‐Domiquez and Reyes, 2009; Ouyang et al., 2009]. These have been expertly reviewed elsewhere [Dasso 2008; Bergink and Jentsch, 2009; Stehmeier and Muller, 2009; Gareau and Lima, 2010; Nagai et al., 2011; Praefcke et al., 2012]. This review covers emerging evidence suggesting that SUMO regulates the cytoskeleton, an idea that has not been widely recognized previously.
The cytoskeleton is commonly considered to be comprised of three polymer networks; the microtubules, the actin cytoskeleton also known as microfilaments, and intermediate filaments. However, a fourth polymeric network, the septins, should be considered as part of the cytoskeleton as well [Gladfelter, 2010; Mostowy and Cossart, 2012] and we include a discussion of its sumoylation in this review. Microtubules are well recognized as a major component of the mitotic spindle that separates genetic information found in chromosomes. Actin plays important roles in a large variety of cellular processes including cytokinesis, muscle contraction, cell motility, endocytosis, and phagocytosis. Both microtubules and microfilaments can serve as railway‐like transit systems that allow the movement of various cargoes by motor proteins along these tracks to specific destinations within the cell. In contrast, intermediate filament networks and septins do not serve as tracks for motor proteins. Instead, intermediate filaments provide structural integrity to the cell, and septins promote cytokinesis by forming a filamentous collar around the neck of dividing cells [Beise and Trimble, 2011].
It has recently been established that SUMO modifies elements of all four of the cytoskeletal networks. While some functions of the SUMO modifications are clear, a plethora of questions remain concerning the when, how, why and by what mechanism SUMO signaling controls the various elements of the cytoskeleton. Elucidating the similarities and differences in SUMO's control of the different cytoskeletal networks will assuredly illuminate new communication circuits within the cell.
A better understanding of the regulation of the cytoskeleton by SUMO is likely to provide significant impacts on human health. As the cytoskeleton is intimately involved in numerous disease processes, new knowledge of its regulation will undoubtedly lead to new ways to intervene when its function is impaired. Sumoylation is well known to transduce signals from multiple types of stress to influence various cellular processes [Tempe et al., 2008]. The idea that a similar signaling paradigm could also modulate the cytoskeleton is just beginning to emerge. Currently however, the mechanisms by which the cytoskeleton deals with these stresses are poorly understood. Indeed, sumoylation is implicated in several neurodegenerative diseases including Alzheimer's [Zhang et al., 2008; McMillan et al., 2011; Hoppe et al., 2013], Parkinson's disease [Kim et al., 2011; Krumova et al., 2011; Weetman et al., 2013], as well as cancer [Lee et al., 2006; Liu et al., 2011; Bettermann et al., 2012], and other diseases [Dorval and Fraser, 2007; Sarge and Park‐Sarge, 2011]. However, how sumoylation affects the cytoskeleton in these diseases remains unclear. Clarification of these pathways could ultimately lead to new paths for therapy development, including new targets for drug screening.
While it is already widely accepted that the cytoskeleton is regulated by a multitude of different post‐translational modifications, these often transduce signals from a variety of inputs and thus produce a variety of outputs. The sumoylation system may provide a single molecular mechanism to signal to the multiple polymers of the cytoskeleton simultaneously. Thus, it is possible that a particular input could result in a coordinated output for multiple cytoskeletal polymers.
The Sumoylation Machinery
SUMO is about 100 amino acids in size [Johnson, 2004]. Although SUMO and ubiquitin share only ∼18% sequence identity, they are structurally quite similar [Vijay‐Kumar et al., 1987; Bayer et al., 1998] (Fig. 1). Like ubiquitin, the tertiary structure of SUMO contains a β‐grasp fold, which is a common characteristic of the ubiquitin protein family [Bayer et al., 1998]. However, there are some differences between the two molecules. SUMO has an amino‐terminal extension approximately 20 amino acids long that is absent in ubiquitin. Both are processed to yield a terminal glycine‐glycine motif that is used in conjugation to target proteins [Ozkaynak et al., 1987; Wilkinson, 1997; Larsen et al., 1998; Li and Hochstrasser, 1999; Fang and Weissman, 2004; Li and Ye, 2008].
Figure 1.
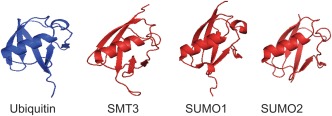
Structure of ubiquitin and SUMO proteins. Ribbon drawing of ubiquitin, Smt3, SUMO1, and SUMO2. These molecules share a common secondary structure ββααββαβ that assembles into a ubiquitin‐like fold. Renderings were developed using the crystallography coordinates available from the Protein Data Bank with the following accession numbers: ubiquitin (1UBQ), Smt3 (3V60), SUMO1 (2UYZ), and SUMO2 (1WM3). The structures for the above molecules were analyzed using the PyMOL Molecular Graphics System, Version 1.3 Schrödinger, LLC.
Classically, SUMO is conjugated to a lysine residue lying within the consensus sequence ΨΚxE/D, where Ψ is a large hydrophobic residue and x is any amino acid [Melchior, 2000; Johnson, 2004]. However, about half of known conjugation events occur within non‐consensus or incomplete consensus sites [Blomster et al., 2009; Matic et al., 2010; Teng et al., 2012].
There are four SUMO paralogs in humans, SUMO1‐4; but only one in the budding yeast, Saccharomyces cerevisiae (Smt3p); and one in the fission yeast, Schizosaccharomyces pombe (Pmt3) [Meluh and Koshland, 1995; Tanaka et al., 1999]. In humans, SUMO1, SUMO2, and SUMO3 can be found in multiple tissues, whereas SUMO4 mRNA expression is most pronounced in lymph nodes and kidney [Citro and Chiocca, 2013]. SUMO2 and SUMO3 are 97% identical in sequence and are considered redundant with each other. Thus, they are often referred to as SUMO 2/3. SUMO1 shares ∼50% sequence identity with SUMO2/3 [Saitoh and Hinchey, 2000]. SUMO1 is most similar to the yeast Smt3p, sharing 50% amino acid sequence identity and a longer N‐terminal extension [Schwarz et al., 1998; Sheng and Liao, 2002]. For any of the SUMO paralogs, SUMO is often conjugated to only a small population of the target protein at any given time [Johnson, 2004; Klug et al., 2013]. Although SUMO interacting motifs (SIMs) play a role, it still remains an outstanding question of what factors specify the conjugation of a particular paralog to a particular cytoskeletal element [Citro and Chiocca, 2013].
The enzyme cascade of the sumoylation pathway is analogous with the ubiquitination pathway, but the enzymes are distinct for each [reviewed in Ulrich, 2009]. Three different classes of enzymes are required for SUMO conjugation to the target protein: an activating enzyme (E1), a conjugating enzyme (E2), and a ligating enzyme (E3), which enhances the efficiency of conjugation and specificity for SUMO targets [Hochstrasser, 2001; Johnson, 2004] (Fig. 2).
Figure 2.
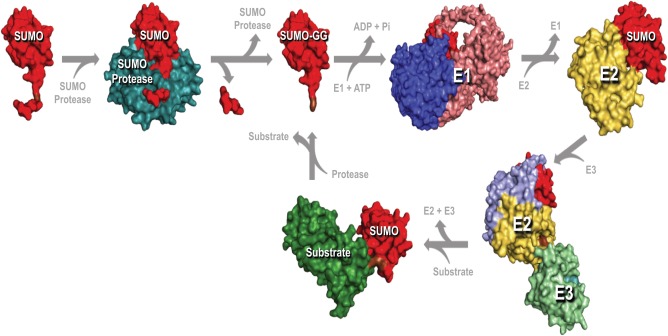
SUMOylation Pathway: To portray each state in the sumoylation pathway, surface maps were developed using crystallography coordinates available from PDB with the following accession numbers: SUMO1 and Senp1 (2IY1), E1 complex (3KYC), E2 complex (2UYZ), E3 complex (3UIP), and sumoylated PCNA (3V60). The orientation of SUMO is maintained throughout the sumoylation processes depicted above.
For both moieties, conjugation consists of isopeptide bond formation between the carboxyl group of the terminal glycine of SUMO to the epsilon amino group of a lysine residue within the target protein, thus forming an isopeptide bond (Fig. 3A). SUMO can either be attached to one lysine residue (mono‐sumoylation), multiple lysine residues (multi‐sumoylation), or form SUMO chains on the target lysine residue (poly‐sumoylation) [Bencsath et al., 2002; Hickey et al. 2012].
Figure 3.
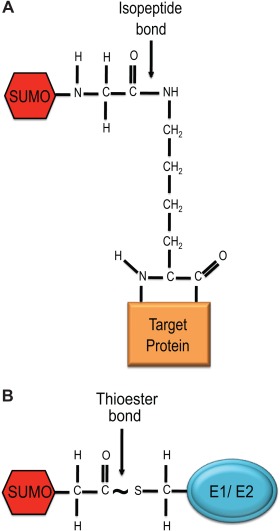
Chemical bonds in the sumoylation pathway. (A) Isopeptide bond. SUMO is conjugated to the target protein via an isopeptide bond linkage between the terminal glycine residue of SUMO and the epsilon amino group of the lysine in the target. (B) Thioester Bond. Chemical linkage is highlighted between the terminal glycine carboxy group of SUMO and the active cysteine in the SUMO activating, and conjugating enzymes.
The conjugation of SUMO to its target substrate requires ATP. The activation of SUMO is initiated with the adenylation of the C‐terminal carboxyl group of SUMO in an ATP dependent reaction. The process continues with the SUMO‐activating enzyme, an E1. This enzyme consists of a heterodimer of Aos1 and Uba2 and is conserved from yeast to human [Dohmen et al., 1995; Johnson et al., 1997; Desterro et al., 1999]. The thiol group of cysteine within the active site of Aos1‐Uba2 attacks the adenylated SUMO, forming a high‐energy thioester bond between the Aos1‐Uba2 heterodimer and the C‐terminus of SUMO [Olsen et al., 2010]. Next, the activated SUMO is transferred to a cysteine within the active site of the E2 SUMO‐conjugating enzyme, Ubc9p, forming a new thioester bond [Johnson and Blobel, 1997] (Fig. 3B).
The sole E2 SUMO‐conjugating enzyme is Ubc9p, which is also highly conserved from yeast to humans [Johnson et al., 1997; Schwarz et al., 1998]. Ubc9 is regulated by multiple post‐translational modifications, including sumoylation, acetylation, and phosphorylation [Ho et al., 2011; Su et al., 2012; Hsieh et al., 2013]. Phosphorylation of Ubc9p by the cyclin‐dependent kinase, CDK1, implies that sumoylation is coordinated with the cell cycle [Su et al., 2012]. This has significant ramifications for control of the cytoskeleton with its myriad layers of cell‐cycle input.
SUMO conjugation can take place in the absence of a SUMO E3, however the E3 is thought to bring the Ubc9p into close proximity with the target substrate to enhance SUMO conjugation and its specificity [Desterro et al., 1999; Okuma et al., 1999; Takahashi et al., 2001]. SUMO E3 enzymes share similar features with the RING‐domain found in the ubiquitin E3s [Hochstrasser, 2001; Johnson and Gupta, 2001] (Fig. 4). There are several classes of SUMO E3s; including the protein inhibitor of activated STAT, known as the PIAS family [Shuai, 2000], polycomb group protein Pc2 [Kagey et al., 2003], and the nuclear pore protein complex RanBP2/Nup358 [Pichler et al., 2002]. In budding yeast, there are four SUMO E3 ligases, Siz1p, Siz2p/Nfi1p, Mms21p/Nse2p, and Cst9p/Zip3p [Johnson and Gupta, 2001; Reindle et al., 2006; Duan et al., 2011; Heideker et al., 2011; Stephan et al., 2011]. Siz1p and Siz2p are responsible for the majority of SUMO conjugation in vivo, with Siz1p having the larger effect on global sumoylation levels [Johnson and Gupta, 2001; Takahashi et al., 2001].
Figure 4.
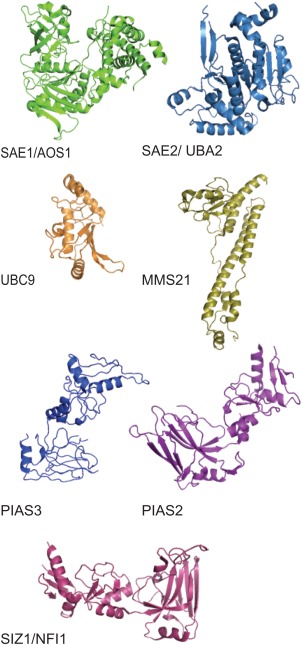
Structure of SUMO E1, E2, E3 enzymes. Tertiary ribbon structure of the SUMO activating enzyme dimers Sae1 and Sae2, SUMO conjugating enzyme Ubc9, and SUMO ligating enzymes Mms21, PIAS3, PIAS2, and Siz1. These renderings were developed using the crystallography coordinates associated with the following PDB accession numbers: Sae1 (1Y8Q), Sae2 (1Y8Q), Ubc9 (2GRR), Mms2 (3HTK), PIAS3 (4MVT), PIAS2 (4FO9), and Siz1 (3I2D).
Like ubiquitin, SUMO can form chains, known as polysumoylation [Johnson and Gupta, 2001]. SUMO chains occur mainly through SUMO's flexible N‐terminal extension containing a ψKxE sequence [Tatham et al., 2001]. SUMO2/3 is more likely to form chains than SUMO1, which lacks the needed lysines [Tatham et al., 2001]. SUMO1 can also cap the end of a SUMO 2/3 chain, limiting its length [Matic et al., 2008]. The budding yeast SUMO, Smt3p, although it displays similarity to SUMO1, also forms chains [Bylebyl et al., 2003].
Two Models for Regulation by SUMO: Conjugation and Non‐covalent Binding
SUMOylation can regulate cellular processes via two major mechanisms. SUMO can become covalently cross‐linked to a target protein or it can interact non‐covalently with a binding partner [reviewed in Kerscher, 2007]. This latter type of interaction typically occurs through SUMO interaction motifs (SIMs) on the interacting protein [Minty et al., 2000; Song et al., 2004; Kroetz and Hochstrasser, 2009]. These are short stretches of the branched hydrophobic amino acids, isoleucine, leucine, valine, in the pattern (I/L/V) X (I/L/V) (I/L/V) with x being any amino acid [Kroetz and Hochstrasser, 2009; Yang and Sharrocks, 2010]. This motif is sometimes flanked on one side by acidic residues, and this enhances binding to SUMO [Hannich et al., 2005; Hecker et al., 2006; Kerscher, 2007; Uzunova et al., 2007]. Some proteins like the kinetochore kinesin, CENP‐E, can interact both ways, covalently and non‐covalently [Zhang et al., 2008]. Very little is known currently about the extent to which various cytoskeletal elements interact non‐covalently with SUMO.
Proteases Make Sumoylation a Reversible Process
Unlike traditional proteases, SENPs/Ulps do not degrade either SUMO or the targets. These enzymes remove SUMO from its targets by cleavage of the isopeptide bond between the glycine of SUMO and the target lysine. This allows the SUMO moiety to be recycled. The deconjugating enzymes responsible for this specialized clipping are termed ULPs in yeast for ubiquitin‐like‐specific protease [Li and Hochstrasser, 1999] and SENPs in plants and metazoans for SUMO/sentrin‐specific proteases. Sentrin was an early name for SUMO [Kamitani et al., 1997]. Several insightful reviews have been written recently on SENPs and Ulps [Mukhopadhyay and Dasso, 2007; Drag and Salvesen, 2008; Su and Hochstrasser, 2010; Gillies and Hochstrasser, 2012; Hickey et al., 2012]. Although the different cytoskeletal polymers themselves display varying degrees of dynamic subunit turnover [Cleveland, 1982; Yoon et al., 2001; Vorobjev et al., 1999], very little is known about the rates of reversible attachment of SUMO on each of the cytoskeletal polymers.
In mammals, there are six SUMO‐cleaving enzymes, SENP1, SENP2, SENP3, SENP5, SENP6, and SENP7, in addition to the recently described DeSumoylating Isopeptidase 1 (DESI) protease [Mukhopadhyay and Dasso, 2007; Shin et al., 2012; Suh et al., 2012]. In Saccharomyces cerevisiae, there are only three SUMO proteases, Ulp1p, Ulp2p, and Wss1p, each belonging to a distinct class [Li and Hochstrasser, 1999; Li and Hochstrasser, 2000; Bylebyl et al., 2003; Gillies and Hochstrasser, 2012].
SENPs
SENP/Ulp enzymes can possess two related cleavage activities, endopeptidase and isopeptidase activity. Whereas both the Ulp1p and Ulp2p families of SENPs desumoylate substrates by cleaving the isopeptide bond located between SUMO and the target, the Ulp1p class (but not the Ulp2 group) can also act as an endopeptidase [Li and Hochstrasser, 1999; Mikolajczyk et al., 2007; Drag and Salvesen, 2008; Lima and Reverter, 2008]. This activity processes the full‐length pro‐SUMO to a conjugatable form by cleaving several amino acids from the carboxy‐terminus to expose the terminal‐glycine used in conjugation [Drag and Salvesen, 2008]. In Saccharomyces cerevisiae, this removes three amino acids, ATY; but for mammalian SUMOs, two to eleven amino acids are removed depending on the SUMO paralog [Hickey et al., 2012].
The Ulp1 and Ulp2 classes display distinct substrate specificities [Li and Hochstrasser, 2000] as evidenced by the fact that when either of the two proteases is absent, different sets of sumoylated products accumulate [Johnson and Blobel, 1999; Li and Hochstrasser, 1999, 2000; Schwienhorst et al., 2000]. These two proteases also display different subcellular localizations and virtually non‐overlapping interactomes [Panse et al., 2003; Cubenas‐Potts et al., 2013; Srikumar et al., 2013]. Yet surprisingly, only a few cytoskeletal substrates are known for each [Hickey et al., 2012]. The Kerscher lab and others have shown that Ulp1p in yeast desumoylates the septins [Takahashi et al., 2000; Elmore et al., 2011]. The sumoylation of septins is described more fully below. Ulp1p also de‐modifies two proteins important for spindle positioning, Kar9p and Pac1p [Leisner et al., 2008; Alonso et al., 2012]. These are described below. We are not aware of any functional evidence that physically links Ulp2p to the major cytoskeletal polymers.
Wss1p
Wss1p is predicted to be a zinc‐dependent metalloprotease, the original member of a distinct class of SUMO proteases termed the WLM family of proteases (Wss1‐Like Metalloproteases) [Iyer et al., 2004; Mullen et al., 2010]. WSS1 was originally identified as a weak suppressor of smt3‐1, a temperature sensitive allele of SUMO [Biggins et al., 2001], clearly implicating it in SUMO‐related functions. Wss1p contains two SIMs (SUMO interacting motifs) within its extreme carboxyl‐terminal domain [Uzunova et al., 2007; Mullen et al., 2010], but it also has significant conservation with deubiquitinating enzymes (DUBs) [Mullen et al., 2010]. Recent work from the Brill lab suggests the possibility that while Wss1p may have both SUMO protease and DUB types of activity, it is a much better SUMO‐cleaving enzyme than a ubiquitin‐cleaving one [Mullen et al., 2010].
In addition to its role in sister chromatid recombination, a type of double‐ strand DNA break repair, Wss1p has recently been linked to another SUMO‐utilizing process, microtubule biology. Two‐hybrid analysis showed that Wss1p interacts with four distinct classes of microtubule‐binding proteins, Kar9p, Bim1p, Bik1p and Pac1p [Meednu et al., 2008; Alonso et al., 2012]. What makes this finding remarkable is that these different classes of MAPs carry out a divergent set of functions for microtubules [Berlin et al., 1990; Schwartz et al., 1997; Miller et al., 1999; Miller et al., 2000; Gundersen and Bretscher, 2003; Hwang et al., 2003; Lee et al., 2003; Sheeman et al., 2003; Miller et al., 2006; Blake‐Hodek et al., 2010; Huang et al., 2012]. The effect of Wss1p on microtubule binding proteins has been examined only for the Pac1p adaptor of the dynein motor protein [Alonso et al., 2012]. These experiments show that deletion of WSS1 results in higher molecular weight forms of Pac1p. This is consistent with the hypothesis from the Brill lab that Wss1p helps direct sumoylated proteins to the proteasome [Mullen et al., 2010; Alonso et al., 2012]. Further work is in progress to determine whether Wss1p alters the levels of ubiquitin on Pac1p.
A portion of Wss1p also localizes to foci in the cytoplasm [van Heusden and Steensma, 2008]. Curiously, this localization is dependent upon the actin‐related component of the dynactin complex, Arp1p, but not on another dynactin component Jnm1p [van Heusden and Steensma, 2008]. Wss1p is reported to localize only in the mother cell [van Heusden and Steensma, 2008]. The punctate pattern is consistent with it also localizing on the ends of cytoplasmic microtubules in the mother cell, but this is not known definitively.
SUMO‐Targeted Ubiquitin Ligases (STUbLs)
Owing that sumoylation is a reversible process, the levels of SUMO on a protein are critical and need to be maintained at an optimal homeostasis [Prudden et al., 2007; Kim and Baek, 2009; Bawa‐Khalfe and Yeh, 2010]. As discussed above, this can be accomplished by cleaving SUMO from targets. Another way to remove excess poly‐sumoylation is to degrade the entire sumoylated protein at the proteasome. For many years, sumoylation and ubiquitination were viewed as distinct modification systems with limited cross talk [Ulrich, 2005]. In one paradigm, ubiquitin and SUMO modify the same lysine at different times, in a competitive relationship [Desterro et al., 1998; Hoege et al., 2002; Steffan et al., 2004]. In this model, SUMO protects the protein from ubiquitin‐mediated degradation. Another type of cross talk employs cooperation between the two modifications in which the target is first modified by SUMO and then by ubiquitin [Huang et al., 2003].
In 2007, a new class of enzyme was described, the SUMO‐targeted ubiquitin ligase (STUbL). With this, communication between ubiquitin and SUMO became more interesting [Prudden et al., 2007; Sun et al., 2007; Uzunova et al., 2007; Xie et al., 2007]. A STUbL is an enzyme with ubiquitin ligase activity that recognizes a sumoylated protein and poly‐ubiquitinates it [reviewed in Perry et al., 2008; Praefcke et al., 2012]. Poly‐ubiquitination then targets that protein for degradation via the proteasome. Thus, sumoylation can be an indirect, upstream signal for protein degradation (Fig. 5).
Figure 5.
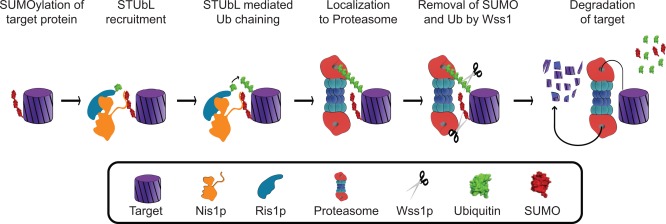
STUbL Pathway Model. The Ris1p‐Nis1p STUbL complex can direct a sumoylated target protein to the proteasome for degradation (Adapted from Alanso et al. 2012).
Three STUbL families have been characterized, Uls1p‐Nis1p and Slx5p‐Slx8p/RNF4, and Rad18p. Both Uls1p‐Nis1p and Slx5p‐Slx8p function as heterodimers [Yang et al. 2006). While both Slx5p and Slx8p contain RING domains, Slx5p is the subunit that targets the complex to substrates via its two SIMs [Xie et al., 2007; Cook et al., 2009; Szymanski and Kerscher, 2013]. Slx5p‐Slx8p is the yeast homologue of the human RNF4, [Sun et al., 2007; Uzunova et al., 2007; Xie et al., 2007]. Little information is presently known about Uls1p targets, with only a few currently identified. These include the microtubule associated protein Pac1p and the DNA binding protein Rap1p [Grunstein, 1997; Jain and Cooper, 2010; Alonso et al., 2012; Zhang et al., 2012].
STUbLs also play an important role in cancer. In one of the best characterized examples, RNF4 functions in the degradation of PML in nuclear bodies [reviewed in de The et al., 2012; Hay, 2013]. In acute promyelocytic leukemia, the PML protein forms an in frame fusion with the retinoic acid receptor alpha (RAR α), forming an oncoprotein that initiates this blood cancer [Tatham et al., 2008]. Arsenic, the major treatment for acute promyelocytic leukemia, causes the sumoylation of PML‐RARα by SUMO2. RNF4 then polyubiquitinates these SUMO chains, resulting in degradation of the aberrant PML by the proteasome [Tatham et al., 2008; Liu et al., 2012; Maroui et al., 2012; Rojas‐Fernandez et al., 2014]. Recently, the novel STUbL, Arkadia, was found to function similarly in PML degradation [Erker et al., 2013]. The elegant work describing PML cell biology and its relationship to effective therapeutic interventions for this disease gives hope to the idea that cytoskeletal accumulation diseases might one day be treated by targeting the SUMO system.
In just a few short years, the number of targets for STUbL enzymes and processes governed by STUbLs has simply exploded, with STUbLs playing critical roles in almost as many cellular processes as SUMO itself. It is perhaps not surprising that STUbLs have now been linked to the cytoskeleton, including interactions with several microtubule‐associated proteins.
SUMO and the Cytoskeleton
Septins
Septins were originally identified in yeast using screens searching for cell division cycle (CDC) genes [Hartwell, 1971; Hartwell et al., 1974]. This work identified four of the five mitotic septins, Cdc3p, Cdc10p, Cdc11p, and Cdc12p, which are essential. The name septin was later coined to describe the role of these genes in cell septation in yeast [Mostowy and Cossart, 2012]. A fifth mitotic septin, Sep7p/Shs1p was later identified as the seventh homolog of a septin [Mino et al., 1998]. Shs1p is not essential. Septins are highly conserved, found in a wide range of organisms ranging from yeast to human. However, no evidence has been found for septins in plants [Field et al., 1996; Nguyen et al., 2000; Gladfelter et al., 2001; Nishihama et al., 2011]. The reader is referred to a comprehensive review of septins that was published recently [Mostowy and Cossart, 2012].
In Saccharomyces cerevisiae, septins form the filaments that encircle the mother‐bud neck, the site of cytokinesis in this yeast [Byers and Goetsch, 1976; Haarer and Pringle, 1987; Ford and Pringle, 1991; Bertin et al., 2012]. A septin patch is formed initially on the cortex of the unbudded cell, just before bud emergence [reviewed in Chen et al., 2011]. As the growing bud emerges, the septins then reorganize to form an hourglass‐like collar that is positioned on both sides of the mother‐bud neck [Longtine and Bi, 2003; Kozubowski et al., 2005; Vrabioiu and Mitchison, 2006]. Electron microscopy studies reveal a gauze‐like meshwork of filaments at the bud neck [Rodal et al., 2005] consisting of filaments running circumferentially around the neck and axial filaments running along the mother‐bud axis [Garcia et al., 2011; Bertin et al., 2012; Bertin and Nogales, 2012]. At cytokinesis, the hourglass collar splits into two rings via rearrangement and reassembly mechanisms, with one ring facing the mother cell and the other facing the bud (Fig. 6) [Garcia et al., 2011; Bertin et al., 2012; Ong et al., 2014].
Figure 6.
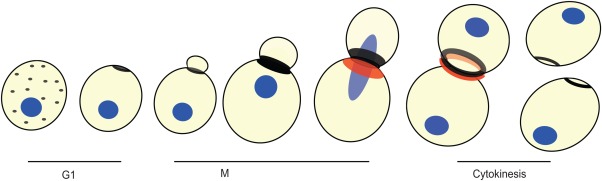
Sumoylation of septins. During G1, a septin patch forms at the site of bud formation. As the bud emerges through the patch, the septins form a collar around the mother‐bud neck. The five septin proteins involved in this process are Cdc3p, Cdc10p, Cdc11p, Cdc12p, and Shs1p. Prior to cytokinesis, three of these septin proteins are sumoylated, but only on the mother side of the mother‐bud neck. Cdc3p is sumoylated at lysines 4, 11, 30, and 63. Cdc11p is sumoylated at lysine 412. Shs1p is sumoylated at lysines 426, and 437 [Johnson and Blobel, 1999; Takahashi et al., 1999]. The sumoylation event is color coded in red. During cytokinesis, the septin “hourglass” collar splits into two rings as the cells divide. After cytokinesis the septin rings dissociates, and the process starts again.
Notably, the two sides of the hourglass collar are not symmetric, and distinct sets of proteins are localized with the ring on the mother side and the ring on the bud side. Still other proteins localize between the two rings [Kozubowski et al., 2005]. Thus, the septins serve as scaffolds for proteins functioning in cytokinesis, bringing in and organizing components of the actomyosin constriction ring and the enzymes needed for cell wall synthesis [Gladfelter et al., 2001; McMurray et al., 2011; Kang et al., 2013].
Septins also play a role in several other cellular processes that are closely associated with membranes. These include spindle alignment and the establishment of the diffusion barrier [Kusch et al., 2002; Dobbelaere and Barral, 2004; Caudron and Barral, 2009]. Diffusion barriers block molecules in one membrane compartment from diffusing through the lipid bilayer into another compartment.
The paired filaments formed by septins are approximately 10 nm in diameter, which can also self assemble in vitro [Byers and Goetsch, 1976; Bertin et al., 2012]. In yeast, electron microscopy studies demonstrate that the basic building block of the septin filament is comprised of the four essential septins arranged into a hetero‐octomer in the order of Cdc11p‐Cdc12p‐Cdc3p‐Cdc10p‐Cdc10p Cdc3p‐Cdc12p‐Cdc11p [Bertin et al., 2008]. The two halves of the octomer fit together around the two‐fold rotational symmetry in the Cdc10p‐Cdc10p homophillic interaction of Cdc10, creating a symmetric rod‐shaped subunit that is the building block of septins [Bertin et al., 2008]. In mammalian systems, the basic building block is a hetero‐hexamer, rather than an octomer, and it also has a rod‐like shape [Sirajuddin et al., 2007; Garcia et al., 2011]. An alternative hetero‐octomer containing Shs1p rather than Cdc11p is important for the bundling of filaments and ring formation in vitro and formation of the septin collar in vivo [Garcia et al., 2011].
Sumoylation of the Septins
Septins were the first substrates of SUMO identified in yeast [Johnson and Blobel, 1999; Takahashi et al., 1999]. Indeed, they are some of the most abundant sumoylated proteins in the cell [Johnson and Blobel, 1999; Wohlschlegel et al., 2004]. Sumoylation serves as one of the markers for the asymmetry of the two septin rings; only the septin ring on the mother side of the bud neck is sumoylated [Johnson and Blobel, 1999; Takahashi et al., 1999; Martin and Konopka, 2004]. This sumoylation occurs during mitosis, with SUMO addition occurring just before anaphase and SUMO removal occurring abruptly at cytokinesis [Johnson and Blobel, 1999]. Consistent with this, the E3 enzyme Siz1p localizes to the septin ring on the mother side of the neck at the same point in the cell cycle as the SUMO modification occurs during mitosis [Johnson and Gupta, 2001]. Additional amounts of GFP‐Siz1p are found inside the nucleus as puncta [Johnson and Gupta, 2001].
Septin sumoylation has been seen to play a role in maintaining the polymerization state of septins as mutants lacking sumoylation sites display a modest delay in the disassembly of the septin rings at cytokinesis [Johnson and Blobel, 1999]. The molecular mechanism of this remains an avenue for future investigations, as the mutation of these lysines could alter other aspects of the septin proteins such as folding and stability. Several questions still remain concerning the roles of septin sumoylation [Oh and Bi, 2011]. While septins are required for cytokinesis, their sumoylation is not [Johnson and Blobel, 1999; Dobbelaere and Barral, 2004]. Septins are also not the essential substrate of sumoylation during the cell cycle, because when all the septin sumoylation sites are mutated and combined into one cell, the cells grow and do not display the cell‐cycle arrest observed in SUMO deficient mutants [Johnson and Blobel, 1999].
It is notable that only a subset of the septins is sumoylated [Johnson and Blobel, 1999]. In yeast, only Cdc3p, Cdc11p, and Shs1p are modified by SUMO [Johnson and Blobel, 1999]. Cdc3p is sumoylated at four sites, Cdc11p at one site, and Shs1p at two sites [Johnson and Blobel, 1999]. These modifications are absent in cellular extracts from a siz1Δ strain but not nfi1Δ [Johnson and Gupta, 2001]. Siz1p also enhances the in vitro sumoylation of septins. Together, these findings suggest that Siz1p is the E3 responsible for septin sumoylation [Johnson and Gupta, 2001].
Two of the sumoylated septins, Shs1p and Cdc11p, occupy the terminal position in the octomeric building‐block for filament assembly. This prompts one to wonder whether this modification may modulate the specialized role of Shs1p in promoting ring formation and filament bundling [Garcia et al., 2011]. One might also speculate whether the high levels of phosphorylation on Shs1p might influence its sumoylation [Egelhofer et al., 2008; Meseroll et al., 2013]. While the precise function of septin sumoylation has been evasive, considering that Cdc11p and Cdc3p are essential, sumoylation is unlikely to play a critical role for these septins. However, the septin Shs1p is not essential, and considering that the phenotypes of Shs1p are milder than mutations in the other two septins, it is possible that the function of sumoylation is tied to this less critical septin.
Sumoylation of the septins is regulated by signals passing through the E2, Ubc9p. The sumoylation levels on the septins are inversely proportional to the levels of Ubc9p auto‐phosphorylation [Ho et al., 2011]. Determining the extent to which Ubc9p phosphorylation by CDKs and other post‐translational modifications affect septin sumoylation should prove to be a worthwhile avenue of future investigation [Su et al., 2012].
Deregulation of septins has been linked to several major diseases, including multiple cancers and neurological diseases, including Parkinson's and Alzheimer's [Ihara et al., 2007]. In Parkinson's disease, the septin SEPT4 has been shown to modulate the neurotoxity of alpha‐synuclein, but it remains to be determined whether the sumoylation of alpha‐synuclein is a part of this modulation [Ihara et al., 2007]. In Alzheimer's, septins have been seen to colocalize in neurofibrillary tangles, an aberrant structure containing the MAP, Tau [Kinoshita et al., 1998]. In several types of cancer, human SEPT9 serves as a biomarker for colon cancer [reviewed in Cerveira et al., 2011; Connolly et al., 2011]. As septins are dynamic structures [Gladfelter, 2010], it is possible that sumoylation may affect their solubility and thus influence their dynamicity.
Microtubules
Microtubules are proteinaceous polymers comprised of alpha‐beta tubulin dimers that make key contributions to intracellular motility and cell division [reviewed in Desai and Mitchison, 1997; Valiron et al., 2001; Howard and Hyman, 2003; Conde and Caceres, 2009; Etienne‐Manneville, 2013]. They serve as tracks along which motors move various cargoes throughout the cell. As a major structural element of the mitotic spindle, they are often referred to as “ropes” owing to their ability to generate pulling forces on chromosomes. Microtubules are highly dynamic, continuously growing and shrinking [Cassimeris et al., 1988; Sammak and Borisy, 1988; Schulze and Kirschner, 1988; Chretien et al., 1995; Akhmanova and Steinmetz, 2008; Gardner et al., 2008]. The faster growing end is referred to as the plus‐end. The less dynamic end is referred to as the minus‐end [Allen and Borisy, 1974; Bergen and Borisy, 1980]. In many cell types, the centrosome serves as a major microtubule‐organizing center (MTOC) and stabilizes the minus‐ends of microtubules embedded within it [Mitchison and Kirschner, 1984]. In yeast, the spindle pole body serves as the MTOC [reviewed in Rout and Kilmartin, 1990; Kahana et al., 1995; Jaspersen and Winey, 2004].
SUMO and Microtubules
Currently, sumoylation is not widely recognized as a post‐translational modification of either tubulin or microtubules [Janke, 2014; Song and Brady, 2015]. To date, there is only limited evidence that tubulin itself may be modified by SUMO. Alpha and/or beta tubulins have been identified as candidates in several global sumoylation screens employing proteomics [Panse et al., 2004; Wohlschlegel et al., 2004; Rosas‐Acosta et al., 2005]. However, only immunoblotting with monoclonal anti‐tubulin was used to confirm the Rosas‐Acosta finding of sumoylated alpha tubulin. The anti‐tubulin reacted with a larger 70 kDa band in the TAP purified samples, but with only the standard 50 kDa size tubulin in the corresponding parental‐control strain [Rosas‐Acosta et al., 2005]. The shifted form was only observed in the SUMO3 TAP purification, but not the SUMO1 purification, indicating that SUMO3 may be responsible for modifying alpha tubulin [Rosas‐Acosta et al., 2005]. As alternative explanations for these findings are possible, additional confirmatory studies are needed before other questions can be answered about how sumoylation might alter the many properties of microtubules and their dynamics.
MAPs
Many classes of microtubule‐associated proteins (MAPs) modify and regulate a multitude of microtubule behaviors. Some of these functions include directing microtubules towards distinct subcellular locations, cross‐linking microtubules, mediating protein‐protein interaction, and either stabilizing or destabilizing microtubules. Some classes of MAPs bind directly to tubulin dimers to help regulate their addition to the microtubule polymer [Etienne‐Manneville, 2010; Gupta et al., 2013; Cheerambathur and Desai, 2014; Ferreira et al., 2014]. Other MAPs, like tau, bind along the sides of microtubules [Al‐Bassam et al., 2002], whereas other classes of MAPs bind at the plus‐end (+TIPs) [Akhmanova and Steinmetz, 2008].
Recently several classes of MAPs have been shown to be modified by SUMO (Table 1) and several other classes interact with SUMO either physically or by two‐hybrid analysis. The MAPs that can be covalently modified include the dynein adapter Pac1p (Lis1), Bik1p (CLIP‐170), the spindle positioning protein Kar9p, the Alzheimer's MAP Tau, and the kinetochore attachment protein Ndc80p [Dorval and Fraser, 2006, 2007; Montpetit et al., 2006; Leisner et al., 2008; Meednu et al., 2008; Alonso et al., 2012]. The kinetochore kinesin CENP‐E is both modified by SUMO and interacts non‐covalently with it [Zhang et al., 2008]. Interaction with the SUMO machinery has also been seen with Bim1p, the EB1 homologue in yeast, but it is not known whether this interaction occurs through conjugation or non‐covalent interactions [Meednu et al., 2008]. This growing list leads us to speculate that sumoylation may control multiple facets of microtubule biology via regulation of its MAPs.
Table 1.
Sumoylation Targets of the Cytoskeleton
| Polymer system | Type of modification | Site of modifcation | Evidence | Reference |
|---|---|---|---|---|
| Septins | ||||
| Cdc3p | Covalent | K4, K11, K30, and K63 | Pulldown | Johnson and Blobel [1999] |
| Cdc11p | Covalent | K412 | Pulldown | Johnson and Blobel [1999] |
| Shslp | Covalent | K426, K437 | Pulldown | Johnson and Blobel [1999] |
| Microtubules | ||||
| Alpha‐beta tubulin | Covalent | N/A | Proteomic screens, pulldown | Panse et al. [2004]; Wohlschlegel et al. [2004]; Rosas‐Acosta et al. [2005] |
| CENP‐E | Covalent and non‐covalent | N/A | Pulldown | Zhang et al. [2008] |
| Ndc80 | Covalent | K231 | Proteomic screens, pulldown | Panse et al. [2004]; Zhou et al. [2004]; Wykoff and O'Shea [2005]; Montpetit et al. [2006] |
| Tau | Covalent | K340 | Pulldown | Dorval and Fraser, [2012, 2006]; Takahashi et al. [2008] |
| Pac1p | Covalent | N/A | Pulldown, Y2H | Alonso et al. [2012] |
| Kar9p | Covalent | K301, K333, K381, and K529 | Pulldown, Y2H | Leisner et al. [2008]; Meednu et al. [2008] |
| Bik1p | Covalent | N/A | In vitro, Y2H | Alonso et al. [2012] |
| Bim1p | N/A | N/A | Y2H | Meednu et al. [2008] |
| La | Covalent | K41 | Pulldown | van Niekerk et al. [2007] |
| Microfilaments | ||||
| Actin | Covalent | K68, and K284 | Proteomic screens, pulldown | Panse et al. [2004]; Vertegaal et al. [2004]; Wohlschlegel et al. [2004]; Rosas‐Acosta et al. [2005]; Hofmann et al. [2009] |
| Rac1 | Covalent | K188, K183, K184, and K186 | Pulldown | Castillo‐Lluva et al. [2010] |
| RhoGDI | Covalent | K138 | Pulldown | Liu et al. [2011]; Yu et al. [2012] |
| Arc35p | Covalent | N/A | Proteomic screen, pulldown | Wohlschlegel et al. [2004]; Sung et al. [2013] |
| Arc40p | Covalent | N/A | Proteomic screen | Wohlschlegel et al. [2004] |
| Arc19p | Covalent | N/A | Proteomic screen | Nie et al. [2012] |
| Arc15p | Covalent | N/A | Proteomic screen | Nie et al. [2012] |
| Intermediate filaments | ||||
| Vimentin | Covalent | N/A | In vitro, pulldown | Wang et al. [2010], Snider et al. [2011] |
| Keratin 8 | Covalent | K285, K364 | In vitro, pulldown | Snider et al. [2011] |
| Keratin 18 | Covalent | K207, K373 | In vitro, pulldown | Snider et al. [2011] |
| Keratin 19 | Covalent | K208 | In vitro, pulldown | Snider et al. [2011] |
| Lamin A | Covalent | K201, K420, and K486 | Pulldown, Y2H | Zhang et al. [2008], Galisson et al. [2011], Simon et al. [2013] |
Kar9p
Kar9p is required for correct orientation of the mitotic spindle and is important for nuclear migration in both mating and mitotic cells [Kurihara et al., 1994; Miller and Rose, 1998]. KAR9 was discovered in a screen for bilateral karyogamy mutants, [Kurihara et al., 1994] and is thought to be analogous to the mammalian adenomatous polyposis coli protein (APC) [Bienz, 2001], which is mutated in a large percentage of human colorectal cancers [Groden et al., 1991; Markowitz and Bertagnolli, 2009]. APC and Kar9p share a number of functional similarities, albeit they have limited homology at the amino acid level [Bienz, 2001; Gundersen, 2002]. At the protein level, Kar9p consists of an N‐terminal acidic domain, a central coil‐coil domain, and a C‐terminal basic domain [Miller and Rose, 1998].
Kar9p plays a key role in positioning the mitotic spindle by orienting the cytoplasmic microtubule into the bud [Miller and Rose, 1998]. Kar9p links the actin and microtubule networks through a bridging complex that contains Bim1p‐Kar9p‐Myo2p [Beach et al., 2000; Hwang et al., 2003]. Bim1p is a microtubule‐binding protein and the yeast homologue of EB1. Myo2p is a type V myosin. The EB1‐like C‐terminus of Bim1p binds the C‐terminal domain of Kar9p [Miller et al., 2000; Moore and Miller, 2007]. Kar9p binds to the tail of Myo2p in a region that overlaps with other cargo‐binding sites [Eves et al., 2012]. When this connection is formed, the myosin walks up the actin cable. The resulting pulling‐force guides the end of the cytoplasmic microtubule into the yeast bud, thus orienting the mitotic spindle. The myosin motor then pulls the spindle up to the bud neck [Beach et al., 2000; Korinek et al., 2000; Miller et al., 2000; Yin et al., 2000].
In orienting the mitotic spindle, it is important that Kar9p is localized on just one of the poles of the spindle. In other words, its localization on the two poles needs to be asymmetric. Otherwise, both poles of the spindle would be pulled into the bud. Kar9p binds to the “old” or original spindle pole body that will be transferred to the daughter cell, whereas the “new” SPB lacking Kar9p is retained in the mother yeast cell [Liakopoulos et al., 2003; Moore et al., 2006; Moore and Miller, 2007].
Kar9p and Sumoylation
Several lines of evidence suggest that Kar9p is sumoylated. Kar9p interacts with SUMO by two‐hybrid analysis. It also interacts with the E2 enzyme Ubc9p and the E3 Nfi1p [Meednu et al., 2008]. Kar9p has been shown to be sumoylated both in vitro and in vivo [Leisner et al., 2008; Meednu et al., 2008]. Four lysines are required for the sumoylation shift of Kar9p, lysines 301, 333, 381, and 529 [Leisner et al., 2008].
Sumoylation is important for multiple aspects of Kar9p function. It is important for the asymmetric localization of Kar9p on SPBs. Mutation of lysines 301, 333, 381, and 529 to arginine (4K→R) results in the mis‐localization of Kar9p on both SPBs, rather than it being restricted to just the old SPB [Leisner et al., 2008]. Similar results were observed with Kar9p mutations at lysine 304, which resides within the sumoylation consensus site of K301 [Meednu et al., 2008]. Functionally, sumoylation is also important for spindle positioning [Leisner et al., 2008; Meednu et al., 2008]. Inhibition of SUMO with a temperature‐sensitive SUMO allele, smt3‐331, results in mispositioning of the mitotic spindle [Leisner et al., 2008; Meednu et al., 2008]. Both the Liakopoulos and Miller labs showed that kar9 mutants lacking the ability to be sumoylated display defects in the position of the mitotic spindle [Leisner et al., 2008; Meednu et al., 2008]. The Kar9‐L340P mutant results in a short‐bipolar spindle that is positioned farther away from the mother‐bud neck compared to wild type [Meednu et al., 2008]. Similarly, the Kar9‐4K→R mutant also shows spindle‐positioning defects, displaying increases in both the angle of spindle alignment and the distance to the bud neck. It is interesting to note, however, that the defect seen in the Kar9‐4K→R mutant is not as severe as that seen in the smt3‐331 mutant of SUMO itself [Leisner et al., 2008]. This suggests that other components required for spindle positioning are also regulated by SUMO. Alonso et al. [2012] posit that at least one of these other components resides within the dynein pathway [Alonso et al., 2012]. Alternatively, the difference could be attributed to activation of the spindle assembly checkpoint (SAC) by the smt3‐331 mutant [Leisner et al., 2008].
The interaction between Kar9p and Bim1p is regulated by both sumoylation and phosphorylation [Huls et al., 2012]. Sumoylation of Kar9p promotes the interaction, with lysine 381 having the most prominent effect. In contrast, phosphorylation of Bim1p by the Ipl1p kinase impedes the interaction [Huls et al., 2012].
Phosphorylation of substrates is one mechanism by which sumoylation can be regulated. This can be either a positive influence or a negative one [Yang et al., 2003; Hietakangas et al., 2006]. Kar9p is one example that illustrates this type of regulation. Cdc28p phosphorylates Kar9p at serine 197 and 496 [Liakopoulos et al., 2003]. Disruption of these phosphorylation sites causes Kar9p to mislocalize to both old and new spindle pole bodies [Liakopoulos et al., 2003; Moore et al., 2006; Moore and Miller, 2007]. Phospho‐mimetic mutations at one of these sites, Kar9p‐A196E S197E, does not interact with SUMO by two‐hybrid analysis, suggesting that phosphorylation at serine 197 blocks the interaction of Kar9p with Smt3p [Meednu et al., 2008]. Consistent with this idea, the Liakpoulos lab showed that the phospho‐inhibited Kar9p‐S197A S496A mutant was still able to be sumoylated [Leisner et al., 2008].
In addition to phosphorylation and sumoylation, Kar9p is also regulated by ubiquitination [Maekawa et al., 2003; Moore et al., 2006; Moore and Miller, 2007; Leisner et al., 2008; Meednu et al. 2008; Kammerer et al., 2010]. Ubiquitination of Kar9p regulates the interaction of astral microtubules with the bud neck, appearing to be involved in the proteasomal degradation of the subset of Kar9p molecules interacting with the bud neck [Kammerer et al., 2010]. The relationship between sumoylation and ubiquitination in this context remains unexplored. However, Kar9p interacts with the STUbL, Uls1p‐Nis1p, and Wss1p by two‐hybrid analysis [Meednu et al., 2008].
Dynein
Dynein is the major motor protein that walks toward the minus‐end of microtubules. Dynein participates in a wide range of cellular functions. Dynein plays several roles in the mitotic spindle and at the kinetochore [Kardon and Vale, 2009]. Dynein is important in chromosome capture and alignment, as well as silencing the spindle assembly checkpoint [Howell et al., 2001; Bader and Vaughan, 2010; Mao et al., 2010]. Together with NuMa, dynein plays a critical role in focusing the poles of the mitotic spindle, helping to generate its cone‐shaped geometry [Gaglio et al., 1997]. Errors in any of these processes can lead to increases in abnormally segregated chromosomes, a condition known as aneuploidy. When dynein is anchored at the cell surface, it can participate in spindle orientation or nuclear migration by pulling on microtubules that are attached to the MTOC [Lee et al., 2005; Collins et al., 2012; Kotak and Gonczy, 2013; Kotak et al., 2014]. Dynein also carries a variety of different cargoes to specific destinations within the cell. Cargoes include endocytic vesicles, viral particles, organelles in retrograde axonal transport, melanosomes, and ER to Golgi transport vesicles [Holzbaur and Vallee, 1994; LaMonte et al., 2002; Watson et al., 2005; Johansson et al., 2007; Rocha et al., 2009; Scherer and Vallee, 2011; Tan et al., 2011; Moughamian and Holzbaur, 2012]. However, regulation of the attachment of cargo to dynein is still poorly understood.
There are two forms of cytoplasmic dynein DHC1a (dynein 1) and DHC1b (dynein 2), both of which are distinct from flageller dynein [Paschal et al., 1987; Gibbons, 1995]. Dynein 1 is the major form of cytoplasmic dynein and is found in all eukaryotes, from fungi to human [King et al., 2002]. Dynein 2, is a less well characterized form of cytoplasmic dynein that is found in most ciliated eukaryotic cells, where it functions in intraflagellar transport and golgi organization [Pazour et al., 1999; Signor et al., 1999; Grissom et al., 2002; Helfand et al., 2002; Mikami et al., 2002]. Mutations in the dynein 2 complex result in a number of ciliopathies [Schmidts et al., 2013a, 2013b]. Dynein 2 associates with different intermediate and light chains than those associated with dynein 1. It also does not interact with other known regulators of dynein, including dynactin, LIS1, and BICD2 [Asante et al., 2014].
Crystallography work from the Vale lab provides detailed insight into how the structure of the dynein 1 motor couples ATP hydrolysis within the main AAA ATPase domain to allosteric changes that result in movement [Carter et al., 2008, 2011; Bhabha et al., 2014].
The dynein heavy chain is complexed with several accessory proteins. These are the intermediate chains, the light chains, and the light intermediate chains [Vaughan and Vallee, 1995; Waterman‐Storer et al., 1995; Ma et al., 1999; Lo et al., 2001; Mok et al., 2001]. Two adaptors for dynein are Lis1/Pac1p and the dynein‐activating complex, better known as the dynactin complex [Vaughan et al., 1999; Faulkner et al., 2000; Tai et al., 2002; Schroer, 2004; Levy and Holzbaur, 2006]. The dynactin complex consists of two sub‐domains, a short actin‐like filament connected to a shoulder‐sidearm projection [Eckley et al., 1999; Quintyne et al., 1999]. The short actin‐like filament consists of Arp1, CapZ, p62, Arp11, p27, and p25. The shoulder‐sidearm projection consists of 150Glued, dynamitin, and p24 [Eckley et al., 1999; Garces et al., 1999]. Both dynactin and Lis1/Pac1p are involved in attaching cargo to the dynein motor, but the mechanisms that regulate cargo attachment to dynein are unclear [Kardon and Vale, 2009; McKenney et al., 2011]. To date, no evidence suggests that the accessory chains, the dynactin complex, or dynein itself are SUMO substrates. However, two reports currently connect SUMO to dynein. One report investigates the dynein cargo, La; and the other examines the adaptor, Pac1p [van Niekerk et al., 2007; Alonso et al., 2012]. Alonso et al. postulate that sumoylation of adaptors could be a new mode of regulation for dynein [Alonso et al., 2012].
La
La is an RNA‐binding protein that is transported by dynein [van Niekerk et al., 2007]. La is also an antigen found in the autoimmune diseases, systemic lupus erythematosus and Sjorgren's syndrome [Kumar et al., 2013], and it can enhance mRNA translation as well as viral replication [Trotta et al., 2003; Kumar et al., 2013].
The Twiss lab demonstrated that La is sumoylated at a unique site, K41 [van Niekerk et al., 2007]. A non‐sumoylatable form of La fails to immunoprecipitate with dynein. The non‐sumoylatable La also moves down the axon in the anterograde direction, but not toward the cell body in the retrograde direction. Together these observations suggests that sumoylation of La promotes its interaction with dynein and is required for its retrograde transport in neurons by dynein. However, several questions remain. Does La transport involve the interaction with other dynein adaptors? Where in the neuron is La sumoylated and does desumoylation regulate the un‐loading of La cargo?
Lis1/Pac1p
Pac1p is the yeast homologue of the Lis1 protein, occasionally referred to as PAFAH1B1 [Hattori et al., 1994]. Mutations in the LIS1 gene are responsible for the severe brain disease, Type 1 lissencephaly, or “smooth brain.” Lissencephaly is a rare brain formation disorder caused by dysfunction in neuronal migration, leading to severe mental disorders and early death [Sapir et al., 1999; Kato and Dobyns, 2003; Reiner et al., 2006; Liu, 2011]. The hallmark of the disease is a drastic decrease in convolutions of the cerebral cortex [Reiner and Sapir, 2013]. While Lis1 is perhaps best known for its role in neurons, it is also important in desmosome stability and cortical microtubule organization in the epidermis. Loss of Lis1 results in fragile desmosomes, where it also localizes [Sumigray and Lechler, 2011; Sumigray et al., 2011]. Lis1 is also critical in the development of hematopoietic stem cells, where it controls the positioning of the mitotic spindle during cell division and the inheritance of cell fate determinants [Zimdahl et al., 2014].
The structure of Pac1p/Lis1 provides clues as to how it serves as a critical regulator of the dynein motor protein. Pac1p is composed of three regions: a LisH domain, a coiled‐coil domain, and a series of highly conserved WD40 repeats. Alone, none of the domains are sufficient for microtubule binding or tracking the plus‐end of the microtubule in vivo [Markus et al., 2011]. In contrast, the WD40 repeats of Pac1p/Lis1 are thought to bind across the intersection of the AAA3 and AAA4 ATPase motifs of dynein [Faulkner et al., 2000; Vallee et al., 2001; McKenney et al., 2011; Huang et al., 2012; Wang et al., 2013; Toropova et al., 2014]. Lis1 also promotes dynein's interaction with certain cargo [Sitaram et al., 2012; Splinter et al., 2012]. The direct binding of Lis1/Pac1p to dynein can regulate several properties of the motor itself. These include its velocity, the load carried, and “processivity.” By inducing a conformational change in the motor, Lis1/Pac1p also increases the “heaviness” of the load that the motor can carry [McKenney et al., 2010]. Dynein bound to Lis1 walks at a slower speed than unbound dynein [McKenney et al., 2010; Markus et al., 2011; Torisawa et al., 2011; Huang et al., 2012; Toropova et al., 2014]. The binding of Lis1 also increases its “processivity” which is the distance that a motor travels before stepping off the track. All of these parameters can be influenced by the time of attachment of dynein to the microtubule [Huang et al., 2012; Toropova et al., 2014]. Thus, the binding of Lis1 to dynein can be thought of as transforming it into a more powerful diesel engine, one in low gear.
In yeast, Pac1p functions in the dynein pathway by working with Bik1p to recruit dynein to the plus‐end of the microtubule before dynein is off‐loaded to the bud cortex [Sheeman et al., 2003; Lee et al., 2005; Li et al., 2005; Markus et al., 2011]. Bik1p is the yeast homologue of mammalian CLIP‐170. In the absence of Pac1p or Bik1p, dynein fails to be recruited to the plus‐end of microtubules, resulting in spindle positioning defects [Sheeman et al., 2003].
Several approaches were employed to show that SUMO is linked to Lis1/Pac1p [Alonso et al., 2012]. First, two‐hybrid analysis was used to show that Pac1p interacts with SUMO and several other members of the sumoylation pathway, including the E2 enzyme, Ubc9p, and the E3, Nfi1p. Second, inhibition of the SUMO protease Ulp1p resulted in multiple higher molecular weight forms of Pac1p, suggesting that Ulp1p removes SUMO from Pac1p [Alonso et al., 2012]. Third, the co‐immunoprecipitation of Pac1p with SUMO strongly suggested that Pac1p is a SUMO substrate. Fourth and also consistent with Pac1p being sumoylated, Pac1p interacted with both components of the STUbL enzyme, Uls1p‐Nis1p, by two‐hybrid analysis [Alonso et al., 2012]. Pac1p shift was increased in strains deleted for the STUbL Uls1p, and in strains where the proteasome was inhibited with the drug MG132 [Alonso et al., 2012]. These data support a model in which the Uls1p‐Nis1p STUbL recognizes a sumoylated Pac1p and thus targets it to the proteasome. Depending on the localization of the STUbL, this could represent a mechanism to degrade a subcellular pool of Pac1p, perhaps on the set of microtubules directed into the bud.
As Pac1p is one of the few examples known for substrates of the STUbL, Uls1p‐Nisp1p, many questions remain about its sumoylation. Additional work is needed to see if this modification is conserved in the mammalian homologue, Lis1. It is also not known how sumoylation of Pac1p might regulate either the cargo selection of dynein or the motor properties of dynein. Work is currently in progress in the Miller lab to identify the sites of modification and determine the function of this modification.
Bik1p/CLIP‐170
Bik1p is the yeast homologue of CLIP‐170, a family of CAP‐Gly proteins that track microtubule plus‐ends [reviewed in Miller et al., 2006; Gupta et al., 2014]. These are often referred to as a member of the a “+TIP” family of proteins [Akhmanova and Steinmetz, 2008]. CLIP‐170 binds the growing ends of microtubules, whereas Bik1p binds microtubules that are both growing and shrinking [Carvalho et al., 2004]. Bik1p also stabilizes microtubules against catastrophe. When Bik1p is absent from the cell, microtubules are very short [Berlin et al., 1990].
Structurally, Bik1p/CLIP‐170 is comprised of an amino‐terminal head domain, a central coiled‐coil domain, and a carboxy‐terminal domain that contains metal‐binding “zinc knuckle” motif. This domain is sometimes referred to as the “cargo‐binding domain” [Miller et al., 2006; Gupta et al., 2010]. In contrast to the yeast Bik1p, the head domain of the mammalian CLIP‐170 contains two CAP‐Gly domain and several serine rich domains [Miller et al., 2006]. Early work suggested that microtubule binding occurred through the CAP‐Gly domains, but recent work demonstrates that the serine rich regions also make substantial contributions to microtubule binding [Gupta et al., 2010]. In addition to binding the microtubule polymer, CLIP‐170 also possesses a significant affinity for tubulin dimers [Folker et al., 2005]. This interaction may play a role in a “co‐polymerization” mechanism by which CLIP‐170 tracks the plus‐end of the growing microtubule [Folker et al., 2005]. The interaction of Pac1p with Bik1p occurs though the carboxy‐terminal domain of Bik1p [Sheeman et al., 2003].
The functions of both CLIP‐170 and Bik1p are closely connected to those of dynein [Vaughan et al., 1999; Tai et al., 2002; Goodson et al., 2003; Sheeman et al., 2003; Caudron et al., 2008]. Bik1p, together with Lis1/Pac1p and Ndl1p, the yeast homologue of nuclear distribution factor E, recruits dynein to the plus‐end of the microtubule, prior to dynein's off‐loading to the cortex [Sheeman et al., 2003; Lee et al., 2005; Markus et al., 2011]. Bik1p also interacts with Kar9p, providing a link between the Kar9p and dynein spindle positioning pathways [Moore et al., 2006].
Bik1p displays several interactions with the sumoylation machinery. Bik1p interacts with SUMO; the SUMO E2 conjugating enzyme Ubc9p, and the E3 Nfi1p by two‐hybrid analysis [Alonso et al., 2012]. Interestingly, the carboxy‐terminal domain of Bik1p, the domain that interacts with Pac1p, is also required for Pac1p's interaction with SUMO. In the reciprocal direction, Pac1p is required for Bik1p's interaction with SUMO in the two‐hybrid assay. These findings suggest the possibility that a mutual‐association of both proteins is required for their modification by SUMO [Alonso et al., 2012]. Bik1p can also be sumoylated using an in vitro assay, resulting in two and possibly three shifted bands. It is not known whether Pac1p might enhance this in vitro sumoylation, which would be consistent with the two‐hybrid data. Sumoylated forms of Bik1p have also been observed in vivo when overexpressed Bik1p and overexpressed SUMO were employed [Alonso et al., 2012]. Ulp1p is one of the major SUMO proteases in the cell that cleaves SUMO from target proteins. In a somewhat surprising finding, inactivation of Ulp1p with a temperature‐sensitive allele did not reveal SUMO‐shifted forms of Bik1p [Alonso et al., 2012]. Thus, identification of a sumoylated form of Bik1p at endogenous levels has remained elusive.
What hypotheses could reconcile this apparent discrepancy? Perhaps Bik1p is not actually conjugated by SUMO and the putative SUMO connection occurs via a non‐covalent interaction. Perhaps SUMO only attaches to Bik1p when the cell is stressed. Another possibility is based on the finding that Bik1p interacts by two‐hybrid analysis with the STUbL enzyme, Uls1p‐Nis1p, and the SUMO isopeptidase, Wss1p. While Bik1p's interaction with this enzyme implies that it is sumoylated at some point, the difficulty of “catching” SUMO on Bik1p is nevertheless perplexing. Perhaps Bik1p's interaction with the STUBL results in its rapid demise by the proteasome. As Bik1p and CLIP‐170 have critical functions for microtubules, further research into the SUMO‐Bik1p connection is anticipated.
Various +TIPs interact with each other to form a web of interactions at the plus‐end of the microtubule [Akhmanova and Steinmetz, 2008]. However, the function of these interactions has remained a mystery [Gupta et al., 2014]. Considering that a growing list of +TIPs are seen to interact with SUMO, we postulate that sumoylation may help in the assembly of higher order molecular structures of +TIP assemblies. This may involve the SIMs of one MAP binding the sumoylated form of an adjacent MAP.
Tau
Tau, tubulin‐associated unit, is a microtubule‐associated protein that helps stabilize microtubules and is highly conserved in higher eukaryotes [Goedert et al., 1989a, 1989b, 1996; Maccioni et al., 1995]. Tau is found mainly in neurons, where it stabilizes microtubules and promotes their polymerization [Cleveland et al., 1977; Binder et al., 1985; Drubin and Kirschner, 1986; Drechsel et al., 1992]. Tau also has the ability to bundle microtubules [Kanai et al., 1992]. Tau is a hydrophilic protein that consists of four regions; an acidic region, a proline‐rich region, a microtubule‐binding region consisting of four repeats of conserved residues, and a basic C‐terminal region. The extreme variation in charge between the N‐terminus and the C‐terminus region of tau can be modulated by various post‐translational modifications. Tau shares homology with other MAPs including MAP2 and MAP3/4 [Chapin and Bulinski, 1991]. Mutations in tau are associated with several neurodegenerative disorders including Alzheimer's, Pick's disease and several tauopathies [reviewed in Goedert, 2001]. Alzheimer's is a neurodegenerative disease characterized by neurofibrillary tangles and senile plaques. The neurofibrillary tangles are intracellular aggregates containing abnormally phosphorylated tau, whereas senile plaques are extracellular deposits of amyloid β‐peptides [Grundke‐Iqbal et al., 1986; Ihara et al., 1986; Delacourte et al., 1999]. In models for tau's role in Alzheimer's, tau first dissociates from microtubules in a phosphorylation‐dependent manner, leading to destabilization of the microtubules. Subsequently, unbound tau oligomerizes to form the paired helical filaments found in neurofibrillary tangles [reviewed in Meraz‐Rios et al., 2010]. As various forms of tau are found in cerebrospinal fluid, it is now being developed as biomarker for Alzheimer's disease to speed early diagnosis [reviewed in Blennow et al., 2012; Kopeikina et al., 2012].
Tau can be tagged by numerous post‐translational modifications, including phosphorylation, glycosylation, glycation, prolyl‐isomerization, nitration, polyamination, ubiquitination, oxidation, and sumoylation [Grundke‐Iqbal et al., 1986; Schweers et al., 1995; Wang et al., 1996; Nacharaju et al., 1997; Murthy et al., 1998; Takahashi et al., 1999; Zhou et al., 2000; David et al., 2002; Horiguchi et al., 2003; Landino et al., 2004; Necula and Kuret, 2004; Zhang et al., 2005; Dorval and Fraser, 2006, 2007; Kuhla et al., 2007; Takahashi et al., 2008; Wang et al., 2008; Arnaud et al., 2009; Bulbarelli et al., 2009; Liu et al., 2009]. Tau has as many as thirty phosphorylation sites that can alter its structure, function, and localization [Grundke‐Iqbal et al., 1986; Litersky et al., 1996; Fischer et al., 2009]. In general, an increase in tau phosphorylation reduces its affinity for microtubules and thus its ability to stabilize microtubules [Drewes et al., 1995].
The relationship between SUMO and ubiquitin on tau is a noteworthy example of one type of crosstalk between two ubiquitin family members. Tau can be ubiquitinated both in vitro and in vivo [David et al., 2002; Petrucelli et al., 2004; Zhang et al., 2005; Arnaud et al., 2009; Liu et al., 2009]. Tau is sumoylated mainly by SUMO1, but in some cases by SUMO2 and SUMO3 [Dorval and Fraser, 2006, 2007; Takahashi et al., 2008]. Mutational analysis showed that the primary attachment site for SUMO is lysine 340, which is located within a microtubule‐binding repeat. Tau has been seen shown to be heavily ubiquitinated in mature tangles of Alzheimer's patients whereas the sumoylation levels in the mature tangles are low [Bancher et al., 1991; Dorval and Fraser, 2006]. It is speculated that ubiquitin and SUMO compete for the same lysine residue. In this case, if one modification is upregulated, the other would be down regulated [Dorval and Fraser, 2006]. Consistent with this model, inhibition of the proteasome causes a decrease on tau sumoylation, while increasing tau ubiquitination [Dorval and Fraser, 2006]. Therefore, the sumoylation of tau could be one mechanism to modulate its turnover rate by blocking the ubiquitination that sends it to the proteasome [Dorval and Fraser, 2006]. The diminished sumoylation of tau observed in Alzheimer's patients is consistent with the diminished proteasome function that is commonly found in many neurodegenerative diseases [Pountney et al., 2003; Dorval and Fraser, 2006].
Tau sumoylation is also partly dependent on phosphorylation. Treatment of cells with the phosphatase inhibitor, okadaic acid, promotes tau sumoylation [Dorval and Fraser, 2006, 2007]. Sumoylation of tau is also increased by treatment of cells with the microtubule‐depolymerizing drug, colchicine, which also releases tau from the microtubule. This finding is consistent with the sumoylation site being located inside the microtubule‐binding region [Dorval and Fraser, 2006]. These findings raise questions about the extent to which sumoylation may control tau solubility. Since tau is implicated in various human diseases, the levels of tau sumoylation should also be examined in other tauopathies. This information could provide insight into our understanding of the role of sumoylation in human disease pathogenesis.
Kinetochore MAPs
Numerous proteins of the kinetochore are sumoylated [Mukhopadhyay and Dasso, 2010; Cubenas‐Potts et al., 2013]. Indeed, SUMO/Smt3p in yeast was identified as the third Suppressor of Mif Two, which is a protein located at the centromere‐kinetochore interface [Lampert and Westermann, 2011]. While the sumoylation of centromere and kinetochore proteins is itself an emerging field of interest, this section focuses on the kinetochore proteins that are also bona fide microtubule‐binding proteins.
Ndc80p
Ndc80p is a conserved part of the kinetochore‐associated Ndc80 complex, also refered to as Hec1p. Ndc80p is also a microtubule‐associated protein. Ndc80p consists of a N‐terminal microtubule‐binding domain, which is negatively regulated by the kinase Aurora B, and a C‐terminal coiled‐coiled domain, which interacts with other components of the kinetochore‐associated Ndc80 complex [Cheeseman et al., 2006; Guimaraes et al., 2008; Miller et al., 2008]. The kinetochore consists of a collection of proteins that assembles on centromere DNA, to which the microtubules then attach. Ndc80p helps organize and stabilize kinetochore‐microtubule interaction in order to facilitate proper chromosome segregation [Wei et al., 2011]. Ndc80p forms a “dumbbell‐like” heterotetramer with Nuf2p, Spc24p, and Spc25p to form the Ndc80 complex [Cheeseman et al., 2006; Tien et al., 2013]. The Ndc80 complex also helps localize spindle assembly checkpoint proteins to the kinetochore [Gillett et al., 2004; Maiato et al., 2004].
In budding yeast, Ndc80p was identified as a sumoylated protein in several SUMO proteomes [Panse et al., 2004; Zhou et al., 2004; Wykoff and O'Shea, 2005]. Later, it was confirmed that Ndc80p is sumoylated in vivo at a lysine residing at position 231 [Montpetit et al., 2006]. Mutation of lysine 231 to arginine completely abolished the higher molecular forms of Ndc80p. It is unlikely that lysine 231 contributes to SUMO chain formation since the laddering effect remains the same in a strain in which SUMO chain formation is blocked [Montpetit et al., 2006]. Instead, the abrogation of the multiple higher molecular weight forms of Ndc80p in the K231R mutant suggests that this amino acid is required for the sumoylation of other lysines. Ndc80p sumoylation levels remain relatively constant over the cell cycle. Its sumoylation is also not affected by the depolymerization of microtubules by nocodazole treatment or by activation of the spindle assembly checkpoint. This is unlike other sumoylated kinetochore proteins, Ndc10p, Bir1p, and Cep3p. This suggests that Ndc80p is regulated differently than these proteins [Montpetit et al., 2006]. Although the evidence shows that Ndc80p is sumoylated in vivo, there are no phenotypes described as yet for the K231R mutant.
CENP‐E
CENP‐E is both a centromere‐associated protein located in the outer plate of the kinetochore and a plus end‐directed microtubule motor from the kinesin family [Yen et al., 1991]. CENP‐E is required for cell‐cycle progression from metaphase to anaphase by helping align chromosomes at the metaphase plate [Yen et al., 1991; Liu et al., 2007]. CENP‐E localization at the kinetochore is crucial for spindle checkpoint activation, which prevents defects in chromosome segregation [Liu et al., 2007]. CENP‐E has been shown to promote plus‐end microtubule elongation in vitro by stabilizing the microtubule as it walks towards the plus‐end [Sardar et al., 2010].
CENP‐E is both a SUMO substrate and a SUMO‐binding protein [Zhang et al., 2008]. The important role that SUMO plays in CENP‐E function was demonstrated by inhibition of sumoylation using overexpression of SENP2, a SUMO‐specific protease. This resulted in cell‐cycle arrest at prometaphase and the mislocalization of CENP‐E from the kinetochore [Zhang et al., 2008]. Overexpression of SENP2 also caused a decrease in sumoylation of other kinetochore‐associated proteins that are needed for proper CENP‐E localization to the kinetochore, since they bind CENP‐E non‐covalently [Zhang et al., 2008]. CENP‐E has also been shown to be a SUMO2/3 binding protein. Disruption of the SIMs in CENP‐E also causes its mislocalization from the kinetochore [Zhang et al., 2008].
In summary, two classes of microtubule motors are linked to SUMO, but by different mechanisms. The kinetochore kinesin, CENP‐E, both binds to and is conjugated by SUMO [Zhang et al., 2008]. The dynein motor is speculated to be regulated by SUMO, but indirectly, through conjugation of its adaptor, Pac1p [Alonso et al., 2012].
Actin
A third major cytoskeletal system is comprised of actin, also known as microfilaments [Chesarone et al., 2010; Ydenberg et al., 2011]. Actin is highly abundant, and can constitute as much as 5% of total cellular protein in some cell types. Actin is found in both the cytoplasm and in the nucleus, and actively shuttles between the two compartments [Dopie et al., 2012; Belin and Mullins, 2013]. Many of the cytoplasmic functions of actin are well characterized. In addition to serving as the cellular tracks on which myosin transports its cargo, the many roles of actin include maintaining cellular shape, formation of the cytokinesis furrow, cellular locomotion, scaffolding sites for signaling proteins, and roles in endocytosis and exocytosis [Pollard and Cooper, 2009; Gardel et al., 2010; Pollard, 2010; Mishra et al., 2014]. The nuclear functions of actin however are less well understood [Hendzel, 2014], but include roles in transcription and chromatin remodeling [Louvet and Percipalle, 2009; Kapoor et al., 2013; Percipalle, 2013]. Nuclear actin also interacts with each of the RNA polymerases, as well as nuclear export and import factors [Hofmann et al., 2004; Hu et al., 2004; Philimonenko et al., 2004; Dopie et al., 2012].
Recent reviews of actin and actin binding proteins have discussed their various post‐translational modifications including acetylation, methylation, phosphorylation, and ubiquitination [reviewed in dos Remedios et al., 2003; Terman and Kashina, 2013]. This review focuses on the effect that the SUMO modification exerts on actin.
Four proteomic studies identified actin as a likely target for SUMO conjugation [Panse et al., 2004; Vertegaal et al., 2004; Wohlschlegel et al., 2004; Rosas‐Acosta et al., 2005]. Hofmann et al. [2009] confirmed these studies, showing that SUMO 2 and 3 are the preferential isoforms of SUMO that modify actin. Lysine to arginine mutagenesis established that two lysines, one at position 68 and another at position 284, are required for actin's sumoylation. However computer modeling predicts that only K284 is conjugated by SUMO and that salt bridges between lysine 68 and SUMO help to stabilize the actin‐SUMO interaction, allowing K284 to be sumoylated.
Cellular fractionation experiments showed that it was predominately the actin in the nuclear fraction that is modified by SUMO. The current model suggests that sumoylation on K284 blocks access to a nuclear export sequence, NES‐1, resulting in sumoylated actin being retained in the nucleus. This idea is supported by the finding that non‐sumoylatable actin mutants are rapidly exported out of the nucleus back to the cytoplasm through an CRM1/exportin‐1 dependent pathway. This export was blocked by leptomycin B, a compound that modifies CRM1, inhibiting its function and nuclear export [Kudo et al., 1998, 1999; Hofmann et al., 2009]. Recent reports also implicate Exp6 in the nuclear export of actin [Dopie et al., 2012].
The import of actin into the nucleus was previously linked to the actin binding protein, cofilin, which contains a nuclear localization signal motif. Early models suggested that actin could “piggy‐back” on cofilin to gain entry into the nucleus [Nishida et al., 1987]. However, Dopie et al. [2012] recently showed that the import factor Ipo‐9 is also critical for actin transport into the nucleus. Informing both models, Hoffman et al.'s finding that non‐sumoylatable actin can easily enter the nucleus suggests that this modification may not be required for either of these import‐dependent interactions [Hofmann et al., 2009].
Structurally, the position of SUMO on actin at lysine K284 suggests that sumoylation would physically block the formation of classical actin filaments. This provides a plausible explanation for the absence of classical actin filaments in the nucleus [Hofmann et al., 2009]. It is also possible that sumoylation provides a mechanism by which actin could adopt alternative structures within the nucleus [Schoenenberger et al., 2005; Jockusch et al., 2006]. This hypothesis is especially intriguing considering that SUMO expression is strongly influenced by stress, and cellular stresses like heat shock and DMSO treatment induce the formation of a type of actin bundle known as actin rods within the nucleus of Xenopus oocytes [Welch and Suhan, 1985; Iida and Yahara, 1986; Iida et al., 1986]. Additional work is warranted to determine the exact role that sumoylation plays in governing the functions of nuclear actin and the types of structures formed, as little is known on this topic [Belin and Mullins, 2013].
Actin Regulatory Proteins and SUMO
The function, dynamics, and interactions of actin in both the cytoplasm and the nucleus are regulated by numerous actin‐binding proteins [Higgs and Pollard, 2001]. In addition to nuclear forms of myosin [Vreugde et al., 2006], several actin‐binding proteins have been shown to be present in the nucleus, such as filamin A, members of the Arp2/3 complex, and thymosin β4 [Vartiainen, 2008]. Their role in the regulation of nuclear actin is less clear [Dopie et al., 2012]. While it is known that actin‐binding proteins undergo several types of post‐translational modification including phosphorylation [Arber et al., 1998; Yang et al., 1998] and ubiquitination [Hao et al., 2013], actin‐binding proteins and actin regulatory proteins are now emerging as new categories of SUMO substrates.
RhoA, and Rac1 are two members of the Rho family of GTPases that play significant regulatory roles for the actin cytoskeleton, and have also been linked to SUMO. They regulate the formation of stress fibers, membrane ruffles, and filopodia [Nobes and Hall, 1995]. In the cell, Rho family GTPases function as molecular switches that toggle between GDP‐bound (inactive) and GTP‐bound (active) forms. This switching is regulated by two other groups of proteins, GAPs (GTPase‐activating proteins) and GEFs (guanine‐nucleotide exchange factors). GAPs facilitate the hydrolysis of GTP to GDP, returning the GTPase to its “inactive” form, whereas GEFs help facilitate the exchange of GDP for GTP, returning the GTPase to an “active” state [reviewed in Cherfils and Zeghouf, 2013].
Ran is another small GTPase, which is central to the regulation of nuclear transport. It also interacts with the nuclear pore protein Ran binding protein (RanBP2), which is a SUMO E3 ligase [Azuma and Dasso, 2002]. The GAP for Ran, RanGAP1, is conjugated by SUMO1 [Joseph et al., 2002]. Ran influences the interaction between microtubules and the kinetochore [Joseph et al., 2004] and this aspect of its function has been expertly reviewed elsewhere [Dasso, 2008; Flotho and Werner, 2012].
In 2010, Rac1 was shown to co‐purify with the SUMO E3 ligase PIAS3, prompting further investigation. Castillo‐Lluva et al. [2010] showed that the Rac1 is sumoylated and that this SUMOylation event promotes cellular migration. These researchers identified four lysine residues in Rac1 to which SUMO‐1 could conjugate. These lysines were identified by using in vitro sumoylated Rac1 and mass spectrometry to reveal the branched “gly‐gly stubs” that are left behind after trypsinsation. This approach is based on the assumption that the fidelity of in vitro sumoylation is quite high. Indeed, they found that mutation of these four lysines to arginine resulted in the loss of the shifted SUMO bands in vitro and in vivo. The four sumoylated lysines reside in the C‐terminal polybasic region of Rac1, a domain that is important for the binding of several effectors of Rac1. Surprisingly however, the non‐sumoylatable Rac1 did not display altered binding to several known effectors. Instead, sumoylation appeared to be important for optimal GTP binding to Rac1. Defects were also observed in lamellipodia‐membrane ruffling. Further, the E3‐SUMO ligase PIAS3 preferentially sumoylated the GTP‐bound activated form of Rac1 over the GDP‐bound form of Rac1. Castillo‐Lluva et al. [2010] postulate that while not all active Rac1 is SUMOylated, the percentage that is could be enough to boost Rac1 activity over a certain threshold that is required for lamelliopodia formation and cellular migration.
RhoGDI
Rho family GTPases are regulated in part by RhoGDIs (Rho GDP‐dissociation inhibitors). RhoGDI can both remove and prevent the binding of Rho‐GTPases to cell membranes [Isomura et al., 1991; Dovas and Couchman, 2005], thereby controlling their cytosol‐membrane cycling. Thus, RhoGDI regulates the activation state of Rho‐GTPases from an active state that is membrane bound to an inactive state in the cytoplasm [Olofsson, 1999]. Rho‐GTPases are known to regulate actin and a variety of cellular events including cellular morphology, cellular adhesion and aggregation, cellular motility, and ruffling of the plasma membrane, as well as formation of stress fibers and focal adhesions [Paterson et al., 1990; Ridley and Hall, 1992; Tominaga et al., 1993; Nishiyama et al., 1994; Takaishi et al., 1994]. Thus, the regulation of RhoGDI has the potential to control many downstream effects.
The RhoGDI can be regulated by multiple mechanisms. The RhoGDI‐RhoGTPase complex can be post‐translationally regulated by the phosphorylation of RhoA and Cdc42 [Forget et al., 2002; Tu et al., 2003]. RhoGDI can itself be post‐translationally modified by phosphorylation, causing the RhoGDI‐RhoGTPase complex to dissociate [Price et al., 2003; DerMardirossian et al., 2006]. RhoGDI can also be modified by sumoylation at lysine 138 [Liu et al., 2011; Yu et al., 2012]. This acts as a switch to activate RhoGDI activity [Yu et al., 2012]. The active sumoylated form of RhoGDI inhibits Rho‐GTPase activity, resulting in the down regulation of actin polymerization and cell motility by decreasing the recruitment of Arp2/3 complex to the cytoskeleton [Yu et al., 2012].
The sumoylation of RhoGDI can be regulated by the RING domain of X‐linked inhibitor of apoptosis protein (XIAP) [Liu et al., 2011; Yu et al., 2012]. The RING domain of XIAP binds RhoGDI and blocks RhoGDI sumoylation [Yu et al., 2012]. By blocking the sumoylation site, XIAP reduces the sumoylation levels of RhoGDI, therefore increasing the recruitment of Arp2/3 to the cytoplasm causing an increase in actin polymerization and cell motility [Yu et al., 2012]. XIAP overexpression has been associated with malignant cancer progression in various types of cancer [Yamazaki et al., 1999; Nemoto et al., 2004; Akyurek et al., 2006; Kleinberg et al., 2007; Kluger et al., 2007; Nagi et al., 2007; Burstein et al., 2008]. However the molecular mechanism for how this occurs remains unknown. Thus, this finding suggests a possible molecular mechanism for how overexpression of XIAP can down regulate RhoGDI, leading to increased actin polymerization and cell motility of cancer cells.
Arp2/3 Complex
Another group of actin binding proteins is the Arp2/3 complex. The principal function of the Arp2/3 complex is to create branches in the elongating actin network near the protruding edge of the plasma membrane [dos Remedios et al., 2003; Firat‐Karalar and Welch, 2011; Rotty et al., 2013]. This complex is conserved from yeast to mammals and consists of seven proteins: Arp2, Arp3, and five smaller proteins (Arcs) [Goley and Welch, 2006].
At least three proteomic studies have identified components of the Arp2/3 complex as potential SUMO targets. Arc35p and Arc40p were identified in a proteomics screen that combined nickel purification of his6‐Smt3p with mass spectrometry [Wohlschlegel et al., 2004]. A second proteomic study using Schizosaccharomyces pombe also identified several Arcs as potential SUMO targets. These included Arc34p, which is the S. pombe ortholog to S. cerevisae Arc35p; Arc5p, which is an ortholog to S. cerevisiae Arc15p; and Arc4p which is an ortholog to S. cerevisiae Arc19p [Nie et al., 2012]. Recently, Sung et al. [2013] also identified Arc35p as a sumoylation candidate by using a bi‐molecular fluorescence complementation assay. This assay is based on the principle that a fluorescent complex will form when two proteins fused to fragments of a fluorescent protein interact with each other. This allows for direct visualization within the cell of the location of a protein‐protein interaction. Arc35p was one of several proteins chosen to validate this approach. In a pull‐down assay, Arc35p co‐fractionated with Smt3p, and anti‐Smt3p reactive bands matched the shifted forms of Arc35p [Sung et al., 2013]. While proteomic studies have consistently identified Arc35p of the Arp2/3 complex as a likely sumoylated protein, follow‐up studies providing more detail are currently lacking. For instance, it is not known whether Arc sumoylation activates or inactivates Arp2/3 for its ability to form actin branches.
Intermediate Filaments (IF)
Intermediate filaments (IF) are the fourth polymeric network of the cytoskeleton, and include six classes of proteins [Eriksson et al., 2009]. The pattern of expression for the various classes of IF is cell‐type specific. For example, the type I and type II IF are the acidic and basic keratins. These are coexpressed in tissues of epithelial origin [Fuchs, 1995]. Type III IF are found in cells of mesenchymal origin, which include cells such as fibroblasts [Franke et al., 1978]. Vimentin is perhaps the best characterized type III IF. The type IV class of IF include synemin, nestin, and the neurofilament proteins of which there are the high, medium, and low molecular weight (H, M, L) forms [Jing et al., 2007; Lepinoux‐Chambaud and Eyer, 2013]. Lamins are type V intermediate filament proteins that line the periphery of the inner membrane of the nuclear envelope [Eriksson et al., 2009]. Members of the type VI IF family include filensin and phakinin, which are present in the fiber cells of the lens [Oka et al., 2008]. Mutations in these proteins result in cataracts [Szeverenyi et al., 2008]. Certain cell types can express more than one class of IF and expression patterns can also be controlled developmentally. Several excellent reviews have been written recently on intermediate filaments [Eriksson et al., 2009; Goldman et al., 2012; Snider and Omary, 2014].
General Structure of IF
Intermediate filaments are 10 nm in diameter, thus giving them the name “intermediate” because they are intermediate in size between the 25 nm microtubules and 7 nm microfilaments [Ishikawa et al., 1968]. IF have three major domains. At the N‐terminus, there is a non‐alpha‐helical head domain. The central part of the protein is composed of an alpha‐helical rod domain containing heptad repeats. These allow the formation of the dimeric coiled‐coil architecture characteristic of IF. The coiled nature of the rod domain is disrupted by three conserved short non‐helical linker regions. The C‐terminus in lamins, also known as the tail domain, forms a β‐fold similar to that seen in immunoglobulins [Herrmann and Aebi, 2004]. Several types of IF organize into homodimers, whereas other types can form heterodimers [Parry et al., 1985; Herrmann and Aebi, 2004; Goldman et al., 2008].
As discussed in more detail below, four of the six classes of IF are modified by SUMO in some capacity. These are the type I and II keratins, type III vimentin, and type V lamins [Zhang et al., 2008; Wang et al., 2010; Snider et al., 2011]. The C. elegans IF protein, IFB‐1, which displays several structural and functional similarities to keratin but has a lamin‐like tail, is also sumoylated [Carberry et al., 2009; Kaminsky et al., 2009]. This raises the question of whether sumoylation of IF is a conserved modification. Will members of the other classes of IF someday be found to be sumoylated?
Keratin
Approximately 30 different keratins have been catalogued and these are classified as either type I or type II IF based on their isoelectric points and sequence homologies. Type I keratins have an acidic isoelectric point, whereas type II keratins are neutral‐basic. As obligate heteropolymers, keratin filaments can only form when a type I keratin forms a heterodimer with a type II keratin.
Keratin IF are specifically expressed in epithelial cells, where they play several vital roles. First, keratins confer structural support and mechanical durability to epithelial cells [Fuchs and Cleveland, 1998]. A second role of keratins is to modulate cell signaling processes through a variety of mechanisms including the recruitment of multiple kinases, phosphatases, and 14‐3‐3 proteins [Eriksson et al., 2009]. Keratins also play a role in the function of organelles and cell migration [Kim and Coulombe, 2007]. Mutations that disrupt the filament forming ability of keratins result in several diseases, including the blistering skin diseases, epidermal bullosa simplex (EBS) and epidermolytic hyperkeratosis (EHS) [Bonifas et al., 1991; Coulombe et al., 1991; Vassar et al., 1991; Coulombe and Fuchs, 1993; Letai et al., 1993; Chipev et al., 1994; Yang et al., 1994, 1996; Fuchs and Cleveland, 1998; Arin et al., 1999, 2000). Mutations in keratin 8 and 18 can predispose patients and mice to liver disease [Ku et al., 2005; Ku and Omary, 2006; Strnad et al., 2012]. Keratin expression can also modulate the invasive nature of some cancers [Chung et al., 2013; Seltmann et al., 2013].
Keratins K8, K18, and K19, which are found in simple epithelia, were recently shown by the Omary laboratory to be sumoylated [Snider et al., 2011]. SUMO 2/3 is used preferentially over SUMO1 [Snider et al., 2011]. Four sites were identified for K8, three for K18, and one for K19. The sites of sumoylation appear to lie within the coiled‐coil alpha‐helical rod domain, similar to the study from the Sarge lab for lamins A (see below) [Zhang et al., 2008]. Due to the geometry of packing of dimers and tetramers, it seems improbable that these sites would be available for sumoylation in the fully formed filament [Snider et al., 2011]. In contrast, an IF from C. elegans, IFB‐1, was found to be sumoylated in the C‐terminal tail domain [Kaminsky et al., 2009].
Keratin sumoylation is regulated by oxidative and other stresses. In cells treated with hydrogen peroxide, as well as compounds used in liver injury models, the sumoylation levels on K8, K18, and K19 increased dramatically [Snider et al., 2011]. Concomitantly, the levels of SUMO co‐localizing with the keratin network also increased significantly [Snider et al., 2011]. It will be useful to know from future studies how keratin sumoylation transduces signals to downstream stress response pathways. It will be equally important to determine whether sumoylation is necessary for the “stress protection” conferred by keratins against various liver diseases.
Studies from two systems, human cells and C. elegans, show that sumoylation regulates the solubility of keratins. Using human keratins 8 and 18, mono‐sumoylation was found to increase the solubility of keratins. In contrast, hyper‐sumoylation decreases the solubility [Snider et al., 2011]. These findings have significant implications for the regulation of IF dynamics [Kaminsky et al., 2009; Snider et al., 2011]. In comparison to other cytoskeletal networks, keratins and other IF have small cytosolic pools of subunits, which are represented by a small number of biochemically soluble subunits [Soellner et al., 1985]. This contributed to an early but inaccurate viewpoint that keratin networks were static, rigid structures [reviewed in Goldman et al., 2012]. Work then began to emerge showing that keratin subunits did in fact exchange with the keratin polymer, albeit the rates for dynamic exchange of keratin were slower than for vimentin, microtubules, or microfilaments [Soellner et al., 1985; Miller et al., 1991, 1993; Yoon et al., 2001, 1998].
These findings are consistent with those made with the C. elegans IF protein, IFB‐1A, which forms epidermal attachment structures. A sumoylation‐deficient mutant of IFB‐1 displayed decreased cytoplasmic staining and disrupted IF formation in vivo in comparison to wild type. Similarly, inhibition of the C. elegans SUMO gene itself with RNAi‐feeding also showed decreased cytoplasmic staining of the IFB‐1 and thicker and shorter filament bundles. These findings support the notion that sumoylation mediates the amount of keratin subunits available for incorporation into newly assembled bundles [Kaminsky et al., 2009]. To investigate the extent to which SUMO‐mediated solubility correlates with IF dynamics, fluorescence recovery after photobleaching (FRAP) experiments were carried out. Both non‐sumolatable IFB‐1 and SUMO mutants exhibited much slower rates of recovery of photobleached IF filaments, suggesting that subunit exchange within the filaments is impaired by the lack of sumoylation in vivo [Kaminsky et al., 2009]. Overexpression of SUMO increased the amount of IF at apparent IF nucleation sites. Together, these findings are consistent with the hypothesis that sumoylation mediates the cytosolic pool of keratin subunits [Kaminsky et al., 2009]. In the future, it will be interesting to find out whether extra sumoylation can induce the disassembly of pre‐formed filaments and/or modulate their interaction with desmosomes and hemi‐desmosomes. If so, what is the extracellular stimuli to which sumoylation of IF responds? This information may provide insight into the function of sumoylation on mechanisms of cell motility.
In polymer science, it is commonly accepted that only very low concentrations of non‐functional subunits need be present to terminate the polymerization of polymer, resulting in much shorter chain lengths [Odian, 1991]. This principle of polymer science leads one to speculate whether a SUMO‐modified IF subunit may serve as a chain‐terminating element for this family of biopolymers. Considering that the vast majority of cytoskeletal protein resides in the polymeric state, sumoylation in this capacity would also be consistent with the low level of sumo‐modified subunits found within the entire population of molecules for a particular cytoskeletal network.
Phosphorylation of the target protein is a common paradigm by which sumoylation can be regulated [Hietakangas et al., 2003]. Phosphorylation of a serine or threonine can create a negatively charged residue that is functionally equivalent to the aspartic (D) or glutamic acid (E) within a canonical consensus site for sumoylation [Hietakangas et al., 2003; Yang et al., 2006; Blomster et al., 2009]. Such a phosphorylation dependent sumoylation motif (PDSM) paradigm is also seen with keratin. Several lines of evidence suggest that keratin 8 sumoylation is regulated in part by phosphorylation [Snider et al., 2011]. Inhibition of phosphatases with okadaic acid results in an increase in keratin 8 sumoylation seen by western blotting and an increase in the amount of SUMO‐2/3 colocalizing with the keratin network seen by immunofluorescence [Snider et al., 2011]. Liver‐injury agents such as porphyrinogenic compound DCC are known to increase keratin phosphorylation. These also result in increased sumoylation of keratin [Snider et al., 2011]. In cells transfected with the phospho‐inhibitory keratin 8‐S74A mutation, there is a moderate decrease in sumoylation of keratin 8 [Snider et al., 2011]. While the decrease was not dramatic, it leaves open the possibility that other uncharacterized phosphorylation sites could be regulating keratin sumoylation. Together these data support the hypothesis that phosphorylation regulates the sumoylation of keratin. Additional knowledge on this topic has the potential to be hugely important in understanding the molecular mechanisms of diseases involving keratins, such as liver disease.
Vimentin
Vimentin is the intermediate filament protein that typifies mesenchymal tissue and its expression characterizes the epithelial to mesenchymal transition in development [Mendez et al., 2010]. In several epithelial cancers, vimentin expression correlates with an increase in cell migration and poorer cancer prognosis [Hendrix et al., 1997; Lepekhin et al., 2001; Mendez et al., 2010; Liang et al., 2014; Niwa et al., 2014]. However in astrocytomas, the correlation of vimentin expression and survival is less clear [Skalli et al., 2013]. The regulation of vimentin disassembly is an important step in the formation of lamelliapodia, a key cellular structure at the leading edge of the cell that is needed for cell migration [Helfand et al., 2011]. Adding to its complexity, vimentin can co‐polymerize with several other types of type III subunits to form IF co‐polymers [Eliasson et al., 1999]. One use for this co‐polymerization is to help assemble the glial fibrillary acidic protein (GFAP) network within astrocytes [Galou et al., 1996]. In addition, vimentin plays a role in anchoring mitochondria and thus modulating their intracellular migration [Nekrasova et al., 2011].
Recently SUMO has been linked to a mutant form of vimentin in an aggressive form of brain cancer, glioblastoma multiforme. In a model system for this, U373 cells displayed inhibited cell migration with overexpression of PIAS, a SUMO ligase [Wang et al., 2010]. In an effort to identify potential targets of SUMO that might play a role in the inhibition of cell migration, a pull down experiment of SUMO1 was carried out. A truncated version of vimentin was co‐isolated from a nuclear fraction and identified by mass spectrometry and western blotting as a candidate [Wang et al., 2010]. Yet, many questions remain. Can full‐length vimentin be sumoylated? Is cytoplasmic vimentin modified by SUMO? Do the other isoforms of SUMO modify vimentin? What is the fate of sumoylated vimentin?
Lamins
Lamins are the major element of a meshwork that provides structural support and shape for the nucleus [Aebi et al., 1986; Belmont et al., 1993; Houben et al., 2007; Dechat et al., 2008]. A fraction of the lamins A/C population is also present in the interior of the nucleus as “speckles” [Jagatheesan et al., 1999; Kumaran et al., 2002; Adhikari et al., 2004]. Lamins participate is a variety of processes, including DNA replication, DNA repair, and transcriptional regulation owing to their ability to segregate heterochromatic domains to the inner edge of the nuclear envelope [Kumaran and Spector, 2008; Shimi et al., 2010]. Lamins also have functions in cell signaling, cell proliferation, development, and differentiation [reviewed in Dechat et al., 2008; Eriksson et al., 2009]. Mutations in the genes encoding lamins result in a class of devastating diseases called laminopathies, which include the premature aging disorder Hutchison‐Gilford Progeria, Emery‐Dreifuss muscular dystrophy, and cardiomyopathies [Sullivan et al., 1999; Sylvius and Tesson, 2006; Eriksson et al., 2009; Schreiber and Kennedy, 2013; Burke and Stewart, 2014].
There are two types of nuclear lamins, A‐type and B‐type. The A‐type lamins are encoded by a single gene in mammals, LMNA. Alternative splicing of this gene leads to expression of different proteins including lamin A and lamin C [Broers et al., 2006]. Mammals express three different B‐type lamins, which are encoded by two different genes, LNMB1 and LNMB2 [Burke and Stewart, 2014].
The first hint that lamins were sumoylated came from two‐hybrid experiments showing that lamin A interacted with the SUMO E2 conjugating enzyme, Ubc9p [Zhang et al., 2008]. Subsequent studies have since confirmed this finding, but with several noteworthy differences between them [Zhang et al., 2008; Boudreau et al., 2012; Simon et al., 2013].
The first study by Zhang and Sarge suggests that the lamin A conjugation occurs predominately with SUMO2, but not SUMO1. These authors identified lysine 201, which is located in the rod domain of lamin A as a sumoylation site [Zhang and Sarge, 2008a, 2008b]. Residing near this SUMO site are two mutations that are associated with familial dilated cardiomyopathy, E203G and E203K. Considering that the acidic residue in the canonical sumoylation consensus sequence (ψKX D/E) is important for the efficiency of sumoylation, E203G and E203K were tested for their effect on sumoylation. Both mutants exhibited significantly decreased levels of sumoylation. GFP fusions with both mutants also displayed abnormal sub‐cellular localization patterns, which were consistent with the K201R mutant. These findings are consistent with those of Boudreau et al. [2012], who also found that several lamin mutants associated with dilated cardiac myopathy were modified by SUMO1, whereas wild type lamin A or C were not. The lamin A of these myopathies was mislocalized into aggregates that also sequestered SUMO1 [Boudreau et al., 2012]. Combined, these results suggest that sumoylation plays an important role in lamin A function and implicate sumoylation in the pathology of cardiomyopathies associated with lamin malfunction.
In the second study from the Hoffman and Wilson labs, Simon et al. [2013] also demonstrated that lamin A is modified by SUMO, but the modification they observed employed SUMO1 preferentially over SUMO2. In a second point of contrast to the Sarge study, these authors showed that sumoylation of lamin A occurred on K420 and K486 in the IgG globular tail domain. Independent mass spectrometry confirmed modification at lysine 420 [Galisson et al., 2011]. These findings are in contrast to the Zhang study in which sumoylation was found in the coiled‐coil domain. The position of such a mutation in the tail domain is noteworthy because as many as 21 different lamin A binding partners interact with lamin A through its tail domain [Simon et al., 2013]. These findings led Simon et al. to propose a model where modification in the tail by SUMO1 regulates the binding of lamin A to its known partners to control the assembly of lamin A filaments.
Why the differences in these two studies? The differences in the types of SUMO used and in the sites of conjugation could be attributed to the origin of cells studied. The Sarge lab used HeLa cells and myocytes, whereas the Wilson and Hoffmann labs used Cos‐7 cells, a SV40 transformed kidney cell line derived from African green monkey. It has also been noted previously that the pattern and number of SUMO modifications can change with cell activation [Galisson et al., 2011], so the growth state of the cells used in these lamin studies could easily play a role in these differences.
In the Simon et al. study, the second site of sumoylation, K486, in the tail was somewhat unexpected because it did not lie within a canonical consensus site. After further examination of the three‐dimensional structure of the IgG fold of the lamin A tail, it was apparent that two acidic residues, E460 and D461, are positioned directly below K486 [Krimm et al., 2002]. Two other acidic residues E536 and E537 were close to K420, but on a different side. Mutation of E460 and D461, or the double mutant E460 D461, diminished sumoylation by 65–80% compared to wild type. Mutations of E536 and E537 reduced sumoylation by 30–50% showing they contribute to sumoylation of K486, but are less important than E460 and D461. These findings suggest the hypothesis that canonical consensus sites can be “conformational” in nature. These results highlight the need for the development of algorithms that identify sumoylation sites not only by linear amino acid sequences, but also by the three‐dimensional structure of the folded protein.
Laminopathies and Sumoylation
It is significant to note that several studies report abnormalities in the sumoylation patterns of patients afflicted with a range of different laminopathies. Lamin A mutations of G465 and K486 are known to cause familial partial lipodystrophy (FPLD), an adipose tissue disease characterized by decreased levels of adipose tissue. As mentioned above, the sumoylation of these mutants is decreased [Simon et al., 2013], leading these investigators to postulate that FPLD in patients with G465 or K486 mutations could arise from deficiencies in sumoylation levels. Lamin A mutations are also frequently seen in patients with dilated cardiomyopathy [Sylvius and Tesson, 2006]. In dilated cardiomyopathy disease, patients carrying a lamin C‐D192G mutation displayed a number of aberrant nuclear phenotypes and greatly reduced SUMO1 patterns [Sylvius and Tesson, 2006]. As noted above, two lamin A mutations E203G and E203K are linked with familial dilated cardiomyopathy. These mutant lamins display altered subcellular localization that match those seen in the sumoylation‐deficient lamin A mutants [Zhang et al., 2008]. Consistent with this pattern, patients with the premature aging disease, Progeria, have a defective nuclear rim staining and also display a disrupted pattern of Ubc9 localization [Kelly et al., 2011]. However, not all laminopathy mutations result in decreased levels of sumoylation. In mouse myoblasts cell lines and mouse muscle tissue, laminopathy mutations associated with dilated cardiac myopathy and Emery‐Dreisfuss muscular dystrophy resulted in intracellular lamin aggregates that had higher levels of sumoylation, as well as increased Ubc9 co‐localization [Boudreau et al., 2012].
Together, these finding are consistent with sumoylation playing a vital role in the physiology of various cell types important for human heath. The differences in these studies highlight the need for additional research into this transformative area of cell biology, while providing critical insight into the cause of human diseases associated with mutations in lamin A. Many laminopathies are known [Eriksson et al., 2009; Boudreau et al., 2012; Burke and Stewart, 2014], but the sumoylation status in most of these diseases remains unknown. This represents an important avenue for future investigations that will likely lead to a better understanding of these diseases. For instance in these various laminopathies, does sumoylation modulate the solubility of lamins as it does for keratin? Does sumoylation promote the removal of damaged lamins by a sumo‐mediated proteasome pathway? Could such a system be harnessed for the treatment of these diseases? [Liu and Zhou, 2008].
Conclusions and Perspectives
Many Questions Remain
A plethora of questions remain about the relationship between sumoylation and the cytoskeleton. How extensive is SUMO's control of cytoskeletal function? Are other cytoskeletal elements controlled by sumoylation? Could sumoylation regulate desmosomes, adherens junctions, tight junctions, or focal adhesions? Microtubule motors CENP‐E and dynein have been linked to SUMO. Are the myosin motors walking along microfilaments also controlled by sumoylation? How wide spread is the altered sumoylation status of cytoskeletal proteins in disease?
An emerging theme is the role that sumoylation plays in controlling the structure of cytoskeletal systems. Sumoylation of actin is found on just the nuclear fraction of actin. It has been speculated that this could lead to a different configuration for actin assembly since the site of sumoylation would obstruct the formation of a classical actin filament. Sumoylation also influences intermediate filament solubility. Yet, a significant gap exists in knowing how this translates into control of other aspects of filament dynamics. Can sumoylation influence the disassembly of pre‐assembled intermediate filament networks? To what extent is the assembly/disassembly or solubility of other cytoskeletal polymers regulated by sumoylation?
Network Connectors
Much of this review discusses the four polymer networks of the cytoskeleton as separate entities, but in fact, several connections are known between these networks. These connections allow for inter‐network communication that could, in theory, be modulated by sumoylation. The sumoylation of Kar9p, a linker between the actin and microtubule networks, supports this contention. The possibility of other linkers being regulated by sumoylation is just now beginning to emerge. For instance, plectin is a very large protein of the plakin family that plays a major role in cytoskeletal organization by providing linkages between the three major cytoskeletal networks; actin, microtubules and intermediate filaments [Svitkina et al., 1996; Wiche, 1998; Sonnenberg and Liem, 2007; Winter and Wiche, 2013; Bouameur et al., 2014]. In skin cells, plectin is an essential part of the hemidesmosome, a junction that plays a major role in anchoring the outer epithelial layer of the skin to the underlying dermal layer [Andra et al., 2003]. It does this in part by attaching keratin intermediate filaments to the basal membrane of cells in the basal cell layer [Wiche, 1998]. Plectin is also a component of desmosomes, which form junctions between neighboring cells [Huber, 2003]. It is intriguing that a recent sumoylation proteomics screen recently identified plectin as a potential substrate for SUMO2 [Wen et al., 2014]. Although further research is needed to confirm this interaction, this opens up a new research avenue that could impact multiple cytoskeletal systems simultaneously. This would provide needed insight into how signaling between the cytoskeletal networks might be coordinated.
Crosstalk With Other Signal Transduction Systems
While many unknowns exist about the relationship of sumoylation with the cytoskeleton, perhaps the biggest unknown is how cytoskeletal sumoylation is integrated with other signal transduction pathways. Can sumoylation be a mechanism of transmitting information between different cell cycle checkpoints, DNA repair, stress response pathways, and transcription to the cytoskeleton?
Sumoylation of many targets can be either increased or decreased with a variety of cellular stresses [Golebiowski et al., 2009; Ren et al., 2014]. Does cellular stresses alter the sumoylation of the entire cytoskeleton? And if so, which stresses? Some work on cellular stress affecting cytoskeletal sumoylation has been done for the intermediate filaments, but little has been done in this regard for the other networks. Does cytoskeletal sumoylation generate crosstalk with other signaling cascades? What other signals are responsible for changes in the sumoylation of the cytoskeleton? A clear understanding these questions is still in its infancy, and sumoylation of the cytoskeleton will certainly be an exciting chapter of new research for years to come.
Acknowledgments
This work was supported in part by funding to R.K.M. from the National Science Foundation (#MCB‐1052174), the Oklahoma Health Research Program of the Oklahoma Center for the Advancement of Science and Technology (OCAST #HR09‐150S), and funds from O.S.U. and the Oklahoma Agricultural Experiment Station, project numbers OKL02715 and OKL02961. A.A. is supported in part by a fellowship from the Sloan Foundation. K.J.D. was supported at O.S.U. by a Niblack Research Scholarship and a Wentz Research Scholarship. We thank Drs. Omar Skalli, Amy Gladfelter, Holly Goodson, and Oliver Kerscher for their critical reading of this manuscript. We also thank the reviewers for their helpful comments. The authors declare that they have no conflict of interest to report.
Monitoring Editor: Bruce Goode
References
- Adhikari AS, Sridhar Rao K, Rangaraj N, Parnaik VK, Mohan Rao CH. 2004. Heat stress‐induced localization of small heat shock proteins in mouse myoblasts: intranuclear lamin A/C speckles as target for alphaB‐crystallin and Hsp25. Exp Cell Res 299:393–403. [DOI] [PubMed] [Google Scholar]
- Aebi U, Cohn J, Buhle L, Gerace L. 1986. The nuclear lamina is a meshwork of intermediate‐type filaments. Nature 323:560–564. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Steinmetz MO. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 9:309–322. [DOI] [PubMed] [Google Scholar]
- Akyurek N, Ren Y, Rassidakis GZ, Schlette EJ, Medeiros LJ. 2006. Expression of inhibitor of apoptosis proteins in B‐cell non‐Hodgkin and Hodgkin lymphomas. Cancer 107:1844–1851. [DOI] [PubMed] [Google Scholar]
- Al‐Bassam J, Ozer RS, Safer D, Halpain S, Milligan RA. 2002. MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J Cell Biol 157:1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C, Borisy GG. 1974. Structural polarity and directional growth of microtubules of Chlamydomonas flagella. J Mol Biol 90:381–402. [DOI] [PubMed] [Google Scholar]
- Alonso A, D'Silva S, Rahman M, Meluh PB, Keeling J, Meednu N, Hoops HJ, Miller RK. 2012. The yeast homologue of the microtubule‐associated protein Lis1 interacts with the sumoylation machinery and a SUMO‐targeted ubiquitin ligase. Mol Biol Cell 23:4552–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andra K, Kornacker I, Jorgl A, Zorer M, Spazierer D, Fuchs P, Fischer I, Wiche G. 2003. Plectin‐isoform‐specific rescue of hemidesmosomal defects in plectin (‐/‐) keratinocytes. J Invest Dermatol 120:189–197. [DOI] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. 1998. Regulation of actin dynamics through phosphorylation of cofilin by LIM‐kinase. Nature 393:805–809. [DOI] [PubMed] [Google Scholar]
- Arin MJ, Longley MA, Anton‐Lamprecht I, Kurze G, Huber M, Hohl D, Rothnagel JA, Roop DR. 1999. A novel substitution in keratin 10 in epidermolytic hyperkeratosis. J Invest Dermatol 112:506–508. [DOI] [PubMed] [Google Scholar]
- Arin MJ, Longley MA, Epstein EH, Jr. , Rothnagel JA, Roop DR. 2000. Identification of a novel mutation in keratin 1 in a family with epidermolytic hyperkeratosis. Exp Dermatol 9:16–19. [DOI] [PubMed] [Google Scholar]
- Arnaud LT, Myeku N, Figueiredo‐Pereira ME. 2009. Proteasome‐caspase‐cathepsin sequence leading to tau pathology induced by prostaglandin J2 in neuronal cells. J Neurochem 110:328–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asante D, Stevenson NL, Stephens DJ. 2014. Subunit composition of the human cytoplasmic dynein‐2 complex. J Cell Sci 127:4774–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Dasso M. 2002. A new clue at the nuclear pore: RanBP2 is an E3 enzyme for SUMO1. Dev Cell 2:130–131. [DOI] [PubMed] [Google Scholar]
- Bader JR, Vaughan KT. 2010. Dynein at the kinetochore: timing, interactions and functions. Semin Cell Dev Biol 21:269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancher C, Grundke‐Iqbal I, Iqbal K, Fried VA, Smith HT, Wisniewski HM. 1991. Abnormal phosphorylation of tau precedes ubiquitination in neurofibrillary pathology of Alzheimer disease. Brain Res 539:11–18. [DOI] [PubMed] [Google Scholar]
- Bawa‐Khalfe T, Yeh ET. 2010. SUMO losing balance: SUMO proteases disrupt SUMO homeostasis to facilitate cancer development and progression. Genes Cancer 1:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Becker J. 1998. Structure determination of the small ubiquitin‐related modifier SUMO‐1. J Mol Biol 280:275–286. [DOI] [PubMed] [Google Scholar]
- Beach DL, Thibodeaux J, Maddox P, Yeh E, Bloom K. 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr Biol 10:1497–1506. [DOI] [PubMed] [Google Scholar]
- Beise N, Trimble W. 2011. Septins at a glance. J Cell Sci 124:4141–4146. [DOI] [PubMed] [Google Scholar]
- Belin BJ, Mullins RD. 2013. What we talk about when we talk about nuclear actin. Nucleus 4:291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS, Zhai Y, Thilenius A. 1993. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol 123:1671–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencsath KP, Podgorski MS, Pagala VR, Slaughter CA, Schulman BA. 2002. Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J Biol Chem 277:47938–47945. [DOI] [PubMed] [Google Scholar]
- Bergen LG, Borisy GG. 1980. Head‐to‐tail polymerization of microtubules in vitro. Electron microscope analysis of seeded assembly. J Cell Biol 84:141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Jentsch S. 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458:461–467. [DOI] [PubMed] [Google Scholar]
- Berlin V, Styles CA, Fink GR. 1990. BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiae, colocalizes with tubulin. J Cell Biol 111:2573–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, Nogales E. 2012. Septin filament organization in Saccharomyces cerevisiae . Commun Integr Biol 5:503–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Grob P, Park SS, Garcia G, 3rd , Patanwala I, Ng HL, Alber T, Thorner J, Nogales E. 2008. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci USA 105:8274–8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Pierson J, Thai L, McDonald KL, Zehr EA, Garcia G, 3rd , Peters P, Thorner J, Nogales E. 2012. Three‐dimensional ultrastructure of the septin filament network in Saccharomyces cerevisiae . Mol Biol Cell 23:423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettermann K, Benesch M, Weis S, Haybaeck J. 2012. SUMOylation in carcinogenesis. Cancer Lett 316:113–125. [DOI] [PubMed] [Google Scholar]
- Bhabha G, Cheng HC, Zhang N, Moeller A, Liao M, Speir JA, Cheng Y, Vale RD. 2014. Allosteric communication in the dynein motor domain. Cell 159:857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M. 2001. Spindles cotton on to junctions, APC and EB1. Nat Cell Biol 3:E67–E68. 6 [DOI] [PubMed] [Google Scholar]
- Biggins S, Bhalla N, Chang A, Smith DL, Murrary AW. 2001. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae . Genetics 159:453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder LI, Frankfurter A, Rebhun LI. 1985. The distribution of tau in the mammalian central nervous system. J Cell Biol 101:1371–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake‐Hodek KA, Cassimeris L, Huffaker TC. 2010. Regulation of microtubule dynamics by Bim1 and Bik1, the budding yeast members of the EB1 and CLIP‐170 families of plus‐end tracking proteins. Mol Biol Cell 21:2013–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blennow K, Zetterberg H, Fagan AM. 2012. Fluid biomarkers in Alzheimer disease. Cold Spring Harb Perspect Med 2:a006221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomster HA, Hietakangas V, Wu J, Kouvonen P, Hautaniemi S, Sistonen L. 2009. Novel proteomics strategy brings insight into the prevalence of SUMO‐2 target sites. Mol Cell Proteomics 8:1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifas JM, Rothman AL, Epstein EH, Jr. 1991. Epidermolysis bullosa simplex: evidence in two families for keratin gene abnormalities. Science 254:1202–1205. [DOI] [PubMed] [Google Scholar]
- Bouameur JE, Favre B, Borradori L. 2014. Plakins, a versatile family of cytolinkers: roles in skin integrity and in human diseases. J Invest Dermatol 134:885–894. [DOI] [PubMed] [Google Scholar]
- Boudreau E, Labib S, Bertrand AT, Decostre V, Bolongo PM, Sylvius N, Bonne G, Tesson F. 2012. Lamin A/C mutants disturb sumo1 localization and sumoylation in vitro and in vivo. PLoS One 7:e45918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. 2006. Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev 86:967–1008. [DOI] [PubMed] [Google Scholar]
- Bulbarelli A, Lonati E, Cazzaniga E, Gregori M, Masserini M. 2009. Pin1 affects tau phosphorylation in response to Abeta oligomers. Mol Cell Neurosci 42:75–80. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. 2014. Functional architecture of the cell's nucleus in development, aging, and disease. Curr Top Dev Biol 109:1–52. [DOI] [PubMed] [Google Scholar]
- Burstein DE, Idrees MT, Li G, Wu M, Kalir T. 2008. Immunohistochemical detection of the X‐linked inhibitor of apoptosis protein (XIAP) in cervical squamous intraepithelial neoplasia and squamous carcinoma. Ann Diagn Pathol 12:85–89. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. 1976. A highly ordered ring of membrane‐associated filaments in budding yeast. J Cell Biol 69:717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylebyl GR, Belichenko I, Johnson ES. 2003. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem 278:44113–44120. [DOI] [PubMed] [Google Scholar]
- Carberry K, Wiesenfahrt T, Windoffer R, Bossinger O, Leube RE. 2009. Intermediate filaments in Caenorhabditis elegans . Cell Motil Cytoskeleton 66:852–864. [DOI] [PubMed] [Google Scholar]
- Carter AP, Garbarino JE, Wilson‐Kubalek EM, Shipley WE, Cho C, Milligan RA, Vale RD, Gibbons IR. 2008. Structure and functional role of dynein's microtubule‐binding domain. Science 322:1691–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. 2011. Crystal structure of the dynein motor domain. Science 331:1159–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Gupta MLJ, Hoyt MA, Pellman D. 2004. Cell cycle control of kinesin‐mediated transport of Bik1(CLIP170) regulates microtubule stability and dynein activation. Dev Cell 6:815–829. [DOI] [PubMed] [Google Scholar]
- Cassimeris L, Pryer NK, Salmon ED. 1988. Real‐time observations of microtubule dynamic instability in living cells. J Cell Biol 107:2223–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo‐Lluva S, Tatham MH, Jones RC, Jaffray EG, Edmondson RD, Hay RT, Malliri A. 2010. SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat Cell Biol 12:1078–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron F, Barral Y. 2009. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell 16:493–506. [DOI] [PubMed] [Google Scholar]
- Caudron F, Andrieux A, Job D, Boscheron C. 2008. A new role for kinesin‐directed transport of Bik1p (CLIP‐170) in Saccharomyces cerevisiae . J Cell Sci 121:1506–1513. [DOI] [PubMed] [Google Scholar]
- Cerveira N, Bizarro S, Teixeira MR. 2011. MLL‐SEPTIN gene fusions in hematological malignancies. Biol Chem 392:713–724. [DOI] [PubMed] [Google Scholar]
- Chapin SJ, Bulinski JC. 1991. Non‐neuronal 210 x 10(3) Mr microtubule‐associated protein (MAP4) contains a domain homologous to the microtubule‐binding domains of neuronal MAP2 and tau. J Cell Sci 98:27–36. [DOI] [PubMed] [Google Scholar]
- Cheerambathur DK, Desai A. 2014. Linked in: formation and regulation of microtubule attachments during chromosome segregation. Curr Opin Cell Biol 26:113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson‐Kubalek EM, Desai A. 2006. The conserved KMN network constitutes the core microtubule‐binding site of the kinetochore. Cell 127:983–997. [DOI] [PubMed] [Google Scholar]
- Chen H, Howell AS, Robeson A, Lew DJ. 2011. Dynamics of septin ring and collar formation in Saccharomyces cerevisiae . Biol Chem 392:689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. 2013. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 93:269–309. [DOI] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. 2010. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol 11:62–74. [DOI] [PubMed] [Google Scholar]
- Chipev CC, Yang JM, DiGiovanna JJ, Steinert PM, Marekov L, Compton JG, Bale SJ. 1994. Preferential sites in keratin 10 that are mutated in epidermolytic hyperkeratosis. Am J Hum Genet 54:179–190. [PMC free article] [PubMed] [Google Scholar]
- Chretien D, Fuller SD, Karsenti E. 1995. Structure of growing microtubule ends: two‐dimensional sheets close into tubes at variable rates. J Cell Biol 129:1311–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BM, Rotty JD, Coulombe PA. 2013. Networking galore: intermediate filaments and cell migration. Curr Opin Cell Biol 25:600–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citro S, Chiocca S. 2013. Sumo paralogs: redundancy and divergencies. Front Biosci (Schol Ed) 5:544–553. [DOI] [PubMed] [Google Scholar]
- Cleveland DW. 1982. Treadmilling of tubulin and actin. Cell 28:689–691. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Hwo SY, Kirschner MW. 1977. Purification of tau, a microtubule‐associated protein that induces assembly of microtubules from purified tubulin. J Mol Biol 116:207–225. [DOI] [PubMed] [Google Scholar]
- Collins ES, Balchand SK, Faraci JL, Wadsworth P, Lee WL. 2012. Cell cycle‐regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol Biol Cell 23:3380–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C, Caceres A. 2009. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci 10:319–332. [DOI] [PubMed] [Google Scholar]
- Connolly D, Abdesselam I, Verdier‐Pinard P, Montagna C. 2011. Septin roles in tumorigenesis. Biol Chem 392:725–738. [DOI] [PubMed] [Google Scholar]
- Cook CE, Hochstrasser M, Kerscher O. 2009. The SUMO‐targeted ubiquitin ligase subunit Slx5 resides in nuclear foci and at sites of DNA breaks. Cell Cycle 8:1080–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe PA, Fuchs E. 1993. Epidermolysis bullosa simplex. Semin Dermatol 12:173–190. [PubMed] [Google Scholar]
- Coulombe PA, Hutton ME, Letai A, Hebert A, Paller AS, Fuchs E. 1991. Point mutations in human keratin 14 genes of epidermolysis bullosa simplex patients: genetic and functional analyses. Cell 66:1301–1311. [DOI] [PubMed] [Google Scholar]
- Cubenas‐Potts C, Goeres JD, Matunis MJ. 2013. SENP1 and SENP2 affect spatial and temporal control of sumoylation in mitosis. Mol Biol Cell 24:3483–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M. 2008. Emerging roles of the SUMO pathway in mitosis. Cell Div 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DC, Layfield R, Serpell L, Narain Y, Goedert M, Spillantini MG. 2002. Proteasomal degradation of tau protein. J Neurochem 83:176–185. [DOI] [PubMed] [Google Scholar]
- de The H, Le Bras M, Lallemand‐Breitenbach V. 2012. The cell biology of disease: acute promyelocytic leukemia, arsenic, and PML bodies. J Cell Biol 198:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. 2008. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev 22:832–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacourte A, David JP, Sergeant N, Buee L, Wattez A, Vermersch P, Ghozali F, Fallet‐Bianco C, Pasquier F, Lebert F, Petit H, Di Menza C. 1999. The biochemical pathway of neurofibrillary degeneration in aging and Alzheimer's disease. Neurology 52:1158–1165. [DOI] [PubMed] [Google Scholar]
- DerMardirossian C, Rocklin G, Seo JY, Bokoch GM. 2006. Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol‐membrane cycling. Mol Biol Cell 17:4760–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. 1997. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13:83–117. [DOI] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT. 1998. SUMO‐1 modification of IkappaBalpha inhibits NF‐kappaB activation. Mol Cell 2:233–239. [DOI] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Kemp GD, Hay RT. 1999. Identification of the enzyme required for activation of the small ubiquitin‐like protein SUMO‐1. J Biol Chem 274:10618–10624. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Barral Y. 2004. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science 305:393–396. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. 1995. An essential yeast gene encoding a homolog of ubiquitin‐activating enzyme. J Biol Chem 270:18099–18109. [DOI] [PubMed] [Google Scholar]
- Dopie J, Skarp KP, Rajakyla EK, Tanhuanpaa K, Vartiainen MK. 2012. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci USA 109:E544–E552. 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorval V, Fraser PE. 2006. Small ubiquitin‐like modifier (SUMO) modification of natively unfolded proteins tau and alpha‐synuclein. J Biol Chem 281:9919–9924. [DOI] [PubMed] [Google Scholar]
- Dorval V, Fraser PE. 2007. SUMO on the road to neurodegeneration. Biochim Biophys Acta 1773:694–706. [DOI] [PubMed] [Google Scholar]
- dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. 2003. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol Rev 83:433–473. [DOI] [PubMed] [Google Scholar]
- Dovas A, Couchman JR. 2005. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem J 390:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drag M, Salvesen GS. 2008. DeSUMOylating enzymes–SENPs. IUBMB Life 60:734–742. [DOI] [PubMed] [Google Scholar]
- Drechsel DN, Hyman AA, Cobb MH, Kirschner MW. 1992. Modulation of the dynamic instability of tubulin assembly by the microtubule‐associated protein tau. Mol Biol Cell 3:1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G, Trinczek B, Illenberger S, Biernat J, Schmitt‐Ulms G, Meyer HE, Mandelkow EM, Mandelkow E. 1995. Microtubule‐associated protein/microtubule affinity‐regulating kinase (p110mark). A novel protein kinase that regulates tau‐microtubule interactions and dynamic instability by phosphorylation at the Alzheimer‐specific site serine 262. J Biol Chem 270:7679–7688. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Kirschner MW. 1986. Tau protein function in living cells. J Cell Biol 103:2739–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Holmes WB, Ye H. 2011. Interaction mapping between Saccharomyces cerevisiae Smc5 and SUMO E3 ligase Mms21. Biochemistry 50:10182–10188. [DOI] [PubMed] [Google Scholar]
- Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. 1999. Analysis of dynactin subcomplexes reveals a novel actin‐related protein associated with the arp1 minifilament pointed end. J Cell Biol 147:307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhofer TA, Villen J, McCusker D, Gygi SP, Kellogg DR. 2008. The septins function in G1 pathways that influence the pattern of cell growth in budding yeast. PLoS One 3:e2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson C, Sahlgren C, Berthold CH, Stakeberg J, Celis JE, Betsholtz C, Eriksson JE, Pekny M. 1999. Intermediate filament protein partnership in astrocytes. J Biol Chem 274:23996–24006. [DOI] [PubMed] [Google Scholar]
- Elmore ZC, Donaher M, Matson BC, Murphy H, Westerbeck JW, Kerscher O. 2011. SUMO‐dependent substrate targeting of the SUMO protease Ulp1. BMC Biol 9:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson JE, Dechat T, Grin B, Helfand B, Mendez M, Pallari HM, Goldman RD. 2009. Introducing intermediate filaments: from discovery to disease. J Clin Invest 119:1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erker Y, Neyret‐Kahn H, Seeler JS, Dejean A, Atfi A, Levy L. 2013. Arkadia, a novel SUMO‐targeted ubiquitin ligase involved in PML degradation. Mol Cell Biol 33:2163–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne‐Manneville S. 2010. From signaling pathways to microtubule dynamics: the key players. Curr Opin Cell Biol 22:104–111. [DOI] [PubMed] [Google Scholar]
- Etienne‐Manneville S. 2013. Microtubules in cell migration. Annu Rev Cell Dev Biol 29:471–499. [DOI] [PubMed] [Google Scholar]
- Eves PT, Jin Y, Brunner M, Weisman LS. 2012. Overlap of cargo binding sites on myosin V coordinates the inheritance of diverse cargoes. J Cell Biol 198:69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Weissman AM. 2004. A field guide to ubiquitylation. Cell Mol Life Sci 61:1546–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O'Connell CB, Wang YL, Vallee RB. 2000. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol 2:784–791. [DOI] [PubMed] [Google Scholar]
- Ferreira JG, Pereira AL, Maiato H. 2014. Microtubule plus‐end tracking proteins and their roles in cell division. Int Rev Cell Mol Biol 309:59–140. [DOI] [PubMed] [Google Scholar]
- Field CM, al‐Awar O, Rosenblatt J, Wong ML, Alberts B, Mitchison TJ. 1996. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J Cell Biol 133:605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, Raman N, Muller S. 2011. SUMO routes ribosome maturation. Nucleus 2:527–532. [DOI] [PubMed] [Google Scholar]
- Firat‐Karalar EN, Welch MD. 2011. New mechanisms and functions of actin nucleation. Curr Opin Cell Biol 23:4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D, Mukrasch MD, Biernat J, Bibow S, Blackledge M, Griesinger C, Mandelkow E, Zweckstetter M. 2009. Conformational changes specific for pseudophosphorylation at serine 262 selectively impair binding of tau to microtubules. Biochemistry 48:10047–10055. [DOI] [PubMed] [Google Scholar]
- Flotho A, Werner A. 2012. The RanBP2/RanGAP1*SUMO1/Ubc9 complex: a multisubunit E3 ligase at the intersection of sumoylation and the RanGTPase cycle. Nucleus 3:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker ES, Baker BM, Goodson HV. 2005. Interactions between CLIP‐170, tubulin, and microtubules: implications for the mechanism of CLIP‐170 plus‐end tracking behavior. Mol Biol Cell 16:5373–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford SK, Pringle JR. 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev Genet 12:281–292. [DOI] [PubMed] [Google Scholar]
- Forget MA, Desrosiers RR, Gingras D, Beliveau R. 2002. Phosphorylation states of Cdc42 and RhoA regulate their interactions with Rho GDP dissociation inhibitor and their extraction from biological membranes. Biochem J 361:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke WW, Schmid E, Osborn M, Weber K. 1978. Different intermediate‐sized filaments distinguished by immunofluorescence microscopy. Proc Natl Acad Sci USA 75:5034–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. 1995. Keratins and the skin. Annu Rev Cell Dev Biol 11:123–153. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Cleveland DW. 1998. A structural scaffolding of intermediate filaments in health and disease. Science 279:514–519. [DOI] [PubMed] [Google Scholar]
- Gaglio T, Dionne MA, Compton DA. 1997. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J Cell Biol 138:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galisson F, Mahrouche L, Courcelles M, Bonneil E, Meloche S, Chelbi‐Alix MK, Thibault P. 2011. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol Cell Proteomics 10:M110.004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galou M, Colucci‐Guyon E, Ensergueix D, Ridet JL, Gimenez y Ribotta M, Privat A, Babinet C, Dupouey P. 1996. Disrupted glial fibrillary acidic protein network in astrocytes from vimentin knockout mice. J Cell Biol 133:853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces JA, Clark IB, Meyer DI, Vallee RB. 1999. Interaction of the p62 subunit of dynactin with Arp1 and the cortical actin cytoskeleton. Curr Biol 9:1497–1500. [DOI] [PubMed] [Google Scholar]
- Garcia G, 3rd , Bertin A, Li Z, Song Y, McMurray MA, Thorner J, Nogales E. 2011. Subunit‐dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. J Cell Biol 195:993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Domiquez M, Reyes JC. 2009. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta 1789:451–459. [DOI] [PubMed] [Google Scholar]
- Gardel ML, Schneider IC, Aratyn‐Schaus Y, Waterman CM. 2010. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol 26:315–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Hunt AJ, Goodson HV, Odde DJ. 2008. Microtubule assembly dynamics: new insights at the nanoscale. Curr Opin Cell Biol 20:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau JR, Lima CD. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR. 1995. Dynein family of motor proteins: present status and future questions. Cell Motil Cytoskeleton 32:136–144. [DOI] [PubMed] [Google Scholar]
- Gillett ES, Espelin CW, Sorger PK. 2004. Spindle checkpoint proteins and chromosome‐microtubule attachment in budding yeast. J Cell Biol 164:535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies J, Hochstrasser M. 2012. A new class of SUMO proteases. EMBO Rep 13:284–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter AS. 2010. Guides to the final frontier of the cytoskeleton: septins in filamentous fungi. Curr Opin Microbiol 13:720–726. [DOI] [PubMed] [Google Scholar]
- Gladfelter AS, Pringle JR, Lew DJ. 2001. The septin cortex at the yeast mother‐bud neck. Curr Opin Microbiol 4:681–689. [DOI] [PubMed] [Google Scholar]
- Goedert M. 2001. The significance of tau and alpha‐synuclein inclusions in neurodegenerative diseases. Curr Opin Genet Dev 11:343–351. [DOI] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. 1989a. Multiple isoforms of human microtubule‐associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron 3:519–526. [DOI] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG, Potier MC, Ulrich J, Crowther RA. 1989b. Cloning and sequencing of the cDNA encoding an isoform of microtubule‐associated protein tau containing four tandem repeats: differential expression of tau protein mRNAs in human brain. EMBO J 8:393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Baur CP, Ahringer J, Jakes R, Hasegawa M, Spillantini MG, Smith MJ, Hill F. 1996. PTL‐1, a microtubule‐associated protein with tau‐like repeats from the nematode Caenorhabditis elegans . J Cell Sci 109:2661–2672. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Grin B, Mendez MG, Kuczmarski ER. 2008. Intermediate filaments: versatile building blocks of cell structure. Curr Opin Cell Biol 20:28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Cleland MM, Murthy SN, Mahammad S, Kuczmarski ER. 2012. Inroads into the structure and function of intermediate filament networks. J Struct Biol 177:14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. 2009. System‐wide changes to SUMO modifications in response to heat shock. Sci Signal 2:ra24. [DOI] [PubMed] [Google Scholar]
- Goley ED, Welch MD. 2006. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol 7:713–726. [DOI] [PubMed] [Google Scholar]
- Goodson HV, Skube SB, Stalder R, Valetti C, Kreis TE, Morrison EE, Schroer TA. 2003. CLIP‐170 interacts with dynactin complex and the APC‐binding protein EB1 by different mechanisms. Cell Motil Cytoskeleton 55:156–173. [DOI] [PubMed] [Google Scholar]
- Grissom PM, Vaisberg EA, McIntosh JR. 2002. Identification of a novel light intermediate chain (D2LIC) for mammalian cytoplasmic dynein 2. Mol Biol Cell 13:817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio, L , Robertson M, et al. 1991. Identification and characterization of the familial adenomatous polyposis coli gene. Cell 66:589–600. [DOI] [PubMed] [Google Scholar]
- Grundke‐Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. 1986. Abnormal phosphorylation of the microtubule‐associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci USA 83:4913–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. 1997. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol 9:383–387. [DOI] [PubMed] [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. 2008. Kinetochore‐microtubule attachment relies on the disordered N‐terminal tail domain of Hec1. Curr Biol 18:1778–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG. 2002. Evolutionary conservation of microtubule‐capture mechanisms. Nat Rev Mol Cell Biol 3:296–304. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Bretscher A. 2003. Microtubule asymmetry. Science 300:2040. [DOI] [PubMed] [Google Scholar]
- Gupta KK, Joyce MV, Slabbekoorn AR, Zhu ZC, Paulson BA, Boggess B, Goodson HV. 2010. Probing interactions between CLIP‐170, EB1, and microtubules. J Mol Biol 395:1049–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KK, Li C, Duan A, Alberico EO, Kim OV, Alber MS, Goodson HV. 2013. Mechanism for the catastrophe‐promoting activity of the microtubule destabilizer Op18/stathmin. Proc Natl Acad Sci USA 110:20449–20454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KK, Alberico EO, Nathke IS, Goodson HV. 2014. Promoting microtubule assembly: a hypothesis for the functional significance of the +TIP network. Bioessays 36:818–826. [DOI] [PubMed] [Google Scholar]
- Haarer BK, Pringle JR. 1987. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10‐nm filaments in the mother‐bud neck. Mol Cell Biol 7:3678–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M. 2005. Defining the SUMO‐modified proteome by multiple approaches in Saccharomyces cerevisiae . J Biol Chem 280:4102–4110. [DOI] [PubMed] [Google Scholar]
- Hao YH, Doyle JM, Ramanathan S, Gomez TS, Jia D, Xu M, Chen ZJ, Billadeau DD, Rosen MK, Potts PR. 2013. Regulation of WASH‐dependent actin polymerization and protein trafficking by ubiquitination. Cell 152:1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH. 1971. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res 69:265–276. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. 1974. Genetic control of the cell division cycle in yeast. Science 183:46–51. [DOI] [PubMed] [Google Scholar]
- Hattori M, Adachi H, Tsujimoto M, Arai H, Inoue K. 1994. Miller‐Dieker lissencephaly gene encodes a subunit of brain platelet‐activating factor acetylhydrolase [corrected]. Nature 370:216–218. [DOI] [PubMed] [Google Scholar]
- Hay RT. 2013. Decoding the SUMO signal. Biochem Soc Trans 41:463–473. [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. 2006. Specification of SUMO1‐ and SUMO2‐interacting motifs. J Biol Chem 281:16117–16127. [DOI] [PubMed] [Google Scholar]
- Heideker J, Prudden J, Perry JJ, Tainer JA, Boddy MN. 2011. SUMO‐targeted ubiquitin ligase, Rad60, and Nse2 SUMO ligase suppress spontaneous Top1‐mediated DNA damage and genome instability. PLoS Genet 7:e1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand BT, Mikami A, Vallee RB, Goldman RD. 2002. A requirement for cytoplasmic dynein and dynactin in intermediate filament network assembly and organization. J Cell Biol 157:795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand BT, Mendez MG, Murthy SN, Shumaker DK, Grin B, Mahammad S, Aebi U, Wedig T, Wu YI, Hahn KM, Inagaki M, Herrmann H, Goldman RD. 2011. Vimentin organization modulates the formation of lamellipodia. Mol Biol Cell 22:1274–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Seftor RE, Trevor KT. 1997. Experimental co‐expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am J Pathol 150:483–495. [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ. 2014. The F‐act's of nuclear actin. Curr Opin Cell Biol 28C:84–89. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. 2004. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu Rev Biochem 73:749–789. [DOI] [PubMed] [Google Scholar]
- Hickey CM, Wilson NR, Hochstrasser M. 2012. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 13:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, Sistonen L. 2003. Phosphorylation of serine 303 is a prerequisite for the stress‐inducible SUMO modification of heat shock factor 1. Mol Cell Biol 23:2953–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. 2006. PDSM, a motif for phosphorylation‐dependent SUMO modification. Proc Natl Acad Sci USA 103:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD. 2001. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu Rev Biochem 70:649–676. [DOI] [PubMed] [Google Scholar]
- Ho CW, Chen HT, Hwang J. 2011. UBC9 autosumoylation negatively regulates sumoylation of septins in Saccharomyces cerevisiae . J Biol Chem 286:21826–21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. 2001. SP‐RING for SUMO: new functions bloom for a ubiquitin‐like protein. Cell 107:5–8. [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfaner b, Moldovan GL, Pyrowolakis G, Jentsch S. 2002. RAD6‐dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135–141. [DOI] [PubMed] [Google Scholar]
- Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, Hozak P, de Lanerolle P. 2004. Actin is part of pre‐initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol 6:1094–1101. [DOI] [PubMed] [Google Scholar]
- Hofmann WA, Arduini A, Nicol SM, Camacho CJ, Lessard JL, Fuller‐Pace FV, de Lanerolle P. 2009. SUMOylation of nuclear actin. J Cell Biol 186:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzbaur EL, Vallee RB. 1994. DYNEINS: molecular structure and cellular function. Annu Rev Cell Biol 10:339–372. [DOI] [PubMed] [Google Scholar]
- Hoppe JB, Rattray M, Tu H, Salbego CG, Cimarosti H. 2013. SUMO‐1 conjugation blocks beta‐amyloid‐induced astrocyte reactivity. Neurosci Lett 546:51–56. [DOI] [PubMed] [Google Scholar]
- Horiguchi T, Uryu K, Giasson BI, Ischiropoulos H, LightFoot R, Bellmann C, Richter‐Landsberg C, Lee VM, Trojanowski JQ. 2003. Nitration of tau protein is linked to neurodegeneration in tauopathies. Am J Pathol 163:1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben F, Ramaekers FC, Snoeckx LH, Broers JL. 2007. Role of nuclear lamina‐cytoskeleton interactions in the maintenance of cellular strength. Biochim Biophys Acta 1773:675–686. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. 2003. Dynamics and mechanics of the microtubule plus end. Nature 422:753–758. [DOI] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol 155:1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YL, Kuo HY, Chang CC, Naik MT, Liao PH, Ho CC, Huang TC, Jeng JC, Hsu PH, Tsai MD, Huang TH, Shih HM. 2013. Ubc9 acetylation modulates distinct SUMO target modification and hypoxia response. EMBO J 32:791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Wu S, Hernandez N. 2004. A role for beta‐actin in RNA polymerase III transcription. Genes Dev 18:3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Roberts AJ, Leschziner AE, Reck‐Peterson SL. 2012. Lis1 acts as a “clutch” between the ATPase and microtubule‐binding domains of the dynein motor. Cell 150:975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TT, Wuerzberger‐Davis SM, Wu ZH, Miyamoto S. 2003. Sequential modification of NEMO/IKKgamma by SUMO‐1 and ubiquitin mediates NF‐kappaB activation by genotoxic stress. Cell 115:565–576. [DOI] [PubMed] [Google Scholar]
- Huber O. 2003. Structure and function of desmosomal proteins and their role in development and disease. Cell Mol Life Sci 60:1872–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huls D, Storchova Z, Niessing D. 2012. Post‐translational modifications regulate assembly of early spindle orientation complex in yeast. J Biol Chem 287:16238–16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang ES, Kusch J, Barral Y, Huffaker TC. 2003. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J Cell Biol 161:483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M, Yamasaki N, Hagiwara A, Tanigaki A, Kitano A, Hikawa R, Tomimoto H, Noda M, Takanashi M, Mori H, Hattori N, Miyakawa T, Kinoshita M. 2007. Sept4, a component of presynaptic scaffold and Lewy bodies, is required for the suppression of alpha‐synuclein neurotoxicity. Neuron 53:519–533. [DOI] [PubMed] [Google Scholar]
- Ihara Y, Nukina N, Miura R, Ogawara M. 1986. Phosphorylated tau protein is integrated into paired helical filaments in Alzheimer's disease. J Biochem 99:1807–1810. [DOI] [PubMed] [Google Scholar]
- Iida K, Yahara I. 1986. Reversible induction of actin rods in mouse C3H‐2K cells by incubation in salt buffers and by treatment with non‐ionic detergents. Exp Cell Res 164:492–506. [DOI] [PubMed] [Google Scholar]
- Iida K, Iida H, Yahara I. 1986. Heat shock induction of intranuclear actin rods in cultured mammalian cells. Exp Cell Res 165:207–215. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Bischoff R, Holtzer H. 1968. Mitosis and intermediate‐sized filaments in developing skeletal muscle. J Cell Biol 38:538–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isomura M, Kikuchi A, Ohga N, Takai Y. 1991. Regulation of binding of rhoB p20 to membranes by its specific regulatory protein, GDP dissociation inhibitor. Oncogene 6:119–124. [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. 2004. Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle 3:1440–1450. [DOI] [PubMed] [Google Scholar]
- Jagatheesan G, Thanumalayan S, Muralikrishna B, Rangaraj N, Karande AA, Parnaik VK. 1999. Colocalization of intranuclear lamin foci with RNA splicing factors. J Cell Sci 112:4651–4661. [DOI] [PubMed] [Google Scholar]
- Jain D, Cooper JP. 2010. Telomeric strategies: means to an end. Annu Rev Genet 44:243–269. [DOI] [PubMed] [Google Scholar]
- Janke C. 2014. The tubulin code: molecular components, readout mechanisms, and functions. J Cell Biol 206:461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. 2004. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol 20:1–28. [DOI] [PubMed] [Google Scholar]
- Jing R, Wilhelmsson U, Goodwill W, Li L, Pan Y, Pekny M, Skalli O. 2007. Synemin is expressed in reactive astrocytes in neurotrauma and interacts differentially with vimentin and GFAP intermediate filament networks. J Cell Sci 120:1267–1277. [DOI] [PubMed] [Google Scholar]
- Jockusch BM, Schoenenberger CA, Stetefeld J, Aebi U. 2006. Tracking down the different forms of nuclear actin. Trends Cell Biol 16:391–396. [DOI] [PubMed] [Google Scholar]
- Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, Neefjes J. 2007. Activation of endosomal dynein motors by stepwise assembly of Rab7‐RILP‐p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol 176:459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES. 2004. Protein modification by SUMO. Annu Rev Biochem 73:355–382. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. 1997. Ubc9p is the conjugating enzyme for the ubiquitin‐like protein Smt3p. J Biol Chem 272:26799–26802. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. 1999. Cell cycle‐regulated attachment of the ubiquitin‐related protein SUMO to the yeast septins. J Cell Biol 147:981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA. 2001. An E3‐like factor that promotes SUMO conjugation to the yeast septins. Cell 106:735–744. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. 1997. The ubiquitin‐like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J 16:5509–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M. 2002. SUMO‐1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol 156:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. 2004. The RanGAP1‐RanBP2 complex is essential for microtubule‐kinetochore interactions in vivo . Curr Biol 14:611–617. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Melhuish TA, Wotton D. 2003. The polycomb protein Pc2 is a SUMO E3. Cell 113:127–137. [DOI] [PubMed] [Google Scholar]
- Kahana JA, Schnapp BJ, Silver PA. 1995. Kinetics of spindle pole body separation in budding yeast. Proc Natl Acad Sci USA 92:9707–9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky R, Denison C, Bening‐Abu‐Shach U, Chisholm AD, Gygi SP, Broday L. 2009. SUMO regulates the assembly and function of a cytoplasmic intermediate filament protein in C. elegans . Dev Cell 17:724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani T, Nguyen HP, Yeh ETH. 1997. Preferential modification of nuclear proteins by a novel ubiquitin‐like molecule. J Biol Chem 272:14001–14004. [DOI] [PubMed] [Google Scholar]
- Kammerer D, Stevermann L, Liakopoulos D. 2010. Ubiquitylation regulates interactions of astral microtubules with the cleavage apparatus. Curr Biol 20:1233–1243. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Chen J, Hirokawa N. 1992. Microtubule bundling by tau proteins in vivo: analysis of functional domains. EMBO J 11:3953–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang PJ, Hood‐DeGrenier JK, Park HO. 2013. Coupling of septins to the axial landmark by Bud4 in budding yeast. J Cell Sci 126:1218–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P, Chen M, Winkler DD, Luger K, Shen X. 2013. Evidence for monomeric actin function in INO80 chromatin remodeling. Nat Struct Mol Biol 20:426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon JR, Vale RD. 2009. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol 10:854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Dobyns WB. 2003. Lissencephaly and the molecular basis of neuronal migration. Hum Mol Genet 12 (Suppl 1):R89–R96. [DOI] [PubMed] [Google Scholar]
- Kelly JB, Datta S, Snow CJ, Chatterjee M, Ni L, Spencer A, Yang CS, Cubeñas‐Potts C, Matunis JJ, Paschal BM. 2011. The defective nuclear lamina in Hutchinson‐gilford progeria syndrome disrupts the nucleocytoplasmic Ran gradient and inhibits nuclear localization of Ubc9. Mol Cell Biol 31:3375–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O. 2007. SUMO junction‐what's your function? New insights through SUMO‐interacting motifs. EMBO Rep 8:550–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Baek SH. 2009. Emerging roles of desumoylating enzymes. Biochim Biophys Acta 1792:155–162. [DOI] [PubMed] [Google Scholar]
- Kim S, Coulombe PA. 2007. Intermediate filament scaffolds fulfill mechanical, organizational, and signaling functions in the cytoplasm. Genes Dev 21:1581–1597. [DOI] [PubMed] [Google Scholar]
- Kim YM, Jang WH, Quezado MM, Oh Y, Chung KC, Junn E, Mouradian MM. 2011. Proteasome inhibition induces alpha‐synuclein SUMOylation and aggregate formation. J Neurol Sci 307:157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Bonilla M, Rodgers ME, Schroer TA. 2002. Subunit organization in cytoplasmic dynein subcomplexes. Protein Sci 11:1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Kinoshita M, Akiyama H, Tomimoto H, Akiguchi I, Kumar S, Noda M, Kimura J. 1998. Identification of septins in neurofibrillary tangles in Alzheimer's disease. Am J Pathol 153:1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberg L, Lie AK, Florenes VA, Nesland JM, Davidson B. 2007. Expression of inhibitor‐of‐apoptosis protein family members in malignant mesothelioma. Hum Pathol 38:986–994. [DOI] [PubMed] [Google Scholar]
- Klug H, Xaver M, Chaugule VK, Koidl S, Mittler G, Klein F, Pichler A. 2013. Ubc9 sumoylation controls SUMO chain formation and meiotic synapsis in Saccharomyces cerevisiae . Mol Cell 50:625–636. [DOI] [PubMed] [Google Scholar]
- Kluger HM, McCarthy MM, Alvero AB, Sznol M, Ariyan S, Camp RL, Rimm DL, Mor G. 2007. The X‐linked inhibitor of apoptosis protein (XIAP) is up‐regulated in metastatic melanoma, and XIAP cleavage by phenoxodiol is associated with carboplatin sensitization. J Transl Med 5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopeikina KJ, Hyman BT, Spires‐Jones TL. 2012. Soluble forms of tau are toxic in Alzheimer's disease. Transl Neurosci 3:223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek WS, Copeland MJ, Chaudhuri A, Chant J. 2000. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science 287:2257–2259. [DOI] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gonczy P. 2014. NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. EMBO J 33:1815–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Gonczy P. 2013. Mechanisms of spindle positioning: cortical force generators in the limelight. Curr Opin Cell Biol 25:741–748. [DOI] [PubMed] [Google Scholar]
- Kozubowski L, Larson JR, Tatchell K. 2005. Role of the septin ring in the asymmetric localization of proteins at the mother‐bud neck in Saccharomyces cerevisiae . Mol Biol Cell 16:3455–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimm I, Ostlund C, Gilquin B, Couprie J, Hossenlopp P, Mornon JP, Bonne G, Courvalin JC, Worman HJ, Zinn‐Justin S. 2002. The Ig‐like structure of the C‐terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure 10:811–823. [DOI] [PubMed] [Google Scholar]
- Kroetz MB, Hochstrasser M. 2009. Identification of SUMO‐interacting proteins by yeast two‐hybrid analysis. Methods Mol Biol 497:107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumova P, Meulmeester E, Garrido M, Tirard M, Hsiao HH, Bossis G, Urlaub H, Zweckstetter M, Kugler S, Melchior F, Bahr M, Weishaupt JH. 2011. Sumoylation inhibits alpha‐synuclein aggregation and toxicity. J Cell Biol 194:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku NO, Omary MB. 2006. A disease‐ and phosphorylation‐related nonmechanical function for keratin 8. J Cell Biol 174:115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku NO, Lim JK, Krams SM, Esquivel CO, Keeffe EB, Wright TL, Parry DA, Omary MB. 2005. Keratins as susceptibility genes for end‐stage liver disease. Gastroenterology 129:885–893. [DOI] [PubMed] [Google Scholar]
- Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. 1998. Leptomycin B inhibition of signal‐mediated nuclear export by direct binding to CRM1. Exp Cell Res 242:540–547. [DOI] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA 96:9112–9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhla B, Haase C, Flach K, Luth HJ, Arendt T, Munch G. 2007. Effect of pseudophosphorylation and cross‐linking by lipid peroxidation and advanced glycation end product precursors on tau aggregation and filament formation. J Biol Chem 282:6984–6991. [DOI] [PubMed] [Google Scholar]
- Kumar A, Ray U, Das S. 2013. Human La protein interaction with GCAC near the INitiator AUG enhances hepatitis C virus RNA replication by promoting linkage between 5' and 3' untranslated regions. J Virol 87:6713–6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. 2008. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol 180:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Muralikrishna B, Parnaik VK. 2002. Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J Cell Biol 159:783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Beh CT, Latterich M, Schekman R, Rose MD. 1994. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J Cell Biol 126:911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch J, Meyer A, Snyder MP, Barral Y. 2002. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev 16:1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMonte BH, Wallace KE, Holloway BA, Shelly SS, Ascano J, Tokito M, Van Winkle T, Howland DS, Holzbaur EL. 2002. Disruption of dynein/dynactin inhibits axonal transport in motor neurons causing late‐onset progressive degeneration. Neuron 34:715–727. [DOI] [PubMed] [Google Scholar]
- Lampert F, Westermann S. 2011. A blueprint for kinetochores ‐ new insights into the molecular mechanics of cell division. Nat Rev Mol Cell Biol 12:407–412. [DOI] [PubMed] [Google Scholar]
- Landino LM, Skreslet TE, Alston JA. 2004. Cysteine oxidation of tau and microtubule‐associated protein‐2 by peroxynitrite: modulation of microtubule assembly kinetics by the thioredoxin reductase system. J Biol Chem 279:35101–35105. [DOI] [PubMed] [Google Scholar]
- Larsen CN, Krantz BA, Wilkinson KD. 1998. Substrate specificity of deubiquitinating enzymes: ubiquitin C‐terminal hydrolases. Biochemistry 37:3358–3368. [DOI] [PubMed] [Google Scholar]
- Lee MH, Lee SW, Lee EJ, Choi SJ, Chung SS, Lee JI, Cho JM, Seol JH, Baek SH, Kim KI, Chiba T, Tanaka K, Bang OS, Chung CH. 2006. SUMO‐specific protease SUSP4 positively regulates p53 by promoting Mdm2 self‐ubiquitination. Nat Cell Biol 8:1424–1431. [DOI] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA. 2003. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol 160:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Kaiser MA, Cooper JA. 2005. The offloading model for dynein function: differential function of motor subunits. J Cell Biol 168:201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner C, Kammerer D, Denoth A, Britschi M, Barral Y, Liakopoulos D. 2008. Regulation of mitotic spindle asymmetry by SUMO and the spindle‐assembly checkpoint in yeast. Curr Biol 18:1249–1255. [DOI] [PubMed] [Google Scholar]
- Lepekhin EA, Eliasson C, Berthold CH, Berezin V, Bock E, Pekny M. 2001. Intermediate filaments regulate astrocyte motility. J Neurochem 79:617–625. [DOI] [PubMed] [Google Scholar]
- Lepinoux‐Chambaud C, Eyer J. 2013. Review on intermediate filaments of the nervous system and their pathological alterations. Histochem Cell Biol 140:13–22. [DOI] [PubMed] [Google Scholar]
- Letai A, Coulombe PA, McCormick MB, Yu QC, Hutton E, Fuchs E. 1993. Disease severity correlates with position of keratin point mutations in patients with epidermolysis bullosa simplex. Proc Natl Acad Sci USA 90:3197–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JR, Holzbaur EL. 2006. Cytoplasmic dynein/dynactin function and dysfunction in motor neurons. Int J Dev Neurosci 24:103–111. [DOI] [PubMed] [Google Scholar]
- Li J, Lee WL, Cooper JA. 2005. NudEL targets dynein to microtubule ends through LIS1. Nat Cell Biol 7:686–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M. 1999. A new protease required for cell‐cycle progression in yeast. Nature 398:246–251. [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M. 2000. The yeast ULP2(SMT4) gene encodes a novel protease specific for the ubiquitin‐like Smt3 protein. Mol Cell Biol 20:2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ye Y. 2008. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci 65:2397–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. 2003. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112:561–574. [DOI] [PubMed] [Google Scholar]
- Liang W, Hao Z, Han JL, Zhu DJ, Jin ZF, Xie WL. 2014. CAV‐1 contributes to bladder cancer progression by inducing epithelial‐to‐mesenchymal transition. Urol Oncol 32:855–863. [DOI] [PubMed] [Google Scholar]
- Lima CD, Reverter D. 2008. Structure of the human SENP7 catalytic domain and poly‐SUMO deconjugation activities for SENP6 and SENP7. J Biol Chem 283:32045–32055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litersky JM, Johnson GV, Jakes R, Goedert M, Lee M, Seubert P. 1996. Tau protein is phosphorylated by cyclic AMP‐dependent protein kinase and calcium/calmodulin‐dependent protein kinase II within its microtubule‐binding domains at Ser‐262 and Ser‐356. Biochem J 316:655–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zhou Z. 2008. Lamin A/C, laminopathies and premature ageing. Histol Histopathol 23:747–763 [DOI] [PubMed] [Google Scholar]
- Liu D, Ding X, Du J, Cai X, Huang Y, Ward T, Shaw A, Yang Y, Hu R, Jin C, Yao X. 2007. Human NUF2 interacts with centromere‐associated protein E and is essential for a stable spindle microtubule‐kinetochore attachment. J Biol Chem 282:21415–21424. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang D, Luo W, Yu Y, Yu J, Li J, Zhang X, Zhang B, Chen J, Wu XR, Rosas‐Acosta G, Huang C. 2011. X‐linked inhibitor of apoptosis protein (XIAP) mediates cancer cell motility via Rho GDP dissociation inhibitor (RhoGDI)‐dependent regulation of the cytoskeleton. J Biol Chem 286:15630–15640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JS. 2011. Molecular genetics of neuronal migration disorders. Curr Neurol Neurosci Rep 11:171–178. [DOI] [PubMed] [Google Scholar]
- Liu JX, Zhou GB, Chen SJ, Chen Z. 2012. Arsenic compounds: revived ancient remedies in the fight against human malignancies. Curr Opin Chem Biol 16:92–98. [DOI] [PubMed] [Google Scholar]
- Liu YH, Wei W, Yin J, Liu GP, Wang Q, Cao FY, Wang JZ. 2009. Proteasome inhibition increases tau accumulation independent of phosphorylation. Neurobiol Aging 30:1949–1961. [DOI] [PubMed] [Google Scholar]
- Lo KW, Naisbitt S, Fan JS, Sheng M, Zhang M. 2001. The 8‐kDa dynein light chain binds to its targets via a conserved (K/R)XTQT motif. J Biol Chem 276:14059–14066. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Bi E. 2003. Regulation of septin organization and function in yeast. Trends Cell Biol 13:403–409. [DOI] [PubMed] [Google Scholar]
- Louvet E, Percipalle P. 2009. Transcriptional control of gene expression by actin and myosin. Int Rev Cell Mol Biol 272:107–147. [DOI] [PubMed] [Google Scholar]
- Ma S, Trivinos‐Lagos L, Graf R, Chisholm RL. 1999. Dynein intermediate chain mediated dynein‐dynactin interaction is required for interphase microtubule organization and centrosome replication and separation in Dictyostelium . J Cell Biol 147:1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccioni RB, Tapia L, Cambiazo V. 1995. Functional organization of tau proteins during neuronal differentiation and development. Braz J Med Biol Res 28:827–841. [PubMed] [Google Scholar]
- Maekawa H, Usui T, Knop M, Schiebel E. 2003. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J 22:438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, DeLuca J, Salmon ED, Earnshaw WC. 2004. The dynamic kinetochore‐microtubule interface. J Cell Sci 117:5461–5477. [DOI] [PubMed] [Google Scholar]
- Mao Y, Varma D, Vallee R. 2010. Emerging functions of force‐producing kinetochore motors. Cell Cycle 9:715–719. [DOI] [PubMed] [Google Scholar]
- Markowitz SD, Bertagnolli MM. 2009. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med 361:2449–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Plevock KM, St Germain BJ, Punch JJ, Meaden CW, Lee WL. 2011. Quantitative analysis of Pac1/Lis1‐mediated dynein targeting: implications for regulation of dynein activity in budding yeast. Cytoskeleton 68:157–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroui MA, Kheddache‐Atmane S, El Asmi F, Dianoux L, Aubry M, Chelbi‐Alix MK. 2012. Requirement of PML SUMO interacting motif for RNF4‐ or arsenic trioxide‐induced degradation of nuclear PML isoforms. PLoS One 7:e44949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SW, Konopka JB. 2004. SUMO modification of septin‐interacting proteins in Candida albicans . J Biol Chem 279:40861–40867. [DOI] [PubMed] [Google Scholar]
- Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC. 2008. In vivo identification of human small ubiquitin‐like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics 7:132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, Gnad F, Mann M, Vertegaal AC. 2010. Site‐specific identification of SUMO‐2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell 39:641–652. [DOI] [PubMed] [Google Scholar]
- McKenney RJ, Vershinin J, Kunwar A, Vallee RB, Gross SP. 2010. Lis1 and NudE induce a persistent dynein force‐producing state. Cell 141:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Weil SJ, Scherer J, Vallee RB. 2011. Mutually exclusive cytoplasmic dynein regulation by NudE‐Lis1 and dynactin. J Biol Chem 286:39615–39622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan LE, Brown JT, Henley JM, Cimarosti H. 2011. Profiles of SUMO and ubiquitin conjugation in an Alzheimer's disease model. Neurosci Lett 502:201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray MA, Bertin A, Garcia G, 3rd , Lam L, Nogales E, Thorner J. 2011. Septin filament formation is essential in budding yeast. Dev Cell 20:540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meednu N, Hoops H, D'Silva S, Pogorzala L, Wood S, Farkas D, Sorrentino M, Sia E, Meluh P, Miller RK. 2008. The spindle positioning protein Kar9p interacts with the sumoylation machinery in Saccharomyces cerevisiae . Genetics 180:2033–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F. 2000. SUMO‐nonclassical ubiquitin. Annu Rev Cell Dev Biol 16:591–626. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP‐C. Mol Biol Cell 6:793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez MG, Kojima S, Goldman RD. 2010. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J 24:1838–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraz‐Rios MA, Lira‐De Leon KI, Campos‐Pena V, De Anda‐Hernandez MA, Mena‐Lopez R. 2010. Tau oligomers and aggregation in Alzheimer's disease. J Neurochem 112:1353–1367. [DOI] [PubMed] [Google Scholar]
- Meseroll RA, Occhipinti P, Gladfelter AS. 2013. Septin phosphorylation and coiled‐coil domains function in cell and septin ring morphology in the filamentous fungus Ashbya gossypii . Eukaryot Cell 12:182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami A, Tynan SH, Hama T, Luby‐Phelps K, Saito T, Crandall JE, Besharse JC, Vallee RB. 2002. Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J Cell Sci 115:4801–4808. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk J, Drag M, Bekes M, Cao JT, Ronai Z, Salvesen GS. 2007. Small ubiquitin‐related modifier (SUMO)‐specific proteases: profiling the specificities and activities of human SENPs. J Biol Chem 282:26217–26224. [DOI] [PubMed] [Google Scholar]
- Miller RK, Rose MD. 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol 140:377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Vikstrom K, Goldman RD. 1991. Keratin incorporation into intermediate filament networks is a rapid process. J Cell Biol 113:843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Khoun S, Goldman RD. 1993. Dynamics of keratin assembly: exogenous type I keratin rapidly associates wit type II keratin in vivo . J Cell Biol 122:123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Matheos D, Rose MD. 1999. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J Cell Biol 144:963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Cheng SC, Rose MD. 2000. Bim1p/Yeb1p mediates the Kar9p‐dependent cortical attachment of cytoplasmic microtubules. Mol Biol Cell 11:2949–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Moore JK, D'Silva S, Goodson HV. 2006. The CLIP‐170 orthologue Bik1p and positioning the mitotic spindle in yeast. Curr Top Dev Biol 76:49–87. [DOI] [PubMed] [Google Scholar]
- Miller SA, Johnson ML, Stukenberg PT. 2008. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1). Curr Biol 18:1785–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino A, Tanaka K, Kamei T, Umikawa M, Fujiwara T, Takai Y. 1998. Shs1p: a novel member of septin that interacts with spa2p, involved in polarized growth in Saccharomyces cerevisiae . Biochem Biophys Res Commun 251:732–736. [DOI] [PubMed] [Google Scholar]
- Minty A, Dumont X, Kaghad M, Caput D. 2000. Covalent modification of p73alpha by SUMO‐1. Two‐hybrid screening with p73 identifies novel SUMO‐1‐interacting proteins and a SUMO‐1 interaction motif. J Biol Chem 275:36316–36323. [DOI] [PubMed] [Google Scholar]
- Mishra M, Huang J, Balasubramanian MK. 2014. The yeast actin cytoskeleton. FEMS Microbiol Rev 38:213–227. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. 1984. Microtubule assembly nucleated by isolated centrosomes. Nature 312:232–237. [DOI] [PubMed] [Google Scholar]
- Mok YK, Lo KW, Zhang M. 2001. Structure of Tctex‐1 and its interaction with cytoplasmic dynein intermediate chain. J Biol Chem 276:14067–14074. [DOI] [PubMed] [Google Scholar]
- Montpetit B, Hazbun TR, Fields S, Hieter P. 2006. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol 174:653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Miller RK. 2007. The cyclin‐dependent kinase Cdc28p regulates multiple aspects of Kar9p function in yeast. Mol Biol Cell 18:1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, D'Silva S, Miller RK. 2006. The CLIP‐170 homologue Bik1p promotes the phosphorylation and asymmetric localization of Kar9p. Mol Biol Cell 17:178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, Cossart P. 2012. Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol 13:183–194. [DOI] [PubMed] [Google Scholar]
- Moughamian AJ, Holzbaur EL. 2012. Dynactin is required for transport initiation from the distal axon. Neuron 74:331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. 2007. Modification in reverse: the SUMO proteases. Trends Biochem Sci 32:286–295. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. 2010. The fate of metaphase kinetochores is weighed in the balance of SUMOylation during S phase. Cell Cycle 9:3194–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Chem CF, Brill SJ. 2010. Wss1 is a SUMO‐dependent isopeptidase that interacts genetically with the Slx5‐Slx8 SUMO‐targeted Ub ligase. Mol Cell Biol 30:3737–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy SN, Wilson JH, Lukas TJ, Kuret J, Lorand L. 1998. Cross‐linking sites of the human tau protein, probed by reactions with human transglutaminase. J Neurochem 71:2607–2614. [DOI] [PubMed] [Google Scholar]
- Nacharaju P, Ko L, Yen SH. 1997. Characterization of in vitro glycation sites of tau. J Neurochem 69:1709–1719. [DOI] [PubMed] [Google Scholar]
- Nagai S, Davoodi N, Gasser SM. 2011. Nuclear organization in genome stability: SUMO connections. Cell Res 21:474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagi C, Xiao GQ, Li G, Genden E, Burstein DE. 2007. Immunohistochemical detection of X‐linked inhibitor of apoptosis in head and neck squamous cell carcinoma. Ann Diagn Pathol 11:402–406. [DOI] [PubMed] [Google Scholar]
- Necula M, Kuret J. 2004. Pseudophosphorylation and glycation of tau protein enhance but do not trigger fibrillization in vitro. J Biol Chem 279:49694–49703. [DOI] [PubMed] [Google Scholar]
- Nekrasova OE, Mendez MG, Chernoivanenko IS, Tyurin‐Kuzmin PA, Kuczmarski ER, Gelfand VI, Goldman RD, Minin AA. 2011. Vimentin intermediate filaments modulate the motility of mitochondria. Mol Biol Cell 22:2282–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto T, Kitagawa M, Hasegawa M, Ikeda S, Akashi T, Takizawa T, Hirokawa K, Koike M. 2004. Expression of IAP family proteins in esophageal cancer. Exp Mol Pathol 76:253–259. [DOI] [PubMed] [Google Scholar]
- Nguyen TQ, Sawa H, Okano H, White JG. 2000. The C. elegans septin genes, unc‐59 and unc‐61, are required for normal postembryonic cytokineses and morphogenesis but have no essential function in embryogenesis. J Cell Sci 113:3825–3837. [DOI] [PubMed] [Google Scholar]
- Nie M, Aslanian A, Prudden J, Heideker J, Vashisht AA, Wohlschlegel JA, Yates JR, 3rd , Boddy MN. 2012. Dual recruitment of Cdc48 (p97)‐Ufd1‐Npl4 ubiquitin‐selective segregase by small ubiquitin‐like modifier protein (SUMO) and ubiquitin in SUMO‐targeted ubiquitin ligase‐mediated genome stability functions. J Biol Chem 287:29610–29619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida E, Iida K, Yonezawa N, Koyasu S, Yahara I, Sakai H. 1987. Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc Natl Acad Sci U S A 84:5262–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R, Onishi M, Pringle JR. 2011. New insights into the phylogenetic distribution and evolutionary origins of the septins. Biol Chem 392:681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Sasaki T, Takaishi K, Kato M, Yaku H, Araki K, Matsuura Y, Takai Y. 1994. rac p21 is involved in insulin‐induced membrane ruffling and rho p21 is involved in hepatocyte growth factor‐ and 12‐O‐tetradecanoylphorbol‐13‐acetate (TPA)‐induced membrane ruffling in KB cells. Mol Cell Biol 14:2447–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Yamada S, Koike M, Kanda M, Fujii T, Nakayama G, Sugimoto H, Nomoto S, Fujiwara M, Kodera Y. 2014. Epithelial to mesenchymal transition correlates with tumor budding and predicts prognosis in esophageal squamous cell carcinoma. J Surg Oncol 110:764–769. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81:53–62. [DOI] [PubMed] [Google Scholar]
- Odian G. 1991. Principles of Polymerization. New York: Wiley. [Google Scholar]
- Oh Y, Bi E. 2011. Septin structure and function in yeast and beyond. Trends Cell Biol 21:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka M, Kudo H, Sugama N, Asami Y, Takehana M. 2008. The function of filensin and phakinin in lens transparency. Mol Vis 14:815–822. [PMC free article] [PubMed] [Google Scholar]
- Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H. 1999. In vitro SUMO‐1 modification requires two enzymatic steps, E1 and E2. Biochem Biophys Res Commun 254:693–698. [DOI] [PubMed] [Google Scholar]
- Olofsson B. 1999. Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling. Cell Signal 11:545–554. [DOI] [PubMed] [Google Scholar]
- Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. 2010. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature 463:906–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong K, Wloka C, Okada S, Svitkina T, Bi E. 2014. Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nat Commun 5:5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Valin A, Gill G. 2009. Regulation of transcription factor activity by SUMO modification. Methods Mol Biol 497:141–152. [DOI] [PubMed] [Google Scholar]
- Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. 1987. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J 6:1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG, Kuster B, Gerstberger T, Hurt E. 2003. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat Cell Biol 5:21–27. [DOI] [PubMed] [Google Scholar]
- Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. 2004. A proteome‐wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem 279:41346–41351. [DOI] [PubMed] [Google Scholar]
- Parry DA, Steven AC, Steinert PM. 1985. The coiled‐coil molecules of intermediate filaments consist of two parallel chains in exact axial register. Biochem Biophys Res Commun 127:1012–1018. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Shpetner HS, Vallee RB. 1987. MAP 1C is a microtubule‐activated ATPase which translocates microtubules in vitro and has dynein‐like properties. J Cell Biol 105:1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson HF, Self AJ, Garrett MD, Just I, Aktories K, Hall A. 1990. Microinjection of recombinant p21rho induces rapid changes in cell morphology. J Cell Biol 111:1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Witman GB. 1999. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol 144:473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P. 2013. Co‐transcriptional nuclear actin dynamics. Nucleus 4:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JJ, Tainer JA, Boddy MN. 2008. A SIM‐ultaneous role for SUMO and ubiquitin. Trends Biochem Sci 33:201–208. [DOI] [PubMed] [Google Scholar]
- Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, Kim J, Dillmann WH, Browne SE, Hall A, Voellmy R, Tsuboi Y, Dawson TM, Wolozin B, Hardy J, Hutton M. 2004. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet 13:703–714. [DOI] [PubMed] [Google Scholar]
- Philimonenko VV, Zhao J, Iben S, Dingova H, Kysela K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozak P, Grummt I. 2004. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol 6:1165–1172. [DOI] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109–120. [DOI] [PubMed] [Google Scholar]
- Pinder JB, McQuaid ME, Dobson MJ. 2013. Deficient sumoylation of yeast 2‐micron plasmid proteins Rep1 and Rep2 associated with their loss from the plasmid‐partitioning locus and impaired plasmid inheritance. PLoS One 8:e60384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. 2010. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol 22:50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. 2009. Actin, a central player in cell shape and movement. Science 326:1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pountney DL, Huang Y, Burns RJ, Haan E, Thompson PD, Blumbergs PC, Gai WP. 2003. SUMO‐1 marks the nuclear inclusions in familial neuronal intranuclear inclusion disease. Exp Neurol 184:436–446. [DOI] [PubMed] [Google Scholar]
- Praefcke GJ, Hofmann K, Dohmen RJ. 2012. SUMO playing tag with ubiquitin. Trends Biochem Sci 37:23–31. [DOI] [PubMed] [Google Scholar]
- Price LS, Langeslag M, Ten Klooster JP, Hordijk PL, Jalink K, Collard JG. 2003. Calcium signaling regulates translocation and activation of Rac. J Biol Chem 278:39413–39421. [DOI] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN. 2007. SUMO‐targeted ubiquitin ligases in genome stability. EMBO J 26:4089–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Perry JJ, Nie M, Vashisht AA, Arvai AS, Hitomi C, Geunther G, Wohlschlegel JA, Tainer JA, Boddy MN. 2011. DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes. Mol Cell Biol 31:2299–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. 1999. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol 147:321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reindle A, Belichenko I, Bylebyl GR, Chen XL, Gandhi N, Johnson ES. 2006. Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J Cell Sci 119:4749–4757. [DOI] [PubMed] [Google Scholar]
- Reiner O, Sapir T. 2013. LIS1 functions in normal development and disease. Curr Opin Neurobiol 23:951–956. [DOI] [PubMed] [Google Scholar]
- Reiner O, Sapoznik S, Sapir T. 2006. Lissencephaly 1 linking to multiple diseases: mental retardation, neurodegeneration, schizophrenia, male sterility, and more. Neuro Mol Med 8:547–565. [DOI] [PubMed] [Google Scholar]
- Ren YH, Liu KJ, Wang M, Yu YN, Yang K, Chen Q, Yu B, Wang W, Li QW, Wang J, Hou ZY, Fang JY, Yeh ET, Yang J, Yi J. 2014. De‐SUMOylation of FOXC2 by SENP3 promotes the epithelial‐mesenchymal transition in gastric cancer cells. Oncotarget 5:7093–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. 1992. The small GTP‐binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70:389–399. [DOI] [PubMed] [Google Scholar]
- Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J. 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7‐RILP‐p150 glued and late endosome positioning. J Cell Biol 185:1209–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Kozubowski L, Goode BL, Drubin DG, Hartwig JH. 2005. Actin and septin ultrastructures at the budding yeast cell cortex. Mol Biol Cell 16:372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas‐Fernandez A, Plechanovova A, Hattersley N, Jaffray E, Tatham MH, Hay RT. 2014. SUMO chain‐induced dimerization activates RNF4. Mol Cell 53:880–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas‐Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. 2005. A universal strategy for proteomic studies of SUMO and other ubiquitin‐like modifiers. Mol Cell Proteomics 4:56–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. 2013. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol 14:7–12. [DOI] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. 1990. Components of the yeast spindle and spindle pole body. J Cell Biol 111:1913–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J. 2000. Functional heterogeneity of small ubiquitin‐related protein modifiers SUMO‐1 versus SUMO‐2/3. J Biol Chem 275:6252–6258. [DOI] [PubMed] [Google Scholar]
- Sammak PJ, Borisy GG. 1988. Direct observation of microtubule dynamics in living cells. Nature 332:724–726. [DOI] [PubMed] [Google Scholar]
- Sapir T, Eisenstein M, Burgess HA, Horesh D, Cahana A, Aoki J, Hattori M, Arai H, Inoue K, Reiner O. 1999. Analysis of lissencephaly‐causing LIS1 mutations. Eur J Biochem 266:1011–1020. [DOI] [PubMed] [Google Scholar]
- Sardar HS, Luczak VG, Lopez MM, Lister BC, Gilbert SP. 2010. Mitotic kinesin CENP‐E promotes microtubule plus‐end elongation. Curr Biol 20:1648–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Park‐Sarge OK. 2011. SUMO and its role in human diseases. Int Rev Cell Mol Biol 288:167–183. [DOI] [PubMed] [Google Scholar]
- Scherer J, Vallee RB. 2011. Adenovirus recruits dynein by an evolutionary novel mechanism involving direct binding to pH‐primed hexon. Viruses 3:1417–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidts M, Arts HH, Bongers EM, Yap Z, Oud MM, Antony D, Duijkers L, Emes RD, Stalker J, Yntema JB, Plagnol V, Hoischen A, Gilissen C, Forsythe E, Lausch E, Veltman JA, Roeleveld N, Superti‐Furga A, Kutkowska‐Kazmierczak A, Kamsteeg EJ, Elcioglu N, van Maarle MC, Graul‐Neumann LM, Devriendt K, Smithson SF, Wellesley D, Verbeek NE, Hennekam RC, Kayserili H, Scambler PJ, Beales PL, Knoers NV, Roepman R, Mitchison HM. 2013a. Exome sequencing identifies DYNC2H1 mutations as a common cause of asphyxiating thoracic dystrophy (Jeune syndrome) without major polydactyly, renal or retinal involvement. J Med Genet 50:309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidts M, Vodopiutz J, Christou‐Savina S, Cortes CR, McInerney‐Leo AM, Emes RD, Arts HH, Tuysuz B, D'Silva J, Leo PJ, Giles TC, Oud MM, Harris JA, Koopmans M, Marshall M, Elcioglu N, Kuechler A, Bockenhauer D, Moore AT, Wilson LC, Janecke AR, Hurles ME, Emmet W, Gardiner B, Streubel B, Dopita B, Zankl A, Kayserili H, Scambler PJ, Brown MA, Beales PL, Wicking C, Duncan EL, Mitchison HM. 2013b. Mutations in the gene encoding IFT dynein complex component WDR34 cause Jeune asphyxiating thoracic dystrophy. Am J Hum Genet 93:932–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenenberger CA, Buchmeier S, Boerries M, Sütterlin R, Aebi U, Jockusch BM. 2005. Conformation‐specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J Struct Biol 152:157–168. [DOI] [PubMed] [Google Scholar]
- Schreiber KH, Kennedy BK. 2013. When lamins go bad: nuclear structure and disease. Cell 152:1365–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA. 2004. Dynactin. Annu Rev Cell Dev Biol 20:759–779. [DOI] [PubMed] [Google Scholar]
- Schulze E, Kirschner M. 1988. New features of microtubule behaviour observed in vivo. Nature 334:356–359. [DOI] [PubMed] [Google Scholar]
- Schwartz K, Richards K, Botstein D. 1997. BIM1 encodes a microtubule‐binding protein in yeast. Mol Biol Cell 8:2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz SE, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. 1998. The ubiquitin‐like proteins SMT3 and SUMO‐1 are conjugated by the UBC9 E2 enzyme. Proc Natl Acad Sci USA 95:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers O, Mandelkow EM, Biernat J, Mandelkow E. 1995. Oxidation of cysteine‐322 in the repeat domain of microtubule‐associated protein tau controls the in vitro assembly of paired helical filaments. Proc Natl Acad Sci USA 92:8463–8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwienhorst I, Johnson ES, Dohmen RJ. 2000. SUMO conjugation and deconjugation. Mol Gen Genet 263:771–786. [DOI] [PubMed] [Google Scholar]
- Seltmann K, Fritsch AW, Kas JA, Magin TM. 2013. Keratins significantly contribute to cell stiffness and impact invasive behavior. Proc Natl Acad Sci USA 110:18507–18512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. 2003. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol 13:364–372. [DOI] [PubMed] [Google Scholar]
- Sheng W, Liao X. 2002. Solution structure of a yeast ubiquitin‐like protein Smt3: the role of structurally less defined sequences in protein‐protein recognitions. Protein Sci 11:1482–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Butin‐Israeli V, Adam SA, Goldman RD. 2010. Nuclear lamins in cell regulation and disease. Cold Spring Harb Symp Quant Biol 75:525–531. [DOI] [PubMed] [Google Scholar]
- Shin EJ, Shin HM, Nam E, Kim WS, Kim JH, Oh BH, Yun Y. 2012. DeSUMOylating isopeptidase: a second class of SUMO protease. EMBO Rep 13:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K. 2000. Modulation of STAT signaling by STAT‐interacting proteins. Oncogene 19:2638–2644. [DOI] [PubMed] [Google Scholar]
- Signor D, Wedaman KP, Orozco JT, Dwyer ND, Bargmann CI, Rose LS, Scholey JM. 1999. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans . J Cell Biol 147:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, Domaradzki T, Hofmann WA, Wilson KL. 2013. Lamin A tail modification by SUMO1 is disrupted by familial partial lipodystrophy‐causing mutations. Mol Biol Cell 24:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Farkasovsky M, Hauer F, Kuhlmann D, Macara IG, Weyand M, Stark H, Wittinghofer A. 2007. Structural insight into filament formation by mammalian septins. Nature 449:311–315. [DOI] [PubMed] [Google Scholar]
- Sitaram P, Anderson MA, Jodoin JN, Lee E, Lee LA. 2012. Regulation of dynein localization and centrosome positioning by Lis‐1 and asunder during Drosophila spermatogenesis. Development 139:2945–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalli O, Wilhelmsson U, Orndahl C, Fekete B, Malmgren K, Rydenhag B, Pekny M. 2013. Astrocytoma grade IV (glioblastoma multiforme) displays 3 subtypes with unique expression profiles of intermediate filament proteins. Hum Pathol 44:2081–2088. [DOI] [PubMed] [Google Scholar]
- Snider NT, Omary MB. 2014. Post‐translational modifications of intermediate filament proteins: mechanisms and functions. Nat Rev Mol Cell Biol 15:163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider NT, Weerasinghe SV, Iniguez‐Lluhi JA, Herrmann H, Omary MB. 2011. Keratin hypersumoylation alters filament dynamics and is a marker for human liver disease and keratin mutation. J Biol Chem 286:2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soellner P, Quinlan RA, Franke WW. 1985. Identification of a distinct soluble subunit of an intermediate filament protein: tetrameric vimentin from living cells. Proc Natl Acad Sci USA 82:7929–7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. 2004. Identification of a SUMO‐binding motif that recognizes SUMO‐modified proteins. Proc Natl Acad Sci USA 101:14373–14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Brady ST. 2015. Post‐translational modifications of tubulin: pathways to functional diversity of microtubules. Trends Cell Biol 25:125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg A, Liem RK. 2007. Plakins in development and disease. Exp Cell Res 313:2189–2203. [DOI] [PubMed] [Google Scholar]
- Splinter D, Razafsky DS, Schlager MA, Serra‐Marques A, Grigoriev I, Demmers J, Keijzer N, Jiang K, Poser I, Hyman AA, Hoogenraad CC, King SJ, Akhmanova A. 2012. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol Biol Cell 23:4226–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikumar T, Lewicki MC, Raught B. 2013. A global S. cerevisiae small ubiquitin‐related modifier (SUMO) system interactome. Mol Syst Biol 9:668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V. 2003. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J Cell Biol 163:729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, Pandolfi PP, Thompson LM, Marsh JL. 2004. SUMO modification of Huntingtin and Huntington's disease pathology. Science 304:100–103. [DOI] [PubMed] [Google Scholar]
- Stehmeier P, Muller S. 2009. Regulation of p53 family members by the ubiquitin‐like SUMO system. DNA Repair (Amst) 8:491–498. [DOI] [PubMed] [Google Scholar]
- Stephan AK, Kliszczak M, Morrison CG. 2011. The Nse/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability. FEBS Lett 585:2907–2913. [DOI] [PubMed] [Google Scholar]
- Strnad P, Kucukoglu O, Lunova M, Guldiken N, Lienau TC, Stickel F, Omary MB. 2012. Non‐coding keratin variants associate with liver fibrosis progression in patients with hemochromatosis. PLoS One 7:e32669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Hochstrasser M. 2010. A WLM protein with SUMO‐directed protease activity. Mol Cell Biol 30:3734–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YF, Yang T, Huang H, Liu LF, Hwang J. 2012. Phosphorylation of Ubc9 by Cdk1 enhances SUMOylation activity. PLoS One 7:e34250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh HY, Kim JH, Woo JS, Ku B, Shin EJ, Yun Y, Oh BH. 2012. Crystal structure of DeSI‐1, a novel deSUMOylase belonging to a putative isopeptidase superfamily. Proteins 80:2099–2104. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalante‐Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. 1999. Loss of A‐type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol 147:913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumigray KD, Lechler T. 2011. Control of cortical microtubule organization and desmosome stability by centrosomal proteins. Bioarchitecture 1:221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumigray KD, Chen H, Lechler T. 2011. Lis1 is essential for cortical microtubule organization and desmosome stability in the epidermis. J Cell Biol 194:631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Leverson JD, Hunter T. 2007. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 26:4102–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MK, Lim G, Yi DG, Chang YJ, Yang EB, Lee K, Huh WK. 2013. Genome‐wide bimolecular fluorescence complementation analysis of SUMO interactome in yeast. Genome Res 23:736–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, Borisy GG. 1996. Plectin sidearms mediate interaction of intermediate filaments with microtubules and other components of the cytoskeleton. J Cell Biol 135:991–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvius N, Tesson F. 2006. Lamin A/C and cardiac diseases. Curr Opin Cardiol 21:159–165. [DOI] [PubMed] [Google Scholar]
- Szeverenyi I, Cassidy AJ, Chung CW, Lee BT, Common JE, Ogg SC, Chen H, Sim SY, Goh WL, Ng KW, Simpson JA, Chee LL, Eng GH, Li B, Lunny DP, Chuon D, Venkatesh A, Khoo KH, McLean WH, Lim YP, Lane EB. 2008. The Human Intermediate Filament Database: comprehensive information on a gene family involved in many human diseases. Hum Mutat 29:351–360. [DOI] [PubMed] [Google Scholar]
- Szymanski EP, Kerscher O. 2013. Budding yeast protein extraction and purification for the study of function, interactions, and post‐translational modifications. J Vis Exp 80:e50921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai CY, Dujardin DL, Faulkner NE, Vallee RB. 2002. The role of dynein, dynactin, and CLIP‐170 interactions in LIS1 kinetochore function. J Cell Biol 156:959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Ishida M, Komano H, Takahashi H. 2008. SUMO‐1 immunoreactivity co‐localizes with phospho‐tau in APP transgenic mice but not in mutant tau transgenic mice. Neurosci Lett 441:90–93. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Tsujioka Y, Yamada T, Tsuboi Y, Okada H, Yamamoto T, Liposits Z. 1999. Glycosylation of microtubule‐associated protein tau in Alzheimer's disease brain. Acta Neuropathol 97:635–641. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Iwase M, Konishi M, Tanaka M, Toh‐e A, Kikuchi Y. 1999. Smt3, a SUMO‐1 homolog, is conjugated to Cdc3, a component of septin rings at the mother‐bud neck in budding yeast. Biochem Biophys Res Commun 259:582–587. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Mizoi J, Toh EA, Kikuchi Y. 2000. Yeast Ulp1, an Smt3‐specific protease, associates with nucleoporins. J Biochem 128:723–725. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Kahyo T, Toh EA, Yasuda H, Kikuchi Y. 2001. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J Biol Chem 276:48973–48977. [DOI] [PubMed] [Google Scholar]
- Takaishi K, Sasaki T, Kato M, Yamochi W, Kuroda S, Nakamura T, Takeichi M, Takai Y. 1994. Involvement of Rho p21 small GTP‐binding protein and its regulator in the HGF‐induced cell motility. Oncogene 9:273–279. [PubMed] [Google Scholar]
- Tan SC, Scherer J, Vallee RB. 2011. Recruitment of dynein to late endosomes and lysosomes through light intermediate chains. Mol Biol Cell 22:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, Murakami Y. 1999. Characterization of a fission yeast SUMO‐1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol 19:8660–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. 2001. Polymeric chains of SUMO‐2 and SUMO‐3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 276:35368–35374. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. 2008. RNF4 is a poly‐SUMO‐specific E3 ubiquitin ligase required for arsenic‐induced PML degradation. Nat Cell Biol 10:538–546. [DOI] [PubMed] [Google Scholar]
- Tempe D, Piechaczyk M, Bossis G. 2008. SUMO under stress. Biochem Soc Trans 36:874–878. [DOI] [PubMed] [Google Scholar]
- Teng S, Luo H, Wang L. 2012. Predicting protein sumoylation sites from sequence features. Amino Acids 43:447–455. [DOI] [PubMed] [Google Scholar]
- Terman JR, Kashina A. 2013. Post‐translational modification and regulation of actin. Curr Opin Cell Biol 25:30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien JF, Fong KK, Umbreit NT, Payen C, Zelter A, Asbury CL, Dunham MJ, Davis TN. 2013. Coupling unbiased mutagenesis to high‐throughput DNA sequencing uncovers functional domains in the Ndc80 kinetochore protein of Saccharomyces cerevisiae . Genetics 195:159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T, Sugie K, Hirata M, Morii N, Fukata J, Uchida A, Imura H, Narumiya S. 1993. Inhibition of PMA‐induced, LFA‐1‐dependent lymphocyte aggregation by ADP ribosylation of the small molecular weight GTP binding protein, rho. J Cell Biol 120:1529–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torisawa T, Nakayama A, Furuta K, Yamada M, Hirotsune S, Toyoshima YY. 2011. Functional dissection of LIS1 and NDEL1 towards understanding the molecular mechanisms of cytoplasmic dynein regulation. J Biol Chem 286:1959–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck‐Peterson SL, Leschziner AE. 2014. Lis1 regulates dynein by sterically blocking its mechanochemical cycle. Elife 3, doi: 10.7554/eLife.03372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta R, Vignudelli T, Candini O, Intine RV, Pecorari L, Guerzoni C, Santilli G, Byrom MW, Goldoni S, Ford LP, Caligiuri MA, Maraia RJ, Perrotti D, Calabretta B. 2003. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell 3:145–160. [DOI] [PubMed] [Google Scholar]
- Tu S, Wu WJ, Wang J, Cerione RA. 2003. Epidermal growth factor‐dependent regulation of Cdc42 is mediated by the Src tyrosine kinase. J Biol Chem 278:49293–49300. [DOI] [PubMed] [Google Scholar]
- Ulrich HD. 2005. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol 15:525–532. [DOI] [PubMed] [Google Scholar]
- Ulrich HD. 2009. The SUMO system: an overview. Methods Mol Biol 497:3–16. [DOI] [PubMed] [Google Scholar]
- Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, Praefcke GJ, Dohmen RJ. 2007. Ubiquitin‐dependent proteolytic control of SUMO conjugates. J Biol Chem 282:34167–34175. [DOI] [PubMed] [Google Scholar]
- Valiron O, Caudron N, Job D. 2001. Microtubule dynamics. Cell Mol Life Sci 58:2069–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, Tai C, Faulkner NE. 2001. LIS1: cellular function of a disease‐causing gene. Trends Cell Biol 11:155–160. [DOI] [PubMed] [Google Scholar]
- van Heusden GP, Steensma HY. 2008. The Saccharomyces cerevisiae Wss1 protein is only present in mother cells. FEMS Microbiol Lett 282:100–104. [DOI] [PubMed] [Google Scholar]
- van Niekerk EA, Willis DE, Chang JH, Reumann K, Heise T, Twiss JL. 2007. Sumoylation in axons triggers retrograde transport of the RNA‐binding protein La. Proc Natl Acad Sci USA 104:12913–12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen MK. 2008. Nuclear actin dynamics–from form to function. FEBS Lett 582:2033–2040. [DOI] [PubMed] [Google Scholar]
- Vassar R, Coulombe PA, Degenstein L, Albers K, Fuchs E. 1991. Mutant keratin expression in transgenic mice causes marked abnormalities resembling a human genetic skin disease. Cell 64:365–380. [DOI] [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. 1995. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J Cell Biol 131:1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT, Tynan SH, Faulkner NE, Echeverri CJ, Vallee RB. 1999. Colocalization of cytoplasmic dynein with dynactin and CLIP‐170 at microtubule distal ends. J Cell Sci 112:1437–1447. [DOI] [PubMed] [Google Scholar]
- Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, Mann M, Lamond AI. 2004. A proteomic study of SUMO‐2 target proteins. J Biol Chem 279:33791–33798. [DOI] [PubMed] [Google Scholar]
- Vijay‐Kumar S, Bugg CE, Cook WJ. 1987. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol 194:531–544. [DOI] [PubMed] [Google Scholar]
- Vorobjev IA, Rodionov VI, Maly IV, Borisy GG. 1999. Contribution of plus and minus end pathways to microtubule turnover. J Cell Sci 112:2277–2289. [DOI] [PubMed] [Google Scholar]
- Vrabioiu AM, Mitchison TJ. 2006. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature 443:466–469. [DOI] [PubMed] [Google Scholar]
- Vreugde S, Ferrai C, Miluzio A, Hauben E, Marchisio PC, Crippa MP, Bussi M, Biffo S. 2006. Nuclear myosin VI enhances RNA polymerase II‐dependent transcription. Mol Cell 23:749–755. [DOI] [PubMed] [Google Scholar]
- Wang DS, Dickson DW, Malter JS. 2008. Tissue transglutaminase, protein cross‐linking and Alzheimer's disease: review and views. Int J Clin Exp Pathol 1:5–18. [PMC free article] [PubMed] [Google Scholar]
- Wang JZ, Grundke‐Iqbal I, Iqbal K. 1996. Glycosylation of microtubule‐associated protein tau: an abnormal posttranslational modification in Alzheimer's disease. Nat Med 2:871–875. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang J, Banerjee S, Barnes L, Barnes L, Sajja V, Liu Y, Guo B, Du Y, Agarwal MK, Wald DN, Wang Q, Yang J. 2010. Sumoylation of vimentin354 is associated with PIAS3 inhibition of glioma cell migration. Oncotarget 1:620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ketcham SA, Schon A, Goodman B, Wang Y, Yates J, 3rd , Freire E, Schroer TA, Zheng Y. 2013. Nudel/NudE and Lis1 promote dynein and dynactin interaction in the context of spindle morphogenesis. Mol Biol Cell 24:3522–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YE, Pernet O, Lee B. 2012. Regulation of the nucleocytoplasmic trafficking of viral and cellular proteins by ubiquitin and small ubiquitin‐related modifiers. Biol Cell 104:121–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman‐Storer CM, Karki S, Holzbaur EL. 1995. The p150Glued component of the dynactin complex binds to both microtubules and the actin‐related protein centractin (Arp‐1). Proc Natl Acad Sci USA 92:1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P, Forster R, Palmer KJ, Pepperkok R, Stephens DJ. 2005. Coupling of ER exit to microtubules through direct interaction of COPII with dynactin. Nat Cell Biol 7:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman J, Wong MB, Sharry S, Rcom‐H'cheo‐Gauthier A, Gai WP, Meedeniya A, Pountney DL. 2013. Increased SUMO‐1 expression in the unilateral rotenone‐lesioned mouse model of Parkinson's disease. Neurosci Lett 544:119–124. [DOI] [PubMed] [Google Scholar]
- Wei R, Ngo B, Wu G, Lee WH. 2011. Phosphorylation of the Ndc80 complex protein, HEC1, by Nek2 kinase modulates chromosome alignment and signaling of the spindle assembly checkpoint. Mol Biol Cell 22:3584–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch WJ, Suhan JP. 1985. Morphological study of the mammalian stress response: characterization of changes in cytoplasmic organelles, cytoskeleton, and nucleoli, and appearance of intranuclear actin filaments in rat fibroblasts after heat‐shock treatment. J Cell Biol 101:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D, Xu Z, Xia L, Liu X, Tu Y, Lei H, Wang W, Wang T, Song L, Ma C, Xu H, Zhu W, Chen G, Wu Y. 2014. Important role of SUMOylation of spliceosome factors in prostate cancer cells. J Proteome Res 13:3571–3582. [DOI] [PubMed] [Google Scholar]
- Wiche G. 1998. Role of plectin in cytoskeleton organization and dynamics. J Cell Sci 111:2477–2486. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD. 1997. Regulation of ubiquitin‐dependent processes by deubiquitinating enzymes. FASEB J 11:1245–1256. [DOI] [PubMed] [Google Scholar]
- Winter L, Wiche G. 2013. The many faces of plectin and plectinopathies: pathology and mechanisms. Acta Neuropathol 125:77–93. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Johnson ES, Reed SI, Yates JR, 3rd . 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae . J Biol Chem 279:45662–45668. [DOI] [PubMed] [Google Scholar]
- Wykoff DD, O'Shea EK. 2005. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol Cell Proteomics 4:73–83. [DOI] [PubMed] [Google Scholar]
- Xie Y, Kerscher O, Kroetz MB, McConchies HF, Sung P, Hochstrasser M. 2007. The yeast Hex3*Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem 282:34176–34184. [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Gong Z, Fukuchi‐Mizutani M, Fukui Y, Tanaka Y, Kusumi T, Saito K. 1999. Molecular cloning and biochemical characterization of a novel anthocyanin 5‐O‐glucosyltransferase by mRNA differential display for plant forms regarding anthocyanin. J Biol Chem 274:7405–7411. [DOI] [PubMed] [Google Scholar]
- Yang JM, Chipev CC, DiGiovanna JJ, Bale SJ, Marekov LN, Steinert PM, Compton JG. 1994. Mutations in the H1 and 1A domains in the keratin 1 gene in epidermolytic hyperkeratosis. J Invest Dermatol 102:17–23. [DOI] [PubMed] [Google Scholar]
- Yang JM, Nam K, Park KB, Kim WS, Moon KC, Koh JK, Steinert PM, Lee ES. 1996. A novel H1 mutation in the keratin 1 chain in epidermolytic hyperkeratosis. J Invest Dermatol 107:439–441. [DOI] [PubMed] [Google Scholar]
- Yang L, Mullen JR, Brill SJ. 2006. Purification of the yeast Slx5‐Slx8 protein complex and characterization of its DNA‐binding activity. Nucleic Acids Res 34:5541–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. 1998. Cofilin phosphorylation by LIM‐kinase 1 and its role in Rac mediated actin reorganization. Nature 393:809–812. [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD. 2010. The SUMO E3 ligase activity of Pc2 is coordinated through a SUMO interaction motif. Mol Cell Biol 30:2193–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Jaffray E, Senthinathan B, Hay RT, Sharrocks AD. 2003. SUMO and transcriptional repression: dynamic interactions between the MAP kinase and SUMO pathways. Cell Cycle 2:528–530. [DOI] [PubMed] [Google Scholar]
- Yang SH, Galanis A, Witty J, Sharrocks AD. 2006. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J 25:5083–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ydenberg CA, Smith BA, Breitsprecher D, Gelles J, Goode BL. 2011. Cease‐fire at the leading edge: new perspectives on actin filament branching, debranching, and cross‐linking. Cytoskeleton 68:596–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, Cleveland DW. 1991. CENP‐E, a novel human centromere‐associated protein required for progression from metaphase to anaphase. EMBO J 10:1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Pruyne D, Huffaker TC, Bretscher A. 2000. Myosin V orientates the mitotic spindle in yeast. Nature 406:1013–1015. [DOI] [PubMed] [Google Scholar]
- Yong‐Gonzales V, Hang LE, Castellucci F, Branzei D, Zhao X. 2012. The Smc5‐Smc6 complex regulates recombination at centromeric regions and affects kinetochore protein sumoylation during normal growth. PLoS One 7:e51540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KH, Yoon M, Moir RD, Khuon S, Flitney FW, Goldman RD. 2001. Insights into the dynamic properties of keratin intermediate filaments in living epithelial cells. J Cell Biol 153:503–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M, Moir RD, Prahlad V, Goldman RD. 1998. Motile properties of vimentin intermediate filament networks in living cells. J Cell Biol 143:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhang D, Liu J, Li J, Yu Y, Wu XR, Huang C. 2012. RhoGDI SUMOylation at Lys‐138 increases its binding activity to Rho GTPase and its inhibiting cancer cell motility. J Biol Chem 287:13752–13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Liu SJ, Li HL, Wang JZ. 2005. Microtubule‐associated protein tau is a substrate of ATP/Mg(2+)‐dependent proteasome protease system. J Neural Transm 112:547–555. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yu Q, Olsen L, Bi X. 2012. Functions of protosilencers in the formation and maintenance of heterochromatin in Saccharomyces cerevisiae . PLoS One 7:e37092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Goeres J, Zhang H, Yen TJ, Porter ACG, Matunis MJ. 2008. SUMO‐2/3 modification and binding regulate the association of CENP‐E with kinetochores and progression through mitosis. Mol Cell 29:729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Sarge KD. 2008a. Sumoylation of amyloid precursor protein negatively regulates Abeta aggregate levels. Biochem Biophys Res Commun 374:673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Sarge KD. 2008b. Sumoylation regulates lamin A function and is lost in lamin A mutants associated with familial cardiomyopathies. J Cell Biol 182:35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ryan JJ, Zhou H. 2004. Global analyses of sumoylated proteins in Saccharomyces cerevisiae: induction of protein sumoylation by cellular stresses. J Biol Chem 279:32262–32268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, Kullertz G, Stark M, Fischer G, Lu KP. 2000. Pin1‐dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell 6:873–883. [DOI] [PubMed] [Google Scholar]
- Zimdahl B, Ito T, Blevins A, Bajaj J, Konuma T, Weeks J, Koechlein CS, Kwon HY, Arami O, Rizzieri D, Broome HE, Chuah C, Oehler VG, Sasik R, Hardiman G, Reya T. 2014. Lis1 regulates asymmetric division in hematopoietic stem cells and in leukemia. Nat Genet 46:245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]


