Summary
The phytohormones cytokinin and auxin orchestrate the root meristem development in angiosperms by determining embryonic bipolarity. Ferns, having the most basal euphyllophyte root, form neither bipolar embryos nor permanent embryonic primary roots but rather an adventitious root system. This raises the questions of how auxin and cytokinin govern fern root system architecture and whether this can tell us something about the origin of that root.
Using Azolla filiculoides, we characterized the influence of IAA and zeatin on adventitious fern root meristems and vasculature by Nomarski microscopy. Simultaneously, RNAseq analyses, yielding 36 091 contigs, were used to uncover how the phytohormones affect root tip gene expression.
We show that auxin restricts Azolla root meristem development, while cytokinin promotes it; it is the opposite effect of what is observed in Arabidopsis. Global gene expression profiling uncovered 145 genes significantly regulated by cytokinin or auxin, including cell wall modulators, cell division regulators and lateral root formation coordinators.
Our data illuminate both evolution and development of fern roots. Promotion of meristem size through cytokinin supports the idea that root meristems of euphyllophytes evolved from shoot meristems. The foundation of these roots was laid in a postembryonically branching shoot system.
Keywords: apical development, auxin, cytokinin, fern, meristem regulation, RNAseq, root development, root evolution
Introduction
Roots are a key innovation of vascular plants. They probably arose twice, once in the fern lineage and once in lycopods (Pires & Dolan, 2012). Hence, the emergence of the complex sporophyte body was accompanied by the emergence of roots (Graham et al., 2000; Doyle, 2013). Phylogenetically early‐branching bryophytes also form root‐like rhizoids. Regardles of whether they are uni‐ or multicellular (Jones & Dolan, 2012), rhizoids are elementary structures that grow through tip growth in the same manner as root hairs (Menand et al., 2007). Gametophytes of ferns form rhizoids, too (Banks, 1999), but in contrast to the bryophytes, their life cycle is dominated by the sporophytic stage (as in angiosperms), in which they build proper roots (Gunning et al., 1978; Banks, 1999). During the evolutionary transition to a sporophyte‐dominated plant life cycle, regulators of gametophyte development were recruited for sporophyte development (Menand et al., 2007; Frank & Scanlon, 2015). Gametophyte and sporophyte development in basal land plants share control mechanisms (Landberg et al., 2013; Bennett et al., 2014; Viaene et al., 2014), but we know little about complex roots of basal vascular plants beyond that.
Two major apical meristems determine complex plant growth: the shoot apical meristem (SAM) and the root apical meristem (RAM). Exploring the root's origin is therefore exploring the origin of the RAM. Apical development through a SAM‐like structure precedes the emergence of the RAM (Graham et al., 2000; Prigge & Bezanilla, 2010) and regulation of the SAM and RAM show striking similarity (Stahl & Simon, 2010). It has therefore been speculated that the initial RAM evolved from the SAM (Stahl & Simon, 2010), a transition that must have happened in the early vascular plants. To understand this transition, we explored the molecular regulation of the fern RAM in comparison to that of angiosperms.
The Arabidopsis RAM has become the model system to study angiosperm meristem function (Petricka et al., 2012). While Arabidopsis forms a taproot system (allorhizy) that is dominated by the embryonic primary root, other plants such as grasses develop a fibrous root system (secondary homorhizy). Regardless of the mature root system architecture, angiosperms establish a primary root (and RAM) during early embryogenesis, serving angiosperm seedlings of both allorhizic and homorhizic species (Bellini et al., 2014). Angiosperm embryos establish the apical–basal polarity already at the two‐cell stage, a process orchestrated by polar auxin flow (Friml et al., 2003; Petrasek & Friml, 2009). Next to auxin, root identity in embryogenesis is determined by master regulators such as WUSCHEL RELATED HOMEOBOX (WOX) genes (Haecker et al., 2004; Schlereth et al., 2010; Radoeva & Weijers, 2014). Angiosperm roots develop longitudinally in three different zones: the meristematic zone (MZ), from which all root tissues arise, the elongation zone (EZ), where the cells reach their final size, and the differentiation zone (DZ), where elongated cells reach full maturation according to their respective fate (Dolan et al., 1993).
All tissues of the root originate from the stem cell niche's (SCN) initials (Petricka et al., 2012) that line the quiescent centre (QC), which itself remains mitotically inactive but maintains the meristematic activity of its surroundings (Clowes, 1954; van den Berg et al., 1997). Activity and establishment of the SCN are orchestrated through a few key regulators such as PLETHORA (PLT; Aida et al., 2004), SHORTROOT (SHR) and SCARECROW (SCR; Scheres et al., 1995; Sabatini et al., 2003). PLT is a further key determinant of the meristem size (Galinha et al., 2007). Activity and size of the RAM are regulated by the antagonists auxin and cytokinin (Blilou et al., 2005; Dello Ioio et al., 2007, 2008). While auxin promotes the MZ size through cell proliferation (Blilou et al., 2005), cytokinin restricts it by promoting earlier cell differentiation (Dello Ioio et al., 2007, 2008). The Aux/IAA auxin signalling repressor (Tian & Reed, 1999) SHORT HYPOCOTYL 2 (SHY2) (that is also targeted by the cytokinin signalling components RESPONSE REGULATOR 1 (ARR1) and ARR12; Sakai et al., 2001; Ishida et al., 2008; Moubayidin et al., 2010) determines the position of the transition zone (TZ; marking the border between MZ and EZ) and therefore the size of the MZ (Dello Ioio et al., 2008; Moubayidin et al., 2010; Perilli et al., 2012). The same appears true for the formation of adventitious roots in angiosperms, where auxin promotes and cytokinin hinders adventitious rooting (Werner et al., 2003; Gutierrez et al., 2012; Bellini et al., 2014).
Fern embryos do not develop in a bipolar manner (Hou & Hill, 2002). The embryonic RAM is only briefly present and the permanent fern roots arise adventitiously from a shoot, a process termed primary homorhizy (Goebel, 1930; Schneider, 2013) that is clearly distinguished from (secondary) homorhizic angiosperms with long‐living embryonic roots (Hou & Hill, 2002; Bellini et al., 2014). Instead of bearing a QC‐centred SCN, fern roots have a single root apical cell (RAC) that gives rise to root tissues (Gunning et al., 1978; Hou & Blancaflor, 2009). Our knowledge about the molecular mechanisms underlying the function of the fern RAC is limited, but Nardmann & Werr (2012) could show that WUSCHEL‐related transcription factors act in the RAC of Ceratopteris, demonstrating them to be ancient master regulators of plant development that radiated even before the angiosperm–gymnosperm split (Hedman et al., 2013). Studies on Ceratopteris showed that auxin did not alter lateral root formation, but inhibited parent root growth (Hou et al., 2004). Yet the molecular framework that underpins fern meristem regulation remained hidden. We used Azolla filiculoides, famous for its unique symbiosis with a nitrogen‐fixing cyanobacterium (Rai et al., 2000; Carrapiço, 2010), to characterize how the key developmental regulators auxin and cytokinin influence fern root development. While the longitudinal organization of the Azolla root resembles that of angiosperms, we found that the phytohormonal meristem regulation through auxin and cytokinin is reversed. Our results shed light on the evolutionary origin of the RAM in euphyllophytes.
Materials and Methods
Azolla culture and phytohormone treatment
Azolla filiculoides Lam. was cultivated in floating culture in a beaker containing 250 ml filtered water (pH 7.0) under 450 μmol quanta m−2 s−1 16 h 24°C : 8 h 20°C, light: dark, day : night, and 75% relative humidity. For phenotypic analysis, 2 d after root removal (hereafter defined as 2 dpc) roots were treated with 2.7 μl EtOH (mock), 0.1 μM IAA (Carl‐Roth; IAA, Karlsruhe, Germany) and 0.5 μM trans‐Zeatin (Sigma‐Aldrich; CK) for 24 h.
Phenotypic analysis
For differential interference contrast (DIC) analyses, roots were mounted in a chloral hydrate solution (5 : 2 : 1, chloral hydrate : glycerol : H2O) and bleached (i.e. tissues were cleared) for several days. Images were taken on a Nikon Ti Eclipse microscope (Nikon DS‐Qi1Mc camera). Root lengths (entire 3 dpc root) were determined via dissection microscopy (Nikon SMZ 745T; Nikon DS‐Ri1 camera). Images were processed using the NIS‐Elements BR 4.20.00 software (Nikon, Tokyo, Japan) and Adobe Photoshop and Illustrator CS6. Statistical analysis was performed using R 3.0.2 (R Core Development Team 2013), normality was tested using a Shapiro–Wilk test (Shapiro & Wilk, 1965) and, accordingly, a Mann–Whitney U‐test (Mann & Whitney, 1947) was performed. Amyloplasts were stained using 5% Lugol's iodine and pictures were taken on a Zeiss Axiophat microscope with an AxioCam ICc 5 camera, using the ZEN 2012 software (Zeiss). Dissection micrographs were generated using a SteREO Discovery V8 (Zeiss) microscopy with a AxioCAM ICc 5 and processed using the ZEN 2012 software (Zeiss).
Global gene expression analysis
For RNAseq analysis, 66 hpc roots were treated as described above but for 6 h; 3–4 mm root tips were collected and extracted using the Spectrum™ Plant Total RNA Kit (Sigma) according to the manufacturer's instructions. RNA was extracted for each treatment in triplicate (n ≥ 100 root tips each), always at the same time of the day. RNA quality was assessed using a formamide gel and shipped on dry ice to BGI Tech Solutions (Hong Kong), where RNA quality assessment by BioAnalyzer (Agilent Technologies, Waldbronn, Germany), library preparation using the TruSeq kit (Illumina, San Diego, CA, USA), and 100 bp paired‐end sequencing via the Illumina HiSeq2000 system were performed. After removal of low‐quality reads, we obtained a total of 270 866 324 reads (Supporting Information Table S1). The Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GBTV00000000 and the National Center for Biotechnology Information (NCBI) Single Read Archive (PRJNA264391). The version described in this paper is the first version, GBTV01000000.
Read quality was assessed using Fastqc v.0.10.1 (FASTQC 2012) and trimmed using Trimmomatic 0.32 (Bolger et al., 2014; settings: ILLUMINACLIP:TruSeq3‐PE.fa:2:30:10; HEADCROP:10; TRAILING:3; SLIDINGWINDOW:4:20; MINLEN:36). All reads were paired‐end assembled using Trinity r20131110 (Grabherr et al., 2011; Haas et al., 2013), resulting in 153 400 contigs (≥ 300 bp). Reads were mapped onto the contigs using the CLC Genomics Workbench 7 (CLC Bio). Contigs were annotated based on a BLASTx approach (Altschul et al., 1997) against the TAIR 10 peptide release (Lamesch et al., 2011); based on the false discovery rate, the e‐value cutoff was set to 10−7. Filtering for contigs with ≥ 150 mapped reads yielded 36 091 contigs (Table S2). Potential contamination was omitted by retaining only contigs that had best BLASTx hits (vs RefSeq; Pruitt et al., 2012) to streptophytes.
Gene onthology (GO) term analysis was performed based on the Arabidopsis annotations (e‐ value < 10−7) using the Gene Ontology enRIchment anaLysis and visuaLizAtion tool (GOrilla; P < 10−3) (Eden et al., 2009).
Differential regulation and statistical analysis were performed based on negative binomial probability distribution and Benjamini–Hochberg correction using edgeR (Robinson et al., 2010). Wordles were generated using wordle.net. Phytohormone signalling pathways were manually curated based on Arabidopsis homologues (Liscum & Reed, 2002; Lamesch et al., 2011; Brenner & Schmüllig, 2012). Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) was performed using the Power SYBR™ Green kit and a StepOnePlus™ (Applied Biosystems) system and analysed according to Pfaffl (2001). AfTufA (AzfiRT00021) and AfCAM5 (AzfiRT00154) were selected as reference genes based on their steady expression (confirmed during qPCR analysis on equal amounts of RNA) in the RNAseq data (see Table S3 for primer sequences).
Protein family matrix
Protein data were extracted from current genome releases (The Arabidopsis Genome Initiative, 2000; Merchant et al., 2007; Rensing et al., 2008; Schnable et al., 2009; The International Brachypodium Initiative, 2010; Banks et al., 2011; The Potato Genome Sequencing Consortium, 2011; Nystedt et al., 2013; Amborella Genome Project, 2013) from NCBI and Joint Genome Institute (JGI) and the longest open reading frames (start to stop; ≥ 100 aa; in silico translated using Emboss v.6.6.0; Rice et al., 2000) of RNAseq and expressed sequence tag (EST) data from Spirogyra pratensis (Timme & Delwiche, 2010), Marchantia polymorpha (Nagai et al., 1999), Ceratopteris richardii (Bushart et al., 2013; same filtering as for the Azolla RNAseq dataset) and Pteridium aquilinum (Der et al., 2011). Orthologous protein families were determined using a bidirectional best hit (Tatusov et al., 1997) all‐against‐all BLASTp v.2.2.29+ approach (e‐value ≤ 10−10; global sequence identity ≥ 30% calculated by Emboss v.6.6.0 Needle; Rice et al., 2000) and clustered using the Markov Cluster Algorithm (MCL) (Enright et al., 2002). Results were displayed in a Matlab R2014a (MathWorks, Natick, MA, USA)‐generated matrix.
Hierarchical clustering of expansin expression and phylogeny
An expansin dataset was generated based on best‐BLASTx hits against the Azolla root transcriptome and sequences from Sampedro & Cosgrove (2005), Li et al. (2002), the NCBI and the Arabidopsis Information Resource (TAIR; Table S4). Protein sequences were generated using the sequence manipulation suite (Stothard, 2000), aligned using Multiple Alignment using Fast Fourier Transform (Mafft) v.7.127b L‐INS‐I (Katoh & Standley, 2013) and Mega5.2.2 (Tamura et al., 2011) and modified to include only the conserved middle region of the protein sequences. Sequences containing only parts of the conserved region were removed. A maximum‐likelihood phylogeny was computed using WAG + G + I (best‐fit substitution model determined by Mega5.2.2; five discrete gamma categories, 1000 bootstrap replicates, partial deletion site coverage cutoff: 99%). Sequences were annotated based on best BLAST hits to Arabidopsis expansins, unless annotation was given. Hierarchical clustering on the normalized expression data was performed by using log2(FC) data. Data for 3 h CK‐ and IAA‐treated Arabidopsis thaliana seedlings were extracted from The Bio‐Analytic Resource for Plant Biology (BAR) (Toufighi et al., 2005; Winter et al., 2007). Clusters were generated using the CLC main workbench 7.0 (CLCbio, Qiagen) and Euclidean distance measures with single linkage.
Results
Azolla has adventitiously emerging shoot‐borne roots
Azolla roots emerge adventitiously as shoot‐borne roots (cf. Groff & Kaplan, 1988) from a node that contains a ventral and a dorsal leaf lobe (Fig. 1a). Roots of angiosperms can be longitudinally separated into the DZ, EZ and MZ (Dolan et al., 1993), a partitioning that we also used for the roots of Azolla. The largest and uppermost segment of the Azolla root is made up by the DZ, where the cells reached their final size, the xylem is differentiated and root hairs are prominent. The EZ begins with cells that are approximately twice as long as those of the MZ (from 8.7 ± 2.5 μm of the three outer cortex cells before the doubling to 16.8 ± 4.5 μm of the three outer cortex cells after the doubling, marking the TZ; Fig. 2a). At the tip lies the MZ, housing the RAC. Trichoblasts, from which the root hairs later emerge, are already distinguishable in the MZ as a result of their triangular appearance; proper differentiation of the root hairs occurs at the end of the MZ and subtending the inner root cap (Fig. S1). Azolla roots bear an outer (Fig. 2a green arrowhead) and an inner root cap (Figs 1b, 2a yellow arrowhead). The outer root cap ends approximately at the TZ, while the inner root cap ends approximately at the transition between EZ and DZ. Potassium iodide (KI) staining revealed no statocytes where one would expect a columella, but instead several basipetal amyloplasts lining the stele (Fig. S2). In contrast to the fern Ceratopteris (Hou et al., 2004; Hou & Blancaflor, 2009), Azolla does not have lateral roots.
Figure 1.

Emergence of shoot‐borne Azolla filiculoides roots. (a) Stereoscopic micrographs of Azolla filiculoides sporophytes in floating culture, showing a dorsal (upper panel), ventral (lower left panel) and detailed view (lower right panel). Arrows mark the shoot‐borne, adventitious emergence of roots. The detailed view shows the nodes containing a dorsal (D) and a ventral (V) leaf lobe. (b) Schematic drawing of the Azolla root that highlights the outer root cap (brown), the inner root cap (yellow), the apical cell (A), and differentiation of the xylem strands in dashed lines (outer thin lines represent protoxylem, and inner thick lines represent metaxylem).
Figure 2.

Azolla root meristem cell number is elevated upon cytokinin and decreased upon auxin treatment. (a) Nomarski interference contrast micrographs of Azolla filiculoides roots treated with solvent (control), 0.5 μM trans‐zeatin (CK) and 0.1 μM IAA. Open arrows mark the root apical cell (RAC) and closed arrows mark the end of the outer cortex meristematic zone (MZ). Inserts show an enlarged view of the outer cortex transition zone (TZ), marking the end of the MZ and giving the average length of the last three cells of the MZ and the first three cells of the elongation zone (EZ) in µm (± SD; cells are retraced by dashed lines). Numbers at the apex give the average length of the first 10 outer cortex cells in μm (± SD). The green arrowhead marks the outer root cap, the yellow arrowhead marks the inner root cap. (b) Quantification of the outer cortex MZ cell number in mock (control)‐, 0.5 μM CK‐ and 0.1 μM IAA‐treated 3 d post‐cut (dpc) roots; significance groups a–c (P < 0.001) were determined using Mann‐Whitney U‐statistics. (c) Quantification of the root length in mock (control)‐, 0.5 μM CK‐ and 0.1 μM IAA‐treated 3 dpc roots; significance groups a and b (P < 0.001) were determined using Mann–Whitney U‐statistics. Box‐plots in (b) and (c) display the interquartile range (IQR; 50 ± 25%) of the data; horizontal lines in each box mark the median (50%), whiskers extend to the furthest data points within the 1.5 × IQR range, and circles mark outliers. (d) Photographs of 3 dpc A. filiculoides sporophytes in floating culture upon mock (control), 0.5 μM CK and 0.1 μM IAA treatment. All presented data points are derived from evaluation of at least 40 roots per treatment (n = 40).
Size of the Azolla root MZ is increased by cytokinin and decreased by auxin
Auxin and cytokinin are key regulators of the MZ's activity in angiosperms and thus we analysed their effect on the Azolla root's MZ. To make sure that only meristems of the same age and developmental stage were analysed, roots were first removed. At 1 dpc, Azolla roots had replaced the removed roots but showed nonuniformal phenotypes. We used the distinct TZ to evaluate the Azolla MZ (starting from first emergence, cells of the outer cortex file were counted until they had doubled in size) and determined that 3 dpc roots showed the steadiest MZ size (Fig. S3), which were thereafter used for all analyses. Ferns were kept in floating culture for 2 dpc and transferred to a new culture containing either solvent (mock treatment) or 0.1 μM IAA or 0.5 μM trans‐zeatin (CK) for 24 h. The concentrations were chosen based on previous studies on meristem size in Arabidopsis (Růžička et al., 2009; Dello Ioio et al., 2012; Moubayidin et al., 2013), as well as preliminary experiments determining the minimal concentrations resulting in clear phenotypes. Mock‐treated roots (3 dpc) had an MZ cell number of 77 ± 12 (Fig. 2a,b). Exogenous 0.1 μM IAA application led to roots with a significant reduction (P < 0.001) in MZ size (62 ± 12; Fig. 2a,b). By contrast, application of 0.5 μM CK increased the MZ size (111 ± 19, P < 0.001; Fig. 2a,b). Application of the phytohormones also influenced the lengths of the MZ cells (Fig. 2a). In mock‐treated roots, the first 10 outer cortex cells had an average length of 5.4 ± 1.4 μm; IAA treatment increased this to 6.9 ± 1.4 μm (P < 0.001), while CK treatment decreased cell size to 4.7 ± 1.7 μm (P < 0.001). Control 3 dpc roots were 6.81 ± 3.1 mm in length. While 0.5 μM CK – although changing the meristem cell number – led to no significant change in root length (6.81 ± 2.7 mm; n = 40), 0.1 μM IAA resulted in a significant decrease (P < 0.001), to 3.73 ± 1.6 mm (Fig. 2c,d).
Auxin and cytokinin affect xylem formation
Auxin and cytokinin are also key regulators of root vasculature in Arabidopsis (Bishopp et al., 2011) and we therefore analysed their influence on xylem morphology under the same conditions as applied earlier (n = 40). Azolla roots bear two narrow protoxylem and two wide metaxylem strands (Fig. 1b). The protoxylem is c. 4 μm in diameter, progressing from scalariform ornamentation to annular ornamentation (Fig. 3). Protoxylem differentiates, on average, seven stele cells above the apex. One protoxylem strand differentiates before the other (approx. two cells), and differentiating either earlier on IAA or later on CK treatment (Fig. 3d). Metaxylem shows a scalariform ornamentation. First metaxylem strands differentiate as wide cell files (c. 8 μm in diameter) more tipwards, and second metaxylem strands differentiate as even wider cell files (c. 13 μm in diameter) more shootwards, than the rim of the inner root cap (Fig. 1b). Upon either phytohormone treatment, both metaxylem strands differentiated more towards the tip than in the untreated roots. The relative appearance of the second compared with the first metaxylem strand was also altered such that both strands appeared to start closer to each other (CK and IAA), or even simultaneously (IAA; Fig. 3f). This was accompanied by a change in the perforation plate angle, resulting in orthogonal instead of rhomboid‐shaped metaxylem cells (Fig. 3g). In addition, IAA caused the secondary cell wall deposition leading to the scalariform ornamentation being closer to each other (Fig. 3h). Both phytohormes resulted in a widening of the second metaxylem strand (Fig. 3i). Although varying marginally in size along the root, the second metaxylem strand was always wider than the first.
Figure 3.
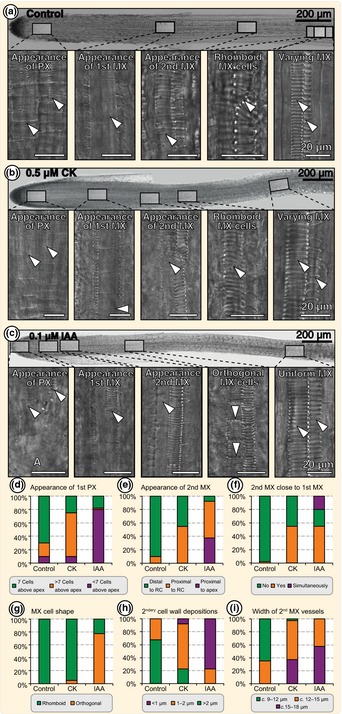
Azolla root xylem development upon cytokinin and auxin treatments. (a–c) Nomarski interference contrast micrographs of Azolla filiculoides roots treated with solvent (control), 0.5 μM trans‐zeatin (CK) and 0.1 μM IAA; blow‐ups of marked positions along the root show details of the xylem phenotypes, marked with arrowheads. A, root apical cell; PX, protoxylem; MX, metaxylem. (d–i) Quantification of observed alterations in the xylem phenotype after control, 0.5 μM CK and 0.1 μM IAA treatment. Data points are derived from evaluation of at least 40 roots per experiment (n = 40). Root cap (RC).
Embryophyte developmental regulators of the Azolla root
Plants colonized land at least 470 million yr ago (Gensel, 2008) and many regulators that shape plant bodies are ancient (Pires & Dolan, 2012). To understand which growth regulators support fern root development, > 270 million paired‐end reads of mRNA isolated from 3‐mm‐long A. filiculoides root tips – and under three different conditions (see later) – were sequenced. The de novo transcriptome data were assembled into 190 000 contigs, then filtered for those that were supported by ≥ 150 reads and an assembled length of ≥ 300 bp, resulting in 36 091 contigs (Fig. 4a; Table S1). To test for the depth and breadth of key developmental regulators among Azolla transcripts, we clustered all proteins derived from available genome, RNAseq and EST data (≥ 100 amino acids long) of a variety of streptophytes (Fig. 4) and Chlamydomonas reinhardtii as an outgroup using a bidirectional BLASTp approach. Screening this dataset for clusters that contained orthologues to 353 known A. thaliana developmental denominators resulted in 161 clusters that contained at least one orthologue from another species, 40 of which contained homologues expressed in the A. filiculoides root. In comparison, proteins from the genomes of Selaginella moellendorffii and Picea abies were detected in 36 and 58 clusters, respectively, and proteins from the transcriptomes of C. richardii and Pteridium aquilinum were detected in only 10 and 14 clusters, respectively. Thus, sequence depth of our de novo transcriptome was sufficient to detect developmental master regulators, such as homologues of WOX13 (AzfiRT13282, AzfiRT23190, AzfiRT23355; e‐value < 10−47), bearing the characteristic ancestral NVYNWFQN peptide sequence at the second turn of the recognition helix (Nardmann & Werr, 2012). Key denominators of true root meristems known from angiosperms, such as three homologues of PLT (AzfiRT20391, AzfiRT31772, AzfiRT33493; e‐value < 10−95), were identified (Table S5), accounting for the readout of the meristem regulon. But does differential expression of these key regulators translate into the different phenotypes observed upon phytohormone treatment?
Figure 4.
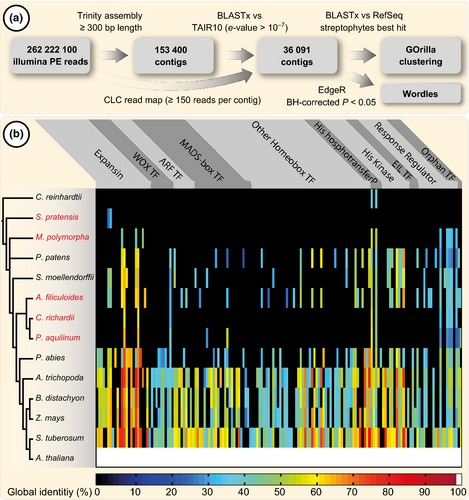
RNAseq of Azolla root tips and identification of orthologues to known developmental regulators across 14 species of Chloroplastida. (a) Workflow of the RNAseq analysis of Azolla filiculoides root tips starting with the filtered reads obtained by illumina paired‐end (PE) sequencing followed by assembly using the Trinity pipeline, annotation and filtering via BLASTx and read mapping using the CLC workbench. For downstream analyses (protein clustering, expression wordles of significantly regulated genes or gene ontology (GO) term enrichment via GO rilla), only contigs that had their best BLASTx hit to streptophytes were used. (b) Protein data were extracted from nine genomes (black species names) and five RNAseq and expressed sequence tag (EST) libraries (red species names), and clustered; Chlamydomonas reinhardtii serves as an outgroup. Clusters were generated from all‐against‐all bidirectional best BLASTp hits. Those clusters were screened for 353 Arabidopsis thaliana proteins from 10 gene families (top row) and 161 clusters that contained at least one other species were extracted. The global identity of the identified homologues is displayed as a gradient colour. Note the high number of identified homologues in the A. filiculoides root tip transcriptome. The phylogeny on the left is based on the National Center for Biotechnology Information taxonomy database.
To connect the phenotypic observations to molecular mechanism, RNA was sequenced in triplicate from 3 mm root tips treated for 6 h with 0.1 μM IAA, 0.5 μM CK or solvent (mock). The 6 h treatment was chosen to detect the established readout over initial signalling components of the phytohormone response (Goda et al., 2004). Observed trends in expression were confirmed by two‐step qRT‐PCR (Fig. S4). To our knowledge, this is the first global gene expression profiling on a fern root, so we first ran a GO term enrichment analysis (P < 10−3) on the top 2000 highest expressed genes from all root datasets and compared this with sequence data available from the Azolla sporophyte (Brouwer et al., 2014) based on homology to Arabidopsis proteins (e‐value < 10−7). The most evident difference is the expression of photosynthesis and pigment metabolism GO terms, detected only in the photosynthesising sporophyte system (Fig. 5a; Table S6). The root expression profiles were enriched for various kinds of transport GO terms, including multifaceted ion transport. Intriguingly, photorespiratory genes were highly expressed in the Azolla root, a phenomenon that was recently discussed for angiosperm roots as well (Nunes‐Nesi et al., 2014).
Figure 5.
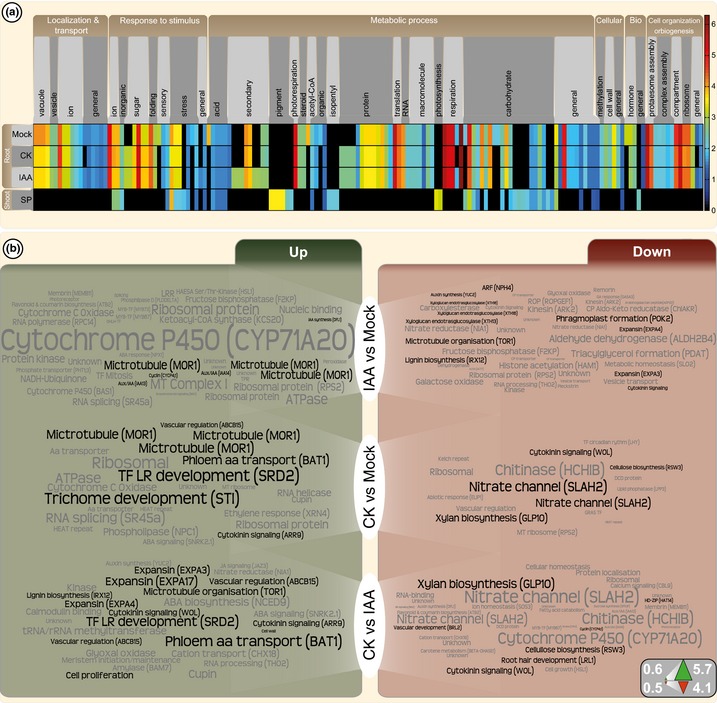
Gene expression changes in the Azolla root tip after auxin and cytokinin treatments. (a) Gene ontology (GO) enrichment analysis (P < 10−3) based on the Arabidopsis annotation (e‐value < 10−7) of the top 2000 expressed genes in 3 d post‐cut (dpc) roots 6 h after treatment with solvent (mock), 0.1 μM IAA and 0.5 μM trans‐zeatin (CK) and in the shoots. The fold enrichment of the GO terms is depicted as a colour gradient and GO terms were manually sorted into umbrella categories (top). (b) Significantly differentially regulated (P < 0.05) homologues (e‐value < 10−7) to Arabidopsis expressed in 0.3 mm of the root tip of Azolla filiculoides, represented as a word cloud. The bigger the word, the stronger the differential regulation comparing 0.1 μM IAA with mock (IAA vs mock), 0.5 μM CK with mock, and 0.5 μM CK with 0.1 μM IAA treatment. Words in black are of special interest with regard to the main text. Homologues are annotated based on A. thaliana; if applicable, the A. thaliana gene symbol is provided in brackets. Up‐regulated homologues are shown on the left, down‐regulated homologues on the right. Word sizes are based on log2(fold change); see the key in the right corner. For all individual values, see Supporting Information Table S6.
Using a ranked GO term enrichment analysis highlighted the differential responses towards IAA and CK (Fig. S5). GO terms for RNA biosynthesis were prominent, indicating that both treatments triggered strong and specific transcriptional responses by the fern. Performing a ranked comparison of genes up‐ and down‐regulated by CK in comparison to IAA highlighted the antagonistic action of the phytohormones. Not only were GO terms for regulation of meristem growth up‐regulated upon CK, but so too were cell cycle regulation and inositol metabolism, which are well‐known growth regulators (Stevenson et al., 2000). Importantly, no GO terms associated with stress responses were highlighted. Hence, the reduction of meristem size observed upon auxin treatment is governed by developmental control and not stress.
Auxin and cytokinin differentially shape Azolla roots through cell wall modification
To detect key players and candidate genes for future studies involved in governing Azolla root development upon IAA and CK, we pursued a top‐down approach. Screening among contigs with homology to nuclear land plant genes, and which were significantly regulated (Benjamini–Hochberg corrected P‐value < 0.05), identified several homologues (e‐value < 10−7) to Arabidopsis growth regulators (Fig. 5b; Table S6). As indicated by the GO terms (Fig. S5) in particular, ‘information processing’ was up‐regulated upon treatment. This included RNA processing such as the RNA splicing SR45 homologue (e‐value 1.74 × 10−09) and ribosomal proteins in general. Notable was the 54‐fold up‐regulation of a CYTOCHROME 450 71A20 (CYP71A20) homologue, whose orthologue in A. thaliana is down‐regulated upon auxin treatment (Goda et al., 2004).
Both phytohormone treatments resulted in pronounced expression changes of the cell wall‐modifying expansins (e.g. EXPA4 CK vs mock, 0.3‐fold up in contrast to 2.1‐fold down in IAA vs mock; P < 0.05). The α‐expansins, in particular, were enriched among Azolla root transcripts, whereas only one β‐expansin and one α‐expansin‐like were detected. Conserved regions of Azolla expansins (Fig. S6b) match well with those of higher embryophytes (Li et al., 2002; Sampedro & Cosgrove, 2005), including the α‐ and β‐specific insertions (Li et al., 2002). Phylogenetic analysis using 93 protein sequences from 10 embryophytes show a clear distinction between the α‐ and β‐expansins, the expansin‐like sequences clustered with the β‐expansins (Fig. S6). By contrast, other expansin phylogenies (Sampedro & Cosgrove, 2005), expansin sequences of Azolla filiculoides, Physcomitrella patens, Marsilea quadrifolia and Regnellidium diphyllum formed lower plant‐specific clades instead of being equally dispersed throughout the α‐sequences. We hierarchically clustered normalized expression data of α‐expansins upon auxin and cytokinin treatment in A. thaliana and A. filiculoides. Four of the 10 clusters obtained were species‐specific, and only one for Azolla (Fig. 6a). The remaining six clusters were dominated by one species, four out of six by A. filiculoides. To analyse putative subfunctionalization after species‐specific duplications, we identified potential orthologues, co‐orthologues and paralogues in A. filiculoides and A. thaliana and estimated pairwise distances within those groups using OrthoMCL (Li et al., 2003). In agreement with our phylogenetic analysis (Fig. S6) we obtained only a few orthologues and a larger group of co‐orthologues. When plotting the pairwise distances of each pair against its absolute expression differences, there was little correlation (Fig. 6b). All in all, the data support the conclusion that, based on a common expansin set in all land plants, ferns – perhaps also mosses – have advanced their expansin repertoire, governing a differential use.
Figure 6.
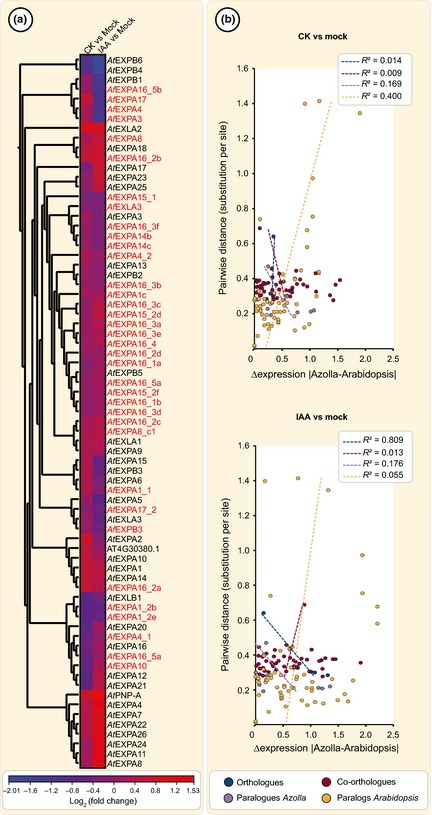
Comparison of Azolla filiculoides and Arabidopsis thaliana expansin expression patterns in relation to their relatedness. (a) Relative expression of the expansin gene family after trans‐zeatin (CK) vs mock treatment (left column) and IAA vs mock treatment (right column) in A. thaliana seedlings and A. filiculoides roots (in red letters) was clustered hierarchically using Euclidean distance measures and single linkage. Expansin expression is shown in red for up und blue for down and was calculated as log2(fold change). Both species‐specific and mixed clusters were obtained, but clusters rarely formed among evolutionarily related expansins. To confirm this relatedness of expansins to each other, a substitution rate per site, estimated with JTT + G + I with five discrete gamma categories (pairwise differences), was correlated to the absolute value of difference in expression between each two expansin genes (Δexpression). (b) No correlation between the pairwise differences and the absolute value of expression differences was observed in any tested phylogenetic category (purple and yellow, paralogues; blue, orthologues; red, co‐orthologues) under CK vs mock treatment. Under IAA vs mock treatment, only a negative correlation for orthologous expansin genes was observed, indicating that, sequence‐wise, highly similar orthologous genes are used differently in ferns and dicots.
Other cell wall modifiers were specifically regulated by the phytohormones as well. A homologue of the cellulose biosynthesis‐associated gene RADIAL SWELLING3 (RSW3; e‐value 0) was down‐regulated significantly (P < 0.05), when comparing CK with IAA treatment (3.6‐fold down), and Arabidopsis rsw3 mutants display a severe root phenotype (Baskin et al., 1992; Burn et al., 2002). An IRX12 (IRREGULAR XYLEM 12, an enzyme involved in lignin biosynthesis) homologue (e‐value 1.74 × 10−166) was significantly (P < 0.05) down‐regulated upon IAA (2.6‐fold) and only marginally down‐regulated upon CK. Arabidopsis irx12 mutants have strong alterations in xylem formation, displaying collapsing vessel elements (Brown et al., 2005), and regulation of Azolla IRX12 homologues might therefore be correlated with the observed xylem phenotypes.
Key players for the correct xylem cell differentiation are cellulose deposition‐guiding microtubules (Oda & Fukuda, 2012). The expression of several MICROTUBULE ORGANISATION 1 homologues (MOR1; e‐values < 1.18 × 10−107) increased upon IAA and CK treatment (4.4‐, 4.1‐ and 3.9‐fold up and 4.1‐, 3.8‐ and 3.7‐fold up, respectively; Fig. 5b). A TORTIFOLIA1 homologue (TOR1; e‐value 1.02 × 10−24) that mediates microtubule‐directed cell wall development (Buschmann et al., 2004) was significantly down‐regulated by auxin (2.8‐fold down; P < 0.05). Gene expression changes associated with vascular function were also detected. This included regulation of the ABC transporter ABCB15 (AzfiRT08887; e‐value 0; 1.9‐fold down upon IAA treatment) that is expressed in developing vasculature and associated with auxin transport and secondary cell wall development (Kaneda et al., 2011) and the phloem amino acid transporter BAT1 homologue (AzfiRT35714; e‐value 0), which was 3.7‐fold up‐regulated upon CK treatment (P < 0.05). Intriguingly, nitrate transport‐associated homologues were up to eightfold down‐regulated upon CK treatment (SLAH2; AzfiRT18705, AzfiRT19565), which could be explained by the link between CK biosynthesis and nitrogen availability (Takei et al., 2004). IAA and CK, therefore, shape the root of Azolla radially and longitudinally by the differential regulation of cell wall‐loosening and ‐reinforcing enzymes.
Auxin and cytokinin activate specific signalling components in Azolla roots
Root meristems are subject to a tight regulation by auxin and cytokinin. CYTOKININ RESPONSE 1 (CRE1) is a key component in cytokinin signalling (Inoue et al., 2001; Fig. 7; Table S6) and Arabidopsis cre1 mutants have severe root phenotypes (Mähönen et al., 2000; Ueguchi et al., 2001). Three homologues of CRE1 (e‐values 2.51 × 10−72, 4.13 × 10−39 and 3.82 × 10−39) were found to be regulated upon CK treatment, two up‐ (1.8‐ and 2.0‐fold), one down‐regulated (3.4‐fold; P < 0.05). A total of 14 contigs with homology to CRE1 (e‐values < 10−14) were detected in total, only seven of which are predicted to represent isoforms. No other cytokinin signalling component showed a strong response, except for an A‐type RESPONSE REGULATOR 9 (ARR9) homologue (AzfiRT12218; e‐value 4.33 × 10−50; Figs 5, 7) that was up‐regulated 2.2‐fold (P < 0.001) upon CK treatment.
Figure 7.
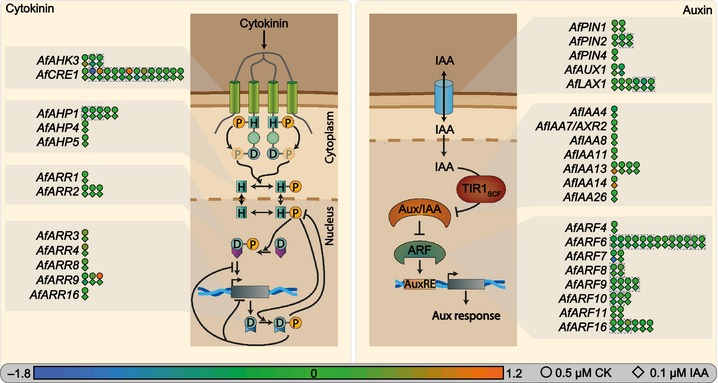
Cytokinin and auxin trigger specific components of their signalling pathways in the Azolla root tip. The figure shows the number of detected Arabidopsis thaliana cytokinin (left) and auxin (right) signalling pathway components expressed in 0.3 mm root tips of A. filiculoides, based on homology (e‐value < 10−7). Differential expression of each contig is represented by a circle (response to 0.5 μM trans‐zeatin, CK) and a rectangle (response to 0.1 μM IAA) which are coloured in a gradient that labels log2(fold change) from orange (1.2) to green (0) to blue (−1.8). Although many homologues for the canonical auxin signalling pathway were detected, no homologue of the SCFTIR 1 complex could be identified. Grey boxes with dotted lines encase potential isoforms. For all individual values, see Supporting Information Table S6. Aux, auxin.
Expression of three known auxin signalling factors was significantly altered upon auxin treatment (Figs 5, 7). Two Aux/IAA homologues were detected, grouping with the B‐group Aux/IAA proteins of Arabidopsis (Fig. S7; Remington et al., 2004) that are known for their importance in primary and lateral root development. One Azolla Aux/IAA homologue showed its highest identity to IAA13 (AzfiRT01636; e value 6.64 × 10−35; up‐regulated almost two‐fold upon auxin treatment), known to be important for root initiation during embryogenesis and later expressed in the stele (Weijers et al., 2005). Besides its phylogenetic placement in the B‐group, the other homologue showed highest identity to IAA14 (Solitary root; AzfiRT00243; e value 1.84 × 10−35; up‐regulated almost two‐fold upon auxin treatment). IAA14 is a known regulator of lateral root formation and Arabidopsis iaa14 mutants do not generate any lateral roots (Fukaki et al., 2002). Finally, a transcript of an AUXIN RESPONSE FACTOR was found to be down‐regulated upon IAA treatment, showing highest identity to ARF7 that is known to be a crucial regulator of lateral root formation (Okushima et al., 2007). Also a homologue of the transcription factor SHOOT REDIFFERENTIATION DEFECTIVE 2 (SRD2; e value 1.40 × 10−10) was found to be up‐regulated upon CK treatment (5.4‐fold; P < 0.05) and Arabidopsis srd2 mutants are known to have impaired lateral and primary root growth (Ohtani & Sugiyama, 2005). Thus, while several components of the CK and IAA signalling pathway are expressed by the Azolla root, only specific factors showed a strong reaction after 6 h of the respective phytohormone treatment, many of which are associated with lateral root formation in Arabidopsis.
Discussion
Understanding meristem regulation is key to understanding plant development. The RAM of Arabidopsis has become a priced model to uncover the basic principles of angiosperm development (Petricka et al., 2012), but our knowledge about the evolutionary earlier fern root development is rather scarce (Hou & Blancaflor, 2009). Our results highlight the fundamental differences in the control of the Azolla RAM by the key regulators cytokinin and auxin.
Auxin, cytokinin and the reciprocal regulation of the root system in Azolla
Similar to Arabidopsis (Dolan et al., 1993), we found that Azolla has a DZ, EZ and MZ. While auxin is known to increase Arabidopsis MZ cell number (Blilou et al., 2005), cytokinin is known to reduce it (Dello Ioio et al., 2007). Similar applies to Arabidopsis’ adventitious roots, which one can assume constitute the more obvious analogue to the Azolla root. Auxin is a known inductor and promoter of adventitious roots, while cytokinin acts antagonistically (Bellini et al., 2014). Surprisingly, application of IAA and CK to the Azolla adventitious roots resulted in the opposite effect. Root growth inhibition through auxin is well known in the fern Ceratopteris (Hou et al., 2004) and also in Arabidopsis, where it acts on cell elongation (Rahman et al., 2007). Yet the exogenous auxin naphthaleneacetic acid (NAA; acting in a similar manner to IAA on root meristems; Rahman et al., 2007) increases MZ cell number in the latter and shows inhibitory effects only at very high concentrations (Růžička et al., 2009). While decreasing MZ size through earlier differentiation (Dello Ioio et al., 2007), cytokinin affects root length in Arabidopsis only at high concentrations (Růžička et al., 2009). In the case of Azolla, CK did not alter root growth but it did alter MZ cell number. This might be explained by a CK‐induced delay in differentiation and a simultaneous restraint on cell length.
In summary, IAA influenced both cell elongation (in MZ and TZ) and MZ cell number. This resulted in its significant impact on root length, whereas phenotypes triggered by CK were mainly governed by changes in cells of the MZ and not the elongation in Azolla roots. This is further supported by the xylem phenotypes that displayed either a delayed or a premature differentiation of proto‐ and metaxylem. More densely ornamented xylem and orthogonal instead of rhomboid metaxylem cells observed upon IAA treatment might be the simple consequence of a delayed or impaired elongation. All of these are linked to the relative position to the apex and are thus probably a direct cause of longitudinal alterations. Our information is limited with regard to understanding the upstream molecular regulation that elicits these phenotypic alterations. For example, the RNAseq analysis revealed CYP71A20 to be differently regulated upon auxin treatment in comparison to its A. thaliana homologue (Goda et al., 2004). This might suggest a possible difference in the upstream perception of auxin in the Azolla root, but it is just as likely that auxin and cytokinin signalling acts downstream on very specific factors. Azolla is currently not accessible for genetic studies. Nevertheless, based on sequence homology, a set of genes were detected that can be associated with the observed phenotypes induced by the phytohormones; that provide information about the Azolla root developmental regulon and identity; and that are valuable candidate genes for future studies on fern root development.
Regulon of Azolla roots
The rigid cell walls of plants pose a challenge when a change in phenotype is required. We found that the most strongly regulated genes in Azolla roots were those involved in cell wall modification, a major group of which were the expansins that are involved in virtually all processes during plant cell remodelling and growth (Sampedro & Cosgrove, 2005). Of all expansins, the α‐expansins are thought to be the most gene‐rich group in the common ancestor of embryophytes (Sampedro & Cosgrove, 2005). The same is true for the transcriptome of the Azolla root. We did not find many orthologous α‐expansin genes between A. thaliana and A. filiculoides, but we did find co‐orthologous ones. This suggests that after an initial round of expansin family inflation in the common ancestor of embryophytes, several subsequent rounds of lineage‐specific radiations occurred. Expansins influence cell wall loosening, which is linked to the auxin‐mediated acid‐growth hypothesis (Rayle & Cleland, 1992; Link & Cosgrove, 1998; Catalá et al., 2000; Cosgrove, 2000). Yet exogenous auxin is known to reduce root length in Arabidopsis through altering the cell elongation (Rahman et al., 2007; Růžička et al., 2009). IAA‐induced reduction in root length and down‐regulation of expansins suggest that the same occurs in Azolla roots. Cell wall modification and cellulose deposition are further mediated through microtubule organization (Wasteneys, 2004). Intriguingly, both CK and IAA lead to similar changes with regard to xylem phenotypes and up‐regulation of MOR1 homologues. Owing to the direct link between xylem phenotype and microtubule organization (Oda & Fukuda, 2012), both phenotypes could be explained through the same mechanism: genetically controlled cell wall modification that shapes the Azolla root.
Fern root systems are governed by adventitiously arising roots (primary homorhizy); their primary RAM and resulting primary root are only short‐lived (Hou & Blancaflor, 2009). This stands in clear contrast to (secondary) homorhizy in angiosperms, where the primary root is long‐living. Yet, as in angiosperms, each of these adventitious roots has, similar to lateral roots (De Smet et al., 2006), its own postembryonic apical meristem in form of a RAC. Regulators of secondarily formed meristems (De Smet et al., 2006) are thus a likely component of the Azolla root's meristematic signature, not least because the control of adventitious and lateral root formation seems to largely overlap (Gutierrez et al., 2009; Bellini et al., 2014). Indeed, although Azolla does not have any lateral roots, a salient amount of differentially expressed Azolla root genes were homologous to lateral root developmental regulators. In angiosperms the determination of the lateral root founder cell identity is largely regulated by auxin (Fukaki et al., 2005). Yet, in Ceratopteris, auxin affects only the adventitious parent root, but not lateral root development (Hou & Hill, 2002; Hou et al., 2004). Consistent with the canonical Arabidopsis auxin signalling pathway (Liscum & Reed, 2002), expression of b‐group Aux/IAAs was induced upon IAA, but apparently did not promote the activity of the adventitiously formed RAC in a manner that would result in a larger MZ. Yet, cytokinin can also regulate lateral root development. Homozygous Arabidopsis wol mutants display no lateral root emergence from the primary root, but bear only hypocotyl‐borne roots (Scheres et al., 1995). We found WOL to be one of the most ubiquitous cytokinin signalling components in the Azolla root. In addition, a possible homologue of BREVIS RADIS (AzfiRT34406; e‐value 1.01 × 10−06; Table S5) was also detected, which is thought to be crucial for cytokinin's regulation of lateral root formation (Li et al., 2008). What could this imply for the evolutionary origin of the euphyllophyte root?
From thallus to cormus and from adventious to primary roots
Apical growth took place in the common ancestor of all land plants and precedes the evolution of roots (Graham et al., 2000; Prigge & Bezanilla, 2010; Pires & Dolan, 2012). Even the most basal plants, including some streptophyte algae (Kenrick & Crane, 1997; Wodniok et al., 2011), develop through the action of an apical cell (Graham et al., 2000). Liverworts, or hornworts, are often discussed to be the most ancient land plants (Gensel, 2008; Wickett et al., 2014) and their thalli grow through marginal meristems (Hagemann, 1999), but also by the activity of apical cells (Goffinet & Buck, 2013). Both sporophytes and gametophytes of ferns (and mosses) bear apical cells mediating their growth (Hagemann, 1999; Prigge & Bezanilla, 2010; Schneider, 2013). Although the roots of lycophytes and euphyllophytes most probably stem from two independent origins, both develop through an RAC (Prigge & Bezanilla, 2010; Pires & Dolan, 2012). Sanders & Langdale (2013) found that auxin and cytokinin act antagonistically on branching in the lycophyte Selaginella kraussiana; in that case, auxin led to a higher degree of root branching. It was concluded that this mechanism could be homologous to lateral root development of angiosperms, in which auxin promotes lateral root formation. In the fern Ceratopteris, however, lateral root development was not influenced by auxin (Hou et al., 2004). This means that while the antagonism of auxin and cytokinin is conserved in lycophytes and euphyllophytes, its outcome for the respective organs is not.
A shoot‐like origin for euphyllophyte roots?
Fossil records show that early vascular plant roots are strikingly similar to shoots. Roots were therefore predicted to have evolved through dichotomous branching from shoots (Graham et al., 2000; Jiang & Feldman, 2005). If we now recall that moss gametophytes and sporophytes share developmental regulators (Menand et al., 2007; Bennett et al., 2014; Viaene et al., 2014), it seems plausible that for the development of the basal fern‐sporophytic body, regulators also found in moss gametophore development were recruited. This process would have then occurred in shoot‐like structures governing a shoot‐first morphology. It has been hypothesized that the RAM arose from the SAM (Jiang & Feldman, 2005). Yet, plant development is often governed by the same regulators, which can act in very different tissues, leading to different outcomes (Prigge et al., 2005). We detected contigs with homologies to Arabidopsis genes that could be characteristic for either root or shoot. Whether these indicate a more root‐like or shoot‐like regulation of the Azolla RAC regulon cannot be stated, not least because of the similarity in RAM and SAM regulation (Stahl & Simon, 2010). Only genetic studies will be able to elucidate the detailed framework that regulates fern RAMs.
The observed phytohormone effect, however, provides intriguing implications for the evolution of root identity. Cytokinin is known as a growth inductor of shoot branches, while auxin inhibits this process (Müller & Leyser, 2011). Both shoot branches and adventitious roots arise laterally from the shoot during postembryonic development. If we entertain the idea of a shoot‐first origin of roots, there are two lessons to be learned from Azolla root development with regard to the basal sporophyte root system in euphyllophytes: their origin is adventitious and postembryonic, and they branch off as an altered shoot supported by cytokinin. Whether these adventitious roots are homologous to the embryonic primary roots of seed plants remains to be proven. Yet, it is clear that auxin's role in the dominant organs of the Arabidopsis root system does not apply in the same way to the fern Azolla. Our data indicate that it might be cytokinin instead. The key will be to determine the identity of the transiently present RAM of the fern embryo. It further raises the question at what point during euphyllophyte evolution the first permanent embryonic roots emerged, and whether this process was the advent of auxin's dominance during (lateral) root developmental regulation.
Author contributions
J.d.V and S.B.G. planned and designed the research. A.M.F. and J.d.V. performed experiments. J.d.V., M.R. and S.R. performed bioinformatic analyses. All authors discussed the results and commented on the manuscript that J.d.V. and S.B.G drafted. The final version of the manuscript was written by J.d.V., H.S., A.B., A.C. and S.B.G. and approved by all authors .
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Root hair development in Azolla.
Fig. S2 Amyloplasts line the Azolla stele.
Fig. S3 Regeneration of the meristematic zone.
Fig. S4 Conformation of global gene expression trends through two‐step quantitative reverse‐transcription PCR.
Fig. S5 Ranked gene ontology analysis of global gene expression in Azolla roots.
Fig. S6 Evolution of expansins.
Fig. S7 Phylogenetic analysis of Arabidopsis and regulated Azolla Aux/IAA proteins.
Table S1 Overview of the RNAseq analysis
Table S2 Annotations and expression values of the Azolla filiculoides root tip transcriptome
Table S3 Oligonucleotides used for two‐step qRT‐PCR
Table S4 Sequences used for the alignment of land plant expansins
Table S5 Known developmental regulators expressed in Azolla root tips
Table S6 Significantly regulated contigs upon auxin and cytokinin treatment and GOterm enrichment analysis depicted in Figs 5 and 7
Acknowledgements
This work was supported by the SFF (Heinrich Heine University) and the DFG (1825/4‐1) to S.B.G. We thank Elke Wieneke for superb technical assistance, Steffen Köhler (CAi, HHU) for excellent photography, Michael Hewera (Population Genetics, HHU) for help with statistical analysis, and Ulla Rasmussen, John Larsson and Birgitta Bergman (Botany, Stockholm University) for plant material.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh Y‐S, Amasino R, Scheres B. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amborella Genome Project . 2013. The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Banks JA. 1999. Gametophyte development in ferns. Annual Review of Plant Physiology and Plant Molecular Biology 50: 163–186. [DOI] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, Bowman JL, Girbskov M, de Pamphilis C, Albert VA, Aono N, Aoyaama T, Ambrose BA et al 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI, Betzner AS, Hoggart R, Cork A, Williamson RE. 1992. Root morphology mutants in Arabidopsis thaliana . Australian Journal of Plant Physiology 19: 427–437. [Google Scholar]
- Bellini C, Pacurar DI, Perrone I. 2014. Adventitious roots and lateral roots: similarities and differences. Annual Review of Plant Biology 65: 639–666. [DOI] [PubMed] [Google Scholar]
- Bennett TA, Liu MM, Aoyama T, Bierfreund NM, Braun M, Coudert Y, Dennis RJ, O'Connor D, Wang XY, White CD et al 2014. Plasma membrane‐targeted PIN proteins drive shoot development in a moss. Current Biology 24: 2776–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. 1997. Short‐range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287–289. [DOI] [PubMed] [Google Scholar]
- Bishopp A, Help H, El‐Showk S, Weijers D, Scheres B, Friml J, Benková E, Mähönen AP, Helariutta Y. 2011. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Current Biology 21: 917–926. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Schmüllig TS. 2012. Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largelysimilar but also organ‐specific responses. BMC Plant Biology 12: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P, Bräutigam A, Külahoglu C, Tazelaar AOE, Kurz S, Nierop KGJ, van der Werf A, Weber APM, Schluepmann H. 2014. Azolla domestication towards a biobased economy? New Phytologist 3: 1069–1082. [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turnera SR. 2005. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn JE, Hurley UA, Birch RJ, Arioli T, Cork A, Williamson RE. 2002. The cellulose‐deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N‐glycans during ER quality control. Plant Journal 32: 949–960. [DOI] [PubMed] [Google Scholar]
- Buschmann H, Fabri CO, Hauptmann M, Hutzler P, Laux T, Lloyd CW, Schäffner AR. 2004. Helical growth of the Arabidopsis mutant tortifolia1 reveals a plant‐specific microtubule‐associated protein. Current Biology 14: 1515–1521. [DOI] [PubMed] [Google Scholar]
- Bushart TJ, Cannon AE, Ul Haque A, San Miguel P, Mostajeran K, Clark GB, Porterfield DM, Roux SJ. 2013. RNA‐seq analysis identifies potential modulators of gravity response in spores of Ceratopteris (Parkeriaceae): evidence for modulation by calcium pumps and apyrase activity. American Journal of Botany 100: 161–174. [DOI] [PubMed] [Google Scholar]
- Carrapiço F. 2010. Azolla as a superorganism. Its implication in symbiotic studies In: Seckbach J, Grube M, eds. Symbioses and stress: joint ventures in biology, cellular origin, life in extreme habitats and astrobiology, vol 17. Berlin, Germany: Springer Science & Business Media SA, 225–241. [Google Scholar]
- Catalá C, Rose JK, Bennett AB. 2000. Auxin‐regulated genes encoding cell wall‐modifying proteins are expressed during early tomato fruit growth. Plant Physiology 122: 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes F. 1954. The promeristem and the minimal constructional centre in grass root apices. New Phytologist 53: 108–116. [Google Scholar]
- Cosgrove DJ. 2000. Loosening of plant cell walls by expansins. Nature 407: 321–326. [DOI] [PubMed] [Google Scholar]
- De Smet I, Vanneste S, Inzé D, Beeckman T. 2006. Lateral root initiation or the birth of a new meristem. Plant Molecular Biology 60: 871–887. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana‐Martinez E, Heidstra R, Costantino P, Sabatini S. 2007. Cytokinins determine Arabidopsis root‐meristem size by controlling cell differentiation. Current Biology 17: 678–682. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Galinha C, Fletcher AG, Grigg SP, Molnar A, Willemsen V, Scheres B, Sabatini S, Baulcombe D, Maini PK et al 2012. A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Current Biology 22: 1699–1704. [DOI] [PubMed] [Google Scholar]
- Der JP, Barker MS, Wickett NJ, dePamphilis CW, Wolf PG. 2011. De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum . BMC Genomics 12: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84. [DOI] [PubMed] [Google Scholar]
- Doyle JA. 2013. Phylogenetic analyses and morphological innovations in land plants. Annual Plant Reviews 45: 1–50. [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large‐scale detection of protein families. Nucleic Acids Research 30: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FASTQC . 2012. A quality control tool for high throughput sequence data. [WWW document] URL http://www. bioinformatics.babraham.ac.uk/projects/fastqc [accessed 7 April 2014]
- Frank MH, Scanlon MJ. 2015. Transcriptomic evidence for the evolution of shoot meristem function in sporophyte‐dominant land plants through concerted selection of ancestral gametophytic and sporophytic genetic programs. Molecular Biology and Evolution 32: 355–367. [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. 2003. Efflux‐dependent auxin gradients establish the apical–basal axis of Arabidopsis . Nature 426: 147–153. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. 2005. Tissue‐specific expression of stabilized SOLITARY‐ROOT/IAA14 alters lateral root development in Arabidopsis . Plant Journal 44: 382–395. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. 2002. Lateral root formation is blocked by a gain‐of‐function mutation in the SOLITARY‐ROOT/IAA14 gene of Arabidopsis . Plant Journal 29: 153–168. [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. 2007. PLETHORA proteins as dose‐dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Gensel PG. 2008. The earliest land plants. Annual Review of Ecology, Evolution, and Systematics 39: 459–477. [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. 2004. Comprehensive comparison of auxin‐regulated and brassinosteroid‐regulated genes in Arabidopsis . Plant Physiology 134: 1555–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel K. 1930. Organographie der Pflanzen, vol. 3. Berlin, Germany: Gustav Fischer Verlag. [Google Scholar]
- Goffinet B, Buck WR. 2013. The evolution of body form in bryophytes. Annual Plant Reviews 45: 51–90. [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q et al 2011. Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nature Biotechnology 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham LE, Cook ME, Busse JS. 2000. The origin of plants: body plan changes contributing to a major evolutionary radiation. Proceedings of the National Academy of Sciences, USA 97: 4535–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groff PA, Kaplan DR. 1988. The relation of root systems to shoot systems in vascular plants. The Botanical Review 54: 387–422. [Google Scholar]
- Gunning B, Hughes JE, Hardham AR. 1978. Formative and proliferative cell divisions, cell differentiation, and developmental changes in the meristem of Azolla roots. Planta 143: 121–144. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C. 2009. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21: 3119–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Mongelard G, Flokova K, Pacurar DI, Novak O, Staswick P, Kowalczyk M, Pacurar M, Demailly H, Geiss G et al 2012. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 24: 2515–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M et al 2013. De novo transcript sequence reconstruction from RNA‐seq using the Trinity platform for reference generation and analysis. Nature Protocols 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A, Gross‐Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. 2004. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana . Development 131: 657–668. [DOI] [PubMed] [Google Scholar]
- Hagemann W. 1999. Towards an organismic concept of land plants: the marginal blastozone and the development of the vegetation body of selected frondose gametophytes of liverworts and ferns. Plant Systematics and Evolution 216: 81–133. [Google Scholar]
- Hedman H, Zhu T, von Arnold S, Sohlberg JJ. 2013. Analysis of the WUSCHEL‐RELATED HOMEOBOX gene family in the conifer Picea abies reveals extensive conservation as well as dynamic patterns. BMC Plant Biology 13: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou G, Blancaflor EB. 2009. Fern root development. Annual Plant Reviews 37: 192–208. [Google Scholar]
- Hou GC, Hill JP. 2002. Heteroblastic root development in Ceratopteris richardii (Parkeriaceae). International Journal of Plant Sciences 163: 341–351. [Google Scholar]
- Hou G, Hill JP, Blancaflor EB. 2004. Developmental anatomy and auxin response of lateral root formation in Ceratopteris richardii . Journal of Experimental Botany 55: 685–693. [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. 2001. Identification of CRE1 as a cytokinin receptor from Arabidopsis . Nature 409: 1060–1063. [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T. 2008. Three type‐B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana . Plant Cell and Physiology 49: 47–57. [DOI] [PubMed] [Google Scholar]
- Jiang K, Feldman LJ. 2005. Regulation of root apical meristem development. Annual Review of Cell and Developmental Biology 21: 485–509. [DOI] [PubMed] [Google Scholar]
- Jones VAS, Dolan L. 2012. The evolution of root hairs and rhizoids. Annals of Botany 110: 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Schuetz M, Lin BSP, Chanis C, Hamberger B, Western TL, Ehlting J, Samuels AL. 2011. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. Journal of Experimental Botany 62: 2063–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. 1997. The origin and early evolution of plants on land. Nature 389: 33–39. [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia‐Hernandez M et al 2011. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Research 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landberg K, Pederson ERA, Viaene T, Bozorg B, Friml J, Jönsson H, Thelander M, Sundberg E. 2013. The moss Physcomitrella patens reproductive organ development is highly organized, affected by the two SHI/STY genes and by the level of active auxin in the SHI/STY expression domain. Plant Physiology 162: 1406–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Mo X, Wang J, Chen N, Fan H, Dai C, Wu P. 2008. BREVIS RADIX is involved in cytokinin‐mediated inhibition of lateral root initiation in Arabidopsis . Planta 229: 593–603. [DOI] [PubMed] [Google Scholar]
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen‐Mason SJ. 2002. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiology 128: 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Research 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link BM, Cosgrove DJ. 1998. Acid‐growth response and alpha‐expansins in suspension cultures of bright yellow 2 tobacco. Plant Physiology 118: 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. 2002. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Molecular Biology 49: 387–400. [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. 2000. A novel two‐component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes and Development 14: 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann HB, Whitney DR. 1947. On a test of whether one of two random variables is stochastically larger than the other. Annals of Mathematical Statistics 1: 50–60. [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L. 2007. An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz‐Laylin LK, Marechal‐Drouard L et al 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Ioio Dello R, Di Mambro R, Costantino P, Sabatini S. 2010. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Current Biology 20: 1138–1143. [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Di Mambro R, Sozzani R, Pacifici E, Salvi E, Terpstra I, Bao D, van Dijken A, Dello Ioio R, Peril S et al 2013. Spatial coordination between stem cell activity and cell differentiation in the root meristem. Developmental Cell 26: 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D, Leyser O. 2011. Auxin, cytokinin and the control of shoot branching. Annals of Botany 107: 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai J, Yamato KT, Sakaida M, Yoda H, Fukuzawa H, Ohyama K. 1999. Expressed sequence tags from immature female sexual organ of a liverwort, Marchantia polymorpha . DNA Research 6: 1–11. [DOI] [PubMed] [Google Scholar]
- Nardmann J, Werr W. 2012. The invention of WUS‐like stem cell‐promoting functions in plants predates leptosporangiate ferns. Plant Molecular Biology 78: 123–134. [DOI] [PubMed] [Google Scholar]
- Nunes‐Nesi A, Florian A, Howden A, Jahnke K, Timm S, Bauwe H, Sweetlove L, Fernie AR. 2014. Is there a metabolic requirement for photorespiratory enzyme activities in heterotrophic tissues? Molecular Plant 7: 248–251. [DOI] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin Y‐C, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A et al 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584. [DOI] [PubMed] [Google Scholar]
- Oda Y, Fukuda H. 2012. Secondary cell wall patterning during xylem differentiation. Current Opinion in Plant Biology 15: 38–44. [DOI] [PubMed] [Google Scholar]
- Ohtani M, Sugiyama M. 2005. Involvement of SRD2‐mediated activation of snRNA transcription in the control of cell proliferation competence in Arabidopsis . Plant Journal 43: 479–490. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis . Plant Cell 19: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Di Mambro R, Sabatini S. 2012. Growth and development of the root apical meristem. Current Opinion in Plant Biology 15: 17–23. [DOI] [PubMed] [Google Scholar]
- Petrasek J, Friml J. 2009. Auxin transport routes in plant development. Development 136: 2675–2688. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN. 2012. Control of Arabidopsis root development. Annual Review of Plant Biology 63: 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Research 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires ND, Dolan L. 2012. Morphological evolution in land plants: new designs with old genes. Philosophical Transactions of the Royal Society B 367: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Bezanilla M. 2010. Evolutionary crossroads in developmental biology: Physcomitrella patens . Development 137: 3535–3543. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. 2005. Class III homeodomain‐leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Brown GR, Maglott DR. 2012. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Research 40: D130–D135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Development Team . 2013. R: a language and environment for statistical computing, versions 3.0.2. [WWW document] URL http://www.R-project.org [accessed 7 May 2014].
- Radoeva T, Weijers D. 2014. A roadmap to embryo identity in plants. Trends in Plant Science 19: 709–716. [DOI] [PubMed] [Google Scholar]
- Rahman A, Bannigan A, Sulaman W, Pechter P, Blancaflor EB, Baskin TI. 2007. Auxin, actin and growth of the Arabidopsis thaliana primary root. Plant Journal 50: 514–528. [DOI] [PubMed] [Google Scholar]
- Rai AN, Söderbäck E, Bergman B. 2000. Cyanobacterium‐plant symbiosis. New Phytologist 147: 449–481. [DOI] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. 1992. The acid growth theory of auxin‐induced cell elongation is alive and well. Plant Physiology 99: 1271–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. 2004. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiology 135: 1738–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud P‐F, Lindquist EA, Kamisugi Y et al 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends in Genetics 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Šimášková M, Duclercq J, Petrášek J, Zažímalová E, Simon S, Friml J, Van Montagu MCE, Benková E. 2009. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proceedings of the National Academy of Sciences, USA 106: 4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. 2003. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes and Development 17: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A. 2001. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. 2005. The expansin superfamily. Genome Biology 6: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders HL, Langdale JA. 2013. Conserved transport mechanisms but distinct auxin responses govern shoot patterning in Selaginella kraussiana . New Phytologist 198: 419–428. [DOI] [PubMed] [Google Scholar]
- Scheres B, Di Laurenzio L, Willemsen V, Hauser M‐T, Janmaat K, Weisbeek P, Benfey PN. 1995. Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62. [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. 2010. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916. [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA et al 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Schneider H. 2013. Evolutionary morphology of ferns (monilophytes). Annual Plant Reviews 45: 115–140. [Google Scholar]
- Shapiro SS, Wilk MB. 1965. An analysis of variance test for normality (complete samples). Biometrika 52: 591–611. [Google Scholar]
- Stahl Y, Simon R. 2010. Plant primary meristems: shared functions and regulatory mechanisms. Current Opinion in Plant Biology 13: 53–58. [DOI] [PubMed] [Google Scholar]
- Stevenson J, Perera I, Heilmann I, Persson S, Boss W. 2000. Inositol signaling and plant growth. Trends in Plant Science 5: 357. [DOI] [PubMed] [Google Scholar]
- Stothard P. 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 28: 1102–1104. [DOI] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H. 2004. AtIPT3 is a key determinant of nitrate‐dependent cytokinin biosynthesis in Arabidopsis . Plant Cell Physiolgy 45: 1053–1062. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278: 631–637. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative . 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- The International Brachypodium Initiative . 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- The Potato Genome Sequencing Consortium . 2011. Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195. [DOI] [PubMed] [Google Scholar]
- Tian Q, Reed J. 1999. Control of auxin‐regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711–721. [DOI] [PubMed] [Google Scholar]
- Timme RE, Delwiche CF. 2010. Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biology 10: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. 2005. The Botany Array Resource: e‐Northerns, expression angling, and promoter analyses. Plant Journal 43: 153–163. [DOI] [PubMed] [Google Scholar]
- Ueguchi C, Sato S, Kato T, Tabata S. 2001. The AHK4 gene involved in the cytokinin‐signaling pathway as a direct receptor molecule in Arabidopsis thaliana . Plant Cell Physiology 42: 751–755. [DOI] [PubMed] [Google Scholar]
- Viaene T, Landberg K, Thelander M, Medvecka E, Pederson E, Feraru E, Cooper ED, Karimi M, Delwiche CF, Ljung K et al 2014. Directional auxin transport mechanisms in early diverging land plants. Current Biology 24: 2786–2791. [DOI] [PubMed] [Google Scholar]
- Wasteneys GO. 2004. Progress in understanding the role of microtubules in plant cells. Current Opinion in Plant Biology 7: 651–660. [DOI] [PubMed] [Google Scholar]
- Weijers D, Benková E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. 2005. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO Journal 24: 1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. 2003. Cytokinin‐deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA et al 2014. Phylotranscriptomic analysis oft he origin and early diversification of land plants. Proceedings of the National Academy of Sciences, USA 111: E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large‐scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodniok S, Brinkmann H, Glöckner G, Heidel AJ, Philippe H, Melkonian M, Becker B. 2011. Origin of land plants: do conjugating green algae hold the key? BMC Evolutionary Biology 11: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Root hair development in Azolla.
Fig. S2 Amyloplasts line the Azolla stele.
Fig. S3 Regeneration of the meristematic zone.
Fig. S4 Conformation of global gene expression trends through two‐step quantitative reverse‐transcription PCR.
Fig. S5 Ranked gene ontology analysis of global gene expression in Azolla roots.
Fig. S6 Evolution of expansins.
Fig. S7 Phylogenetic analysis of Arabidopsis and regulated Azolla Aux/IAA proteins.
Table S1 Overview of the RNAseq analysis
Table S2 Annotations and expression values of the Azolla filiculoides root tip transcriptome
Table S3 Oligonucleotides used for two‐step qRT‐PCR
Table S4 Sequences used for the alignment of land plant expansins
Table S5 Known developmental regulators expressed in Azolla root tips
Table S6 Significantly regulated contigs upon auxin and cytokinin treatment and GOterm enrichment analysis depicted in Figs 5 and 7


