Abstract
Natural product discovery efforts have focused primarily on microbial biosynthetic gene clusters (BGCs) containing large multi-modular PKSs and NRPSs; however, sequencing of fungal genomes has revealed a vast number of BGCs containing smaller NRPS-like genes of unknown biosynthetic function. Using comparative metabolomics, we show that a BGC in the human pathogen Aspergillus fumigatus named fsq, which contains an NRPS-like gene lacking a condensation domain, produces several novel isoquinoline alkaloids, the fumisoquins. These compounds derive from carbon-carbon bond formation between two amino acid-derived moieties followed by a sequence that is directly analogous to isoquinoline alkaloid biosynthesis in plants. Fumisoquin biosynthesis requires the N-methyltransferase FsqC and the FAD-dependent oxidase FsqB, which represent functional analogs of coclaurine N-methyltransferase and berberine bridge enzyme in plants. Our results show that BGCs containing incomplete NRPS modules may reveal new biosynthetic paradigms and suggest that plant-like isoquinoline biosynthesis occurs in diverse fungi.
Introduction
One major source of medicinally important small molecules are microbial BGCs that encode polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs), two classes of multi-functional mega-enzymes that are usually accompanied by sets of tailoring enzymes. The recent surge in the sequencing of fungal genomes has revealed large numbers of BGCs that do not appear to encode enzymes involved in the production of any known metabolites (“orphan BGCs”) and thus may harbor new biosynthetic capabilities1–3.
In search of new biosynthetic mechanisms, we focused on orphan BGCs that contain small NRPS-like genes that diverge from canonical NRPSs in their domain structure. Canonical NRPSs include, at a minimum, one adenylation domain that selects and activates an amino acid (or related building block), one thiolation domain for covalent attachment of the activated amino acid, and a condensation domain that catalyzes formation of a peptide bond between the tethered amino acid and another substrate4–6. However, sequencing of fungal genomes has additionally revealed numerous genes that encode only a subset of these domains.1,2 Such non-canonical, NRPS-like genes may feature adenylation and thiolation domains, but lack condensation domains, and thus likely do not function as peptide synthetases7,8. We hypothesized that analysis of BGCs including small NRPS-like genes encoding incomplete modules would likely reveal novel biosynthetic functions.
Here we show that the fsq gene cluster in A. fumigatus, which features an NRPS-like gene, fsqF, produces a series of novel isoquinoline alkaloids, the fumisoquins. We then demonstrate that FsqF, which lacks a canonical condensation domain, is required for carbon-carbon bond formation between L-serine and L-tyrosine-derived building blocks in the fumisoquin biosynthetic pathway. Two additional enzymes encoded by the fsq cluster, the N-methyltransferase FsqC and the FAD-dependent oxidase FsqB, catalyze the formation of the isoquinoline ring system in the fumisoquins via a sequence that we show is directly analogous to the biosynthesis of a prominent group of plant isoquinoline alkaloids, one of the largest families of medicinally important natural products9,10.
Results
FsqF encodes an incomplete NRPS module
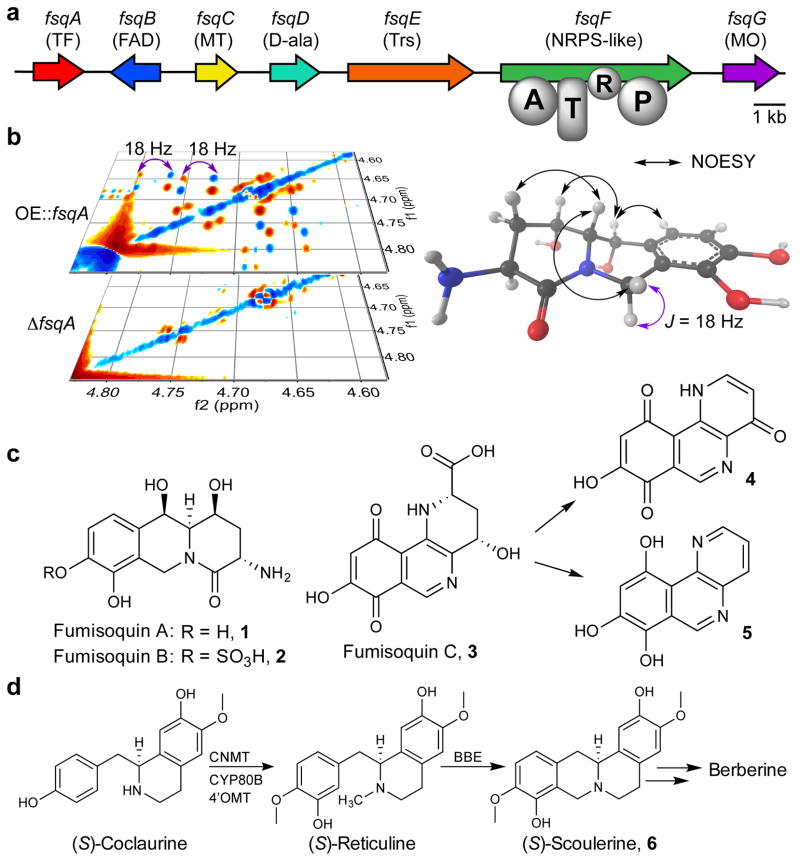
Expression of many fungal orphan BGCs is under the control of the nuclear protein LaeA, a global regulator of morphogenesis and virulence factor in A. fumigatus and other pathogenic fungi11,12. Among LaeA-regulated orphan BGCs in the human pathogen A. fumigatus, the fsq cluster features a small NRPS-like gene, fsqF, which encodes an adenylation, thiolation, reductase and pyridoxal-phosphate dependent aminotransferase domain (Fig. 1a). Notably, FsqF lacks the condensation domain that is essential for canonical peptide bond formation, and bioinformatic analysis of the vicinity of fsqF did not reveal any genes that could code for a second NRPS or free-standing condensation domain13. We found that expression of fsqF is regulated by the Zn(II)2Cys6-type transcriptional activator FsqA (Fig. 1a and Supplementary Results, Supplementary Fig. 1). Zn(II)2Cys6 transcription factors are unique to fungi and have been shown to regulate many metabolic pathways, e.g. aflatoxin biosynthesis in A. flavus14. In addition to fsqF, the transcription factor FsqA was found to regulate expression of five adjacent genes, annotated as ABC transporter (fsqE), fructosyl amino acid oxidase (fsqB), N-methyl-transferase (fsqC), phenol 2-monooxygenase (fsqG), and ATP-grasp enzyme (fsqD), defining the boundaries of the fsq cluster (Supplementary Fig. 1)13.
Figure 1. Analysis of the fsq gene cluster and metabolite production.
(a) fsq gene cluster and putative assignments of encoded proteins. TF: transcription factor; FAD: FAD-binding domain protein; MT: N-methyltransferase; D-ala: ATP-grasp enzyme (D-alanine ligase); Trs: transporter; NRPS: nonribosomal peptide synthetase (A = adenylation, T = thiolation, R = short-chain dehydrogenase/reductase domain, P: pyridoxal phosphate binding domain); MO: phenol 2-monooxygenase. (b) Section of the dqfCOSY spectra of OE::fsqA and ΔfsqA metabolite extracts used for comparative metabolomics (DANS). (c) Identified fsq-dependent compounds fumisoquin A and B (1 and 2), including stereochemical assignments via NOESY for 1 (see Supplementary Note), as well as structure of fumisoquin C (3), which decomposes to 4 and 5. (d) Biosynthesis of structurally related isoquinoline alkaloids in plants via coclaurine N-methyltransferase (CNMT), N-methylcoclaurine 3′-monooxygenase (CYP80B), 3′-hydroxy-N-methyl-(S)-coclaurine 4′-O-methyltransferase (4′OMT) and berberine bridge enzyme (BBE).
Identification of fsq cluster-derived metabolites
Overexpression of fsqA was associated with accumulation of a characteristic brown pigment in the surrounding media (Supplementary Fig. 2), suggesting secretion of a colored metabolite(s). For the identification of fsq-derived metabolites we employed 2D NMR-based comparative metabolomics (differential analysis by 2D-NMR spectroscopy, DANS), which facilitates comprehensive and largely unbiased detection of new metabolites associated with gene cluster overexpression15,16. DANS-based comparison of whole metabolome extracts from overexpression (OE::fsqA) and deletion (ΔfsqA) strains revealed several differential signals, most prominently two sets of OE-dependent doublets between 4.6 and 5.0 ppm with large coupling constants of 18–20 Hz (Fig. 1b), suggesting a diastereotopic benzylic CH2 group within a bi- or oligocyclic structure. The OE::fsqA-dependent metabolites were characterized further following partial purification via reversed-phase chromatography, using a standard suite of 2D NMR spectra and high-resolution mass spectrometry (HRMS) (Supplementary Table 1 and Supplementary Note). These analyses revealed two novel tricyclic isoquinoline derivatives, the pyrido[1,2-b]isoquinolines, fumisoquin A (1, Fig. 1c) and fumisoquin B (2, Fig. 1c) as major metabolites associated with fsq-overexpression. Fumisoquin A and B decompose gradually during chromatography, likely as a result of the oxidation-prone hydroquinone moiety. We further noted a deeply purple OE-dependent metabolite that was not captured by DANS because of decomposition during sample preparation. Optimization of extraction conditions and reverse-phase fractionation, followed by 2D NMR spectroscopic and HRMS analysis allowed to identify a third isoquinoline, fumisoquin C (3, Fig 1c), as the deeply purple colored metabolite (Supplementary Fig. 3, and Supplementary Note). Upon standing or during chromatography, fumisoquin C decomposes into compounds 4 and 5, which are more stable and were isolated (Fig. 1c, Supplementary Fig. 4a, and Supplementary Note). Treatment of partially purified fumisoquin C with (trimethylsilyl)diazomethane furnished the corresponding dimethyl derivative (Supplementary Fig. 4b), which is much less prone to decomposition and provided additional NMR spectroscopic data to confirm structure and relative configuration of fumisoquin C (Supplementary Note).
A plant-analogous isoquinoline formation pathway
Among known fungal natural products, isoquinoline alkaloids are rare, and the tricyclic ring systems of the fumisoquins, to the best of our knowledge, are unprecedented from fungi. However, the identified dihydroxylated isoquinolines are strikingly reminiscent of a large and diverse family of plant-derived isoquinoline alkaloids (pyrido[1,2-b]isoquinolines, e.g. scoulerine, 6, Fig. 1d), whose biosynthetic pathways have been studied extensively17–19. Plant pyrido[1,2-b]isoquinoline biosynthesis proceeds via N-methylation of a tyrosine-derived catechol precursor, followed by oxidative cyclization via the FAD-dependent berberine bridge enzyme (BBE, Fig. 1d)17, a member of a large group of flavin-dependent amine oxidases20,21. The cyclization likely proceeds via a two-step mechanism, beginning with FAD-dependent oxidation of the methyl group to an iminium species followed by electrophilic attack on the deprotonated phenol, although earlier studies suggested an alternative, concerted mechanism17,19. Because of the manifold pharmaceutical uses of plant isoquinoline alkaloids, there is considerable interest in developing microbial-based production approaches22–24, and yeast-based expression systems for several groups of plant isoquinoline alkaloids have been developed, including the berberines25–30. We noted that the fsq cluster features a set of genes that may encode functional analogs of the berberine biosynthetic enzymes, including the putative phenol 2-monooxygenase FsqG, N-methyltransferase FsqC, and FAD-dependent oxidase FsqB. Bioinformatic analysis revealed several orphan gene clusters in fungi that feature homologous sets of genes (Supplementary Table 2 and Supplementary Fig. 5). Moreover, the only other known fungal isoquinolines have been linked to a gene cluster containing homologs for these three genes31.
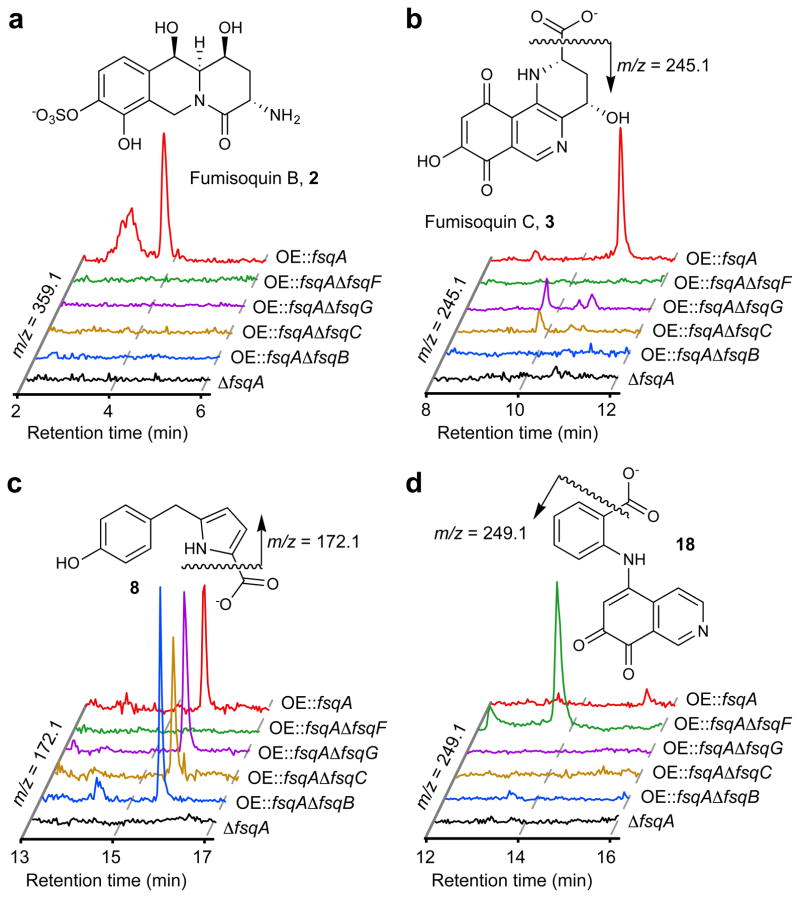
To investigate the biosynthesis of the fumisoquins, we created deletion strains in the OE::fsqA background for all genes in the fsq cluster, except for fsqD and the putative transporter fsqE, which we were unable to delete, which could be due to a protective function of these proteins as has been found in several other fungal BGCs1. We then compared the metabolomes of each of these deletion strains with those of OE::fsqA and ΔfsqA via DANS and HPLC-MS (Fig. 2a–d). We found that deletion of the N-methyltransferase FsqC, phenol 2-monooxygenase FsqG, or FAD-dependent oxidase FsqB in OE::fsqA background led to complete abolishment of isoquinoline alkaloid production and accumulation of a series of benzyl pyrroles (7–8, Fig. 2c and Fig. 3), including the previously reported fumipyrrole, 7 (where the fsq cluster was referred to as fmp32), smaller amounts of which were also produced in the OE::fsqA control strain (Fig. 2c). Comparison of the carbon skeletons of the pyrroles (7–8, Fig. 3) with those of the fumisoquins provided support for the predicted N-methyltransferase and phenol 2-monooxygenase functions of FsqC and FsqG, respectively, and suggested that the fumisoquins form from an open-chain precursor such as 9 (Fig. 3), in a manner very similar to that of plant benzophenanthridine alkaloid biosynthesis18.
Figure 2. Comparative metabolomics of fsq gene cluster mutants by HPLC-MS.
(a–d) HPLC-MS analysis of fsq-gene deletion strains in OE::fsqA background, shows ion chromatograms representing major products and shunt metabolites.
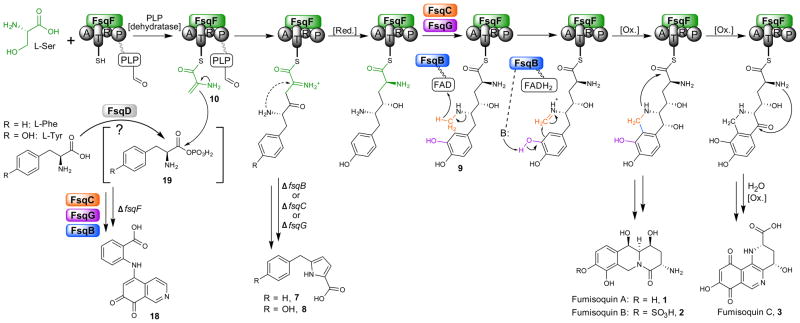
Figure 3. Model for fumisoquin biosynthesis.
(A = adenylation, T = thiolation, R = short-chain dehydrogenase/reductase domain, P: pyridoxal phosphate binding domain), [Red.] = reduction, [Ox.] = oxidation. The proposed mechanism for FsqB-mediated isoquinoline formation is based on functional similarity to plant BBE17–19, see Fig. 5 for details. The role of the ATP-grasp enzyme FsqD in activating tyrosine to form 19 is putative.
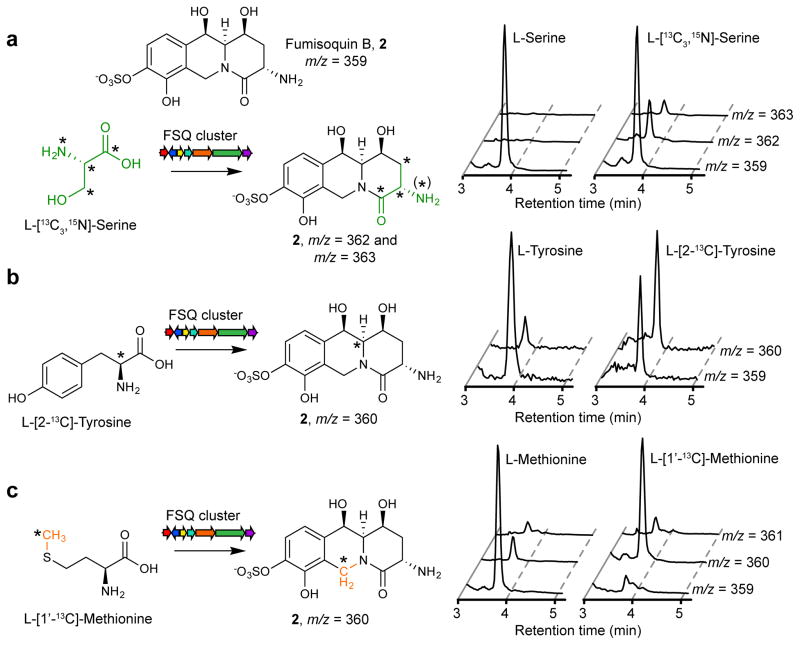
The structure of the putative open-chain precursor 9 suggested that it may be derived from tyrosine and a dehydroalanine equivalent (10, Fig. 3), which could be produced from serine or cysteine via the pyridoxal-phosphate dependent aminotransferase domain of the NRPS-like protein FsqF10,33. Growing the OE::fsqA strain in media supplemented with different stable-isotope labeled amino acids, we found that L-tyrosine and L-serine are incorporated into the fumisoquins and pyrroles (1–3 and 8) in a manner consistent with the biosynthetic model shown in Fig. 3, whereas L-cysteine is not incorporated (Fig. 4a,b and Supplementary Fig. 6, 7b–d, also see Online Methods). Furthermore, we showed that the L-methionine methyl group is incorporated into the fumisoquin ring systems at the predicted position, in agreement with the putative function of FsqC as an S-adenosyl methionine-dependent N-methyltransferase (Fig. 4c and Supplementary Fig. 8).
Figure 4. The fumisoquins incorporate L-serine, L-tyrosine, and an L-methionine-derived methyl group.
Shown are HPLC-MS ion chromatograms derived from fungal cultures grown with (a) L-serine or L-[13C3,15N]-serine, (b) L-tyrosine or L-[2-13C]-tyrosine and (c) L-methionine or L-[1′-13C]-methionine. In contrast, L-cysteine is not incorporated (Supplemental Fig. 6). See Supplementary Fig. 7 for mass spectra. (*) denotes labeled isotopes. In the case of L-[13C3,15N]-serine, loss of 15N due to partial deamination/amination contributes to increased m/z = 362.
FsqB is a functional analog of plant BBE
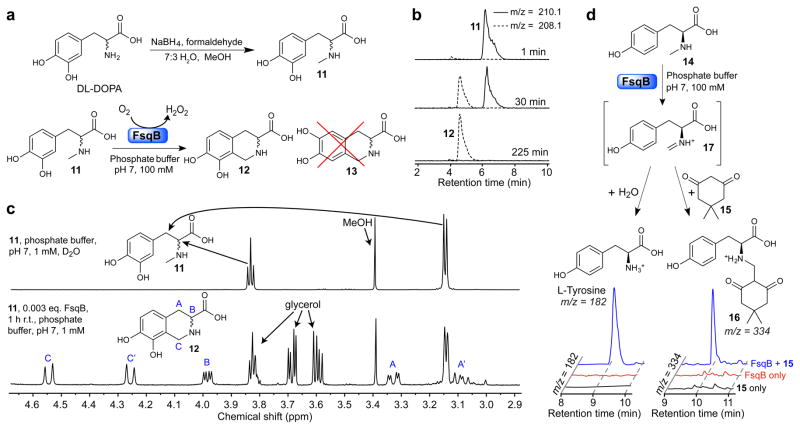
To test whether FsqB catalyzes isoquinoline ring formation in 1–3 and thus acts as a functional ortholog of plant BBE, we recombinantly produced polyhistidine-tagged FsqB in E. coli (Supplementary Fig. 9). We then assayed its activity on a range of potential substrates that mimicked the tyrosine-derived portion of putative intermediate 9. Whereas N-methyl dopamine did not react, we found that FsqB converts N-methyl DOPA 11, prepared via a simple one-step procedure, directly into isoquinoline 12 (Fig. 5a and Supplementary Fig. 10, 12a). We first tested FsqB activity on 11 on a small scale suitable for LCMS, which yielded near quantitative conversion to 12 within 3–4 h (100 mM phosphate buffer, pH 7, KM = 142.5 ± 42.9 μM) (Fig. 5b, Supplementary Fig. 11). Next we demonstrated isoquinoline formation from 11 on a milligram scale by conducting the reaction in an NMR tube using deuterated phosphate buffer (Fig. 5c and Supplementary Note). Notably, FsqB-catalyzed formation of 12 proceeds with complete regioselectivity, as the alternative cyclization product, 6,7-dihydroxy-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid, 13, which does form alongside 12 in a non-enzymatic reaction of DOPA with formaldehyde, was not observed (Supplementary Fig. 10 and Supplementary Note). In contrast, we observed no cyclized products when incubating FsqB with N-methyl-L-tyrosine, N,N-dimethyl DOPA, or the BBE-substrate (S)-reticuline, suggesting that both a catechol moiety and a secondary β-N-methylamine are required for FsqB activity (Supplementary Fig. 12b–d).
Figure 5. Enzymatic activity of recombinant FsqB.
(a) Synthesis of N-methyl-3′,4′-dihydroxy-DL-phenylalanine 11 (top) and regioselective conversion of 11 into 12 by purified recombinant FsqB (bottom). The isomer 13 does not form. (b) Ion chromatograms at indicated timepoints for the FsqB-catalyzed in vitro conversion of 11 to 12 (100 mM phoshate buffer, pH 7, 100:1 substrate to enzyme). (c) 1H NMR spectra of a sample of 11 (1 mM phosphate buffer in D2O) prior to FsqB addition (top) and 1 h post FsqB addition (333:1 substrate to enzyme), showing selective conversion of 11 into 12 (bottom) (d) Ion chromatograms showing formation of tyrosine and 16 as a result of capture of intermediate 17 by H2O or dimedone, 15 (1.5 μM FsqB and 400 μM N-methyl-L-tyrosine in 100 mM phoshate buffer, pH 7).
Next we asked whether the mechanism of isoquinoline formation as catalyzed by FsqB is directly analogous to that of the extensively studied plant enzyme, BBE9,17,19,34. Fluorescence analysis of recombinant FsqB showed excitation and emission maxima at 461 nm and 530 nm, respectively (Supplementary Fig. 9), suggesting FsqB utilized a flavin cofactor, as predicted35. Further proteomic analysis of FsqB confirmed the presence of FAD, which was found to be covalently bound to cysteine-414 (Supplementary Fig. 13a and Supplementary Table 3). FAD is also part of BBE where it is bound to a histidine residue17. Flavin reduction during catalytic cycling of BBE is recovered with molecular oxygen generating hydrogen peroxide17, a characteristic that was also observed with FsqB utilizing an Amplex Red assay (Supplementary Fig. 13b). Isoquinoline formation as catalyzed by BBE likely proceeds via an imine intermediate in a two-step mechanism19. To test whether isoquinoline formation via FsqB also involves an imine intermediate, we incubated FsqB with N-methyl-L-tyrosine 14, which we hypothesized may represent an oxidation-competent substrate for FsqB but, as shown above, does not get cyclized, in the presence of the electrophile trapping agent dimedone, 15. This reaction produced the adduct 16 as well as free tyrosine, consistent with capture of an imine intermediate, 17, by dimedone or water (Fig. 5d). Therefore, it appears that the fungal enzyme FsqB catalyzes isoquinoline formation via a two-step mechanism that is analogous to that of plant-derived BBE.
Carbon-carbon bond formation requires FsqF
Next we recombinantly produced the FsqF adenylation domain to test whether it activates L-tyrosine or L-serine, as suggested by the stable isotope labeling study, using the ATP-[32P]pyrophosphate exchange activity36. However, neither enantiomer of serine or tyrosine showed any activity in the in vitro assay nor did any other tested standard amino acid (Supplementary Fig. 14). These results may indicate that the recombinantly expressed FsqF adenylation domain was not functional or that the true substrate is not a standard amino acid. Based on the homology of the PLP-dependent domain in FsqF to other serine dehydrogenases13,33, we hypothesized that L-serine may first be converted into dehydroalanine, which is then activated and tethered to the thiolation domain (Fig. 3).
To further investigate the role of the NRPS-like enzyme FsqF in fumisoquin biosynthesis, we analyzed the effect of fsqF deletion on the OE::fsqA metabolome. As anticipated, production of the fumisoquins (1–3) as well as the pyrroles (7–8) was completely abolished in OE::fsqA-ΔfsqF (Fig. 2d). Significantly, we also observed production of a single, brightly red shunt metabolite, the anthranilic acid-substituted isoquinoline 18 (Fig. 2d and Fig. 3). Feeding experiments of the OE::fsqA-ΔfsqF mutant with stable-isotope labeled L-tyrosine confirmed that the isoquinoline ring system in 18 is derived from tyrosine, as in the case of the fumisoquins (Supplementary Fig. 15). Incubation of 12, obtained in vitro from recombinant FsqB, with anthranilic acid (19) led to formation of shunt metabolite 18, consistent with a model in which shunt metabolite 18 forms non-enzymatically from 12 via oxidation to the corresponding o-quinone, subsequent capture of anthranilic acid, and decarboxylation (Supplementary Fig. 16). Compound 12 was not observed in any of the analyzed OE::fsqA-ΔfsqF extracts, likely as a result of this compound’s high susceptibility to oxidize to the reactive o-quinone, which is rapidly captured by anthranilic acid (Supplementary Fig. 16).
The identification of shunt metabolite 18 indicates that the NRPS-like protein FsqF is only required for addition of a serine-derived dehydroalanine moiety to activated tyrosine, but is not essential for the subsequent steps leading to isoquinoline formation, and that a different enzyme, likely the ATP-grasp enzyme FsqD, is responsible for activation of tyrosine (Fig. 3). The production of shunt metabolite 7 would then indicate that FsqD is also capable of activating phenylalanine; however, phenylalanine-derived intermediates only lead to the production of 7, as expected based on the predicted function of FsqG as a phenol 2-monooxygenase13 and given the observation that recombinantly produced FsqB requires a catechol as substrate. FsqD has homology to carboxylate-amine ligases that furnish aminoacyl phosphate from ATP and amino acid precursors37,38. In a recent example, the FsqD homolog PGM1 was shown to activate non-proteinogenic amino acids for peptide-bond formation in pheganomycin biosynthesis39. In contrast, FsqD appears to activate tyrosine (or phenylalanine) for subsequent condensation with serine-derived dehydroalanine (Fig. 3), providing a first example for a novel carbon-carbon bond forming strategy in fungi.
Discussion
Our analysis of the fsq cluster revealed a plant-like strategy for the biosynthesis of oligocyclic alkaloids in fungi. The biosynthetic steps of phenol-hydroxylation, N-methylation, and oxidative cyclization appears to be the same in plants and fungi, and thus the corresponding enzymes can be considered functionally equivalent. Sequence analysis does not suggest homology for these proteins, though it is notable that cyclization proceeds via analogous two-step mechanisms that are catalyzed by FAD-dependent oxidases in both fungi and plants19, presenting a striking case of convergent evolution of specialized metabolic pathways. Given that analysis of available fungal genomes13 revealed many co-occurrences of homologs of the N-methyltransferase fsqC and FAD-dependent oxidase fsqB (Supplementary Fig. 5), it seems likely that fungi are capable of producing a diverse range of yet undiscovered isoquinoline alkaloids. For example, in addition to Aspergillus spp., the genomes of plant pathogenic Fusarium spp. feature BGCs that include homologues for most fsq genes40.
Together with other recent examples7,8, the identification of the fumisoquins shows that analysis of gene clusters containing incomplete NRPS modules can reveal intriguing new structures and biosynthetic strategies. It remains unclear, however, whether the biosynthetic roles of NRPS-like enzymes can be predicted, as current examples, though few, hint at considerable functional diversity. For example, two NRPS-like enzymes in A. flavus, named LnaA and LnaB, have been shown to produce a series of tyrosine-derived piperazines, pyridines and morpholines7, whereas an NRPS-like enzyme from A. terreus was shown to reduce products of an accompanying non-reducing PKS8. The example of the fsq pathway demonstrates that NRPS-like enzymes lacking condensation domains may nonetheless contribute to ligating amino acids, albeit via formation of carbon-carbon bonds. Characterization of BGCs featuring small NRPS-like genes may thus reveal new types of alliances of NRPS-like enzymes with other amino acid-activating proteins, e.g. ATP-grasp enzymes such as FsqD.
This work supports a mining strategy of assessing fungal natural products, beyond not only canonical NRPS but also PKS-like genes and even those fungal clusters lacking NRPS or PKS-like domain genes all together. Recent examples have focused on cyclic ribosomal peptides41 but a bioinformatic scan of fungal genomes and expression data shows the existence of yet more alternative biosynthetic clusters – lacking NRPS-like, PKS-like and canonical ribosomal peptides - suggestive of unexplored chemical diversity42. Efforts to characterize such alternative clusters may yield even more unique fungal chemistry.
Online Methods
Strains, media, and growth conditions
The fungal strains used in this study are listed in Supplementary Table 4. All strains were grown at 37 °C on glucose minimal medium (GMM)43 and, when appropriate, were supplemented with 0.56 g/L uracil, 1.26 g/L uridine, 1.0 g/L arginine and maintained as glycerol stocks at −80 °C. Escherichia coli strain DH5α was propagated in LB medium with appropriate antibiotics for plasmid DNA.
Gene cloning, plasmid construction, and genetic manipulations
The plasmids utilized in this study are listed in Supplementary Table 4. The oligonucleotide sequences for PCR primers are given in Supplementary Table 5. PCR amplification was carried out on a C1000TM Thermal Cycler (Bio-Rad). For creation of the fsqA overexpression (OE) strain (TJES1.18) the OE cassette was constructed by amplifying the fsqA open reading frame (ORF) from Af293 genomic DNA using primers Afu6g03430_NcoI_For and Afu6g03430_NotI_Rev, which introduced a 5′ NcoI restriction site and a 3′ NotI restriction site. This PCR product was purified with a QIAquick gel extraction kit (Qiagen), quantified, and digested with the appropriate restriction enzymes before being cloned into pJMP8.1244, resulting in plasmid pJES 1.2. Subsequently, A. fumigatus argB was transferred from pJMP4.145 by digesting with EcoRI and introduced into pJES1.2, resulting in plasmid pJES2.7 (A. fumigatus argB::gpdA(p)::fsqA). All PCR steps were carried out with Pfu Ultra II DNA Polymerases (Agilent) and all digestion reactions were carried out with NEB enzymes (New England BioLabs). Correctness of the inserted DNA was then confirmed by sequencing.
Construction of the fsq gene knock-out cassettes were generated using standard double-joint PCR procedures46. Briefly, A. parasiticus pyrG (A.ppyrG) was amplified from pJMP9.1. Then, an approximately 1 kb fragment upstream and downstream of each fsq gene was amplified from genomic DNA of A. fumigatus Af293 using designated primers, respectively. These three amplified PCR products were purified with a QIAquick gel extraction kit, quantified, and fused using standard double-joint PCR procedures. The final PCR product was amplified with the bottom primer pairs, gene_5′F_flank and _3′R_flank, confirmed with endonuclease digestion, and purified for fungal transformation. The first two rounds of PCR were done with Pfu Ultra II DNA Polymerases (Agilent), and the final PCR step used Expand long template PCR system (Roche) according to the manufacturer’s instructions. Af293.6 (double auxotroph A. fumigatus, pyrG−, argB−) was used to make the OE::fsqA, pyrG− auxotroph, TJES1.18. This strain was used as the recipient host strain for subsequent deletions of fsq genes as well as the ectopic complementation of pyrG1 with plasmid pJMP9.1 (containing A.ppyrG) resulting in prototrophic strain TJES3.1 (OE::fsqA, A.ppyrG). Similarly ΔfsqA auxotrophic strains (TJES2.20) were made by transforming Af293.6 with deletion cassettes and complemented with ectopically integrated pJMP4 to make ΔfsqA prototroph TJES8.2.
For the creation of deletion mutants in OE::fsqA TJES1.18 background, a deletion cassette of each cluster gene (fsqB to fsqG) was constructed by using double-joint PCR with A. parasiticus pyrG (A.ppyrG) for replacement of target genes. The A.ppyrG gene was amplified from the plasmid pJMP9.1 as template. Five μg of the double-joint PCR cassette were used to delete each fsq cluster gene by using TJES1.18 (A.fargB::gpdA(p):: fsqA, pyrG1) as the recipient host. After transformation, transformants were grown on minimal media plates without supplements for screening. All strains were verified by PCR and Southern blot analysis (Supplementary Fig. 17. Multiple confirmed strains for each mutant were stored at −80 °C with 33 % glycerol for future use.
Heterologous protein production and biochemical assays
The expression vectors for the fsqF (NRPS-like) adenylation domain (pJES13.2) were constructed by amplifying bases 1-1968 of the coding DNA sequence (CDS) of the gene Afu6g03480 from Af293 using primers fqsF_Adomain_xp_5′F and fqsF_Adomain_xp_3′R introducing 5′ NotI and 3′ XhoI restriction sites, respectively. These products were gel purified and digested with the appropriate enzymes before being cloned into pET30-a(+) (EMD Biosciences) to create an expression plasmid with a 6xHis tagged fsqF adenylation domain. The plasmid was introduced into chemically competent E.coli DH5α, selected on LB agar with Kanamycin and confirmed by sequencing. Subsequently, they were extracted from DH5α cells and used to transform expression host E.coli BL21. Protein purification was carried out as described47. All reactions were carried out in triplicate. Reaction parameters for the ATP-[32P]pyrophosphate exchange assay were: total assay volume of 100 μL at 25°C in 100 mM phosphate buffer, 5 mM MgCl2, 125 nM EDTA, 5 mM ATP, 100 nM purified FsqF adenylation domain, 0.1 μM [32P]pyrophosphate (50 Ci/mmol), and 1 mM amino acid substrate. The reaction proceeded for 30 min before it was stopped (1 % (w/v) activated charcoal, 4.5 % (w/v) tetrasodium pyrophosphate, 3.5 % (v/v) perchloric acid) and further processed as described48. Pyrophosphate exchange was quantified on a scintillation counter (PerkinElmer TriCarb 2910TR).
The expression vector for fsqB (fructosyl amino acid oxidase) was constructed using the CDS of the gene Afu6g03440 from OE::fsqA using primers pet28_fsqB_3′_fwd and fsqB_5′_pet28_rev. These products were gel purified and cloned into pET28-b(+) using ligase-free PCR cloning to create an expression plasmid with a 6xHis tagged FsqB. The plasmid was introduced into chemically competent E. coli DH5α, selected on LB agar with Kanamycin and confirmed by sequencing. Subsequently, they were extracted from DH5α cells and used to transform expression host E. coli BL21(DE3) (New England BioLabs). BL21(DE3) pET28-b(+) fsqB containing E. coli were grown in LB containing 50 μg/mL Kanamycin at 37 °C to an OD of ~0.6, cooled to 18 °C and expression was induced with 250 μM IPTG for 15 hrs. 4 L of LB containing E. coli were harvested at 6000 G for 20 min yielding 10.6 g of wet weight pellet and stored at −80 °C. Protein purification was carried out at 4 °C by resuspending the pellet in lysis buffer containing 100 mM phosphate buffer pH 7.8, 10 mM imidazole, 10 % (v/v) glycerol, 1 mM PMSF, 0.1 mg/mL lysozyme, 0.1 mg/mL Benzonase (EMD Millipore) and sonicated. All following steps were performed in low light to avoid detrimental flavin mediated reactions. Lysed cells were centrifuged at 20000 G for 20 min, and loaded onto TALON affinity resin (Clontech) pre-equilibrated with lysis buffer and incubated for 30 min with stirring. Resin was loaded onto a column and washed with 2 column volumes of lysis buffer without lysosome or Benzonase. FsqB was eluted with 100 mM phosphate buffer pH 7.8, 150 mM imidazole, 10 % (v/v) glycerol with 8 column volumes, concentrated and buffer exhanged with Amicon Ultra-15 centrifugal filter 30 kDa cutoff with 100 mM phosphate buffer pH 7.8, 25 % (v/v) glycerol and stored at −80°C. Activity assays were performed in total assay volumes of 100 μL at 37 °C in 100 mM phosphate buffer pH 7.0 in the dark. Reaction products were identified on a Thermo Scientific-Dionex Ultimate3000 UHPLC system equipped with a diode array detector and connected to a Thermo Scientific Q Exactive Orbitrap operated in electrospray negative (ESI-) ionization mode. UV-Vis absorbance spectra were obtained on a Cary 5000 UV-Vis-NIR spectrophotometer, and fluorescence emission was obtained on a HORIBA Nanolog Spectrofluorometer.
FsqB Steady-state kinetic analysis and imine capture
Steady-state kinetics were carried out at 25 °C in 100 mM phosphate buffer pH 7.0 with 100 nM FsqB at final volumes of 500 or 1000 μL at various initial concentrations of N-methyl-3,4-dihydroxy-DL-phenylalanine (11). Imine capture was carried out at 25 °C in 100 mM phosphate buffer pH 7.0 with 1.5 μM FsqB, 400 μM N-methyl-L-tyrosine (14), and 1 mM dimedone (15) (Sigma Aldrich) with <0.5% (v/v) methanol for dimedone solubility. Both kinetics and imine capture were monitored with low-resolution HPLC-MS performed on an Agilent 1100 series HPLC system equipped with a diode array detector and connected to a Quattro II mass spectrometer (Micromass/Waters) operated in electrospray negative ionization (ESI−) mode for kinetic analysis and positive electrospray ionization (ESI+) mode for imine capture. Kinetic data analysis was performed using GraphPad Prism version 6.00 for Windows (GraphPad Software).
FsqB Amplex Red assay
100 μM N-methyl-DL-DOPA or DL-DOPA, either with or without 15 μM FsqB, were incubated in 100 mM pH 7.0 potassium phosphate buffer at 25 °C in 100 μL total reaction volume in the dark in a 96-well plate with gentle shaking. After 1.5 h of incubation, 100 μL of hydrogen peroxide detection mixture (containing 50 mM potassium phosphate pH 7.0, 100 μM Amplex Red (Thermo Scientific), 0.2 units/mL HRP) was added and incubated at 25 °C in the dark for 10 minutes. Readings were performed on a BioTek Synergy 2 96-well plate reader using a 500±27 nm excitation filter and a 615±15 nm emission filter and normalized to 0 and 100 μM hydrogen peroxide standards. All conditions were performed in triplicate.
FsqB trypsinization analysis
80 μg of recombinant FsqB was buffer exchanged into 55 μL of 50 mM ammonium bicarbonate. 3 μL of 0.33 μg/μL of sequencing grade porcine trypsin (Promega) was added to FsqB and incubated at 37 °C for 12 h. 5 μL 50 mM TCEP was added and the mixture was further incubated at 37 °C for 10 min. The solution was cooled to 25 °C and 5 uL of 100 mM iodoacetamide was added and incubated at 25 °C in the dark for 1 h. Formic acid was added to 1 % (v/v), and peptides were prepared with 100 μL Pierce C18 Tips (Thermo Scientific) per manufacturers protocol utilizing 100 μL elution solution. Peptides were identified via UHPLC-MS/MS using a Thermo Scientific Dionex Ultimate 3000 UHPLC system equipped with a diode array detector and connected to a Thermo Scientific Q Exactive Orbitrap operated in positive electrospray (ESI+) ionization mode.
Nucleic acid analysis
Preparation of plasmids, restriction enzyme digestions, gel electrophoresis, blotting, probe preparation and hybridization were carried out by standard protocols. Aspergillus DNA was extracted using a previously described method49. Sequence data were analyzed using the LASERGENE software package from DNASTAR.
Northern analysis
50 mL of liquid GMM43 were inoculated with 1.0×106 spores (asexual) mL−1 of all appropriate strains in this study and incubated with shaking at 250 rpm at 25 °C. After 48 h, the mycelium was collected and total RNA was extracted by using Isol-RNA Lysis Reagent according to the manufacturer’s instructions (5 Prime).
In order to determine the boundaries of the fsq gene cluster, gene fragments of potential cluster genes used as probes were amplified individually from Af293 genomic DNA with appropriate primers. WT, OE::fsqA (TJES3.1), and ΔfsqA (TJES8.2) were used for this experiment. About 40 μg of total RNA were used for RNA blot analysis. RNA blots were hybridized with designated DNA fragments. All experiments were performed in duplicate. Detection of signals was carried out with a Storm 860 phosphoimager (Molecular Dynamics).
Fermentation and metabolome extraction for comparative metabolomics by DANS and HPLC-UV-MS
Preparation for NMR spectroscopic analysis: A. fumigatus strains were inoculated (1.0×106 spores mL−1) into 1 L GMM43 in a 2 L Erlenmeyer flask at 37 °C with shaking at 220 rpm. After 4 days, liquid fungal cultures including fungal tissue and media were frozen using a dry ice acetone bath, and lyophilized. The lyophilized residues were extracted with 500 mL of 10 % methanol in ethyl acetate for 3.5 h with vigorous stirring. Extracts were filtered over cotton, evaporated to dryness, and stored in 8 mL glass vials at −20 °C. Prior to NMR spectroscopic analysis, the crude extracts (~30–50 mg) were suspended in 0.15 mL of methanol-d4. The resulting suspension was evaporated to dryness, the residue re-suspended in 0.6 mL of methanol-d4, centrifuged to remove insoluble materials, and the supernatant was transferred into a 5 mm NMR tube.
Preparation for HPLC-MS analysis: A. fumigatus strains were inoculated (1.0×106 spores mL−1) into 50 mL GMM43 in a 125 mL Erlenmeyer flask at 37 °C with shaking at 220 rpm. After 4 days, liquid fungal cultures including fungal tissue and media were frozen using a dry ice acetone bath, and lyophilized. The lyophilized residues were extracted with 30 mL of MeOH for 1.5 h with vigorous stirring. Extracts were filtered over cotton, evaporated to dryness, and stored in 4 mL vials. Crude extracts were suspended in 0.5 mL of MeOH and centrifuged to remove insoluble materials, and the supernatant was subjected to HPLC-MS analysis.
Fermentation and metabolome extraction for compounds 3–5
A. fumigatus strains were inoculated (1.0×106 spores mL−1) into 1 L GMM43 in a 2 L Erlenmeyer flask at 37 °C with shaking at 220 rpm. After 3.5 days, liquid fungal cultures including fungal tissue and media were frozen using a dry ice acetone bath, and lyophilized. The lyophilized residues were extracted with 500 mL of MeOH for 1 h with vigorous stirring. Extracts were filtered over cotton and evaporated on Celite-545 (Acros Organics) to dryness. The dry Celite was then loaded into a 25 gram solid phase cartridge (Isco) and subjected to large-scale reverse phase chromatography (see Chromatographic enrichment section below for details). Compounds 4 and 5 are also minor components of the culture media.
Analytical methods and equipment overview
(a) NMR spectroscopy: NMR spectroscopic instrumentation: a Varian INOVA 600 MHz NMR spectrometer (600 MHz 1H reference frequency, 151 MHz for 13C) equipped with an HCN indirect-detection probe or a Bruker AvanceIII HD (800 MHz 1H reference frequency, 201 MHz for 13C) equipped with a 5 mm CPTCL 1H-13C/15N cryo probe. Non-gradient phase-cycled dqfCOSY spectra were acquired using the following parameters: 0.6 s acquisition time, 400–600 complex increments, 8, 16 or 32 scans per increment. Non-gradient HSQC, HMQC, and HMBC spectra were acquired with these parameters: 0.25 s acquisition time, 200–500 increments, 8–64 scans per increment. 1H, 13C-HMBC spectra were optimized for JH,C = 6 Hz. HSQC spectra were acquired with or without decoupling. Susceptibility-matched NMR tubes (Shigemi) were used for sample amounts smaller than 1 mg. NMR spectra were processed and baseline corrected using MestreLabs MNOVA software packages. (b) Mass spectrometry: high-resolution UHPLC-MS was performed on a Thermo Scientific-Dionex Ultimate3000 UHPLC system equipped with a diode array detector and connected to a Thermo Scientific Q Exactive Orbitrap operated in electrospray positive (ESI+) or electrospray negative (ESI−) ionization mode. Low-resolution HPLC-MS was performed on an Agilent 1100 series HPLC system equipped with a diode array detector and connected to a Quattro II mass spectrometer (Micromass/Waters) operated in ESI+ or ESI− mode. Data acquisition and processing for the LC-HRMS was controlled by Thermo Scientific Xcalibur software. Data acquisition and processing for the HPLC-MS was controlled by Waters MassLynx software. (c) Chromatography: flash chromatography was performed using a Teledyne ISCO CombiFlash system. For semi-preparative HPLC Agilent Zorbax Eclipse XDB-C18 or -C8 columns (25 cm × 10 mm, 5 μm particle diameter) were used. An Agilent Zorbax RRHD Eclipse XDB-C18 column (2.1 × 100 mm, 1.8 μm particle diameter) was used in the LC-HRMS A. fumigatus mutant profiling analysis. An Agilent Zorbax Eclipse XDB-C18 column (4.6 × 250 mm, 5 μm particle diameter) was used in the HPLC-MS A. fumigatus mutant profiling analysis.
Chromatographic enrichment of compounds 1, 3, dimethyl-3, 7–8, 11–13, and 18
Methanol extracts derived from 1–2 L of A. fumigatus cultures were fractionated using large-scale reverse-phase flash chromatography on a Teledyne ISCO CombiFlash with a Teledyne C18 gold (100 gram) column with acetonitrile (organic phase) and 0.1 % acetic acid in water (aqueous phase) as solvents at a flow rate of 60 mL/min. A linear ramp from 0 % organic to 100 % organic over 30 min was used and fractions containing compounds of interest were collected, evaporated to dryness, and stored in 8 mL glass vials at −20 °C. Fractions containing compounds 1, 3, dimethyl-3, 7–8, 11–13, and 18 were further purified via semi-preparative HPLC using an Agilent XDB C-18 or C-8 column (25 cm × 10 mm, 5 μm particle diameter) acetonitrile (organic phase) and 0.1 % acetic acid in water (aqueous phase) as solvents at a flow rate of 3.6 mL/min. A solvent gradient scheme was used, starting at 5 % organic for 3 min, followed by a linear increase to 100 % organic over 27 min, holding at 100 % organic for 5 min, then decreasing back to 5 % organic for 0.1 min, and holding at 5 % organic for the final 4.9 min, for a total of 40 min.
In vivo stable isotope labeling experiments
Deuterium- and 13C-labeled amino acids were purchased from Cambridge Isotopes Inc. Each amino acid (labeled or unlabeled control) was added to 50 mL (2 mM) of GMM43 media in 125 mL Erlenmeyer flasks. Each flask was inoculated with OE::fsqA or OE::fsqA-ΔfsqF (1.0 × 106 spores mL−1) and grown for 4 days at 37 °C with shaking at 220 rpm. After 4 days, liquid fungal cultures including fungal tissue and media were frozen using a dry ice acetone bath and lyophilized. The lyophilized residues were extracted with MeOH (25 mL) for 1.5 h with vigorous stirring. Extracts were filtered over cotton, evaporated to dryness, and stored in 4 mL vials at −20 °C. Crude extracts were suspended in 0.5 mL of MeOH and centrifuged to remove insoluble materials, and the supernatant was subjected to HPLC-MS analysis.
Conversion of 3 into dimethyl-3 for NMR spectroscopic analysis
To a solution of a partially purified fumisoquin C (3) in 3:2 toluene/MeOH (2 mL) was added (trimethylsilyl)diazomethane (Sigma-Aldrich) (20 μL, 0.04 mmol) as a 2.0 M diethyl ether solution. After stirring at room temperature for 30 min, the mixture was quenched with glacial acetic acid (10 μL) and dried by rotary evaporation. The mixture was resuspended in MeOH (100 μL) and dimethyl-3 was purified by semi-preparative HPLC (see section “chromatographic enrichment” above for details).
Synthesis of FsqB substrates
All reagents were purchased and used as is from Sigma-Aldrich. All solvents were purchased and used as is from Fischer Scientific. N-methyl-3,4-dihydroxy-DL-phenylalanine (11). A 100 mL Schlenk flask under argon at room temperature was charged with 3,4-dihydroxy-DL-tyrosine (394 mg, 2.0 mmol), dissolved in 50 mL of a 7:3 mixture of water and MeOH. To this NaBH4 (228 mg, 6.6 mmol) was added, followed by immediate addition of formaldehyde (0.5 mL of a 37 % solution in H2O, 6.0 mmol). The reaction was stirred for 1 h at room temperature then quenched by addition of glacial acetic acid (10 mL). The solvents were removed by rotary evaporation, and the product isolated by reverse-phase HPLC. See Supplementary Note for full NMR spectroscopic data of (12). N-methyl-dopamine. This compound was prepared according to the literature procedure50.
Supplementary Material
Acknowledgments
This research was funded in part by NIH R01GM112739-01 (to N.P.K and F.C.S.), an NIH CBI Training grant T32GM008500 (to J.A.B.), and an NSF Graduate Research Fellowship under grant no. DGE-1256259 (to J.E.S.). A.A.B. and D.H. were supported by the Deutsche Forschungsgemeinschaft (CRC ChemBioSys 1127). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of General Medical Sciences, National Institutes of Health, or National Science Foundation. The authors would like to thank David Kiemle for his kind assistance in operating the Bruker AvanceIII HD 800 MHz NMR spectrometer.
Footnotes
Author Contributions
J.A.B. and F.C.S. characterized and identified metabolites and biosynthetic pathways. J.E.S. and J.W.B. created all A. fumigatus mutants used. J.A.B. and C.G. performed stable-isotope labeling experiments. H.H.L., E.B., and D.H. contributed biochemical assays. J.M. and A.A.B. contributed to the identification of new metabolites. J.A.B., J.E.S., D.H., N.P.K., and F.C.S. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Contributor Information
Joshua A. Baccile, Email: jab826@cornell.edu.
Joseph E. Spraker, Email: jspraker@wisc.edu.
Henry H. Le, Email: hhl43@cornell.edu.
Eileen Brandenburger, Email: Eileen.Brandenburger@leibniz-hki.de.
Christian Gomez, Email: cg454@cornell.edu.
Jin Woo Bok, Email: jwbok@wisc.edu.
Juliane Macheleidt, Email: Juliane.Macheleidt@leibniz-hki.de.
Axel A. Brakhage, Email: Axel.Brakhage@leibniz-hki.de.
Dirk Hoffmeister, Email: Dirk.Hoffmeister@leibniz-hki.de.
Nancy P. Keller, Email: npkeller@wisc.edu.
Frank C. Schroeder, Email: schroeder@cornell.edu.
References
- 1.Keller NP. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat Chem Biol. 2015;11:671–677. doi: 10.1038/nchembio.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez JF, Somoza AD, Keller NP, Wang CC. Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat Prod Rep. 2012;29:351–371. doi: 10.1039/c2np00084a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brakhage AA. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 4.Fischbach MA, Walsh CT. Assembly-Line Enzymology for Polyketide and Nonribosomal Peptide Antibiotics: Logic, Machinery, and Mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 5.Jenke-Kodama H, Dittmann E. Bioinformatic perspectives on NRPS/PKS megasynthases: advances and challenges. Nat Prod Rep. 2009;26:874–883. doi: 10.1039/b810283j. [DOI] [PubMed] [Google Scholar]
- 6.Walsh CT, O’Brien RV, Khosla C. Nonproteinogenic Amino Acid Building Blocks for Nonribosomal Peptide and Hybrid Polyketide Scaffolds. Angew Chem Int Ed Engl. 2013;52:7098–7124. doi: 10.1002/anie.201208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forseth RR, et al. Homologous NRPS-like Gene Clusters Mediate Redundant Small-Molecule Biosynthesis in Aspergillus flavus. Angew Chem Int Ed Engl. 2013;52:1590–1594. doi: 10.1002/anie.201207456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang M, Beissner M, Zhao H. Aryl-aldehyde formation in fungal polyketides: Discovery and characterization of a distinct biosynthetic mechanism. Chem Biol. 2014;21:257–263. doi: 10.1016/j.chembiol.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagel JM, Facchini PJ. Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant Cell Physiol. 2013;54:647–672. doi: 10.1093/pcp/pct020. [DOI] [PubMed] [Google Scholar]
- 10.Bentley KW. β-phenylethylamines and the isoquinoline alkaloids. Nat Prod Rep. 2006;23:444–463. doi: 10.1039/b509523a. [DOI] [PubMed] [Google Scholar]
- 11.Bok JW, et al. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot Cell. 2005;4:1574–1582. doi: 10.1128/EC.4.9.1574-1582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Research. 2015;43:222–226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forseth RR, Schroeder FC. NMR-spectroscopic analysis of mixtures: from structure to function. Curr Opin Chem Biol. 2011;15:38–47. doi: 10.1016/j.cbpa.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forseth RR, Schroeder FC. Correlating secondary metabolite production with genetic changes using differential analysis of 2D NMR spectra. Methods Mol Biol. 2012;944:207–219. doi: 10.1007/978-1-62703-122-6_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutchan TM, Dittrich H. Characterization and Mechanism of the Berberine Bridge Enzyme, a Covalently Flavinylated Oxidase of Benzophenanthridine Alkaloid Biosynthesis in Plants. J Biol Chem. 1995;270:24475–24481. doi: 10.1074/jbc.270.41.24475. [DOI] [PubMed] [Google Scholar]
- 18.Winkler A, et al. A concerted mechanism for berberine bridge enzyme. Nat Chem Biol. 2008;4:739–741. doi: 10.1038/nchembio.123. [DOI] [PubMed] [Google Scholar]
- 19.Gaweska HM, Roberts KM, Fitzpatrick PF. Isotope effects suggest a stepwise mechanism for Berberine Bridge Enzyme. Biochemistry. 2012;51:7342–7347. doi: 10.1021/bi300887m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scrutton NS. Chemical aspects of amine oxidation by flavoprotein enzymes. Nat Prod Rep. 2004;21:722–730. doi: 10.1039/b306788m. [DOI] [PubMed] [Google Scholar]
- 21.Dunn RV, Munro AW, Turner NJ, Rigby SE, Scrutton NS. Tyrosyl radical formation and propagation in flavin dependent monoamine oxidases. Chembiochem. 2010;11:1228–1231. doi: 10.1002/cbic.201000184. [DOI] [PubMed] [Google Scholar]
- 22.Leonard E, Runguphan W, O’Connor S, Prather KJ. Opportunities in metabolic engineering to facilitate scalable alkaloid production. Nat Chem Biol. 2009;5:292–300. doi: 10.1038/nchembio.160. [DOI] [PubMed] [Google Scholar]
- 23.Glenn WS, Runguphan W, O’Connor SE. Recent progress in the metabolic engineering of alkaloids in plant systems. Curr Opin Biotechnol. 2013;24:354–365. doi: 10.1016/j.copbio.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakagawa A, et al. A bacterial platform for fermentative production of plant alkaloids. Nat Commun. 2011;2:326. doi: 10.1038/ncomms1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLoache WC, et al. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat Chem Biol. 2015;11:465–471. doi: 10.1038/nchembio.1816. [DOI] [PubMed] [Google Scholar]
- 26.Hawkins KM, Smolke CD. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat Chem Biol. 2008;4:564–573. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fossati E, et al. Reconstitution of a 10-gene pathway for synthesis of the plant alkaloid dihydrosanguinarine in Saccharomyces cerevisiae. Nat Commun. 2014;5:3283. doi: 10.1038/ncomms4283. [DOI] [PubMed] [Google Scholar]
- 28.Thodey K, Galanie S, Smolke CD. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat Chem Biol. 2014;10:837–844. doi: 10.1038/nchembio.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galanie S, Smolke CD. Optimization of yeast-based production of medicinal protoberberine alkaloids. Microb Cell Fact. 2015;14:144. doi: 10.1186/s12934-015-0332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imamura K, et al. Identification of a Gene Involved in the Synthesis of a Dipeptidyl Peptidase IV Inhibitor in Aspergillus oryzae. Appl Environ Microbiol. 2012;78:6996–7002. doi: 10.1128/AEM.01770-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macheleidt J, et al. Transcriptome analysis of cyclic AMP-dependent protein kinase A–regulated genes reveals the production of the novel natural compound fumipyrrole by Aspergillus fumigatus. Mol Microbiol. 2015;96:148–162. doi: 10.1111/mmi.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada T, et al. Crystal Structure of Serine Dehydratase from Rat Liver. Biochemistry. 2003;42:12854–12865. doi: 10.1021/bi035324p. [DOI] [PubMed] [Google Scholar]
- 34.Wallner S, et al. Catalytic and Structural Role of a Conserved Active Site Histidine in Berberine Bridge Enzyme. Biochemistry. 2012;51:6139–6147. doi: 10.1021/bi300411n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee A, Walker J, Weyant KB, Schroeder CM. Characterization of flavin-based fluorescent proteins: an emerging class of fluorescent reporters. PLoS One. 2013;8:e64753. doi: 10.1371/journal.pone.0064753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider P, Weber M, Rosenberger K, Hoffmeister D. A One-Pot Chemoenzymatic Synthesis for the Universal Precursor of Antidiabetes and Antiviral Bis-Indolylquinones. Chem Biol. 2007;14:635–644. doi: 10.1016/j.chembiol.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 37.Fawaz MV, Topper M, Firestine SM. The ATP-Grasp Enzymes. Bioorg Chem. 2011;39:185–191. doi: 10.1016/j.bioorg.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan C, Moews PC, Shi Y, Walsh CT, Knox JR. A common fold for peptide synthetases cleaving ATP to ADP: glutathione synthetase and D-alanine:d-alanine ligase of Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:1172–1176. doi: 10.1073/pnas.92.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noike M, et al. A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin. Nat Chem Biol. 2015;11:71–76. doi: 10.1038/nchembio.1697. [DOI] [PubMed] [Google Scholar]
- 40.Tobiasen C, et al. Nonribosomal peptide synthetase (NPS) genes in Fusarium graminearum, F. culmorum and F. pseudograminearium and identification of NPS2 as the producer of ferricrocin. Current Genetics. 2007;51:43–58. doi: 10.1007/s00294-006-0103-0. [DOI] [PubMed] [Google Scholar]
- 41.Nagano N, et al. Class of cyclic ribosomal peptide synthetic genes in filamentous fungi. Fungal Genet Biol. 2016;86:58–70. doi: 10.1016/j.fgb.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 42.Wiemann P, et al. Perturbations in small molecule synthesis uncovers an iron-responsive secondary metabolite network in Apsergillus fumigatus. Front Microbiol. 2014;5:530. doi: 10.3389/fmicb.2014.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimizu K, Keller NP. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics. 2001;157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sekonyela R, et al. RsmA regulates Aspergillus fumigatus gliotoxin cluster metabolites including cyclo(L-Phe-L-Ser), a potential new diagnostic marker for invasive aspergillosis. PLoS ONE. 2013;8:e62591. doi: 10.1371/journal.pone.0062591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer JM, et al. Loss of CclA, required for histone 3 lysine 4 methylation, decreases growth but increases secondary metabolite production in Aspergillus fumigatus. PeerJ. 2013;1:e4. doi: 10.7717/peerj.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu JH, et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Kalb D, Lackner G, Hoffmeister D. Functional and phylogenetic divergence of fungal adenylate-forming reductases. Appl Environ Microbiol. 2014;80:6175–6183. doi: 10.1128/AEM.01767-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider P, Bouhired S, Hoffmeister D. Characterization of the atromentin biosynthesis genes and enzymes in the homobasidiomycete Tapinella panuoides. Fungal Genet Biol. 2008;45:1487–1496. doi: 10.1016/j.fgb.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 49.Bok JW, et al. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carpenter JF. An improved synthesis of 5,6-diacetoxy-N-methylindole and of epinine. J Org Chem. 1993;58:1607–1609. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.