Abstract
Bacterial vaccines can reduce carriage rates. Colonization is usually a binary endpoint. Real time quantitative PCR (qPCR) can quantify bacterial DNA in mucosal samples over a wide range. Using culture and single-gene species-specific qPCRs for Streptococcus pneumoniae (lytA), Streptococcus pyogenes (ntpC), Moraxella catarrhalis (ompJ), Haemophilus influenzae (hdp) and Staphylococcus aureus (nuc) and standard curves against log-phase reference strain broth cultures we described frequency and peak density distributions of carriage in nasopharyngeal swabs from 161 healthy 2–4 y old children collected into STGG broth. In general, detection by qPCR and culture was consistent. Discordance mostly occurred at lower detection thresholds of both methods, although PCR assays for S. pyogenes and S. aureus were less sensitive. Density varied across 5-7 orders of magnitude for the 5 species with the abundant species skewed toward high values (modes: S. pneumoniae log3-4, M. catarrhalis & H. influenzae log4-5 CFU/ml broth). Wide ranges of bacterial DNA concentrations in healthy children carrying these bacteria could mean that different individuals at different times vary greatly in infectiousness. Understanding the host, microbial and environmental determinants of colonization density will permit more accurate prediction of vaccine effectiveness.
Keywords: colonization, quantitative PCR, bacterial density, children
Introduction
The bacterial species Streptococcus pneumoniae, Streptococcus pyogenes (group A streptococcus; GAS), Moraxella catarrhalis, Haemophilus influenzae and Staphylococcus aureus are common inhabitants of the mucosal surfaces of the human nasopharynx (NP). Children routinely become colonised by these species although the age distribution and the duration of colonization varies.1-5 An example of this is colonization with S. pneumoniae, rates of which increase in infancy, peak at 1–2 y of age and then decline. GAS colonization on the other hand tend to be lower in infants with gradual increases during the first decade of life.6 Several capsular types of bacteria have been targeted by infant conjugate vaccine programmes which not only induce direct protection in recipients but can also reduce carriage rates and so transmission, thereby reducing the incidence of symptomatic infection in the population at large.7 Thus improved methodologies for the study of colonization have the potential to improve our understanding of the pathogenesis of disease, the mechanisms of vaccine-induced herd protection, the potential for inter-species interactions8 and ecological changes over time. Until recently most studies have reported the detection of these organisms using cultures from upper respiratory tract swabs.9 We recently reported associations between both rhinitis symptoms and respiratory viral infections and nasal bacterial density ascertained by attributing semi-quantitative scores to numbers of colonies visualised on bacterial culture plates10,11 but this approach only allows resolution of differences in bacterial numbers over a narrow dynamic range. Improved and broadened quantitation of bacterial carriage over a wider range would facilitate the study of the biology of colonization and exploration of the determinants of transmission. This can be achieved using real time PCR which is now widely used in clinical practice and has great potential utility in research studies by virtue of high levels of automation, reproducibility, rapidity, sensitivity and specificity and low unit staff-time and consumables costs.12,13 Whereas in clinical practice the capacity of PCR to detect bacterial DNA in the absence of viable organisms, for example following antibiotic treatment,14,15 may complicate interpretation of results, in studies of bacterial colonization it can be an advantage as this footprint is of potential relevance. Finally, provided that appropriate species-specific DNA sequences are targeted, PCR is less subject to the potential biases and errors that can result during bacterial culture and the phenotypic identification of resulting bacterial colonies.
In this study we employed qPCR monoplex assays using previously published gene targets for S. pneumoniae (lytA), GAS (ntpC), M. catarrhalis (ompJ), H. influenzae (hdp) and S. aureus (nuc) and serial dilutions of culture-quantified log-phase broth cultures of reference strains of these species, permitting conversion of qPCR cycle threshold (Ct) values into equivalent bacterial concentrations measured in colony forming units (CFU)/ml of culture broth. We validated these qPCR assays against conventional agar plate cultures using a randomly selected subset of 149 NP swab samples and then used them to describe the rates and density distributions of nasal carriage of these 5 bacterial species in 161 healthy 2–4 y old children each sampled up to 5 times over the single winter season 2011-2012.
Results
Peak density distributions of bacterial carriage in young healthy children
From 619 samples collected from 161 healthy children and analyzed by qPCR, 79.5% were positive for S. pneumoniae, 4.4% for GAS, 86.1% for M. catarrhalis, 85.3% for H .influenzae and 6.6% for S. aureus. The respective percentages of children in whom each of these species were detected at least once during the study were S. pneumoniae 97.5%, GAS 14.8%, M. catarrhalis 98.1%, H. influenzae 98.7% and S. aureus 21.6%. For each species, the result from each colonised child which had the highest density (peak value) was selected for presentation and these data are summarised as frequency histograms in Figure 1. The mode densities and the distributions around them look quite distinct for the different species with the 3 most frequently carried species (S. pneumoniae, M. catarrhalis and H. influenzae) being, on average, present at much higher density than the 2 less commonly carried species (GAS and S. aureus).
Figure 1.
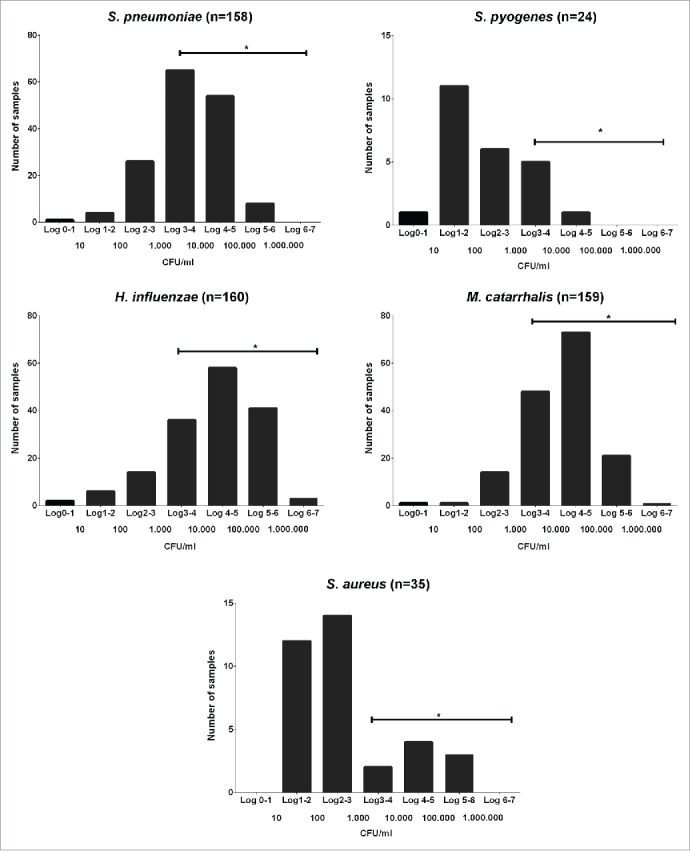
Bacterial peak density range. Peak winter time nasal density distributions of the 5 studied species in pre-school children determined by qPCR. *:Horizontal lines indicate the range of densities that could be reliably quantified by qPCR but not by semi-quantitative culture.
Comparison of RT-PCR against conventional cultures
In order to compare detection with qPCR and conventional cultures, 149 NP swab samples were selected randomly and both cultured and subjected to qPCR analysis for S. pneumoniae, GAS, M. catarrhalis, H. influenzae and S. aureus (Table 1). For S. pneumoniae, M. catarrhalis and H. influenzae, species for which the large majority of samples were positive, detection rates were 9.4–26.2% higher using qPCR than culture and the large majority of culture-positive samples also tested PCR-positive (Table 1). For GAS and S. aureus, which were much less commonly found, detection rates were lower by PCR than culture and culture-positive PCR-negative samples were much more abundant relative to samples positive by both methods. In all species tested, the mean density was lower in samples only positive by either method alone than in those positive by both.
Table 1.
Culture and qPCR results in 149 clinical NP swabs from 2–4 y old healthy children attending day care centers. Mean bacterial density in PCR positive samples is shown in CFU/ml and in culture positive samples as means of density scores (DS)(0:0 1:1–5 CFU/ml broth 2:6-20 3:21-50 4:51-100 5:>100)(10).*: One sample failed the GAS PCR and was omitted from the comparison
| S. pneumoniae | GAS* | M. catarrhalis | H. influenzae | S. aureus | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Culture + qPCR+ | 85 (57%) | DS: 4.1 | 9 (6.1%) | DS: 3.3 | 106 (71.1%) | DS: 3.8 | 61 (40.9%) | DS: 4.1 | 6 (4.0%) | DS: 2.6 |
| 645 CFU/ml | 825 CFU/ml | 1330 CFU/ml | 4840 CFU/ml | 1495 CFU/ml | ||||||
| Culture + qPCR- | 2 (1.3%) | DS: 2.5 | 5 (3.4%) | DS: 1.6 | 10 (6.7%) | DS: 2.2 | 3 (2.0%) | DS: 2.3 | 10 (6.7%) | DS: 2.1 |
| N/A | N/A | N/A | N/A | N/A | ||||||
| Culture - qPCR+ | 14 (9.4%) | N/A | 0 (0%) | N/A | 19 (12.8%) | N/A | 39 (26.2%) | N/A | 7 (4.7%) | N/A |
| 17 CFU/ml | N/A | 27 CFU/ml | 205 CFU/ml | 195 CFU/ml | ||||||
| Culture - qPCR- | 48 (32.2%) | N/A | 134 (90.5%) | N/A | 14 (9.4%) | N/A | 46 (30.9%) | N/A | 126 (84.6%) | N/A |
| N/A | N/A | N/A | N/A | N/A | ||||||
PCR specificity panel
In every case, all 3 dilutions of all the bacterial strains in the specificity panel yielded appropriate positive or negative results by PCR.
Liquid cultures, dilutions and standard curves
Growth curves for the liquid cultures of the 5 reference strains are shown in Figure 2. Dilutions yielded pure growths of the respective species and appropriate colony counts for the 10-fold dilutions across the 3.5 log range (3-4 dilutions) in which colonies were both detectable and countable (data not shown).
Figure 2.
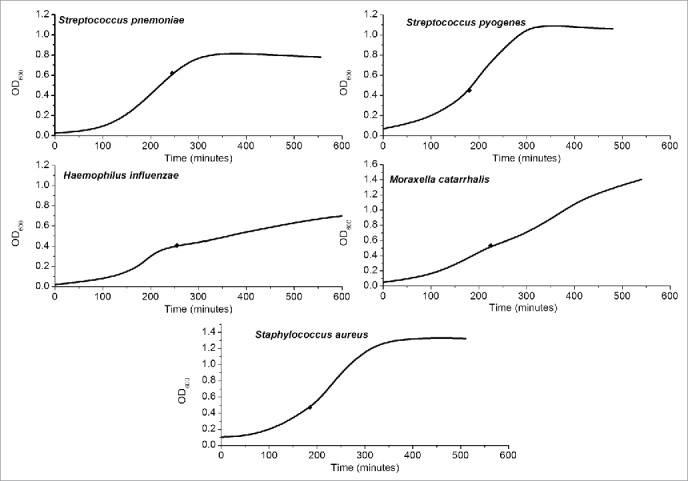
Liquid cultures. Optical density at 600 nm (OD600) of the 5 species studied in liquid culture over time. The black diamonds represent the points of harvest for quantification experiments.
The qPCR CT values for the heat inactivated 10-fold serial dilutions from each species were used to plot standard curves (twice each) against observed and calculated CFU/ml counts for corresponding 10-fold dilutions of the liquid cultures (Fig. 3) providing for each a conversion equation from one to the other, taking into account the volumes used for culture and PCR. These were used to generate CFU/ml broth equivalent values from qPCR CT results from clinical samples processed in the same assays. qPCR assays for S. aureus and GAS did not reliably detect DNA from these standard strains from broth dilutions with low density growth on agar plates (Fig. 3).
Figure 3.
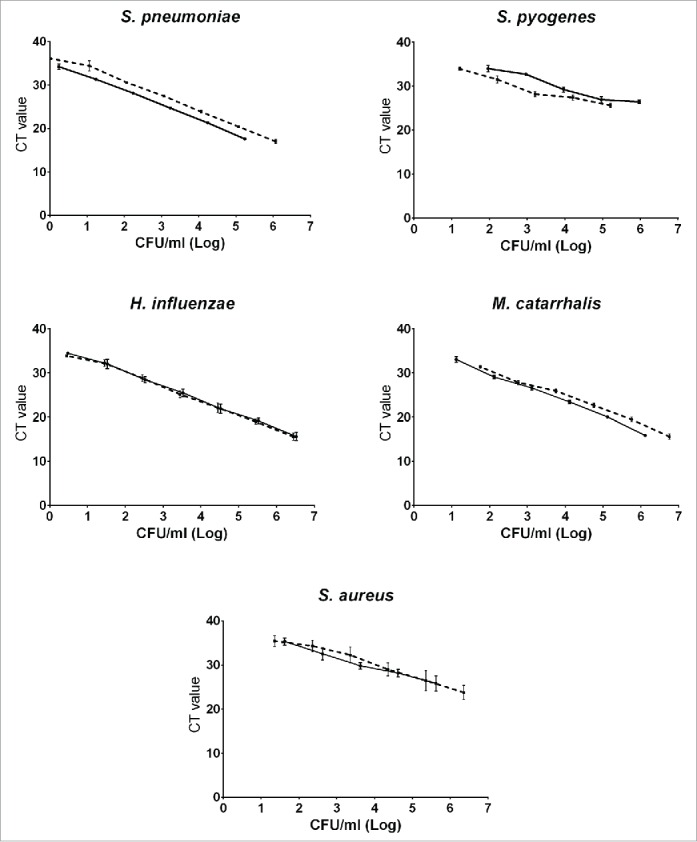
qPCR standard curves. Standard curves (culture results (real and calculated) in CFU/ml broth vs cycle threshold values). Each curve represents the average of the results of 3 PCR runs performed on a dilution series of cultures of which 2 (both shown, solid line and dashed line) were done for each species. CT: Cycle Threshold. CFU: colony forming units.
Discussion
In this study, we describe, for the first time, the distributions of peak colonization density of the common culturable bacterial species that inhabit the nasopharynges of healthy young children. We found somewhat higher rates of carriage for pneumococci, M. catarrhalis and H. influenzae than previous reports.16-19 These high rates by PCR may represent DNA detection from organisms previously present but no longer viable and so undetectable by culture while relatively high rates by culture in our population may reflect high rates of wintertime intercurrent viral infections and intense transmission in these young children in close contact with one another in the day care setting. Marked differences in the relationship between colonization frequency and density between species are immediately obvious (Fig. 1) which are likely to affect the efficiency with which they are transmitted within this population and to other age groups. This analysis may therefore more accurately predict spread than the simple rates of detection reported in previous studies.20,21 Perhaps more importantly, large differences in maximal colonization density are evident between individual children. We propose that children with high density colonization will transmit bacterial carriage more efficiently than those with low density colonization. Given the evident importance of indirect effects on transmission, for the effectiveness of bacterial vaccine programmes22-24 we propose that future attempts to measure and predict such effects should take account of the effect of vaccination on colonization density and not just its frequency.
We used qPCR to detect and quantify bacteria obtained by nasopharyngeal swabbing rather than conventional culture methods, principally because of the wide range of bacterial density over several orders of magnitude25 that can be measured simply and accurately using this technique, but also because of its expected greater efficiency and lower cost. However, with the methodology we used, sensitivity was poor for GAS and S. aureus particularly in low density samples. This may have been due to inefficient DNA extraction due to the robust structure of the cell wall of these organisms26-28 so that more efficient lysis of samples prior to DNA extraction might increase the sensitivity of these assays. Alternatively or additionally reducing the extraction eluate volume to increase DNA concentration in samples might do likewise. The efficiency of these 2 qPCR assays was also lower than the others, leading to more than the expected 3.3 cycle difference of amplification signal between successive 10-fold serial dilutions (Fig. 3). Different combinations of primer concentrations (50, 300 and 900 nM) were tested without improvement (data not shown) and the presence of inhibitors could possibly explain this.29 Finally, since, in particular, experience with the ntpC gene target for PCR detection of GAS is still very limited, it may be that other gene targets will provide better assay performance. Development and validation of a multiplex assay for the detection of these organisms will increase efficiency and reduce costs even further although the potential pitfalls of reduced sensitivity, accuracy and inter-assay competition30 will need to be addressed before implementation. A cut-off level of 35 cycles is widely used in PCR assays and we have adopted that in our work. More individually adapted levels of detection for each primer set could possibly be adapted for increased accuracy.
The creation of standard curves which permit translation of qPCR cycle threshold numbers for detection into absolute values of bacterial density in CFU/ml makes the meaning of assay results more accessible but also makes direct comparison between studies of colonization density feasible even if different assays or methods are used. Replicate dilution series done for each bacterial species were closely concordant for M. catarrhalis and H. influenzae and similar for S. pneumoniae but showed more variation for the 3 less abundant species (Fig. 3).
Unsurprisingly, since PCR can detect DNA from non-viable bacteria or bacterial fragments, although the large majority of the 149 randomly selected nasal swab samples generated concordant positive or negative culture and qPCR results for all 6 bacterial species, all apart from GAS yielded some that were PCR-positive but culture-negative (Table 1). Of particular note, about one in 4 swabs were positive for H. influenzae by qPCR but not culture. The lower detection rates of H. influenzae by culture may partly be explained by failure reliably to observe the small colonies on agar plate cultures and/or inaccurate results from biological identification tests which are often ambiguous. For the rest, PCR-positive culture-negative samples may represent samples with DNA but in which viable bacteria are not present. Discrepancies could also be due to genetic variation in primer or probe recognition sequences. However, in the context of colonization studies, in contrast to clinical diagnostics, if this represents detection of the footprint of previous carriage, these positive results are of interest and importance. As might be expected, for all 5 species for which such samples were found, they had substantially lower average of gene copies detected than samples in which culture was also positive.
Among the 149 samples, there were also between 2 and 10 PCR-negative culture-positive samples for each species. Given the observed low sensitivity of the GAS and S. aureus qPCR assays, for these, such samples were to be expected. However, since there is potential to detect even a single viable bacterium manifest as a single colony from a 50-100 µl inoculum of broth on agar, whereas even a highly efficient PCR reaction may need multiple copies of a single gene in DNA extracted from 350 µl of broth, in order reliably to detect their presence above background, small numbers of such discrepant results are also unsurprising. Once again, for all species tested, these samples had lower average density by culture than samples positive both by culture and PCR.
It will be of interest to compare these findings made in a UK pre-school population with bacterial carriage density distributions in other places, in other age groups and in children who are unwell.12 The other important question that arises is how, exactly, colonization density equates to efficiency of transmission. Is there a density below which it becomes very rare or, conversely, is there a density above which there is no further increase in what is effectively maximal transmission? Between these extremes, is the relationship between density and successful transfer linear or, more likely, otherwise? Finally, what other determinants of bacterial phenotype also affect the capacity to transmit successfully (so that 2 children with similar colonization density might have very different infectiousness)? The interplay between respiratory viral infections, upper respiratory tract-colonising bacteria and host innate and specific anti-viral and anti-bacterial mucosal immune responses will need to be elucidated more fully before this ecosystem, its occasional punctuation by episodes of clinically significant infectious illness and the ways it is affected by both bacterial and viral vaccines can be properly understood. It is important to find ways to answer such questions in order that future studies of carriage, conducted to evaluate the indirect effects of vaccines, can be interpreted correctly.
Subjects, Materials and Methods
Human subjects and sampling
The nasopharyngeal swab samples used were collected in clinical studies of bacterial colonization in healthy 2–4 y old children attending 9 day care centers in Bristol, UK between 2011 and 2013. Studies were approved by the National Research Ethics Service – Central Bristol Committee and sponsored by the University of Bristol. The children were all fully vaccinated according to the national UK schedule, were all attending the DCC and had no signs of systemic illness at the time of sampling. Fine tip pediatric nasopharyngeal swabs with rayon buds (Peel Pouch Dryswab™ Medical Wire & Equipment, Wiltshire, UK) were inserted horizontally into the nasopharynx until resistance was met then retracted with a slight twisting motion and tips were inserted into 2 ml tubes containing 1.5 ml Skim milk-Tryptone-Glucose-Glycerol (STGG) broth. Each child was sampled up to 5 times during a single winter season. Swabs were marked with a random number from 1–700 using pre-printed non-consecutive numbered labels, and stored at −80° C in boxes containing up to 100 samples each.
Bacterial strains and plate cultures
Reference bacterial strains used for PCR assay development and standardisation were: S. pneumoniae (ATCC 6303), GAS (ATCC 19615), M. catarrhalis (ATCC 25240), H. influenzae (ATCC 10211) and S. aureus (ATCC 25923), all from LGC Standards Laboratories (Teddington, UK). M. catarrhalis and S. aureus strains were cultured overnight for 16-18 hours on a Columbia Blood Agar Base (CBA) supplemented plate with 5% horse defibrinated blood (Oxoid, Basingstoke, UK). S. pneumoniae and GAS strains were cultured for 16-18 hours overnight on a COBA plate (CBA plate with 0.25 mg colistin and 0.125 mg oxalinic acid). H. influenzae was cultured for 16-18 hours overnight on a bacitracin chocolate agar plate. All cultures were performed at incubation temperature of 37°C. 50 μl broth from STGG clinical swab samples were likewise cultured on this set of plates and under these conditions. For comparison of culture and qPCR, given the high colonization rates, 150 samples were considered a representative sample size and they were randomly selected by choosing the first 22 samples from each sample box until 150 was reached. One sample was destroyed during the process and was not replaced. A semi-quantitative score was allocated for each cultured species from 0 through 5 (0 = no growth, 1 = 1-5 colonies per plate (thus 20-100 CFU/ml broth), 2 = 6-20, 3 = 21-50, 4 = 51-100 and 5 = >100).10
Cultures in liquid medium
3-4 colonies of each standard strain from plate cultures were used to inoculate 20 ml liquid media as follows: Todd-Hewitt broth (Media services, School of Cellular and Molecular Medicine, University of Bristol, UK) with 0.5% yeast extract (S. pneumoniae & GAS), Brain-Heart Infusion (Oxoid, Basingstoke, UK) alone (S. aureus), with 10% defibrinated horse blood (M. catarrhalis) or with 5% Fildes (haemin and nicotinamide adenine dinucleotide from Oxoid, Basingstoke, UK) (H. influenzae) and cultured at 37°C in 5% CO2 static (S. pneumoniae & GAS) or at 50 (H. influenzae & S. aureus) or 100 (M. catarrhalis) rotations per minute in 25 ml glass universals with the caps loosely fitted. After 1 hour, and subsequently at regular intervals until the stationary phase was reached, optical density (OD) at 600 nm was measured (Thermo Spectronic Genesys 6, Thermo Electron Scientific Instruments LLC, WI, USA). When the OD600 was 0.4–0.6, 1 ml each broth culture was harvested and 10 ten-fold serial dilutions prepared in STGG broth. 100 μl aliquots of each dilution were plated out (as above) and aerobically incubated for 16-18 hours at 37°C/5% CO2 prior to colony counts being performed up to a maximum of 750 CFU per plate. For the purposes of PCR, 350 μl aliquots of each liquid culture dilution were also heat inactivated at 100°C for 10 minutes using a digital block heater (Grant Boekel BBD, Grant instruments, Cambridge, UK). Successful inactivation was confirmed by appropriate plate cultures for each species. For Sa, 90 minutes of heat inactivation were necessary.
Preparation for and conduct of PCR assays
Target genes and corresponding primer selection were informed by the published literature.31-36 Primers and probes (Table 2) were assessed and optimised using Primer Express version 3.0 (Life Technologies). The specificity of the primers for the target species was assessed in silico using the National Center for Biotechnology Information (NCBI) database and the Basic Local Alignment Search Tool (BLAST).
Table 2.
Target genes and primer and probe sequences used for qPCR assays. BHQ: Black Hole Quencher. *:Primer design from this study
| Organism | Gene | Forward Primer Sequence 5′ - 3′ | Reverse Primer Sequence 5′ - 3′ | [Dye]ProbeSequence 5′ - 3′ |
|---|---|---|---|---|
| S. pneumoniae31 | lytA | ACGCAATCTAGCAGATGAAGCA | TCGTGCGTTTTAATTCCAGCT | [Cy5]GCCGAAAACGCTTGATACAGGGAG[BHQ2] |
| S. pyogenes* | ntpC | TGCACCAGTCAGTCTATCACC | AGCGATAGGTCTTGGTTAGCTC | [6FAM]TGACGATCTCAACGATTTGGATCGG[BHQ1] |
| M. catarrhalis36 | ompJ | CAGCCTAGCAGGCGGTGTT | TTGCTTCAACGCCCACATT | [VIC]TTGGCTTTAAACCATTAGC[MGB] |
| H. influenzae* | hdp | TTGGCCCAGGTTGGTATATG | TTACGCACGGTGTAAGGATG | [6FAM]CACTCCGTTGGTAAAAGAACTTGCACA[BHQ1] |
| S. aureus33 | nuc | GTTGCTTAGTGTTAACTTTAGTTGTA | AATGTCGCAGGTTCTTTATGTAATTT | [6FAM]AAGTCTAAGTAGCTCAGCAAATGCA[BHQ1] |
Automated extraction of nucleic acids from samples was performed (QIAsymphony® QIAGEN, CA, USA) using DSP Virus/Pathogen Mini Kit version 1 (QIAGEN, CA, USA). In brief, after vortexing, 350-μl of each sample underwent bacterial cell lysis, and total nucleic acids were captured on magnetic beads and eluted into 110 μl elution buffer. DNA extract eluates were dispensed into 96-well elution microtubes (QIAGEN, CA, USA). After extraction the plates were sealed and stored frozen in −80°C. Plates were thawed and centrifuged prior to PCR. A QIAagility pipetting robot and software (QIAGEN, CA, USA) was used to prepare PCR plates in reaction mixes containing Applied Biosystems Fast Universal Master Mix (10 µl), primers (300 nM), probe (100 nM) and DNA extracts (5 µl) - to a total reaction volume of 20 µl.
The Applied Biosystems ViiA 7™ real time PCR system (Life Technologies, USA) was used for amplification and detection of DNA using MicroAmp optical 384-well reaction plates (Life Technologies, USA). Reaction conditions for DNA amplification were a 20 second hold stage at 95°C followed by 50 cycles of 95°C for 3 seconds and 60°C for 1 min. Samples with CT values ≤ 35 were considered positives. ViiA™ 7 Software version 1.2.2 (Life Technologies, USA) was used for data analysis.
The bacteriophage T4 was used as an internal amplification control (successful DNA extraction and absence of PCR inhibition) in all PCR assays. Two dilutions of DNA extracts of each target reference strain were used as positive controls and DNA extracts of STGG broth and molecular grade water as negative reagent controls. A specificity panel (Table 3) containing DNA extracts of three dilutions (2-, 4- and 6-fold dilution with STGG broth of pure culture isolates) of heat-inactivated cultures of the same and other species was also tested.
Table 3.
Bacterial strains used to prepare a panel of specificity controls
| Organism | Strain |
|---|---|
| Streptococcus agalactiae | ATCC® 12403 |
| Streptococcus mitis | NCTC 10712 |
| Staphylococcus epidermidis | NCTC 11047 |
| Enterococcus faecalis | JH2–2 |
| Streptococcus oralis | NCTC 11427 |
| Staphylococcus aureus | ATCC® 25923 |
| Streptococcus pneumoniae | ATCC® 6303 |
| Streptococcus pyogenes | ATCC BAA-1064 |
| Streptococcus pseudopneumoniae | ATCC® BAA-960 |
| Streptococcus parasanguinis | ATCC® 15912 |
| Escherichia coli | ATCC 25922 |
| Neisseria lactamica | From other carriage studies |
| Neisseria gonorrhoea | ATCC® 31426 |
| Pseudomonas aeruginosa | From other carriage studies |
| Haemophilus parainfluenzae | From other carriage studies |
| Neisseria meningitidis A | ATCC® 53417 |
| Neisseria meningitidis B | ATCC® BAA-335 |
| Neisseria meningitidis C | ATCC® 53414 |
| Neisseria meningitidis D | ATCC® 53419 |
| Neisseria meningitidis W135 | ATCC® 35559 |
| Neisseria meningitidis X | ATCC® 35560 |
| Neisseria meningitidis Y | ATCC® 35561 |
| Klebsiella pneumoniae | From other carriage studies |
Disclosure of Potential Conflicts of Interest
Prof Finn has received research grants from GSK, AstraZeneca, Alios, Pfizer, Novartis and SPSMD and consultancy fees and speaking honoraria, all paid to his employers, from the same companies as well as Takeda. He is a member of the WHO European Technical Advisory Group of Experts on Immunisation and the UK Department of Health's Joint Committee on Vaccination and Immunisation and subcommittees. Other authors report no conflicts of interest.
Acknowledgments
The authors thank Howard Jenkinson & Darryl Hill for advice and bacterial strains and Paulina Sikora-Liszka for assistance with the laboratory work.
Funding
This work was supported by an ESPID research fellowship.
References
- 1.Syrjänen RK, Kilpi TM, Kaijalainen TH, Herva EE, Takala AK. Nasopharyngeal carriage of Streptococcus pneumoniae in Finnish children younger than 2 years old. J Infect Dis 2001; 184(4):451-9; PMID:11471103; http://dx.doi.org/ 10.1086/322048 [DOI] [PubMed] [Google Scholar]
- 2.Christenson B, Sylvan SP, Noreen B. Carriage of multiresistant Streptococcus pneumoniae among children attending day-care centres in the Stockholm area. Scand J Infect Dis 1997; 29(6):555-8; PMID:9571733; http://dx.doi.org/ 10.3109/00365549709035893 [DOI] [PubMed] [Google Scholar]
- 3.García-Rodríguez JA, Fresnadillo Martínez MJ. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J Antimicrob Chemother 2002; 50 Suppl S2:59-73; PMID:12556435; http://dx.doi.org/ 10.1093/jac/dkf506 [DOI] [PubMed] [Google Scholar]
- 4.Leach AJ, Boswell JB, Asche V, Nienhuys TG, Mathews JD. Bacterial colonization of the nasopharynx predicts very early onset and persistence of otitis media in Australian aboriginal infants. Pediatr Infect Dis J 1994; 13(11):983-9; PMID:7845752; http://dx.doi.org/ 10.1097/00006454-199411000-00009 [DOI] [PubMed] [Google Scholar]
- 5.Le Polain de Waroux O, Flasche S, Prieto-Merino D, Edmunds WJ. Age-dependent prevalence of nasopharyngeal carriage of streptococcus pneumoniae before conjugate vaccine introduction: a prediction model based on a meta-analysis. PLoS One 2014; 9(1):e86136; PMID:24465920; http://dx.doi.org/ 10.1371/journal.pone.0086136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yagupsky P, Landau D, Beck A, Dagan R. Carriage of Streptococcus pyogenes among infants and toddlers attending day-care facilities in closed communities in southern Israel. Eur J Clin Microbiol Infect Dis 1995; 14(1):54-8; PMID:7729455; http://dx.doi.org/ 10.1007/BF02112621 [DOI] [PubMed] [Google Scholar]
- 7.Seale A, Finn A. What is the best way to use conjugate vaccines? Curr Opin Infect Dis 2011; 24(3):219-24; PMID:21522065; http://dx.doi.org/ 10.1097/QCO.0b013e3283468996 [DOI] [PubMed] [Google Scholar]
- 8.Spijkerman J, Prevaes SM, van Gils EJ, Veenhoven RH, Bruin JP, Bogaert D, Wijmenga-Monsuur AJ, van den Dobbelsteen GP, Sanders EA. Long-term effects of pneumococcal conjugate vaccine on nasopharyngeal carriage of S. pneumoniae, S. aureus, H. influenzae and M. catarrhalis. PLoS One 2012; 7(6):e39730; PMID:22761879; http://dx.doi.org/ 10.1371/journal.pone.0039730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satzke C, Turner P, Virolainen-Julkunen A, Adrian PV, Antonio M, Hare KM, Henao-Restrepo AM, Leach AJ, Klugman KP, Porter BD, et al.. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: Updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 2013; 32(1):165-79; PMID:24331112; http://dx.doi.org/ 10.1016/j.vaccine.2013.08.062 [DOI] [PubMed] [Google Scholar]
- 10.Rodrigues F, Foster D, Nicoli E, Trotter C, Vipond B, Muir P, Gonçalves G, Januário L, Finn A. Relationships between rhinitis symptoms, respiratory viral infections and nasopharyngeal colonization with Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in children attending daycare. Pediatr Infect Dis J 2013; 32(3):227-32; PMID:23558321; http://dx.doi.org/ 10.1097/INF.0b013e31827687fc [DOI] [PubMed] [Google Scholar]
- 11.Wolter N, Tempia S, Cohen C, Madhi SA, Venter M, Moyes J, Walaza S, Malope-Kgokong B, Groome M, du Plessis M, et al.. High nasopharyngeal pneumococcal density, increased by viral coinfection, is associated with invasive pneumococcal pneumonia. J Infect Dis 2014; 210(10):1649-57; PMID:24907383; http://dx.doi.org/ 10.1093/infdis/jiu326 [DOI] [PubMed] [Google Scholar]
- 12.Vu HT, Yoshida LM, Suzuki M, Nguyen HA, Nguyen CD, Nguyen AT, Oishi K, Yamamoto T, Watanabe K, Vu TD. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J 2011; 30(1):11-8; PMID:20686433; http://dx.doi.org/ 10.1097/INF.0b013e3181f111a2 [DOI] [PubMed] [Google Scholar]
- 13.Albrich WC, Madhi SA, Adrian PV, van Niekerk N, Mareletsi T, Cutland C, Wong M, Khoosal M, Karstaedt A, Zhao P, et al.. Use of a rapid test of pneumococcal colonization density to diagnose pneumococcal pneumonia. Clin Infect Dis 2012; 54(5):601-9; PMID:22156852; http://dx.doi.org/ 10.1093/cid/cir859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaschke AJ, Heyrend C, Byington CL, Obando I, Vazquez-Barba I, Doby EH, Korgenski EK, Sheng X, Poritz MA, Daly JA, et al.. Molecular analysis improves pathogen identification and epidemiologic study of pediatric parapneumonic empyema. Pediatr Infect Dis J 2011; 30(4):289-94; PMID:21057372; http://dx.doi.org/ 10.1097/INF.0b013e3182002d14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kais M, Spindler C, Kalin M, Ortqvist A, Giske CG. Quantitative detection of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in lower respiratory tract samples by real-time PCR. Diagn Microbiol Infect Dis 2006; 55(3):169-78; PMID:16626914; http://dx.doi.org/ 10.1016/j.diagmicrobio.2006.01.007 [DOI] [PubMed] [Google Scholar]
- 16.Shiri T, Nunes MC, Adrian PV, Van Niekerk N, Klugman KP, Madhi SA. Interrelationship of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus colonization within and between pneumococcal-vaccine naïve mother-child dyads. BMC Infect Dis 2013; 13:483; PMID:24134472; http://dx.doi.org/ 10.1186/1471-2334-13-483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Bergh MR, Spijkerman J, Swinnen KM, François NA, Pascal TG, Borys D, Schuerman L, Ijzerman EP, Bruin JP, van der Ende A, et al.. Effects of the 10-valent pneumococcal nontypeable Haemophilus influenzae protein D-conjugate vaccine on nasopharyngeal bacterial colonization in young children: a randomized controlled trial. Clin Infect Dis 2013; 56(3):e30-9; PMID:23118268; http://dx.doi.org/ 10.1093/cid/cis922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosikowska U, Korona-Głowniak I, Niedzielski A, Malm A. Nasopharyngeal and adenoid colonization by haemophilus influenzae and haemophilus parainfluenzae in children undergoing adenoidectomy and the ability of bacterial isolates to biofilm production. Med (Baltimore) 2015; 94(18):e799; PMID:25950686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jourdain S, Smeesters PR, Denis O, Dramaix M, Sputael V, Malaviolle X, Van Melderen L, Vergison A. Differences in nasopharyngeal bacterial carriage in preschool children from different socio-economic origins. Clin Microbiol Infect 2011; 17(6):907-14; PMID:20977542; http://dx.doi.org/ 10.1111/j.1469-0691.2010.03410.x [DOI] [PubMed] [Google Scholar]
- 20.Mackenzie GA, Leach AJ, Carapetis JR, Fisher J, Morris PS. Epidemiology of nasopharyngeal carriage of respiratory bacterial pathogens in children and adults: cross-sectional surveys in a population with high rates of pneumococcal disease. BMC Infect Dis 2010; 10:304; PMID:20969800; http://dx.doi.org/ 10.1186/1471-2334-10-304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oikawa J, Ishiwada N, Takahashi Y, Hishiki H, Nagasawa K, Takahashi S, Watanabe M, Chang B, Kohno Y. Changes in nasopharyngeal carriage of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis among healthy children attending a day-care centre before and after official financial support for the 7-valent pneumococcal conjugate vaccine and H. influenzae type b vaccine in Japan. J Infect Chemother 2014; 20(2):146-9; PMID:24582389; http://dx.doi.org/ 10.1016/j.jiac.2013.10.007 [DOI] [PubMed] [Google Scholar]
- 22.Isaacman DJ, McIntosh ED, Reinert RR. Burden of invasive pneumococcal disease and serotype distribution among Streptococcus pneumoniae isolates in young children in Europe: impact of the 7-valent pneumococcal conjugate vaccine and considerations for future conjugate vaccines. Int J Infect Dis 2010; 14(3):e197-209; PMID:19700359; http://dx.doi.org/ 10.1016/j.ijid.2009.05.010 [DOI] [PubMed] [Google Scholar]
- 23.Trotter CL, Andrews NJ, Kaczmarski EB, Miller E, Ramsay ME. Effectiveness of meningococcal serogroup C conjugate vaccine 4 years after introduction. Lancet 2004; 364(9431):365-7; PMID:15276396; http://dx.doi.org/ 10.1016/S0140-6736(04)16725-1 [DOI] [PubMed] [Google Scholar]
- 24.Loo JD, Conklin L, Fleming-Dutra KE, Knoll MD, Park DE, Kirk J, Goldblatt D, O'Brien KL, Whitney CG. Systematic review of the indirect effect of pneumococcal conjugate vaccine dosing schedules on pneumococcal disease and colonization. Pediatr Infect Dis J 2014; 33 Suppl 2:S161-71; PMID:24336058; http://dx.doi.org/ 10.1097/INF.0000000000000084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein D. Quantification using real-time PCR technology: applications and limitations. Trends Mol Med 2002; 8(6):257-60; PMID:12067606; http://dx.doi.org/ 10.1016/S1471-4914(02)02355-9 [DOI] [PubMed] [Google Scholar]
- 26.Al-Talib H, Yean Yean C, Al-Khateeb A, Ravichandran M. Comparative evaluation of three different methods of genomic DNA extraction for staphylococcus aureus. World Appl Sci J 2013; 21(3):424-7 [Google Scholar]
- 27.van Belkum A, Bax R, Peerbooms P, Goessens WH, van Leeuwen N, Quint WG. Comparison of phage typing and DNA fingerprinting by polymerase chain reaction for discrimination of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol 1993; 31(4):798-803; PMID:8463389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rantakokko-Jalava K, Jalava J. Optimal DNA isolation method for detection of bacteria in clinical specimens by broad-range PCR. J Clin Microbiol 2002; 40(11):4211-7; PMID:12409400; http://dx.doi.org/ 10.1128/JCM.40.11.4211-4217.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors - occurrence, properties and removal. J Appl Microbiol 2012; 113(5):1014-26; PMID:22747964; http://dx.doi.org/ 10.1111/j.1365-2672.2012.05384.x [DOI] [PubMed] [Google Scholar]
- 30.Edwards MC, Gibbs RA. Multiplex PCR: advantages, development, and applications. PCR Methods Appl 1994; 3(4):S65-75; PMID:8173510; http://dx.doi.org/ 10.1101/gr.3.4.S65 [DOI] [PubMed] [Google Scholar]
- 31.Carvalho Mda G, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, Steigerwalt A, Whaley M, Facklam RR, Fields B, et al.. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 2007; 45(8):2460-6; PMID:17537936; http://dx.doi.org/ 10.1128/JCM.02498-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung GC, Nagamine K, Li B, Lo SC. Identification of DNA signatures suitable for use in development of real-time PCR assays by whole-genome sequence approaches: use of Streptococcus pyogenes in a pilot study. J Clin Microbiol 2012; 50(8):2770-3; PMID:22593599; http://dx.doi.org/ 10.1128/JCM.01155-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chien YW, Vidal JE, Grijalva CG, Bozio C, Edwards KM, Williams JV, Griffin MR, Verastegui H, Hartinger SM, Gil AI, et al.. Density interactions among Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children. Pediatr Infect Dis J 2013; 32(1):72-7; PMID:22935873; http://dx.doi.org/ 10.1097/INF.0b013e318270d850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Binks MJ, Temple B, Kirkham LA, Wiertsema SP, Dunne EM, Richmond PC, Marsh RL, Leach AJ, Smith-Vaughan HC. Molecular surveillance of true nontypeable Haemophilus influenzae: an evaluation of PCR screening assays. PLoS One 2012; 7(3):e34083; PMID:22470516; http://dx.doi.org/ 10.1371/journal.pone.0034083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hays JP, van Selm S, Hoogenboezem T, Estevao S, Eadie K, van Veelen P, Tommassen J, van Belkum A, Hermans PW. Identification and characterization of a novel outer membrane protein (OMP J) of Moraxella catarrhalis that exists in two major forms. J Bacteriol 2005; 187(23):7977-84; PMID:16291671; http://dx.doi.org/ 10.1128/JB.187.23.7977-7984.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stol K, Diavatopoulos DA, Graamans K, Engel JA, Melchers WJ, Savelkoul HF, Hays JP, Warris A, Hermans PW. Inflammation in the middle ear of children with recurrent or chronic otitis media is associated with bacterial load. Pediatr Infect Dis J 2012; 31(11):1128-34; PMID:22668804; http://dx.doi.org/ 10.1097/INF.0b013e3182611d6b [DOI] [PubMed] [Google Scholar]


