Abstract
Background
Recent meta-analyses show that individuals with high risk variants in CHRNA5 on chromosome 15q25 are likely to develop lung cancer earlier than those with low-risk genotypes. The same high-risk genetic variants also predict nicotine dependence and delayed smoking cessation. It is unclear whether smoking cessation confers the same benefits in terms of lung cancer risk reduction for those who possess CHRNA5 risk variants versus those who do not.
Methods
Meta-analyses examined the association between smoking cessation and lung cancer risk in 15 studies of individuals with European ancestry who possessed varying rs16969968 genotypes (N = 12,690 ever smokers, including 6988 cases of lung cancer and 5702 controls) in the International Lung Cancer Consortium.
Results
Smoking cessation (former vs. current smokers) was associated with a lower likelihood of lung cancer (OR = 0.48, 95%CI = 0.30–0.75, p = 0.0015). Among lung cancer patients, smoking cessation was associated with a 7-year delay in median age of lung cancer diagnosis (HR = 0.68, 95%CI = 0.61–0.77, p = 4.9 ∗ 10–10). The CHRNA5 rs16969968 risk genotype (AA) was associated with increased risk and earlier diagnosis for lung cancer, but the beneficial effects of smoking cessation were very similar in those with and without the risk genotype.
Conclusion
We demonstrate that quitting smoking is highly beneficial in reducing lung cancer risks for smokers regardless of their CHRNA5 rs16969968 genetic risk status. Smokers with high-risk CHRNA5 genotypes, on average, can largely eliminate their elevated genetic risk for lung cancer by quitting smoking- cutting their risk of lung cancer in half and delaying its onset by 7 years for those who develop it. These results: 1) underscore the potential value of smoking cessation for all smokers, 2) suggest that CHRNA5 rs16969968 genotype affects lung cancer diagnosis through its effects on smoking, and 3) have potential value for framing preventive interventions for those who smoke.
Keywords: Smoking cessation, Genetics, Meta-analysis, Lung cancer
Highlights
-
•
CHRNA5 rs16969968 confers risk for earlier lung cancer diagnosis, but quitting produces benefit regardless of genotype.
-
•
Smokers can cut their risk of lung cancer in half and delay its onset by 7 years among those diagnosed.
-
•
Precision prevention allows clinicians to provide personalized health benefits of smoking cessation.
This is a report on whether smoking cessation confers the same benefits in terms of lung cancer risk reduction for those who possess CHRNA5 risk variants versus those who do not. We determined that quitting smoking is highly beneficial in reducing lung cancer risk levels for smokers regardless of their CHRNA5 rs16969968 genetic risk status. Although CHRNA5 rs16969968 increases risk for earlier lung cancer by 4 years, quitting produces essentially the same benefit for smokers with either high or low genetic risks. Smokers can cut their risk of lung cancer in half and delay its onset by 7 years among those diagnosed. These results are important for smokers to prevent cancer. On average, smokers at all genetic risk levels can largely eliminate their elevated risk for lung cancer by quitting smoking.
1. Introduction
Cigarette smoking is the leading modifiable risk for cancer and multiple major chronic illnesses. Growing evidence identifies that genetic variation in the α5 nicotinic cholinergic receptor subunit (CHRNA5) gene plays a key role in both heavy smoking and nicotine dependence (Bierut et al., 2007, Bierut et al., 2008a). Multiple large meta-analyses demonstrated the association with smoking quantity, defined by number of cigarettes smoked per day (CPD) (Liu et al., 2010, Saccone et al., 2010, TAG, 2010, Thorgeirsson et al., 2010), with the most robust signal for two highly correlated genetic variants, rs16969968 and rs1051730 in subjects of European ancestry (TAG, 2010). The coding variant, rs16969968, results in an amino acid change in the α5 nicotinic cholinergic receptor subunit, and subsequently alters nicotinic receptor conductance in vitro (Bierut et al., 2008b, Kuryatov et al., 2011). Genetic variation in CHRNA5 increases risk for smoking-related disorders such as lung cancer and chronic obstructive pulmonary disease (COPD) (Amos et al., 2008, Pillai et al., 2009, Thorgeirsson et al., 2008). Research suggests that the association between CHRNA5 and lung cancer may be mediated by COPD (Young et al., 2011, Brennan et al., 2011, Wang et al., 2010). Further, CHRNA5 has been reported to predict smoking cessation in both cessation trials (Baker et al., 2009, Bergen et al., 2013, Chen et al., 2012, Munafo et al., 2011, Sarginson et al., 2011) and general population studies (TAG, 2010, Chen et al., 2012, Chen et al., 2015a). The CHRNA5 variant rs16969968 was recently shown to be a marker of delayed smoking cessation in a meta-analysis (Chen et al., 2015a). Smokers with the high-risk genotype had delayed age of smoking cessation compared with those with the low-risk genotype (age 56 versus age 52). Similarly, those with the high-risk genotype had a 4-year earlier age of lung cancer diagnosis compared to those with the low-risk genotypes (age 61 vs. age 65).
Although it is clear that smoking cessation reduces cancer risk (Jha et al., 2013), and that CHRNA5 risk variants affect both smoking heaviness and duration, it is unclear whether CHRNA5 alters the health benefits of smoking cessation. For example, the CHRNA5 high-risk variants may directly affect lung cancer risk or, instead, influence risk via effects on smoking heaviness and cessation (Amos et al., 2008, Thorgeirsson et al., 2008, Ji et al., 2015, Tseng et al., 2014, Wang et al., 2012, Hung et al., 2008a). In the first case, smoking cessation would have negligible effects on lung cancer latency or risk among those with the high-risk variants. Smokers with such risk variants may have smoked so intensively prior to quitting, that cessation confers lesser benefit for them (in terms of lung cancer onset or likelihood). Conversely, smokers with low-risk genotypes may not benefit as much from cessation because they might have low genetic risk for lung cancer in addition to their lower risk for heavy smoking.
Large epidemiologic studies have shown the clear benefits of smoking cessation in reducing mortality and morbidity (Jha et al., 2013), but no studies have examined whether such benefits vary based on a smoker's genotype. Certainly, cessation has the potential to benefit almost any smoker, but discovering that some smokers may especially benefit from cessation could not only elucidate the nature of the mechanism(s) linking CHRNA5 to lung cancer, but might, also, inform personalized prevention efforts. For instance, such findings might encourage directing some smokers to additional preventive interventions and might be used to support “gain framed” messaging for such smokers, an approach to prevention with especially strong supporting evidence (Gallagher and Updegraff, 2012).
The CHRNA5 risk variants were chosen for this study because they are associated with lung cancer risk and onset, with exposure to a primary cancer etiologic factor (smoking duration and heaviness), and with the effects of a preventive action (smoking cessation). Therefore, they have the potential to elucidate the relations among genetic risk, etiologic factors, and preventive actions.
To address the gap in knowledge concerning CHRNA5, smoking cessation, and lung cancer risk, we meta-analyzed results from 15 European ancestry samples in the International Lung Cancer Consortium (ILCCO) and Transdisciplinary Research in Cancer of the Lung (TRICL). We explored two linked aims: 1) Do both CHRNA5 genetic risk and smoking cessation independently affect the risk of lung cancer? 2) Does the effect of smoking cessation on the risk of lung cancer vary with variation in CHRNA5? i.e., does the effect of smoking cessation on lung cancer risk and onset differ as a function of CHRNA5 rs16969968 genotype?
2. Methods
2.1. Samples
This is a collaborative meta-analysis based on the International Lung Cancer Consortium (ILCCO) and Transdisciplinary Research in Cancer of the Lung (TRICL) which were established with the aim of sharing comparable data from ongoing case-control and cohort studies of lung cancer. (Hung et al., 2008b, Timofeeva et al., 2012). To examine CHRNA5, smoking cessation, and lung cancer, we invited all ILCCO and TRICL studies of individuals of European Ancestry, of which, 15 (out of 27 invited studies) participated in the collaborative meta-analysis and pooled analysis with shared individual-level data. Results from 15 case control studies of lung cancer (N = 12,690 unrelated smokers of European ancestry) contributed to the meta-analyses. Informed consent was obtained from participants, and all studies received approval from the appropriate institutional review boards. To be included in analyses, each subject was required to be an ever-smoker (> 100 cigarettes in his or her lifetime). Tables S1, S2, and Text S1 provide additional details for each study.
With the aim of studying smoking cessation, CHRNA5, and lung cancer risk, we invited all ILCCO and TRICL studies of individuals of European Ancestry and 15 (out of 27 invited studies) participated in the collaborative meta-analysis and pooled analysis with shared individual-level data.
2.2. Outcomes
The primary outcomes of the analyses were: 1) case vs. control status for lung cancer in the combined sample of cases and controls, and 2) time (in years) from birth to lung cancer diagnosis among lung cancer cases.
2.3. Variants for Analyses
Because of its biological significance, we targeted the CHRNA5 variant rs16969968 for association testing. The variant rs16969968 was available in all datasets except the Germany Study for which we used a proxy variant (rs1051730, r2 =0.99 estimated based on 1000 Genomes for the EUR samples) for analyses because the imputed genotype was not available (Altshuler et al., 2010, Durbin et al., 2010). In addition, we conducted the meta-analyses with and without the Germany Study and reached similar results.
2.4. Statistical Analyses and Meta-analyses
In each dataset, we used logistic regression and Cox regression models to evaluate the association between rs16969968 and the two primary outcomes: i.e., lung cancer case vs. control, and age of lung cancer diagnosis among the cases, respectively. Age as a continuous variable and sex were included as covariates when appropriate. Additional covariates included smoking quantity and pack years. Smoking quantity when subjects smoked regularly was assessed with cigarettes smoked per day (CPD), defined as a 4-level ordered trait (CPD ≤ 10; 11 ≤ CPD ≤ 20; 21 ≤ CPD ≤ 30; CPD ≥ 31, coded as 0, 1, 2, 3, respectively). The variable “pack years” was defined by the product of smoking duration and CPD, and was modeled via quartiles.
Genotypes were coded additively as the number of minor alleles (A), where the reference allele was defined as the major allele (G) in the European ancestry population (Sherry et al., 2001). Consistency of allelic coding was confirmed by comparing allele labels and frequencies across datasets (Table S2). Analyses were performed at Washington University, as individual-level data were available for both meta-analyses and pooled analyses. Individual SNP analyses were performed using SAS (SAS Institute, Cary, NC).
The R package, rmeta, was used to generate meta-analysis plots (Lumley, 2012). We reported results from random effects models for all meta-analyses. Heterogeneity across studies was assessed with the Cochran Q test and resulting P values were reported for all analyses. There was evidence of heterogeneity across datasets for the analyses of the effect of smoking cessation on lung cancer, but not for the effect of genotypes on lung cancer. This heterogeneity may be due to the varying study designs and ascertainment strategies. Heterogeneity results for all meta-analysis are listed in Table S3.
3. Results
We examined individuals who ever smoked at least 100 cigarettes (5833 current and 6857 former smokers) from 15 case-control studies of lung cancer. CHRNA5 rs16969968 is known to predict risk for lung cancer and delayed smoking cessation based on previous reports of overlapping samples including these studies (Hung et al., 2008a, Chen et al., 2015a). We hypothesized that smoking cessation is associated with a decreased likelihood of lung cancer and delayed onset of lung cancer in this sample. Further, we investigated whether the association of smoking cessation and lung cancer risk varied with rs16969968 genotype risk levels.
3.1. As Previously Reported, CHRNA5 rs16969968 Predicts Lung Cancer and Earlier Diagnosis of Lung Cancer
First, we confirmed a consistent association as previously reported (Hung et al., 2008a, Chen et al., 2015a) between CHRNA5 rs16969968 and 1) increased likelihood of lung cancer when comparing cases and controls of lung cancer (meta-analysis OR = 1.29, 95%CI = 1.21–1.38, p = 3.5 ∗ 10–13, PHeterogeneity = 0.196, see Fig. S1;this genetic association does not vary significantly with smoking quantity), and 2) earlier diagnosis of lung cancer among cases of lung cancer (meta-analysis HR = 1.07, 95%CI = 1.03–1.11, p = 1.1 ∗ 10–4, PHeterogeneity = 0.561, see Fig. S2).
3.2. Quitting Smoking Decreases Risk Of Lung Cancer - Effect does not Vary With rs16969968 Genotypes
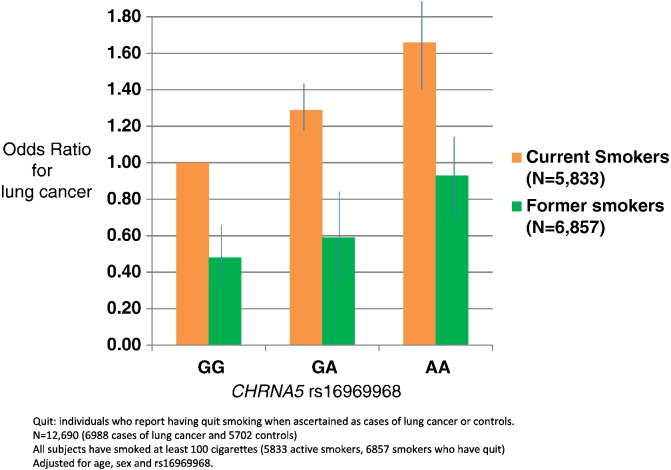
In these case control studies, current smoking status was ascertained simultaneously with case or control status for lung cancer. Ever-smokers who reported having quit smoking > 1 year prior to the cancer diagnosis were defined as former smokers; ever smokers who reported active smoking were defined as current smokers. The effect of smoking cessation was defined as the comparison of former vs. current smokers. Among ever smokers, smoking cessation was associated with a lower likelihood of lung cancer (OR = 0.48, 95%CI = 0.30–0.75, p = 0.0015, PHeterogeneity < 10–16). Both smoking cessation and rs16969968 were significantly associated with lung cancer risk (Fig. 1, Fig. S1).
Fig. 1.
CHRNA5 rs16969968 predicts risk of lung cancer. Smoking cessation decreases probability of lung cancer regardless of CHRNA5 rs16969968 genotype.
Quit: individuals who report having quit smoking when ascertained as cases of lung cancer or controls.
N = 12,690 (6988 cases of lung cancer and 5702 controls).
All participants have smoked at least 100 cigarettes (5833 active smokers, 6857 smokers who have quit).
Adjusted for age, sex and rs16969968 genotype.
The effect of smoking cessation on lung cancer risk remained after additionally adjusting for smoking quantity or pack years, and smoking cessation remained a predictor of lower risk of lung cancer diagnosis (OR = 0.46, 95%CI = 0.29–0.74, p = 0.0013 adjusted for smoking quantity) When adjusting for pack years, the effect size point estimate was similar (OR = 0.47, 95%CI = 0.14–1.52, p = 0.21) although it did not approach statistical significance as data on pack years were available for only a subset of the studies).
Further, we examined whether this association varied in individuals with different rs16969968 genotypes and found that smoking cessation was significantly associated with lower lung cancer risks for all genotypes (GG: OR = 0.48, 95%CI = 0.30–0.77, p = 0.0024, PHeterogeneity < 10–16; GA: OR = 0.46, 95%CI = 0.28–0.76, p = 0.0025, PHeterogeneity < 10–16; AA: OR = 0.56, 95%CI = 0.34–0.91, p = 0.019, PHeterogeneity = 3.36 ∗ 10–6) (Fig. S1). Genotype did not modify the effect of smoking cessation on decreased risk of lung cancer among ever smokers, and there was no interaction (OR = 1.01, 95%CI = 0.86–1.18, p = 0.93, PHeterogeneity = 0.139, Fig. S1). The finding remained robust in the sensitivity analyses when all possible sets of studies (up to N = 10) were excluded from the meta-analysis.
In addition, we obtained similar results in pooled analysis using available individual-level data adjusted for age, sex, rs16969968 genotype, and study, and found that smoking cessation was associated with decreased risk of lung cancer (N = 12,690, OR = 0.78, 95%CI = 0.75–0.91, P = 5.65 ∗ 10–31), and that genotype did not interact with smoking cessation in predicting lung cancer likelihood (i.e., the beneficial effect of smoking cessation did not vary with rs16969968 genotype (OR = 0.99, 95%CI = 0.94–1.05, P = 0.887)).
3.3. Quitting Smoking Delays Lung Cancer and This Effect Does not Vary With CHRNA5 rs16969968 Genotype
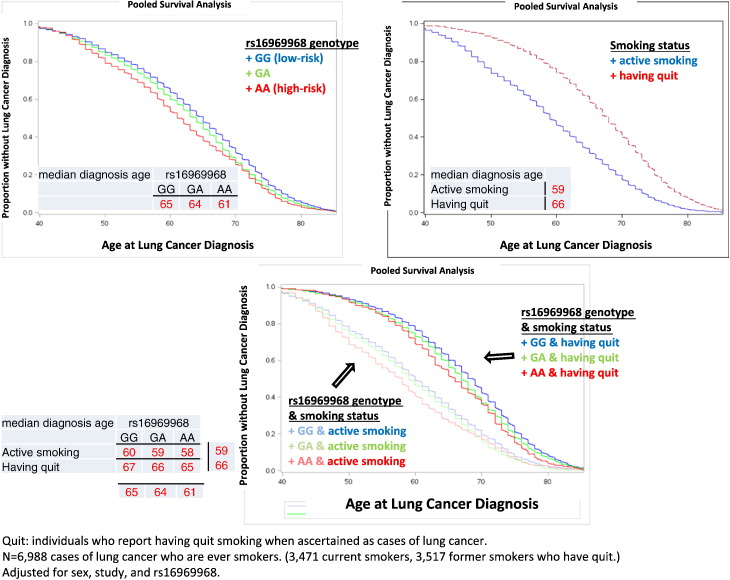
Among ever smokers who were diagnosed with lung cancer, smoking cessation was associated with delayed age of diagnosis, adjusted for sex and rs16969968 (HR = 0.68, 95%CI = 0.61–0.77, p = 4.9 ∗ 10–10, PHeterogeneity = 2.87 ∗ 10–8, see Fig. S2). Both smoking cessation and rs16969968 were independent predictors of age of lung cancer diagnosis. The median age at lung cancer diagnosis was 59 years for current smokers, which was a 7-year earlier onset compared to former smokers, who had a median age of diagnosis of 66 years (Fig. 2).
Fig. 2.
CHRNA5 rs16969968 predicts earlier lung cancer. Smoking cessation delays lung cancer, regardless of CHRNA5 rs16969968 genotype.
Quit: individuals who report having quit smoking when ascertained as cases of lung cancer.
N = 6,988 cases of lung cancer who are ever smokers. (3,471 current smokers, 3,517 former Smokers.
Adjusted for sex, study, and rs16969968 genotype.
The effect of smoking cessation on age of lung cancer diagnosis remained after additionally adjusting for cigarettes smoked per day, or pack years, and smoking cessation remained a significant predictor of a delayed age of lung cancer diagnosis (HR = 0.68, 95%CI = 0.60–0.76, p = 1.4 ∗ 10–10, adjusted for cigarettes smoked per day; HR = 0.62, 95%CI = 0.46–0.85, p = 0.0032, adjusted for pack years for studies with the information).
We examined whether this association varies by genotype and found that smoking cessation was associated with delayed age of lung cancer diagnosis for all genotypes (GG: HR = 0.70, 95%CI = 0.61–0.82, p = 5.2 ∗ 10–6, PHeterogeneity = 0.002; GA: HR = 0.63, 95%CI = 0.55–0.72, p = 7.8 ∗ 10–12, PHeterogeneity = 0.001; AA: HR = 0.66, 95%CI = 0.57–0.76, p = 1.3 ∗ 10–8, PHeterogeneity = 0.344) (Fig. S2). Even though CHRNA5 rs16969968 risk genotypes were associated with delayed quitting and earlier age of lung cancer diagnosis as previously reported (Chen et al., 2015a), genotypes did not modify the effect of smoking cessation on the delayed age of lung cancer diagnosis and there was no interaction (HR = 0.98, 95%CI = 0.91–1.05, p = 0.57, PHeterogeneity = 0.542, Fig. S2). The finding remained robust in the sensitivity analyses when all possible sets (up to N = 10) of studies were excluded from the meta-analysis.
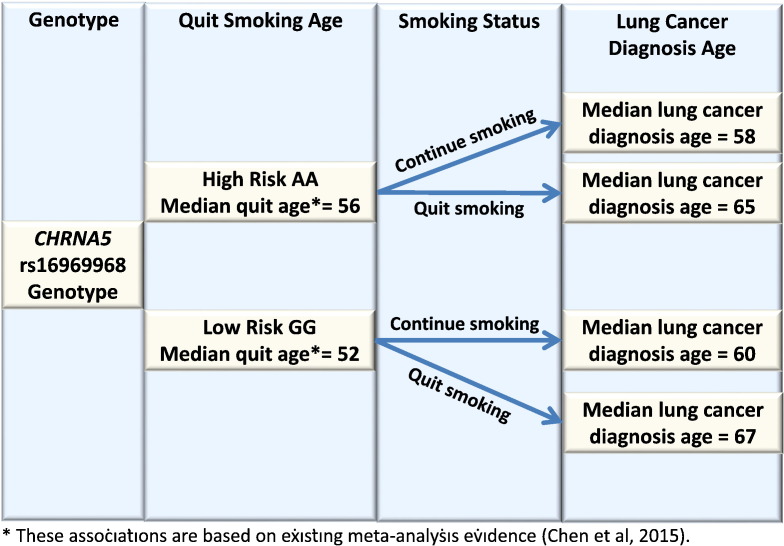
The median ages of lung cancer diagnosis for current and former smokers of different genotypes are in Fig. 3. Smokers with the high-risk AA genotypes, compared with those with the low-risk GG genotypes, have earlier age of lung cancer diagnosis, but this risk is greatly reduced by successful smoking cessation. Individuals with the high-risk genotype who have quit smoking have a mean age of lung cancer diagnosis of 65 vs. a mean age of 58 for those who continue to smoke. Individuals with the low-risk genotype who have quit smoking have a mean age of lung cancer diagnosis of 67 vs. a mean age of 60 for who continue smoking.
Fig. 3.
Summary diagram of CHRNA5 rs16969968 genotypes, smoking cessation, and lung cancer risks.
*These associations are based on existing evidence (Chen et al., 2015a, Chen et al., 2015b).
In addition, we reached similar conclusions in pooled analysis using available individual-level data adjusted for sex, rs16969968, and study; we found that smoking cessation was associated with delayed lung cancer diagnosis (N = 6988,HR = 0.62, 95%CI = 0.59–0.65, P = 1.26 ∗ 10–77), and this association did not vary with rs16969968 genotype (HR = 1.00, 95%CI = 0.94–1·07, P = 0.958).
4. Discussion
Genetic information is increasingly used by both patients and providers to determine health risks. Multiple meta-analyses have shown that the nicotinic receptor variant, CHRNA5 rs16969968, predicts heavy smoking (Liu et al., 2010, Saccone et al., 2010, TAG, 2010, Thorgeirsson et al., 2010), delayed quitting (TAG, 2010, Baker et al., 2009, Bergen et al., 2013, Chen et al., 2012, Munafo et al., 2011, Sarginson et al., 2011, Chen et al., 2015a), and earlier onset and higher probability of lung cancer (Amos et al., 2008, Thorgeirsson et al., 2008, Chen et al., 2015a, Hung et al., 2008a, Lips et al., 2010, Spitz et al., 2008). It is important to determine how malleable these genetic risks are to preventive or treatment efforts. For example, will individuals with high-risk genetic variants for lung cancer benefit from known effective preventive measures such as smoking cessation? In essence, do genetically high-risk and low-risk individuals experience different levels of benefit from cessation? Evidence related to this issue could both further elucidate the influences on cancer, and also serve as a basis for genetically informed preventive intervention. This is a large meta-analysis to examine the benefit of smoking cessation for lung cancer in individuals with different CHRNA5 rs16969968 genotypes. Obviously, these results cannot be extrapolated to the effects of other genotypes or other preventive actions (i.e., different from smoking cessation).
We found that smoking cessation is beneficial in reducing and delaying lung cancer and this beneficial effect does not vary with CHRNA5 rs16969968, which is a genetic marker for nicotine dependence (Liu et al., 2010, Saccone et al., 2010, TAG, 2010, Thorgeirsson et al., 2010), lung cancer (Amos et al., 2008, Thorgeirsson et al., 2008), COPD (Pillai et al., 2009), and delayed quitting (Chen et al., 2015a). Even though the CHRNA5 variant, rs16969968, predicts a 4-year earlier diagnosis of lung cancer among smokers with the high-risk genotypes vs. the low-risk genotypes (Chen et al., 2015a), quitting smoking is a highly effective preventive measure, cutting lung cancer risk approximately in half for individuals with all genotypes. Furthermore, among those who developed lung cancer, quitting smoking delayed diagnosis by 7 years (from 59 years of age for active smokers to 66 years of age, Fig. 2), with the delay not differing by genotype.
By simultaneously studying both genetic risk and preventive action (cessation), this study provides an informative perspective on comparative risk. For instance, it is clear that the effect of smoking cessation is larger than the effect of CHRNA5 rs16969968 genotypes on the risk of lung cancer. Smokers with the high-risk genotype, compared with those with the low-risk genotype, do have an increased risk for lung cancer, but this risk can be greatly reduced if they successfully quit smoking. In fact, smokers with the high-risk genotype who quit have lower risk than those with the low-risk genotypes who continue smoking (i.e., individuals with the high-risk genotype who have quit smoking have a mean age of lung cancer diagnosis of 65 vs. a mean age of 60 for smokers with a low-risk genotype who continue smoking).
The mechanisms through which CHRNA5 increases the risk for, and accelerates the onset age of, lung cancer likely involve multiple direct or indirect pathways (mediated through cigarette smoking (Liu et al., 2010, Saccone et al., 2010, TAG, 2010, Thorgeirsson et al., 2010), deeper inhalation of cigarettes leading to higher carcinogen exposure (Bloom et al., 2014, Le Marchand et al., 2008), and a delay in smoking cessation (Chen et al., 2015a). There has been inconsistent evidence on whether or not the genetic association of CHRNA5 and lung cancer is largely mediated through smoking) (Amos et al., 2008, Thorgeirsson et al., 2008, Ji et al., 2015, Tseng et al., 2014, Wang et al., 2012, Hung et al., 2008a).
Some study suggests that CHRNA5 confers risk of lung and cardiovascular diseases largely through an effect on behavior (Thorgeirsson et al., 2008), while other studies suggest that an increased risk with CHRNA5 and lung cancer in non-smokers would indicate that the disease mechanism with lung cancer is unlikely to be explained by an association with tobacco addiction. (Ji et al., 2015, Hung et al., 2008a) Most studies suggest that CHRNA5 had both a direct effect on overall lung cancer risk and an indirect effect through smoking. (Amos et al., 2008, Tseng et al., 2014, Wang et al., 2012) Our finding that smoking cessation decreases the elevated genetic risk for lung cancer supports the view that smoking at least partially mediates the effect of this genetic risk. This evidence suggests that the increased risk associated with high-risk genotypes for lung cancer is at least partially attributed to the smoker's inability to quit smoking relatively early (Chen et al., 2015a). For those who succeed in quitting, the elevated genetic risk for lung cancer conferred by rs16969968 (AA genotypes: 30% more likely to get lung cancer and 4 year earlier diagnosis) can be reduced drastically, resulting in halving the risk for lung cancer and delaying the age of diagnosis by 7 years.
These results should be interpreted in the context of multiple limitations. First, smoking cessation in the samples was self-reported and not assessed using biochemical confirmation. However, research shows that self-report is a valid indicator of current smoking, especially when there are no strong incentives to deceive (Subcommittee on Biochemical Verification SRNT, 2002). Second, we compared current smokers and former smokers based on the self-reported smoking status when cases and controls for lung cancer were recruited. Caution is needed for results based on the cross-sectional nature of the data. For a small portion of smokers (5.7%) who receive the lung cancer diagnosis and quit smoking in the same year, it is unknown whether they quit before the manifestation of disease or their diagnosis. We have repeated the same analyses by including, excluding, or reclassifying their smoking status and reached similar results. Third, this work analyzed only one genetic variant, and it is clear that multiple genes or variants contribute to lung cancer and smoking behaviors (Uhl et al., 2012). Fourth, we found no significant interaction between smoking cessation and rs16969968 genotype on lung cancer risk, despite the non-linear trend in point estimates of the effect of quitting across genotypes on lung cancer risk or age at diagnosis. With our sample size of 12,690 smokers and the allele frequency of rs16969968, we have sufficient power (0·8) to detect an interaction effect size of 1.12. Clearly a larger sample size would be needed to detect a smaller interaction. Fifth, this work analyzed the association between CHRNA5 and lung cancer, and it is possible that this association is mediated by other factors such as chronic obstructive pulmonary disease (COPD) (Young et al., 2008). Finally, this study included only subjects of European ancestry.
Lung cancer is the most common cancer in US and worldwide (Centers for Disease Control and Prevention and National Cente, American Cancer Society, 2014), and the survival rate is low (Howlader et al., 2013). Most cases (90%) of lung cancer are attributable to smoking (General, 2014). We demonstrate that quitting smoking is highly beneficial in reducing lung cancer risks for smokers regardless of their CHRNA5 rs16969968 genetic risk status. Evidence suggest that relative to individuals with CHRNA5 rs16969968 low-risk genotypes, those with high-risk genotypes are likely to smoke greater quantities, inhale more deeply, have more difficulty quitting, and have higher risk for lung cancer (Bierut et al., 2007, Saccone et al., 2010, TAG, 2010, Thorgeirsson et al., 2010, Thorgeirsson et al., 2008, Chen et al., 2012, Chen et al., 2015a, Hung et al., 2008a, Bloom et al., 2014). We now extend this evidence to show that quitting smoking produces essentially equivalent benefit regardless of genotype for this genetic risk factor. These results have potential value for preventive counseling with smokers; smokers with high-risk CHRNA5 genotypes can be informed that, on average, smokers with their genetic risk can largely eliminate their elevated genetic risk for lung cancer by quitting smoking. They can cut their risk of lung cancer in half and delay its onset by 7 years, if they develop it. These results underscore the potential value of smoking cessation for all smokers, they elucidate the causal path from CHRNA5 risk to lung cancer diagnosis, and they have potential value for framing preventive interventions for those who smoke.
Funding
Dr. Bierut is supported by National Institute on Drug Abuse grant R01DA036583 and National Cancer Institute grant P30CA091842.
Dr. LiShiun Chen was supported by R01DA038076, KL2RR024994 and K08DA030398.
Dr. Robert Culverhouse was supported by National Institute on Drug Abuse grant R21DA038241 and R21DA033827.
Dr. Amos is supported by NIH grants U19CA148127, and P30CA023108, The ILCCO data harmonization is supported by Cancer Care Ontario Research Chair of Population Studies to R. H. and Lunenfeld-Tanenbaum Research Institute, Sinai Health System.
International Lung Cancer Consortium (ILCCO): The data management of ILCCO is supported by Cancer Care Ontario Research Chair awarded to R. Hung, and NIH U19 CA148127.
Transdisciplinary Research in Cancer of the Lung (TRICL). The TRICL study was supported by a grant from the National Institute of Health (U19CA148127). The Toronto study was also supported by Canadian Cancer Society Research Institute (no.020214), Ontario Institute of Cancer and Cancer Care Ontario Chair Award to RH. For the German Lung Cancer Study, funding for MD Anderson Cancer Study was provided by NIH grants (P50 CA70907, R01CA121197, R01 CA127219, U19CA148127, R01CA55769) and CPRIT grant (RP100443). The Harvard Lung Cancer Study was funded by the National Institutes of Health (CA074386, CA092824, CA090578. The National Key Basic Research Program Grant was funded by (2011CB503805) and the National Natural Science Foundation of China (30730080, 30972541, 30901233 and 30872178). MD Anderson was funded by NCI/NIH K07CA160753.
German Lung Cancer Study (Germany).
The German Lung Cancer Study was made of three independent German studies: a) the LUCY study (“LUng Cancer in the Young ”, conducted by the Institute of Epidemiology, Helmholtz Zentrum München, PI Brüske ; and the Department of Genetic Epidemiology, Medical School, University of Göttingen, PI Bickeböller), b) the Heidelberg lung cancer case-control study (conducted by the German Cancer Research Center, PI Risch) and c) the KORA surveys (“Cooperative health research in the Region of Augsburg”, conducted by the Institute of Epidemiology II, Helmholtz Zentrum München).
LUCY was partly funded by the National Genome Research Network (NGFN), the DFG (BI 576/2-1; BI 576/2–2), the HGF and the Federal office for Radiation Protection (BfS: STSch4454). The Heidelberg sample collection was partly supported by the Deutsche Krebshilfe (70–2919). The KORA platform predominantly is financed by public funds allocated to the Helmholtz Zentrum München by the Federal Ministry of Education and Research and the State of Bavaria. Genotyping was performed in the Genome Analysis Center (GAC) of the Helmholtz Zentrum München.
Germany Saarland ESTHER Study. This study was supported in part by the Baden-Württemberg State Ministry of Science, Research and Arts; by the German Federal Ministry of Education and Research.
Greater Toronto Area Lung Cancer study: This study was supported by Canadian Cancer Society Research Institute (no. 020,214) to R. Hung.
Hawaii Case-Control Study: This project was supported by Grant ROI-CA-55874 and Contract NOI-CN-05,223 from the United States National Cancer Institute and by Grant EDT-78 from the American Cancer Society.
Karmanos Cancer Institute, Wayne State University: The Karmanos Cancer Institute contribution was supported by the National Institutes of Health (NIH) (R01CA60691, R01CA87895, N01PC35145, P30CA022453).
Mayo Study, Mayo Clinic, College of Medicine: The contribution was supported by NIH-R01–80127/84354 and Mayo Foundation Fund.
NELCS: The New England Lung Cancer Study was funded by Grant Number P20RR018787 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH).
Netherlands Radboudumc: The study was funded by an investment grant of Radboud university medical center.
Northern California Lung Cancer Study, University of California San Francisco (UCSF):
This work was supported in part by grants from the National Institutes of Health (grants ES06717, R01CA52689). The collection of cancer incidence data was supported by the California Department of Public Health as part of the statewide cancer reporting program; the National Cancer Institute's Surveillance, Epidemiology and End Results Program under contract N01-PC-35,136 awarded to the Northern California Cancer Center; and the Centers for Disease Control and Prevention's National Program of Cancer Registries, under agreement #U55/CCR921930-02 awarded to the Public Health Institute.
Norway: Funding by the Norwegian Cancer Society and the Norwegian Research Council.
The Resource for Lung Cancer in North Trent (ReSoLUCENT)/Sheffield: This study has been supported by Sheffield ECMC (Experimental Cancer Medicine Centre) and Weston Park Hospital Cancer Charity.
Tampa, FL: This study was supported by Public Health Service grants P01-CA68384 and R01-DE13158 from the National Institutes of Health.
UCLA: This study is supported by the Alper Research Program for Environmental Genomics of the UCLA Jonsson Comprehensive Cancer Center and the National Institute of Health (CA90833, DA11386, CA77954, CA09142, CA96134, and ES 011667).
Funders had no role in study design, data collection, data analysis, interpretation, and writing of the report.
Conflict of interest
LJ Bierut is listed as an inventor on Issued U.S. Patent 8,080,371,“Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction.
All other authors have no conflict of interest to disclose.
Contribution to Literature Search
Li-Shiun Chen, Amy Horton.
Contribution to Figures
Li-Shiun Chen, Amy Horton.
Contribution to Study Design
Li-Shiun Chen, Rayjean J. Hung, Robert Culverhouse, Sarah Hartz, Nancy Saccone, Christopher I. Amos, Laura J. Bierut.
Contribution to Data Collection:
Rayjean J. Hung, Heike Bickeböller, Hermann Brenner, Eric J. Duell, Angeline S. Andrew, Aage Haugen, Irene Brüske, Lambertus A. Kiemeney, Philip Lazarus, Loic Le Marchand, Geoffrey Liu, Jose Mayordomo, Angela Risch, Ann G. Schwartz, M. Dawn Teare, Xifeng Wu, John K. Wiencke, Ping Yang, Zuo-Feng Zhang, Margaret R. Spitz, Christopher I. Amos, Margaret Wrensch, Helen Hansen, John Wiencke.
Contribution to data analysis:
Amy Horton, Bo Deng, Younghun Han, Helen M. Hansen, Janet Horsman, Claire Kim, Albert Rosenberger, Katja K. Aben, Angeline S. Andrew, Shen-Chih Chang, Kai-Uwe Saum, Hendrik Dienemann, Mala Pande, Margaret R. Wrensch, John McLaughlin, Vidar Skaug, Erik H. van der Heijden, Jason Wampfler, Angela Wenzlaff, Penella Woll, Shanbeh Zienolddiny.
Contribution to data interpretation:
Li-Shiun Chen, Timothy Baker, Rayjean J. Hung, Robert Culverhouse, Sarah Hartz, Amy Horton, Nancy Saccone, Dorothy K. Hatsukami, Eric O. Johnson, Margaret R. Spitz, Christopher I. Amos, Laura J. Bierut, Iona Cheng.
Contribution to writing:
Li-Shiun Chen, Timothy Baker, Rayjean J. Hung, Amy Horton, Christopher I. Amos, Laura J. Bierut.
Contribution to manuscript scientific review and comments:
Li-Shiun Chen, Timothy Baker, Rayjean J. Hung, Amy Horton, Robert Culverhouse, Sarah Hartz, Nancy Saccone, Iona Cheng, Bo Deng, Younghun Han, Helen M. Hansen, Janet Horsman, Claire Kim, Albert Rosenberger, Katja K. Aben, Angeline S. Andrew, Shen-Chih Chang, Kai-Uwe Saum, Hendrik Dienemann, Dorothy K. Hatsukami, Eric O. Johnson, Mala Pande, Margaret R. Wrensch, John McLaughlin, Vidar Skaug, Erik H van der Heijden, Jason Wampfler, Angela Wenzlaff, Penella Woll, Shanbeh Zienolddiny, Heike Bickeböller, Hermann Brenner, Eric J. Duell, Aage Haugen, Irene Brüske, Lambertus A. Kiemeney, Philip Lazarus, Loic Le Marchand, Geoffrey Liu, Jose Mayordomo, Angela Risch, Ann G. Schwartz, M. Dawn Teare, Xifeng Wu, John K. Wiencke, Ping Yang, Zuo-Feng Zhang, Margaret R. Spitz, Christopher I. Amos, Laura J. Bierut.
All authors reviewed the manuscript for scientific content and approved the final version.
Acknowledgements
The authors thank Sherri Fisher and Nina Smock for editorial support of the manuscript.
International Lung Cancer Consortium (ILCCO): We thank Xuchen Zong for the data management of ILCCO Data Repository.
German Lung Cancer Study (Germany): We thank the subjects who participated in any contributing study, the LUCY-consortium (detail in Sauter et al. 2008), Wiebke Sauter, Martina Mittelstrass, Vera Zietemann and Prof. H.-E. Wichmann, the KORA study group, P. Drings and the staff at the Thoraxklinik Heidelberg.
Greater Toronto Area Study: We thank Dr. Geoffrey Liu's laboratory for the genotyping work related to this manuscript.
Mayo Clinic: We thank the study participants for their dedication and commitment to Epidemiology and Genetics of Lung Cancer (REGLC) and Mayo Foundation for their support.
Tampa, FL: We thank Kathy Eyring and Lindsay Mericle for sample and data collection at the H. Lee Moffitt Cancer Center.
UCLA: We thank the study participants for their dedication and commitment and the Alper Research Program for Environmental Genomics of the UCLA Jonsson Comprehensive Cancer Center for their support.
UCSF: We appreciate the subjects and their families for participating in this study.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.08.012.
Appendix A. Supplementary data
Table S1. Summary of the participating studies; Table S2. Distribution of demographics, Cigarettes Smoked Per Day (CPD), and rs16969968 in participating studies;Table S3. Table of meta-analysis results with heterogeneity test results.
Figure S1. CHRNA5 rs16969968 predicts risk of lung cancer. Smoking cessation decreases probability of lung cancer regardless of CHRNA5 rs16969968 genotype.; Figure S2. CHRNA5 rs16969968 predicts earlier lung cancer. Smoking cessation delays lung cancer, regardless of CHRNA5 rs16969968 genotype.; Figure S3. Meta-analysis: No interaction of smoking cessation and CHRNA5 rs16969968 on risk for lung cancer; Figure S4. Meta-analysis: No interaction of smoking cessation and CHRNA5 rs16969968 on diagnosis age of lung cancer.
: Descriptions of contributing datasets
References
- Altshuler D.M., Gibbs R.A., Peltonen L. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Cancer Society . 2014. Cancer Facts and Figures. [Google Scholar]
- Amos C.I., Wu X., Broderick P. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T.B., Weiss R.B., Bolt D. Human neuronal acetylcholine receptor A5-A3-B4 haplotypes are associated with multiple nicotine dependence phenotypes. Nicotine Tob. Res. 2009;11(7):785–796. doi: 10.1093/ntr/ntp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen A.W., Javitz H.S., Krasnow R. Nicotinic acetylcholine receptor variation and response to smoking cessation therapies. Pharmacogenet. Genomics. 2013;23(2):94–103. doi: 10.1097/FPC.0b013e32835cdabd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut L.J., Madden P.A., Breslau N. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut L.J., Stitzel J.A., Wang J.C. Nicotine dependence and the a5-a3-b4 nicotinic receptor gene cluster: variants in the nicotinic receptors Alter the risk for nicotine dependence. Am. J. Psychiatr. 2008;9(165):1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut L.J., Stitzel J.A., Wang J.C. Variants in nicotinic receptors and risk for nicotine dependence. Am. J. Psychiatry. 2008;165(9):1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom A.J., Hartz S.M., Baker T.B. Beyond cigarettes per day. A genome-wide association study of the biomarker carbon monoxide. Ann. Am. Thorac. Soc. 2014;11(7):1003–1010. doi: 10.1513/AnnalsATS.201401-010OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan P., Hainaut P., Boffetta P. Genetics of lung-cancer susceptibility. Lancet Oncol. 2011;12(4):399–408. doi: 10.1016/S1470-2045(10)70126-1. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention, National Center for Health Statistics . 2013. CDC Wonder Online Database, Compiled From Compressed Mortality File 1999–2010 Series 20 No. 2P. [Google Scholar]
- Chen L.S., Baker T.B., Piper M.E. Interplay of genetic risk factors (CHRNA5-CHRNA3-CHRNB4) and cessation treatments in smoking cessation success. Am. J. Psychiatry. 2012;169(7):735–742. doi: 10.1176/appi.ajp.2012.11101545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.S., Hung R.J., Baker T. CHRNA5 risk variant predicts delayed smoking cessation and earlier lung cancer diagnosis-a meta-analysis. J. Natl. Cancer Inst. 2015;107(5) doi: 10.1093/jnci/djv100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.S., Baker T.B., Jorenby D. Genetic variation (CHRNA5), medication (combination nicotine replacement therapy vs. varenicline), and smoking cessation. Drug Alcohol Depend. 2015;154:278–282. doi: 10.1016/j.drugalcdep.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R.M., Abecasis G.R., Altshuler D.L. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher K.M., Updegraff J.A. Health message framing effects on attitudes, intentions, and behavior: a meta-analytic review. Ann. Behav. Med. 2012;43(1):101–116. doi: 10.1007/s12160-011-9308-7. [DOI] [PubMed] [Google Scholar]
- General S. 2014. The Health Consequences of Smoking. [Google Scholar]
- Howlader N., Noone A.M., Krapcho M. Seer Cancer Statistics Review, 1975–2010. 2013. http://seer.cancer.gov/csr/1975_2010/ (accessed April 2013)
- Hung R.J., McKay J.D., Gaborieau V. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25.[see comment] Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Hung R.J., Christiani D.C., Risch A. International lung cancer consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol. Biomark. Prev. 2008;17(11):3081–3089. doi: 10.1158/1055-9965.EPI-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha P., Ramasundarahettige C., Landsman V. 21st-century hazards of smoking and benefits of cessation in the United States. N. Engl. J. Med. 2013;368(4):341–350. doi: 10.1056/NEJMsa1211128. [DOI] [PubMed] [Google Scholar]
- Ji X., Gui J., Han Y. The role of haplotype in 15q25.1 locus in lung cancer risk: results of scanning chromosome 15. Carcinogenesis. 2015;36(11):1275–1283. doi: 10.1093/carcin/bgv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryatov A., Berrettini W., Lindstrom J. Acetylcholine receptor (AChR) alpha5 subunit variant associated with risk for nicotine dependence and lung cancer reduces (alpha4beta2)alpha5 AChR function. Mol. Pharmacol. 2011;79(1):119–125. doi: 10.1124/mol.110.066357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Marchand L., Derby K.S., Murphy S.E. Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 2008;68(22):9137–9140. doi: 10.1158/0008-5472.CAN-08-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips E.H., Gaborieau V., McKay J.D. Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17 000 individuals. Int. J. Epidemiol. 2010;39(2):563–577. doi: 10.1093/ije/dyp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.Z., Tozzi F., Waterworth D.M. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet. 2010;42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumley T. R Package Version 2.15. 2012. rmeta: Meta-analysis. [Google Scholar]
- Munafo M.R., Johnstone E.C., Walther D., Uhl G.R., Murphy M.F., Aveyard P. CHRNA3 rs1051730 genotype and short-term smoking cessation. Nicotine Tob. Res. 2011;13(10):982–988. doi: 10.1093/ntr/ntr106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S.G., Ge D., Zhu G. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5(3) doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone N.L., Culverhouse R.C., Schwantes-An T.H. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6(8):1–16. doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarginson J.E., Killen J.D., Lazzeroni L.C. Markers in the 15q24 nicotinic receptor subunit gene cluster (CHRNA5-A3-B4) predict severity of nicotine addiction and response to smoking cessation therapy. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2011;156B(3):275–284. doi: 10.1002/ajmg.b.31155. [DOI] [PubMed] [Google Scholar]
- Sherry S.T., Ward M.H., Kholodov M. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz M.R., Amos C.I., Dong Q., Lin J., Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J. Natl. Cancer Inst. 2008;100(21):1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subcommittee on Biochemical Verification SRNT Biochemical verification of tobacco use and cessation. Nicotine Tob. Res. 2002;4(2):149–159. doi: 10.1080/14622200210123581. [DOI] [PubMed] [Google Scholar]
- TAG Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson T.E., Geller F., Sulem P. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(3):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson T.E., Gudbjartsson D.F., Surakka I. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeeva M.N., Hung R.J., Rafnar T. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum. Mol. Genet. 2012;21(22):4980–4995. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng T.S., Park J.Y., Zabaleta J. Role of nicotine dependence on the relationship between variants in the nicotinic receptor genes and risk of lung adenocarcinoma. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl G.R., Walther D., Musci R. Smoking quit success genotype score predicts quit success and distinct patterns of developmental involvement with common addictive substances. Mol. Psychiatry. 2012 doi: 10.1038/mp.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Spitz M.R., Amos C.I., Wilkinson A.V., Wu X., Shete S. Mediating effects of smoking and chronic obstructive pulmonary disease on the relation between the CHRNA5-A3 genetic locus and lung cancer risk. Cancer. 2010;116(14):3458–3462. doi: 10.1002/cncr.25085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Spitz M.R., Amos C.I. Method for evaluating multiple mediators: mediating effects of smoking and COPD on the association between the CHRNA5-A3 variant and lung cancer risk. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0047705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.P., Hopkins R.J., Hay B.A., Epton M.J., Black P.N., Gamble G.D. Lung cancer gene associated with COPD: triple whammy or possible confounding effect? Eur. Respir. J. 2008;32(5):1158–1164. doi: 10.1183/09031936.00093908. [DOI] [PubMed] [Google Scholar]
- Young R.P., Hopkins R.J., Whittington C.F., Hay B.A., Epton M.J., Gamble G.D. Individual and cumulative effects of GWAS susceptibility loci in lung cancer: associations after sub-phenotyping for COPD. PLoS One. 2011;6(2) doi: 10.1371/journal.pone.0016476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of the participating studies; Table S2. Distribution of demographics, Cigarettes Smoked Per Day (CPD), and rs16969968 in participating studies;Table S3. Table of meta-analysis results with heterogeneity test results.
Figure S1. CHRNA5 rs16969968 predicts risk of lung cancer. Smoking cessation decreases probability of lung cancer regardless of CHRNA5 rs16969968 genotype.; Figure S2. CHRNA5 rs16969968 predicts earlier lung cancer. Smoking cessation delays lung cancer, regardless of CHRNA5 rs16969968 genotype.; Figure S3. Meta-analysis: No interaction of smoking cessation and CHRNA5 rs16969968 on risk for lung cancer; Figure S4. Meta-analysis: No interaction of smoking cessation and CHRNA5 rs16969968 on diagnosis age of lung cancer.
: Descriptions of contributing datasets