Abstract
Aspirin prevents cardiovascular disease and colon cancer; however aspirin's inhibition of platelet COX-1 only partially explains its diverse effects. We previously identified an aspirin response signature (ARS) in blood consisting of 62 co-expressed transcripts that correlated with aspirin's effects on platelets and myocardial infarction (MI). Here we report that 60% of ARS transcripts are regulated by RUNX1 – a hematopoietic transcription factor - and 48% of ARS gene promoters contain a RUNX1 binding site. Megakaryocytic cells exposed to aspirin and its metabolite (salicylic acid, a weak COX-1 inhibitor) showed up regulation in the RUNX1 P1 isoform and MYL9, which is transcriptionally regulated by RUNX1. In human subjects, RUNX1 P1 expression in blood and RUNX1-regulated platelet proteins, including MYL9, were aspirin-responsive and associated with platelet function. In cardiovascular disease patients RUNX1 P1 expression was associated with death or MI. RUNX1 acts as a tumor suppressor gene in gastrointestinal malignancies. We show that RUNX1 P1 expression is associated with colon cancer free survival suggesting a role for RUNX1 in aspirin's protective effect in colon cancer. Our studies reveal an effect of aspirin on RUNX1 and gene expression that may additionally explain aspirin's effects in cardiovascular disease and cancer.
Keywords: Aspirin, Systems pharmacogenomics, Cardiovascular disease, Colorectal cancer, Gene expression profiling, RUNX1
Highlights
-
•
Aspirin regulates RUNX1 gene expression and the expression of RUNX1-regulated platelet proteins
-
•
RUNX1 gene expression in blood is associated with death and myocardial infarction in patients with cardiovascular disease
-
•
RUNX1 expression in colon cancers is associated with cancer free survival
Aspirin is among the most commonly used medications to prevent cardiovascular disease and colon cancer. Here we describe a property of aspirin that affects a gene named RUNX1 that is important in regulating platelets – the cells that form blood clots that cause heart attack and stroke – and in colon cancer. We go on to show that levels of RUNX1 are informative of which patients with cardiovascular disease will have future heart attacks and which patients with colon cancer will die from their cancers. This property of aspirin helps to explain how a drug like aspirin can produce such diverse effects.
1. Introduction
Aspirin is commonly used to prevent cardiovascular disease and colon cancer – leading causes of death and disease worldwide. There are large knowledge gaps in how this drug produces such diverse effects. Aspirin inhibits platelet function through platelet COX-1 suppression (Burch et al., 1978). However, this “on-target” mechanism is insufficient to explain aspirin's full effects on platelets since aspirin resistance (defined as aspirin's ability to inhibit platelet COX-1) is rare (Grosser et al., 2012) and uninhibited COX-1 does not explain variation in aspirin's effects on platelet function (Voora et al., 2012). Further, this mechanism is not implicated in cancer (Guillem-Llobat et al., 2014). In our prior work (Voora et al., 2013) we used aspirin as a probe in an experimental human system and assayed aspirin's pharmacodynamic effects using several assays of platelet function to comprehensively identify the effects of aspirin on platelet function. In 50 healthy volunteers administered 325 mg/day aspirin and simultaneous peripheral blood gene expression profiling before and after aspirin exposure, we identified an aspirin response signature (ARS) as a set of 62 tightly co-expressed genes in peripheral blood RNA that was correlative of the effects of aspirin on ADP, epinephrine, and collagen induced platelet aggregation (Voora et al., 2013). We subsequently validated the ARS in an independent cohort of 53 healthy volunteers and 132 patients with cardiovascular disease (Voora et al., 2013). That we did not observe a correlation between the ARS and platelet function in the absence of aspirin exposure suggested that part of the biologic effect of aspirin may be to modify platelet function via changes in gene expression. Here, we show that RUNX1, a key regulator of numerous genes governing both platelet and megakaryocyte biology, is an aspirin-responsive transcription factor and this represents an effect of aspirin on platelet/megakaryocyte biology and protection against cardiovascular events. RUNX1 is also a tumor suppressor gene in the colon (Fijneman et al., 2012) and we show that RUNX1 and aspirin-responsive genes regulated by RUNX1 are dysregulated in colon cancer and are associated with cancer-free survival.
2. Materials and Methods
2.1. Megakaryocytes
Human erythroleukemia (HEL) cells (ATCC, Manassas,VA) were grown in RPMI-1640 medium (Mediatech,Flemington, NJ) supplemented with 10% fetal bovine serum (HyClone, ThermoFisher Scientific, Pittsburgh, PA) and 1% of antibiotics (penicillin and streptomycin from ATCC, Manassas, VA). To induce megakaryocytic transformation, HEL cells were grown in the presence of phorbol 12-myristate 13-acetate (10 nM PMA) for 24 h (Jalagadugula et al., 2010). Megakaryocytic HEL cells (1 × 10^6) were treated with acetylsalicylic acid (ASA, Sigma, St. Louis, MO) or sodium salicylic acid (SS, Thermo Fisher Scientific, Pittsburgh, PA) (0.1, 0.5 and 1 mM) for 24, 48 and 72 h. In control experiments cells were treated with DMSO (Dimethylsulfoxide). In 48 and 72 h experiments, medium was replenished with fresh ASA or SS every 24 h. Cells were harvested and centrifuged (at 2000 rpm for 2 min on a table top centrifuge) and the pellets were resuspended in RNAprotect Cell Reagent (Qiagen, Venlo, Limburg) in 1:5 volume.
2.2. Description of Human Cohorts
Healthy volunteers (n = 53) were exposed to 325 mg/day aspirin for four weeks according to a previously published experimental protocol (Voora et al., 2012, Voora et al., 2013). Peripheral blood for RNA analysis and purified platelets for proteomic analyses were banked before and after aspirin exposure as previously described (Voora et al., 2013).
Two cohorts of patients with cardiovascular disease, baseline clinical, medication, and peripheral blood microarray data, and long-term clinical outcomes from the Duke Catheterization Genetics (CATHGEN) cohort have been previously described (Voora et al., 2013).
All human subject participants provided signed informed consent and protocols were approved by the Duke University Institutional Review Board according to standards indicated by the Declaration of Helsinki.
2.3. RNA Purification
RNA was isolated from HEL cells from cultured megakaryocytes using the QIAgen miRNAeasy Mini Kit (cat.no. 217004) following the manufacturer's QIAcube protocol and from peripheral blood as previously described (Voora et al., 2013).
2.4. qPCR
After determining sufficient RNA quality and quantity, cDNA was synthesized using SuperScript VILO Master Mix (Life Technologies; Grand Island, NY) in 20ul reactions using 500 ng of RNA. The reactions were run according to the manufacturer's recommended protocol. Aliquots of RNA samples were diluted to matching concentrations prior to cDNA synthesis. This reaction generated cDNA at a final estimated concentration of 25 ng/μl. PCR was performed in 10ul triplicate wells with 2.5 ng cDNA/well with the following kit: BioLine Lo-Rox SYBR #BIO-94020 and the following protocol: 95° 2 min, 95° 5 s, 55° 20 s, 95° 15 s, 55° 1 min. Gene expression for targets of interest were normalized to the geometric mean expression of normalizing genes (FPGS and GAPDH) as previously described (Voora et al., 2013). Primers used in this study are available upon request.
2.5. Statistical Analysis
2.5.1. Enrichment Analyses
ChIP – seq peaks for RUNX1 were obtained from the supplemental data of Tijssen et al. (2011). The coordinates of the peaks were converted from hg18 to hg19 using liftOver (https://genome.ucsc.edu/cgi-bin/hgLiftOver) and gene annotation was determined using Homer (http://homer.salk.edu/homer/ngs/index.html). Peaks that were annotated as intergenic were assigned to genes with transcription start sites (TSS) within 100 kb of the peak. HUGO gene symbols where then used to match ChIP-seq peaks with Affymetrix gene expression probe identifiers. A one-sided Fisher's exact test was used to test enrichment of RUNX1 binding sites (ChIP-seq peaks) and predicted combinations of transcription factors among genes in the Aspirin Response Signature compared to all other expressed genes. The enrichment p-value was verified though permutations testing random gene sets.
2.5.2. PCR Analysis
Normalized expression data as previously described (Voora et al., 2013). The fold change relative to vehicle control was calculated by dividing the expression for each time point by the average expression of the vehicle control from that experiment and that time point. Unpaired t-tests were used to compare fold changes in expression at each time point vs. vehicle control.
2.5.3. Platelet Proteomic Analyses
Platelet proteomic data was normalized as previously described (Voora et al., 2013) and filtered to focus on those protein products of genes that we identified as down-regulated in a platelets of a patient with RUNX1 haploinsufficiency (Sun et al., 2007).
Healthy Volunteer expression analysis – platelet function score and microarray gene expression data from health volunteers (available through the Database of Genotypes and Phenotypes [dbGaP] phs000551.v1.p1) before and after exposure and the aspirin response signature (ARS) was calculated as previously described (Voora et al., 2013). Correlations between individual probe sets, aspirin response signature (ARS), and platelet function score were performed using Spearman correlation.
2.5.4. CATHGEN RUNX1 Expression Analysis
Microarray gene expression data was normalized using RMA separately for the two cohorts making up the CATHGEN study: observational (N = 190) and case-control (N = 397) (Voora et al., 2013) available through dbGaP (phs000551.v1.p1). In each cohort, logistic regression was used to evaluate the effect of each RUNX1 probe expression on myocardial infarction (MI) or death events. Two covariate models were evaluated: 1) the reduced covariate model controlling for the effects of age on time to event, and 2) the full covariate model controlling for the effects of age, sex, race, smoking status, hyperlipidemia, hypertension, diabetes, platelet function, aspirin treatment, and the projected aspirin response gene expression signature. RUNX1 probes were tested for a probe-set effect on MI or death events using the score-based Global Test (Goeman et al., 2005). Two nested logistic regression models were contrasted, the alternative model including all RUNX1 probes and covariates listed above and the null model including only the covariates listed above. Meta-analysis was used to combine evidence of RUNX1 expression on survival in each cohort for both the probe-level and probe-set analyses. For the probe-level analyses, the log odds ratios and their standard errors were pooled as the weighted average of the log odds ratios using inverse variance weights, assuming a fixed effect model. For the probe-set analysis, Fisher's method was used to combine p-values from the cohort level Global Test results. All data processing and statistical analyses were conducted in R version 3.2 (http://www.r-project.org/) using packages affy, globaltest, and meta for normalization, probe-set tests, and meta-analyses respectively.
2.5.5. Colon Cancer Gene Expression Analysis
Microarray gene expression datasets (GEO accessions GSE14333 [N = 290] (Jorissen et al., 2009) and GSE17536 [N = 177] (Smith et al., 2010)) were downloaded and each experiment normalized using RMA. Probe-level analyses were conducted in each experiment using the Cox proportional hazards model, modeling disease free survival time as a function of probe expression while controlling for cancer stage. The ARS probe-set was tested for an effect on disease-free survival using the score-based approach Global Test (Goeman et al., 2005). Results from each experiment were combined using meta-analysis. The probe-level results were pooled assuming a fixed effects model using the weighted average of the log hazards ratios with inverse variance weights. For the probe-set analysis, Fisher's method was used to combine the Global Test p-values from each experiment. All data processing and statistical analyses were conducted in R version 3.2 (http://www.r-project.org/) using packages affy, survival, globaltest, and meta for normalization, survival analyses, probe-set tests, and meta-analyses respectively.
Differential expression analysis was conducted in GSE14333 and GSE17536 using a moderated t-test (Smyth, 2004) to contrast probe expression between cancer stages 1 and 2, stages 1 and 3, and stages 1 and 4. Meta-analysis was conducted using the weighted average of the log2 fold change with inverse variance weights. All statistical analyses were conducted in R version 3.2 (http://www.r-project.org/) using packages limma differential expression analysis and meta for meta-analyses.
3. Results
3.1. RUNX1 Regulates Aspirin Response Signature Genes
To identify potential transcription factor (s) that may be directly regulated by aspirin and as a consequence affect aspirin response signature (ARS) gene expression, we used chromatin immunopreciptation sequencing (ChIP-seq) data (Tijssen et al., 2011) generated by using RUNX1, GATA 1 or 2, FLI1, and SCL as probes in megakaryocytic cell lines to identify genes with putative binding sites for these transcription factors. Each transcription factor had binding sites that were significantly (p-value ≤ 0.001) enriched in ARS gene promoters demonstrating that each of these transcription factors – either alone or in combination - in megakaryocytes may transcriptionally regulate ARS genes. Loss of function mutations in RUNX1 in humans (an autosomal dominant disorder named Familial Platelet Disorder with associated Myeloid Malignancy (Song et al., 1999a)) lead to defects in platelet function including agonist-induced aggregation, secretion, protein phosphorylation, and activation of GPIIb and GPIIIa, thrombocytopenia, and down regulation in several platelet genes including ALOX12, MYL9, and PF4 (Sun et al., 2004, Song et al., 1999b, Songdej and Rao, 2015), thus providing a useful model to study human RUNX1 loss of function. In a human subject with a truncating mutation in the RUNX1 DNA binding RUNT domain leading to haplodeficiency, we identified over 200 platelet genes that are down-regulated (Sun et al., 2007). A majority (60%, 37/62) of ARS genes identified in our prior study (Voora et al., 2013) are down regulated in RUNX1 haplodeficiency and 48% (28/62) of ARS genes contain putative RUNX1 binding sites based on ChIP-seq data, with several (Jalagadugula et al., 2010, Kaur et al., 2010, Aneja et al., 2011) shown to be direct transcriptional targets of RUNX1. Therefore a large proportion of ARS genes appear to be regulated, in part, by RUNX1.
3.2. Aspirin Affects RUNX1 Expression in Megakaryocytic Cells
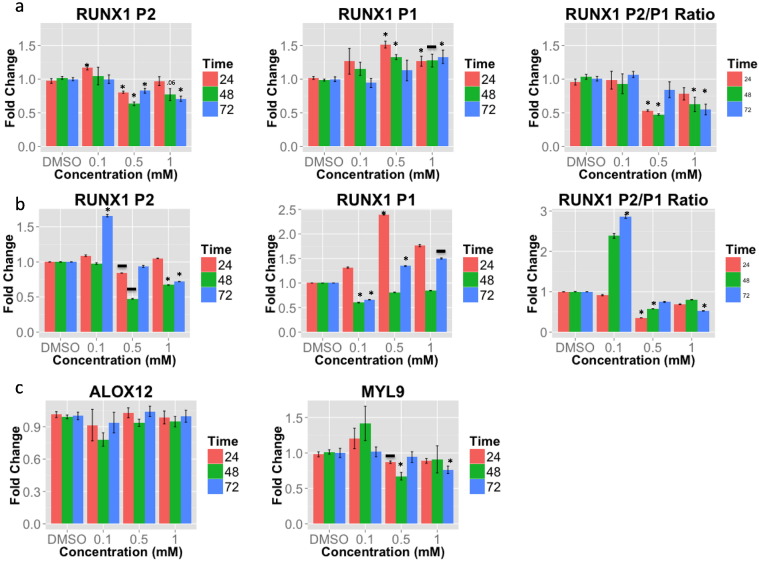
To test the hypothesis that aspirin directly regulates RUNX1 (and as a consequence, ARS gene expression), we exposed megakaryocytic cells to aspirin. RUNX1 expression is known to initiate at two, alternate (Levanon and Groner, 2004) promoter start sites (P1, distal, and P2, proximal, Supplementary Data) that produces isoforms that have nonredundant (Bee et al., 2010) roles in hematopoiesis and whose ratio is important for megakaryocyte vs. erythrocyte differentiation (Draper et al., 2016). We exposed megakaryocytic cell lines to aspirin for up to 72 h and found that aspirin concentrations ≥ 0.5 mM down-regulated RUNX1 P2 expression (Fig. 1A) and up regulated RUNX1 P1 expression (Fig. 1A) with a suppression of the P2/P1 ratio (Fig. 1A). When we tested the effects of aspirin's main metabolite, salicylic acid - a weak COX-1 inhibitor - we found similar changes in RUNX1 expression as with aspirin (Fig. 1B). Thus both aspirin and its primary metabolite have similar effects on RUNX1 expression. To test the hypothesis that aspirin-induced changes in RUNX1 expression alters RUNX1 function and regulation of downstream genes, we examined ALOX12 and MYL9 - two genes that we have shown are direct transcriptional targets of RUNX1 (Kaur et al., 2010, Jalagadugula et al., 2010) and are members of the ARS gene set (Voora et al., 2013). We found that aspirin down-regulated MYL9, which encodes myosin light chain, a component of the motor protein myosin. ALOX12 expression was not decreased (Fig. 1C), suggesting that not all RUNX1 targets genes are affected. Overall, a pharmacodynamic effect of aspirin and its metabolite salicylate are to down regulate P2 and upregulate P1 RUNX1 expression in megakaryocytic cells and as a consequence, RUNX1 function, thus altering the expression of a downstream gene known to influence platelet function.
Fig. 1.
Aspirin and salicylate effects on RUNX1 P2 and P1. Megakaryocytic HEL cells were exposed to escalating concentrations and durations of aspirin (panel a) or salicylic acid (panel b). Changes in RUNX1 P1, P2, and the P2/P1 ratio in response to aspirin or salicylic acid are displayed relative to DMSO. Effects of aspirin on two RUNX1 regulated genes, MYL9 and ALOX12 (panel c). Data are mean ± standard error of the mean. (*) = p-values < 0.05 and (−) = p-values for ≤ 0.08 for unpaired parametric two-tailed t-tests of fold change of aspirin or salicylate compared to DMSO for each time point. Each bar represents the average of 4–5 experiments.
3.3. RUNX1 Regulated Genes Are Aspirin Responsive
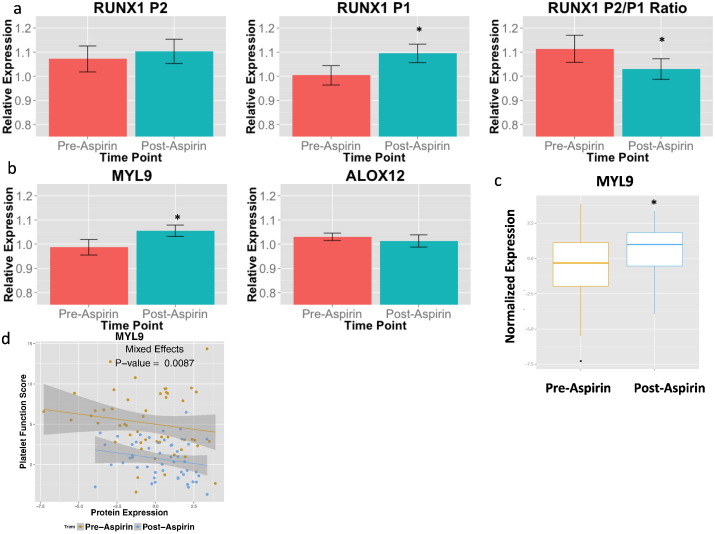
To test the effects of aspirin on the RUNX1 pathway in vivo, we administered 325 mg/day non-enteric coated aspirin for four weeks to healthy volunteers in an experimental setting where compliance was ensured (Voora et al., 2012). Because, very low RUNX1 expression in purified platelets (Ct values > 30 in over half of samples) precluded accurate quantification for comparative purposes of gene expression analysis by PCR, we assayed for RUNX1 expression in peripheral blood. To study RUNX1 regulated genes and proteins we analyzed purified platelets before and after aspirin exposure. Similar to the response observed in HEL cells, in peripheral blood from healthy volunteers, aspirin exposure resulted in an up-regulation in RUNX1 P1 and down-regulation in the P2/P1 ratio (Fig. 2A). Therefore, RUNX1 gene expression in peripheral blood may represent a pharmacodynamic biomarker for the gene expression modulating effects of aspirin. In purified platelets from the same subjects, aspirin exposure was associated with higher MYL9 gene expression in platelets (Fig. 2B), indicating that aspirin alters the expression of this gene. ALOX12 levels were not altered (Fig. 2B). We performed open label proteomics on platelet protein (Voora et al., 2013) and filtered this dataset of 235 proteins to those 22 genes that are differentially expressed in the patient with RUNX1 haplodeficiency (Sun et al., 2007). Nearly all proteins (21/22) were responsive to aspirin exposure, including MYL9 (Fig. 2C); the majority (19/21) were up-regulated.
Fig. 2.
Effects of aspirin exposure on RUNX1 function in humans. a, In peripheral blood RNA, RUNX1 P1, P2, and the P2/P1 ratio is displayed at baseline and after 4 weeks of 325 mg/day aspirin exposure in healthy volunteers (n = 45). Data are mean ± standard error of the mean (SEM, y-axis, scaled 0.75–1.25). Panels b and c, in purified platelet RNA (B) and platelet protein (C) from healthy volunteers exposed to 325 mg/day aspirin, MYL9 expression before and after aspirin exposure is displayed. Data are SEM (y-axis, scaled 0.75–1.25). * = p-values < 0.05 for unpaired parametric two-tailed t-tests compared to DMSO at for each time point. Panel d, Using a previously described (Voora et al., 2013) composite metric of nine platelet function measures termed the Platelet Function Score, MYL9 protein expression (x-axis) is plotted versus the platelet function score (y-axis) before and after aspirin exposure in healthy volunteers. P-value represents ANOVA p-value comparing mixed-effects models with vs. without MYL9 protein expression. Shaded area represents 95% confidence interval around regression line.
3.4. RUNX1 Regulated Platelet Proteins Are Associated With Platelet Function
To test the hypothesis that aspirin-responsive, RUNX1-regulated proteins underlie variation in platelet function, we correlated platelet protein expression with a composite metric (named the platelet function score (Voora et al., 2012)) of ADP, epinephrine, and collagen induced platelet aggregation before and after aspirin exposure. All 21 aspirin-responsive proteins, including MYL9, were correlative of platelet function (Fig. 2D and Table 2) regardless of aspirin exposure. Aspirin exposure did not change the platelet count (data not shown). The consistency of effects across RUNX1-regulated proteins demonstrates that aspirin affects the RUNX1 pathway in vivo and provide an additional mechanism for aspirin's effects on modulating platelet function.
Table 2.
Correlation with platelet function and change in response to aspirin exposure for RUNX1 regulated platelet proteins.
| Protein symbol | Regression coefficient for correlation with platelet function | Regression p-value | Mean difference (pre-post) | Paired t test p-value |
|---|---|---|---|---|
| FLNA | 0.11 | 0.01 | 3.91 | 0.01 |
| F13A | 0.20 | 0.01 | 1.83 | 0.02 |
| PDE5A | − 0.35 | 0.34 | − 0.06 | 0.78 |
| SH3L2 | − 1.31 | 0.01 | − 0.26 | 0.03 |
| ILEU | − 1.10 | 0.02 | − 0.28 | 0.03 |
| PGH1 | − 1.36 | 0.00 | − 0.30 | 0.03 |
| VDAC3 | − 1.11 | 0.02 | − 0.33 | 0.01 |
| JAM1 | − 0.84 | 0.03 | − 0.36 | 0.02 |
| 1433F | − 1.04 | 0.01 | − 0.39 | 0.02 |
| NP1L1 | − 0.70 | 0.02 | − 0.46 | 0.02 |
| CALD1 | − 0.65 | 0.01 | − 0.69 | 0.02 |
| PRDX6 | − 0.55 | 0.01 | − 0.70 | 0.02 |
| LEGL | − 0.58 | 0.01 | − 0.72 | 0.01 |
| TBA4A | − 0.48 | 0.01 | − 0.80 | 0.01 |
| SDPR | − 0.44 | 0.01 | − 0.84 | 0.01 |
| LTBP1 | − 0.48 | 0.01 | − 0.85 | 0.01 |
| MYL9 | − 0.45 | 0.01 | − 0.89 | 0.01 |
| TAGL2 | − 0.32 | 0.01 | − 0.99 | 0.06 |
| PLF4 | − 0.33 | 0.03 | − 1.05 | 0.01 |
| TPM4 | − 0.28 | 0.01 | − 1.40 | 0.01 |
| TBB1 | − 0.28 | 0.01 | − 1.58 | 0.01 |
| ACTN1 | − 0.20 | 0.01 | − 2.22 | 0.01 |
3.5. RUNX1 Expression Is Associated With Cardiovascular Events
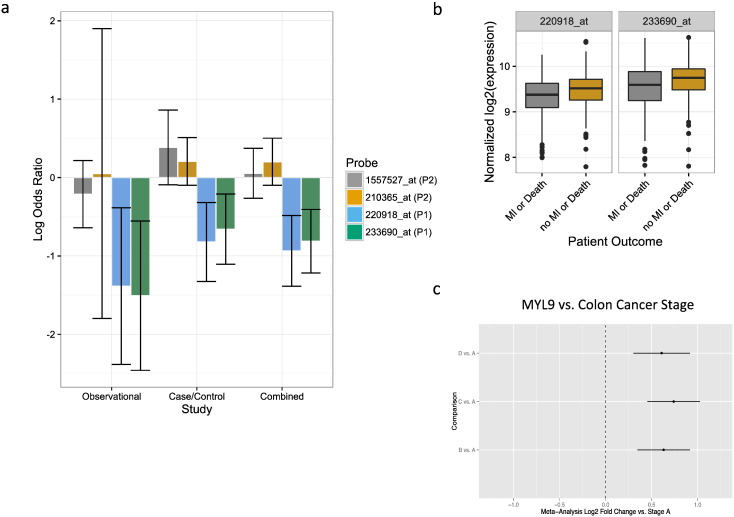
Aspirin prevents cardiovascular death and myocardial infarction (MI). Based on our findings above in megakaryocytes and humans, we hypothesized that this aspirin-responsive pathway is associated with death or MI. To test this hypothesis we utilized data from two independent cohorts (case-control and observational) of patients with cardiovascular disease who were treated with aspirin for which we have previously demonstrated an association between ARS gene expression in peripheral blood with death or MI·(Voora et al., 2013) To explore the relation between aspirin, platelet function, and cardiovascular events with RUNX1 gene expression we used RUNX1 probe sets that were available on the U133 microarray and that map to the RUNX1 locus on chromosome 21 (Supplementary Data). To ensure that these probe sets could represent RUNX1 expression and distinguish between RUNX1 P1 vs. P2 expression we applied two criteria: 1) inclusion of those probe sets that uniquely map to a single gene (i.e. exclusion of probe sets with ‘s_at’ or ‘x_at’ suffixes) and 2) correlation with PCR-based RUNX1 P1 or P2 expression in healthy volunteers, respectively. Based on these criteria four probe sets (Supplementary Data) met our criteria and were used to represent RUNX1 P1 (233690_at and 220918_at) and P2 (1557527_at and 210365_at) expression.
To test the hypothesis that RUNX1 probe set expression was correlated with either ARS gene expression or platelet function we used healthy volunteer data before and after aspirin exposure (Voora et al., 2013). The RUNX1 P2 probe set 210365_at was associated with ARS expression at baseline and after aspirin exposure; neither P2 probe set was associated with platelet function (Table 1). In contrast, P1 probe set 220918_at was associated with ARS gene expression and platelet function after aspirin exposure. Therefore, peripheral blood RUNX1 microarray data are consistent with the hypothesis that the P1 RUNX1 isoform regulates ARS gene expression and platelet function in an aspirin dependent manner. To test the hypothesis that RUNX1 expression was associated with cardiovascular events we again focused on these four probe sets (Fig. 3A). The RUNX1 P2 probe sets were not associated with death or MI, however, both P1 probe sets were consistently protective for death or MI (Fig. 3A). This finding extends the pharmacologic effect of aspirin on upregulating RUNX1 P1 expression to the antiplatelet effects of aspirin on platelet function and cardiovascular events.
Table 1.
Correlation of RUNX1 probe sets with Aspirin Response Signature expression and platelet function before and after aspirin exposure in healthy volunteers.
| Probe set ID | Correlation coefficients |
||||
|---|---|---|---|---|---|
| Pre-aspirin ARS | Post-aspirin ARS | Change in ARS (post-pre) | Pre-aspirin platelet function score | Post-aspirin platelet function scorea | |
| P2 RUNX1 probe sets | |||||
| 210365_at | 0.31⁎ | 0.39⁎⁎ | 0.30⁎⁎ | − 0.01 | − 0.07 |
| 1557527_at | − 0.12 | − 0.12 | − 0.01 | 0.16 | 0.15 |
| P1 RUNX1 probe sets | |||||
| 233690_at | − 0.11 | − 0.05 | 0.13 | − 0.15 | 0.10 |
| 220918_at | − 0.18 | − 0.45⁎⁎ | − 0.27⁎ | − 0.03 | 0.34⁎⁎ |
p ≤ 0.05.
p ≤ 0.01.
The platelet function score (11) (PFS) was used to quantify platelet function before and after aspirin exposure.
Fig. 3.
RUNX1 and MYL9 association with cardiovascular events or colon cancer stage Panel a, Using whole blood RNA microarray data collected from patients at the time of cardiac catheterization and long-term follow up for death and myocardial infarction (MI), we assembled case-control (n = 190) and observational patient cohorts (n = 397) where we have previously shown (Voora et al., 2013) an association between an Aspirin Response Signature (ARS) and death/MI events. The associations of P2 and P1 RUNX1 probe sets (see text for definitions) with death or MI in the observational and case-control individually and combined through meta-analysis are displayed as log Odds Ratios and 95% confidence intervals. Log odds ratios less than zero correspond to protective effects (i.e. higher expression associated with lower risk) and vice versa. All analyses are adjusted for age and sex. Panel b, Normalized expression for RUNX1 P1 probe sets are plotted vs. death or MI outcomes in the combined observational and case-control cohorts. Panel c, using two publically available datasets of colorectal cancer tumors (Jorissen et al. (2009), n = 290 and Smith et al. (2010), n = 177), a meta-analysis of differential MYL9 expression for stages B–D vs. A was conducted and the log2 fold change and 95% confidence intervals for each pairwise comparison is presented.
3.6. Aspirin Response Signature Genes Are Associated With Colorectal Cancer Stage and Survival
Beyond its use in cardiovascular disease, aspirin use is recommended to prevent GI malignancies (Bibbins-Domingo, 2016) and is being tested (NCT02467582, NCT02301286, and NCT00565708) as adjuvant therapy in patients treated for colon cancer. RUNX1 is a known tumor suppressor gene in acute myeloid leukemia (AML (Ichikawa et al., 2013)) and gastrointestinal (GI) malignancies (Fijneman et al., 2012, Dulak et al., 2012). RUNX1 (Sakakura et al., 2005) and several ARS genes (MYL9 (Yan et al., 2012), CTTN (Gardina et al., 2006), CALD1 (Gardina et al., 2006), SPARC (Chew et al., 2011)) are dysregulated in GI malignancies. To test the hypothesis that RUNX1 and its downstream ARS genes represent a mechanistic link for the chemoprotective effects of aspirin on GI cancers, we tested the association of the microarray expression of RUNX1 and ARS genes with colorectal cancer stage and cancer free survival in two independent cohorts (Smith et al., 2010, Jorissen et al., 2009) not previously studied for these genes. We found that RUNX1 and the majority of ARS probe sets are expressed in normal colon tissue (53/72) as well as in colon cancer samples (68/72). To test the hypothesis that aspirin responsive RUNX1 was associated with disease-free survival, we tested the P2 and P1 RUNX1 probe sets and found that higher expression of RUNX1 P1 probe sets were associated with survival (meta-analysis hazard ratio for P1 probe sets 233690_at and 220918_at, 95% confidence intervals, and meta-analysis p-values were 1.6 [1.1 – 2.3], 0.01 and 1.6 [0.9 – 2.7], 0.09, respectively). The remaining RUNX1 probe sets were not associated with survival or with cancer stage (data not shown). Further, the RUNX1 responsive, ARS gene set was differentially expressed in advanced colon cancer stage (B-D) compared to stage A cancers (genesettest p-values = 1.34e − 06 and 0.001). In particular, several ARS genes including MYL9, were up-regulated in stages B–D compared to stage A (Fig. 3B, Supplementary Data). Furthermore, independent of cancer stage, several ARS genes including SPARC, CTTN, CALD1, and MYL9 in colorectal cancer tumors were associated with cancer-free survival (meta-analysis odds ratios = 1.3–2.7, meta-analysis p-values < 0.01, Supplementary Data). To account for potential confounding by clinical characteristics and treatments, we included age, sex, adjuvant chemotherapy, and adjuvant radiation therapy as covariates in the Jorissen et al. analysis. Aspirin use was not available in either. To test for potential confounding effects, we compared effect sizes for each ARS and RUNX1 probeset in the survival analysis with and without inclusion of these covariates and found a strong correlation of effect sizes (r = 0.94, slope = 1.02, Supplementary Data). These data suggest that the aspirin responsive RUNX1 – ARS pathway that we identified in blood may be active in colon cancers and illuminates a potential additional mechanism for aspirin's chemoprotective role following colon cancer diagnosis.
4. Discussion
Aspirin is widely used to prevent cardiovascular disease and is recommended (Bibbins-Domingo, 2016) to prevent colon cancer. To study the effects of aspirin, we used a systems pharmacogenomics approach that combines systems biology, genetics, and pharmacology. Previously, we used serial gene expression profiling in humans exposed to aspirin (Voora et al., 2013). We identified an aspirin response signature (ARS) as a set of peripheral blood genes, that correlated with the effects of aspirin on platelet function, suggesting that part of the biologic effect of aspirin on platelet function may be to modify platelet gene expression (Voora et al., 2013). Because of the co-expressed nature of the ARS and their platelet origin we tested the hypothesis that aspirin produces its effects on gene expression through modulation of a megakaryocytic transcription factor (s). Our findings demonstrate that 1) ARS genes are RUNX1 responsive 2) aspirin and salicylate directly regulate RUNX1 in a promoter specific (P1 versus P2) fashion; 3) aspirin influences ARS gene expression in megakaryocytes and platelets; 4) RUNX1 P1 gene expression is associated with improved long-term clinical outcomes in patients with cardiovascular disease and colon cancer. Together, these findings provide evidence for an effect for aspirin on RUNX1 gene expression and function. This aspirin-responsive mechanism is implicated in cardiovascular disease and colon cancer outcomes.
It is well known that aspirin's mechanism of action is to inhibit platelet COX-1. However, aspirin likely has additional mechanisms for inhibiting platelet function because aspirin doses in excess of 81 mg/day do not result in additional COX-1 suppression but produce additional platelet inhibition as assessed by ADP and collagen (Gurbel et al., 2007). Although aspirin and salicylate, are well-known to inhibit NFKB (Kopp and Ghosh, 1994, Frantz et al., 1995), IkB phosphorylation (Pierce et al., 1996), and other cellular kinases (Stevenson et al., 1999, Schwenger et al., 1997), many of these effects are observed at concentrations not achieved (Roberts et al., 1984) with cardioprotective dosages (≤ 325 mg/day). In support of our hypothesis that aspirin alters gene expression, Massimi et al. identified that aspirin can upregulate expression of a single gene, MRP4, in megakaryocytes and platelets – an effect that was attributed to variation in platelet function in response to aspirin (Massimi et al., 2014). In this study we link the effects of aspirin and salicylate to RUNX1, a well-known transcription factor critical for normal hematopoiesis, megakaryocyte differentiation, and platelet function and a number of genes that it regulates (Elagib et al., 2003). That we observed similar effects with salicylate compared to aspirin on megakaryocyte gene expression suggests that aspirin's ability to inhibit COX-1 may not be required for this effect because salicylate has a ~ 20-fold lower potency for inhibiting COX-1 (Riendeau et al., 1997). Therefore in addition to the well-known effects of aspirin on platelet COX-1, our findings identify an effect of aspirin (or its metabolite salicylate) on RUNX1 and downstream gene expression.
RUNX1 is critical for normal hematopoiesis and megakaryocyte differentiation. Deletion of RUNX1 leads to embryonic lethality in mice due to hemorrhage (Wang et al., 1996), and humans with mutations leading to RUNX1 haploinsufficiency (Song et al., 1999a) are characterized by thrombocytopenia, defects in multiple aspects of platelet function, and a predisposition to leukemia (Sun et al., 2004, Song et al., 1999b, Ho et al., 1996). These prior studies in humans demonstrate that reductions in RUNX1 expression can lead to profound changes in platelet function. Our data demonstrate that aspirin can produce ~ 2 fold changes in RUNX1 in the megakaryocyte and in MYL9 - a gene that we have shown is directly regulated by RUNX1 (Jalagadugula et al., 2010). MYL9 plays a major role in platelet function and formation (Gilles et al., 2009) and in megakaryocyte biology (Jalagadugula et al., 2010). Therefore the observed effects of aspirin on RUNX1 and downstream gene expression provides an additional mechanism by which aspirin modulates platelet function and prevents cardiovascular disease.
Colorectal cancer is among the most common malignancy in the US and developed countries. Despite several lines of evidence implicating COX-2 (Algra and Rothwell, 2012, Rothwell et al., 2010, Rothwell et al., 2011, Clevers, 2006, Rothwell et al., 2012, Bertagnolli et al., 2006, Arber et al., 2006) as the mechanism by which aspirin prevents colon cancer, low-dose (< 100 mg/day) aspirin, which is associated with a reduced risk of colon cancer (Cook et al., 2013, Chan et al., 2008), is a weak COX-2 inhibitor (Mitchell et al., 1993). Therefore additional mechanisms linking aspirin to colon cancer prevention are likely. Our findings demonstrate that RUNX1 and several ARS genes, which are RUNX1-responsive, are correlated with disease free survival in two independent datasets of colon cancer patients. Therefore, ARS genes, which we have shown are aspirin- and RUNX1-responsive in the hematopoietic system, are also associated with colon cancer outcomes.
There are some limitations to our findings that deserve consideration. First, in megakaryocytic cells, we observed a down regulation in MYL9 compared to an upregulation in human platelets. The different directions of change in MYL9 expression may be attributed to 1) differences in duration of aspirin exposure (72 h [in vitro] vs. 4 weeks [in vivo]); the longer duration may permit compensatory mechanisms to develop; 2) the impact of cell specific transcriptional regulators (HEL-derived early stage megakaryocytic cells vs. native megakaryocytes), or 3) MYL9 measured in megakaryocytic cells vs. platelets. Second, while aspirin changed MYL9 expression there was no change in ALOX12. One explanation is that blocking COX-1 with aspirin may shunt arachidonic acid into the lipoxygenase pathways and thus cause a compensatory increase in ALOX12 that balances any effects of aspirin on RUNX1. Third, the dose- and time-dependent effects of aspirin or salicylate were not linear (Fig. 1). These non-linearities will likely be understood as part of future research around the mechanism of aspirin on RUNX1 expression. Fourth, the salicylate concentrations in this study may be higher than those achieved clinically. Because aspirin is quickly metabolized to salicylate, there is no systemic accumulation of aspirin. However, there is significant accumulation of salicylate (which has a half-life of 3–4 h) in the 30–100 μM range (Nagelschmitz et al., 2014, Charman et al., 1993) for 75–100 mg aspirin and 400–800 μM (Crome et al., 1991, Nagelschmitz et al., 2014) with 300 mg aspirin dosing based on area under the concentration-time curve analyses. These data, combined with the finding that we see similar effects of aspirin on RUNX1 with 325 mg/day dosing suggests that the concentrations used are physiologic. Last, we used an aspirin dose that is at the upper limit of the dosage range for cardiovascular prevention (75–325 mg/day). We chose this dose because 325 mg/day remains dosage commonly used in clinical practice (Hall et al., 2014) and one that is currently being tested in randomized clinical trials in cardiovascular disease (theaspirinstudy.org - NCT02697916) and colon cancer (www.capp3.org - NCT02497820). We recognize the importance of understanding the dose-response relationship between aspirin and RUNX1. Future studies will examine the extent to which changes in gene expression seen in this study extend to lower aspirin doses. Despite these limitations, we show that RUNX1 is an aspirin responsive transcription factor, leading to alteration in numerous downstream platelet genes and proteins that regulate platelet function and whose levels are correlated with known platelet physiology.
Our work on aspirin represents a proof-of-concept application of the systems pharmacogenomics approach to identify novel effects for aspirin that were not predicted based on aspirin's known mechanism of COX-1 inhibition. Future work focused on identifying the mechanism underlying this effect may lead to novel molecules that exploit this property for cardiovascular disease and cancer prevention. RUNX1 pathway biomarkers may be useful to individualize and optimize antiplatelet therapy or identify novel conditions that may be treated by aspirin. By using a systems pharmacogenomics approach we identify an off-target mechanism of action for one of the most commonly used medications worldwide and implicate this mechanism to cardiovascular disease and colorectal cancer.
Competing Interest Statement
DV, TLO, and GSG are listed as co-inventors on a patent application using RUNX1 as a biomarker for antiplatelet therapy.
Funding Sources
Grant funding for this work provided by RC1GM091083 (GSG), R01HL109568 (AKR), and R01HL118049 (DV).
Author Contributions
DV, EH, and GSG designed the human subjects' studies, analyzed the data, and wrote the manuscript. DV, GJ, and AKR designed the in vitro studies and analyzed the data. RM and DV designed and conducted statistical analyses related to Chip-Seq and microarray data. TLO supervised all platelet function testing. All authors discussed the results and commented on the manuscript.
Acknowledgements
We thank the staff of the Duke Clinical Research Unit and the Duke Hemostasis and Thrombosis Core Laboratory.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.08.021.
Contributor Information
Deepak Voora, Email: deepak.voora@duke.edu.
Geoffrey S. Ginsburg, Email: geoffrey.ginsburg@duke.edu.
Appendix A. Supplementary Data
Supplementary material.
References
- Algra A.M., Rothwell P.M. Effects of regular aspirin on long-term cancer incidence and metastasis: a systematic comparison of evidence from observational studies versus randomised trials. Lancet Oncol. 2012;13:518–527. doi: 10.1016/S1470-2045(12)70112-2. [DOI] [PubMed] [Google Scholar]
- Aneja K., Jalagadugula G., Mao G., Singh A., Rao A.K. Mechanism of platelet factor 4 (PF4) deficiency with RUNX1 haplodeficiency: RUNX1 is a transcriptional regulator of PF4. J. Thromb. Haemost. 2011;9:383–391. doi: 10.1111/j.1538-7836.2010.04154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber N., Eagle C.J., Spicak J., Racz I., Dite P., Hajer J., Zavoral M., Lechuga M.J., Gerletti P., Tang J., Rosenstein R.B., Macdonald K., Bhadra P., Fowler R., Wittes J., Zauber A.G., Solomon S.D., Levin B., Pre S.A.P.T.I. Celecoxib for the prevention of colorectal adenomatous polyps. N. Engl. J. Med. 2006;355:885–895. doi: 10.1056/NEJMoa061652. [DOI] [PubMed] [Google Scholar]
- Bee T., Swiers G., Muroi S., Pozner A., Nottingham W., Santos A.C., Li P.-S., Taniuchi I., De Bruijn M.F.T.R. Nonredundant roles for RUNX1 alternative promoters reflect their activity at discrete stages of developmental hematopoiesis. Blood. 2010;115:3042–3050. doi: 10.1182/blood-2009-08-238626. [DOI] [PubMed] [Google Scholar]
- Bertagnolli M.M., Eagle C.J., Zauber A.G., Redston M., Solomon S.D., Kim K., Tang J., Rosenstein R.B., Wittes J., Corle D., Hess T.M., Woloj G.M., Boisserie F., Anderson W.F., Viner J.L., Bagheri D., Burn J., Chung D.C., Dewar T., Foley T.R., Hoffman N., Macrae F., Pruitt R.E., Saltzman J.R., Salzberg B., Sylwestrowicz T., Gordon G.B., Hawk E.T., Investigators, A. P. C. S. Celecoxib for the prevention of sporadic colorectal adenomas. N. Engl. J. Med. 2006;355:873–884. doi: 10.1056/NEJMoa061355. [DOI] [PubMed] [Google Scholar]
- Bibbins-Domingo K. Aspirin use for the primary prevention of cardiovascular disease and colorectal cancer: U.S. preventive services task force recommendation statement aspirin use for the primary prevention of CVD and CRC. Ann. Intern. Med. 2016 doi: 10.7326/M16-0577. (N/A, N/A-N/A) [DOI] [PubMed] [Google Scholar]
- Burch J.W., Stanford N., Majerus P.W. Inhibition of platelet prostaglandin synthetase by oral aspirin. J. Clin. Invest. 1978;61:314–319. doi: 10.1172/JCI108941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.T., Giovannucci E.L., Meyerhardt J.A., Schernhammer E.S., Wu K., Fuchs C.S. Aspirin dose and duration of use and risk of colorectal cancer in men. Gastroenterology. 2008;134:21–28. doi: 10.1053/j.gastro.2007.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charman W.N., Charman S.A., Monkhouse D.C., Frisbee S.E., Lockhart E.A., Weisman S., Fitzgerald G.A. Biopharmaceutical characterisation of a low-dose (75 mg) controlled-release aspirin formulation. Br. J. Clin. Pharmacol. 1993;36:470–473. doi: 10.1111/j.1365-2125.1993.tb00399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew A., Salama P., Robbshaw A., Klopcic B., Zeps N., Platell C., Lawrance I.C. SPARC, FOXP3, CD8 and CD45 correlation with disease recurrence and long-term disease-free survival in colorectal cancer. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Colon cancer–understanding how NSAIDS work. N. Engl. J. Med. 2006;354:761–763. doi: 10.1056/NEJMcibr055457. [DOI] [PubMed] [Google Scholar]
- Cook N.R., Lee I.M., Zhang S.M., Moorthy M.V., Buring J.E. Alternate-day, low-dose aspirin and cancer risk: long-term observational follow-up of a randomized trial. Ann. Intern. Med. 2013;159:77–85. doi: 10.7326/0003-4819-159-2-201307160-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crome P., Wijayawardhana P., Guy L., Flanagan R.J., Streete P.J. Salicylate pharmacokinetics after single and multiple doses of Nu-Seals® aspirin in young and elderly male and female volunteers. Drug Investigation. 1991;3:252–257. [Google Scholar]
- Draper J.E., Sroczynska P., Tsoulaki O., Leong H.S., Fadlullah M.Z., Miller C., Kouskoff V., Lacaud G. Runx1b expression is highly heterogeneous and distinguishes megakaryocytic and erythroid lineage fate in adult mouse hematopoiesis. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1005814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak A.M., Schumacher S.E., Van Lieshout J., Imamura Y., Fox C., Shim B., Ramos A.H., Saksena G., Baca S.C., Baselga J., Tabernero J., Barretina J., Enzinger P.C., Corso G., Roviello F., Lin L., Bandla S., Luketich J.D., Pennathur A., Meyerson M., Ogino S., Shivdasani R.A., Beer D.G., Godfrey T.E., Beroukhim R., Bass A.J. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012;72:4383–4393. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagib K.E., Racke F.K., Mogass M., Khetawat R., Delehanty L.L., Goldfarb A.N. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- Fijneman R.J., Anderson R.A., Richards E., Liu J., Tijssen M., Meijer G.A., Anderson J., Rod A., O'sullivan M.G., Scott P.M., Cormier R.T. RUNX1 is a tumor suppressor gene in the mouse gastrointestinal tract. Cancer Sci. 2012;103:593–599. doi: 10.1111/j.1349-7006.2011.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz B., O'Neill E.A., Ghosh S., Kopp E. The effect of sodium salicylate and aspirin on Nf-Κb. Science. 1995;270:2017–2019. doi: 10.1126/science.270.5244.2017. [DOI] [PubMed] [Google Scholar]
- Gardina P.J., Clark T.A., Shimada B., Staples M.K., Yang Q., Veitch J., Schweitzer A., Awad T., Sugnet C., Dee S., Davies C., Williams A., Turpaz Y. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles L., Bluteau D., Boukour S., Chang Y., Zhang Y., Robert T., Dessen P., Debili N., Bernard O.A., Vainchenker W., Raslova H. Mal/Srf complex is involved in platelet formation and megakaryocyte migration by regulating Myl9 (Mlc2) and Mmp9. Blood. 2009;114:4221–4232. doi: 10.1182/blood-2009-03-209932. [DOI] [PubMed] [Google Scholar]
- Goeman J.J., Oosting J., Cleton-Jansen A.M., Anninga J.K., Van Houwelingen H.C. Testing association of a pathway with survival using gene expression data. Bioinformatics. 2005;21:1950–1957. doi: 10.1093/bioinformatics/bti267. [DOI] [PubMed] [Google Scholar]
- Grosser T., Fries S., Lawson J.A., Kapoor S.C., Grant G.R., Fitzgerald G.A. Drug resistance and pseudoresistance: an unintended consequence of enteric coating aspirin. Circulation. 2012 doi: 10.1161/CIRCULATIONAHA.112.117283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillem-Llobat P., Dovizio M., Alberti S., Bruno A., Patrignani P. Platelets, cyclooxygenases, and colon cancer. Semin. Oncol. 2014;41:385–396. doi: 10.1053/j.seminoncol.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Gurbel P.A., Bliden K.P., Dichiara J., Newcomer J., Weng W., Neerchal N.K., Gesheff T., Chaganti S.K., Etherington A., Tantry U.S. Evaluation of dose-related effects of aspirin on platelet function: results from the aspirin-induced platelet effect (aspect) study. Circulation. 2007;115:3156–3164. doi: 10.1161/CIRCULATIONAHA.106.675587. [DOI] [PubMed] [Google Scholar]
- Hall H.M., De Lemos J.A., Enriquez J.R., Mcguire D.K., Peng S.A., Alexander K.P., Roe M.T., Desai N., Wiviott S.D., Das S.R. Contemporary patterns of discharge aspirin dosing after acute myocardial infarction in the United States: results from the National Cardiovascular Data Registry (NCDR) Circulation: Cardiovascular Quality and Outcomes. 2014 doi: 10.1161/CIRCOUTCOMES.113.000822. [DOI] [PubMed] [Google Scholar]
- Ho C., Otterud B., Legare R., Varvil T., Saxena R., Dehart D., Kohler S., Aster J., Dowton S., Li F., Leppert M., Gilliland D. Linkage of a familial platelet disorder with a propensity to develop myeloid malignancies to human chromosome 21q22.1-22.2. Blood. 1996;87:5218–5224. [PubMed] [Google Scholar]
- Ichikawa M., Yoshimi A., Nakagawa M., Nishimoto N., Watanabe-Okochi N., Kurokawa M. A role for RUNX1 in hematopoiesis and myeloid leukemia. Int. J. Hematol. 2013;97:726–734. doi: 10.1007/s12185-013-1347-3. [DOI] [PubMed] [Google Scholar]
- Jalagadugula G., Mao G., Kaur G., Goldfinger L.E., Dhanasekaran D.N., Rao A.K. Regulation of platelet myosin light chain (MYL9) by RUNX1: implications for thrombocytopenia and platelet dysfunction in RUNX1 haplodeficiency. Blood. 2010;116:6037–6045. doi: 10.1182/blood-2010-06-289850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorissen R.N., Gibbs P., Christie M., Prakash S., Lipton L., Desai J., Kerr D., Aaltonen L.A., Arango D., Kruhoffer M., Orntoft T.F., Andersen C.L., Gruidl M., Kamath V.P., Eschrich S., Yeatman T.J., Sieber O.M. Metastasis-associated gene expression changes predict poor outcomes in patients with dukes stage B and C colorectal cancer. Clin. Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G., Jalagadugula G., Mao G., Rao A.K. RUNX1/core binding factor A2 regulates platelet 12-lipoxygenase gene (ALOX12): studies in human RUNX1 haplodeficiency. Blood. 2010;115:3128–3135. doi: 10.1182/blood-2009-04-214601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp E., Ghosh S. Inhibition of NF-kappa B by sodium salicylate and aspirin. Science. 1994;265:956–959. doi: 10.1126/science.8052854. [DOI] [PubMed] [Google Scholar]
- Levanon D., Groner Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23:4211–4219. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- Massimi I., Guerriero R., Lotti L.V., Lulli V., Borgognone A., Romani F., Barillà F., Gaudio C., Gabbianelli M., Frati L., Pulcinelli F.M. Aspirin influences megakaryocytic gene expression leading to upregulation of multidrug resistance protein-4 in human platelets. Br. J. Clin. Pharmacol. 2014 doi: 10.1111/bcp.12432. (N/A-N/A) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.A., Akarasereenont P., Thiemermann C., Flower R.J., Vane J.R. Selectivity of nonsteroidal antiinflammatory drugs as inhibitors of constitutive and inducible cyclooxygenase. Proc. Natl. Acad. Sci. 1993;90:11693–11697. doi: 10.1073/pnas.90.24.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagelschmitz J., Blunck M., Kraetzschmar J., Ludwig M., Wensing G., Hohlfeld T. Pharmacokinetics and pharmacodynamics of acetylsalicylic acid after intravenous and oral administration to healthy volunteers. Clinical Pharmacology: Advances and Applications. 2014;6:51–59. doi: 10.2147/CPAA.S47895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce J.W., Read M.A., Ding H., Luscinskas F.W., Collins T. Salicylates inhibit I kappa B-alpha phosphorylation, endothelial-leukocyte adhesion molecule expression, and neutrophil transmigration. J. Immunol. 1996;156:3961–3969. [PubMed] [Google Scholar]
- Riendeau D., Charleson S., Cromlish W., Mancini J.A., Wong E., Guay J. Comparison of the cyclooxygenase-1 inhibitory properties of nonsteroidal anti-inflammatory drugs (NSAIDS) and selective COX-2 inhibitors, using sensitive microsomal and platelet assays. Can. J. Physiol. Pharmacol. 1997;75:1088–1095. [PubMed] [Google Scholar]
- Roberts M.S., Mcleod L.J., Cossum P.A., Vial J.H. Inhibition of platelet function by a controlled release acetylsalicylic acid formulation–single and chronic dosing studies. Eur. J. Clin. Pharmacol. 1984;27:67–74. [PubMed] [Google Scholar]
- Rothwell P.M., Fowkes F.G., Belch J.F., Ogawa H., Warlow C.P., Meade T.W. Effect of daily aspirin on long-term risk of death due to cancer: analysis of individual patient data from randomised trials. Lancet. 2011;377:31–41. doi: 10.1016/S0140-6736(10)62110-1. [DOI] [PubMed] [Google Scholar]
- Rothwell P.M., Price J.F., Fowkes F.G., Zanchetti A., Roncaglioni M.C., Tognoni G., Lee R., Belch J.F., Wilson M., Mehta Z., Meade T.W. Short-term effects of daily aspirin on cancer incidence, mortality, and non-vascular death: analysis of the time course of risks and benefits in 51 randomised controlled trials. Lancet. 2012;379:1602–1612. doi: 10.1016/S0140-6736(11)61720-0. [DOI] [PubMed] [Google Scholar]
- Rothwell P.M., Wilson M., Elwin C.E., Norrving B., Algra A., Warlow C.P., Meade T.W. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet. 2010;376:1741–1750. doi: 10.1016/S0140-6736(10)61543-7. [DOI] [PubMed] [Google Scholar]
- Sakakura C., Hagiwara A., Miyagawa K., Nakashima S., Yoshikawa T., Kin S., Nakase Y., Ito K., Yamagishi H., Yazumi S., Chiba T., Ito Y. Frequent downregulation of the runt domain transcription factors RUNX1, RUNX3 and their cofactor CBFB in gastric cancer. Int. J. Cancer. 2005;113:221–228. doi: 10.1002/ijc.20551. [DOI] [PubMed] [Google Scholar]
- Schwenger P., Bellosta P., Vietor I., Basilico C., Skolnik E.Y., Vilček J. Sodium salicylate induces apoptosis via P38 mitogen-activated protein kinase but inhibits tumor necrosis factor-induced c-Jun N-terminal kinase/stress-activated protein kinase activation. Proc. Natl. Acad. Sci. 1997;94:2869–2873. doi: 10.1073/pnas.94.7.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.J., Deane N.G., Wu F., Merchant N.B., Zhang B., Jiang A., Lu P., Johnson J.C., Schmidt C., Bailey C.E., Eschrich S., Kis C., Levy S., Washington M.K., Heslin M.J., Coffey R.J., Yeatman T.J., Shyr Y., Beauchamp R.D. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Song W.J., Sullivan M.G., Legare R.D., Hutchings S., Tan X., Kufrin D., Ratajczak J., Resende I.C., Haworth C., Hock R., Loh M., Felix C., Roy D.C., Busque L., Kurnit D., Willman C., Gewirtz A.M., Speck N.A., Bushweller J.H., Li F.P., Gardiner K., Poncz M., Maris J.M., Gilliland D.G. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- Song W.J., Sullivan M.G., Legare R.D., Hutchings S., Tan X., Kufrin D., Ratajczak J., Resende I.C., Haworth C., Hock R., Loh M., Felix C., Roy D.C., Busque L., Kurnit D., Willman C., Gewirtz A.M., Speck N.A., Bushweller J.H., Li F.P., Gardiner K., Poncz M., Maris J.M., Gilliland D.G. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- Songdej N., Rao A.K. Hematopoietic transcription factor mutations and inherited platelet dysfunction. F1000prime Rep. 2015;7:66. doi: 10.12703/P7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson M.A., Zhao M.-J., Asea A., Coleman C.N., Calderwood S.K. Salicylic acid and aspirin inhibit the activity of RSK2 kinase and repress RSK2-dependent transcription of cyclic amp response element binding protein- and NF-κB-responsive genes. J. Immunol. 1999;163:5608–5616. [PubMed] [Google Scholar]
- Sun L., Gorospe J.R., Hoffman E.P., Rao A.K. Decreased platelet expression of myosin regulatory light chain polypeptide (MYL9) and other genes with platelet dysfunction and CBFA2/RUNX1 mutation: insights from platelet expression profiling. J. Thromb. Haemost. 2007;5:146–154. doi: 10.1111/j.1538-7836.2006.02271.x. [DOI] [PubMed] [Google Scholar]
- Sun L., Mao G., Rao A.K. Association of CBFA2 mutation with decreased platelet PKC-theta and impaired receptor-mediated activation of GPIIb-IIIa and pleckstrin phosphorylation: proteins regulated by CBFA2 play a role in GPIIb-IIIa activation. Blood. 2004;103:948–954. doi: 10.1182/blood-2003-07-2299. [DOI] [PubMed] [Google Scholar]
- Tijssen, Marloes R., Cvejic A., Joshi A., Hannah R.L., Ferreira R., Forrai A., Bellissimo D.C., Oram S.H., Smethurst P.A., Wilson N.K., Wang X., Ottersbach K., Stemple D.L., Green A.R., Ouwehand W.H., Göttgens B. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev. Cell. 2011;20:597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voora D., Cyr D., Lucas J., Chi J.-T., Dungan J., Mccaffrey T.A., Katz R., Newby L.K., Kraus W.E., Becker R.C., Ortel T.L., Ginsburg G.S. Aspirin exposure reveals novel genes associated with platelet function and cardiovascular events. J. Am. Coll. Cardiol. 2013;62:1267–1276. doi: 10.1016/j.jacc.2013.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voora D., Ortel T.L., Lucas J.E., Chi J.T., Becker R.C., Ginsburg G.S. Time-dependent changes in non-COX-1-dependent platelet function with daily aspirin therapy. J. Thromb. Thrombolysis. 2012;33:246–257. doi: 10.1007/s11239-012-0683-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Stacy T., Binder M., Marin-Padilla M., Sharpe A.H., Speck N.A. Disruption of the CBFA2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. U. S. A. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Li J., Xiong Y., Xu W., Zheng G. Identification of candidate colon cancer biomarkers by applying a random forest approach on microarray data. Oncol. Rep. 2012;28:1036–1042. doi: 10.3892/or.2012.1891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.