Abstract
The architecture and regulation of Saccharomyces cerevisiae metabolic network are among the best studied owing to its widespread use in both basic research and industry. Yet, several recent studies have revealed notable limitations in explaining genotype–metabolic phenotype relations in this yeast, especially when concerning multiple genetic/environmental perturbations. Apparently unexpected genotype–phenotype relations may originate in the evolutionarily shaped cellular operating principles being hidden in common laboratory conditions. Predecessors of laboratory S. cerevisiae strains, the wild and the domesticated yeasts, have been evolutionarily shaped by highly variable environments, very distinct from laboratory conditions, and most interestingly by social life within microbial communities. Here we present a brief review of the genotypic and phenotypic peculiarities of S. cerevisiae in the context of its social lifestyle beyond laboratory environments. Accounting for this ecological context and the origin of the laboratory strains in experimental design and data analysis would be essential in improving the understanding of genotype–environment–phenotype relationships.
Keywords: Saccharomyces cerevisiae, microbial community, genotype–phenotype relation, species interaction, ecological context
Could complex genotype–phenotype observations in Saccharomyces cerevisiae originate from its evolutionary history rich with social life?
INTRODUCTION
In laboratories, Saccharomyces cerevisiae is usually grown in isolation and under well-defined conditions. Laboratory experiments therefore only faintly represent the challenges in the natural ecological habitats of wild and domesticated (i.e. strains adapted to human use for food and beverage fermentation already thousands of years ago) S. cerevisiae. Furthermore, in a natural habitat, the yeast metabolism needs to adapt and respond to the presence of other species. The absence of interspecies social life and unrepresentative growth conditions in laboratory experiments may thus hide the evolutionarily shaped operating principles of S. cerevisiae metabolic and regulatory networks. Furthermore, minimal spatial variation in liquid laboratory cultures hardly supports the phenotypic heterogeneity arising due to chemical gradients and physical proximity (Campbell et al. 2015; Campbell, Vowinckel and Ralser 2016). The laboratory studies of S. cerevisiae, with the above-mentioned limitations, are also generally limited to few strains. All these factors may have unforeseen and fundamental effects on the interpretation of experimental data and thereby present a challenge for building a quantitative understanding of the genotype–phenotype relationship.
Saccharomyces cerevisiae metabolic responses are yet difficult to predict
Genome-scale metabolic models can be used to predict the phenotype dependence on the status of metabolic genes (Forster et al. 2003; Herrgard et al. 2008). These models are a gene-annotated stoichiometric representation of the full metabolic potential of a species augmented with thermodynamic and reaction capacity constraints. Despite the wealth of knowledge represented by these models, the prediction of gene essentiality is not yet flawless (Heavner and Price 2015), and the ability to predict the dependence of the metabolic flux distribution on the gene status (presence/absence) has been found to be poor (Pereira, Nielsen and Rocha 2016). In addition, the prediction accuracy of conditional gene essentialities in prototrophic deletion mutants on different carbon and nitrogen source has been found to be weak (VanderSluis et al. 2014). Furthermore, synthetic lethal phenotypes arising from genetic interactions can hardly be explained using metabolic models (Szappanos et al. 2011; Brochado et al. 2012). In auxotrophic laboratory strains grown on supplemented media, the interpretation of phenotypes is further complicated by yeast metabolizing the supplements, and the cessation of pathways because of the end product availability. This has shown to lead to complex molecular phenotypes through epistatic effects (Brem et al. 2002; Mulleder et al. 2012; Campbell et al. 2015).
Saccharomyces cerevisiae laboratory genotypes
A majority of laboratory experiments are performed with only a few strains of S. cerevisiae which may not represent the full genetic potential of the species (Steinmetz et al. 2002; Carreto et al. 2008; Ehrenreich et al. 2010; Warringer et al. 2011; Strope et al. 2015). S. cerevisiae strains from genotypically different population origins exhibit large trait divergence in terms of growth characteristics on various substrates, in the presence of toxins or effectors, and mineral and vitamin limitations (Warringer et al. 2011). A laboratory strain may even lack evolutionary streamlining of the genotype–phenotype relation if its genome has not undergone the evolutionary selection under the corresponding growth conditions. Indeed, Qian et al. (2012) observed that in a common laboratory culture media, a laboratory strain of S. cerevisiae expresses genes that are rather deleterious than beneficial, indicating antagonistic pleiotropy that has not been resolved by adaptation to the corresponding environment (Qian et al. 2012). Relaxation of natural selection pressures has also been found to enrich, possibly through genetic drift, population specific alleles (Warringer et al. 2011; Zorgo et al. 2012). Additionally, auxotrophies commonly present in laboratory strains have been shown to affect the expression of a large number of genes and metabolite levels even when the growth medium is adequately supplemented (Brem et al. 2002; Mulleder et al. 2012). The supplementation thus causes effects beyond the stoichiometric fulfillment of the nutrients corresponding to the auxotrophies.
Habitats of Saccharomyces cerevisiae
The ecology of the wild S. cerevisiae is relatively poorly understood (Boynton and Greig 2014), mainly because of early domestication (Sicard and Legras 2011) and widespread use of commodity strains. S. cerevisiae has been used for food and beverage fermentation for several thousand years due to its unique metabolic properties: fermentative metabolism, resistance to high sugar and ethanol concentrations, and production of specific aroma compounds. Humans have therefore significantly facilitated dispersal of the yeast (Goddard et al. 2010). For instance, the strains used for wine fermentation in Australia, Chile and New Zealand have shared recent ancestors with European wine strains (Legras et al. 2007; Liti et al. 2009; Goddard et al. 2010; Dunn et al. 2012). Through a large-scale population history study the genotypes of S. cerevisiae were found to fit to five primary lineages with shared ancestor populations (i.e. Malaysian, West African, North American, European and Sake) (Liti et al. 2009; Liti 2015). The genetic variations found in strains in a lineage were unique and equally distributed in the genome (Liti et al. 2009). However, a separate investigation of Chinese wild S. cerevisiae isolates revealed a larger and hitherto unknown reservoir of genetic variation (Wang et al. 2012). The natural history of S. cerevisiae including the known genetic variation is comprehensively reviewed by Liti (Liti 2015).
While S. cerevisiae is very abundant in human-made environments, such as wineries (Ciani et al. 2004), it appears to be rather rare in natural reservoirs (Goddard and Greig 2015). Thus, investigations of wild isolates are hindered by small population sizes (Liti 2015). In a search for the natural S. cerevisiae habitats, it has been isolated from plants (Wang et al. 2012), and the bark and leaves of oak trees and oak-associated soil (Sniegowski, Dombrowski and Fingerman 2002; Sampaio and Goncalves 2008; Zhang et al. 2010). Recently, Kowallik and Greig (2016) observed that yeast was more abundant in oak leaf litter than in the oak bark and that the leaf litter provides a refuge all year round (Kowallik and Greig 2016). Consistently, it has also been confirmed that S. cerevisiae can sporulate in soil and survive in this stress-resistant state until more nutritious conditions arise (Knight and Goddard 2016). S. cerevisiae indeed seems to respond to lignocellulosic solids from Birch tree by activating stress tolerance mechanisms—an observation that we suggest could be due to its evolutionary linkage to the bark niche (Koppram et al. 2016). Despite the common belief that yeasts are naturally found on the surface of grapes, a study finds that only one in thousand grapes are positive for S. cerevisiae (Mortimer and Polsinelli 1999). In cases of damaged fruit or berries, on the other hand, the occurrence and cell counts of S. cerevisiae were found to be higher (Mortimer and Polsinelli 1999). Interestingly, insects serve also as natural reservoirs and vectors that promote yeast dispersal: S. cerevisiae can be found associated with flies (Chandler, Eisen and Kopp 2012), social wasps (Stefanini et al. 2012) and bees (Goddard et al. 2010). Given the hitherto focus on prokaryotes in microbial diversity analysis due to sample preparation constraints, further natural reservoirs of the yeast might not have been discovered yet.
The known natural reservoirs of S. cerevisiae are usually nutrient poor with occasional periods of rich resource availability (e.g. after a transfer from oak bark to a faulty fruit by an insect) (Liti 2015). Therefore, unlike human-associated yeasts, wild strains most likely spend the most of their life in a dormant state. It has been argued that S. cerevisiae does not show adaptations to any particular habitat, but rather an ability to survive in a wide range of conditions (such as temperature, pH, nutrient concentrations and osmolarity) (Goddard and Greig 2015). The tolerance to a variety of environmental perturbations is consistent with the lifestyle of nomadic generalist that inhabits diverse niches at low abundance. High adaptability of yeast is supported by a remarkable chromosomal number plasticity (Pavelka et al. 2010; Liu et al. 2015; Selmecki et al. 2015). Furthermore, yeast is capable of sexual reproduction, which also facilitates rapid adaptation (Goddard 2016). Nevertheless, variation in the natural ecological niches is shown to be reflected in trait divergence within S. cerevisiae strains associated with different population origins (Warringer et al. 2011).
Upon sudden exposure to excess glucose, even under aerobic conditions, S. cerevisiae exhibits high glycolytic and fermentative fluxes (Pronk, Steensma and vanDijken 1996)—a complex trait called short-term Crabtree effect. Several traits that contribute to the short-term Crabtree effect have appeared along the evolutionary history of Saccharomyceta (Hagman et al. 2013). Thus, S. cerevisiae exhibits an evolutionarily shaped trait to tolerate or even benefit from a sudden change in glucose availability.
In contrast to the natural reservoirs, usual laboratory growth medium is either a defined medium optimized for short generation times or a rich medium like in food and beverage fermentation applications of S. cerevisiae. Thus, the metabolism of S. cerevisiae is best understood in the fast growing states of fermentation. Wild strains from natural environments generally show lower glucose utilization rate than the domesticated strains of S. cerevisiae that have been selected in conditions of high glucose availability (Spor et al. 2009). Modeling results by Nidelet et al. (2016) accordingly suggest that the intracellular metabolic fluxes vary among S. cerevisiae strains from different ecological origins (bread, rum, wine, flour, Mediterranean and American oak) (Nidelet et al. 2016). Their results suggest that while the glycolytic and fermentative fluxes are similar between strains, the flux through the pentose phosphate pathway is strain dependent. Surprisingly, the domesticated (i.e. for food and beverage fermentation) strains show generally higher phenotypic diversity, in terms of growth characteristics under different environmental conditions, than the investigated wild strains of S. cerevisiae, even though the wild strains are genetically more diverse and show stronger geographical patterning (Warringer et al. 2011). The phenotypic diversity of the domesticated strains may result from selection for traits associated with specific fermentation substrates (e.g. rice fermentation for sake, grape for wine and barley for beer) (Liti et al. 2009; Boynton and Greig 2014; Gallone et al. 2016). For example, wine fermentations present challenging conditions such as high sugar concentration and low pH. Due to the equal abundance of glucose and fructose in the grape must, the total sugar concentration is extremely high creating challenging conditions of high osmolarity. The low pH of grape must originates from the presence of organic acids. When exposed to wine fermentation mimicking conditions, S. cerevisiae strains from different sources (i.e. laboratory strains, wild strains, clinical isolates, vineyard isolates, bakery strains, commercial wine strains, strains domesticated for other fermentation processes) showed distinct fermentation characteristics (Camarasa et al. 2011). While commercial wine yeasts were able to perform as they have been selected to, i.e. to complete the fermentation, strains from other sources commonly grew poorly and could not ferment the sugars completely. Notably, under the challenging conditions laboratory strains diverged from all the rest of the strains in their poor biomass formation and fermentation capability and in their product profile. Furthermore, since wine fermentations are commonly unsterilized, the domesticated S. cerevisiae wine strains have been simultaneously exposed to the extreme abiotic conditions in grape must and challenged with social life with other species.
Social life of Saccharomyces cerevisiae—symbionts impact yeast metabolism
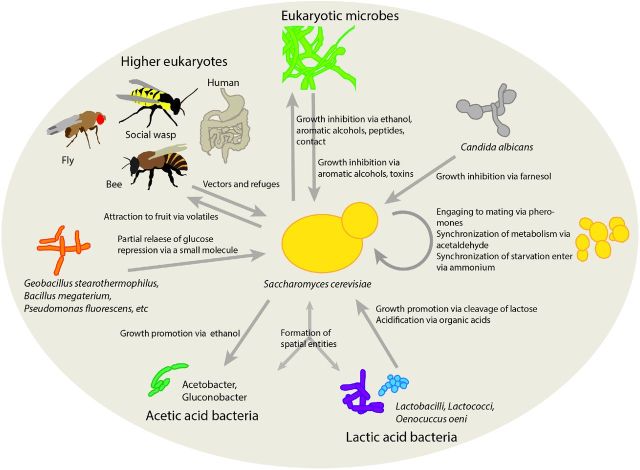
In addition to a dynamic abiotic environment, co-habiting organisms constitute another important ecological dimension shaping S. cerevisiae metabolism (Fig. 1). This social dimension also applies, despite being removed from their original ecological context, to domesticated yeasts growing in human created/controlled environments. Wine fermentation is perhaps the best-studied environment of S. cerevisiae with respect to the interspecies interactions (Mortimer and Polsinelli 1999; Fleet 2003; Alexandre et al. 2004; Comitini et al. 2005; Ciani et al. 2010; Barata, Malfeito-Ferreira and Loureiro 2012; Branco et al. 2014; Jolly, Varela and Pretorius 2014; Ramirez et al. 2015; Ciani et al. 2016; Liu et al. 2016; Wang, Mas and Esteve-Zarzoso 2016). Since grape must is typically not sterilized for wine fermentations, they represent a multispecies ecosystem. The yeast spectrum in the ecosystems includes over 40 different species that have been isolated from grape must (Jolly, Varela and Pretorius 2014). Understanding the interactions between fermenting yeasts and other microorganisms in wine has a distinct economic and gustatory incentive (Fleet 2003). More recently, social life of yeast has been observed also in other environments such as kefir (i.e. fermented milk product with originating from Caucasian mountains) (Simova et al. 2002; Farnworth 2005), sourdough (De Vuyst et al. 2014) and biofuel production cultures (Watanabe, Nakamura and Shima 2008; Lucena et al. 2010; Tiukova, Eberhard and Passoth 2014). Lactic acid bacteria (LAB) are commonly found together with S. cerevisiae in the domestic applications (e.g. wine, sourdough and kefir), but also in nature in overripened or faulty fruits (Barata, Malfeito-Ferreira and Loureiro 2012). Furthermore, LAB are common spoilage organisms in open S. cerevisiae fermentations used for biofuel production (Watanabe, Nakamura and Shima 2008; Lucena et al. 2010; Tiukova, Eberhard and Passoth 2014). In bioethanol production, Lactobacillus fermentum and L. brevis have been reported as the most common contaminants (Watanabe, Nakamura and Shima 2008). Tiukova, Eberhard and Passoth (2014) observed that L. vini in co-cultures with S. cerevisiae or Dekkera bruxellensis causes bacteria–yeast aggregation, thereby decreasing the ethanol production by yeast (Tiukova, Eberhard and Passoth 2014). S. cerevisiae can in turn benefit from the presence of LAB, for example, in lactose-rich environments. While S. cerevisiae is not able to utilize lactose, LAB are able to cleave it into glucose and galactose, which can be readily metabolized by the yeast (Mendes et al. 2013) (Fig. 1). Organic acids produced by fermentative microbes may also benefit yeast. S. cerevisiae is capable of consuming lactate during cheese ripening, simultaneously producing valuable aroma compounds (Kagkli et al. 2006). Though active transport of other acids such as malate and succinate across the cell membrane is not known in S. cerevisiae, these may diffuse into the cell contributing to the metabolism (Barnett and Kornberg 1960). The laboratory strains of S. cerevisiae show specific regulatory responses towards weak acids (Mira, Teixeira and Sa-Correia 2010), but the dependence of this response on the natural habitat or the origin of S. cerevisiae strains is yet unexplored.
Figure 1.
The social life of S. cerevisiae. Yeast shows numerous interactions with bacteria, other fungi and higher eukaryotes. Niche engineering is performed by the yeast and interacting organisms. S. cerevisiae cells communicate also within species by modifying the chemical environment. Yeast–plant interactions have not been considered here.
In addition to organic acids, other compounds secreted by coexisting species may affect S. cerevisiae phenotypes by directly triggering regulatory networks. Recently, some bacteria were found to partially release glucose repression in S. cerevisiae by inducing a prion-like element through secretion of a yet unidentified small molecule(Jarosz et al. 2014a,b) (Fig. 1). Several different bacteria were shown to relieve glucose repression when co-cultured with S. cerevisiae. This was found to benefit both the bacterial and yeast symbionts. While lower ethanol production reduced the toxic effect on bacteria, the yeast gained an ability to utilize other carbon sources, which was found particularly advantageous in mixed carbon source media (e.g. molasses).
Fungal symbionts of yeast can also impact its growth and metabolism through toxins and signaling molecules (Fig. 1). For instance, a toxin-secreting Torulaspora delbrueckii strain is able to kill S. cerevisiae (Ramirez et al. 2015). Another example is growth impairment of S. cerevisiae by Candida albicans through secretion of farnesol, a quorum-sensing molecule (Machida et al. 1998). Farnesol is an isoprenoid alcohol that interferes with the cell cycle signaling in S. cerevisiae in particular, but also affects the mitochondrial function through increased generation of reactive oxygen species (Machida et al. 1998, 1999). Aromatic alcohols tyrosol, tryptophol and phenylethanol are further fungal quorum-sensing molecules, which S. cerevisiae is also able to produce (Bruce et al. 2004) and can induce changes in growth morphology (Cottier and Muhlschlegel 2012).
Certain volatiles produced by yeast, commonly higher alcohols and their esters, attract flies, social wasps and bees. These insects can thereby transport S. cerevisiae to new habitats (Chandler, Eisen and Kopp 2012) (Fig. 1). Yeasts in turn can benefit its host through establishing a mutualistic interaction. While S. cerevisiae that attract flies better get an advantage of effective dispersal (Gilbert 1980; Chandler, Eisen and Kopp 2012; Hoang, Kopp and Chandler 2015) and outbreeding (Reuter, Bell and Greig 2007), Drosophila populated with more attractive yeast species demonstrate higher fecundity (Buser et al. 2014). Larvae of flies also benefit from yeast symbiont, mainly as a dietary supplement facilitating development and survival (Anagnostou, Dorsch and Rohlfs 2010).
Social life of Saccharomyces cerevisiae—yeast impacts community composition
Metabolites and peptides secreted by yeast can have a substantial impact on its co-habitants. Metabolites produced by S. cerevisiae have been found to reduce the cultivability of a number of non-Saccharomyces yeasts during co-fermentation or in conditioned medium (Wang, Mas and Esteve-Zarzoso 2016). S. cerevisiae produces high concentrations of ethanol that are toxic for many other microbial species. Saccharomyceta have gained traits contributing to the high fermentative capacity along their evolutionary history (Hagman et al. 2013). Only more recently S. cerevisiae with its closely related species gained a further increased ethanol production capability through a trait in which respiration becomes repressed in a high glucose environment even when oxygen is available (i.e. long-term Crabtree effect; Pronk, Steensma and vanDijken 1996) (Hagman and Piskur 2015). The high fermentative capacity allows to convert sugar to ethanol at a fast rate, and later shift to a respiratory metabolic phenotype to consume the ethanol (diauxic shift). The yeast thus seems to apply a make-accumulate-consume strategy in high glucose environments (Piskur et al. 2006).
While ethanol has considered to be the toxic product which S. cerevisiae benefits from the most, it has recently become evident that it may not be the most efficient of the weapons of S. cerevisiae. S. cerevisiae employs a 2-fold strategies to inhibit the growth of bacteria and fungi: secretion of small molecules and proteinaceous compounds such as peptides and physical cell–cell contact (Nissen, Nielsen and Arneborg 2003; Piskur et al. 2006; Branco et al. 2014; Wang, Mas and Esteve-Zarzoso 2016) (Fig. 1). In addition to ethanol, S. cerevisiae produces volatile compounds such as aromatic alcohols, which are implicated in inhibition of other fungi (Bruce et al. 2004; Cottier and Muhlschlegel 2012). S. cerevisiae secretes antimicrobial peptides that have either fungistatic (e.g. against Lachanchea thermotolerans (Kluyveromyces thermotolerans) and T. delbrueckii) or even fungisidic (e.g. against Kluyveromyces marxianus) effects on other fungal microbes (Albergaria et al. 2010). When exposed to S. cerevisiae's antimicrobial peptides, Hanseniaspora guilliermondii suffers from alterations in membrane permeability leading to a severe loss of intracellular pH homeostasis (Branco et al. 2015). Interestingly, it was recently found that anti-fungal and anti-bacterial killer peptides secreted by S. cerevisiae during wine fermentation include peptides of glyceraldehyde dehydrogenase (GAPDH), a glycolytic enzyme (Branco et al. 2014). The polypeptides of the GAPDH isoenzymes have been observed to be associated also with the cell wall (Delgado et al. 2001) where they are amenable for interactions with other species.
Cell–cell contact with S. cerevisiae has been found to contribute to the death of La. thermotolerans (K. thermotolerans) and T. delbrueckii in wine fermentations (Nissen, Nielsen and Arneborg 2003; Kemsawasd et al. 2015). This phenomenon could possibly be mediated by the anti-fungal peptides residing attached to the cell membrane.
Besides producing toxic compounds to fight competitors, S. cerevisiae can also provide benefits to other species. Already in 1965, commensalism of bacterium Proteus vulgaris with S. cerevisiae was observed (Shindala et al. 1965). The growth benefit of co-culturing with the yeast was found dependent on a niacin-like nutrient secreted by S. cerevisiae and essential for P. vulgaris. Megee et al. (1972) reported that they had been able to create various relationships (i.e. commensalism, competition and mutualism) between S. cerevisiae and Lactobacillus casei by varying the concentrations of glucose and riboflavin in the medium (Megee et al. 1972).
The stationary state survival of Pseudomonas putida increases substantially in co-culture with S. cerevisiae, which consumes glucose rapidly and lowers the pH (Romano and Kolter 2005). Saccharomyces cerevisiae has also been shown to support the growth of L. delbrueckii ssp. bulgaricus in lactose media by supplying L-Alanine and CO2 to the bacteria (Mendes et al. 2013). In return, the bacteria provided galactose, as a cleavage product of lactose, for S. cerevisiae growth.
In the late stages of wine fermentation, yeast can either promote or inhibit the growth of LAB involved in malolactic fermentation (Liu et al. 2016). The yeast inhibiting bacteria secretes sulfur-containing peptides, in distinction to the yeast phenotype creating a supporting niche for LAB. Together with LAB, acetic acid bacteria (AAB) commonly co-occur with S. cerevisiae (Farnworth 2005; Camu et al. 2007). Acetobacter, Gluconobacter and Gluconacetobacter species of AAB are the most prevalent co-occurring taxons. S. cerevisiae together with LAB and AAB species has been found for instance in spontaneous cocoa bean (Camu et al. 2007), kefir (Farnworth 2005) and kombucha (Jayabalan et al. 2014) fermentations. In spontaneous cocoa bean fermentations, yeasts, including S. cerevisiae, together with LAB engineer a niche for AAB. LAB consume citrate initially present, thus, increasing pH that favors AAB. Mainly Acetobacter pasteurianus oxidizes ethanol produced by the yeasts (Fig. 1). Oxygen-dependent conversion of ethanol to acetate and/or acetoin increases the temperature until the microbial activities cease. Ethanol and acetate diffuse into the cocoa beans creating the desired flavor and color characteristics. In addition to ethanol, AAB oxidize lactic acid secreted by LAB and are also able to oxidize glucose to gluconic acid and mannitol into fructose, at least in the presence of ethanol (Moens, Lefeber and De Vuyst 2014). Thus, ethanol produced by yeast enables AAB to oxidize a wider range of substrates.
Phenotypic heterogeneity and cross-feeding in yeast populations
In a yeast colony, the cells encounter a variable chemical environment depending on their location. This may induce phenotypic heterogeneity within species, possibly enhanced by intraspecies communication by secreted metabolites. While complementary auxotrophic strains of S. cerevisiae have been found to fail to cross-feed sufficiently for survival, a viable complementary cross-feeding community is formed when the auxotrophs emerge from a self-supporting cell during colony formation (Campbell et al. 2015; Campbell, Vowinckel and Ralser 2016). This phenomenon was discovered in a system where the cells carried in separate plasmids the genes for the synthesis of four compounds essential for growth (i.e. histidine, leucine, uracil and methionine). During colony development, prototrophy was progressively lost leading to phenotypic heterogeneity, but the community could survive by exchanging the essential nutrients.
In colony development, S. cerevisiae cells behave periodically with acidic and alkali phases (Palkova et al. 1997). When the cells switch from an acidic phase to an alkaline phase, they produce volatile ammonia as a pulse, which triggers ammonia production in surrounding colonies. The released ammonia signals for the synchronization of nutrient starvation response. Ammonium secreted by S. cerevisiae could potentially be sensed by other yeasts as well (Gori et al. 2007). Other small molecules can also mediate communication between yeast cells (Fig. 1). At high cell densities in liquid cultures, S. cerevisiae cells synchronize their metabolism with secreted acetaldehyde (Richard et al. 1996). Other fungi can thus also influence S. cerevisiae metabolism through acetaldehyde (Cheraiti, Guezenec and Salmon 2005). Other examples of communication include bicarbonate for synchronization of the onset of sporulation or meiosis (Hayashi, Ohkuni and Yamashita 1998; Ohkuni, Hayashi and Yamashita 1998), small peptide pheromones used to facilitate partner finding during sexual reproduction (Jones and Bennett 2011) and secreted Hsp12p protein as a ‘danger signal’ in order to activate the stress response in the surrounding yeast cells (Rivero et al. 2015).
CONCLUSIONS
Most of the current knowledge of S. cerevisiae metabolism pertains to (or is interpreted in the context of) its life as a single entity, devoid of species interactions, and in a limited set of laboratory conditions. The limited understanding of the challenges and possibilities that have evolutionarily shaped the metabolic and regulatory systems of yeast may be the major factor hindering us from explaining complex genotype–environment–phenotype interactions. Accounting for the ecological context of yeast could also help us to assign functions to the uncharacterized 10% of genes (669 uncharacterized open reading frames, www.yeastgenome.org, on 15 August 2016) in the yeast genome, and to identify moonlighting functions of metabolic enzymes. Mainly pioneered by enological research, phenotypic peculiarities arising from the social life of yeast are increasingly being revealed. These findings present an excellent opportunity towards building more accurate models of metabolic and regulatory networks. The full phenotypic potential to be revealed will also increase the application possibilities of the already widely used industrial production host.
FUNDING
This work was supported by in the context of the EU-funded initiative ERA-Net for Applied Systems Biology ERASysAPP [WineSys to KRP and PJ].
Conflict of interest. None declared.
REFERENCES
- Albergaria H, Francisco D, Gori K, et al. Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl Microbiol Biot. 2010;86:965–72. doi: 10.1007/s00253-009-2409-6. [DOI] [PubMed] [Google Scholar]
- Alexandre H, Costello PJ, Remize F, et al. Saccharomyces cerevisiae - Oenococcus oeni interactions in wine: current knowledge and perspectives. Int J Food Microbiol. 2004;93:141–54. doi: 10.1016/j.ijfoodmicro.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Anagnostou C, Dorsch M, Rohlfs M. Influence of dietary yeasts on Drosophila melanogaster life-history traits. Entomol Exp Appl. 2010;136:1–11. [Google Scholar]
- Barata A, Malfeito-Ferreira M, Loureiro V. The microbial ecology of wine grape berries. Int J Food Microbiol. 2012;153:243–59. doi: 10.1016/j.ijfoodmicro.2011.11.025. [DOI] [PubMed] [Google Scholar]
- Barnett JA, Kornberg HL. The utilization by yeasts of acids of the tricarboxylic acid cycle. J Gen Microbiol. 1960;23:65–82. doi: 10.1099/00221287-23-1-65. [DOI] [PubMed] [Google Scholar]
- Boynton PJ, Greig D. The ecology and evolution of non-domesticated Saccharomyces species. Yeast. 2014;31:449–62. doi: 10.1002/yea.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco P, Francisco D, Chambon C, et al. Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl Microbiol Biot. 2014;98:843–53. doi: 10.1007/s00253-013-5411-y. [DOI] [PubMed] [Google Scholar]
- Branco P, Viana T, Albergaria H, et al. Antimicrobial peptides (AMPs) produced by Saccharomyces cerevisiae induce alterations in the intracellular pH, membrane permeability and culturability of Hanseniaspora guilliermondii cells. Int J Food Microbiol. 2015;205:112–8. doi: 10.1016/j.ijfoodmicro.2015.04.015. [DOI] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, et al. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–5. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Brochado AR, Andrejev S, Maranas CD, et al. Impact of stoichiometry representation on simulation of genotype-phenotype relationships in metabolic networks. PLoS Comput Biol. 2012;8:e1002758. doi: 10.1371/journal.pcbi.1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A, Verrall S, Hackett CA, et al. Identification of volatile organic compounds (VOCs) from bacteria and yeast causing growth inhibition of sapstain fungi. Holzforschung. 2004;58:193–8. [Google Scholar]
- Buser CC, Newcomb RD, Gaskett AC, et al. Niche construction initiates the evolution of mutualistic interactions. Ecol Lett. 2014;17:1257–64. doi: 10.1111/ele.12331. [DOI] [PubMed] [Google Scholar]
- Camarasa C, Sanchez I, Brial P, et al. Phenotypic landscape of Saccharomyces cerevisiae during wine fermentation: evidence for origin-dependent metabolic traits. PLoS One. 2011;6:e25147. doi: 10.1371/journal.pone.0025147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K, Vowinckel J, Mülleder M, et al. Self-establishing communities enable cooperative metabolite exchange in a eukaryote. Elife. 2015;4:e09943. doi: 10.7554/eLife.09943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K, Vowinckel J, Ralser M. Cell-to-cell heterogeneity emerges as consequence of metabolic cooperation in a synthetic yeast community. Biotechnol J. 2016;11:1169–78. doi: 10.1002/biot.201500301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camu N, De Winter T, Verbrugghe K, et al. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl Environ Microb. 2007;73:1809–24. doi: 10.1128/AEM.02189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreto L, Eiriz MF, Gomes AC, et al. Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics. 2008;9:524. doi: 10.1186/1471-2164-9-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JA, Eisen JA, Kopp A. Yeast communities of diverse Drosophila species: comparison of two symbiont groups in the same hosts. Appl Environ Microb. 2012;78:7327–36. doi: 10.1128/AEM.01741-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheraiti N, Guezenec S, Salmon JM. Redox interactions between Saccharomyces cerevisiae and Saccharomyces uvarum in mixed culture under enological conditions. Appl Environ Microb. 2005;71:255–60. doi: 10.1128/AEM.71.1.255-260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani M, Capece A, Comitini F, et al. Yeast Interactions in Inoculated Wine Fermentation. Front Microbiol. 2016;7:555. doi: 10.3389/fmicb.2016.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani M, Comitini F, Mannazzu I, et al. Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking. Fems Yeast Res. 2010;10:123–33. doi: 10.1111/j.1567-1364.2009.00579.x. [DOI] [PubMed] [Google Scholar]
- Ciani M, Mannazzu I, Marinangeli P, et al. Contribution of winery-resident Saccharomyces cerevisiae strains to spontaneous grape must fermentation. Anton Leeuw Int J G. 2004;85:159–64. doi: 10.1023/B:ANTO.0000020284.05802.d7. [DOI] [PubMed] [Google Scholar]
- Comitini F, Ferretti R, Clementi F, et al. Interactions between Saccharomyces cerevisiae and malolactic bacteria: preliminary characterization of a yeast proteinaceous compound(s) active against Oenococcus oeni. J Appl Microbiol. 2005;99:105–11. doi: 10.1111/j.1365-2672.2005.02579.x. [DOI] [PubMed] [Google Scholar]
- Cottier F, Muhlschlegel FA. Communication in fungi. Int J Microbiol. 2012;2012:351832. doi: 10.1155/2012/351832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vuyst L, Van Kerrebroeck S, Harth H, et al. Microbial ecology of sourdough fermentations: diverse or uniform? Food Microbiol. 2014;37:11–29. doi: 10.1016/j.fm.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Delgado ML, O'Connor JE, Azorin I, et al. The glyceraldehyde-3-phosphate dehydrogenase polypeptides encoded by the Saccharomyces cerevisiae TDH1, TDH2 and TDH3 genes are also cell wall proteins. Microbiol-UK. 2001;147:411–7. doi: 10.1099/00221287-147-2-411. [DOI] [PubMed] [Google Scholar]
- Dunn B, Richter C, Kvitek DJ, et al. Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 2012;22:908–24. doi: 10.1101/gr.130310.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Torabi N, Jia Y, et al. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2010;464:1039–42. doi: 10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnworth ER. Kefir—A complex probiotic. Food Sci Technol Bull. 2005;2:1–17. [Google Scholar]
- Fleet GH. Yeast interactions and wine flavour. Int J Food Microbiol. 2003;86:11–22. doi: 10.1016/s0168-1605(03)00245-9. [DOI] [PubMed] [Google Scholar]
- Forster J, Famili I, Fu P, et al. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13:244–53. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallone B, Steensels J, Prahl T, et al. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell. 2016;166:1397–410. doi: 10.1016/j.cell.2016.08.020. e1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DG. Dispersal of yeasts and bacteria by Drosophila in a temperate forest. Oecologia. 1980;46:135–7. doi: 10.1007/BF00346979. [DOI] [PubMed] [Google Scholar]
- Goddard MR. Molecular evolution: sex accelerates adaptation. Nature. 2016;531:176–7. doi: 10.1038/nature17304. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Anfang N, Tang R, et al. A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ Microbiol. 2010;12:63–73. doi: 10.1111/j.1462-2920.2009.02035.x. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Greig D. Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res. 2015;15:fov009. doi: 10.1093/femsyr/fov009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori K, Mortensen HD, Arneborg N, et al. Ammonia production and its possible role as a mediator of communication for Debaryomyces hansenii and other cheese-relevant yeast species. J Dairy Sci. 2007;90:5032–41. doi: 10.3168/jds.2006-750. [DOI] [PubMed] [Google Scholar]
- Hagman A, Piskur J. A study on the fundamental mechanism and the evolutionary driving forces behind aerobic fermentation in yeast. PLoS One. 2015;10:e0116942. doi: 10.1371/journal.pone.0116942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman A, Sall T, Compagno C, et al. Yeast "make-accumulate-consume" life strategy evolved as a multi-step process that predates the whole genome duplication. PLoS One. 2013;8:e68734. doi: 10.1371/journal.pone.0068734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Ohkuni K, Yamashita I. An extracellular meiosis-promoting factor in Saccharomyces cerevisiae. Yeast. 1998;14:617–22. doi: 10.1002/(SICI)1097-0061(199805)14:7<617::AID-YEA265>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Heavner BD, Price ND. Comparative analysis of yeast metabolic network models highlights progress, opportunities for metabolic reconstruction. PLoS Comput Biol. 2015;11:e1004530. doi: 10.1371/journal.pcbi.1004530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrgard MJ, Swainston N, Dobson P, et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol. 2008;26:1155–60. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang D, Kopp A, Chandler JA. Interactions between Drosophila and its natural yeast symbionts-Is Saccharomyces cerevisiae a good model for studying the fly-yeast relationship? Peerj. 2015;3:e1116. doi: 10.7717/peerj.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Brown JC, Walker GA, et al. Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell. 2014a;158:1083–93. doi: 10.1016/j.cell.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Lancaster AK, Brown JC, et al. An evolutionarily conserved prion-like element converts wild fungi from metabolic specialists to generalists. Cell. 2014b;158:1072–82. doi: 10.1016/j.cell.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayabalan R, Malbasa RV, Loncar ES, et al. A review on kombucha tea—microbiology, composition, fermentation, beneficial effects, toxicity, and tea fungus. Compr Rev Food Sci F. 2014;13:538–50. doi: 10.1111/1541-4337.12073. [DOI] [PubMed] [Google Scholar]
- Jolly NP, Varela C, Pretorius IS. Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. Fems Yeast Res. 2014;14:215–37. doi: 10.1111/1567-1364.12111. [DOI] [PubMed] [Google Scholar]
- Jones SK, Jr, Bennett RJ. Fungal mating pheromones: choreographing the dating game. Fungal Genet Biol. 2011;48:668–76. doi: 10.1016/j.fgb.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagkli DM, Tache R, Cogan TM, et al. Kluyveromyces lactis and Saccharomyces cerevisiae, two potent deacidifying and volatile-sulphur-aroma-producing microorganisms of the cheese ecosystem. Appl Microbiol Biot. 2006;73:434–42. doi: 10.1007/s00253-006-0461-z. [DOI] [PubMed] [Google Scholar]
- Kemsawasd V, Branco P, Almeida MG, et al. Cell-to-cell contact and antimicrobial peptides play a combined role in the death of Lachanchea thermotolerans during mixed-culture alcoholic fermentation with Saccharomyces cerevisiae. FEMS Microbiol Lett. 2015;362:fnv103. doi: 10.1093/femsle/fnv103. [DOI] [PubMed] [Google Scholar]
- Knight SJ, Goddard MR. Sporulation in soil as an overwinter survival strategy in Saccharomyces cerevisiae. FEMS Yeast Res. 2016;16:fov102. doi: 10.1093/femsyr/fov102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppram R, Mapelli V, Albers E, et al. The presence of pretreated lignocellulosic solids from birch during Saccharomyces cerevisiae fermentations leads to increased tolerance to inhibitors—a proteomic study of the effects. PLoS One. 2016;11:e0148635. doi: 10.1371/journal.pone.0148635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowallik V, Greig D. A systematic forest survey showing an association of Saccharomyces with oak leaf litter. Environ Microbiol Rep. 2016 doi: 10.1111/1758-2229.12446. [DOI] [PubMed] [Google Scholar]
- Legras JL, Merdinoglu D, Cornuet JM, et al. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- Liti G. The fascinating and secret wild life of the budding yeast S. cerevisiae. Elife. 2015;4:e05835. doi: 10.7554/eLife.05835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–41. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GW, Yong MYJ, Yurieva M, et al. Gene Essentiality is a quantitative property linked to cellular evolvability. Cell. 2015;163:1388–99. doi: 10.1016/j.cell.2015.10.069. [DOI] [PubMed] [Google Scholar]
- Liu YZ, Forcisi S, Harir M, et al. New molecular evidence of wine yeast-bacteria interaction unraveled by non-targeted exometabolomic profiling. Metabolomics. 2016;12:unsp69. [Google Scholar]
- Lucena BT, dos Santos BM, Moreira JL, et al. Diversity of lactic acid bacteria of the bioethanol process. BMC Microbiol. 2010;10:298. doi: 10.1186/1471-2180-10-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida K, Tanaka T, Fujita K, et al. Farnesol-induced generation of reactive oxygen species via indirect inhibition of the mitochondrial electron transport chain in the yeast Saccharomyces cerevisiae. J Bacteriol. 1998;180:4460–5. doi: 10.1128/jb.180.17.4460-4465.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida K, Tanaka T, Yano Y, et al. Farnesol-induced growth inhibition in Saccharomyces cerevisiae by a cell cycle mechanism. Microbiol-UK. 1999;145:293–9. doi: 10.1099/13500872-145-2-293. [DOI] [PubMed] [Google Scholar]
- Megee RD, 3rd, Drake JF, Fredrickson AG, et al. Studies in intermicrobial symbiosis. Saccharomyces cerevisiae and Lactobacillus casei. Can J Microbiol. 1972;18:1733–42. doi: 10.1139/m72-269. [DOI] [PubMed] [Google Scholar]
- Mendes F, Sieuwerts S, de Hulster E, et al. Transcriptome-Based Characterization of Interactions between Saccharomyces cerevisiae and Lactobacillus delbrueckii subsp bulgaricus in Lactose-Grown Chemostat Cocultures. Appl Environ Microb. 2013;79:5949–61. doi: 10.1128/AEM.01115-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira NP, Teixeira MC, Sa-Correia I. Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: a genome-wide view. OMICS. 2010;14:525–40. doi: 10.1089/omi.2010.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens F, Lefeber T, De Vuyst L. Oxidation of Metabolites Highlights the Microbial Interactions and Role of Acetobacter pasteurianus during Cocoa Bean Fermentation. Appl Environ Microb. 2014;80:1848–57. doi: 10.1128/AEM.03344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R, Polsinelli M. On the origins of wine yeast. Res Microbiol. 1999;150:199–204. doi: 10.1016/s0923-2508(99)80036-9. [DOI] [PubMed] [Google Scholar]
- Mulleder M, Capuano F, Pir P, et al. A prototrophic deletion mutant collection for yeast metabolomics and systems biology. Nat Biotechnol. 2012;30:1176–8. doi: 10.1038/nbt.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nidelet T, Brial P, Camarasa C, et al. Diversity of flux distribution in central carbon metabolism of S. cerevisiae strains from diverse environments. Microb Cell Fact. 2016;15:58. doi: 10.1186/s12934-016-0456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Nielsen D, Arneborg N. Viable Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-Saccharomyces yeasts in mixed cultures by a cell-cell contact-mediated mechanism. Yeast. 2003;20:331–41. doi: 10.1002/yea.965. [DOI] [PubMed] [Google Scholar]
- Ohkuni K, Hayashi M, Yamashita I. Bicarbonate-mediated social communication stimulates meiosis and sporulation of Saccharomyces cerevisiae. Yeast. 1998;14:623–31. doi: 10.1002/(SICI)1097-0061(199805)14:7<623::AID-YEA264>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Palkova Z, Janderova B, Gabriel J, et al. Ammonia mediates communication between yeast colonies. Nature. 1997;390:532–6. doi: 10.1038/37398. [DOI] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–5. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira R, Nielsen J, Rocha I. Improving the flux distributions simulated with genome-scale metabolic models of Saccharomyces cerevisiae. Metab Eng Commun. 2016;3:153–63. doi: 10.1016/j.meteno.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskur J, Rozpedowska E, Polakova S, et al. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006;22:183–6. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Pronk JT, Steensma HY, vanDijken JP. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–33. doi: 10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Qian W, Ma D, Xiao C, et al. The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Rep. 2012;2:1399–410. doi: 10.1016/j.celrep.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M, Velazquez R, Maquedal M, et al. A new wine Torulaspora delbrueckii killer strain with broad antifungal activity and its toxin-encoding double-stranded RNA virus. Front Microbiol. 2015;6:983. doi: 10.3389/fmicb.2015.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Bell G, Greig D. Increased outbreeding in yeast in response to dispersal by an insect vector. Curr Biol. 2007;17:R81–3. doi: 10.1016/j.cub.2006.11.059. [DOI] [PubMed] [Google Scholar]
- Richard P, Bakker BM, Teusink B, et al. Acetaldehyde mediates the synchronization of sustained glycolytic oscillations in populations of yeast cells. Eur J Biochem. 1996;235:238–41. doi: 10.1111/j.1432-1033.1996.00238.x. [DOI] [PubMed] [Google Scholar]
- Rivero D, Berna L, Stefanini I, et al. Hsp12p and PAU genes are involved in ecological interactions between natural yeast strains. Environ Microbiol. 2015;17:3069–81. doi: 10.1111/1462-2920.12950. [DOI] [PubMed] [Google Scholar]
- Romano JD, Kolter R. Pseudomonas-saccharomyces interactions: Influence of fungal metabolism on bacterial physiology and survival. J Bacteriol. 2005;187:940–8. doi: 10.1128/JB.187.3.940-948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio JP, Goncalves P. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and are sympatric with S. cerevisiae and S. paradoxus. Appl Environ Microb. 2008;74:2144–52. doi: 10.1128/AEM.02396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Maruvka YE, Richmond PA, et al. Polyploidy can drive rapid adaptation in yeast. Nature. 2015;519:349–52. doi: 10.1038/nature14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindala A, Bungay HR, 3rd, Krieg NR, et al. Mixed-culture interactions. I. Commensalism of proteus vulgaris with Saccharomyces cerevisiae in continuous culture. J Bacteriol. 1965;89:693–6. doi: 10.1128/jb.89.3.693-696.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard D, Legras JL. Bread, beer and wine: yeast domestication in the Saccharomyces sensu stricto complex. C R Biol. 2011;334:229–36. doi: 10.1016/j.crvi.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Simova E, Beshkova D, Angelov A, et al. Lactic acid bacteria and yeasts in kefir grains and kefir made from them. J Ind Microbiol Biot. 2002;28:1–6. doi: 10.1038/sj/jim/7000186. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Dombrowski PG, Fingerman E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Spor A, Nidelet T, Simon J, et al. Niche-driven evolution of metabolic and life-history strategies in natural and domesticated populations of Saccharomyces cerevisiae. BMC Evol Biol. 2009;9:296. doi: 10.1186/1471-2148-9-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanini I, Dapporto L, Legras JL, et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. P Natl Acad Sci USA. 2012;109:13398–403. doi: 10.1073/pnas.1208362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz LM, Sinha H, Richards DR, et al. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 2002;416:326–30. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- Strope PK, Skelly DA, Kozmin SG, et al. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 2015;25:762–74. doi: 10.1101/gr.185538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szappanos B, Kovacs K, Szamecz B, et al. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nat Genet. 2011;43:656–62. doi: 10.1038/ng.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiukova I, Eberhard T, Passoth V. Interaction of Lactobacillus vini with the ethanol-producing yeasts Dekkera bruxellensis and Saccharomyces cerevisiae. Biotechnol Appl Bioc. 2014;61:40–4. doi: 10.1002/bab.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderSluis B, Hess DC, Pesyna C, et al. Broad metabolic sensitivity profiling of a prototrophic yeast deletion collection. Genome Biol. 2014;15:R64. doi: 10.1186/gb-2014-15-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CX, Mas A, Esteve-Zarzoso B. The interaction between Saccharomyces cerevisiae and non-saccharomyces yeast during alcoholic fermentation is species and strain specific. Front Microbiol. 2016;7:502. doi: 10.3389/fmicb.2016.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QM, Liu WQ, Liti G, et al. Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol Ecol. 2012;21:5404–17. doi: 10.1111/j.1365-294X.2012.05732.x. [DOI] [PubMed] [Google Scholar]
- Warringer J, Zorgo E, Cubillos FA, et al. Trait variation in yeast is defined by population history. PLos Genet. 2011;7:e1002111. doi: 10.1371/journal.pgen.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe I, Nakamura T, Shima J. A strategy to prevent the occurrence of Lactobacillus strains using lactate-tolerant yeast Candida glabrata in bioethanol production. J Ind Microbiol Biot. 2008;35:1117–22. doi: 10.1007/s10295-008-0390-1. [DOI] [PubMed] [Google Scholar]
- Zhang HY, Skelton A, Gardner RC, et al. Saccharomyces paradoxus and Saccharomyces cerevisiae reside on oak trees in New Zealand: evidence for migration from Europe and interspecies hybrids. FEMS Yeast Res. 2010;10:941–7. doi: 10.1111/j.1567-1364.2010.00681.x. [DOI] [PubMed] [Google Scholar]
- Zorgo E, Gjuvsland A, Cubillos FA, et al. Life history shapes trait heredity by accumulation of loss-of-function alleles in yeast. Mol Biol Evol. 2012;29:1781–9. doi: 10.1093/molbev/mss019. [DOI] [PubMed] [Google Scholar]