Abstract
The thylakoid proton motive force (pmf) generated during photosynthesis is the essential driving force for ATP production; it is also a central regulator of light capture and electron transfer. We investigated the effects of elevated pmf on photosynthesis in a library of Arabidopsis thaliana mutants with altered rates of thylakoid lumen proton efflux, leading to a range of steady-state pmf extents. We observed the expected pmf-dependent alterations in photosynthetic regulation, but also strong effects on the rate of photosystem II (PSII) photodamage. Detailed analyses indicate this effect is related to an elevated electric field (Δψ) component of the pmf, rather than lumen acidification, which in vivo increased PSII charge recombination rates, producing singlet oxygen and subsequent photodamage. The effects are seen even in wild type plants, especially under fluctuating illumination, suggesting that Δψ-induced photodamage represents a previously unrecognized limiting factor for plant productivity under dynamic environmental conditions seen in the field.
DOI: http://dx.doi.org/10.7554/eLife.16921.001
Research Organism: <i>A. thaliana</i>
Introduction
The thylakoid proton motive force (pmf), the transmembrane electrochemical gradient of protons generated during the light reactions of photosynthesis, is a fundamental entity of bioenergetics, coupling light-driven electron transfer reactions to the phosphorylation of ADP via the ATP synthase (Avenson et al., 2004; Kramer and Evans, 2011). In oxygenic photosynthesis, light energy is captured by pigments in light-harvesting complexes and transferred to a subset of chlorophylls in photosystem I (PSI) and photosystem II (PSII), where it drives the extraction of electrons from water and their transfer through redox cofactors to ultimately reduce NADP+. The vectorial transfer of electrons across the membrane is tightly coupled with the generation of the pmf, composed of both electric field (Δψ) and pH (ΔpH) gradients.
In addition to its role in energy conservation, the pmf is also critical for feedback regulation of photosynthesis (Cruz et al., 2005). Acidification of the thylakoid lumen activates the photoprotective energy-dependent exciton quenching (qE) process, which dissipates excess absorbed light energy in the photosynthetic antenna complexes by the activation of violaxanthin deepoxidase (Demmig-Adams and Adams, 1992) and protonation of PsbS (Li et al., 2004). Lumen acidification also regulates the oxidation of plastoquinol by the cytochrome b6f complex, slowing electron transfer from PSII and preventing the accumulation of electrons on PSI, which can otherwise lead to photodamage (Nishio and Whitmarsh, 1993; Hope et al., 1994). In vitro work has shown that excessive lumen acidification can inactivate PSII (Krieger and Weis, 1993), decrease the stability of plastocyanin (Gross et al., 1994) and severely restrict electron flow through the cytochrome b6f complex (Kramer et al., 1999). Taken together, the in vivo and in vitro evidence of the susceptibility of photosynthetic components to acidification has led to the proposal that the extent of pmf and its partitioning into Δψ and ΔpH components is regulated to maintain the lumen pH above about 5.8, where it can regulate photoprotection. However, under environmental stresses the regulation of photosynthesis may become overwhelmed, leading to PSII damage, or photoinhibition, from lumen over-acidification, although this has not been shown to occur in vivo (Kramer et al., 1999).
PSII photoinhibition can be a major contributor to loss of photosynthetic productivity (Raven, 2011), particularly under rapid fluctuations in environmental conditions experienced in the field (Kulheim et al., 2002). However, the mechanisms and regulation of photoinhibition remain highly debated (Keren and Krieger-Liszkay, 2011; Tyystjärvi, 2013). Though several mechanisms have been proposed for the photodamage process, it is not known which of these operate in vivo under diverse environmental conditions. In addition, there are differing views on whether the extent of photoinhibition is governed by the rate of photodamage to PSII or by regulation of PSII repair (Takahashi et al., 2007). Answering these questions is essential to understanding how plants respond to rapidly changing conditions and thus of critical importance to improving plant productivity.
The extent of the pmf can be modulated by altering the light-driven influx of protons into the lumen, i.e. by changing the rates of linear electron flow (LEF) or cyclic electron flow, or the efflux of protons through the ATP synthase (reviewed in Kramer et al., 2004). This latter mode of regulation is important for co-regulation of the light reactions with downstream metabolic processes. For example, under CO2 limitations (Kanazawa and Kramer, 2002) as well as during stromal Pi limited conditions (Takizawa et al., 2008) the chloroplast ATP synthase activity is strongly down-regulated, decreasing the proton conductivity (gH+) of the thylakoid, leading to buildup of pmf and activation of qE. Similar decreases in gH+ and increases in pmf are seen when ATP synthase protein content is decreased (Rott et al., 2011) or in mutants with an altered ATP synthase γ-subunit (Kohzuma et al., 2012).
Results
We took advantage of the effects of the ATP synthase on the photosynthetic proton circuit to probe how the pmf influences photoinhibition in vivo. We constructed a series of Arabidopsis thaliana mutants, which we termed minira (minimum recapitulation of ATPC2), in which we complemented a γ1-subunit (ATPC1) T-DNA knockout line (Dal Bosco et al., 2004) with γ-coding sequences containing site-directed mutations to specifically incorporate amino acid changes around the redox regulatory cysteines present in ATPC2 into ATPC1 (Figure 1A) (Inohara et al., 1991). Each minira line is designated numerically based on amino acid position and independent transformation events; for example minira 4–1, minira 4–2, and minira 4–3 represent three independent transformation events of the same I201V mutation. The minira library was originally developed as part of an on-going 'domain swapping' approach to assess functional differences in the two ATPC paralogs in Arabidopsis, ATPC1 and ATPC2, but the minira mutants also shows a range of ATP synthase activities useful to the present work. As described below, we also confirmed key aspects of the work using previously characterized ATPC1 tobacco antisense lines (Rott et al., 2011). However, variations in ATP synthase suppression in the antisense lines between leaves and plant generations limited their experimental utility. The current mutagenesis approach was preferable as it allowed repeatable analyses of multiple photosynthetic parameters in stable, identical genetic backgrounds within each mutant line.
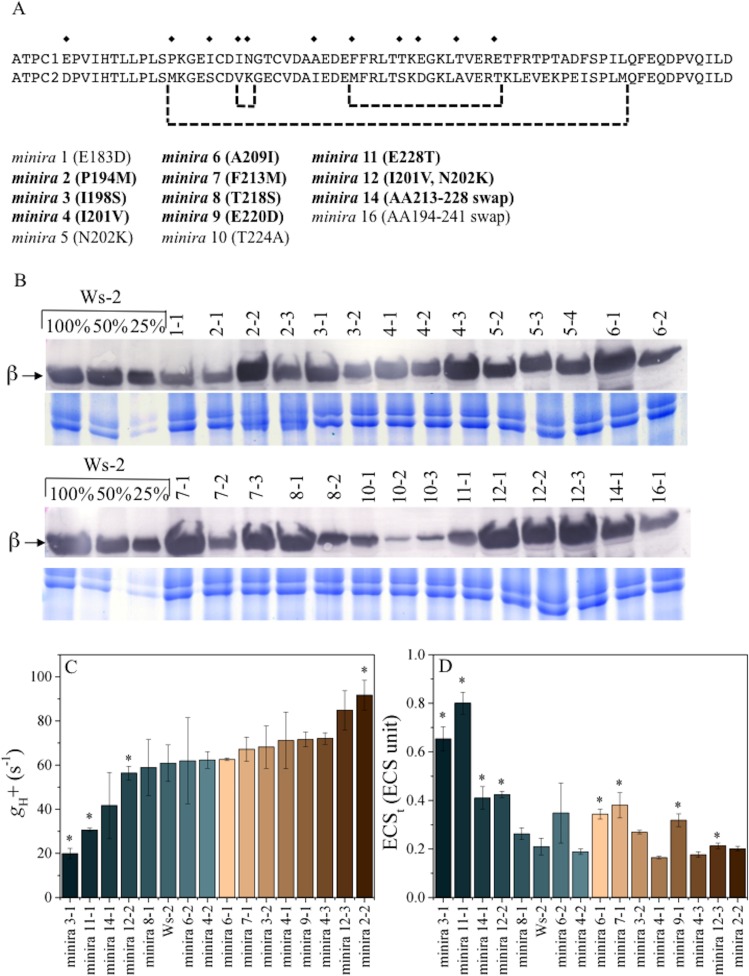
Figure 1. γ-subunit mutations alter photosynthetic proton efflux.
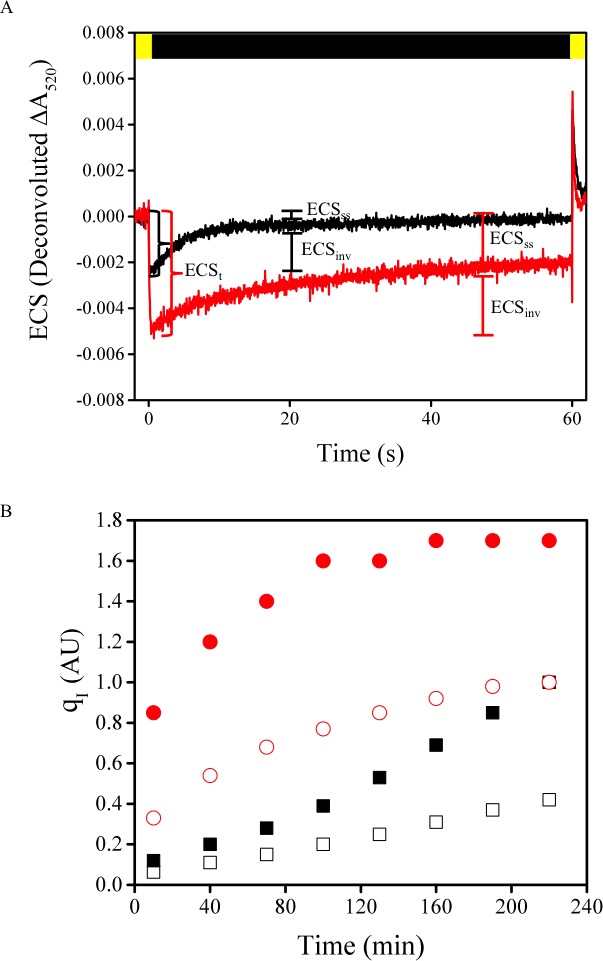
Sequence alignment of Arabidopsis ATPC1 and ATPC2 regulatory region (A). Amino acid differences incorporated into ATPC1 to generate minira are indicated by symbols (♦). Amino acid numbers are based on standard spinach positions, which minira numeric designations are based upon position within the amino acid primary sequence. Regions where multiple changes were incorporated from ATPC2 are outlined in brackets. Bold: mutations resulting in successful transgenic plants with stable phenotypes utilized for experiments. Accumulation of chloroplast ATP synthase complexes was verified across resulting transformant lines (B). Total leaf protein was probed for chloroplast ATP synthase β-subunit compared to a titration of wild type (Ws-2) accumulation. Gels run with identical samples stained with Coomassie Brilliant Blue are shown below to ensure equal loading to the 100% wild type samples. The conductivity of the ATP synthase for protons (gH+, C) and the light-driven pmf (ECSt, D), calculated from the decay of the electrochromic shift at 100 μmol photons m−2s−1 actinic light (mean ± s.d, n = 3). Statistically significant differences (*p<0.05) from wild type were determined using a t-test. ECS units were defined as the deconvoluted ΔA520 μg chlorophyll−1 cm2.
We assessed the effects of minira modifications on gH+ and the extent of the light-driven pmf in vivo based on the decay kinetics of the electrochromic shift (ECS), which reports changes in the thylakoid electric field (Sacksteder and Kramer, 2000). The minira lines displayed a range of ATP synthase activities (Figure 1C), from about 30–120% that of wild type (Wassilewskija-2, Ws-2), resulting in similar variations in light-driven pmf (Figure 1D). Multiple independent transformations were utilized for the same minira mutation, as the gH+ changes likely reflect both intrinsic ATP synthase activity changes due to the mutations, as well as changes due to protein expression level or stability of the mutated subunit within the complex (Figure 1B). While some minira mutants display an increase in gH+ relative to the wild type and will acidify the lumen at a slower rate, others have a large decrease in total ATP synthase content. For the purpose of understanding how a high pmf impacts photosynthesis we have primarily focused on those mutants that modified gH+ while maintaining an ATP synthase content similar to wild type levels.
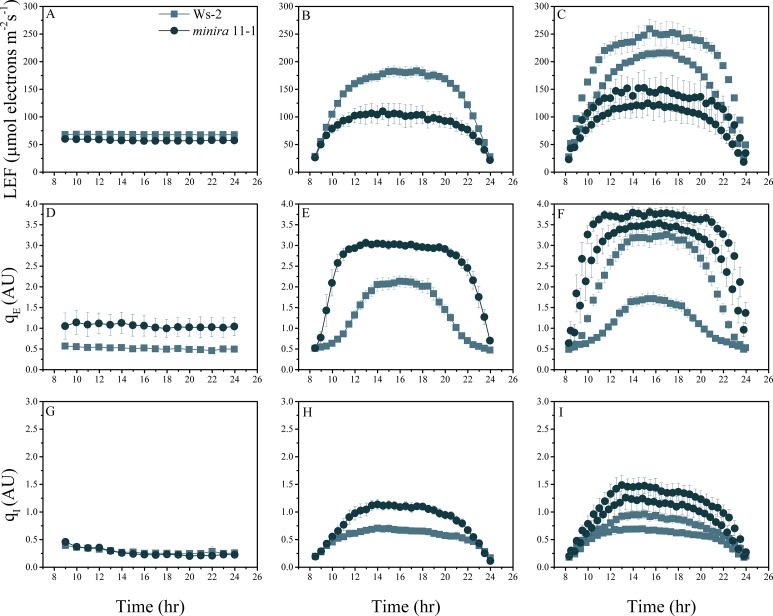
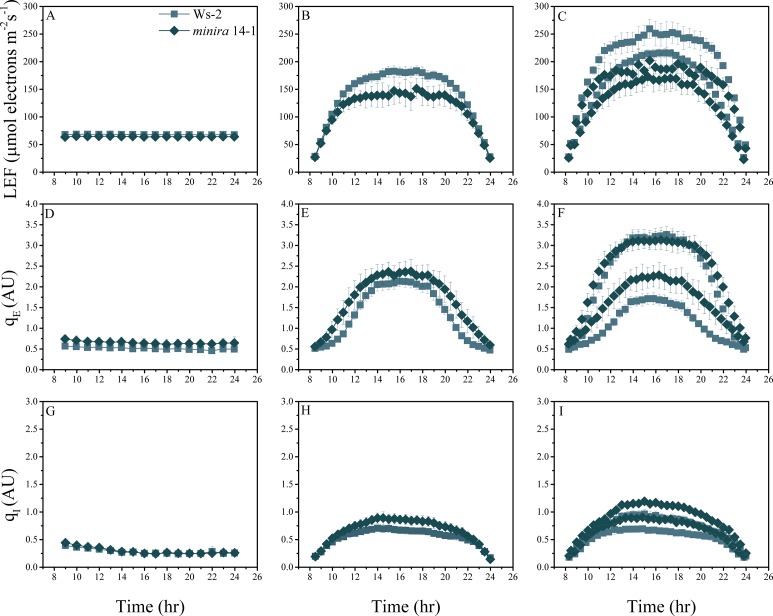
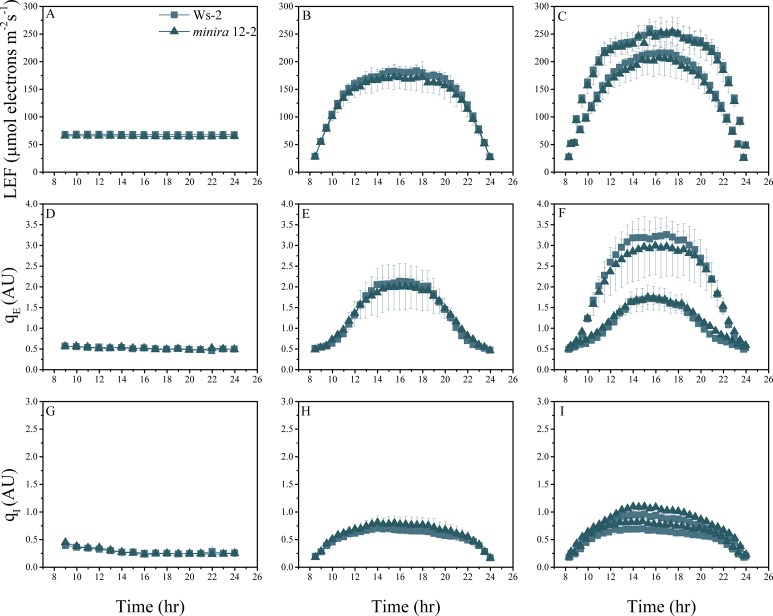
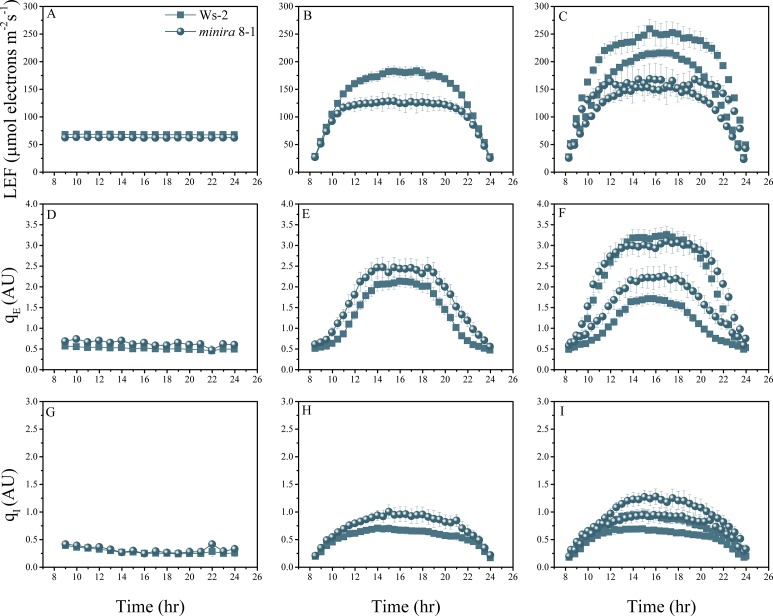
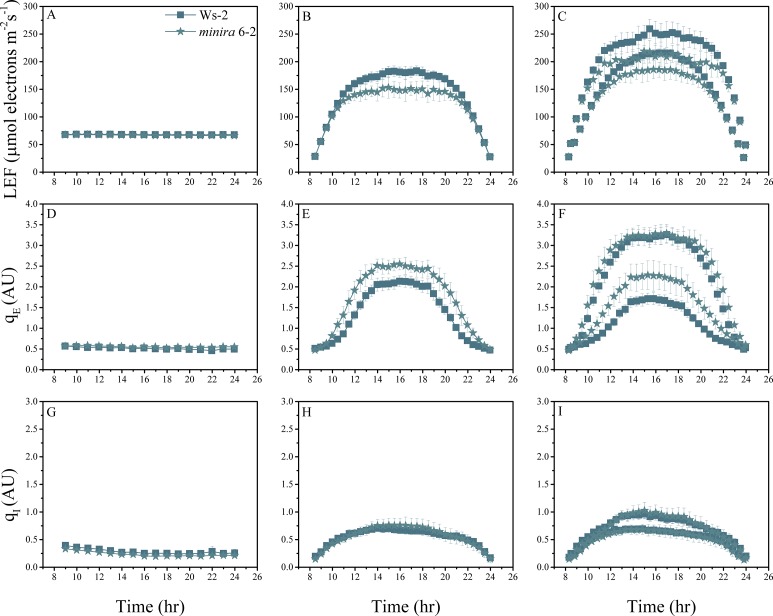
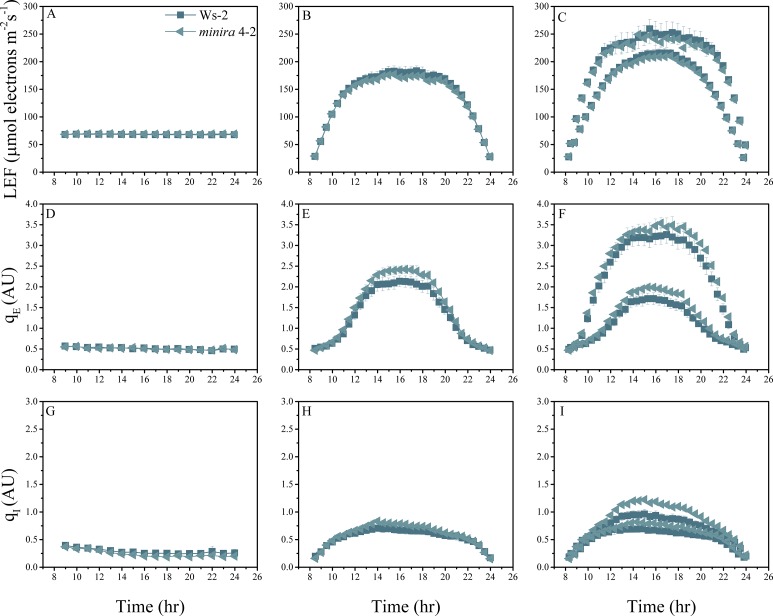
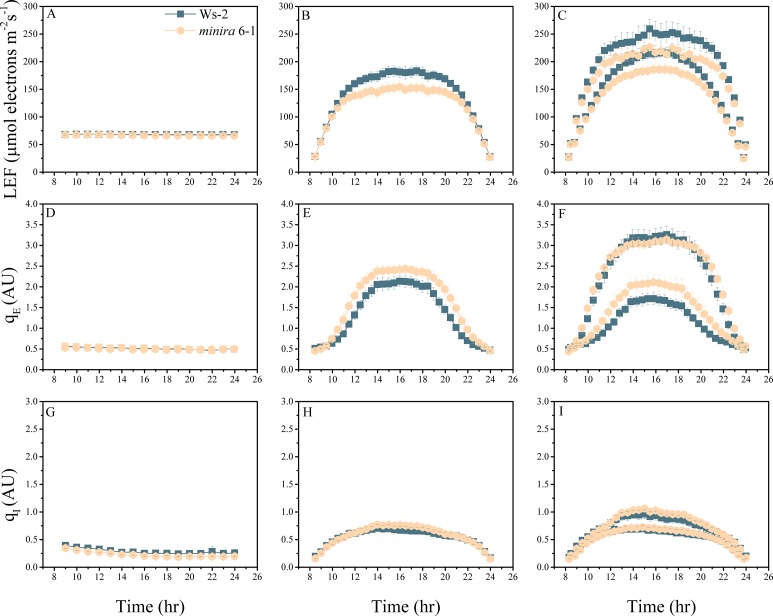
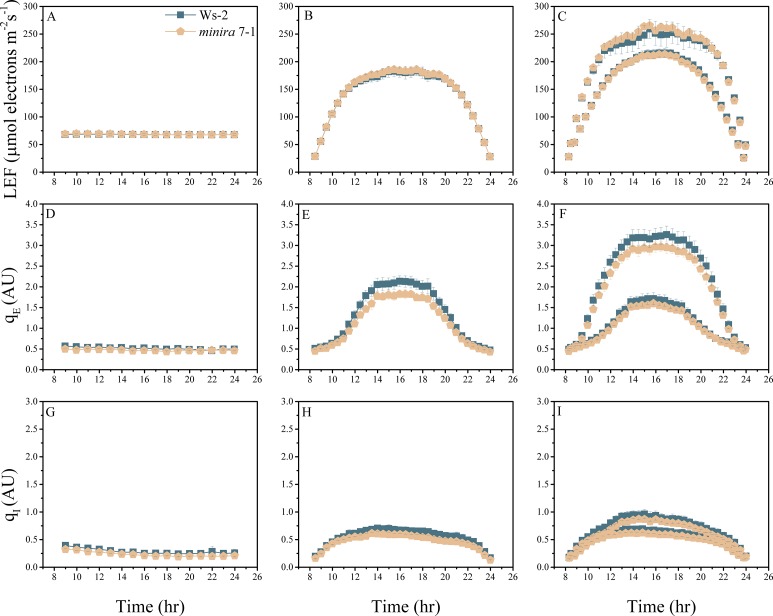
The minira library was then screened for photosynthetic phenotypes using whole plant chlorophyll fluorescence imaging (Cruz et al., 2016) over a consecutive three-day photoperiod (Figure 2A). Photosynthetic parameters were calculated from these images (videos are shown in videos 1–9) and shown as kinetic traces (Figure 2—figure supplements 1–15) or as log-fold changes compared to wild type (Figure 2B–D). Under 'standard' laboratory growth chamber lighting on day one, most minira lines showed relatively small differences from wild type in LEF (Figure 2B), qE (Figure 2C), and photoinhibitory quenching (qI) (Figure 2D), with the exceptions of minira 3–1 and 11–1, which also showed the most severe decreases in gH+ (Figure 1C). Stronger photosynthetic phenotypes appeared on days two and three, implying that the decreased gH+ and pmf effects were enhanced by intense or fluctuating illumination.
Video 1. (phi2_movie_labeled_day1): Whole plant PSII quantum efficiency (ΦII) during constant illumination.
False-colored chlorophyll fluorescence images of whole plants obtained during constant, 100 μmol photons m−2 s−1 actinic illumination over a 16 hr photoperiod. Measurements and calculations were performed as described in Materials and methods. Images were false colored according to calculated values, from magenta (high) to blue (low).
Video 2. (phi2_movie_labeled_day2): Whole plant PSII quantum efficiency (ΦII) during sinusoidal illumination.
False-colored chlorophyll fluorescence images of whole plants obtained during sinusoidal actinic illumination over a 16 hr photoperiod. Measurements and calculations were performed as described in Materials and methods. Images were false colored according to calculated values, from magenta (high) to blue (low).
Video 3. (phi2_movie_labeled_day3): Whole plant PSII quantum efficiency (ΦII) during fluctuating sinusoidal illumination.
False-colored chlorophyll fluorescence images of whole plants obtained during fluctuating sinusoidal actinic illumination over a 16 hr photoperiod. Measurements and calculations were performed as described in Materials and methods. Images were false colored according to calculated values, from magenta (high) to blue (low).
Video 4. qE_movie_labeled_day1): Whole plant pH-dependent quenching (qE) during constant illumination.
False-colored chlorophyll fluorescence images of whole plants obtained during constant, 100 μmol photons m−2 s−1 actinic illumination over a 16 hr photoperiod. Measurements and calculations were performed as described in Materials and methods. Images were false colored according to calculated values, from magenta (high) to blue (low).
Video 5. (qE_movie_labeled_day2): Whole plant pH-dependent quenching (qE) during sinusoidal illumination.
False-colored chlorophyll fluorescence images of whole plants obtained during sinusoidal actinic actinic illumination over a 16 hr photoperiod. Measurements and calculations were performed as described in Materials and methods. Images were false colored according to calculated values, from magenta (high) to blue (low).
Video 6. (qE_movie_labeled_day3): Whole plant pH-dependent quenching (qE) during fluctuating sinusoidal illumination.
False-colored chlorophyll fluorescence images of whole plants obtained during fluctuating sinusoidal actinic illumination over a 16 hr photoperiod. Measurements and calculations were performed as described in Materials and methods. Images were false colored according to calculated values, from magenta (high) to blue (low).
Video 7. (qI_movie_labeled_day1): Whole plant photoinhibitory quenching (qI) during constant illumination.
False-colored chlorophyll fluorescence images of whole plants obtained during constant, 100 μmol photons m−2 s−1 actinic illumination over a 16 hr photoperiod. Measurements and calculations were performed as described in Materials and methods. Images were false colored according to calculated values, from magenta (high) to blue (low).
Video 8. (qI_movie_labeled_day2): Whole plant photoinhibitory quenching (qI) during sinusoidal illumination.
False-colored chlorophyll fluorescence images of whole plants obtained during sinusoidal actinic actinic illumination over a 16 hr photoperiod. Measurements and calculations were performed as described in Materials and methods. Images were false colored according to calculated values, from magenta (high) to blue (low).
Video 9. (qI_movie_labeled_day3): Whole plant photoinhibitory quenching (qI) during fluctuating sinusoidal illumination.
False-colored chlorophyll fluorescence images of whole plants obtained during fluctuating sinusoidal actinic illumination over a 16 hr photoperiod. Measurements and calculations were performed as described in Materials and methods. Images were false colored according to calculated values, from magenta (high) to blue (low).
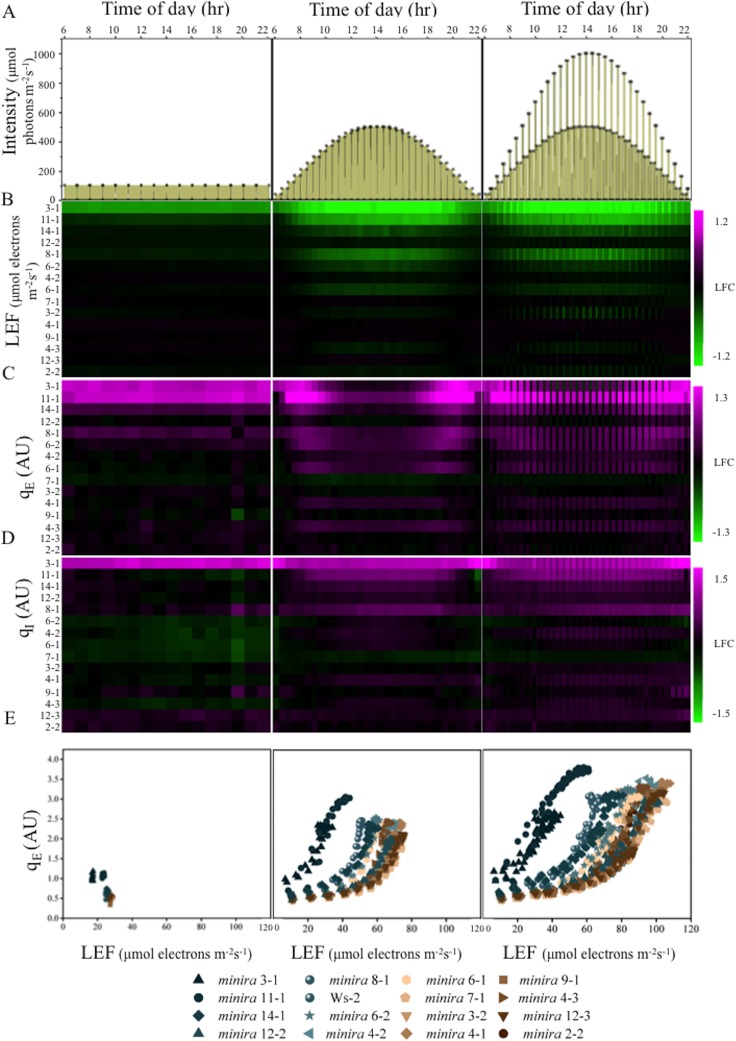
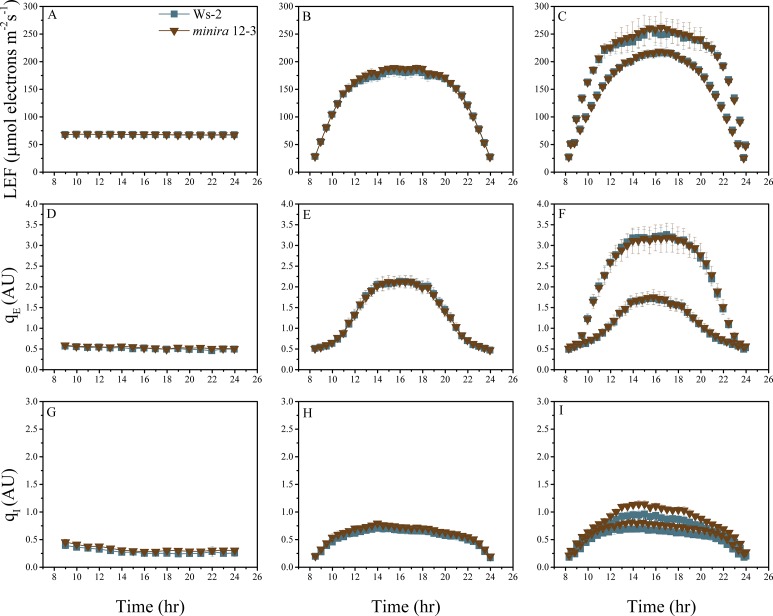
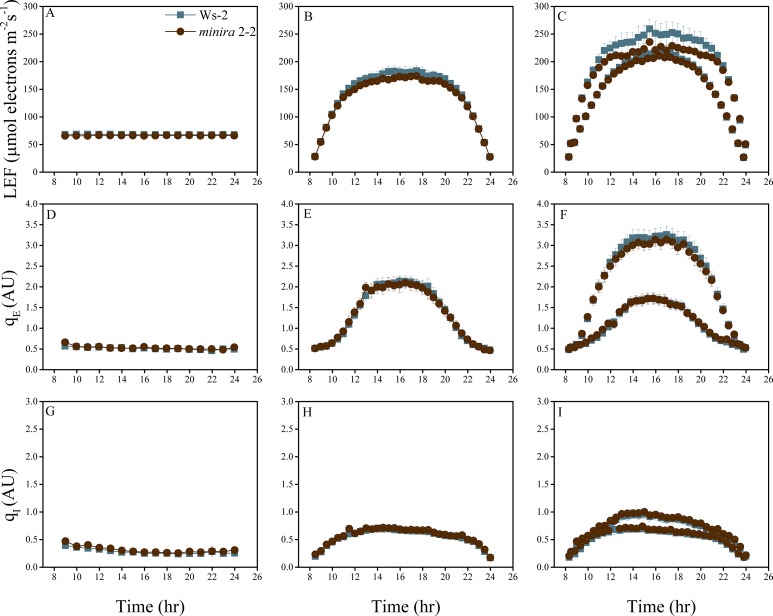
Figure 2. Dynamic light conditions enhance pmf dependent phenotypes.
Whole plant fluorescent images were captured over three days under the illumination conditions displayed in Panel A and listed in Supplementary file 2. Plants were illuminated over the 16-hr photoperiod, shown as yellow filled areas representing the light intensity when present in A, under either a constant light intensity (day one), a sinusoidal photoperiod (day two), or a sinusoidal photoperiod interrupted by fluctuations in light intensity (day three). Square symbols in Panel A indicate each light intensity change. Steady-state fluorescence parameters were captured for each plant at the end of each light condition. Panels B,C and D represent the responses of LEF, qE and qI respectively (n ≥ 3). Data are shown as log2-fold changes compared to the wild type. Kinetic data including wild type are shown in Figure 2—figure supplements 1–15. The rows were sorted in order of ascending gH+ values measured as in Figure 1C. Panel E plots the dependence of the mean (n ≥ 3) of qE against the linear electron flow (LEF) for each time point measured for the day. For visualization purposes the error bars have been omitted from E.
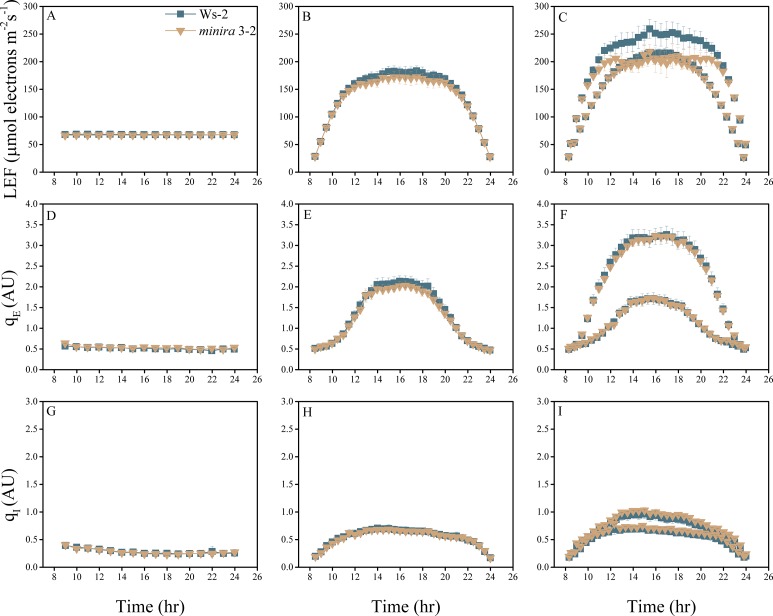
Figure 2—figure supplement 1. Whole plant fluorescence imaging phenotyping of minira 3–1 mutant.
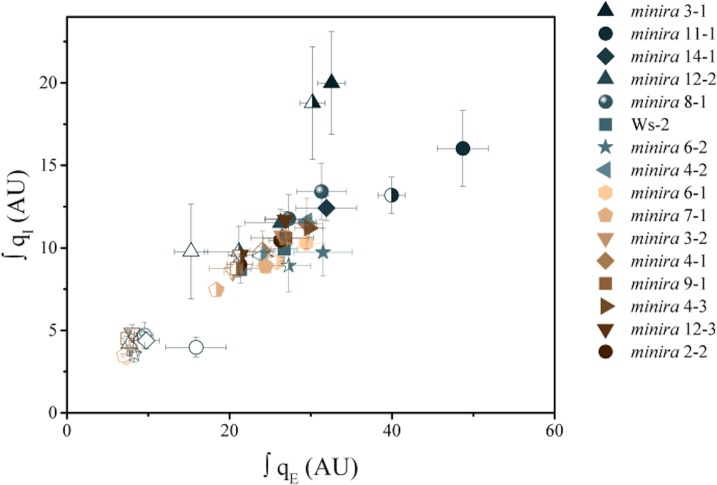
Figure 2—figure supplement 2. Increased pH-dependent quenching correlates with increased photoinhibitory quenching.
Figure 2—figure supplement 3. Whole plant fluorescence imaging phenotyping of minira 11–1 mutant.
Figure 2—figure supplement 4. Whole plant fluorescence imaging phenotyping of minira 14–1 mutant.
Figure 2—figure supplement 5. Whole plant fluorescence imaging phenotyping of minira 12–2 mutant.
Figure 2—figure supplement 6. Whole plant fluorescence imaging phenotyping of minira 8–1 mutant.
Figure 2—figure supplement 7. Whole plant fluorescence imaging phenotyping of minira 6–2 mutant.
Figure 2—figure supplement 8. Whole plant fluorescence imaging phenotyping of minira 4–2 mutant.
Figure 2—figure supplement 9. Whole plant fluorescence imaging phenotyping of minira 6–1 mutant.
Figure 2—figure supplement 10. Whole plant fluorescence imaging phenotyping of minira 7–1 mutant.
Figure 2—figure supplement 11. Whole plant fluorescence imaging phenotyping of minira 3–2 mutant.
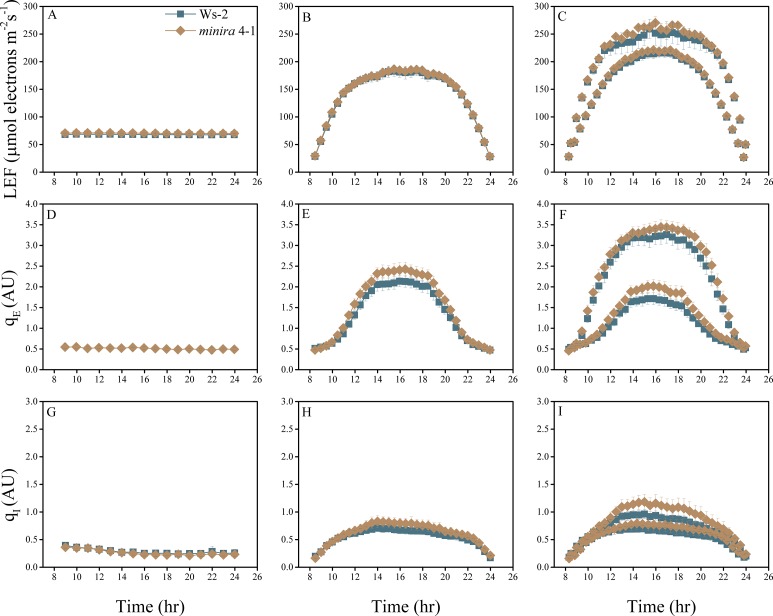
Figure 2—figure supplement 12. Whole plant fluorescence imaging phenotyping of minira 4–1 mutant.
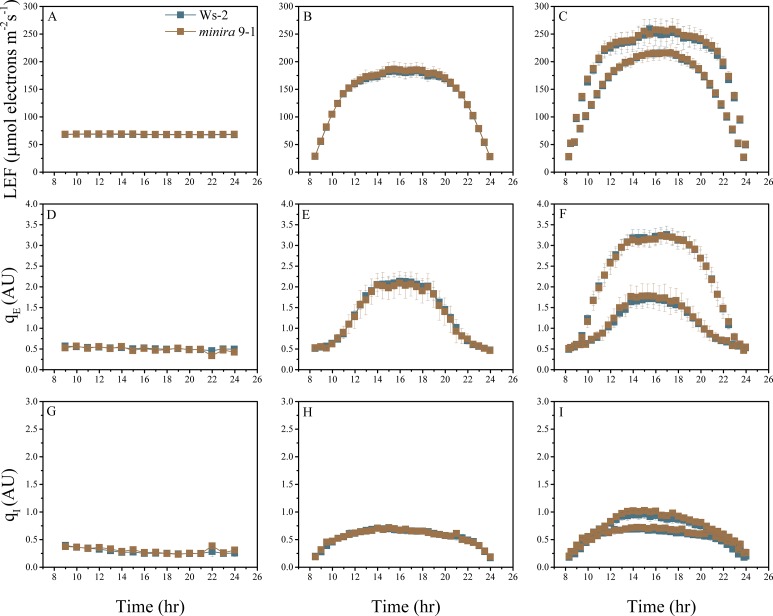
Figure 2—figure supplement 13. Whole plant fluorescence imaging phenotyping of minira 9–1 mutant.
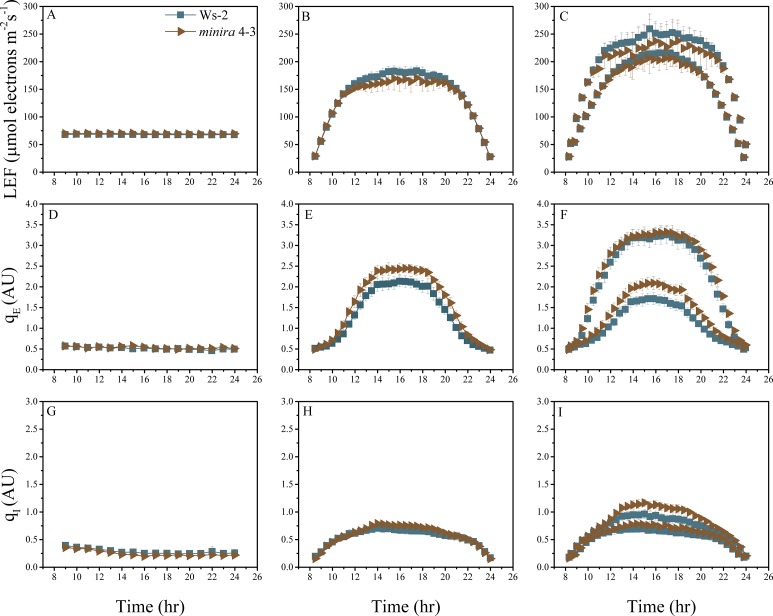
Figure 2—figure supplement 14. Whole plant fluorescence imaging phenotyping of minira 4–3 mutant.
Figure 2—figure supplement 15. Whole plant fluorescence imaging phenotyping of minira 12–3 mutant.
Figure 2—figure supplement 16. Whole plant fluorescence imaging phenotyping of minira 2–2 mutant.
In general, LEF decreased with decreasing gH+ (Figure 2B, rows are ordered by increasing gH+) while qE increased (Figure 2C). The increases in qE were especially pronounced at higher light intensities on days two and three. These effects can be explained by slowing of proton efflux through the ATP synthase in the mutants that results in increased pmf for a given LEF. This is reflected in the higher lumen pH-sensitive qE response for a given LEF (Figure 2E), which is strikingly similar to that attributed to ATP synthase regulation in wild type plants during limitations in carbon fixation (Kanazawa and Kramer, 2002) or decreases in ATP synthase content (Rott et al., 2011). The qE sensitivities for the mutants remained similar throughout the experiments, i.e. the data for each mutant followed similar curves, implying that the ATP synthase activities were relatively constant within a particular line, consistent with a lack of light-dependent modulation of gH+ that has previously been observed (Avenson et al., 2005). The extents of qE did not exceed about 3.5 units in any of the lines, suggesting that either the lumen pH was restricted to a moderate acidity or that the capacity of the qE response was saturated as light intensities increased.
We also observed a strong correlation between increased qE and qI, (for quantitative comparisons, see Figure 2—figure supplement 2), implying that decreases in ATP synthase activity in the mutants led to not only higher photoprotection but also higher rates of PSII photoinhibition. This result appears counterintuitive, in that we would expect the photoprotective qE response to prevent photoinhibition. Particularly striking was the relative loss in qE near the peak light intensities on days two and three in the most strongly affected minira lines (Figure 2C). We attribute this effect to strong accumulation of PSII photoinhibition (Figure 2D) leading to the loss of photosynthetic capacity (see Figure 2B) that limited acidification of the thylakoid lumen.
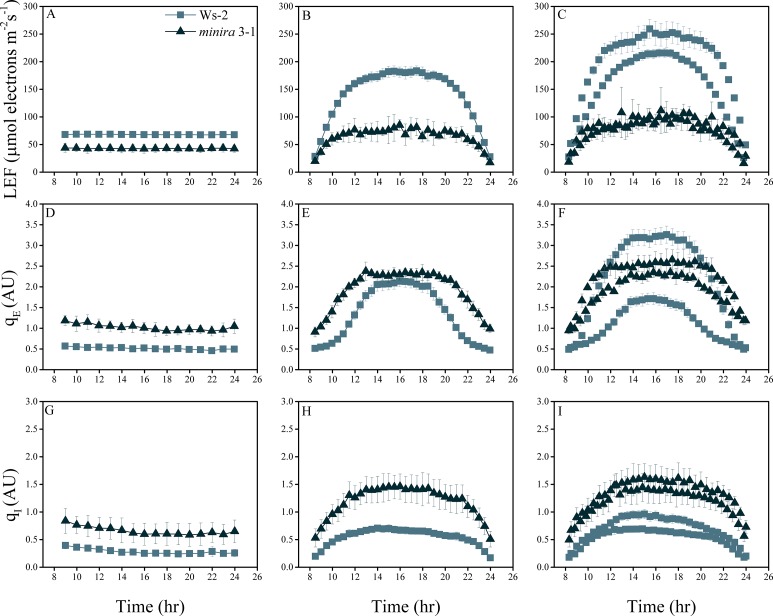
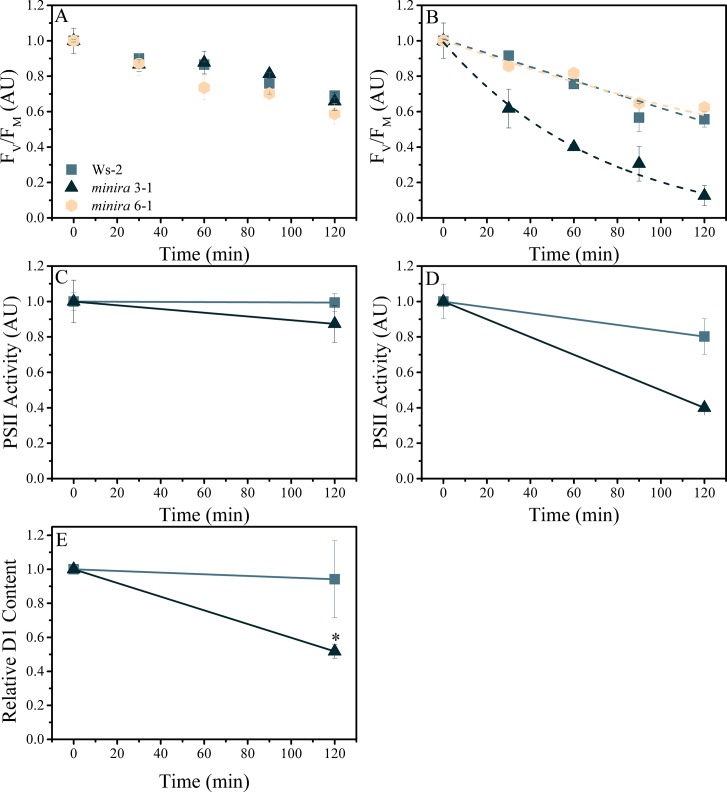
The observed increases in photoinhibition at high pmf could be caused by several mechanisms. It has been proposed that photoinhibition is primarily controlled by modulating the rate of PSII repair, i.e. the rate of damage is dependent solely on light intensity but repair being inhibited by stress-induced ROS production (Nishiyama and Murata, 2014). However, blocking PSII repair with the chloroplast translation inhibitor lincomycin (Tyystjarvi and Aro, 1996) revealed that, when compared to wild type or minira lines with wild type like gH+ (e.g. minira 6–1), minira lines with low gH+ and high pmf (e.g. minira 3–1, Figure 1C, Figure 3) had higher rates of photodamage as reflected in both decreased maximal PSII quantum efficiency and loss of t capacity to perform charge separation in PSII (Joliot and Delosme, 1974; Joliot et al., 1977; Bailleul et al., 2010) and significantly decreased levels of D1 protein (Figure 3C–E). Thus, decreasing ATP synthase activity led to increased PSII photodamage rather than decreased rates of repair, in contradiction with strict control of photoinhibition by repair (Takahashi et al., 2007).
Figure 3. Elevated pmf leads to PSII photodamage.
Detached leaves were infiltrated with either water (A,C) or a 3 mM solution of lincomycin (B,D) and treated with 1000 μmol photons m−2s−1 red light for the times indicated. Following dark adaptation, FV/FM values were obtained (A,B) for treated leaves of wild type (square, initial FV/FM 0.75 ± 0.006 and 0.76 ± 0.012 for water and lincomycin, respectively), minira 3–1 (triangle, initial FV/FM 0.60 ± 0.07 and 0.54 ± 0.10 for water and lincomycin, respectively), and minira 6–1 (hexagon, initial FV/FM 0.72 ± 0.004 and 0.73 ± 0.01 for water and lincomycin, respectively) (mean ± s.d., n ≥ 3). Data were normalized to the initial dark-adapted FV/FM values to remove intrinsic differences between the three lines. In panel B, dashed lines represent the best fit curves for a single exponential decay. The ability of photoinhibited leaves to perform PSII charge separation was determined in Ws-2 and minira 3–1 by measuring the ECS absorbance changes following two consecutive single-turnover saturating flashes in the presence of DCMU (C,D). Leaves infiltrated with water (C, initial amplitudes of 8.88 × 10−4 ± 9.0 × 10−5 and 9.59 × 10−5 ± 3.6 × 10−5 for wild type and minira 3–1, respectively) or 3 mM lincomycin (D, initial amplitudes of 8.59 × 10−4 ± 1.2 × 10−4 and 1.63 × 10−4 ± 3.1 × 10−5 for wild type and minira 3–1, respectively) were infiltrated with DCMU following the indicated light treatment time and dark adaptation. PSII activity was determined by subtracting the ECS amplitude induced by the second flash from the ECS amplitude induced by the first flash (mean ± s.d., n ≥ 4). Loss of the PSII reaction center D1 protein over the time course of illumination in lincomycin treated wild type and minira 3–1 leaves (E). Leaves treated as in panel B were analyzed by western blot (n = 4) using an α-PsbA antibody and the 32 kDa band was quantified. Band intensities were normalized to the time zero point for each genotype within a single blot to control for differences in development intensities. Statistically significant differences (*p<0.05) from wild type were determined using a t-test.
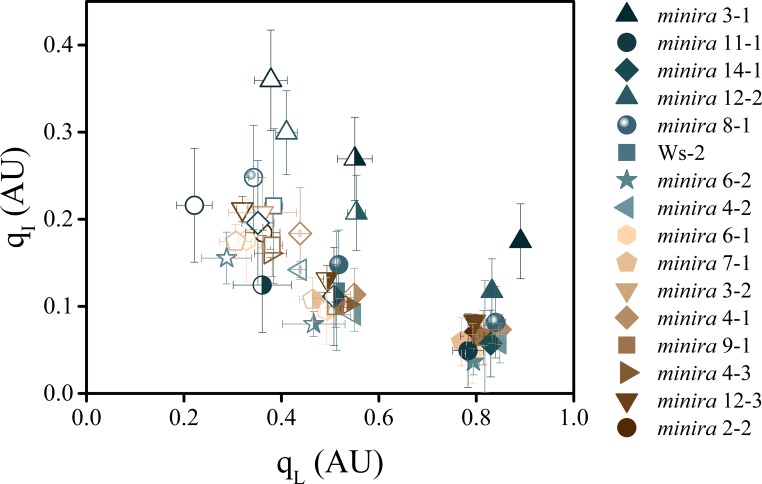
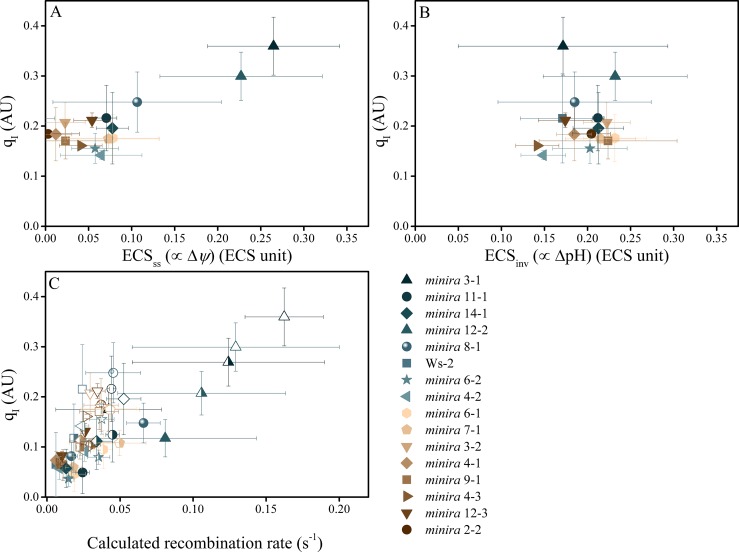
Photodamage can be induced in vitro by excitation of PSII centers with previously reduced primary quinone acceptor (QA) leading to the formation of the doubly-reduced QAH2 state (Keren and Krieger-Liszkay, 2011). This situation might be expected if a high pmf slowed electron transfer through the b6f complex, resulting in the accumulation of electrons on PSII acceptors. When data at a range of light intensities are compared, the relationship between the QA redox state (qL) (Kramer et al., 2004) and qI is statistically significant (ANOVA, p=2 × 10−16) (Figure 4). However, both the light intensity (p=3 × 10−5) and qE (p=7 × 10−3) are significant interacting factors. At any one light intensity, qL was relatively stable, whereas qI was strongly dependent upon the mutant background and underlining pmf changes, indicating that while QA reduction may be a contributing factor, it cannot by itself explain the observed extents of photoinhibition in the minira lines. On the other hand, this dependence is also consistent with an alternative model, proposed below, that involves effects on PSII recombination rates.
Figure 4. The dependence of photoinhibition on the redox state of QA.
The redox state of the primary electron acceptor QA was assayed using the qL fluorescence parameter concurrently with photoinhibitory quenching qI at 100 (solid symbols), 300 (half filled symbols), and 500 μmol photons m−2s−1 (open symbols, mean ± s.d., n = 3). Plants were exposed to at least 10 min of actinic illumination prior to qL measurement, and qI measured after 10 min of dark relaxation. While the extent of qI varies between plants, the relative redox state of QA remains similar between all plants within each actinic light intensity.
Changes in chlorophyll content have also been correlated with increases in PSII photoinhibition (Pätsikkä et al., 2002), likely due to less light being absorbed at the leaf surface and the subsequent increased light penetration into the leaf reaching more PSII centers. While the leaf chlorophyll content was altered in the minira mutants from wild type levels (Supplementary file 3), the leaf chlorophyll content does not fall below where (Pätsikkä et al., 2002) observed correlations between a lack of chlorophyll content and photoinhibition.
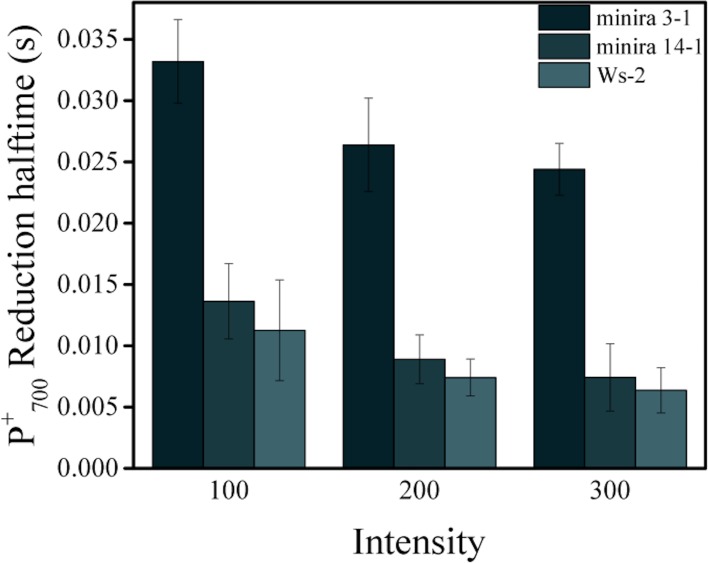
We next hypothesized that the most probable explanation for the increased photoinhibition is direct sensitization of PSII to photodamage by pmf. In the 'acid-damage' model (Kramer et al., 1999), it was proposed that excessive lumen acidification at high ΔpH (i.e. low lumen pH) could sensitize PSII centers to photodamage. To test this possibility, we compared the rates of photoinhibition with the extents of the ΔpH and Δψ components of the pmf by measuring the relaxation kinetics of the ECS signal (Takizawa et al., 2007). Surprisingly, increased extents of photoinhibition in low gH+ lines were not correlated with ΔpH, but were with Δψ (Figure 5), contrary to what was expected with the acid-damage model. Consistent with this result, the rates of P700+ reduction, which reflect the lumen pH-sensitive turnover of the cytochrome b6f complex remained in a range consistent with a lumen pH above or near the pKa for b6f down-regulation, i.e. above about 6.0 (Figure 5—figure supplement 1).
Figure 5. Photoinhibition is strongly correlated with Δψ but not ΔpH in minira lines.
Photoinhibition, estimated by the qI fluorescence parameter, is plotted against either the ΔpH or Δψ components of pmf, estimated by the ECSss (A) and ECSinv (B) parameters, as described in Materials and methods. Measurements shown were taken during exposure to 500 μmol photons m−2s−1 actinic light (mean ± s.d., n = 3). Two-way analysis of variance (ANOVA) of all combined data, 15 minira lines and wild type, showed a stronger correlation between qI and Δψ (F = 9.5, p=0.003) than ΔpH (F = 4.05, p=0.05). This correlation is also seen with the expected pH-dependent alterations of P700+ reduction for the observed partitioning differences from wild type (Figure 5—figure supplement 1) and an increase in the fraction of total pmf stored as Δψ at the expense of ΔpH over multiple light intensities (Figure 5—figure supplement 2). Increased storage of pmf as Δψ is also observed in tobacco ATP synthase knock-down plants (Figure 5—figure supplement 3). ECS units were defined as the deconvoluted ΔA520 μg chlorophyll−1 cm2. The influence of Δψ on the rate of PSII recombination was estimated based on the change in the equilibrium constant for the sharing of electrons between pheophytin and QA (described in Materials and methods) (C). The influence of Δψ on the calculated recombination rate taking into account the fraction of reduced QA using the equations described in Materials and methods and described in the main text. Data were obtained at 100 (solid symbols), 300 (half filled symbols), and 500 μmol photons m−2s−1 (open symbols) (mean ± s.d., n = 3).
Figure 5—figure supplement 1. Reduction kinetics of P700+.
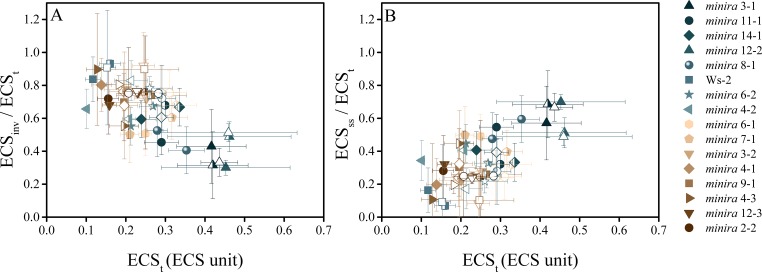
Figure 5—figure supplement 2. The electric field component of the pmf dominates under high pmf conditions.
Figure 5—figure supplement 3. Tobacco ATPC1 antisense knockdown increase Δψ partitioning under high pmf conditions.
These results are consistent with an increase in the partitioning of pmf into Δψ as the total pmf increased (Figure 5—figure supplement 2), as have been observed previously (Avenson et al., 2004), and are likely caused by alterations in ion movements across both the thylakoid and chloroplast envelope membranes (Avenson et al., 2004; Armbruster et al., 2014; Kunz et al., 2014). Our results suggest that pmf partitioning acts to maintain a permissible lumen pH during large pmf increases.
We observed similar results throughout the range of minira mutants with low gH+ as well as with tobacco γ-subunit antisense plants (Rott et al., 2011) with reduced ATP synthase complexes, finding that a large increase in total pmf was accompanied by increased partitioning of pmf into Δψ (Figure 5—figure supplement 3), and increased rates of photodamage in the presence and absence of lincomycin (Figure 5—figure supplement 3). These results suggest that low gH+ or high pmf-related PSII damage is a more general phenomenon, which is related to excess Δψ and likely to be independent of such factors as changes in the protein or supercomplex content (see also discussion in Rott et al., 2011).
As described in Figure 8, we hypothesize that high Δψ accelerates photodamage by favoring recombination reactions within PSII (de Grooth and van Gorkom, 1981; Diner and Joliot, 1976; Satoh and Katoh, 1983; Rappaport et al., 1999) that lead to the formation of chlorophyll triplet states that in turn generate 1O2 (Johnson et al., 1995).
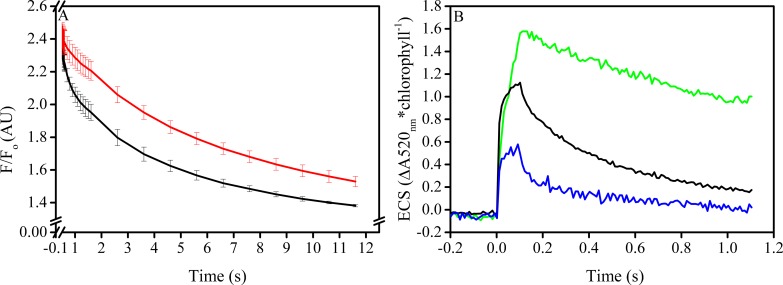
To test this model, we studied the relationship between elevated Δψ and PSII charge recombination in isolated spinach thylakoids (Figure 6), which unlike Arabidopsis can be isolated as highly intact chloroplasts and tightly coupled thylakoids. Consistent with our model, we found that elevated Δψ, produced by artificial decyl-ubiquinol mediated cyclic electron flow through PSI, increased the rate of charge recombination from the S2QA− state compared to samples treated with gramicidin to dissipate Δψ (Figure 6A). This recombination reaction can also occur in vivo during normal turnover when the S2QA− state is formed given that the equilibrium constant for sharing of electrons between QA and QB is small (Robinson and Crofts, 1983). The extent of Δψ generated in these experiments was similar to that observed in minira leaves under photoinhibitory conditions (Figure 6B), suggesting similar increases in recombination rates should occur in vivo.
Figure 6. In vitro manipulation of the pmf Δψ alters PSII S2QA− recombination rates.
Isolated spinach thylakoids in the presence of 5 μM spinach ferredoxin and 10 μm sodium ascorbate were treated with 3–3,4-dichlorophenyl 1,1-dimethylurea (DCMU) to block PSII forward electron transfer, and a trans-thylakoid pmf generated utilizing decyl-ubiquinol mediated PSI cyclic electron transfer (A). Recombination from the S2QA− state was probed by observing the decrease in the high fluorescence state associated with QA− following a short (100 ms) actinic flash to dark-adapted thylakoids (black line). Depletion of the pmf, which under these conditions is stored almost exclusively as Δψ, in the presence of 25 μM gramicidin (red line) resulted in an approximate 5-fold increase in the initial rate of decay when Δψ was largest, and an overall 2-fold increase in the lifetime of the high fluorescence state. The extent of Δψ generated by the 100 ms light (B), estimated by the ECS signal measured at 520 nm and normalized to chlorophyll content. Thylakoids were assayed in the absence of inhibitors (green line), in the presence of DCMU (blue line), and in the presence of both DCMU and 50 μM decyl-ubiquinol (black line) to generate pmf through PSI turnover, corresponding to the condition used in panel A.
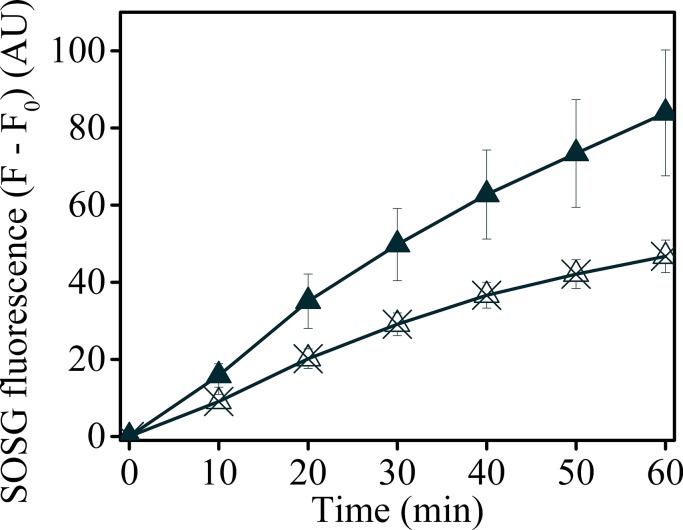
We next tested the predicted connection between elevated Δψ and singlet oxygen (1O2) generation (Figure 7D). In wild type leaves, moderate illumination (30 min of 300 μmol photons m−2s−1) resulted in no detectable light dependent changes in 1O2 (Figure 7D) using Singlet Oxygen Sensor Green (SOSG) dye fluorescence (Dall'Osto et al., 2012; Ramel et al., 2013; Shumbe et al., 2016). In contrast, the low gH+minira 3–1 line showed a strong induction of SOSG fluorescence within the first 10 min of illumination, which saturated by about 30 min. Infiltration of leaves with valinomycin, a potassium ionophore that decreases the Δψ component of pmf (Satoh and Katoh, 1983), partially inhibited the rise in in SOSG fluorescence (Figure 7—figure supplement 1). While care must be taken making quantitative estimates of 1O2 from SOSG fluorescence (Koh and Fluhr, 2016), within the limits of these experiments the Δψ-dependence of the SOSG fluorescence increases strongly support a role for Δψ-induced 1O2 production from photosynthesis.
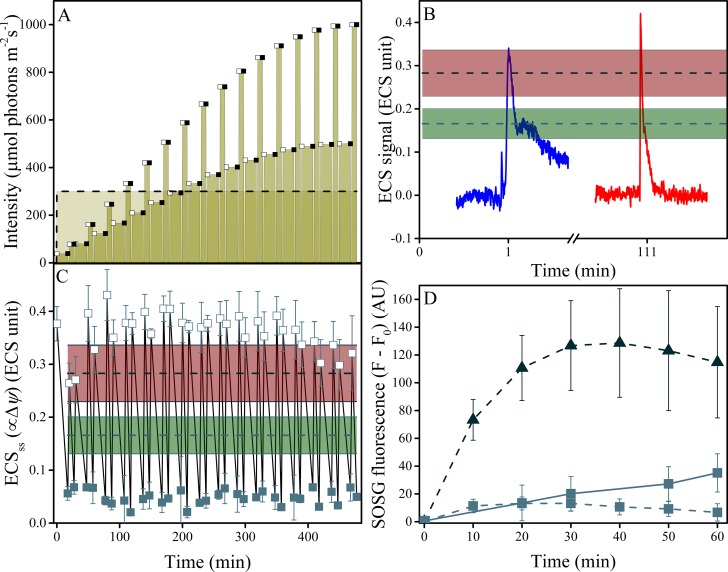
Figure 7. Induction of Δψ and 1O2 production under fluctuating light in wild type plants.
(A) Illumination conditions and measurement points used in the experiments. Fluctuating light conditions (replicating Figure 2 day three) are shown as connected points, with open squares representing measurements obtained 10 s after the light transition and closed squares the end of steady-state illumination. Constant illumination of 300 μmol m−2s−1 is represented as a dotted line. (B) Representative traces of the light-fluctuation induced ECS signals resulting in transient ECS 'spikes' are shown for the first fluctuation (dark to 39 μmol m−2s−1, blue) and the fluctuation from 167 to 333 μmol m−2s−1, red). A full set of ECS kinetic 'spikes' following increased light fluctuations can be found in Figure 7—figure supplement 2. The extents of light-induced Δψ in wild type (green line and shaded box, indicating mean ± s.d) and minira 3–1 (red lines and box, indicating mean ± s.d) at 300 μmol m−2s−1 are shown for comparison. (C) The extents of light-induced Δψ, estimated using the ECSss parameter over the time-course of the fluctuating light experiment, compared to those obtained under continuous illumination in wild type and minira 3–1 (green and red lines and boxes, respectively, as in Panel B). Open and closed squares correspond to the ECSss measurements taken at the timing designated in panel A. (D) Time-course of SOSG fluorescence changes for wild type (squares) and minira 3–1 (triangles) during exposure to constant 300 μmol m−2s−1 (dotted lines)and wild type leaves under the first hour of fluctuating light (solid line). A decrease in SOSG fluorescence occurs when Δψ is collapsed with the addition of the ionophore valinomycin (Figure 7—figure supplement 1). All data in A, B, and C represent mean (n ≥ 3) ± s.d. ECS units were defined as the deconvoluted ΔA520 μg chlorophyll−1 cm2.
Figure 7—figure supplement 1. Uncoupling Δψ decreases SOSG fluorescence in minira 3–1.
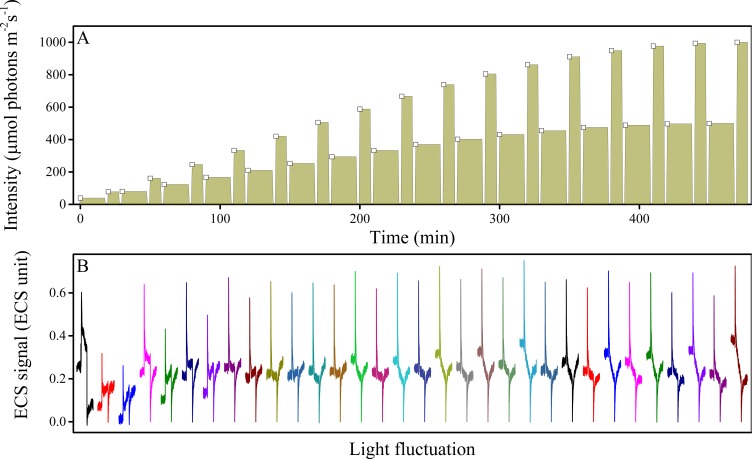
Figure 7—figure supplement 2. Fluctuations in light intensity result in transient ECS spikes.
Singlet oxygen is produced by the interaction of O2 with triplet excited states of pigments, most likely from the 3P680 chlorophyll within the PSII reaction centers generated by recombination reactions (Krieger-Liszkay et al., 2008; Telfer, 2014), as further discussed below. It is unlikely that such triplets could be generated in the bulk light harvesting pigments because high NPQ in the mutants will decrease the lifetime of antenna excited states, and chlorophyll triplets generated in light harvesting complexes are efficiently quenched by carotenoids (Telfer et al., 2008). We thus propose that elevated Δψ induces 1O2 production by accelerating PSII recombination in low gH+ mutants when pmf is large. Keren et al. (Keren et al., 1997) suggested that recombination-induced triplet formation could explain the photoinhibitory effects of very low light, when PSII charge recombination is preferred over the forward electron transfer reactions. Our work implies that this type of phenomenon is greatly accelerated by high Δψ, potentially making it relevant to photosynthesis under growth light conditions.
The effects of high Δψ would be expected to alter the rates of PSII recombination through P+Pheo− (where P+ is the oxidized primary chlorophyll donor and Pheo the D1 subunit pheophytin) in an increasing dependence upon the fraction of QA−, consistent with the observed correlation between qI and qL as the light intensity increased (Figure 4). To test this relationship, we estimated the recombination rates from S2QA− through the P+Pheo− pathway, considering QA redox state (estimated by 1-qL) and the expected impact of Δψ on the equilibrium constant for sharing electrons between Pheo and QA (based on ECSss and the position of QA in the structure relative to the membrane dielectric). The basis of this estimate is described in more detail in Materials and methods. As shown in Figure 5C, we see a positive correlation between qI and estimated recombination through P+Pheo− over both mutant variants and light intensities, indicating that the combined effects of Δψ and QA redox state can explain a large fraction of the observed extents of photoinhibition. While it is likely that multiple mechanisms of photoinhibition exist, which may also explain some of the qI variation, overall the greatest impact upon photoinhibition under these conditions can be explained by Δψ-mediated changes in PSII electron recombination.
The obvious questions are: does Δψ-induced photoinhibition occur in wild type plants and if so under what conditions? Photosynthesis is known to be particularly sensitive to rapid fluctuations in light intensity (Kulheim et al., 2002), at least some of this sensitivity is associated with photoinhibition of PSI, especially in cyanobacteria (Allahverdiyeva et al., 2015). However, such fluctuations should also result in large transient changes in Δψ, as the thylakoid membrane has a low electrical capacitance and low permeability to counter-ions, while the lumen and stroma have high proton buffering capacity (Cruz et al., 2001). The slow onset of qE and other down-regulatory processes in photosynthesis should exacerbate these effects and allow for large fluxes of electrons when light levels are rapidly increased, resulting in large, transient Δψ 'pulses'.
We therefore hypothesized that Δψ-induced photodamage may contribute to the increased photodamage seen under fluctuating light. To test this possibility, we measured light-driven pmf (Δψ and ΔpH) and 1O2 generation in wild type Arabidopsis under fluctuating light conditions (Figure 7A, replicating the first 8 hr of Figure 2A day three). The initial dark-to-light transition resulted in an immediate, transient 'spike'in Δψ, even though the light intensity was low (39 μmol photons m−2s−1, Figure 7B). The spike was transient, and decreased to steady-state levels within tens of seconds, as shown in the blue trace in Figure 7B. Spikes in Δψ of similar amplitudes were also seen upon each increase in light at the onset of each fluctuation, though the recovery kinetics tended to be more rapid than those seen at the first dark-light transient. An example of these transients, taken at the transition between 167 and 333 μmol photons m−2 s−1 is shown in (Figure 7B). A more complete set of transient kinetics, over the entire course of the experiment is presented in Figure 7—figure supplement 2. The amplitudes of these Δψ transients were similar to or larger than those seen in the minira 3–1 line under constant 300 μmol photons m−2 s−1 light, which also induced 1O2 generation (see red horizontal bars in Figure 7B). These spikes reflect increases in Δψ above that already produced by steady-state photosynthesis, so the true extent of Δψ is likely considerably higher, and based on estimates of the calibration of the ECS signal likely range between 150–260 mV (see Figure 7—figure supplement 2).
By contrast, wild type plants under constant 300 μmol photons m−2 s−1 produced much lower Δψ extents (Figure 7B green bars) and had no detectible 1O2 generation (Figure 7D). These results suggest that even low amplitude light fluctuations are capable of inducing Δψ large enough to produce 1O2 and PSII photodamage. Supporting this interpretation, wild type leaves under these fluctuating light conditions produced substantial amounts of 1O2 during the first hour of fluctuating conditions compared to higher intensity, but constant illumination (Figure 7D).
The above results lead us to conclude that fluctuating light likely induces strong effects through Δψ-induced recombination reactions in PSII (see below for considerations of potential contributions to PSI). In addition to direct damage to the enzymes of photosynthesis, 1O2 can also activate plant light stress-related gene expression and programmed cell death (Zhang et al., 2014), suggesting a possible physiological linkage between pmf-enhanced recombination and plant regulatory pathways that may result in long-term acclimation to fluctuating light.
At a mechanistic level, we propose that imposing a large Δψ across the PSII complex will decrease the standard free energy gap between the vectorial electron transfer steps and thus the back-reaction will be accelerated in competition with forward (energy-storing) reactions (de Grooth and van Gorkom, 1981; Vos et al., 1991; Johnson et al., 1995) (Figure 8). It is known that decreasing the energy gap between Pheo and QA favors recombination from P+QA− via Pheo− (de Grooth and van Gorkom, 1981; Johnson et al., 1995) rather than directly from QA− to P+ (Johnson et al., 1995; Krieger-Liszkay and Rutherford, 1998; Rutherford et al., 2012). The observed increased recombination rates in vitro (Figure 6) when Δψ is present, combined with 1O2 production under high Δψ conditions in vivo suggest that the fraction of electrons recombining to P+ through a Pheo− intermediate is greatly increased, as the recombination pathway via the Pheo− has a high yield for formation of the triplet state of P (3P) that in turn can interact with O2 to produce 1O2 (Johnson et al., 1995; Keren and Krieger-Liszkay, 2011; Rutherford et al., 2012).
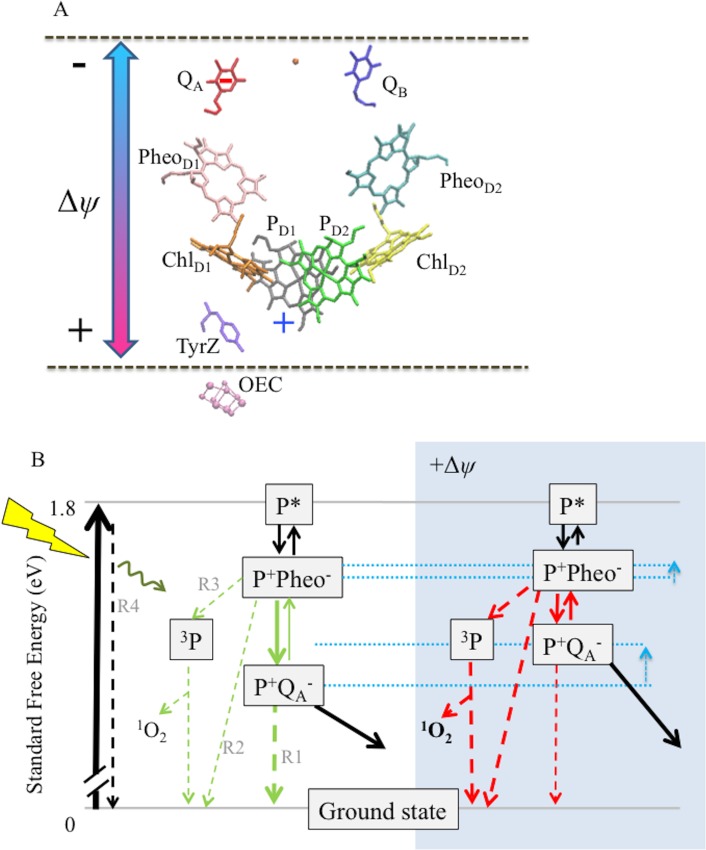
Figure 8. Schemes for the trans-thylakoid Δψ-induced acceleration of recombination reactions in PSII and subsequent production of 1O2.
(A) The relative positions of PSII electron transfer cofactors with respect to the electric field (double-headed arrow) imposed across the thylakoid membrane (dotted lines). The red and blue arrows indicate the Δψ-induced changes in the equilibrium constant for the sharing of electrons (−) between QA and Pheo, and electron holes (+) among P+ and the oxygen evolving complex. Excitation of PSII by light leads to formation of excited chlorophyll states (P*), the excitation is shared over the 4 chlorophylls and the 2 pheophytins. Charge separation occurs between more than one pair of pigments, so at short times the situation is not well defined, but ChlD1+Pheo D1− appears to be the dominant radical pair. Secondary electron transfer events occur forming PD1+PheoD1−, the second radical pair, which is present in nearly all centers. This radical pair is stabilized by electron transfer from Pheo− to QA forming PD1+QA−. This radical pair is further stabilized by electron transfer from D1Tyr161 (TyrZ) forming a neutral tyrosyl radical, which oxidizes the Mn cluster of the oxygen evolving complex to form the state Sn+1QA−. Finally, QA− reduces QB to form Sn+1QB-. Upon a second PSII turnover the double reduced and protonated QB plastohydroquinone becomes protonated and is exchanged with an oxidized plastoquinone from the membrane pool (black arrows). The illustration was based on crystal structure 3WU2 (Umena et al., 2011). (B) The charge separation states described above are unstable and recombination competes with the energy-storing reactions. When PD1+ is present, recombination reactions can occur by several pathways as indicated by the dashed lines: (1) direct electron transfer from QA− to P+ (R1); (2) by the back reaction to form the P+Pheo− state, which can then recombine directly (R2) or, (3) when the P+Pheo− radical pair is present as a triplet state, 3[P+Pheo−], the dominant state when formed by the back-reaction, 3[P+Pheo−] charge recombination forms 3P (R3), a long lived chlorophyll triplet that can easily interact with O2 to form 1O2; (4) complete reversal of electron transfer can also occur, repopulating P* (R4), which can return to the ground state by emitting fluorescence (luminescence) or heat. Route 3, the triplet generating pathway, is the dominant recombination route in fully functional PSII. For simplicity the 3[P+Phe-] is not distinguised from the singlet form in this scheme. A Δψ across the membrane should destabilize P+QA− relative to the other states (see dotted blue lines), affecting the rates of reactions indicated in the green versus red. A Δψ across the membrane should also destabilize P+Pheo−, but because of the smaller dielectric span across the membrane, to a lesser extent than P+QA−. Thus, the buildup of Δψ should shift the equilibrium constant for sharing electrons between QA− and Pheo, favoring the formation of Pheo− and thus increasing the rate of recombination through R3 (as well as the R2 and R4), resulting in increased production of 3P and 1O2. Destabilization of P+QA− will also increase the driving force for P+QA− recombination via R1, however this recombination is already driven by 1.4eV and it is thus likely to be in the Marcus inverted region. Thus increasing the driving force will slow recombination by this route.
Discussion
This proposed mechanism of Δψ-mediated photoinhibition has broad implications for the energy limitations of photosynthesis. Although the two components of pmf are energetically equivalent for driving ATP synthesis (Hangarter and Good, 1982), they have distinct effects on the regulation of photosynthesis (Kramer et al., 1999; 2004; Finazzi et al., 2015). It has thus been proposed that the partitioning of pmf into Δψ and ΔpH is regulated to maintain a balance between efficient energy storage and regulation of light capture (Cruz et al., 2005). We demonstrate here another important constraint on this balance: the avoidance of photodamage caused by recombination reactions in PSII, and this may explain the need for complex ion balancing systems in chloroplasts (Cruz et al., 2001; Armbruster et al., 2014; Kunz et al., 2014).
The effect of Δψ on recombination and 1O2 production may, at least in part, explain the severe effects of fluctuating light on photosynthesis, and thus could constitute a significant limitation to photosynthetic productivity. In the absence of a large Δψ, the energy gap between the P+Pheo− and P+QA− appears to be sufficient to keep detrimental recombination to a manageable level when the usual regulatory mechanisms are functional (Rutherford et al., 2012). During photosynthesis, and especially under fluctuating light, though, the energy gaps between the photo-generated radical pairs vary dynamically under the influence of the pmf, so that high Δψ renders the charge-separated states in the photosystems considerably less stable. The Δψ is also expected to influence the trap depth (i.e. the energy level between P* and the first radical pair(s), the most relevant probably being P+Pheo−) and this could potentially affect the quantum yield of charge separation and the yield of radiative recombination (luminescence).
Transthylakoid electric fields also influence recombination reactions in PSI reaction centers (van Gorkom, 1996). It is thought that P700 triplet formation is minimized in PSI by the presence of the higher potential quinone in PsaA, making this side of the reaction center the safe charge recombination pathway (Rutherford et al., 2012). In light of the present findings, it is worth considering whether a transiently large Δψ could make this protective mechanism less efficient, though this question has yet to be addressed experimentally.
Over evolutionary time scales, the Δψ-effect may have constrained other bioenergetics features of photosynthesis. It has long been known that oxygenic photosynthesis is limited, in most organisms, to wavelength ranges shorter than about 700 nm (the 'red limit'), resulting in the loss of a large fraction of the light energy hitting the plant (Blankenship et al., 2011). (Gust et al., 2008) proposed that the red limit may be the result of certain limitations imposed in part by key biochemical properties of life (e.g. the properties of energy storage molecules NAD(P)H and ATP, the use of certain biochemical pathways, etc.) that evolved before the advent of photosynthesis. Milo (Milo, 2009) came to a different conclusion based on estimates of the theoretical wavelength dependence of energy conversion efficiency for plant photosynthesis, based on the Shockley and Queisser equation (Shockley and Queisser, 1961). The maximum efficiency was about 700 nm, similar to the red limit of oxygenic photosynthesis, suggesting that evolution has selected for photosynthetic energetics based on this fundamental limit, rather than any biological imperative. However, the predicted wavelength-dependence of the energy efficiency is very broad, with only about a 15% decrease from the peak at wavelengths out to 800 nm, far beyond the red limit, even in organisms with red-shifted reaction centers. Marosvolgyi and van Gorkom (Marosvolgyi and van Gorkom, 2010) drew a similar conclusion but with additional restraints and suggested a narrower maximum more closely overlapping with the red absorption of chlorophyll a.
Rutherford et al. (Rutherford et al., 2012) took a different view based on an analysis of the bioenergetics of reaction centers. They noted that PSII was in a uniquely difficult situation in energy terms: (1) it does multi-electron chemistry (at both sides of the reaction center) and so cannot prevent back reactions by kinetic control, (2) it has a very energy demanding reaction to do: water oxidation and quinone reduction with a ∆E ~920 meV in functional conditions, and requires a significant over-potential not just for attaining a high quantum yield of photochemistry but also for achieving water oxidation and quinol release, and (3) its chlorophyll cation chemistry is uniquely oxidizing and thus it cannot use carotenoids to protect itself from chlorophyll triplet formation at the heart of the reaction center. This situation means that unlike other type-II reaction centers, PSII is unable to prevent electrons from the bound semiquinones getting back to the Pheo and then recombining with P+ and forming 3P.
This lack of energy 'headroom' was seen not only as a major factor in PS II’s susceptibility to photodamage but also as a reason why oxygenic photosynthesis is pinned to chlorophyll a photochemistry at around 680 nm as the red limit. The existence of efficient oxygenic photosynthesis at longer wavelengths seemed to question that view (Miyashita et al., 1996; Kuhl et al., 2005; Gan et al., 2014; Behrendt et al., 2015). However, it was pointed out that these species seem to exist in very stable environments that have very little variation in light conditions (Cotton et al., 2015). Under such a narrow range of illumination conditions it is not unreasonable that less energy 'headroom' is required (Cotton et al., 2015).
Clearly, these specific energy limitations of PSII will be exacerbated by spikes in the Δψ reported here. Indeed, it seems reasonable to suggest that the existing 'energy headroom' postulated in normal chlorophyll a-containing PSII, while too small to avoid photodamage altogether, exists quite specifically to mitigate the extra photodamage from back-reactions enhanced by spikes in Δψ due to variable light intensities. In this way, the extent of the variable light-induced Δψ may be considered to contribute to the position of the red limit of oxygenic photosynthesis.
The need to prevent Δψ-induced recombination may also have guided the evolution of other photosynthetic components. Recent mechanistic models of the ATP synthase suggest that the ratio of protons passed through the ATP synthase per ATP synthesized depends on the number of subunits in the c-ring (Silverstein, 2014). The chloroplasts of green plants and algae thus far studied possess ATP synthase complexes with larger c-ring stoichiometries than their mitochondrial and bacterial homologues (Seelert et al., 2000), imposing higher fluxes of protons to generate ATP and necessitating the engagement of additional bioenergetic processes, including cyclic electron flow, to make up the ATP deficit needed to sustain photosynthesis (Kramer et al., 2004; Avenson et al., 2005). While this increased H+ demand is often viewed as a bioenergetic limitation to photosynthetic electron and proton transfer, a high H+/ATP ratio decreases the pmf needed to maintain a given ATP free energy state (ΔGATP), thus allowing photosynthesis to operate at a decreased steady-state pmf. In this context, the deleterious electron recombination effects of a high pmf may have favored the evolution of ATP synthase complexes with high H+/ATP ratios in chloroplasts.
Materials and methods
Plant materials and growth conditions
Wild type Arabidopsis thaliana (ecotype Wassilewskija-2) and ATP synthase γ-subunit mutants were germinated on Murashige and Skoog medium supplemented with 2% (w/v) sucrose, and 10 mg L−1 sulfadiazine for selection of transgenic minira lines (Hadi et al., 2002). Following germination plants were grown on soil under a 16 hr photoperiod at 100 μmol photons m−2 s−1 at 22°C for three weeks.
Nicotiana tabacum wild type (cv Samsun NN) and ATPC1 antisense lines were germinated and grown as in (Rott et al., 2011) under a 16 hr photoperiod at 300 μmol photons m−2 s−1. Measurements were performed at the onset of flowering on the youngest, fully expanded leaves.
Generation of chloroplast ATP synthase γ-subunit minira (minimum recapitulation of ATPC2) mutants
A T-DNA insertion mutant for ATPC1 (dpa1) (Dal Bosco et al., 2004) was used to introduce the mutated constructs as a complemented allele as in (Kohzuma et al., 2012). Site-directed mutations in the redox-regulatory domain of ATPC1 were designed to incorporate amino acid differences from the redox inactive ATPC2 into the redox regulated ATPC1 (Figure 1A). The Arabidopsis thaliana AtpC1 gene was excised from binary vector pSex001 as a SmaI/XbaI fragment and cloned into SmaI/XbaI digested pBluescript plasmid. The resulting plasmid was named pDA15. The mutations were introduced into ATPC1 using a combination of three approaches.
The first approach used an adaptor ligation strategy. Oligonucleotides were designed to introduce desired mutations. Adaptors representing the 5’ and 3’ strand of DNA targeting specific mutations were obtained independently from Sigma Aldrich. A total of 5 ml of each oligonucleotide pair (10 mM) were mixed and denatured at 95°C for 10 min in a boiling water bath. The oligonucleotides were allowed to reach room temperature over two hours in the water bath allowing for efficient annealing of complementary strands. The adaptors thus obtained were utilized for adaptor ligation.
The target region from ATPC1 cDNA was removed using BglII/HpaI restriction enzymes. The larger linearized backbone of pDA15 was used for ligation with the adaptors (Supplementary file 1). The resultant vector was double digested with SmaI and XbaI to excise the mutated ATPC1 gene, which was then ligated back into the SmaI/XbaI digested binary vector, pSex001.
A second set of mutations was introduced by first digesting pDA15 with BglII followed by a partial digestion with TatI. The resulting plasmid backbone was used for ligation with the adaptors having the desired mutation (Supplementary file 1b). As in the first adaptor ligation-mediated mutagenesis approach, the resultant vector was double digested with SmaI and XbaI to excise the respective mutated atpC1 gene and ligated into SmaI/XbaI digested binary vector, pSex001.
A second mutagenesis strategy used splicing by overlap extension (SOE) PCR. For each desired mutation, two sets of primers were designed to produce two overlapping fragments during amplification of atpC1 from pDA15 such that the mutation was generated in the region of overlap (Supplementary file 1c). The two fragments were mixed together and the resulting DNA solution was used as a template for a subsequent PCR using primers that amplify the complete AtpC1 gene (DMP 45 and DMP 46). This led to the amplification of the entire atpC1 gene with the desired mutation. The mutated atpC1 was digested with SmaI and XbaI and was sub-cloned into SmaI/XbaI digested pSex001.
The third mutagenesis approach involved swapping of target domains (delete swaps) using synthetic gene fragments synthesized at GenScript USA (Supplementary file 1). The synthetic gene was used to replace the ATPC1 redox regulatory domain in the native gene. To swap the domains, pDA15 was digested with BsrGI and XbaI to remove the native domain and ligated with the synthetic fragment derived from the synthetic gene construct after digestion with BsrGI and XbaI. To introduce a synthetic gene with a single nucleotide mutation, the synthetic gene construct for minira 3 and pDA15 were double digested with BsrGI and XbaI and the synthetic BsrGI/XbaI fragment with the mutation was ligated into pDA15 where the original BsrGI/XbaI fragment had been removed The resultant intermediate plasmid was digested with SmaI/XbaI to obtain the gene with swapped C domain. This gene was then sub-cloned into SmaI/XbaI digested pSex001. All of the introduced mutations were confirmed by Sanger sequencing of individual plasmids.
Following successful mutagenesis, minira constructs were mobilized into the binary vector pSEX001-VS under control of the Cauliflower mosaic virus 35S promoter. A single minira construct was transformed into heterozygous dpa1 plants via Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998). The resulting transgenic plants were screened for the minira insertion via PCR using the forward primer 5’ -GGTAATATCCGGAAACCTCC- 3’ and the reverse primer 5’ -GTACAAGAGCTCGACTTTCTCG- 3’ followed by dpa1 screening using the forward primer 5’ -CACATCATCTCATTGATGCTTGG- 3’ and the reverse primer 5’ -GTACAAGAGCTCGACTTTGTCG- 3’. Transgenic plants containing both minira and dpa1 insertions were self-pollinated until plants were homozygous for both dpa1 (Δatpc1) and the correct minira mutation, which was subsequently confirmed by sequencing.
Isolation of tightly coupled chloroplasts and intact thylakoids
Chloroplasts were extracted from market spinach with modifications to the method described in Seigneurin-Berny et al. (Seigneurin-Berny et al., 2008). All centrifugation steps were carried out at 4°C and exposure to light was kept to a minimum. Briefly, approximately 20 g of spinach leaves were homogenized in a blender for 10 s with ice cold homogenization buffer of 50 mM HEPES (pH 7.6), 330 mM sorbitol, 5 mM MgCl2, 2 mM EDTA and supplemented with 0.1% BSA for grinding. The homogenate was filtered through three layers of wetted Miracloth and one layer of wetted muslin followed by centrifugation at 4000 x g for 10 min. The pellet was resuspended in homogenization buffer and layered on top of a single step 80%–40% Percoll gradient. Intact chloroplasts were recovered after centrifugation for 20 min at 3000 x g in a swinging bucket rotor. The intact chloroplasts were diluted approximately 4-fold with homogenization buffer and centrifuged for 5 min at 4000 xg. The chloroplast pellet was resuspended in a minimal amount of homogenization buffer (<1 mL) and chlorophyll quantified in 80% acetone (Porra et al., 1989).
Spectroscopic measurements
Near simultaneous chlorophyll fluorescence and electrochromic shift (ECS) measurements were performed on a custom made spectrophotometer (Hall et al., 2013). For in vivo spectroscopic measurements, following a 10 min dark acclimation the maximal PSII quantum efficiency, linear electron flow (LEF), energy-dependent exciton quenching (qE), and photoinhibitory quenching (qI) were estimated using saturation pulse chlorophyll a fluorescence as described previously (Livingston et al., 2010). The extent of the qI component of NPQ was determined following at least 10 min dark relaxation to eliminate the residual effects of qE type quenching. Red actinic illumination was used for all measurements to prevent incorrect assessment of chloroplast movement as qI, as red light is ineffective in inducing chloroplast movements (Cazzaniga et al., 2013). A Stern-Volmer derivation of qE (qE(SV)) was used to minimize the contribution of qI in the determination of qE (Krause and Jahns, 2003). Estimates of the relative redox status of QA (qL) were performed as described in (Kramer et al., 2004) after at least 10 min of actinic illumination. The relative extents of steady state pmf (ECSt) and the conductivity of ATP synthase to protons (gH+) were measured using the dark interval relaxation kinetics of absorbance changes associated with the electrochromic shift (ECS) fit to a first-order exponential decay (Sacksteder and Kramer, 2000). Partitioning of the pmf was determined from deconvolution of the absorbance change at three wavelengths (505, 520 and 535 nm) around 520 nm during the dark interval ECS changes and the ECS steady-state (Δψ) and ECS inverse (ΔpH) were determined as in (Takizawa et al., 2007). Briefly, the total amplitude of the deconvoluted ECS signal following a rapid light/dark transition was used to estimate the total light-induced pmf (ECSt). The steady-state Δψ component was determined from the extent to which the inverted ECS signal during the dark interval decreased from the steady-state baseline. The steady-state ΔpH component was determined as the amplitude of the inverted ECS signal during the dark interval. The ECS measurements were corrected for pigment variations by normalizing to chlorophyll content determined from acetone extraction as above. For tobacco measurements, the ECS measurements were normalized to the xenon-flash induced extent of the ΔA520 nm ECS rise. P700+ reduction kinetics were measured from the dark interval relaxation kinetics of the absorbance change at 810 nm after subtracting the 930 absorbance change (Baker et al., 2007).
In vitro chloroplast measurements were performed on a similar instrument described above modified to measure a cuvette held sample. Chloroplasts were osmotically shocked on ice in buffer containing 10 mM HEPES (pH 7.8) and 10 mM MgCl2 to a final chlorophyll concentration of 20 μg ml−1 supplemented with 5 μM spinach ferredoxin and 10 μM ascorbate. Where noted, thylakoids were treated with 50 μM decyl-ubiquinol to catalyze PSI cyclic electron transfer and generate a pmf, 50 μM 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) to block PSII forward electron transfer, and 25 μM gramicidin to decouple the pmf. Fluorescence measurements were performed as above, with variable fluorescence measured after a 20 min dark adaptation after which the thylakoids were excited with a single 100 ms subsaturating actinic pulse. From a dark-adapted state, the application of a single turnover pulse will lead to rapid accumulation of Δψ, due to the high buffering capacity of the thylakoid lumen as well as the low capacitance of the thylakoid membrane, allowing gramicidin to decouple Δψ, as the Δψ primarily composes the pmf under these conditions (Cruz et al., 2001). The F0 measurements for all samples were taken from the first measured point to avoid any actinic effects due to the measuring pulses themselves.
Estimation of recombination rate
The Δψ-induced enhancement of the rate of recombination from the S2QA− state was estimated based on the change in the equilibrium constant for the sharing of electrons in the presence of Δψ. Other states will also recombine (e.g. the S3QA− state) but we use S2QA− as a proxy because we expect most of these states to respond to changes in QA and Δψ in similar ways. The rate of recombination from S2QA− was calculated as:
| (1) |
where [S2QA−] is the concentration of PSII centers with reduced QA, kr the intrinsic rate of recombination from S2QA−, and ΔEstab is the free energy for stabilization of the charge separated state, expressed in eV. In the absence of a field and in the presence of DCMU where all QA is reduced, vr is measured to be about 0.3 s−1 (Figure 5), but in the uninhibited complex under steady state photosynthesis, this rate will be decreased proportionally by oxidation of QA, while ΔEstab will be decreased by Δψ so that:
| (2) |
where 1−qL is an estimation of the fraction of QA in the reduced form (Kramer et al., 2004), and Δψlight-dark is the light-dark difference in electric field in mV. From Takizawa et al. (Takizawa et al., 2007) we obtained a factor for estimating Δψ from the extents of ECS, and correcting for the chlorophyll content as performed here, we obtain:
| (3) |
where ECSss is the normalized ECS signal representing the light-dark difference in Δψ normalized to the chlorophyll content.
Chlorophyll fluorescence imaging
In vivo whole plant chlorophyll a fluorescence imaging was performed in an imaging chamber equipped with 50W Bridgelux White LEDs (BXRA-56C5300, Bridgelux Inc., Livermore, California) for white actinic illumination (Cruz et al., 2016). Pre-illumination values for F0 and FM were captured just prior to the beginning of the photoperiod and subsequent fluorescence parameters obtained using a 300 ms saturating actinic pulse at ~25,000 μmol photons m−2 s−1 and a Red LED matrix (Luxeon Rebel SMT High Power LED Red, LXM2-PD01-0050, Philips Lumiled, San Jose, California) to measure chlorophyll which was then captured by a CCD camera (AVT Manta 145 M) equipped with a near infrared long pass filter (RT-830, Hoya Glass). Plants were imaged over three consecutive 24 hr photoperiods (Figure 2A, videos 1–9, timing and light intensities are described in Supplementary file 2). During the first day, the actinic light intensity remained constant at 100 μmol photons m−2 s−1 to collect growth chamber conditions. Days two and three represented ramped lighting perturbations. The photoperiod for day two was sinusoidal, beginning at 39 μmol photons m−2 s−1 and increasing in intensity by approximately 1.2 times every 30 min until midday where it peaked at 500 μmol photons m−2 s−1, after which the light intensity decreased at the same rate every 30 min. The photoperiod for day three was sinusoidal with brief fluctuations in light intensity. Starting at 39 μmol photons m−2 s−1, the light intensity was doubled after 15 min followed by 1.5-fold increase for 12 min and the cycle repeated until peak intensities of 1000 and 500 μmol photons m−2 s−1 were cycled through at midday, after which the sinusoidal fluctuations decreased at the same rate as the increases. Steady state values for NPQ parameters of chlorophyll fluorescence were captured prior to the ramp to the next light intensity on days two and three, or hourly on day one and calculated as noted above for the fluorescence spectroscopy. Sequences of images were captured with a 60 ms delay between images for a 15 frame total for each measurement pre- during and post-saturation flash followed by images taken to correct for artifacts due to residual electrons in the CCD array. Images were analyzed using open source software (ImageJ, NIH) modified in house to allow calculations of photosynthetic fluorescence parameters across selected regions of interest.
Photoinhibition of detached leaves
Plant leaves were excised and incubated in the dark for 3 hr with their petioles submerged in either water or 3 mM lincomycin to inhibit chloroplast protein translation (Tyystjarvi and Aro, 1996). The leaves were then illuminated with red light at 1000 μmol photons m−2 s−1 using a red actinic light for indicated periods of time. During illumination, the petioles of the leaves remained submerged in the treatment solution. Following illumination, leaves were allowed to dark adapt for 20 min, after which the F0 and FM values of chlorophyll fluorescence were measured in order to determine FV/FM. For tobacco plants, leaf discs were soaked in a lincomycin solution and fluorescence parameters determined at 600 μmol photons m−2 s−1.
To determine PSII activity, following photoinhibitory treatment, performed as above, leaves were dark adapted for 20 min and then vacuum infiltrated with a 50 μM DCMU solution. Analysis of PSII activity was determined from the amplitude of the ΔA520 nm ECS signal using two saturating single-turnover flashes provided by a xenon lamp spaced 200 ms apart. The amplitude of the second flash, corresponding to PSI centers capable of charge separation, was subtracted from the amplitude of the first flash, corresponding to both PSI and PSII centers capable of charge separation, to obtain the relative PSII photosystems capable of activity both before and after photoinhibition. Xenon flashes were judged to be fully saturating by ensuring that essentially identical results were obtained with a 50% weaker intensity.
Protein analysis
Photoinhibited leaf samples were collected as described above and total leaf proteins were extracted as described in Livingston et al. (Livingston et al., 2010). Analysis of chloroplast ATP synthase complexes was carried out on 20 μg of protein using leaves collected from 3–4 plants taken from the growth chamber. Analysis of PsbA (D1) protein levels during photoinhibitory treatment was carried out on 30 μg of total protein from 3–4 leaves sampled during the photoinhibitory treatment time points as described above. Proteins were separated by SDS-PAGE and the ATPB (β-subunit) and PsbA (D1) proteins detected using commercially purchased antibodies (Agrisera, Vannas, Sweden).
1O2 detection
Plant leaves were excised and incubated in the dark for 3 hr in 250 μM Singlet Oxygen Sensor Green (SOSG, Life Technologies) prepared according to manufacturer’s instructions. Petioles were maintained below the liquid’s surface during the infiltration and were wrapped in a Kimwipe soaked in water during imaging to prevent drying. For valinomycin treatments, leaves were vacuum infiltrated with either SOSG solution or with SOSG supplemented with 50 μM valinomycin and subsequently measured. Successful penetration of leaf epidermal and mesophyll cells, as well as SOSG penetrance throughout the cells was confirmed via confocal microscopy. The leaves were imaged in a chamber equipped with the same lighting as above. Qualitatively similar data were obtained for minira 3–1 leaves under both white and red (650 nm) LED illumination, ensuring that photosensitization of SOSG was not responsible for the signals obtained (Ragas et al., 2009). Images were captured with a cooled CCD camera (AVT Bigeye G 132B-NIR) equipped with a 555 nm 10 nm band pass filter. Fluorescence excitation was provided via 458 nm LEDS (Cree Inc). Images were analyzed using ImageJ software.
Data analysis
All spectroscopic data were analyzed and figures generated using Origin 9.0 software (Microcal Software). Statistical analyses of data were performed in R package, utilizing two-way ANOVA to test for significant effects on photoinhibition (qI) from the interaction with either the ΔpH or Δψ component of the pmf.
Acknowledgements
This work was supported by the U.S. Department of Energy (DOE), Office of Science, Basic Energy Sciences (BES) under Award number DE-FG02-91ER20021 and the MSU Center for Advanced Algal and Plant Phenotyping (CAAPP). AWR was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) grant (BB/K002627/1) and the Royal Society Wolfson Research Merit Award.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grants:
Basic Energy Sciences DE-FG02-91ER20021 to Geoffry A Davis, Atsuko Kanazawa, Kaori Kohzuma, John E Froehlich, Mio Satoh-Cruz, David M Kramer.
Biotechnology and Biological Sciences Research Council BB/K002627/1 to A William Rutherford.
Royal Society Wolfson Research Merit Award to A William Rutherford.
Michigan State University Center for Advanced Algal and Plant Phenotyping to David M Kramer.
Michigan State University AgBioResearch to David M Kramer.
Additional information
Competing interests
DMK: Founder and stock holder in Phenometrics, Inc. and a founder of the PhotosynQ.org project, these entities manufacture and distribute instrumentation for plant and algal phenotyping. However, neither of these technologies are used in the current work.
The other authors declare that no competing interests exist.
Author contributions
GAD, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
AK, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
MAS, Conception and design, Analysis and interpretation of data, Drafting or revising the article.
KK, Conception and design, Drafting or revising the article.
JEF, Acquisition of data, Analysis and interpretation of data.
AWR, Analysis and interpretation of data, Drafting or revising the article.
MS-C, Conception and design, Analysis and interpretation of data.
DM, Conception and design, Acquisition of data, Drafting or revising the article.
ST, Acquisition of data, Analysis and interpretation of data.
AD, Conception and design, Drafting or revising the article.
DMK, Conception and design, Analysis and interpretation of data, Drafting or revising the article.
Additional files
(a) Oligonucleotide sequences utilized for adapter ligation mutagenesis. (b) Oligonucleotide sequences utilized for adapter ligation mutagenesis to introduce secondary mutations. (c) Oligonucleotide sequences utilized for splicing by overlap extension PCR. (d) Synthetic gene constructs incorporating multiple ATPC2 mutations into ATPC1.
(a) Timing and light profile of imaging day one. (b) Timing and light profile of imaging day two. (c) Timing and light profile of imaging day three.
All measurements were performed on three-week old leaves following in vivo spectroscopic measurements as described in Materials and methods. Data represent the mean ± s.d. for n ≥ 3 leaves. Statistically significant differences (*p<0.05) from wild-type were determined using a t-test.
References
- Allahverdiyeva Y, Suorsa M, Tikkanen M, Aro EM. Photoprotection of photosystems in fluctuating light intensities. Journal of Experimental Botany. 2015;66:2427–2436. doi: 10.1093/jxb/eru463. [DOI] [PubMed] [Google Scholar]
- Armbruster U, Carrillo LR, Venema K, Pavlovic L, Schmidtmann E, Kornfeld A, Jahns P, Berry JA, Kramer DM, Jonikas MC. Ion antiport accelerates photosynthetic acclimation in fluctuating light environments. Nature Communications. 2014;5:e16921. doi: 10.1038/ncomms6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avenson TJ, Cruz JA, Kanazawa A, Kramer DM. Regulating the proton budget of higher plant photosynthesis. PNAS. 2005;102:9709–9713. doi: 10.1073/pnas.0503952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avenson TJ, Cruz JA, Kramer DM. Modulation of energy-dependent quenching of excitons in antennae of higher plants. PNAS. 2004;101:5530–5535. doi: 10.1073/pnas.0401269101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul B, Cardol P, Breyton C, Finazzi G. Electrochromism: a useful probe to study algal photosynthesis. Photosynthesis Research. 2010;106:179–189. doi: 10.1007/s11120-010-9579-z. [DOI] [PubMed] [Google Scholar]
- Baker NR, Harbinson J, Kramer DM. Determining the limitations and regulation of photosynthetic energy transduction in leaves. Plant, Cell & Environment. 2007;30:1107–1125. doi: 10.1111/j.1365-3040.2007.01680.x. [DOI] [PubMed] [Google Scholar]
- Behrendt L, Brejnrod A, Schliep M, Sørensen SJ, Larkum AW, Kühl M, Kuhl M. Chlorophyll f-driven photosynthesis in a cavernous cyanobacterium. ISME Journal. 2015;9:2108–2111. doi: 10.1038/ismej.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship RE, Tiede DM, Barber J, Brudvig GW, Fleming G, Ghirardi M, Gunner MR, Junge W, Kramer DM, Melis A, Moore TA, Moser CC, Nocera DG, Nozik AJ, Ort DR, Parson WW, Prince RC, Sayre RT. Comparing photosynthetic and photovoltaic efficiencies and recognizing the potential for improvement. Science. 2011;332:805–809. doi: 10.1126/science.1200165. [DOI] [PubMed] [Google Scholar]
- Cazzaniga S, Dall' Osto L, Kong SG, Wada M, Bassi R. Interaction between avoidance of photon absorption, excess energy dissipation and zeaxanthin synthesis against photooxidative stress in Arabidopsis. Plant Journal. 2013;76:568–579. doi: 10.1111/tpj.12314. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cotton CA, Douglass JS, De Causmaecker S, Brinkert K, Cardona T, Fantuzzi A, Rutherford AW, Murray JW. Photosynthetic constraints on fuel from microbes. Frontiers in Bioengineering and Biotechnology. 2015;3:e16921. doi: 10.3389/fbioe.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JA, Avenson TJ, Kanazawa A, Takizawa K, Edwards GE, Kramer DM. Plasticity in light reactions of photosynthesis for energy production and photoprotection. Journal of Experimental Botany. 2005;56:395–406. doi: 10.1093/jxb/eri022. [DOI] [PubMed] [Google Scholar]
- Cruz JA, Sacksteder CA, Kanazawa A, Kramer DM. Contribution of Electric Field (Δψ) to Steady-State Transthylakoid Proton Motive Force ( pmf ) in Vitro and in Vivo. Control of pmf Parsing into Δψ and ΔpH by Ionic Strength †. Biochemistry. 2001;40:1226–1237. doi: 10.1021/bi0018741. [DOI] [PubMed] [Google Scholar]
- Cruz JA, Savage LJ, Zegarac R, Hall CC, Satoh-Cruz M, Davis GA, Kovac WK, Chen J, Kramer DM. Dynamic environmental photosynthetic imaging reveals emergent phenotypes. Cell Systems. 2016;2:365–377. doi: 10.1016/j.cels.2016.06.001. [DOI] [PubMed] [Google Scholar]
- Dal Bosco C, Lezhneva L, Biehl A, Leister D, Strotmann H, Wanner G, Meurer J. Inactivation of the chloroplast ATP synthase gamma subunit results in high non-photochemical fluorescence quenching and altered nuclear gene expression in Arabidopsis thaliana. Journal of Biological Chemistry. 2004;279:1060–1069. doi: 10.1074/jbc.M308435200. [DOI] [PubMed] [Google Scholar]
- Dall'Osto L, Holt NE, Kaligotla S, Fuciman M, Cazzaniga S, Carbonera D, Frank HA, Alric J, Bassi R. Zeaxanthin protects plant photosynthesis by modulating chlorophyll triplet yield in specific light-harvesting antenna subunits. Journal of Biological Chemistry. 2012;287:41820–41834. doi: 10.1074/jbc.M112.405498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Grooth BG, Van Gorkom HJ. External electric field effects on prompt and delayed fluorescence in chloroplasts. Biochimica Et Biophysica Acta. 1981;635:445–456. doi: 10.1016/0005-2728(81)90104-3. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW. Photoprotection and other responses of plants to high light stress. Annual Review of Plant Physiology and Plant Molecular Biology. 1992;43:599–626. doi: 10.1146/annurev.pp.43.060192.003123. [DOI] [Google Scholar]
- Diner B, Joliot P. Effect of the transmembrane electric field on the photochemical and quenching properties of Photosystem II in vivo. Biochimica Et Biophysica Acta . 1976;423:479–498. doi: 10.1016/0005-2728(76)90202-4. [DOI] [PubMed] [Google Scholar]
- Finazzi G, Petroutsos D, Tomizioli M, Flori S, Sautron E, Villanova V, Rolland N, Seigneurin-Berny D. Ions channels/transporters and chloroplast regulation. Cell Calcium. 2015;58:86–97. doi: 10.1016/j.ceca.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Gan F, Zhang S, Rockwell NC, Martin SS, Lagarias JC, Bryant DA. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science. 2014;345:1312–1317. doi: 10.1126/science.1256963. [DOI] [PubMed] [Google Scholar]
- Gross EL, Pan B, Li B, Brown L. Stability of plastocyanin to acid pH. Biophysical Journal. 1994;66:e16921 [Google Scholar]
- Gust D, Kramer D, Moore A, Moore TA, Vermaas W. Engineered and artificial photosynthesis: human ingenuity enters the game. MRS Bulletin. 2008;33:383–387. doi: 10.1557/mrs2008.78. [DOI] [Google Scholar]
- Hadi MZ, Kemper E, Wendeler E, Reiss B. Simple and versatile selection of Arabidopsis transformants. Plant Cell Reports. 2002;21:130–135. doi: 10.1007/s00299-002-0473-9. [DOI] [Google Scholar]
- Hall CC, Cruz J, Wood M, Zegarac R, DeMars D, Carptenter J, Kramer DM. Photosynthesis Research for Food, Fuel and Future (15th International Conference on Photosynthesis) Springer Berlin Heidelberg; 2013. Photosynthetic measurements with the Idea Spec: an integrated diode emitter array spectrophotometer/fluorometer; pp. 184–188. [Google Scholar]
- Hangarter RP, Good NE. Energy thresholds for ATP synthesis in chloroplasts. Biochimica Et Biophysica Acta . 1982;681:397–404. doi: 10.1016/0005-2728(82)90181-5. [DOI] [Google Scholar]
- Hope AB, Valente P, Matthews DB. Effects of pH on the kinetics of redox reactions in and around the cytochromebf complex in an isolated system. Photosynthesis Research. 1994;42:111–120. doi: 10.1007/BF02187122. [DOI] [PubMed] [Google Scholar]
- Inohara N, Iwamoto A, Moriyama Y, Shimomura S, Maeda M, Futai M. Two genes, atpC1 and atpC2, for the gamma subunit of Arabidopsis thaliana chloroplast ATP synthase. Journal of Biological Chemistry. 1991;266:7333–7338. [PubMed] [Google Scholar]
- Johnson GN, Rutherford AW, Krieger A. A change in the midpoint potential of the quinone QA in Photosystem II associated with photoactivation of oxygen evolution. Biochimica Et Biophysica Acta. 1995;1229:202–207. doi: 10.1016/0005-2728(95)00003-2. [DOI] [Google Scholar]
- Joliot P, Delosme R, Joliot A. 515nm absorption changes in Chlorella at short times (4–100 μs) after a flash. Biochimica Et Biophysica Acta . 1977;459:47–57. doi: 10.1016/0005-2728(77)90007-X. [DOI] [PubMed] [Google Scholar]
- Joliot P, Delosme R. Flash-induced 519 nm absorption change in green algae. Biochimica Et Biophysica Acta . 1974;357:267–284. doi: 10.1016/0005-2728(74)90066-8. [DOI] [PubMed] [Google Scholar]
- Kanazawa A, Kramer DM. In vivo modulation of nonphotochemical exciton quenching (NPQ) by regulation of the chloroplast ATP synthase. PNAS. 2002;99:12789–12794. doi: 10.1073/pnas.182427499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N, Berg A, van Kan PJM, Levanon H, Ohad I. Mechanism of photosystem II photoinactivation and D1 protein degradation at low light: The role of back electron flow. PNAS. 1997;94:1579–1584. doi: 10.1073/pnas.94.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N, Krieger-Liszkay A. Photoinhibition: molecular mechanisms and physiological significance. Physiologia Plantarum. 2011;142:1–5. doi: 10.1111/j.1399-3054.2011.01467.x. [DOI] [PubMed] [Google Scholar]
- Koh E, Fluhr R. Singlet oxygen detection in biological systems: Uses and limitations. Plant Signaling & Behavior. 2016;11:e16921. doi: 10.1080/15592324.2016.1192742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohzuma K, Dal Bosco C, Kanazawa A, Dhingra A, Nitschke W, Meurer J, Kramer DM. Thioredoxin-insensitive plastid ATP synthase that performs moonlighting functions. PNAS. 2012;109:3293–3298. doi: 10.1073/pnas.1115728109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Avenson TJ, Edwards GE. Dynamic flexibility in the light reactions of photosynthesis governed by both electron and proton transfer reactions. Trends in Plant Science. 2004;9:349–357. doi: 10.1016/j.tplants.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Kramer DM, Evans JR. The importance of energy balance in improving photosynthetic productivity. Plant Physiology. 2011;155:70–78. doi: 10.1104/pp.110.166652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Johnson G, Kiirats O, Edwards GE. New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynthesis Research. 2004;79:209–218. doi: 10.1023/B:PRES.0000015391.99477.0d. [DOI] [PubMed] [Google Scholar]
- Kramer DM, Sacksteder CA, Cruz JA. How acidic is the lumen? Photosynthesis Research. 1999;60:151–163. doi: 10.1023/A:1006212014787. [DOI] [Google Scholar]
- Krause GH, Jahns P. Pulse amplitude modulated chlorophyll fluorometry and its application in plant science. In: Green BR, Parson WW, editors. Advances in Photosynthesis and Respiration, Vol 13. Dordrecht, The Netherlands: Kluwer Academic; 2003. pp. 373–399. [Google Scholar]
- Krieger A, Weis E. The role of calcium in the pH-dependent control of Photosystem II. Photosynthesis Research. 1993;37:117–130. doi: 10.1007/BF02187470. [DOI] [PubMed] [Google Scholar]
- Krieger-Liszkay A, Fufezan C, Trebst A. Singlet oxygen production in photosystem II and related protection mechanism. Photosynthesis Research. 2008;98:551–564. doi: 10.1007/s11120-008-9349-3. [DOI] [PubMed] [Google Scholar]
- Krieger-Liszkay A, Rutherford AW. Influence of herbicide binding on the redox potential of the quinone acceptor in photosystem II: relevance to photodamage and phytotoxicity. Biochemistry. 1998;37:17339–17344. doi: 10.1021/bi9822628. [DOI] [PubMed] [Google Scholar]
- Kühl M, Chen M, Ralph PJ, Schreiber U, Larkum AW. Ecology: a niche for cyanobacteria containing chlorophyll d. Nature. 2005;433:e16921. doi: 10.1038/433820a. [DOI] [PubMed] [Google Scholar]
- Külheim C, Agren J, Jansson S. Rapid regulation of light harvesting and plant fitness in the field. Science. 2002;297:91–93. doi: 10.1126/science.1072359. [DOI] [PubMed] [Google Scholar]
- Kunz H-H, Gierth M, Herdean A, Satoh-Cruz M, Kramer DM, Spetea C, Schroeder JI. Plastidial transporters KEA1, -2, and -3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. PNAS. 2014;111:7480–7485. doi: 10.1073/pnas.1323899111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Gilmore AM, Caffarri S, Bassi R, Golan T, Kramer D, Niyogi KK. Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. Journal of Biological Chemistry. 2004;279:22866–22874. doi: 10.1074/jbc.M402461200. [DOI] [PubMed] [Google Scholar]
- Livingston AK, Cruz JA, Kohzuma K, Dhingra A, Kramer DM. An Arabidopsis mutant with high cyclic electron flow around photosystem I (hcef) involving the NADPH dehydrogenase complex. Plant Cell. 2010;22:221–233. doi: 10.1105/tpc.109.071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston AK, Kanazawa A, Cruz JA, Kramer DM. Regulation of cyclic electron flow in C₃ plants: differential effects of limiting photosynthesis at ribulose-1,5-bisphosphate carboxylase/oxygenase and glyceraldehyde-3-phosphate dehydrogenase. Plant, Cell & Environment. 2010;33:1779–1788. doi: 10.1111/j.1365-3040.2010.02183.x. [DOI] [PubMed] [Google Scholar]
- Marosvölgyi MA, van Gorkom HJ. Cost and color of photosynthesis. Photosynthesis Research. 2010;103:105–109. doi: 10.1007/s11120-009-9522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R. What governs the reaction center excitation wavelength of photosystems I and II? Photosynthesis Research. 2009;101:59–67. doi: 10.1007/s11120-009-9465-8. [DOI] [PubMed] [Google Scholar]
- Miyashita H, Ikemoto H, Kurano N, Adachi K, Chihara M, Miyachi S. Chlorophyll d as a major pigment. Nature. 1996;383:e16921. doi: 10.1038/383402a0. [DOI] [Google Scholar]
- Nishio JN, Whitmarsh J. Dissipation of the proton electrochemical potential in intact chloroplasts (II. The pH gradient monitored by cytochrome f reduction kinetics) Plant Physiology. 1993;101:89–96. doi: 10.1104/pp.101.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Murata N. Revised scheme for the mechanism of photoinhibition and its application to enhance the abiotic stress tolerance of the photosynthetic machinery. Applied Microbiology and Biotechnology. 2014;98:8777–8796. doi: 10.1007/s00253-014-6020-0. [DOI] [PubMed] [Google Scholar]
- Pätsikkä E, Kairavuo M, Sersen F, Aro EM, Tyystjärvi E. Excess copper predisposes photosystem II to photoinhibition in vivo by outcompeting iron and causing decrease in leaf chlorophyll. Plant Physiology. 2002;129:1359–1367. doi: 10.1104/pp.004788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica Et Biophysica Acta. 1989;975:384–394. doi: 10.1016/S0005-2728(89)80347-0. [DOI] [Google Scholar]
- Ragàs X, Jiménez-Banzo A, Sánchez-García D, Batllori X, Nonell S. Singlet oxygen photosensitisation by the fluorescent probe Singlet Oxygen Sensor Green. Chemical Communications. 2009;20:2920–2922. doi: 10.1039/b822776d. [DOI] [PubMed] [Google Scholar]
- Ramel F, Ksas B, Akkari E, Mialoundama AS, Monnet F, Krieger-Liszkay A, Ravanat JL, Mueller MJ, Bouvier F, Havaux M. Light-induced acclimation of the Arabidopsis chlorina1 mutant to singlet oxygen. Plant Cell. 2013;25:1445–1462. doi: 10.1105/tpc.113.109827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport F, Finazzi G, Pierre Y, Bennoun P. A new electrochemical gradient generator in thylakoid membranes of green algae. Biochemistry. 1999;38:2040–2047. doi: 10.1021/bi982351k. [DOI] [PubMed] [Google Scholar]
- Raven JA. The cost of photoinhibition. Physiologia Plantarum. 2011;142:87–104. doi: 10.1111/j.1399-3054.2011.01465.x. [DOI] [PubMed] [Google Scholar]
- Robinson HH, Crofts AR. Kinetics of the oxidation-reduction reactions of the photosystem II quinone acceptor complex, and the pathway for deactivation. FEBS Letters. 1983;153:221–226. doi: 10.1016/0014-5793(83)80152-5. [DOI] [Google Scholar]
- Rott M, Martins NF, Thiele W, Lein W, Bock R, Kramer DM, Schöttler MA. ATP synthase repression in tobacco restricts photosynthetic electron transport, CO2 assimilation, and plant growth by overacidification of the thylakoid lumen. Plant Cell. 2011;23:304–321. doi: 10.1105/tpc.110.079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford AW, Osyczka A, Rappaport F. Back-reactions, short-circuits, leaks and other energy wasteful reactions in biological electron transfer: redox tuning to survive life in O(2) FEBS Letters. 2012;586:603–616. doi: 10.1016/j.febslet.2011.12.039. [DOI] [PubMed] [Google Scholar]
- Sacksteder CA, Kramer DM. Dark-interval relaxation kinetics (DIRK) of absorbance changes as a quantitative probe of steady-state electron transfer. Photosynthesis Research. 2000;66:145–158. doi: 10.1023/A:1010785912271. [DOI] [PubMed] [Google Scholar]
- Satoh K, Katoh S. Induction kinetics of millisecond-delayed luminescence in intact Bryopsis chloroplasts. Plant and Cell Physiology. 1983;24:953–962. [Google Scholar]
- Seelert H, Poetsch A, Dencher NA, Engel A, Stahlberg H, Müller DJ. Structural biology. Proton-powered turbine of a plant motor. Nature. 2000;405:418–419. doi: 10.1038/35013148. [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Salvi D, Joyard J, Rolland N. Purification of intact chloroplasts from Arabidopsis and spinach leaves by isopycnic centrifugation. Current Protocols in Cell Biology. 2008;3:3–30. doi: 10.1002/0471143030.cb0330s40. [DOI] [PubMed] [Google Scholar]
- Shockley W, Queisser HJ. Detailed balance limit of efficiency of p-n junction solar cells. Journal of Applied Physics. 1961;32:e16921. doi: 10.1063/1.1736034. [DOI] [Google Scholar]
- Shumbe L, Chevalier A, Legeret B, Taconnat L, Monnet F, Havaux M. Singlet oxygen-induced cell death in Arabidopsis under high-light stress is controlled by OXI1 kinase. Plant Physiology. 2016;170:1757–1771. doi: 10.1104/pp.15.01546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein TP. An exploration of how the thermodynamic efficiency of bioenergetic membrane systems varies with c-subunit stoichiometry of F₁F₀ ATP synthases. Journal of Bioenergetics and Biomembranes. 2014;46:229–241. doi: 10.1007/s10863-014-9547-y. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Bauwe H, Badger M. Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiology. 2007;144:487–494. doi: 10.1104/pp.107.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa K, Cruz JA, Kanazawa A, Kramer DM. The thylakoid proton motive force in vivo. Quantitative, non-invasive probes, energetics, and regulatory consequences of light-induced pmf. Biochimica Et Biophysica Acta. 2007;1767:1233–1244. doi: 10.1016/j.bbabio.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Takizawa K, Kanazawa A, Kramer DM. Depletion of stromal P(i) induces high 'energy-dependent' antenna exciton quenching (q(E)) by decreasing proton conductivity at CF(O)-CF(1) ATP synthase. Plant, Cell & Environment. 2008;31:235–243. doi: 10.1111/j.1365-3040.2007.01753.x. [DOI] [PubMed] [Google Scholar]
- Telfer A, Pascal A, Gall A. Carotenoids in Photosynthesis. In: Britton G, Liaaen-Jensen S, Pfander H, editors. Carotenoids, Vol 4 . Birkhäuser Basel; 2008. pp. 265–308. [Google Scholar]
- Telfer A. Singlet oxygen production by PSII under light stress: mechanism, detection and the protective role of β-carotene. Plant and Cell Physiology. 2014;55:1216–1223. doi: 10.1093/pcp/pcu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyystjarvi E, Aro EM. The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity. PNAS. 1996;93:2213–2218. doi: 10.1073/pnas.93.5.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyystjärvi E. Photoinhibition of Photosystem II. International Review of Cell and Molecular Biology. 2013;300:243–303. doi: 10.1016/B978-0-12-405210-9.00007-2. [DOI] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature. 2011;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- van Gorkom HJ. Electroluminescence. Photosynthesis Research. 1996;48:107–116. doi: 10.1007/BF00041001. [DOI] [PubMed] [Google Scholar]
- Vos MH, van Gorkom HJ, van Leeuwen PJ. An electroluminescence study of stabilization reactions in the oxygen-evolving complex of Photosystem II. Biochimica Et Biophysica Acta . 1991;1056:27–39. doi: 10.1016/S0005-2728(05)80069-6. [DOI] [Google Scholar]
- Zhang S, Apel K, Kim C. Singlet oxygen-mediated and EXECUTER-dependent signalling and acclimation of Arabidopsis thaliana exposed to light stress. Philosophical Transactions of the Royal Society. 2014;369:e16921. doi: 10.1098/rstb.2013.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]