Abstract
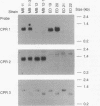
The cyclophilins are a family of ubiquitous eukaryotic proteins first identified by high affinity for cyclosporin A (CsA). The immunosuppressant and cytotoxic effects of CsA are thought to result from formation of a toxic complex between cyclophilin and CsA rather than from inhibition of cyclophilin function. The physiological role(s) of the cyclophilins is unknown. Cyclophilins have in vitro peptidylprolyl cistrans isomerase (PPIase) activity, and thus may be involved in protein folding in vivo. We have isolated a yeast cyclophilin gene, CPR3, which encodes a presumptive mitochondrial isoform. While CPR3 disruption mutants lack any phenotype at 30 degrees C, they are unable to grow on L-lactate at 37 degrees C. Disruptions of two other cyclophilin genes (CPR1, CPR2) and of FPR1, the gene encoding an FK506 binding protein with PPIase activity, do not affect growth on L-lactate at 37 degrees C. L-Lactate metabolism requires transcriptional induction of CYB2, the gene encoding flavocytochrome b2; cpr3 mutants induce transcription of this gene normally. This result demonstrates a conditional lethal phenotype for a cyclophilin mutation and presents a system for genetic and biochemical analysis of cyclophilin function.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- APPLEBY C. A., MORTON R. K. Crystalline cytochrome b2 and lactic dehydrogenase of yeast. Nature. 1954 Apr 24;173(4408):749–752. doi: 10.1038/173749a0. [DOI] [PubMed] [Google Scholar]
- Attardi G., Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Caroni P., Rothenfluh A., McGlynn E., Schneider C. S-cyclophilin. New member of the cyclophilin family associated with the secretory pathway. J Biol Chem. 1991 Jun 15;266(17):10739–10742. [PubMed] [Google Scholar]
- Cheng M. Y., Hartl F. U., Martin J., Pollock R. A., Kalousek F., Neupert W., Hallberg E. M., Hallberg R. L., Horwich A. L. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 1989 Feb 16;337(6208):620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- Danielson P. E., Forss-Petter S., Brow M. A., Calavetta L., Douglass J., Milner R. J., Sutcliffe J. G. p1B15: a cDNA clone of the rat mRNA encoding cyclophilin. DNA. 1988 May;7(4):261–267. doi: 10.1089/dna.1988.7.261. [DOI] [PubMed] [Google Scholar]
- Fischer G., Wittmann-Liebold B., Lang K., Kiefhaber T., Schmid F. X. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature. 1989 Feb 2;337(6206):476–478. doi: 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]
- Friedman J., Weissman I. Two cytoplasmic candidates for immunophilin action are revealed by affinity for a new cyclophilin: one in the presence and one in the absence of CsA. Cell. 1991 Aug 23;66(4):799–806. doi: 10.1016/0092-8674(91)90123-g. [DOI] [PubMed] [Google Scholar]
- Gething M. J., Sambrook J. Protein folding in the cell. Nature. 1992 Jan 2;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Guiard B. Structure, expression and regulation of a nuclear gene encoding a mitochondrial protein: the yeast L(+)-lactate cytochrome c oxidoreductase (cytochrome b2). EMBO J. 1985 Dec 1;4(12):3265–3272. doi: 10.1002/j.1460-2075.1985.tb04076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haendler B., Hofer-Warbinek R., Hofer E. Complementary DNA for human T-cell cyclophilin. EMBO J. 1987 Apr;6(4):947–950. doi: 10.1002/j.1460-2075.1987.tb04843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haendler B., Keller R., Hiestand P. C., Kocher H. P., Wegmann G., Movva N. R. Yeast cyclophilin: isolation and characterization of the protein, cDNA and gene. Gene. 1989 Nov 15;83(1):39–46. doi: 10.1016/0378-1119(89)90401-0. [DOI] [PubMed] [Google Scholar]
- Handschumacher R. E., Harding M. W., Rice J., Drugge R. J., Speicher D. W. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984 Nov 2;226(4674):544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- Hasel K. W., Glass J. R., Godbout M., Sutcliffe J. G. An endoplasmic reticulum-specific cyclophilin. Mol Cell Biol. 1991 Jul;11(7):3484–3491. doi: 10.1128/mcb.11.7.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Movva N. R., Hiestand P. C., Hall M. N. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1948–1952. doi: 10.1073/pnas.88.5.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochgeschwender U., Sutcliffe J. G., Brennan M. B. Construction and screening of a genomic library specific for mouse chromosome 16. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8482–8486. doi: 10.1073/pnas.86.21.8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koser P. L., Bergsma D. J., Cafferkey R., Eng W. K., McLaughlin M. M., Ferrara A., Silverman C., Kasyan K., Bossard M. J., Johnson R. K. The CYP2 gene of Saccharomyces cerevisiae encodes a cyclosporin A-sensitive peptidyl-prolyl cis-trans isomerase with an N-terminal signal sequence. Gene. 1991 Dec 1;108(1):73–80. doi: 10.1016/0378-1119(91)90489-x. [DOI] [PubMed] [Google Scholar]
- Liu J., Farmer J. D., Jr, Lane W. S., Friedman J., Weissman I., Schreiber S. L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991 Aug 23;66(4):807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- Liu J., Walsh C. T. Peptidyl-prolyl cis-trans-isomerase from Escherichia coli: a periplasmic homolog of cyclophilin that is not inhibited by cyclosporin A. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4028–4032. doi: 10.1073/pnas.87.11.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer P. E., Krieg U. C., Hwang S. T., Vestweber D., Schatz G. A precursor protein partly translocated into yeast mitochondria is bound to a 70 kd mitochondrial stress protein. EMBO J. 1990 Dec;9(13):4315–4322. doi: 10.1002/j.1460-2075.1990.tb07880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh B. H., Stamnes M. A., Seavello S., Harris G. L., Zuker C. S. The ninaA gene required for visual transduction in Drosophila encodes a homologue of cyclosporin A-binding protein. Nature. 1989 Mar 2;338(6210):67–70. doi: 10.1038/338067a0. [DOI] [PubMed] [Google Scholar]
- Spik G., Haendler B., Delmas O., Mariller C., Chamoux M., Maes P., Tartar A., Montreuil J., Stedman K., Kocher H. P. A novel secreted cyclophilin-like protein (SCYLP). J Biol Chem. 1991 Jun 15;266(17):10735–10738. [PubMed] [Google Scholar]
- Stamnes M. A., Shieh B. H., Chuman L., Harris G. L., Zuker C. S. The cyclophilin homolog ninaA is a tissue-specific integral membrane protein required for the proper synthesis of a subset of Drosophila rhodopsins. Cell. 1991 Apr 19;65(2):219–227. doi: 10.1016/0092-8674(91)90156-s. [DOI] [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Hayano T., Suzuki M. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature. 1989 Feb 2;337(6206):473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- Tropschug M., Barthelmess I. B., Neupert W. Sensitivity to cyclosporin A is mediated by cyclophilin in Neurospora crassa and Saccharomyces cerevisiae. Nature. 1989 Dec 21;342(6252):953–955. doi: 10.1038/342953a0. [DOI] [PubMed] [Google Scholar]
- Tropschug M., Nicholson D. W., Hartl F. U., Köhler H., Pfanner N., Wachter E., Neupert W. Cyclosporin A-binding protein (cyclophilin) of Neurospora crassa. One gene codes for both the cytosolic and mitochondrial forms. J Biol Chem. 1988 Oct 5;263(28):14433–14440. [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]