Abstract
In the past decades ancient DNA research has brought numerous insights to archaeological research where traditional approaches were limited. The determination of sex in human skeletal remains is often challenging for physical anthropologists when dealing with incomplete, juvenile or pathological specimens. Molecular approaches allow sexing on the basis of sex-specific markers or by calculating the ratio of DNA derived from different chromosomes. Here we propose a novel approach that relies on the ratio of X chromosome-derived shotgun sequencing data to the autosomal coverage, thus establishing the probability of an XX or XY karyotype. Applying this approach to the individuals of the Upper Paleolithic triple burial of Dolní Věstonice reveals that all three skeletons, including the individual DV 15, whose sex has long been debated due to a pathological condition, were male.
Introduction
Sexing of human skeletal material is traditionally performed in archeology by assessing sexually dimorphic traits of the pelvis and skull and, where possible, taking into account typically gendered burial positions and grave-goods. However, the expression of these skeletal traits falls on a continuum and is population-dependent, while gendered burial positions and goods were only common in some cultures and might have reflected different attitudes towards gender and biological sex that we are not aware of. Determination of sex is further complicated if the skeleton is fragmented or incomplete, the individual is sub-adult or when a pathological condition affects the morphology. However, the biological sex of an individual can be assessed by determining the sex chromosomal karyotype. In the case of DNA derived from ancient human remains, a method has been proposed that identifies sex by considering the number of reads in shotgun DNA sequencing data that align to the X and Y chromosomes [1], which is advantageous over previous PCR-based approaches that targeted sex-specific markers [2–4] and that can easily be compromised by modern contamination [5]. However, this method relies on at least 100,000 sequences mapping to the human genome for accurate assignment, a prohibitive requirement for many badly preserved ancient remains. For the majority of prehistoric skeletons a loss of DNA due to post-mortem decay results in a fraction of lower than 0.5% endogenous human DNA in most parts of the skeleton with the rest being mostly comprised of a mix of microbial DNA from bacteria and fungi that settled the body after the individual died [6]. Furthermore, contamination by modern humans, e.g. the excavators or lab technicians, renders the analyses of authentic ancient human DNA difficult.
The Upper Paleolithic triple burial of Dolní Věstonice, site II, (part of the Dolní Věstonice-Pavlov-Milovice site complex in Moravia; Fig 1), dated to 26,640±110 BP (31,155 ± 85 calBP; GrN-14831) [7], has notoriously been difficult to interpret in regard to anthropological sexing. While the two flanking individuals, DV 13 and DV 14, could be identified as a 17–19 and a 16–17 year old male, respectively [8], the individual DV 15 in the middle position of the burial, about 20 years of age, evades osteological sexing due to a pathological, possibly congenital deformation affecting symmetry and proportion of limbs as well as tooth and pelvis morphology [9, 10]. It has been ascribed to both male [11, 12] and female sex [7, 13] and diagnosis of the pathology as the X-linked dominant form of chondrodysplasia calcificans punctata (CCP) has been put forth [14], which is lethal in most cases in males end would thus correspond with a female assignment. A previous study has found a close maternal relationship between DV 14 and DV 15, who carry identical haplotypes for the mitochondrial DNA [15] and kinship between all three individuals has been suggested based on odontological and other non-metric traits [16]. Besides this triple burial and a single male burial DV 16, excavations at Dolní Věstonice II since 1985 have unearthed a structured settlement with a number of stone and bone tools, decorative objects, as well as fragmented remains of associated human individuals making it one of the most important sites of the Central European Gravettian. As extensive rituals seem to have accompanied the burials, the site is especially intriguing for understanding the ideology and social structures of Paleolithic communities [17].
Fig 1. The triple burial of Dolní Věstonice, Moravia, dated to around 31,000 years before present.
From left to right: DV 13, DV 15, DV 14.
Applying a novel method for genetic sex determination that uses shotgun sequencing data to calculate the ratio of endogenous DNA assigned to autosomes to that assigned to the X chromosomes we are able to establish male sex for all three individuals despite a low fraction of endogenous DNA and the presence of modern contamination.
Results
Shotgun sequencing resulted in 7,641,368 to 11,902,891 merged and quality filtered reads, of which 2,788 to 16,099 unique reads of 30 bp or longer mapped to the human genome (Table 1). The endogenous DNA content was 0.21%, 0.08% and 0.03% for DV 13, DV 14 and DV 15, respectively. Mapped reads of the samples showed elevated levels of deamination towards the ends (Table 1 and S1 Fig) and a read length distribution shifted toward short reads (S2 Fig), both characteristics of ancient DNA.
Table 1. Summary of sequencing results.
| Sample | Merged, quality filtered reads | Unique human reads | % endogenous DNA | Cluster factor | % deamination at 5’-end | Contamination on mtDNA15 |
|---|---|---|---|---|---|---|
| DV 13 | 7641368 | 16099 | 0.21 | 1.77 | 27.8 | 0.9%–2.4% |
| DV 14 | 11902891 | 8945 | 0.08 | 1.01 | 6.4 | 1.9%–9.2% |
| DV 15 | 10298290 | 2788 | 0.03 | 1.83 | 5.9 | 0%–3.9% |
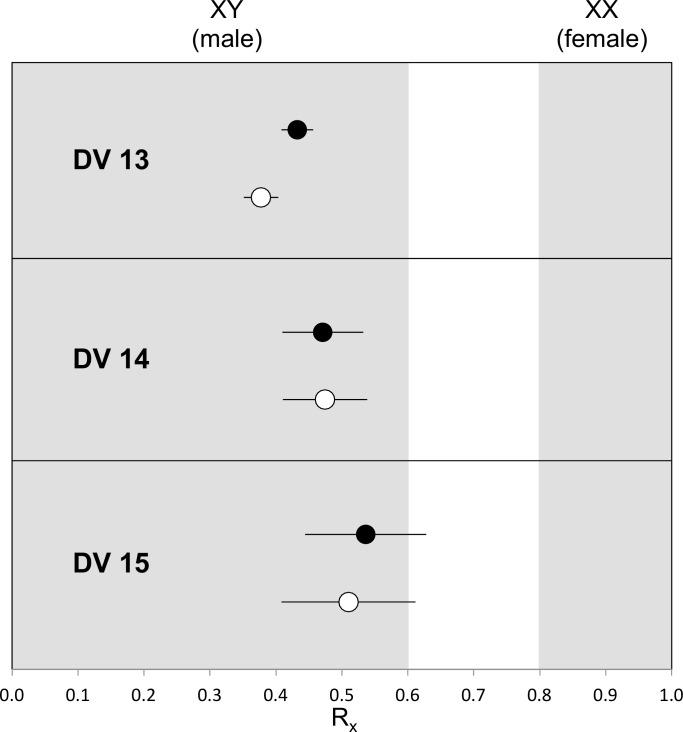
We first calculated Ry following Skoglund et al.’s method [1] (S1 Table). The chromosomal sexes of DV 13 and DV 14 are consistent with XY but not XX. But Ry could not give a sex assignment for DV 15, as there are not enough reads mapped onto the Y chromosome. Rx, defined as the ratio of the alignments to chromosome X to the alignments to autosomes, all normalized against the overall number of alignments to the reference genome, falls below 0.6 for all three individuals indicating male sex (Fig 2, S2 Table). It has been previously established that all three samples exhibit an amount of modern mtDNA contamination that ranges from a maximum of 2.4% in DV 13 to up to 9.2% in DV 14 [15] (Table 1). It can therefore be assumed that modern contamination is also present in the nuclear DNA. After excluding all sequencing reads that did not show any evidence of post-mortem damage in the form of cytosine deamination towards the 5’-end of the molecule, which is a characteristic of authentic ancient DNA [18], filtered reads were consistent with unfiltered reads in giving an Rx below 0.6 and thus a male assignment for all three individuals (Fig 2).
Fig 2. Ratio of alignments to chromosome X compared to ratio of alignments of all autosomes (Rx).
An Rx of below 0.6 indicates male sex, error bars represent the 95% CI. Male assignment is consistent for all three samples for both unfiltered sequences (black dots) and sequences that show evidence of post-mortem deamination (white dots).
However, there are only 400 reads left after the above 30bp filtration and an additional post-mortem damage filtration for sample DV15. We caution that the Rx approach is not able to handle well with a sample of only a few hundred reads. To confirm the XY karyotype in DV 15, reads mapping to the Y chromosome were checked. In total 7 sequences securely map to chromosome Y, two of which show cytosine deamination and are thus likely authentic, indicating the presence of ancient Y chromosomal DNA in those remains.
The results obtained here through low coverage shotgun sequencing data are in concordance with the ratio of the number of SNPs mapped on X chromosome and Y chromosome to those mapped on autosomes using genome-wide capture data for those three samples from Dolní Věstonice. They have all been classified as males [19].
Discussion
While reconstruction and interpretation of pre-historic events is a contentious exercise for archeologists, valuable insights into past social behaviour and attitudes toward death and afterlife can nevertheless be gained through the study of ancient burials.
The triple burial of Dolní Věstonice is especially intriguing due to the peculiarity of the individuals’ positioning–DV 13 on his side facing the central DV 15 with hands reaching the pubic region of the latter, and DV 14 laying face-down. The prominent central position of DV 15 is even more highlighted due to his pathological deformations. The fact that his sex was undeterminable by means of bone morphology and metrics suggests a unique character of this person and such individuals may have received a specific status in egalitarian societies. In addition, the three skeletons were covered partially in ochre and the whole situation was protected by burnt spruce logs and branches, obviously remains of a larger structure.
The sexing of the middle individual DV 15 as male gives us clues to the relationship the dead had to each other. The maternal kinship of DV 14 and DV 15 established by study of the mtDNA [15] as well as their close age raises the possibility of them being brothers. A sibling relationship within the triple burial has been proposed before on the basis of skeletal variants present in all three individuals, however this type of analysis cannot be conclusive, as traits that are rare in extant populations might appear at higher frequency in smaller pre-historic communities due to endogamy [16]. While a close maternal relationship of DV 13 to the other two individuals can be ruled out, a relationship to the degree of paternal half-brother cannot.
The causes of death cannot be established at this point, however, the male assignment of DV 15 rules out some proposed scenarios such as death during childbirth [13]. This individual’s severe pathological condition might have led to his early death. The differential diagnosis as the X-linked form of CCP [14] can now be considered less likely, as male individuals with the condition usually die in early childhood, however, adult male survivors have been reported in the literature and genetic mechanisms for this have been characterized [20, 21].
The molecular sexing of these badly preserved DNA samples was made possible with a novel approach that takes into account the ratio of sequence alignments to chromosome X compared to the autosomes and which gives accurate results with as little as several thousands of reads mapping to the human genome. This method is therefore suitable for light shotgun sequencing data even of samples that contain only a small percentage of endogenous DNA or that are contaminated by modern human DNA. We also see an application of an adapted version of this approach to detect major chromosomal anomalies such as trisomy 21, conditions that were undoubtedly present in pre-historic populations but that can only tentatively be diagnosed in skeletal remains through anthropological methods [22].
Methods
DNA extraction and library preparation
All pre-amplification procedures took place in the clean-room facilities at the Max-Planck-Institute for Evolutionary Anthropology in Leipzig, Germany, (DV 13, DV 14) and at the University of Tübingen, Germany, (DV 15) where procedures to minimize contamination with modern DNA are implemented [23, 24]. Sampling on the long bones of the individuals was carried out using a sterile dentistry drill. DNA was extracted from 30–160 mg per sample as previously described [23]. Negative extraction controls were included (S3 Table). From an aliquot of the extract, DNA libraries were made following a modified protocol as described [25]. Each DNA fragment was extended by individual DNA tag combinations corresponding exclusively to one sample to prevent contamination from other sequencing libraries [26]. After amplification of aliquots of these DNA libraries, shotgun sequencing was performed.
Sequencing and data analysis
High-throughput shotgun sequencing for the libraries of DV 13 and DV 14 was carried out on the Illumina Genome Analyzer IIx platform using 2 x 76 + 7 cycles, for DV 15 on the Illumina MiSeq platform for 2 x 150 + 8 + 8 cycles and for all three libraries on a HiSeq 2500 with RapidRun mode for 2 x 101 + 8 + 8 according to the manufacturer’s instructions for multiplex. Raw sequencing reads were pooled per individual and processed together with a custom pipeline that performs adapter-clipping and merging of reads overlapping at 11 or more bases as well as mapping [27]. The sequences were mapped to the complete human reference genome (hg19/GRCh37/1000Genomes) with BWA 0.6.1 [28].
Sex assignment
We first used the Ry approach [1] to infer the biological sex. Ry was performed by computing the number of reads mapped to Y chromosome as a fraction of the total number of alignments to both sex chromosomes.
We here propose a different approach to calculate the averaged normalized ratio of the X chromosome. Let f1, f2,…, f22, and fx denote the ratio of the alignments to each chromosome to the total number of alignments to autosomes and sex chromosomes, respectively. {fi}(i = 1, 2,…, 22, x) can be estimated directly from the sequenced individual. We then calculated the normalized ratio of each chromosome {ρi}(i = 1, 2,…, 22, x) by dividing {fi} by the corresponding chromosome ratio of the reference genome used for alignment.
Let Rx be the averaged normalized ratio of X chromosome:
| (1) |
If the shotgun sequencing is completely random, the Rx should be around 0.5 for male samples and 1.0 for females. We tested this approach by assigning several individuals with previous known sex assignments [29–32]. We also follow the approach of Ry [1] to use extreme values as the sex dividing lines. We first performed a linear regression to test if the numbers of sequenced and mapped reads on each chromosome are correlated with the number of reference reads. The assignment result will be invalid if there is no correlation, which means the sequencing is insufficient. We assigned a sample as male if its 95% confidence interval (CI) upper bound for Rx was lower than 0.60 and assigned a sample as female if its Rx 95% CI lower bound was higher than 0.80 (Table 2). The 95% CI was computed as Rx±1.96SE. SE is the standard error measuring the amount of variability in the Rx mean compared with 22 autosomes.
Table 2. Test of Rx approach for individuals with known sex assignments.
| Sample | Nseq | NchrX | NchrY | p-value | Rx | 95% CI | Assignment |
|---|---|---|---|---|---|---|---|
| HG00096 | 129336187 | 3404137 | 338394 | 5.857e-13 | 0.5313 | 0.5029–0.5596 | XY |
| HG00099 | 215786533 | 10712732 | 7026 | 2.200e-16 | 1.0191 | 0.9684–1.0699 | XX |
| HG00100 | 351555180 | 18015697 | 12776 | 2.200e-16 | 1.0590 | 1.0024–1.1156 | XX |
| HG00101 | 185089582 | 4997300 | 499215 | 3.813e-13 | 0.5523 | 0.5174–0.5873 | XY |
| Vi33.16 | 16648258 | 794453 | 2813 | 1.458e-15 | 0.9551 | 0.9092–1.0011 | XX |
| Vi33.25 | 15431136 | 767200 | 2124 | 2.699e-16 | 1.0042 | 0.9541–1.0542 | XX |
| Vi33.26 | 15051507 | 750642 | 2098 | 2.200e-16 | 1.0109 | 0.9587–1.0630 | XX |
| Mezmaiskaya-E733 | 23589975 | 1114078 | 3640 | 1.193e-14 | 0.9334 | 0.8864–0.9803 | XX |
| Ajv52 | 4084279 | 110151 | 9607 | 5.128e-13 | 0.5478 | 0.5131–0.5825 | XY |
| Ajv53 | 861535 | 44341 | 119 | 2.200e-16 | 1.0636 | 1.0025–1.1247 | XX |
| Ajv58 | 95232858 | 2466677 | 208840 | 1.158e-12 | 0.5159 | 0.4897–0.5422 | XY |
| Ajv59 | 214849 | 5690 | 505 | 9.797e-13 | 0.5306 | 0.5030–0.5583 | XY |
| Ajv70 | 7189980 | 181985 | 15666 | 3.529e-12 | 0.4981 | 0.4749–0.5214 | XY |
| Gok2 | 53548001 | 2429279 | 2785 | 1.754e-12 | 0.8972 | 0.8382–0.9562 | XX |
| Gok4 | 1769314 | 46570 | 3938 | 8.424e-13 | 0.5315 | 0.4992–0.5638 | XY |
| Gok5 | 770044 | 34904 | 26 | 6.264e-13 | 0.9016 | 0.8460–0.9572 | XX |
| Gok7 | 562131 | 24580 | 26 | 6.143e-12 | 0.8685 | 0.8092–0.9278 | XX |
| Ire8 | 2274888 | 60465 | 4891 | 7.349e-13 | 0.5441 | 0.5060–0.5823 | XY |
| Denisova_4 | 38626 | 1021 | 80 | 1.491e-12 | 0.5519 | 0.5054–0.5984 | XY |
| Denisova_8 | 828216 | 21641 | 1854 | 1.157e-12 | 0.5286 | 0.4939–0.5633 | XY |
Nseq: number of total alignments; NchrX: number of alignments on X chromosome; NchrY: number of alignments on Y chromosome; p-value: F-statistic p-value in linear regression of the number of reference reads with number of mapped reads.
To further test the minimum number of reads required for Rx sex assignment, we down-sampled reads with a mapping quality higher than 30 from 16 ancient individuals, and found all their sex could be confidently identified down to about 1000 reads (S4 Table).
Patterns of deamination towards read ends were analyzed and plotted with mapDamapge [33]. To account for possible modern DNA contamination the reads were additionally filtered to exclude those that did not show deamination towards the end of the molecule using PMDtools [34] with parameter—threshold 3 (S2 Table). Assignment was also performed with reads with a mapping quality higher than 30.
We provide an R script to compute Rx and assign sex (S1 File).
Supporting Information
(PDF)
(PDF)
(R)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Michael Bolus for helpful discussions and our anonymous reviewers for suggestions.
Data Availability
Data are available on the Short Read Archive, http://www.ncbi.nlm.nih.gov/sra (Accession numbers: SAMN04457495, SAMN04457496, SAMN04457497).
Funding Statement
This research was funded by the Deutsche Forschungsgemeinschaft (KR 4015/1-1 to JK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Skoglund P, Storå J, Götherström A, Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J Archaeol Sci. 2013;40(12):4477–82. [Google Scholar]
- 2.Hummel S, Herrmann B. Y-Chromosome-Specific DNA Amplified in Ancient Human Bone. Naturwissenschaften. 1991;78(6):266–7. [DOI] [PubMed] [Google Scholar]
- 3.Gibbon V, Paximadis M, Strkalj G, Ruff P, Penny C. Novel methods of molecular sex identification from skeletal tissue using the amelogenin gene. Forensic Sci Int-Gen. 2009;3(2):74–9. [DOI] [PubMed] [Google Scholar]
- 4.Daskalaki E, Anderung C, Humphrey L, Gotherstrom A. Further developments in molecular sex assignment: a blind test of 18th and 19th century human skeletons. J Archaeol Sci. 2011;38(6):1326–30. [Google Scholar]
- 5.Quincey D, Carle G, Alunni V, Quatrehomme G. Difficulties of sex determination from forensic bone degraded DNA: A comparison of three methods. Sci Justice. 2013;53(3):253–60. 10.1016/j.scijus.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 6.Allentoft ME, Sikora M, Sjogren KG, Rasmussen S, Rasmussen M, Stenderup J, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522(7555):167–72. 10.1038/nature14507 [DOI] [PubMed] [Google Scholar]
- 7.Vlček E. Die Mammutjäger von Dolni Vestonice. Archäologie und Museum. 1991;22. [Google Scholar]
- 8.Sládek V, Trinkaus E, Hillson SW, Holliday TW. The people of the Pavlovian Skeletal catalogue and osteometrics of the Gravettian fossil hominids from Dolni Vestonice and Pavlov. Svoboda J, editor. Brno: Academy of Sciences of the Czech Republic; 2000. 244 p. [Google Scholar]
- 9.Novotny V. Pelves and the sexual dimorphism in hunters of Dolni Vestonice. Sbornik Narodniho Muzea v Praze Rada B Prirodni Vedy. 1992;48(1–4):152–64. [Google Scholar]
- 10.Cerny M. Sex Determination on the Humeri and Femora of Skeletons of Dolni Vestonice. Sbornik Narodniho Muzea v Praze Rada B Prirodni Vedy. 1992;48(1–4):130–5. [Google Scholar]
- 11.Jelinek J. New Upper Paleolithic burials from Dolní Věstonice In: Toussaint M, editor. L'Aventure Humaine: 5 Millions d'Années: Études et Recherches Achéologiques de l'Université de Liège; 1992. p. 207–28. [Google Scholar]
- 12.Bruzek J, Franciscus RG, Novotny V, Trinkaus E. The Assessment of Sex In: Trinkaus E, Svoboda J, editors. Early Modern Human Evolution in Central Europe: The People of Dolní Vĕstonice and Pavlov. Dolni Vestonice Studies 12. New York: Oxford University Press; 2006. p. 46–62. [Google Scholar]
- 13.Klima B. A triple burial from the Upper Paleolithic of Dolní Věstonice, Czechoslovakia. J Hum Evol. 1987;16(7–8):831–5. [Google Scholar]
- 14.Formicola V, Pontrandolfi A, Svoboda J. The Upper Paleolithic triple burial of Dolni Vestonice: Pathology and funerary behavior. American journal of physical anthropology. 2001;115(4):372–9. [DOI] [PubMed] [Google Scholar]
- 15.Fu Q, Mittnik A, Johnson PL, Bos K, Lari M, Bollongino R, et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Current biology: CB. 2013;23(7):553–9. 10.1016/j.cub.2013.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alt KW, Pichler S, Vach W, Klima B, Vlcek E, Sedlmeier J. Twenty-five thousand-year-old triple burial from Dolni Vestonice: An ice-age family? American journal of physical anthropology. 1997;102(1):123–31. [DOI] [PubMed] [Google Scholar]
- 17.Svoboda J. The Burials: Ritual and Taphonomy In: Trinkaus E, Svoboda J, editors. Early Modern Human Evolution in Central Europe: The People of Dolní Vĕstonice and Pavlov. Dolni Vestonice Studies 12. New York: Oxford University Press; 2006. p. 15–26. [Google Scholar]
- 18.Sawyer S, Krause J, Guschanski K, Savolainen V, Paabo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PloS one. 2012;7(3):e34131 10.1371/journal.pone.0034131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Q, Posth C, Hajdinjak M, Petr M, Mallick S, Fernandes C, et al. The genetic history of Ice Age Europe. Nature. 2016; 534:200–5. 10.1038/nature17993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aughton DJ, Kelley RI, Metzenberg A, Pureza V, Pauli RM. X-linked dominant chondrodysplasia punctata (CDPX2) caused by single gene mosaicism in a male. Am J Med Genet A. 2003;116A(3):255–60. [DOI] [PubMed] [Google Scholar]
- 21.Milunsky JM, Maher TA, Metzenberg AB. Molecular, biochemical, and phenotypic analysis of a hemizygous male with a severe atypical phenotype for X-linked dominant Conradi-Hunermann-Happle syndrome and a mutation in EBP. Am J Med Genet A. 2003;116A(3):249–54. [DOI] [PubMed] [Google Scholar]
- 22.Rivollat M, Castex D, Hauret L, Tillier AM. Ancient Down syndrome: An osteological case from Saint-Jean-des-Vignes, northeastern France, from the 5–6th century AD. Int J Paleopathol. 2014;7:8–14. [DOI] [PubMed] [Google Scholar]
- 23.Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nature protocols. 2007;2(7):1756–62. [DOI] [PubMed] [Google Scholar]
- 24.Green RE, Briggs AW, Krause J, Prüfer K, Burbano HA, Siebauer M, et al. The Neandertal genome and ancient DNA authenticity. The EMBO journal. 2009;28(17):2494–502. 10.1038/emboj.2009.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor protocols. 2010;2010(6):pdb prot5448. [DOI] [PubMed] [Google Scholar]
- 26.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic acids research. 2012;40(1):e3 10.1093/nar/gkr771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peltzer A, Jäger G, Herbig A, Seitz A, Kneip C, Krause J, et al. EAGER: Efficient Ancient Genome Reconstruction. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.1000 genomes project consortium, 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skoglund P, Malmström H, Raghavan M, Storå J, Hall P, Willerslev E, et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336 (6080): 466–469. 10.1126/science.1216304 [DOI] [PubMed] [Google Scholar]
- 31.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, et al. A draft sequence of the Neandertal genome. Science. 2010; 328: 710–722. 10.1126/science.1188021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawyer S, Renaud G, Viola B, Hublin JJ, Gansauge MT, Shunkov MV, et al. Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc Natl Acad Sci U S A. 2015; 112(51):15696–700. 10.1073/pnas.1519905112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skoglund P, Northoff BH, Shunkov MV, Derevianko AP, Päabo S, Krause J, et al. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. PNAS. 2014;111(6):2229–34. 10.1073/pnas.1318934111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ginolhac A, Rasmussen M, Gilbert MT, Willerslev E, Orlando, L. mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics. 2011;27(15): 2153–55. 10.1093/bioinformatics/btr347 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(R)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
Data are available on the Short Read Archive, http://www.ncbi.nlm.nih.gov/sra (Accession numbers: SAMN04457495, SAMN04457496, SAMN04457497).