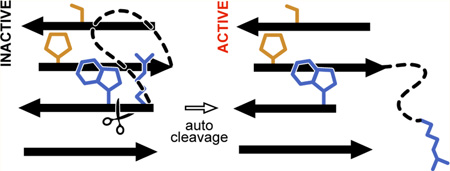
Abstract
Enterotoxigenic Bacteroides fragilis produces a secreted metalloprotease known as B. fragilis toxin (BFT), which contributes to anaerobic sepsis, colitis, and colonic malignancy in mouse models of disease. A C11 family cysteine protease, fragipain (Fpn), directly activates BFT in the B. fragilis cell by removing the BFT prodomain. Fpn is itself a proenzyme and is autoactivated upon cleavage at an arginine residue in its activation loop. We have defined the proteolytic active site of Fpn, demonstrated that Fpn autoactivation can occur by an in trans loop cleavage mechanism, and characterized structural features of the Fpn activation loop that control peptidase activity against several substrates, including BFT. An arginine residue at the autocleavage site determines the fast activation kinetics of Fpn relative to the homologous C11 protease, PmC11, which is cleaved at lysine. Arginine to alanine substitution at the cleavage site ablated peptidase activity, as did partial truncation of the Fpn activation loop. However, complete truncation of the activation loop yielded an uncleaved, pro form of Fpn that was active as a peptidase against both Fpn and BFT substrates. Thus, Fpn can be transformed into an active peptidase in the absence of activation loop cleavage. This study provides insight into the mechanism of fragipain activation and, more generally, defines the role of the C11 activation loop in the control of peptidase activity and substrate specificity.
Graphical abstract
Bacteroides spp. are the predominant anaerobes in the human gut, where they maintain a generally beneficial relationship with the host.1–3 However, these bacteria can escape the gastrointestinal tract and cause disease.1,3–8 Bacteroides fragilis is the leading cause of anaerobic sepsis and bacteremia in humans4–6,9 and possesses virulence factors that allow survival outside the host digestive tract and determine pathogenicity.1 Among the primary B. fragilis virulence determinants is a metalloprotease toxin known as fragilysin, or BFT, which is expressed by enterotoxigenic strains.10–14 BFT contributes to sepsis and pro-inflammatory disease within the colon by cleaving cadherin family proteins and thereby compromising tissue barrier integrity.15–18 BFT is synthesized as an inactive zymogen that consists of an N-terminal prodomain, a flexible linker, and a C-terminal catalytic domain.13 It is activated through cleavage of an arginine-containing site in its flexible linker11,13 by a peptidase known as fragipain (Fpn),18 a member of the C11 clostripain family.19–22
An X-ray crystal structure of Fpn revealed a cysteine-histidine catalytic dyad with cleavage specificity for Arg–Xaa bonds.18 Notably, Fpn contains its own arginine-containing cleavage site (R147/W148) within a flexible loop and, like clostripains23–25 and related proteases,22,26–29 is self-cleaving. The crystal structure of Fpn captured the protein in a trans-cleaved state with the loop arginine projecting into the active site of an adjacent Fpn molecule in the crystal lattice.18 As observed in related cysteine proteases,29–33 autocleavage is presumed to enhance Fpn peptidase activity by changing active site accessibility and/or restructuring catalytic residues. The goal of this study was to assess the trans autocleavage mechanism suggested by the Fpn crystal structure and to characterize the role of the cleavage loop in control of Fpn peptidase activity.
We expressed and purified a set of Fpn loop and active site mutants and evaluated their autopeptidase activities and their ability to process BFT. We demonstrate that an intact Cys-His active site is required for autocleavage and BFT cleavage, and that Fpn can cut itself in trans. Substitution of arginine with lysine at the Fpn cleavage site reduced autopeptidase activity, while arginine to alanine substitution ablated Fpn autocleavage and BFT processing. Truncating the Fpn cleavage loop by five residues resulted in a total loss of peptidase activity, while complete removal of this same loop yielded an enzyme that was uncleaved yet active as a peptidase against Fpn and BFT substrates. Thus, loop removal results in a pro form of Fpn that is enzymatically active, albeit less active than wild-type Fpn. Our data provide evidence of a model in which the Fpn cleavage loop functions to regulate peptidase activity both by steric occlusion of the Fpn active site and by controlling the active site conformation.
MATERIALS AND METHODS
Recombinant Expression Plasmids
Strains expressing wild-type Fpn, FpnH135A+C180A, and BFT have been described previously.18 Primers used to introduce deletions and produce FpnR147K, FpnR147A, FpnLΔ5, FpnLΔ7, and FpnLΔ7+R147A mutant proteins are listed in Table S1. pET-upstream and T7-term primers were used to amplify the different inserts, and the pET28-fpn plasmid was used as a template. The 19 N-terminal amino acids of Fpn, corresponding to the lipoprotein signal sequence, were absent from the tagged recombinant proteins. After purification, the different polymerase chain reaction products were digested with NcoI–NotI restriction enzymes and ligated into the pET28c expression plasmid (KanR). Plasmids were then transformed into Escherichia coli Top10. After sequencing, plasmids with the different inserts were purified and transformed into the E. coli Rosetta (DE3) pLysS strain for protein expression (see Table S2 for strain information).
Protein Expression and Purification
A 50 mL overnight Luria broth (LB, Fisher Scientific) culture inoculated with the protein expression strains (see Table S2 for strain information) was used to inoculate 300 mL of LB medium supplemented with the appropriate antibiotics. Overexpression of C-terminally His-tagged Fpn and BFT proteins was induced at an OD600 of ~0.8 (37 °C, 220 rpm) by adding 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG, GoldBio). After being induced for 5 h, cells were harvested by centrifugation at 11000g for 20 min at 4 °C. Cell pellets were resuspended in 10 mL of 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 10 mM imidazole buffer supplemented with 5 µg/mL DNase I. Cells were disrupted by one passage in a microfluidizer (Microfluidics LV1). The resulting cell lysate was clarified by centrifugation at 39000g for 20 min at 4 °C.
Purification of His-tagged proteins was performed using nickel affinity chromatography (NTA resin, GE Healthcare). After the clarified lysate samples had been bound to the column, three washing steps were performed using 10, 75, and 200 mM imidazole Tris-NaCl buffers followed by elution with 500 mM imidazole Tris-NaCl buffer. All purification steps were performed at 4 °C. The protein purity of the different samples was assessed with a 14% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel stained with Coomassie Blue. For every construct, protein concentrations were estimated by UV absorption spectroscopy (280 nm) using a NanoDrop 1000 spectrophotometer (Thermo Scientific).
Protease Cleavage Assays
After purification, the different Fpn mutant proteins as well as BFT were dialyzed overnight against a Tris-NaCl buffer containing 10 mM Tris (pH 7.4) and 150 mM NaCl. To assess peptidase digestion patterns, 5 µL of the different Fpn mutant proteins (at 20 µM) was mixed with 5 µL of wild-type Fpn or FpnH135A+C180A proteins (at 20 µM) or with 5 µL of the BFT toxin (at 20 µM). These 10 µL reaction mixtures were then incubated at 37 °C for 45 min. After incubation, 5 µL of SDS–PAGE loading buffer was added to the reaction mixture and boiled for 2 min at 95 °C. These 15 µL samples were then completely loaded on a 14% SDS–PAGE gel, resolved, and Coomassie stained.
Nα-Benzoyl-l-arginine Ethyl Ester (BAEE) Assay
One milliliter of phosphate buffer [67 mM NaH2PO4 (pH 7.5)] containing 200 nM wild-type or mutant Fpn proteins was mixed with 1.25 mM BAEE. The absorbance at 253 nm was measured using a UV–vis spectrophotometer (Shimadzu UV-1650pc) and a quartz cuvette. Experiments were conducted in triplicate.
Protease Kinetic Assay
After purification and dialysis, 100 µL of a solution containing 5 µM wild-type Fpn or FpnLΔ7+R147A and 5 µM BFT or FpnH135+C180A was incubated at 37 °C. Aliquots (10 µL) were taken at regular time intervals, mixed with 5 µL of SDS–PAGE loading buffer, and boiled for 2 min at 95 °C. The 15 µL samples were then loaded and resolved on a 14% SDS–PAGE gel and Coomassie stained. After destaining, intensities of the bands corresponding to cleaved BFT or FpnH135+C180A were quantified using ImageJ34 and converted into a percent cleaved value. Data were plotted and fitted using Prism. Digestion kinetic assays were conducted in triplicate with three independent purifications of each protein.
Generation of B. fragilis Strains
To introduce fpn mutant alleles into a Δfpn B. fragilis strain, the replicating pAH2 plasmid (CliR) was used (gift of A. Hecht); cloning details were previously described.18
Western Blot Analysis
Preparation of B. fragilis cell lysates and supernatants for Western blot analysis was performed as previously described,18 using the following primary and secondary antibodies: α-Fpn rabbit polyclonal antisera (1:2500), α-BFT rabbit polyclonal antisera (1:2000), α-RpoA (1:20000), and α-rabbit Alexa Fluor 680.
Protein Structure Alignments
Structural alignments and root-mean-square deviation (rmsd) calculations were performed using PyMOL (PyMOL Molecular Graphics System, version 1.7.4, Schrödinger, LLC). Residue substitutions in the Fpn active site were performed using Coot.35
RESULTS
The Cysteine Protease Fpn Can Autocleave in Trans
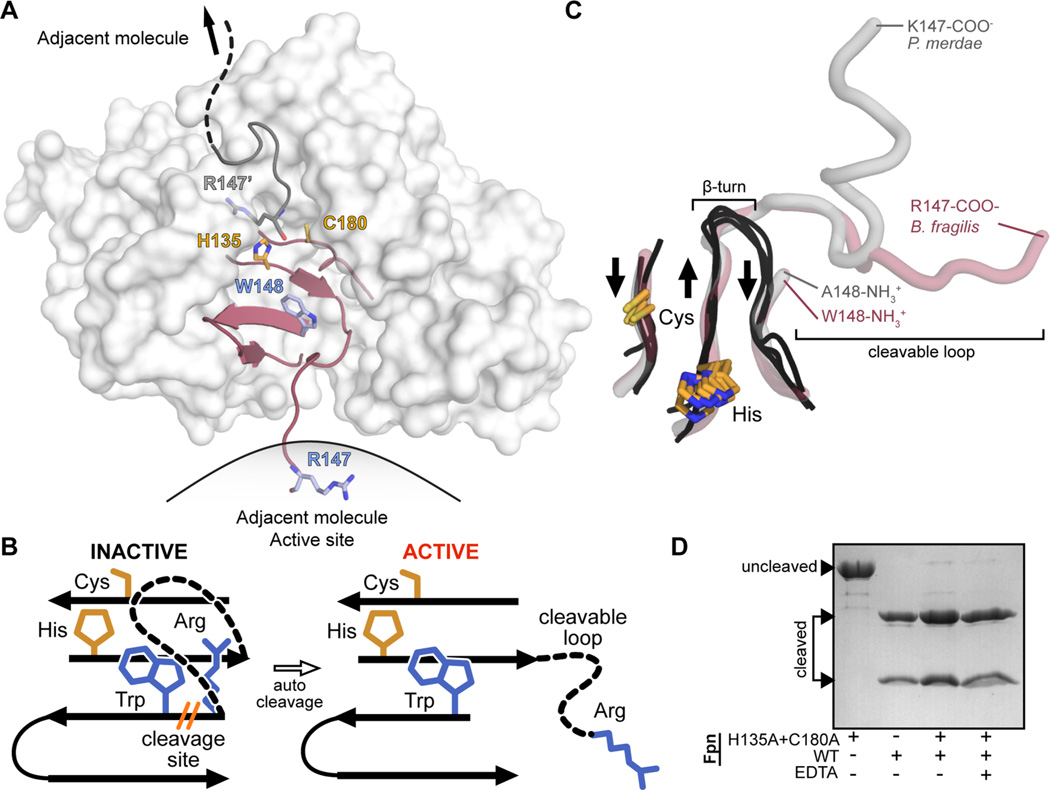
In a previous study,18 we crystallized and determined the structure of the Fpn cysteine protease [Protein Data Bank (PDB) entry 5DYN (http://dx.doi.org/10.15785/SBGRID/225)]. This structure uniquely captured the active site bound to the cleaved loop of an adjacent Fpn molecule in trans (Figure 1A). The site of Fpn autocleavage is contained in a flexible loop region of eight residues that connect β-strand 6 to β-strand 7. Within β-strand 6 is a histidine residue (H135) that forms the protein active site with a proximal cysteine (C180) (Figure 1A,B). The structure of Fpn is similar to that of the clostripain-like protein PmC11 of Parabacteroides merdae,36 which has an identical fold, and a closely related active site and loop cleavage site (Figure 1C). Though PmC11 and Fpn have a low level of structural homology to cysteine proteases such as legumain (PDB entry 4AW9),30 caspase-3 (PDB entry 1NMS),37 and gingipain (PDB entry 4RBM),38 the active sites are related (Figure 1C). However, cleavable activation loops are not present at equivalent positions in these proteases. This suggests that the activation loop present in the C11 peptidases Fpn and PmC11 may serve a unique structural role in controlling peptidase activity.
Figure 1.
B. fragilis Fpn active site and cleavable activation loop. Residue numbering includes the 19-amino acid predicted signal peptide present at the N-terminus of Fpn. (A) Surface representation of Fpn (white) with the β-strands constituting the active site colored maroon (PDB entry 5DYN). B. fragilis Fpn was crystallized in a cleaved form (cleavage site R147/W148, side chains colored light blue, catalytic dyad H135 and C180, side chains colored orange), with the cleaved region trapped in the active site of the neighboring molecule. (B) Cartoon representation of the B. fragilis Fpn active site and cleavable loop. When Fpn is inactive, the cleavable loop is intact and inhibits the active site composed of the catalytic dyad, H135 and C180 (orange sticks). Fpn is activated upon cis or trans loop cleavage between residues R147 and W148 (blue sticks), which modifies active site accessibility. (C) Superposition of the active site regions of legumain (PDB entry 4AW9), caspase-3 (PDB entry 1NMS), gingipain (PDB entry 4RBM), PmC11 (PDB entry 3UWS), and Fpn structures (PDB entry 5DYN). β-Strands composing the active sites of legumain, caspase-3, and gingipain are shown as black loops, and the equivalent regions in PmC11 and Fpn are show as transparent gray and red tubes, respectively. The aligned catalytic dyad side chains of cysteine and histidine are shown as orange sticks. The β-turn present in legumain, caspase-3, and gingipain structures and the cleaved loops present at the equivalent position in PmC11 and Fpn structures are highlighted. Residues forming the termini of the cleavage sites are annotated. (D) Wild-type Fpn trans cleavage assay, in which Fpn was incubated with the active site mutant FpnH135A+C180A, in the presence (+) or absence (−) of EDTA.
The Fpn crystal structure provided evidence of a model in which the loop is cut in trans. To test this model, we first generated and purified Fpn active site mutant protein (FpnH135A+C180A). Purified FpnH135A+C180A resolved as a single polypeptide by SDS–PAGE, confirming that this mutant has no autocleavage activity (Figure 1D). Wild-type Fpn resolved as two bands (at ≈14 and 28 kDa), consistent with autocleavage at a single site. It was not known whether wild-type Fpn autocleaves in cis, in trans, or both. As such, we tested whether wild-type Fpn could cut FpnH135A+C180A in trans. We mixed the two proteins, incubated them for 45 min at 37 °C, and resolved the mixture by SDS–PAGE. The high-molecular weight band at ≈42 kDa corresponding to uncleaved and inactive FpnH135A+C180A disappeared, and we observed only the two cut bands on a gel (Figure 1D). This provides evidence that FpnH135A+C180A is cut by wild-type Fpn in trans and supports the trans-cleavage activation mechanism evidenced by our crystal structure. We further tested the concentration dependence of this cleavage reaction by mixing 5 µL of inactive FpnH135A+C180A (20 µM) with 5 µL of wild-type Fpn at various concentrations (from 20 to 0.01 µM). After being incubated at 37 °C for 5 min, samples were resolved by SDS–PAGE. Cleavage of FpnH135A+C180A by wild-type Fpn is concentration-dependent; 0.125 µM wild-type Fpn was not sufficient to completely cleave FpnH135A+C180A (Figure S1).
We next tested whether Fpn activity required divalent cations as described for some cysteine proteases.22 We performed the same cleavage experiment between wild-type Fpn and the FpnH135A+C180A active site mutant in the presence of 50 mM EDTA. After incubation for 45 min at 37 °C, the band corresponding to uncleaved FpnH135A+C180A was absent, providing evidence that Fpn peptidase activity does not require divalent cations (Figure 1D). This confirms what has been described for Parabacteroides merdae PmC11.36
Fpn and PmC11 Have Related Structures and Active Sites but Differing Autocleavage Kinetics
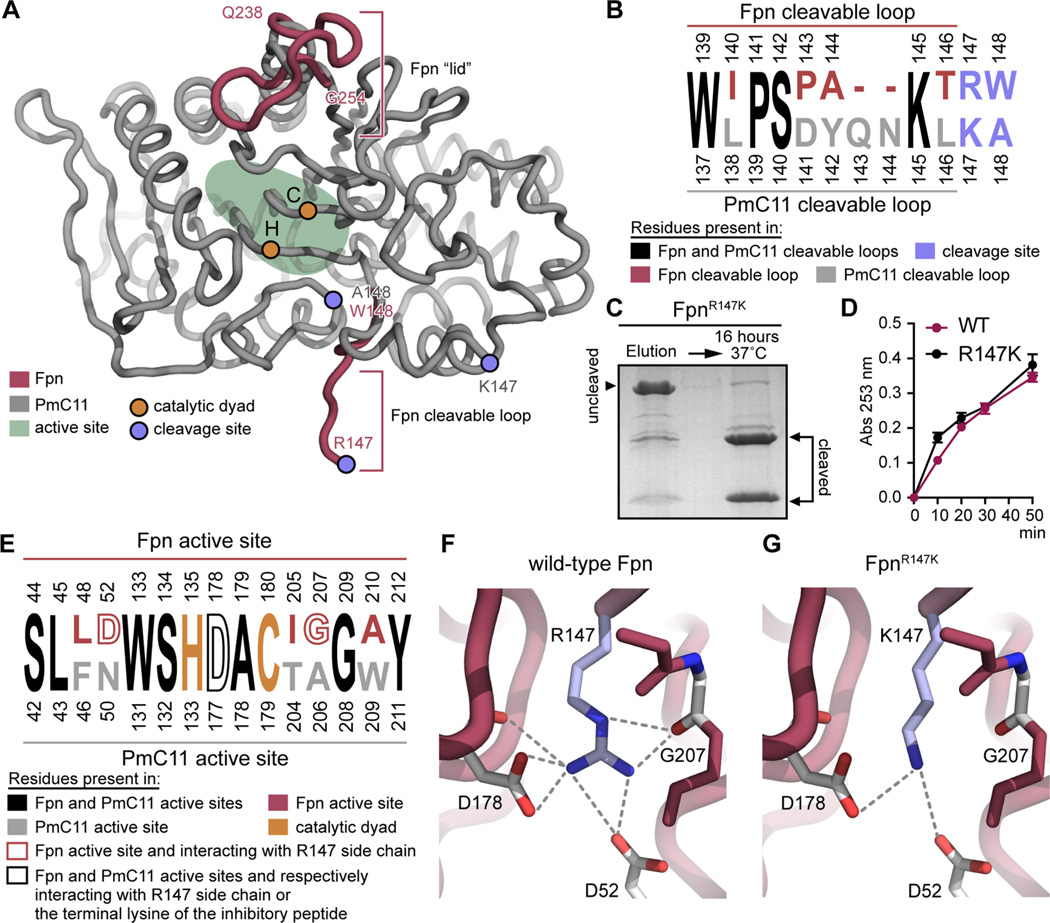
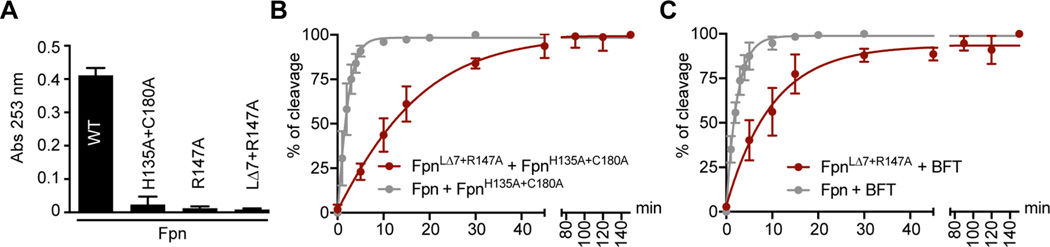
P. merdae PmC11 (PDB entry 3UWS)36 and B. fragilis Fpn (PDB entry 5DYN)18 have 35% identical sequences and a high degree of overall tertiary structural homology (rmsd of Cα coordinates = 0.81 Å) (Figure 2A). Differences in structure are evident in the cleavable loop. An additional loop (Q238–G254) is present in only Fpn and acts like a lid, partially covering the active site. In the Fpn crystal structure, the cleavable loop is eight residues long, cut, and unfolded and terminates at an arginine residue (R147). The corresponding loop in PmC11 is longer (10 amino acids) and helical and terminates at a lysine residue (K147) (Figures 1C and 2A,B). We tested whether substitution of R147 with lysine (as observed in PmC11) affects autocleavage. Like PmC11,36 FpnR147K was purified as a partially cleaved protein that resolved as three bands as determined by SDS–PAGE (at ≈42, 28, and 14 kDa) (Figure 2C). A 16 h incubation at 37 °C resulted in complete cleavage of the protein, providing evidence that a lysine-containing cleavage site is not as efficient a substrate as an arginine-containing site for this class of peptidase. When we compared the ability of wild-type Fpn and fully processed FpnR147K mutant protein to cleave the synthetic substrate Nα-benzoyl-l-arginine ethyl ester (BAEE), both proteins showed equivalent activities. Thus, FpnR147K has specifically reduced cleavage kinetics for the lysine-containing polypeptide in the cleavage loop but not for the small substrate BAEE (Figure 2D).
Figure 2.
Comparison of B. fragilis Fpn and P. merdae PmC11 structures. Residue numbering includes the 19-amino acid predicted signal peptide present at the N-terminus of Fpn. (A) Structural alignment of P. merdae PmC11 (PDB entry 3UWS) (gray) and B. fragilis Fpn (PDB entry 5DYN) (maroon). Only regions of Fpn that are structurally distinct from PmC11 are presented. Positions of catalytic dyad residues are colored orange; positions of cleavage site residues are colored blue, and the active site is delimited in light green. (B) Alignment of the amino acid residues present in P. merdae PmC11 and B. fragilis Fpn cleavable loops. Numbered residues are colored black when they are present in both PmC11 and Fpn, maroon when they are present in only Fpn, and gray when they are present in only PmC11. Basic residues present at the PmC11 and Fpn cleavage sites are colored light blue. (C) Autocleavage of FpnR147K immediately after purification and 16 h after purification at 37 °C. (D) BAEE cleavage assay. Cleavage of 1250 µM BAEE by 200 nM wild-type Fpn or FpnR147K at 37 °C monitored at 253 nm. The experiment was conducted in triplicate; error bars represent standard deviations (mean ± standard deviation). (E) Alignment of residues present in P. merdae PmC11 and B. fragilis Fpn active sites. Numbered residues are colored black when they are present in both PmC11 and Fpn active sites, maroon when they are present in only the Fpn active site, and gray when they are present in only the PmC11 active site. Residues constituting the catalytic dyad are colored orange, and residues interacting with the arginine side chain in the Fpn structure (PDB entry 5DYN) and with the lysine side chain of the inhibitory peptide in the PmC11 structure (PDB entry 4YEC) are shown in empty maroon and black letters, respectively. (F) Polar interactions between the R147 side chain (blue stick) and the residues present in the Fpn active site (white sticks). (G) Model of the lysine side chain (blue stick) at position 147 in the Fpn active site and prediction of the polar interactions. Polar interactions were determined using PyMOL with a distance cutoff of 3.6 Å.
We then compared the structures of the Fpn and PmC11 active sites. Six residues differ between Fpn and PmC11. Only residues D52, D178, and G207 interact with the R147 side chain in Fpn (Figure 2E,F). In PmC11 cocrystallized with a peptide-like inhibitor (PDB entry 4YEC), the side chain of the terminal lysine residue interacts with D177 only (Figure 2E). Given these structural data, substitution of R147 with lysine is predicted to result in fewer polar contacts with the surrounding residues present in the Fpn active site (Figure 2G). This may explain the difference in autocleavage kinetics between Fpn and PmC11.
Loop Cleavage Activates Fpn Peptidase
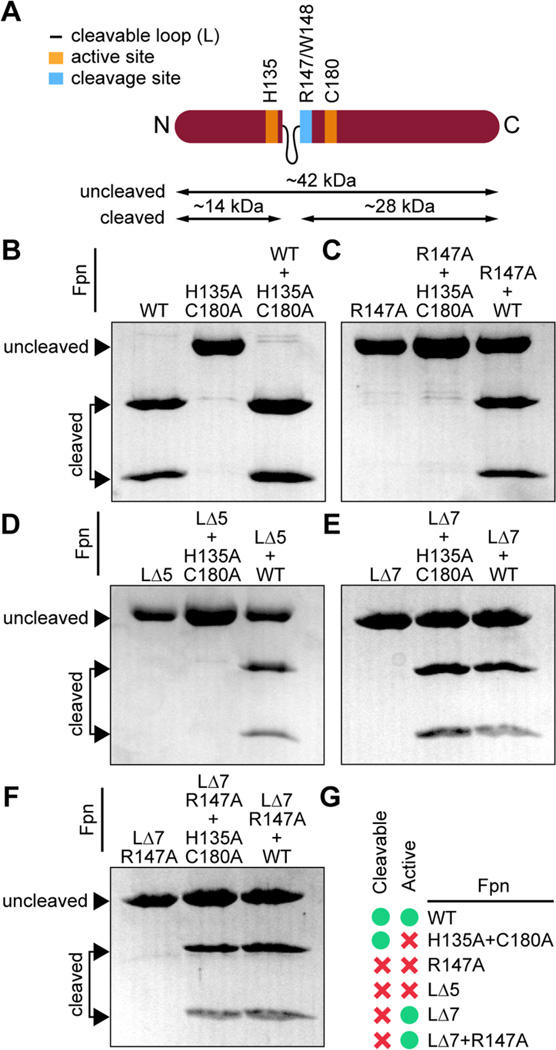
We tested whether an Fpn mutant with an intact active site but missing the arginine at the cleavage site (FpnR147A) would cut in trans. A cleaved protein runs as two fragments (≈14 and 28 kDa) on a SDS–PAGE gel (Figure 3A,B). Like FpnH135A+C180A, an FpnR147A mutant was purified as a single polypeptide with a molecular weight of ≈42 kDa, demonstrating that it was uncleaved (Figure 3C). Mixing FpnR147A with FpnH135A+C180A yielded no cleaved product. This provides evidence that Fpn loop cleavage is a required step in activation of Fpn peptidase function. As a positive control, we assessed FpnH135A+C180A cleavage by wild-type Fpn at the same time interval and observed trans cutting (Figure 3B). Wild-type Fpn failed to cleave FpnR147A, demonstrating that mutation of the arginine at the cleavage site ablates cutting by wild-type Fpn in trans (Figure 3C).
Figure 3.
Protease activity of Fpn active site and cleavage loop mutants. (A) Cartoon representation of the Fpn protein with the catalytic site (H135+C180) colored orange, the cleavage site (R147/W148) colored blue, and the cleavable loop represented as a thin black line. Sizes of the uncleaved protein (42 kDa) and of the two cleavage products (14 and 28 kDa) are delimited. The N- and C-termini are annotated. (B–F) Resolution of cleaved and uncleaved products by 14% SDS–PAGE. (B) Left lane, wild-type Fpn; middle lane, FpnH135A+C180A (active site mutant); right lane, FpnH135A+C180A cleaved by wild-type Fpn. (C) Left lane, FpnR147A (cleavage site mutant); middle lane, FpnH135A+C180A in the presence of inactive FpnR147A; right lane, FpnR147A in the presence of cleaved wild-type Fpn. (D) Left lane, FpnLΔ5 (short deletion of the cleavable loop); middle lane, FpnH135A+C180A in the presence of FpnLΔ5; right lane, FpnLΔ5 in the presence of cleaved wild-type Fpn. (E) Left lane, FpnLΔ7 (full deletion of the cleavable loop); middle lane, FpnH135A+C180A in the presence of FpnLΔ7; right lane, FpnLΔ7 in the presence of cleaved wild-type Fpn. (F) Left lane, FpnLΔ7+R147A (full deletion of the cleavable loop and cleavage site mutant); middle lane, FpnH135A+C180A in the presence of active FpnLΔ7+R147A; right lane, FpnLΔ7+R147A in the presence of cleaved wild-type Fpn. (G) Tabulation of Fpn active site and loop mutant peptidase activity and cleavability (green dot for activity and/or cleavability observed, red cross for no activity and/or cleavability observed).
Partial Truncation of the Cleavage Loop Results in Lower in Trans Peptidase Activity
As described above, the Fpn cleavage site (R147/W148) is positioned at the end of a eight-residue flexible loop (…WIPSPAKTR/W…). We hypothesized that this loop physically occludes the accessibility of the active site to the substrate. Loop cleavage may serve to increase active site accessibility and permit in trans peptidase activity. To test this hypothesis, we engineered a small loop (L) mutant from which five amino acids (ΔS142–T146) were deleted. Purified FpnLΔ5 resolved as a single 42 kDa band by SDS–PAGE (Figure 3D). This protein was incapable of cleaving the FpnH135A+C180A active site mutant in trans and could not be cleaved by wild-type Fpn in trans (Figure 3D). Thus, removing these five residues of the Fpn cleavage loop either blocks accessibility of the active site or introduces structural constraints on Fpn that are incompatible with peptidase activity.
Complete Removal of the Fpn Cleavage Loop Yields an Enzyme That Is Active in the Pro Form
To further explore the functional role of the Fpn cleavage loop, we engineered an additional Fpn mutant with the entire loop region deleted (ΔI140–T146). On the basis of published cysteine protease structures, complete loop deletion should result in an active site that is closer in geometry to the legumain, gingipain, or caspase-3 active sites (Figure 1C). This “loopless” mutant (FpnLΔ7) resolved as an uncleaved 42 kDa polypeptide by SDS–PAGE. Though FpnLΔ7 was evidently in the pro form, it was capable of cleaving FpnH135A+C180A in trans. Wild-type Fpn did not cleave loopless Fpn in trans (Figure 3E). To rule out the possibility that the observed peptidase activity of FpnLΔ7 was due to a small amount of cleaved protein present in the sample, we engineered a mutant with an alanine at the cleavage site. This mutant (FpnLΔ7+R147A) had the same characteristics as FpnLΔ7: it was purified as a single polypeptide, cleaved FpnH135A+C180A, and could not be processed by wild-type Fpn (Figure 3F). We conclude that complete removal of the cleavable loop of Fpn results in a pro form of the enzyme that is enzymatically active; i.e., FpnLΔ7 does not require loop cleavage for activity. A table summarizing the activity and cleavability of each Fpn mutant is presented in Figure 3G.
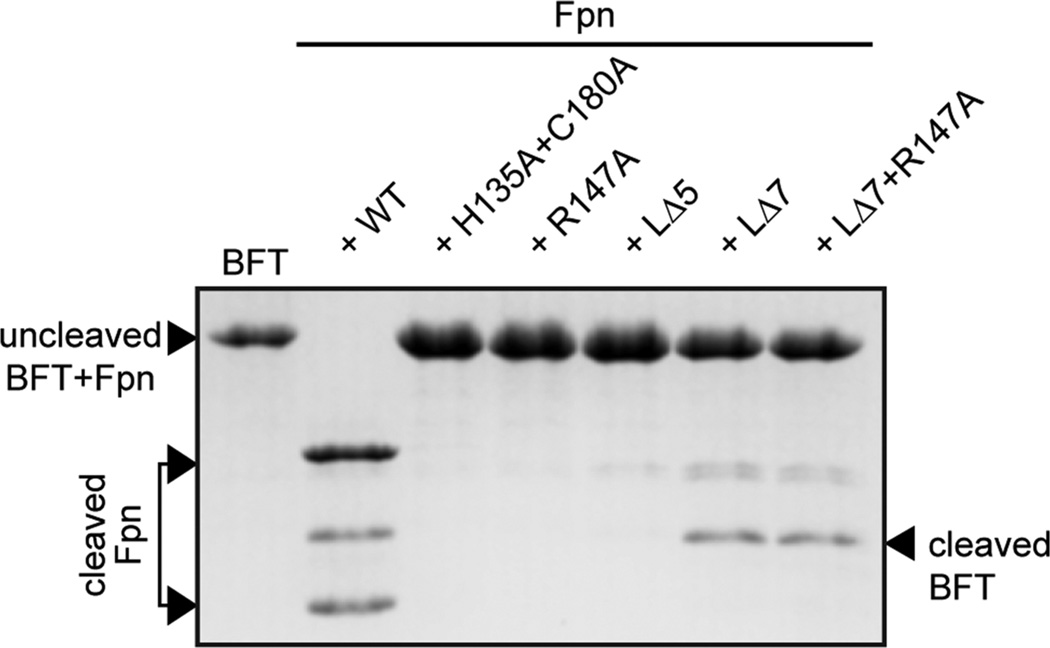
Loopless Fpn Processes the Metalloprotease Toxin BFT
In B. fragilis, a target of Fpn is the metalloprotease toxin BFT.18 Cleavage of pro-BFT by Fpn activates BFT, which proteolyzes E-cadherin and VE-cadherin in the mammalian host. We tested the ability of our different Fpn mutants to cleave pro-BFT. The primary Fpn cut site in BFT is located after residue R211; an alternative but minor cut site is present after residue R162.18 As expected, BFT was cut by wild-type Fpn, which resulted in a 17 kDa BFT fragment on a SDS–PAGE gel. The active site mutant protein (FpnH135A+C180A), the cleavage site mutant protein (FpnR147A), or the “short” deletion loop mutant protein (FpnLΔ5) had no observable enzymatic activity against BFT after incubation for 45 min at 37 °C (Figure 4). However, the loopless mutant proteins (FpnLΔ7 and FpnLΔ7+R147A) cleaved BFT. This demonstrates that the loopless pro form mutants of Fpn can process BFT as a substrate (Figure 4).
Figure 4.
Testing Fpn active site and loop mutants as proteases of the B. fragilis toxin, BFT. Wild-type Fpn cleavage of BFT results in a 17 kDa band on a SDS–PAGE gel. The active site mutant (FpnH135A+C180A), the cleavage site mutant (FpnR147A), and the loop mutant (FpnLΔ5) do not process BFT after incubation at 37 °C for 45 min. A loopless mutant of Fpn (FpnLΔ7) and a loopless mutant of Fpn harboring a substitution of the arginine residue at the loop cleavage site (FpnLΔ7+R147A) cleave BFT.
Loopless Pro-Fpn Has a Peptidase Activity Lower Than That of Wild-Type Fpn
We next measured activities of the different Fpn mutants using BAEE as a substrate. As expected, wild-type Fpn had high activity while FpnH135A+C180A and FpnR147A were inactive (Figure 5A). Surprisingly, the loopless mutant, FpnLΔ7+R147A, was also unable to process BAEE (Figure 5A). This is inconsistent with our result showing that FpnLΔ7+R147A cleaves FpnH135A+C180A and BFT as substrates. We hypothesize that complete loop removal may have introduced some structural constraints in or near the active site that reduce activity against the smaller substrate, BAEE.
Figure 5.
Enzymatic activities of wild-type and mutant Fpn. (A) Absorbance at 253 nm representing the cleavage activities of 200 nM wild-type Fpn, FpnH135A+C180A, FpnR147A, or FpnLΔ7+R147A mutant proteins in the presence of 1250 µM synthetic substrate BAEE after incubation at 37 °C for 45 min. (B) Cleavage rate of purified FpnH135A+C180A used as a substrate in the presence of wild-type Fpn (gray) or FpnLΔ7+R147A (maroon). (C) Cleavage rate of purified BFT used as a substrate in the presence of wild-type Fpn (gray) or FpnLΔ7+R147A (maroon). In panels A and B, each cleavage experiment was performed three times; error bars represent standard deviations (SD) (mean ± SD).
The absence of detectable enzymatic activity of FpnLΔ7+R147A against BAEE suggested that the FpnLΔ7+R147A cleavage rate of FpnH135A+C180A and BFT may be slow in comparison to that of the wild type. To test this hypothesis, we evaluated the FpnLΔ7+R147A cleavage rate using FpnH135A+C180A or BFT as the substrate. Briefly, loopless FpnLΔ7+R147A was mixed with FpnH135A+C180A or BFT and incubated at 37 °C. Samples were taken at different time points and visualized by SDS–PAGE. Intensities of cleaved FpnH135A+C180A and BFT bands were quantified and plotted (representative SDS–PAGE gels are shown in Figure S2). Compared to that of the wild type, loopless FpnLΔ7+R147A had a lower apparent activity. FpnLΔ7+R147A took approximately 45 min to process FpnH135A+C180A (Figure 5B) or BFT (Figure 5C). In comparison, wild-type Fpn processed FpnH135A+C180A (Figure 5B) or BFT (Figure 5C) in approximately 5 min. Thus, loopless FpnLΔ7+R147A is less efficient at processing Fpn and BFT substrates.
To assess whether the activities of Fpn mutants measured in vitro reflect the activity in B. fragilis cells, we measured B. fragilis BFT levels by Western blotting in fpn mutant backgrounds. Consistent with our in vitro results, a B. fragilis strain expressing only uncleaved, loopless fpn (Δfpn/pAH2-fpnLΔ7) processed BFT, but with an efficiency lower than that of a B. fragilis Δfpn/pAH2-fpn strain. Control strains B. fragilis Δfpn and Δfpn/pAH2-fpnH135A+C180A did not show any cleaved BFT by Western blotting (Figure S3).
DISCUSSION
Cysteine proteases are regulated in multiple ways, including interaction with protein inhibitors, or by inhibitory domains in cis.33,39,40 The B. fragilis cysteine protease Fpn contains an inhibitory loop domain that is autocleaved, which results in enzyme activation. We previously reported a crystal structure of a cleaved form of Fpn in which this loop occupies a conformation that is compatible with an in trans autocleavage mechanism.18 In this structure, Fpn is cut at an arginine residue that is positioned between the histidine and cysteine of the catalytic dyad of an adjacent molecule in the crystal lattice and forms a covalent bond with the cysteine side chain (Figure 1A). To the best of our knowledge, an equivalent configuration in a cysteine protease has only been observed in a structure of cathepsin L, in which the cleaved region was trapped in the active site of a neighboring molecule.41 Biochemical data presented in this study support a trans model of Fpn cleavage but do not rule out the possibility that Fpn can also autocleave in cis.
The closest homologue of Fpn in the PDB is P. merdae PmC11.36 Fpn and PmC11 are 35% identical at the primary structure level and exhibit a high degree of tertiary structural similarity (rmsd of Cα coordinates of <1 Å). A comparison of the active sites and the cleavage sites of these structures reveals some differences (Figure 2A,B,E) that we propose have important consequences in Fpn and PmC11 autocleavage kinetics. Briefly, Fpn expressed in a heterologous E. coli system is purified as a fully cleaved active enzyme that resolves as two bands by SDS–PAGE (Figures 1D and 3B), while PmC11 is purified as a population of cleaved and uncleaved (i.e., active and inactive, respectively) protein that is processed to completion after 16 h at 37 °C.36 The difference in autocleavage kinetics between these related proteins can be attributed to the lysine residue (K147) in the PmC11 cleavable loop (Figure 2B). Weakened molecular interaction of K147 relative to that of R147 (Figure 2F,G) is predicted to reduce stability of the active site–loop interaction and slow the cleavage reaction. This is supported by our data showing that replacement of R147 with lysine reduces the rate of Fpn autocleavage (Figure 2C) and by the published observation that PmC11 has a stronger preference for substrates containing arginine over those containing lysine.36 Slower autoprocessing kinetics may have some physiological importance in P. merdae, though the in vivo substrate of PmC11 remains undefined. P. merdae does not encode a homologue of BFT.
The extended loop conformation captured in the Fpn crystal structure (Figure 1A) suggests a minimum loop length requirement for Fpn to be cut in trans. We have shown that a loop truncated by five residues cannot be cut in cis or in trans, even if the proper residue identity of cleavage site is intact (Figure 3D). However, complete removal of the loop results in an active site that more closely resembles caspases, legumain, or gingipain (Figure 1C). This loopless Fpn mutant has clear peptidase activity even though the enzyme remains in an uncleaved pro form (Figures 3E,F and 4). Together, these results suggest that the cleavage loop may function both to limit access to the active site and to control the conformation of active site residues required for peptide bond cleavage. Structures of uncleaved Fpn mutants will provide deeper insight into the molecular basis of loop inhibition.
In conclusion, we provide new structural insight into how the cleavable loop of C11 family peptidases controls activity. We demonstrate that Fpn can be activated by loop cleavage in trans. We successfully engineered an active pro form of Fpn, though this loopless mutant (FpnLΔ7+R147A) processed substrates more slowly than wild-type Fpn did (Figure 5A–C). We postulate that β-strands 6 and 7, which contain the catalytic dyad, are more flexible in the cleaved wild-type form of Fpn and can thus accommodate and process a wider range of substrates more efficiently than the constrained, loopless Fpn can. Nevertheless, the fact that we are able to produce an active version of pro-Fpn by loop removal suggests that modifications of the loop region may be a general approach to engineering C11 family cysteine proteases with variable substrate specificities and peptidase activities.
Supplementary Material
Acknowledgments
Funding
This work was supported in part by a Pilot and Feasibility Award from the Digestive Diseases Research Core Center at the University of Chicago (National Institute of Diabetes and Digestive and Kidney Diseases Grant P30DK42086). V.M.C. received support from the National Institutes of Health Medical Scientist Training Program at the University of Chicago (GM007281).
The authors thank members of the Crosson lab for useful discussion.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Three figures and two tables (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karlsson FH, Ussery DW, Nielsen J, Nookaew I. A closer look at bacteroides: phylogenetic relationship and genomic implications of a life in the human gut. Microb. Ecol. 2011;61:473–485. doi: 10.1007/s00248-010-9796-1. [DOI] [PubMed] [Google Scholar]
- 3.Wick EC, Sears CL. Bacteroides spp. and diarrhea. Curr. Opin. Infect. Dis. 2010;23:470–474. doi: 10.1097/QCO.0b013e32833da1eb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redondo MC, Arbo MD, Grindlinger J, Snydman DR. Attributable mortality of bacteremia associated with the Bacteroides fragilis group. Clin. Infect. Dis. 1995;20:1492–1496. doi: 10.1093/clinids/20.6.1492. [DOI] [PubMed] [Google Scholar]
- 5.Ngo JT, Parkins MD, Gregson DB, Pitout JD, Ross T, Church DL, Laupland KB. Population-based assessment of the incidence, risk factors, and outcomes of anaerobic bloodstream infections. Infection. 2013;41:41–48. doi: 10.1007/s15010-012-0389-4. [DOI] [PubMed] [Google Scholar]
- 6.Snydman DR, Jacobus NV, McDermott LA, Golan Y, Hecht DW, Goldstein EJ, Harrell L, Jenkins S, Newton D, Pierson C, Rihs JD, Yu VL, Venezia R, Finegold SM, Rosenblatt JE, Gorbach SL. Lessons learned from the anaerobe survey: historical perspective and review of the most recent data (2005–2007) Clin. Infect. Dis. 2010;50(Suppl. 1):S26–S33. doi: 10.1086/647940. [DOI] [PubMed] [Google Scholar]
- 7.Rodrigues C, Siciliano RF, Zeigler R, Strabelli TM. Bacteroides fragilis endocarditis: a case report and review of literature. Braz. J. Infect. Dis. 2012;16:100–104. [PubMed] [Google Scholar]
- 8.McHenry MC, Wellman WE, Martin WJ. Bacteremia due to Bacteroides. Review of 11 cases. Arch. Intern. Med. 1961;107:572–577. doi: 10.1001/archinte.1961.03620040098011. [DOI] [PubMed] [Google Scholar]
- 9.Robert R, Deraignac A, Le Moal G, Ragot S, Grollier G. Prognostic factors and impact of antibiotherapy in 117 cases of anaerobic bacteraemia. Eur. J. Clin. Microbiol. Infect. Dis. 2008;27:671–678. doi: 10.1007/s10096-008-0487-5. [DOI] [PubMed] [Google Scholar]
- 10.Moncrief JS, Obiso R, Jr, Barroso LA, Kling JJ, Wright RL, Van Tassell RL, Lyerly DM, Wilkins TD. The enterotoxin of Bacteroides fragilis is a metalloprotease. Infect. Immun. 1995;63:175–181. doi: 10.1128/iai.63.1.175-181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiryaev SA, Aleshin AE, Muranaka N, Kukreja M, Routenberg DA, Remacle AG, Liddington RC, Cieplak P, Kozlov IA, Strongin AY. Structural and functional diversity of metalloproteinases encoded by the Bacteroides fragilis pathogenicity island. FEBS J. 2014;281:2487–2502. doi: 10.1111/febs.12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franco AA, Mundy LM, Trucksis M, Wu S, Kaper JB, Sears CL. Cloning and characterization of the Bacteroides fragilis metalloprotease toxin gene. Infect. Immun. 1997;65:1007–1013. doi: 10.1128/iai.65.3.1007-1013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goulas T, Arolas JL, Gomis-Ruth FX. Structure, function and latency regulation of a bacterial enterotoxin potentially derived from a mammalian adamalysin/ADAM xenolog. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1856–1861. doi: 10.1073/pnas.1012173108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Tassell RL, Lyerly DM, Wilkins TD. Purification and characterization of an enterotoxin from Bacteroides fragilis. Infect. Immun. 1992;60:1343–1350. doi: 10.1128/iai.60.4.1343-1350.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu S, Lim KC, Huang J, Saidi RF, Sears CL. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14979–14984. doi: 10.1073/pnas.95.25.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, Shin J, Zhang G, Cohen M, Franco A, Sears CL. The Bacteroides fragilis toxin binds to a specific intestinal epithelial cell receptor. Infect. Immun. 2006;74:5382–5390. doi: 10.1128/IAI.00060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sears CL. The toxins of Bacteroides fragilis. Toxicon. 2001;39:1737–1746. doi: 10.1016/s0041-0101(01)00160-x. [DOI] [PubMed] [Google Scholar]
- 18.Choi VM, Herrou J, Hecht AL, Teoh WP, Turner JR, Crosson S, Bubeck Wardenburg J. Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice. Nat. Med. 2016;22:563–567. doi: 10.1038/nm.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kocholaty W, Weil L, Smith L. Proteinase secretion and growth of Clostridium histolyticum. Biochem. J. 1938;32:1685–1690. doi: 10.1042/bj0321685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kembhavi AA, Buttle DJ, Rauber P, Barrett AJ. Clostripain: characterization of the active site. FEBS Lett. 1991;283:277–280. doi: 10.1016/0014-5793(91)80607-5. [DOI] [PubMed] [Google Scholar]
- 22.McLuskey K, Mottram JC. Comparative structural analysis of the caspase family with other clan CD cysteine peptidases. Biochem. J. 2015;466:219–232. doi: 10.1042/BJ20141324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dargatz H, Diefenthal T, Witte V, Reipen G, von Wettstein D. The heterodimeric protease clostripain from Clostridium histolyticum is encoded by a single gene. Mol. Gen. Genet. 1993;240:140–145. doi: 10.1007/BF00276893. [DOI] [PubMed] [Google Scholar]
- 24.Labrou NE, Rigden DJ. The structure-function relationship in the clostripain family of peptidases. Eur. J. Biochem. 2004;271:983–992. doi: 10.1111/j.1432-1033.2004.04000.x. [DOI] [PubMed] [Google Scholar]
- 25.Witte V, Wolf N, Diefenthal T, Reipen G, Dargatz H. Heterologous expression of the clostripain gene from Clostridium histolyticum in Escherichia coli and Bacillus subtilis: maturation of the clostripain precursor is coupled with self-activation. Microbiology. 1994;140(Part 5):1175–1182. doi: 10.1099/13500872-140-5-1175. [DOI] [PubMed] [Google Scholar]
- 26.Klaiman G, Champagne N, LeBlanc AC. Self-activation of Caspase-6 in vitro and in vivo: Caspase-6 activation does not induce cell death in HEK293T cells. Biochim. Biophys. Acta, Mol. Cell Res. 2009;1793:592–601. doi: 10.1016/j.bbamcr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Dall E, Brandstetter H. Activation of legumain involves proteolytic and conformational events, resulting in a context-and substrate-dependent activity profile. Acta Crystallogr., Sect. F. Struct. Biol. Cryst. Commun. 2012;68:24–31. doi: 10.1107/S1744309111048020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JM, Fortunato M, Barrett AJ. Activation of human prolegumain by cleavage at a C-terminal asparagine residue. Biochem. J. 2000;352(Part2):327–334. [PMC free article] [PubMed] [Google Scholar]
- 29.Mallorqui-Fernandez N, Manandhar SP, Mallorqui-Fernandez G, Uson I, Wawrzonek K, Kantyka T, Sola M, Thogersen IB, Enghild JJ, Potempa J, Gomis-Ruth FX. A new autocatalytic activation mechanism for cysteine proteases revealed by Prevotella intermedia interpain A. J. Biol. Chem. 2008;283:2871–2882. doi: 10.1074/jbc.M708481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dall E, Brandstetter H. Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:10940–10945. doi: 10.1073/pnas.1300686110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan AR, Khazanovich-Bernstein N, Bergmann EM, James MN. Structural aspects of activation pathways of aspartic protease zymogens and viral 3C protease precursors. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10968–10975. doi: 10.1073/pnas.96.20.10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27:6194–6206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- 33.Rzychon M, Chmiel D, Stec-Niemczyk J. Modes of inhibition of cysteine proteases. Acta Biochim. Pol. 2004;51:861–873. [PubMed] [Google Scholar]
- 34.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLuskey K, Grewal JS, Das D, Godzik A, Lesley SA, Deacon AM, Coombs GH, Elsliger MA, Wilson IA, Mottram JC. Crystal Structure and Activity Studies of the C11 Cysteine Peptidase from Parabacteroides merdae in the Human Gut Microbiome. J. Biol. Chem. 2016;291:9482–9491. doi: 10.1074/jbc.M115.706143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erlanson DA, Lam JW, Wiesmann C, Luong TN, Simmons RL, DeLano WL, Choong IC, Burdett MT, Flanagan WM, Lee D, Gordon EM, O’Brien T. In situ assembly of enzyme inhibitors using extended tethering. Nat. Biotechnol. 2003;21:308–314. doi: 10.1038/nbt786. [DOI] [PubMed] [Google Scholar]
- 38.de Diego I, Veillard F, Sztukowska MN, Guevara T, Potempa B, Pomowski A, Huntington JA, Potempa J, Gomis-Ruth FX. Structure and mechanism of cysteine peptidase gingipain K (Kgp), a major virulence factor of Porphyromonas gingivalis in periodontitis. J. Biol. Chem. 2014;289:32291–32302. doi: 10.1074/jbc.M114.602052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grzonka Z, Jankowska E, Kasprzykowski F, Kasprzykowska R, Lankiewicz L, Wiczk W, Wieczerzak E, Ciarkowski J, Drabik P, Janowski R, Kozak M, Jaskolski M, Grubb A. Structural studies of cysteine proteases and their inhibitors. Acta Biochim. Pol. 2001;48:1–20. [PubMed] [Google Scholar]
- 40.Otlewski J, Jelen F, Zakrzewska M, Oleksy A. The many faces of protease-protein inhibitor interaction. EMBO J. 2005;24:1303–1310. doi: 10.1038/sj.emboj.7600611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sosnowski P, Turk D. Caught in the act: the crystal structure of cleaved cathepsin L bound to the active site of Cathepsin L. FEBS Lett. 2016;590:1253–1261. doi: 10.1002/1873-3468.12140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.