Abstract
Thrombocytes are nucleated platelets involved in immune functions such as pathogen recognition and release of pro-inflammatory bioactive compounds when exposed to bacterial and viral molecules. However, the complete role of these cells in innate and adaptive immune responses is not understood, and little is known about their biology at the molecular-genetic level. Highly sensitive RNA-sequencing technologies were used to analyze the complete transcriptome of thrombocytes for the first time with analytical resolution focused on cell-based components of the immune system/response. Amongst all the genes listed in the current chicken genome assembly, 10,041 gene transcripts were found in the chicken thrombocyte. After 1-hour in vitro stimulation with lipopolysaccharide (LPS, Salmonella minnesota), 490 genes were upregulated and 359 genes were downregulated, respectively, with at least a 1-fold change relative to unexposed thrombocytes. Additionally, by constructing a de novo assembly, we were able to identify a total of 3,030 novel genes in the thrombocyte transcriptome. The information generated here is useful in development of novel solutions to lower the economic burden and zoonotic threat that accompanies infectious diseases for birds and fish. In addition, the resources created here have translational utility as a model system to find orthologous genes and genes related to its enucleated counterpart, the platelet.
Background
Thrombocytes are smallest of the blood cells and are found in lower vertebrates such as reptiles, amphibians, fish, and birds [1]. These cells are nucleated and considered the first cells to evolve that specialize in hemostasis [1–3]. Nonnucleated thrombocytes or platelets are only found in mammals [1]; and thrombocytes are functionally comparable and primarily involved in hemostatic functions/wound healing. In recent years, these cells have been shown to have roles in inflammation, anti-microbial host defense, and overall immune response [4–8].
The role of thrombocytes in immunity was shown with evidence of phagocytic ability, followed by a role in the inflammatory response. Thrombocytes have been shown to express, produce or release a variety of mediators of inflammation, antimicrobial activity and other immune modulating activity [6]. The discovery of Pathogen Recognition Receptors (PRRs) such as Toll-like Receptors (TLRs) on these cells has led to a new understanding of the thrombocyte role in immune responses [4,9–14]. Thrombocytes respond to lipopolysaccharide (LPS) [12,15], and this stimulation takes place through TLR4, mitogen-activated (protein (MAP) kinase (ERK, MEK1 and p38 MAPK) and nuclear factor-κ light-chain-enhancer of activated B cells (NF-κB) pathways [12].
Thrombocytes have been suggested to be the hemostatic homologue of the mammalian platelet due to combined morphologic, immunologic and functional evidence; and conservation of major hemostatic pathways involved in platelet function and blood coagulation [16,17]. Due to the importance of platelets in human medicine, a substantial amount of research has been conducted to study the role of these cells in physiological function [18–20], and the capability to be involved in the immune response [4,5,21–23] is better understood compared to the thrombocyte. However, the full capability of the platelet to influence overall physiology would be better revealed through experimentation with the thrombocyte. Thrombocytes could serve as a nucleated model to provide new insights into platelet hemostasis, thrombosis and even bleeding disorders.
Our lab has studied in vitro stimulation of chicken thrombocytes with bacterial and viral Toll-like receptor ligands for several years [12,24] to establish the proper role of this cell in immunity. Here, we have used RNAseq technology to characterize the global transcriptome of the chicken thrombocyte and its in vitro response to stimulation with Salmonella-derived endotoxin (LPS) in order to expand our knowledge about these cells. The long-term aim of our research is to generate an essential genomic resource that will have translational utility in the medical world as a model system to find orthologous genes and genes related to platelet disorders. In addition, such resources will be useful in development of novel solutions to lower the economic burden and zoonotic threat that accompany infectious diseases for birds and fish. Such solutions include the identification of biomarkers for elite disease resistance genes expressed by thrombocytes. Determination of biomarkers by examining the gene expression profile of thrombocytes, stimulated with pathogenic agents, may be used as an early detection of zoonotic/infectious agents affecting young or even breeding-age poultry.
Materials and Methods
Chickens
Three female Single Comb White Leghorn (SCWL) chickens (16-weeks old) were randomly selected for blood collection in this study. The chickens were housed at the Clemson University Morgan Poultry Center, Clemson, SC, which is an Institutional Animal Care and Use Committee (IACUC) approved animal facility operating under standard management practices adhering to the Association for Assessment and Accreditation of Laboratory Animal Care International criteria.
Thrombocyte Isolation and In Vitro Stimulation
Syringes fitted with needles were used to collect 3 mL of whole blood from the wing vein of each chicken into 0.1 mL of 10% ethylenediaminetetraacetic acid (EDTA) solution. The collected blood samples were stored on ice until brought back to the laboratory. Each blood sample was diluted (1:1) with calcium and magnesium free Hank’s balanced salt solution (HBSS) (Cambrex Bio Sciences Walkersville Inc., Walkersville, MD). Diluted blood samples were then layered on a lymphocyte separation medium (Density 1.077–1.080 g/mL, Mediatech. Inc., Herdon, VA) and centrifuged at 1700 x g for 30 min at 23°C to collect the thrombocyte-rich band as previously described by Scott and Owens [12]. The isolated thrombocyte enriched cell suspension routinely is 99% positive for the thrombocyte specific marker CD41/61 [14,25]. Trypan blue solution (0.4% w/v in normal saline) was used for quantification of viable cell numbers on a SPolite® Hemacytometer (Baxter Healthcare, McGaw Park, IL) with the aid of an upright light microscope. The isolated thrombocytes from each chicken were incubated with 1 μg/mL of ultra pure LPS from Salmonella minnesota (InvivoGen, San Diego, CA). The control samples were incubated with only HBSS and no LPS. The cell suspensions with and without LPS were incubated in sterile 1.5 mL microcentrifuge tubes (1 x 107 cells per tube) on a rocking platform (VWR, Suwanee, GA) at 41°C for 60 min. The concentration of LPS used and stimulation length was chosen based on previous experiments performed in our laboratory.
RNA Isolation, Quantification, and Quality Assessment
For RNA isolation after thrombocyte stimulation, cells were centrifuged at 5000 x g for 2 min to pellet. The pellets were stored in 100 μL of RNAlater™ (Qiagen Inc., Valencia, CA), an RNA stabilizing solution. After 24 hr at 4°C in RNAlater™, the cells were centrifuged again to remove the supernatant and stored at -20°C until thawed for RNA isolation. The RNeasy® Kit (Qiagen Inc., Valencia, CA) was used according to the manufacturer’s protocol to isolate the total RNA from these samples. The RNA samples were treated with an on-column DNase (Qiagen Inc., Valencia, CA) to remove any possible contamination from chicken genomic DNA. Isolated RNA samples were quantified and integrity validated on a Nano Drop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA) and Bioanalyzer 2100 (Agilent Technologies).
Illumina Library Construction, RNA Sequencing, and Analysis
Each thrombocyte sample was normalized to a standard input concentration (1 μg of Total RNA) and an Illumina compatible sequencing library was prepared robotically on a Microlab STAR (Hamilton) with the TruSeq stranded total RNA library prep kit (Illumina) following the manufacturer’s recommended procedures (Illumina). The resulting sequencing libraries were assessed for size on a 2100 Bioanalyzer (Agilent) and sequence data collected on 1 lane of an Illumina HiSeq2500 with a 2x125 bp PE read on high-output mode. Raw sequence reads were assessed for run quality with the FastQC analysis package [26], and then preprocessed to remove adapter and low quality bases with the Trimmomatic software package [27]. Processed reads were mapped to the Gallus_gallus-4.0 reference assembly (GenBank Assembly ID GCA_000002315.2) [28] with the BWA [29]. Each replicate transcriptome was plotted together as a multidimensional scaling plot to observe global sample variation (S1 Fig). The control replicate Number 2, and the LPS stimulated replicate Number 2 were removed due to high variability in the first dimension. Further analysis were performed with an n = 2 for both unstimulated and LPS stimulated conditions. Read abundance counts per exon were determined with the Subread [30] and differential gene expression determined with EDGER [31]. Gene Ontology enrichment and analysis was performed with the Panther suite of analytical tools [32]. We utilized the Panther derived gene ontology (GO-slim terms) used for broad classification of molecular function, biological processes, and cellular components[32,33]. De novo transcriptome assembly of the thrombocyte was performed with the Trinity [34].
Results
Thrombocyte Transcriptome
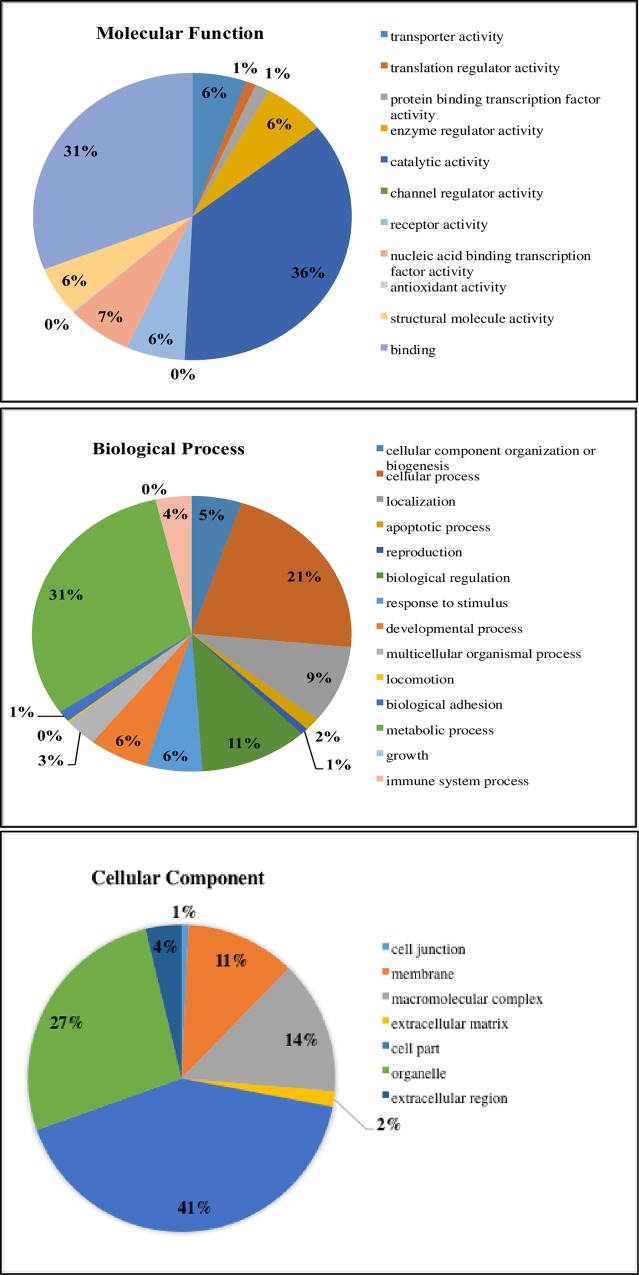
A total of 10,041 transcripts were detected in unstimulated control chicken thrombocytes compared to the 17,108 total annotated gene sequences identified in the reference chicken annotation. In order to decipher the functional aspects of the thrombocyte genes, we organized the transcripts based on their Gene Ontology (GO) functional categories. The results indicated that these cells have a role in a broad range of different biological activities and functions (Fig 1). Within the molecular function (MF) category, the most abundant terms observed include catalytic activity, binding, and nucleic acid binding transcription factor activity, representing close to 75% of all the MF terms. Within the biological processes (BP) category, the most abundant terms detected were metabolic processes, cellular processes, biological regulation, localization, response to stimulus, and immune system processes. Within the cellular component (CC) category, 41% of the terms were related to cell part, 27% to organelle, 14% to macromolecular complex and 11% to membranes.
Fig 1.
Distribution of the chicken thrombocyte transcripts in unstimulated cells categorized as cellular processes according to the Gene Ontology (GO)-slim categories of molecular function (MF), biological process (BP), and cellular component (CC).
Pathways and Genes Involved with Immune Function
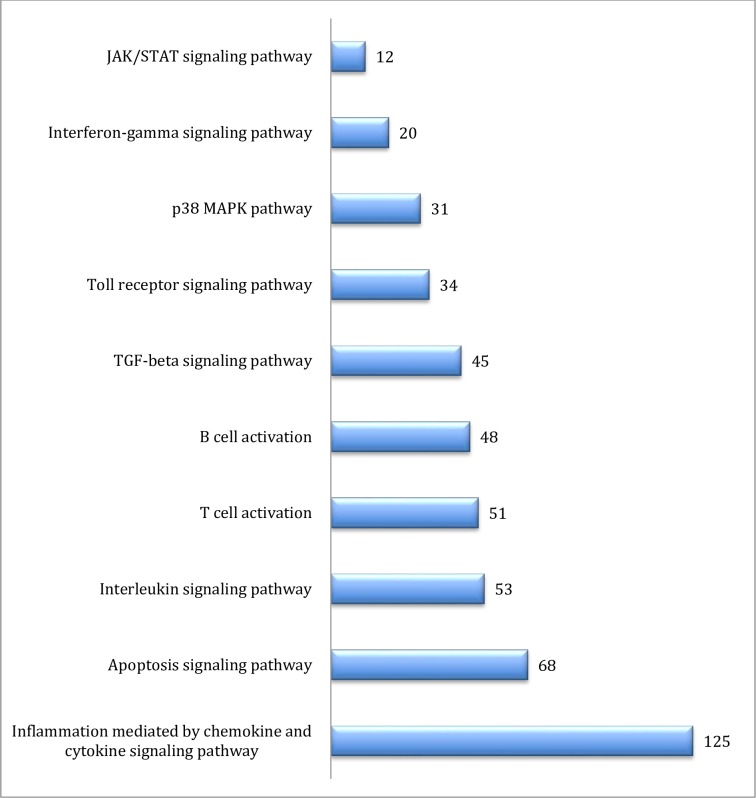
In order to give a broad classification of gene product function, we categorized the gene ontology content as respective biochemical pathways according to selected GO-slim terms; the cellular processes, pathways, and the number of genes associated with each of the assigned terms are listed in S1 Table. In order to further expand the knowledge regarding the role of thrombocytes in the immune system, the results presented here are directed toward the identification of biochemical pathways and genes that are involved in immune function. Upon examination of the GO-slim biochemical pathways, we identified ten pathways associated with immune signaling. We identified a total of 453 genes with roles in inflammation mediated chemokine and cytokine signaling (125), apoptosis signaling (68), interleukin signaling (53), T-cell activation (51), B-cell activation (48), transforming growth factor beta (TGF-β) signaling (45), TLR signaling (34), p38 mitogen-activated protein kinase (MAPK) signaling (31), interferon-gamma (IFN-γ) signaling (20) and Janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling (12, Fig 2).
Fig 2. Representative number of genes in the top 10 immune-related GO-slim biochemical pathways.
The numbers beside the blue bars represent the number of genes in that category. The complete list of GO-slim biochemical pathways detected in unstimulated thrombocytes are listed in S1 Table.
In addition to TLR signaling genes, we detected gene transcripts for nucleotide binding oligomerization domain (NOD)-like receptors (NLRs) such as NLR Family Member X1 (NLRX1), NLR Family CARD Domain Containing 3 and 5 (NLRC3, NLRC5) in the thrombocyte transcriptome. According to our RNAseq data, thrombocytes expressed MHC class I alpha chain 2 (such as BFIV21), and MHC class II genes (such as BLB1, BLB2, B-MA2). Many genes associated with major histocompatibility complex I and II, antigen processing and presentation were found in thrombocytes (S2 Table) including transcripts for accessory molecules CD40 and CD80 that are found on antigen presenting cells (APCs) in unstimulated thrombocytes. Furthermore, a broad search of the GO terms for “immune” revealed 244 genes with this assigned annotation in both the biological process and molecular function categories (S3 Table).
Transcriptional Response to LPS
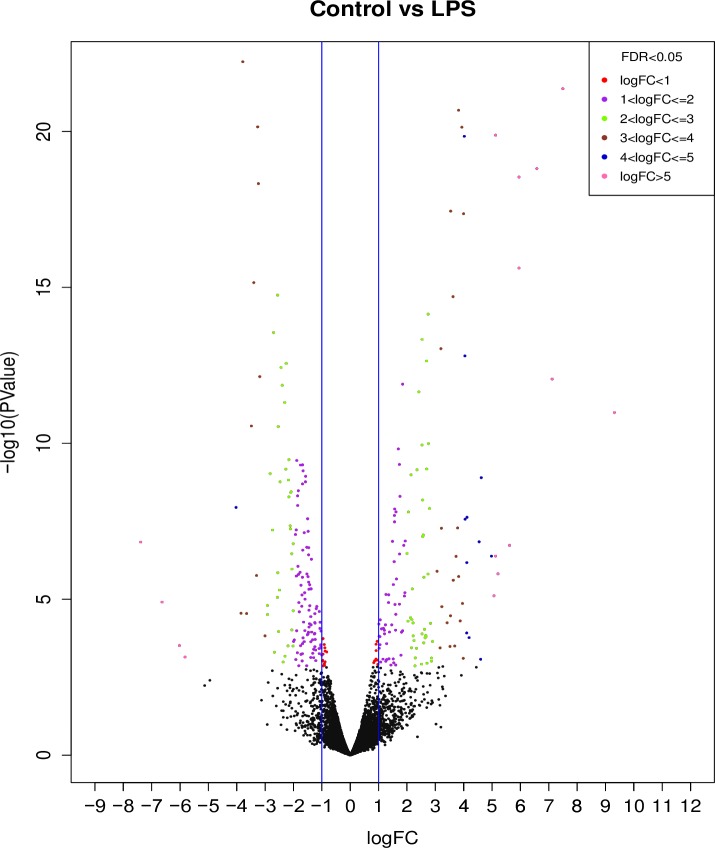
Upon stimulation with LPS, we detected evidence for a total of 10,148 genes being transcribed, of which, 354 are unique to this treatment relative to control thrombocytes (S4 Table). Differential expression profiling yielded upregulation of transcription for 490 genes and downregulation of 359 transcripts relative to the unstimulated control cells (Fig 3, S5 Table). The top 10 biochemical pathways up and downregulated in response to LPS stimulation are shown in Table 1. The upregulated pathways include Wnt signaling, inflammation mediated by chemokine and cytokine, cholecystokinin signaling map, angiogenesis, interleukin signaling, integrin signaling, platelet-derived growth factor signaling, cadherin signaling while the downregulated pathways include inflammation mediated by chemokine and cytokine, TGF-β signaling, angiogenesis, Huntington disease, Wnt signaling and others (complete list can be found in S6 Table).
Fig 3. A volcano plot of genes with differential expression profiles after a 1 hr treatment with LPS.
Positive values on the X-axis indicate genes with increased transcript abundance, and negative values indicate genes with decreased transcript abundance. Black dots indicate genes that are greater than the false discovery rate (fdr) of 0.05, and the vertical blue bars delineate a threshold of 1 fold change.
Table 1. Top ten categories by biochemical pathway using up/down regulated genes in response to LPS stimulation.
(The complete list can be found in S6 Table.)
| Upregulated | Downregulated |
|---|---|
| Wnt signaling pathway | Inflammation mediated by chemokine and cytokine signaling pathway |
| Inflammation mediated by chemokine and cytokine signaling pathway | TGF-beta signaling pathway |
| CCKR signaling map | Angiogenesis |
| Angiogenesis | Huntington disease |
| Interleukin signaling pathway | Wnt signaling pathway |
| Integrin signaling pathway | Parkinson disease |
| PDGF signaling pathway | PDGF signaling pathway |
| Cadherin signaling pathway | Gonadotropin releasing hormone receptor pathway |
| Axon guidance mediated by semaphorins | Metabotropic glutamate receptor group III pathway |
| Alzheimer disease-presenilin pathway | Ionotropic glutamate receptor pathway |
The PANTHER overrepresentation test shows that among all three categories (GO biological process, PANTHER protein class and GO molecular function) immune related processes or proteins are upregulated (Table 2). Under GO biological processes, neutrophil and granulocyte migration and chemotaxis have the highest fold enrichment. Chemokine and chemokine activity are the most upregulated in terms of fold enrichment in PANTHER protein class and GO molecular function, respectively.
Table 2. Panther Over Representation Test.
| Fold Enrichment | P value | |
|---|---|---|
| GO Biological Process | ||
| neutrophil migration | 8.98 | 2.45E-02 |
| neutrophil chemotaxis | 8.98 | 2.45E-02 |
| granulocyte chemotaxis | 8.44 | 3.85E-02 |
| granulocyte migration | 8.44 | 3.85E-02 |
| defense response | 2.84 | 7.88E-04 |
| immune response | 2.78 | 2.87E-03 |
| PANTHER Protein Class | ||
| chemokine | 14.33 | 1.65E-04 |
| cytokine | 4.62 | 6.08E-05 |
| signaling molecule | 2.17 | 1.60E-04 |
| GO Molecular Function | ||
| chemokine activity | 14.5 | 5.06E-03 |
| cytokine activity | 3.66 | 2.08E-03 |
| receptor binding | 2.29 | 7.71E-05 |
The top 10 upregulated and downregulated genes are listed in Table 3. Among the upregulated transcripts, IL-6 and IL-1β have roles in inflammation; IL-8 has roles in mediating cell activation and migration, and CSF3 influences granulocyte production. Among the genes with extreme downregulation profiles, we identified Glycerol-3-phosphate dehydrogenase, Rho GTPase Activating Protein 20, Semaphorin VIB, von Willebrand Factor A Domain Containing 1, Glutamate Decarboxylase 1, RasGEF Domain Family, Member 1B, Growth Arrest-specific-2, Apolipoprotein A-I, and several other genes with diverse functional roles (S5 Table). In addition to genes shown in Table 3, there were many more genes that were up or downregulated when thrombocytes were stimulated with LPS (S5 Table).
Table 3. Top 10 up- and down-regulated genes in response to in vitro stimulation of chicken thrombocyte.
| ENSGALG ID | Gene | Transcript Name | logFc• | Known or possible function |
|---|---|---|---|---|
| ENSGALG00000010915 | IL6 | Interleukin 6 | 9.63 | Inflammatory response and b cell maturation |
| ENSGALG00000026420 | CSF3 | Colony Stimulating Factor 3 | 9.31 | Cytokine that controls various aspects of granulocyte production |
| ENSGALG00000005616 | ARSI | Arylsulfatase Family, Member | 7.49 | Hormone biosynthesis, cell signaling |
| ENSGALG00000011668 | K60/ IL8L1 | Interleukin-8 precursor/ interleukin 8-like 1 | 7.25 | |
| ENSGALG00000005619 | F3 | Coagulation Factor III | 7.12 | Cell surface glycoprotein, affinity receptor for F7 |
| ENSGALG00000026098 | IL8 | Interleukin-8 | 6.74 | Mediates activation and migration of neutrophils |
| ENSGALG00000006106 | TNFRSF6B | Tumor Necrosis Factor Receptor Superfamily, Member 6b | 6.58 | Suppression of cell death |
| ENSGALG00000000534 | IL1B | Interleukin 1, Beta | 6.54 | Induction of COX2, mediator of the inflammatory response |
| ENSGALG00000021025 | LOC419276 | Bactericidal permeability-increasing protein-like | 5.95 | Antibacterial response |
| ENSGALG00000016919 | IRG1 | Immunoresponsive 1 Homolog | 5.95 | Negative regulator of TLR mediated responses |
| ENSGALG00000012061 | GPD1 | Glycerol-3-phosphate dehydrogenase | -7.38 | a component of glycerophospholipids. |
| ENSGALG00000004827 | Uncharacterized | -6.63 | ||
| ENSGALG00000017150 | ARHGAP20 | Rho GTPase Activating Protein 20 | -6.31 | related pathways are Signaling by GPCR and Signaling by Rho GTPases. |
| ENSGALG00000001171 | SEMA68 | Semaphorin VIB | -6.02 | may be involved in both peripheral and central nervous system development |
| ENSGALG00000001527 | VWA1 | Von Willebrand Factor A Domain Containing 1 | -5.82 | appears to play a role in cartilage structure and function |
| ENSGALG00000024466 | CC17 | Uncharacterized protein | -5.13 | |
| ENSGALG00000009589 | GAD67 | Glutamate Decarboxylase 1 (Brain, 67kDa) | -4.95 | responsible for catalyzing the production of gamma-aminobutyric acid from L-glutamic acid |
| ENSGALG00000010903 | RASGEF1B | RasGEF Domain Family, Member 1B | -4.18 | Guanine nucleotide exchange factor |
| ENSGALG00000028373 | GAS2L3 | Growth Arrest-Specific 2 Like 3 | -4.02 | Cytoskeletal linker protein |
| ENSGALG00000007114 | APOA1 | Apolipoprotein A-I | -3.85 | Participates in the reverse transport of cholesterol from tissues to the liver for excretion by promoting cholesterol efflux from tissues and by acting as a cofactor for the lecithin cholesterol acyltransferase |
*(logFC indicates the log2 of the fold-change between the treated and untreated samples)
Thrombocyte Transcriptome de novo Assembly
In an effort to search for novel genes expressed in the thrombocyte transcriptome that are not currently annotated in the Gal gal 4 reference assembly, we performed a de novo transcriptome assembly and removed known annotated chicken genes (See Methods). After strict assembly and filtering criteria, we identified a total of 3,030 putative new coding transcripts (S7 Table). A blastx alignment to the SwissProt database revealed that only 780 of these putative genes do not produce a hit at a 1e-05 threshold. Alignment to the Gene Ontology produced a total of 1,857 hits (only 308 unique terms) (S8 Table). Molecular function included genes with roles in RNA/DNA binding, aspartic-type endopeptidase activity, and protein binding. The biological process category contained genes with roles in transmembrane kinase signaling, cellular process, and nucleobase-containing compounds (S8 Table). Moreover, a search of the GO terms for “immune” revealed a total of 80 genes (S7 Table). Among these, we identified a B-cell antigen receptor complex associated protein, several complement decay-accelerating factors, interleukins, T-cell receptors, and numerous transcription factors and other signaling molecules (S7 Table). Of the 3,030 putative new gene sequences, a total of 83 genes displayed an increase of at least a fold change of 2, and only 53 displayed a decrease in transcriptional abundance with a fold change of at least 2 when assayed for expression changes under LPS stimulation (S9 Table). Among the putative novel genes that appear to be upregulated, we identified genes with homology to interleukins, RAS-GTPase, Filamin-C, and other growth factor like genes (S9 Table). Novel genes that appear to have a decrease in expression profiles include snRNA-like, methyltransferase-like genes, among others.
Discussion
This is the first analysis of the complete transcriptome of the thrombocyte. Analysis of GO functional categories demonstrated that these cells have a role in a broad range of different biological activities and functions. For this paper, we focused on processes, pathways, and genes related to immune response; particularly those affected by LPS exposure.
Among all the gene transcripts detected, GO-slim biological processes showed 466 genes related to immune system processes (S1 Table). Other biological process categories such as response to stimulus, and biological regulation may also have genes indirectly related to immune response. Among all biochemical pathways shown by GO-slim analysis (Fig 2), the greatest number of genes (125) was associated with inflammation mediated by chemokine and cytokine signaling pathways. These genes are primarily associated with the metabolic process, cell communication, response to stimulus, immune response, and inflammation. Apoptosis and p38 MAPK signaling genes generally participate in the signaling cascade that controls cellular responses to cytokines and stress. The TGF-β signaling pathway is commonly involved in regulation of fundamental cell processes such as proliferation, differentiation, death, cytoskeletal organization, adhesion, and migration [35]. Cellular effects of IFN-γ include up-regulation of pathogen recognition, antigen processing and presentation, the antiviral state, inhibition of cellular proliferation and effects on apoptosis, activation of microbicidal effector functions, immunomodulation, leukocyte trafficking and integration of signaling and response with other cytokines [36]. The JAK/STAT pathway is the principal signaling mechanism for a wide array of cytokines and growth factors [37]. TLR signaling is also among the GO-slim analysis of biochemical pathways observed in the thrombocyte transcriptome. TLR signaling is activated by pathogen associated molecular pattern leads to immediate innate immune responses preventing spread of infection and in the potentiation and direction of the later responses of acquired immunity [38]. A preliminary evaluation of transcripts for TLR pathway components linked to LPS stimulation of thrombocytes via TLR4 provides a characteristic set of signals leading to gene expression of pro-inflammatory mediators (i.e., IL-1β, IL-6, IL-8).
In addition to genes related to TLR signaling, we detected transcripts for NLRs. NLRs along with TLRs and others (such as mannose receptors, C-type lectin receptors, RIG-I-like receptors) are involved in the innate pathogen pattern recognition system. Among NLRs detected in the thrombocyte, NLRX1 is known to be a regulator of mitochondrial antivirus responses [39], and NLRC3 is a cytosolic negative regulator of innate immunity [40]. NLR5 is a critical regulator of MHC class I-dependent immune responses [41]. In mice, deficiency of NLR5 expression has been associated with impaired MHC class I expression, and impaired CD8+ T-cell activation [42]. Human NLR5 has a role in anti-viral innate immune responses[43]. NLRs have been shown to respond to intracellular pathogens and play important roles in distinct biological processes ranging from regulation of antigen presentation, sensing metabolic changes in the cell, modulation of inflammation, embryo development, cell death, and differentiation of the adaptive immune response [44].
Based on our experience in working with thrombocytes and previous studies done in our laboratory [12,14,15,24], stimulation with 1 μg/mL of LPS for 1 hr is more than sufficient to induce these cells. The most upregulated gene transcript is IL-6 when thrombocytes are exposed to LPS for 1 hr. IL-6 is a pro-inflammatory cytokine that also induces the synthesis of acute phase response proteins, terminal differentiation of B cells to antibody producing plasma cells, differentiation of monocytes to macrophages, and growth of hematopoietic stem cells [45]. Colony stimulating factor (CSF) 3 is the next most upregulated gene. CSF3 controls the production, differentiation, and function of mature granulocytes [46]. IL-8 precursor and IL-8, which is a chemoattractant, were also among the upregulated genes. IL-8 is a proinflammatory cytokine that is involved in activation and migration of neutrophils (heterophils) during inflammation [47]. Bactericidal permeability-increasing protein-like (LOC4192760) and immunoresponsive 1 homolog (IRG1) genes were also among the top upregulated genes involved in immune response. Arylsulfatase Family (ARSI) was one of the genes in this list that is not directly involved in immune response. ARSI hydrolyzes sulfate esters from sulfated steroids, carbohydrates, proteoglycans, and glycolipids and is known to be involved in hormone biosynthesis and cell signaling [48]. Coagulation factor III, known to initiate the blood coagulation cascades, was the only gene from blood coagulation that was in the list of top upregulated genes.
Depending on the cytokines and other expressed cellular markers, thrombocytes may be able to activate and affect naïve T cells to differentiate into effector T cell types. Here, we were able to identify genes in thrombocytes that indicate involvement of thrombocytes in more than just innate immunity. Expression of MHC II genes and molecules is a unique finding for nucleated thrombocytes [13,14,49] since mammalian platelets are devoid of MHC class II molecules [4,14]. It is interesting to detect MHC II transcripts in unstimulated thrombocytes since this is a feature limited to true APCs such as dendritic cells. Generally, dendritic cells (DCs, professional APCs) express both class I and class II MHC molecules while macrophages and B cells must be activated to express class II. Expression of MHC II is important as the first signal for stimulation of the T cell. We have observed that thrombocytes also express co-stimulatory molecules such as CD40 and CD80 on control unstimulated thrombocytes. Co-stimulatory molecules on APCs generally bind to CD28 on T cells and act as a second signal to activate T cells. We also observed upregulation of some cytokines like IL-6 (Table 3) and IL-1β (S5 Table). We have previously reported that LPS stimulation leads to upregulation of gene expression for IL-6 and IL-12 in chicken thrombocytes [15] providing polarizing signals for T-cell activation. IL-6 promotes Th2 differentiation and simultaneously inhibits Th1 polarization through two independent molecular mechanisms [50]. Likewise, IL-12 can positively influence differentiation to a Th1 state when conditions favor more cytotoxicity. This ability to influence T cells is a hallmark of adaptive immunity in which thrombocytes most likely share with professional antigen presenting cells.
Although mammalian enucleated platelets are devoid of MHC II, the role of these cells in adaptive immunity is fairly well established[51–53]. Platelets have been shown to activate DCs in vitro and promote T-cell responses via CD40L [4]. Platelet-derived CD40L has been reported to support B-cell differentiation and immunoglobulin class switching in mice [53]. Platelet-derived CD40L also has been shown to augment CD8+ T-cell responses, both in vitro and in vivo, and to promote protective T-cell responses following infection with Listeria monocytogenes [54]. Among cytokines that generally affect T-cell activation and proliferation[55], we were able to detect transcripts for IL-15. We have also detected transcripts for the components of the IL-2 receptor (IL2RA, IL2RB and IL2RG), which may be used by a number of cytokines for stimulating T-cell proliferation including IL-15. We also detected IL-16 that can function as a chemoattractant and modulator of T-cell activation.
Since thrombocytes we isolated for this study were from chickens, in addition to discussing what we have observed in our dataset, we compared our thrombocyte RNAseq data with published datasets for other chicken immune cells. This will be valuable for understanding the overall role of this particular cell in the immune system of chickens. Upon performing an extensive literature search, we were able to find RNAseq data of chicken heterophil, macrophage and dendritic cells. The publicly available RNAseq data for bone marrow-derived dendritic cells and macrophages, and heterophils isolated from blood downloaded for comparative analysis were both unstimulated and LPS stimulated for 24 hr [56]. Although this varies in terms of length of stimulation time, this dataset was most similar to the overall format of our study.
Our objective was to uncover common markers for sentinel and antigen presenting cells to compare with the thrombocyte transcriptome (S10 Table). We generated a semi-quantitative table based on raw counts. This comparison provided a side-by-side examination of some key molecules such as TLRs, TLR associated molecules, costimulatory markers, MHC and cytokines in all of these cell types. Overall, gene transcripts for most of these markers were present in each cell type making the inclusion of thrombocytes relevant with important immune cells. TLR2-2, TLR5, and IRF6 transcripts were not observed in thrombocytes, Cytokine transcripts for IL-6, IL-8 and IL-12β were not seen in unstimulated control but were observed in LPS treated cells. The expression of gene transcripts for TLR, TLR associated molecules, cytokines, co-stimulatory makers and MHC (at least BFIV21 and BLB1) were similar among the four cell types. It is essential to note that this comparison was done with publicly available RNAseq data and some of the genes that may or may not be present here could be due to an incomplete chicken genome database or to point/time of sample collections.
The potential of these cells to be involved in adaptive immunity, or at least as a bridge between innate and adaptive immune responses, has been indicated previously by some researchers [13,14,57]. Tregaskes and his colleagues demonstrated, among other biologically active surface molecules and receptors, avian thrombocytes also express CD40L [57]. The discovery of functional CD40L is of vital importance in the potential modulatory capacity of thrombocytes in bridging innate immunity to the adaptive side of immune responsiveness. CD40/CD40L is a receptor-ligand pair with a central role in promoting interactions between lymphocytes and APCs such as DCs. Therefore, thrombocytes appear to be more than innate effector cells. In addition to CD40L, it has been shown that CD40, CD80, CD86 and MHC II (molecules that are generally associated with antigen presentation) were detected on thrombocytes using flow cytometry [14]. Chicken thrombocytes should not only have the ability to interact with APC, but also have the potential to play a role in antigen presentation.
Conclusions
To the extent that we know, this is the first report on the entire identifiable transcriptome of the nucleated thrombocyte from chicken. Global transcriptome information on these cells is important due the comparative aspect for other species to provide acceptable and valuable biomedical models for platelet physiology studies in mammals. In addition, defining the role of these cells in immune responses will be useful for economically important agriculture species such as poultry and fish. Databases generated from studies like this will be useful to discover biomarkers for assessing overall animal health.
Supporting Information
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
The raw data for this paper are available from NCBI GenBank Sequence Read Archive under the following SRA Accessions: SAMN05818716-SAMN05818721 under the BioProject Accession: PRJNA34407.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Levin J. The Evolution of Mammalian Platelets In: Michelson AD, Coller BS, editors. Platelets. Amsterdam: Elsevier; 2007. pp. 3–22. [Google Scholar]
- 2.Ratnoff OD. The evolution of hemostatic mechanisms. Perspect Biol Med. 1987;31: 4–33. 10.1353/pbm.1987.0003 [DOI] [PubMed] [Google Scholar]
- 3.Schneider W, Gattermann N. Megakaryocytes: origin of bleeding and thrombotic disorders. Eur J Clin Invest. 1994;24: 16–20. 10.1111/j.1365-2362.1994.tb02420.x [DOI] [PubMed] [Google Scholar]
- 4.Semple JW, Italiano JE Jr, Freedman J. Platelets and the immune continuum. Nat Rev Immunol. 2011;11: 264–274. 10.1038/nri2956 [DOI] [PubMed] [Google Scholar]
- 5.Morrell CN, Aggrey AA, Chapman LM, Modjeski KL. Emerging roles for platelets as immune and inflammatory cells. Blood. 2014;123: 2759–2767. 10.1182/blood-2013-11-462432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferdous F, Scott TR. A comparative examination of thrombocyte/platelet immunity. Immunol Lett. 2015;163: 32–39. 10.1016/j.imlet.2014.11.010. 10.1016/j.imlet.2014.11.010 [DOI] [PubMed] [Google Scholar]
- 7.Ware J, Corken A, Khetpal R. Platelet function beyond hemostasis and thrombosis. Curr Opin Hematol. 2013;20: 451–456. 10.1097/MOH.0b013e32836344d3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeaman MR. Bacterial-platelet interactions: virulence meets host defense. Future Microbiol. 2010;5: 471–506. 10.2217/fmb.09.112 [DOI] [PubMed] [Google Scholar]
- 9.Aslam R, Speck ER, Kim M, Crow AR, Bang KW, Nestel FP, et al. Platelet Toll-like receptor expression modulates lipopolysaccharide-induced thrombocytopenia and tumor necrosis factor-alpha production in vivo. Blood. 2006;107: 637–641. 10.1182/blood-2005-06-2202 [DOI] [PubMed] [Google Scholar]
- 10.Shiraki R, Inoue N, Kawasaki S, Takei A, Kadotani M, Ohnishi Y, et al. Expression of Toll-like receptors on human platelets. Thromb Res. 2004;113: 379–385. 10.1016/j.thromres.2004.03.023 [DOI] [PubMed] [Google Scholar]
- 11.Cognasse F, Hamzeh H, Chavarin P, Acquart S, Genin C, Garraud O. Evidence of Toll-like receptor molecules on human platelets. Immunol Cell Biol. 2005;83: 196–198. 10.1111/j.1440-1711.2005.01314.x [DOI] [PubMed] [Google Scholar]
- 12.Scott T, Owens MD. Thrombocytes respond to lipopolysaccharide through Toll-like receptor-4, and MAP kinase and NF-kappaB pathways leading to expression of interleukin-6 and cyclooxygenase-2 with production of prostaglandin E2. Mol Immunol. 2008;45: 1001–1008. 10.1016/j.molimm.2007.07.035 [DOI] [PubMed] [Google Scholar]
- 13.St Paul M, Paolucci S, Barjesteh N, Wood RD, Schat KA, Sharif S. Characterization of chicken thrombocyte responses to Toll-like receptor ligands. PLoS One. 2012;7: e43381 10.1371/journal.pone.0043381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferdous F. The avian thrombocyte is a specialized immune cell. Ph.D, Clemson University. 2014. Available: http://tigerprints.clemson.edu/all_dissertations/1289.
- 15.Ferdous F, Maurice D, Scott T. Broiler chick thrombocyte response to lipopolysaccharide. Poult Sci. 2008;87: 61–63. 10.3382/ps.2007-00356 [DOI] [PubMed] [Google Scholar]
- 16.Jagadeeswaran P, Liu YC, Sheehan JP. Analysis of hemostasis in the zebrafish. Method Cell Biol. 1999;59: 337–357. [DOI] [PubMed] [Google Scholar]
- 17.Jagadeeswaran P, Sheehan JP, Craig FE, Troyer D. Identification and characterization of zebrafish thrombocytes. Br J Haematol. 1999;107: 731–738. 10.1046/j.1365-2141.1999.01763.x [DOI] [PubMed] [Google Scholar]
- 18.Vieira-de-Abreu A, Campbell RA, Weyrich AS, Zimmerman GA. Platelets: versatile effector cells in hemostasis, inflammation, and the immune continuum. Semin Immunopathol. 2012;34: 5–30. 10.1007/s00281-011-0286-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GC White I. Platelet physiology and function. Blood Coagulation Fibrinol. 2000;11: S53 10.1097/00001721-200004001-00012 [DOI] [PubMed] [Google Scholar]
- 20.George JN. Platelets. Lancet. 2000;355: 1531–1539. 10.1016/S0140-6736(00)02175-9 [DOI] [PubMed] [Google Scholar]
- 21.Duerschmied D, Bode C, Ahrens I. Immune functions of platelets. Thromb Haemost. 2014;112: 678–691. 10.1160/TH14-02-0146 [DOI] [PubMed] [Google Scholar]
- 22.Garraud O, Cognasse F. Platelet Toll-like receptor expression: the link between "danger" ligands and inflammation. Inflamm Allergy Drug Targets. 2010;9: 322–333. 10.2174/187152810793937991 [DOI] [PubMed] [Google Scholar]
- 23.Lam FW, Vijayan KV, Rumbaut RE. Platelets and their interactions with other immune cells. 2015. [DOI] [PMC free article] [PubMed]
- 24.Ferdous F, Scott T. Bacterial and viral induction of chicken thrombocyte inflammatory responses. 2015;49: 225–230. 10.1016/j.dci.2014.11.019. 10.1016/j.dci.2014.11.019 [DOI] [PubMed] [Google Scholar]
- 25.Lacoste-Eleaume A, Bleux C, Quere P, Coudert F, Corbel C, Kanellopoulos-Langevin C. Biochemical and functional characterization of an avian homolog of the integrin GPIIb-IIIa present on chicken thrombocytes. Exp Cell Res. 1994;213: 198–209. 10.1006/excr.1994.1191 [DOI] [PubMed] [Google Scholar]
- 26.Babraham Bioinofrmatics. FastQC. Available: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 27.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432: 695–716. 10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41: e108 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44: D336–42. 10.1093/nar/gkv1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss A, Attisano L. The TGFbeta superfamily signaling pathway. 2013;2: 47–63. 10.1002/wdev.86 [DOI] [PubMed] [Google Scholar]
- 36.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75: 163–189. 10.1189/jlb.0603252 [DOI] [PubMed] [Google Scholar]
- 37.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117: 1281–1283. 10.1242/jcs.00963 [DOI] [PubMed] [Google Scholar]
- 38.Brownlie R, Allan B. Avian toll-like receptors. Cell Tissue Res. 2011;343: 121–130. 10.1007/s00441-010-1026-0 [DOI] [PubMed] [Google Scholar]
- 39.Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451: 573–577. 10.1038/nature06501 [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Mo J, Swanson KV, Wen H, Petrucelli A, Gregory SM, et al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity. 2014;40: 329–341. 10.1016/j.immuni.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi KS, van den Elsen, Peter J. NLRC5: a key regulator of MHC class I-dependent immune responses.. 2012;12: 813–820. 10.1038/nri3339 [DOI] [PubMed] [Google Scholar]
- 42.Biswas A, Meissner TB, Kawai T, Kobayashi KS. Cutting edge: impaired MHC class I expression in mice deficient for Nlrc5/class I transactivator. J Immunol. 2012;189: 516–520. 10.4049/jimmunol.1200064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neerincx A, Lautz K, Menning M, Kremmer E, Zigrino P, Hosel M, et al. A role for the human nucleotide-binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses. J Biol Chem. 2010;285: 26223–26232. 10.1074/jbc.M110.109736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Motta V, Soares F, Sun T, Philpott DJ. NOD-like receptors: versatile cytosolic sentinels. Physiol Rev. 2015;95: 149–178. 10.1152/physrev.00009.2014 [DOI] [PubMed] [Google Scholar]
- 45.Hirano T. Interleukin 6 and its receptor: ten years later. Int Rev Immunol. 1998;16: 249–284. 10.3109/08830189809042997 [DOI] [PubMed] [Google Scholar]
- 46.Panopoulos AD, Watowich SS. Granulocyte colony-stimulating factor: molecular mechanisms of action during steady state and ‘emergency’hematopoiesis. Cytokine. 2008;42: 277–288. 10.1016/j.cyto.2008.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammond ME, Lapointe GR, Feucht PH, Hilt S, Gallegos CA, Gordon CA, et al. IL-8 induces neutrophil chemotaxis predominantly via type I IL-8 receptors. J Immunol. 1995;155: 1428–1433. [PubMed] [Google Scholar]
- 48.Sardiello M, Annunziata I, Roma G, Ballabio A. Sulfatases and sulfatase modifying factors: an exclusive and promiscuous relationship. Hum Mol Genet. 2005;14: 3203–3217. ddi351 [pii]. 10.1093/hmg/ddi351 [DOI] [PubMed] [Google Scholar]
- 49.Nagasawa T, Nakayasu C, Rieger AM, Barreda DR, Somamoto T, Nakao M. Phagocytosis by thrombocytes is a conserved innate immune mechanism in lower vertebrates. Front Immunol. 2014;5 10.3389/fimmu.2014.00445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diehl S, Rincón M. The two faces of IL-6 on Th1/Th2 differentiation. Mol Immunol. 2002;39: 531–536. 10.1016/S0161-5890(02)00210-9. [DOI] [PubMed] [Google Scholar]
- 51.Elzey BD, Sprague DL, Ratliff TL. The emerging role of platelets in adaptive immunity. Cell Immunol. 2005;238: 1–9. 10.1016/j.cellimm.2005.12.005 [DOI] [PubMed] [Google Scholar]
- 52.Elzey BD, Ratliff TL, Sowa JM, Crist SA. Platelet CD40L at the interface of adaptive immunity. Thromb Res. 2011;127: 180–183. 10.1016/j.thromres.2010.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elzey BD, Tian J, Jensen RJ, Swanson AK, Lees JR, Lentz SR, et al. Platelet-mediated modulation of adaptive immunity: a communication link between innate and adaptive immune compartments. Immunity. 2003;19: 9–19. 10.1016/S1074-7613(03)00177-8 [DOI] [PubMed] [Google Scholar]
- 54.Elzey BD, Schmidt NW, Crist SA, Kresowik TP, Harty JT, Nieswandt B, et al. Platelet-derived CD154 enables T-cell priming and protection against Listeria monocytogenes challenge. Blood. 2008;111: 3684–3691. 10.1182/blood-2007-05-091728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ku CC, Murakami M, Sakamoto A, Kappler J, Marrack P. Control of homeostasis of CD8+ memory T cells by opposing cytokines. Science. 2000;288: 675–678. 8464 [pii]. 10.1126/science.288.5466.675 [DOI] [PubMed] [Google Scholar]
- 56.ArrayExpress. EMTAB-2996—RNA-seq of coding RNA from bone marrow derived dendritic cells, bone marrow derived macrophages and heterophils isolated from blood of day-old chicks which were control or LPS stimulated. 2013. Available: http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-2996/.
- 57.Tregaskes CA, Glansbeek HL, Gill AC, Hunt LG, Burnside J, Young JR. Conservation of biological properties of the CD40 ligand, CD154 in a non-mammalian vertebrate. Dev Comp Immunol. 2005;29: 361–374. 10.1016/j.dci.2004.09.001. 10.1016/j.dci.2004.09.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The raw data for this paper are available from NCBI GenBank Sequence Read Archive under the following SRA Accessions: SAMN05818716-SAMN05818721 under the BioProject Accession: PRJNA34407.