Abstract
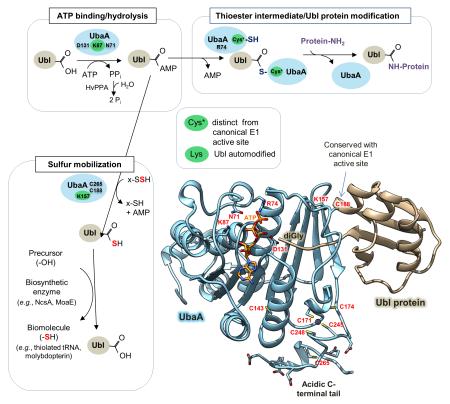
Here we provide the first detailed biochemical study of a noncanonical E1-like enzyme with broad specificity for cognate ubiquitin-like (Ubl) proteins that mediates Ubl protein modification and sulfur mobilization to form molybdopterin and thiolated tRNA. Isothermal titration calorimetry and in vivo analyses proved useful in discovering that environmental conditions, ATP binding and Ubl type controlled the mechanism of association of the Ubl protein with its cognate E1-like enzyme (SAMP and UbaA of the archaeon Haloferax volcanii, respectively). Further analysis revealed ATP hydrolysis triggered the formation of thioester and peptide bonds within the Ubl:E1-like complex. Importantly, the thioester was an apparent precursor to Ubl protein modification but not sulfur mobilization. Comparative modeling to MoeB/ThiF guided the discovery of key residues within the adenylation domain of UbaA that were needed to bind ATP as well as residues that were specifically needed to catalyze the downstream reactions of sulfur mobilization and/or Ubl protein modification. UbaA was also found to be Ubl-automodified at lysine residues required for early (ATP binding) and late (sulfur mobilization) stages of enzyme activity revealing multiple layers of auto-regulation. Cysteine residues, distinct from the canonical E1 ‘active site’ cysteine, were found important in UbaA function supporting a model that this non-canonical E1 is structurally flexible in its active site to allow Ubl~adenylate, Ubl~E1-like thioester and cysteine persulfide(s) intermediates to form.
Keywords: post-translational modification, molydopterin biosynthesis, sulfur relay, tRNA thiolation, ubiquitin, proteasomes, archaea
Illustrated Abstract
Here we report the first detailed biochemical study of an E1-like enzyme (UbaA) that forms diverse ubiquitin-like (Ubl) protein conjugates and mobilizes sulfur to generate thiolated tRNA and molybdopterin. Importantly, UbaA is found autoregulated by Ubl-protein modification and structurally flexible to allow Ubl~adenylate, Ubl~E1 thioester and persulfide(s) intermediates to form in the absence of a canonical E1 active site cysteine.
INTRODUCTION
Ubiquitylation and related protein modification systems are important in regulating cell function including proteasome-mediated proteolysis, cell division, subcellular localization of proteins, signal transduction, and DNA repair [1]. In eukaryotes, E1-E2-E3 enzymes catalyze the covalent attachment of ubiquitin (Ub) to protein targets. At the apex of the process is the E1, which adenylates the C-terminus of Ub at the expense of ATP [2]. The Ub adenylate (Ub~AMP) undergoes nucleophilic attack by a conserved E1 ‘active site’ cysteine to form a Ub~E1 thioester [2]. E1 next recruits cognate E2s, and the Ub is transferred from the E1 catalytic cysteine to the E2 catalytic cysteine to form an Ub~E2 thioester [2]. The charged Ub~E2 combines with an array of E3 ligases to ensure proteins are properly ubiquitylated. In this process, the C-terminus of Ub is attached by isopeptide bonds to the ε-amino group of lysine residues and by linear peptide bonds to the α-amino group of protein N-termini [3, 4].
Ancestral relatives of Ub/E1 are identified that mobilize sulfur to form sulfur-containing biomolecules (e.g., thiamine, molybdopterin (MPT), thiolated tRNA, thiol-functionalized siderophores, and 2-thiosugars) [5, 6]. In bacteria, these E1-like enzymes are well studied and found to adenylate and activate the C-terminus of the Ub-like (Ubl) protein by hydrolyzing ATP and releasing PPi [7, 8]. E2 and E3 enzymes are not used in the sulfur relay, instead the E1-like enzyme converts the Ubl~AMP (adenylate) to a Ubl protein thiocarboxylate with the sulfur derived from the cysteinyl persulfide group of a rhodanese domain (RHD) protein or cysteine desulfurase [9-13]. The sulfur is directed to biosynthesis by association of the thiocarboxylated Ubl protein with a protein partner dedicated to the synthesis of a particular sulfur-containing biomolecule (e.g., thiocarboxylated MoaD associates with MoaE to form MPT [14]).
Ubl/E1-like cognate pairs are recently described to serve in Ubl protein modification and the mobilization of sulfur to form biomolecules [15]. Examples of dual function E1-/Ubl pairs include Uba4p/Urm1p of yeast [16], UbaA/SAMPs of archaea [5] and TtuC/TtuB of thermophilic bacteria [17, 18]. While downstream protein partners are identified for sulfur mobilization (e.g., in archaea SAMP1 partners with MoaE to form MPT and SAMP2 associates with NcsA to thiolate tRNA)[19, 20], E2- and E3-type enzymes have yet to be identified for the Ubl protein modification activities of these dual function E1-/Ubl pairs. Furthermore, the molecular mechanism of how a single E1-like enzyme can function to activate an Ubl protein for sulfur mobilization and protein modification is poorly understood.
Here an insight into non-canonical E1-/Ubl pairs that are of dual function is provided by determining biochemical properties and structure-function relationships of the archaeal UbaA/SAMPs. In particular, UbaA was defined in terms of its nucleotide ligands, thermodynamics of Ubl protein binding, thioester intermediate, regulation by auto-modification, and amino acid residues important for catalytic function. Site-directed mutagenesis was found critical for refining 3D-homology models and for identifying amino acid residues important for UbaA function.
RESULTS
Nucleotide ligands of UbaA
The noncanonical E1-like enzyme UbaA, of the archaeon Hfx. volcanii, was purified as a homodimer (Fig. 1) and analyzed for nucleotide binding by differential scanning fluorimetry (DSF) (Table 1). Sypro Orange was used detect an increase in the stability and associated melting temperature (Tm) of UbaA when bound to its nucleotide ligands, as this dye displays increase in fluorescence when bound to hydrophobic residues exposed during protein thermal denaturation [21]. Of the ligands tested, ATP and its non-hydrolyzable analog AMP-PNP were found to have the greatest influence on the Tm of UbaA (with an increase of 8.2 °C and 7.4 °C, respectively) (Table 1). ADP also increased the Tm of UbaA (by 4.4 °C); whereas, AMP, CTP, GTP, UTP, and TTP had no detectable effect in these assays (Table 1). Thus, UbaA was found to bind the nucleotide ligands AMP-PNP, ATP and ADP but not AMP, CTP, GTP, UTP, and TTP. These results are supported by our recent finding that PPi is released when UbaA is incubated with SAMPs and ATP, but not the other nucleoside triphosphates [22].
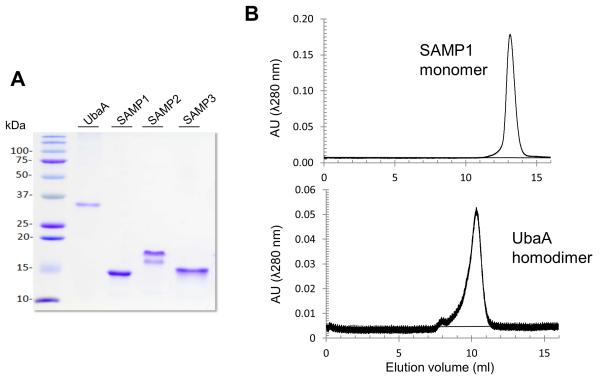
Figure 1. UbaA and SAMP proteins analyzed by reducing SDS-PAGE and size exclusion chromatography.
(A) UbaA and SAMP1/3 were found to migrate as single protein bands and SAMP2 as two distinct protein bands, when separated by reducing SDS-PAGE and stained with Coomassie brilliant blue. The flexible β-hinge region of SAMP2 [27] was attributed to account for its unusual migration by SDS-PAGE, based on analysis of samples by electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS) that suggested protein homogeneity (Table S1). (B) Representative SEC elution profiles are presented which reveal UbaA and SAMP1 to be a homodimer (~ 55 kDa) and monomer (~10 kDa), respectively. Similar results were obtained for the other UbaA and SAMP proteins purified including the site-directed variants.
Table 1.
Thermal stability of UbaA and its variants determined by differential scanning fluorimetry (DSF) in the presence and absence of nucleotides.
| UbaA Variant | Ligand | Tm (°C) |
Tm Shift (°C) |
|---|---|---|---|
| wt | -a | 58.60 ± 1.13 | - |
| ATP | 66.80 ± 3.96 | 8.20 ± 4.12 | |
| AMP | 59.90 ± 0.42 | 1.30 ± 1.21 | |
| ADP | 63.00 ± 0.85 | 4.40 ± 1.41 | |
| AMP-PNP | 66.00 ± 3.39 | 7.40 ± 3.58 | |
| CTP | 57.60 ± 1.13 | -1.00 ± −1.60 | |
| GTP | 59.60 ± 1.41 | 1.00 ± 1.81 | |
| TTP | 59.70 ± 2.12 | 1.10 ± 2.40 | |
| UTP | 59.70 ± 0.99 | 1.10 ± 1.50 | |
|
| |||
| K87R | - | 59.68 ± 1.42 | - |
| ATP | 60.12 ± 0.46 | 0.44 ± 1.61 | |
|
| |||
| D131N | - | 61.12 ± 1.85 | - |
| ATP | 60.92 ± 3.02 | −0.20 ± 3.54 | |
|
| |||
| C188A | - | 56.08 ± 2.04 | - |
| ATP | 63.20 ± 1.11 | 7.12 ± 2.32 | |
-, no ligand control. Means of thermal shift difference and standard deviations were calculated (n = 2 for the nucleotide ligands, n = 6 for the UbaA variants). See Methods for assay details.
Influence of ATP ligand on UbaA binding to SAMP1/2
Thermodynamic parameters of the UbaA homodimer and its binding to the monomeric SAMPs were determined by isothermal titration calorimetry (ITC) analysis (Table 2, supporting data Fig. S1). To assess Ubl:E1-like complex formation in the absence of ATP hydrolysis, ITC analysis was performed with and without added AMP-PNP and metal-free ATP (EDTA-treated). By this approach, UbaA homodimers were found to bind SAMP1/2 monomers in 1:2 molar ratio with the C-terminal diglycine (diGly) residues of the SAMPs required for this association (Table 2). UbaA associated with SAMP1 independent of nucleotide (Kd of 4.1 μM); however, docking of ATP promoted a tighter binding of UbaA to SAMP1 based on a 2- to 3-fold reduction in the dissociation constant observed upon addition of metal-free ATP (Kd of 1.2 μM) or AMP-PNP (Kd of 1.7 μM) to the assay (Table 2). AMP and PPi had little, if any, influence on SAMP1 binding to UbaA (Kd of 4.4 μM) compared to no nucleotide (Table 2). The binding of SAMP1 to UbaA was entropically driven (ΔS>0), suggesting that the association of these protein partners caused hydrophobic side chains to become buried and water to be released with a minimal loss of conformational degrees of freedom. In contrast to SAMP1, ATP binding was required for UbaA to associate with SAMP2 with an observed Kd of 4.3 μM that was not detected in the absence of metal-free ATP. The binding of SAMP2 to UbaA was enthalpically favored (ΔH<0), revealing hydrogen bonding and van der Waals interactions likely dominated the binding interface. Overall, these results demonstrate significant differences in the thermodynamic properties of UbaA binding to the SAMPs that are dependent upon the SAMP-type.
Table 2.
Thermodynamic parameters derived from the calorimetric titration of UbaA with SAMPs in the presence or absence of nucleotides.
| Protein in ITC Cell |
Injectant Protein |
Nucleotide |
KD (μM) |
N |
KA (μM-1) |
ΔH ( kJ/mol) |
ΔS (J/mol/K) |
|---|---|---|---|---|---|---|---|
| UbaA | SAMP1 | -a | 4.1 ± 0.8 | 1.02 ± 0.03 | 0.25 ± 0.05 | 16.0 ± 0.6 | 157 |
| UbaA | SAMP1 | AMP + PPi | 4.4 ± 0.8 | 0.95 ± 0.03 | 0.23 ± 0.05 | 20.7 ± 0.8 | 172 |
| UbaA | SAMP1 | Metal-free ATPb | 1.2 ± 0.2 | 0.98 ± 0.01 | 0.85 ± 0.13 | 12.2 ± 0.2 | 154 |
| UbaA | SAMP1 | AMP-PNP | 1.7 ± 0.1 | 0.95 ± 0.01 | 0.59 ± 0.06 | 36.2 ± 0.4 | 232 |
| UbaA | SAMP1ΔGG | - | UDc | UD | UD | UD | UD |
| UbaA | SAMP1ΔGG | Metal-free ATP | UD | UD | UD | UD | UD |
| UbaA | SAMP2 | - | UD | UD | UD | UD | UD |
| UbaA | SAMP2 | Metal-free ATP | 4.3 ± 0.8 | 0.99 ± 0.03 | 0.23 ± 0.05 | -69 ± 3 | -129 |
| UbaA | SAMP2ΔGG | - | UD | UD | UD | UD | UD |
-, no nucleotide.
Metal-free ATP generated by EDTA treatment.
UD, undetectable. Raw isothermal titration calorimetry (ITC) data and enthalpy plots are provided in Fig. S1. Means and standard deviations were calculated (n = 2). See Methods for assay details.
UbaA associates with the SAMPs in vivo
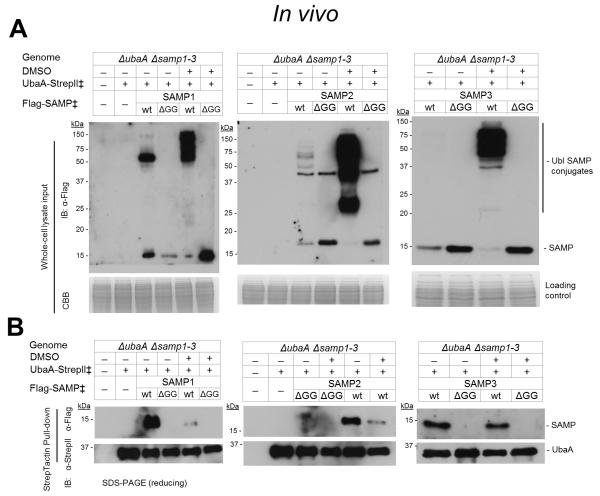
The binding of UbaA to SAMPs was assessed in vivo including an evaluation of the impact of culture conditions. To accomplish this, the UbaA and SAMPs were co-expressed as functional epitope-tag fusions (C-terminal StrepII and N-terminal Flag, respectively) [23-25]. The SAMPs devoid of their C-terminal diGly residues (ΔGG) were included for comparison. Protein partners of UbaA were captured by StrepTactin chromatography, with buffers supplemented with 2 M salt to maintain non-covalent bonds (e.g., haloarchaeal 20S proteasomes disassemble at salt concentrations less than 1 M) [26]. Hfx. volcanii strains were grown in the absence and presence of DMSO, with the latter used to induce Ubl bond formation [23]. Analysis of the whole-cell lysate input revealed SAMP1-3 formed Ubl bonds on diverse protein targets; whereas, SAMP1/3 ΔGG did not form Ubl bonds and SAMP2 ΔGG formed a Ubl conjugate of 44 kDa (Fig. 2A). SAMP2 has a long and flexible C-terminal tail compared to SAMP1/3 [27], which may explain why SAMP2 ΔGG can form a Ubl bond with its C-terminal lysine. Further analysis of the pull-down fractions revealed UbaA bound all three SAMPs in non-covalent complexes and required the C-terminal diGly residues to detect this association (Fig. 2B). The levels of SAMP1/2 non-covalently bound to UbaA were significantly decreased when cells were grown in the presence of DMSO, while SAMP3 bound UbaA irrespective of the growth conditions examined (Fig. 2B). Taken together, these results demonstrate that UbaA can associate with all three SAMPs in non-covalent complexes in vivo and that the association of UbaA with SAMP1/2 is modulated by environmental conditions (DMSO).
Figure 2. UbaA associates with SAMPs in vivo in non-covalent complexes that are modulated by environmental growth conditions.
(A) Ubl-modifications detected in the cell lysate of Hfx. volcanii strains expressing UbaA and SAMPs with epitope tags grown aerobically to stationary phase with and without the addition of 100 mM DMSO as indicated. (B) UbaA-StrepII and associated proteins are purified from cell lysate by StrepTactin chromatography. ‡, expressed in trans. See Methods for details.
Modeling the 3D structure of UbaA
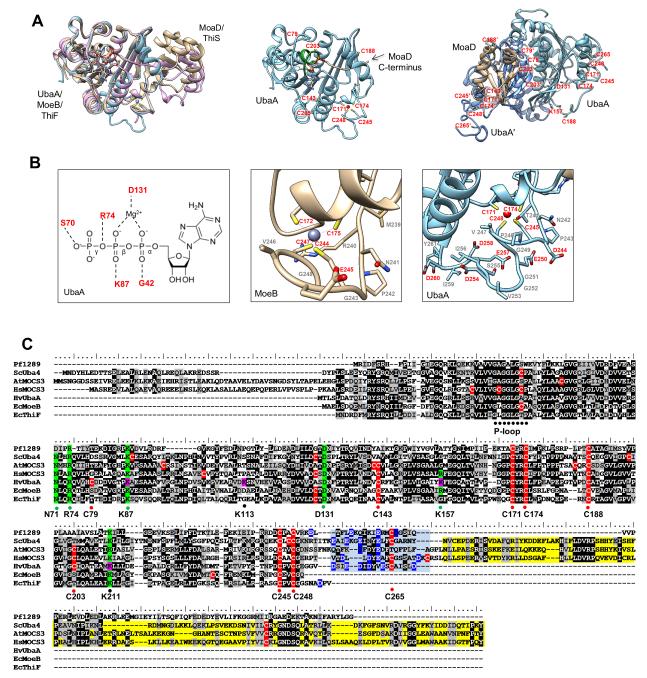
The UbaA adenylation domain could be modeled with high (> 90%) confidence by comparison to bacterial MoeB/ThiF crystal structures (Fig. 3A). Within the adenylation domain, the phosphate-binding loop (P-loop) (39-GAGGLGAP-46) of UbaA was found to differ from the classical Walker A motif of G-X4-GK[T/S] (where X represents any amino acid) by the absence of the conserved lysine residue required for nucleotide binding. Instead, UbaA K87, G42, R74 and S70 were modeled to form direct H-bonds to the oxygen atoms of the nucleotide triphosphate groups of ATP, and D131 was predicted to coordinate the Mg2+ ion associated with the nucleotide (Fig. 3B). UbaA residues that would interface and activate the Ubl SAMPs and coordinate a structural Zn2+ ion were not as clearly defined by homology modeling. The majority of cysteine residues appeared on the surface of UbaA. The conserved ‘active site’ cysteine (C188) of UbaA was modeled to a unstructured region (V166 to G195) that appeared to contact the Ubl MoaD, yet was at a distance from the site of activation (the C-terminal diGly) (Fig. 3A). UbaA C79 and C203 were positioned within the homodimer interface at a substantial distance that would prohibit disulfide bonding. UbaA C171, C174, C245, and C248 aligned with the cysteine residues of a tetrad motif that coordinates Zn2+ in MoeB/ThiF crystal structures (Fig. 3B-C). However, a C-terminal extension was also identified in UbaA that was rich in acidic amino acid residues (Asp/Glu) as well as C265 that could theoretically coordinate Zn2+ (Fig. 3B-C). This C-terminal extension was found to be widely conserved in archaeal E1 homologs, yet absent in bacterial MoeB/ThiF proteins and fused to an additional rhodanese domain (RHD) in Uba4p/MOCS3 (Fig. 3C). Thus, while the model of UbaA appeared robust in predicting residues that could coordinate ATP, the remaining portions of the protein were not as well defined.
Figure 3. UbaA 3D-structural model and multiple amino acid sequence alignment.
(A) Ribbon diagram UbaA (blue) predicted structure and its superimposition onto the crystal structures of E. coli covalent acyl-adenylate form of the MoeB:MoaD complex (gold) (1JWB) and monomeric half of the ThiF:ThiS complex (purple) (1ZUD). Coordinating AMP, Zn2+ ligands and UbaA P-loop (green) are highlighted. UbaA homodimer is designated by light (UbaA) and dark (UbaA’) blue ribbon diagram with cysteine and other residues highlighted. (B) From left to right: i) schematic representation showing conserved residues of the UbaA adenylation domain predicted to coordinate Mg-ATP; ii) ribbon diagram of MoeB C-terminal region including the Cys tetrad residues (C172-X2-C175-Xn-C244-X2-C247) that coordinate Zn2+; and iii) ribbon diagram of 3D-model of UbaA C-terminal region that includes Cys tetrad residues (C171-X2-C174-Xn-C245-X2-C248) and acidic residues (D244, E250, D254, E257, D258, and D260) which could coordinate Zn2+. (C) Multiple amino acid sequence alignment of UbaA with homologs of the E1/MoeB/ThiF superfamily. Predicted catalytic cysteine (C188), conserved Cys-tetrad (C171, C174, C245, C248), and less conserved cysteine residues (C79, C143, C203, and C265) of UbaA are indicated in red. UbaA residues of the adenylation domain found important for ATP binding (K87 and D131) and other activities (R74 and N71) are highlighted in green. UbaA lysine residues modified by Ubl bonds are indicated in pink, with K87 and K157 further highlighted in green to indicate their important role in catalytic activity. Highlighted in blue is the acidic C-terminal tail common to archaeal UbaA homologs and linker to the rhodanese domain of eukaryotic Uba4/MOCS3 homologs (the latter highlighted in yellow). Abbreviations with UniProt accession numbers: Hfx. volcanii HvUbaA (sp∣D4GSF3∣), E. coli EcMoeB (sp∣P12282∣), E. coli EcThiF (sp∣P30138∣), P. furiosus Pf1289 (tr∣Q8U1C8∣), yeast ScUba4 (sp∣P38820∣), human HsMOCS3 (sp∣O95396∣), Arabidopsis thaliana AtMOCS3 (sp∣Q9ZNW0∣).
UbaA residues required for nucleotide binding
Amino acid exchange analysis was used to define UbaA residues within the adenylation domain that could bind ATP. Residues predicted to coordinate the β-phosphate group (K87) and the Mg2+ ion (D131) of Mg-ATP were targeted for site-directed modification. The conserved ‘active site’ cysteine (C188), previously reported to be important in sulfur mobilization and Ubl bond formation (sampylation) [24], was included for comparison. The amino acid exchange variants of UbaA (K87R, D131N and C188A) were purified and the DSF-derived Tm values were compared to wild type in the presence and absence of ATP (Table 1). All three UbaA variants were found to purify as homodimers similarly to wild type and to have Tm values comparable to wild type when assayed in the absence of nucleotide ligand (Table 1). The C188A variant had an increase in Tm of 7.12 °C when assayed in the presence of ATP that was similar to wild type and indicative of ATP binding. By contrast, ATP did not significantly alter the Tm values of UbaA K87R and D131N, with thermal shifts of less than 0.5 °C. Thus, UbaA K87 and D131 were important for ATP binding, while C188 was not needed to coordinate this ligand.
Key residues within the adenylation domain are required for UbaA activity
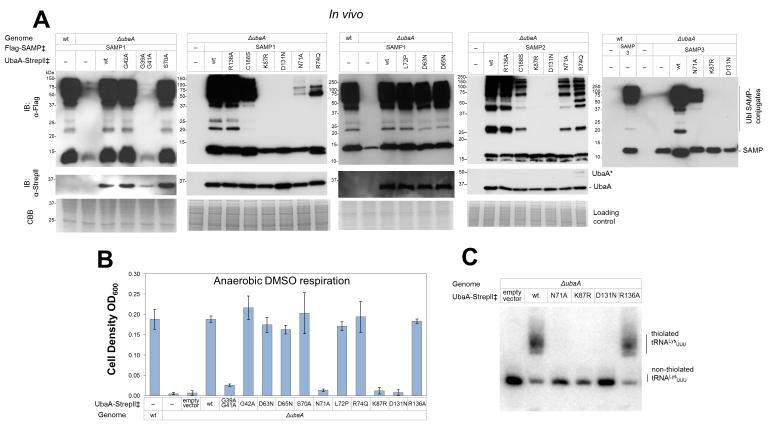
Site-directed variants of the UbaA adenylation domain were next analyzed for E1-like activities downstream of ATP binding. By this approach, the residues (K87 and D131) needed to bind ATP were also found important for all of UbaA activities including Ubl bond formation (Fig. 4A), sulfur mobilization to form MPT (Fig. 4B), and tRNALysUUU thiolation (Fig. 4C). By contrast, UbaA residues in close proximity to K87 and the ATP β/γ-phosphate region had a differential effect; N71 and R74 were important for Ubl bond formation, but only N71 was essential for sulfurtransferase activity (MPT biosynthesis and tRNA thiolation) (Fig. 4). Substitutions of glycine residues within the P-loop were inconclusive, as the single G42A had no effect and the double G39A G41A substitution inactivated UbaA but also reduced the levels of this E1-like protein in the cell (Fig. 4). No apparent difference from wild type was observed in strains expressing UbaA D63N, D65N, S70A, and L72P (Fig. 4). Together these results demonstrate that the UbaA residues needed to bind ATP (D131 and K87) are required for all UbaA activities and, thus, are positioned at the early stage of Ubl protein activation. Furthermore, the results reveal that UbaA harbors residues within the β/γ-phosphate binding region that are important in Ubl bond formation but not necessarily needed for sulfur mobilization.
Figure 4. UbaA residues within the adenylation domain analyzed for function.
UbaA residues of the adenylation domain were analyzed for their role in: (A) forming Ubl bonds, (B) mobilizing sulfur to form molybdopterin for DMSO respiration, and (C) thiolating tRNALysUUU. ‡, expressed in trans. Error bars represent standard deviations (n = 3). See Methods for details.
Thiol-dependent linkage of UbaA to SAMPs
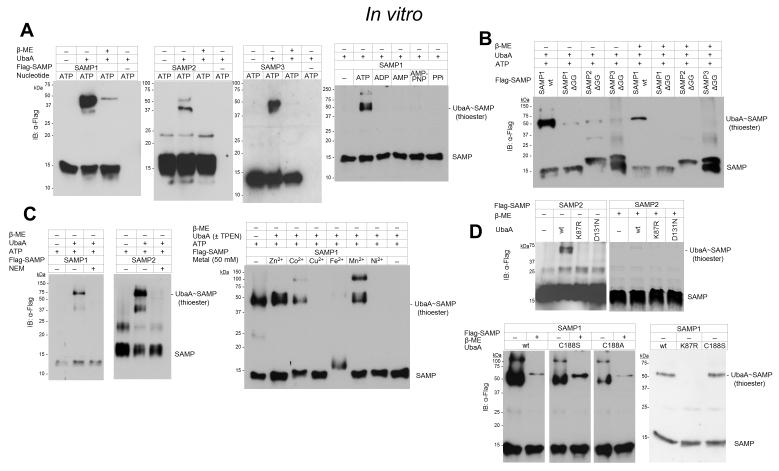
We next examined whether ATP hydrolysis could drive the formation of covalent bonds between UbaA and SAMP by gel mobility shift assay. After incubating UbaA and SAMP with various nucleotides, the purified proteins were boiled in SDS buffer under reducing (β-mercaptoethanol, β-ME) and non-reducing conditions, separated by PAGE and analyzed for complex formation by immunoblotting. All three SAMPs were found to associate with UbaA in covalent complexes that were sensitive to reducing reagent suggesting formation of a thioester intermediate (UbaA~SAMP) (Fig. 5A). Under these brief incubation periods, a small portion of the complexes were resistant to β-ME indicating that Ubl covalent bonds were also formed on UbaA (see Autosampylation section for detailed analysis). The non-hydrolyzable ATP analog, AMP-PNP, as well as ADP, AMP, PPi, GTP, CTP, TTP and UTP could not substitute for ATP in formation of these covalent bonds (Fig. 5A). The C-terminal diGly residues of the SAMPs were also found important in the covalent linkages (Fig. 5B). Preincubation of UbaA with the sulfhydryl-modifying NEM or the heavy metal chelator TPEN inhibited complex formation, with addition of Zn2+, Co2+ and Mn2+ (but not Cu2+, Fe2+, Ni2+ or Mg2+ of the Mg-ATP) to TPEN treated UbaA restoring complex formation (Fig. 5C). The heavy metal ion needed to form the UbaA~SAMP complex was most likely a non-catalytic Zn2+ ion based on comparison to other enzymes of the E1/MoeB/ThiF superfamily [28, 29]. Thus, an NEM-sensitive cysteine residue(s) of UbaA is suggested to form a thioester intermediate with the SAMPs in the presence of hydrolyzable ATP that requires UbaA to be bound to a structural Zn2+ ion.
Figure 5. UbaA~SAMP thioester intermediate formed in vitro.
(A) In the presence of hydrolyzable ATP, all three SAMPs associate with UbaA in covalent complexes that are sensitive to reducing reagent (β-ME). (B) SAMP C-terminal diGly residues are required to form the thiol-dependent UbaA~SAMP complexes. (C) The UbaA~SAMP thioester intermediate is not formed when UbaA is pre-incubated with N-ethylmaleimide (NEM), a modifier of cysteine thiols or the metal chelator TPEN. Addition of Zn2+ (Co2+ and Mn2+) to the TPEN treated UbaA restores formation of the thioester bond. (D) UbaA K87 and D131 of the adenylation domain, but not the conserved ‘active site cysteine’ C188, are required to form the UbaA~SAMP thioester. *UbaA, covalently modified. See Methods for details.
UbaA residues required for thiol-dependent linkage to the SAMPs
Investigation was next carried out to determine the amino acid residues required to form the UbaA~SAMP thioester. Our focus was on the UbaA residues that were found important for ATP binding/hydrolysis (K87 and D131) as well as the conserved cysteine (C188) positioned within the central α-helix of the UbaA model. UbaA K87R, D131N and C188[A/S] proteins were assayed by gel mobility shift assay with SAMP1. With this approach, UbaA K87 and D131 were found crucial for the formation of the thiol intermediate (Fig. 5D), most likely due to the requirement of these residues for ATP binding. To our surprise, the conserved ‘active-site’ C188 of UbaA was not needed to form the thiol intermediate, yet the complex was sensitive to NEM (Fig. 5D). Thus, UbaA harbored one or more cysteine residues, distinct from C188, which formed a thioester intermediate with the SAMPs.
UbaA cysteine residues important for catalysis
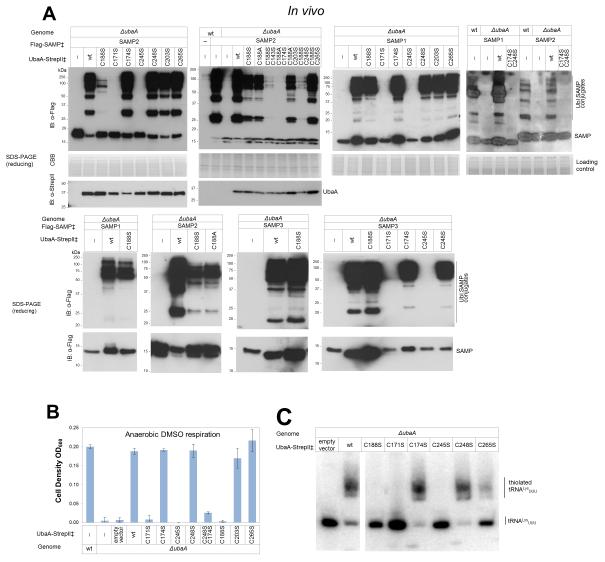
Alanine/serine scanning mutagenesis was used to determine cysteine residue(s) which may be responsible for UbaA activities. UbaA has nine cysteine residues including those of a Cys-tetrad motif (C171, C174, C245 and C248) and an ‘active site’ cysteine (C188) conserved among members of the E1/MoeB/ThiF superfamily (Fig. 3C). Interestingly, UbaA C188[A/S], which formed a thioester bond with SAMP1/2 in vitro, also functioned to modify proteins with Ubl bonds under optimized in vivo assay conditions (Fig. 6A), but was not found to mobilize sulfur to form MPT or thiolated tRNA (Fig. 6B-C). While the analogous ‘active site’ cysteine of canonical E1s forms a Ub/Ubl thioester intermediate, this cysteine residue is not needed for bacterial MoeB function and has important/unimportant roles in yeast Uba4p/human MOCS3 depending upon the assay conditions (see Table 3 for comparison). Further individual amino acid substitution of the Cys-tetrad of UbaA revealed the first cysteine residue (C171 and C245) of each CX2C pair was essential for Ubl bond formation and sulfur mobilization, while the second cysteine of the pair (C174 and C248) was not needed for function (Fig. 6). Thus, UbaA differed from its bacterial MoeB counterpart, which requires all Cys-tetrad motif residues for sulfurtransferase activity (Table 3). Further analysis of the other UbaA variants revealed C203 was nonessential, while C265 near the C-terminus was exceptional in its requirement for the thiolation of tRNA but not for Ubl bond or MPT formation (Fig. 6). Double amino acid substitution of C188S with other cysteine variants that alone had little to no impact on UbaA activity resulted in a significant reduction in the level of Ubl bonds formed but did not impair UbaA protein levels (Fig. 6A). These findings support a model in which UbaA uses cysteine residue(s) distinct from C188 to form a Ubl thioester intermediate(s) and subsequent Ubl bonds. Our results suggest UbaA may have a significant amount of flexibility to accommodate multiple cysteine residues in Ubl protein activation during Ubl bond formation, which is consistent with the disordered region overlap of MoeB/ThiF (Fig. 3A). When compared to MoeB/ThiF, one structural difference of UbaA is its extended C-terminal tail that is rich in Cys/Glu/Asp residues that could bind Zn2+ and free up the Cys-tetrad for use in forming the thioester intermediate observed between SAMP and UbaA C188[S/A] (Fig. 3C). Further deliberation on the noncanonical E1-like mechanism is in the Discussion section.
Figure 6. UbaA cysteine residues in Ubl bond formation and the mobilization of sulfur to form MPT and thiolated tRNA.
UbaA cysteine residues were analyzed for their role in: (A) forming Ubl bonds, (B) mobilizing sulfur to form molybdopterin for DMSO respiration, and (C) thiolating tRNALysUUU. ‡, expressed in trans. Error bars represent standard deviations (n = 3). See Methods for details.
Table 3.
Comparison of E1/MoeB/ThiF superfamily proteins by amino acid exchange studiesa.
| Description | Archaeal UbaA | E. coli MoeB | Yeast Uba4p | Mammalian MOCS3 | E1 enzymes |
|---|---|---|---|---|---|
| Ubl protein partner |
SAMP1, SAMP2 and SAMP3 |
MoaD | Urm1 | MOCS2A |
e.g., Ub, SUMO and NEDD8 |
| Ub/Ubl bond formation |
Yes | No | Yes | Yes | Yes |
| MPT biosynthesis |
Yes | Yes | No | Yes | No |
| Thiolation of tRNA |
Yes | No | Yes | Yes | No |
| Thiol- dependent intermediate |
Yes | No [7] | Yes [58] and No [59] | Not determined | Yes |
| PDB 3D structure |
No | 1JW9, 1JWA, 1JWB |
No | 3I2V (RHD domain only) |
4NNJ (and other examples) |
| Conserved ‘active site’ cysteine |
C188 critical for sulfurtransferase activity, but not needed to form thiol- intermediate or Ubl bonds |
C187 nonessential [7] |
C225 essential in vivo in Ubl bond formation [58] and the thiolation of wobble uridine tRNA [60, 61]; not essential in vitro in the thiocarboxylation or adenylation of Ubl proteins [59] |
C239 essential in vitro in the thiocarboxylation of MOCS2A [62] |
forms thioester intermediate with the Ub/Ubl protein needed form Ub/Ubl bonds [37, 63] |
| Cys-tetrad (CX2C-Xn- CX2C) |
C171 and C245 essential for all activities; C174 and C248 not essential unless combined with other Cys variants |
C172, C175, C244 and C247 essential for binding structural Zn2+ and sulfurtransferase activity [7] |
Conserved | Conserved | Zn2+ motif suggested to be important in binding and preventing Ubl protein adenylate from dissociating [64] |
| C-terminal extension/linker |
C265 needed for thiolation of wobble uridine tRNA |
- | Conserved | C316 and C324 not needed in the thiocarboxylation of MOCS2A in vitro [62] |
- |
| ATP coordinating residues |
R74 not important as sulfurtransferase (at least MPT biosynthesis); important in Ubl bond formation N71 required for sulfurtransferase (MPT and thiolated tRNA); important in Ubl bond formation K87 and D131 needed for all UbaA activities including ATP binding; K87 is autosampylated |
R73 nonessential [28] Conserved Conserved |
Conserved Conserved Conserved |
Conserved Conserved Conserved |
Generally needed for Ubl bond formation |
| R/K at end of β-sheet within docking site of Ubl protein C- terminus |
K157 autosampylated; required for MPT biosynthesis but not Ubl bond formation |
Conserved | - | Conserved | - |
| Rhodanese (RHD) domain |
- | - | C397 needed in vitro to form Ubl protein thiocarboxylates but not adenylation [59]; essential for thiolation of wobble uridine tRNA in vivo [60, 61] |
C412 persulfide detected by MS [65] and found essential in thiocarboxylation of MOCS2A [62]; D417[R/T] increases sulfurtransferase activity of RHD domain [66] |
- |
-
E1/MoeB/ThiF proteins serve as adenylyltransferases in Ubl bond formation and as sulfurtransferases in MPT biosynthesis and the thiolation of tRNA. Due to space limitation only a few citations are provided for E1 enzymes. -, not conserved.
Autosampylation
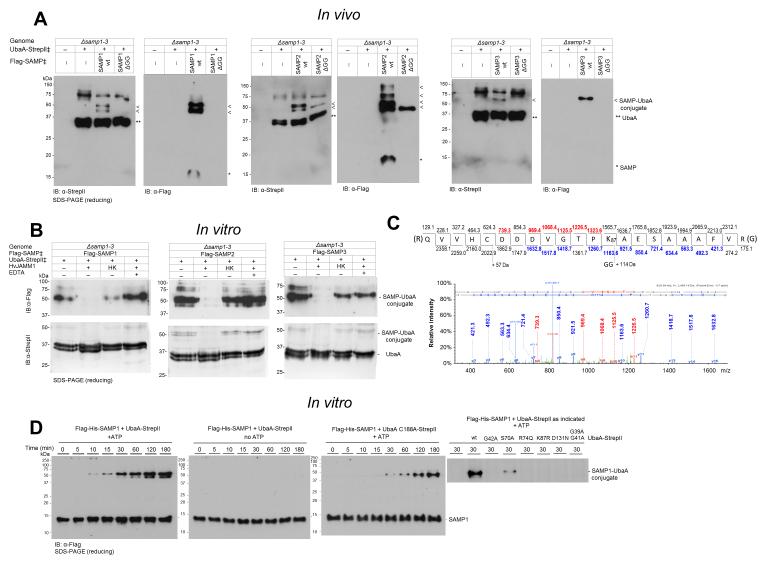
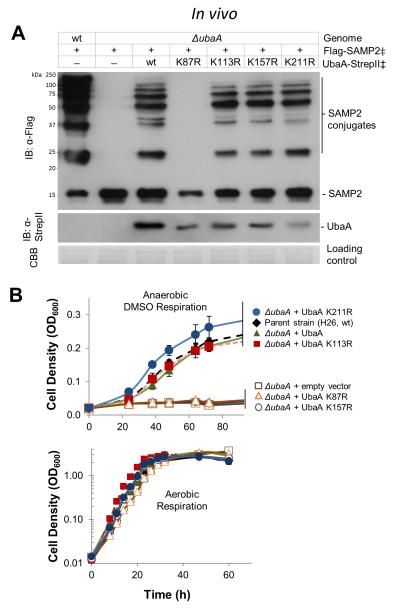
Several lines of evidence support autosampylation of UbaA (i.e., the sole E1/MoeB/ThiF superfamily member of Hfx. volcanii forms Ubl linkages on itself). First, UbaA is found covalently linked to the Ubl SAMPs. Of its four lysine residues, UbaA is sampylated at two lysines, K157 (modified by SAMP1) and K113 (modified by SAMP1/2), based on previous study [25, 30]. In addition, in this study, UbaA was found to be covalently linked to all three SAMPs (SAMP1/2/3) by Ubl bonds that were resistant to reducing SDS-PAGE (Fig. 7A) and hydrolyzed by HvJAMM1 (Fig. 7B), a Zn2+ dependent metalloprotease that cleaves Ubl bonds [19]. Further analysis by LC-MS/MS, revealed UbaA K87 to be linked to SAMP2 by an Ubl bond (Fig. 7C). In addition to finding Ubl bonds on UbaA, in vitro assay with purified components demonstrated UbaA to catalyze autosampylation. Covalent complexes of UbaA-SAMP (recalcitrant to β-ME) were detected when UbaA homodimers and SAMP1 monomers were incubated at 1:2 molar ratio in the presence of ATP (Fig. 7D). The Ubl linked products were evident after the SAMPs were incubated with UbaA wt (10 min) or C188A (30 min), but were not detected when ATP was omitted from the assay (Fig. 7D). UbaA residues important in ATP binding (K87 and D131) and other residues in the P-loop region (G39, G41, G42 and R74) were required to detect the autosampylation activity (Fig. 7D). Further analysis of UbaA lysine residue function revealed that, while K87 was essential for all enzyme activities, K157 was required for only sulfur mobilization (not Ubl bond formation) and K113 and K211 were unessential (Fig. 8). Overall, these results reveal UbaA is autosampylated through an ATP-dependent mechanism that is catalyzed independent of E2/E3 enzymes.
Figure 7. UbaA is autosampylated.
(A) >UbaA is covalently modified by all three SAMP types. SAMP-modified and unmodified UbaA purified by StrepTactin affinity chromatography from DMSO grown Hfx. volcanii strains (as indicated) and normalized based on total protein of the cell lysate applied to each StrepTactin column. (B) UbaA-SAMP conjugates cleaved in vitro by HvJAMM1 but not the heat-killed (HK) protease. (C) UbaA K87 is covalently modified by SAMP2. Ubl modification sites were identified by CID LC-MS/MS analysis of tryptic peptides of UbaA purified as in panel A. The diGly remnant (+114 Da) on K87 indicated SAMP2 modification. D) UbaA is autosampylated in vitro. Purified SAMP1 and UbaA (wt and variant) proteins were incubated with and without ATP as indicated. ‡, expressed in trans. See Methods for details.
Figure 8. Autosampylation of K87 and K157 likely regulates UbaA function.
UbaA lysine residues were analyzed for their role in: (A) forming Ubl bonds and (B) mobilizing sulfur to form molybdopterin for DMSO respiration. ‡, expressed in trans. Means and standard deviations were calculated (n = 3). See Methods for details.
DISCUSSION
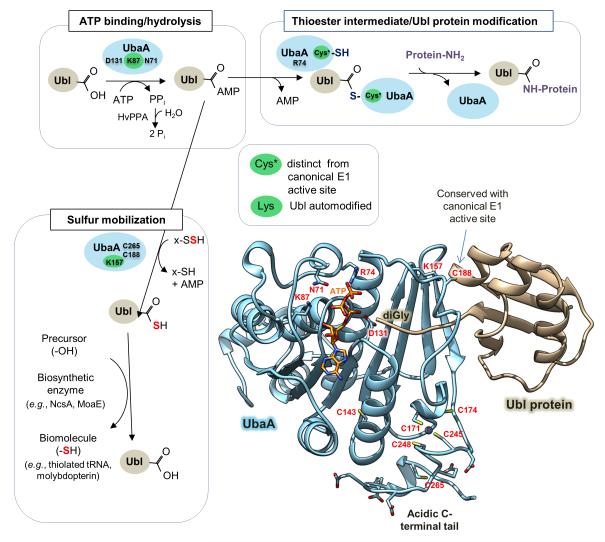
Here we summarize the major findings of the structure-function study a non-canonical E1-like enzyme with broad specificity for cognate Ubl proteins that mediates Ubl-protein modification and sulfur mobilization (to form MPT and thiolated tRNA). In particular, we highlight key findings regarding the archaeal E1-like UbaA and Ubl SAMPs (Fig. 9, Table 3).
Figure 9. Schematic representation of UbaA and its function in Archaea.
Highlighted are UbaA residues important in ATP binding/hydrolysis (D131, K87 and N71), Ubl protein modification (R74 and Cys*) and sulfur transfer (K157 and C265) (where Cys* represents a cysteine residue that is required to form a UbaA~Ubl thioester intermediate distinct from the canonical ‘active site’ cysteine residue (C188). The thioester is an apparent intermediate of Ubl bond formation (not sulfur mobilization). X, represents a rhodanese domain (RHD) protein speculated to be required for sulfur transfer to the Ubl proteins. E2/E3 analogs required for Ubl protein modification pathway remain undetermined. HvPPA inorganic pyrophosphatase hydrolyzes the PPi byproduct of SAMP adenylation catalyzed by UbaA [22]. UbaA is regulated by Ubl automodification (autosampylation) of lysine residues highlighted in green, where K87 is required for all UbaA activities, and K157 is required for UbaA sulfurtransferase activity. See Discussion for details.
UbaA binding to its cognate SAMP is influenced by environmental condition
Compared to oxygen alone, the non-covalent binding of UbaA to SAMP1/2 is reduced when cells are grown to stationary phase with added DMSO (a mild oxidant and terminal electron acceptor). One possible explanation for these findings is that SAMP1/2 are not only Ubl protein modifiers but are also sulfur-carriers [20, 24]. The abundance of thiocarboxylated SAMP1/2 is likely to be increased with added DMSO to generate the MPT needed for DMSO respiration and the thiolated tRNA for stress resistance and long-term survival. Thiocarboxylated SAMP1/2 is likely to have reduced affinity for UbaA binding to allow for association with protein partners (MoaE/NcsA) that direct the sulfur to the appropriate biomolecule (MPT biosynthesis/ tRNA thiolation) [20, 24]. SAMP3 binding to UbaA is unaffected by DMSO but has no known role in sulfur mobilization.
The type of Ubl protein has a major influence on the order of substrate binding to UbaA
SAMP1/2 are Ubl fold proteins with distinct structural features that are likely to be influential factors in binding UbaA. SAMP1, while not a close homolog, does share 3D structural elements [31] and C-terminal amino acid sequence (−GDE[L/V]ALFPPV[S/T]GG) with bacterial MoaD. Much like the bacterial MoaD binding to MoeB [28, 32], the archaeal SAMP1 binds UbaA in the absence of ATP, with this association stabilized by ATP binding. By contrast, SAMP2, which has an extended C-terminal tail and unique β-hinge region [27, 33], binds UbaA like eukaryotic Ub binds E1 [34], through an ordered mechanism in which ATP binding precedes Ub/Ubl protein binding to the E1/E1-like enzyme. Interestingly, the downstream consequences of SAMP2 binding to UbaA are also eukaryotic-like (SAMP2 tags proteins for destruction by proteasomes, whereas, SAMP1-modified proteins can be quite stable) [19, 35]. The importance of the Ub/Ubl C-terminal diGly motif in E1/E1-like binding is evolutionarily conserved in that the archaeal SAMP diGly is needed to bind UbaA, the bacterial MoaD G81 (of the diGly motif) is needed to bind MoeB [32], and the eukaryotic Ub diGly is needed to form the Ub adenylate (Ub~AMP) and E1~Ub thioester [36-39] implying that the diGly is also needed for E1 binding. Whether archaeal Ub-fold proteins devoid of C-terminal diGly residues, as predicted by genomics [40], use different binding determinants or do not bind E1-like enzymes remains to be determined.
Residues within the UbaA adenylation domain bind/hydrolyze ATP and direct SAMPs to either Ubl bond formation or sulfur mobilization
In particular, K87 and D131 (residues modeled to coordinate the β-phosphate and Mg2+ ion of Mg-ATP, respectively) are found important for all UbaA functions including forming Ubl bonds and mobilizing sulfur to form MPT and thiolated tRNA. UbaA deficiency in these diverse activities is explained by the inability of the K87R and D131N variants to bind ATP (the first step in the pathway). By contrast, residues modeled near the ATP γ-phosphate binding pocket are generally important in Ubl bond formation but have differential roles in sulfur mobilization (e.g., N71 is crucial in formation of MPT and thiolated tRNA, while R74 is unimportant in these activities).
UbaA forms a thioester intermediate with the SAMPs
Previous work demonstrates UbaA adenylates SAMPs (an early step in the pathway) [22, 41]. Here, we find UbaA forms a thioester intermediate with the SAMPs (a later step in the pathway). Thiol-dependent intermediates are commonly reported for the canonical Ub~E1s of eukaryotes [42, 43] and the dual function TtuC~TtuB of hyperthermophilic bacteria [18], but are not observed for systems associated solely with sulfur mobilization, such as the bacterial MoeB:MoaD [7]. We argue that UbaA cysteines and not cysteine persulfides act as the apparent nucleophile in formation of the UbaA~SAMP thioester intermediate based on the following. First, a thiol-dependent bond was detected between SAMP and UbaA that required the input of ATP but not a sulfur donor (e.g., thiosulfate or cysteine). Second, the SAMPs and UbaA used for the in vitro reactions were purify to homogeneity and did not have a +16 Da mass increase indicative of added sulfur as determined by ESI-TOF MS analysis (Table S1). The thioester bond is argued to be the C-terminal residue of SAMP linked to an UbaA cysteine residue(s), based on finding that the bond is sensitive to SAMPΔGG and NEM treatment, which would alkylate active site thiol groups present in UbaA (not the cysteine-free SAMPs).
UbaA activates the SAMP in formation of a thioester by a cysteine residue that is distinct from the conserved ‘active site’ cysteine
Stunningly, the conserved ‘active site’ cysteine of UbaA is only needed for sulfur mobilization and is not required to activate the SAMP in formation of the thioester intermediate or subsequent Ubl bonds. Candidate UbaA cysteines that would form the thioester reside within the Cys-tetrad motif. Unlike bacterial MoeB which requires the Cys-tetrad for binding a structural Zn2+ ion [7], the UbaA residues of the Cys-tetrad are in close proximity to an extended C-terminal tail rich in Cys/Asp/Glu residues that could in theory contribute to electrostatic coordination of a structural Zn2+. If so, the UbaA Cys-tetrad residues would be free to form a thioester intermediate with the SAMPs. This added flexibility would also allow UbaA to form and/or associate with cysteine persulfides during the mobilization of sulfur, a process that would require the attack of another cysteine residue, leading to disulfide bonds that require input of reductant for enzyme recycling. The implication of such a mechanism would be that at least one other cysteine must reach into the region where the SAMP adenylate needs to be attacked. A good candidate is C265 within the C-terminal tail of UbaA, since this cysteine residue is uniquely required in the thiolation of tRNA, but is not needed to form Ubl bonds. Even the canonical eukaryal E1s display a high degree of plasticity within the active site [2]. In the initial stages of binding Mg-ATP and Ub, the E1 active site cysteine is relatively far from the adenylation active site [2]. Once Ub is adenylated and PPi is released, the E1 active site undergoes a network of complementary conformation changes to allow for thioester bond formation (including a 130° rotation of the domain harboring the active site cysteine) [2].
UbaA is autosampylated at lysine residues important to its function
UbaA is the sole member of the E1/MoeB/ThiF superfamily in many archaea including Hfx. volcanii, thus, autosampylation of UbaA and its homologs could have a significant impact on archaeal Ubl biology. In Hfx. volcanii, UbaA is modified at K87 by SAMP2, K113 by SAMP1/2, and K157 by SAMP1 (this study and [25, 30]). Likewise, in Sulfolobus acidocaldarius, the UbaA homolog ELSA is autoconjugated by the SAMP1 structural homolog Urm1 [44], although the site and function of this modification remains to be determined. Here we find UbaA K87 is critical for ATP binding (an early step in Ubl bond formation and sulfur mobilization). Thus, autosampylation of K87 is likely to downregulate all UbaA activities that require ATP hydrolysis and to occur through an intersubunit mechanism (within the UbaA homodimer that binds SAMP at a 1:2 molar ratio). Our finding that K157 is needed in sulfur mobilization but not Ubl bond formation, suggests the Ubl SAMP1 linkage at this site is a specialized form of autoregulation, having little to no impact on Ubl bond formation, but instead is likely to reduce the rate of sulfur mobilization. The positioning of K157 near the unstructured C188 loop in the UbaA model (Fig. 3A) is consistent with this lysine residue having an influential role in UbaA activity. Members of E1-E2-E3 systems are auto-regulated through Ub/Ubl modification in eukaryotic cells [45-51]; thus, the archaeal Ubl system is no exception.
Materials and Methods
Materials
Biochemicals were purchased from Sigma-Aldrich (St. Louis, MO). Other organic and inorganic analytical-grade chemicals were from Fisher Scientific (Atlanta, GA) and Bio-Rad (Hercules, CA). Desalted oligonucleotides were from Integrated DNA Technologies (Coralville, IN). Phusion and Taq DNA polymerases, restriction enzymes, T4 polynucleotide kinase and T4 DNA ligase were from New England Biolabs (Ipswich, MA).
Strains, media and growth conditions
Strains, plasmids and primers used in this study are summarized in Tables S2-S3. Hfx. volcanii strains were grown aerobically by rotary shaking (200 rpm) at 42 °C in ATCC 974 medium supplemented with novobiocin (Nv, 0.2 μg·ml−1). Escherichia coli strains were grown in LB medium with ampicillin (Ap, 100 μg·ml−1), chloramphenicol (Cm, 30 μg·ml−1) and/or kanamycin (Km, 50 μg·ml−1) as needed. Anaerobic growth by DMSO respiration was assessed as previously described [24]. Growth of cell cultures was monitored by optical density at 600 nm (OD600).
Protein purification
SAMPs (N-term Flag-His6 tag) and UbaA (N-term His6- or C-term StrepII tag) were expressed and purified from recombinant E. coli Rosetta (DE3) and Hfx. volcanii HM1052 or NH02. Cells were lysed by French Press (24000 psi; chilled on ice) in Tris-salt buffer (2 M NaCl, 50 mM Tris-HCl, pH 7.5) (supplemented with 40 mM imidazole for His6-proteins). Lysate was clarified by centrifugation and filtration (0.45 μm). Clarified lysate of cells expressing His6-proteins was applied to a HisTrap HP Ni2+-affinity chromatography (GE Healthcare) equilibrated and washed in Tris-salt buffer with 40 mM imidazole; proteins were eluted in Tris-salt buffer with 500 mM imidazole. For StrepII-proteins, clarified cell-free extract was applied to a Strep-Tactin column (Qiagen) equilibrated and washed in Tris-salt buffer, and proteins were eluted in Tris-salt buffer supplemented with d-desthiobiotin (5 mM). Samples were concentrated to 0.5-2.0 mg·ml−1 protein by Ultracel-10 centrifugal filtration (Millipore). Proteins (at 500 μl per run) were separated by size exclusion chromatography (SEC) using a Superdex 75 10/300 GL column (GE Healthcare) equilibrated in HEPES-salt buffer (2 M NaCl, 1 mM DTT, 50 mM HEPES, pH 7.5) at 0.3 ml per min. NaCl in the SEC buffer was reduced from 2 M to 150 mM for separation of the molecular mass standards: blue dextran (void volume), bovine serum albumin (66 kDa), horseradish peroxidase (44 kDa), bovine erythrocyte carbonic anhydrase (29 kDa), and equine heart cytochrome C (12.4 kDa) (Sigma Aldrich). Chromatography steps were aerobic at room temperature, and samples were stored on ice or at 4 °C. Protein concentration was determined by bicinchoninic acid (BCA) protein assay (Thermo Scientific, Rockville, IL) using bovine serum albumin (Thermo Scientific, Rockville, IL) as the protein standard. Proteins were determined to be homogenous by SDS-PAGE (Fig. 1) and electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS) analyses (Table S1). The homodimeric and monomeric configurations of the respective UbaA and SAMPs were based on SEC analysis (Fig. 1). UbaA-StrepII proteins were purified from Hfx. volcanii strains by StrepTactin chromatography as previously described [52].
Immunoblotting
Proteins were separated by SDS-PAGE and transferred onto Hybond PVDF membrane (0.45 μM, Amersham). Samples were normalized for loading by total protein and stained with Coomassie brilliant blue (CBB) to confirm equal sample loading. Affinity tagged proteins were detected on membranes by immunoblotting using alkaline phosphatase-linked anti-Flag M2 monoclonal antibody (Sigma) or a combination of anti-StrepII polyclonal antibody (Qiagen) and alkaline phosphatase-linked goat anti-rabbit IgG antibody (SouthernBiotech). Chemiluminescent signals were visualized on X-ray film (Research Products Intl. Corp.) by alkaline phosphatase activity using CDP-Star (Applied Biosystems) as the substrate.
3D homology structural modeling
UbaA was comparatively modeled by Phyre2 (Protein Homology/AnalogY Recognition Engine) [53] using the E. coli complexes MoeB-MoaD (1JW9) and ThiF-ThiS (1ZUD) from the Structural Classification of Proteins (SCOP) and the Protein Data Bank as fold templates (where, UbaA shares a 42/56 and 39/57 % amino acid sequence identity/similarity over a 90% query coverage of MoeB and ThiF, respectively). The disordered N- and C-termini (25 residues total) were modelled by ab initio simulation using the simplified protein-folding simulator Poing, available through Phyre2 [53]. Ligand-binding sites were determined using the 3DLigandSiteligand binding prediction server [54].
Differential scanning fluorimetry (DSF)
Purified UbaA-StrepII (20 μM) was mixed with 5 mM nucleotides (ATP, GTP, CTP, TTP, ADP, AMP, AMP-PNP) or PPi in DSF buffer (50 mM HEPES pH 7.5, 2 M NaCl, 5 mM MgCl2, 1 mM DTT and 50 μM ZnCl2) and 1× Sypro Orange (Invitrogen). Mixtures (40 μl) were incubated at room temperature for 10 min in a 96-well PCR microplate in the dark. The thermal stability of UbaA was determined from protein melting curves generated by increasing temperature from 22 to 95 °C at 1 °C•min−1 using a C1000 thermal cycler (BioRad CX96 Realtime System). Fluorescence was scanned for 5 s in temperature increments of 0.2 °C. Protein melting temperatures were calculated by melting curve fitting using CFX Manager 2.1 (Bio-Rad). The general approach was adapted from [21].
Isothermal titration calorimetry (ITC)
ITC experiments were carried out on a MicroCal iTC200 microcalorimeter (MicroCal Inc., Northampton, MA) using the iTC200 v 1.24.2 software for instrument control and data acquisition. Protein samples (SAMPs and UbaA) separately contained in 200-μl dialysis tube, were co-dialyzed against Tris-salt buffer with or without 1 mM nucleotides (ATP, AMP-PNP or AMP and PPi) supplemented with 1 mM EDTA at 4 °C to minimize buffer mismatch and prevent formation of the SAMP adenylates and PPi. SAMP (600 μM) in a 40-μl syringe was titrated in 20 subsequent injections (0.4-μl pre-injection followed by nineteen 2-μl injections) to 201.9 μl of 60 μM UbaA in the sample cell to reach the final molar ratio of SAMP to UbaA of about 2:1. Injection duration was set to 7.82 s with a 3-min interval between injections into the sample cell. Mixing was carried out at 25 °C with reference power and rotational stirring set at 5 μcal/s and 1000 rpm, respectively. The change in thermal power as a function of each injection was recorded and the raw data were fully processed using MicroCal ORIGIN 7.0 software. The binding isotherms of heat determined from the integral of the calorimetric signal were plotted as a function of molar ratio of SAMP to UbaA. To extract observed affinity (Kobs) and observed enthalpy (Hobs), the binding isotherms were iteratively fit via non-linear least squares regression analysis to the following equation:
where q(i) is the heat released or absorbed (kJ/mol) for the ith injection, N is the binding stoichiometry, Vo is the cell volume, U is the UbaA concentration (μM) and S is the concentration of SAMP (μM). The equation above is derived from law of mass action assuming a one-site binding model in a macromolecule. The observed free energy of binding (ΔGobs ) was calculated from the following equation:
where R is the universal molar gas constant (8.315 J/mol/K) and T is the absolute temperature (298K). Observed entropic contribution (ΔSobs) to binding was calculated from the relationship:
Calorimetric data were corrected from background signals generated by heat effects of injection, mixing, hydration, and dilution. Background controls include buffer to buffer, SAMP to buffer, and buffer to UbaA titrations.
Gel shift assay
UbaA and SAMP interactions were examined by gel shift assay as follows. Reaction mixtures (20 μl) containing 5 μM UbaA-StrepII, 5 μM Flag-His6-SAMP, 10 mM nucleotide (ATP, ADP, AMP-PNP, GTP, CTP, TTP or UTP) in SAMP-activation assay buffer (50 μM ZnCl2, 10 mM MgCl2, 0.5 mM DTT, 2 M NaCl, 20 mM HEPES, pH 7.5) were incubated at 50 °C for 30 min. Reactions were stopped by adding equal volume of 2× SDS-PAGE loading buffer with and without 71 mM β-mercaptoethanol. Samples were boiled for 5 min, separated by 12 % SDS-PAGE and visualized by anti-Flag immunoblotting. To determine the importance of cysteine thiol groups, UbaA was pre-treated with 5 mM N-ethylmaleimide (NEM) for 2 min prior to addition of SAMP and ATP to the assay. To chelate Zn2+ from UbaA prior to assay, a 100-μl mixture containing 10 μM UbaA was incubated with 100 μM N,N,N’,N’-tetrakis (2-pyridylmethyl) ethylenediamine (TPEN) in high-salt HEPES buffer (2 M NaCl, 20 mM HEPES, pH 7.5) at room temperature for 15 min. Formation of thiol-dependent UbaA~SAMP complexes was carried out in 20-μl reaction mixtures, containing 1 μM TPEN-pre-treated UbaA, 1 μM SAMP, 5 mM MgCl2, 50 mM of divalent cation (Zn2+, Co2+, Cu2+, Fe2+, Mn2+, or Ni2+) in high-salt HEPES buffer.
Detection of thiolated tRNA
Total RNA was isolated from log-phase cells of Hfx. volcanii as described previously [20]. Purified RNA was precipitated in 250 mM sodium acetate (pH 5.0) with two volumes of pre-chilled 95% (v/v) ethanol and washed thoroughly with 70% (v/v) ethanol. The RNA pellet was air-dried and resuspended in 30 μl DEPC-treated H2O. RNA integrity was assessed by agarose gel electrophoresis, and the concentration was subsequently determined spectrophotometrically (A260 nm). [(N-acryloylamino)phenyl] mercuric chloride polyacrylamide gel electrophoresis (APM–PAGE) was used to ascertain the thiolation of tRNA as previously described [24]. An oligonucleotide specific for tRNALysUUU (5′-CGGGCTGGGAGGGACTTGAACCCCC-3′) (5′ end-labeled with [γ-32P]ATP and T4 polynucleotide kinase) was used as the probe for analysis of the APM gels by Northern blotting.
In vitro cleavage of SAMP-UbaA conjugates by HvJAMM1 desampylase
N-terminal His6-tagged HvJAMM1 was purified as previously described [55]. Desampylation of SAMP-UbaA conjugates was carried out in 20-μl reaction mixture containing 5 mM HvJAMM1 and 5 μg SAMP-UbaA conjugates in 20 mM HEPES buffer, pH 7.5, with 50 mM ZnCl2 and 2 M NaCl. Reactions were carried out at 50°C for 2-3 h. Separate mixtures without the enzyme, with the heat-killed enzyme (pre-boiled for 15 min), and with addition of 50 mM EDTA served as negative controls. Desampylation products were visualized by anti-Flag and anti-StrepII immunoblotting.
Mapping ubiquitin-like bond formation on UbaA
Conjugation site of SAMP2 on UbaA was determined by electrospray ionization, collision-induced dissociation and tandem mass spectrometry (ESI-CID-MS/MS) analysis. UbaA-StrepII, purified from Hfx. volcanii HM1052-pJAM957, was digested with trypsin. Tryptic digests were injected onto a capillary trap column (LC Packings PepMap) and desalted for 5 min with a flow rate of 3 μl•min−1 of 0.1% v/v acetic acid. The samples were loaded onto an LC Packing C18 Pep Map nanoflow HPLC column. The elution gradient of the HPLC column started at 3% solvent A, 97% solvent B and finished at 60% solvent A, 40% solvent B for 30 min for protein identification. Solvent A consisted of 0.1% v/v acetic acid, 3% v/v acetonitrile (ACN), and 96.9% v/v H2O, while solvent B contained 0.1% v/v acetic acid, 96.9% v/v ACN, and 3% v/v H2O. LC-MS/MS analysis was carried out on a hybrid quadrupole-TOF mass spectrometer (QSTAR Elite, Applied Biosystems, Framingham, MA). The focusing potential and ion spray voltage were set to 225 V and 2400 V, respectively. The information-dependent acquisition (IDA) mode of operation was employed in which a survey scan from m/z 400–1800 was acquired followed by collision induced dissociation (CID) of the four most intense ions. Survey and MS/MS spectra for each IDA cycle were accumulated for 1 and 3 s, respectively. For protein search algorithm, the tandem mass spectra were extracted by ABI Analyst version 2.0. All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.2.07). Mascot was set up to search Haloferax_032511 database assuming trypsin digestion, with fragment ion and parent ion mass tolerances both set at 0.50 Da. Variable modifications including iodoacetamide derivative of Cys, deamidation of Asn and Gln, and oxidation of Met were specified in the software. Scaffold (Proteome Software Inc., Portland, OR; version 4.3.4) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if probability was >95.0% cutoff specified by the Peptide Prophet algorithm [56] or >99.0% probability of at least two identified unique peptides, as assigned by the Protein Prophet algorithm [57].
Supplementary Material
Acknowledgements
The authors would like to thank M. C. Dancel at the Mass Spectrometry Facility of UF Department of Chemistry for ESI-TOF-MS, R. Zheng at the UF ICBR Proteomics and Mass Spectrometry Core for LC-MS/MS, and S. Shanker at the UF ICBR Sanger Sequencing Core for DNA sequencing.
Funding information. This study was funded in part by the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences and Biosciences, Physical Biosciences Program (DE-FG02-05ER15650 to J.M-F.), the National Institute of General Medical Sciences (NIH R01 GM57498-15 to J.M-F. and GM22854-42 to D. Söll), the USDA National Institute of Food and Agriculture (Hatch 1005900 to J. M-F.), and the Ford Foundation International Fellowships Program (to N.H). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Abbreviations
- SAMP
small ubiquitin-like archaeal modifier protein
- Ub
ubiquitin
- Ubl
ubiquitin-like
- E1
ubiquitin-activating enzyme
- UbaA
E1-like SAMP-activating enzyme of Archaea
- MoaD
molybdopterin converting factor subunit 1
- MoeB
molybdopterin synthase adenyltransferase
- MPT
molybdopterin
- DMSO
dimethylsulfoxide
- DSF
differential scanning fluorimetry
- ITC
isothermal titration calorimetry
- APM
acryloylaminophenylmercuric chloride
- TPEN
N,N,N’,N’-tetrakis (2-pyridylmethyl) ethylenediamine
- NEM
N-ethylmaleimide
- diGly
C-terminal diglycine residues of SAMPs
- ΔGG
C-terminal diGly residues deleted
- SEC
size exclusion chromatography
- wt
wild type
- CBB
Coomassie brilliant blue
Footnotes
The authors do not have a conflict of interest to declare.
Author Contributions. N.H., I.V, S.C., X.F., Y.W., S.U., N.C., M.E., and D.Su performed the research. N.H., I.dV, D.Söll, D.K. and J. M-F. designed the research, analyzed data, wrote and revised the paper. All authors approve of the paper.
Table S1. Theoretical and observed masses of SAMP and UbaA proteins
Table S2. List of strains and plasmids used in this study.
Table S3. List of primers used in this study.
Figure S1. Raw isothermal titration calorimetry (ITC) data and corresponding enthalpy plots of UbaA:SAMP1/2 binding at 25 °C.
References
- 1.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–9. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Streich FC, Lima CD. Structural and functional insights to ubiquitin-like protein conjugation. Annu Rev Biophys. 2014;43:357–79. doi: 10.1146/annurev-biophys-051013-022958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kravtsova-Ivantsiv Y, Sommer T, Ciechanover A. The lysine48-based polyubiquitin chain proteasomal signal: not a single child anymore. Angew Chem Int Ed Engl. 2013;52:192–8. doi: 10.1002/anie.201205656. [DOI] [PubMed] [Google Scholar]
- 4.Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–6. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Maupin-Furlow JA. Ubiquitin-like proteins and their roles in archaea. Trends Microbiol. 2013;21:31–8. doi: 10.1016/j.tim.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki E, Zhang X, Sun HG, Lu MY, Liu TL, Ou A, Li JY, Chen YH, Ealick SE, Liu HW. Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis. Nature. 2014;510:427–31. doi: 10.1038/nature13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leimkühler S, Wuebbens MM, Rajagopalan KV. Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J Biol Chem. 2001;276:34695–701. doi: 10.1074/jbc.M102787200. [DOI] [PubMed] [Google Scholar]
- 8.Park JH, Dorrestein PC, Zhai H, Kinsland C, McLafferty FW, Begley TP. Biosynthesis of the thiazole moiety of thiamin pyrophosphate (vitamin B1) Biochemistry. 2003;42:12430–8. doi: 10.1021/bi034902z. [DOI] [PubMed] [Google Scholar]
- 9.Dahl JU, Urban A, Bolte A, Sriyabhaya P, Donahue JL, Nimtz M, Larson TJ, Leimkühler S. The identification of a novel protein involved in molybdenum cofactor biosynthesis in Escherichia coli. J Biol Chem. 2011;286:35801–12. doi: 10.1074/jbc.M111.282368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutzke G, Fischer B, Mendel RR, Schwarz G. Thiocarboxylation of molybdopterin synthase provides evidence for the mechanism of dithiolene formation in metal-binding pterins. J Biol Chem. 2001;276:36268–74. doi: 10.1074/jbc.M105321200. [DOI] [PubMed] [Google Scholar]
- 11.Taylor SV, Kelleher NL, Kinsland C, Chiu HJ, Costello CA, Backstrom AD, McLafferty FW, Begley TP. Thiamin biosynthesis in Escherichia coli. Identification of ThiS thiocarboxylate as the immediate sulfur donor in the thiazole formation. J Biol Chem. 1998;273:16555–60. doi: 10.1074/jbc.273.26.16555. [DOI] [PubMed] [Google Scholar]
- 12.Marelja Z, Mullick Chowdhury M, Dosche C, Hille C, Baumann O, Löhmannsröben HG, Leimkühler S. The L-cysteine desulfurase NFS1 is localized in the cytosol where it provides the sulfur for molybdenum cofactor biosynthesis in humans. PLoS One. 2013;8:e60869. doi: 10.1371/journal.pone.0060869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Gomez NC, Palmer LD, Vivas E, Roach PL, Downs DM. The rhodanese domain of ThiI is both necessary and sufficient for synthesis of the thiazole moiety of thiamine in Salmonella enterica. J Bacteriol. 2011;193:4582–7. doi: 10.1128/JB.05325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudolph MJ, Wuebbens MM, Rajagopalan KV, Schindelin H. Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation. Nat Struct Biol. 2001;8:42–6. doi: 10.1038/83034. [DOI] [PubMed] [Google Scholar]
- 15.Maupin-Furlow JA. Prokaryotic ubiquitin-like protein modifiers. Annu Rev Microbiol. 2014;68:155–175. doi: 10.1146/annurev-micro-091313-103447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, Liu M, Qiu R, Ji C. The dual role of ubiquitin-like protein Urm1 as a protein modifier and sulfur carrier. Protein Cell. 2011;2:612–9. doi: 10.1007/s13238-011-1074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shigi N. Posttranslational modification of cellular proteins by a ubiquitin-like protein in bacteria. J Biol Chem. 2012;287:17568–77. doi: 10.1074/jbc.M112.359844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shigi N, Sakaguchi Y, Asai S, Suzuki T, Watanabe K. Common thiolation mechanism in the biosynthesis of tRNA thiouridine and sulphur-containing cofactors. EMBO J. 2008;27:3267–78. doi: 10.1038/emboj.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao S, Hepowit N, Maupin-Furlow JA. Ubiquitin-like protein SAMP1 and JAMM/MPN+ metalloprotease HvJAMM1 constitute a system for reversible regulation of metabolic enzyme activity in Archaea. PLoS One. 2015;10:e0128399. doi: 10.1371/journal.pone.0128399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chavarria NE, Hwang S, Cao S, Fu X, Holman M, Elbanna D, Rodriguez S, Arrington D, Englert M, Uthandi S, Söll D, Maupin-Furlow JA. Archaeal Tuc1/Ncs6 homolog required for wobble uridine tRNA thiolation is associated with ubiquitin-proteasome, translation, and RNA processing system homologs. PLoS One. 2014;9:e99104. doi: 10.1371/journal.pone.0099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 22.McMillan LJ, Hepowit NL, Maupin-Furlow JA. Archaeal inorganic pyrophosphatase displays robust activity under high-salt conditions and in organic solvents. Appl Environ Microbiol. 2015;82:538–48. doi: 10.1128/AEM.03055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miranda HV, Antelmann H, Hepowit N, Chavarria NE, Krause DJ, Pritz JR, Bäsell K, Becher D, Humbard MA, Brocchieri L, Maupin-Furlow JA. Archaeal ubiquitin-like SAMP3 is isopeptide-linked to proteins via a UbaA-dependent mechanism. Mol Cell Proteomics. 2014;13:220–39. doi: 10.1074/mcp.M113.029652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miranda H, Nembhard N, Su D, Hepowit N, Krause D, Pritz J, Phillips C, Söll D, Maupin-Furlow J. E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc Natl Acad Sci U S A. 2011;108:4417–22. doi: 10.1073/pnas.1018151108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humbard M, Miranda H, Lim J, Krause D, Pritz J, Zhou G, Chen S, Wells L, Maupin-Furlow J. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson H, Aldrich H, Maupin-Furlow J. Halophilic 20S proteasomes of the archaeon Haloferax volcanii: Purification, characterization, and gene sequence analysis. J Bacteriol. 1999;181:5814–5824. doi: 10.1128/jb.181.18.5814-5824.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Maciejewski MW, Martin J, Jin K, Zhang Y, Maupin-Furlow JA, Hao B. Crystal structure of the ubiquitin-like small archaeal modifier protein 2 from Haloferax volcanii. Protein Sci. 2013;22:1206–17. doi: 10.1002/pro.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414:325–9. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 29.Lehmann C, Begley TP, Ealick SE. Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry. 2006;45:11–9. doi: 10.1021/bi051502y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dantuluri S, Wu Y, Hepowit NL, Chen H, Chen S, Maupin-Furlow JA. Proteome targets of ubiquitin-like samp1ylation are associated with sulfur metabolism and oxidative stress in Haloferax volcanii. Proteomics. 2016;16:1100–10. doi: 10.1002/pmic.201500153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeong YJ, Jeong BC, Song HK. Crystal structure of ubiquitin-like small archaeal modifier protein 1 (SAMP1) from Haloferax volcanii. Biochem Biophys Res Commun. 2011;405:112–7. doi: 10.1016/j.bbrc.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz J, Wuebbens MM, Rajagopalan KV, Leimkühler S. Role of the C-terminal Gly-Gly motif of Escherichia coli MoaD, a molybdenum cofactor biosynthesis protein with a ubiquitin fold. Biochemistry. 2007;46:909–16. doi: 10.1021/bi062011w. [DOI] [PubMed] [Google Scholar]
- 33.Liao S, Zhang W, Fan K, Ye K, Zhang X, Zhang J, Xu C, Tu X. Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP2) from Haloferax volcanii. Sci Rep. 2013;3:2136. doi: 10.1038/srep02136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang DT, Walden H, Duda D, Schulman BA. Ubiquitin-like protein activation. Oncogene. 2004;23:1958–71. doi: 10.1038/sj.onc.1207393. [DOI] [PubMed] [Google Scholar]
- 35.Fu X, Liu R, Sanchez I, Silva-Sanchez C, Hepowit NL, Cao S, Chen S, Maupin-Furlow J. Ubiquitin-like proteasome system represents a eukaryotic-like pathway for targeted proteolysis in archaea. mBio. 2016;7:e00379–16. doi: 10.1128/mBio.00379-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haas AL, Rose IA. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J Biol Chem. 1982;257:10329–37. [PubMed] [Google Scholar]
- 37.Haas AL, Warms JV, Hershko A, Rose IA. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J Biol Chem. 1982;257:2543–8. [PubMed] [Google Scholar]
- 38.Haas AL, Warms JV, Rose IA. Ubiquitin adenylate: structure and role in ubiquitin activation. Biochemistry. 1983;22:4388–94. doi: 10.1021/bi00288a007. [DOI] [PubMed] [Google Scholar]
- 39.Pickart CM, Kasperek EM, Beal R, Kim A. Substrate properties of site-specific mutant ubiquitin protein (G76A) reveal unexpected mechanistic features of ubiquitin-activating enzyme (E1) J Biol Chem. 1994;269:7115–23. [PubMed] [Google Scholar]
- 40.Makarova KS, Koonin EV. Archaeal ubiquitin-like proteins: functional versatility and putative ancestral involvement in tRNA modification revealed by comparative genomic analysis. Archaea. 2010;2010 doi: 10.1155/2010/710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ranjan N, Damberger FF, Sutter M, Allain FH, Weber-Ban E. Solution structure and activation mechanism of ubiquitin-like small archaeal modifier proteins. J Mol Biol. 2011;405:1040–55. doi: 10.1016/j.jmb.2010.11.040. [DOI] [PubMed] [Google Scholar]
- 42.Kumar A, Ito A, Hirohama M, Yoshida M, Zhang KY. Identification of sumoylation activating enzyme 1 inhibitors by structure-based virtual screening. J Chem Inf Model. 2013;53:809–20. doi: 10.1021/ci300618e. [DOI] [PubMed] [Google Scholar]
- 43.Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–12. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anjum RS, Bray SM, Blackwood JK, Kilkenny ML, Coelho MA, Foster BM, Li S, Howard JA, Pellegrini L, Albers SV, Deery MJ, Robinson NP. Involvement of a eukaryotic-like ubiquitin-related modifier in the proteasome pathway of the archaeon Sulfolobus acidocaldarius. Nat Commun. 2015;6:8163. doi: 10.1038/ncomms9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amemiya Y, Azmi P, Seth A. Autoubiquitination of BCA2 RING E3 ligase regulates its own stability and affects cell migration. Mol Cancer Res. 2008;6:1385–96. doi: 10.1158/1541-7786.MCR-08-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen A, Kleiman FE, Manley JL, Ouchi T, Pan ZQ. Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase. J Biol Chem. 2002;277:22085–92. doi: 10.1074/jbc.M201252200. [DOI] [PubMed] [Google Scholar]
- 47.Truong K, Lee TD, Chen Y. Small ubiquitin-like modifier (SUMO) modification of E1 Cys domain inhibits E1 Cys domain enzymatic activity. J Biol Chem. 2012;287:15154–63. doi: 10.1074/jbc.M112.353789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32:767–77. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 49.Cooper HJ, Tatham MH, Jaffray E, Heath JK, Lam TT, Marshall AG, Hay RT. Fourier transform ion cyclotron resonance mass spectrometry for the analysis of small ubiquitin-like modifier (SUMO) modification: identification of lysines in RanBP2 and SUMO targeted for modification during the E3 autoSUMOylation reaction. Anal Chem. 2005;77:6310–9. doi: 10.1021/ac058019d. [DOI] [PubMed] [Google Scholar]
- 50.Lai Z, Yang T, Kim YB, Sielecki TM, Diamond MA, Strack P, Rolfe M, Caligiuri M, Benfield PA, Auger KR, Copeland RA. Differentiation of Hdm2-mediated p53 ubiquitination and Hdm2 autoubiquitination activity by small molecular weight inhibitors. Proc Natl Acad Sci U S A. 2002;99:14734–9. doi: 10.1073/pnas.212428599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho CW, Chen HT, Hwang J. UBC9 autosumoylation negatively regulates sumoylation of septins in Saccharomyces cerevisiae. J Biol Chem. 2011;286:21826–34. doi: 10.1074/jbc.M111.234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Humbard MA, Zhou G, Maupin-Furlow JA. The N-terminal penultimate residue of 20S proteasome α1 influences its Nα acetylation and protein levels as well as growth rate and stress responses of Haloferax volcanii. J Bacteriol. 2009;191:3794–803. doi: 10.1128/JB.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wass MN, Kelley LA, Sternberg MJ. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010;38:W469–73. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hepowit NL, Uthandi S, Miranda HV, Toniutti M, Prunetti L, Olivarez O, De Vera IM, Fanucci GE, Chen S, Maupin-Furlow JA. Archaeal JAB1/MPN/MOV34 metalloenzyme (HvJAMM1) cleaves ubiquitin-like small archaeal modifier proteins (SAMPs) from protein-conjugates. Mol Microbiol. 2012;86:971–987. doi: 10.1111/mmi.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 57.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 58.Furukawa K, Mizushima N, Noda T, Ohsumi Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J Biol Chem. 2000;275:7462–5. doi: 10.1074/jbc.275.11.7462. [DOI] [PubMed] [Google Scholar]
- 59.Schmitz J, Chowdhury MM, Hänzelmann P, Nimtz M, Lee EY, Schindelin H, Leimkühler S. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry. 2008;47:6479–89. doi: 10.1021/bi800477u. [DOI] [PubMed] [Google Scholar]
- 60.Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–32. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 61.Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37:1335–52. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matthies A, Rajagopalan KV, Mendel RR, Leimkühler S. Evidence for the physiological role of a rhodanese-like protein for the biosynthesis of the molybdenum cofactor in humans. Proc Natl Acad Sci U S A. 2004;101:5946–51. doi: 10.1073/pnas.0308191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schäfer A, Kuhn M, Schindelin H. Structure of the ubiquitin-activating enzyme loaded with two ubiquitin molecules. Acta Crystallogr D Biol Crystallogr. 2014;70:1311–20. doi: 10.1107/S1399004714002910. [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Chen Y. Role of the Zn2+ motif of E1 in SUMO adenylation. J Biol Chem. 2010;285:23732–8. doi: 10.1074/jbc.M110.114660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matthies A, Nimtz M, Leimkühler S. Molybdenum cofactor biosynthesis in humans: identification of a persulfide group in the rhodanese-like domain of MOCS3 by mass spectrometry. Biochemistry. 2005;44:7912–20. doi: 10.1021/bi0503448. [DOI] [PubMed] [Google Scholar]
- 66.Krepinsky K, Leimkühler S. Site-directed mutagenesis of the active site loop of the rhodanese-like domain of the human molybdopterin synthase sulfurase MOCS3. Major differences in substrate specificity between eukaryotic and bacterial homologs. FEBS J. 2007;274:2778–87. doi: 10.1111/j.1742-4658.2007.05811.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.