Abstract
The three-dimensional organization of the genome plays important roles in regulating the functional output of the genome, and even the maintenance of epigenetic inheritance and genome stability. Here, we review and compare a number of newly developed methods that enables the direct visualization of specific, endogenous DNA sequences in living cells, especially that utilizing the CRISPR-Cas9 system. We further discuss the practical considerations in implementing the CRIPSR imaging technique in order to achieve sufficient signal-to-background level, high specificity and high labeling efficiency. With these DNA labeling methods enabling the tracking of the copy number, localization and movement of genomic elements, we discuss their potential applications in understanding the searching and targeting mechanism of Cas9-sgRNA complex, investigating chromosome organization, as well as visualizing genome instability and rearrangement.
Keywords: microscopy, CRISPR, Cas9, fluorescence, chromatin, nucleus
Introduction
The maintenance of the genome and the regulation of its functional output are one of the most fundamental processes in living organisms. It is now more and more clear that the regulation of genome function goes far beyond the linear sequence. The organization of the genome in the physical space has particularly emerged as an important but still elusive mechanism. In order to understand how the genome is packaged in the nucleus, chromosome conformation capture (3C) and its derived methods such as Hi-C have now provided new insights into the spatial organization principles for the genome such as the existence of topologically associated domains (TADs). On the other hand, microscopy observation has long been the classic approach to directly provide the morphological information of cellular structures as well as localization information of molecules. For example, fluorescence in situ hybridization (FISH) allows sequence-specific visualization of DNA and RNA in fixed cells. Chromosome level FISH have revealed chromosome territories (14), and FISH has now been extended to high-throughput imaging (128) and super-resolution imaging (6).
Complementary to structural mapping in fixed systems, live observation reveals the dynamic behavior. Live cell labeling of all chromatins can be achieved with cell-permeable DNA-intercalating dyes such as Hoechst 33342 and DRAQ5 (8, 93, 160), introduction of modified nucleotides (3) or with histone fused to fluorescent proteins (FPs) (e.g. H2B-GFP) (7, 31, 65, 68, 99). Labeling of specific genomic loci, however, was constrained by the availability of sequence-specific DNA-binding proteins. Certain genomic elements such as telomeres and centromeres can be labeled by fluorescently tagging their corresponding binding proteins (46, 95, 120, 121). For other loci, Lac or Tet operator repeats were inserted into random positions or a specific genomic region by genome engineering. These artificial sites can then be traced with Lac or Tet repressor proteins, respectively, fused to FPs (46, 95, 120, 121). This system, however, relies on artificially introduced sequences and genome engineering tools. The ParB/INT system can similarly label a specific genomic region by inserting ~ 1 kb INT element, along which ParB protein can spread by oligomerization to amplify the signal. The small insertion does not interfere chromatin dynamics and transcription, providing a tool to investigate chromatin positioning and dynamics (122).
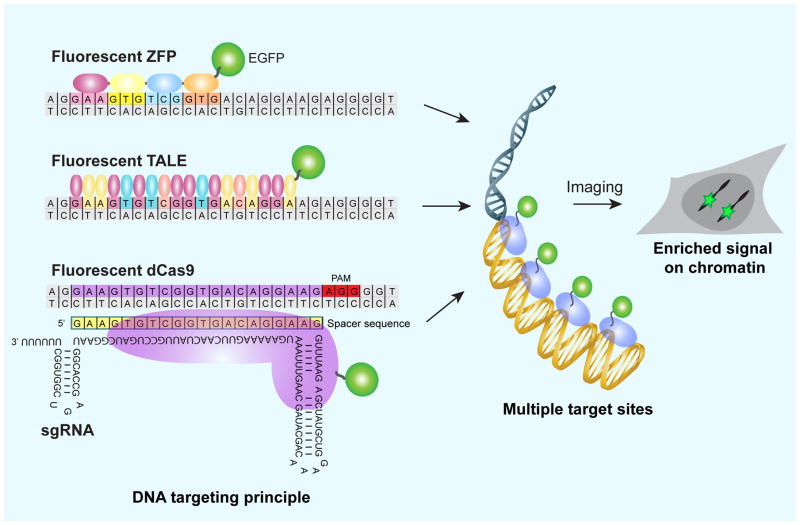
Efforts to label and image endogenous genomic element have benefited from the series of modular proteins with specific DNA recognition which has been developed in the past years for gene editing, starting with Zinc finger modules, then transcription activator-like effector (TALE), and more recently, the CRISPR (clustered regularly interspaced short palindromic repeat) - Cas (CRISPR-associated) system. These system provide a module that can recognize a specific DNA sequence. When fused with fluorescent proteins, they can guide the fluorescence signal to a specific sequence within the complex genome (Figure 1). This reviewer will discuss the imaging of specific genomic DNA sequences in living cells using these methods, especially about CRISPR-Cas9-mediated genomic imaging, termed “CRISPR imaging”. We have summarized all live DNA imaging methods in Table 1.
Fig. 1.
Scheme of live cell DNA labeling using ZFP, TALE and the CRISPR-Cas9 system.
Table 1.
Comparison of different live DNA detection methods
| Method | Endogenous | Specific sequence | Other advantages | Other disadvantages | References |
|---|---|---|---|---|---|
| DNA dye | Yes | No | Easy to perform | Bulk labeling | (93) |
| Fluorescent nucleotide | Yes | No | Bulk labeling;Requires single cell injection or ‘rubbing’ | (3) | |
| Histones | Yes | No | Stable fluorescence;Any cell type;Easy to perform; | Bulk labeling | (7, 31, 65, 68, 99) |
| Repressor operator array | No | Yes | Stable fluorescence | Artificial locus, insertion of 5–10 kb of DNA arrays;Difficult in metazoans;Potential chromatin disruption | (46, 95, 120, 121) |
| ParB-INT system | No | Yes | Strong signal through ParB oligomerization;Insertion dose not interfere chromatin dynamics and transcription | Artificial locus, insertion of ~1kb unique sequence | (122) |
| ZFP | Yes | Yes | Stable fluorescence;Potentially any cell type | Restricted to repetitive sequences; Assembly and screening of ZFPs is challenging; Costly, laborious and time-consuming | (19, 81) |
| TALE | Yes | Yes | Stable fluorescence;Potentially any cell type;Potentially any sequence | Restricted to repetitive sequences; Costly, laborious and time-consuming | (84, 98, 140, 153) |
| CRISPR-Cas9 | Yes | Yes | Stable fluorescence;Potentially any cell type;Easy to perform;Affordable and scalable | Restricted to sequences locating upstream of a specific PAM;Local DNA unwinding/DNA-RNA triple helix formation | (1, 24, 72, 83) |
Methods for sequence-specific imaging of endogenous DNA in living cells
Fluorescent zinc-finger proteins
The Cys2-His2 zinc finger, originally discovered in Xenopus (96), is the most common type of DNA-binding motifs found in eukaryotes (143). An individual zinc finger consist of 30 amino acids in a conserved ββα configuration (5). Each zinc finger can be engineered to recognize a nucleotide triplet (71, 110) and several fingers can be arranged in tandem as a zinc-finger protein (ZFP) to recognize a broad spectrum of DNA sequences with high specificity (29, 107). Engineered ZFP-based DNA binding domains with novel specificities have been extensively applied in vivo to target various effector domains, such as the cleavage domain of FokI, to produce a zinc-finger nuclease (ZFN) for genome editing (10, 11, 115, 145). ZFP fused with fluorescent proteins have been used for tracing specific DNA sequences in living cells (19, 81). GFP-tagged ZFN was first constructed to label specific DNA sequence in Arabidopsis and mouse (81). A ZFP-GFP protein was designed to specifically recognize a 9-bp sequence within centromeric 180-bp repeat and monitor centromeres in living roots. Similarly, the major satellite repeat was also visualized by a designed ZFP-GFP in mouse, demonstrating that this approach can be potentially applied in different organisms. Despite the numerous successful uses of engineered zinc finger proteins for editing, regulating and imaging genome, the full potential of this technology has not yet been fulfilled. It has been shown that ZFP exhibit context-dependent binding preference due to crosstalk between adjacent modules when assemble into a larger array (86). On the other hand, although several strategies have been developed to overcome these limitations, assembly of functional ZFPs for targeting specific sequences still remains a major challenge that requires an extensive screening process.
Fluorescent TALEs
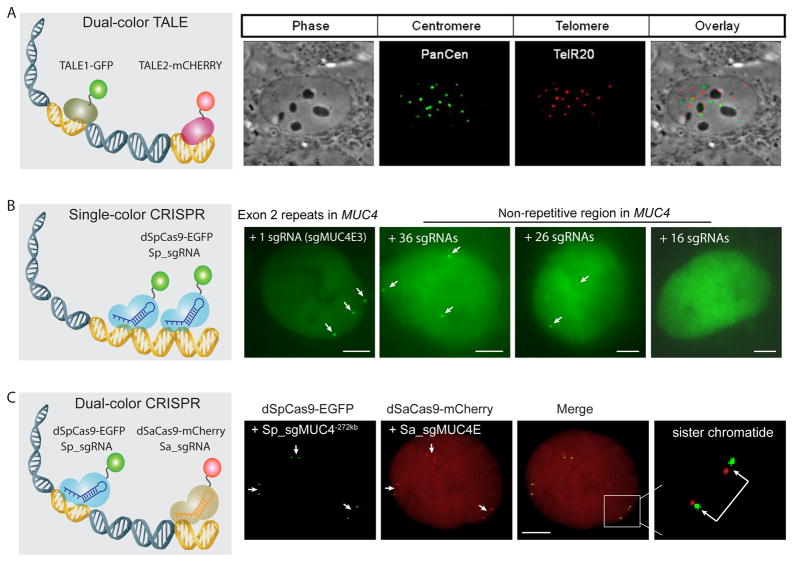
TALEs, originally discovered from the plant pathogenic bacteria Xanthomonas (13, 101), contain a DNA binding domain that can be engineered to bind specific DNA sequences (18, 87, 97, 100, 157). TALEs recognize the target DNA through tandemly arranged 33–35 amino acid repeats, with each repeat binding one base. The base preference of individual repeats is defined by amino acids 12 and 13, referred to as repeat-variable diresidues (RVDs). This simple code allows the efficient generation of DNA-binding modules for targeting any specific sequences. Synthetic TALEs, fusion of TALEs with specific functionality such as nucleases and transcriptional modulators, have been successfully applied in genome editing and transcriptional regulation (85, 113, 137, 157). TALEs fused with fluorescent proteins have been developed to visualize endogenous repetitive sequences in culture cells and living organisms (84, 98, 140, 153). This approach was first developed to visualize centromeres and telomeres in live mouse embryonic stem (ES) cells. 15 nt or longer sequences were selected for targets of TALEs. By fusing TALEs with different fluorescent proteins, centromeres and telomeres were detected in the same cell, demonstrating the capability of TALEs for imaging multiple loci in one cell (Figure 2A) (84). The high specificity of TALEs allows distinguishing 1-nt or 2-nt differences on the target sequence. Fluorescent TALEs were thus able to distinguish chromosomes in a parent-of-origin manner by recognition of single-nucleotide polymorphisms (SNPs) (98). Long-term live cell imaging demonstrated that fluorescent TALEs enables DNA sequence tracing during the cell cycle. Depending on the target sequences, fluorescent TALEs can associate with DNA through the whole cell cycle (98) or disassociate in mid to late prophase but reassociate in early telophase (140). These observations are consistent with the hypothesis that chromatin environment can influence TALE binding and activity (18, 35). Yuan and Colleagues have utilized this approach to follow the replication behavior of particular satellite sequences during a major embryonic transition in Drosophila and reported that developmental signals can separately influence the timing of different satellite sequences (153). This novel application of TALEs open new perspectives to identify and elucidate cell cycle and development-specific changes in genome organization and chromatin dynamics.
Fig. 2.
Imaging of genomic loci using the TALE and the CRISPR-Cas9 methods. (A) Two color TALE imaging of telomere and centromere repeats (84). (B) CRISPR imaging of tandem repeats and non-repetitive sequences in the MUC4 gene in a trisomy human cell line (24). (C) Two color CRISPR imaging of two tandem repeats separated by 272 kb at G2 phase in the same cell line using SpCas9 and SaCas9. Scale bars for (B) and (C): 5 μm.
Fluorescent TALEs, though conceptually similar to fluorescent zinc-finger proteins (19, 81), offers several advantages including ease of design, higher affinity, its potential ability to be applied to any sequences and simpler optimization. However, it is still challenging to extend this approach to label non-repetitive sequences. To gain sufficient signal for detection, it will be necessary to construct multiple TALEs over the target sequence. Because of their repetitive nature, it is difficult to construct TALEs with definite specificity. To overcome this technical difficulty, several cloning strategies have been developed. These methods include “Golden Gate” molecular cloning (21), high-throughput solid-phase assembly (16, 119), and ligation-independent cloning techniques (126). In addition, delivery of multiple TALEs into one cell is another factor that has to be considered. The number of targeted TALEs that is necessary to achieve non-repetitive sequence labeling needs to be further optimized.
Fluorescent dCas9 proteins
The CRISPR-Cas system provides prokaryotes with adaptive immunity to invading viruses and plasmids (4, 56, 139, 150). In the type II CRISPR-Cas system, short foreign DNA fragments, named “spacers” are integrated into CRISPR loci and then transcribed and processed into short CRISPR RNAs (crRNAs), which in turn pair with a trans-activating crRNA (tracrRNA) and direct sequence-specific silencing of foreign nucleic acids by Cas proteins (17, 45). Biochemical characterizations showed that purified Cas9 from Streptococcus pyogenes can be guided by crRNA-tracrRNA to cleave target DNA in vitro (61). A single guide RNA (sgRNA) generated by fusing crRNA and tracrRNA sequences can replace the two RNAs to mediate the DNA cleavage. This finding created a simple two-component system, sgRNA and Cas9 protein, in which changes in the guide sequence of the sgRNA can direct Cas9 to target any DNA sequence of interest. Target recognition, binding and subsequent cleavage by the Cas9-sgRNA complex requires both Watson-Crick-base paring between the spacer and the target ‘protospacer’ sequence as well as the presence of an appropriate protospacer-adjacent motif (PAM) sequence at the 3′ end of the target sequence (59, 60, 133). Many type II systems have differing PAM requirements, which can limit their ease of targeting (11). The most commonly engineered system thus far, which comes from Streptococcus pyogenes, requires a PAM with sequence NGG, where N is any nucleotide (61). The CRISPR-Cas9 system have been engineered for robust RNA-guided genome editing in mammalian cells (30, 62, 91) as well as in many model organisms (91, 125).
In addition to genome editing, the ability of Cas9 to bind to DNA at sites defined by the guide RNA sequence and the PAM has allowed many more applications. By fusing regulatory domains to a catalytically deactivated Cas9 (dCas9), CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) has been developed to regulate endogenous gene expression (50, 85, 90, 112, 116). The programmable binding capability of dCas9 was also adapted for imaging specific endogenous DNA sequence in living cells. A fluorescent protein tagged dCas9 protein and a structurally optimized sgRNA were shown to image both repetitive and non-repetitive elements in living human cells (Figure 2B) (24). For tandem repetitive sequences, a single sgRNA may be sufficient to enrich GFP signal for detection, whereas labeling non-repetitive sequences may need the simultaneous expression of at least 30 sgRNAs. This labeling approach has been successfully applied in living mouse embryonic stem cells (153) as well as a variety of cultured human cells (25, 72, 83).
Multicolor imaging using TALE or the CRISPR-Cas9 system
Elucidating how the genome is spatiotemporally organized inside the nucleus is imperative to understanding how genes are regulated during normal development and dysregulated in various disease states (12, 76). Mapping the functional organization of the genome can be greatly assisted with methods to directly visualize the interactions between different genomic elements (e.g. enhancers and promoters) in living cells. To simultaneously image and track multiple genomic loci, multicolor imaging approach would be indispensable.
Because TALE recognizes DNA in a protein-only manner, it is straightforward to implement multicolor imaging with TALE simply by tagging different TALE proteins with different fluorophores (84). In contrast, the target specificity of CRISRP-Cas9 system is mainly determined by the sgRNA, thus enabling the following two alternative approaches to achieve multicolor CRISPR imaging:
The first strategy is to use fluorescent Cas9 orthologs from different bacteria species. Except the Cas9 from Streptococcus pyogenes (SpCas9), two more orthogonal Cas9 proteins from Neisseria meningitidisn (NmCas9) and Streptococcus thermophilus (St1Cas9) have been repurposed for multicolor CRISPR imaging. Using pairs of differently colored dCas9-sgRNAs, it becomes possible to determine the intranuclear distance between loci on different chromosomes or the spatial resolution between two loci on the same chromosome (83). However, both NmCas9 and St1Cas9 require complex PAM sequences, which restricts the range of accessible targets (30, 41, 57). Moreover, their targeting activity need more characterizations. Recently, an additional Cas9 ortholog, SaCas9, has been characterized and effectively used for in vivo genome editing using single guide RNAs (117). This new Cas9 enzyme is small and can be used for broadly targeting with a PAM of “NNGRRT”. Thus, SaCas9 has the potential to be combined with SpCas9 for simultaneously tracking multiple genomic loci in one cell with high efficiency and robustness (Figure 2C). Recent efforts in altering the PAM specificity of SpCas9 (69, 70) and the discovery of the Cas9-like activities of the Cpf1 protein (155) may further expand the palette of multicolor CRIPSR imaging.
The second strategy is to tether fluorescent RNA-binding protein to the sgRNA though aptamer fusions which converts the sgRNA into a scaffold RNA (scRNA) that encodes both information about the target locus and the fluorescent color. This approach has been successfully used for genomic regulatory programming by recruiting regulatory domains to target loci through scRNAs (74, 90, 154).
Comparisons of TALE and CRISPR imaging
Although both TALE and the CRISPR-Cas9 system can be programmed to image repetitive genomic elements effectively, their different DNA binding mechanisms and characteristics may affect the choice between these two approaches.
Flexibility in target site selection
CRISPR-Cas9 targets must immediately precede a PAM sequence (such as “NGG” for SpCas9) and are suggested to start with “G” (61), while the only limitation of TALEs recognition seems to be the requirement for a “T” at the 5′ end of the target sequence (89). Although it is usually not difficult to locate GG sites for targeting, this constraint do limit the choices of target sites. In contrast, a TALE-FP can in principle target almost any given site in a genome.
The ease of design and construction
Typically, TALEs are designed to recognize 15 to 20 DNA base-pairs at a target site, corresponding to a custom-designed TALE protein of ~ 500–700 amino acids in size. In contrast, the specificity of RNA-guided Cas9 system can be customized by replacing a short synthetic RNA molecule without changing the protein component, making it easier and more cost-effective to design and producing the labeling constructs. Moreover, to label a non-repetitive genomic region, multiple sites must be labeled simultaneously to enrich the signal over the background for detection. For this application as well as co-labeling of many loci, the CRISPR-Cas9 system is thus the easier choice.
Targeting efficiency and specificity
For genome editing, TALE nuclease (TALEN) and CRISPR-Cas9 both exhibit high variabilities in efficiency: 1% to ~ 60% mutation rate for TALEN in cultured mammalian cells (67, 119), and 2.3–79% editing efficiency for CRISPR (28, 30, 36, 62, 90). So far, there are no reliable rules to predict their targeting efficiency before experimental validation (66). Therefore, it is almost required try multiple target sequences for a given loci to ensure successful labeling, for which CRISPR has the advantage over TALE.
Although a major concern for genome editing, the off-target effect is not as critical an issue for imaging, because it require simultaneous, stable binding to many site at the target loci to obtain a detectable signal. However, with future developments that can reduce the number of binding sites, off-targeting may have to be taken into consideration. The specificity of TALEs can be modulated by changing the number of TALE modules. To avoid off-target DNA cleavage, genome editing applications typically use a TALEN pair that bind on opposite sides of the target site. Such an extremely long (approximately 36 bp) DNA binding site is expected to be found rarely in genomes. However, when perform TALE imaging, TALE-FP molecules function as monomers, which may increase the chance of off-target labeling. In comparison, Cas9/sgRNA can sometimes cleave DNA sequences with up to five mismatches with the sgRNA protospacer region (42), and Cas9 has been shown to physically associate with many off-target sites in the genome (75, 152). It was suggested that shortening sgRNAs to as few as 17 nucleotides can reduce off-targeting for editing (43).
Perturbation to the target site
An ideal DNA labeling system should not interfere with the function of the target loci. The association of Cas9/sgRNA complex with the target DNA induces local dsDNA unwinding (133), which may affect the localization of histones and other DNA-binding proteins. It was suggested that CRISPR imaging may perturb gene expression in a target site-dependent manner, which could be minimized by targeting the far downstream region, or upstream region of the promoter while avoiding the enhancers (24, 49, 50). On the other hand, it was shown that dCas9 labeled telomeres displayed similar movement dynamics as those labeled by telomere-binding proteins, such as TRF-1, nor does dCas9 binding to telomeres affect integrity of telomere shelterin complex (24). Different from CRISPR, TALE directly binds to target dsDNA without inducing its unwinding. Histone H3 occupancy on satellite repeats and a histone modication of pericentromeric heterochromatin, trimethylated histone H3K9 (H3K9me3), remained unchanged upon expression of TALE against satellite repeats, thus suggesting that the binding of TALE to its target DNA does not result in a detectable change in chromatin configuration (98). So far, TALE has not been used for labeling protein-coding genes yet. Therefore, its affect in gene expression remains to be characterized.
Collectively, TALE and CRISPR imaging tools should be more complementary than competitive approaches in labeling repetitive genomic region. The major limitation of TALE imaging will be the labeling of non-repetitive genomic region. Multiple-sites targeting requires not only engineering multiple TALE-FPs, but also co-delivery multiple proteins into one cell. The highly repetitive nature of TALE-coding sequences creates barriers to their delivery using certain viral vectors, such as lentiviruses (55). Additionally, every individual TALE is fused with a fluorescent protein, which will result in very high background signal because of co-expressing multiple TALE-FPs. Different from TALE imaging, a clonal cell line expressing suitable level of dCas9-FP can be easily maintained and transduced with multiple sgRNAs for labeling non-repetitive loci. This is a major advantage of CRISPR imaging system.
Practical considerations for implementing CRISPR imaging technology
Signal-to-background ratio
The intensity of CRISPR signal from the target locus is determined by the number of fluorophores bound to this target. Due to the background signal arising from the free dCas9-FP in the nucleoplasm and cell autofluorescence, it is challenging to detect a single FP in the nucleus of a living cell. Therefore, multiple dCas9-FP is needed at the same locus to generate detectable signal. For tandem repeats, such as telomeres and satellite DNA, a single sgRNA can achieve successful labeling (1, 24, 83). Although tandem repeats are widely present in many genomes, it is not always possible to find tandem repeats near the locus of interest. In these cases that require the labeling of a non-repetitive locus, multiple sgRNAs are needed. Generally, creating create a reliably detectable fluorescent puncta over the background signal may need at least 10 FPs at the same locus. Considering the high variability in the targeting efficiency of sgRNAs (90), a group of at least 30 sgRNAs are usually required to effectively label a non-repetitive region, unless they are individually validated. For example, a non-repetitive region in MUC4 locus has been labeled by targeting at least 26 sites (24).
SunTag, a repetitive peptide array, has been recently developed for recruiting multiple (e.g. 24x) copies of a single-chain antibody (scFv) fused to FP or other proteins (138). Telomere labeling with dCas9-(SunTag)24x and scFv-GFP have demonstrated about 20 fold signal enhancement, with no perturbation of telomere mobility. Such signal amplification should greatly help long-term live imaging, which often suffers from photobleaching.
sgRNA efficacy
It is important to note that the efficiency of Cas9 targeting for any genomic locus can be dramatically influenced by the guide RNA(s) used. For example, three spacers targeting neighboring sequences of MUC4 locus resulted in very different labeling efficiencies (24) despite that these sites may share similar chromatin structures and DNA modifications, which have been suggested to be strong determinants of dCas9 targeting efficiency (72, 75, 152). A number of studies to date have tried to elucidate the connection between the target sequence features, the PAM and the sgRNA architecture to the sgRNA activity for genome editing or gene expression regulation (23, 38, 91, 149). Although imaging is a different application of the CRISPR-Cas9 system, guidelines from these studies would still be beneficial to sgRNA design for CRISPR imaging.
A pooled loss-of-function genetic screening revealed that target sequence with very high or low GC content were less effective against their targets and sgRNAs targeting the transcribed strand were less effective than those targeting the non-transcribed strand. In addition, a dCas9-binding assay indicated that Cas9 preferentially bound sgRNAs containing purines in the last four nucleotides (149). Another high-throughput tiling screen using CRISPRi as readout indicated that sgRNAs with protospacer lengths of 18–21 nt were significantly more active than sgRNAs containing longer protospacers (49). Furthermore, this screening revealed that nucleotide homopolymers had a strongly negative effect on sgRNA activity. However, neither the DNA strand that was targeted nor the GC content of sgRNA strongly correlated with sgRNA activity, which is inconsistent with the other study (149) using genome editing as readout. A third study, creating a pool of sgRNAs tiling across all possible target sites of nine endogenous genes to produce null alleles, indicated no preference of targeted DNA strands and sgRNAs with low or high GC content did tend to be less active (38). Notably, they also observed a preference in the variable nucleotide of the PAM, where cytosine was favored and thymine was disfavored. The preference for cytosine at this position has also recently been observed in zebrafish (44).
Collectively, the sequence features within and surrounding the target site do strongly influence sgRNA activity. Rigorous quantification of all these influences directly on dCas9 DNA-binding activity would be critical to maximize sgRNA efficacy for effective CRISPR labeling, especially when targeting non-repetitive sequences, which requires many sgRNAs to function simultaneously. It is also worth noting that not all guidelines obtained from genome editing are applicable to imaging. For example, although efficient DNA cleavage require a protospacer length of ~ 20 nt (75, 117, 152), pairing of the ~ 11 nt PAM-proximal seed sequence region is sufficient for dCas9 binding (152). Thus, CRISPR imaging can use shorter protospacer lengths without affecting the efficiency.
Targeting specificity
A series of studies have assessed issues related to the CRISPR-Cas9 specificity, which has been recognized as the major constraint that limits Cas9-mediated genome engineering applications (42, 53, 58, 90, 109, 117, 141, 142). To assess Cas9 targeting specificity, several groups have generated variant sgRNAs bearing one to four nucleotide mismatches in the complementary region and then examined the abilities of these molecules to direct Cas9 nuclease activity in human cells (42, 58). These studies revealed that perfect base-paring within 8–12 bp directly 5′ of the PAM (seed sequence) determines SpCas9 cleavage specificity, whereas multiple PAM-distal mismatches can be tolerated (58, 60, 61, 127, 151). In addition, not all nucleotide substitutions at a given position necessarily have equivalent effects on activity (58). Overall, for any given target site, it is not currently possible to predict how many mismatches can be tolerated. However, different from genome engineering, targeting specificity is not a major concern for CRISPR imaging due to the need of signal enrichment for detection. The off-target effects may be directly identified if the karyotype of the cell line used is known. Additionally, the labeling specificity can be further validated by FISH (25). In contrast with genome editing and regulation, mismatches within PAM-distal region can be considered as potential targets, especially when there are no many choices of target sites. To more precisely extract the true underlying mismatching features that affect dCas9 DNA-binding activity, CRISPR labeling efficiency should be directly examined as the readout to address the mismatch effects.
Delivery of CRISPR-Cas components
In cultured mammalian cells, electroporation, Nucleofection or Lipofecamine-mediated transfection have been used to transiently express Cas9 and sgRNAs (30, 36, 43, 90), which is sufficient to induce efficient genome editing. More recent studies have started to use lentiviral delivery system to do genome-scale screening with relatively higher efficiency (49, 63, 73, 129, 149, 159). This delivery system helped CRISPR-Cas9-based multiplex genome engineering in diverse cell types (63). For imaging purposes, lentiviral vectors have been used to constitutively express Cas9 and/or sgRNAs in cultured cells, which is convenient for the maintenance of stable cell line for downstream experiments (24). dCas9-FP and sgRNA are suggested to be delivered into the cells sequentially. Clonal cell lines with variable dCas9-FP expression levers are better to be first generated and then screened by looking for a suitable dCas9 expression level with active sgRNAs. This selection is determined by labeling efficiency, signal-to-background ratio and cell state. In general, a healthy cell line with best dCas9-FP expression level can be used for targeting any locus. This cell line can be maintained for imaging any target of interest. Lentivirus expressing sgRNA can then be tansduced to the cells as a separate step.
It has been found that sgRNA expression dosage is strongly correlate with CRISPR efficiency (24). Thus, any approach that can increase the sgRNA expression would help improve the labeling efficiency. In particular, non-repetitive locus labeling is largely limited by the difficulty to deliver many sgRNAs into the same cell. Two new strategies, developed to deliver multiple sgRNAs from a single vector for multiplex genome engineering, may be potentially helpful to non-repetitive locus labeling and the labeling of multiple loci. One strategy is to use a single lentiviral system to express up to four sgRNAs from independent RNA polymerase III promoters (63). The limitation, though, is that the identical sequences in the scaffold part of the four sgRNAs may cause rearrangement in the vector. Although this problem can be minimized by the cloning strategy, it is indeed time-consuming to generate the vector. The other strategy is to utilize RNA processing (105). The type III CRISPR/Cas associated Csy4 protein from Psedomonas aeruginosa cleaves RNA following a 28-nt sequence. By bearing this sequence upstream of the sgRNA, a single transcript can be processed into multiple sgRNAs from the same Pol II promoter (105, 141). The drawback of this method is that Csy4 RNA endonuclease must also be expressed which might bring toxicity to the cell.
Applications of CRISPR imaging
Understanding the searching and targeting mechanism of Cas9-sgRNA complex
Being able to image and track fluorescent Cas9 molecules offers the opportunity to understand the mechanism of searching and binding of Cas9 protein to its DNA target. For example, in vitro single molecule tracking of Cas9 on the DNA curtain platform (133) (parallel strands of extended DNA whose two ends are anchored to a cover glass surface) have revealed the search kinetics and bias. More recently, single molecule tracking of dCas9 in live cells have further elucidated the effect of nuclear environment, such as heterochromatin, on the kinetics of target searching (72).
Investigating chromosome organization
The spatial organization of genomes is increasingly recognized as important for maintaining and regulating cell functions such as gene expressions, gene replications, and proper partitions of its genetic materials through cell divisions. Below we suggest open questions regarding the regulation and role of chromosome organizations in various biological processes that CRISPR imaging might help address.
Chromatin loops and domains
Chromatin loops and domains are revealed recently by emerging sequencing-based techniques such as Hi-C methods, which detect physical “contacts” in a chromosome between two DNA sequences that are far away in one-dimensional genomic distance but close in three-dimensional space (32, 34, 80, 118, 146). These studies suggest that chromatin loops and domains are highly regulated: preferential contacts are common in eukaryotic organisms ranging from yeast to human (12, 39, 48, 80); the sequence of contacts is largely conserved between cell types (37); change in gene expression is accompanied by change in the contacts in specific chromosome domains (20, 106, 148).
Although the sequencing-based techniques have the high-throughput to perform genomic scale mapping, direct CRISPR imaging of specific genomic loci in single living cells would yield complementary information such as when and how these contacts form and how persistent or labile they are. Moreover, as large heterogeneity is seen at the single cell level that cannot be simply extrapolated from the ensemble measurement (102), CRISPR imaging can study the heterogeneity by directly imaging single cells. A third advantage enabled by CRISPR imaging is that the spatial information is directly measured by imaging loci positions rather than modeled from satisfying contact constraints (51, 64, 102) and therefore open up new possibilities in chromosome conformation studies. For example, in embryonic development, thousands of pairs of contact between distal enhancer and promoter region through chromatin looping are tightly regulated to control gene expressions in a coordinated fashion in the signaling network (26, 54, 111, 124). Direct imaging of a distal enhancer and promoter pair in development might reveal the timing of the contact in development and how strong the contact is. Some long range looping interactions such as those in the sonic hedgehog pathway (on the order of 1 Mb) have indeed been resolved using conventional optical microscopy (123, 158).
CRISPR imaging has the promise to image chromosome conformation in live cells if multiple specific loci are imaged on a single chromosome, say, a hundred fiducial markers along a chromosome chain such that the each locus can be tracked with high spatial resolution. As chromosome conformation has been primarily investigated by sequencing methods or staining fixed cells, the ability to image chromosome conformation in live cells has broad applications. First, chromosomes undergo global scale condensation and de-condensation through mitosis (79). Whereas interphase chromosomes adopt conformation with preferential contacts, mitotic chromosomes tightly pack in a way that seems independent of the genomic sequence (103, 136). How a chromosome alternates between the two drastically different conformations is largely unknown. The reverse process, how cells re-establish preferential chromosome contacts and topologically associated domains (TADs) after mitotic exit is equally elusive (33).
Spatial organization of chromosomes
FISH on interphase nuclei in fixed cells reveals chromosome territories – each chromosome occupies a space where other chromosomes are excluded (14). Chromosomes show preferential spatial distribution within the nucleus with the proper nuclear alignment (9). The spatial organization and relative position of chromosomes is highly conversed and has important consequences (108). It is related to coordinated gene expression as proteins might bring chromosomes to vicinity to each other to coordinate gene expression (94, 132). It is also related to recurrent chromosomal translocations in tumors as translocations preferentially form between neighboring chromosomes (121, 131).
Curiously, the spatial organization of interphase chromosomes is inherited to a great extent from mother to daughter cells despite large chromosome movement and spatial rearrangement through mitosis (40, 135). How the spatial organization is inherited is unknown. An interesting model (47) proposes that different chromosomes might separate at slightly different time due to slightly different centromere strength according to chromosome identity. The ones that separate early will end up far away from the cell middle whereas the late ones will be near the cell midplane. Therefore, a spatial distribution is dictated by the chromosome identity and could be inherited. CRISPR imaging allows tagging multiple specific chromosomes simultaneously through delivery of multiple guide RNA species corresponding to specific chromosomes. Therefore multiple specific chromosomes could be tracked and how their relative positions are regulated through cell division could be directly imaged.
Furthermore, other cyclic changes of chromosome organization happen through cell cycles. One example is that the active transcription centers of a cell, nucleoli, disintegrate during mitosis and reform afterwards, and small nucleoli fuse gradually to form large ones in daughter cells (144). CRISPR imaging can track ribosomal DNA segments distributed on multiple chromosomes that participate in the dynamic cyclic formation of nucleoli. Another example is to probe the dynamics of genomic loci that are periodically expressed and repressed such as cyclins. By labeling these genes, CRISPR imaging might reveal periodic changes in the chromosome organization as a way to regulate gene expression.
In X chromosome inactivation, direct imaging shows the pairing and physical tether between two X chromosomes and the subsequent silencing of one X chromosome during stem cell differentiation (94). It is unclear when and how cells count their X chromosomes. Evidence suggests that cells compare the number of X chromosome to that of autosomes to apply dosage compensation accordingly (2, 104). Therefore, it is possible that similar counting and pairing mechanism exists for autosomes which might be pre-requisite for X chromosome pairing and silencing. CRISPR imaging can monitor X chromosomes and multiple specific autosomes simultaneously to observe whether there is similar autosome pairing in differentiation program and how that correlates with the dynamics and silencing of X chromosomes.
Visualizing genome instability and rearrangement
Genome instability and rearrangement occurs in diverse biological processes and its consequences vary from being extremely helpful to catastrophic to the cells. Although key enzymes are extensively studied, recent studies with direct imaging of the genome suggest that dynamics of chromosomes and genomic loci are critical to these processes.
Chromosome mis-segregation and aneuploidy
Although aneuploidy, where a cell has abnormal number of chromosomes, is recurrent in birth defects and tumors (134, 147), how it arises and its consequences are unclear partly due to a lack of live-cell aneuploidy reporter. CRISPR imaging can faithfully report the number of a specific chromosome by tagging a sequence unique to that chromosome and therefore accurately detect chromosome mis-segregation and aneuploidy. For example, aneuploidy is commonly thought to arise directly from mis-segregation in mitosis. However, live-cell imaging suggests that mis-segregation in mammalian cells often leads to binuclei fusion and tetraploid progenies instead of aneuploidy (104). Another seeming paradox is that aneuploidy seems to be detrimental at the single cell level but confer growth advantage in tumors (130). Some hypothesizes that aneuploidy leads to higher genome instability and more mutations and growth advantage is acquired through step-wise mutations. Using CRISPR imaging and lineage tracking, one might monitor the growth kinetics of aneuploidy with high temporal resolution in a population of cells. This imaging platform might be further adapted to study therapeutic effects to curb tumor growth by selecting normal cells against cells with aneuploidy (27, 130).
DNA damage repair
Cells constantly experience DNA damage and have to employ various mechanisms to efficiently carry out DNA repair. Although little is known about the chromosome dynamics during DNA repair, direct imaging of chromosomes suggests intriguing chromosome behavior and repair kinetics. For instance, studies using transgenes to tag the broken ends of double stranded breaks show that instead of freely dangling and instant separation as previously thought, broken ends are typically held together and exhibit restricted motion for hours after DNA damage (121, 131); distant sister chromatids can undergo large movement to find each other in homology-directed repair (HDR) in bacteria cells (78).
CRISPR imaging can conveniently label specific genomic loci and therefore the DNA damage site might be precisely tracked during repair. Additionally, CRISPR imaging label simultaneously homologous sequences (from both homologous chromosomes and sister chromatids) and therefore might be particularly suited to monitoring HDR process. A systematic study to monitor chromosome motion during DNA repair and read out the corresponding repair result afterwards would be ideal to reveal the underlying connection and might help predict and influence the outcome of DNA repair (15, 22). A method to combine CRISPR imaging and in-situ sequencing (77) might serve this purpose.
One extreme case of DNA damage and repair is chromothripsis where extensive chromosomal rearrangements (up to thousands) occur in a single event in one chromosome (156). Using CRISPR imaging to tag multiple loci on a single chromosome, one might directly observe this catastrophic event to see how a chromosome is broken into pieces, how it is re-assembled, and what affects the outcome of the re-assembly process.
Gene recombination
A given set of genome produce limited protein species whereas many circumstances require a large repertoire of diverse proteins. Nature solves this problem in a simple and efficient way, through gene segment recombination in specific cell lineages in a highly regulated fashion (88, 114). For example, during B cell maturation, one variable (V) gene segment, one diversity (D) gene segment, and one joining (J) gene segment is randomly chosen from the pool of 44 variable gene segments, 27 diversity gene segments and 6 joining gene segments on one chromosome to produce one heavy chain protein in the so-called V(D)J recombination. Similar recombination processes are involved in light chain production, class switching, T cell surface receptor diversification, olfactory cell receptor production, etc.
Although key enzymes involved in these gene recombination processes have been identified through decades of work, relatively little is known about the timing and kinetics of gene recombination or the dynamics of the relevant gene segments. Live cell imaging of gene segments using transgene method shows complex dynamics that suggest interesting loci searching mechanisms (82), but no recombination events was direct imaged possibly due to uncontrolled transgene insertion site. CRISPR imaging can follow the recombination process to reveal how the loci accurately find their respective joining segments, and the duration and kinetics of the recombination. CRISPR imaging might be particularly suited here given the repetitive sequences endogenously present in these systems.
Recent studies demonstrate genome rearrangement and recombination are influenced by the genome spatial organization. For example, chromosomal translocations preferentially happen between neighboring chromosomes (121). Human immunodeficiency virus (HIV) preferentially targets the part of the genome close to the nuclear envelope for integration and lead to sequence hotspots during HIV infection (92). This discovery gives new insight on how our genomes evolve as virus genomes are integrated into ours through millions of years’ evolution. The bias created by the genome spatial organization might be responsible for other processes of genome rearrangements such as in transposons through the course of evolution (52). These possibilities highlight a great need to acquire spatial information of specific DNA sequences in live cells and CRISPR imaging can provide such information in the context of 3D genome.
Conclusions and future development of CRISPR imaging technology
Chromatin structures and dynamics are increasingly recognized as important in diverse cellular functions. Emerging techniques of chromatin imaging show the promise to bridge the long-standing gap between sequencing studies that give genomic information and imaging studies that provide spatial and temporal information. Although technical challenges need to be overcome, the potential of CRISPR imaging will help solve many genome and chromatin mysteries through direct live-cell imaging. With its unprecedented flexibility and precision in genomic sequence targets, we believe the best is yet to come.
Future research directions to improve the technology will include further optimizing sgRNA scaffold which is crucial for the targeting efficiency through its binding affinity for Cas9. More well designed experiments using CRISPR imaging as readout are required to explore the target sequence features that determine the sgRNA activity. To achieve full potential of this technology for labeling single copy genes, development of new sgRNA delivery systems will be a critical area of future research. Additional studies will be also required to optimize the signal-to-background ratio, possibly by further amplifying the CRISPR signal. Alternatively, using cutting-edge microscopy with high sensitivity of signal detection will greatly assist the imaging efficiency. Direct engineering of the Cas9 proteins from different bacterial species should offer a path toward PAM independence, and generating even more efficient Cas9 proteins. These achievements will allow developing more efficient multicolor CRISPR imaging method for simultaneously tracking many loci in one cell. Finally, CRISPR imaging method should be also applied in living organism, which will truly unravel the function of genome spatial-temporal organization. Together, all these technologies promise to expand our ability to uncover the mystery of the dynamics of complex genome organization.
Acknowledgments
This work is supported by the W. M. Keck Foundation and NIH Single Cell Analysis Program (R33EB019784).
Contributor Information
Baohui Chen, Email: baohui.chen@ucsf.edu.
Juan Guan, Email: juan.guan@ucsf.edu.
Bo Huang, Email: bo.huang@ucsf.edu.
Literature Cited
- 1.Anton T, Bultmann S, Leonhardt H, Markaki Y. Visualization of specific DNA sequences in living mouse embryonic stem cells with a programmable fluorescent CRISPR/Cas system. Nucleus. 2014;5:163–72. doi: 10.4161/nucl.28488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avner P, Heard E. X-chromosome inactivation: counting, choice and initiation. Nat Rev Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- 3.Bancaud A, Huet S, Daigle N, Mozziconacci J, Beaudouin J, Ellenberg J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009;28:3785–98. doi: 10.1038/emboj.2009.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 5.Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–41. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 6.Beliveau BJ, Boettiger AN, Avendano MS, Jungmann R, McCole RB, et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat Commun. 2015;6:7147. doi: 10.1038/ncomms8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belmont AS. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 2001;11:250–7. doi: 10.1016/s0962-8924(01)02000-1. [DOI] [PubMed] [Google Scholar]
- 8.Belmont AS, Braunfeld MB, Sedat JW, Agard DA. Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma. 1989;98:129–43. doi: 10.1007/BF00291049. [DOI] [PubMed] [Google Scholar]
- 9.Berger AB, Cabal GG, Fabre E, Duong T, Buc H, et al. High-resolution statistical mapping reveals gene territories in live yeast. Nat Methods. 2008;5:1031–7. doi: 10.1038/nmeth.1266. [DOI] [PubMed] [Google Scholar]
- 10.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 12.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–84. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–12. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 14.Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. Plos Biology. 2005;3:826–42. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 16.Briggs AW, Rios X, Chari R, Yang L, Zhang F, et al. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40:e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bultmann S, Morbitzer R, Schmidt CS, Thanisch K, Spada F, et al. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 2012;40:5368–77. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casas-Delucchi CS, Becker A, Bolius JJ, Cardoso MC. Targeted manipulation of heterochromatin rescues MeCP2 Rett mutants and re-establishes higher order chromatin organization. Nucleic Acids Res. 2012;40:e176. doi: 10.1093/nar/gks784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–9. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 23.Chari R, Mali P, Moosburner M, Church GM. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods. 2015;12:823–6. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–91. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen B, Huang B. Imaging genomic elements in living cells using CRISPR/Cas9. Methods Enzymol. 2014;546:337–54. doi: 10.1016/B978-0-12-801185-0.00016-7. [DOI] [PubMed] [Google Scholar]
- 26.Chen CY, Morris Q, Mitchell JA. Enhancer identification in mouse embryonic stem cells using integrative modeling of chromatin and genomic features. BMC Genomics. 2012;13:152. doi: 10.1186/1471-2164-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G, Mulla WA, Kucharavy A, Tsai HJ, Rubinstein B, et al. Targeting the adaptability of heterogeneous aneuploids. Cell. 2015;160:771–84. doi: 10.1016/j.cell.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–2. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 29.Choo Y, Sanchez-Garcia I, Klug A. In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature. 1994;372:642–5. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- 30.Cong L, Ran FA, Cox D, Lin S, Barretto R, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das T, Payer B, Cayouette M, Harris WA. In vivo time-lapse imaging of cell divisions during neurogenesis in the developing zebrafish retina. Neuron. 2003;37:597–609. doi: 10.1016/s0896-6273(03)00066-7. [DOI] [PubMed] [Google Scholar]
- 32.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes & Development. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dekker J. Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture. Epigenetics Chromatin. 2014;7:25. doi: 10.1186/1756-8935-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng D, Yan C, Pan X, Mahfouz M, Wang J, et al. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335:720–3. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393–4. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–7. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–7. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Essers J, van Cappellen WA, Theil AF, van Drunen E, Jaspers NG, et al. Dynamics of relative chromosome position during the cell cycle. Mol Biol Cell. 2005;16:769–75. doi: 10.1091/mbc.E04-10-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–21. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–6. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–84. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gasser SM. Visualizing chromatin dynamics in interphase nuclei. Science. 2002;296:1412–6. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- 47.Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–64. doi: 10.1016/s0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- 48.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–82. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–61. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–51. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, et al. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–63. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Green B, Bouchier C, Fairhead C, Craig NL, Cormack BP. Insertion site preference of Mu, Tn5, and Tn7 transposons. Mob DNA. 2012;3:3. doi: 10.1186/1759-8753-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–82. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19:541–9. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holkers M, Maggio I, Liu J, Janssen JM, Miselli F, et al. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41:e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 57.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110:15644–9. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–32. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang F, Doudna JA. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol. 2015;30:100–11. doi: 10.1016/j.sbi.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–81. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- 61.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–8. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–85. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 66.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–34. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 67.Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, et al. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013;31:251–8. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- 68.Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–53. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015 doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–U249. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klug A. Co-chairman’s remarks: protein designs for the specific recognition of DNA. Gene. 1993;135:83–92. doi: 10.1016/0378-1119(93)90052-5. [DOI] [PubMed] [Google Scholar]
- 72.Knight SC, Xie L, Deng W, Guglielmi B, Witkowsky LB, et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350:823–6. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- 73.Koike-Yusa H, Li Y, Tan EP, del Velasco-Herrera MC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–73. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 74.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–8. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–83. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 76.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–15. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 77.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–3. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lesterlin C, Ball G, Schermelleh L, Sherratt DJ. RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature. 2014;506:249-+. doi: 10.1038/nature12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li G, Sudlow G, Belmont AS. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J Cell Biol. 1998;140:975–89. doi: 10.1083/jcb.140.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lindhout BI, Fransz P, Tessadori F, Meckel T, Hooykaas PJ, van der Zaal BJ. Live cell imaging of repetitive DNA sequences via GFP-tagged polydactyl zinc finger proteins. Nucleic Acids Res. 2007;35:e107. doi: 10.1093/nar/gkm618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lucas JS, Zhang Y, Dudko OK, Murre C. 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell. 2014;158:339–52. doi: 10.1016/j.cell.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci U S A. 2015;112:3002–7. doi: 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma H, Reyes-Gutierrez P, Pederson T. Visualization of repetitive DNA sequences in human chromosomes with transcription activator-like effectors. Proc Natl Acad Sci U S A. 2013;110:21048–53. doi: 10.1073/pnas.1319097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–9. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mahfouz MM, Li L, Piatek M, Fang X, Mansour H, et al. Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein. Plant Mol Biol. 2012;78:311–21. doi: 10.1007/s11103-011-9866-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maizels N. Immunoglobulin gene diversification. Annu Rev Genet. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- 89.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–9. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–8. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–63. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marini B, Kertesz-Farkas A, Ali H, Lucic B, Lisek K, et al. Nuclear architecture dictates HIV-1 integration site selection. Nature. 2015;521:227–31. doi: 10.1038/nature14226. [DOI] [PubMed] [Google Scholar]
- 93.Martin RM, Leonhardt H, Cardoso MC. DNA labeling in living cells. Cytometry A. 2005;67:45–52. doi: 10.1002/cyto.a.20172. [DOI] [PubMed] [Google Scholar]
- 94.Masui O, Bonnet I, Le Baccon P, Brito I, Pollex T, et al. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell. 2011;145:447–58. doi: 10.1016/j.cell.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matzke AJ, Huettel B, van der Winden J, Matzke M. Use of two-color fluorescence-tagged transgenes to study interphase chromosomes in living plants. Plant Physiol. 2005;139:1586–96. doi: 10.1104/pp.105.071068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–14. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–8. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 98.Miyanari Y, Ziegler-Birling C, Torres-Padilla ME. Live visualization of chromatin dynamics with fluorescent TALEs. Nat Struct Mol Biol. 2013;20:1321–4. doi: 10.1038/nsmb.2680. [DOI] [PubMed] [Google Scholar]
- 99.Mora-Bermudez F, Ellenberg J. Measuring structural dynamics of chromosomes in living cells by fluorescence microscopy. Methods. 2007;41:158–67. doi: 10.1016/j.ymeth.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 100.Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790–9. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 102.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, et al. Organization of the mitotic chromosome. Science. 2013;342:948–53. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 105.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–85. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. 2001;70:313–40. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 108.Parada LA, McQueen PG, Munson PJ, Misteli T. Conservation of relative chromosome positioning in normal and cancer cells. Curr Biol. 2002;12:1692–7. doi: 10.1016/s0960-9822(02)01166-1. [DOI] [PubMed] [Google Scholar]
- 109.Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–43. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–17. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 111.Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: five essential questions. Nat Rev Genet. 2013;14:288–95. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–6. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, et al. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Methods. 2013;10:239–42. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Petrie HT, Livak F, Burtrum D, Mazel S. T cell receptor gene recombination patterns and mechanisms: cell death, rescue, and T cell production. J Exp Med. 1995;182:121–7. doi: 10.1084/jem.182.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 116.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–83. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–91. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–5. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, et al. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T. Spatial dynamics of chromosome translocations in living cells. Science. 2013;341:660–4. doi: 10.1126/science.1237150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Saad H, Gallardo F, Dalvai M, Tanguy-le-Gac N, Lane D, Bystricky K. DNA dynamics during early double-strand break processing revealed by non-intrusive imaging of living cells. PLoS Genet. 2014;10:e1004187. doi: 10.1371/journal.pgen.1004187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sagai T, Amano T, Tamura M, Mizushina Y, Sumiyama K, Shiroishi T. A cluster of three long-range enhancers directs regional Shh expression in the epithelial linings. Development. 2009;136:1665–74. doi: 10.1242/dev.032714. [DOI] [PubMed] [Google Scholar]
- 124.Sakabe NJ, Savic D, Nobrega MA. Transcriptional enhancers in development and disease. Genome Biol. 2012;13:238. doi: 10.1186/gb-2012-13-1-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–55. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schmid-Burgk JL, Schmidt T, Kaiser V, Honing K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol. 2013;31:76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011;108:10098–103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shachar S, Pegoraro G, Misteli T. HIPMap: A High-Throughput Imaging Method for Mapping Spatial Gene Positions. Cold Spring Harb Symp Quant Biol. 2015 doi: 10.1101/sqb.2015.80.027417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Siegel JJ, Amon A. New Insights into the Troubles of Aneuploidy. Ann Rev Cell Dev Biol. 2012;28:189–214. doi: 10.1146/annurev-cellbio-101011-155807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–82. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 133.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–7. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Streblow RC, Dafferner AJ, Nelson M, Fletcher M, West WW, et al. Imbalances of chromosomes 4, 9, and 12 are recurrent in the thecoma-fibroma group of ovarian stromal tumors. Cancer Genet Cytogenet. 2007;178:135–40. doi: 10.1016/j.cancergencyto.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Strickfaden H, Zunhammer A, van Koningsbruggen S, Kohler D, Cremer T. 4D chromatin dynamics in cycling cells: Theodor Boveri’s hypotheses revisited. Nucleus. 2010;1:284–97. doi: 10.4161/nucl.1.3.11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Strukov YG, Belmont AS. Mitotic chromosome structure: reproducibility of folding and symmetry between sister chromatids. Biophys J. 2009;96:1617–28. doi: 10.1016/j.bpj.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sun N, Zhao H. Transcription activator-like effector nucleases (TALENs): a highly efficient and versatile tool for genome editing. Biotechnol Bioeng. 2013;110:1811–21. doi: 10.1002/bit.24890. [DOI] [PubMed] [Google Scholar]
- 138.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell. 2014;159:635–46. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–7. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thanisch K, Schneider K, Morbitzer R, Solovei I, Lahaye T, et al. Targeting and tracing of specific DNA sequences with dTALEs in living cells. Nucleic Acids Res. 2014;42:e38. doi: 10.1093/nar/gkt1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–76. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–97. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–3. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- 144.Tuzel E. Organelle dynamics: a tale of fusing nucleoli. Curr Biol. 2011;21:R395–7. doi: 10.1016/j.cub.2011.03.068. [DOI] [PubMed] [Google Scholar]
- 145.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–51. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 146.van Berkum NL, Lieberman-Aiden E, Williams L, Imakaev M, Gnirke A, et al. Hi-C: a method to study the three-dimensional architecture of genomes. J Vis Exp. 2010 doi: 10.3791/1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vasuri F, Magrini E, Foschini MP, Eusebi V. Trisomy of chromosome 6 in Merkel cell carcinoma within lymph nodes. Virchows Arch. 2008;452:559–63. doi: 10.1007/s00428-008-0599-4. [DOI] [PubMed] [Google Scholar]
- 148.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–4. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 151.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A. 2011;108:10092–7. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–6. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yuan K, Shermoen AW, O’Farrell PH. Illuminating DNA replication during Drosophila development using TALE-lights. Curr Biol. 2014;24:R144–5. doi: 10.1016/j.cub.2014.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–50. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–71. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, et al. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–84. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–53. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang Y, Wong CH, Birnbaum RY, Li G, Favaro R, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–10. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhou Y, Zhu S, Cai C, Yuan P, Li C, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–91. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 160.Zink D, Sadoni N, Stelzer E. Visualizing chromatin and chromosomes in living cells. Methods. 2003;29:42–50. doi: 10.1016/s1046-2023(02)00289-x. [DOI] [PubMed] [Google Scholar]