Abstract
Short tandem repeats (STRs) are short tandemly repeated DNA sequences that involve a repetitive unit of 1–6 bp. Because of their polymorphisms and high mutation rates, STRs are widely used in biological research. Strand-slippage replication is the predominant mutation mechanism of STRs, and the stepwise mutation model is regarded as the main mutation model. STR mutation rates can be influenced by many factors. Moreover, some trinucleotide repeats are associated with human neurodegenerative diseases. In order to deepen our knowledge of these diseases and broaden STR application, it is essential to understand the STR mutation process in detail. In this review, we focus on the current known information about STR mutation.
Key words: short tandem repeats, mutation
Introduction
Short tandem repeats (STRs), also known as microsatellites or simple sequence repeats, are shorl tandemly repeated DNA sequences that involve a repetitive unit of 1-6 bp (1), forming series with lengths of up to 100 nucleotides (nt). STRs are widely found in prokaryotes and eukaryotes, including humans. They appear scattered more or less evenly throughout the human genome, accounting for about 3% of the entire genome. However, their distribution within chromosomes is not quite uniform—they appear less frequently in subtelomeric regions (2). Most STRs are found in the noncoding regions, while only about 8% locate in the coding regions (3). Moreover, their densities vary slightly among chromosomes. In humans, chromosome 19 has the highest density of STRs (4). On average, one STR occurs per 2,000 bp in the human genome (5). The most common STRs in humans are A-rich units: A, AC, AAAN, AAN, and AG 5., 6.. The STR locus is named as, for example, D3S1266, where D represents DNA, 3 means chromosome 3 on which the STR locus locates, S stands for STR, and 1266 is the unique identifier.
On the basis of different repeat units, STRs can be classified into different types. On the one hand, according to the length of the major repeat unit, STRs are classified into mono-, di-, tri-, tetra-, penta-, and hexanucleotide repeats. The total number of each type decreases as the size of the repeat unit increases. The most common STRs in the human genome are dinucleotide repeats (5). On the other hand, according to the repeat structure, STRs are classified into perfect repeats (simple repeats), containing only one repetitive unit, and imperfect repeats (compound repeats), consisting of different composition repeats (7).
Since the last decade of the 20th century, scientists have been interested in the direct functions of STRs in some of their host organisms. Although STRs widely exist in organisms, most of them are thought to have no biological uses at all and are regarded as “junk DNA”. However, several interesting hypotheses suggest that STRs actually play an important role in many organisms. In many disease-causing bacteria, some “contingency genes” reside in STR sequences. The STRs in such genes could cause frameshift mutations, which will change the expression of some proteins. These proteins are not necessary for the viability of the bacteria, but they can help bacteria to evade the human immune system. Some STRs may take part in regulating the transcription. From yeasts to humans, many proteins involved in transcriptional regulation contain glutamine-rich domains and trinucleotide repeats encoding series of polyglutamine (8). Moreover, researches have shown that some STRs can regulate the transcription of the epidermal growth factor gene (9), the tyrosine hydroxylase gene (10), and the PIG3 gene (11). In addition, some STRs may influence the regulation of gene expression. For example, in mammalian genomes, (CA)n and (CT)n near particular genes can affect the expression of these genes 12., 13.. STRs may also affect recombination (14), generation of nucleosome positioning signals (15), and maintenance of chromatin spatial organization (16).
Although currently more and more biological functions of STRs are being discovered, most of them remain unknown. Therefore, further study of the mutation and variability of STRs is required to understand their biological functions.
High Mutation Rates of STRs
Unique DNA sequences in a genome exhibit a very low mutation rate (approximately 10−9 nt per generation), whereas the mutation rates in STR sequences are several orders of magnitude higher (3), ranging from 10−6 to 10−2 nt per generation. STR mutation rates are specific for organisms in vivo. For instance, the STR mutation rate in yeast (16) and human 17., 18. is 10−5 nt and 10−3−10−5 nt per cell division, respectively.
There is apparently great variation in mutation rates among loci. Chakraborty et al. (19) showed that in human nonpathogenic STR loci, dinucleotide repeats display the highest mutation rate, while those of tetranucleotide STRs are 50% lower. However, the mutation rates of disease-associated trinucleotide repeats exceed the normal value by four to seven times.
Several approaches have been developed for elucidating the mutation rates of STRs, such as the familial approach (17), biological model approach, population approach (20), and germline cell approach. The familial approach is the most direct one, where both the mutation rate and mutation type can be directly examined during STR transmission from the parents to the offspring (17). In the biological model approach, an STR is cloned into a vector and propagated in its host, then the spontaneous rates of STR mutations can be evaluated and the effects of various factors on STR mutation can be estimated. Using the population approach, the common evolutionary origin of STRs can be detected and the mutation events can be traced back many generations (21). STR mutation rates can also be directly analyzed in germline cells by the germline cell approach, especially in sperms (22).
Mechanisms of STR Mutation
STRs were identified in eukaryotic DNA at the beginning of the 1970s. However, for the past decades, the mechanisms of STR mutation remain poorly understood. Up to date, three possible mechanisms have been proposed: (1) unequal crossing over in meiosis; (2) retrotransposition mechanism; (3) strand-slippage replication. Among these mechanisms, strand-slippage replication appears to be widely regarded as the main pattern of STR mutation.
Unequal crossing over in meiosis
This is a well-known mechanism generating large blocks of satellite DNA. It is associated with the exchange of repeat units between homologous chromosomes. However, this process involves different chromosomes, thus it plays a restricted role in STR mutation. Nevertheless, this mechanism may be responsible for STR multistep mutations (17), which will be mentioned later.
Retrotransposition mechanism
This mechanism speculates that A-rich STRs are generated by a 3’ extension of retrotranscripts, similar to the polyadenylation of mRNA. Evidence has shown the association between the most common human STRs with A-rich content and transposable elements (6). However, a high density of transposable elements does not always coincide with a high density of STRs. Further studies are needed to elucidate whether it is really a mechanism for STR mutation.
Strand-slippage replication
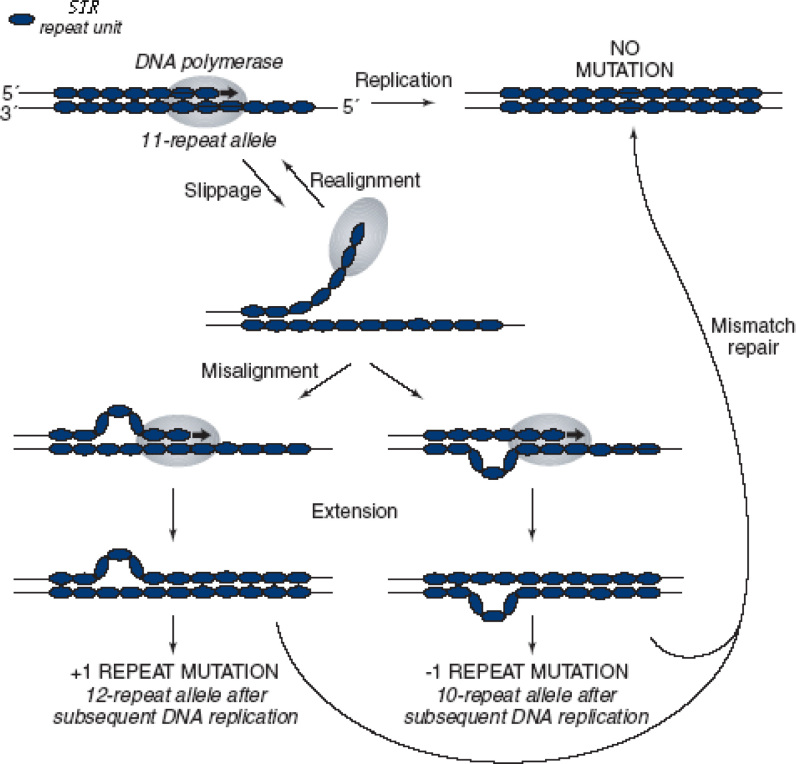
This model was first proposed by Kornberg et al. in 1964 (23), which has also been called DNA slippage, polymerase slippage, or slipped strand mispairing. At present, it appears to be widely accepted as the main explanation of the STR mutation process. The slippage occurs during DNA replication, with a consequence of mispairing (by one or more repeat units) between the nascent and template strands. Next, the repeated DNA fragment is forced to “loop out” at the mismatch site. If DNA synthesis continues on this molecule, then the repeat number of the STR is altered (Figure 1) (24).
Fig. 1.
Schematic illustration of the strand-slippage replication at STR (24).
However, the slippage rate is not the same as the apparent mutation rate of STR. Experiments in vitro have demonstrated that DNA slippage occurs at very high rates (25). But in vivo, most of the DNA loops are recognized and eliminated by the mismatch repair system. It has been shown that a functional mismatch repair system reduces the STR mutation rate between 100 and 1,000 folds (26). Thus, the observed STR mutation rate depends on the rate of slippage and the efficiency of the repair system correcting the mismatches.
Several factors can affect the rate of slippage events, among which the repeat unit is the most important factor. A negative correlation was suggested between the length of the repeat unit and the rate of slippage (25). Kruglyak et al. (27) showed that the rate of slippage was the highest in dinucleotide STRs and the lowest in tetranucleotide STRs. This is consistent with the observation that the longer the repeat unit, the less the total amount of STRs. Probably the longer repeat units would require the strand to slip further before the bases could pair correctly again, and then they become less common in the genomes.
Besides the repeat unit, other factors such as the number, location, and sequence of repeats are also likely to affect the rate and direction of slippage (28). For instance, in humans, the rate of slippage events exponentially increases with the increasing repeat number (29).
The genesis of STRs assumes that the generation of STRs requires short “proto-STR”, which is subsequently extended by DNA slippage (30). Once proto-STR arises, the repeat sequence acquires the ability of mutation. The minimum number of repeats needed for further expansion is four to five repeats for a dinucleotide STR and two repeats for a tetranucleotide STR (31).
Models of STR Mutation
Infinite alleles model (IAM)
Kimura and Crow proposed this model in 1964 (32) based on an assumption that each new mutation produces a new allele and all mutations are equiprobable. Therefore, it can involve any number of tandem repeats and always results in a new allele state not previously existing in the population. However, many studies on STR mutation showed that this model was incompatible with real mutation processes.
Stepwise mutation model (SMM)
This model was developed by Ota and Kimura in 1973 (33). It was originally used to describe the changes of charged proteins inferred from electrophoretic mobility, which was later shown to be ineffective. However, it proved to be suitable for describing STR mutations. SMM is based on the following assumptions: (1) small changes in repeat number; (2) equal probabilities of increasing and decreasing repeat number; (3) unlimited allele size; (4) independence of the rate and size of mutations from the repeat number. SMM is in agreement with the strand-slippage replication mechanism, which is currently accepted as the main mechanism for STR mutation.
In this model, alleles can mutate up or down by one or a small number of repeat units (25). The model that only changes one repeat unit each time is called strict (single-step) SMM. When the change is more than one unit each time, it is called two-phase mutation model (TPM), which is also termed as generalized or multistep mutation model (MMM). In general, SMM refers to the strict SMM. Some reports suggested that SMM was consistent with the distribution of alleles at STR loci (34). But many other studies demonstrated that strict SMM might not be sufficient to account for allele frequency distributions at STR alleles. These studies imply that the more complex the repeat structure is, the lower the likelihood that strict stepwise mutational events will happen (35).
TPM was developed by Di Rienzo et al. in 1994 (35), which predicts the expected variance in repeat number under different mutational processes and demographic histories. It incorporates the mutational process of SMM, but allows for mutations of a larger magnitude to occur. As the variance in repeat number increases, the frequency of multistep mutations increases. If the distribution of STR alleles corresponds to MMM, such STR mutations could be caused by unequal crossing over. TPM is found in various organisms including Primates (36). Huang et al. reported that the proportion of multistep mutations in human dinucleotide repeats is 62.9% (17). Other researchers reported a much lower value of the average frequency of multistep human STR mutations, with an average of 23.8% 18., 37..
Factors Influencing STR Mutation
Repeat number
One of the key effective factors influencing STR mutation is repeat number. Studies using different methods, such as familial approach (38) and population approach (39), have strongly suggested that STR mutation rate increases with repeat number. Some studies displayed a positive association between mutation rate and repeat number in many vertebrate species (40) including humans (17). The direction of mutation may be different for alleles of different sizes within a locus (41). An expansion occurs more frequently in short STRs, while a reduction of repeat number exists in longer ones (29).
Repeat unit
The mutation rate of dinucleotide repeats is higher than that of trinucleotide repeats 19., 42.. These results agree with the slippage studies mentioned above.
Repeat structure
In autosomes, Y chromosome, and tumor cells, it was found that the mutation frequency was appreciably higher in heterozygotes with large allele span (43), indicating that the repeat structure could have some contributions to the STR mutation process.
Base composition of repeat unit
Sequences of the unit with a high AT content mutate faster than those with a high GC content (19), suggesting that the template stability could influence the mutation rate. Perhaps the sequences with high GC content could reduce the frequency of strand-slippage events.
Flanking sequence
Glenn et al. observed a significant negative correlation between allelic diversity and GC content of flanking sequences (44). However, others did not agree with it (45). Further studies are required to find out the true role of GC content of STR flanking sequences in the STR mutation process.
Recombination
The published research results were controversial on whether recombination is associated with STR mutation. Some detected a correlation, while others found no correlation. In humans, genome-wide analyses provided no evidence for a strong positive correlation between recombination rate and STR mutation 17., 46.. The STR loci from a non-recombining region of the human Y chromosome display the same mutation rate to that of autosomal loci, suggesting that recombination is not the predominant mechanism generating STR variability.
Sex
The mutation rates at most loci in germ cells are higher in males than in females (47). It is widely accepted that sperms undergo more DNA replication cycles than eggs. The more replication cycles a cell experiences, the higher frequency of mutation is.
Age
The mean age of the men who carry STR mutations is significantly greater than that of the men lacking of these mutations 18., 38.. This is probably because sperms experience more mitoses and thus have higher mutation possibilities. A sperm of men undergoes about 380 and 540 mitoses by the age of 28 and 35, respectively. Therefore, in a way, the mutation rate of STRs depends on the age of men.
Interruptions in STR
A special mutation in STR is slippage, which induces insertion or deletion of one or several repeats. Besides, STRs also display “nonspecific” mutations, such as transitions/transversions, single-nucleotide insertions/deletions, and other events. Their frequency is low as compared to that of specific mutations, but they disrupt the STR nucleotide sequences and change their mutability. STRs can be stabilized by insertions of a different nucleotide composition.
STR Mutation and Diseases
The interest in STR mutation comes from the discovery that some trinucleotide repeats are involved in human neurodegenerative diseases. Trinucleotide repeat associated diseases are known to include many rare, dominant, and mainly neurological disorders, such as fragile X syndrome, Huntington’s disease, myotonic dystrophy, and certain types of spinocerebellar ataxia. To date, trinucleotide repeat associated diseases have only been identified in humans. This has led to the hypothesis that the presence of trinucleotide repeats within certain brain related genes may contribute to the evolution of brain function.
Trinucleotide repeat associated diseases are characterized by the trinucleotide repeats that expand far outside of their “normal” polymorphic ranges. Such trinucleotide repeats are usually inside genes, most of which encode clusters of glutamine residues; others, which reside outside the genes, are currently close enough to disrupt the genes’ functions. In general, disease severity often appears to correlate with the extent of abnormal expansion. For instance, the CGG repeat, which encodes runs of arginine, resides on the 5’ end in the fragile X mental retardation-I syndrome. Usually the number of repeats ranges from 6 to 46, with an average of 29. When the repeats overrun 52 times, the STR region will be unstable during meiosis, with a consequence of rapid expanding. The CGG repeat number in the carrier without any symptoms ranges from 60 to 200. However, the patient with obvious symptoms carries more than 230 repeats of CGG (48). Therefore, in order to better understand these human neurodegenerative diseases, it is important to understand the particular mutation process of STRs.
Application of STRs in Population Genetics
STRs have such properties as abundant, codominant, highly polymorphic, and nearly selectively neutral. Besides, STRs contain DNA fragments that are small enough to be amplified by polymerase chain reaction and separated in high-resolution media like polyacrylamide. With the availability of high-throughout capillary sequencers or mass spectrography, the sizing of alleles is no longer a bottleneck in STR analysis. Thus STRs are widely used in scientific and applied research.
STRs are extremely useful in applications such as the construction of genetic maps (49), gene location, genetic linkage analysis, identification of individuals, paternity testing, as well as disease diagnosis 50., 51.. STR analysis has also been employed in population genetics. Nevertheless, the application of STRs to population genetics requires a more detailed understanding of the STR mutation process.
We can apply STRs to reconstruct the history of migration and evolution of the species, as well as to assess biological diversity at various levels of biological organization (52). A method of absolute genetic dating uses mutation rates as molecular clocks. Such a molecular clock based on STR, whose mutation rate is very high, can be applied to human evolution. Therefore, STRs are likely to reflect relatively recent divergence (53).
The difference in size between two different STR alleles might be informative: the larger the difference, the more the number of mutation events. Thus there is a “memory” of past mutation events (54). That is, when a mutation occurs, the new mutant is related to the allele from which it was derived. In this case, the difference in length between alleles contains phylogenetic information (55). However, the prevalence of different mutational events may vary dramatically among groups. Ignoring the possibility that the same allelic type found in different individuals or populations may be derived from different evolutionary processes, it might lead to biased estimates of genetic structure. Consequently, it is very important to know the mutation process of STRs in detail before they are applied to population genetics studies.
Mutation models for the evolutionary process of STRs are needed in order to estimate phylogenetic relationships, population differentiation measures, and genetic distances from STR data. Different kinds of estimators based on IAM have been developed, such as DAS (shared allele distance), DCH (Cavalli-Sforza and Edwards chord distance), and DS (Nei’s standard genetic distance). On SMM/TPM, estimators include (δμ)2, DSW (stepwise weighted genetic distance), and RST. Different estimators can be effective in different situations. Goldstein et al. concluded that for a relatively short period of time, DAS or DS is a better measure, but as time increases, the estimator based on SMM such as (δμ)2 becomes superior (55).
In 1995, Goldstein et al. predicted that STR loci would ultimately allow a high-resolution description of the human evolutionary history (56). Many researchers have studied the history of human evolution and migration by using STR loci 57., 58., 59., 60.. Mountain et al. developed a new combination polymorphism, namely SNPSTRs (61), in which each such segment includes one or more single nucleotide polymorphisms (SNPs) and exactly one STR locus, providing insights into population history. At present, STR loci are employed to reveal the relationship of populations in different regions, as well as the route of migration of ancient peoples.
Perspective
With the development of the third generation genetic markers, SNP will replace STR for some applications like genome mapping. However, a comprehensive understanding of STR mutation and its high informative characteristics will increase the application of STR analysis in many more fields of science.
References
- 1.Tautz D. Notes on the defunction and nomenclature of tandemly repetitive DNA sequences. In: Pena S.D.J., editor. DNA Fingerprinting: State of the Science. Birkhauser Verlag; Basel, Switzerland: 1993. pp. 21–28. [Google Scholar]
- 2.Koreth J. Microsatellites and PCR genomic analysis. J. Pathol. 1996;178:239–248. doi: 10.1002/(SICI)1096-9896(199603)178:3<239::AID-PATH506>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 3.Ellegren H. Heterogeneous mutation processes in human microsatellite DNA sequences. Nat. Genet. 2000;24:400–402. doi: 10.1038/74249. [DOI] [PubMed] [Google Scholar]
- 4.Subramanian S. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Nadir E. Microsatellite spreading in the human genome: evolutionary mechanisms and structural implications. Proc. Natl. Acad. Sci. USA. 1996;93:6470–6475. doi: 10.1073/pnas.93.13.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urquhart A. Variation in short tandem repeat sequences: a survey of twelve microsatellite locifor use as forensic identification markers. Int. J. Legal Med. 1994;107:13–20. doi: 10.1007/BF01247268. [DOI] [PubMed] [Google Scholar]
- 8.Escher D. Conservation of glutamine-rich transactivation function between yeast and humans. Mol. Cell. Biol. 2000;20:2774–2782. doi: 10.1128/mcb.20.8.2774-2782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gebhardt F. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J. Biol. Chem. 1999;274:13176–13180. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- 10.Meloni R. A tetranucleotide polymorphic microsatellite, located in the first intron of the tyrosine hydroxylase gene, acts as a transcription regulatory element in vitro. Hum. Mol. Genet. 1998;3:423–428. doi: 10.1093/hmg/7.3.423. [DOI] [PubMed] [Google Scholar]
- 11.Contente A. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat. Genet. 2002;30:315–320. doi: 10.1038/ng836. [DOI] [PubMed] [Google Scholar]
- 12.Hamada H. Enhanced gene expression by the poly(dT-dG).poly(dC-dA) sequence. Mol. Cell. Biol. 1984;12:2622–2630. doi: 10.1128/mcb.4.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naylor L.H., Clark E.M. d(TG)n.d(CA)n sequences upstream of the rat prolactin gene form Z-DNA and inhibit gene transcription. Nucleic Acids Res. 1990;18:1595–1601. doi: 10.1093/nar/18.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahls W.P. The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol. Cell. Biol. 1990;10:785–793. doi: 10.1128/mcb.10.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y.H., Griffin J. Expanded CTG triplet blocks from myotonic dystrophy gene create the strongest known natural nucleosome positioning elements. Genomics. 1995;25:570–573. doi: 10.1016/0888-7543(95)80061-p. [DOI] [PubMed] [Google Scholar]
- 16.Heale S.M., Petes T.D. The stabilization of repetitive tracts of DNA by variant repeats requires a functional DNA mismatch repair system. Cell. 1995;83:539–545. doi: 10.1016/0092-8674(95)90093-4. [DOI] [PubMed] [Google Scholar]
- 17.Huang Q.Y. Mutation patterns at dinucleotide microsatellite loci in humans. Am. J. Hum. Genet. 2002;70:625–634. doi: 10.1086/338997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayser M. Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am. J. Hum. Genet. 2000;66:1580–1588. doi: 10.1086/302905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakraborty R. Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc. Natl. Acad. Sci. USA. 1997;94:1041–1046. doi: 10.1073/pnas.94.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Rienzo A. Heterogeneity of microsatellite mutations within and between loci, and implications for human demographic histories. Genetics. 1998;148:1269–1284. doi: 10.1093/genetics/148.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hastbacka J. Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nat. Genet. 1992;2:204–211. doi: 10.1038/ng1192-204. [DOI] [PubMed] [Google Scholar]
- 22.Holtkemper U. Mutation rates at two human Y-chromosomal microsatellite loci using small pool PCR techniques. Hum. Mol. Genet. 2001;10:629–633. doi: 10.1093/hmg/10.6.629. [DOI] [PubMed] [Google Scholar]
- 23.Kornberg A. Enzymatic Synthesis of deoxyribonucleic Acid. XVI. Oligonucleotides as Templates and the Mechanisms of Their replication. Proc. Natl. Acad. Sci. USA. 1964;51:315–323. doi: 10.1073/pnas.51.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jobling M.A. The diversity of the human genome. In: Jobling M.A., editor. Human Evolutionary Genetics: Origins, Peoples & Disease. Garland Science; New York, USA: 2004. p. 67. [Google Scholar]
- 25.Schlötterer C., Tautz D. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 1992;20:211–215. doi: 10.1093/nar/20.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strand M. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 27.Kruglyak S. Equilibrium Distributions of Microsatellite Repeat Length resulting from a Balance between Slippage Events and Point Mutations. Proc. Natl. Acad. Sci. USA. 1998;95:10774–10778. doi: 10.1073/pnas.95.18.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlötterer C. Genome evolution: are microsatellites really simple sequences? Curr. Biol. 1998;8:132–134. doi: 10.1016/s0960-9822(98)70989-3. [DOI] [PubMed] [Google Scholar]
- 29.Lai Y., Sun F. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol. Biol. Evol. 2003;20:2123–2131. doi: 10.1093/molbev/msg228. [DOI] [PubMed] [Google Scholar]
- 30.Rose O., Falush D. A threshold size for microsatellite expansion. Mol. Biol. Evol. 1998;15:613–615. doi: 10.1093/oxfordjournals.molbev.a025964. [DOI] [PubMed] [Google Scholar]
- 31.Messier W. The birth of microsatellites. Nature. 1996;381:483. doi: 10.1038/381483a0. [DOI] [PubMed] [Google Scholar]
- 32.Kimura M., Crow J.F. The number of alleles that can be maintained in a finite population. Genetics. 1964;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ota T., Kimura M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet. Res. 1973;22:201–204. doi: 10.1017/s0016672300012994. [DOI] [PubMed] [Google Scholar]
- 34.Shriver M.D. VNTR allele frequency distributions under the stepwise mutation model: a computer simulation approach. Genetics. 1993;134:983–993. doi: 10.1093/genetics/134.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Rienzo A. Mutational processes of simple sequence repeat loci in human populations. Proc. Natl. Acad. Sci. USA. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garza J.C., Freimer N.B. Homoplasy for size at microsatellite loci in humans and chimpanzees. Genome Res. 1996;6:211–217. doi: 10.1101/gr.6.3.211. [DOI] [PubMed] [Google Scholar]
- 37.Sajantila A. Experimentally observed germline mutations at human micro- and minisatellite loci. Eur. J. Hum. Genet. 1999;7:263–266. doi: 10.1038/sj.ejhg.5200257. [DOI] [PubMed] [Google Scholar]
- 38.Brinkmann B. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am. J. Hum. Genet. 1998;62:1408–1415. doi: 10.1086/301869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldstein D.B., Clark A.G. Microsatellite variation in North American populations of Drosophila melanogaster. Nucleic Acids Res. 1995;23:3882–3886. doi: 10.1093/nar/23.19.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neef B.D., Gross M.R. Microsatellite evolution in vertebrates: inference from AC dinucleotide repeats. Evolution. 2001;55:1717–1733. doi: 10.1111/j.0014-3820.2001.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 41.Lee J.S. Relative stabilities of dinucleotide and tetranucleotide repeats in cultured mammalian cells. Hum. Mol. Genet. 1999;8:2567–2572. doi: 10.1093/hmg/8.13.2567. [DOI] [PubMed] [Google Scholar]
- 42.Zhivotovsky L.A. Biased mutations and microsatellite variation. Mol. Biol. Evol. 1997;14:926–933. doi: 10.1093/oxfordjournals.molbev.a025835. [DOI] [PubMed] [Google Scholar]
- 43.Amos W. Microsatellites show mutational bias and heterozygote instability. Nat. Genet. 1996;13:390–391. doi: 10.1038/ng0896-390. [DOI] [PubMed] [Google Scholar]
- 44.Glenn T.C. Allelic diversity in alligator microsatellite loci is negatively correlated with GC content of flanking sequences and evolutionary conservation of PCR amplifiability. Mol. Biol. Evol. 1996;13:1151–1154. doi: 10.1093/oxfordjournals.molbev.a025678. [DOI] [PubMed] [Google Scholar]
- 45.Balloux F. Microsatellite conservation, polymorphism, and GC content in shrews of the genus Sorex (Insectivora, Mammalia) Mol. Biol. Evol. 1998;15:473–475. doi: 10.1093/oxfordjournals.molbev.a025945. [DOI] [PubMed] [Google Scholar]
- 46.Payseur B.A., Nachman M.W. Microsatellite variation and recombination rate in the human genome. Genetics. 2000;156:1285–1298. doi: 10.1093/genetics/156.3.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimmin L.C. Male-driven evolution of DNA sequences. Nature. 1993;362:745–747. doi: 10.1038/362745a0. [DOI] [PubMed] [Google Scholar]
- 48.Liu Q.Z. Chromosomal diseases. In: Li P., editor. Medical Genetics. second edition. Peking Union Medical College Press; Beijing, China: 2004. pp. 153–155. [Google Scholar]
- 49.Dib C. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 50.Bowcock A. High Resolution of human evolutionary Trees with polymorphic microsatellites. Nature. 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 51.Hammond H.A. Evaluation of 13 short tandem repeat loci for use in personal identification applications. Am. J. Hum. Genet. 1994;55:175–189. [PMC free article] [PubMed] [Google Scholar]
- 52.Edwards A. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics. 1992;12:241–253. doi: 10.1016/0888-7543(92)90371-x. [DOI] [PubMed] [Google Scholar]
- 53.Romualdi C. Patterns of human diversity, within and among continents, inferred from biallelic DNA polymorphisms. Genome Res. 2002;12:602–612. doi: 10.1101/gr.214902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardy O.J. Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics. 2002;163:1467–1482. doi: 10.1093/genetics/163.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldstein D.B. An evaluation of genetic distances for use with microsatellite loci. Genetics. 1995;139:463–471. doi: 10.1093/genetics/139.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldstein D.B. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc. Natl. Acad. Sci. USA. 1995;92:6723–6727. doi: 10.1073/pnas.92.15.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nei M., Takezaki N. The root of the phylogenetic tree of human populations. Mol. Biol. Evol. 1996;13:170–177. doi: 10.1093/oxfordjournals.molbev.a025553. [DOI] [PubMed] [Google Scholar]
- 58.Calafell F. Short tandem repeat polymorphism evolution in humans. Eur. J. Hum. Genet. 1998;6:38–40. doi: 10.1038/sj.ejhg.5200151. [DOI] [PubMed] [Google Scholar]
- 59.Jorde L.B. The distribution of human genetic diversity: a comparison of mitochondrial, autosomal, and Y-chromosome data. Am. J. Hum. Genet. 2000;66:979–988. doi: 10.1086/302825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenberg N.A. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 61.Mountain J.L. SNPSTRs: empirically derived, rapidly typed, autosomal haplotypes for inference of population history and mutational processes. Genome Res. 2002;12:1766–1772. doi: 10.1101/gr.238602. [DOI] [PMC free article] [PubMed] [Google Scholar]