Abstract
It is believed that in the RNA world the operational (ribozymes) and the informational (riboscripts) RNA molecules were created with only three (adenosine, uridine, and guanosine) and two (adenosine and uridine) nucleosides, respectively, so that the genetic code started uncomplicated. Ribozymes subsequently evolved to be able to cut and paste themselves and riboscripts were acceptive to rigorous editing (adenosine to inosine); the intensive diversification of RNA molecules shaped novel cellular machineries that are capable of polymerizing amino acids—a new type of cellular building materials for life. Initially, the genetic code, encoding seven amino acids, was created only to distinguish purine and pyrimidine; it was later expanded in a stepwise way to encode 12, 15, and 20 amino acids through the relief of guanine from its roles as operational signals and through the recruitment of cytosine. Therefore, the maturation of the genetic code also coincided with (1) the departure of aminoacyl-tRNA synthetases (AARSs) from the primordial translation machinery, (2) the replacement of informational RNA by DNA, and (3) the co-evolution of AARSs and their cognate tRNAs. This model predicts gradual replacements of RNA-made molecular mechanisms, cellular processes by proteins, and informational exploitation by DNA.
Key words: genetic code, codon, aminoacyl-tRNA synthase, GC content
Introduction
The evolution of artificial codes depends upon human intelligence (1), whereas the genetic code is believed to evolve through very lengthy and very ancient selection processes that began in the RNA world (2) and subsequently optimized and maturated in the modern world after DNA finally replaced one of RNA’s major roles—bearing and passing on the genetic information in a robust way. The birth of life as its primordial form—RNA—was proposed to take place about 3.5 billion years ago around a time window of a few hundred million years 3., 4., 5.. Although it is impossible to reconstruct real cellular processes of the two early yet brilliant transitions of life: from the RNA life first to the RNA–protein life and then to the RNA–protein–DNA life (6), a description of plausible scenarios for the processes is of importance in understanding stepwise creations of many molecular mechanisms and their basic machineries. In this short paper, we attempt to propose a theoretical framework for such transitions to better understand their impact on the maturity of the genetic code. This proposition is certainly not free of loopholes but should be able to stimulate further contemplation and imagination. Whether a model becomes popular or not relies entirely on its predicting power and its insights into molecular details yet to be revealed.
Model
The RNA world and its early code
The evolution of the genetic code began in the early phase of the RNA world where RNA molecules started to be built as simple nucleotide repeats or polymers. These de novo-synthesized polymers had to survive somehow for millions and millions of years in order to allow life to get started with structurally and functionally divergent RNA molecules that provide complexity and perform sufficient functions. Although template-directed synthesis might not be initially necessary since protocells certainly had to fight for life’s “seed components” among themselves, these RNA molecules could either be cut and pasted at the molecular level or be chemically modified to turn into other similar structures at the building-block level for structural and functional diversities. RNA editing was obviously a molecular mechanism as part of the RNA polymerization machinery aside from splicing. Once engulfing wars among the protocells started going, RNA molecules and their complexes had to be consistently synthesized, chemically modified, spliced, and assembled into two essential classes, operational and informational. Up to a point, template-directed synthesis of RNAs might have exhibited advantages over simple undirected polymerization. The operational RNAs or ribozymes resembled the modern proteinaceous molecules and their complexes, whereas the informational RNAs or riboscripts were functionally equivalent to messenger RNAs in the contemporary biological world (CB world). In the CB world, the latter is called RNA splicing, which is either catalyzed by a “-some” (usually a complex formed by proteins and RNA as well as DNA sometimes; Table 1)—the spliceosome—or self-spliced. Of course, we have made here a bold assumption that life may start as a prototype of eukaryotic organism rather than prokaryote-like before recruitment of DNA, and eukaryotes are known to have preserved some of the critical molecular mechanisms such as RNA splicing through the spliceosomal pathway and complex organelles generated from intermediates of engulfing each other.
Table 1.
Basic cellular machinery for the RNA world and the CB world
| Cellular machinery | Phase | World and function | Substrate * |
||
|---|---|---|---|---|---|
| R | P | D | |||
| Editosome | I | The RNA world I: RNA synthesis and editing | + | ||
| Spliceosome | I | The RNA world I: RNA splicing | + | ||
| Translatosome | II | The RNA world II: RNA–peptide and protein synthesis | + | + | |
| Reverse-transcriptosome | III | The CB world: RNA-based DNA synthesis | + | + | |
| Transcriptosome | III | The CB world: DNA-based RNA synthesis | + | + | |
| Replisome | III | The CB world: DNA replication | + | + | |
| Repairosome | III | The CB world: DNA repair | + | + | |
R, P, and D stand for RNA, protein, and DNA, respectively. The “+” signs indicate the presence of a particular molecular mechanism and its corresponding substrates.
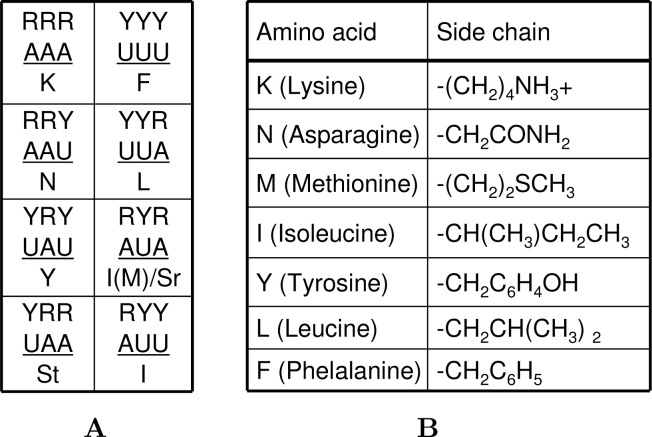
The primitive genetic code would not be considered necessary until early versions of the RNA-built translatosome were invented, which made primitive life forms leap into the late phase (Phase II) of the RNA world—the RNA–protein life (Table 1). Once requisite polypeptides were synthesized according to a ciphertext, genetic codes came into the play. If we assume that the early life forms and their shared genetic code did not use cytidine (C) before the involvement of DNA, since it seems not stable enough to join primitive organisms 7., 8., the first set of codons was simple and purine-sensitive at the third codon position (cp3) 9., 10.. The codons were mostly made of adenosine (A) and uridine (U), formed by a binary code that only distinguished purine from pyrimidine (Figure 1A). If we assume that the modern code became universal in life’s early history or inherited the RNA code with faith, it encoded possibly seven amino acids (here we assume isoleucine and methionine are exchangeable and functionally equivalent; both are capable of starting peptide synthesis) as well as possessed both start and stop signals. These amino acids have rather impressive physicochemically diversified side chains, albeit relatively devoid of small and acidic amino acids (Figure 1B).
Fig. 1.
The R–Y (A–U) code (A) and its encoded amino acids (B). R and Y stand for purine and pyrimidine, respectively. Start and stop codons are indicated with Sr and St, respectively. RYR or AUA is assumed as start codon rather than encodes isoleucine since RYY or AUU already does so.
Since primitive translatosomes were made to be simple, there was a possibility that the first aminoacyl-tRNA synthetases (AARSs) might have started as a permanent part of this protein-manufacturing machinery and fell off from it, together with tRNAs, as the genetic code forged ahead for creating peptide complexity. The first batch of RNA-encoded proteins was mostly protective for integrity of primordial cells and their cellular components, and undoubtedly included those for RNA binding and membrane stability, constituted by basic, aromatic, and hydrophobic amino acids. The first division of AARSs was predicted to ensure protein diversity so that they must distinguish the two polar amino acids, asparagine and tyrosine, as well as the two aromatic amino acids, phenylalanine and tyrosine. In contrast, it might not be necessary to tell leucine, isoleucine, and methionine apart.
The first expansion of the early code
The expansion of the early code relies on the recruitment of new building blocks. There are at least two possible scenarios: one concerns the limited recruitment of guanine (G) and the other assumes editing mechanisms that convert adenosine (A) to inosine (I). Both scenarios should be able to provide significant structural diversity and coding capacity for ribozymes and riboscripts, respectively. Base or nucleoside conversions between the two purine-containing nucleosides—A and I—as well as between the two pyrimidine-containing nucleosides—U and C, have been carried over to the CB world. Inosine is capable of forming double hydrogen bonds with U, G, and C. Although the two scenarios may not be mutually exclusive, that is, they might have evolved independently or co-existed, we discuss them separately just for simplicity.
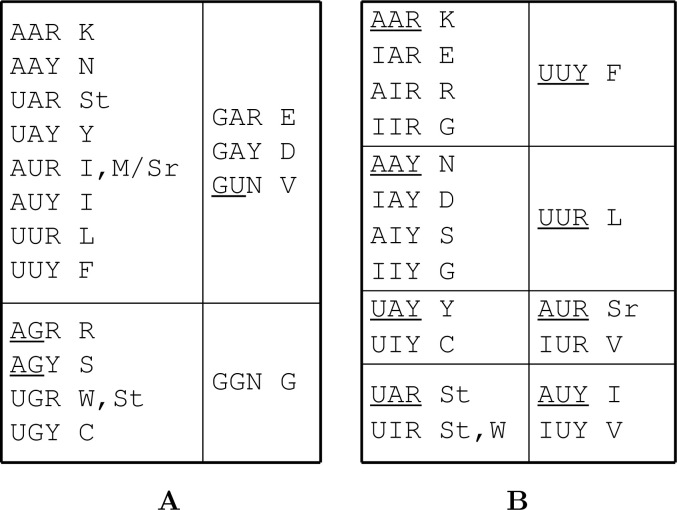
In one scenario, we assume that G was recruited by riboscripts in a limited way in addition to serving as a divergent building block and processing signals for ribozymes (Figure 2A). Although a ribozyme without G and C was proven functional (11), structural and functional diversities provide advantages for life forms to compete for survival. Since dinucleotides AG and GU are designated as signals for splice sites, the expansion of the codons in this scenario might be limited to tryptophane, glutamic acid, aspartic acid, cysteine, and glycine. These five new recruits are very impressive: the largest (tryptophane), the negatively charged (glutamic acid and aspartic acid), the disulfide-bond-forming (cysteine), and the smallest (glycine) amino acids.
Fig. 2.
The extended early code in two scenarios: (A) incorporation of G but avoiding AG and GU (both dinucleotides were used as splicing signals) and (B) extended through base editing from A to I. The codons overlapping with splice signals and the original codons are underlined.
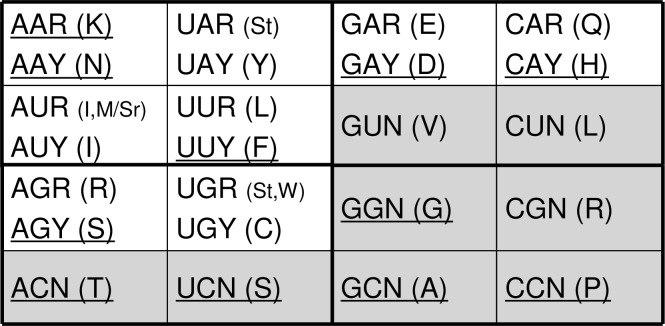
In the other scenario, we assume that A was selectively and constantly edited into I in riboscripts in a context of A and I co-existence, so codons were extended to match more AA-tRNAs. The result of this extension became identical to the first scenario (Figure 2B). This scenario is strongly supported by the distribution of AARS classes (Figure 3) as the expansion of amino acids and their corresponding AARSs follow the class rule largely (12). In addition, similar roles of nucleotide modifications have been inherited by all the extant life forms, such as wobble pairing between anticodons of tRNA molecules and codons of mRNA (13). For instance, AAY (N) and its “sibling codons”, IAY (D), AIY (S), and IIY (G), share the same class of AARSs. The K group (AAR, AIR, IAR, and IIR) has a little complication, as there are two Lys-RSs belonging to classes I and II. Correspondingly, lysine’s “sibling codons” can certainly go with class I (Glu-RS and Arg-RS) except glycine that was defined by its Y-ending codons. An alternative explanation is that Gly-RS may have an unusual history since its active form is a unique tetramer. The consensus of the two scenarios suggests an early-expanded genetic code that encodes twelve amino acids other than start and stop codons.
Fig. 3.
The organization of the genetic code and AARSs. The code is divided into two halves, pro-diversity (unshaded area) and pro-robustness (shaded area), according to sensitivity of the codons to purine (AG) content changes. AARSs are also divided into two types and the Type II enzymes are underlined. There are both types of AARSs for lysine although it is underlined in this figure. After C was recruited as an essential building block, the code was extended to include more amino acids with its C-containing codons. The rule of extension for AARSs followed a G–C conversion trend except the six-fold codons (L, R, and S). For instance, the pairs, such as CAR and GAR, CAY and GAY, GCN and GGN, share the same class of AARSs.
The second expansion of the genetic code
The second recruitment of the early genetic code has to be arginine, serine, and valine after dinucleotides GU and AG were finally freed from serving as sequences of splice sites since spliceosomes became more sophisticated. The new addition that makes a set of fifteen amino acids was a subtle extension of the existing amino acids considering both the physicochemical property and the secondary structure: arginine was an alternative of lysine; serine was a smaller version of tyrosine; and valine added another variation to the hydrophobic amino acids—leucine, isoleucine, methionine, and phenylalanine 14., 15., 16., 17..
The most puzzling feature of the code is its unusual redundancy where only three amino acids, arginine, leucine, and serine, are encoded with six codons; they by now have all been recruited and later expanded to acquire their quadruplets when cytosine joined the genetic code. Let us first make a few observations on leucine in comparison to the other two amino acids. First, although they are all among the most abundant amino acids in the extant genomes, leucine is always the most abundant in all three kingdoms of modern life forms. Serine comes to the second among eukaryotic genomes, such as in the human and Arabidopsis genomes. Arginine is the least abundant among the three, which barely makes it to the top ten among some of the bacterial genomes. Second, leucine has the easiest codon conversion between the doublet and the quadruplet among all three amino acids: a simple base transition between U and C results in a change from UUR to CUR. This suggests that leucine is capable of playing essential structural roles for most proteins and maintains their integrity when GC content increases. Similarly, to keep arginine and serine unchanged, transversions have to be introduced; a single transversion has to take place to change AGR to CGR for arginine, and double transversions, AGY to UCY for serine, are indispensable. Their changes are not as easy as what is seen for leucine. Third, leucine has dimensions most similar to four other amino acids with side chains that have rather diverse physicochemical properties from it: isoleucine, histidine, methionine, and lysine 14., 15.. All three observations support the notion that leucine should be the most abundant amino acids for all major life forms. By the same token, serine ranks the second. It has two counterparts, threonine and tyrosine. Serine differs from leucine and arginine in forming protein secondary structures; it prefers turns as compared to leucine that favors alpha-helix and arginine that is rather neutral to all three major secondary structures. Arginine also has two counterparts, histidine and lysine. It is unique in forming protein secondary structures—the only amino acid that is indiscriminately honored by alpha-helix, beta-sheet, and turn. These observations lead to a hypothesis that the additional codons for these three amino acids were tailored to balance the abundant amino acids when DNA nucleotide composition changes, such as GC content or AG (purine) content increases. The corresponding codons are organized in such a way that they balance between pro-diversity and pro-robustness halves of the genetic code 9., 10.. The result of such a balancing power is the stability of amino acid composition and its subtle effect on protein conformations when mutations bombard the coding sequence over evolutionary time scales. By now, the genetic code is good enough for directing protein synthesis, and the sophistication of proteinaceous cellular machineries have made life more diverged, robust, and complex.
The final expansion of the genetic code
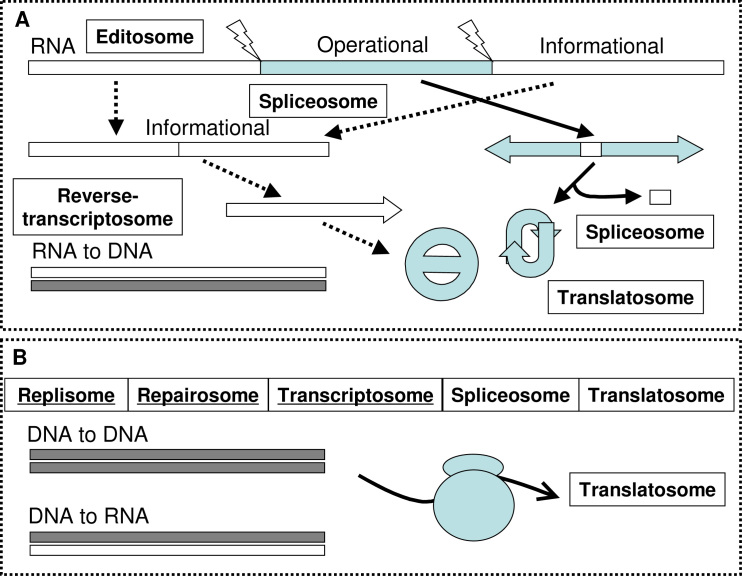
The final or the third recruitment of the code had to happen when DNA replaced RNA as the informational molecule for better precision and stability. It was the invention of the most critical cellular mechanism—reverse-transcription—that made this a reality, and the template-directed DNA replication marked the beginning of the new world. The evolution of many new cellular mechanisms, such as DNA replication, repair, and DNA-directed transcription, made the new world having achieved its perfection almost immediately (Figure 4). The contemporary genetic code was born and fixed after cytosine and its deoxyl derivative joined in as one of the four building blocks for RNA and DNA, respectively.
Fig. 4.
Hypothetical schemes illustrating how RNAs were spliced into informational and operational molecules in the RNA world with the involvement of cellular machineries, such as spliceosome and reverse-transcriptosome (A). In the DNA–RNA–protein world, replisome and repairosome were created for managing DNA processes (B).
The code had to be filled up with new recruits as the coding capacity increased. Histamine and glutamine filled in instantly due to their contributions to catalytic properties and similarities to the two existing basic amino acids, respectively. Threonine extended the function of serine but added subtlety in protein structures. Alanine has almost identical size and volume parameters as serine but is hydrophobic 14., 15.. This new recruit plays a very crucial role in protein structure and function diversity: swapping between a hydroxyl group with hydrophilic property and hydrophobic side chain if the size change is tolerable for essential functions of a protein. Proline is undoubtedly the last addition. On the one hand, it distorts the protein backbone in a unique way that no other amino acid does; on the other hand, it fits in with its hydrophobicity and modest size, resulting minimal changes when replacing other amino acids, such as aspartic acid, glutamine, and threonine.
The corresponding expansion pattern in AARS classes also supports the simple extension hypothesis. Aside from the six-fold degenerate codons, there are six sets of codons involved in the final expansion, which encode six amino acids. They are all in the same class of AARSs as those of the closest (or neighboring) G-containing or I-pairing codons. For instance, AARSs for two doublet-encoding amino acids, histidine (CAR) and glutamine (CAY), are the same as those for glutamic acid (GAR) and aspartic acid (GAY), respectively. The rest are CCN to GGN as well as ACN to GCN/GGN.
Evolution has certainly been involved in shaping up the genetic code. First, it shaped up the code through a long creation and optimization process so that the code finally adapted to a format that minimized the damage power of nucleotide changes on RNA in the RNA world or on both DNA and RNA in the CB world. Second, the code has organized in such a way that changes in DNA composition alter protein composition in a very distinct direction—from the AU-rich quarter to the GC-rich quarter, a shift emphasizing amino acids in favorite of either the catalytic moiety or the structural moiety, respectively. Third, while minimizing damage through a well-organized code, evolution also took the advantage of sequence variation at the third codon position; variations of the transversion type (between R and Y) at this position alter the encoded amino acids. There are 15 amino acids (75%) in the pro-diversity half of the canonical genetic code, which are sensitive to transversional changes. Finally, the relic of the changing code has still been observed in yeast and some organellar genomes, involving especially amino acids with six-fold degenerate codes—arginine, leucine, and serine (18).
Evolution also worked on molecular mechanisms. Making multiple copies of RNA molecules must have been the first molecular mechanism invented in the RNA world. Since replication as a biological term is dedicated to describe the process of making copies of a DNA molecule, we have to invent another word for making RNA copies, namely editosome, which is capable of both replicating a RNA molecule and editing it to change its minor content individually. The second major molecular mechanism in the RNA world has to be the spliceosome that cut and paste RNA fragments. It remains active in the CB world. The third one is the translatosome that manufactures proteins directly; it marks the transition of a primitive RNA world to a mature RNA world where a transition to the modern world or the CB world started. The key contribution of proteins to this transition is the accuracy of physicochemical activities of active proteins such as enzymes and receptors. Another key molecular mechanism invented in the transition time was the reverse-transcriptosome. DNA was finally introduced to life by this protein–RNA complex, so did the CB world thereafter by the invention of replisome, repairosome, and transcriptosome; all of them are DNA-dependent. If we say the translatosome marked the ending of the RNA world, the reverse-transcriptosome declared the birth of the CB world where new inventions continued until the rest of the “-somes” were made to work (Table 1).
Evolution works on genes and their variants that are borne by individuals within a species. This is largely true for multicellular organisms but not true for most of the unicellular organisms, especially prokaryotes where horizontal gene transfer is a major cellular process for exchanging genes and their variants. Individuals carry gene variations distinct from the rest of the same species and survive within a breeding population. Selection will only work on the variations of genes and DNA elements in germlines for multicellular organisms where they may result in advantage in survival for the variation-bearing individuals. Speciation depends on the degree and accumulation of such variations. Therefore, evolution starts from alterations of DNA sequences, filtered through the genetic code, reaches protein sequences, and the result is tested by fitness and survival at the individual level.
Exemplified predictions based on the codon expansion model
Whether a theoretical model becomes popular or not depends on its predicting power and subsequent validation of its predictions. Although it is extremely hard for a model that attempts to predict the almost unpredictable—what had happened in the RNA world, we can still make some of the most obvious predictions. We would like to give a few examples here. First, the codon expansion model predicts that some of the protein domains may be created with the early codons and their corresponding amino acids so they are transversion-sensitive at cp3. The idea can be extended to expect that most of the protein domains in DNA-related machineries may be built by the fully expanded codons so they were able to recruit the full set of amino acids for functional intricacies. However, it is very difficult to re-establish the initial composition of the assumed codon-biased domains since evolution has been taking its toll of altering them constantly for billions of years. Second, the model predicts that the splicing and editing machineries are invented earlier for building a viable ancestral life form so that the prokaryotes might have lost most of them, if not all. Since heavy compartmentalization, such as building organelles and nucleus, had to come after proteins replaced most of the operational RNAs, we believe that a true eukaryote might have been born from an eukaryote-like precursor rather than its function-striped forms—prokaryotes or prokaryote-like organisms. The final example is the prediction that certain groups of prokaryotes may keep significantly low GC content for maintaining a biased purine content, and these organisms should use more ancient protein domains in their proteomes dominated by purine-sensitive amino acids 19., 20., 21..
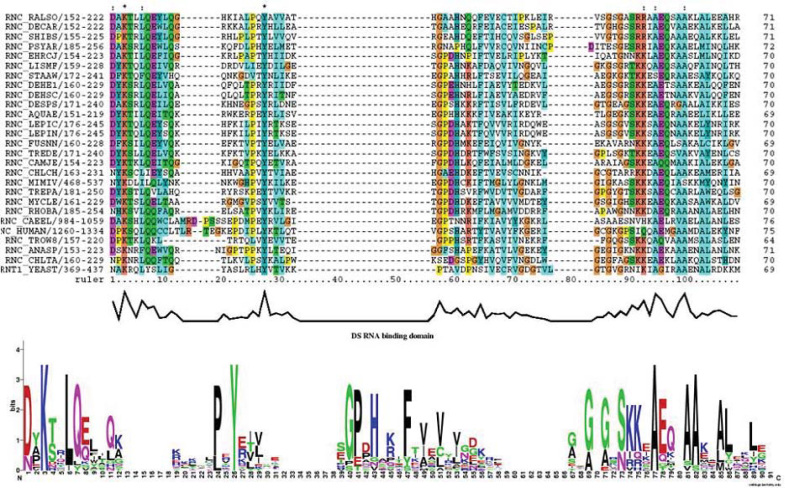
We did try to validate some of our predictions by examining some ancient proteins that are believed to be created in the RNA world. For instance, some of the RNA-binding proteins must be among the first to be invented for the protection of functional RNA molecules, including single-strand or double-strand binding proteins as well as their binding domains: the single-stranded RNA-binding domain (ssRBD) and double-stranded RNA-binding domain (dsRBD). Since evolution has done its job to check the essentiality of every amino acid for a given protein domain, we need only to align the sequences over a diverged panel and look for the decisive or highly conserved amino acids in the domain. Taking the dsRBD of ribonuclease 3 as an example, we demonstrate a two-parameter method to identify the most essential amino acids for the domain based on the physicochemical properties of amino acid side chains. The single parameters are simple physicochemical property measures, such as polarity, surface area, size, charge, hydrophobicity, and disulfide-linkage. The double parameters are various combinations of the single parameters, such as size–polarity and surface area–hydrophobicity. In the alignment of dsRBD with four subdomains from various ribonuclease 3 proteins, we can easily recognize a few amino acids that are either strictly conserved or less strictly conserved across wide taxonomic groups (Figure 5). Lysine is firmly restricted in both size and charge for RNA binding through electronic interaction. In contrast, aspartic acids (asparagine) and leucine (phenylalanine) in the subdomains are less conserved, perhaps only polarity and hydrophobicity are important for their RNA-binding functions, respectively, that is, they are restricted only by a single parameter. Tyrosine is another strictly conserved amino acid among the four subdomains; it is constraint by both shape and hydrophobicity, which are important factors for RNA binding through the π–π interaction (specific to aromatic amino acids). The highly conserved lysine and tyrosine in ribonuclease 3 RNA-binding domains suggest an early invention.
Fig. 5.
Multiple sequence alignments of the RNA-binding domain in ribonuclease 3. The sequences were retrieved from public databases from the top to the bottom as abbreviated names of the following: Pseudomonas solanacearum, Dechloromonas aromatic, Shigella boydii serotype 4, Psychrobacter arcticum, Ehrlichia canis, Listeria monocytogenes serotype 4b, Staphylococcus aureus, Dehalococcoides ethenogenes, Dehalococcoides sp., Desulfotalea psychrophila, Aquifex aeolicus, Leptospira interrogans, Fusobacterium nucleatum, Treponema denticola, Campylobacter jejuni, Chlorobium chlorochromatii, Mimivirus, Treponema pallidum, Mycobacterium leprae, Rhodopirellula baltica, Caenorhabditis elegans, human, Tropheryma whipplei, Anabaena sp., Chlamydia trachomatis, and yeast. Strictly conserved and less strictly conserved amino acids are indicated with stars and solid doubled dots, respectively.
Conclusion
Primordial life has been evolving from simple to complex as the genetic code expanded. A primordial code was composed of A and U rather than all four nucleotides—A, U, G, and C. Early in the RNA world, G served as one of the three essential building blocks of the operational RNA molecules but not part of the genetic code. If interactions among molecules started easy, these interactions should be less intimate, which leads to our second assumption for the first set of amino acids: they might be the larger and more diversified in physicochemical properties. Each of the new additions was added stepwisely with justification on subtle to significant alterations with minimal functional damage for proteins. As the molecular mechanisms evolved, the genetic code eventually became mature and fixed to a large extent in the CB world. We may never be able to prove the history and maturation process of the genetic code, but a meaningful scenario will stimulate our thoughts and give us a logical way to understand the possible arrangement of the genetic code. New ideas will come soon, agree or disagree with us, leading to an active forum for fruitful discussions on other scenarios on the origin of the genetic code.
Acknowledgements
This work was supported by the “100 Talents” grant from the Chinese Academy of Sciences awarded to JY.
References
- 1.Singh S. Anchor Books; New York, USA: 1999. The Code Book. [Google Scholar]
- 2.Gesteland R.F. second edition. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, USA: 1999. The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA. [Google Scholar]
- 3.Joyce G.F. The rise and fall of the RNA world. New Biol. 1991;3:399–407. [PubMed] [Google Scholar]
- 4.Joyce G.F. The antiquity of RNA-based evolution. Nature. 2002;418:214–221. doi: 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- 5.Orgel L.E. The origin of life—a review of facts and speculations. Trends Biochem. Sci. 1998;23:491–495. doi: 10.1016/s0968-0004(98)01300-0. [DOI] [PubMed] [Google Scholar]
- 6.Forterre P. The two ages of the RNA world, and the transition to the DNA world: a story of viruses and cells. Biochimie. 2005;87:793–803. doi: 10.1016/j.biochi.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Levy M., Miller S.L. The stability of the RNA bases: implications for the origin of life. Proc. Natl. Acad. Sci. USA. 1998;95:7933–7938. doi: 10.1073/pnas.95.14.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro R. Prebiotic cytosine synthesis: a critical analysis and implications for the origin of life. Proc. Natl. Acad. Sci. USA. 1999;96:4396–4401. doi: 10.1073/pnas.96.8.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J. A content-centric organization of the genetic code. Genomics Proteomics Bioinformatics. 2007;5:1–6. doi: 10.1016/S1672-0229(07)60008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu J. An evolutionary scenario for the origin of the genetic code. Communications of Chinese-American Chemical Society. 2007;2007(Fall):3–7. [Google Scholar]
- 11.Reader J.S., Joyce G.F. A ribozyme composed of only two different nucleotides. Nature. 2002;420:841–844. doi: 10.1038/nature01185. [DOI] [PubMed] [Google Scholar]
- 12.O’Donoghue P., Luthey-Schulten Z. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol. Mol. Biol. Rev. 2003;67:550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crick F.H. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 14.Chothia C. The nature of the accessible and buried surfaces in proteins. J. Mol. Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 15.Zamyatnin A.A. Protein volume in solution. Prog. Biophys. Mol. Biol. 1972;24:107–123. doi: 10.1016/0079-6107(72)90005-3. [DOI] [PubMed] [Google Scholar]
- 16.Chou P.Y., Fasman G.D. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 17.Chou P.Y., Fasman G.D. Empirical predictions of protein conformation. Annu. Rev. Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 18.Söll D., RajBhandary U.L. The genetic code—thawing the ‘frozen accident’. J. Biosci. 2006;31:459–463. doi: 10.1007/BF02705185. [DOI] [PubMed] [Google Scholar]
- 19.Hu J. Replication-associated purine asymmetry may contribute to strand-biased gene distribution. Genomics. 2007;90:186–194. doi: 10.1016/j.ygeno.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Hu J. Compositional dynamics of guanine and cytosine content in prokaryotic genomes. Res. Microbiol. 2007;158:363–370. doi: 10.1016/j.resmic.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Zhao X. GC content variability of eubacteria is governed by the pol III alpha subunit. Biochem. Biophys. Res. Commun. 2007;356:20–25. doi: 10.1016/j.bbrc.2007.02.109. [DOI] [PubMed] [Google Scholar]