Abstract
Plants are colonized by a diverse community of microorganisms, the plant microbiota, exhibiting a defined and conserved taxonomic structure. Niche separation based on spatial segregation and complementary adaptation strategies likely forms the basis for coexistence of the various microorganisms in the plant environment. To gain insights into organism-specific adaptations on a molecular level, we selected two exemplary community members of the core leaf microbiota and profiled their proteomes upon Arabidopsis phyllosphere colonization. The highly quantitative mass spectrometric technique SWATH MS was used and allowed for the analysis of over two thousand proteins spanning more than three orders of magnitude in abundance for each of the model strains. The data suggest that Sphingomonas melonis utilizes amino acids and hydrocarbon compounds during colonization of leaves whereas Methylobacterium extorquens relies on methanol metabolism in addition to oxalate metabolism, aerobic anoxygenic photosynthesis and alkanesulfonate utilization. Comparative genomic analyses indicates that utilization of oxalate and alkanesulfonates is widespread among leaf microbiota members whereas, aerobic anoxygenic photosynthesis is almost exclusively found in Methylobacteria. Despite the apparent niche separation between these two strains we also found a relatively small subset of proteins to be coregulated, indicating common mechanisms, underlying successful leaf colonization. Overall, our results reveal for two ubiquitous phyllosphere commensals species-specific adaptations to the host environment and provide evidence for niche separation within the plant microbiota.
Higher multicellular organisms live in close association with a remarkable diversity of microorganisms, the microbiota (1–4). The vast majority of community members are nonpathogenic commensal bacteria, but the host microbiota has been associated with beneficial traits ranging from disease susceptibility to host nutrition, inflammatory responses, host immunity and growth promotion (5–7). Rapid advances in next-generation sequencing technologies accelerated the detailed understanding of the phylogenetic composition of the microbiota of various organisms, including the economically highly relevant associations of microbes with terrestrial plants (8–13).
The phyllosphere environment comprises the above ground parts of plants and offers an excellent system to study ecological concepts of microbial communities, because gnotobiotic model systems are available and colonization as well as community structure can be linked to spatial information recorded at various scales (14, 15). The predominant component of the phyllosphere are leaves, which represent a harsh, oligotrophic and rapidly changing microbial environment that is exposed to the diurnal cycle and to extreme conditions, including UV radiation, frequent changes in nutrient and water availability as well as wide temperature gradients (13, 16). Nonetheless, a diverse community of microorganisms inhabits the different niches on leaves (10, 12, 17). Cultivation-independent studies revealed that mainly the four phyla Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria constitute the phyllosphere community of different host plants. In particular, the class Alphaproteobacteria predominates in abundance and some of the observed genera, e.g. Sphingomonas and Methylobacterium are conserved between phylogenetically distinct host plants and geographic regions (12, 13, 18, 19). For both genera, beneficial plant-microbe interactions have been reported, either conferring a protection against pathogen infection or an increase in biomass production to the plant (20–23). Unraveling molecular mechanisms of these plant-microbe interactions will be crucial, to develop and implement applications to further improve host fitness.
Several studies addressing microbial adaptation to the plant habitat indicated conserved strategies as well as species-specific mechanisms by which the phyllosphere inhabitants adapt to their niches and cope with the harsh conditions they encounter (13, 24–26). However, to explain coexistence and stable community formation of core community members a more profound understanding of their physiology as well as their interactions with the plant host will be required.
Earlier studies gave first insights into the physiology of different community members by combining metaproteomics with metagenomic shotgun sequencing of natural phyllosphere communities (10, 12). However, because proteome coverage per species was low in studies on complex natural communities, complementary approaches are required for an in-depth understanding of bacterial fitness traits on leaves. The recently developed Sequential Windowed Aquisition of all THeoretical fragment ions Mass Spectrometry (SWATH MS)1 technique allows for the reproducible quantification of thousands of proteins in a single run, overcoming technical limitations of previous studies and proteomic strategies and enabling systems level insights into each strain's physiology (27–29). In short, the SWATH MS approach generates a complete map of all detectable fragment ions derived from peptide precursors in a given sample using data independent acquisition (DIA). In a targeted analysis strategy, known m/z and retention time coordinates for each peptide (collected in a spectral library) are then used to extract quantitative signals for the peptides (and by inference, the corresponding proteins) of interest (27, 30, 31). Here, we applied SWATH MS to study proteomic changes of two commensal model strains of the core leaf microbiota, Methylobacterium extorquens PA1 and Sphingomonas melonis Fr1, upon plant colonization.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rational
For each bacterial strain and growth condition (see Fig. 1A), we generated three biological replicates. The samples on minimal media agar plates were all generated at the same time, whereas the plant samples were generated and measured at different times because of the large scale of the experiments. We chose three replicate experiments for each condition to be able to assess biological variation. The log-normalized transition group intensities (from OpenSWATH output) were approximately normally distributed and were used for all statistical tests. The statistical methods used at each step are described in detail in the corresponding paragraph describing that experimental/analysis procedure.
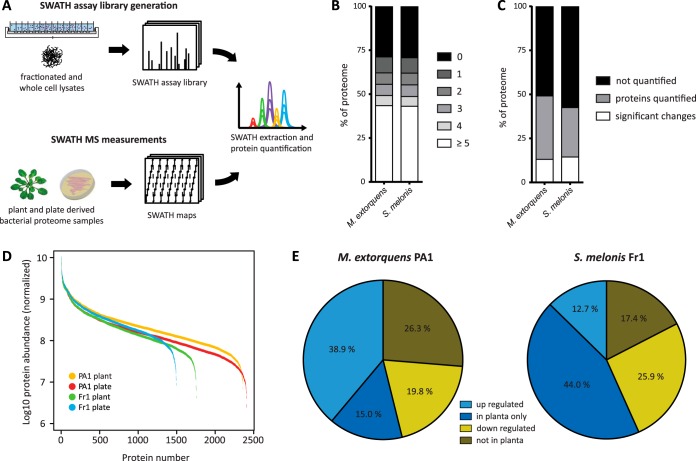
Fig. 1.
Experimental setup and characteristics of SWATH assay libraries and SWATH measurements. Comprehensive SWATH assay libraries were generated for M. extorquens and S. melonis based on various sample types and proteomic changes during growth on the leaf surface was subsequently studied by SWATH MS (A). SWATH assay library coverage of M. extorquens PA1 and S. melonis Fr1 (B). Gray shadings indicate the number of unambiguous peptide identifications per protein. Percentage of quantified and differentially regulated target proteins per strain (C). For (B) and (C), 100% corresponds to all annotated proteins within the genome. Dynamic quantification range of the two sample types analyzed by SWATH MS measurements of both strains (D). The abundance of all identified proteins was estimated using the best flyer approach (see Methods). Overview of differentially regulated proteins of M. extorquens and S. melonis during colonization of leaves or minimal media plates (E). 100% corresponds to the total number of differentially regulated proteins per strain.
Bacterial Strains and Growth Conditions
Methylobacterium extorquens PA1 (19) and Sphingomonas melonis Fr1 (21) were routinely grown in phosphate buffered minimal media (32) at pH 6.5 supplemented with 25 mm d-glucose and 125 mm methanol as carbon sources at 28 °C. For generation of SWATH assay libraries, strains were grown in liquid media in baffled flasks until early exponential, exponential and stationary growth phase. In addition, strains were grown on minimal media agar plates containing 1.5% agar at 22 °C in plant growth chambers (ATC26, Conviron, Winnipeg, Canada) exposed to the diurnal cycle with a photoperiod of 9 h per day.
Plant Growth Conditions and Inoculation of Phyllosphere Bacteria
Arabidopsis thaliana ecotype Col-0 plants were grown in microboxes (Combiness, Nazareth, Belgium) under gnotobiotic conditions as describes previously (21). Sterilized Lumox Film 25 (Sarstedt, Nümbrecht, Germany) containing eight holes were applied to the agar surface before sowing of surface sterilized seeds. The film prevents leaves touching the agar surface and therefore cross-contamination. For inoculation of phyllosphere bacteria ∼1 × 106 CFU were pipetted onto each seed immediately after sowing and microboxes were sealed with covers containing XXL filters for gas exchange. Plants were incubated in standard growth chambers (ATC26, Conviron) at 22 °C under long day conditions (16 h photoperiod) for 7 d before conditions were changed to short day conditions (9 h photoperiod) until harvest. All plants grew comparably to un-inoculated control plants, were harvested before flowering and did not show disease symptoms during the time course of the experiment. We did not detect bacterial growth on noninoculated control plants on minimal or complex media (nutrient broth (NB) without additional NaCl, pH 6.9, Sigma-Aldrich Chemie GmbH, Buchs, Switzerland).
Harvest of Plants and Recovery of Phyllosphere Bacteria
Plants were harvested after 28 d of incubation, above ground parts were carefully separated from roots using sterilized razor blades and plants of two microboxes were transferred into 50 ml Falcon tubes containing 25 ml ice cold TE-P buffer containing 10 mm Tris-HCl, 1 mm EDTA, 20% Percoll, 0.1% Silwett l-77, 0.02% Pefabloc SC (Roche Diagnostics, Rotkreuz, Switzerland) at pH 7.5. Phyllosphere bacteria were washed off leaves by three consecutive cycles of intense mixing and sonication before bacterial cells were separated from leaf material by filtration through a nylon mesh of 200 μm pore size (Spectrum Europe B.V., Breda, Netherlands). Cells were subsequently collected by centrifugation at 3220 × g for 10 min, transferred to 2 ml microcentrifuge tubes and washed twice with TE buffer, before cells of one experiment were pooled and stored at −80 °C until further processing.
Microscopy
Visualization of phyllosphere bacteria on cuticle tape lifts and infrared autofluorescence of anoxygenic photosynthesis was done as described previously (14). In brief, double-sided adhesive tape was glued onto microscopy slides and turgescent Arabidopsis leaves were flattened on the upper sticky layer using sterilized glass rods. Removal of the leaf material by tweezers results in a leaf imprint of the phylloplane, which was visualized by phase contrast microscopy using an AxioObserver D1 epifluorescence microscope (Carl Zeiss GmbH, Oberkochen, Germany) connected to a X-Cite 120Q (Lumen Dynamics Group, Mississauga, ON, Canada) light source. Infrared autofluorescence was visualized using a custom filter set consisting of a 320–650 nm excitation filter (BG39, Schott AG, Mainz, Germany), a 650 nm dichroic mirror (5650dcxru, Chroma, Bellows Falls, VT) and a 850 nm longpass emission filter (RG840, Schott AG). All pictures were acquired using an AxioCam Mrm (Carl Zeiss GmbH) and software AxioVision 4.8 (Carl Zeiss GmbH).
Preparation of Proteins Samples for MS
Bacterial cell pellets were dissolved in lysis buffer containing 100 mm ammonium bicarbonate, 8 m urea and 0.1% RapiGest (Waters Corporation, Milford, MA) (33) and bacterial cells were lysed by a combinatory approach of sonication (10 times for 10s, Intensity 50%, Mode 50, Vibra Cell, Sonics & Materials Inc., Danbury, CT) and bead beating (10 times for 30s, CapMix, 3 m ESPE AG, Seefeld, Germany) using 0.1 mm silica beads (BioSpec Products, Bartlesville, OK). Protein concentration of lysates was determined using the BCA assay kit according to the manufacturer's instructions (Thermo Fisher Scientific, Reinach, Switzerland). The buffer of all samples was subsequently exchanged using Zeba Spin Desalting Columns (7K MWCO, 5 ml, Thermo Fisher Scientific) and samples were concentrated to a final protein concentration of 1 mg/ml using Amicon Ultra centrifugal filters (0.5 ml, 3K MWCO, Merck Millipore Ltd., Schaffhausen, Switzerland). Protein disulfide bonds were reduced by addition of 5 mm tris(2-carboxylethyl)phosphine (TCEP) and incubation for 30 min at 37 °C, before cysteine residues were alkylated by adding 10 mm iodoacetamide (IAA) and incubation for 1 h in the dark at 25 °C. Samples were subsequently diluted 1:5 with freshly prepared 50 mm ammonium bicarbonate buffer to achieve urea concentrations below 2 m and sequencing grade modified trypsin (Promega AG, Dübendorf, Switzerland) was added at an enzyme to protein ratio of 1:100. The mixture was incubated over night at 37 °C shaking at 300 rpm for digestion (trypsin specifically cleaves C-terminally of lysine and arginine residues, unless followed by a proline). Subsequently, 50% trifluoroacetic acid (TFA) was added to an approximate concentration of 1% to reduce the pH value below 3 and stop the trypsin digest as well as to precipitate RapiGest. The solution was incubated at 37 °C shaking at 500 rpm for 30 min before insoluble particles were removed by centrifugation at 20,000 × g for 10 min. The supernatant was subsequently desalted using Sep-Pak Vac C18 (Waters Corporation, Milford, MA) reversed phase columns. At first, columns were activated using buffer A containing 80% acetonitrile (ACN) and 0.1% TFA and equilibrated with buffer B containing 2% ACN and 0.1% TFA. After loading of samples columns were washed five times using buffer B before samples were eluted by gravity flow with buffer C containing 50% ACN and 0.1% TFA. Eluted samples were dried under vacuum and re-solubilized in 2% ACN, 0.1% formic acid (FA) to a final concentration of 0.2–1.0 mg/ml. Three independent biological replicates were generated for each condition.
SWATH Assay Library Generation
A SWATH assay library was built for each organism separately. Liquid cultures at different growth stages (early exponential, mid-exponential and stationary phase) were harvested and peptide samples were prepared as described above. To increase library coverage, we then pooled and fractionated the peptide samples of the three different growth stages by isoelectric focusing as described before (33). Several of the resulting 24 fractions were pooled, resulting in 10 fractions. To avoid missing proteins specifically present on plant leaves, we included whole cell lysates obtained from cultures grown on leaves for the generation of the SWATH assay library. We performed the OGE fractionation only on the samples from liquid cultures, where biological material was not limiting and it was readily feasible to obtain the relatively large amounts needed for the fractionation approach. supplemental Fig. S1 shows the contribution of the respective samples to the combined library as well as the fraction of the libraries that contributed to the quantification of proteins by SWATH MS. However, we cannot exclude that the deeper coverage of proteins from liquid culture-derived bacteria compared with plant-derived bacteria in the library, because of OGE fractionation, may slightly favor the identification and quantification of proteins classified as “detectable on plate only” as compared with proteins classified as “detectable on plant only” in the SWATH MS measurements. To each of the samples, iRT peptides (RT-kit WR, Biognosys, AG, Schlieren, Switzerland) for retention time alignment of different LC-MS/MS runs were added (34). Mass spectra were acquired on a TripleTOF 5600 in data/information-dependent acquisition (IDA) mode: The TripleTOF 5600 mass spectrometer (AB Sciex, Concord, Canada) was coupled to a nanoLC 1Dplus system (Eksigent, Technologies, Dublin, CA) and the chromatographic separation of the peptides was performed on a 20-cm emitter (75 μm inner diameter, #PF360–75-10-N-5, New Objective, Inc., Woburn, MA) packed in-house with C18 resin (Magic C18 AQ 3 μm diameter, 200 Å pore size, Michrom BioResources, Inc., Auburn, CA). A linear gradient from 2–35% solvent B (98% ACN/0.1% FA) was run over 120 min at a flow rate of 300 nl/min. The mass spectrometer was operated in IDA mode with a 500 ms survey scan from which up to 20 ions exceeding 250 counts per second were isolated with a quadrupole resolution of 0.7 Da, using an exclusion window of 20 s. Rolling collision energy was used for fragmentation and an MS2 spectrum was recorded after an accumulation time of 150 ms.
Raw data files (wiff) were centroided and converted into mzML format using the AB Sciex converter (version 1.2, 111102 beta release) and subsequently converted into mzXML using openMS (version 1.9), followed by indexing with indexmzXML (part of TPP 4.3) (35). The converted data files were searched using the search engines X!Tandem (k-score, TPP 4.6.0) and OMSSA (version 2.1.9), against the corresponding protein sequence database of the 4829 annotated proteins of M. extorquens PA1 (http://www.ncbi.nlm.nih.gov/nuccore/NC_010172.1) or the 3857 annotated proteins of S. melonis Fr1 (IMG database; https://img.jgi.doe.gov/cgi-bin/m/main.cgi; Taxon ID 2517093015). Each protein sequence database contained the sequences of the 11 iRT peptides and for every target protein a corresponding decoy protein based on the reversed protein sequence. Tryptic or semi-tryptic peptides with up to one missed cleavages were allowed for the database search. The tolerated mass errors were 50 ppm on MS1 level and 0.05 Da on MS2 level. Carbamidomethylation of cysteines was defined as a fixed modification and methionine oxidation as a variable modification. The search results were processed with PeptideProphet (36) and iProphet (37) as part of the TPP 4.6.0 (35). To determine the iProphet cut-off corresponding to a 1% protein FDR, the software tool MAYU (38) was applied. The SWATH assay libraries were constructed from the iProphet results with an iProphet cut-off of 0.952195 for M. extorquens PA1 and 0.96029 for S. melonis Fr1, corresponding to a 1% FDR on protein level. The raw and consensus spectral libraries were built with SpectraST (version 4.0) (39, 40) using the -cICID_QTOF option for high resolution and high mass accuracy. Retention times were converted to iRT units using the retention time information of the spiked-in iRT peptides. The 6 most intense y and b fragment ions of charge state 1, 2 and 3 between 400 and 2000 m/z were extracted from the consensus spectral library using spectrast2tsv.py from msproteomicstools https://pypi.python.org/pypi/msproteomicstools). Fragment ions falling into the swath window of the precursor were excluded as the resulting signals are often highly interfered. Neutral loss fragment ions were included if they were among the 6 most intense fragment ions: −17 (NH3), −18 (H2O), −64 (typical for oxidized methionines). The library was converted into TraML format using the OpenMS tool ConvertTSVToTraML (development branch of OpenMS 1.10; commit 4caef80). Decoy transition groups were generated based on shuffled sequences (decoys similar to targets were excluded) by the OpenMS tool OpenSwathDecoyGenerator (development branch of OpenMS 1.10; commit 4caef80) and appended to the final SWATH library in TraML format. The data of the spectral libraries is summarized in Suppl. Data 1.
SWATH Data Acquisition
The TripleTOF 5600 mass spectrometer was set up as described above, but operated in SWATH mode (27) using the following parameters: For the liquid chromatography, the following two solvents, buffer A (2% acetonitrile and 0.1% formic acid in HPLC-grade water) and buffer B (2% water and 0.1% formic acid in acetonitrile) were used. A linear gradient from 2–35% solvent B, complemented to 100% with corresponding amounts of buffer A, was run over 120 min at a flow rate of 300 nl/min. Acquisition of a 100-ms survey scan was followed by acquisition of 32 fragment ion spectra from 32 precursor isolation windows (swaths) of 26 m/z each. The swaths were overlapping by 1 m/z and thus cover a range of 400–1200 m/z. The SWATH MS2 spectra were recorded with an accumulation time of 100 ms and cover 100–2000 m/z. The collision energy for each window was determined according to the calculation for a charge 2+ ion centered upon the window with a spread of 15.
SWATH Data Analysis with OpenSWATH
Raw SWATH data files (wiff) were converted into mzXML format using ProteoWizard (version 3.0.3316) (41) without centroiding. For the SWATH data analysis, the assay libraries of both strains were combined. The SWATH data was analyzed using OpenSWATH (version 29995b387c238fdc58b195b0390aadcb2b355aa6) (31) with the following parameters: Chromatograms were extracted with 50 ppm around the expected mass of the fragment ions and with an extraction window of ±5 min around the expected retention time after iRT alignment. The best model to separate true from false positives (per run) was determined by pyprophet (version 0.9.1) with 10 cross-validation runs (42). The pyprophet software re-implements the mProphet algorithm (43), which uses target and decoy signals to compute an optimal discriminant score which is then applied to all peak groups. Next, it estimates the false discovery rate (FDR) based on the Storey-Tibshirani method and computes a q-value for each peak group. The runs were subsequently aligned with the TRIC (TRansfer of Identification Confidence) algorithm which selected a corrected experiment-wide identification q-value cutoff of 0.00159 across the complete experiment to achieve 1% FDR for peptide identification. Next, TRIC used nonlinear retention time correction to align all runs and computed a narrow retention time window for each run where the analyte was expected to elute. If a suitable peak group was found inside the window, it was selected for quantification (a manually selected score cut-off of 0.05 was used for quantification) (Röst et al., unpublished). The openSWATH output data is summarized in Suppl. Data 2.
Relative quantification by ANOVA
The data matrix produced by the OpenSWATH pipeline was compared for each pair of conditions separately using the R programming language. First, peptide analytes detected in less than 3 LC-MS/MS for any condition were removed from the data analysis and the filtered data was normalized by its median. An analysis of variance (ANOVA) model was used for pairwise comparison of the conditions and computation of the effect size as well as significance. The resulting p values were corrected using the Benjamini-Hochberg correction. The results of the relative quantification and statistical analysis are summarized in Suppl. Data 3.
Protein Inference by aLFQ
To obtain a protein intensity value that correlates with its actual abundance, we ran the aLFQ R package (version 1.3.1) (44). Beforehand, the OpenSWATH output was filtered to contain only features below an mScore of 0.01 (1% FDR). The sum of the five most intense transitions per peptide averaged over the three most intense peptides per protein was used as the protein intensity for all proteins identified by SWATH MS (29, 45). Also proteins with only one or two peptides were included. Inferred protein intensities are provided in Suppl. Data 4.
Proteome Comparison of Different Conditions
For the proteome comparison of samples derived from individual strain colonization of plates and plants (three independent biological replicates per condition (n = 3); each plate sample consisted of three individual plates pooled to form one replicates, whereas plant samples consisted of 30 independent plant growth containers, each containing eight plants, pooled to form one biological replicate) all proteins of the ANOVA output table with a fold change greater two (log2 fold change ≥ 1 or ≤ −1) and a p value below 0.05 were considered significantly differentially regulated between the two conditions. To determine which proteins were only detectable in planta, the aLFQ output was filtered for proteins which are detectable in at least two out of three plant derived samples and in no plate derived sample. The same was subsequently done to identify proteins only detectable on plates. Genome annotation files from the Integrated Microbial Genomes (IMG) database of the Joint Genome Institute of the United States Department of Energy (46) were used to obtain KO terms of these proteins. KO terms were subsequently grouped into functional categories based on the KEGG BRITE database (http://www.genome.jp/kegg/brite.html; functional categories are based on hierarchy level B and categories irrelevant for microbial physiology, e.g. Cancers or Neurodegenerative diseases, were excluded). Functional enrichment analysis was performed using custom input files and the Cytoscape plugin BiNGO v 2.44 (47) with all proteins included in the spectral library as reference set of proteins for enrichment analyses.
Protein Comparison with Other Leaf Microbiota Members
We used blastp of the BLAST standalone software (v. 2.2.31+) to identify homologous proteins in the genomes of Methylobacteria of a recently established leaf microbiota strain collection (48). A protein was considered a hit, if the alignment covered at least 70% of the M. extorquens PA1 query protein sequence with at least 70% sequence identity. To identify similar proteins in more distantly related strains we downloaded all protein sequences of the assigned KO terms from the UniProt database (on November 2, 2015) and used the HMMER toolkit (www.hmmer.org; v. 3.1b2) to first build Hidden Markov Models based on sequence alignments (Clustal Omega) of each KO term and subsequently query our leaf strain collection database with these models (for the KO term K08928 of the photosynthetic reaction center subunit L, two proteins annotated as subunit M were removed from the downloaded sequence list prior to aligning the sequences). Proteins were considered similar if the alignment covered at least 70% of the model with an e-value threshold of 10e-05.
Data Availability
The SWATH assay libraries for M. extorquens PA1 and S. melonis FR1 are available through the SWATHAtlas database (www.SWATHAtlas.org). All SWATH MS raw data have been made available through the PeptideAtlas database (http://www.peptideatlas.org/PASS/PASS00686).
RESULTS
SWATH Assay Library Construction
To reliably quantify the proteome of the two commensal bacteria, M. extorquens PA1 and S. melonis Fr1, under different environmental conditions, we applied the targeted proteomic technique SWATH MS (27). In SWATH MS, identification and quantification of proteins requires knowledge on specific mass spectrometric coordinates for every protein of interest, which are typically compiled in an assay library (27) (Fig. 1A). For every protein, these coordinates consist of a number of representative peptides, their mass-over-charge ratio (m/z) and charge state, the most intense fragment ions formed during fragmentation of the peptide, as well as the normalized chromatographic retention time of the peptide (30). We initially built a comprehensive SWATH assay library for each of the two commensal strains M. extorquens PA1 and S. melonis Fr1 to support the subsequent quantitative proteome measurements. In order to cover the vast majority of proteins produced during colonization of the phyllosphere environment, we performed shotgun mass spectrometry of digested protein extracts of leaf washes as well as extensively fractionated, trypsin-digested protein extracts of liquid cultures grown to early exponential, mid-exponential and stationary growth phase (Fig. 1A). In total, the SWATH assay libraries generated for the two species comprise 3440 (71% of predicted ORFs) and 2729 (71%) proteins, for M. extorquens PA1 and S. melonis Fr1 respectively, whereas 2685 (56%) and 2132 (55%) proteins are represented by three or more peptides (Fig. 1B). These are the first assay libraries for core plant microbiota members allowing the comprehensive and accurate quantification of their proteomes by targeted mass spectrometry. Such near-complete SWATH assay libraries covering over 70% proteome coverage are to date only available for few other organisms, i.e. Saccharomyces cerevisiae, Mycobacterium tuberculosis, and Streptococcus pyogenes (28, 29, 31). The two SWATH assay libraries are available for download through the SWATHAtlas database (www.SWATHAtlas.org) as a public resource for future proteomic studies of these two model strains of the plant microbiota.
Overview of SWATH Measurements
To analyze the in planta physiology of M. extorquens and S. melonis under controlled laboratory conditions, we modified a protocol which has been successfully used to examine the in planta proteome of M. extorquens AM1 as well as the metaproteome of complex natural phyllosphere communities (12, 26). In combination with SWATH MS this procedure allowed for the quantification of 2373 proteins (49.1% of predicted ORFs) and 1610 (42.5%) proteins for M. extorquens PA1 and S. melonis Fr1, respectively (Fig. 1C). Estimated protein intensities were spanning more than three orders of magnitude for cells grown on solidified minimal media in agar plates as well as for phyllosphere-derived bacterial proteome samples (Fig. 1D). For M. extorquens we identified 635 candidate proteins being significantly regulated when growing on leaves compared with growth on minimal media (247 up- and 95 down-regulated proteins; fold change ≥ 2, p value ≤ 0.05) or only detectable in one condition (126 in planta only and 167 not detected in planta) (Fig. 1E). For S. melonis, 545 proteins were significantly regulated when growing on leaves compared with growth on minimal media, with 69 up- and 141 down-regulated proteins (fold change ≥ 2, p value ≤ 0.05), 240 proteins being only detectable on plants and 95 proteins only detectable on minimal media (Fig. 1E). In summary, we obtained deep proteome coverage for both members of the Arabidopsis microbiota in their natural habitat, the phyllosphere, as well as on minimal media, allowing us to study plant-specific physiology and thus, infer evolved adaptive mechanisms of each of the strains in a comprehensive manner.
Adaptation of S. melonis Fr1 to the Arabidopsis Phyllosphere
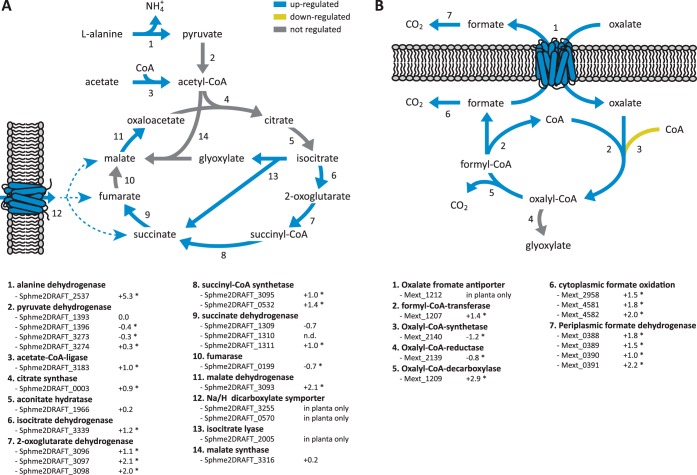
S. melonis is an organo-heterotrophic bacterium that is colonizing the phyllosphere of various host plants. To identify molecular processes representing the adaptation of S. melonis to the leaf compartment we mined the proteome of cells colonizing Arabidopsis plants for induced proteins as compared with growth on minimal media. Colonization of leaves by S. melonis Fr1 led to a significantly higher production of TCA cycle proteins, including isocitrate dehydrogenase (Sphme2DRAFT_3339), three different subunits of the α ketoglutarate dehydrogenase complex (E1 component Sphme2DRAFT_3096, E2 component Sphme2DRAFT_3097, E3 component Sphme2DRAFT_3098), the alpha and beta subunit of the succinyl-CoA synthetase (Sphme2DRAFT_3095, Sphme2DRAFT_0532) as well as a flavoprotein subunit of the succinate dehydrogenase (Sphme2DRAFT_1311), and malate dehydrogenase (Sphme2DRAFT_3093) (Fig. 2A, Table I). Up-regulation of these proteins might reflect higher flux through the TCA cycle and utilization of available substrates feeding into this central metabolic cycle compared with the glycolytic growth on minimal media. The most strongly up-regulated protein on plants was alanine dehydrogenase (Sphme2DRAFT_2537), converting l-alanine into pyruvate and subsequently feeding into the TCA cycle via oxidative decarboxylation to acetyl-CoA (Fig. 2A, Table I). Besides l-alanine conversion, two proteins associated with degradation of arginine (N-carbamoylputrescine amidase Sphme2DRAFT_1690 and succinylglutamic semialdehyde dehydrogenase Sphme2DRAFT_0117) and one protein involved in valine degradation (methylmalonic acid semialdehyde dehydrogenase Sphme2DRAFT_1054) were among the most strongly up-regulated proteins during plant colonization and additionally indicated utilization of available amino acids (Table I). Furthermore, two subunits of the 2-oxoisovalerate dehydrogenase (Sphme2DRAFT_2061, Sphme2DRAFT_2063), responsible for the degradation of branched-chain amino acids, were induced in S. melonis grown on plants.
Fig. 2.
Metabolic responses of S. melonis and M. extorquens during growth on plants. Up-regulation of TCA cycle proteins during leaf colonization of S. melonis Fr1 (A). Several pathways feeding into this central metabolic cycle indicate which plant derived substrates might be utilized during phyllosphere growth. Pathway and involved proteins of oxalate metabolism by M. extorquens PA1 (B). Arrows are shown in blue (up-regulated) and yellow (down-regulated) if corresponding proteins are significantly regulated (fold change ≥ 2, p value ≤ 0.05). Fold changes indicated below the figures are logarithmized (log2); a star indicates a p value ≤ 0.05.
Table I. Most strongly induced proteins during phyllosphere colonization of S. melonis Fr1 and M. extorquens PA1. Only significantly regulated proteins (Benjamini-Hochberg corrected p value ≤ 0.05) are shown.
| Gene | Locus-Tag | Annotation | Log2 FC | p valuea |
|---|---|---|---|---|
| Sphingomonas melonis Fr1 | ||||
| 2517158111 | Sphme2DRAFT_2537 | alanine dehydrogenase | 5.3 | <0.01 |
| 2517158262 | Sphme2DRAFT_2688 | 3-oxoacid CoA-transferase, B subunit | 3.4 | <0.01 |
| 2517158089 | Sphme2DRAFT_2515 | Protease subunit of ATP-dependent Clp proteases | 2.6 | <0.01 |
| 2517158671 | Sphme2DRAFT_3097 | 2-oxoglutarate dehydrogenase complex E2 component | 2.1 | 0.0273 |
| 2517158667 | Sphme2DRAFT_3093 | malate dehydrogenase, NAD-dependent | 2.1 | <0.01 |
| 2517158672 | Sphme2DRAFT_3098 | dihydrolipoamide dehydrogenase | 2.0 | <0.01 |
| 2517159474 | Sphme2DRAFT_3904 | hypothetical protein | 2.0 | <0.01 |
| 2517156663 | Sphme2DRAFT_1089 | d-3-phosphoglycerate dehydrogenase | 1.9 | <0.01 |
| 2517156254 | Sphme2DRAFT_0680 | Predicted outer membrane lipoprotein | 1.9 | <0.01 |
| 2517158622 | Sphme2DRAFT_3048 | hypothetical protein | 1.9 | <0.01 |
| 2517158626 | Sphme2DRAFT_3052 | Predicted sugar kinase | 1.9 | 0.0182 |
| 2517157004 | Sphme2DRAFT_1430 | Sporulation related domain. | 1.8 | <0.01 |
| 2517157264 | Sphme2DRAFT_1690 | N-carbamoylputrescine amidase | 1.7 | 0.0179 |
| 2517157334 | Sphme2DRAFT_1760 | Multidrug resistance efflux pump | 1.6 | <0.01 |
| 2517157234 | Sphme2DRAFT_1660 | Predicted periplasmic/secreted protein | 1.6 | <0.01 |
| 2517158635 | Sphme2DRAFT_3061 | Membrane proteins related to metalloendopeptidases | 1.6 | <0.01 |
| 2517158142 | Sphme2DRAFT_2568 | Deoxyhypusine synthase | 1.6 | <0.01 |
| 2517156139 | Sphme2DRAFT_0565 | pyruvate kinase | 1.6 | <0.01 |
| 2517158432 | Sphme2DRAFT_2858 | NAD-dependent aldehyde dehydrogenases | 1.6 | <0.01 |
| 2517158634 | Sphme2DRAFT_3060 | tyrosyl-tRNA synthetase | 1.6 | <0.01 |
| 2517156628 | Sphme2DRAFT_1054 | methylmalonic acid semialdehyde dehydrogenase | 1.5 | <0.01 |
| 2517157977 | Sphme2DRAFT_2403 | Glutathione S-transferase | 1.5 | <0.01 |
| 2517155766 | Sphme2DRAFT_0192 | ribosomal protein L21 | 1.5 | <0.01 |
| 2517158541 | Sphme2DRAFT_2967 | Peroxiredoxin | 1.5 | <0.01 |
| 2517156538 | Sphme2DRAFT_0964 | Glycine cleavage system protein P (pyridoxal-binding), C-terminal domain | 1.5 | <0.01 |
| 2517158824 | Sphme2DRAFT_3250 | Cysteine synthase | 1.5 | 0.0213 |
| 2517156106 | Sphme2DRAFT_0532 | succinyl-CoA synthetase, beta subunit | 1.4 | <0.01 |
| 2517156919 | Sphme2DRAFT_1345 | Short-chain alcohol dehydrogenase of unknown specificity | 1.4 | 0.0147 |
| 2517158295 | Sphme2DRAFT_2721 | Glucose/sorbosone dehydrogenases | 1.4 | 0.0106 |
| 2517155691 | Sphme2DRAFT_0117 | succinylglutamic semialdehyde dehydrogenase | 1.4 | 0.0115 |
| 2517156060 | Sphme2DRAFT_0486 | Domain of unknown function (DUF3576) | 1.3 | <0.01 |
| 2517158171 | Sphme2DRAFT_2597 | hypothetical protein | 1.3 | <0.01 |
| 2517155822 | Sphme2DRAFT_0248 | RND family efflux transporter, MFP subunit | 1.3 | <0.01 |
| 2517158649 | Sphme2DRAFT_3075 | Long-chain fatty acid transport protein | 1.3 | <0.01 |
| 2517156431 | Sphme2DRAFT_0857 | argininosuccinate lyase | 1.3 | <0.01 |
| 2517156535 | Sphme2DRAFT_0961 | glycine cleavage system T protein | 1.3 | <0.01 |
| 2517156271 | Sphme2DRAFT_0697 | Beta-lactamase class A | 1.3 | <0.01 |
| 2517158162 | Sphme2DRAFT_2588 | Ribosomal protein L10 | 1.3 | <0.01 |
| 2517158942 | Sphme2DRAFT_3368 | ATP-binding cassette protein, ChvD family | 1.2 | <0.01 |
| 2517158985 | Sphme2DRAFT_3411 | Cell shape-determining protein | 1.2 | <0.01 |
| Methylobacterium extorquens PA1 | ||||
| NC_010172.1_cdsid_YP_001642205.1 | Mext_4766 | hypothetical protein | 4.5 | <0.01 |
| NC_010172.1_cdsid_YP_001640200.1 | Mext_2738 | photosynthetic reaction center cytochrome c subunit (PufC) | 4.3 | 0.022 |
| NC_010172.1_cdsid_YP_001640859.1 | Mext_3402 | hypothetical protein | 4.2 | <0.01 |
| NC_010172.1_cdsid_YP_001642131.1 | Mext_4692 | protease Do | 3.9 | <0.01 |
| NC_010172.1_cdsid_YP_001641409.1 | Mext_3967 | hypothetical protein | 3.7 | <0.01 |
| NC_010172.1_cdsid_YP_001638811.1 | Mext_1339 | methanol/ethanol family PQQ-dependent dehydrogenase | 3.2 | <0.01 |
| NC_010172.1_cdsid_YP_001640093.1 | Mext_2630 | hypothetical protein | 3.1 | <0.01 |
| NC_010172.1_cdsid_YP_001638682.1 | Mext_1209 | putative oxalyl-CoA decarboxylase | 2.9 | 0.013 |
| NC_010172.1_cdsid_YP_001640767.1 | Mext_3309 | molybdopterin binding oxidoreductase | 2.9 | <0.01 |
| NC_010172.1_cdsid_YP_001641647.1 | Mext_4207 | triple helix repeat-containing collagen | 2.9 | <0.01 |
| NC_010172.1_cdsid_YP_001640025.1 | Mext_2560 | phasin (Gap20) | 2.8 | <0.01 |
| NC_010172.1_cdsid_YP_001642168.1 | Mext_4729 | CsbD family protein | 2.7 | <0.01 |
| NC_010172.1_cdsid_YP_001639679.1 | Mext_2213 | hypothetical protein | 2.7 | <0.01 |
| NC_010172.1_cdsid_YP_001637566.1 | Mext_0067 | hypothetical protein | 2.7 | <0.01 |
| NC_010172.1_cdsid_YP_001639614.1 | Mext_2148 | hypothetical protein | 2.6 | <0.01 |
| NC_010172.1_cdsid_YP_001641262.1 | Mext_3817 | hypothetical protein | 2.6 | <0.01 |
| NC_010172.1_cdsid_YP_001641592.1 | Mext_4152 | hypothetical protein | 2.6 | <0.01 |
| NC_010172.1_cdsid_YP_001638148.1 | Mext_0663 | putative DNA-binding protein | 2.6 | <0.01 |
| NC_010172.1_cdsid_YP_001637886.1 | Mext_0393 | hypothetical protein | 2.6 | <0.01 |
| NC_010172.1_cdsid_YP_001638358.1 | Mext_0882 | PfpI family intracellular peptidase | 2.6 | <0.01 |
| NC_010172.1_cdsid_YP_001639279.1 | Mext_1809 | methanol/ethanol family PQQ-dependent dehydrogenase (XoxF) | 2.5 | <0.01 |
| NC_010172.1_cdsid_YP_001642090.1 | Mext_4651 | aromatic amino acid beta-eliminating lyase/threonine aldolase | 2.5 | <0.01 |
| NC_010172.1_cdsid_YP_001639237.1 | Mext_1767 | hypothetical protein | 2.5 | <0.01 |
| NC_010172.1_cdsid_YP_001638912.1 | Mext_1440 | hypothetical protein | 2.4 | 0.025 |
| NC_010172.1_cdsid_YP_001640238.1 | Mext_2776 | hypothetical protein | 2.4 | <0.01 |
| NC_010172.1_cdsid_YP_001638796.1 | Mext_1324 | hypothetical protein | 2.4 | <0.01 |
| NC_010172.1_cdsid_YP_001637885.1 | Mext_0392 | hypothetical protein | 2.4 | <0.01 |
| NC_010172.1_cdsid_YP_001638850.1 | Mext_1378 | hypothetical protein | 2.4 | <0.01 |
| NC_010172.1_cdsid_YP_001639962.1 | Mext_2496 | hypothetical protein | 2.4 | <0.01 |
| NC_010172.1_cdsid_YP_001640222.1 | Mext_2760 | NAD(P) transhydrogenase subunit PntAB | 2.4 | <0.01 |
| NC_010172.1_cdsid_YP_001640865.1 | Mext_3408 | alkanesulfonate monooxygenase | 2.3 | <0.01 |
| NC_010172.1_cdsid_YP_001642123.1 | Mext_4684 | hypothetical protein | 2.3 | <0.01 |
| NC_010172.1_cdsid_YP_001637681.1 | Mext_0184 | hypothetical protein | 2.3 | <0.01 |
| NC_010172.1_cdsid_YP_001639333.1 | Mext_1863 | hypothetical protein | 2.2 | <0.01 |
| NC_010172.1_cdsid_YP_001640504.1 | Mext_3044 | hypothetical protein | 2.2 | 0.023 |
| NC_010172.1_cdsid_YP_001637884.1 | Mext_0391 | formate dehydrogenase subunit gamma (Fdh3) | 2.2 | <0.01 |
| NC_010172.1_cdsid_YP_001642264.1 | Mext_4825 | gluconate 2-dehydrogenase | 2.2 | 0.021 |
| NC_010172.1_cdsid_YP_001640221.1 | Mext_2759 | NAD(P)(+) transhydrogenase | 2.1 | <0.01 |
| NC_010172.1_cdsid_YP_001640937.1 | Mext_3481 | hypothetical protein | 2.1 | <0.01 |
| NC_010172.1_cdsid_YP_001641211.1 | Mext_3766 | hypothetical protein | 2.1 | <0.01 |
a Benjamini-Hochberg corrected for multiple testing.
Other differentially regulated proteins involved in acetate utilization (acetate-CoA ligase, Sphme2DRAFT_3183), oxidation of 3-oxoacids (3-oxoacid CoA-transferase subunit B, Sphme2DRAFT_2688), and fatty acid oxidation (long chain fatty acid transport protein, Sphme2DRAFT_3075, 3-hydroxyacyl-CoA dehydrogenase, Sphme2DRAFT_3077, acyl-CoA synthetase Sphme2DRAFT_3079, acyl-CoA dehydrogenases Sphme2DRAFT_0054, Sphme2DRAFT_1323, Sphme2DRAFT_2248 and enoyl-CoA hydratase Sphme2DRAFT_1289, Sphme2DRAFT_2065) indicate that fatty acids, or potentially long chain alkanes, which are part of the cuticle, might serve as additional nutrients during colonization of leaves. In agreement with the utilization of compounds yielding acetyl-CoA, isocitrate lyase (Sphme2DRAFT_2005), the key enzyme of the glyoxylate shunt, was only detectable in planta. Furthermore, dicarboxylates such as malate, succinate or fumarate might be taken up by the Na+/H+-dicarboxylate symporter proteins (Sphme2DRAFT_3255 and Sphme2DRAFT_0570), which were also only detectable during phyllosphere colonization (Fig. 2A, supplemental Table S1). Other differentially regulated transport proteins comprise iron uptake and efflux pump systems for detoxification and resistance (supplemental Table S1). Overall, our data indicate that growth of S. melonis on leaves is driven by plant-derived amino acids, small molecules like acetate and fatty acids or other hydrocarbon compounds, catabolized through the TCA cycle.
Adaptation of M. extorquens PA1 to the Arabidopsis Phyllosphere
M. extorquens PA1 is a facultative methylotroph and capable of utilizing plant derived methanol in the phyllosphere, a byproduct of plant cell wall biosynthesis (22, 49, 50). To identify molecular processes underlining the adaptation of M. extorquens to plant surfaces we looked for significantly induced proteins during growth on leaves as compared with growth on solidified minimal media, as described for S. melonis Fr1 above. This analysis confirmed the catalytic subunit of methanol dehydrogenase MxaF as the most abundant protein under both growth conditions. In addition, we found two other methanol dehydrogenase-like proteins, Mext_1809 (XoxF) and Mext_1339, to be up-regulated during plant colonization (supplemental Table S2, supplemental Fig. S2). Furthermore, a large number of proteins involved in formate oxidation (Mext_4581 and Mext_4582, two subunits of the cytoplasmic Fdh1; Mext_0389, Mext_0390; Mext_0391, three subunits of the periplasmic Fdh3) were significantly more produced during plant colonization (Fig. 2B and supplemental Table S2). Interestingly, most proteins of the serine cycle and the ethylmalonyl-CoA pathway, important for assimilation of one-carbon (C1) compounds (51), were present at a significantly lower level during growth on plants, indicating that other multi-carbon compounds are used preferentially for assimilatory processes (supplemental Fig. S2, supplemental Table S2).
The detection of enzymes involved in oxalate metabolism suggests that the plant-derived two-carbon (C2) compound might be metabolized by the methylotroph M. extorquens PA1. Formyl-CoA transferase and oxalyl-CoA decarboxylase (Mext_1207 and Mext_1209, respectively), needed for oxalate conversion, were up-regulated on leaves and the oxalate formate antiporter (Mext_1212) was only detectable during phyllosphere colonization (Fig. 2B, supplemental Table S3). Exported formate can subsequently be oxidized by the periplasmic dehydrogenase Fdh3, which was up-regulated during plant colonization. Previous characterization of the oxalate metabolism in M. extorquens AM1 revealed that reduction of oxalyl-CoA to glyoxylate is the preferred route of oxalate assimilation and that NAD(P)+ transhydrogenase for redox balance was highly expressed in cells grown on oxalate (52). In the present study, NAD(P)+ transhydrogenase was induced upon plant colonization in M. extorquens PA1, however, the corresponding proteins for assimilatory reduction to glyoxylate were down-regulated, indicating that oxalate oxidation is rather used for energy conservation than for carbon assimilation (supplemental Table S3).
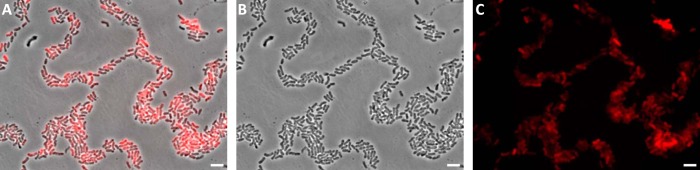
Besides oxidation of organic carbon, utilization of sunlight may act as accessory mode of energy conservation for phyllosphere bacteria (53, 54). The M. extorquens PA1 genome encodes for proteins of aerobic anoxygenic photosynthesis and photosynthetic cytochrome PufC (Mext_2738) was among the most up-regulated proteins in planta (Table I). In addition, the subunits L and H of the photosynthetic reaction center (Mext_2736 and Mext_4810, respectively) were only detectable in samples from in planta conditions, whereas two proteins potentially involved in bacteriochlorophyll biosynthesis (HemY domain containing protein and magnesium-protoporphyrin IX monomethyl ester cyclase, Mext_0735 and Mext_4806, respectively) were up-regulated during plant colonization. In agreement with this finding, infrared autofluorescence, indicative of anoxygenic photosynthesis was detectable in M. extorquens PA1 cells colonizing the A. thaliana phyllosphere (Fig. 3).
Fig. 3.
False color images of M. extorquens cells showing infrared autofluorescence indicative of aerobic anoxygenic photosynthesis during leaf colonization. Overlay (A) of pictures acquired from the same field of view in phase contrast (B) and infrared fluorescence channels (C). Scale bar corresponds to 5 μm. Methylobacterium cells grown on minimal medium agar plates in the dark do not show background infrared autofluorescence indicative of anoxygenic photosynthesis (supplemental Fig. S3).
Another phyllosphere-specific response of M. extorquens PA1 was the up-regulation of genes for alkanesulfonate monooxygenases. In total, the bacterial genome encodes four alkanesulfonate monooxygenases genes including homologs of SsuD and SfnG. Three of these genes are predicted to be part of transcriptional units including genes for uptake of sulfonate substrates and indeed all 16 genes were induced on plants (supplemental Table S4). Alkanesulfonate monooxygenases are generally involved in sulfur acquisition during sulfur limiting conditions (55). Up-regulation of other genes involved in sulfate uptake (sulfate ABC transport permease and sulfate ABC transporter ATPase, Mext_0583 and Mext_0584, respectively) and sulfate assimilation (sulfate adenylyltransferase, Mext_2232 and Mext_2233) further support the notion of sulfur limitation on leaves.
Other regulated transport proteins in our data set comprise uptake proteins of unknown specificity (e.g. general substrate transporter Mext_4685) as well as efflux systems to lower the concentration of toxic compounds (e.g. RND family efflux transporter MFP subunit, Mext_2832 among others) (supplemental Table S1). Notably, many proteins up-regulated during plant colonization (Table I) are not yet characterized and might be involved in yet unknown adaptive processes.
In conclusion, our results underline the importance of methanol oxidation during plant colonization by M. extorquens and identify anoxygenic photosynthesis and oxalate metabolism as accessory modes of energy conversation during growth on leaves.
Metabolic Specialization of Methylobacterium Strains
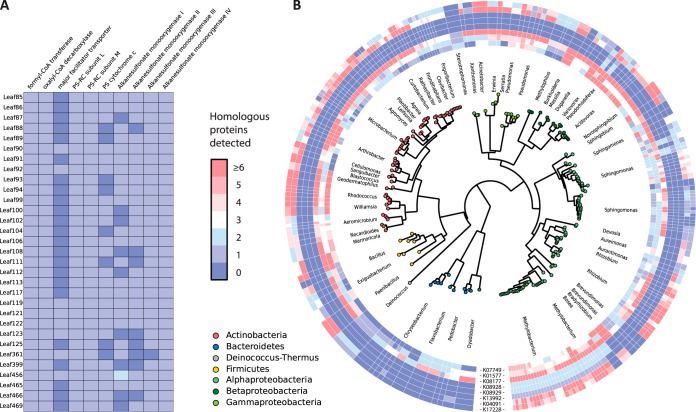
Because some traits induced by M. extorquens during plant colonization suggest niche specialization, we analyzed whether anoxygenic photosynthesis, oxalate metabolism and utilization of organic sulfonates are common features of leaf colonizing Methylobacteria. Using BLAST search against genomes of a recently established A. thaliana microbiota strain collection (48) we found that homologs of these proteins are encoded in the genomes of essentially all 31 Methylobacterium strains analyzed (Fig. 4A). The data suggests that the processes we identified in M. extorquens PA1 might be conserved among plant associated Methylobacterium strains. To identify similar proteins in more distantly related strains, we built profile Hidden Markov Models based on the assigned KEGG Orthology (KO) terms and found that genes of oxalate and alkanesulfonate metabolism are widespread among leaf microbiota members, whereas proteins of aerobic anoxygenic photosynthesis are almost exclusively found in Methylobacteria (Fig. 4B).
Fig. 4.
Number and distribution of homologs of the identified target proteins in other leaf colonizing Methylobacteria and leaf microbiota members. We identified homologous proteins (Protein Blast; >70% query sequence coverage, >70% sequence identity) of the target proteins involved in phyllosphere colonization by M. extorquens PA1 in the genomes of 31 leaf colonizing Methylobacteria (Methylobacterium sp. Leaf85 to Methylobacterium sp. Leaf469) of a recently established bacterial phyllosphere strain collection (48) (A). Hidden Markov Models based on assigned KEGG Orthology (KO) terms were used to identify functionally similar proteins in more distantly related leaf microbiota members (B). The color scheme indicates the number of putative homologs identified within the queried genomes of the distinct target strains. Proteins are formyl-CoA transferase (KO term: K07749), oxalyl-CoA decarboxylase (K01577) and the major facilitator transporter (K08177) of the oxalate metabolism, subunits L, M and the cytochrome c subunit of the photosynthesis apparatus (K08928, K08929 and K13992, respectively) as well as the alkanesulfonate monooxygenase homologs SsuD (K04091) and SfnG (K17228).
Overlap of Regulated Proteins Between M. extorquens PA1 and S. melonis Fr1
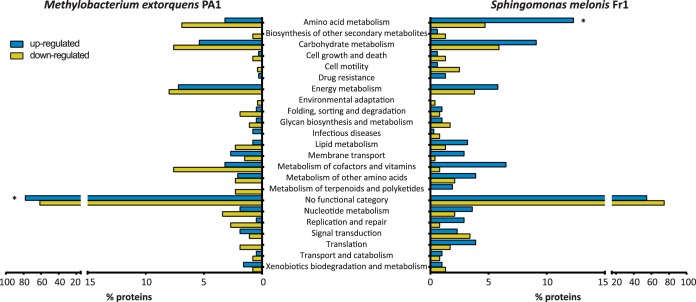
The analysis of the in planta proteomes of M. extorquens PA1 and S. melonis Fr1 suggest species-specific adaptations, nonetheless common adaptation mechanisms upon plant colonization may also exist. To check if a subset of proteins or protein functions is shared between the two commensals we compared the assigned KO terms of all differentially regulated proteins. In total, genome annotation of M. extorquens PA1 contained KO terms for 263 of all significantly regulated proteins (41%) with 125 (20%) and 138 (22%) showing induced or reduced expression on plants, respectively. On the other hand, 317 S. melonis proteins (58%) are assigned KO terms and 201 (37%) of them were induced on leaves, whereas 116 proteins (21%) were present at lower levels. We first grouped all KO terms according to their functional category. Subsequent enrichment analysis revealed that the category “amino acid metabolism” is significantly enriched in S. melonis Fr1 on leaves (Fig. 5, p value = 0.00275, Hypergeometric test, Benjamini-Hochberg FDR correction), underlining our earlier observations regarding the induced utilization of amino acids. In M. extorquens PA1, proteins without assigned functional category were enriched (Fig. 5, p = 1.24 × 10−4, Hypergeometric test, Benjamini-Hochberg FDR correction) indicating that many potentially important plant adaptive processes are not yet characterized. In total, we found a relatively small subset of 12 KO terms shared among the up-regulated proteins and 7 KO terms shared among the down-regulated proteins of the two strains (supplemental Table S5). Notably, one of the proteins induced on leaves in both strains is the alkanesulfonate monooxygenase subunit SsuE (K00299), possibly indicating widespread utilization of sulfonates for sulfur acquisition during plant colonization. Other proteins induced in planta by both commensals include ABC type transporter proteins, a polysaccharide export protein, the phosphoenolpyruvate carboxykinase, phosphoglycerate dehydrogenase, threonine aldolase as well as a number of hypothetical proteins (K09796; Mext_1378, Mext_1379, Sphme2DRAFT_0994) only containing a conserved domain of unknown function (supplemental Table S5). Overall, the relatively small subset of proteins found to be coregulated in M. extorquens PA1 and S. melonis Fr1 during growth on plant leaves underlines the apparent niche separation via distinct metabolic capacities and strategies for host colonization.
Fig. 5.
Proteome comparison of M. extorquens PA1 and S. melonis Fr1. KO term assignments of all differentially regulated proteins of M. extorquens PA1 and S. melonis Fr1 and their functional categories based on the KEGG Brite database. Blue and yellow bars represent the percentage of up and down-regulated proteins, respectively for each functional category. Asterisks indicate significant enrichments (p ≤ 0.05; Hypergeometric test, Benjamini-Hochberg FDR correction) of up- or down-regulated proteins of each functional category.
DISCUSSION
Investigating molecular adaptations in members of microbial communities when they colonize their host is crucial to understand stable community patterns observed under environmental conditions and to unravel the molecular basis of host-microbe interactions. Here we extended earlier proteomic analyses of microbial plant community members (10, 56, 57) by applying the massively parallel targeted proteomic technique SWATH MS to reliably quantify the proteomic changes of two ubiquitous plant commensals upon colonization of A. thaliana leaves. Methylobacterium and Sphingomonas are two genera commonly inhabiting the phyllosphere of different host plants and previous studies suggested distinct metabolic capabilities to be important drivers for niche separation (12).
In the present study we confirmed the importance of methanol oxidation for energy conservation of M. extorquens during plant colonization including the methanol dehydrogenase-like protein XoxF. Previous studies have shown that XoxF is among the most abundant proteins in natural phyllosphere communities and that a xoxF mutant of M. extorquens is affected in plant colonization under competitive conditions (12, 58). Based on the high proteome coverage achieved with SWATH MS, we found that enzymes of the linear methanol oxidation pathway are induced during growth on plants, however, proteins involved in the metabolic pathways for C1 assimilation were down-regulated, indicating that additional carbon compounds might be used preferentially for assimilatory purposes during plant colonization by the facultative methylotroph. In this context, also our observation of up-regulation of proteins for oxalate dissimilation is notable (Fig. 2B), indicating utilization of the plant-derived C2 compound by Methylobacterium. Oxalate is produced by a wide range of plants and is involved in various processes, including calcium storage, detoxification of heavy metals as well as plant protection. Generally, oxalotrophy was reported to be a rare bacterial trait, but it has frequently been found in microbes living in close association with plants (59). Based on assigned KO terms we identified the key enzymes in oxalate conversion in a wide range of phylogenetically distinct leaf microbiota members (Fig. 4B) confirming this tendency. Besides oxalate biosynthesis by the plant host, some phytopathogenic fungi and bacteria are known to produce oxalate as a functionally diverse virulence factor. Therefore, oxalate degradation can be considered a plant protective feature and oxalotrophy was shown to be involved in recruiting beneficial Burkholderia strains to the plant root (59, 60). Methylobacteria and other microbiota members may act as sink of oxalate on leaves and therefore lower the risk of infectious diseases, for example caused by the oxalate-secreting broad host pathogen Botrytis cinerea.
Besides proteins related to carbon metabolism, we found a conspicuous overrepresentation of proteins involved in uptake and utilization of sulfonates for sulfur acquisition, e.g. the 16 genes of predicted transcriptional units containing alkanesulfonate monooxygenase genes, were induced upon colonization of the leaf surface. The SsuE subunit of the alkanesulfonate monooxygenase was not only up-regulated in M. extorquens, but also in S. melonis and it was further shown to be induced during epiphytic growth of the plant pathogen Pseudomonas syringae B728a (61, 62). Generally, sulfur metabolism in the phyllosphere is poorly understood, but our findings indicate broader utilization of organic sulfonates by the plant microbiota.
S. melonis Fr1 is known to confer a protective effect to the plant host A. thaliana against the foliar pathogen Pseudomonas syringae pv. tomato DC3000. The detailed molecular mechanism of this interaction remains elusive, but several traits might contribute incrementally to plant host protection, including stimulation of plant host immunity and competition for nutrients, especially during the first phase of pathogen invasion into the host-associated microbiota (20, 21). The in planta proteome of S. melonis Fr1 indicates utilization of substrates feeding into the TCA cycle including acetate, fatty acid compounds, dicarboxylates and the amino acid alanine. Alanine dehydrogenase was the most strongly up-regulated protein during phyllosphere colonization of S. melonis, being approximately forty times more abundant compared with growth on minimal media. Interestingly, the importance of alanine catabolism for successful in vivo proliferation has been demonstrated for another Pseudomonad, the human pathogen Pseudomonas aeruginosa during infection of the lungs (63). Besides alanine dehydrogenase, enzymes for arginine and valine degradation were among the most up-regulated proteins, in line with the recently acquired metabolic footprint by MALDI-TOF imaging of S. melonis, demonstrating arginine utilization on Arabidopsis leaves (64). We furthermore found a set of transport proteins involved in multidrug resistance, efflux of toxic compounds as well as iron uptake induced in S. melonis Fr1 upon plant colonization, which might contribute to its perseverance in the phyllosphere, also with regard to pathogenic strains invading the habitat.
The comparison of the differential proteomes of M. extorquens PA1 and S. melonis Fr1 underlines the apparent niche separation via distinct metabolic capacities and strategies for host colonization, as only a relatively small subset of proteins was found to be coregulated between both strains. Common traits include sulfonate metabolism, phosphate uptake and secretion of polysaccharides. We also found hypothetical proteins containing a conserved domain of unknown function to be coregulated between the two strains, indicating that other conserved responses might be hidden among proteins for which no functional annotations are yet available (59 and 42% of candidate proteins for M. extorquens and S. melonis, respectively).
In summary, our study shows that SWATH MS is a suitable tool to study physiological responses of the indigenous microbiota to their natural habitat within the respective host. Our findings indicate that the two ubiquitous commensals M. extorquens and S. melonis have evolved strain-specific and fundamentally different ways to adapt to the plant environment. Although S. melonis is utilizing a variety of more general organic substrates like acetate, dicarboxylates, amino acids, or potentially hydrocarbon compounds, M. extorquens is highly specified in using C1 and C2 plant compounds as well as anoxygenic photosynthesis to support growth on leaves. Despite the apparent niche separation, a subset of shared proteins between the two strains indicate common adaptive processes to the phyllosphere, and identify organic sulfonates as potentially widespread sulfur source on leaves.
Supplementary Material
Acknowledgments
We would like to thank David Campbell for making our SWATH libraries available through the SWATHAtlas database.
Footnotes
Author contributions: D.B.M., O.T.S., H.R., R.A., and J.A.V. designed research; D.B.M. and O.T.S. performed research; H.R. contributed new reagents or analytic tools; D.B.M., O.T.S., H.R., R.A., and J.A.V. analyzed data; D.B.M., O.T.S., and J.A.V. wrote the paper.
* The work was supported by ETH Research Grant ETH-41 14-2 and an ERC Advanced grant (PhyMo).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- SWATH MS
- Sequential Windowed Acquisition of all THeoretical fragment ions Mass Spectrometry
- MWCO
- molecular weight cut-off
- IDA
- information dependent acquisition
- ORF
- open reading frame.
REFERENCES
- 1. Russell J. A., Dubilier N., and Rudgers J. A. (2014) Nature's microbiome: introduction. Mol. Ecol. 23, 1225–1237 [DOI] [PubMed] [Google Scholar]
- 2. Bulgarelli D., Schlaeppi K., Spaepen S., Ver Loren van Themaat E., and Schulze-Lefert P. (2013) Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838 [DOI] [PubMed] [Google Scholar]
- 3. Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R., and Gordon J. I. (2007) The Human Microbiome Project. Nature 449, 804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ley R. E., Hamady M., Lozupone C., Turnbaugh P. J., Ramey R. R., Bircher J. S., Schlegel M. L., Tucker T. A., Schrenzel M. D., Knight R., and Gordon J. I. (2008) Evolution of mammals and their gut microbes. Science 320, 1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Backhed F., Fraser C. M., Ringel Y., Sanders M. E., Sartor R. B., Sherman P. M., Versalovic J., Young V., and Finlay B. B. (2012) Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe 12, 611–622 [DOI] [PubMed] [Google Scholar]
- 6. Abt M. C., and Pamer E. G. (2014) Commensal bacteria mediated defenses against pathogens. Curr. Opinion Immunol. 29, 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lugtenberg B., and Kamilova F. (2009) Plant-Growth-Promoting Rhizobacteria. Annu. Rev. Microbiol. 63, 541–556 [DOI] [PubMed] [Google Scholar]
- 8. Bulgarelli D., Rott M., Schlaeppi K., Ver Loren van Themaat E., Ahmadinejad N., Assenza F., Rauf P., Huettel B., Reinhardt R., Schmelzer E., Peplies J., Gloeckner F. O., Amann R., Eickhorst T., and Schulze-Lefert P. (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95 [DOI] [PubMed] [Google Scholar]
- 9. Lundberg D. S., Lebeis S. L., Paredes S. H., Yourstone S., Gehring J., Malfatti S., Tremblay J., Engelbrektson A., Kunin V., del Rio T. G., Edgar R. C., Eickhorst T., Ley R. E., Hugenholtz P., Tringe S. G., and Dangl J. L. (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Knief C., Delmotte N., Chaffron S., Stark M., Innerebner G., Wassmann R., von Mering C., and Vorholt J. A. (2012) Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 6, 1378–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zarraonaindia I., Owens S. M., Weisenhorn P., West K., Hampton-Marcell J., Lax S., Bokulich N. A., Mills D. A., Martin G., Taghavi S., van der Lelie D., and Gilbert J. A. (2015) The soil microbiome influences grapevine-associated microbiota. mBio 6, e02527–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delmotte N., Knief C., Chaffron S., Innerebner G., Roschitzki B., Schlapbach R., von Mering C., and Vorholt J. A. (2009) Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 16428–16433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vorholt J. A. (2012) Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840 [DOI] [PubMed] [Google Scholar]
- 14. Remus-Emsermann M. N., Lücker S., Müller D. B., Potthoff E., Daims H., and Vorholt J. A. (2014) Spatial distribution analyses of natural phyllosphere-colonizing bacteria on Arabidopsis thaliana revealed by fluorescence in situ hybridization. Environ. Microbiol. 16, 2329–2340 [DOI] [PubMed] [Google Scholar]
- 15. Meyer K. M., and Leveau J. H. J. (2012) Microbiology of the phyllosphere: a playground for testing ecological concepts. Oecologia 168, 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lindow S. E., and Brandl M. T. (2003) Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bodenhausen N., Horton M. W., and Bergelson J. (2013) Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PloS One 8, e56329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Knief C., Frances L., Cantet F., and Vorholt J. A. (2008) Cultivation-independent characterization of Methylobacterium populations in the plant phyllosphere by automated ribosomal intergenic spacer analysis. Appl. Environ. Microbiol. 74, 2218–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Knief C., Frances L., and Vorholt J. A. (2010) Competitiveness of diverse Methylobacterium strains in the phyllosphere of Arabidopsis thaliana and identification of representative models, including M. extorquens PA1. Microbial Ecol. 60, 440–452 [DOI] [PubMed] [Google Scholar]
- 20. Vogel C., Innerebner G., Zingg J., Guder J., and Vorholt J. A. (2012) Forward genetic in planta screen for identification of plant-protective traits of Sphingomonas sp. strain Fr1 against Pseudomonas syringae DC3000. Appl. Environ. Microbiol. 78, 5529–5535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Innerebner G., Knief C., and Vorholt J. A. (2011) Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 77, 3202–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abanda-Nkpwatt D., Musch M., Tschiersch J., Boettner M., and Schwab W. (2006) Molecular interaction between Methylobacterium extorquens and seedlings: growth promotion, methanol consumption, and localization of the methanol emission site. J. Exp. Botany 57, 4025–4032 [DOI] [PubMed] [Google Scholar]
- 23. Koenig R. L., Morris R. O., and Polacco J. C. (2002) tRNA is the source of low-level trans-zeatin production in Methylobacterium spp. J. Bacteriol. 184, 1832–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Monier J. M., and Lindow S. E. (2003) Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl. Acad. Sci. U.S.A. 100, 15977–15982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jacobs J. L., Carroll T. L., and Sundin G. W. (2005) The role of pigmentation, ultraviolet radiation tolerance, and leaf colonization strategies in the epiphytic survival of phyllosphere bacteria. Microbial Ecol. 49, 104–113 [DOI] [PubMed] [Google Scholar]
- 26. Gourion B., Rossignol M., and Vorholt J. A. (2006) A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc. Natl. Acad. Sci. U.S.A. 103, 13186–13191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gillet L. C., Navarro P., Tate S., Rost H., Selevsek N., Reiter L., Bonner R., and Aebersold R. (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Selevsek N., Chang C. Y., Gillet L. C., Navarro P., Bernhardt O. M., Reiter L., Cheng L. Y., Vitek O., and Aebersold R. (2015) Reproducible and consistent quantification of the Saccharomyces cerevisiae proteome by SWATH-mass spectrometry. Mol. Cell. Proteomics 14, 739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schubert O. T., Ludwig C., Kogadeeva M., Zimmermann M., Rosenberger G., Gengenbacher M., Gillet L. C., Collins B. C., Rost H. L., Kaufmann S. H., Sauer U., and Aebersold R. (2015) Absolute proteome composition and dynamics during dormancy and resuscitation of Mycobacterium tuberculosis. Cell Host Microbe 18, 96–108 [DOI] [PubMed] [Google Scholar]
- 30. Schubert O. T., Gillet L. C., Collins B. C., Navarro P., Rosenberger G., Wolski W. E., Lam H., Amodei D., Mallick P., MacLean B., and Aebersold R. (2015) Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat. Protocols 10, 426–441 [DOI] [PubMed] [Google Scholar]
- 31. Röst H. L., Rosenberger G., Navarro P., Gillet L., Miladinovic S. M., Schubert O. T., Wolskit W., Collins B. C., Malmstrom J., Malmstrom L., and Aebersold R. (2014) OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 32, 219–223 [DOI] [PubMed] [Google Scholar]
- 32. Peyraud R., Kiefer P., Christen P., Massou S., Portais J. C., and Vorholt J. A. (2009) Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proc. Natl. Acad. Sci. U.S.A. 106, 4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schubert O. T., Mouritsen J., Ludwig C., Rost H. L., Rosenberger G., Arthur P. K., Claassen M., Campbell D. S., Sun Z., Farrah T., Gengenbacher M., Maiolica A., Kaufmann S. H. E., Moritz R. L., and Aebersold R. (2013) The Mtb proteome library: A resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe 13, 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Escher C., Reiter L., MacLean B., Ossola R., Herzog F., Chilton J., MacCoss M. J., and Rinner O. (2012) Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics 12, 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., and Aebersold R. (2010) A guided tour of the trans-proteomic pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keller A., Nesvizhskii A. I., Kolker E., and Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 37. Shteynberg D., Deutsch E. W., Lam H., Eng J. K., Sun Z., Tasman N., Mendoza L., Moritz R. L., Aebersold R., and Nesvizhskii A. I. (2011) iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reiter L., Claassen M., Schrimpf S. P., Jovanovic M., Schmidt A., Buhmann J. M., Hengartner M. O., and Aebersold R. (2009) Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell. Proteomics 8, 2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., King N., Stein S. E., and Aebersold R. (2007) Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7, 655–667 [DOI] [PubMed] [Google Scholar]
- 40. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., Stein S. E., and Aebersold R. (2008) Building consensus spectral libraries for peptide identification in proteomics. Nature Meth. 5, 873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M. Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., and Mallick P. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nature Biotech. 30, 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Teleman J., Rost H. L., Rosenberger G., Schmitt U., Malmstrom L., Malmstrom J., and Levander F. (2015) DIANA-algorithmic improvements for analysis of data-independent acquisition MS data. Bioinformatics 31, 555–562 [DOI] [PubMed] [Google Scholar]
- 43. Reiter L., Rinner O., Picotti P., Huttenhain R., Beck M., Brusniak M. Y., Hengartner M. O., and Aebersold R. (2011) mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nature Meth. 8, 430–435 [DOI] [PubMed] [Google Scholar]
- 44. Rosenberger G., Ludwig C., Rost H. L., Aebersold R., and Malmstrom L. (2014) aLFQ: an R-package for estimating absolute protein quantities from label-free LC-MS/MS proteomics data. Bioinformatics 30, 2511–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ludwig C., Claassen M., Schmidt A., and Aebersold R. (2012) Estimation of absolute protein quantities of unlabeled samples by selected reaction monitoring mass spectrometry. Mol. Cell. Proteomics 11, M111.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Markowitz V. M., Chen I. M., Palaniappan K., Chu K., Szeto E., Grechkin Y., Ratner A., Jacob B., Huang J., Williams P., Huntemann M., Anderson I., Mavromatis K., Ivanova N. N., and Kyrpides N. C. (2012) IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 40, D115–D122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maere S., Heymans K., and Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 48. Bai Y., Müller D. B., Srinivas G., Garrido-Oter R., Potthoff E., Rott M., Dombrowski N., Münch P. C., Spaepen S., Remus-Emsermann M., Hüttel B., McHardy A. C., Vorholt J. A., and Schulze-Lefert P. (2015) Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369 [DOI] [PubMed] [Google Scholar]
- 49. Sy A., Timmers A. C., Knief C., and Vorholt J. A. (2005) Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl. Environ. Microbiol. 71, 7245–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fall R., and Benson A. A. (1996) Leaf methanol - The simplest natural product from plants. Trends Plant Sci. 1, 296–301 [Google Scholar]
- 51. Ochsner A. M., Sonntag F., Buchhaupt M., Schrader J., and Vorholt J. A. (2015) Methylobacterium extorquens: methylotrophy and biotechnological applications. Appl. Microbiol. Biotech. 99, 517–534 [DOI] [PubMed] [Google Scholar]
- 52. Schneider K., Skovran E., and Vorholt J. A. (2012) Oxalyl-Coenzyme A reduction to glyoxylate Is the preferred route of oxalate assimilation in Methylobacterium extorquens AM1. J. Bacteriol. 194, 3144–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Atamna-Ismaeel N., Finkel O., Glaser F., von Mering C., Vorholt J. A., Koblizek M., Belkin S., and Beja O. (2012) Bacterial anoxygenic photosynthesis on plant leaf surfaces. Env. Microbiol. Rep. 4, 209–216 [DOI] [PubMed] [Google Scholar]
- 54. Stiefel P., Zambelli T., and Vorholt J. A. (2013) Isolation of optically targeted single bacteria by application of fluidic force microscopy to aerobic anoxygenic phototrophs from the phyllosphere. Appl. Environ. Microbiol. 79, 4895–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ellis H. R. (2011) Mechanism for sulfur acquisition by the alkanesulfonate monooxygenase system. Bioorg. Chem. 39, 178–184 [DOI] [PubMed] [Google Scholar]
- 56. Knief C., Delmotte N., and Vorholt J. A. (2011) Bacterial adaptation to life in association with plants - A proteomic perspective from culture to in situ conditions. Proteomics 11, 3086–3105 [DOI] [PubMed] [Google Scholar]
- 57. Afroz A., Zahur M., Zeeshan N., and Komatsu S. (2013) Plant-bacterium interactions analyzed by proteomics. Front. Plant Sci. 4, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schmidt S., Christen P., Kiefer P., and Vorholt J. A. (2010) Functional investigation of methanol dehydrogenase-like protein XoxF in Methylobacterium extorquens AM1. Microbiology 156, 2575–2586 [DOI] [PubMed] [Google Scholar]
- 59. Kost T., Stopnisek N., Agnoli K., Eberl L., and Weisskopf L. (2014) Oxalotrophy, a widespread trait of plant-associated Burkholderia species, is involved in successful root colonization of lupin and maize by Burkholderia phytofirmans. Front. Microbiol. 4, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schoonbeek H. J., Jacquat-Bovet A. C., Mascher F., and Metraux J. P. (2007) Oxalate-degrading bacteria can protect Arabidopsis thaliana and crop plants against Botrytis cinerea. Mol. Plant Microbe Interact. 20, 1535–1544 [DOI] [PubMed] [Google Scholar]
- 61. Yu X., Lund S. P., Scott R. A., Greenwald J. W., Records A. H., Nettleton D., Lindow S. E., Gross D. C., and Beattie G. A. (2013) Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc. Natl. Acad. Sci. U.S.A. 110, E425–E434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Marco M. L., Legac J., and Lindow S. E. (2005) Pseudomonas syringae genes induced during colonization of leaf surfaces. Environ. Microbiol. 7, 1379–1391 [DOI] [PubMed] [Google Scholar]
- 63. Boulette M. L., Baynham P. J., Jorth P. A., Kukavica-Ibrulj I., Longoria A., Barrera K., Levesque R. C., and Whiteley M. (2009) Characterization of alanine catabolism in Pseudomonas aeruginosa and its importance for proliferation in vivo. J. Bacteriol. 191, 6329–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ryffel F., Helfrich E. J., Kiefer P., Peyriga L., Portais J. C., Piel J., and Vorholt J. A. (2015) Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. ISME J. 10, 632–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SWATH assay libraries for M. extorquens PA1 and S. melonis FR1 are available through the SWATHAtlas database (www.SWATHAtlas.org). All SWATH MS raw data have been made available through the PeptideAtlas database (http://www.peptideatlas.org/PASS/PASS00686).