Abstract
The initiation of B-cell ligand recognition is a critical step for the generation of an immune response against foreign bodies. We sought to identify the biochemical pathways involved in the B-cell ligand recognition cascade and sets of ligands that trigger similar immunological responses. We utilized several comparative approaches to analyze the gene coexpression networks generated from a set of microarray experiments spanning 33 different ligands. First, we compared the degree distributions of the generated networks. Second, we utilized a pairwise network alignment algorithm, BiNA, to align the networks based on the hubs in the networks. Third, we aligned the networks based on a set of KEGG pathways. We summarized our results by constructing a consensus hierarchy of pathways that are involved in B cell ligand recognition. The resulting pathways were further validated through literature for their common physiological responses. Collectively, the results based on our comparative analyses of degree distributions, alignment of hubs, and alignment based on KEGG pathways provide a basis for molecular characterization of the immune response states of B-cells and demonstrate the power of comparative approaches (e.g., gene coexpression network alignment algorithms) in elucidating biochemical pathways involved in complex signaling events in cells.
Keywords: Ligand recognition, B-cells, Gene coexpression network alignment
Introduction
B-cell ligand recognition plays a large role in various immune responses ranging from the recognition of foreign invaders such as viruses and bacteria to the recognition of cancerous cells. B-cells act as the body’s most effective line of defense to invaders [1]. Several types of responses may be induced in naïve mature B-cells through the activation of different receptors (e.g., cytokine and chemokine receptors) [2], [3]. Recognition of ligands by the B-cell Ag receptor (BCR) begins with the activation of an array of intracellular effector molecules and ends with phenotypic modifications that define the cell’s response to the stimulus [3]. As more and more players in this process are uncovered, the current schematic of BCR signal transduction has become a “labyrinth” of interconnecting pathways [4]. Despite the complicated events that occur during this process, the resultant reaction is very ordered and precise. The activation of various signal transduction pathways in mature B cells is influenced by the combination of ligands presented to the B-cells. The presence of different ligands may trigger cell-proliferation, activation, differentiation, migration, isotype switching and apoptosis [1], [5], [6]. Of particular interest in this area is the elucidation of the regulatory mechanisms that are involved in B-cell recognition of various ligands. These data provide a detailed look at the finite states that B-cells can enter upon exposure to ligands. Understanding the genetic interactions that are required for this process allows the design of drugs that are capable of triggering a specific immune response at a given time point, identifying the mechanisms that underly different auto-immune diseases, and allowing for the detection of key molecules involved in the regulation of B-cell function.
Several studies [7], [8], [9] have examined the changes in expression patterns of B-cells in response to exposure to different ligands. These studies used differential gene expression analysis of microarray data, such as Significance Analysis of Microarrays (SAM) [10] and Gene Ontology (GO) [11] terms, to detect genes that were significantly differentially expressed and whose pathway annotations shared significant GO terms. This approach, although well developed and widely used, suffers from an important limitation: it focuses on differences in expression patterns of individual genes across the different treatments or time points rather than differences between specific pathways/modules based on prior information of pathway relationships. It is of note that although software such as Gene Set Enrichment Analysis (GSEA) [12] conducts analysis based on pathways or selected groups of genes of interest, such methods do not account for the topology of networks or connectivity/relationships within the genes of interest.
Gene coexpression networks in which the nodes represent genes and the weighted links between pairs of nodes encode the correlations in expression patterns of the corresponding genes offer a useful way to represent cellular responses to each of the different treatments (e.g., exposure to different ligands). Network alignment methods are available to overcome the limitation of differentially expressed gene analysis and GO enrichment analysis [13], [14], [15], [16], [17], [18], [19], [20]. The advantage of using these methods is that they account for the connectivity of genes rather than focusing on single gene regulation. Hence, we utilized a pairwise network alignment algorithm, BiNA [21], to align 33 gene coexpression networks generated from a set of microarray experiments spanning 33 different ligands (see Table 1 for a complete list of the ligands) [8]. A network alignment (analogous to a sequence alignment) compares two input networks and returns a set of common pathways across the networks with a score denoting the similarity between the networks being compared. By constructing a symmetric 33 × 33 distance matrix using the alignment scores across the 33 networks, a hierarchical cluster was constructed based on the distance matrix to visualize relationships across the networks representing the gene expression changes due to exposure to different ligands. The common pathways detected across the most similar networks were examined and the pathways were annotated according to KEGG [22]. Using this approach, we examined the regulation mechanisms specific to certain groups of ligands. Based on this method, we identified a set of specific genes and pathways that appear to be involved in BCR-mediated ligand capture, vesicle function and vesicle trafficking during B-cell antigen processing and presentation for the set of 33 ligands we examined.
Table 1.
Full list of the ligands and their abbreviations examined in the current study
| Ligand abbreviation | Ligand name |
|---|---|
| 2MA | 2-Methyl-thio-ATP |
| AIG | Antigen (Anti-Ig) |
| BAF | BAFF (B-cell activating factor) |
| BLC | BLC (B-lymphocyte chemoattractant) |
| BOM | Bombesin |
| 40L | CD40 ligand |
| 70L | CD70/CD27 ligand |
| CGS | CGS-21680 hydrochloride (2-p-[2-Carboxyethyl]phenethylamino-5′-N-ethylcarboxamidoadenosine) |
| CPG | CpG-containing oligonucleotide |
| DIM | Dimaprit |
| ELC | ELC (Epstein Barr Virus-induced molecule-1 ligand chemokine) |
| FML | fMLP (formyl-Met-Leu-Phe) |
| GRH | Growth hormone-releasing hormone |
| IGF | Insulin-like growth factor 1 |
| IFB | Interferon-beta |
| IFG | Interferon-gamma |
| I10 | Interleukin 10 |
| IL4 | Interleukin 4 |
| LPS | Lipopolysaccharide |
| LB4 | Leukotriene B4 (LTB4) |
| LPA | Lysophosphatidic acid |
| M3A | MIP3-alpha (Macrophage inflammatory protein-3) |
| NEB | Neurokinin B |
| NPY | Neuropeptide Y |
| NGF | Nerve growth factor |
| PAF | Platelet activating factor |
| PGE | Prostaglandin E2 |
| SDF | SDF1 alpha (Stromal cell derived factor-1) |
| SLC | Secondary lymphoid-organ chemokine |
| S1P | Sphingosine-1-phosphate |
| TER | Terbutaline |
| TNF | Tumor necrosis factor-alpha |
| TGF | Transforming growth factor-beta 1 |
Note: This list was adapted from Lee et al. [7].
Results and discussion
Cells respond to stimuli through a myriad of pathways. However, they deploy similar modules in their response to distinct ligands. The major objective of this study was to explore the space of signaling responses of B-cells to naturally occurring stimuli and identify the commonality and differences in the ligand response. Such analysis will provide an insight into the space of responses of B-cells in native physiology and provide pathway motifs that can be explored through further experimentation.
We utilized several different approaches for comparing and aligning gene coexpression networks constructed from microarray data obtained from B-cells treated with different ligands. These include comparison of degree distributions of networks using Kolmogorov-Smirnoff statistic, and alignment of the networks based on the top 2000 highly connected nodes and based on KEGG pathways that were enriched with high intensity probes.
Clustering based on degree distribution
In order to determine the relationships of the ligand networks based on the network topology, we computed the degree distribution (Figure S1) for each of the 33 ligand networks. The degree distribution plots show the relationship between the degree of a node and the frequency of nodes with that degree (P(Degree)). We show that it is possible to get a reasonable estimate of the relationships between networks by utilizing only the degree distributions of the networks.
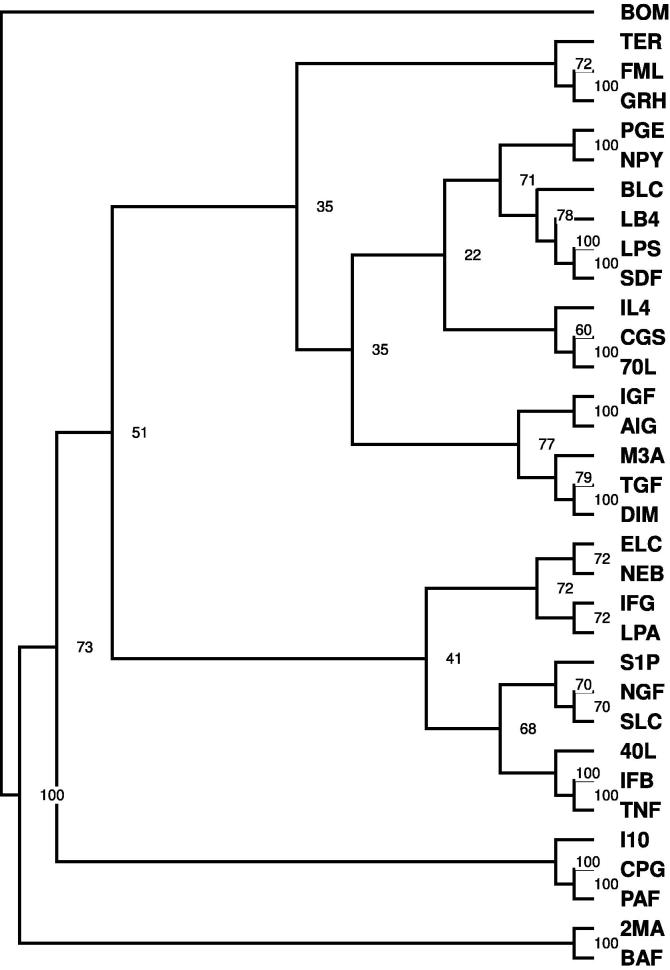
We compared the resulting 33 distributions using the two-sample Kolmogorov–Smirnov statistic [23]. Specifically, we used the Kolmogorov–Smirnov statistic to compute the 33 × 33 pairwise distances from the 33 degree distributions. Thus, we constructed a 33 × 33 matrix Dtoplogical where the entry in the i-th row and j-th column in the matrix corresponds to the distance between the degree distributions of the i-th and j-th networks as determined by the Kolmogorov–Smirnov statistic. The Dtoplogical matrix was then fed into a hierarchical neighbor-joining algorithm to construct the hierarchical cluster. Figure 1 shows the relationships between the ligand networks obtained by the topological comparison of the networks based on their degree distributions. Ligand networks with high number of (at least 100) differentially expressed genes at the 4 h time point relative to untreated samples, based on the classification of Lee et al. [7] using the SAM [10] tool, have been highlighted in the figure. As shown in Figure 1, ligand networks with a high number of differentially expressed genes relative to untreated samples share the same subtree/clade in the hierarchical network (P = 0.032, see “Hierarchical clustering” section in Methods). This result indicates that the network structure that was measured by the degree distribution and compared by the Kolmogorov–Smirnov statistic (similarly utilized in [24], [25], [26]) can be used to detect ligands that elicit similar responses upon exposure to B-cells.
Figure 1.
Network clustering based on degree distribution The figure shows the result of hierarchically clustering of the networks based on Kolmogorov–Smirnov test statisitic between degree distributions of the networks as distance measure of network similarity. Ligand networks with a high number of differentially expressed genes relative to untreated samples (as indicated in [7] have been highlighted in the figure (LPS, I04, BOM, 2MA, AIG, GRH, IFB, CGS, 40L, CPG). The clade with an asterisk (*) is highly enriched (P = 0.032 in ligand-response networks that induced a high number of differentially expressed genes).
Although topological comparison of gene coexpression networks based on their degree distributions is simple, intuitive, and computationally inexpensive, it fails to take into account the node labels or the biological annotation for the nodes in the networks. In order to compare the networks based on both the network topology and the node labels/biological annotation (e.g., signaling pathways, metabolic pathways…etc.) for the nodes, we utilized a network alignment algorithm implemented in the Biomolecular Network Alignment (BiNA) toolkit [21], [27].
Clustering based on alignment of high degree nodes in ligand networks
The network alignment algorithm implemented in BiNA allows the comparison of gene coexpression networks based on not only the extent to which they share similar topologies, but also the weights on the links (e.g., similarities in gene coexpression patterns) and the similarities of node and/or edge labels (biological annotations). We used the BiNA toolkit to run all-vs-all comparisons between all 33 ligand networks and construct a 33 × 33 distance matrix Dhubs whose entries signify the similarity score between ligands. Initially, we reduced the comparison to an alignment of the neighborhood around the top 2000 highly connected nodes (hubs) between all 33 ligand networks. Although we started aligning all nodes in the network, we quickly noticed that the total alignment score between two networks saturated after 2000 hubs (Figure S4). Specifically, to construct Dhubs, the output of a pairwise alignment between two networks (e.g., between ligand network 1, L1(V1, E1) and ligand network 2, L2(V2, E2)) is considered as a set of matched nodes S1 (for ligand network 1, where ) and S2 (for ligand network 2, where ) with a corresponding score set M. The corresponding entries , and Mi signify matching k-hop neighborhoods around the nodes and with a similarity score Mi (where 1 ⩽ i ⩽ 2000 since we are considering 2000 hubs). The overall pairwise similarity score between the two ligand networks is calculated by summing the scores across all matched neighborhoods (see Alignment subsection in Methods for more information on how neighborhood scores are calculated). The overall similarity scores between all 33 ligand networks were assembled into a similarity matrix Dhubs with each entry in the matrix signifying the similarity score between the ligand networks (e.g., entry in Dhubs contains the similarity score between ligand network 1 and ligand network 2 as determined by BiNA). The Dhubs matrix was then fed into a hierarchical neighbor-joining algorithm to construct the hierarchical cluster representing the similarity between the ligand networks.
Finally, in order to calculate confidence measures on the branches of the hierarchical clusters produced by the alignment, the tree produced by hierarchical clustering was bootstrapped [28], [29] by sampling randomly (with replacement) from the top 2000 hubs 100 times. This random resampling on the M set, followed by summing the scores of the resampled set for each cell in Dhubs results in 100 distance matrices which are fed into the same hierarchical neighbor-joining algorithm to construct 100 hierarchical similarity trees. The consensus tree of the hierarchical clusters based on the bootstrapped trees is produced using the Phylip [30] “consense” tool. Figure 2 shows the bootstrapped tree resulting from this method.
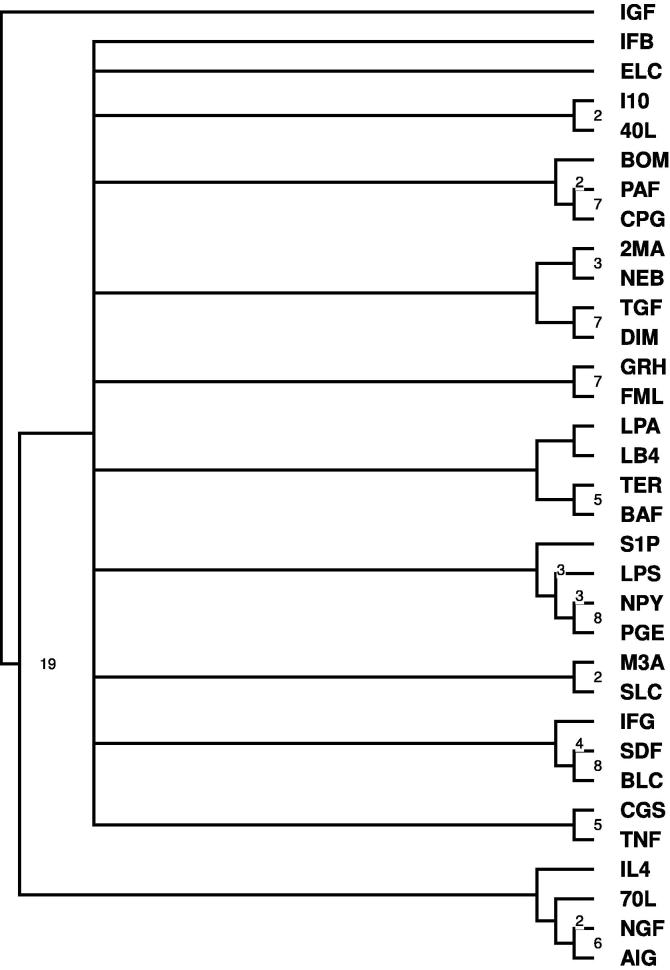
Figure 2.
Bootstrapped tree showing the relationship between all 33 ligand networks The tree was constructed using the network alignment score to measure the distance between networks. This tree shows that ligands with similar induced reaction (e.g., LPS and SDF, both affect pathways involved in cell migration) are clustered together.
Figure 2 shows that ligands with a similar induced reaction (e.g., LPS and SDF, both affect pathways involved in cell migration) are clustered together. It is important to note that the pathways necessary for migration would still be activated regardless of whether migration was the end point phenotypic response of B-cells to migratory ligands such as LPS and SDF, thus clustered together in our analysis. Such an analysis yields not only general similarity relationships between the ligand networks, but also provides specific gene and pathway information as seen from clustering based on signaling pathways (see below). The cluster shown in Figure 2 describes the similarity of expression based on node labels as well as correlation between the genes in the ligand networks. However, the hierarchical cluster from Figure 2 does not provide specific information as to which sets of pathways are shared/similarly regulated across ligand networks that fall under the same clade/subtree in the hierarchical cluster. KEGG [22] annotation of pathways was used to link the node labels in the networks to biological pathways (such as metabolism or signal processing). The additional pathway annotation can be used to determine the specific biological pathways that are involved in B-cell ligand recognition, and how those pathways are regulated based on exposure to each ligand. This procedure is described in detail in the next section.
Clustering based on ligand similarity across signaling pathways
We wanted to choose pathways based on the highly regulated genes in the microarray dataset rather than relying on a priori knowledge from the literature. The reasons for this choice are: (i) a choice of pathways that is unbiased by what is currently known in the literature can help identify novel pathways involved in B-cell ligand recognition (ii) if the list of pathways determined to be highly regulated based on the microarray data happens to share a high degree of overlap with the list generated based on literature surveys, it helps establish the utility of the approach in settings where prior knowledge available in the literature is quite sparse.
We choose pathways according to the following procedure. Firstly (step 1), in the fully normalized dataset (all 422 microarray samples), we search for genes that meet the following criteria (referred to as “high intensity” genes in what follows). Briefly, we wanted to maximize the sensitivity of detection of genes that are differentially regulated upon exposure of B-cells to ligands compared to untreated B-cells. This procedure maximizes sensitivity at the cost of specificity. The list of genes generated by this approach will be further reduced by comparing the neighborhoods in the ligand networks using network alignments. To do this, we (a) calculate the fold difference between the average probe expression level and the expression level for all probes in each sample (see Methods section); (b) select probes whose fold-difference is higher than 1 in at least one of the 422 samples and (c) of the probes selected in step (b), find probes that are expressed at least 1-fold higher compared to the same probes from the untreated samples. Secondly (step 2), once the high intensity probes are selected from (c), the probe IDs are mapped back to their respective gene IDs. Lastly (step 3), among all the pathways in KEGG, we count the number of genes from step 2 that show up in each KEGG pathway.
The results of the preceding steps are summarized in Table 2. As shown in Table 2, many of the pathways enriched in high-intensity genes are known to be implicated in the development of the immune system and processing of ligands. It should be noted that although KEGG considers the immune system pathways (KEGG category 5.1) to be a part of organismal system (KEGG category 5), we considered the immune system pathways separately (Table 2) since we wanted to specifically examine the immune system pathways.
Table 2.
List of pathways detected based on high-intensity probes from the microarray data
| KEGG pathway category | No. of subpathways | KEGG subpathway ID |
|---|---|---|
| Cellular processes | 10 | mmu04142, mmu04144, mmu04145, mmu04520, mmu04540, mmu04810, mmu04110, mmu04114, mmu04115, mmu04140 |
| Environmental information processing | 2 | mmu04150, mmu04310 |
| Organismal system | 6 | mmu04962, mmu04964, mmu04966, mmu04260, mmu04722, mmu04910 |
| Genetic information processing | 15 | mmu03020, mmu03022, mmu03030, mmu03040, mmu03050, mmu03060, mmu03410, mmu03420, mmu03430, mmu03440, mmu04120, mmu04130, mmu00970, mmu03010, mmu03018 |
| Human diseases | 12 | mmu05100, mmu05210, mmu05212, mmu05214, mmu05215, mmu05216, mmu05219, mmu05222, mmu05010, mmu05012, mmu05014, mmu05016 |
| Immune system | 4 | mmu04623, mmu04662, mmu04666, mmu04622 |
| Metabolism | 19 | mmu00020, mmu00030, mmu00051, mmu00072, mmu00100, mmu00130, mmu00190, mmu00230, mmu00240, mmu00260, mmu00290, mmu00460, mmu00510, mmu00511, mmu00563, mmu00630, mmu00670, mmu00740, mmu00900 |
Note: This table with pathway names and relative number of genes enriched in the pathway based on the data. Please see Table S1 for more detail.
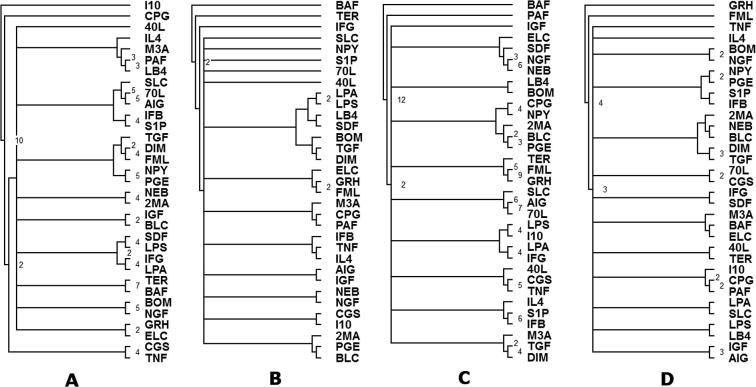
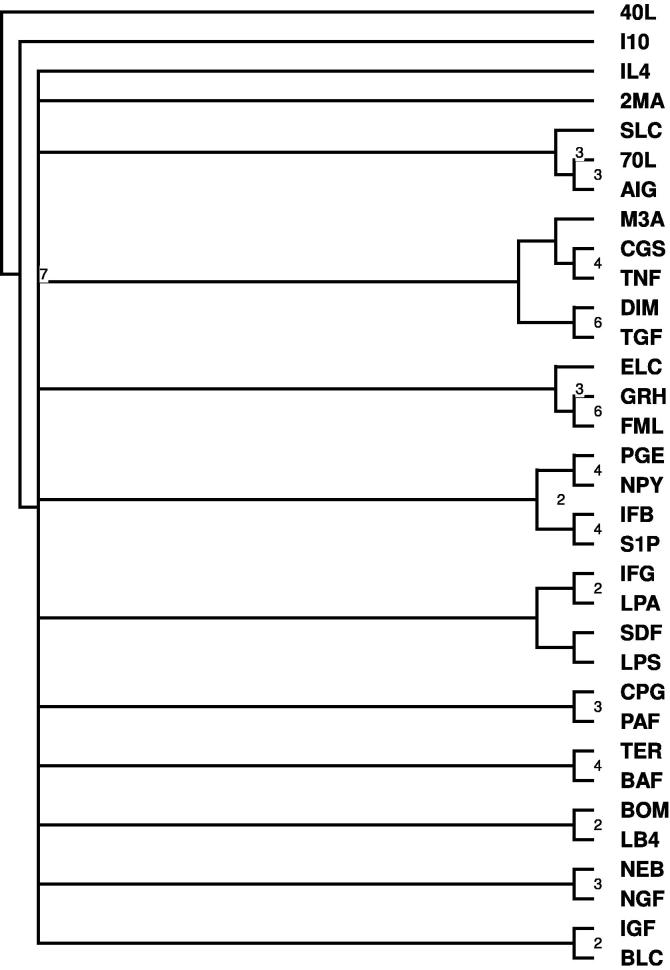
After considering all pathways of each of the seven general KEGG categories summarized in Table 2, we constructed a clustering tree for each pathway across each of the subcategories, a consensus network across each of the subcategories (Figure 3, Figure 4, Figure 6 and S3) and a consensus network based on all the networks in Table 2 (shown in Figure 5).
Figure 3.
Consensus tree constructed based on all metabolism pathways inTable 2 The tree was constructed using the network alignment score to measure the distance between networks. The values on the branches indicate the total number of times that the branch appeared across all networks (total of 19). If no value is indicated, the branch appeared only once.
Figure 4.
Consensus tree constructed based on all Genetic Information Processing Pathways inTable 2 The tree was constructed using the network alignment score to measure the distance between networks. The values on the branches indicate the total number of times that the branch appeared across all networks (total of 15). If no value is indicated, the branch appeared only once.
Figure 6.
Consensus trees constructed based on other pathways inTable 2 Consensus tree was constructed based on other pathways in Table 2 including all cellular processes pathways (A), all environmental information processing pathways (B), all human diseases pathways (C) and all immune system pathways (D), respectively. The values on the branches indicate the total number of times that the branch appeared across all networks (totals of 10, 2, 12, and 4 for A, B, C, and D, respectively). If no value is indicated, the branch appeared only once.
Figure 5.
Consensus of all pathway categories inTable 2 The values on the branches indicate the total number of times that the branch appeared across all networks (total of 7). If no value is indicated, the branch appeared only once.
Figure 3, Figure 4 present examples of the alignment based on the KEGG metabolism and Genetic Information Processing pathways. The numbers on the branches signify the number of similarly regulated subpathways between any two ligands. It was shown that some ligand networks (e.g., TER/BAF and FML/GRH) fall under the same clade/subtree in the two pathways, signifying general similarity in the regulation/signaling of pathways by such ligands. Differences between the trees show that the ligands may have different effects depending on the pathway being observed.
Figure 5 shows a consensus tree based on all seven general pathway categories highlighted in Table 2. GRH and FML, for example, fall under the same clade/subtree in the consensus tree in Figure 2 and the consensus tree constructed based on differentially expressed pathways (Table 2) shown in Figure 5. Overall, this shows that the results of the alignment is consistent across the different pathways chosen to ascertain the similarity hierarchy between the overall networks. The numbers on the branches can also serve as confidence measures for grouping certain leaves/networks with each other.
We also utilized specific signaling pathways highlighted in the literature [7], [8] (Table S1) to align the networks and constructed a cladogram (Figure S2) describing the relationship between the ligands. The results from the alignments showed that some ligands tend to have similar expression patterns based on the KEGG pathways used to anchor the pairwise all-vs-all alignments for the 33 ligand networks. Table 3 presents a detailed list of ligands that induce similar expression cascades in the KEGG pathways highlighted in Table 2. Several of the matched ligands (Figure 5) are actually known to induce similar reactions in B-cells based on a literature search we conducted. It is important to point out that the algorithm is detecting expression patterns that are similar in B-cells across different ligands, though not all such patterns may necessarily be important for cell function.
Table 3.
Top matched ligands based on expression patterns in the consensus tree shown in Figure 5
| Matched ligands | Conserved KEGG pathway categories | Conserved KEGG subpathways |
|---|---|---|
| 70L/AIG/SLC | Cellular processes, human diseases, organismal system | Cell cycle, p53 signaling pathway, phagosome, Parkinson’s disease, Huntington’s disease |
| LPA/IFG | Cellular processes, human diseases | p53 signaling pathway, bacterial invasion of epithelial cells |
| GRH/FML | Cellular processes, environmental information processing, genetic information processing, Human diseases, metabolism, organismal system | Cell cycle, regulation of autophagy, Aminoacyl-tRNA biosynthesis, ribosome, RNA degradation, RNA polymerase, DNA replication, ubiquitin mediated proteolysis, Parkinson’s disease, Huntington’s disease, thyroid cancer, TCA cycle, oxidative phosphorylation, pyrimidine metabolism, glyoxylate and dicarboxylate metabolism |
| PGE/NPY | Cellular processes, immune system, metabolism, organismal system | Oocyte meiosis, cytosolic DNA-sensing pathway, Fc gamma R-mediated phagocytosis, TCA cycle, ubiquinone and other terpenoid-quinone biosynthesis, oxidative phosphorylation, pyrimidine metabolism, riboflavin metabolism, terpenoid backbone biosynthesis |
| IFB/S1P | Cellular processes, human diseases, immune system, organismal system | Cell cycle, oocyte meiosis, p53 signaling pathway, Parkinson’s disease, Huntington’s disease, bacterial invasion of epithelial cells, Fc gamma R-mediated phagocytosis |
| BOM/LB4 | Human diseases, organismal system | Colorectal cancer, Glioma, Cardiac muscle contraction |
| NEB/NGF | Environmental information processing, human diseases, organismal system | mTOR signaling pathway, Parkinson’s disease, Amyotrophic lateral sclerosis, Colorectal cancer, Glioma, Neurotrophin signaling pathway |
| TNF/CGS | Cellular processes, genetic information processing, human diseases, metabolism | Cell cycle, p53 signaling pathway, ribosome, DNA replication, mismatch repair, SNARE interactions in vesicular transport, Parkinson’s disease, bacterial invasion of epithelial cells, steroid biosynthesis, oxidative phosphorylation, glyoxylate and dicarboxylate metabolism |
| PAF/CPG | Environmental information processing, immune system, metabolism | RIG-I-like receptor signaling pathway, cytosolic DNA-sensing pathway, pyrimidine metabolism, cyanoamino acid metabolism, one carbon pool by folate, riboflavin metabolism |
| TER/BAF | Cellular processes, environmental information processing, genetic information processing, metabolism | Cell cycle, oocyte meiosis, p53 signaling pathway, endocytosis, aminoacyl-tRNA biosynthesis, RNA degradation, spliceosome, ubiquitin mediated proteolysis, TCA cycle, pentose phosphate pathway, cyanoamino acid metabolism |
| DIM/TGF | Environmental information processing, genetic information processing, human diseases, immune system, metabolism, organismal system | Aminoacyl-tRNA biosynthesis, ribosome, RNA polymerase, basal transcription factors, spliceosome, protein export, mismatch repair, bacterial invasion of epithelial cells, colorectal cancer, RIG-I-like receptor signaling pathway, cytosolic DNA-sensing pathway, B-cell receptor signaling pathway, TCA cycle, pentose phosphate pathway, steroid biosynthesis, oxidative phosphorylation |
For example, lipopolysaccharide (LPS) and stromal cell derived factor-1 (SDF) are known to affect cellular migration, interferon-gamma (IFG) and lysophosphatidic acid (LPA) are known to trigger changes in isotype switching [7], [8]. Macrophage inflammatory protein-3 (M3A)/dimaprit (DIM)/transforming growth factor-beta 1 (TGF) have several effects: M3A is strongly chemotactic for lymphocytes, DIM, an analog to histamine, activates immune response, while TGF provides a chemotactic gradient for leukocytes and down-regulates the activity of immune cells [31]. Neurokinin B (NEB) and nerve growth factor (NGF) have both been shown to be involved in the growth and development of neurons [32], [33]. Furthermore, tumor necrosis factor-alpha (TNF) has been shown to be highly involved in mediating inflammatory and immune responses [34], similar to what has been recently observed using CGS (CGS-21680 hydrochloride) [35].
In addition, the relationship between each of the above ligands as to exactly which ligands trigger similar expression patterns in the selected KEGG subpathways is also shown (Tables 3 and S3, the expanded version of Table 3). We can see that several major pathways are regulated in B-cells in response to the exposure to the 33 ligands shown in Table 1. First, human disease pathways (e.g., cancer and asthma) are the most prevalent pathways triggered by over half the ligands: 70L, AIG, SLC, LPA, IFG, GRH, FML, IFB, S1P, BOM, LB4, NEB, NGF, TNF, CGS, DIM and TGF. Those ligands constitute a set of molecules that trigger a wide variety of responses in B-cells and can be used to further ascertain the conditions under which B-cells activate under certain situations in human diseases. Second, cellular process pathways (e.g., endocytosis and apoptosis) seem to be also over-represented among the pathways that significantly change in expression across upon exposure to ligands. Some of the ligands (70L, AIG, SLC, LPA, IFG, GRH, FML, IFB, S1P, TNF and CGS) seem to trigger both human disease and cellular process pathways, while other ligands (PGE, NPY, TER and BAF) only trigger cellular pathways. Such ligands constitute a set of molecules that trigger changes in B-cells that may affect their growth and proliferation. The third major pathway commonly regulated in B-cells upon ligand exposure is metabolism with a sizable number of ligands (GRH, FML, PGE, NPY, TNF, CGS, PAF, CPG, TER, BAF, DIM and TGF) triggering pathways in that category. Ligands that only triggered pathways in B-cells related to metabolism but not “human diseases” or “cellular processes” are PGE, NPY, PAF, CPG. Since those ligands are known to affect inflammation and antibody production, the metabolic pathways expressed as a result of B-cell exposure to those ligands may be important indicators of B-cell immune response.
Conclusion
Identifying sets of ligands that trigger similar B-cell responses provides a basis for elucidating the specific genetic interactions that play a role in the recognition of ligands by B-cells. To achieve this goal, we constructed 33 gene coexpression networks that represented the genetic interactions in B-cells after exposure to each of the 33 ligands. Each network represents the response of normal splenic B-cells to a specific ligand across four different time points with three replicates per time point. We then utilized several comparative approaches to identify shared subnetworks/pathways among the 33 networks. Based on those pathways (Table 2), we were able to identify ligands that trigger similar expression changes in each of the pathways (Table 3, Figure 5, Figure 6, and Supplementary materials).
Aligning the 33 ligand networks allowed the detection of the specific relationships between the ligands in terms of the pathways that they regulate in B-cells. Additionally, the alignment pointed out specific pathways that share expression patterns across ligands and are involved in BCR activation. We have been able to validate some of the relationships we uncovered based on the immune responses described in the literature in the case of some of the ligands in our dataset. The computation tools and methods we utilized for constructing the alignments and analyzing the results are available online as part of the BiNA (Biomolecular Network Alignment) toolkit http://www.cs.iastate.edu/~ftowfic. An analysis pipeline based on network alignment such as the one used in this study may also serve as a general template for identifying pathways with conserved expression patterns across different conditions in other types of experiments. Some promising directions for further work include integrating additional types of information (e.g., protein–protein interaction networks) in our analyses and overlaying our pathways with already known protein–protein interactions to detect specific proteins that are responsible for triggering the signaling cascades for each ligand. Such information can aid in narrowing down the list of pathways to their core protein interactions.
Materials and methods
Microarray data
The microarray data [7], [8] were collected from the Alliance for Cell Signaling (AfCS) site (http://www.signaling-gateway.org/) [36]. Briefly, the experiments were designed to examine gene expression changes induced by the 33 single ligands.
Mouse splenic B-cells were cultured with ligands in serum-free medium for 0.5, 1, 2, and 4 h. cDNA synthesized from the RNA of B-cells was labeled with Cy5 and hybridized onto custom-made two-color Agilent cDNA arrays (Containing 16273 probes) with a Cy3-labeled cDNA prepared from the RNA of total splenocytes. There were a total of 424 Agilent chips hybridized in this study [7], [8].
The data was processed using MatLab® Bioinformatics toolbox. The background corrected intensity values were used for each chip. Some of the background corrected intensities were negative and made it difficult to take the logarithm of the data. To circumvent this problem, a very low positive value (10, a value that was 500 times below the mean intensity of all chips) was assigned to these probes. Each chip was also normalized to its mean intensity. Chip-to-chip normalization was performed via the LOWESS normalization method to allow for adequate analysis between chips [37]. After the normalization, the replicate chips were averaged. To remove the outliers each replicated probe was subjected to an outlier test. The outlier test was as follows: First, we calculate the mean and standard deviation (SD) for all replicates of each probe. Second, select the probes in the range of mean ± 1.2 SD for the calculation of a new mean and SD. Third, we discard the probes out of the range of the new mean ± 2 new SD. Finally, we calculate the fold change as ligand treated divided by control (untreated) samples for each probe on the chip. The log fold-change was calculated using R’s [38] BioConductor [39] package.
Construction of gene coexpression networks
After obtaining the expression matrices for each of the 33 ligands (33 expression matrices total), we merged expression levels from probesets that mapped onto the same gene. This was done by averaging the log(FC) values across the probesets that mapped to the same gene as indicated by the microarray chip annotation information provided by Agilent. After obtaining a single expression matrix per ligand (where rows in the matrix are genes and columns are the replicates/timepoints for that particular ligand), Pearson correlation was used to obtain the gene coexpression matrices. We obtained 33 gene coexpression matrices (E1…33), one for each ligand, then applied a correlation cutoff of ⩾0.8 to sparsify the matrices. Entries in the matrix Ek were set to 0 whenever for 1 ⩽ k ⩽ 33 and 1 ⩽ i,j ⩽ n where n is the number of genes/rows in the matrix Ek. Remaining entries signified edges in the networks that connected genes whose expression patterns were correlated above our chosen cutoff. It is important to note that when a gene does not change in treatment samples (distribution of expression follows a normal distribution) relative to control (also a normal distribution due to normalization), the correlation is 0. As such, the edge does not exist in the graph. Additionally, we did not disregard any nodes in the networks explicitly based on a strict cutoff of differential expression since we did not want to bias the network analysis based on network size. As a result, all genes were considered in our analysis. The resulting networks were treated as undirected, weighted graphs with an average of 10,000 nodes (genes) and 1 million edges ( million possible edges in a fully connected graph). We varied the threshold cutoff around our chosen value (0.8) from [0.78, 0.82] in 0.01 increments and the distances between the degree distributions (see Figure S1 for example) of the ligand networks did not significantly (P < 0.01) differ as measured by the Friedman test. We also removed edges whose P value (calculated using Student’s t-distribution for a transformation of the correlation as implemented in Matlab) did not pass a significance threshold of P < 0.05. The percentage of edges removed using the correlation significance procedure is indicated in Table S4. Removing such edges did not influence the results as measured by comparing the degree distributions of the networks with and without such edges using the Friedman test.
Gene coexpression network alignment
Given two gene coexpression networks (graphs 1 and 2), the graphs are treated as weighted (where the weights on the edges denote the pairwise correlation in the expression of the corresponding genes). A k-hop neighborhood-based approach to alignment is used [21], [27]. The k-hop neighborhood of a vertex of the graph G1(V1, E1) is simply a subgraph of G1 that connects with the vertices in V1 that are reachable in k-hops from using the edges in E1. Given two graphs G1(V1, E1) and G2(V2, E2), a mapping matrix P that associates each vertex in V1 with zero or more vertices in V2 (the matrix P can be constructed based on BLAST matches or gene IDs. In our analysis, using a 1-to-1 mapping between expression networks based on gene IDs and a user-specified parameter k, we construct for each vertex its corresponding k-hop neighborhood Cx in G1. We then use the mapping matrix P to obtain the set of matches for vertex among the vertices in V2 and construct the k-hop neighborhood Zy for each matching vertex in G2 and . Let be the resulting collection of k-hop neighborhoods in G2 associated with the vertex in G1. We compare each k-hop subgraph Cx in G1 with each member of the corresponding collection to identify the k-hop subgraph of G2 that is the best match for Cx (based on a chosen similarity measure). We utilized a k-hop value of 1 for the analysis we discussed in this paper. The analysis was conducted on eight nodes from the San Diego Supercomputer Center’s Triton cluster with eight cores and 24 GB of memory per node.
Shortest path graph kernel score
The shortest path graph kernel was first described by Borgwardt and Kriegel [40]. The kernel acts as a scoring function that compares the length of the shortest paths between any two nodes in a graph based on a pre-computed shortest-path distance. The shortest path distances for each graph may be computed using the Floyd–Warshall algorithm. We modified the Shortest-Path Graph Kernel to take into account the labels of the nodes being compared as computed by BLAST [41] or as a mapping in the mapping matrix P. The shortest path graph kernel for subgraphs and (e.g., k-hop subgraphs) is given by:
where and are the lengths of the shortest paths between and computed by the Floyd–Warshall algorithm. For gene coexpression networks, the Floyd–Warshall algorithm takes into account the weight of the edges (correlations) in the graphs. The runtime of the Floyd–Warshall Algorithm is O(n3). The shortest path graph kernel has a runtime of O(n4) (where n is the maximum number of nodes in the larger of the two graphs being compared).
Hierarchical clustering
A set of symmetric 33 × 33 distance matrices using the alignment scores across the 33 networks was constructed. Each matrix was constructed based on a specific subset of genes on the microarray chip (e.g., all genes involved in Calcium Signaling Pathway, all genes involved in Notch Signaling Pathway…etc. Please see Table 2, S1 and S2 for a full list of pathways utilized for comparing the networks). For each matrix, the diagonals contained the sum of the rows in the matrix and the off diagonals contained the alignment score comparing the network from row i with the network in column j where 1 ⩽ i,j ⩽ 33. The hierarchical cluster was constructed using a neighbor-joining method based on the distance matrix in Matlab. The hierarchical cluster can be used to visualize the relationship across the networks representing the gene expression changes due to exposure to different ligands. TreeView [42] was used to visualize the hierarchical clusters and the “consense” program of Phylip [30] was used to merge hierarchical clusters and to compute majority-rule consensus trees. The majority rule consensus approach has been shown to minimize the number of false groupings and provides a good summary of the posterior distribution over the trees that were used to construct the consensus tree [43]. Significance of clusters was computed using a hypergeometric distribution using the simple scheme:
where d is the number of ligands that had a high number of differentially expressed genes (10, as highlighted in Figure 1). c is the number of ligands in the cluster (17, which includes TFR, SLC, IGF, TGF, IFG, CPG, M3A, S1P, 40L, CGS, IFB, 70L, GRH, AIG, 2MA, ELC, BOM). l is the number of ligands in the experiment (namely 33), and r is the number of ligands that had a high number of differentially expressed genes in the cluster (eight from Figure 1, namely: CPG, 40L, CGS, IFB, GRH, AIG, 2MA, and BOM).
Authors’ contributions
FT and SG assembled and verified the datasets for the analysis. FT wrote the algorithms, FT and SG ran the experiments and drafted the manuscript. SS and VH supervised the analysis, the algorithm design and manuscript revisions. All authors read and approved the final manuscript.
Competing interests
The authors declared that they have no competing interests.
Acknowledgments
This research was supported in part by a Cornette Fellowship award and an Integrative Graduate Education and Research Training (IGERT) fellowship to FT, funded by the National Science Foundation (NSF) Grant (DGE 0504304) to Iowa State University and NSF Grants 0939370, 0835541 and 0641037 awarded to SS. We also thank Raj Srikrishnan for his help in data processing. The work of VH was supported by the NSF, while working at the Foundation. Any opinion, finding, and conclusions contained in this article are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Supplementary material associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.gpb.2012.03.001.
Supplementary material
Supplementary Figure 1.
Number of clusters/k-hop neighborhoods needed for the network alignment This plot shows that we need around 638 nodes with the highest degree to accurately approximate the total alignment score if we had used up to 5,000 k-hops. In other words, one can reach within 0.9 or 90th percentile of the total score (that results from utilizing 5000 k-hop neighborhoods) by utilizing only the k-hop neighborhoods around the top 638 highly connected nodes. In summary, this plot shows that we only need to compute the alignment of the top 638 nodes if we are simply interested in the distance between two networks. The full alignment may later be computed after determining which networks are closest.
Example of degree distributions used for Kolmogorov-Smirnov test for initial clustering of the ligands based on network topology As seen from the figure, the expression networks exhibit scale-free like behavior as described by Barabsi and Oltavai [1].
Consensus tree constructed based on all KEGG pathways in Table S1 The values on the branches indicate the total number of times that the branch appeared across all networks (total of 11). If no value is indicated, the branch appeared only once.
Consensus tree constructed based on all organismal system pathways in Table S2 The values on the branches indicate the total number of times that the branch appeared across all networks (total of 6). If no value is indicated, the branch appeared only once.
Schematic of the experiments utilized to construct the gene coexpression networks for the 33 Ligands B cells were stimulated with 33 possible ligands (listed in Table 1) and harvested at 0.5,1, 2 and 4 hours (hr) post stimulation. Gene expression was assayed using Agilent cDNA arrays. Gene coexpression networks were then constructed by assembling the time point expression data from each stimulation and the correlation between all probes was then measured across all 4 time points (and 3 replicates per time point). A correlation magnitude cutoff of 0.8 was utilized to finally yield the networks used for this study (positive correlations are indicated by solid edges, while negative correlations are indicated by dashed edges in the figure). The networks were compared using a network alignment approach that took into account the weight on the edges around each respective node. Networks with similar topologies and weights around respective genes had higher score than those with a good toplogical match, but poor weight matches around matching genes.
Supplementary Tables
References
- 1.Chaplin D.D. Overview of the immune response. J Allergy Clin Immunol. 2010;125:S3–23. doi: 10.1016/j.jaci.2009.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeFranco A.L. Molecular aspects of B-lymphocyte activation. Annu Rev Cell Biol. 1987;3:143–178. doi: 10.1146/annurev.cb.03.110187.001043. [DOI] [PubMed] [Google Scholar]
- 3.Hsueh R.C., Scheuermann R.H. Tyrosine kinase activation in the decision between growth, differentiation, and death responses initiated from the B cell antigen receptor. Adv Immunol. 2000;75:283–316. doi: 10.1016/s0065-2776(00)75007-3. [DOI] [PubMed] [Google Scholar]
- 4.Dal Porto J.M., Gauld S.B., Merrell K.T., Mills D., Pugh-Bernard A.E., Cambier J. B cell antigen receptor signaling 101. Mol Immunol. 2004;41:599–613. doi: 10.1016/j.molimm.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Saitoh T., Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010;189:925–935. doi: 10.1083/jcb.201002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harwood N.E., Batista F. Early events in B cell activation. Annu Rev Immunol. 2009;28:185–210. doi: 10.1146/annurev-immunol-030409-101216. [DOI] [PubMed] [Google Scholar]
- 7.Lee J.A., Sinkovits R.S., Mock D., Rab E.L., Cai J., Yang P. Components of the antigen processing and presentation pathway revealed by gene expression microarray analysis following B cell antigen receptor (BCR) stimulation. BMC Bioinformatics. 2006;7:237. doi: 10.1186/1471-2105-7-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu X., Hart R., Chang M.S., Kim J.W., Lee S.Y., Cao Y.A. Analysis of the major patterns of B cell gene expression changes in response to short-term stimulation with 33 single ligands. J Immunol. 2004;173:7141–7149. doi: 10.4049/jimmunol.173.12.7141. [DOI] [PubMed] [Google Scholar]
- 9.Murn J., Mlinaric-Rascan I., Vaigot P., Alibert O., Frouin V., Gidrol X. A Myc-regulated transcriptional network controls B-cell fate in response to BCR triggering. BMC Genomics. 2009;10:323. doi: 10.1186/1471-2164-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Pro Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koyutürk M., Kim Y., Topkara U., Subramaniam S., Szpankowski W., Grama A. Pairwise alignment of protein interaction networks. J Comput Biol. 2006;13:182–199. doi: 10.1089/cmb.2006.13.182. [DOI] [PubMed] [Google Scholar]
- 14.Tian W., Samatova N.F. Pairwise alignment of interaction networks by fast identification of maximal conserved patterns. Pac Symp Biocomput. 2009;14:99–110. [PubMed] [Google Scholar]
- 15.Flannick J., Novak A., Srinivasan B.S., McAdams H.H., Batzoglou S. Graemlin: General and robust alignment of multiple large interaction networks. Genome Res. 2006;16:1169–1181. doi: 10.1101/gr.5235706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalaev M., Smoot M., Ideker T., Sharan R. NetworkBLAST: comparative analysis of protein networks. Bioinformatics. 2008;24:594–596. doi: 10.1093/bioinformatics/btm630. [DOI] [PubMed] [Google Scholar]
- 17.Kelley B.P., Yuan B., Lewitter F., Sharan R., Stockwell B.R., Ideker T. PathBLAST: a tool for alignment of protein interaction networks. Nucleic Acids Res. 2004;32:W83–W88. doi: 10.1093/nar/gkh411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott J., Ideker T., Karp R.M., Sharan R. Efficient Algorithms for Detecting Signaling Pathways in Protein Interaction Networks. J Comput Biol. 2006;13:133–144. doi: 10.1089/cmb.2006.13.133. [DOI] [PubMed] [Google Scholar]
- 19.Sharan R., Ideker T. Modeling cellular machinery through biological network comparison. Nat Biotechnol. 2006;24:427–433. doi: 10.1038/nbt1196. [DOI] [PubMed] [Google Scholar]
- 20.Liao C.S., Lu K., Baym M., Singh R., Berger B. IsoRankN: spectral methods for global alignment of multiple protein networks. Bioinformatics. 2009;25:i253–i258. doi: 10.1093/bioinformatics/btp203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Towfic F, Heather M, Greenlee W, Honavar V. Aligning biomolecular networks using modular graph kernels. In: Salzberg SL, Warnow T, editors. WABI’09 Proceedings of the 9th international conference on Algorithms in bioinformatics. Springer-Verlag Berlin Heidelberg 2009; LNBI Vol. 5724, p. 345–61.
- 22.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M. Kegg for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsaglia G., Tsang W.W., Wang J. Evaluating Kolmogorov’s distribution. J Stat Softw. 2003;8:1–4. [Google Scholar]
- 24.Goñi J., Esteban F.J., de Mendizábal N.V., Sepulcre J., Ardanza-Trevijano S., Agirrezabal I. A computational analysis of protein-protein interaction networks in neurodegenerative diseases. BMC Syst Biol. 2008;2:52. doi: 10.1186/1752-0509-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altay G., Emmert-Streib F. Structural influence of gene networks on their inference: analysis of C3NET. Biol Direct. 2011;6:31. doi: 10.1186/1745-6150-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kugler K.G., Mueller L.A., Graber A., Dehmer M. Integrative network biology: Graph prototyping for co-expression cancer networks. PloS One. 2011;6:e22843. doi: 10.1371/journal.pone.0022843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Towfic F., VanderPlas S., Oliver C.A., Couture O., Tuggle C.K., West Greenlee M.H. Detection of gene orthology from gene co-expression and protein interaction networks. BMC Bioinformatics. 2010;11(Suppl 3):S7. doi: 10.1186/1471-2105-11-S3-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Efron B. The jackknife, the bootstrap and other resampling plans, vol. 38. Society for Industrial Mathematics; 1982.
- 29.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 30.Felsenstein, J. P HYLIP (phylogeny inference package) version 3.6. Distributed by the author. Seattle: Department of Genome Sciences, University of Washington; 2005.
- 31.Letterio J.J., Roberts A.B. Regulation of immune responses by TGF-beta. Annu Rev Immunol. 1998;16:137–161. doi: 10.1146/annurev.immunol.16.1.137. [DOI] [PubMed] [Google Scholar]
- 32.Fiore M., Chaldakov G.N., Aloe L. Nerve growth factor as a signaling molecule for nerve cells and also for the neuroendocrine-immune systems. Rev Neurosci. 2009;20:133–145. doi: 10.1515/revneuro.2009.20.2.133. [DOI] [PubMed] [Google Scholar]
- 33.Topaloglu A.K., Reimann F., Guclu M., Yalin A.S., Kotan L.D., Porter K.M. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2008;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pradervand S., Maurya M.R., Subramaniam S. Identification of signaling components required for the prediction of cytokine release in RAW 264.7 macrophages. Genome Biol. 2006;7:R11. doi: 10.1186/gb-2006-7-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vuaden F.C., Savio L.E., Bastos C.M., Bogo M.R., Bonan C.D. Adenosine A(2A) receptor agonist (CGS-21680) prevents endotoxin-induced effects on nucleotidase activities in mouse lymphocytes. Eur J Pharmacol. 2011;651:212–217. doi: 10.1016/j.ejphar.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Dinasarapu A.R., Saunders B., Ozerlat I., Azam K., Subramaniam S. Signaling gateway molecule pages—a data model perspective. Bioinformatics. 2011;27:1736–1738. doi: 10.1093/bioinformatics/btr190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32 Suppl:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 38.R Development Core Team. R: a language and environment for statistical computing. http://www.R-project.org; 2010. ISBN 3-900051-07-0.
- 39.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borgwardt K, Kriegel H. Shortest-path kernels on graphs. In: Proceedings of the fifth IEEE international conference on data mining; 2005. p. 74–81.
- 41.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Page R.D. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 43.Holder M.T., Sukumaran J., Lewis P.O. A justification for reporting the majority-rule consensus tree in Bayesian phylogenetics. Syst Biol. 2008;57:814–821. doi: 10.1080/10635150802422308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Example of degree distributions used for Kolmogorov-Smirnov test for initial clustering of the ligands based on network topology As seen from the figure, the expression networks exhibit scale-free like behavior as described by Barabsi and Oltavai [1].
Consensus tree constructed based on all KEGG pathways in Table S1 The values on the branches indicate the total number of times that the branch appeared across all networks (total of 11). If no value is indicated, the branch appeared only once.
Consensus tree constructed based on all organismal system pathways in Table S2 The values on the branches indicate the total number of times that the branch appeared across all networks (total of 6). If no value is indicated, the branch appeared only once.
Schematic of the experiments utilized to construct the gene coexpression networks for the 33 Ligands B cells were stimulated with 33 possible ligands (listed in Table 1) and harvested at 0.5,1, 2 and 4 hours (hr) post stimulation. Gene expression was assayed using Agilent cDNA arrays. Gene coexpression networks were then constructed by assembling the time point expression data from each stimulation and the correlation between all probes was then measured across all 4 time points (and 3 replicates per time point). A correlation magnitude cutoff of 0.8 was utilized to finally yield the networks used for this study (positive correlations are indicated by solid edges, while negative correlations are indicated by dashed edges in the figure). The networks were compared using a network alignment approach that took into account the weight on the edges around each respective node. Networks with similar topologies and weights around respective genes had higher score than those with a good toplogical match, but poor weight matches around matching genes.
Supplementary Tables