Abstract
In eukaryotes, the messenger RNA (mRNA), the blueprint of a protein‐coding gene, is processed and packaged into a messenger ribonucleoprotein particle (mRNP) by mRNA‐binding proteins in the nucleus. The steps of mRNP formation – transcription, processing, packaging, and the orchestrated release of the export‐competent mRNP from the site of transcription for nuclear mRNA export – are tightly coupled to ensure a highly efficient and regulated process. The importance of highly accurate nuclear mRNP formation is illustrated by the fact that mutations in components of this pathway lead to cellular inviability or to severe diseases in metazoans. We hypothesize that efficient mRNP formation is realized by a molecular mRNP packaging station, which is built by several recruitment platforms and coordinates the individual steps of mRNP formation.
Keywords: gene expression, molecular mRNP packaging station, mRNP formation, mRNA processing, nuclear mRNA export, transcription, TREX
Abbreviations
- CTD
C‐terminal domain of Rpb1
- CTR
C‐terminal region of Spt5
- mRNA
messenger RNA
- mRNP
messenger ribonucleoprotein particle
- Paf1C
Paf1 complex
- PRP19C
PRP19 complex
- RNAPII
RNA polymerase II
- S. cerevisiae
Saccharomyces cerevisiae
Introduction
Gene expression in eukaryotic cells is a multi‐step process starting with the transcription of protein‐coding genes by RNA polymerase II (RNAPII) synthesizing the mRNA. As an important feature of eukaryotic cells, the nascent mRNA is matured into an export‐competent mRNP, which is exported through the nuclear pore to the cytoplasm, where translation of the mRNA takes place 1. During this nuclear mRNP formation, the mRNA is processed (capped, spliced, and polyadenylated) and bound by multiple RNA‐binding proteins. Throughout this review, we will refer to mRNA processing and packaging as mRNP formation and to the proteins mediating, these processes as mRNP formation factors. mRNP formation heavily impacts downstream processes of gene expression, such as alternative splicing, leading to diversity and regulation of the produced proteins 2. It also determines the stability of the mRNA and thus its half‐life, potentially affecting final protein levels. Moreover, nuclear mRNP formation also has the potential to change the transcript's cellular localization after mRNP export. Taken together, nuclear mRNP formation is important for the regulation of gene expression.
In this review, we give an overview of nuclear mRNP formation and the coordination of its different steps leading to a mature, export‐competent mRNP with a focus on Saccharomyces cerevisiae (S. cerevisiae). We hypothesize that the coordination of the single steps of nuclear mRNP formation is implemented in form of a molecular mRNP packaging station that ensures a highly efficient process.
Nuclear mRNP formation
mRNP formation starts with the synthesis of the mRNA by RNAPII. After transcription initiation, RNAPII undergoes extensive structural changes and switches to productive elongation 3. As soon as the 5′‐end of the nascent RNA emerges from RNAPII, the first step of mRNP formation takes place: the mRNA is capped at its 5′‐end by the capping enzyme (Fig. 1). The m7‐G‐cap is made by removal of the 5′‐phosphate group by Cet1, addition of a GTP by Ceg1 under the loss of a pyrophosphate resulting in a 5′‐5′‐triphosphate linkage 4, and methylation of N7 of the GTP by Abd1. The 5′ cap of the mRNA is then bound by the cap‐binding complex (Cbp80 and Cbp20 in yeast). The cap is important to protect the mRNA from exonucleolytic degradation and promotes several downstream processes of gene expression: splicing, mRNA export, and translation (reviewed in 4).
Figure 1.
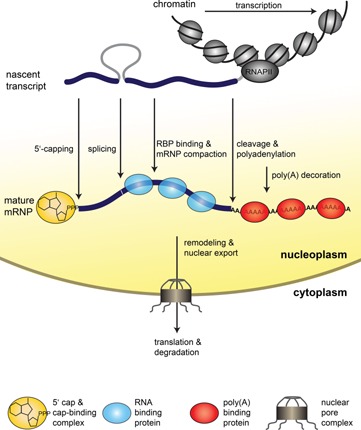
Steps of nuclear mRNP formation. The mRNA is synthesized from the protein coding gene by RNAPII as a nascent transcript. The nascent transcript is processed while transcription elongation still takes place. It is capped, spliced, cleaved, and polyadenylated. In addition, mRNA‐binding proteins bind to the mRNA and package it into an mRNP. These mRNA‐binding proteins influence mRNA stability and are necessary for nuclear export. After a likely remodeling of the mRNP, it is exported to the cytoplasm through the nuclear pore complex (NPC). In the cytoplasm the mRNA is translated by the ribosomes, which synthesize the encoded protein. Eventually, the mRNA is degraded.
As the nascent mRNA grows, the mRNA is spliced, i.e. the introns are removed and the exons are ligated (Fig. 1). This process, two trans‐esterification reactions per intron, is catalyzed by the spliceosome 5. The spliceosome is a multiprotein RNA complex and assembles on the mRNA in a stepwise, highly ordered fashion, which ensures correct splice site recognition and precise intron removal and exon ligation 5.
In parallel, the mRNA is bound by several mRNA‐binding proteins and packaged into an mRNP. mRNA‐binding proteins have multiple functions such as stabilization of the mRNA against degradation, nuclear mRNA export, and mRNP compaction 1, 6. The TREX complex is involved in transcription and couples transcription to mRNA export (also see below) and consists of the heteropentameric THO complex (consisting of Tho2, Hpr1, Mft1, Thp2, Tex1), the mRNA export factors Sub2 and Yra1, and the SR‐like proteins Gbp2 and Hrb1 7, 8, 9. The SR‐like protein Npl3 functions in mRNA export and antagonizes premature 3′‐end processing by competing with 3′‐end processing factors for binding to the mRNA 10, 11. Nab2, a nuclear poly(A)‐binding protein, functions in poly(A) tail length control, mRNA export, and compaction of the mRNA 6, 12, 13. TREX components and Npl3 recruit the mRNA export receptor Mex67‐Mtr2 to the mRNA 14, 15, 16. Mex67‐Mtr2 directly binds to the mRNA and to the nuclear pore complex and exports the mRNP to the cytoplasm 14, 17, 18.
The last co‐transcriptional step of mRNP formation takes place at the poly(A) site: the nascent pre‐mRNA is released from the transcription machinery by cleavage at its 3′‐end. The cleavage and polyadenylation machinery consists of the subcomplexes CFIA, CFIB, and CPF and also polyadenylates the mRNA. The poly(A) tail of the mRNA has several functions: it marks mature transcripts and regulates its half‐life. In vivo, poly(A) tails are heavily decorated by poly(A)‐binding proteins 19. After completion of 3′‐end processing, the mRNP is most likely remodeled to a mature mRNP that is released from the site of transcription and competent for nuclear export. In vivo, mRNP formation is thought to be a highly interconnected and continuous process with several steps being carried out in parallel.
Three recruitment platforms are important for co‐transcriptional mRNP formation
The C‐terminal domain of RNA polymerase II
One of the key players for the recruitment of proteins involved in mRNP formation is the C‐terminal domain (CTD) of Rpb1, the largest subunit of RNAPII. It serves as a landing pad for most of the mRNP formation factors during transcription elongation 20 (Fig. 2). Depending on the organism, the CTD consists of 26–52 heptapeptide repeats with the consensus sequence YSPTSPS. These repeats are highly conserved in the N‐terminal part and show more variation in the C‐terminal part of the CTD 20. As RNAPII traverses through the transcription cycle it is selectively phosphorylated. During transcription initiation, the CTD is phosphorylated at several of its S5 and S7 positions. As RNAPII transits to elongation, S5 and S7 phosphorylations decrease, while S2 and Y1 phosphorylations increase. Y1 phosphorylation levels decrease at the poly(A) site and S2 phosphorylation levels at the termination site (Fig. 2) 21, 22, 23.
Figure 2.
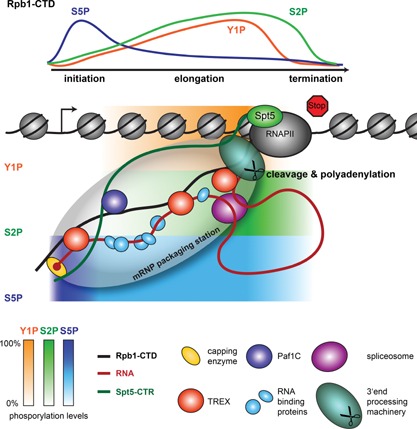
The mRNP packaging station. As the mRNA emerges from RNAPII during transcription elongation, it is capped, spliced, cleaved, and polyadenylated (processing). In addition, a multitude of RNA‐binding proteins binds to the mRNA, packaging it in an mRNP. We hypothesize that these single steps of mRNP formation are coordinated and controlled within an mRNP packaging station. This mRNP packaging station is assembled by three recruitment platforms and possibly additional coordinating proteins such as the TREX complex, which ensure spatial proximity of all processes. The three recruitment platforms for mRNA processing and mRNA‐binding proteins are indicated by different colors (red: RNA, black: Rpb1‐CTD, and green: Spt5‐CTR). The different mRNA processing and mRNA‐binding proteins are defined in the legend below the figure. The capping enzyme is exchanged for the cap binding complex consisting of Cbc20 and Cbc80 after capping is complete. The differential phosphorylation of the CTD (C‐terminal domain of Rpb1, the largest subunit of RNAPII) is indicated at the top as well as below the chromatin (blue: S5P, orange: Y1P, green: S2P). The phosphorylation status is indicated by a gradient from 0–100% as indicated by the bars in the bottom left hand corner. Zero per cent reflects no detectable phosphorylation while 100% reflects the maximum signal observed at an average gene in a ChIP experiment.
The CTD phosphorylation pattern coordinates transcription with mRNP formation by the timely recruitment of mRNP formation factors that selectively bind to a specifically phosphorylated CTD (Table 1). The capping enzyme binds to the S5 phosphorylated CTD 24, 25 and is thus recruited during transcription initiation. Splicing is largely co‐transcriptional in S. cerevisiae and higher eukaryotes 26, 27. Consistently, several subunits of the spliceosome interact directly with the phosphorylated CTD, e.g. Prp40, a subunit of the U1 snRNP complex of the spliceosome 28. Additional factors are thought to interact indirectly with the CTD via CTD‐binding proteins, such as the splicing factor TCERG1 (aka CA150) 29 and the RNA‐binding proteins PSF and p54nrb 30, both of which are implicated in splicing. The yeast SR‐like protein Npl3 is involved in splicing and binds directly to the S2 phosphorylated CTD 31, 32. Hence, the spliceosome is thought to be recruited to the site of transcription by binding to the CTD. Corroborating the importance of the CTD for splicing, splicing reactions are more efficient in vitro in the presence of a recombinant CTD 33, and in vivo splicing is promoted by the CTD via recruitment of a U2AF65‐Prp19 34.
Table 1.
S. cerevisiae proteins and protein complexes with a function in nuclear mRNP formation
| Complex | Protein | Function | Interaction with phospho‐CTD | RNA | Spt5 |
|---|---|---|---|---|---|
| Capping enzyme | Ceg1 | Guanyltransferase | S5‐P 24, 25 | 5′ Cap | pCTRa 45, 52 |
| Cet1 | 5′ Triphosphatase, homodimer with Ceg1 | 5′ Cap | pCTRa 45, 52 | ||
| Abd1 | Methyltransferase | pCTD 106 | 5′ Cap | pCTRa 45, 52 | |
| Cap binding complex | Cbc1 and Cbc2 | 5′ Cap | Via S2 kinases 107 | 5′ Cap 108 | ND |
| Spliceosome | Prp40 | Splicing | pCTD 28 | Via U1 snRNA 109 | ND |
| TREX complex, THO subcomplex | Tho2, Hpr1, Mft1, Thp2, Tex1 | Transcription and mRNA export | S2S5‐P 35 | General 110 | ND |
| TREX | Yra1 | mRNA export, Mex67 recruitment | S2S5‐P 73 | General 14 | Spt5 45 |
| Sub2 | mRNA export, DEAD‐box helicase | ND | DEAD‐box helicase | ND | |
| Gbp2 | mRNA export, splicing control | ND | Motif 42 | ND | |
| Hrb1 | mRNA export, splicing control | ND | Motif 42 | ND | |
| THSC/TREX2 | Sus1 | mRNA export/chromatin modification | S5‐P 111 | General 112 | ND |
| hnRNP | Tho1 | mRNA export | ND | General 113 | Spt5 45 |
| Export receptor Mex67‐Mtr2 | Mex67 | Nuclear mRNA export | ND | General 14 (via Yra1 14, Nab2 76, and Npl3 16) | ND |
| Nuclear poly(A) binding protein | Nab2 | Poly(A) binding and export | ND | PolyA 36, motif 13, 42 | ND |
| SR‐like‐protein | Npl3 | Promotes elongation/prevents polyadenylation | S2‐P 32 | General, motif 10, b | ND |
| Cleavage Factor I (CFI) | Pcf11 | Cleavage/polyadenylation | S2‐P 37 | 114 | ND |
| Rna14 | Cleavage/polyadenylation | pCTD 38 | 115 | pCTR 45, 49 | |
| Rna15 | Cleavage/polyadenylation | pCTD 38 | pCTR 49 | ||
| Hrp1 | Cleavage/polyadenylation | ND | UA rich sites 115 | pCTR 115 | |
| Cleavage and polyadenylation factor (CPF) | Ydh1 | Cleavage/polyadenylation | pCTD 39 | Via Yhh1 | ND |
| Yhh1 | Cleavage/polyadenylation | pCTD 40 | Motif 40 | ND | |
| Pta1 | Cleavage/polyadenylation | S5‐P 116 | Via Yhh1 | ND | |
| Nrd1‐Nab3‐Sen1 complex | Nrd1 | Transcription termination/processing | S5‐P 117 | Motif 42, 118 | Via Sen1 |
| Nab3 | Transcription termination/processing | Via Nrd1 | Motif 118 | Via Sen1 | |
| Sen1 | Transcription termination/processing | Via Nrd1 | General 118 | Spt5 45 | |
| Rtt103 | Termination | S5‐P 119 | Exonuclease | ND | |
| Poly(A) binding protein | Pab1 | Poly(A) binding | Indirect via interaction with RNA15 120 | Motif 42 | Spt5 45 |
ND, not determined; S2‐P, CTD phosphorylated on serine 2; S5‐P, CTD phosphorylated on serine 5; S2S5‐P, CTD phosphorylated on serine 2 and serine 5; pCTD, phosphorylated CTD, phosphorylated residue not known; pCTR, phosphorylated C‐terminal region (of Spt5).
Binding to the pCTR shown only indirectly by ChIP.
Competition with RNA15.
Several of the mRNA‐binding proteins that package the mRNA into an mRNP are also recruited to the site of transcription by the phosphorylated CTD (Table 1). THO, a subcomplex of the TREX complex, binds directly to the S2–S5 phosphorylated CTD 35. The SR‐protein Npl3 is also recruited to the nascent mRNA by direct binding to the phospho‐CTD 32. Furthermore, Nab2 binds to RNA 36 and is recruited to the transcription site 35. Finally, increasing S2 phosphorylation, decreasing S5 phosphorylation, and decreasing Y1 phosphorylation approximately at the cleavage and polyadenylation site mark the next event in mRNP formation: the recruitment of the 3′‐end processing machinery. The recruitment of Pcf11, a subunit of the cleavage factor IA (CFIA), to the CTD is inhibited by Y1 and S2 di‐phosphorylated repeats and binds to the S2 phosphorylated CTD only after Y1 dephosphorylation 23, 37. Other 3′‐end processing factors also bind to the S2 phosphorylated CTD: Rna14 (CFIA), Rna15 (CFIA), Ydh1 (CPF), and Yhh1 (CPF) 38, 39, 40. Taken together, the CTD is a landing platform for many mRNA processing and packaging factors, and its differential phosphorylation coordinates mRNP formation with transcription.
The nascent RNA
Not surprisingly, another recruitment platform for mRNP formation factors is the nascent mRNA itself (Fig. 2). Several mRNA‐binding proteins recognize specific RNA sequence motifs. Examples for this are proteins involved in 3′‐end processing, which e.g. recognize the positioning element and the efficiency element (reviewed in 19). The spliceosome also recognizes motifs within the RNA, although not by protein–RNA interactions, but by base‐pairing of spliceosomal RNA components with the mRNA 5. RNA sequence motifs are thought to provide higher specificity: e.g. during splicing several consecutive base‐specific binding events ensure that the trans‐esterification reaction is carried out at exactly the right position of the mRNA. Interestingly, a similar mechanism is used to control pervasive transcription: Nrd1 carries out its function in terminating transcription of anti‐sense transcripts after being transferred from the CTD to its RNA motif. As these motifs occur randomly in RNA but are massively depleted in sense transcripts, Nrd1 is able to control pervasive transcription 41. Both processes have in common that their recruitment to the CTD ensures availability of the protein and its binding to an RNA motif ensures specificity.
Many proteins of the mRNP bind the mRNA without a well‐defined RNA sequence motif. Gbp2 and Hrb1 have only a preference for highly degenerate sequence motifs 42, 43. Besides binding to polyA tracks Nab2 also binds to degenerated A‐rich motifs 42, 43. Npl3 shows a low specificity for GU‐rich sequences but also binds unspecifically to RNA 11. These proteins might be recruited mainly by protein‐protein interactions and then transferred to the mRNA. For example, efficient THO complex recruitment to the site of transcription depends on the two recruitment platforms RNA and CTD 35. The non‐ or low specificity of RNA‐binding by these mRNP proteins might be necessary to enable “even” packaging and compaction of the mRNA into an mRNP since each mRNA – depending on the protein it encodes – has a different sequence. Taken together, the RNA – whether with or without specific motifs – is an important platform in the recruitment of mRNP formation factors.
The C‐terminal region of the transcription elongation factor Spt5
A third platform for mRNP formation factor recruitment is the general transcription elongation factor Spt5, which binds to the body of RNAPII 44 (Table 1). A mass spectrometry study revealed that several proteins involved in mRNP biogenesis copurify with Spt5 in yeast, among them the capping enzymes, 3′‐end processing factors, and mRNA‐binding proteins such as poly(A)‐binding protein Pab1 and the TREX component Yra1 45. Similar to Rpb1, Spt5 also has a repetitive C‐terminal region (CTR). In yeast, the CTR of Spt5 consists of 16 hexa‐repeats, which were shown to be phosphorylated on their S1 residues during transcription elongation by the kinase Bur1 in yeast and its homolog P‐TEFb in humans 46, 47, 48. The CTR is needed for efficient recruitment of the Cleavage Factor I (CFI) to the transcription site 49. Additionally, the Paf1 complex, which is implicated in several processes like chromatin remodeling and 3′‐end processing (reviewed in 50), was shown to depend on correct CTR phosphorylation for efficient recruitment 51. Moreover, the capping enzyme needs the CTR for stable recruitment 52. Taken together, these results show a general role for the Spt5‐CTR in the recruitment of mRNP formation factors.
Chromatin influences mRNP formation
mRNPs form co‐transcriptionally and therefore in a chromatin environment. Chromatin is well known to influence transcription, but its function in mRNP formation emerged only recently. Several reports connect the chromatin state to the recruitment of mRNP formation factors, suggesting that chromatin might serve as a regulating platform similar to the Rpb1‐CTD, the Spt5‐CTR, and the RNA.
The best‐investigated co‐transcriptional mRNP formation process influenced by the local chromatin state is splicing. H3K36me3 together with methylation of H3K79 contributes to exon definition, a central process in splicing, by accumulation of both methylation marks at exons. Consistently, alternative exons have lower H3K36me3 levels than constitutive exons 53, 54. Nevertheless, precisely how the particular chromatin modifications regulate splicing is not yet understood, but to date two main – not mutually exclusive – models exist: the kinetic and the recruitment model (reviewed in 55). According to the kinetic model, the chromatin is thought to locally slow down the elongating RNAPII and lead to selection of weaker splice sites due to a prolonged window of opportunity for the splicing reaction. This hypothesis is supported by the finding that a “slower” RNAPII mutant influences splice site selection 56. H3K36me3 slows down RNAPII by HDAC1 recruitment, which removes acetylations from the chromatin leading to a more repressive state 57. Furthermore, H3K9me3 is enriched over a specific subset of alternative exons and recruits Cbx3, which in turn reduces RNAPII elongation rate 58. The second model is based on the recruitment of splicing factors and the spliceosome components by epigenetic marks: H3K36me3 was shown to stimulate recruitment of splicing factors, e.g. MRG15 59 and Psip1 60. Moreover, H3K4me3 interacts with Chd1 which is also able to interact with several spliceosome components. Knockdown of Chd1 or reduced H3K4me3 levels lead to reduced association of U2 snRNP components with the chromatin and altered splicing efficiency in vivo 61. In brief, chromatin promotes recruitment of splicing factors, contributes to exon definition and potentially regulates alternative splicing.
In addition to splicing, mRNA export is influenced by the local chromatin state. In higher eukaryotes, Aly, the mammalian homolog of Yra1, interacts with IWS1 (Spt6), a protein involved in chromatin modification 62, and with the H3K36 methylase KMT4 63. Moreover, the ubiquitinylated histone H2B (ubH2B) controls formation of export competent mRNPs by its ability to promote ubiquitinylation of Swd2 (ubSwd2) at the 5′‐end of genes 64. Swd2 is a subunit of the chromatin modifying Set1/COMPASS complex and the cleavage and polyadenylation factor in yeast. ubSwd2 is required for proper mRNP formation reflected by decreased levels of the export receptor adaptor proteins Yra1 and Nab2 at the mRNP in a non‐ubiquitinylatable Swd2 mutant 64, 65. Furthermore, Npl3 genetically interacts with H2B and binds to Bre1, which ubiquitinylates H2B 66.
As a third step in mRNP formation, 3′‐end processing is influenced by the local chromatin structure. Poly(A) sites are strongly depleted of nucleosomes while their downstream regions are enriched for nucleosomes. Additionally, weak alternative poly(A) sites are less depleted of nucleosomes than strong sites 67. Thus, chromatin probably contributes as recruitment platform to mRNP formation, but also seems to be involved in its regulation.
Importantly, the influence of the chromatin state on mRNP formation is not unidirectional; instead a crosstalk between chromatin and mRNP formation exists. This is of great importance since a feedback of mRNP formation processes on chromatin enables the establishment of a stable chromatin state. The first example for this is H3K36me3 and splicing: as outlined above H3K36me3 can stimulate spliceosome recruitment, and splicing in turn stimulates H3K36 trimethylation. Mutation of the splice site or depletion of the splicing factor SAP130 leads to decreased Kmt3a recruitment and H3K36me3 levels in vivo 68, 69, 70. Most likely, other mRNP formation factors might also contribute to feedback to the chromatin to ensure stable propagation of the chromatin state, but have yet to be found.
In summary, local chromatin impacts and modulates mRNP biogenesis in several ways, and mRNP formation seems to be part of a positive feedback loop stabilizing the present chromatin state. This is a typical type of regulation in epigenetics, which ensures constant propagation of a specific state and raises the question, how this state is changed. As the first examples of the impact of chromatin on mRNP formation are uncovered, further research is needed to understand the function of chromatin in mRNP formation and the crosstalk between chromatin and mRNP formation.
The orchestration of mRNP formation within a molecular mRNP packaging station
Several platforms are implicated in recruiting mRNP formation factors to the nascent RNA: the Rpb1‐CTD, the nascent RNA itself, Spt5‐CTR, and the local chromatin state. Most of the known mRNP formation factors are recruited by one or more of these recruitment platforms (Fig. 2). In addition, several factors bind to each other such as the capping enzyme, which binds to the two main CTD‐S2 kinases Bur1 and Ctk1, and Yra1, which is part of the TREX complex and interacts with the 3′‐end processing machinery and Nab2. Such mutual interactions between mRNP formation factors most likely ensure the recruitment of all proteins by forming additional recruitment interfaces.
Furthermore, the estimated in vivo elongation rate of RNAPII is approximately 2,000 RNA bases per minute in yeast 71, which corresponds to a length of 560 nm. Thus, the nascent mRNA grows very quickly and the window of opportunity for each mRNP formation step is very narrow. Hence, the following questions arise: how does the cell ensure that all factors are available in a timely manner? How are all steps of this complicated, multistep process carried out correctly in the short time that is available for each of them? Finally, how is the premature release of incomplete mRNPs prevented to make mRNP formation efficient?
We hypothesize that mRNP formation is coordinated within a molecular mRNP packaging station, which keeps all functionalities needed for proper mRNP formation in spatial proximity to the nascent RNA. We propose that such an efficient molecular mRNP packaging station is realized by a combination of two functional divisions (Fig. 2): (i) the recruitment platforms, which ensure the timely recruitment of the mRNP formation factors and (ii) regulatory factors, which ensure that each modification of the pre‐mRNA is carried out at the right time point and that incomplete mRNPs are not prematurely released.
In addition to the recruitment platforms, Rpb1‐CTD, Spt5‐CTR, chromatin, and RNA described above, additional mRNP formation factors might facilitate the spatial proximity of the RNA to the recruitment platform(s) and thus to mRNP formation factors (Fig. 2). This is most likely achieved by bi‐functional binding to the RNA and to at least one of the recruitment platforms. Second, such (a) hypothetical regulatory factor(s) could orchestrate all steps of mRNP formation and should therefore interact with all other key players of mRNP formation. Ideally, it/they should also control successful completion of each step. Third, (a) central factor(s) coordinating mRNP formation should control the last step(s) of mRNP formation to prevent premature release of the mRNP from the transcription site and provide, if required, additional time for the mRNP to mature. Last, such (a) factor(s) ideally would also be involved in nuclear mRNA surveillance and promote degradation of faulty mRNPs. In summary, we propose that nuclear mRNP formation takes place in an mRNP packaging station, and suggest that (an) additional factor(s) integrate(s) and control(s) mRNP formation and thus largely improve(s) efficiency of mRNP formation.
The TREX complex could be a central factor of the mRNP packaging station
Considering the importance of effective mRNP formation, existence of a factor bundling mRNP formation within an mRNP packaging station would be advantageous, but was not shown to date. We hypothesize that the TREX complex fulfills this function. TREX couples transcription to mRNA export and thus participates in mRNP formation from beginning to end. TREX consists of the THO subcomplex (as described above), which promotes efficient transcription elongation 7, 71 and is recruited to the phosphorylated CTD 35. Further TREX components are Yra1, Sub2, Hrb1, and Gbp2 8. Hrb1 and Gbp2 are two SR‐like proteins involved in control of correct splicing prior to mRNP export (72, also see above). Sub2, a DExD‐box helicase, interacts directly with Yra1, which in turn binds directly to the export receptor Mex67, probably attracting it to the mRNP 14. Yra1 likely plays a central role in mRNP formation, which is underlined by its alternative recruitment by TREX and the phosphorylated Rpb1‐CTD 73. Yra1 also interacts with Pcf11 74, a subunit of the cleavage and polyadenylation machinery, and modulates poly(A)‐site choice 75. To facilitate mRNA export, Yra1 forms a ternary complex with Nab2 and Mex67 76. Upon ubiquitination, Yra1 dissociates from this ternary complex, and the mRNP is released for nuclear export. Yra1 dissociation is most likely the last quality control point prior to export. Thus, by interaction with key players of mRNP formation Yra1/TREX could coordinate mRNP formation and control release of the mature mRNP.
Moreover, the phenotypes of TREX knockout or mutation underline the regulatory role of TREX in mRNP formation. THO components were initially identified to prevent transcription‐dependent hyper‐recombination: the not properly packed mRNA hybridizes with the DNA forming so called R‐loops 77, 78. Furthermore, upon TREX mutation or knock‐out, 3′‐end processing malfunctions resulting in failure of mRNP maturation 79, 80. A dense chromatin fraction is formed, which contains the mRNP, 3′‐end processing machinery and even nuclear pore proteins 80. This clearly shows that TREX is necessary for mRNP formation and correct release of mature mRNPs.
Interestingly, TREX recruitment to the site of transcription increases from the 5′ to the 3′‐end of the gene, i.e. as RNA length increases 35. A tho2 recruitment mutant, in which TREX is recruited to the 5′ end but the 5′ to 3′ increase is abolished, causes reduced expression of long transcripts, which most likely need more TREX complexes for full expression 35. This suggests – in combination with the direct binding of TREX to the CTD – that TREX bridges between the mRNAs and the CTD, i.e. to keep the RNA in spatial proximity to the recruitment platforms 35. Hypothetically, this interaction could be important to ensure efficient and correct processing and packaging of the mRNA, which is consistent with the finding that a continuous transcript is needed for efficient mRNA processing 81. Taken together, TREX could play a pivotal role in the coordination of mRNP formation within the mRNP packaging station.
Hypothetical spatial organization of an mRNP packaging station
Tethering of the RNA to the recruitment platforms would be an elegant solution to promote mRNP formation. Even short periods of time, during which the RNA is in close proximity to the CTD and CTR, could extend the window of opportunity and thus enhance efficiency and prevent defective mRNP formation. How RNA tethering is organized on a molecular basis, i.e. whether the RNA is tethered to the Rpb1‐CTD over its whole length or only partially by several copies of TREX or other factors (Fig. 2), remains an open question. In yeast, a fully extended CTD is approximately 700 Å long and thus corresponds to the length of a 2.5 kb long extended mRNA. Since the median length of an mRNA in S. cerevisiae is 1,436 nucleotides 82, the CTD is able to span the entire length of an average mRNA. However, it seems unlikely that the CTD or the mRNA exist in a fully extended form in vivo. The RNA could e.g. form several loops such that only several points of the RNA are associated with the CTD. This model is compatible with the fact that in higher eukaryotes, exon definition is facilitated by the indirect binding of the two exonic sequences to the CTD in close proximity to each other and that the (large) introns form non‐CTD‐associated loops 83 (Fig. 2). Additionally, the human CTD is twice as long as the yeast CTD, thus harboring more recruitment capacity. Spatial proximity of the RNA and the CTD could contribute to the compaction of the mRNP, since mRNA‐binding proteins that package the mRNA are present at the CTD.
Instead of or in addition to TREX, other factors could promote mRNP formation by similar mechanisms. A very interesting complex essential for mRNA export and mRNP formation is THSC/TREX‐2 1. THSC is also needed for the relocation of Gal genes to the nuclear pore upon their induction and is thought to thereby modulate gene expression 1. This process is known as gene gating. However, its impact on gene expression is not completely understood, as on the one hand mRNA export might be enhanced at the nuclear pore but on the other hand localization of chromatin to the nuclear periphery might regulate their expression.
Npl3, which is also recruited co‐transcriptionally by binding to the phosphorylated CTD 32, is an interesting candidate. It is involved in splicing, controls 3′‐end processing by competition with the recruitment of CFI, binds RNA, and promotes mRNA export 10, 16. As a constraint, Npl3 recruitment does not increase from 5′ to 3′ of genes and, consequently, Npl3 does not offer CTD‐RNA contacts that increase with RNA length. To date, TREX is the only complex with a 5′–3′ increase turning it into one of the most promising candidates for a regulatory factor within the mRNP packaging station 35.
The protein components of the mRNP packaging station are most likely held together by reciprocal protein interactions. The Paf1 complex (Paf1C), for example, might ensure spatial proximity between the two recruitment platforms Spt5‐CTR and Rpb1‐CTD, as Paf1C is able to bind both during transcription elongation 51. Additionally, Paf1C might function as an interface between chromatin and mRNP formation as it is involved in ubiquitylation of H2B 84 and several other histone modifications such as H3K4‐me3 and H3K36‐me3 (reviewed in 50). The PRP19 complex (PRP19C) functions in splicing and – independently – in transcription elongation by ensuring TREX occupancy 85, 86. Hence, PRP19C might well hold the mRNP packaging station together. Taken together, the mRNP packaging station is most likely formed by multiple interactions between the recruitment platforms (Rpb1‐CTD, Spt5‐CTR, RNA, and chromatin) and mRNP formation factors.
Conservation and implications for higher eukaryotes
Although overall S. cerevisiae is very similar to higher eukaryotes, differences exist, e.g. in transcription and mRNP formation. One potentially important difference is that most genes in baker's yeast do not contain introns, although highly transcribed yeast genes often do harbor introns. In higher eukaryotes, it is thought that the TREX complex is not recruited by the transcription machinery as in yeast, but instead by the spliceosome during splicing 87. Consistently, human THO/TREX subunits were found to co‐purify with the spliceosome 5, 88, 89 and to be needed for release of spliced mRNA from the nuclear speckles domains 90. Additionally, human TREX is recruited to the 5′‐end of the mRNA by the interaction of Aly (Yra1) with the cap binding complex protein Cbp80 91. Nevertheless, TREX is also recruited independently of splicing by binding to specific RNA sequence elements 92, 93, 94. Thus, in higher eukaryotes, TREX is most likely also recruited co‐transcriptionally. Recently, a function of human Aly in transcription was demonstrated adding further evidence that human TREX could be recruited co‐transcriptionally 95. Furthermore, it is known that the Rpb1‐CTD and its functions are conserved in yeast and higher eukaryotes. Although the human CTD is approximately twice as long as the yeast CTD, the phospho‐code is highly conserved. The increased complexity of the CTD in higher eukaryotes might account for the greater flexibility of transcription and mRNP biogenesis in more complex multicellular organisms. In this light, an integrator and bridging protein appears to be even more important to orchestrate the mRNP packaging station. Moreover, almost all factors contributing to mRNP formation are conserved in higher eukaryotes and show similar interactions with the recruitment platforms. Besides the CTD, the other recruitment platforms are also present in higher eukaryotes: the phosphorylated Spt5‐CTR (by P‐TEFb) 47, the RNA, and chromatin. In several cases, the proteins differ in their architecture, e.g. in the capping enzyme of higher eukaryotes the enzymatic function of Ceg1 and Cet1 is present in a single protein 4, or sequence specificity, e.g. in 3′‐end processing, where some sequence elements differ in sequence and positioning 19. However, the underlying principle of coupling between the recruitment platforms is conserved for all steps of mRNP biogenesis. Additionally, the nuclear speckles, which are not known to exist in yeast, but are found in higher eukaryotes might be related and more elaborated forms of the here proposed mRNP packaging station. For further information on nuclear speckles and other nuclear suborganelles, see Box 1.
Box 1. Nuclear bodies: Hot spots of RNP formation.
Most nuclear biogenesis pathways consist of multiple steps that are catalyzed by a multitude of proteins and RNA‐protein complexes in a highly ordered fashion. Often, the involved proteins are spatially organized forming suborganelles to enhance efficiency and facilitate regulation. These suborganelles, which are often called bodies, do not have a membrane but are composed of larger protein and nucleic acid assemblies. This increases the local concentration of reactants, ensures availability of the proteins, and shortens the diffusion time of the proteins to the reaction site. One of these bodies is the nucleolus, where the chromatin encoding rRNA is located and transcribed, and the first steps of rRNA maturation are carried out. Further nuclear bodies are Cajal bodies, nuclear speckles, and histone locus bodies 102. Cajal bodies are involved in the biogenesis of small nuclear RNPs (snRNPs), subunits of the spliceosome, and in the assembly of small nucleolar RNPs (snoRNPs), needed for the processing of rRNA transcripts. Additionally, the telomerase RNP is assembled in Cajal bodies (reviewed in 103). Nuclear speckles are thought to serve as storage and modification sites for the splicing machinery (snRNPs and SR‐proteins) (reviewed in 104). A recent study indicated that 80–85% of the splicing takes place co‐transcriptionally in the periphery of nuclear speckles and that only the remaining 15–20% of the active spliceosomes reside inside nuclear speckles for post‐transcriptional splicing 105. In line with this observation, nuclear speckles are located in proximity to transcription factories (see below) and were found in spatial proximity to highly induced genes. Reflecting that yeast genes more rarely contain introns and that nuclear speckles are important for the regulation of splicing, they are not known to exist in yeast. Another nuclear body are transcription factories. They are relatively small protein assemblies comprising several RNAPII molecules and transcription factors and are considered to be the active transcription units. Moreover, histone locus bodies have been identified, which contain genes coding for the major histones. Those transcripts have very unique features: they do not contain introns, and instead of cleavage and polyadenylation they carry a 3′‐extension that is cleaved before export with the help of the U7 snRNP. Additionally, histone mRNPs are exported by a stem loop binding protein. Histone locus bodies underline the importance of effective mRNP formation: important, specialized genes like the ones coding for histones might even have their own specialized mRNP packaging station tailored to their needs. Taken together, inside the nucleus several suborganelles or larger assemblies are used as common design principle to ensure a locally high concentration of factors to make multistep processes more efficient, similar to the mRNA packaging station proposed here.
The great importance of efficient mRNP formation is underlined by observations connecting mRNP biogenesis factors with various diseases. TREX was shown to be deregulated in various primary cancers and to be a target of several oncogenic kinases 96. Additionally, TREX is essential for stem cell maintenance and growth factor/cytokine‐mediated differentiation and proliferation 97. Furthermore, defects in REF/Aly can cause a severe form of myotonic dystrophy, which is thought to be caused by aberrant nuclear retention of CUG repeat transcripts 98. Moreover, mutations affecting splicing were shown to cause diseases and to be connected to cancer (for review see 99, 100). Such mutations often affect splice site selection, as e.g. for the tumor suppressor gene LKB1. Additionally, changes in the choice of alternative poly(A) sites lead to changed 3′ UTRs, affecting inclusion of miRNA‐binding sites into the transcript, which can activate oncogenes 101. In summary, efficient mRNP formation is of great importance for all eukaryotes as illustrated by the high conservation of the mRNP formation machinery and by the impact of mutations in mRNP formation factors, which lead to inviability or disease.
Conclusion
We hypothesize that the highly complex and multistep process of mRNP formation is executed within a molecular mRNP packaging station (Figs. 1 and 2). This mRNP packaging station comprises recruitment platforms and probably one or more additional factors such as TREX (Fig. 2). In the future, it will be important to prove the presence of such a molecular mRNP packaging station and to address further questions concerning the events taking place in this factory: how is mRNP formation chronologically organized? Does the mRNP packaging station form a compact or a loose assembly, which is flexibly adjusted to the need of each gene/mRNP? Is there any specificity for single transcripts? How is the mRNP remodeled? What are the consequences of mRNP formation on downstream processes such as nuclear export, translation, and mRNA degradation? In summary, an mRNP packaging station would be another good example, of how a pathway is provided with efficiency and surveillance by spatially bundling together several steps of this pathway in a highly concerted way.
The authors have declared no conflicts of interest.
Acknowledgements
We thank Sabine Meinel and members of the lab for critical reading of the manuscript. KS was supported by a Starting Grant of the European Research Council (ERC).
References
- 1. Rondon AG, Jimeno S, Aguilera A. 2010. The interface between transcription and mRNP export: from THO to THSC/TREX‐2. Biochim Biophys Acta 1799: 533–8. [DOI] [PubMed] [Google Scholar]
- 2. Tian B, Manley JL. 2013. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci 38: 312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hahn S. 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol 11: 394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. 2011. Cap and cap‐binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA 2: 277–98. [DOI] [PubMed] [Google Scholar]
- 5. Will CL, Luhrmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Batisse J, Batisse C, Budd A, Bottcher B, et al. 2009. Purification of nuclear poly(A)‐binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J Biol Chem 284: 34911–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chavez S, Beilharz T, Rondon AG, Erdjument‐Bromage H, et al. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae . EMBO J 19: 5824–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Strasser K, Masuda S, Mason P, Pfannstiel J, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–8. [DOI] [PubMed] [Google Scholar]
- 9. Hurt E, Luo MJ, Rother S, Reed R, et al. 2004. Cotranscriptional recruitment of the serine‐arginine‐rich (SR)‐like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci USA 101: 1858–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bucheli ME, Buratowski S. 2005. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J 24: 2150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deka P, Bucheli ME, Moore C, Buratowski S, et al. 2008. Structure of the yeast SR protein Npl3 and Interaction with mRNA 3′‐end processing signals. J Mol Biol 375: 136–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Green DM, Marfatia KA, Crafton EB, Zhang X, et al. 2002. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J Biol Chem 277: 7752–60. [DOI] [PubMed] [Google Scholar]
- 13. Hector RE, Nykamp KR, Dheur S, Anderson JT, et al. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J 21: 1800–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Strasser K, Hurt E. 2000. Yra1p, a conserved nuclear RNA‐binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J 19: 410–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gwizdek C, Iglesias N, Rodriguez MS, Ossareh‐Nazari B, et al. 2006. Ubiquitin‐associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci USA 103: 16376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gilbert W, Guthrie C. 2004. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell 13: 201–12. [DOI] [PubMed] [Google Scholar]
- 17. Santos‐Rosa H, Moreno H, Simos G, Segref A, et al. 1998. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol 18: 6826–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Segref A, Sharma K, Doye V, Hellwig A, et al. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J 16: 3256–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mandel CR, Bai Y, Tong L. 2008. Protein factors in pre‐mRNA 3′‐end processing. Cell Mol Life Sci 65: 1099–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang DW, Rodriguez‐Molina JB, Tietjen JR, Nemec CM, et al. 2012. Emerging views on the CTD code. Genet Res Int 2012: 347214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tietjen JR, Zhang DW, Rodriguez‐Molina JB, White BE, et al. 2010. Chemical‐genomic dissection of the CTD code. Nat Struct Mol Biol 17: 1154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mayer A, Lidschreiber M, Siebert M, Leike K, et al. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–8. [DOI] [PubMed] [Google Scholar]
- 23. Mayer A, Heidemann M, Lidschreiber M, Schreieck A, et al. 2012. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 336: 1723–5. [DOI] [PubMed] [Google Scholar]
- 24. Cho EJ, Takagi T, Moore CR, Buratowski S. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy‐terminal domain. Genes Dev 11: 3319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fabrega C, Shen V, Shuman S, Lima CD. 2003. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy‐terminal domain of RNA polymerase II. Mol Cell 11: 1549–61. [DOI] [PubMed] [Google Scholar]
- 26. Carrillo Oesterreich F, Bieberstein N, Neugebauer KM. 2011. Pause locally, splice globally. Trends Cell Biol 21: 328–35. [DOI] [PubMed] [Google Scholar]
- 27. Brugiolo M, Herzel L, Neugebauer KM. 2013. Counting on co‐transcriptional splicing. F1000Prime Rep 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morris DP, Greenleaf AL. 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl‐terminal domain of RNA polymerase II. J Biol Chem 275: 39935–43. [DOI] [PubMed] [Google Scholar]
- 29. Carty SM, Goldstrohm AC, Sune C, Garcia‐Blanco MA, et al. 2000. Protein‐interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc Natl Acad Sci USA 97: 9015–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emili A, Shales M, McCracken S, Xie W, et al. 2002. Splicing and transcription‐associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8: 1102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kress TL, Krogan NJ, Guthrie C. 2008. A single SR‐like protein, Npl3, promotes pre‐mRNA splicing in budding yeast. Mol Cell 32: 727–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, et al. 2008. Unphosphorylated SR‐like protein Npl3 stimulates RNA polymerase II elongation. PLoS ONE 3: e3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Millhouse S, Manley JL. 2005. The C‐terminal domain of RNA polymerase II functions as a phosphorylation‐dependent splicing activator in a heterologous protein. Mol Cell Biol 25: 533–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. David CJ, Boyne AR, Millhouse SR, Manley JL. 2011. The RNA polymerase II C‐terminal domain promotes splicing activation through recruitment of a U2AF65‐Prp19 complex. Genes Dev 25: 972–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meinel DM, Burkert‐Kautzsch C, Kieser A, O'Duibhir E, et al. 2013. Recruitment of TREX to the transcription machinery by its direct binding to the phospho‐CTD of RNA polymerase II. PLoS Genet 9: e1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anderson JT, Wilson SM, Datar KV, Swanson MS. 1993. NA B2: a yeast nuclear polyadenylated RNA‐binding protein essential for cell viability. Mol Cell Biol 13: 2730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Noble CG, Hollingworth D, Martin SR, Ennis‐Adeniran V, et al. 2005. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat Struct Mol Biol 12: 144–51. [DOI] [PubMed] [Google Scholar]
- 38. Barilla D, Lee BA, Proudfoot NJ. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl‐terminal domain of RNA polymerase II in Saccharomyces cerevisiae . Proc Natl Acad Sci USA 98: 445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kyburz A, Sadowski M, Dichtl B, Keller W. 2003. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre‐mRNA 3′‐end formation. Nucleic Acids Res 31: 3936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dichtl B, Blank D, Sadowski M, Hubner W, et al. 2002. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J 21: 4125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schulz D, Schwalb B, Kiesel A, Baejen C, et al. 2013. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell 155: 1075–87. [DOI] [PubMed] [Google Scholar]
- 42. Riordan DP, Herschlag D, Brown PO. 2011. Identification of RNA recognition elements in the Saccharomyces cerevisiae transcriptome. Nucleic Acids Res 39: 1501–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tuck AC, Tollervey D. 2013. A transcriptome‐wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell 154: 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klein BJ, Bose D, Baker KJ, Yusoff ZM, et al. 2011. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci USA 108: 546–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lindstrom DL, Squazzo SL, Muster N, Burckin TA, et al. 2003. Dual roles for Spt5 in pre‐mRNA processing and transcription elongation revealed by identification of Spt5‐associated proteins. Mol Cell Biol 23: 1368–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu Y, Warfield L, Zhang C, Luo J, et al. 2009. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol 29: 4852–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamada T, Yamaguchi Y, Inukai N, Okamoto S, et al. 2006. P‐TEFb‐mediated phosphorylation of hSpt5 C‐terminal repeats is critical for processive transcription elongation. Mol Cell 21: 227–37. [DOI] [PubMed] [Google Scholar]
- 48. Zhou K, Kuo WH, Fillingham J, Greenblatt JF. 2009. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci USA 106: 6956–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mayer A, Schreieck A, Lidschreiber M, Leike K, et al. 2012. The spt5 C‐terminal region recruits yeast 3' RNA cleavage factor I. Mol Cell Biol 32: 1321–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jaehning JA. 2010. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta 1799: 379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qiu H, Hu C, Gaur NA, Hinnebusch AG. 2012. Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR‐independent recruitment of Paf1 complex. EMBO J 31: 3494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lidschreiber M, Leike K, Cramer P. 2013. Cap completion and C‐terminal repeat domain kinase recruitment underlie the initiation‐elongation transition of RNA polymerase II. Mol Cell Biol 33: 3805–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hon G, Wang W, Ren B. 2009. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol 5: e0000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dhami P, Saffrey P, Bruce AW, Dillon SC, et al. 2010. Complex exon‐intron marking by histone modifications is not determined solely by nucleosome distribution. PLoS ONE 5: e12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brown SJ, Stoilov P, Xing Y. 2012. Chromatin and epigenetic regulation of pre‐mRNA processing. Hum Mol Genet 21: R90–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. La Mata M, Alonso CR, Kadener S, Fededa JP, et al. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–32. [DOI] [PubMed] [Google Scholar]
- 57. Jelinic P, Pellegrino J, David G. 2011. A novel mammalian complex containing Sin3B mitigates histone acetylation and RNA polymerase II progression within transcribed loci. Mol Cell Biol 31: 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Saint‐Andre V, Batsche E, Rachez C, Muchardt C. 2011. Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat Struct Mol Biol 18: 337–44. [DOI] [PubMed] [Google Scholar]
- 59. Luco RF, Pan Q, Tominaga K, Blencowe BJ, et al. 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pradeepa MM, Sutherland HG, Ule J, Grimes GR, et al. 2012. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet 8: e1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sims RJ, 3rd , Millhouse S, Chen CF, Lewis BA, et al. 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre‐mRNA splicing. Mol Cell 28: 665–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yoh SM, Cho H, Pickle L, Evans RM, et al. 2007. The Spt6 SH2 domain binds Ser2‐P RNAPII to direct Iws1‐dependent mRNA splicing and export. Genes Dev 21: 160–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Park G, Gong Z, Chen J, Kim JE. 2010. Characterization of the DOT1L network: implications of diverse roles for DOT1L. Protein J 29: 213–23. [DOI] [PubMed] [Google Scholar]
- 64. Vitaliano‐Prunier A, Menant A, Hobeika M, Geli V, et al. 2008. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol 10: 1365–71. [DOI] [PubMed] [Google Scholar]
- 65. Vitaliano‐Prunier A, Babour A, Herissant L, Apponi L, et al. 2012. H2B ubiquitylation controls the formation of export‐competent mRNP. Mol Cell 45: 132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Moehle EA, Ryan CJ, Krogan NJ, Kress TL, et al. 2012. The yeast SR‐like protein Npl3 links chromatin modification to mRNA processing. PLoS Genet 8: e 1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Spies N, Nielsen CB, Padgett RA, Burge CB. 2009. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yuan W, Xie J, Long C, Erdjument‐Bromage H, et al. 2009. Heterogeneous nuclear ribonucleoprotein L Is a subunit of human KMT3a/Set2 complex required for H3 Lys‐36 trimethylation activity in vivo. J Biol Chem 284: 15701–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. de Almeida SF, Grosso AR, Koch F, Fenouil R, et al. 2011. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol 18: 977–83. [DOI] [PubMed] [Google Scholar]
- 70. Kim S, Kim H, Fong N, Erickson B, et al. 2011. Pre‐mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci USA 108: 13564–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mason PB, Struhl K. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17: 831–40. [DOI] [PubMed] [Google Scholar]
- 72. Hackmann A, Wu H, Schneider UM, Meyer K, et al. 2014. Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nat Commun 5: 3123. [DOI] [PubMed] [Google Scholar]
- 73. MacKellar AL, Greenleaf AL. 2011. Cotranscriptional association of mRNA export factor Yra1 with C‐terminal domain of RNA polymerase II. J Biol Chem 286: 36385–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Johnson SA, Cubberley G, Bentley DL. 2009. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell 33: 215–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Johnson SA, Kim H, Erickson B, Bentley DL. 2011. The export factor Yra1 modulates mRNA 3′ end processing. Nat Struct Mol Biol 18: 1164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, et al. 2010. Ubiquitin‐mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev 24: 1927–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Huertas P, Aguilera A. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription‐associated recombination. Mol Cell 12: 711–21. [DOI] [PubMed] [Google Scholar]
- 78. Dominguez‐Sanchez MS, Barroso S, Gomez‐Gonzalez B, Luna R, et al. 2011. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet 7: e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Saguez C, Schmid M, Olesen JR, Ghazy MA, et al. 2008. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol Cell 31: 91–103. [DOI] [PubMed] [Google Scholar]
- 80. Rougemaille M, Dieppois G, Kisseleva‐Romanova E, Gudipati RK, et al. 2008. THO/Sub2p functions to coordinate 3′‐end processing with gene‐nuclear pore association. Cell 135: 308–21. [DOI] [PubMed] [Google Scholar]
- 81. Fong N, Ohman M, Bentley DL. 2009. Fast ribozyme cleavage releases transcripts from RNA polymerase II and aborts co‐transcriptional pre‐mRNA processing. Nat Struct Mol Biol 16: 916–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nagalakshmi U, Wang Z, Waern K, Shou C, et al. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zeng C, Berget SM. 2000. Participation of the C‐terminal domain of RNA polymerase II in exon definition during pre‐mRNA splicing. Mol Cell Biol 20: 8290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kim J, Roeder RG. 2009. Direct Bre1‐Paf1 complex interactions and RING finger‐independent Bre1‐Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem 284: 20582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chanarat S, Seizl M, Strasser K. 2011. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev 25: 1147–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chanarat S, Burkert‐Kautzsch C, Meinel DM, Strasser K. 2012. Prp19C and TREX: interacting to promote transcription elongationand mRNA export. Transcription 3: 8–12. [DOI] [PubMed] [Google Scholar]
- 87. Masuda S, Das R, Cheng H, Hurt E, et al. 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 19: 1512–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhou Z, Licklider LJ, Gygi SP, Reed R. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–5. [DOI] [PubMed] [Google Scholar]
- 89. Chen YI, Moore RE, Ge HY, Young MK, et al. 2007. Proteomic analysis of in vivo‐assembled pre‐mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res 35: 3928–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dias AP, Dufu K, Lei H, Reed R. 2010. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat Commun 1: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cheng H, Dufu K, Lee CS, Hsu JL, et al. 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127: 1389–400. [DOI] [PubMed] [Google Scholar]
- 92. Nojima T, Hirose T, Kimura H, Hagiwara M. 2007. The interaction between cap‐binding complex and RNA export factor is required for intronless mRNA export. J Biol Chem 282: 15645–51. [DOI] [PubMed] [Google Scholar]
- 93. Katahira J, Inoue H, Hurt E, Yoneda Y. 2009. Adaptor Aly and co‐adaptor Thoc5 function in the Tap‐p15‐mediated nuclear export of HSP70 mRNA. EMBO J 28: 556–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lei H, Zhai B, Yin S, Gygi S, et al. 2013. Evidence that a consensus element found in naturally intronless mRNAs promotes mRNA export. Nucleic Acids Res 41: 2517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Stubbs SH, Conrad NK. 2015. Depletion of REF/Aly alters gene expression and reduces RNA polymerase II occupancy. Nucleic Acids Res 43: 504–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Siddiqui N, Borden KL. 2012. mRNA export and cancer. Wiley Interdiscip Rev RNA 3: 13–25. [DOI] [PubMed] [Google Scholar]
- 97. Tran DD, Koch A, Tamura T. 2014. THOC5, a member of the mRNA export complex: a novel link between mRNA export machinery and signal transduction pathways in cell proliferation and differentiation. Cell Commun Signal 12: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Garcia‐Lopez A, Monferrer L, Garcia‐Alcover I, Vicente‐Crespo M, et al. 2008. Genetic and chemical modifiers of a CUG toxicity model in Drosophila. PLoS ONE 3: e1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ward AJ, Cooper TA. 2010. The pathobiology of splicing. J Pathol 220: 152–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Srebrow A, Kornblihtt AR. 2006. The connection between splicing and cancer. J Cell Sci 119: 2635–41. [DOI] [PubMed] [Google Scholar]
- 101. Mayr C, Bartel DP. 2009. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Morimoto M, Boerkoel CF. 2013. The role of nuclear bodies in gene expression and disease. Biology (Basel) 2: 976–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Machyna M, Heyn P, Neugebauer KM. 2013. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA 4: 17–34. [DOI] [PubMed] [Google Scholar]
- 104. Spector DL, Lamond AI. 2011. Nuclear speckles. Cold Spring Harb Perspect Biol 3: pii: a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Girard C, Will CL, Peng J, Makarov EM, et al. 2012. Post‐transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat Commun 3: 994. [DOI] [PubMed] [Google Scholar]
- 106. McCracken S, Fong N, Rosonina E, Yankulov K, et al. 1997. 5′‐Capping enzymes are targeted to pre‐mRNA by binding to the phosphorylated carboxy‐terminal domain of RNA polymerase II. Genes Dev 11: 3306–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hossain MA, Chung C, Pradhan SK, Johnson TL. 2013. The yeast cap binding complex modulates transcription factor recruitment and establishes proper histone H3K36 trimethylation during active transcription. Mol Cell Biol 33: 785–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mazza C, Segref A, Mattaj IW, Cusack S. 2002. Large‐scale induced fit recognition of an m(7) GpppG cap analogue by the human nuclear cap‐binding complex. EMBO J 21: 5548–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kao HY, Siliciano PG. 1996. Identification of Prp40, a novel essential yeast splicing factor associated with the U1 small nuclear ribonucleoprotein particle. Mol Cell Biol 16: 960–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jimeno S, Rondon AG, Luna R, Aguilera A. 2002. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Pascual‐Garcia P, Govind CK, Queralt E, Cuenca‐Bono B, et al. 2008. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev 22: 2811–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ellisdon AM, Dimitrova L, Hurt E, Stewart M. 2012. Structural basis for the assembly and nucleic acid binding of the TREX‐2 transcription‐export complex. Nat Struct Mol Biol 19: 328–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Jimeno S, Luna R, Garcia‐Rubio M, Aguilera A. 2006. Tho1, a novel hnRNP, and Sub2 provide alternative pathways for mRNP biogenesis in yeast THO mutants. Mol Cell Biol 26: 4387–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhang Z, Fu J, Gilmour DS. 2005. CTD‐dependent dismantling of the RNA polymerase II elongation complex by the pre‐mRNA 3’‐end processing factor, Pcf11. Genes Dev 19: 1572–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Noble CG, Walker PA, Calder LJ, Taylor IA. 2004. Rna14‐Rna15 assembly mediates the RNA‐binding capability of Saccharomyces cerevisiae cleavage factor IA. Nucleic Acids Res 32: 3364–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Rodriguez CR, Cho EJ, Keogh MC, Moore CL, et al. 2000. Kin28, the TFIIH‐associated carboxy‐terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol 20: 104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Vasiljeva L, Kim M, Mutschler H, Buratowski S, et al. 2008. The Nrd1‐Nab3‐Sen1 termination complex interacts with the Ser5‐phosphorylated RNA polymerase II C‐terminal domain. Nat Struct Mol Biol 15: 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Creamer TJ, Darby MM, Jamonnak N, Schaughency P, et al. 2011. Transcriptome‐wide binding sites for components of the Saccharomyces cerevisiae non‐poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet 7: e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kim M, Ahn SH, Krogan NJ, Greenblatt JF, et al. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Amrani N, Minet M, Le Gouar M, Lacroute F, et al. 1997. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol 17: 3694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]


