Abstract
A vast network of cellular circadian clocks regulates 24‐hour rhythms of behavior and physiology in mammals. Complex environments are characterized by multiple, and often conflicting time signals demanding flexible mechanisms of adaptation of endogenous rhythms to external time. Traditionally this process of circadian entrainment has been conceptualized in a hierarchical scheme with a light‐reset master pacemaker residing in the hypothalamus that subsequently aligns subordinate peripheral clocks with each other and with external time. Here we review new experiments using conditional mouse genetics suggesting that resetting of the circadian system occurs in a more “federated” and tissue‐specific fashion, which allows for increased noise resistance and plasticity of circadian timekeeping under natural conditions.
Keywords: circadian clock, clock genes, entrainment, light, mammals, peripheral clocks, SCN
Introduction
The circadian system provides a first‐rate example illustrating in what way changes in the environment shape behavior and physiology. The 24‐hour periodicity of axial rotation of our planet has favored the evolution of circadian clocks, endogenous cellular pacemakers (terms in italics throughout the text are defined in Box 1) that allow organisms to adapt to the natural light‐dark rhythm. Circadian clocks are present in unicellular organisms such as dinoflagellates 1, in plants 2, 3, in insects 4, and in vertebrates 5. The first gene encoding a critical component of a circadian clock (Period) was discovered in Drosophila by Konopka and Benzer in 1971, showing that circadian clocks are genetically encoded 6. In mammals, circadian clocks are found in nearly all cells and tissues. At the biochemical level, they consist of coupled feedback loops that collectively establish a self‐sustained, yet adjustable molecular oscillator that controls, via transcriptional programs, a wide spectrum of cellular and organismal processes (Fig. 1). A critical feature of circadian clocks is their ability to sustain circadian oscillations for days or even weeks, even in the absence of an external periodic stimulus. This property is maintained even in tissue explants and cell cultures 7, 8.
Figure 1.
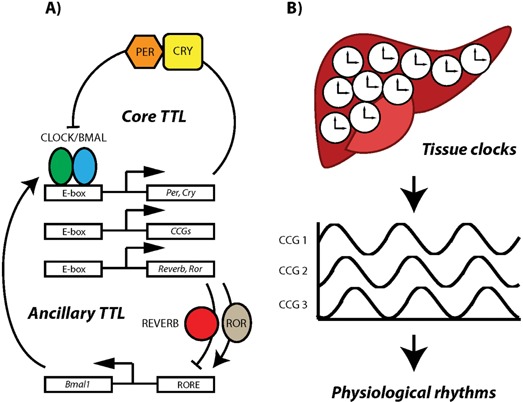
The circadian molecular clockwork. A: At the molecular level circadian clocks are based on interlocked transcriptional/translational feedback loops (TTLs). In the circadian core TTL the transcription factors CLOCK and BMAL1 activate the expression of Period (Per1–3) and Cryptochrome (Cry1/2) genes during the day. PER and CRY proteins accumulate in the cytoplasm and toward the night translocate to the nucleus where they inhibit CLOCK/BMAL1‐mediated transcription. Accompanied by modifications of histone tails around the DNA‐binding sites of CLOCK/BMAL1, PER/CRY binding leads to a dissociation of the transcription complex and suppresses production of Per and Cry mRNAs. Gradual degradation of PER/CRY complexes toward the end of the night releases the CLOCK/BMAL1 dimer from PER/CRY suppression, thus re‐initiating the clock cycle by induction of Per and Cry transcription. A number of ancillary feedback loops 73, 74, 75 as well as post‐translational mechanisms 76 fine‐tune and stabilize the oscillation of this core TTL. In addition to creating a self‐sustaining clockwork, core clock proteins also control transcription of a large number of tissue‐specific clock‐controlled genes (CCGs). B: Typically each cell of an organ such as the liver houses its own clock which drives the circadian expression of CCGs to create organ‐specific rhythmic physiological responses. Although the clock components may differ between species, this principle organization of circadian timekeeping is preserved in all animals and plants.
Box 1. Abbreviations and terminology.
DD: Constant darkness; used to analyze free‐running rhythms of the circadian clock.
Entrainment: The coordination of a self‐sustained oscillator via rhythmic signals from a pacemaking oscillator (i.e. a zeitgeber). In a loose way, this refers to the process of alignment of the internal clock with external time.
Free‐run: Behavior of internal clocks under Zeitgeber‐free conditions. When free‐running, circadian rhythms cycle with their endogenous period which usually deviates from 24 hours. For example, in the C57BL/6 mouse strain, the free‐running period in DD is roughly 23.7 hours. However, this value differs with strain, species, and light intensity.
Jetlag: Misalignment of internal and external time after rapid crossing of several time zones. Re‐entrainment of circadian clocks to local time ends jetlag after a number of days depending on the magnitude of the time shift.
LD: Rhythmic light:dark conditions. While the time intervals can differ depending on experimental setup, in this paper, we refer to standard 12 hours light:12 hours dark conditions, which synchronize central and peripheral clocks when no conflicting Zeitgeber signals are present.
Masking: Masking refers to a direct regulation of an overt rhythm while over‐riding entrainment, or enforcement of periodicity independent of circadian control. Masking does not affect clock function and does also not depend on a functional circadian system.
Pacemaker: A central clock (i.e. the SCN) that coordinates rhythmic output of peripheral clocks and aligns them to external time.
Peripheral clocks: Cellular oscillators outside the SCN; found in peripheral tissues, but also in various brain areas. An alternative designation is subordinate clocks, emphasizing their dependence on SCN input.
Phase angle: Refers to the temporal position of a landmark point of an expressed internal rhythm with respect to an external reference, e.g. the time of sleep onset relative to the time of “lights off.”
RF: Time‐restricted feeding; a potent zeitgeber for peripheral tissue clocks.
SCN: Suprachiasmatic nucleus, a hypothalamic nucleus above the optic chiasm considered to be the circadian pacemaker in mammals.
TTL: Transcriptional‐translational feedback loop; the core organizational unit of the circadian clock (see Fig. 1A).
VIP: Vasointestinal polypeptide; peptide mediator of the hypothalamus and the gut.
Zeitgeber: A rhythmic signal capable of entraining circadian clocks.
Organization of the circadian system
Because of the widespread presence of clocks throughout the body, the circadian system of an animal resembles a clock shop rather than a single clock. Therefore, the important question arises of how rhythms of so many clocks are efficiently synchronized. There are two obvious ways of circadian rhythm coordination. One assumes a master pacemaker that instructs all subordinate, i.e. peripheral, clocks (hierarchical system). The other mechanism would depend on coupling of a multitude of essentially equivalent oscillators (“federated” system). In mammals, the discovery of a central circadian pacemaker in the suprachiasmatic nucleus (SCN) of the hypothalamus long suggested a strict top‐down control 9, 10. Because the SCN receives direct input from the retina via the retino‐hypothalamic tract, this nucleus serves as a direct window looking into the outside world. The neurons within the SCN are coupled and as an ensemble produce electrical and molecular circadian rhythms of a robustness not seen in isolated neurons and other types of cells 11. The SCN is connected to several other hypothalamic and extra‐hypothalamic nuclei thereby becoming part of an intricate neuronal network 12. A vast body of literature supports a hierarchical control of circadian synchronization, in which the rhythm of the master pacemaker in the SCN is entrained by an external zeitgeber: the rhythmic change of light and dark. The SCN subsequently synchronizes peripheral clocks with each other and thus aligns the entire circadian system to the external light‐dark cycle. The detailed nature of the synchronizing signals is still being investigated, but relies on a combination of humoral factors and the peripheral autonomic nervous system (ANS) 16, 17.
This model of a strictly SCN‐centric circadian control has been challenged. Timing signals other than light, such as rhythmically timed food intake, can potently reset peripheral clocks directly without affecting clock rhythms in the SCN 13, 14. Ishida and co‐workers showed that light exposure can acutely activate clock gene expression and glucocorticoid release in the adrenal gland via sympathetic innervation by the splanchnic nerve 15, and this effect appears to be independent of photic responses at the level of the SCN 16. A more “federated” organization of independent oscillators offers the opportunity of different zeitgeber signals acting independently on different peripheral clocks. For example, timed food uptake could primarily reset the liver clock and thereby regulate liver metabolism. The light‐dark cycle could primarily entrain the SCN and its output such as rhythmic locomotor activity 17. The challenge for the “federated” control will be to identify the signaling routes that ensure that all the clocks of the clock shop are properly coordinated.
It should be recalled that under natural conditions, many zeitgeber signals are tied to the day‐night cycle (Fig. 2). This potent photic zeitgeber evokes (in most species) a 24‐hour sleep‐wake rhythm. Sleep precludes both food intake and locomotor activity. Thus, the sleep‐wake rhythm effectively, yet indirectly, drives food intake and body temperature cycles. Nonetheless, one can uncouple light and food: when sleep rhythms are disrupted (e.g. in the case of jetlag) or food intake is restricted to the natural sleeping phase, alignment of different zeitgeber signals is impaired, leading to an uncoupling of SCN and peripheral clocks 14, 18. Many insights into the circadian mechanisms stem from such experimental interference studies.
Figure 2.
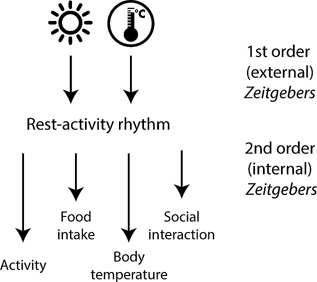
Different zeitgeber signals synchronize the circadian clock. As the endogenous period of the circadian clock is only approximately 24 hours, it has to be entrained to the external 24‐hour cycle every day. Zeitgeber signals are stimuli that can reset the circadian clock. The major external zeitgeber signals (first order signals) are the light‐dark cycle and external temperature changes. In addition, internal zeitgeber signals (second order signals) ensure the synchronization of different body clocks with each other and with external time. The most important internal zeitgeber signals are locomotor activity, the timing of food intake, body temperature, and social interaction.
Excellent reviews on the molecular mechanism of circadian timekeeping have recently appeared 5, 19, 20, 21, 22, some of which also address the clock coordination problem (e.g. 20). In this paper, we discuss to which extent the circadian clock of the SCN is required for peripheral clock synchronization with an emphasis on experimental strategies in which the SCN clock was genetically perturbed, an approach complementary to classical SCN lesion and transplantation. In addition, we review work in which the SCN was ablated and the response of peripheral clocks was investigated by non‐invasive analyses. We discuss these data in relation to the hierarchical and “federated” models. It is worth noting that recent work in Drosophila suggests a revision of a hitherto widely accepted dual‐oscillator model in which pigment‐dispersing factor (PDF) positive neurons were thought to serve as master pacemakers controlling PDF‐negative output neurons. The new findings suggest that rhythms result from an interaction between multiple independent oscillators 23.
New genetic tools enable functional manipulation of the SCN circadian clock
Complete bilateral lesions of the SCN evoke arrhythmicity of locomotor activity, drinking behavior, hormone release, and body temperature 9, 10, 24. Tissues from SCN‐lesioned animals show abolished clock gene rhythmicity under both LD and DD conditions 25, 26, 27, 28, 29, 30. These lesion experiments, in combination with SCN transplantation studies 31, 32, thus show that the SCN controls coherent behavioral and molecular rhythmicity in intact organisms. A significant limitation of lesions is that they not only ablate the structure in question, but also interrupt the associated neuronal networks. Since the SCN is also part of such a network, it is necessary that classical SCN surgical experiments are complemented by alternative approaches 33 such as SCN‐specific deletion of clock genes that preserves the neuronal circuit.
Light can synchronize peripheral clocks in mice with Syt10‐driven deletion of Bmal1 in the SCN
Husse and colleagues recently developed a Syt10‐driven CRE line to delete – in a floxed Bmal1 mouse 34 – the essential clock gene Bmal1 predominantly in neurons of the SCN while leaving Bmal1, and hence clocks, unaffected in peripheral tissues 35. BMAL1 immunohistochemistry, in situ hybridization of clock and clock output genes, as well as PER2::LUC explant recordings from control and mutant SCN all showed that experimental parameters were near base level in mutant SCN supporting the notion that the SCN clock was strongly impaired 36. In line with this, mutant mice did not exhibit circadian rhythms of locomotor activity in DD conditions 35. In LD conditions, however, 24‐hour activity rhythms were preserved, indicating that photic masking was by and large maintained – which is not the case in SCN‐lesioned mice. After SCN lesioning peripheral clock rhythms are abolished. In mice in which solely the SCN clock function was impaired high‐amplitude rhythms of clock gene expression were maintained, and rhythms were synchronized to the external light‐dark cycle and with each other 36. This indicates that SCN clock function is dispensable for the synchronization of peripheral clocks under these conditions. Circadian profiling of clock gene expression and PER2::LUC explant rhythms revealed that peripheral clocks initially were capable of sustaining synchronized rhythmicity under zeitgeber‐free conditions in the absence of the SCN pacemaker. However, when PER2::LUC rhythms were again assessed after 7 days, overt rhythms were lost due to desynchronization between free‐running tissue oscillators. Husse et al. monitored fecal glucocorticoid levels, a robust endocrine clock marker that can be tracked non‐invasively over several days. In LD, glucocorticoid levels were strongly circadian, but upon release into DD a gradual dampening of rhythms was observed resulting in a complete loss of circadian rhythmicity after 7 days in the zeitgeber‐free environment. When the circadian system was challenged by subjecting SCN clock‐deficient mice to an abrupt shift in the LD cycle (jetlag paradigm), the absence of the SCN clock resulted in accelerated resetting of PER2::LUC rhythms in peripheral tissue explants. Such hastened light adaptation of peripheral clock rhythms in the absence of SCN pacemaker function suggests that, in a wild‐type animal, the SCN clock stabilizes the phase of the peripheral circadian system against external perturbation. In summary, these findings suggest that under LD, but not DD, conditions the SCN clock is dispensable for the synchronization of peripheral clocks. Importantly, while putting into perspective the coordinating function of the SCN in circadian rhythm regulation, the mouse genetic findings immediately propose an important function for the central pacemaker: if the circadian system is challenged by an absence of zeitgeber information (DD) or when conflicting zeitgeber signals interfere (jetlag), the SCN clock becomes essential to steady the circadian cellular oscillator network.
It is debatable whether such alignment of peripheral clocks with the external light‐dark cycle qualifies as entrainment in the classical sense (see 37 for a detailed discussion of entrainment theory), which requires the existence of two independent and self‐sustained oscillators. The fact that peripheral clock gene rhythms are sustained at least for some time under DD conditions suggests that these can be seen as oscillators of limited self‐sustainment, comparable to a pendulum that due to friction will eventually cease swinging, unless it is periodically deflected. One also has to keep in mind that the loss of circadian rhythmicity at the organ level does not necessarily imply that individual cells are arrhythmic but might also be due to a loss of intercellular synchrony.
Light can synchronize peripheral clocks in mice with CamK2‐driven deletion of Bmal1 in the SCN
The findings of Husse et al. were buttressed by a study using a CamK2‐CRE driver line to abolish Bmal1 throughout the forebrain, including the SCN, while leaving peripheral tissues unaffected 38. Bmal1 deletion in the SCN was efficient, as shown by attenuated clock gene rhythms as well as a loss of circadian locomotor activity in DD. Mutant mice were rhythmic in LD; the locomotor activity onset, however, was more variable and appeared slightly phase‐advanced when compared to controls. The authors also investigated the masking response in more detail and found an impairment of light‐induced suppression of behavioral activity in mutant mice. Clock gene expression in peripheral tissues of mutants exhibited well‐phased rhythms in LD conditions, indicating that internal synchrony was maintained. In DD, peripheral clock gene expression profiles showed dispersed phasing as well as reduced oscillatory amplitudes. Using high‐resolution bioluminescence imaging of heart tissue, Izumo and co‐workers demonstrated that the decreased amplitude of peripheral clocks was due to a combination of decreased cellular rhythmicity and increased phase desynchrony between cellular clocks 38. In an additional set of experiments, time‐restricted feeding (RF) in DD was used to rescue peripheral clock regulation in forebrain clock‐deficient mice. Feeding cues synchronized circadian clocks in liver and kidney, while peripheral clocks in heart, lung, and spleen were less responsive to timed food intake. The oscillatory amplitude was restored to a level comparable to control mice only in liver, while all other peripheral tissues showed lower amplitude oscillations even after 2 weeks of RF. The authors concluded that internal synchrony and high‐amplitude peripheral rhythms vanish in the absence of a functional SCN pacemaker. However, rhythms can be restored by the external zeitgeber light or, at least partially, by timed feeding 38.
In summary, both studies conclude that despite a genetic ablation of the SCN clock peripheral clock rhythmicity and synchrony is largely maintained in the presence of an external zeitgeber. While light resets all tissues, feeding cues only partially restore internal synchrony in SCN clock‐deficient mice. In the absence of any zeitgeber peripheral rhythms are attenuated and internal synchrony is markedly impaired. Together these findings provide strong genetic evidence for the notion that in addition to the sequential pathway, in which light synchronizes the SCN clock, and the SCN clock in turn transmits the timing information to peripheral clocks (Fig. 3, pathway 1), there exist additional pathways by which peripheral clocks are synchronized independently of the SCN clock (Fig. 3, pathway(s) 2). At this point, the nature of these pathways remains to be explored. Light‐driven activation of SCN neurons may lead to rhythmic autonomic, endocrine, or behavioral outputs that then translate into rhythmic clock gene activation in peripheral tissues. Alternatively, various hypothalamic and some extra‐hypothalamic regions outside the SCN receive retinal input and might influence endocrine, autonomic, or behavioral outputs that regulate the periphery 39. Some of these areas may even house endogenous, light‐entrainable circadian clocks. SCN ablation studies seemingly have missed out on such additional pathways.
Figure 3.
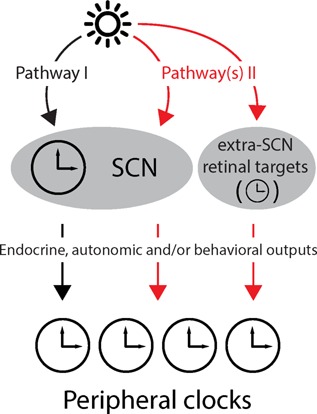
Parallel pathways synchronize peripheral clocks to the LD cycle. Light is the most important zeitgeber for circadian clocks and can reach peripheral clocks via several routes. In pathway 1, light from the retina reaches the SCN through the retino‐hypothalamic tract and entrains the SCN clock that then transmits the timing information to peripheral clocks along neuronal and/or endocrine pathways. Pathway 2 depicts additional routes for transmission of photic information that bypass the SCN clock and directly or indirectly reset peripheral clocks.
It is important to realize that despite the use of different CRE driver lines to delete Bmal1 in overlapping, but not identical, regions of the brain (compare 42 and 37), both studies gave very similar results. It is well known that CRE driver lines have limitations with regard to tissue specificity and recombination efficiency. Therefore, the use of two independent drivers targeting the same gene (Bmal1) strengthens the conclusions that can be drawn from this genetic approach.
In vivo PER2::LUC recordings in anesthetized mice reveal SCN‐independent peripheral clock resetting
Recent advances were also made in the use of real‐time, non‐invasive analysis of the output of mammalian circadian clocks. High‐resolution time series have long been available for locomotor activity 40 and for secretion of glucocorticoids 41, 42, but directly following molecular clocks inside the body was achieved just recently 43, 44.
Tahara et al. 44 used in vivo bioluminescence monitoring in PER2::LUC mice bred onto an albino ICR background. For measurements of luminescence animals were repeatedly anesthetized with isoflurane, and luciferin was infused to elicit light emission through luciferase‐mediated oxidation. Kidneys, livers, and submandibular glands were monitored. In all three tissues PER2::LUC activity was rhythmic with peaks in the first half of the dark phase. In tissues of Bmal1‐LUC mice, in which luciferase expression is driven by the Bmal1 promoter, luminescence rhythms were shifted by 8 hours relative to those observed for PER2::LUC. Apparently, the reporters recapitulate the naturally occurring anti‐phasic expression profiles of endogenous PER2 and Bmal1. After SCN lesion, peripheral tissue rhythms were rhythmic but greatly dampened, and normal phase relationships between tissues were abolished. This contrasts with previous data obtained from real‐time quantitative PCR (qPCR) experiments with SCN‐lesioned animals where peripheral clock gene rhythms were completely lost 27. Such discrepancy may be due to the fact, that qPCR profiles usually combine average data from different animals 27. While these findings do not devaluate the old studies, they emphasize the necessity of time series from individual animals to overcome inter‐animal variations in gene activity levels.
To assess free‐running periods of different tissue clocks, Tahara et al. adapted SCN‐lesioned mice to RF under LD conditions and then subjected them to ad libitum food access and DD 44. Under RF/LD conditions PER2::LUC rhythms in kidney, liver, and submandibular gland were reset by 12 hours compared to ad libitum food/LD consistent with what had been observed from sequential sampling studies before 14. Under free‐run conditions, rhythmicity in peripheral clock activity was maintained at a period of slightly less than 24 hour, reminiscent of what is observed for locomotor activity in wild‐type mice. However, rhythm amplitudes became dampened soon after release into ad libitum food/DD, indicating that a functional SCN pacemaker coordinates and maintains peripheral rhythmicity in the absence of an external zeitgeber.
PER2::LUC recordings in freely moving mice reveal accelerated resetting of the liver clock in SCN‐lesioned mice
Saini et al. 43 have invented a device that houses an individual mouse that has access to food and water, whose locomotion can be monitored by infrared sensors and whose bioluminescence photons emitted (e.g. from liver) are collimated and recorded by highly sensitive photomultiplier technology. Bioluminescence in liver was evoked by transducing hepatocytes with an adenoviral Bmal1‐LUC reporter introduced by tail vein injection. Luciferase substrate was delivered continuously with an osmotic mini pump implant. Saini and co‐workers used this set‐up to investigate the regulation of hepatocyte clocks by SCN and food entrainment signals 43. Animals maintained under night‐restricted feeding were shifted to daytime‐only feeding, and the phase shifting of liver bioluminescence rhythms was recorded for several days at 1‐minute intervals. Animals with an intact or a sham‐operated SCN responded slowly to a shift in the feeding regimen. By contrast, mice with SCN lesions showed a rapid phase‐shift in their hepatic BMAL1‐LUC activity rhythms indicating that SCN function shields the liver clock from food‐induced phase shifting.
As is often the case, new methods and technologies shed new light on long‐standing questions. With regard to the essentiality of a master pacemaker in the SCN, the recent work suggests that a genetic deletion of Bmal1 in the SCN only to some extent affects the amplitude and phase of peripheral clocks, as long as animals are maintained in an LD environment. The rhythmic change of light and dark is sufficient to impose a rhythm of clock gene expression in peripheral organs, even if the SCN clock is completely abolished. If such SCN‐clock deficient mice are maintained in a zeitgeber‐free (e.g. DD) environment, peripheral clock gene expression rhythms become phase‐dispersed and their amplitude is strongly reduced, presumably because cellular synchronization in peripheral tissues is abolished. In mice with an intact SCN, neither timed feeding nor a reversal of feeding time has an immediate consequence on the activity rhythm of a hepatic luciferase reporter. In contrast, in mice in which the SCN was surgically ablated, timed feeding imposes a rapid shift in hepatic luciferase reporter activity rhythms, thus revealing the existence of nutrient‐dependent zeitgeber cues that substitute for the SCN signal. What is the biological relevance of such metabolic cues? Envisage a scenario where a nocturnal rodent has limited access to food at dusk. If metabolic enzymes required for the uptake and processing of nutrients were entirely subject to regulation by the SCN clock, the clock controlled enzymes required for digestion might not be optimally expressed. This problem is resolved, if food uptake itself serves as a zeitgeber to turn on the expression of clock controlled metabolic enzymes. Such decentralized control provides the organism with the opportunity to integrate complex periodic changes (light, food, temperature, humidity, social cues, etc.) that occur in the surroundings over the 24‐hour period and over which the organism has no control.
Organization of the “federated” multi‐clock system
The studies reviewed above, favor a “federated” model of the circadian timing system. This raises a host of questions such as how the myriads of peripheral tissue clocks are synchronized with each other and with external time (Fig. 4). Intercellular and inter‐tissue synchronization is particularly challenging because most tissues contain multiple cell types. Little is known about the mechanisms of intercellular synchronization except for the SCN in which paracrine signals (VIP and others), gap junctions, and electrical signaling appear to play a role in keeping SCN cell rhythms aligned with each other 45, 46, 47, 48. In the case of peripheral clocks, mechanisms of intercellular synchronization are largely elusive. Some studies suggest that cellular coupling is confined to the SCN 11. It should be recalled, however, that while in dispersed cell cultures overt rhythms are lost within 2–3 days, clocks in explants of peripheral tissues retain high‐amplitude circadian oscillations for substantially longer 7. These findings support the existence of short‐range signaling through direct cell‐to‐cell communication, for example via gap junctions in endothelial tissues, or by paracrine factors. In development, such short‐range interactions are mediated by transmembrane signaling based on direct interactions between cell surface receptors and their cognate ligands. Whether this signaling mechanism occurs in the circadian timing system remains to be seen. It is also unclear whether in a multicellular tissue all cellular clocks are equal. It is tempting to speculate that the clock in a specific cell type might set the phase of the clock in neighboring cells as was proposed for Clara cells in the lung epithelium 49.
Figure 4.
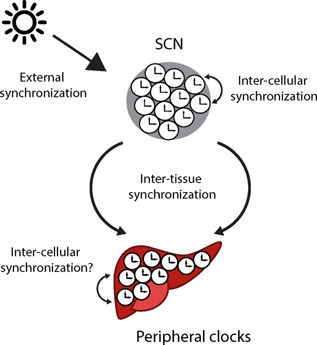
Types of synchronization within the circadian timing system. Several synchronization mechanisms are required to ensure internal and external synchrony of the numerous cellular clocks present in higher organisms. Single cells within a tissue have to be synchronized with each other (inter‐cellular organization) to allow synchronized circadian output of a particular peripheral tissue clock. In addition, different tissue clocks have to be synchronized with each other, to maintain systemic internal synchrony. Lastly, the entire organism has to be synchronized to the external 24‐hour LD cycle to ensure temporal adaptation (external synchronization).
Rhythmically released hormones such as melatonin or glucocorticoids, other blood‐borne signals 50, or even body temperature cycles could, either alone or in combination, sustain synchronization of multiple peripheral clocks. The timing of each of these signals may also be controlled by the autonomic nervous system and perhaps by other tissue clocks, such as the adrenal 51. Additional control of inter‐tissue synchronization may stem from hypothalamic nuclei, many of which contribute to the regulation of physiological functions known to be clock‐controlled 52. Synchronization to environmental zeitgebers are well‐characterized in the case of photic entrainment of the SCN clock 53, but there are only a few studies on non‐SCN tissues 15, 54.
A “federated” clock system increases temporal stability and plasticity
From an evolutionary perspective, the question arises why it would be beneficial for multicellular organisms to maintain a complex “federated” network organization of clocks instead of a seemingly more straightforward hierarchical organization with a master regulator controlling coherent circadian physiology across the body. One possible explanation would be that cellular clocks took over additional adaptive functions such as coordinating tissue‐specialization and cross‐tissue coordination resulting in increased overall fitness. Additional rationales are increased resilience to conflicting zeitgeber signals due to parallel and largely independent routes of synchronization and increased plasticity in response to complex environments and, thus, environmental demands.
The multimodality of resetting mechanisms characterizing the “federated” clock model allows for a more differentiated network response. At the same time, by routing the time signal through different paths, it increases the overall stability of the system, protecting it against conflicting zeitgeber signals (Fig. 5). Supporting this notion, Husse et al. have shown that peripheral clock adaptation to a shifted LD cycle is accelerated if only one resetting pathway is functional 36. Similarly, peripheral clocks in SCN‐lesioned mice reset faster to a shifted feeding schedule 43. Under artificial conditions such as jetlag increased internal phase stability becomes a liability, because one would want clocks to adapt fast. In a more natural and, thus, rather noisy zeitgeber environment, however, it proves highly advantageous, because it prevents the system from unwanted phase shifts in response to stochastic or intermittent zeitgeber signaling.
Figure 5.
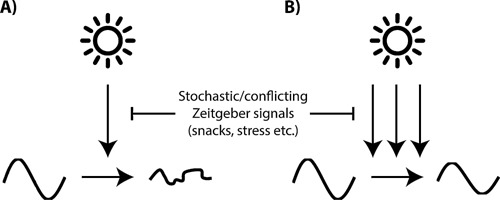
A: Redundant synchronization pathways protect the circadian system against zeitgeber noise. In a scenario where a single pathway resets the circadian clock, external perturbations such as conflicting zeitgeber signals (e.g. from occasional food intake during the rest phase) would potentially have marked effects on the amplitude and phase of the circadian system. B: Redundant synchronization pathways protect the clock network against such sporadic external zeitgeber signalling and allow high‐amplitude and synchronized circadian clock rhythmicity even under complex (and noisy) environmental conditions.
Another potential advantage of a “federated” circadian network is increased network plasticity in the context of complex environmental conditions. As described above, different clocks respond differently to certain timing signals. For example, the liver as a metabolic organ is highly sensitive to RF while the SCN controlling, e.g. locomotion is primarily reset by light 14. A strictly centralized network would result in synchronization of all physiological processes to the same zeitgeber signal. A “federated” network, however, allows for each clock and, therefore, each physiological process to be synchronized to those zeitgeber signals that are most relevant for a particular process, resulting in a tailored response. For example, the liver clock should be synchronized to rhythms in food intake, but it should also respond to changes in energy demands or variations in oxygen supply. A “federated” organization allows for better adaptation to changing environments. For example, seasonal adaptation of circadian rhythms might be facilitated by differential resetting of tissue clocks in response to changes in photoperiod, temperature, humidity, and other parameters that change over the course of a year. It has been suggested that the phase distribution of individual SCN cells contains information about day‐length 55 and that coupling of individual SCN cells facilitates photoperiod adaptation 56.
Future routes of research
It will be of interest to further explore the different synchronization pathways in SCN clock‐deficient mice. Such experiments should focus on modifying the circadian rhythm of secondary zeitgeber signals and observing the responses of the peripheral clock network. For example to approach the role of food as a secondary zeitgeber (Fig. 2) in photic entrainment, SCN clock‐deficient mice could be subjected to a constant feeding paradigm, in which mice are fed every few hours such that the circadian rhythm in food intake is abolished 57. If peripheral clocks remain aligned to the LD cycle under such conditions, it can be concluded that food signals are not necessary to synchronize peripheral clocks in SCN clock‐deficient mice. Another zeitgeber candidate, glucocorticoid rhythms, could be abolished by subcutaneous implants that release corticosterone constantly over the course of the day in surgically or chemically adrenal‐ablated mice 58. To test the role of autonomic signals, optogenetic approaches may be used to manipulate circadian ANS activity and investigate the effects on peripheral clock function. Of note, with experiments such as described above, only the necessity for a single zeitgeber signal can be tested. It is likely that a real‐life situation is more complicated with several, partially redundant signaling pathways contributing to synchronizing peripheral clocks. Moreover, depending on the external condition, the contribution of different pathways to peripheral synchronization may vary. Experiments in which mice are subjected to conflicting or noisy zeitgeber signals will be very informative in this context.
An alternative approach to analyze circadian network connectivity would be to abolish clock function in several tissues at the same time and investigate the response of the remaining clock network. Given the role of glucocorticoids in tissue clock regulation it might be of particular interest to disrupt, in addition to the SCN pacemaker, the adrenal clock or adrenal clock outputs in SCN clock‐deficient mice, e.g. by adrenal‐targeted disruption of the essential clock gene Bmal1 51, 59. One hypothesis is that adrenal and SCN clocks together play a key role in synchronizing the overall circadian network, thus expecting further deterioration of internal synchrony in adrenal and SCN clock double‐deficient mice.
It should be noted that so far most genetic experiments targeting the SCN clock use a deletion of Bmal1. This is understandable since Bmal1 is the only single clock gene discovered so far whose deletion results in complete behavioral arrhythmicity under constant conditions. However, combinatorial deletions of other genes – e.g. Cry1/2 60 or Per1/2 61 – also result in a loss of circadian rhythmicity. Thus, in order to clearly distinguish between (Bmal1) gene and circadian clock loss effects one would need to test whether manipulation of these gene pairs specifically in the SCN has similar effects. Moreover, current conditional genetics never target all cells within a tissue of interest and in, both, the Husse and the Izumo papers not all cells in the SCN are affected, thus offering the possibility that residual rhythms are retained in the mutant SCN. In line with this, Lee et al. 62, using a similar approach targeting neuromedin S‐positive neurons in the SCN, have shown that even when 80% of SCN neurons lack Bmal1 residual behavioral rhythmicity is observed for some time after mutant mice are released into DD. Finally, even in conventional – and thus complete – Bmal1 mutant SCN slice cultures residual oscillations are observed, although with different periodicity and reduced robustness 63.
While cell culture data suggest that Bmal1 deletion disrupts clock function at the molecular level, an alternative explanation would be that Bmal1 knock out in the SCN may mainly affect intercellular synchrony within the SCN neuronal network. In line with this, residual oscillations of at least some clock genes are still observed in the SCN of mice with Syt10‐CRE driven deletion of Bmal1 64. Importantly, similar effects can be obtained by impairing synaptic activity in the SCN by tetanus toxin 62. Further, a common feature of all these mouse models is an accelerated phase shifting under jetlag conditions, which is also observed when SCN intercellular coupling is affected by different means 65, 66, 67. Taken together, further complementary studies are needed to distinguish between cellular network and molecular clock disruption in the SCN and its effects on peripheral clock gene regulation.
It will further be of importance to study the molecular endpoints of resetting signals in target tissues. For example, it has been shown that some rhythmic liver transcripts are regulated by signals derived from outside this tissue, whereas other transcripts are controlled by the local clock gene machinery 68, 69. Studies on SCN clock‐deficient mice published so far only looked at a handful of canonical clock genes in a limited number of tissues. In order to obtain a more comprehensive picture of synchronization mechanisms and physiological outputs it will be necessary to study a broader spectrum of readouts. Transcriptome studies from peripheral tissues could identify those genes that are reset independently of the SCN clock and those that require SCN pacemaker function. In combination with the above‐described experiments aimed at dissecting the internal routes of entrainment such approaches will allow to specifically associate transcriptional – and, ultimately, physiological – rhythms with different synchronization signals. It is intriguing to speculate that different zeitgeber signals may act via different mechanisms (in different tissues). For example photic signals have been shown to affect the clockwork via induction of Per gene expression whereas metabolic cues may signal via Bmal1, Rev‐erba 70, or Clock 71. Even others may exert their effect through epigenetic mechanisms 72.
Conclusions and prospects
Recent data suggest that the organization of the mammalian multi‐clock system is not strictly hierarchical. Peripheral clocks can be aligned with and, likely, entrained to external conditions in a largely “federated” fashion that conveys plasticity to the system. In the absence of periodic zeitgeber information, the clock system relies on coordinating signals from the SCN master clock, thus switching to a more hierarchical organization. This mixed modality may represent a compromise between a need for fine‐tuned adaptation to complex environmental signatures and a need for circadian stability under less reliable zeitgeber conditions. The relative contribution of either mechanism might depend on the situation. With the development of more sophisticated genetic tools, we are finally in a position to start to functionally dissect circadian network organization in the living mammal and devise more specific means of manipulating clock function – for example during jetlag or in the context of various diseases.
Acknowledgments
GE and JH are supported by the Max Planck Society. HO is a Lichtenberg fellow of the Volkswagen Foundation.
The authors have declared no conflict of interest.
References
- 1. Hastings JW. 2007. The Gonyaulax clock at 50: translational control of circadian expression. Cold Spring Harb Symp Quant Biol 72: 141–4. [DOI] [PubMed] [Google Scholar]
- 2. McClung CR. 2013. Beyond Arabidopsis: the circadian clock in non‐model plant species. Semin Cell Dev Biol 24: 430–6. [DOI] [PubMed] [Google Scholar]
- 3. Nakamichi N. 2011. Molecular mechanisms underlying the Arabidopsis circadian clock. Plant Cell Physiol 52: 1709–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boothroyd CE, Young MW. 2008. The in(put) s and out(put) s of the Drosophila circadian clock. Ann NY Acad Sci 1129: 350–7. [DOI] [PubMed] [Google Scholar]
- 5. Partch CL, Green CB, Takahashi JS. 2014. Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24: 90–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Konopka RJ, Benzer S. 1971. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA 68: 2112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, et al. 2004. PERIOD2::LUCIFERASE real‐time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA 101: 5339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balsalobre A, Damiola F, Schibler U. 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93: 929–37. [DOI] [PubMed] [Google Scholar]
- 9. Stephan FK, Zucker I. 1972. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci USA 69: 1583–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moore RY, Eichler VB. 1972. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res 42: 201–6. [DOI] [PubMed] [Google Scholar]
- 11. Liu AC, Welsh DK, Ko CH, Tran HG, et al. 2007. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129: 605–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brancaccio M, Enoki R, Mazuski CN, Jones J, et al. 2014. Network‐mediated encoding of circadian time: the suprachiasmatic nucleus (SCN) from genes to neurons to circuits, and back. J Neurosci 34: 15192–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buijs R, Salgado R, Sabath E, Escobar C. 2013. Peripheral circadian oscillators: time and food. Prog Mol Biol Transl Sci 119: 83–103. [DOI] [PubMed] [Google Scholar]
- 14. Damiola F, Le Minh N, Preitner N, Kornmann B, et al. 2000. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14: 2950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishida A, Mutoh T, Ueyama T, Bando H, et al. 2005. Light activates the adrenal gland: timing of gene expression and glucocorticoid release. Cell Metab 2: 297–307. [DOI] [PubMed] [Google Scholar]
- 16. Kiessling S, Sollars PJ, Pickard GE. 2014. Light stimulates the mouse adrenal through a retinohypothalamic pathway independent of an effect on the clock in the suprachiasmatic nucleus. PLoS ONE 9: e92959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Challet E, Caldelas I, Graff C, Pevet P. 2003. Synchronization of the molecular clockwork by light‐ and food‐related cues in mammals. Biol Chem 384: 711–9. [DOI] [PubMed] [Google Scholar]
- 18. Barclay JL, Husse J, Bode B, Naujokat N, et al. 2012. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS ONE 7: e37150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Buhr ED, Takahashi JS. 2013. Molecular components of the mammalian circadian clock. Handb Exp Pharmacol 217: 3–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Granados‐Fuentes D, Herzog ED. 2013. The clock shop: coupled circadian oscillators. Exp Neurol 243: 21–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Minami Y, Ode KL, Ueda HR. 2013. Mammalian circadian clock: the roles of transcriptional repression and delay. Handb Exp Pharmacol 217: 359–77. [DOI] [PubMed] [Google Scholar]
- 22. Richards J, Gumz ML. 2013. Mechanism of the circadian clock in physiology. Am J Physiol Regul Integr Comp Physiol 304: R1053–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yao Z, Shafer OT. 2014. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343: 1516–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Refinetti R, Kaufman CM, Menaker M. 1994. Complete suprachiasmatic lesions eliminate circadian rhythmicity of body temperature and locomotor activity in golden hamsters. J Comp Physiol A 175: 223–32. [DOI] [PubMed] [Google Scholar]
- 25. Akhtar RA, Reddy AB, Maywood ES, Clayton JD, et al. 2002. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12: 540–50. [DOI] [PubMed] [Google Scholar]
- 26. Sakamoto K, Nagase T, Fukui H, Horikawa K, et al. 1998. Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. J Biol Chem 273: 27039–42. [DOI] [PubMed] [Google Scholar]
- 27. Terazono H, Mutoh T, Yamaguchi S, Kobayashi M, et al. 2003. Adrenergic regulation of clock gene expression in mouse liver. Proc Natl Acad Sci USA 100: 6795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iijima M, Nikaido T, Akiyama M, Moriya T, et al. 2002. Methamphetamine‐induced, suprachiasmatic nucleus‐independent circadian rhythms of activity and mPer gene expression in the striatum of the mouse. Eur J Neurosci 16: 921–9. [DOI] [PubMed] [Google Scholar]
- 29. Guo H, Brewer JM, Champhekar A, Harris RB, et al. 2005. Differential control of peripheral circadian rhythms by suprachiasmatic‐dependent neural signals. Proc Natl Acad Sci USA 102: 3111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo H, Brewer JM, Lehman MN, Bittman EL. 2006. Suprachiasmatic regulation of circadian rhythms of gene expression in hamster peripheral organs: effects of transplanting the pacemaker. J Neurosci 26: 6406–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ralph MR, Foster RG, Davis FC, Menaker M. 1990. Transplanted suprachiasmatic nucleus determines circadian period. Science 247: 975–8. [DOI] [PubMed] [Google Scholar]
- 32. Silver R, LeSauter J, Tresco PA, Lehman MN. 1996. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature 382: 810–3. [DOI] [PubMed] [Google Scholar]
- 33. Morin LP. 2013. Neuroanatomy of the extended circadian rhythm system. Exp Neurol 243: 4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Storch KF, Paz C, Signorovitch J, Raviola E, et al. 2007. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell 130: 730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Husse J, Zhou X, Shostak A, Oster H, et al. 2011. Synaptotagmin10‐Cre, a driver to disrupt clock genes in the SCN. J Biol Rhythms 26: 379–89. [DOI] [PubMed] [Google Scholar]
- 36. Husse J, Leliavski A, Tsang AH, Oster H, et al. 2014. The light‐dark cycle controls peripheral rhythmicity in mice with a genetically ablated suprachiasmatic nucleus clock. FASEB J 28: 4950–60. [DOI] [PubMed] [Google Scholar]
- 37. Roenneberg T, Daan S, Merrow M. 2003. The art of entrainment. J Biol Rhythm 18: 183–94. [DOI] [PubMed] [Google Scholar]
- 38. Izumo M, Pejchal M, Schook AC, Lange RP, et al. 2014. Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. eLife 3: e04617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Canteras NS, Ribeiro‐Barbosa ER, Goto M, Cipolla‐Neto J, et al. 2011. The retinohypothalamic tract: comparison of axonal projection patterns from four major targets. Brain Res Rev 65: 150–83. [DOI] [PubMed] [Google Scholar]
- 40. Albrecht U, Oster H. 2001. The circadian clock and behavior. Behav Brain Res 125: 89–91. [DOI] [PubMed] [Google Scholar]
- 41. Tsang AH, Barclay JL, Oster H. 2014. Interactions between endocrine and circadian systems. J Mol Endocrinol 52: R1–16. [DOI] [PubMed] [Google Scholar]
- 42. Leliavski A, Dumbell R, Ott V, Oster H. 2015. Adrenal clocks and the role of adrenal hormones in the regulation of circadian physiology. J Biol Rhythms 30: 20–34. [DOI] [PubMed] [Google Scholar]
- 43. Saini C, Liani A, Curie T, Gos P, et al. 2013. Real‐time recording of circadian liver gene expression in freely moving mice reveals the phase‐setting behavior of hepatocyte clocks. Genes Dev 27: 1526–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tahara Y, Kuroda H, Saito K, Nakajima Y, et al. 2012. In vivo monitoring of peripheral circadian clocks in the mouse. Curr Biol 22: 1029–34. [DOI] [PubMed] [Google Scholar]
- 45. Maywood ES, Chesham JE, O'Brien JA, Hastings MH. 2011. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci USA 108: 14306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamaguchi S, Isejima H, Matsuo T, Okura R, et al. 2003. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302: 1408–12. [DOI] [PubMed] [Google Scholar]
- 47. Welsh DK, Takahashi JS, Kay SA. 2010. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol 72: 551–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rash JE, Olson CO, Pouliot WA, Davidson KG, et al. 2007. Connexin36 vs. connexin32, “miniature” neuronal gap junctions, and limited electrotonic coupling in rodent suprachiasmatic nucleus. Neurosci 149: 350–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gibbs J, Ince L, Matthews L, Mei J, et al. 2014. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat Med 20: 919–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gerber A, Esnault C, Aubert G, Treisman R, et al. 2013. Blood‐borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell 152: 492–503. [DOI] [PubMed] [Google Scholar]
- 51. Oster H, Damerow S, Kiessling S, Jakubcakova V, et al. 2006. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab 4: 163–73. [DOI] [PubMed] [Google Scholar]
- 52. Guilding C, Hughes AT, Brown TM, Namvar S, et al. 2009. A riot of rhythms: neuronal and glial circadian oscillators in the mediobasal hypothalamus. Mol Brain 2: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Challet E. 2007. Minireview: entrainment of the suprachiasmatic clockwork in diurnal and nocturnal mammals. Endocrinology 148: 5648–55. [DOI] [PubMed] [Google Scholar]
- 54. Cailotto C, Lei J, van der Vliet J, van Heijningen C, et al. 2009. Effects of nocturnal light on (clock) gene expression in peripheral organs: a role for the autonomic innervation of the liver. PLoS ONE 4: e5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. VanderLeest HT, Houben T, Michel S, Deboer T, et al. 2007. Seasonal encoding by the circadian pacemaker of the SCN. Curr Biol 17: 468–73. [DOI] [PubMed] [Google Scholar]
- 56. vanderLeest HT, Rohling JH, Michel S, Meijer JH. 2009. Phase shifting capacity of the circadian pacemaker determined by the SCN neuronal network organization. PLoS ONE 4: e4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cailotto C, La Fleur SE, Van Heijningen C, Wortel J, et al. 2005. The suprachiasmatic nucleus controls the daily variation of plasma glucose via the autonomic output to the liver: are the clock genes involved? Eur J Neurosci 22: 2531–40. [DOI] [PubMed] [Google Scholar]
- 58. Kiessling S, Eichele G, Oster H. 2010. Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J Clin Invest 120: 2600–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Son GH, Chung S, Choe HK, Kim HD, et al. 2008. Adrenal peripheral clock controls the autonomous circadian rhythm of glucocorticoid by causing rhythmic steroid production. Proc Natl Acad Sci USA 105: 20970–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. van der Horst GT, Muijtjens M, Kobayashi K, Takano R, et al. 1999. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398: 627–30. [DOI] [PubMed] [Google Scholar]
- 61. Zheng B, Albrecht U, Kaasik K, Sage M, et al. 2001. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105: 683–94. [DOI] [PubMed] [Google Scholar]
- 62. Lee IT, Chang AS, Manandhar M, Shan Y, et al. 2015. Neuromedin s‐producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 85: 1086–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ko CH, Yamada YR, Welsh DK, Buhr ED, et al. 2010. Emergence of noise‐induced oscillations in the central circadian pacemaker. PLoS Biol 8: e1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Husse J, Leliavski A, Tsang AH, Oster H, et al. 2014. The light‐dark cycle controls peripheral rhythmicity in mice with a genetically ablated suprachiasmatic nucleus clock. FASEB J 28: 4950–60. [DOI] [PubMed] [Google Scholar]
- 65. An S, Harang R, Meeker K, Granados‐Fuentes D, et al. 2013. A neuropeptide speeds circadian entrainment by reducing intercellular synchrony. Proc Natl Acad Sci USA 110: E4355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, et al. 2013. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 342: 85–90. [DOI] [PubMed] [Google Scholar]
- 67. Mieda M, Ono D, Hasegawa E, Okamoto H, et al. 2015. Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 85: 1103–16. [DOI] [PubMed] [Google Scholar]
- 68. Kornmann B, Schaad O, Bujard H, Takahashi JS, et al. 2007. System‐driven and oscillator‐dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hughes ME, Hong HK, Chong JL, Indacochea AA, et al. 2012. Brain‐specific rescue of Clock reveals system‐driven transcriptional rhythms in peripheral tissue. PLoS Genet 8: e1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu C, Li S, Liu T, Borjigin J, et al. 2007. Transcriptional coactivator PGC‐1alpha integrates the mammalian clock and energy metabolism. Nature 447: 477–81. [DOI] [PubMed] [Google Scholar]
- 71. Chaves I, van der Horst GT, Schellevis R, Nijman RM, et al. 2014. Insulin‐FOXO3 signaling modulates circadian rhythms via regulation of clock transcription. Curr Biol 24: 1248–55. [DOI] [PubMed] [Google Scholar]
- 72. Aguilar‐Arnal L, Sassone‐Corsi P. 2013. The circadian epigenome: how metabolism talks to chromatin remodeling. Curr Opin Cell Biol 25: 170–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Preitner N, Damiola F, Lopez‐Molina L, Zakany J, et al. 2002. The orphan nuclear receptor REV‐ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251–60. [DOI] [PubMed] [Google Scholar]
- 74. Sato TK, Panda S, Miraglia LJ, Reyes TM, et al. 2004. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43: 527–37. [DOI] [PubMed] [Google Scholar]
- 75. Ukai H, Ueda HR. 2010. Systems biology of mammalian circadian clocks. Annu Rev Physiol 72: 579–603. [DOI] [PubMed] [Google Scholar]
- 76. Maywood ES, Chesham JE, Meng QJ, Nolan PM, et al. 2011. Tuning the period of the mammalian circadian clock: additive and independent effects of CK1epsilonTau and Fbxl3Afh mutations on mouse circadian behavior and molecular pacemaking. J Neurosci 31: 1539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]


