Abstract
Temozolomide (TMZ) is approved for use as first-line treatment for glioblastoma multiforme (GBM). However, GBM shows chemoresistance shortly after the initiation of treatment. In order to detect whether silencing of human protein phosphatase 1D magnesium dependent (PPM1D) gene could increase the effects of TMZ in glioma cells, glioma cells U87-MG were infected with lentiviral shRNA vector targeting PPM1D silencing. After PPM1D silencing was established, cells were treated with TMZ. The multiple functions of human glioma cells after PPM1D silencing and TMZ chemotherapy were detected by flow cytometry and MTT assay. Significantly differentially expressed genes were distinguished by microarray-based gene expression profiling and analyzed by gene pathway enrichment analysis and ontology assessment. Western blotting was used to establish the protein expression of the core genes. PPM1D gene silencing improves TMZ induced cell proliferation and induces cell apoptosis and cell cycle arrest. When PPM1D gene silencing combined with TMZ was performed in glioma cells, 367 genes were upregulated and 444 genes were downregulated compared with negative control. The most significant differential expression pathway was pathway in cancer and IGFR1R, PIK3R1, MAPK8 and EP300 are core genes in the network. Western blotting showed that MAPK8 and PIK3R1 protein expression levels were upregulated and RB1 protein expression was decreased. It was consistent with that detected in gene expression profiling. In conclusion, PPM1D gene silencing combined with TMZ eradicates glioma cells through cell apoptosis and cell cycle arrest. PIK3R1/AKT pathway plays a role in the multiple functions of glioma cells after PPM1D silencing and TMZ chemotherapy.
Keywords: glioma, protein phosphatase 1D magnesium dependent, temozolomide, PIK3R1
Introduction
Malignant gliomas account for close to 50% of all CNS tumors. The median survival of patients with glioblastoma multiforme (GBM) remains close to one year from the time of diagnosis in spite of surgical resection followed by radiotherapy and chemotherapy (1). Such poor outcome has led to the exploration of a wide variety of novel therapies, and some of them have been incorporated as a standard treatment for patients with this GBM. Temozolomide (TMZ) is approved to use as first-line treatment for patients with primary and recurrent high-grade gliomas. It has been shown that TMZ combined with radiotherapy can improved the 2-year survival rate from 10.4% with radiotherapy alone to 26.5% in patients with GBM (1). Whereas the efficacy of TMZ is encouraging, additional prolongation of survival remains a challenge. GBM shows chemoresistance shortly after the initiation of treatment. Additionally, recent studies suggested that 60–75% of patients with GBM derive no benefit from treatment with TMZ (2). There is a critical need for means to overcome this drug resistance and expand the limited therapeutic benefit of TMZ.
TMZ is an alkylating agent which binds to DNA and interferes with replication, resulting in the insertion of DNA strand breaks and, ultimately, cell death (3). p53 status in addition to MGMT plays a role in chemotherapy resistance to TMZ (4,5). GBM patients with low mutant p53 expression have higher progression-free survival time and may have longer life expectancy in comparison with the high mutant p53 expression group (6). p53 facilitates favorable antitumor drug response through a variety of key cellular functions, including cell cycle arrest, senescence, and apoptosis (5). Wip1 (wild-type p53-induced phosphatase 1, or PPM1D), initially identified as a p53-regulated allele located on 17q23-24, is a member of the protein phosphatase 2C (PP2C) family and expressed in a p53-dependent manner (7). PPM1D is frequently amplified and overexpressed in many cancers, including gliomas, breast cancers, neuroblastomas, ovarian clear cell adenocarcinomas, and medulloblastomas (8–13). In addition, it has been shown that PPM1D may serve as an oncogene important to astrocytoma progression, especially in astrocytomas with wild-type p53 (8). PPM1D overexpression inhibits p53 functions and reduces selection for p53 mutations during cancer progression. However, whether PPM1D has a role in chemotherapy resistance to TMZ through regulating p53 functions remains uncertain.
In our previous studies, the lentiviral shRNA expression vector capable of stable PPM1D gene silencing at both mRNA and protein levels in glioma cells was constructed (14). In the present study, we demonstrated PPM1D silencing can improve the effect of TMZ on inhibiting the growth and inducing cell apoptosis in glioma cells. The possible mechanisms were also detected.
Materials and methods
Cell culture
The human glioma cell line U87-MG cells were cultured in complete medium consisting of DMEM (Gibco, Grand Island, NY, USA) containing high glucose and pyruvate, supplemented with 10% FBS, 2 mmol/l L-glutamine, 100 U/ml penicillin G and 100 ng/ml streptomycin. Cells were maintained at 37°C in a humidified 5% CO2 atmosphere. The cells were dissociated using 0.25% trypsin and 0.02% EDTA solution and subcultured once in 3 days.
Lentiviral vector infection and PPM1D silencing
The target shRNAs against human PPM1D gene (GenBank accession NM_003620) for RNAi were designed as: 5′-GCATAGACGAAATGGCTTA-3′. The sequence 5′-TTCTCCGAACGTGTCACGT-3′, which had no significant homology to any known human or mouse genes, was used as a negative control. Lentiviral vector infection, assessment of infection rate and silencing efficacy determination were performed as previously described (14). Cells treated with PBS instead of lentiviral vector infected were used as control. PPM1D mRNA expression levels in U87-MG cells were detected by real-time quantitative PCR. Primer sequences used to amplify PPM1D mRNA for RT-qPCR were as follows: forward 5′-CTACACCACCAGTCAAGTCAC-3′, and reverse 5′-AGAAGGCATTGCTACGAACC-3′; these amplified a 93-bp product. Primer sequences of the control amplification of GAPDH mRNA were as follows: forward 5′-TGACTTCAACAGCGACACCCA-3′ and reverse 5′-CACCCTGTTGCTGTAGCCAAA-3′; these amplified a fragment of 121 bp. The relative mRNA level was calculated comparing normalized gene expression values in treated versus untreated control U87-MG cells with the 2−ΔΔCt method.
Drug treatment
Three days after cells were infected with lentiviral vector and the silencing efficacy of PPM1D was verified, cells were harvested and approximately 1.0×105 cells were plated in 60-mm petri dishes 24 h before treatment with 0.1 mmol/l TMZ (Schering-Plough, Madison, NJ, USA). The IC50 value of TMZ, defined as the concentration that reduces the global growth of cells by 50%, was previously determined to be approximately 200 mmol/l (15). Therefore, the concentration of TMZ used in this study was set at 0.1 mmol/l, which was considered to be approximately similar to the plasma concentration found in human subjects (16) and was the same as published data of TMZ used for glioma cells (17,18). TMZ stocks were prepared by dissolving the drug in DMSO, diluting with sterile H2O, filtering, and storing at −80°C. After treatment, cells were gently washed, incubated in fresh media at 37°C, and harvested at various time points. Cells were divided into 3 groups: CON (infection with PBS alone+DMSO), NC (infection with negative control vector+TMZ) and KD (infection with PPM1D RNAi lentiviral vector+TMZ).
Cell proliferation assay
Cell proliferation assay was performed with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT). In brief, when cells were treated with TMZ and incubated for 1, 2, 3, 4 and 5 days, 10 μl of the 5 mg/ml MTT was added. The liquid was disgarded and 100 μl DMSO was added. The absorbance (A) at 490 nm was measured using a microplate reader after the plate was incubated. Then a calibration curve was prepared using the data obtained from the wells that contained known numbers of viable cells. Experiments were performed three times with representative data presented. The experiment was performed in triplicate and repeated three times.
Cell cycle analysis
Cells were trypsinized and collected together with the cells floating in the media 4 days after treated with TMZ. These cells were washed in PBS, fixed in 70% (v/v) ethanol for more than 1 h. Then the cells were washed and resuspended in PBS containing 50 mg/ml propidium iodine (PI, Sigma Chemical Co.) and 100 μg/ml RNase A (Fermentas Co., Glen Burnie, MD, USA) for 1 h at room temperature in the dark. Then a total of 20,000 PI-stained nuclei were analyzed using a Becton Dickinson FACScan (BD Biosciences, San Jose, CA, USA). Analysis was used DNA content as a measure of progression in the cell cycle.
Apoptosis assay
After infected with lentiviral vector and treated with TMZ, both floating and adherent cells were collected 4 days after TMZ treatment. The cell aliquots were combined, washed with ice-cold PBS, resuspended in 1X binding buffer, and stained using the Annexin V-APC apoptosis detection kit (BD Biosciences), following the protocols provided. Annexin V-APC staining cells were quantified by using a Becton Dickinson FACScan, according to the manufacturer's instructions.
Microarray-based gene expression profiling
Total RNA was extracted from cells with TRIzol (Invitrogen), according to the manufacturer's instructions. After RNA yield and quality were assessed by Thremo Nanodrop 2000 and Agilent 2100 Bioanalyzer, amplified RNA (aRNA) was obtained with GeneChip 3′IVT Express kit (Affymetrix), following the instructions provided. Then purified aRNA was fragmented and hybridized with hybridization reaction mixture for 16 h at 45°C to human cDNA microarrays. Preparation of microarrays for scanning was conducted with Affymetrix wash protocols appropriately matched to the specific chip type on a Model 450 Fluidics station. Microarrays were scanned on a GeneChip Scanner 3000 (Affymetrix) after the last wash. The Affymetrix GeneChip operating Software v1.3 operating system controlled data acquisition functions and maintained the mediated first-level data analysis and desktop data management for the entire GeneChip System.
Data selection
Primary data collection was done and analyzed by GeneChip Analysis Software (Affymetrix). The fold changes were calculated relative to baseline controls. Data from three independent replicate experiments were used to perform a paired two-sample t-test for each gene. Regulated genes used to define genes as significantly differentially expressed as fold change of ≥1.5 or ≤ −1.5, which signifies changes in the expression level between KD and NC cells. A change must be in the P-value of <0.05, which describes the likelihood of change of expression for each transcript, where P-values indicated the level of significance of the difference based on the paired two-sample t-test. Pathway Architect Software (Stratagene) was used for gene pathway enrichment analysis and ontology assessment.
Western blotting
After treated with TMZ for 4 days, the cells were lysed using M-PER lysis buffer. Protein was extracted and quantified using a BCA protein assay kit (Pierce Biotechnology, Inc., Rockford, IL, USA). A total of 30 μg of each sample was heated at 95°C for 10 min and loaded into 10% gel. Samples were electrophoresed at 110 V for 60–90 min and then transferred to PVDF membranes at 300 mA for 150 min using a semi-dry transfer apparatus. Membranes were blocked in 5% non-fat dry milk for 2 h, incubated with primary antibodies [anti-AKT3, anti-RB1, anti-MAPK8 and anti-PIK3R1 (Cell Signaling Technology, Inc., Danvers, MA, USA)] overnight, washed with TBS containing 0.05% Triton-X 100 (TBST) followed by an incubation of 1 h in goat anti-rabbit secondary antibody (1:5,000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) conjugated with HRP. After final washing with TBST, the membranes were developed using chemiluminescence and exposed to X-ray film. The immunoblots were quantified with software quantity one version 4.6.2. The expression levels of AKT3, RB1, MAPK8 and PIK3R1 in each sample were internally normalized to GAPDH and levels were given relative to expression in NC group.
Statistical analysis
Statistical analyses were performed using SPSS software version 16.0. The relationships between various parameters were analyzed statistically by the χ2-test, ANOVA, Fisher's exact test, or Student's t-test as appropriate. P<0.05 was considered to indicate a statistically significant difference.
Results
Lentiviral vector infection and PPM1D silencing
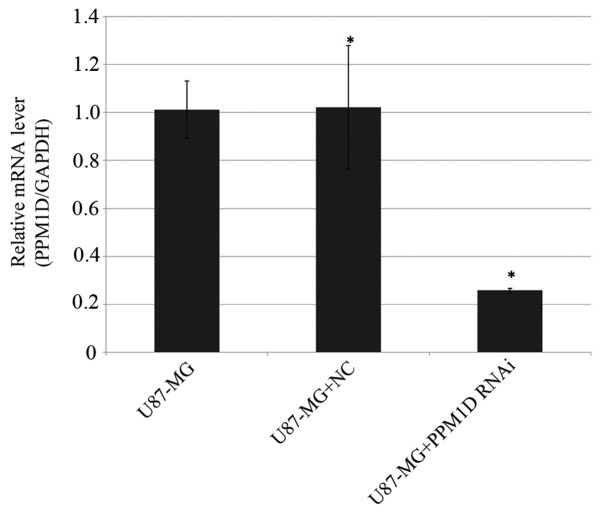
Infection efficiency of lentiviral RNAi vector of PPM1D and the negative control vector in U87-MG cells was >90% (data not shown). The silencing efficacy of PPM1D in U87-MG cells was determined by RT-qPCR. As shown in Fig. 1, the expression level of PPM1D in U87-MG cells was remarkably reduced by 74.3% compared with that in negative control cells (P<0.05).
Figure 1.
PPM1D mRNA expression levels in U87-MG cells. The relative copies of PPM1D mRNA were significantly decreased after infected with RNAi lentivirus compared with negative control. Means ± SD (bars) are shown. *P<0.05.
PPM1D gene silencing improves TMZ induced cell proliferation
The cell numbers of PPM1D silencing and control siRNA transfection cells following TMZ administration were detected by MTT at 24, 48, 72, 96 and 120 h. A statistically significant (P<0.05) reduction in cell growth was seen in KD group comparing with NC group and CON group (Fig. 2). Cell proliferation ability in KD group has reduced by 57.7% comparing with NC group 72 h after TMZ treatment. In contrast, though cell growth in NC group showed a slightly reduced cell proliferation, the differences did not reach statistical significance.
Figure 2.
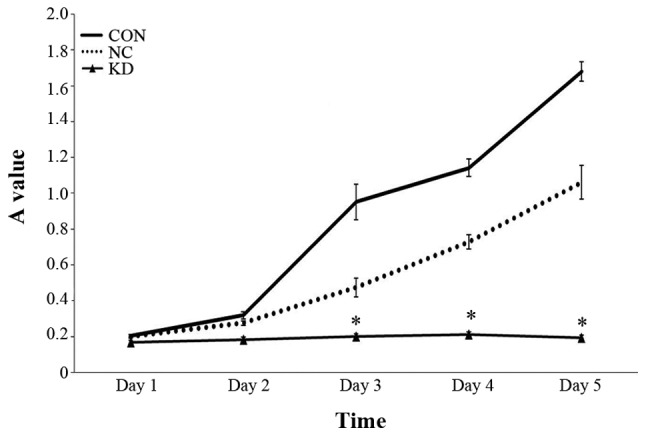
The effect of PPM1D silencing combined with TMZ on proliferation of U87-MG cells. A MTT analysis of the cell viability of PPM1D silencing and negative control cells were treated with TMZ. Cell numbers were counted at 24, 48, 72, 96 and 120 h after TMZ administration. Statistically significant (P<0.05) differences are marked with an asterisk. Error bars indicate ± SD of five replicates.
Combination treatment of TMZ and PPM1D gene silencing induce apoptosis and cell cycle arrest
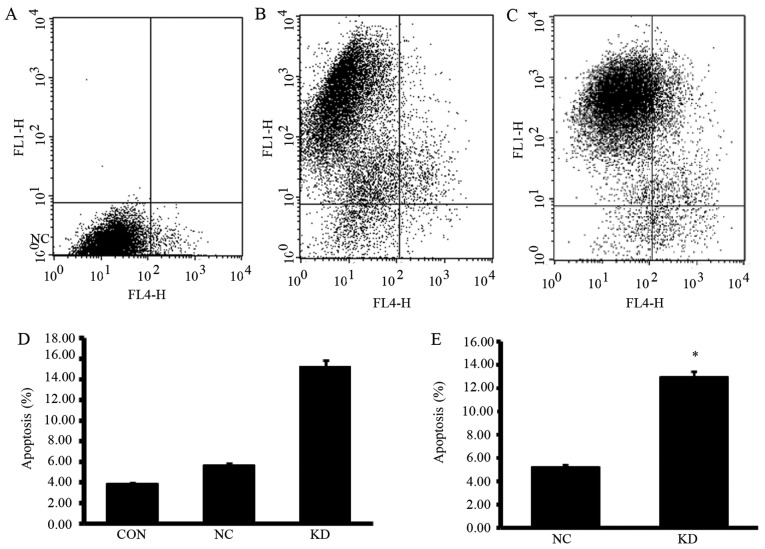
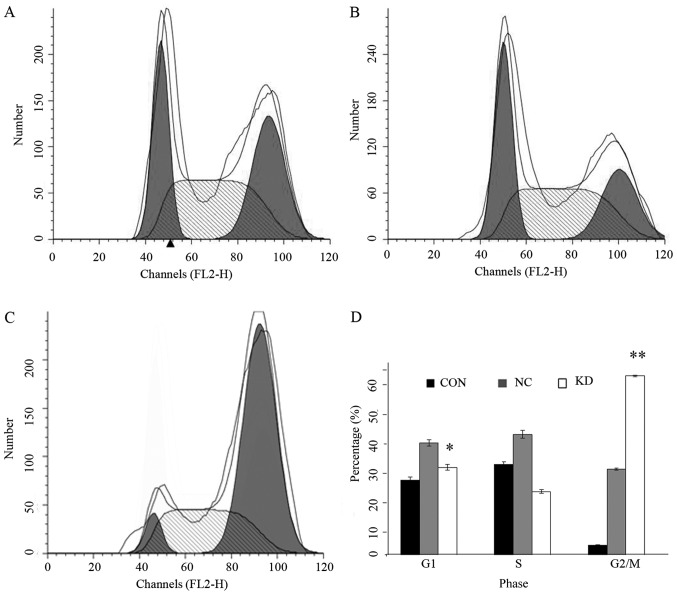
To explore whether the reduction in cell number in KD group was caused by reduced cell proliferation or by increased cell death, we subjected the cells to cell cycle and apoptosis analyses. As shown in Fig. 3, cells in KD group showed a significantly increased G2/M phase and decreased G1 phase cells (P<0.05). Apoptosis analyses showed that PPM1D silencing enhanced the sensitivity to TMZ induced apoptosis inU87-MG cells (Fig. 4). Moreover, NC group cells which were treated with TMZ alone showed slightly increased apoptotic cells compared with CON group cells, but the differences did not reach statistical significance.
Figure 3.
The effect of PPM1D silencing combined with TMZ on apoptosis of U87-MG cells. The number of apoptotic cells was determined on day 4 after TMZ treatment using flow cytometry. (A) CON group, (B) NC group, (C) KD group. (D) Summary of the experiment illustrated in (A–C). (E) To exactly analyze the influences induced by PPM1D silencing, cells in NC and KD groups with positive expression of GFP were analyzed. Apoptotic cells significantly increased in KD group with PPM1D silencing when treated with TMZ. Error bars indicate ± SD of three replicates. *P<0.05 vs. NC.
Figure 4.
The effect of PPM1D silencing combined with TMZ on cell cycle distribution of U87-MG cells. Cell cycle distributions were determined on day 4 after TMZ treatment using flow cytometry. (A) CON group, (B) NC group, (C) KD group. (D) Summary of the experiment illustrated in (A–C). G1 and G2/M cells in KD vs. NC, respectively, *P<0.05, **P<0.01.
Differential gene induction and pathway enrichment analysis in PPM1D gene silencing U-87MG cells treated with TMZ
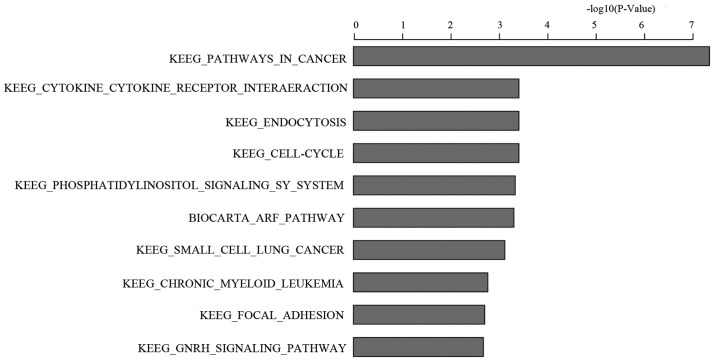
To investigate the mechanisms of changes in cell biology when PPM1D gene silenced U-87MG cells were treated with TMZ, both KD and NC cells were conducted with a whole genome-based transcriptional analysis. Compared with NC group, 367 genes were upregulated and 444 genes were downregulated. Then all the differentially expressed genes were confirmed with gene pathway enrichment analysis, based on all known KEGG pathway genes. The analysis algorithm ranks all genes by their significance of differential expression and then looks for groups of biologically relevant genes that are enriched at either the top or bottom of the ranked list. The statistical significance of a pathway was examined by a permutation procedure and showed as a nominal P-value according to the enrichment score. The ten most significant differential expression pathways and genes were listed in Fig. 5 and Table I, based on an ascending P-value. Cancer in KEGG pathway was the most significant differential expression pathway with a P-value of 4.36E-08.
Figure 5.
Pathway enrichment analysis. The differentially expressed genes were confirmed with gene pathway enrichment analysis. The ten most significant differential expression pathways and genes are listed, based on an ascending P-value. Cancer in KEGG pathway was the most significant differential expression pathway. The level of significance of the difference between the two groups is indicated with a P-value of 4.36E-08 in cancer pathway.
Table I.
The ten most significant differential expression pathways and genes between KD and NC.
| Gene set name | Pathway analysis
|
||
|---|---|---|---|
| No. of genes | P-value | Gene names | |
| KEGG_PATHWAYS_IN_CANCER | 26 | 4.36E-08 | RB1, ABL1, CDK6, SKP2, CBL, CBLB, PIK3R1, RAC1, AKT3, ITGA6, FN1, LAMB2, MAX, RUNX1, MAPK8, MMP2, FGF2, FGF5, FGF14, LEF1, TCF7, WNT5A, FZD6, RALA, GLI2, MMP1 |
| KEGG_CELL_CYCLE | 12 | 3.78E-04 | RB1, ABL1, CDK6, SKP2, CDC25B, MCM2, MCM3, MCM4, CDC20, ESPL1, E2F4, RBL2 |
| KEGG_ENDOCYTOSIS | 14 | 3.78E-04 | CBL, CBLB, PIP5K1C, PIP5K1A, AP2M1, AP2A1, EEA1, SMURF2, NEDD4, PARD6B, ACAP3, LDLRAP1, LDLR, VPS36 |
| KEGG_CYTOKINE_CYTOKINE_ RECEPTOR_INTERAERACTION | 17 | 3.78E-04 | IL1A, IL6R, IL6ST, IL11, LEPR, PRLR, TNFRSF10D, TNFRSF10C, CCL2, CXCL2, CXCL3, BMPR2, TNFRSF9, TNFRSF21, TNFRSF11B, TNFSF14, CD70 |
| KEGG_PHOSPHATIDYLINOSITOL_ SIGNALING_SY_SYSTEM | 9 | 4.51E-04 | PIK3R1, PIP5K1C, PIP5K1A, ITPR1, SYNJ2, INPP5A, OCRL, INPP4B, DGKE |
| BIOCARTA_ARF_PATHWAY | 5 | 4.79E-04 | RB1, ABL1, PIK3R1, RAC1, POLR1A |
| KEGG_SMALL_CELL_LUNG_CANCER | 9 | 7.38E-04 | RB1, CDK6, SKP2, PIK3R1, AKT3, ITGA6, FN1, LAMB2, MAX |
| KEGG_CHRONIC_MYELOID_LEUKEMIA | 8 | 1.66E-03 | RB1, ABL1, CDK6, CBL, CBLB, PIK3R1, AKT3, RUNX1 |
| KEGG_FOCAL_ADHESION | 13 | 1.93E-03 | PIK3R1, RAC1, AKT3, ITGA6, FN1, LAMB2, MAPK8, PIP5K1C, ELK1, THBS1, ARHGAP5, COL1A1, COL5A2 |
| KEGG_GNRH_SIGNALING_PATHWAY | 9 | 2.05E-03 | MAPK8, MMP2, ITPR1, ELK1, MAP2K4, ADCY8, ADCY7, ADCY6, GNA11 |
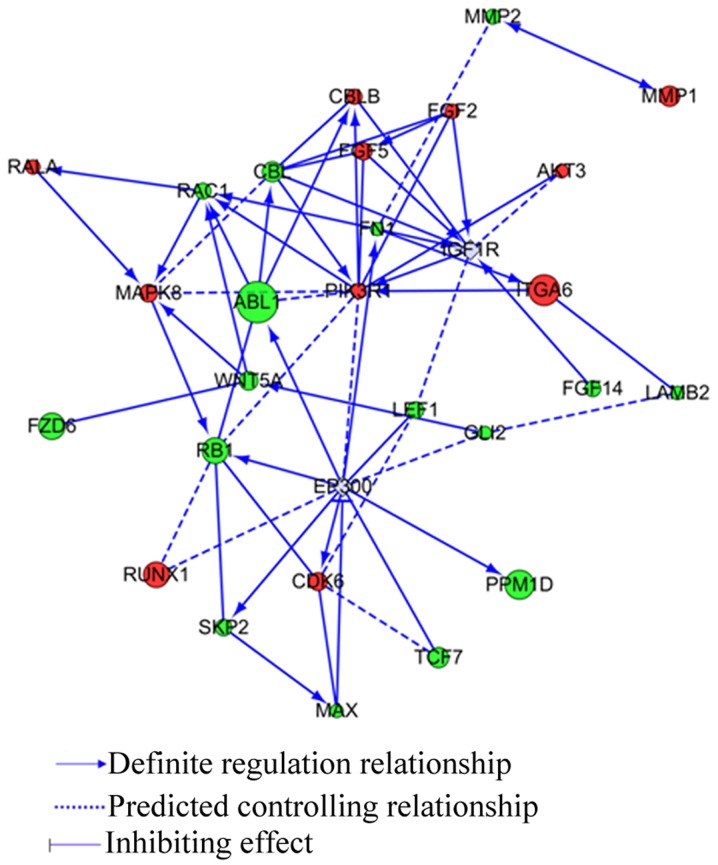
To further analyze the PPM1D effects when U-87MG cells were administered with TMZ, the most significant differential expression pathway in cancer in KEGG pathway was confirmed in gene network construction. The relationship between PPM1D and the 26 differential genes is shown in Fig. 6. IGFR1R, PIK3R1, MAPK8, and EP300 are core genes in the network.
Figure 6.
Gene network construction. Pathway in cancer in KEGG was confirmed in gene network construction. The relationship between PPM1D and the 26 differential genes is shown. Green and red round dots represent downregulated gene and upregulated gene, respectively. The gray represent added gene. The size of the dot represents the significance of the difference.
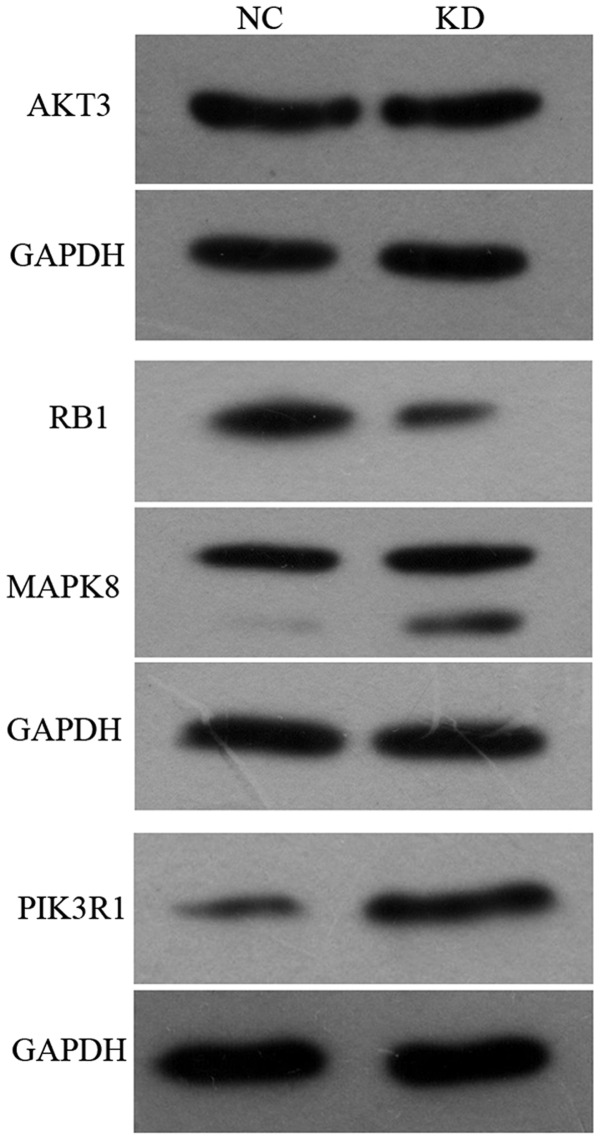
Western blotting
Several target genes deemed biologically interesting because of their differential expression levels in glioma cells in pathways in cancer were validated by western blot analysis. As shown in Fig. 7, MAPK8 and PIK3R1 protein expression levels in KD group were increased by 64.1% and 261.6%, respectively, relative to the expression level in NC group. It was consistent with that detected in gene expression profiling. RB1 protein expression was decreased by 61.5% compared with NC group. The expression of AKT3 was not significantly different.
Figure 7.
Western blot analysis of some differential genes. When administered with TMZ, glioma cells with PPM1D silencing showed significantly increased protein expression levels in MAPK8 and PIK3R1, while RB1 protein expression was decreased. The expression of AKT3 was not significantly different. The immunoblots were quantified with software and the data were repeated for at least three times in different experiments (n=3, P<0.05).
Discussion
Malignant gliomas account for close to 50% of all CNS tumors. The median survival of patients with GBM remains close to 1 year from the time of diagnosis in spite of surgical resection followed by radiotherapy and chemotherapy (1). The average five year survival is less than 3%, leading to the fact that GBM is the most lethal form of brain tumor (19). Such poor outcome has led to the exploration of a wide variety of novel therapies, and some of them have been incorporated as a standard of care for patients with GBM. TMZ is approved for use as first-line treatment for patients with primary and recurrent high-grade gliomas. It has been shown that TMZ combined with radiotherapy can improve the 2-year survival rate from 10.4% with radiotherapy alone to 26.5% in patients with GBM and radiotherapy (1). Whereas the efficacy of TMZ is encouraging, additional prolongation of survival remains a challenge. GBM shows chemoresistance shortly after the initiation of treatment. Additionally, recent studies suggest that 60–75% of patients with GBM derive no benefit from treatment with TMZ (2). Thus, exploring alternative therapeutic strategies is required.
TMZ is an alkylating agent that binds to DNA and interferes with replication, resulting in the insertion of DNA strand breaks and, ultimately, cell death (3). p53 facilitates favorable antitumor drug response through a variety of key cellular functions, including cell cycle arrest, senescence, and apoptosis (4). p53 status in addition to MGMT plays a role in chemotherapy resistance to TMZ. p53 wild-type glioma cells are significantly more sensitive than p53-mutated glioma cells to apoptosis induced by TMZ (18). GBM patients with low mutant p53 expression have higher progression-free survival time and may have longer life expectancy in comparison with the high mutant p53 expression group (6). Initially identified as a p53-regulated allele located on 17q23-24, PPM1D is a member of the protein phosphatase 2C (PP2C) family and expressed in a p53-dependent manner (7). PPM1D can negatively regulate key DNA damage response proteins such as p53, ATM, and p38MAPK and control cell cycle checkpoints in response to DNA damage (20). PPM1D is frequently amplified and overexpressed in many cancers, including gliomas, breast cancers, neuroblastomas, ovarian clear cell adenocarcinomas, and medulloblastomas (8–13). PPM1D is recognized as a novel oncogene and is widely believed to be a promising therapeutic target for cancers (21,22). In addition, we found that PPM1D may serve as an oncogene important to astrocytoma progression, especially in astrocytomas with wild-type p53 (8). However, whether PPM1D plays a role in TMZ-induced DNA damage through regulating p53 functions has not been clarified yet.
In the current study, we demonstrated that PPM1D silencing can increase the antiproliferative activity of TMZ in glioma cells. Cell cycle and apoptosis analyses showed that the decreased proliferative activity was partly due to increased cell apoptosis. It has been reported that U87-MG cells with p53 wild-type are sensitive to apoptosis induced by TMZ (18). However, it was shown that although TMZ-treated cells underwent cell cycle arrest, the apoptotic cells were few and did not significantly increase throughout the 10 days after TMZ treatment (17). In addition, it was reported that there was no significant difference in the rate of apoptosis 72 h after treatment with either DMSO control or 0.1 mmol/l TMZ, though TMZ reduced cell viability and caused cell cycle arrest (16). TMZ induced autophagic, but not apoptotic processes in glioma cells. our results established that apoptotic cells were few when TMZ was administrated alone. However, PPM1D silencing could significantly induce cell apoptosis in TMZ-treated cells. Cell cycle arrest is one of the important mechanisms through which TMZ exerts antitumor effects. In this study, we found that PPM1D silencing combined with TMZ could induce significantly increased G2/M cells and decreased G1 cells. Glioma cells with PPM1D silencing are more sensitive to TMZ induced apoptosis and cell cycle is arrested in G2/M. Therefore, our results established that PPM1D controls cell cycle checkpoints in response to TMZ- induced DNA damage.
To investigate the mechanisms of changes in cell biology when PPM1D gene silencing U-87MG cells were treated with TMZ, cells were assessed with microarray-based gene expression profile. Our results demonstrated that many genes attribute to the different changes when PPM1D silencing glioma cells were treated with TMZ. Pathway in cancer was the most significant pathway. Our results proved that PPM1D is associated with glioma development, which is consistent with the fact that PPM1D is recognized as a novel oncogene in many cancers (21,22). PPM1D was initially identified as a p53-regulated allele and can negatively regulate key DNA damage response proteins such as p53, ATM, and p38MAPK (20). The most significant network arose around the IGFR1R, PIK3R1, MAPK8 and EP300, when glioma cells were treated with combination of PPM1D silencing and TMZ. Accumulating evidence has shown that GBM frequently displays hyperactivation of the AKT pathway (23,24) and endogenous AKT kinase activity can be activated in response to clinically relevant concentrations of TMZ (25,26). Over 80% of GBMs have an acquired alteration in the RTK/PI3K/AKT pathway (27). It has been detected that PPM1D knocked out mesenchymal stem cells may be attributed to increases in the pAKT/AKT ratio and PI3K/AKT signaling axis (28).
Our microarray-based gene expression profile and western blot results detected that PIK3R1 is upregulated and RB1 protein expression was decreased when PPM1D silencing glioma cells were treated with TMZ. However, AKT3 protein expression levels were not different between the two groups. It was proved that Rb was a substrate of PI3K/Akt signaling pathway (29). Activation of PI3K/AKT pathway contributed to promote Rb phosphorylation and degradation (30). It has been previously shown that inhibition of PI3K/Akt/mToR-mediated signaling in glioblastoma cell lines strongly amplifies cell death induced by radiotherapy and a wide range of chemotherapeutics. As a regulatory subunit of phosphoinositide-3-kinase (PI3K), PIK3R1 target the PI3K/AKT pathway. However, our results showed different tendency of change of PIK3R1. Thus, PPM1D may not regulate PI3K/AKT signaling axis through PIK3R1 (31).
It has been demonstrated that downregulated expression of COX-2, Akt1 and PIK3R1 could upregulate p53 and exert proliferation and invasion inhibition effects on U251 cells (32). We infer it should be partly due to the different status of p53. The exact mechanism still remains to be explored.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant no. 81201995), the Natural Science Foundation of Guangdong Province, China (grant no. 2015A030313532) and the Science and Technology Planning Project of Guangdong Province, China (grant no. 2014A020212678).
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group: Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Chamberlain MC. Temozolomide: Therapeutic limitations in the treatment of adult high-grade gliomas. Expert Rev Neurother. 2010;10:1537–1544. doi: 10.1586/ern.10.32. [DOI] [PubMed] [Google Scholar]
- 3.Marchesi F, Turriziani M, Tortorelli G, Avvisati G, Torino F, De Vecchis L. Triazene compounds: Mechanism of action and related DNA repair systems. Pharmacol Res. 2007;56:275–287. doi: 10.1016/j.phrs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Miao W, Liu X, Wang H, Fan Y, Lian S, Yang X, Wang X, Guo G, Li Q, Wang S. p53 upregulated modulator of apoptosis sensitizes drug-resistant U251 glioblastoma stem cells to temozolomide through enhanced apoptosis. Mol Med Rep. 2015;11:4165–4173. doi: 10.3892/mmr.2015.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Rivera M, Siddik ZH. Resistance and gain-of-resistance phenotypes in cancers harboring wild-type p53. Biochem Pharmacol. 2012;83:1049–1062. doi: 10.1016/j.bcp.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, Jiang T, Li G, Wang Z. Impact of p53 status to response of temozolomide in low MGMT expression glioblastomas: Preliminary results. Neurol Res. 2008;30:567–570. doi: 10.1179/174313208X297913. [DOI] [PubMed] [Google Scholar]
- 7.Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, O'Connor PM, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci USA. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, Rao J, Yang H, Zhao H, Yang L. Wip1 overexpression correlated with TP53/p14(ARF) pathway disruption in human astrocytomas. J Surg oncol. 2011;104:679–684. doi: 10.1002/jso.22004. [DOI] [PubMed] [Google Scholar]
- 9.Bulavin DV, Demidov ON, Saito S, Kauraniemi P, Phillips C, Amundson SA, Ambrosino C, Sauter G, Nebreda AR, Anderson CW, et al. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet. 2002;31:210–215. doi: 10.1038/ng894. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KC, Gabriele T, McCurrach ME, Marks JR, Hoey T, et al. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31:133–134. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 11.Saito-Ohara F, Imoto I, Inoue J, Hosoi H, Nakagawara A, Sugimoto T, Inazawa J. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res. 2003;63:1876–1883. [PubMed] [Google Scholar]
- 12.Liang C, Guo E, Lu S, Wang S, Kang C, Chang L, Liu L, Zhang G, Wu Z, Zhao Z, et al. Over-expression of wild-type p53-induced phosphatase 1 confers poor prognosis of patients with gliomas. Brain Res. 2012;1444:65–75. doi: 10.1016/j.brainres.2011.12.052. [DOI] [PubMed] [Google Scholar]
- 13.Hirasawa A, Saito-Ohara F, Inoue J, Aoki D, Susumu N, Yokoyama T, Nozawa S, Inazawa J, Imoto I. Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin Cancer Res. 2003;9:1995–2004. [PubMed] [Google Scholar]
- 14.Wang P, Rao J, Yang H, Zhao H, Yang L. PPM1D silencing by lentiviral-mediated RNA interference inhibits proliferation and invasion of human glioma cells. J Huazhong Univ Sci Technolog Med Sci. 2011;31:94–99. doi: 10.1007/s11596-011-0157-1. [DOI] [PubMed] [Google Scholar]
- 15.Spiro T, Liu L, Gerson S. New cytotoxic agents for the treatment of metastatic malignant melanoma: Temozolomide and related alkylating agents in combination with guanine analogues to abrogate drug resistance. Forum (Genova) 2000;10:274–285. [PubMed] [Google Scholar]
- 16.Shen W, Hu JA, Zheng JS. Mechanism of temozolomideinduced antitumour effects on glioma cells. J Int Med Res. 2014;42:164–172. doi: 10.1177/0300060513501753. [DOI] [PubMed] [Google Scholar]
- 17.Hirose Y, Berger MS, Pieper RO. p53 effects both the duration of G2/M arrest and the fate of temozolomide-treated human glioblastoma cells. Cancer Res. 2001;61:1957–1963. [PubMed] [Google Scholar]
- 18.Batista LF, Roos WP, Kaina B, Menck CF. p53 mutant human glioma cells are sensitive to UV-C-induced apoptosis due to impaired cyclobutane pyrimidine dimer removal. Mol Cancer Res. 2009;7:237–246. doi: 10.1158/1541-7786.MCR-08-0428. [DOI] [PubMed] [Google Scholar]
- 19.Tivnan A, Zakaria Z, O'leary C, Kögel D, Pokorny JL, Sarkaria JN, Prehn JH. Inhibition of multidrug resistance protein 1 (MRP1) improves chemotherapy drug response in primary and recurrent glioblastoma multiforme. Front Neurosci. 2015;9:218. doi: 10.3389/fnins.2015.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu YH, Zhang CW, Lu L, Demidov ON, Sun L, Yang L, Bulavin DV, Xiao ZC. Wipl regulates the generation of new neural cells in the adult olfactory bulb through p53-dependent cell cycle control. Stem Cells. 2009;27:1433–1442. doi: 10.1002/stem.65. [DOI] [PubMed] [Google Scholar]
- 21.Tang YL, Liu X, Gao SY, Feng H, Jiang YP, Wang SS, Yang J, Jiang J, Ma XR, Tang YJ, et al. WIP1 stimulates migration and invasion of salivary adenoid cystic carcinoma by inducing MMP-9 and VEGF-C. Oncotarget. 2015;6:9031–9044. doi: 10.18632/oncotarget.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilmartin AG, Faitg TH, Richter M, Groy A, Seefeld MA, Darcy MG, Peng X, Federowicz K, Yang J, Zhang SY, et al. Allosteric Wip1 phosphatase inhibition through flap-subdomain interaction. Nat Chem Biol. 2014;10:181–187. doi: 10.1038/nchembio.1427. [DOI] [PubMed] [Google Scholar]
- 23.Hirose Y, Katayama M, Mirzoeva OK, Berger MS, Pieper RO. Akt activation suppresses Chk2-mediated, methylating agent-induced G2 arrest and protects from temozolomide-induced mitotic catastrophe and cellular senescence. Cancer Res. 2005;65:4861–4869. doi: 10.1158/0008-5472.CAN-04-2633. [DOI] [PubMed] [Google Scholar]
- 24.Molina JR, Hayashi Y, Stephens C, Georgescu MM. Invasive glioblastoma cells acquire stemness and increased Akt activation. Neoplasia. 2010;12:453–463. doi: 10.1593/neo.10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporali S, Levati L, Starace G, Ragone G, Bonmassar E, Alvino E, D'Atri S. AKT is activated in an ataxia-telangiectasia and Rad3-related-dependent manner in response to temozolomide and confers protection against drug-induced cell growth inhibition. Mol Pharmacol. 2008;74:173–183. doi: 10.1124/mol.107.044743. [DOI] [PubMed] [Google Scholar]
- 26.De Salvo M, Maresca G, D'agnano I, Marchese R, Stigliano A, Gagliassi R, Brunetti E, Raza GH, De Paula U, Bucci B. Temozolomide induced c-Myc-mediated apoptosis via Akt signalling in MGMT expressing glioblastoma cells. Int J Radiat Biol. 2011;87:518–533. doi: 10.3109/09553002.2011.556173. [DOI] [PubMed] [Google Scholar]
- 27.Signore M, Pelacchi F, di Martino S, Runci D, Biffoni M, Giannetti S, Morgante L, De Majo M, Petricoin EF, Stancato L, et al. Combined PDK1 and CHK1 inhibition is required to kill glioblastoma stem-like cells in vitro and in vivo. Cell Death Dis. 2014;5:e1223. doi: 10.1038/cddis.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang Y, Liu L, Sheng M, Xiong K, Huang L, Gao Q, Wei J, Wu T, Yang S, Liu H, et al. Wip1 knockout inhibits the proliferation and enhances the migration of bone marrow mesenchymal stem cells. Exp Cell Res. 2015;334:310–322. doi: 10.1016/j.yexcr.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Gao N, Zhang Z, Jiang BH, Shi X. Role of PI3K/AKT/mTOR signaling in the cell cycle progression of human prostate cancer. Biochem Biophys Res Commun. 2003;310:1124–1132. doi: 10.1016/j.bbrc.2003.09.132. [DOI] [PubMed] [Google Scholar]
- 30.Bao JM, He MY, Liu YW, Lu YJ, Hong YQ, Luo HH, Ren ZL, Zhao SC, Jiang Y. AGE/RAGE/Akt pathway contributes to prostate cancer cell proliferation by promoting Rb phosphorylation and degradation. Am J Cancer Res. 2015;5:1741–1750. [PMC free article] [PubMed] [Google Scholar]
- 31.Westhoff MA, Faham N, Marx D, Nonnenmacher L, Jennewein C, Enzenmüller S, Gonzalez P, Fulda S, Debatin KM. Sequential dosing in chemosensitization: Targeting the PI3K/Akt/mTOR pathway in neuroblastoma. PLoS one. 2013;8:e83128. doi: 10.1371/journal.pone.0083128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Y, Zhang Q, Kang C, Zhang K, Zhang J, Pu P, Wang G, Wang T. Inhibitory effects of adenovirus mediated COX-2, Akt1 and PIK3R1 shRNA on the growth of malignant tumor cells in vitro and in vivo. Int J oncol. 2009;35:583–591. doi: 10.3892/ijo_00000369. [DOI] [PubMed] [Google Scholar]