Researchers report how sulfur deficiency affects glucosinolate biosynthesis in plants.
Keywords: Glucosinolates, sulfur, Arabidopsis thaliana, sulfur deficiency induced, MYB28
Abstract
Glucosinolates (GSLs) in the plant order of the Brassicales are sulfur-rich secondary metabolites that harbor antipathogenic and antiherbivory plant-protective functions and have medicinal properties, such as carcinopreventive and antibiotic activities. Plants repress GSL biosynthesis upon sulfur deficiency (−S); hence, field performance and medicinal quality are impaired by inadequate sulfate supply. The molecular mechanism that links –S to GSL biosynthesis has remained understudied. We report here the identification of the –S marker genes sulfur deficiency induced 1 (SDI1) and SDI2 acting as major repressors controlling GSL biosynthesis in Arabidopsis under –S condition. SDI1 and SDI2 expression negatively correlated with GSL biosynthesis in both transcript and metabolite levels. Principal components analysis of transcriptome data indicated that SDI1 regulates aliphatic GSL biosynthesis as part of –S response. SDI1 was localized to the nucleus and interacted with MYB28, a major transcription factor that promotes aliphatic GSL biosynthesis, in both yeast and plant cells. SDI1 inhibited the transcription of aliphatic GSL biosynthetic genes by maintaining the DNA binding composition in the form of an SDI1-MYB28 complex, leading to down-regulation of GSL biosynthesis and prioritization of sulfate usage for primary metabolites under sulfur-deprived conditions.
INTRODUCTION
Sulfur, as an essential macronutrient, plays a crucial role in plant growth and development (1). Photosynthetic organisms use sulfate (SO42−) as a primary sulfur source to synthesize an array of S-containing metabolites, including the amino acids cysteine (Cys) and methionine (Met), the tripeptide glutathione (GSH), vitamins and cofactors (such as thiamine, biotin, and coenzyme A), and chloroplastic sulfolipids (2, 3). Moreover, primary sulfur assimilation is a prerequisite for synthesizing glucosinolates (GSLs) in Brassicales.
GSLs are nitrogen- and sulfur-containing compounds found in the Brassicaceae family, including several important crops, such as oilseed rape (Brassica napus/Brassica rapa), cabbage (Brassica oleracea var. capitata), broccoli (B. oleracea var. italica), Chinese cabbage (B. rapa), and the model plant Arabidopsis thaliana (4). GSLs are important defense compounds against pathogens and herbivores and also act as S-storage sources (5–7). Moreover, potential health benefits of GSL-rich diets to humans come from the carcinopreventive properties of GSL hydrolysis products, which have been documented in multiple studies (8, 9). For example, sulforaphane, an isothiocyanate derivative of 4-methylsulfinylbutyl GSL, and other isothiocyanates are potential candidates to prevent tumor growth by blocking the cell cycle, and have a potential for treating Helicobacter pylori–caused gastritis and stomach cancer (10).
GSLs are divided into three groups depending on their amino acid precursors: aliphatic, benzenic (or aromatic), and indolic GSLs. In A. thaliana, 40 structurally different GSLs have been detected, most of which are aliphatic and indolic GSLs derived from Met and tryptophan (Trp), respectively (11). Because of their importance in agriculture and for human health, the GSL biosynthetic pathway was extensively investigated, and more than 20 genes involved in GSL biosynthesis have been identified in Arabidopsis to date (11). Despite our knowledge on the GSL biosynthetic pathways, understanding of the regulatory mechanisms and their synthesis upon environmental perturbations, for example, S deficiency (−S), remains fragmentary. Several R2R3 MYB family transcription factors have been identified as positive regulators of GSL synthesis, that is, MYB28, MYB29, and MYB76 as those of aliphatic GSLs (12, 13) and MYB34, MYB51, and MYB122 as those of indolic GSLs (14–16). Among them, MYB28 and MYB34 were identified as the dominant regulators of aliphatic and indolic GSLs, respectively, and are considered the major transcriptional inducers of GSL biosynthetic genes (16, 17). In addition, basic helix-loop-helix (bHLH) transcription factors MYC2 (bHLH06), MYC3 (bHLH05), and MYC4 (bHLH04) have been reported to regulate GSL levels, to some extent, both dependent and independent of the aforementioned MYB transcription factors (18–20).
GSL accumulation is responding to plant development and abiotic factors, such as nitrogen and sulfur supply (21). In plants of the Brassicaceae family, the backbone of GSLs contains three S atoms, which can account for up to 30% of the total sulfur content of the entire plant (22). Thus, GSLs need to be tightly regulated in relation to the S nutritional status (22, 23). Under short-term sulfur starvation (−S), all the major GSL biosynthetic genes, such as MAM, CYP79, and CYP83 families, are down-regulated, and consequently, the GSL levels decrease (22, 24). Concomitantly with the down-regulation of GSL synthesis, up-regulation of GSL catabolic genes under −S is reported by several transcriptomic studies (25–30). The S-storage function of GSL has been shown by the disruption of GSL transporters in Arabidopsis seeds, eliminating seed-borne GSLs and resulting in reduced seedling growth under −S (31). Thus, it can be hypothesized that, under −S, GSLs provide an important sulfur source and that the plant prioritizes protein synthesis and other essential functions above defense (22). However, it is largely unknown how sulfate availability regulates the expression of GSL pathway genes.
In addition to the modification of GSL metabolism, plants increase sulfate uptake and sulfur assimilation capacity in response to −S (2, 3). The EIL family transcription factor SLIM1 has been identified as a regulator of plant −S responses associated with the up-regulation of sulfate uptake, GSL catabolism, and the down-regulation of GSL synthesis (29). This broad range of the metabolic pathways regulated by SLIM1 suggests the existence of other protein factors that mediate −S signals specific to each metabolic pathway. The presence of additional mediators is also suggested based on the fact that the expression levels of the MYB transcription factors do not fully correlate with the decreased GSL levels under –S (23, 32). MYB28 is not down-regulated under –S, and its expression level becomes even higher under long-term sulfur starvation, whereas MYB29 and MYB76 are repressed in both early and late phases of –S (23, 32). This provides an implication that an additional mechanism may be involved in the negative regulation of GSL biosynthesis under –S.
Several transcriptome studies have revealed that a set of functionally unknown genes termed S-marker genes, including sulfur deficiency induced 1 (SDI1; At5g48850) and SDI2 (At1g04770), are up-regulated under sulfur starvation (25–30). A homologous gene of SDI has also been identified by a complementary DNA-based amplified fragment length polymorphism (cDNA-AFLP) analysis of field-grown, S-deficient wheat (Triticum aestivum) (33). SDI1 may play a functional role in the utilization of stored sulfate under −S because Arabidopsis sdi knockout lines accumulate more sulfate than do wild-type (WT) controls (33). SLIM1 appears to down-regulate SDI1 and SDI2 under S-sufficient conditions and the opposite is required for the induction of SDI1 and SDI2 under –S, whereas its functionality is probably modulated through a posttranslational mechanism, which can be more complex and possibly involve additional factors (29). SDI1 and SDI2 levels are correlated to the O-acetylserine (OAS) content under a wide range of stress conditions, among them is sulfate starvation (34). OAS is the precursor of Cys synthesis in the S assimilation pathway and accumulates under –S conditions. Hence, it has been considered to be a signaling compound for −S responses (25–27, 34, 35).
SDI1 and SDI2 responsiveness to sulfate starvation and a genome-wide association study suggesting SDI2 as a candidate that plays a role in GSL accumulation (36) prompted us to investigate the function of SDI proteins in relation to GSL and sulfur metabolism in Arabidopsis. We identified SDI1 and SDI2 as repressors of GSL synthesis by performing metabolome and transcriptome analyses of sdi1 and sdi2 single- and double-knockout lines together with constitutive SDI1- and SDI2-overexpressing lines. Determination of the protein-protein interaction between SDI1 and MYB28 and its effects on DNA binding and transactivation potential of MYB28 revealed the molecular machinery that shows how SDI1 represses GSL synthesis in response to −S. These findings fill the gap in our understanding of how sulfate starvation is linked to the down-regulation of GSL synthesis. This study extends our understanding on plant responses to −S and provides significant implications for designing strategies for the improvement of sulfur use efficiency and production of plant-derived, S-containing beneficial natural products in agriculture.
RESULTS
SDI family proteins in plants
The SDI family consists of five members in A. thaliana (Fig. 1, A and B, and fig. S1). The high similarity between SDI1 and SDI2 and their shorter lengths compared with the other three proteins suggest similar or closely related functions (Fig. 1B and fig. S1). The SDI family proteins contain one tetratricopeptide repeat (TPR) domain, which is known to mediate protein-protein interactions, including both TPR-TPR and TPR–non-TPR interactions (Fig. 1A and fig. S1) (37). TPR proteins are involved in diverse molecular processes, including transcriptional control and protein folding, which may function in biological processes, such as cell cycle regulation and neurogenesis (38). One of the SDI family proteins in Arabidopsis, MS5 (male sterile 5; At4g20900), also known as TDM1 (three division mutant 1), is known to function in pollen development. Disruption of MS5 causes male sterility by impairing the division of unreplicated haploid nuclei after meiosis II (39, 40). Sequence comparison of Arabidopsis SDI family proteins revealed that MS5 contains a TPR domain variant of 25 amino acids rather than 34 amino acids (fig. S1). Among the family members, At5g44330 and SDI1, but not SDI2, contain a putative nuclear localization signal (Fig. 1A and fig. S1).
Fig. 1. SDI1 and SDI2 are TPR proteins induced by sulfur deficiency in Arabidopsis.
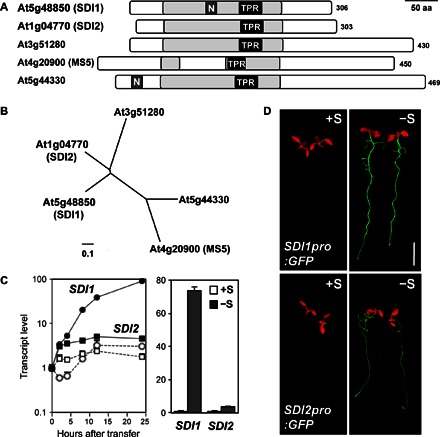
(A) Schematic representation of the TPR domain that exists in Arabidopsis SDI proteins. TPR-like helical domains detected by InterPro scan [pale gray bar (38), TPR motifs; black bar with TPR, TPR1, and PF00515] and nuclear localization signals (dark gray bar with N, NLS_BP, and PS50079) detected by Motif Scan (64) were indicated. Numbers on the right represent amino acid length of each protein. Scale bar, 50 amino acids (aa). (B) Phylogenetic relationships of SDI family proteins in Arabidopsis. Protein sequences of SDI1 (At5g48850), SDI2 (At1g04770), At3g51280, MS5 (At4g20900), and MS5-like (At5g44330) were obtained from The Arabidopsis Information Resources (TAIR) (www.arabidopsis.org). They were aligned using a Clustal W program, and an unrooted phylogenetic tree was drawn by neighbor-joining methods using the Geneious software (Biomatters Ltd.). (C) Induction of SDI1 and SDI2 by sulfur deficiency. (Left) Transcript accumulation of SDI1 (circle) and SDI2 (square) in roots of WT transferred under +S to +S (open markers) or +S to –S (filled markers) conditions detected by previous microarray experiments (28). (Right) Transcript levels of SDI1 and SDI2 in roots of WT grown under +S (1500 μM sulfate, white bars) or –S (15 μM sulfate, gray bars) conditions detected by previous microarray experiments [means ± SE of duplicates (29)]. (D) Green fluorescent protein (GFP) accumulation in SDI1pro:GFP and SDI2pro:GFP plants grown under +S and –S conditions. GFP fluorescence was visualized under an image analyzer, as described in Materials and Methods. Scale bar, 5 mm.
SDI homologs exist in monocots and dicots, including important crop species, such as wheat, rice, and maize (fig. S2). In the PLAZA database (41), we identified 151 SDI family proteins from 29 different species, including a variety of dicot plants, such as Medicago, soybean, strawberry, apple, poplar, rapeseed, tomato, and potato, and in monocot crop species, such as rice, wheat, and maize. On the basis of a phylogenetic analysis of the protein sequences of the SDI homologs from nine major crops, including both monocot and dicot species, these proteins were categorized into four distinct clades: (i) SDI1 and SDI2 clade, (ii) At3G51280 clade, (iii) MS5 and At5G44330 clade, and (iv) a more distant clade. Arabidopsis SDI1 and SDI2 belong to a distinctive subbranch that exclusively contains the proteins of the Brassicaceae family. This observation suggests that SDI1 and SDI2 have a particular function in the Brassicaceae family. Homologs of SDI1 and SDI2 from monocots (rice, wheat, and maize) being present in the neighboring subbranch of the clade may indicate a probable functional relation with homologs in Brassicaceae.
Tissue-specific expression of SDI1 and SDI2
Previously published microarray data indicated a clear induction of SDI1 and SDI2 transcripts in the roots of Arabidopsis during both long-term and short-term −S treatments (Fig. 1C) (28, 29). On the basis of this initial observation, we examined the tissue specificity and –S induction of SDI1 and SDI2 gene expression using Arabidopsis transformed with SDI1pro:GFP/GUS and SDI2pro:GFP/GUS fusion constructs (Fig. 1D and fig. S3). GFP fluorescence was observed using SDI1pro:GFP and SDI2pro:GFP plants grown under sulfur-sufficient (+S; 1500 μM sulfate) and sulfur-deficient (−S; 15 μM sulfate) conditions (Fig. 1D). Both transgenic lines accumulated high GFP signals in the roots in response to −S. Similarly, −S and OAS treatment induced GUS expression in the vascular tissues of both roots and shoots of SDI1pro:GUS and SDI2pro:GUS lines, where SDI2pro:GUS displayed a lower intensity (fig. S3A). Histochemical GUS analysis also depicted the expression patterns of SDI1 and SDI2 in plant tissues grown under long-day conditions (fig. S3B). In SDI1pro:GUS plants, GUS expression was detected in all plant tissues, including shoots (both vascular and cortical cells), stems, flower buds, stigma, and stamens (fig. S3B, a to f). SDI2pro:GUS plants showed an almost identical pattern of GUS expression, with the exception that GUS expression was restricted to the vasculature, whereas in the shoots of SDI1pro:GUS, GUS was expressed in and around the vasculature (fig. S3B, a). SDI2pro:GUS flower buds exhibited lower GUS expression compared to SDI1pro:GUS lines (fig. S3B, i). GUS expression of neither isoform could be detected in mature siliques (fig. S3B, f and l).
Negative correlation between SDI expression and GSL levels in plants
To gain insight into the function of SDI1 and SDI2 in GSL biosynthesis, we isolated homozygous transfer DNA (T-DNA) insertion lines (knockouts), constructed overexpressing lines for both SDI1 and SDI2 genes, and analyzed them under +S and –S conditions. To investigate the phenotypes of the SDI knockouts and SDI-overexpressing lines in response to –S, where SDI1 and SDI2 are strongly induced, Arabidopsis seedlings were grown simultaneously on +S and –S agar medium, respectively. Shoots and roots were harvested separately, and the transcript levels of GSL biosynthetic genes and GSL levels were analyzed (Figs. 2 and 3).
Fig. 2. Disruption of SDI1 and SDI2 enhances the GSL accumulation.
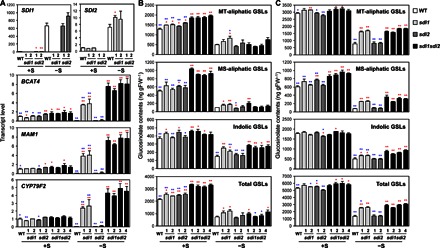
(A) Transcript levels of SDI1, SDI2, and three mGSL biosynthetic genes (BCAT4, MAM1, and CYP79F2) in roots of WT (white bars), sdi1 (pale gray bars), sdi2 (dark gray bars), and sdi1sdi2 (black bars) plants analyzed by qRT-PCR. (B and C) MT-aliphatic, MS-aliphatic, indolic, and total GSL contents in shoot (B) and root (C) tissues of WT, sdi1, sdi2, and sdi1sdi2. Plants were grown under +S or −S conditions. Bars and error bars show means and SE of triplicates. **P < 0.01, *P < 0.05, significant differences detected using Student’s t test between WT and T-DNA insertion mutants (red) and those between four lines of sdi1sdi2 and other plant lines (blue). ng gFW−1, nanogram per gram fresh weight.
Fig. 3. Overexpression of SDI1 and SDI2 represses the GSL accumulation.
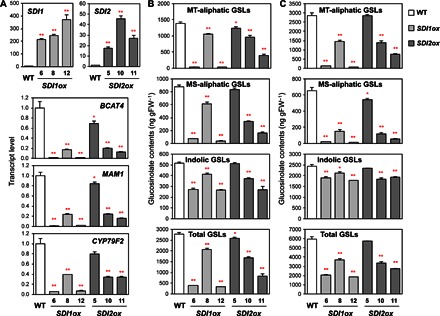
(A) Transcript levels of SDI1, SDI2, and three mGSL biosynthetic genes, BCAT4, MAM1, and CYP79F2, in roots of WT (white bars), SDI1ox (pale gray bars), and SDI2ox (dark gray bars) plants analyzed by qRT-PCR. (B and C) MT-aliphatic, MS-aliphatic, indolic, and total GSL contents in shoot (B) and root (C) tissues of WT, SDI1ox, and SDI2ox plants. Plants were grown under +S conditions. Bars and error bars show means and SE of triplicates. **P < 0.01, *P < 0.05, significant differences detected using Student’s t test between WT and overexpression lines.
In the WT, the transcript levels of several aliphatic GSL biosynthetic genes in roots, namely, BCAT4, MAM1, CYP79F2, CYP79F1, and CYP83A1, were lower under –S relative to +S conditions (Fig. 2A and fig. S4). These results were in accordance with previous observations that suggest a negative impact of sulfur starvation on gene expression of GSL biosynthetic genes (25–30). In contrast, in sdi1 single-knockout and sdi1sdi2 double-knockout lines, the transcript levels of these genes were a few times higher under –S relative to +S conditions (Fig. 2A and fig. S4). These inverse –S responses of gene expression were more evident in sdi1sdi2 double-knockout than in sdi1 single-knockout lines, whereas they were almost absent in sdi2 single-knockout lines that displayed similar –S response profiles as in WT. In sdi1sdi2 double-knockout lines, the transcript levels of BCAT4 and CYP79F1 were higher than those in WT even under +S conditions (Fig. 2A and fig. S4).
GSL levels were also significantly altered by disruption of SDI1 and SDI2 (Fig. 2, B and C, and tables S1 and S2). The total GSL levels in shoots were 1.48 to 1.56 times higher in sdi1sdi2 double-knockout lines than in WT under +S conditions, whereas in the parental sdi1 and sdi2 single-knockout lines, those were about 1.2 times the WT levels (Fig. 2B). The increase in total GSL accumulation in shoots by disruption of SDI1 and SDI2 could mainly be ascribed to accumulations of methylsulfinylalkyl (MS)– and methylthioalkyl (MT)–aliphatic GSLs (Fig. 2B). In contrast to aliphatic GSLs, the amount of indolic GSLs accumulated in shoots was not substantially different among the plant lines grown under +S conditions. However, in plants grown under –S conditions, indolic GSLs were accumulated to relatively higher levels in sdi1 and sdi1sdi2 knockout lines compared with WT, and these changes were reflected in total GSL levels (Fig. 2B). The total GSL content also increased in roots by disruption of SDI1 and SDI2 (Fig. 2C and tables S1 and S2). The increase observed in roots under +S conditions was rather marginal, reflecting the MS-aliphatic GSL content being 1.42 to 1.59 times higher in sdi1sdi2 than in WT (Fig. 2C). In contrast, under –S, the total GSL content in roots of sdi1 and sdi1sdi2 lines was about twice the amount accumulated in WT (Fig. 2C). These changes in total GSL levels were relevant to accumulation of all three GSL species: MT-aliphatic, MS-aliphatic, and indolic GSL contents in sdi1 and sdi1sdi2 were 2.03 to 2.39 times, 3.16 to 4.68 times, and 1.33 to 1.85 times the WT levels, respectively. Generally, under –S, the root GSL contents were significantly higher in sdi1 and sdi1sdi2 compared with WT and sdi2, and the effect of SDI disruption was stronger on aliphatic GSLs than on indolic GSLs.
In contrast to the results shown with the sdi1 and sdi2 knockout lines, the transcript levels of GSL biosynthetic genes decreased in both SDI1- and SDI2-overexpressing lines (SDI1ox and SDI2ox) (Fig. 3A and fig. S4). Three independent homozygous lines of SDI1ox and SDI2ox that display a broad range of SDI expression levels were selected and grown under +S conditions. The transcript levels of SDI1 in SDI1ox lines and SDI2 in SDI2ox lines were about 200- to 400-fold and 20- to 50-fold, respectively, of the endogenous expression levels in WT. Overexpression of SDI1 strongly repressed the expression of GSL biosynthetic genes. Similar but rather moderate effects were observed in SDI2ox lines. The relative transcript levels of BCAT4 in roots of SDI1ox-6, SDI1ox-8, and SDI1ox-12 in comparison with WT were 0.8, 17, and 0.6%, respectively, whereas those in SDI2ox-5, SDI2ox-10, and SDI2ox-11 were 69, 20, and 13%, respectively. Other GSL pathway genes, such as MAM1 and CYP79F2, showed similar expression patterns with significant reduction in their transcript levels in SDI1ox and SDI2ox lines in comparison with WT.
GSL levels were significantly lowered in both shoots and roots in SDI1ox relative to WT but to a lesser extent in SDI2ox lines (Fig. 3, B and C, and tables S3 and S4). Total GSL levels in SDI1ox (lines 6, 8, and 12) were 14, 75, and 12%, respectively, in shoots and 34, 63, and 31%, respectively, in roots of the WT levels. However, in SDI2ox (lines 5, 10, and 11), these percentage values relative to the WT levels were 93, 60, and 30%, respectively, in shoots and 97, 56, and 46%, respectively, in roots. Overexpression of SDI1 and SDI2 strongly affected the accumulation of aliphatic GSLs. In contrast, the effect on indolic GSLs was moderate. The GSL levels were correlated to the expression levels of GSL biosynthetic genes. These results of GSL profiling of SDI gene knockout and overexpressing lines indicate that SDI1 is the major form, whereas SDI2 can be an additively functional form, contributing to the down-regulation of GSL biosynthesis.
Changes in SDI expression levels influencing primary sulfur metabolism
Because GSL metabolism is connected to primary sulfur metabolism, the effect of SDI knockout and overexpression on the sulfur assimilatory pathway was analyzed (Fig. 4, fig. S5, and table S5). The mutant lines sdi1-2, sdi2-2, sdi1sdi2-2, and sdi1sdi2-4 accumulated higher levels of sulfate in both shoots and roots (as well as sdi1sdi2-3 in roots) relative to WT under +S conditions (Fig. 4, A and B, and table S5). Although the levels of sulfate accumulation were strongly reduced when plants were grown under –S, the knockout lines tended to accumulate more sulfate than did WT (Fig. 4A and table S5). In contrast to a slight increase in sulfate accumulation, GSH and Cys levels were decreased by disruption of SDI1 and SDI2. Under +S conditions, the GSH accumulations in shoots and roots of the sdi1sdi2 double-knockout lines were 24 to 35% and 6 to 15%, respectively, of the WT levels (Fig. 4, A and B, and table S5). The GSH levels were also lower in sdi1 and sdi2 single-knockout lines relative to WT, whereas these changes were less significant compared to sdi1sdi2 double knockouts, except for the observation in the roots of sdi2. Similar results were obtained from plants grown under –S conditions. Compared to changes in GSH levels, the effect of SDI1 and SDI2 on Cys accumulation was moderate. The Cys levels in sdi1sdi2 double knockout were 50 to 70% of the WT in shoots and roots under +S conditions (Fig. 4, A and B, and table S5). This decrease in Cys accumulation in sdi1 and sdi2 single-knockout lines was only significant in roots. In contrast, the Cys content remained almost unchanged in both shoot and root tissues under –S conditions (Fig. 4, A and B, and table S5).
Fig. 4. Perturbation of SDI1 and SDI2 influences the accumulation of primary sulfur metabolites.
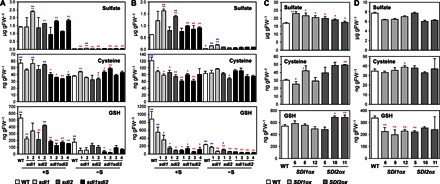
(A and B) Sulfate, Cys, and GSH content in shoot (A) and root (B) tissues of WT (white bars), sdi1 (pale gray bars), sdi2 (dark gray bars), and sdi1sdi2 (black bars) plants grown under +S or −S conditions. **P < 0.01, *P < 0.05, significant differences detected using Student’s t test between WT and T-DNA insertion mutants (red) and those between four lines of sdi1sdi2 and other plant lines (blue). (C and D) Sulfate, Cys, and GSH content in shoot (C) and root (D) tissues of WT (white bars), SDI1ox (pale gray bars), and SDI2ox (dark gray bars) plants grown under +S conditions. Bars and error bars show means and SE of triplicates. **P < 0.01, *P < 0.05, significant differences detected using Student’s t test between WT and overexpression lines.
Sulfate concentrations in shoots of SDI1ox and SDI2ox lines were 1.22 to 1.46 times and 1.03 to 1.18 times, respectively, of the WT levels, whereas in roots, those were not significantly changed or slightly reduced in both SDI-overexpressing lines compared with WT (Fig. 4, C and D, and table S5). The amount of GSH accumulated in shoots was similar between the SDI-overexpressing lines and WT but slightly higher in two of the three SDI2ox lines (lines 10 and 11) (Fig. 4C). In contrast, the GSH levels were lower in roots of SDI1ox and SDI2ox lines compared with WT (Fig. 4D). The Cys contents tended to be higher in SDI2ox than in WT in shoots, whereas they were similar among SDI1ox, SDI2ox, and WT in roots (Fig. 4, C and D, and table S5).
These results indicated that sdi1 and sdi2 knockouts could affect the concentrations of sulfate, Cys, and, to a greater extent, GSH, whereas SDI overexpression only have a marginal effect on these primary metabolites, unlike the accumulations of GSLs that are strongly influenced by both disruption and overexpression of SDI genes in a characteristic inverse manner. In contrast to the metabolite levels, the transcript levels of genes involved in sulfur assimilation, that is, sulfate transporters SULTR1;2 and SULTR2;1 and adenosine 5′-phosphosulfate (APS) reductase APR3, were similarly influenced by SDI overexpression as the GSL biosynthetic genes, suggesting that the sulfate assimilation pathway is reduced concomitantly with GSL biosynthesis. The SULTR1;2, SULTR2;1, and APR3 transcript levels increased under –S in WT, as previously reported, and in the sdi1 and sdi1sdi2 knockout lines, they showed even stronger responses to –S (Fig. 5A and fig. S5) (25–31). The transcript levels of SULTR1;2 and SULTR2;1 were higher in sdi1sdi2 knockout lines relative to WT even under +S (fig. S5A). Upon –S, their transcript levels were significantly higher in sdi1 and sdi1sdi2 relative to WT and sdi2, suggesting that SDI1 is the major component that affects the expression of sulfate assimilatory genes, as it was for GSL biosynthesis. In the SDI1- and SDI2-overexpressing lines, their transcript levels were generally reduced relative to WT (fig. S5B).
Fig. 5. PCA of −S and SDI-regulated genes.
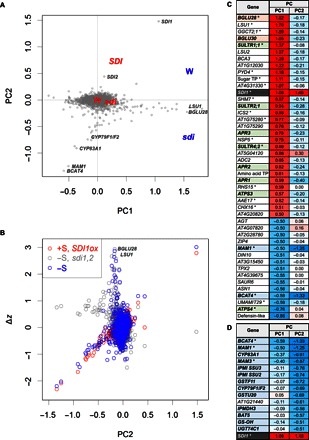
(A) PC biplot of the transcript levels in root tissues of WT (W), sdi1sdi2 (sdi), and SDI1ox (SDI) plants grown under +S (red) and –S (blue) conditions. Scores of sample PC (W, SDI, and sdi) and gene PC (gray spots) are placed on a biplot (44). For each group, means of sample PC are presented; the scores were magnified by eight times. Positions of the gene PC with higher absolute scores in PC1 (BGLU28, LSU1, SDI1, MAM1, and BCAT4) and PC2 (SDI1, CYP79F1/F2, CYP83A1, MAM1, and BCAT4) were indicated. (B) Comparisons between PC2 and expressional changes (Δz) from the control groups. Δz of genes selected for PCA in WT grown under –S (blue), sdi1sdi2 grown under –S (gray), and SDI1ox grown under +S (red) compared to WT grown under +S were spotted. Positions of the genes that take top two PC1 scores, BGLU28 and LSU1, were indicated. The spot located at the upper right is SDI1. (C and D) Heat map of the gene PC with the absolute value of more than 0.5 in PC1 (C) and PC2 (D). Asterisks show SLIM1-regulated genes (29).
SDI1-regulated transcriptome
To obtain further insight into the effect of SDI and –S on the global transcript levels, we performed microarray analyses. RNA samples were prepared from roots of two independent lines of SDI1ox, sdi1sdi2 lines, and WT grown under +S, and the same sdi1sdi2 lines and WT grown were under –S. The probes from these RNA samples were hybridized to ATH1 GeneChip arrays. Raw data were parametrically normalized, and the transcript level of each gene was obtained as z-score (42). The individual and combinatorial effects of –S and SDI genotypes on differential gene expression were defined here by calculating the transcript levels relative to the WT grown under +S and by presenting them as log ratios (Fig. 5 and tables S6 to S10). Using two-way analysis of variance (ANOVA), we selected 6000 genes as those that were differentially regulated among plant lines or conditions; many of the genes showed low P values (fig. S6), negating problems caused by multiple testing (43). These normalized expression data were converted through principal components analysis (PCA) to identify the directions of changes in gene expression at transcriptome levels shared with plant lines or conditions (Fig. 5 and figs. S6 to S9) (44). Four independent principal components (PC) that represent characteristic directions were identified through PCA (fig. S7 and table S10). Among these directions, PC1, PC2, PC3, and PC4 contributed 71, 18, 8, and 3%, respectively, to the variance. The PC1 to PC2 plot, contributing together 89% of the variance within the data sets, conclusively showed a clear separation between SDIox, sdi knockouts, and the –S effect (Fig. 6). PC1 and PC2 were normally distributed with heavy tails (fig. S8). Outlying genes (blue and red in fig. S8) were further selected, and the trends in their gene/protein functions were estimated using Gene Ontology (GO) keywords (fig. S10).
Fig. 6. Transcript profiles of the genes involved in GSL synthesis and sulfur assimilation in sdi1sdi2 and SDI1ox plants.
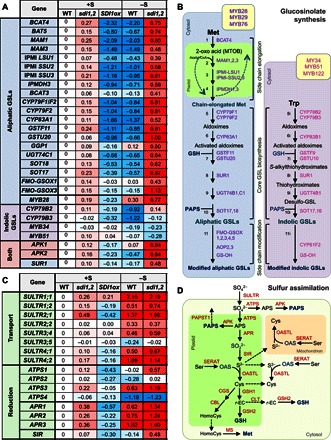
(A) Heat map of the transcript profiles of the key GSL-related genes in sdi1sdi2 and SDI1ox lines. Values indicate log ratios of the transcript levels in root tissues of WT, sdi1sdi2, and SDI1ox grown under +S and –S versus those of WT grown under +S. (B) Heat map of the transcript profiles of the genes involved in the sulfate transport and reduction in sdi1sdi2 and SDI1ox lines. Values are the same with those in (A). (C) Schematic diagram of aliphatic and indolic GSL biosynthetic pathways, including amino acid chain elongation (steps 1 to 4), biosynthesis of core GSL structure (steps 5 to 10), and secondary modification (step 11). Chain elongation of GSLs occurs in the plastid (green), and the rest of the reactions occur in the cytosol (pale blue for aliphatic GSL, pale purple for indolic GSL). Enzymes involved in the aliphatic and indolic GSL synthesis are depicted in purple and magenta, respectively. Transcription factors that control aliphatic and indolic GSL synthesis are depicted in yellow boxes. BCAT, branched-chain amino acid aminotransferase; MAM, methylthioalkylmalate synthase; IPMI, isopropylmalate isomerase; IPMDH, isopropylmalate dehydrogenase; CYP79, cytochrome P450 of the CYP79 family; CYP83, cytochrome P450 of the CYP83 family; GST, glutathione S-transferase; SUR1, C-S lyase; UGT74, glucosyltransferases of the UGT74 family; SOT, sulfotransferase; FMO-GS-OX, S-oxygenase; AOP, 2-oxoglutarate-dependent dioxygenase; GS-OH, 2-oxoacid–dependent dioxygenase. (D) Schematic diagram of sulfate assimilatory pathway. Reactions occur in the plastid (green), cytosol (pale green), and mitochondrion (pale orange). Enzymes are depicted in red. ATPS, adenosine triphosphate sulfurylase; APR, APS reductase; APK, APS kinase; PAPST1, PAPS transporter; SiR, sulfite reductase; SERAT, serine acetyl-transferase; OAS-TL, OAS(thiol)-lyase; GSH1, γ-glutamylcysteine synthase; GSH2, glutathione synthase; CLT, CRT-like transporter; CGS, cystathionine γ-synthase; CBL, cystathionine β-lyase; MS, methionine synthase; r-EC, γ-glutamylcysteine; HomoCys, homocysteine.
The first PC (PC1), which indicates the direction of the maximum variance of the differential gene expression, was associated with the –S responses (Fig. 5A). The direction indicated by PC1 showed high correlations with the –S responses in WT and sdi1sdi2, with the correlation factors of 0.9257 and 0.9071, respectively (fig. S9). Genes with high scores in PC1 were identical to those previously reported as –S-responsive genes, including putative thioglucosidases BGLU28 and BGLU30, several SULTRs and APRs, and −S marker genes LSU1, LSU2, and SDI1 that were displayed toward the positive direction of the PC1 axis, whereas MAM1, BCAT4, and ATPS4 were displayed oppositely toward the negative direction (Fig. 5, A and C, and table S6).
Part of the changes in gene expression caused by –S overlapped with those modulated by overexpression of SDI1, and this direction was represented by the PC2 (Fig. 5, A and B). Comparisons of the effect of SDI1ox (red) and –S (blue) with the combinatorial effect of sdi1sdi2 knockout and –S (gray) suggested that a group of PC2 genes are repressed in common by SDI1 overexpression and by –S in the WT (Fig. 5B). Most of these genes that highly contributed to the negative PC2 scores were clearly the genes involved in aliphatic GSL synthesis (Fig. 5D), and those to the positive PC2 were SDI1 and SDI2 (Fig. 5C and table S7). The GO keywords and metabolic pathways, which represent the genes with negative PC2 scores, provided further supporting evidence that the GSL synthetic process was the major metabolic pathway strongly influenced by SDI expression (fig. S10). In addition to the GSL synthetic process, genes related to amino acid synthesis and auxin [indoleacetic acid (IAA)] synthesis were detected with negative PC2 scores (fig. S10).
Changes in gene expression associated with GSL synthesis and sulfate assimilation are summarized in Fig. 6 and tables S8 and S9. Most of the genes involved in aliphatic GSL synthesis were down-regulated by –S and overexpression of SDI1. Conversely, they were induced by the knockout of SDI1 and SDI2, especially under –S conditions (Fig. 6, A and B); this was consistent with the results obtained by quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis (Figs. 2A and 3A and fig. S4). SDI1 also affected the expression of CYP79B2 and CYP79B3, which are involved in indolic GSL synthesis (Fig. 6, A and B, and table S8). MYB transcription factors were differently regulated between those involved in aliphatic GSL and indolic GSL synthesis; that is, MYB28 was up-regulated by –S and negatively affected by SDI1, but MYB34 and MYB51 were down-regulated by –S and rather positively affected by SDI1, although changes were marginal compared with MYB28 (Fig. 6A and table S8). In contrast to GSL-related genes, sulfate assimilation pathway genes were generally induced by –S in WT and sdi1sdi2, whereas ATPS1, ATPS3, APR1, APR2, APR3, and SIR, in particular, showed higher expression levels in sdi1sdi2 than in WT (Fig. 6, C and D, and table S9). SDI1 overexpression and sdi1sdi2 showed a much weaker but opposite effect on those genes under +S, indicating that SDI1 and SDI2 are inhibitory factors of sulfate assimilation (Fig. 6C).
SDI1 interacts with MYB28 in the nucleus
The effect of SDI on the transcriptome and the prediction of nuclear localization of SDI1 suggested that SDI1 would function possibly as a transcription factor or as a factor in a transcriptional complex. To determine the subcellular localization of SDI1, fusion proteins of GFP to the N and C termini of SDI1, GFP-SDI1, and SDI1-GFP were transiently expressed in Arabidopsis protoplasts and visualized by confocal microscopy (Fig. 7A). GFP signals were detected in the nucleus and the cytosol of the protoplasts (Fig. 7A). Both GFP-SDI1 and SDI1-GFP led to similar observations.
Fig. 7. SDI1 locates to the nucleus and interacts with MYB28.
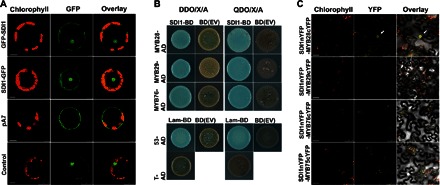
(A) Subcellular localization of SDI1 in Arabidopsis protoplasts. GFP signals were localized in the transformed protoplasts with SDI1-GFP and GFP-SDI1 constructs. pA7 empty vector and NAC transcription factor 104 fused with GFP were used as negative and positive controls, respectively. Scale bars, 75 μm (GFP-SDI1 and pA7), 10 μm (SDI1-GFP), and 25 μm (control). (B) Y2H assay of SDI1-BD (as bait) and MYB28/MYB29/MYB76-AD (as preys). Yeasts have been grown on -Leu/-Trp medium containing X-α-Gal (DDO/X) and -Leu/-Trp/-Ade/-His medium containing X-α-Gal and Aureobasidin (QDO/X/A). Cotransformations of bait empty vector [BD(EV)] with preys were performed as negative controls (brown colonies). The two lower panels show positive and negative controls, respectively, as explained in Materials and Methods. (C) BiFC analysis of interaction between SDI1nYFP and MYB28cYFP, MYB29cYFP, and MYB76cYFP in epidermal cells of N. benthamiana leaves. Arrows denote the interaction of MYB28 and SDI1 in the nucleus (top panel). MYB75cYFP was used as the negative control of the interaction with SDI1nYFP. Scale bar, 50 μm.
Nuclear localization of SDI1 motivated us to investigate how SDI1 represses the expression of GSL biosynthetic genes. Because SDI proteins are assumed to interact with proteins due to their TPR domain, protein-protein interactions between SDI1 and the known transcription factors for aliphatic GSL biosynthesis (MYB28, MYB29, and MYB76) were analyzed using a targeted yeast two-hybrid (Y2H) screening. Y2H screening was performed using the SDI1-bait construct pGBKT7-SDI1 screened against the prey constructs pGADT7-MYB28, pGADT7-MYB29, and pGADT7-MYB76. All the MYB proteins showed interaction with SDI1 in yeast (Fig. 7B). Bimolecular fluorescence complementation (BiFC) was performed to verify in planta interaction of SDI1 with the MYB transcription factors. SDI1::nYFP expressing the fusion protein of SDI1 and N-terminal part of yellow fluorescent protein (YFP) was agroinfiltrated to leaf cells of Nicotiana benthamiana with combinations of MYB28::cYFP, MYB29::cYFP, and MYB76::cYFP constructs expressing the fusion protein of these MYB transcription factors and C-terminal part of YFP. Among all combinations, yellow fluorescence representing the reconstitution of YFP through protein-protein interaction between SDI1 and MYB was detected exclusively in the nuclei of tobacco cells coexpressing SDI1::nYFP and MYB28::cYFP (Fig. 7C), whereas the coexpression of SDI1::nYFP with MYB29::cYFP, MYB76::cYFP, and the negative control MYB75(PAP1)::cYFP did not show any YFP signal (Fig. 7C). These results indicated that SDI1 at least interacts with MYB28 in the nucleus of plant cells.
SDI1 inhibits transcriptional activity of MYB28
To determine how the binding of SDI1 influences MYB28 activity as a transcriptional activator of aliphatic GSL biosynthesis, we conducted transient transactivation assays and electrophoretic mobility shift assays (EMSAs) using SDI1 and MYB28. Cultured A. thaliana cells were transformed with a supervirulent Agrobacterium tumefaciens strain carrying either effector constructs containing MYB28 and/or SDI1 or the reporter constructs expressing the uidA (GUS) gene under the control of the 5′-upstream sequences of CYP79F1 and CYP83A1 (Fig. 8) (12). GUS activity in the cultured cells that contain the reporter constructs was greatly increased by the coexpression with MYB28 (Fig. 8, B and E). When SDI1 was coexpressed with MYB28 as an additional effector, induction of GUS activity by MYB28 was decreased (Fig. 8, B and E). These results, together with our transcriptome data, indicate that SDI1 has a considerable negative impact on the transcriptional activity of MYB28.
Fig. 8. SDI1 inhibits MYB28-mediated transactivation of aliphatic GSL biosynthetic genes CYP79F1 and CYP83A1.
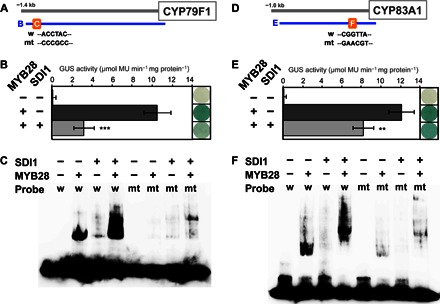
(A and D) Promoter positions of CYP79F1 and CYP83A1 used for transactivation assay (B, −1388 to + 45 bp for CYP79F1; E, −919 to + 63 bp for CYP83A1) and EMSA (C and F). AC element in CYP79F1 promoter [WT probe (w) in (A) (45)], MYB core sequence in CYP83A1 promoter [w in (D) (45)], and the mutated sequences (mt) in the probe used for EMSA were indicated. (B and E) Cotransformation assays for the determination of activation or inhibitory potential of MYB28 and SDI1 toward CYP79F1 and CYP83A1 promoter. A. thaliana cells were inoculated with the supervirulent Agrobacterium strain LBA4404.pBBR1MCS.virGN54D containing either only the reporter construct [CYP79F1:GUS in (B) and CYP83A1:GUS in (E)] or the effector constructs (Pro35S:MYB28 and/or Pro35S:SDI1). Transfection rate for different constructions and biological replicates was controlled by coexpressing the 35S:LUC construct. GUS staining (right) and GUS activity (left) of the cultured cells 5 days after transformation were indicated. Means of GUS activity in micromole monomer unit per minute and milligram protein ± SD, n = 5. ***P < 0.001, **P < 0.01, significant differences detected using Student’s t test between Arabidopsis cells expressing a reporter [CYP79F1:GUS in (B) and CYP83A1:GUS in (E)] and an effector (Pro35S:MYB28) construct with or without Pro35S:SDI1. (C and F) SDI1 does not inhibit the DNA binding of MYB28 to the respective MYB-binding site of CYP79F1 and CYP83A1 promoter. EMSA shows the binding activity of MYB28 to an AC element in the CYP79F1 promoter (C) or a MYB core sequence in the CYP83A1 promoter (F) and the effects of SDI1 on the binding. The WT probe (w) or the mutated probe (mt) labeled with biotin to their 3′ end was incubated in the presence (+) or absence (−) of MYB28 and/or SDI1 proteins. In (C), the probe sequence used for the analysis was the region from −1249 to −1208 from the translational start site of CYP79F1 (5′-ATAAACGTGTTCACCTACCCAAGGTATTATTTATTTATTC-3′). The underlined AC element (45) was mutated as indicated in (A). In (F), the probe sequence used for the analysis was the region from −191 to −125 from the translational start site of CYP83A1 (5′-ACACGTGAGGTTTCGTAAGTAGGTATAAGGGAGTAACCATTTGATTAAAAATTTGATGTCGGTTAAG-3′). The underlined MYB core sequence (45) was mutated as indicated in (D).
The effect of the interaction between SDI1 and MYB28 on DNA binding of MYB28 was analyzed by EMSA (Fig. 8, C and F). AC element and MYB core element are known as R2R3-MYB binding sequences on the basis of results of yeast one-hybrid (Y1H) assays (45). However, direct binding of MYB28 to the target DNA sequence has not been verified yet by EMSA. We used the AC element and MYB core element, present in the 5′-upstream sequences of CYP79F1 and CYP83A1, respectively, as probes for EMSA. Biotin-labeled oligo DNA probes containing AC element (ACCTAC) or MYB core element (CGGTTA) and MYB28 and/or SDI1 proteins produced by wheat germ expression system were used for the experiments. EMSA results revealed that MYB28 bound the respective DNA probes; however, SDI1 did not bind to either of the probes (Fig. 8, C and F). When SDI1 and MYB28 were incubated together, a supershift was observed with both CYP79F1 and CYP83A1 probes, indicating that DNA binding of MYB28 was not impaired by SDI1, whereas the protein complex of MYB28 and SDI1 can still interact with the oligo DNAs. Specific binding of MYB28 to both cis-elements was confirmed by EMSA using mutated probes containing nucleotide substitutions in the AC element and the MYB core element, respectively (Fig. 8, C and F).
DISCUSSION
SDI1 and SDI2 repress GSL synthesis
SDI1 and SDI2 have been described in Arabidopsis and wheat as S-responsive marker genes strongly induced upon –S (25–30, 33). Moreover, SDI1 and SDI2 are induced by OAS, which builds up in plants upon –S, and also due to other stresses (34). Our present study demonstrates that SDI1 and SDI2 are negative regulators of GSL biosynthesis in Arabidopsis. Metabolite data indicate that loss of SDI function leads to an increased aliphatic GSL accumulation, whereas SDI overexpression has the opposite effect, both under +S and –S conditions (Figs. 2 and 3 and tables S1 to S4). The transcript levels of most of the genes involved in the aliphatic GSL pathway increase in sdi1sdi2 double knockouts but decrease in SDI1ox lines (Fig. 6A and table S8). These data suggest that SDI1 and SDI2 negatively regulate GSL biosynthesis at the transcript levels, resulting in reduced GSL levels, especially aliphatic GSLs. Our data further indicate that SDI1 is the major protein component that contributes to this inhibitory action, whereas SDI2 appears to have an additive effect (Figs. 2 and 3). In support of this idea, SDI1 is more abundantly expressed than SDI2 under –S (Fig. 1C and tables S7, S10, and S11), and the sdi1 single knockouts result in highly accumulating GSLs even under –S, whereas the sdi2 single knockouts exhibit GSL profiles similar to WT (Fig. 2, B and C, and table S1).
SDI1 and SDI2 clearly exhibit a control over the biosynthesis of aliphatic GSLs and, to a minor extent, over indolic GSLs in Arabidopsis. The GSL profiles of sdi1sdi2 and SDIox indicate that SDI1 and SDI2 act negatively on both the aliphatic and the indolic GSL pathways (Figs. 2, 3, and 6). The stronger control of SDIs over the aliphatic than the indolic GSLs is obvious because a higher accumulation of aliphatic than indolic GSL is observed in sdi1sdi2 (Fig. 2) and because of the fact that indolic GSL contents are less impaired in SDI1ox lines than in aliphatic GSLs (Fig. 3). Despite these metabolic changes, transcript analyses revealed subtle changes in the transcript levels of the biosynthetic genes of indolic GSLs (Fig. 6A). Expression levels of CYP79B2 and CYP79B3, which are recognized as key genes in indolic GSL biosynthesis (10, 11), are affected by the sdi1sdi2 knockout. Thus, SDI1 and SDI2 impart negative control of expression of CYP79B2 and CYP79B3, which is in line with our observations on the accumulation of indolic GSL in respective mutant and overexpression lines. However, the mechanisms on how exactly SDIs regulate indolic GSL pathway remain to be established.
The PCA of global gene expression provides implications that SDIs are regulators for transcriptional control mechanisms associated with S responses. The PC1 and PC2 obtained from the analysis explain about 90% of the observed variability in the data sets. PC1 is mainly linked to the environmental effect, that is, –S, whereas PC2 mainly correlates to SDI1 inversely with the expression of GSL biosynthetic genes (Fig. 5). This suggests a link between the response to –S and the down-regulation of GSL synthesis mediated by SDIs and further provides evidence that SDI1 is a major regulator of the aliphatic GSL biosynthesis. The more effective and tighter control of the aliphatic versus the indolic GSL corroborates with the fact that aliphatic GSLs show greater sensitivity to –S than do indolic GSL (22, 23, 29). This might be cost-effective for controlling sulfur use under –S condition because aliphatic GSLs synthesized from Met can be a stronger sink for sulfur than indolic GSLs; hence, their tighter control may be essential.
SDI1 acts as a transcriptional repressor via the interaction with MYB28
Aliphatic GSL pathway genes are positively regulated by MYB28, MYB29, and MYB76 (12, 13). Several interacting protein partners have been identified for the other R2R3-MYB type of transcription factors, which act as enhancers or repressors of their activity and provide rapid on-and-off switches for the transcriptional machinery (46). Our present study demonstrates that SDI1 is a novel negative regulator for aliphatic GSL synthesis, which acts via direct physical interaction in the nucleus with MYB28 (Fig. 7C), the major transcription factor that regulates the pathway. Multiple lines of evidence support our conclusion. SDI1 interacts physically by protein-protein interaction with MYB28 both in yeast and in the nucleus of plant cells (Fig. 7, B and C). SDI1 inhibits the MYB28-mediated transactivation of the promoters of two aliphatic GSL biosynthetic genes, CYP79F1 and CYP83A1 (Fig. 8, B and E). Furthermore, SDI1 does not affect binding of MYB28 to the putative MYB binding sites, AC element and MYB core sequence (45), in the promoter fragments of CYP79F1 and CYP83A1 (Fig. 8, C and F). The results from EMSA indicate that SDI1 does not directly bind to these cis-elements but that SDI1 binds to MYB28, maintaining its DNA binding activity, as indicated by the supershift of the signal when both MYB28 and SDI1 are present (Fig. 8, C and F). These results suggest that the SDI1-MYB28 complex formation does not interfere with DNA binding of MYB28. Because SDI1 does not prevent DNA binding of MYB28 to its cis-elements, sterical hindrance of the transcription machinery by the SDI1-MYB28 complex formation can be a mechanism that may possibly block the function of MYB28 as an activator. The roles of the TPR domain in the SDI1-MYB28 interaction and the actual action mechanisms of this interaction remain to be investigated in the future.
Positive regulations between the MYB transcription factors that control aliphatic GSL synthesis have been reported. MYB28 activates the expression of MYB29 and MYB76, whereas MYB28 expression is barely controlled by MYB29 and MYB76 (17, 47). MYB28 further controls its own expression in a positive feedback loop through the interaction with its own promoter (48). Transcript data indicate that SDI1 and SDI2 negatively affect the expression of MYB28 (Fig. 6A and table S8). Therefore, we speculate that protein-protein interaction between SDI1 and MYB28 blocks the action of MYB28 on its target genes, thus rendering it unable to up-regulate the expression of MYB29, MYB76, and MYB28 itself. However, this model needs further verification because the transcripts of MYB29 and MYB76 could not be detected in this study (table S8). Tissues where SDI1 and SDI2 are abundantly expressed are similar to those where MYB28, MYB29, and MYB76 are present (fig. S3) (12, 47). MYB28, MYB29, and MYB76 are preferentially expressed in the vascular tissues of leaves, where many of the GSL synthetic genes are expressed (49, 50), correlating with accumulation of GSLs (17, 51). Hence, the expression of SDI1 and SDI2 in vascular tissues further supports the regulatory connection between SDI1 and MYB28 and aliphatic GSL synthesis (fig. S3).
SDI connecting sulfur availability and regulation of GSL synthesis
Characteristics of SDI1 and SDI2 as repressors of GSL accumulation and their induced expression under –S raise a question on how sulfur availability regulates their expression and, eventually, GSL synthesis. Previously, we identified SLIM1 as a transcription factor, which regulates a broad range of –S responses in plants, including down-regulation of GSL synthesis (29). The SDI1 gene expression is affected by slim1. The transcript levels of SDI1 are higher in slim1 mutants than in WT under +S conditions. In contrast, the SDI1 levels are lower in slim1 than in WT under –S, despite the significant increase observed in both lines (table S11) (29). These gene expression profiles suggest that SLIM1 is a regulatory factor that represses SDI1 expression under +S and assumingly releases this repression in response to S depletion, although it is also possible to consider that the function of SLIM1 is required for further activation of SDI1 expression under –S conditions. In agreement with these gene expression profiles of SDI1, the transcript levels of MYB34 and its downstream genes, including CYP79B2, CYP79B3, and CYP83B1, and some genes of aliphatic GSL biosynthesis, such as BCAT4, CYP79F1, CYP79F2, MAM1, and MAM3, are consistently lower in slim1 mutants relative to WT under +S but higher in slim1 than in WT under −S conditions (29). Because SLIM1 is not transcriptionally regulated in response to –S, posttranslational modifications of SLIM1 may be necessary to modulate the SDI1 gene expression through the SLIM1-mediated pathway (table S11). The relationships between –S and transcriptional regulation of SDI1 and SDI2 and their downstream genes can also be supported by evidence that OAS is an inducer for their gene expression, mimicking the cellular status under –S (29, 34).
Our findings demonstrate a regulatory model that SDI1 interacts with MYB28 to repress aliphatic GSL biosynthesis in plants (Figs. 7 and 8). SDI1 and SDI2 proteins overaccumulate as a result of the –S-driven, SLIM1-mediated regulatory pathway, which brings SDI1 to be assembled into the SDI1-MYB28 complex and subsequently to be in action to repress GSL biosynthesis. In addition to direct evidence demonstrating the function of SDI1 and SDI2 in transcriptional regulation of GSL biosynthetic genes, transcriptome analysis of sdi1sdi2 mutant provides further implication that these repressor proteins also have negative impact on MYB28 gene expression itself under –S (Fig. 6A), extending the idea of SDI1-MYB28 interplay model that can be applied for the promoter-dependent regulation of MYB28 (48). In contrast, previously reported microarray data indicate that SDI1 and SDI2 are up-regulated when MYB28 and MYB29 are overexpressed, whereas both are repressed in myb28 single- and myb28myb29 double-knockout lines (fig. S11) (13, 52). Thus, a MYB28-driven mechanism that feedback controls the levels of SDI1 and SDI2 also appears to be present in the regulatory circuit, allowing the flux of GSL biosynthesis to be lessened when MYB28 overproduces aliphatic GSLs. Although SLIM1 appears to be central for controlling the levels of SDIs, a reciprocal fine-tuning mechanism proposed here based on regulatory relationships between SDI1/SDI2 and MYB28 would also be important for regulation of GSL biosynthesis in response to changes under –S conditions. Given the mechanisms based on protein-protein interaction would allow plants to quickly revert their metabolic status, the SDI1-MYB28 interplay system might be essential for releasing the inhibition of GSL biosynthesis once sulfate becomes available. The mechanisms associated with regulation of SDI1 at protein levels, for example, protein modification or protein turnover, have yet to be identified in the future. Hence, SDI1 and SDI2 are assumingly targets of a multifactorial control of SLIM1 and further, yet unidentified factors.
PCA of the microarray data indicates significant overlaps among the genes regulated by –S, SLIM1, and SDI1 (Fig. 5 and tables S8 and S9); that is, 20 of 45 genes that highly contribute to PC1 (−S signature) and 5 of 15 genes that highly contribute to PC2 (SDI1 signature) are identical to those previously reported as SLIM1-dependent (Fig. 5) (29). These data indicate the overlap between –S response, SLIM1 control, and SDI effects but also suggest multiple signaling pathways to be in action, because these overlaps of regulated genes found in this study are not complete, suggesting that additional players are involved in the response network to sulfate starvation. The transcript levels of genes involved in aliphatic GSL biosynthesis are not completely diminished in either myb28myb29 double or myc2myc3myc4 triple mutants (19), suggesting the existence of other transcription factors that stimulate their expression. On the basis of a large-scale Y1H screening with the 5′-upstream sequences of aliphatic GSL biosynthetic genes and metabolite profiling of gene-disrupted Arabidopsis lines for candidate proteins identified from screening, 29 transcription factors are suggested to modulate GSL biosynthesis (48). These proteins might be additional candidates that play roles in the regulation of GSL biosynthesis in either a SDI1-dependent or SDI1-independent manner.
SDI function on primary sulfur metabolism
Although the effect of SDI1 and SDI2 on the regulation of GSL biosynthesis is a dominant feature in Arabidopsis, they also seem to affect primary sulfur metabolic pathways. Both SDI1 and SDI2 are induced under –S and OAS accumulation (29, 34), suggesting that their effect on primary sulfur metabolism may be fundamental and shared in all plant species, including those unable to synthesize GSLs. The SDI1 and SDI2 orthologs form a unique clade in a phylogenetic tree, indicating their close evolutionary relationships (fig. S2). However, SDI1 and SDI2 from Brassicales plants containing GSLs form a subclade that is separated from others, indicating coevolution due to functional specialization in these plant species.
The metabolite profiles of sdi1sdi2 double-knockout lines indicate that disruption of SDI1 and SDI2 causes significant decrease in tissue GSH levels (Fig. 4). Although these changes are based on observation of steady-state metabolite levels, the lesser accumulation of GSH in sdi1sdi2 double knockout relative to WT can be at least partially attributed to the consumption of GSH as the donor of reduced sulfur in GSL biosynthesis (11, 53). A decrease in GSH levels in sdi1sdi2 knockouts relative to WT might also indicate increased allocation of sulfate to 3′-phosphoadenosine-5′-phosphosulfate (PAPS) biosynthesis. The transcript levels of APK1 and APK2 are higher in sdi1sdi2 knockouts relative to WT (Fig. 6A and table S8), supporting the idea of APK as a key enzyme for S flux control between primary and secondary plant metabolism (54, 55). A reduction in APK activity results in reducing the GSL levels and increasing the concentrations of Cys and GSH (55), which is the exact opposite trend observed in sdi1sdi2 (Figs. 2 and 4). Thus, SDI1 and SDI2 might repress APK gene expression and reduce the amount of sulfate to be used for synthesizing PAPS, which serves as sulfate donors for the synthesis of GSLs in Brassicaceae and various downstream sulfated metabolites in diverse plant species (55). Hence, this feature of SDI1 and SDI2 function mirrors their inhibitory actions on GSL biosynthesis. How SDIs inhibit APK transcription needs to be investigated. However, it has been shown that the MYB transcription factors, which regulate aliphatic and indolic GSL biosynthesis, control the expression of the primary sulfate metabolic genes, such as APK1, APK2, ATPS1, ATPS3, APR isoforms, and SIR (54). Therefore, given the substantial effects of SDI1 on the MYB28 transcript levels, it is reasonable to speculate that the repression of APK gene expression is conveyed through the SDI-mediated MYB28 inactivation.
In addition, it cannot be excluded that an increase in metabolic flux toward GSL synthesis might be driving the activity of sulfate uptake and assimilation in the sdi1sdi2 mutants. The sulfate levels are higher in sdi1sdi2 relative to WT, in agreement with changes in the transcript levels of sulfate transporters in roots (Figs. 4, A and B, and 6C and fig. S5). This result is consistent with similar observations of sulfate accumulation in sdi1 single knockouts in a previous study (33). The sulfate assimilatory pathway appears to be induced concomitantly in sdi1sdi2 mutants, although this induction seems to have not resulted in increasing GSH content because of usage of S for GSL biosynthesis. In an opposite manner, a decrease in sulfur flux toward GSL biosynthesis may have caused repression of sulfate transporters and APS reductase in SDI1- and SDI2-overexpressing lines (Fig. 6C and fig. S5). Thus, an increase in sulfate accumulation in shoots and a decrease in GSH levels in roots of SDI-overexpressing lines in comparison with WT might be consequences of transcriptional repression of the sulfate reduction pathway. Attenuating GSL biosynthesis could have additionally caused the level of nonmetabolized sulfate pool in shoots to increase. The levels of primary sulfur metabolites can be strongly affected by the amount of initial sulfate uptake, internal sulfate distributions (for example, root-to-shoot transport), and changes in sulfur metabolic fluxes. Interpreting the roles of the regulatory proteins SDI1 and SDI2 in the primary sulfur metabolism appears rather complex in contrast to their regulatory-metabolic relationships demonstrated for the regulation of the GSL pathways.
Under sulfate-deprived growth conditions, the de novo biosynthesis of the sulfur-rich GSLs, mainly the aliphatic GSLs, is reduced. SDI controls the biosynthesis of GSLs through the protein-protein interaction with MYB28. In addition to this regulatory pathway, a still mechanistically unclear negative effect of SDIs on the expression of the core indolic GSL biosynthetic genes CYP79B2 and CYP79B3 could be observed under −S, resulting in a reduction of indolic GSL in addition to aliphatic GSL contents. Although SDI1 and SDI2 are major control factors, they are embedded in a complex regulatory network that involves MYB and probably various other transcription factors. Thus, plants have a fine-tuned regulatory mechanism that can reduce the production of secondary S-containing pools of reduced S under –S stress to prevent a flow of S into S-rich compounds, such as aliphatic and indolic GSLs, competing with primary plant metabolism. Moreover, because it has been revealed that GSL production entails a remarkable growth cost, that is, 15% increase in photosynthetic energy (56), regulation of GSLs via SDI can be considered as a cost-effective way, besides GSL turnover, to balance the amount of primary reduced S compounds, such as Cys and GSH, that is allocated to this metabolite pool. Yet, the findings of this study also provide a tool to increase or sustain GSL contents and thus quality in plants, producing health beneficial and defensive S compounds by breeding for reduced leaf SDI1 and SDI2 activities.
MATERIALS AND METHODS
Plant materials and growth conditions
A. thaliana accession Columbia (Col-0) was used as WT. Arabidopsis plants were grown at 22°C under 16-hour light/8-hour dark cycles. Plants were grown on mineral nutrient medium (57) containing 1% sucrose. For preparation of agar medium, agar was washed twice with 1 liter of deionized water and vacuum-filtrated to remove sulfate. +S agar medium contained 1500 μM MgSO4 as the sulfur source. –S agar medium contained 15 μM MgSO4. The Mg concentration of the –S medium was adjusted to 1500 μM by adding MgCl2. Seeds were sown directly on +S and –S medium, and these agar plates were incubated in the same growth chamber under controlled conditions. Shoots and roots were harvested separately.
Isolation of T-DNA insertion mutants of SDI1 and SDI2
Homozygous knockout mutants for SALK_145035 (sdi1-1) and SALK_099766 (sdi1-2) containing T-DNA insertions in the intron regions of SDI1 and those for SALK_091618 (sdi2-1) and SALK_110128 (sdi2-2) containing T-DNA insertions in the exon and intron regions of SDI2, respectively, were obtained by screening based on PCR analysis (58). Confirmation of the T-DNA insertion in these lines was carried out by PCR on genomic DNA using T-DNA left border primer LB-02 and gene-specific primers for SDI1, sdi1-1 LB, sdi1-2 LB, or sdi1-1,2 RB and those for SDI2, sdi2-1 LB, sdi2-1 RB, sdi2-2 LB, or sdi2-2 RB, as described previously (58). Plants were used for further analysis after disruption of transcripts by T-DNA insertions was confirmed by RT-PCR. The primer sequences used are listed in table S13. For generation of sdi1sdi2 double-knockout mutants, the single homozygous knockout mutants were cross-fertilized, and the segregation of T-DNA insertion was analyzed in F2 progenies. F3 progenies without SDI1 and SDI2 expression were named as sdi1sdi2-1, sdi1sdi2-2, sdi1sdi2-3, and sdi1sdi2-4 for sdi1-1sdi2-1, sdi1-1sdi2-2, sdi1-2sdi2-1, sdi1-2sdi2-2, respectively, and were used for further analysis.
SDI-overexpressing plants
SDI overexpression constructs (SDI1ox and SDI2ox) were created by cloning the SDI coding regions under the control of the CaMV 35S promoter in vector pSMAH621 (59) using the In-Fusion HD Cloning Kit (Takara-Clontech) and the primers listed in table S12. Binary plasmids were transferred to A. tumefaciens GV3101 (pMP90) (60, 61) and introduced to Arabidopsis plants according to the floral dip method (62). Transgenic plants were selected on medium containing hygromycin B (25 mg/liter), and T3 progenies were used for the analysis.
Motif search and phylogenetic analysis
Protein sequences of the SDI family proteins in Arabidopsis—SDI1 (At5g48850), SDI2 (At1g04770), At3g51280, MS5 (At4g20900), and MS5-like (At5g44330)—were obtained from TAIR (www.arabidopsis.org). They were aligned using a Clustal W program at the DNA Data Bank of Japan (www.ddbj.nig.ac.jp/search/clustalw-j.html; fig. S1), and the unrooted phylogenetic tree was drawn by neighbor-joining methods using the Geneious software (Biomatters Ltd.). Motifs that existed in SDI family proteins were predicted by InterProScan (www.ebi.ac.uk/interpro/search/sequence-search) (38), Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan) (63), and PROSITE (http://prosite.expasy.org/) (64).
Phylogenetic relationship of SDI family of nine different species was analyzed using the protein sequences of SDI family homologs obtained from the PLAZA website (http://bioinformatics.psb.ugent.be/plaza/versions/plaza_v2_5/) (41). SDI1 wheat sequence was obtained from the National Center for Biotechnology Information. Forty-seven proteins were aligned using Clustal X 2.1 and realigned in MEGA (version 6) by implementing Clustal W (65). The phylogenetic trees were constructed by MEGA using a statistical neighbor-joining method by applying the following parameters: bootstrap method (1000 replicates), Poisson correction, and complete deletion. The phylogenetic tree is shown in fig. S2.
Vector construction of SDI1pro:GFP/GUS and plant transformation
SDI1pro:GFP and SDI2pro:GFP constructs were prepared by cloning the upstream 3.1-kb promoter regions of SDI1 and SDI2 in the pBI101-GFP vector (66), which has a replacement of the β-glucuronidase gene in pBI101 (Clontech) with sGFP. SDI1pro:GUS and SDI2pro:GUS constructs were made using a gateway cassette within a GUS expression vector, pKGWFS7 (Invitrogen) (67). The 5′-upstream 2.0-kb regions of the SDI1 and SDI2 genes were amplified by genomic PCR and cloned into the pKGWFS7 gateway vector. Primer sequences used for the PCR are summarized in table S12. Binary plasmids were transferred to A. tumefaciens GV3101 (pMP90) (60, 61) and introduced to Arabidopsis plants according to the floral dip method (62). Transgenic plants were selected on medium containing kanamycin sulfate (50 mg/liter). We used T2 progenies for GFP analysis and T3 progenies for GUS analysis.
Imaging of GFP expression
The expression of GFP in whole intact Arabidopsis seedlings was visualized by using the image analyzer Typhoon Trio equipped with a 515- to 545-nm band-pass filter and a 488-nm laser (GE Healthcare) (Fig. 1D). Autofluorescence derived from chlorophyll was scanned in parallel using a 610-nm long-pass filter. Localization of GFP, YFP, and autofluorescence of chlorophyll in Arabidopsis protoplasts (Fig. 7A) and BiFC assay of N. benthamiana leaves (Fig. 7C) were determined by a Leica TCS SP5 laser scanning confocal microscope (Leica Microsystems GmbH) using an HCX PL APO CS 63×/1.20 water-corrected objective lens. Excitation wavelengths and emission filters were 488 nm per 505- to 530-nm band pass for GFP, 514 nm per 535/30-nm band pass for YFP, and 488 nm per 650- to 710-nm long pass for chlorophyll autofluorescence. Image analysis was performed with the Leica confocal LAS AF software.
Histochemical GUS assay
Different Arabidopsis plant tissues were immersed in freshly prepared GUS staining solution with a mixture of 100 mM phosphate buffer (pH 7.0), 0.5 mM ferrocyanide, 0.5 mM ferricyanide, 0.1% Triton X-100, and 0.1% X-GlcA. Tissues were incubated overnight at 37°C and destained with several changes of ethanol (68, 69). To analyze the GUS expression, light microscopy was performed using a Leica MZ 12.5 stereomicroscope with a dark-field illumination technique. LAS software version 4.2 (Leica) was used for image processing.
RT-PCR and qRT-PCR
RNA preparation and reverse transcription were performed as reported previously (70). RT-PCR for the determination of SDI1 and SDI2 expression was carried out with Ex Taq (Takara) and GeneAmp 9700 (Applied Biosystems) using the gene-specific primers presented in table S13 (SDI1-F, SDI1-R, SDI2-F, and SDI2-R). qRT-PCR was carried out using SYBR Green Perfect Real Time kit (Takara) or SYBR Select Master Mix (Applied Biosystems) and Thermal Cycler Dice Real Time System (Takara) or ABI PRISM 7900HT Fast Real-Time PCR System (Applied Biosystems). The relative mRNA levels were calculated using ubiquitin 2 (UBQ2) as an internal standard. The primer sequences used are listed in table S14 (29, 32).
Metabolite analysis
Plant tissues were harvested and frozen in liquid nitrogen before extraction of metabolites. Plant extraction was performed as described previously (29). GSL was analyzed by liquid chromatography–mass spectrometry [Acquity UPLC system (Waters) connected to a Q-Tof Premier TOF-MS analyzer (Micromass)], and the amount of each GSL species was determined according to the method described previously (29, 71, 72). Sulfate content was determined by a capillary electrophoresis–photodiode array detection system according to the manufacturer’s protocol (Agilent Technologies). Cys and GSH contents were determined by monobromobimane (Molecular Probes) labeling of the thiols after reduction of the plant extracts by dithiothreitol. The labeled products were analyzed by high-performance liquid chromatography according to the method described previously (29).
GeneChip hybridization and data analysis
WT (Col), two lines of sdi1sdi2 double-knockout mutants (sdi1sdi2-1 and sdi1sdi2-4), and two lines of SDI1 overexpression plants (SDI1ox-6 and SDI1ox-12) were grown for 10 days under +S (1500 μM sulfate) or –S (15 μM sulfate) conditions. Duplicate RNA samples were prepared from root tissues, and hybridization of ATH1-121501.1 array (Affymetrix) was performed according to the manufacturer’s protocol.
Raw cell data were parametrically normalized (42) using the SuperNORM data service (Skylight Biotech Inc.) to obtain z-score for each cell value. Significance of changes in gene expression among the experimental groups was tested by two-way ANOVA (43). The level of significance used for the selection was 0.001. The raw and normalized data were deposited in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81347).
To compare the effects of genetic manipulation and sulfate depletion, we performed PCA on the ANOVA-positive genes (44). Changes in gene expression were displayed as differences in z-scores from the control group, that is, WT grown under +S. To reduce the effects of individual variability among samples, the axes of PCA were obtained on the basis of means of replicated measurements for each group. This methodology allowed us to determine common directions of changes in expression and then settled a set of perpendicular axes that accorded to the directions. Then, it measured PC for each gene and sample, which represented changes in gene expression that were projected on the PCA plot.
Trends of changes in gene expression were estimated in appearance of keywords for particular PC axes. Keywords were found in the “Gene Ontology Biological Process” section of the manufacturer’s annotation sheet (ATH1-121501.na35.annot.csv). Genes specifically expressed toward each PC axis were further selected as follows: distribution of PC scores was checked by using Normal QQ plot, and scale parameter of the distribution was robustly estimated by using median absolute deviation (mad). Genes with either more or less than three times the mad of each PC were selected, and keywords were extracted. For each keyword, frequencies of genes were compared between the selected genes and the whole chip contents. Then, the P value was estimated on binominal distribution model using a null hypothesis; the genes were selected randomly. Keywords of which the P values were less than 0.001 and the frequency in a selected gene greater than 4 (fig. S10A) or 2 (fig. S10B) were listed.
Transient subcellular localization assay
For SDI1:GFP and GFP:SDI1 fusion construct, the full-length coding DNA sequence of SDI1 was amplified with the primers listed in table S12 without including stop or start codons, respectively, and cloned into the TA-pCR2.1 vector (Invitrogen). After the SDI1 coding regions were digested with corresponding enzymes, they were subcloned into the expression vector pA7-GFP (73). GFP fusion constructs were then transiently transformed into isolated Arabidopsis protoplasts (74). Protoplast isolation and transformation were performed via the tape sandwich method (74). Protoplasts were transformed with 20 μg of plasmid DNA and incubated overnight at room temperature until the GFP signal was visible.
BiFC assay
BiFC assays were performed, as previously described, with some modifications (75). Seven constructs were generated in two different GATEWAY-based BiFC vectors, pDEST-VYNE(R)GW and pDEST-GWVYCE (75). Full-length coding cDNA sequences of SDI1, MYB28, MYB29, MYB76, and MYB75 were amplified with the corresponding primers (table S12) and cloned into the pCR8-TOPO TA vector according to the manufacturer’s instructions (pCR8/GW/TOPO TA Cloning Kit, Invitrogen). SDI1 entry vector was subcloned into pDEST-VYNE(R)GW, and MYB28, MYB29, MYB76, and MYB75 were subcloned into pDEST-GWVYCE by performing LR recombination reaction (Invitrogen). A. tumefaciens strain GV3101 harboring either the cassettes of SDI1-nYFP or cYFP-MYB28/MYB29/MYB76/MYB75 vectors was adjusted to equal concentration and volume (optical density at 600 nm = 0.4) in the infiltration medium (pH 5.6; 0.2 mM acetosyringone, 10 mM MgCl2, and 10 mM MES) and, after incubation for 2 hours at room temperature (75-rpm shaking), co-infiltrated into 4-week-old N. benthamiana leaves. Each combination of constructs was infiltrated in separate leaves. Three days after the infiltration, the lower epidermis of N. benthamiana leaves were subjected to confocal microscopy for detection of YFP signal.
Y2H analysis
Y2H screening was performed using the Matchmaker Gold Yeast Two-Hybrid System according to the manufacturer’s instructions (Clontech). To generate the bait and prey constructs, the coding sequences of SDI1, MYB28, MYB29, and MYB76 were PCR-amplified using the primers listed in table S12 and directionally cloned into the pGBKT7 and pGADT7 (Clontech) vectors using the In-Fusion HD Cloning Kit according to the manufacturer’s instructions (Clontech). One hundred nanograms of pGBKT7-SDI1 (SDI1-BD) and pGADT7-MYBs (MYB28/MYB29/MYB76-AD) vectors was introduced into Y2HGold yeast strains using Yeastmaker Yeast Transformation System 2 (Clontech). Negative (pGBKT7-Lam and pGADT7-T) and positive (pGADT7-53 and pGBKT7-Lam) controls were used according to the manufacturer’s instructions. The Y2HGold yeast strains cotransformed with pGBKT7 and MYB28/MYB29/MYB76-AD were used as additional negative controls. Yeast harboring bait and prey vectors were plated on double dropout medium containing X-α-Gal (40 μg/ml; -Leu/Trp/X = DDO/X) and quadruple dropout medium containing X-α-Gal (40 μg/ml) and Aureobasidin A (200 ng/ml; -Leu/Trp/Ade/His/X/A = QDO/X/A). To verify the growth of positive colonies, 3 days after transformation, target colonies were selected and resuspended in double-distilled water and dropped on DDO, QDO, DDO/X, and QDO/X/A.
Transactivation assays in cultured Arabidopsis thaliana cells
The promoter sequences of CYP83A1 [−919 to +63 base pairs (bp)] and CYP79F1 (−1388 to +45 bp) cloned into the binary plant transformation vector pGWB3i (47, 76) were used as reporter constructs. MYB28 and SDI1 that were fused to the CaMV 35S promoter in pGWB2 were used as effector constructs (47). The reporter and effector constructs were transferred to the supervirulent Agrobacterium strain LBA4404 pBBR1MCS.virGN54D (provided by J. Memelink, University of Leiden, Netherlands). For transient expression assays in the cell culture, Agrobacterium containing the effector constructs, the antisilencing 19-K protein, or one of the reporter constructs were taken from fresh yeast extract broth (YEB) plates, grown overnight, and resuspended in 1 ml of AT medium. To control the transfection rate in wells with different technical and biological replicates, the Agrobacterium strain containing the 35S:LUC construct was cocultivated together with the Agrobacterium strains containing the reporter, effector, and antisilencing constructs in cultured plant cells. These Agrobacterium strains were mixed in a 1:1:1:1 ratio, and 100 μl of this suspension was added to 3 ml of cultured A. thaliana root cells, which were then grown for 5 days in the dark and subsequently used for LUC and GUS activity measurements or GUS staining.
Electrophoretic mobility shift assays
His-tagged SDI1 and MYB28 proteins were synthesized with wheat germ cell–free system using the pEU-E01-MCS vector (WEPRO7240H Expression Kit, CellFree Sciences) according to the manufacturer’s instructions. Protein synthesis was performed in a 1.2-ml volume. After purification of the proteins using a Ni Sepharose 6 Fast Flow (GE Healthcare), the solvent [20 mM sodium phosphate (pH 7.5), 0.5 M NaCl, and 0.5 M imidazole] was replaced with storage buffer [10 mM tris-HCl (pH 7.5), 50 mM KCl, 1 mM dithiothreitol, and 10% glycerol] using Amicon Ultra Centrifugal Filters (Ultracel 10K, Millipore) to a final volume of 100 μl.
Oligonucleotide probes were labeled using the Biotin 3′ End DNA Labeling Kit (Thermo Scientific). EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific). Biotin-labeled probes (20 fmol) were incubated in 1× binding buffer, 2.5% glycerol, 50 mM KCl, 5 mM MgCl2, and 10 mM EDTA with or without SDI1 and/or MYB28 proteins at 4°C for 20 min. The probe sequences were as follows: CYP79F1, 5′-ATAAACGTGTTCacctacCCAAGGTATTATTTATTTATTC-3′; CYP79F1mt, 5′-ATAAACGTGTTCcccgccCCAAGGTATTATTTATTTATTC-3′; CYP83A1, 5′-ACACGTGAGGTTTCGTAAGTAGGTATAAGGGAGTAACCATTTGATTAAAAATTTGATGTcggttaAG-3′; CYP83A1mt, 5′-ACACGTGAGGTTTCGTAAGTAGGTATAAGGGAGTAACCATTTGATTAAAAATTTGATGTgaacgtAG-3′.
Supplementary Material
Acknowledgments
We acknowledge the Arabidopsis Biological Resource Center and Salk Institute Genomic Analysis Laboratory for providing the T-DNA insertion lines of SDI1 and SDI2. We thank Y. Okuo, A. Hayashi, A. Onoda, K. Nakabayashi, C. Komori, C. Uchiyama, K. Nojima, and Y. Suzuki for growing the plants; Y. Niwa (University of Shizuoka, Japan) for providing sGFP (S65T) vector; E. Maximova for help with microscopy; M. Rauf for providing the GFP nuclear control construct (NAC104); I. Kasprzyk for the help in Y2H assay; and J. Bergstein for photography. Funding: This research was supported by Japan Society for the Promotion of Science KAKENHI grants 24380040 and 20770044; Kyushu University Interdisciplinary Programs in Education and Projects in Research Development grant 23305 (to A.M.-N.); the NOVARTIS Foundation (Japan) for the Promotion of Science (to A.M.-N.); the Tojuro Iijima Foundation for Food Science and Technology (to A.M.-N.); Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (to H.N.); the Max-Planck Society (to R.H. and A.R.F.); the Ministry of Science, Research and Technology of Iran (to F.A.); NSF grant MCB-1244300; and AgBioResearch grant MICL02313 (to H.T.). Author contributions: A.M.-N., H.T., H.-M.H., F.A., and R.H. designed the research. A.M.-N. isolated single and double knockouts, generated overexpression lines performed qRT-PCR and microarray analysis and prepared plant samples for metabolite analysis and tissue distribution using the Pro-GFP lines. F.A. isolated single and double knockouts, generated overexpression lines, and performed qRT-PCR, subcellular localization assay, Y2H, BiFC, and tissue distribution analysis using the Pro-GUS lines. M.K. and H.N. performed EMSA. T.T., M.W., K.S., and A.R.F. performed GSL analysis. T.K. performed statistical analysis of microarray data. T.G. performed transactivation assay. M.T. isolated double-knockout lines, prepared plant samples, and performed qRT-PCR. Y.S. performed sulfate, Cys, and GSH analysis. H.-M.H. isolated single-knockout lines. F.A., M.K., T.T., T.K., T.G., M.T., Y.S., M.W., H.N., A.R.F., K.S., H.T., H.-M.H., R.H., and A.M.-N. analyzed data. A.M.-N. and F.A. wrote the paper. H.T., A.R.F., and R.H. edited the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The raw and normalized data of microarray analysis were deposited in Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81347). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Plant materials used in this study can be distributed upon request to the corresponding author.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/10/e1601087/DC1
fig. S1. Alignment of SDI family proteins in Arabidopsis.
fig. S2. Phylogenetic relationships of SDI family proteins in some monocot and dicot species.
fig. S3. Histochemical staining of Arabidopsis plants transformed with SDI1pro:GUS and SDI2pro:GUS.
fig. S4. Transcript levels of CYP79F1 and CYP83A1 were influenced by SDI1 and SDI2 similar with other mGSL synthesis genes.
fig. S5. Perturbation of SDI1 and SDI2 influences on the transcript levels of genes involved in primary sulfur metabolism.
fig. S6. Frequency plot of genes according to P values.
fig. S7. Contribution of each PC.
fig. S8. Distribution of PC1 (A) and PC2 (B) scores.
fig. S9. PC1 shows linear correlation with −S-responsive gene expression.
fig. S10. Keywords that frequently appeared in genes with high scores in PC2.
fig. S11. Effects of the manipulation of MYB28, MYB29, and MYB76 on gene expression of SDI1, SDI2, and other factors.
table S1. MT, MS, indolic, and total GSL contents in shoot and root tissues of WT, sdi1, sdi2, and sdi1sdi2.
table S2. Fold changes of GSL contents in shoot and root tissues of sdi1, sdi2, and sdi1sdi2 versus WT.
table S3. MT, MS, indolic, and total GSL contents in shoot and root tissues of WT and SDI1- and SDI2-overexpressing lines.
table S4. Fold changes of GSL contents in shoot and root tissues of SDI1ox and SDI2ox versus WT.
table S5. Sulfate, Cys, and GSH contents in shoot and root tissues of sdi knockouts and overexpressing lines.
table S6. Transcript levels of genes highly contributed to PC1.
table S7. Transcript levels of genes highly contributed to PC2.
table S8. Transcript levels of GSL-related genes in root tissues of WT, sdi1sdi2, and SDI1ox detected by microarray analysis.
table S9. Transcript levels of sulfur assimilatory genes in root tissues of WT, sdi1sdi2, and SDI1ox detected by microarray analysis.
table S10. Transcript levels and gene PCs of genes selected for PCA.
table S11. Transcript levels of GSL-related genes in root tissues of parental, slim1-1, and slim1-2 detected by microarray analysis.
table S12. Oligonucleotides used for the vector construction.
table S13. Oligonucleotides used for the isolation of the T-DNA insertion lines.
table S14. Oligonucleotides used for qRT-PCR analysis.
REFERENCES AND NOTES
- 1.Droux M., Sulfur assimilation and the role of sulfur in plant metabolism: A survey. Photosynth. Res. 79, 331–348 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Saito K., Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 136, 2443–2450 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi H., Kopriva S., Giordano M., Saito K., Hell R., Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184 (2011). [DOI] [PubMed] [Google Scholar]
- 4.de Kraker J.-W., Gershenzon J., From amino acid to glucosinolate biosynthesis: Protein sequence changes in the evolution of methylthioalkylmalate synthase in Arabidopsis. Plant Cell 23, 38–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clay N. K., Adio A. M., Denoux C., Jander G., Ausubel F. M., Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bednarek P., Piślewska-Bednarek M., Svatoš A., Schneider B., Doubský J., Mansurova M., Humphry M., Consonni C., Panstruga R., Sanchez-Vallet A., Molina A., Schulze-Lefert P., A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Fan J., Crooks C., Creissen G., Hill L., Fairhurst S., Doerner P., Lamb C., Pseudomonas sax genes overcome aliphatic isothiocyanate–mediated non-host resistance in Arabidopsis. Science 331, 1185–1188 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., Yao S., Li J., Vegetable-derived isothiocyanates: Anti-proliferative activity and mechanism of action. Proc. Nutr. Soc. 65, 68–75 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Ambrosone C. B., Tang L., Cruciferous vegetable intake and cancer prevention: Role of nutrigenetics. Cancer Prev. Res. 2, 298–300 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Halkier B. A., Gershenzon J., Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Sønderby I. E., Geu-Flores F., Halkier B. A., Biosynthesis of glucosinolates—Gene discovery and beyond. Trends Plant Sci. 15, 283–290 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Gigolashvili T., Yatusevich R., Berger B., Müller C., Flügge U.-I., The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 51, 247–261 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Hirai M. Y., Sugiyama K., Sawada Y., Tohge T., Obayashi T., Suzuki A., Araki R., Sakurai N., Suzuki H., Aoki K., Goda H., Nishizawa O. I., Shibata D., Saito K., Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 6478–6483 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celenza J. L., Quiel J. A., Smolen G. A., Merrikh H., Silvestro A. R., Normanly J., Bender J., The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 137, 253–262 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gigolashvili T., Berger B., Mock H.-P., Müller C., Weisshaar B., Flügge U.-I., The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50, 886–901 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Frerigmann H., Gigolashvili T., MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 7, 814–828 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Sønderby I. E., Burow M., Rowe H. C., Kliebenstein D. J., Halkier B. A., A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiol. 153, 348–363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernández-Calvo P., Chini A., Fernández-Barbero G., Chico J.-M., Gimenez-Ibanez S., Geerinck J., Eeckhout D., Schweizer F., Godoy M., Franco-Zorrilla J. M., Pauwels L., Witters E., Puga M. I., Paz-Ares J., Goossens A., Reymond P., De Jaeger G., Solano R., The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schweizer F., Fernández-Calvo P., Zander M., Diez-Diaz M., Fonseca S., Glauser G., Lewsey M. G., Ecker J. R., Solano R., Reymond P., Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25, 3117–3132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frerigmann H., Berger B., Gigolashvili T., bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 166, 349–369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kliebenstein D. J., Kroymann J., Mitchell-Olds T., The glucosinolate–myrosinase system in an ecological and evolutionary context. Curr. Opin. Plant Biol. 8, 264–271 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Falk K. L., Tokuhisa J. G., Gershenzon J., The effect of sulfur nutrition on plant glucosinolate content: Physiology and molecular mechanisms. Plant Biol. 9, 573–581 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Frerigmann H., Gigolashvili T., Update on the role of R2R3-MYBs in the regulation of glucosinolates upon sulfur deficiency. Front. Plant Sci. 5, 626 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirai M. Y., Klein M., Fujikawa Y., Yano M., Goodenowe D. B., Yamazaki Y., Kanaya S., Nakamura Y., Kitayama M., Suzuki H., Sakurai N., Shibata D., Tokuhisa J., Reichelt M., Gershenzon J., Papenbrock J., Saito K., Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 280, 25590–25595 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Hirai M. Y., Fujiwara T., Awazuhara M., Kimura T., Noji M., Saito K., Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 33, 651–663 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Nikiforova V., Freitag J., Kempa S., Adamik M., Hesse H., Hoefgen R., Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: Interlacing of biosynthetic pathways provides response specificity. Plant J. 33, 633–650 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Maruyama-Nakashita A., Inoue E., Watanabe-Takahashi A., Yarnaya T., Takahashi H., Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 132, 597–605 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maruyama-Nakashita A., Nakamura Y., Watanabe-Takahashi A., Inoue E., Yamaya T., Takahashi H., Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 42, 305–314 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Maruyama-Nakashita A., Nakamura Y., Tohge T., Saito K., Takahashi H., Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18, 3235–3251 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikiforova V. J., Kopka J., Tolstikov V., Fiehn O., Hopkins L., Hawkesford M. J., Hesse H., Hoefgen R., Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol. 138, 304–318 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nour-Eldin H. H., Andersen T. G., Burow M., Madsen S. R., Jørgensen M. E., Olsen C. E., Dreyer I., Hedrich R., Geiger D., Halkier B. A., NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488, 531–534 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Sawada Y., Hirai A., Sato M., Kuwahara A., Yan X., Hirai M. Y., Novel insights into the function of Arabidopsis R2R3-MYB transcription factors regulating aliphatic glucosinolate biosynthesis. Plant Cell Physiol. 54, 1335–1344 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Howarth J. R., Parmar S., Barraclough P. B., Hawkesford M. J., A sulphur deficiency-induced gene, sdi1, involved in the utilization of stored sulphate pools under sulphur-limiting conditions has potential as a diagnostic indicator of sulphur nutritional status. Plant Biotechnol. J. 7, 200–209 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Hubberten H.-M., Klie S., Caldana C., Degenkolbe T., Willmitzer L., Hoefgen R., Additional role of O-acetylserine as a sulfur status-independent regulator during plant growth. Plant J. 70, 666–677 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Hirai M. Y., Yano M., Goodenowe D. B., Kanaya S., Kimura T., Awazuhara M., Arita M., Fujiwara T., Saito K., Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101, 10205–10210 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan E. K. F., Rowe H. C., Corwin J. A., Joseph B., Kliebenstein D. J., Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLOS Biol. 9, e1001125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T. K., Bateman A., Bernard T., Binns D., Bork P., Burge S., de Castro E., Coggill P., Corbett M., Das U., Daugherty L., Duquenne L., Finn R. D., Fraser M., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., McMenamin C., Mi H., Mutowo-Muellenet P., Mulder N., Natale D., Orengo C., Pesseat S., Punta M., Quinn A. F., Rivoire C., Sangrador-Vegas A., Selengut J.D., Sigrist C. J. A., Scheremetjew M., Tate J., Thimmajanarthanan M., Thomas P. D., Wu C. H., Yeats C., Yong S.-Y., InterPro in 2011: New developments in the family and domain prediction database. Nucl. Acids Res. 40, D306–D312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamb J. R., Tugendreich S., Hieter P., Tetratrico peptide repeat interactions: To Tpr or not to Tpr. Trends Biochem. Sci. 20, 257–259 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Ross K. J., Fransz P., Armstrong S. J., Vizir I., Mulligan B., Franklin F. C. H., Jones G. H., Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Res. 5, 551–559 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Bulankova P., Riehs-Kearnan N., Nowack M. K., Schnittger A., Riha K., Meiotic progression in Arabidopsis is governed by complex regulatory interactions between SMG7, TDM1, and the meiosis I–specific cyclin TAM. Plant Cell 22, 3791–3803 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Proost S., Van Bel M., Sterck L., Billiau K., Van Parys T., Van de Peer Y., Vandepoele K., PLAZA: A comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21, 3718–3731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Konishi T., Konishi F., Takasaki S., Inoue K., Nakayama K., Konagaya A., Coincidence between transcriptome analyses on different microarray platforms using a parametric framework. PLOS One 3, e3555 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konishi T., Microarray test results should not be compensated for multiplicity of gene contents. BMC Syst. Biol. 5 (suppl. 2), S6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konishi T., Principal component analysis for designed experiments. BMC Bioinformatics 16 (suppl. 18), S7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelemen Z., Sebastian A., Xu W., Grain D., Salsac F., Avon A., Berger N., Tran J., Dubreucq B., Lurin C., Lepiniec L., Contreras-Moreira B., Dubos C., Analysis of the DNA-binding activities of the Arabidopsis R2R3-MYB transcription factor family by one-hybrid experiments in yeast. PLOS One 10, e0141044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pireyre M., Burow M., Regulation of MYB and bHLH transcription factors: A glance at the protein level. Mol. Plant 8, 378–388 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Gigolashvili T., Engqvist M., Yatusevich R., Müller C., Flügge U.-I., HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 177, 627–642 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Li B., Gaudinier A., Tang M., Taylor-Teeples M., Nham N. T., Ghaffari C., Benson D. S., Steinmann M., Gray J. A., Brady S. M., Kliebenstein D. J., Promoter-based integration in plant defense regulation. Plant Physiol. 166, 1803– 1820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reintanz B., Lehnen M., Reichelt M., Gershenzon J., Kowalczyk M., Sandberg G., Godde M., Uhl R., Palme K., bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13, 351–367 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao C., Craig J. C., Petzold H. E., Dickerman A. W., Beers E. P., The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138, 803–818 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shroff R., Vergara F., Muck A., Svatoš A., Gershenzon J., Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. U.S.A. 105, 6196–6201 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sønderby I. E., Hansen B. G., Bjarnholt N., Ticconi C., Halkier B. A., Kliebenstein D. J., A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLOS One 2, e1322 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlaeppi K., Bodenhausen N., Buchala A., Mauch F., Reymond P., The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis. Plant J. 55, 774–786 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Yatusevich R., Mugford S. G., Matthewman C., Gigolashvili T., Frerigmann H., Delaney S., Koprivova A., Flügge U.-I., Kopriva S., Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana. Plant J. 62, 1–11 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Mugford S. G., Yoshimoto N., Reichelt M., Wirtz M., Hill L., Mugford S. T., Nakazato Y., Noji M., Takahashi H., Kramell R., Gigolashvili T., Flügge U.-I., Wasternack C., Gershenzon J., Hell R., Saito K., Kopriva S., Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21, 910–927 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bekaert M., Edger P. P., Hudson C. M., Pires J. C., Conant G. C., Metabolic and evolutionary costs of herbivory defense: Systems biology of glucosinolate synthesis. New Phytol. 196, 596–605 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Hirai M. Y., Fujiwara T., Chino M., Naito S., Effects of sulfate concentrations on the expression of a soybean seed storage protein gene and its reversibility in transgenic Arabidopsis thaliana. Plant Cell Physiol. 36, 1331–1339 (1995). [PubMed] [Google Scholar]
- 58.Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H. M., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., Parker H., Prednis L., Ansari Y., Choy N., Deen H., Geralt M., Hazari N., Hom E., Karnes M., Mulholland C., Ndubaku R., Schmidt I., Guzman P., Aguilar-Henonin L., Schmid M., Weigel D., Carter D. E., Marchand T., Risseeuw E., Brogden D., Zeko A., Crosby W. L., Berry C. C., Ecker J. R., Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T., Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koncz C., Schell J., The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396 (1986). [Google Scholar]
- 61.Weigel D., Glazebrook J., Transformation of Agrobacterium using electroporation. Cold Spring Harb. Protoc. 2006, 22484681 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Rice P., Longden I., Bleasby A., EMBOSS: The European molecular biology open software suite. Trends Genet. 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- 64.Sigrist C. J. A., Cerutti L., de Castro E., Langendijk-Genevaux P. S., Bulliard V., Bairoch A., Hulo N., PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 38, D161–D166 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahashi H., Watanabe-Takahashi A., Smith F. W., Blake-Kalff M., Hawkesford M. J., Saito K., The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 23, 171–182 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Karimi M., Inzé D., Depicker A., GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002). [DOI] [PubMed] [Google Scholar]
- 68.S. Vitha, K. Beneš, J. Phillips, K. M. A. Gartland, in Agrobacterium Protocols, K. A. Gartland, M. Davey, Eds. (Springer, 1995), vol. 44, pp. 185–193.
- 69.Jefferson R. A., Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405 (1987). [Google Scholar]
- 70.Maruyama-Nakashita A., Nakamura Y., Yamaya T., Takahashi H., A novel regulatory pathway of sulfate uptake in Arabidopsis roots: Implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 38, 779–789 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Tohge T., Fernie A. R., Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protoc. 5, 1210–1227 (2010). [DOI] [PubMed] [Google Scholar]
- 72.Tohge T., Fernie A. R., Web-based resources for mass-spectrometry-based metabolomics: A user’s guide. Phytochemistry 70, 450–456 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Voelker C., Schmidt D., Mueller-Roeber B., Czempinski K., Members of the Arabidopsis AtTPK/KCO family form homomeric vacuolar channels in planta. Plant J. 48, 296–306 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Wu F.-H., Shen S.-C., Lee L.-Y., Lee S.-H., Chan M.-T., Lin C.-S., Tape-Arabidopsis Sandwich - A simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gehl C., Waadt R., Kudla J., Mendel R.-R., Hänsch R., New GATEWAY vectors for high throughput analyses of protein–protein interactions by bimolecular fluorescence complementation. Mol. Plant 2, 1051–1058 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Berger B., Stracke R., Yatusevich R., Weisshaar B., Flügge U.-I., Gigolashvili T., A simplified method for the analysis of transcription factor-promoter interactions that allows high-throughput data generation. Plant J. 50, 911–916 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Maruyama-Nakashita A., Hirai M. Y., Funada S., Fueki S., Exogenous application of 5-aminolevulinic acid increases the transcript levels of sulfur transport and assimilatory genes, sulfate uptake, and cysteine and glutathione contents in Arabidopsis thaliana. Soil Sci. Plant Nutr. 56, 281–288 (2010). [Google Scholar]
- 78.Maruyama-Nakashita A., Watanabe-Takahashi A., Inoue E., Yamaya T., Saito K., Takahashi H., Sulfur-responsive elements in the 3′-nontranscribed intergenic region are essential for the induction of SULFATE TRANSPORTER 2;1 gene expression in Arabidopsis roots under sulfur deficiency. Plant Cell 27, 1279–1296 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/10/e1601087/DC1
fig. S1. Alignment of SDI family proteins in Arabidopsis.
fig. S2. Phylogenetic relationships of SDI family proteins in some monocot and dicot species.
fig. S3. Histochemical staining of Arabidopsis plants transformed with SDI1pro:GUS and SDI2pro:GUS.
fig. S4. Transcript levels of CYP79F1 and CYP83A1 were influenced by SDI1 and SDI2 similar with other mGSL synthesis genes.
fig. S5. Perturbation of SDI1 and SDI2 influences on the transcript levels of genes involved in primary sulfur metabolism.
fig. S6. Frequency plot of genes according to P values.
fig. S7. Contribution of each PC.
fig. S8. Distribution of PC1 (A) and PC2 (B) scores.
fig. S9. PC1 shows linear correlation with −S-responsive gene expression.
fig. S10. Keywords that frequently appeared in genes with high scores in PC2.
fig. S11. Effects of the manipulation of MYB28, MYB29, and MYB76 on gene expression of SDI1, SDI2, and other factors.
table S1. MT, MS, indolic, and total GSL contents in shoot and root tissues of WT, sdi1, sdi2, and sdi1sdi2.
table S2. Fold changes of GSL contents in shoot and root tissues of sdi1, sdi2, and sdi1sdi2 versus WT.
table S3. MT, MS, indolic, and total GSL contents in shoot and root tissues of WT and SDI1- and SDI2-overexpressing lines.
table S4. Fold changes of GSL contents in shoot and root tissues of SDI1ox and SDI2ox versus WT.
table S5. Sulfate, Cys, and GSH contents in shoot and root tissues of sdi knockouts and overexpressing lines.
table S6. Transcript levels of genes highly contributed to PC1.
table S7. Transcript levels of genes highly contributed to PC2.
table S8. Transcript levels of GSL-related genes in root tissues of WT, sdi1sdi2, and SDI1ox detected by microarray analysis.
table S9. Transcript levels of sulfur assimilatory genes in root tissues of WT, sdi1sdi2, and SDI1ox detected by microarray analysis.
table S10. Transcript levels and gene PCs of genes selected for PCA.
table S11. Transcript levels of GSL-related genes in root tissues of parental, slim1-1, and slim1-2 detected by microarray analysis.
table S12. Oligonucleotides used for the vector construction.
table S13. Oligonucleotides used for the isolation of the T-DNA insertion lines.
table S14. Oligonucleotides used for qRT-PCR analysis.


