SUMMARY
The rate of glycolytic metabolism changes during differentiation of human embryonic stem cells (hESCs) and reprogramming of somatic cells to pluripotency. However, the functional contribution of glycolytic metabolism to the pluripotent state is unclear. Here we show that naive hESCs exhibit increased glycolytic flux, MYC transcriptional activity, and nuclear N-MYC localization relative to primed hESCs. This status is consistent with the inner cell mass of human blastocysts, where MYC transcriptional activity and nuclear N-MYC levels are also higher than in primed hESCs. Reduction of glycolysis decreases self-renewal of naive hESCs and feeder-free primed hESCs, but not primed hESCs grown in feeder-supported conditions. Reduction of glycolysis in feeder-free primed hESCs also enhances neural specification. These findings reveal associations between glycolytic metabolism and human naive pluripotency and differences in the metabolism of feeder-/feeder-free cultured hESCs. They may also suggest methods for regulating self-renewal and initial cell fate specification of hESCs.
INTRODUCTION
An association between glycolysis and pluripotency is well-established (Folmes et al., 2012a, 2012b, 2013; Zhang et al., 2011, 2012). Cultured pluripotent human embryonic stem cells (hESCs) exhibit high rates of glycolysis that diminish upon differentiation (Chung et al., 2010; Prigione et al., 2010). Additionally, somatic cells exhibit increased glycolysis and decreased respiration upon reprogramming into pluripotent stem cells (Folmes et al., 2013, 2011; Zhang et al., 2012). However, the role of glycolytic metabolism in the ability of hESCs to undergo self-renewal or differentiation is not well understood.
Recently several methods have been developed to allow for a conversion from the typical hESC state of pluripotency to a more naive state, akin to that found in mouse ESCs (Chan et al., 2013; Gafni et al., 2013; Takashima et al., 2014; Theunissen et al., 2014; Valamehr et al., 2014; Ware et al., 2014). Conventional hESCs, now considered to be in the primed pluripotent state, share molecular and functional properties with epiblast stem cells as described in mouse development, whereas naive hESCs are thought to better represent cells found in the inner cell mass of an embryo (Gafni et al., 2013; Theunissen et al., 2014; Ware et al., 2014). Culturing of naive hESCs represents a major advance to regenerative medicine since the ability to produce “clones” of human pluripotent stem cells is severely hampered in primed hESCs and induced pluripotent stem cells (hiPSCs), limiting the opportunities to perform genomic manipulation by homologous recombination or CRISPR/Cas systems. The naive state of pluripotency is defined by expression of a specific set of pluripotency genes, genome-wide chromatin changes such as DNA hypomethylation, and the ability to survive plating at clonal density. While extensive effort has established the metabolic state of primed hESCs as characterized by enhanced glycolysis and decreased respiration, the metabolic state of naive cells remains less well understood. Recently Takashima et al. showed induction of oxidative phosphorylation pathways and changes in mitochondrial depolarization in human naive cells (Takashima et al., 2014), and Sperber et al. demonstrated naive and primed cells differ significantly in their metabolome, affecting their epigenetic landscapes (Sperber et al., 2015), but neither specifically measured glycolytic rate, utilization of glucose molecules, or the regulation of glycolysis in naive hESCs. Further characterization of naive cell metabolism may reveal additional defining characteristics of the naive state and improve our understanding of the links between metabolism and pluripotency.
Primed hESC lines are made from blastocyst stage embryos, and were first isolated and plated into specialized media conditions that required fibroblast feeders for support (Thomson et al., 1998). However, over the past decade, development of new media compilations involving supra-physiologic amounts of fibroblast growth factor have enabled culturing of human ESCs in “feeder-free” conditions. The ability to maintain and grow hESCs and hiPSCs in feeder-free defined media has substantially improved the consistency and simplicity of both culture and differentiation (Lu et al., 2006; Peiffer et al., 2008; Rajala et al., 2010). However, a complete accounting of physiological differences of hESCs in feeder versus feeder-free culture is currently lacking. While both culture systems appear to maintain the pluripotent state, it is critical to know what physiological differences are prevalent, especially as hESCs and hiPSCs grown in defined feeder-free conditions move toward clinical applications.
Here we investigate glucose metabolism in naive versus primed hESCs, in primed hESCs across culture systems, and the role of glycolytic metabolism in hESC self-renewal capacity, pluripotency, and differentiation capacity. In so doing, we make important insights about the metabolism of cells at different stages of pluripotency, and develop new methods that manipulate metabolism to influence self-renewal and cell fate specification of human pluripotent stem cells.
RESULTS
Naive hESCs exhibit increased glycolysis
Consistent with an association between glycolytic metabolism and the pluripotent state (Folmes et al., 2012b; Varum et al., 2011), we found that retinoic acid-induced differentiation of primed hESCs results in decreased glucose consumption (Figure 1A), decreased lactate production (Figure 1B), and increased oxygen consumption rates (Figure S1A). These results suggest a shift away from glycolytic metabolism and towards oxidative metabolism during retinoic acid-induced differentiation of hESCs into the three primordial germ layers.
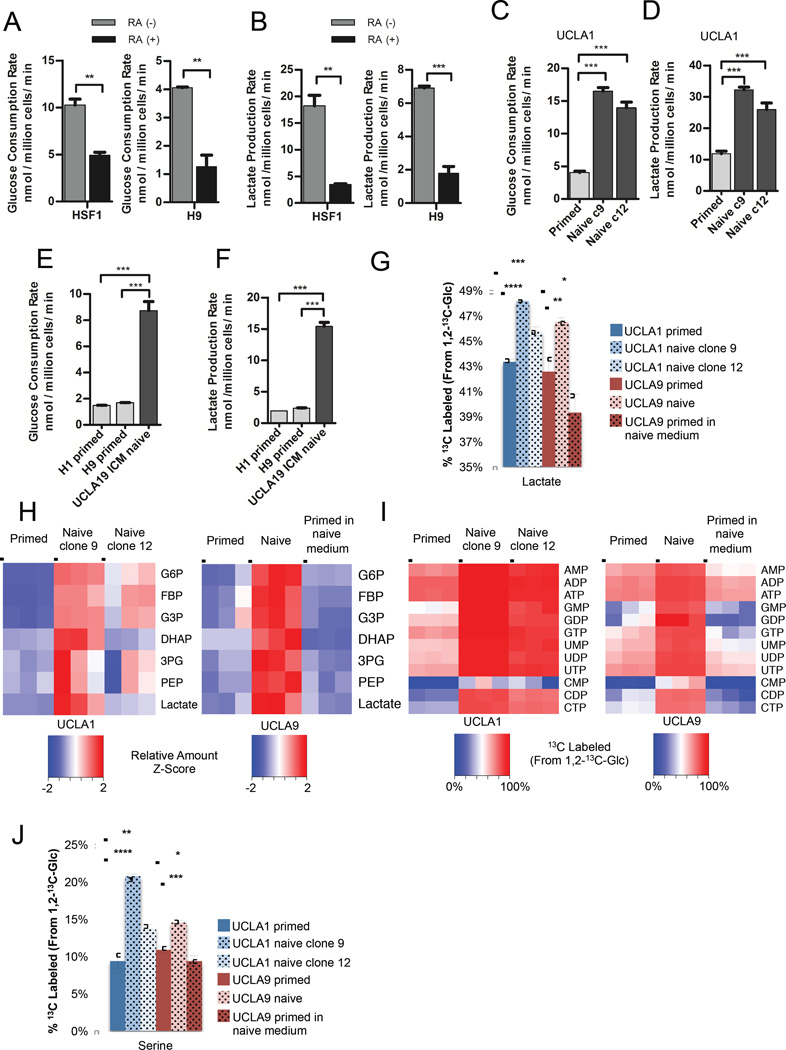
Figure 1. Naive hESCs exhibit increased glycolysis.
Glucose consumption rates (A) and lactate production rates (B) of primed feeder-free HSF1 and H9 hESCs treated for 7 days with DMSO (RA (-)) or 10 µM retinoic acid (RA (+)). Glucose consumption rates (C) and lactate production rates (D) of primed UCLA1 hESCs and naive UCLA1 hESC clone 9 and clone 12 generated by the 5i/LAF method. Glucose consumption rates (E) and lactate production rates (F) of primed H1 and H9 hESCs, and naive UCLA19 hESCs derived from the inner cell mass of a human blastocyst and cultured in 5i/LAF medium. For (G) – (J), primed UCLA1 hESCs, naive UCLA1 hESC clone 9 and clone 12, and primed UCLA9 hESCs, naive UCLA9 hESC, and primed UCLA9 hESCs placed in naive cell medium, were cultured in medium containing 1,2-13C-glucose for 24 hours prior to metabolite extraction and analysis by LC-MS. (G) Percentages of 13C-labeled lactate extracted from the indicated cells. (H) Relative amounts of glycolytic intermediates, glucose-6-phosphate (G6P), fructose bisphosphate (FBP), glyceraldehyde-3-phosphate (G3P), dihydroxyacetone phosphate (DHAP), 3-phosphoglycerate (3PG), phosphoenolpyruvate (PEP), and lactate, from the indicated cells. The heatmap shows standardized amounts of indicated metabolites across samples (Z score). (I) Percentages of 13C-labeled nucleotides extracted from the indicated cells. (J) Percentages of 13C-labeled serine extracted from the indicated cells. For (A) – (G), and (J), error bars indicate ± 1 SEM of biological replicates (n = 3). * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. See also Figure S1.
Since glycolytic metabolism changes at different developmental stages, we hypothesized that glycolytic rate may vary between naive and primed human pluripotent stem cells. To determine whether conversion of primed hESCs to the naive pluripotent state impacts glycolytic rate, we induced primed hESC lines UCLA1 and UCLA9 to a naive state by the 5i/LAF method (Theunissen et al., 2014; Pastor et al., 2016). Sub-clones of naive cells were isolated, and the metabolism of these naive cells was compared to the primed hESCs from which they were derived. Notably, both clones of naive UCLA1 and UCLA9 hESCs exhibit higher glucose consumption and lactate production rates (Figures 1C and 1D and Figure S1B) than the primed cells from which they were derived. To further examine whether increased glycolytic rate is associated with naive pluripotency, we derived a naive cell line UCLA19 directly from the inner cell mass of a human blastocyst in 5i/LAF medium. In this naive line that was never exposed to primed culture conditions, we also found increased glucose consumption and lactate production rates relative to two primed hESC lines, H1 and H9 (Figures 1E and 1F). Additionally, we found that naive/reset hESCs derived using the Takashima et al. method (Takashima et al., 2014) exhibit increased glycolytic rates compared to the primed hESCs from which they were derived (Figure S1C). Consistent with the induction of the naive state by Takashima et al., we also observed an increase in oxygen consumption in our 5i/LAF induced naive lines compared to their primed counterparts (Fig S1D). These results support a strong association between glycolytic metabolism and pluripotency and suggest that modulation of glucose metabolism may be involved in the acquisition of a naive state.
To further characterize how the glucose metabolism of naive hESCs differs from that of primed cells, we labeled UCLA1 and UCLA9 naive and primed hESCs with 1,2-13C-glucose and traced the incorporation of 13C into downstream glucose metabolites using liquid chromatography-mass spectrometry (LC-MS). Consistent with elevated glycolytic flux in naive hESCs, we found increased 13C incorporation into lactate (Figure 1G) and increased levels of most glycolytic intermediates (Figures 1H and S1E) in naive versus primed UCLA1 and UCLA9 hESCs. Importantly, primed UCLA9 hESCs placed in naive cell medium for 24 hours did not exhibit increased lactate production rate (Figure S1B), increased 13C-labeling of lactate (Figure 1G), or increased glycolytic intermediate levels (Figure 1H and Figure S1E). This suggested that the increased glycolysis observed in naive hESCs is associated with acquisition of naive cell identity, and was not a result of factor(s) in the media used to derive the naive hESCs. Notably, we also found that naive hESCs derived from two different hESC lines, UCLA1 and UCLA9, incorporate more glucose carbons into purine and pyrimidine nucleotides (Figure 1I and Figure S1F). The increased M1 isotopologues of nucleotides from naive hESCs labeled with 1,2-13C-glucose suggests increased flux through the oxidative pentose phosphate pathway in naive versus primed hESCs (Figure S1G and S1H). Additionally, we found that UCLA1 and UCLA9 naive hESCs incorporate more glucose carbons into serine than primed hESCs (Figure 1J), an important metabolite for one one-carbon metabolism and purine and glutathione biosynthesis (Locasale, 2013). Unlike naive hESCs, primed UCLA9 hESCs placed in naive cell medium for 24 hours do not exhibit increased 13C-labeling of nucleotides or serine (Figures 1I and 1J and Figure S1F). Collectively, these results suggest that acquisition of the naive state is accompanied by a further increase glucose metabolism and the use of glucose to generate nucleotides and serine.
N-MYC is associated with human naive pluripotency
Given our data that the metabolic phenotype of naive cells is characterized by increased glucose metabolism, we next performed gene set enrichment analysis (GSEA) (Subramanian et al., 2005) on RNA-Seq data from the primed hESCs versus naive cell clones to identify metabolism-related gene sets distinct between these two states of pluripotency. We found that MYC-regulated gene sets are significantly enriched in naive versus primed cells (Figure 2A and Figure S2A). To test whether this enrichment applies to other naive conditions than the 5i/LAF method, we analyzed previously published human blastocyst datasets (Vassena et al., 2011; Yan et al., 2013) and another naive hESC dataset by Takashima et al. (Takashima et al., 2014). We first noticed the expression patterns in KEGG glycolysis pathway genes are very similar between naive hESCs derived by Takashima et al. and those derived using the 5i/LAF method when compared to their respective primed counterparts (Figure S2B–E). More importantly, in multiple independent studies, we found that MYC target genes are enriched in blastocyst versus primed hESCs (Figures 2B and 2C and Figure S2F), and in another naive line versus its primed counterpart (Figure S2G), suggesting that elevated MYC target gene expression is not unique to 5i/LAF-derived naive cells, but is characteristic of naive pluripotency in vivo and likely other naive culture conditions as well. This is notable since MYC can promote increased glucose metabolism and is associated with the pluripotent state (Takahashi and Yamanaka, 2006; Thai et al., 2014).
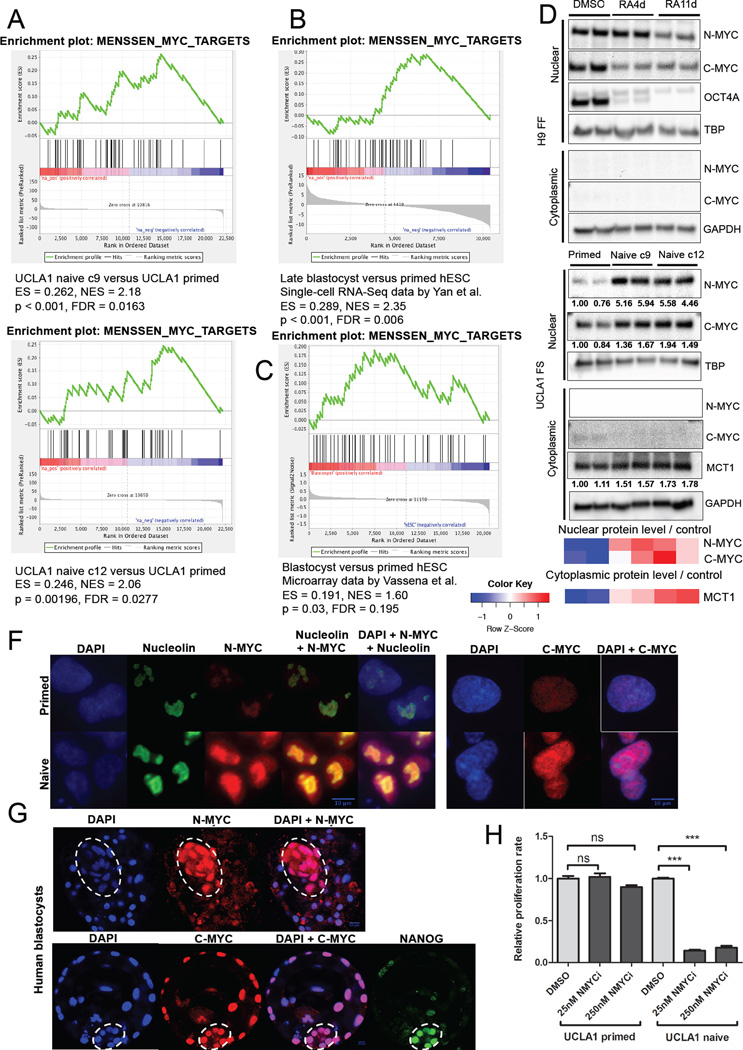
Figure 2. N-MYC is associated with human naive pluripotency.
GSEA mountain plots displaying enrichment of a MYC-regulated gene set in naive UCLA1 clone 9 and naive UCLA1 clone 12 versus primed UCLA1 hESCs (A), human late blastocyst versus primed hESCs (B) using RNA-Seq data by Yan et al., 2013, and human blastocyst versus primed hESCs (C) from mRNA microarray data by Vassena et al., 2011. (D) Immunoblot showing nuclear and cytoplasmic N-MYC and C-MYC levels in primed feeder-free H9 hESCs treated with DMSO, retinoic acid for 4 days (RA4d), or retinoic acid for 11 days (RA11d). Nuclear OCT4A levels are also shown. (E) Immunoblot showing nuclear and cytoplasmic N-MYC and C-MYC levels in primed, naive clone 9, and naive clone 12 feeder-supported UCLA1 hESCs. Cytoplasmic MCT1 levels are also shown. The heatmap shows standardized levels of indicated markers across samples (Z score). For (D) and (E), TBP is used as a loading control for the nuclear lysates. GAPDH is used as a loading control for the cytoplasmic lysates. Lysates were prepared in biological duplicates. (F) Immunofluorescence staining for N-MYC and C-MYC in naive versus primed UCLA1 hESCs. Nucleolin staining is also shown. (G) Immunofluorescence staining for N-MYC and C-MYC in human blastocysts. The contour of inner cell mass is indicated by dashed line. NANOG staining is also shown. For (F) and (G), scale bar = 10µm. (H) Proliferation rates of primed versus naive feeder-supported UCLA1 hESCs treated with DMSO or 250 nM CD532 (NMYCi). Error bars indicate ± 1 SEM of biological replicates (n = 3). ns: not significant; *** p < 0.001. See also Figure S2.
Consistent with an association between MYC activity and pluripotency, we found that nuclear N-MYC and C-MYC levels are decreased upon retinoic acid-induced differentiation of primed hESCs (Figure 2D), while two independently derived naive clones exhibit higher nuclear N-MYC and C-MYC levels than the primed cells from which they were derived (Figure 2E). We confirmed the increased N-MYC and C-MYC levels in naive versus primed hESCs by immunofluorescence staining, which additionally shows differences in sub-nuclear localization of N-MYC: dispersive nuclear N-MYC signal in naive cells, and contained nucleolar N-MYC staining in primed cells (Figure 2F). To test whether the elevated nuclear N-MYC levels we detected in 5i/LAF-derived naive hESCs reflect that of naive pluripotency in vivo, we examined N-MYC and C-MYC levels by immunofluorescence in human blastocysts, and found increased nuclear N-MYC signal in the inner cell mass relative to that found in trophoblasts (Figure 2G). In contrast, we readily detected nuclear C-MYC signal in both the inner cell mass and trophoblasts, suggesting N-MYC may be more specifically associated with naive pluripotency than C-MYC. Given the enrichment of MYC target gene expression and elevation of nuclear N-MYC levels in naive versus primed hESCs, we postulated that N-MYC activity might be important for the maintenance of naive pluripotency. Consistent with this notion, we found that treatment with CD532, a small molecule inhibitor towards N-MYC (Gustafson et al., 2014), dramatically decreases the proliferation of naive hESCs without affecting the proliferation of primed hESCs (Figure 2H), suggesting an important role for N-MYC specifically in naive hESCs. These results further suggest an association between MYC activity, glucose metabolism, and the pluripotent state.
Manipulation of hESC metabolism via MCT1 inhibition
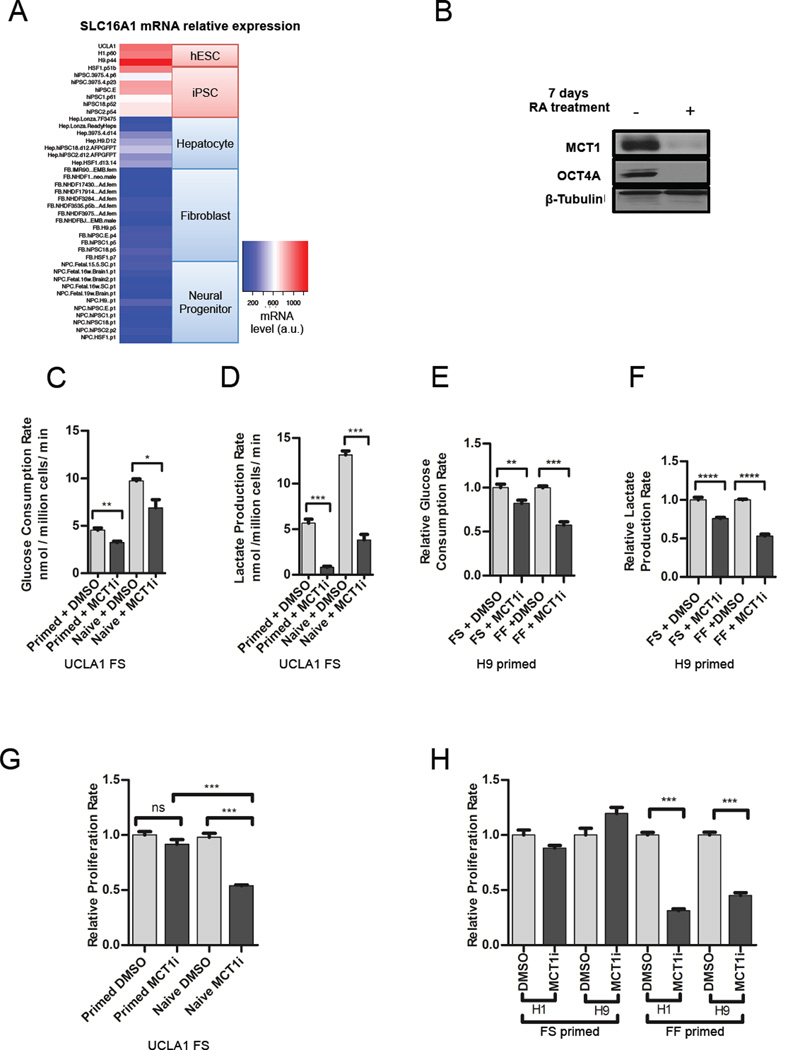
Since our results support a link between MYC-driven glycolytic metabolism and the pluripotent state, we hypothesized that inhibition of glycolysis may impact pluripotency or promote differentiation. However, to identify a glycolysis-related protein that would serve as a good target for hESC metabolism manipulation, we first examined the mRNA expression levels of metabolic genes important for glycolysis towards which small molecule inhibitors have been developed. As shown in Figure 3A, we found that Solute Carrier 16A1 (SLC16A1) mRNA levels significantly correlate with the pluripotent state across a panel of pluripotent cell lines and somatic cell lineages. SLC16A1 is expressed at high levels in both primed hESCs and hiPSCs, but at relatively low levels in non-pluripotent cell lines and tissues (Figure 3A). An important target of MYC-driven glycolysis in cancer, SLC16A1 encodes Monocarboxylate Transporter 1 (MCT1), which transports lactate, pyruvate, and other monocarboxylates across the plasma membrane in a proton-linked bidirectional manner (Doherty et al., 2014; Adijanto and Philp, 2012). An inhibitor of MCT1 activity called AZD3965 is currently undergoing Phase I evaluation for cancer treatment and reduces glycolytic rate and proliferation of cancer cell lines in vitro by decreasing lactate export rate (Polanski et al., 2014). To examine whether the MCT1 protein level is associated with the pluripotent state, we measured MCT1 levels in lysates from primed versus naive hESCs, and in primed hESCs treated with DMSO or treated with retinoic acid for seven days to induce differentiation. MCT1 levels are modestly elevated in naive versus primed hESCs (Figure 2E). Compared to DMSO-treated primed hESCs, MCT1 levels are markedly reduced, and OCT4A levels are absent, in retinoic acid-treated primed hESCs (Figure 3B). Together, these results indicate that high MCT1 expression is associated with the pluripotent state and may serve as a good target for inhibition of glycolysis in hESCs.
Figure 3. Manipulation of hESC metabolism via MCT1 inhibition.
(A) Heatmap depicting SLC16A1 mRNA levels across a panel of pluripotent and differentiated cell lines. Relative mRNA levels are color coded with a gradient from blue for the minimum through red for the maximum reading. (B) Immunoblotting of lysates from H9 primed hESCs treated with DMSO or 10 µM RA treatment for 7 days and probed with antibodies towards MCT1, OCT4A, and beta-tubulin as a loading control. Glucose consumption (C) and lactate production rates (D) of primed versus naive feeder-supported UCLA1 hESCs treated with DMSO or 250 nM AZD3965 (MCT1i) for 24 hours. Glucose consumption (E) and lactate production (F) rates of feeder-supported (FS) versus feeder-free (FF) cultured primed H9 hESCs treated with DMSO or 250 nM AZD3965 (MCT1i) for 24 hours. (G) Proliferation rates of primed versus naive feeder-supported UCLA1 hESCs treated with DMSO or AZD3965 (MCT1i) for 24 hours. (H) Proliferation rates of feeder-supported versus feeder-free H1 and H9 primed hESCs treated with DMSO or 250 nM AZD3965 (MCT1i). For (C) – (H), error bars indicate ± 1 SEM of biological replicates (n = 3). ns: not significant; * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. See also Figure S3.
To test whether MCT1 inhibition reduces hESC glycolytic rate, we compared the glucose consumption and lactate production rates of naive versus primed feeder-supported hESCs, and feeder-supported (FS) versus feeder free-cultured (FF) primed hESCs treated with DMSO or AZD3965. MCT1 inhibition via AZD3965 treatment reduces glucose consumption and lactate production rates in all hESC types tested: naive feeder-supported, primed feeder-supported, and primed feeder-free hESCs (Figures 3C–F and Figure S3A and S3B). To our knowledge, feeder-free naive hESCs cannot be successfully derived using the 5i/LAF method. In addition, MCT1 inhibition increases oxygen consumption rate in FF hESCs (Figure S3C and S3D). These results demonstrate that MCT1 inhibition is a feasible strategy to manipulate hESC glucose metabolism without toxic side-effects.
To determine whether reduction of glycolytic flux impacts hESC self-renewal capacity, we compared the proliferation rates of hESCs treated with DMSO versus AZD3965. Notably, we observed a proliferative impairment in naive FS hESCs, but not in primed FS hESCs (Figure 3G). These results are consistent with our metabolomics results demonstrating increased use of glucose for biosynthesis of nucleotides and serine in naive versus primed hESCs (Figures 1I and 1J). Furthermore, we found that MCT1 inhibition via AZD3965 treatment decreases proliferation rate of FF primed hESCs, but not of FS primed hESCs (Figure 3H). Treatment with another glycolytic inhibitor, dichloroacetic acid (DCA), similarly decreases proliferation rate of FF primed hESCs, but not of FS primed hESCs (Figure S3E–G). These results indicate that primed hESCs react to decreased glycolytic rate differently depending on the presence/absence of mouse embryonic fibroblasts (MEFs), and could suggest that the presence of feeder cells makes primed hESCs less reliant on glycolysis for proliferation.
Feeder free-cultured primed hESCs exhibit increased anabolic glucose metabolism relative to feeder-supported primed hESCs
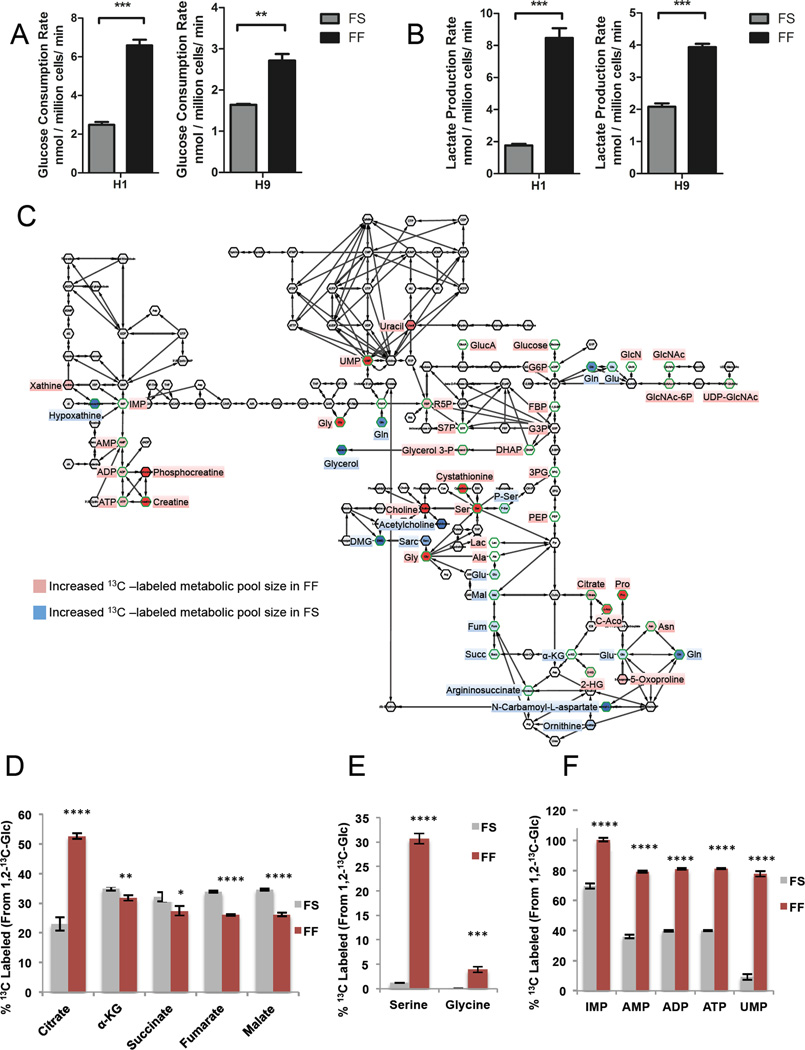
The differential effect of MCT1 inhibition on proliferation of FS primed hESCs versus FF primed hESCs suggests that FS and FF hESCs use glucose differently, and that FF hESCs may use glucose more for biosynthetic pathways that support proliferation. To test this possibility, we compared the glucose consumption and lactate production rates of FS and FF primed H1 and H9 hESCs. We found H1 and H9 FF hESCs consume more glucose and produce more lactate compared to FS cells (Figures 4A and 4B). To further determine whether FS and FF hESCs metabolize glucose differently, we labeled the cells with 1,2-13C-glucose, and traced the incorporation of 13C into downstream glucose metabolites using LC-MS. To control for the presence of feeder cells in the FS conditions, we also labeled a plate of irradiated MEFs without hESCs with 1,2-13C-glucose, and traced the incorporation of 13C into downstream glucose metabolites using LC-MS. Consistent with the small relative number of MEFs compared to hESCs in FS conditions (usually 2–10%), the relative amounts of metabolites as measured by LC-MS from the feeder-only plate were much smaller (0.2–10% depending on the metabolite). We determined that the presence of MEF-derived metabolites in our FS hESC metabolite samples only has a minor effect on the overall labeling pattern (Figure S4A–D). We found that two independent primed hESC lines, H1 and H9, exhibit markedly different patterns of glucose utilization depending on whether they are cultured in FS or FF conditions (Figure 4C and Figure S4E).
Figure 4. Feeder free-cultured primed hESCs exhibit increased anabolic glucose metabolism relative to feeder-supported primed hESCs.
Glucose consumption (A) and lactate production (B) rates of feeder-supported (FS) versus feeder-free cultured (FF) primed H1 and H9 hESCs. For (C) – (F), FS and FF primed H9 hESCs were cultured in medium with 1,2-13C-glucose for 24 hours. Metabolites were extracted and analyzed by LC-MS, and the percentage of 13C-labeled metabolites in FS vs FF primed H9 hESCs was compared. (C) Cytoscape network map showing the ratio of labeled metabolites from 13C glucose in FS versus FF primed H9 hESCs. Metabolites in red indicate increased 13C-labeled metabolic pool sizes in FF hESCs compared to FS hESCs, while metabolites in blue indicate increased 13C-labeled metabolic pool sizes in FS hESCs compared to FF hESCs. Green borders indicate statistically significant (p < 0.05) changes in incorporation of 13C from 1,2-13C-glucose in FS hESCs versus FF hESCs. Metabolites in white indicate similar incorporation of 13C from 1,2-13C-glucose in FS hESCs and FF hESCs. Metabolites in grey were undetected. (D) Percentages of the indicated 13C-labeled TCA cycle metabolites in FS versus FF primed H9 hESCs. (E) Percentages of 13C-labeled serine and glycine in FS versus FF primed H9 hESCs. (F) Percentages of 13C-labeled nucleotides in FS versus FF primed H9 hESCs. For (A), (B), and (D) – (F), error bars indicate ± 1 SEM of biological replicates (n = 3). * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. See also Figure S4.
The consistent metabolic changes in H1 and H9 primed hESCs in FF versus FS conditions suggests increased incorporation of glucose carbons into metabolites used for biosynthesis in FF conditions. For example, FF H1 and H9 hESCs incorporate more glucose carbons into citrate, an important metabolic intermediate for fatty acid, cholesterol, and hexosamine biosynthesis (Figure 4D and Figure S4F). However, FF hESCs incorporate significantly less glucose carbons into other TCA metabolites than citrate, such as αKG, succinate, fumarate, and malate (Figure 4D and Figure S4F), consistent with increased use of the glucose-derived citrate for biosynthesis rather than for maintenance of the TCA cycle. Additionally, FF hESCs incorporate more glucose carbons into serine and glycine, products of the serine synthesis pathway important for purine synthesis, glutathione synthesis, protein synthesis, and synthesis of lipid head groups (Rabinowitz and Vastag, 2012) (Figure 4E and Figure S4G). FF hESCs also incorporate more glucose carbons into nucleotides, including IMP, AMP, ADP, ATP, and UMP (Figure 4F and Figure S4H). These data suggest that FF primed hESCs use glucose more for biosynthesis than FS primed hESCs.
MEF-secreted factors make primed hESCs less reliant on glucose for proliferation
To further investigate the source of metabolic differences between FS and FF culturing methods, we assessed whether fibroblast feeder cell-secreted factors impact the response of primed hESCs to MCT1 inhibition. We incubated FF medium with irradiated MEFs in the absence of hESCs for 24 hours and collected MEF-conditioned medium. We then examined whether MEF-conditioned medium impacted the proliferation response of FF hESCs to MCT1 inhibition. As shown in Figures 5A and 5B, MEF-conditioned medium rescues the proliferation defect caused by MCT1 inhibition of FF H1 and H9 hESCs (Figures 5A and 5B) without causing differentiation (Figure S5A and S5B). To determine if MEF-conditioned medium rescues the proliferation defect of AZD3965-treated hESCs by restoring the glycolytic rate blunted by MCT1 inhibition, we measured glucose consumption and lactate production rates in MEF-conditioned medium treated FF cells treated with DMSO or AZD3965. We found that MEF-conditioned medium does not alter the inhibitory effect of AZD3965 treatment on glycolysis (Figures 5C and 5D). These data suggest that MEFs can impact hESC metabolism, at least partially, by secreting factor(s) that reduce reliance of primed hESCs on glucose for proliferation.
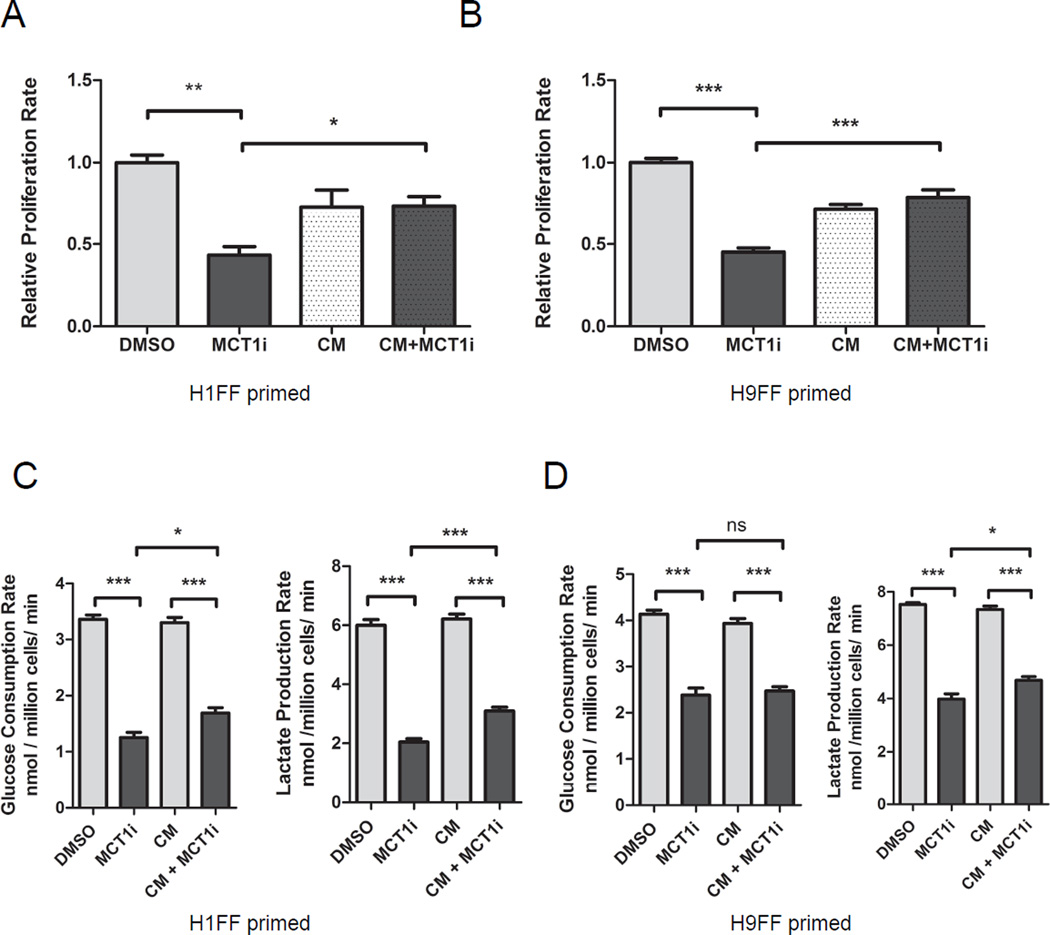
Figure 5. MEF-secreted factors make primed hESCs less reliant on glucose for proliferation.
Proliferation rates of primed feeder-free H1 (A) and H9 (B) hESCs treated with DMSO, AZD3965 (MCT1i), MEF-conditioned medium (CM), or AZD3965 along with MEF-conditioned medium (CM + MCT1i) for 5 days. Glucose consumption and lactate production rates of feeder-free H1 (C) and H9 (D) primed hESCs FF, treated with DMSO, AZD3965 (MCT1i), MEF conditioned medium (CM), or AZD3965 along with MEF conditioned medium (CM + MCT1i) for 5 days. For (A) – (D), error bars indicate ± 1 SEM of biological replicates (n = 3). ns: not significant; * p < 0.05; ** p < 0.01; *** p < 0.001. See also Figure S5.
MYC activity in primed hESCs is modulated by MEF-secreted factors
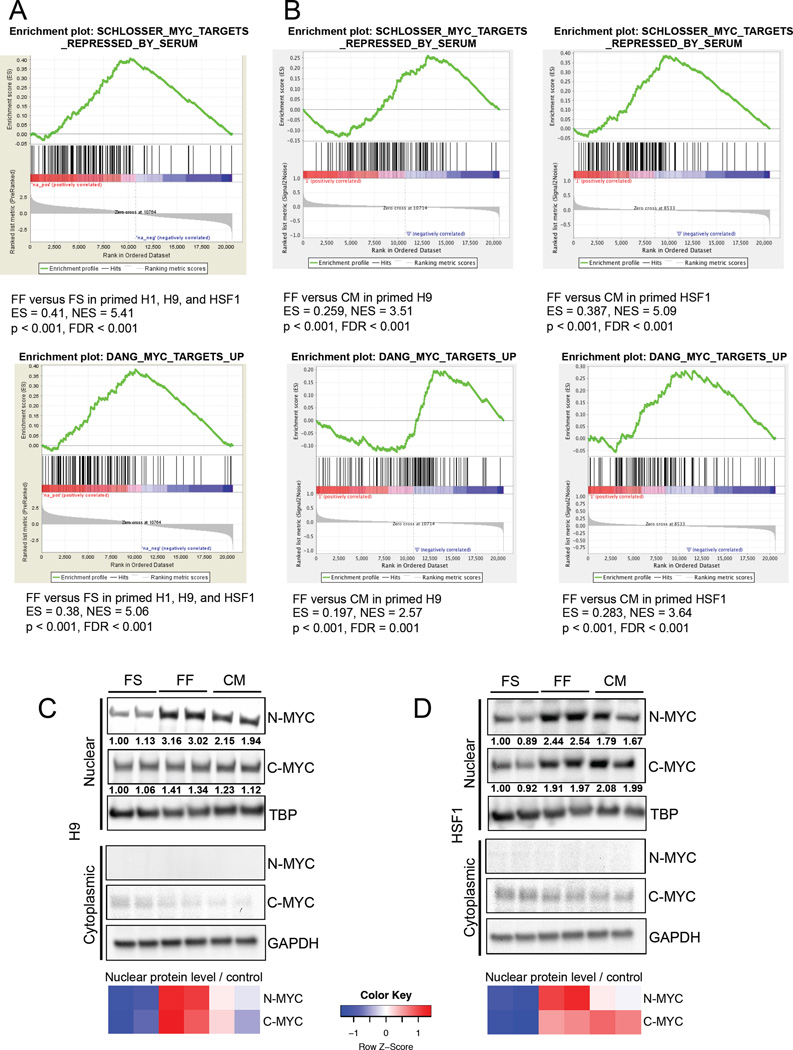
To examine the mechanism by which MEF-secreted factors reprogram primed hESC metabolism, we conducted Gene Set Enrichment Analysis (GSEA) on mRNA microarray data from FS versus FF primed hESCs, and from FF primed hESCs cultured in MEF-conditioned medium. Notably, we found that MYC-regulated gene sets are significantly enriched in FF compared to FS hESCs (Figure 6A). Additionally, MYC-regulated gene sets are significantly enriched in FF hESCs compared to FF hESCs cultured for 24 hours in MEF-conditioned medium (Figure 6B), suggesting that MEF-secreted factor(s) decrease MYC transcriptional activity in FF hESCs. We next examined the levels of nuclear N-MYC and C-MYC in FS, FF, and MEF-conditioned medium-treated FF hESCs. As shown in Figures 6C and 6D, H9 and HSF1 hESCs exhibit elevated nuclear N-MYC levels in FF conditions relative to FS conditions, and MEF-conditioned medium treatment decreases nuclear N-MYC level in FF hESCs. On the other hand, neither cytoplasmic N-MYC level nor cytoplasmic/nuclear C-MYC level is consistently altered in FS, FF, and MEF-conditioned medium-treated H9 and HSF1 hESCs (Figures 6C and 6D). These data suggest that N-MYC may be responsible for the difference in glucose metabolism between FS and FF primed hESCs, and that N-MYC levels and transcriptional activity can be down-regulated by MEF-secreted factors.
Figure 6. MYC activity in hESCs is modulated by MEF-secreted factors.
(A) GSEA mountain plots displaying enrichment of MYC-regulated gene sets in feeder-free (FF) versus feeder-supported (FS) primed H1, H9, and HSF1 hESCs. (B) GSEA mountain plots displaying enrichment of MYC-regulated gene sets in feeder-free (FF) versus MEF-conditioned medium treated FF (CM) primed H9 and HSF1 hESCs. Immunoblot showing nuclear and cytoplasmic N-MYC and C-MYC levels in FS, FF and MEF-conditioned medium (CM) treated primed H9 hESCs (C) and HSF1 hESCs (D). For (C) and (D), the heatmap shows standardized levels of indicated markers across samples (Z score). TBP was used to control for nuclear lysate loading, and GAPDH was used to control for cytoplasmic lysate loading. Lysates were prepared in biological duplicates.
MCT1 inhibition of feeder free-cultured primed hESCs promotes neural lineage specification
Since MCT1 inhibition through AZD3965 treatment reliably reduces glycolytic metabolism of FS and FF primed hESCs (Figures 3E and 3F), we next assessed whether moderate suppression of glycolysis by MCT1 inhibition has a causal effect on the differentiation status of primed hESCs. We first noticed that five days of AZD3965 treatment causes FF hESCs to exhibit altered morphology relative to DMSO-treated hESCs. AZD3965-treated FF hESCs appear elongated, spindle-like, and more light-reflective compared to the round and smooth-edged DMSO-treated cells (Figure 7A).
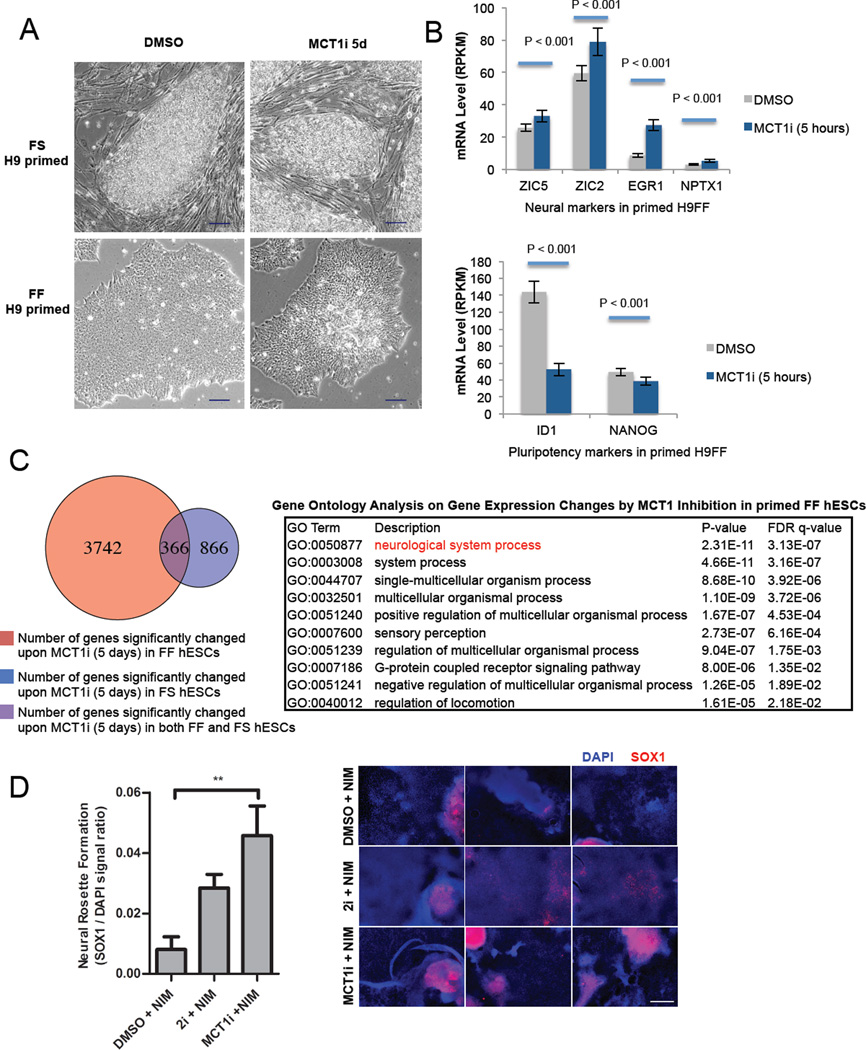
Figure 7. MCT1 inhibition of feeder free-cultured primed hESCs promotes neural lineage specification.
(A) Phase contrast microscopic images of live FS and FF primed H9 hESCs treated with DMSO or 250nM AZD3965 (MCT1i) for 5 days (scale bar = 100 µm). (B) mRNA levels of neural markers (ZIC5, ZIC2, EGR1 and NPTX1) and pluripotency markers (ID1 and NANOG) in primed FF H9 hESCs, treated with DMSO or 250 nM AZD3965 (MCT1i) for five hours. mRNA levels were obtained by RNA sequencing biological duplicates, and expressed in RPKM (Reads Per Kilobase of transcript per Million mapped reads). (C) Table depicting gene ontology analysis of gene expression profiling after five days AZD3965 versus DMSO treatment of primed FF H9, H1, and HSF1 hESCs. (D) Neural rosette formation efficiency of primed FF H9 hESCs treated with neural induction medium (NIM) supplemented with DMSO, 2i dual smad inhibition (10 µM SB-431542 and 100 nM LDN193189), or 250 nM AZD3965 (MCT1i) for 10 days. SOX1 to DAPI ratio was quantified using ImageJ after acquiring immunofluorescent images from fixed cells. Corresponding images of immunofluorescence staining for SOX1 are also shown. Scale bar = 400 µm. For (B), error bars indicate ± 1 SEM of biological duplicates (n = 3). ** p < 0.01. For (D), error bars indicate ± 1 SEM of biological replicates (n = 3). See also Figure S6 and Table S1.
Because we had observed metabolic changes in FF primed hESCs in as little as 30 minutes post MCT1 inhibition, we first measured gene expression changes at five hours to determine whether moderate suppression of glycolysis could affect differentiation status. We sequenced the transcriptome of H9 FF hESCs at five hours post DMSO or AZD3965 treatment, and examined the gene expression differences for patterns of differentiation to particular cell types. While few genes were consistently changed at this short time point of treatment, we did find that MCT1 inhibition promoted elevated expression of the genes involved in early specification of the neural tube including ZIC5, ZIC2, EGR1, and NPTX1, and reduction of the pluripotency markers Inhibitor of Differentiation 1 (ID1) and NANOG (Figure 7B). Gene ontology analysis of gene expression profiling after five days of AZD3965 treatment demonstrated a clear shift in expression that appeared to correlate with neural specification (Figure 7C and Figure S6A and Table S1). Together with data from the five-hour time point, it seems clear that the inhibition of glycolysis with AZD3965 could, at a minimum, lower the threshold for neural specification from primed hESCs.
To assay whether MCT1 inhibition could promote neural specification under differentiation conditions, we subjected FF primed hESCs according to a well described neural specification protocol (Chambers et al., 2009). AZD3965 treatment significantly promotes the formation of neural rosette structures and neural specification compared to DMSO-treated hESCs (Figure 7D), as judged by the increased number of SOX1 positive cells. DCA treatment does not significantly enhance neural rosette structure formation (Figure S6B), likely due to the decreased effectiveness of 5mM DCA treatment compared to 250nM AZD3965 treatment in reducing glycolysis (Figures 3E and 3F and Figure S3E and S3F). Our results collectively demonstrate that glycolytic rate in human pluripotent stem cells is tightly correlated with the differentiation status, from naive to primed to specified cell types.
DISCUSSION
This study confirms previous findings that glycolytic metabolism is associated with the pluripotent state, and extends them significantly to both the naive state and to differences between various primed hESC culture methods. We show that acquisition of a naive state further increases hESC glycolysis, that retinoic acid-induced differentiation decreases glycolytic metabolism, and that abating glycolytic metabolism in hESCs can push them towards specification. The elevated glycolytic metabolism in naive hESCs is accompanied by increased MYC transcriptional activity and increased nuclear N-MYC and C-MYC levels. While C-MYC has previously been associated with pluripotency and has been used to enhance reprogramming of somatic cells into induced pluripotent stem cells (Smith et al., 2010; Takahashi and Yamanaka, 2006), N-MYC has been less studied in hESCs and may be an important contributor to pluripotency-associated glycolytic metabolism. Our findings that naive hESCs exhibit increased glycolysis, together with recent studies showing increased respiration in naive versus primed hESCs (Carbognin et al., 2016; Takashima et al., 2014), supports the notion that glycolysis and oxidative phosphorylation are not mutually exclusive in naive pluripotency. In fact, high glucose uptake has been an established indicator of human blastocyst quality and viability for in vitro fertilization (Gardner et al., 2011), consistent with the increased glucose metabolism found in naive hESCs.
Through LC-MS-based metabolomics, we show that naive hESCs incorporate more glucose carbons into lactate, nucleotides, and serine. Since naive hESCs are thought to be more representative of the inner cell mass of the preimplantation embryo than primed hESCs, which are thought to resemble the postimplantation epiblast (Nichols and Smith, 2009), we hypothesize that variation in nutrient availability to the embryo before and after implantation may contribute to the differences in metabolism between the naive and primed state. Consistent with this notion, we found that MYC target genes are enriched in human embryos at the morula and blastocyst stages relative to hESCs (Figure S2F and Vassena et al., 2011), suggesting that our cultured naive hESCs, which also show enrichment in MYC target genes, may best represent cells from the morula and blastocyst stages.
The increased glycolytic rate we show in human naive versus primed ESCs is different from the glucose metabolism found in mouse naive versus primed ESCs (mESCs versus Epiblast Stem cells (mEpiSCs), respectively). mESCs exhibit lower glycolytic rates compared to mEpiSCs (Zhou et al., 2012 and Figure S7A). This discrepancy may be partially explained by the lower nuclear C-MYC levels in mESCs versus mEpiSCs (Marks et al., 2012 and Figure S7B). In human, the opposite is true: naive hESCs have higher nuclear C-MYC levels than primed hESCs (Figure 2E). Interestingly, nuclear N-MYC levels are elevated in both human and mouse naive ESCs versus primed counterparts, suggesting N-MYC may be more closely associated with naive pluripotency. Of late, there is emerging literature on the differences in early embryo development between human and mouse: lineage and X chromosome dynamics (Petropoulos et al., 2016), dependence on FGF signaling (Roode et al., 2012), and gene expression patterns (Niakan and Eggan, 2013). Considering that our key observations regarding glycolysis in naive hESCs holds true in a naive line directly derived from a human blastocyst, and were corroborated by MYC gene expression studies and immunofluorescence staining in human blastocysts, our results suggest that regulation of glycolytic metabolism may be yet another key aspect of the human-mouse difference during early embryo development.
To our knowledge this is also the first study to describe in detail that glucose metabolism varies significantly between primed hESCs grown in FF versus FS conditions, and to provide evidence that this is at least in part due to MEF-secreted factors. We found that FF cultured primed hESCs exhibit elevated anabolic glucose metabolism and increased reliance on glucose for proliferation. MEF-secreted factor(s) decrease primed hESC reliance on glucose for proliferation. Relative to FS primed hESCs, we show that FF primed hESCs exhibit higher MYC transcriptional activity and nuclear N-MYC levels, both of which are reduced in the presence of MEF-secreted factor(s). These results suggest that N-MYC may promote increased anabolic glucose metabolism in FF versus FS cultured primed hESCs, and that N-MYC is subject to regulation by MEF-secreted factor(s).
Given the difference in glucose metabolism between FS and FF cultured primed hESCs, it will be interesting in the future to determine whether FF versus FS cultured primed hESCs serve to explain distinct biases towards differentiation into different cell fates across labs and pluripotent lines (Osafune et al., 2008). These data could also point towards improved methods for the development of defined media that accurately mimic FS culture as this would facilitate consistent differentiation methods and ease the transition of hESC or hiPSC cultures toward clinical applications.
While significant effort has been applied to characterize the metabolism of human pluripotent stem cells (Prigione et al., 2010; Yoshida et al., 2009; Zhang et al., 2011, 2012), our work sheds light on how to manipulate metabolism to promote a particular cell fate. This study took a clue from gene expression data at an early time point post glycolysis inhibition and focused on studying its effects on neural progenitor specification. It is entirely possible and of great interest that metabolic manipulation might impact and regulate differentiation processes towards other lineages as well. Understanding whether and how the epigenetic state of human pluripotent stem cells is altered by inhibition of glycolysis will be of key importance to determine in future studies. Going forward, it will also be important to determine whether promotion of hESC glycolysis can improve either the efficiency or fidelity of naive human pluripotent stem cell generation. This approach could improve efforts to generate stable human naive culture systems, which are currently plagued by instability of self-renewal, survival, differentiation potential, and even genomic integrity.
EXPERIMENTAL PROCEDURES
Cell Culture
Feeder-supported hESC lines were maintained on radiation inactivated MEFs (GlobalStem). Naive hESCs were maintained using the 5i/LAF condition as described (Pastor et al., 2016; Theunissen et al., 2014). Feeder-free hESCs were cultured on the plates pre-coated with 1:25 diluted Matrigel (BD).
Metabolic Measurements
Culture medium was collected after 24 hours to measure glucose and lactate amounts using a Nova Bioanalyzer. Among different figures, the same cell line might show different values in glucose and lactate readings, which was due to the difference in cell numbers between different experiments. Since plating density of cells can impact metabolism, we always ensured that different treatment groups or cell lines within a given experiment had similar cell counts when glucose and lactate measurements were taken. We accomplished this by plating hESCs at different densities and measuring the media glucose and lactate amounts in the ones with similar cell counts. For metabolomics analysis, cells were incubated in medium containing 1,2-13C-glucose for 24 hours. The analysis was performed on a Q Exactive (Thermo Scientific) mass spectrometer.
Gene Expression Analysis
For RNA-Seq, hESCs were feeder depleted, and RNA samples were submitted to UCLA High-Throughput Sequencing Facility. For microarray, whole-genome expression analysis was performed with the HG-U133 plus 2 array (Affymetrix) at the UCLA Clinical Microarray Core. Gene Set Enrichment Analysis (Subramanian et al., 2005) was performed using the Molecular Signatures Database (MSigDB) C2 collection. A web-based application GOrilla (Eden et al., 2009) was used to identify enriched Gene Ontology terms in a ranked list of all genes according to the differential expression in AZD3965 versus DMSO-treated hESCs.
Statistical Analysis
Data were analyzed by two-sample Student’s t-test, or one-way ANOVA with three or more samples.
Supplementary Material
Acknowledgments
We thank AstraZeneca for use of AZD3965 and members of the Christofk laboratory for helpful discussions. We thank Jennifer Tsoi and Dr. Thomas Graeber for help with GSEA. We thank Drs. Jin Zhang and Michael Teitell for help with the oxygen consumption measurements. We thank Drs. Ziwei Li and Amander Clark for help with the hESC culture. We thank Jessica Cinkornpumin for RNA extraction. We thank Jinghua Tang for culture of hESC lines. W.G. acknowledges the support of California Institute for Regenerative Medicine Training Grant (TG2-01169) and UCLA Dissertation Year Fellowship for funding this project. This study was supported by NIH 5P01GM099134 and by an Innovation Award granted to W.E.L., H.R.C, and Amy Rowat from the UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research, as well as additional funding awarded to H.R.C. from the UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research and the CONCERN Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Please see Supplemental Experimental Procedures for detailed methods.
ACCESSION NUMBERS
The accession number for the data reported in this paper is GEO: GSE83491.
SUPPLEMENTAL INFORMATION
Supplemental information includes Supplemental Experimental Procedures, seven figures, and one table can be found with this article online at
AUTHOR CONTRIBUTIONS
W.G. helped design the study, conducted experiments, analyzed results, and wrote the manuscript. X.G. conducted neural induction experiments, analyzed results, and edited the manuscript. A.S. and R.K. generated the naive hESCs, analyzed results, and edited the manuscript. A.B.C. and C.P.H. helped conduct some of the experiments with AZD3965. D.B. conducted the metabolomics measurements, analyzed metabolomics data, and edited the manuscript. K.P. helped design parts of the study involving naive hESCs, analyzed results, and edited the manuscript. W.E.L. helped design many parts of the study, analyzed results, and wrote the manuscript. H.R.C. designed the study, analyzed results, and wrote the manuscript.
REFERENCES
- Adijanto J, Philp NJ. Chapter Nine - The SLC16A Family of Monocarboxylate Transporters (MCTs)—Physiology and Function in Cellular Metabolism, pH Homeostasis, and Fluid Transport. In: Bevensee MO, editor. In Current Topics in Membranes. Academic Press; 2012. pp. 275–312. [DOI] [PubMed] [Google Scholar]
- Carbognin E, Betto RM, Soriano ME, Smith AG, Martello G. Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J. 2016;35:618–634. doi: 10.15252/embj.201592629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y-S, Göke J, Ng J-H, Lu X, Gonzales KAU, Tan C-P, Tng W-Q, Hong Z-Z, Lim Y-S, Ng H-H. Induction of a Human Pluripotent State with Distinct Regulatory Circuitry that Resembles Preimplantation Epiblast. Cell Stem Cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- Chung S, Arrell DK, Faustino RS, Terzic A, Dzeja PP. Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. J. Mol. Cell. Cardiol. 2010;48:725–734. doi: 10.1016/j.yjmcc.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty JR, Yang C, Scott KEN, Cameron MD, Fallahi M, Li W, Hall MA, Amelio AL, Mishra JK, Li F, et al. Blocking Lactate Export by Inhibiting the Myc Target MCT1 Disables Glycolysis and Glutathione Synthesis. Cancer Res. 2014;74:908–920. doi: 10.1158/0008-5472.CAN-13-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Arrell DK, Zlatkovic-Lindor J, Martinez-Fernandez A, Perez-Terzic C, Nelson TJ, Terzic A. Metabolome and metaboproteome remodeling in nuclear reprogramming. Cell Cycle. 2013;12:2355–2365. doi: 10.4161/cc.25509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CDL, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic Oxidative Bioenergetics Transitions into Pluripotency-Dependent Glycolysis to Facilitate Nuclear Reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CDL, Martinez-Fernandez A, Faustino RS, Yamada S, Perez-Terzic C, Nelson TJ, Terzic A. Nuclear Reprogramming with c-Myc Potentiates Glycolytic Capacity of Derived Induced Pluripotent Stem Cells. J Cardiovasc. Transl. Res. 2012a;6:10–21. doi: 10.1007/s12265-012-9431-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CDL, Nelson TJ, Dzeja PP, Terzic A. Energy metabolism plasticity enables stemness programs. Ann. N. Y. Acad. Sci. 2012b;1254:82–89. doi: 10.1111/j.1749-6632.2012.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Lane M, Stevens J, Schoolcraft WB. Noninvasive assessment of human embryo nutrient consumption as a measure of developmental potential. Fertil. Steril. 2001;76:1175–1180. doi: 10.1016/s0015-0282(01)02888-6. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Wale PL, Collins R, Lane M. Glucose consumption of single post-compaction human embryos is predictive of embryo sex and live birth outcome. Hum. Reprod. Oxf Engl. 2011;26:1981–1986. doi: 10.1093/humrep/der143. [DOI] [PubMed] [Google Scholar]
- Gustafson WC, Meyerowitz JG, Nekritz EA, Chen J, Benes C, Charron E, Simonds EF, Seeger R, Matthay KK, Hertz NT, et al. Drugging MYCN through an Allosteric Transition in Aurora Kinase A. Cancer Cell. 2014;26:414–427. doi: 10.1016/j.ccr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Hou R, Booth CJ, Yang S-H, Snyder M. Defined culture conditions of human embryonic stem cells. Proc. Natl. Acad. Sci. 2006;103:5688–5693. doi: 10.1073/pnas.0601383103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Francis Stewart A, Smith A, et al. The Transcriptional and Epigenomic Foundations of Ground State Pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, Eggan K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev. Biol. 2013;375:54–64. doi: 10.1016/j.ydbio.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and Primed Pluripotent States. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Chen D, Liu W, Kim R, Sahakyan A, Lukianchikov A, Plath K, Jacobsen SE, Clark AT. Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell Stem Cell. 2016;18:323–329. doi: 10.1016/j.stem.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer I, Barbet R, Zhou Y-P, Li M-L, Monier M-N, Hatzfeld A, Hatzfeld JA. Use of Xenofree Matrices and Molecularly-Defined Media to Control Human Embryonic Stem Cell Pluripotency: Effect of Low Physiological TGF-β Concentrations. Stem Cells Dev. 2008;17:519–534. doi: 10.1089/scd.2007.0279. [DOI] [PubMed] [Google Scholar]
- Petropoulos S, Edsgärd D, Reinius B, Deng Q, Panula SP, Codeluppi S, Plaza Reyes A, Linnarsson S, Sandberg R, Lanner F. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell. 2016;165:1012–1026. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanski R, Hodgkinson CL, Fusi A, Nonaka D, Priest L, Kelly P, Trapani F, Bishop PW, White A, Critchlow SE, et al. Activity of the Monocarboxylate Transporter 1 Inhibitor AZD3965 in Small Cell Lung Cancer. Clin. Cancer Res. 2014;20:926–937. doi: 10.1158/1078-0432.CCR-13-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The Senescence-Related Mitochondrial/Oxidative Stress Pathway is Repressed in Human Induced Pluripotent Stem Cells. STEM CELLS. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- Rabinowitz JD, Vastag L. Teaching the design principles of metabolism. Nat. Chem. Biol. 2012;8:497–501. doi: 10.1038/nchembio.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala K, Lindroos B, Hussein SM, Lappalainen RS, Pekkanen-Mattila M, Inzunza J, Rozell B, Miettinen S, Narkilahti S, Kerkelä E, et al. A Defined and Xeno-Free Culture Method Enabling the Establishment of Clinical-Grade Human Embryonic, Induced Pluripotent and Adipose Stem Cells. PLoS ONE. 2010;5:e10246. doi: 10.1371/journal.pone.0010246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roode M, Blair K, Snell P, Elder K, Marchant S, Smith A, Nichols J. Human hypoblast formation is not dependent on FGF signalling. Dev. Biol. 2012;361:358–363. doi: 10.1016/j.ydbio.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Singh AM, Dalton S. Myc Represses Primitive Endoderm Differentiation in Pluripotent Stem Cells. Cell Stem Cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Fischer KA, Devi A, Detraux D, Gu H, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 2015;17:1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al. Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai M, Graham NA, Braas D, Nehil M, Komisopoulou E, Kurdistani SK, McCormick F, Graeber TG, Christofk HR. Adenovirus E4ORF1-Induced MYC Activation Promotes Host Cell Anabolic Glucose Metabolism and Virus Replication. Cell Metab. 2014;19:694–701. doi: 10.1016/j.cmet.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. Systematic Identification of Culture Conditions for Induction and Maintenance of Naive Human Pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Valamehr B, Robinson M, Abujarour R, Rezner B, Vranceanu F, Le T, Medcalf A, Lee TT, Fitch M, Robbins D, et al. Platform for Induction and Maintenance of Transgene-free hiPSCs Resembling Ground State Pluripotent Stem Cells. Stem Cell Rep. 2014;2:366–381. doi: 10.1016/j.stemcr.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CA, IV, Ramalho-Santos J, Van Houten B, Schatten G. Energy Metabolism in Human Pluripotent Stem Cells and Their Differentiated Counterparts. PLoS ONE. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassena R, Boué S, González-Roca E, Aran B, Auer H, Veiga A, Belmonte JCI. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development. 2011;138:3699–3709. doi: 10.1242/dev.064741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, Jimenez-Caliani AJ, Deng X, Cavanaugh C, Cook S, et al. Derivation of naïve human embryonic stem cells. Proc. Natl. Acad. Sci. 2014;111:4484–4489. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia Enhances the Generation of Induced Pluripotent Stem Cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, Wahjudi PN, Setoguchi K, Wang G, Do A, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 2011;30:4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolic Regulation in Pluripotent Stem Cells during Reprogramming and Self-Renewal. Cell Stem Cell. 2012;11:589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C, et al. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.