Abstract
The cellular turnover of adult tissues and injury-induced repair proceed through an exquisite integration of proliferation, differentiation, and survival signals that involve stem/progenitor cell populations, their progeny, and differentiated tissues. GATA factors are DNA binding proteins that control stem cells and the development of tissues by activating or repressing transcription. Here we examined the role of GATA transcription factors in Schmidtea mediterranea, a freshwater planarian that provides an excellent model to investigate gene function in adult stem cells, regeneration, and differentiation. Smed-gata4/5/6, the homolog of the three mammalian GATA-4,-5,-6 factors is expressed at high levels in differentiated gut cells but also at lower levels in neoblast populations, the planarian stem cells. Smed-gata4/5/6 knock-down results in broad differentiation defects, especially in response to injury. These defects are not restricted to the intestinal lineage. In particular, at late time points during the response to injury, loss of Smed-gata4/5/6 leads to decreased neoblast proliferation and to gene expression changes in several neoblast subpopulations. Thus, Smed-gata4/5/6 plays a key evolutionary conserved role in intestinal differentiation in planarians. These data further support a model in which defects in the intestinal lineage can indirectly affect other differentiation pathways in planarians.
1. Introduction
GATA factors form a family of transcription factors containing zinc finger motifs, which bind to the DNA sequence “GATA” (Merika and Orkin, 1993; Patient and McGhee, 2002). In mammals, six GATA family members (GATA1-6) control cellular differentiation and organogenesis during development and in adults (Chlon and Crispino, 2012; Duncan, 2005), including hematopoiesis (Rodrigues et al., 2005; Weiss and Orkin, 1995), cardiac development (Kawamura et al., 2005; Pikkarainen et al., 2004), mammary gland development (Asselin-Labat et al., 2007; Kouros-Mehr et al., 2006), and the differentiation of tissues derived from the endoderm (Aronson et al., 2014; Gao et al., 1998; Zaret, 1999; Zaret et al., 2008). Early during development, GATA factors can control the self-renewal and the differentiation of embryonic stem cells (Capo-Chichi et al., 2010; Serrano et al., 2013; Turbendian et al., 2013), especially differentiation towards the extra-embryonic endoderm (Artus and Chazaud, 2014). GATA factors activity has also been implicated in abnormal proliferation and differentiation in cancer cells (Akiyama et al., 2003; Vicente et al., 2012; Zheng and Blobel, 2010).
GATA factors have been extensively studied in mammalian systems, but the elucidation of their exact roles in stem/progenitor cells and their differentiated progeny is complicated by the overlapping and distinct functions of each family member (Bresnick et al., 2010; Gao et al., 1998; Merika and Orkin, 1993; Patient and McGhee, 2002). Schematically, GATA1, GATA2, and GATA3 are often considered the “hematopoietic” GATA factors, based on their key roles in various aspects of hematopoiesis (Kobayashi-Osaki et al., 2005; Leonard et al., 1993; Orkin, 1992). In contrast, GATA4, GATA5, and GATA6 are expressed in endodermal and mesodermal lineages and have been more implicated in the development of organs derived from these lineages such as the heart, the lung, and the intestine (Bossard and Zaret, 1998; Charron and Nemer, 1999; Liu et al., 2002; Zaret et al., 2008; Zhao et al., 2005).
Planarians are multicellular animals with bilateral symmetry that display a striking capacity to repair injured or lost structures through a robust regeneration process. At any given time, homeostasis is maintained in planarians by dividing cells that generate the cellular progeny that forms adult tissues after terminal differentiation. In amputated or injured animals, a burst of proliferation occurs to form the regenerative blastema, the anatomical place where missing structures are recreated (reviewed in Reddien and Sanchez Alvarado (2004), Sanchez Alvarado and Yamanaka (2014), Tanaka and Reddien (2011)). The planarian stem cells, also known as neoblasts, are the only source of new cells in intact and amputated planarians (Betchaku, 1967; Pedersen, 1959; Scimone et al., 2014; van Wolfswinkel et al., 2014). Heterogeneity exists in neoblast populations, but it is likely that at least one subpopulation acts as a true stem cell while other subsets may have more restricted differentiation capacity (Scimone et al., 2014; van Wolfswinkel et al., 2014; Wagner et al., 2011). Based on these properties, planarians are an exceptional model to decipher fundamental mechanisms of stem cell biology and tissue regeneration.
The different biological functions of each GATA factor in mammals are associated with biochemical and molecular complexity that may involve compensatory functions. Therefore, some of this complexity can be resolved by studying GATA factors in animal species in which the GATA family has not expanded to the levels found in mice or humans. For example, in Caenorhabditis elegans, intestinal development is largely controlled by one GATA factor (McGhee, 2013; McGhee et al., 2007). Schmidtea mediterranea possesses a single homolog for GATA-4, -5, and -6, and phylogenetic analysis has shown Smed-gata4/5/6 falls within the GATA-4,-5, and -6 clade (Wagner et al., 2011). All six mammalian GATA transcription factors contain a highly conserved DNA binding domain consisting of two zinc fingers with a Cys-X 2-Cys-X 17-Cys-X 2-Cys motif that dictates binding to the GATA nucleotide sequence element (Molkentin, 2000): these two key domains are conserved in Smed-gata4/5/6 (Supplemental Fig. S1A), suggesting this GATA factor can function as a transcriptional regulator in planarians. Previous RNA-sequencing (RNA-Seq) studies have shown Smed-gata4/5/6 transcripts are expressed at high levels in the intestine but also in populations of neoblasts (Onal et al., 2012; Resch et al., 2012) (Supplemental Fig. S1B). These observations are consistent with recent studies of single neoblast cells that showed expression of Smed-gata4/5/6 in the gamma subset of neoblasts (van Wolfswinkel et al., 2014; Wurtzel et al., 2015) (Supplemental Fig. S1C) and a previous study showing Smed-gata4/5/6 expression in neoblasts interspersed between the intestinal branches (Wagner et al., 2011).
Here we found that disruption of Smed-gata4/5/6 function in intact and injured worms primarily results in intestinal defects. In addition, however, we observed that the Smed-gata4/5/6(RNAi) phenotype does not exclusively affect the intestinal lineage, indicating that Smed-gata4/5/6 function may play a role in the differentiation of other cell types in planarians. Our data support a model in which intestinal defects due to knock-down of Smed-gata4/5/6 may indirectly affect neoblast populations and the differentiation of non-intestinal lineages.
2. Results
2.1. Loss of Smed-gata4/5/6 function disrupts homeostasis in planarians
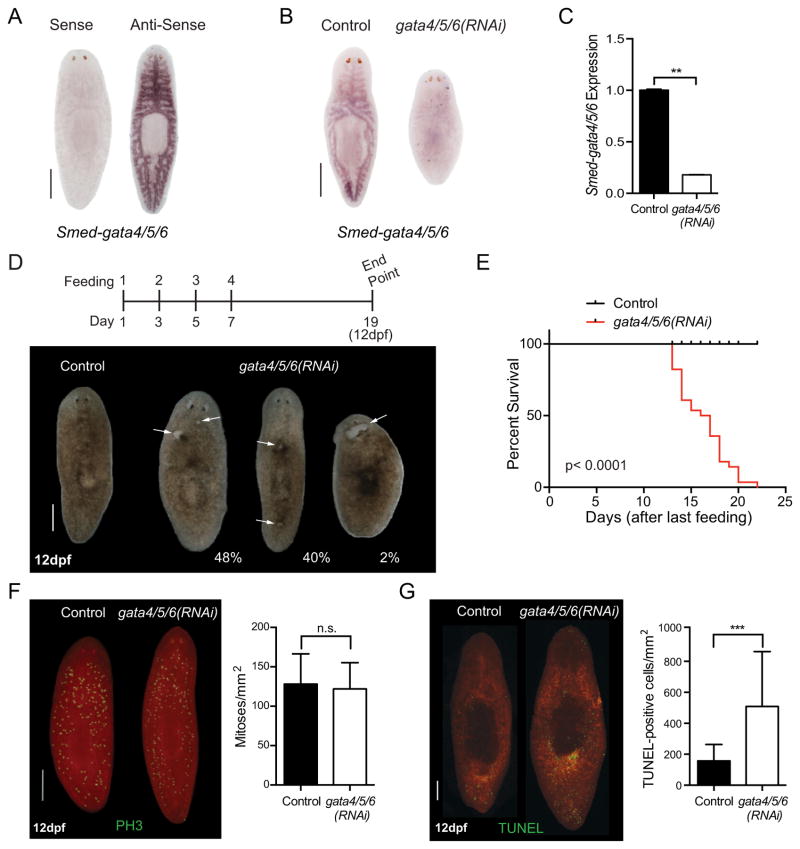
Whole-mount in situ hybridization (WISH) showed high levels of Smed-gata4/5/6 expression in the digestive tract of the worms (Fig. 1A), as previously described (Wagner et al., 2011), and reminiscent of what is seen in the mammalian gut (Beuling et al., 2011; Bossard and Zaret, 1998; Dusing and Wiginton, 2005). These observations suggested that Smed-gata4/5/6 may play a role in the differentiation and/or the maintenance of intestinal structures in S. mediterranea.
Fig. 1.
Smed-gata4/5/6 loss in homeostatic animals increases cell death. (A) Whole-mount in situ hybridization (WISH) for Smed-gata4/5/6 expression (anti-sense probe) in uninjured animal. The sense Smed-gata4/5/6 probe is used as a control. Scale bar: 500 μm. (B) Representative WISH for Smed-gata4/5/6 in a control and Smed-gata4/5/6(RNAi) animal, 12 days after final RNAi feeding (anti-sense probe). Scale bar: 500 μm. (C) RT-qPCR analysis of Smed-gata4/5/6 levels in control and Smed-gata4/5/6(RNAi) animals. Analysis based on 2 biological replicates (n = 10), each replicate containing 5 animals pooled. Ct values are normalized to internal control GAPDH and relative to controls. Two-tailed unpaired Student’s t-test p-value = 0.0091, values represent average and error bars s.e.m. (D) Representative live images of intact RNAi animals 12 days after the final feeding. Smed-gata4/5/6(RNAi) animals develop lesions on both the anterior and posterior as indicated by white arrows. Scale bar: 500 μm. (n > 50) The feeding time line is shown above. (E) Kaplan-Meier survival of Smed-gata4/5/6(RNAi) animals (n = 30, med. survival = 16.5 days). p-value < 0.0001 by log-rank test for significance. (F) Representative images of whole-mount immunostaining using anti-PH3 antibody in intact animals 12 days after final feeding (left) and quantification of mitoses (right) (two-way ANOVA, p-value = 0.5433). Three independent experiments, n = 30. Results represent average and error bars s.e.m. Scale bar: 500 μm. (G) TUNEL assay quantification of control and Smed-gata4/5/6(RNAi) intact animals 12 days after final feeding (two-way ANOVA, ***: p-value = 0.0002). Two independent experiments, n ≥ 19. Results represent average and error bars s.e.m.
To investigate the role of Smed-gata4/5/6, we knocked down its expression in intact animals by RNA interference (RNAi). We developed a feeding schedule that consists of four feedings every 2 days (Fig. 1D). The effective downregulation of Smed-gata4/5/6 mRNA was independently confirmed by WISH and RT-qPCR experiments (Fig. 1B and C). Smed-gata4/5/6(RNAi) animals developed dorsal lesions twelve days after the final feeding (12dpf, Fig. 1D), which eventually led to animal lethality (Fig. 1E). These observations indicate that Smed-gata4/5/6 is required for the long-term maintenance of adult tissue and homeostasis in planarians.
To investigate the cellular basis of these observations, we first examined whether the Smed-gata4/5/6(RNAi) would affect the proliferation of neoblasts. However, under these conditions, we observed no significant differences in the mitotic activity of control and experimental animals as measured by immunostaining for phospho-Histone H3 (PH3) expression, a marker of mitosis (Fig. 1F). Under physiological conditions, cell turnover is a balanced combination of cell division and cell death (Pellettieri et al., 2010). No changes in mitotic activity suggested that tissue loss in Smed-gata4/5/6(RNAi) animals could result from an increase in cell death. Indeed, quantification of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) showed a significant increase in TUNEL-positive cells in Smed-gata4/5/6(RNAi) animals compared to controls at 12dpf (Fig. 1G).
The increase in cell death after Smed-gata4/5/6(RNAi) may be a consequence of dysfunctional neoblast response to cellular turnover demands or structural defects in the intestine where Smed-gata4/5/6 is expressed. Throughout the initial stages of the experiment, we did not observe animal impairment to search for food nor differences in size between Smed-gata4/5/6(RNAi) and control worms (data not shown). Nonetheless, to investigate the possibility of abnormalities in intestinal morphology that develop overtime, we fed control and Smed-gata4/5/6(RNAi) worms with liver paste mixed with fluorescent-conjugated dextran, which labels the intestinal phagocytes and allow intestine visualization in situ (Forsthoefel et al., 2011). This experiment did not reveal any visible difference between the two groups at an early time point (Supplemental Fig. S2A). However, when we performed WISH for the intestinal marker, smedinx-9, at late stages of the experiment (12dpf), when animals were unable to eat, we found a significant loss of expression and intestinal integrity (Supplemental Fig. S2B). These experiments suggest that deterioration of the intestine may be linked to cell death and animal survival as some Smed-gata4/5/6 (RNAi) animals began to die at this time point.
Altogether, these experiments indicate that Smed-gata4/5/6 is necessary for the long-term maintenance of intestinal function and overall survival of planarians under homeostatic conditions. Based on these studies in intact animals, we sought to investigate the role of Smed-gata4/5/6 and to explore its mechanisms of action under conditions where neoblasts are challenged.
2.2. Smed-gata4/5/6 is critical for the regeneration of planarians after amputation
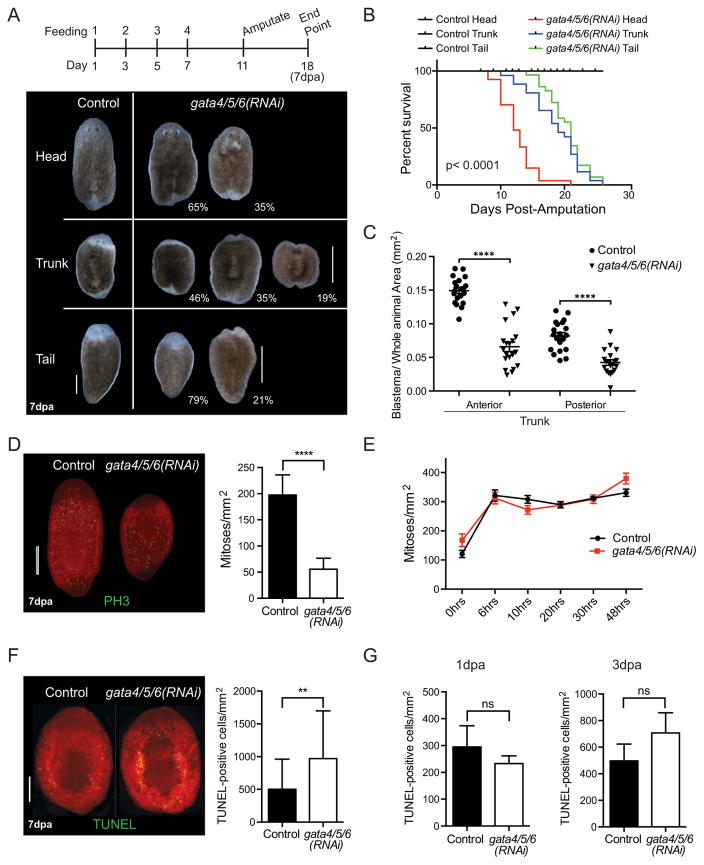
We performed RNAi feeding, 4 feedings every 2 days, and amputated planarians four days after the final feeding. We analyzed the animals 7 days post-amputation (7dpa) and observed similar defects as with the uninjured worms, but exacerbated, including visible epithelial lesions. Head, trunk, and tail fragments all showed some impaired blastema formation and increased mortality upon Smed-gata4/5/6 knock-down (Fig. 2A and B).
Fig. 2.
Smed-gata4/5/6 loss perturbs regeneration, decreases mitotic activity, and increases cell death. (A) Representative live images of regenerating RNAi head, trunk, and tail worms 7 days post-amputation. Scale bars: 500 μm. (n > 50) The feeding time line is shown above. (B) Kaplan-Meier survival of Smed-gata4/5/6(RNAi) animals (n = 30, head-med. survival = 12 days, trunk-med. survival = 19 days, tail-med. survival = 21 days). p-value < 0.0001 by log-rank test for significance. (C) Loss of Smed-gata4/5/6 prevents blastema growth. Represented are the anterior and posterior blastema areas of the trunks. A ratio of the blastema area over the whole animal area was used to take into account the initial size of the regenerating fragment (two-way ANOVA, ****: p-value < 0.0001). Two independent experiments, n = 20. (D) Representative images of whole-mount immunostaining using anti-PH3 antibody in regenerating animals 7 days post-amputation. Quantification of mitoses in the trunk of control and Smed-gata4/5/6(RNAi) animals (two-way ANOVA, p-value < 0.0001). Three independent experiments, n ≥ 25. Results represent average and error bars s.e.m. Scale bar: 500 μm. (E) Graph of early mitosis peaks after amputation. In controls two peaks of mitotic activity occur, first at 6 h and second at 30–48 h post-amputation. Two independent experiments, n = 10 per time point. Results represent average and error bars s.e.m. (F) Representative images of apoptosis (TUNEL-positive cells) in RNAi animals 7 days post-amputation. Quantification of TUNEL-positive cells in control and Smed-gata4/5/6(RNAi) worms 7 days post-amputation (two-way ANOVA, ****: p-value < 0.0001, **: p-value = 0.0010, *: p-value = 0.0239). Two independent experiments, n ≥ 19. Results represent average and error bars s.e.m. Scale bar: 100 μm. (E) Graphs of early apoptosis after amputation. Quantification of TUNEL-positive cells at 1 d and 3 days post-amputation (two-way ANOVA). Two independent experiments, n = 10 per time point. Results represent average and error bars s.e.m.
We decided to focus our analyses on the phenotypes of regenerating trunks because they have to regenerate both a tail and a head. Decreased blastema growth was highly significant in both the anterior and the posterior parts of regenerating trunks in knock-down animals (Fig. 2C). All 7dpa mutant trunk fragments analyzed lacked photoreceptor pigmentation (Fig. 2A and data not shown, see below). Next, to determine the role of Smed-gata4/5/6 in neoblast populations during regeneration, we quantified neoblast mitotic activity with PH3 at 7dpa. We found that cell divisions were significantly decreased in Smed-gata4/5/6(RNAi) animals compared to controls (Fig. 2D), suggesting that disruption of Smed-gata4/5/6 impairs the neoblast response to the demands of tissue regeneration. A previous study has shown that two early bursts of neoblast proliferation take place in planarians just after amputation, a systemic response at ~6h and a local response at ~48 h (Wenemoser et al., 2012). In Smed-gata4/5/6 (RNAi) animals, we found no observable changes in these early proliferative events (Fig. 2E).
At 7dpa we also found a significant increase in TUNEL-positive cells in Smed-gata4/5/6(RNAi) animals compared to controls (Fig. 2F). When we performed the TUNEL assay on 7dpa transverse sections, we observed cell death throughout the animal, and not only in the intestine of Smed-gata4/5/6(RNAi) animals (Supplemental Fig. S3). To determine if Smed-gata4/5/6(RNAi) leads to cell death during the early stages of regeneration, we performed the TUNEL assay at two early time points, 1 and 3 days post-amputation (1dpa and 3dpa, respectively) and found no significant differences in Smed-gata4/5/6(RNAi) animals as compared to controls (Fig. 2G).
Together, the late decrease in proliferation and the late increase in cell death observed in Smed-gata4/5/6(RNAi) worms suggested that Smed-gata4/5/6 may not directly affect neoblasts but that loss of Smed-gata4/5/6 may indirectly affect the differentiation of neoblast populations during the late stages of injury response.
2.3. Smed-gata4/5/6 is critical for the differentiation of specific cell lineages in planarians during regeneration
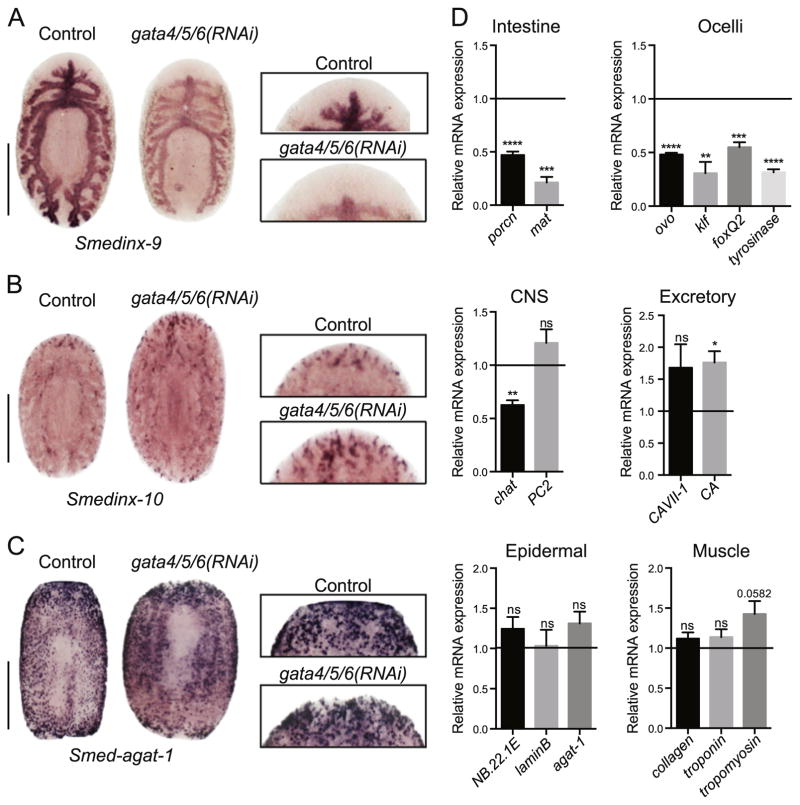
To investigate a potential role for Smed-gata4/5/6 in the differentiation and remodeling of different organs, we examined markers for various lineages. First, we performed WISH for smedinx-9, an intestinal marker (Oviedo and Levin, 2007). Qualitative evaluation of the smedinx-9 staining revealed that the intensity of the signal was diminished 7dpa in Smed-gata4/5/6(RNAi) animals; the gross morphology of the intestine within the pre-existing tissue appeared to remain without changes (Fig. 3A). Strikingly, however, 7dpa Smed-gata4/5/6(RNAi) animals did not develop new intestinal branches within the newly formed blastema, in contrast to control animals (Fig. 3A). These observations provide support for a key role for Smed-gata4/5/6 in intestinal differentiation.
Fig. 3.
Analysis of differentiated cell lineages in Smed-gata4/5/6(RNAi) animals. (A) Representative WISH for smedinx-9, an intestine marker, in control and Smed-gata4/5/6 (RNAi) animals (anti-sense probe). A magnification of the blastema (inset) shows the absence of intestinal branching in the newly formed tissue in knock-down worms 7 days post-amputation. (n = 5) Scale bar: 250 μm. (B) Representative WISH for smedinx-10, an excretory marker (flame cells), in control and Smed-gata4/5/6(RNAi) animals (anti-sense probe). A magnification of the blastema (inset) shows that new flame cells are established in the newly-formed tissue 7 days post-amputation. (n = 5) Scale bar: 250 μm. (C) Representative WISH for Smed-agat-1, a general differentiation marker, in control and Smed-gata4/5/6(RNAi) animals (anti-sense probe). A magnification of the blastema (inset) shows that differentiated cell types are found in the new tissue 7 days post-amputation. (n = 5) Scale bar: 500 μm. (D) RT-qPCR analysis showing mRNA expression changes in differentiation tissue marker transcripts following Smed-gata4/5/6(RNAi) 7 days post-amputation. Bar graphs show fold change in expression relative to controls after normalization to GAPDH. Controls represented by horizontal line set at 1. Two-tailed unpaired Student’s t-test (****: p-value < 0.0001, ***: p-value < 0.001, **: p-value > 0.01*: p-value > 0.05, ns: not significant) values represent average and error bars s.e.m. Analysis for regenerating trunks based on 3 biological replicates (n = 15), each replicate containing 5 animals pooled.
More surprisingly, we found defects after Smed-gata4/5/6(RNAi) in the terminal differentiation of the ocelli, as noted above, with a complete absence of photoreceptor pigmentation in all regenerating tail and trunk fragments, even at late time points in survival studies (Fig. 2A, and data not shown). Using Synapsin as a marker for the differentiation of the central nervous system, we identified differentiated neuronal cells in the regenerating foremost anterior region of trunks and posterior fragments both in controls and knock-down worms but we also observed fusion defects between the two CNS tracts in the knock-down animals in the most severe cases (unresolved cleft in the blastema) (Supplemental Fig. S4). Thus, low levels of Smed-gata4/5/6 prevent the development of photoreceptors and sometimes lead to developmental defects in the CNS.
In contrast, WISH for smedinx-10, a marker for the excretory system (flame cells) (Oviedo et al., 2010), showed no visible defects in the knock-down animals, with clear expression of this marker in the newly regenerated tissue (Fig. 3B). Similarly, when we examined the expression of a L-arginine:glycine amidino-transferase (Smed-AGAT-1), which is expressed broadly in sub-epidermal mesenchymal tissue (Eisenhoffer et al., 2008; Wagner et al., 2011), we found no qualitative difference in the expression of this marker between Smed-gata4/5/6(RNAi) animals and controls 7dpa (Fig. 3C).
To quantitatively assess changes in gene expression in differentiated tissues, we performed RT-qPCR experiments for genes associated with the intestine, the muscles, the epidermis, the nervous and excretory systems. These experiments confirmed a significant inhibition of intestinal and photoreceptor differentiation in Smed-gata4/5/6(RNAi) worms, variable changes in the CNS, and no significant changes in the expression of most markers belonging to the excretory system, muscles, and the epidermis (Fig. 3D).
Together, these experiments identified a key role for Smed-gata4/5/6 in intestinal regeneration and selective roles of this transcription factor in non-intestinal tissues in injured animals during the later stages of regeneration.
2.4. Absence of Smed-gata4/5/6 affects several neoblast populations
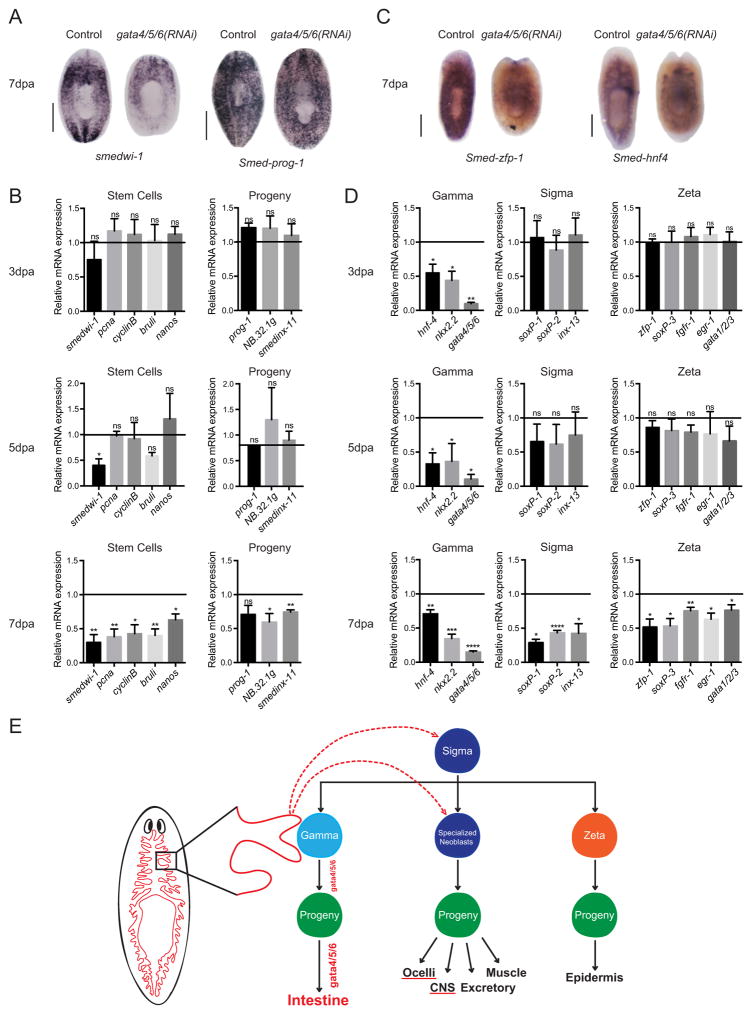
Based on our observations of significantly decreased mitotic cells in regenerating Smed-gata4/5/6(RNAi) animals at late time points (Fig. 2D) and the defects described above in differentiated cell lineages (Fig. 3), we sought to examine the role of Smed-gata4/5/6 in neoblasts and their early progeny. We first compared the expression of neoblasts and early progeny markers (smedwi-1 and Smed-prog1, respectively) by WISH following Smed-gata4/5/6 (RNAi) in 7dpa trunks. We found a decrease in smedwi-1 expression and no visible change in Smed-prog-1 (Fig. 4A). RT-qPCR analysis for general neoblast and early progeny markers showed no or few significant changes at 3dpa and 5dpa but significant decreases in these markers by 7dpa (Fig. 4B); this effect was not observed in uninjured animals (data not shown). Thus, the effects of Smed-gata4/5/6(RNAi) on neoblasts likely begin around day 4–5 of regeneration.
Fig. 4.
Loss of Smed-gata4/5/6 rapidly eliminates the gamma sub-class of neoblasts and affects both the zeta-class and sigma-class. (A) Representative WISH for smedwi-1 and Smed-prog-1, markers for the stem cells and progeny, respectively. (n = 5) Scale bars: 500 μm. (B) Representative WISH for Smed-zfp-1 and Smed-hnf-4, markers for the zeta-class and gamma-class, respectively. (n = 5) Scale bars: 500 μm. (C) RT-qPCR analysis showing mRNA expression changes in stem cell and progeny marker transcripts of control and Smed-gata4/5/6(RNAi) animals. Bar graphs show fold change in expression relative to controls after normalization to GAPDH. Controls represented by horizontal line set at 1. Two-tailed unpaired Student’s t-test (****: p-value < 0.0001,***: p-value < 0.001, **: p-value > 0.01, *: p-value > 0.05, ns: not significant) values represent average and error bars s.e.m. Analysis for 3dpa and 7dpa regenerating trunks based on 3 biological replicates (n = 15), each replicate containing 5 animals pooled. Analysis for 5dpa regenerating trunks based on 2 biological replicates (n = 10), each replicate containing 5 animals pooled. (D) RT-qPCR analysis showing mRNA expression changes in gamma-class, zeta-class, and sigma-class marker transcripts following Smed-gata4/5/6 RNAi. Bar graphs show fold change in expression relative to controls after normalization to GAPDH. Controls represented by horizontal line set at 1. Two-tailed unpaired Student’s t-test (****: p-value < 0.0001, ***: p-value < 0.001, **: p-value > 0.01, *: p-value > 0.05, ns: not significant) values represent average and error bars s.e.m. Analysis for 3dpa and 7dpa regenerating trunks based on 3 biological replicates (n = 15), each replicate containing 5 animals pooled. Analysis for 5dpa regenerating trunks based on 2 biological replicates (n = 10), each replicate containing 5 animals pooled. (E) Model of Smed-gata4/5/6 action in S. mediterranea (based on van Wolfswinkel et al. (2014)). Gamma neoblasts are required for the development and maintenance of the intestine, and Smed-gata4/5/6, which is expressed at low levels in these neoblasts and higher levels in differentiated intestinal cells, is intrinsically essential for this process. The most likely mechanism to explain the late defects in other neoblast populations (e.g. sigma and zeta subtypes) and other differentiation lineages (e.g. ocelli and CNS) in Smed-gata4/5/6 (RNAi) animals would be that intestinal defects disrupt these neoblasts and differentiation pathways indirectly (possible disruption of a niche, or lack of unidentified proliferation/survival factors). See text for a discussion on alternative models.
A recent study of the planarian stem cell compartment identified two major classes of neoblasts, the zeta-class and sigma-class, which are further divided into subclasses (van Wolfswinkel et al., 2014). The gamma subclass, a branch of sigma, was predicted to be involved in the development of the planarian gut, and Smed-gata4/5/6 is expressed in this subpopulation (van Wolfswinkel et al., 2014; Wurtzel et al., 2015) (Supplemental Fig. S1C). The phenotypes of Smed-gata4/5/6(RNAi) worms and our molecular analyses raised the possibility that gamma neoblasts might be affected by loss of Smed-gata4/5/6. Indeed, RT-qPCR analysis of two other markers of the gamma neoblast population showed a significant decrease in expression as early as 3dpa, suggestive of an early loss of these populations in Smed-gata4/5/6(RNAi) worms (Fig. 4D). In contrast, loss of zeta and sigma neoblast expression markers was only significant following Smed-gata4/5/6(RNAi) in regenerating 7dpa trunks (Fig. 4D). Qualitative analysis by WISH following Smed-gata4/5/6(RNAi) in 7dpa trunks also showed a decrease in Smed-hnf4 (gamma) and Smed-zfp-1 (zeta) expression (Fig. 4C). Together, these observations indicate that Smed-gata4/5/6 is a key regulator of intestinal differentiation in Schmidtea mediterranea, from gamma neoblasts to differentiated intestinal cells, and suggest that intestinal defects induced by loss of this intestinal transcription factor may indirectly affect other neoblast populations and the differentiation of other lineages.
3. Discussion
Here we examined the role of the planarian homolog of mammalian GATA-4, -5, and -6 transcription factors in organismal homeostasis and differentiation. Smed-gata4/5/6(RNAi) is detrimental to long-term neoblast maintenance, regeneration, differentiation of specific lineages, and ultimately survival. Our data further indicate that, in addition to its main role in intestinal differentiation from the gamma subclass of neoblasts, Smed-gata4/5/6 may be indirectly implicated in the differentiation of other cell lineages (Fig. 4E).
GATA transcription factors are involved in embryonic development, differentiation, and adult tissue maintenance (Chlon and Crispino, 2012; Duncan, 2005). In vertebrates, six GATA factors are conserved and are separated into two major subfamilies. In contrast, C. elegans and Drosophila have one GATA1/2/3-like factor and multiple endoderm/mesoderm GATA4/5/6 related GATA factors (Aronson et al., 2014). In S. mediterranea we found two GATA factors orthologous to vertebrates, Smed-gata1/2/3 and Smed-gata4/5/6, indicating that this planarian lacks the functionally redundant GATA4/5/6-like transcription factors often seen in lower Metazoa. Intriguingly, both planarian GATA factors maintain complete dual zinc finger domains, unlike C. elegans where all GATA4/5/6 subgroup factors lack the N-terminal zinc finger (Gillis et al., 2008). Initial Smed-gata1/2/3 RNAi experiments did not yield visible phenotypes (data not shown), which led us to focus on Smed-gata4/5/6 for this study; in addition, double RNAi experiments with Smed-gata1/2/3 and Smed-gata4/5/6 did not visibly enhance the phenotypes of Smed-gata4/5/6(RNAi) worms (data not shown), suggesting that Smed-gata1/2/3 does not compensate for the loss of Smed-gata4/5/6. Additionally, Smed-gata1/2/3 is expressed in zeta neoblasts (van Wolfswinkel et al., 2014). The functional role of Smed-gata1/2/3 will need to be investigated in future studies.
Smed-gata4/5/6(RNAi) prevented the development of new intestinal branches in planarians, showing that the intrinsic function of this transcription factor and its vertebrate homologs in intestinal development is conserved (Bossard and Zaret, 1998; Charron and Nemer, 1999; Liu et al., 2002; Zaret et al., 2008; Zhao et al., 2005). These data reveal selective contribution of a transcription factor during simultaneous regeneration of adult tissues, which further validates S. mediterranea as a relevant model organism to study developmental pathways in the context of the whole organism. Recent single-cell transcriptional profiling and RNA-Seq studies indicate that Smed-gata4/5/6 may not be expressed in all intestinal cells and possibly not all gamma neoblasts (Fig. S1C) (van Wolfswinkel et al., 2014; Wurtzel et al., 2015). Further experiments will be required to address whether Smed-gata4/5/6 is responsible for the generation of all cells within the digestive system; it is also possible that the role of Smed-gata4/5/6 may be dependent on whether the animals are under homeostatic conditions or responding to injury.
Photoreceptor pigmentation and, in severe cases, CNS development were also affected by loss of Smed-gata4/5/6. Emerging evidence indicates that GATA4 and GATA6 are expressed in the CNS and that GATA4 may play a role in the proliferation and the survival of astrocytes (Agnihotri et al., 2009; Kamnasaran and Guha, 2005). Similarly, Smed-gata4/5/6(RNAi) caused an increase in cell death, supporting previous reports indicating GATA4 and GATA6 regulate anti-apoptotic signaling (Agnihotri et al., 2009; Rong et al., 2012; Suzuki, 2011). A careful analysis of data from Wurtzel and colleagues (Wurtzel et al., 2015) shows some expression in a “neural” cluster (Fig. S1C); expression of Smed-gata4/5/6 in some “neural” cells might be the cause of some direct effects on neuronal differentiation in the knock-down animals. However, similar RNA-Seq data from Molinaro and colleagues (Molinaro and Pearson, 2016) in cells under homeostatic conditions indicate that a subtype of neoblasts that may contribute specifically to neuronal lineages and these cells do not express Smed-gata4/5/6. These analyses are clearly preliminary and do not exclude a direct effect, but, overall, would suggest that the phenotypes observed in non-intestinal lineages in Smed-gata4/5/6(RNAi) animals are indirect.
Another open question is why the phenotypes we describe here with Smed-gata4/5/6(RNAi) are different from those observed with Smed-nkx2.2(RNAi), even though Smed-nkx2.2 may be another key regulator of intestinal differentiation expressed in gamma neoblasts and differentiated intestinal cells (Forsthoefel et al., 2012). Loss of Smed-nkx2.2 has similar effects on intestinal differentiation as loss of Smed-gata4/5/6 but more rapid effects on overall neoblast proliferation (Forsthoefel et al., 2012). Analysis of single-cell RNA-Seq shows that the pattern of expression of the two genes is similar but not identical, which may explain these differences (for example, only 17/28 intestinal cells express both genes in Wurtzel et al. (2015) – data not shown). It is also possible that different protocols and knock-down efficiency result in different phenotypes.
In conclusion, single-cell RNA-Seq studies strongly indicate that Smed-gata4/5/6 is expressed mostly in the intestinal lineage, from gamma neoblasts to differentiated intestinal cells (Molinaro and Pearson, 2016; Scimone et al., 2016, 2014; van Wolfswinkel et al., 2014; Wurtzel et al., 2015). Together with our observations that phenotypes in non-intestinal lineages arise late during the response to injury, this supports a model in which Smed-gata4/5/6 plays a critical role in intestinal differentiation and wherein the differentiation of other lineages may be affected indirectly by intestinal defects (Fig. 4E). This model would fit with a conserved role of this GATA transcription factor in intestinal development and would provide a simple explanation for its role in other lineages. Other less-likely models may explain our data, including functional interactions between gamma neoblasts and other neoblast subpopulations but the inability to perform lineage-tracing assays in planarians severely limit possible investigations of the functional interactions between neoblast subclasses and different differentiation lineages in this model. In our favored model where the intestine serves as a structural and/or functional niche that normally supports the differentiation of other lineages from neoblasts, we do not understand why some lineages are more affected or more rapidly affected than others. Notably, a primary defect in the CNS has been shown to result in secondary defects in the planarian gut (Cebria and Newmark, 2007), and the converse is therefore possible. Future studies will seek to identify the mechanisms underlying such non-cell autonomous roles for the intestine in the development of other differentiation pathways in planarians.
4. Material and methods
4.1. Protein sequence and phylogenetic analysis
Smed-gata4/5/6 was found annotated in the NCBI database with the use of BLAST (GenBank accession # JF802198). Protein sequence alignments with other species and a predictive evolutionary model were created using CLUSTALW and MEGA6 software (www.megasoftware.net), respectively.
4.2. Planarian culture and RNAi
The asexual CIW4 strain of Schmidtea mediterranea was used in all experiments and maintained as previously described (Oviedo et al., 2008a). For RNA interference assays (RNAi), HT115 bacteria containing cDNA was cloned in to the pPR244 vector to make dsRNA as previously described (Recombinant DNA procedures approved under APB# 712-JS0510) (Reddien et al., 2005). Briefly, bacteria were grown to an OD600 of 0.6 and induced with 1 mM isopropyl-β-thiogalactopyranoside for 2 h, centrifuged and mixed with liver paste. Animals were fed every 2 days for 4 feedings. Amputation was performed four days after the final feeding. The control RNAi plasmid used contains the C. elegans gene unc-22 (Addgene plasmid 1690). For dextran feeding assays, animals were fed a dextran-liver paste mixture 4 days after the last feeding and imaged 3 days later. 100μl of liver paste was mixed with 2 μl (1 mg/mL) 10,000 MW dextran conjugated to Alexa 546 (Molecular Probes), and fed to the planarians (Forsthoefel et al., 2011).
4.3. RNA analysis by RT-qPCR and in situ hybridization
RNA was extracted with TRIzol (Invitrogen). RT-PCR and quantitative Real-Time PCR were performed using the ProtoScript cDNA synthesis kit (New England BioLabs) and the PerfeCTa SYBRGreen FastMix (Quanta Biosciences), respectively. All reactions were performed in triplicates and run on an ABI 7900 HT Fast Real Time PCR System (Applied Biosystems). Fold change in expression of Smed-gata4/5/6(RNAi) animals shown relative to controls (unc-22(RNAi) animals) after all CT values are normalized to the internal control Smed-GAPDH. See Supporting Information Table S1 for primer sequences. Animals were fixed and whole-mount in situ hybridization (WISH) was performed as previously described (King and Newmark, 2013; Pearson et al., 2009).
4.4. Immunostaining, TUNEL assay, and image processing
Planarians were fixed and immunostaining was performed as previously reported (Oviedo et al., 2008b). Antibodies were used at the following dilutions: 1:250 anti-phospho-histone H3 (phosphorylated Serine 10 on histone H3, Millipore), 1:75 anti-SYNORF1 (Synapsin, Developmental Studies Hybridoma Bank), 1:800 anti-rabbit DyLight 594 conjugated and 1:500 anti-mouse HRP. Tyramide development was performed as previously described (Cowles et al., 2012). TUNEL assay on whole worms was performed as previously described (Pellettieri et al., 2010). Counted foci were normalized to the area (mm2) using NIS element software (Nikon). Digital images were captured using a Nikon AZ-100 multizoom microscope and NIS Elements AR 3.2 software. Area measurements and scale bars were calculated on NIS Elements AR 3.2 software.
For TUNEL assay on sections, animals were fixed according to (King and Newmark, 2013). Briefly, animals were killed in 5% N-acetyl cysteine in PBS for 5 min, then fixed in 4% formaldehyde in PBSTx (0.3% Triton X-100) for 30 min, and washed 2 × in PBSTx. Animals were then dehydrated in 50% methanol in PBSTx followed by 100% methanol and stored for < 2weeks at −20 °C in 100% methanol before being processed and embedded in paraffin. Blocks were serially sectioned at 10 μM. For staining the Apop-Tag® Red Apoptosis Detection Kit (Millipore cat. S7165) protocol was followed. Immunostaining for anti-Smed-6G10 (Developmental Studies Hybridoma Bank) was performed as previously reported (Ross et al., 2015). Counted foci were normalized to the area (mm2) using Image J. Digital images were captured using a Keyence All-in-one Fluorescent Microscope BZ-X700 series and BZ-X Analyzer software. Scale bars and area measurements were calculated on BZ-X Analyzer software and Image J, respectively. Adobe Photoshop was used to adjust the brightness and contrast, and merge images.
4.5. Statistical analyses
Statistical analyses, two-way ANOVA and Student’s t-Test, were performed using Prism6 (GraphPad).
Supplementary Material
Acknowledgments
The authors wish to thank members of the Sage and Oviedo labs for helpful advice and discussions, especially T. Harshani Peiris and Daniel Ramirez. Research reported in this manuscript was supported by the National Institutes of Health (CA176114 and GM109372 to N.J.O. and CA09302 to N.F. as part of the Cancer Biology program), and the Lucile Packard Foundation for Children’s Health (J.S.). J.S is the Harriet and Mary Zelencik Scientist in Children’s Cancer and Blood Diseases. The authors declare no conflict of interest.
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2016.08.015.
Footnotes
Author contribution
NF: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing.
NO: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing.
JS: Conception and design, data analysis and interpretation, manuscript writing.
References
- Agnihotri S, Wolf A, Picard D, Hawkins C, Guha A. GATA4 is a regulator of astrocyte cell proliferation and apoptosis in the human and murine central nervous system. Oncogene. 2009;28:3033–3046. doi: 10.1038/onc.2009.159. [DOI] [PubMed] [Google Scholar]
- Akiyama Y, Watkins N, Suzuki H, Jair KW, van Engeland M, Esteller M, Sakai H, Ren CY, Yuasa Y, Herman JG, Baylin SB. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol Cell Biol. 2003;23:8429–8439. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson BE, Stapleton KA, Krasinski SD. Role of GATA factors in development, differentiation, and homeostasis of the small intestinal epithelium. Am J Physiol Gastrointest Liver Physiol. 2014;306:G474–G490. doi: 10.1152/ajpgi.00119.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J, Chazaud C. A close look at the mammalian blastocyst: epiblast and primitive endoderm formation. Cell Mol Life Sci. 2014;71:3327–3338. doi: 10.1007/s00018-014-1630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, Lindeman GJ, Visvader JE. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- Betchaku T. Isolation of planarian neoblasts and their behavior in vitro with some aspects of the mechanism of the formation of regeneration blastema. J Exp Zool. 1967;164:407–433. doi: 10.1002/jez.1401640310. [DOI] [PubMed] [Google Scholar]
- Beuling E, Baffour-Awuah NY, Stapleton KA, Aronson BE, Noah TK, Shroyer NF, Duncan SA, Fleet JC, Krasinski SD. GATA factors regulate proliferation, differentiation, and gene expression in small intestine of mature mice. Gastroenterology. 2011;140(1219–1229):e1211–e1212. doi: 10.1053/j.gastro.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-Chichi CD, Smedberg JL, Rula M, Nicolas E, Yeung AT, Adamo RF, Frolov A, Godwin AK, Xu XX. Alteration of differentiation potentials by modulating GATA transcription factors in murine embryonic stem cells. Stem Cells Int. 2010;2010:602068. doi: 10.4061/2010/602068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebria F, Newmark PA. Morphogenesis defects are associated with abnormal nervous system regeneration following roboA RNAi in planarians. Development. 2007;134:833–837. doi: 10.1242/dev.02794. [DOI] [PubMed] [Google Scholar]
- Charron F, Nemer M. GATA transcription factors and cardiac development. Semin Cell Dev Biol. 1999;10:85–91. doi: 10.1006/scdb.1998.0281. [DOI] [PubMed] [Google Scholar]
- Chlon TM, Crispino JD. Combinatorial regulation of tissue specification by GATA and FOG factors. Development. 2012;139:3905–3916. doi: 10.1242/dev.080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles MW, Hubert A, Zayas RM. A Lissencephaly-1 homologue is essential for mitotic progression in the planarian Schmidtea mediterranea. Dev Dyn. 2012;241:901–910. doi: 10.1002/dvdy.23775. [DOI] [PubMed] [Google Scholar]
- Duncan SA. Generation of embryos directly from embryonic stem cells by tetraploid embryo complementation reveals a role for GATA factors in organogenesis. Biochem Soc Trans. 2005;33:1534–1536. doi: 10.1042/BST0331534. [DOI] [PubMed] [Google Scholar]
- Dusing MR, Wiginton DA. Epithelial lineages of the small intestine have unique patterns of GATA expression. J Mol Histol. 2005;36:15–24. doi: 10.1007/s10735-004-2908-9. [DOI] [PubMed] [Google Scholar]
- Eisenhoffer GT, Kang H, Sanchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel DJ, James NP, Escobar DJ, Stary JM, Vieira AP, Waters FA, Newmark PA. An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Dev Cell. 2012;23:691–704. doi: 10.1016/j.devcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel DJ, Park AE, Newmark PA. Stem cell-based growth, regeneration, and remodeling of the planarian intestine. Dev Biol. 2011;356:445–459. doi: 10.1016/j.ydbio.2011.05.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Sedgwick T, Shi YB, Evans T. Distinct functions are implicated for the GATA-4,-5, and -6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol Cell Biol. 1998;18:2901–2911. doi: 10.1128/mcb.18.5.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis WQ, Bowerman BA, Schneider SQ. The evolution of protostome GATA factors: molecular phylogenetics, synteny, and intron/exon structure reveal orthologous relationships. BMC Evolut Biol. 2008;8:112. doi: 10.1186/1471-2148-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamnasaran D, Guha A. Expression of GATA6 in the human and mouse central nervous system. Brain Res Dev Brain Res. 2005;160:90–95. doi: 10.1016/j.devbrainres.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Ono K, Morimoto T, Wada H, Hirai M, Hidaka K, Morisaki T, Heike T, Nakahata T, Kita T, Hasegawa K. Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. J Biol Chem. 2005;280:19682–19688. doi: 10.1074/jbc.M412428200. [DOI] [PubMed] [Google Scholar]
- King RS, Newmark PA. In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev Biol. 2013;13:8. doi: 10.1186/1471-213X-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi-Osaki M, Ohneda O, Suzuki N, Minegishi N, Yokomizo T, Takahashi S, Lim KC, Engel JD, Yamamoto M. GATA motifs regulate early hematopoietic lineage-specific expression of the Gata2 gene. Mol Cell Biol. 2005;25:7005–7020. doi: 10.1128/MCB.25.16.7005-7020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard M, Brice M, Engel JD, Papayannopoulou T. Dynamics of GATA transcription factor expression during erythroid differentiation. Blood. 1993;82:1071–1079. [PubMed] [Google Scholar]
- Liu C, Morrisey EE, Whitsett JA. GATA-6 is required for maturation of the lung in late gestation. Am J Physiol Lung Cell Mol Physiol. 2002;283:L468–L475. doi: 10.1152/ajplung.00044.2002. [DOI] [PubMed] [Google Scholar]
- McGhee JD. The Caenorhabditis elegans intestine. Wiley interdisciplinary reviews. Dev Biol. 2013;2:347–367. doi: 10.1002/wdev.93. [DOI] [PubMed] [Google Scholar]
- McGhee JD, Sleumer MC, Bilenky M, Wong K, McKay SJ, Goszczynski B, Tian H, Krich ND, Khattra J, Holt RA, Baillie DL, Kohara Y, Marra MA, Jones SJ, Moerman DG, Robertson AG. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev Biol. 2007;302:627–645. doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro AM, Pearson BJ. In silico lineage tracing through single cell transcriptomics identifies a neural stem cell population in planarians. Genome Biol. 2016;17:87. doi: 10.1186/s13059-016-0937-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD. The zinc finger-containing transcription factors GATA-4,-5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- Onal P, Grun D, Adamidi C, Rybak A, Solana J, Mastrobuoni G, Wang Y, Rahn HP, Chen W, Kempa S, Ziebold U, Rajewsky N. Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. EMBO J. 2012;31:2755–2769. doi: 10.1038/emboj.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- Oviedo NJ, Levin M. smedinx-11 is a planarian stem cell gap junction gene required for regeneration and homeostasis. Development. 2007;134:3121–3131. doi: 10.1242/dev.006635. [DOI] [PubMed] [Google Scholar]
- Oviedo NJ, Nicolas CL, Adams DS, Levin M. Establishing and maintaining a colony of planarians. CSH protocols. 2008a;2008 doi: 10.1101/pdb.prot5053. pdb prot5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oviedo NJ, Pearson BJ, Levin M, Sanchez Alvarado A. Planarian PTEN homologs regulate stem cells and regeneration through TOR signaling. Dis Models Mech. 2008b;1:131–143. doi: 10.1242/dmm.000117. discussion 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates) Curr Opin Genet Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sanchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen KJ. Cytological studies on the planarian neoblast. Z Zellforsch Mikrosk Anat. 1959;50:799–817. doi: 10.1007/BF00342367. [DOI] [PubMed] [Google Scholar]
- Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sanchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Dev Biol. 2010;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikkarainen S, Tokola H, Kerkela R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Sanchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Resch AM, Palakodeti D, Lu YC, Horowitz M, Graveley BR. Transcriptome analysis reveals strain-specific and conserved stemness genes in Schmidtea mediterranea. PloS One. 2012;7:e34447. doi: 10.1371/journal.pone.0034447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues NP, Janzen V, Forkert R, Dombkowski DM, Boyd AS, Orkin SH, Enver T, Vyas P, Scadden DT. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106:477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- Rong L, Liu J, Qi Y, Graham AM, Parmacek MS, Li S. GATA-6 promotes cell survival by up-regulating BMP-2 expression during embryonic stem cell differentiation. Mol Biol Cell. 2012;23:3754–3763. doi: 10.1091/mbc.E12-04-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross KG, Omuro KC, Taylor MR, Munday RK, Hubert A, King RS, Zayas RM. Novel monoclonal antibodies to study tissue regeneration in planarians. BMC Dev Biol. 2015;15:2. doi: 10.1186/s12861-014-0050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Alvarado A, Yamanaka S. Rethinking differentiation: stem cells, regeneration, and plasticity. Cell. 2014;157:110–119. doi: 10.1016/j.cell.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Cote LE, Rogers T, Reddien PW. Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. Elife. 2016;5 doi: 10.7554/eLife.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Kravarik KM, Lapan SW, Reddien PW. Neoblast Specialization in Regeneration of the Planarian Schmidtea mediterranea. Stem Cell Rep. 2014;3:339–352. doi: 10.1016/j.stemcr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano F, Calatayud CF, Blazquez M, Torres J, Castell JV, Bort R. Gata4 blocks somatic cell reprogramming by directly repressing Nanog. Stem Cells. 2013;31:71–82. doi: 10.1002/stem.1272. [DOI] [PubMed] [Google Scholar]
- Suzuki YJ. Cell signaling pathways for the regulation of GATA4 transcription factor: implications for cell growth and apoptosis. Cell Signal. 2011;23:1094–1099. doi: 10.1016/j.cellsig.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Dev Cell. 2011;21:172–185. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turbendian HK, Gordillo M, Tsai SY, Lu J, Kang G, Liu TC, Tang A, Liu S, Fishman GI, Evans T. GATA factors efficiently direct cardiac fate from embryonic stem cells. Development. 2013;140:1639–1644. doi: 10.1242/dev.093260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Wagner DE, Reddien PW. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell. 2014;15:326–339. doi: 10.1016/j.stem.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente C, Conchillo A, Garcia-Sanchez MA, Odero MD. The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Crit Rev Oncol/Hematol. 2012;82:1–17. doi: 10.1016/j.critrevonc.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Orkin SH. GATA transcription factors: key regulators of hematopoiesis. Exp Hematol. 1995;23:99–107. [PubMed] [Google Scholar]
- Wenemoser D, Lapan SW, Wilkinson AW, Bell GW, Reddien PW. A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 2012;26:988–1002. doi: 10.1101/gad.187377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Cote LE, Poirier A, Satija R, Regev A, Reddien PW. A generic and cell-type-specific wound response precedes regeneration in planarians. Dev Cell. 2015;35:632–645. doi: 10.1016/j.devcel.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev Biol. 1999;209:1–10. doi: 10.1006/dbio.1999.9228. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Watts J, Xu J, Wandzioch E, Smale ST, Sekiya T. Pioneer factors, genetic competence, and inductive signaling: programming liver and pancreas progenitors from the endoderm. Cold Spring Harb Symp Quant Biol. 2008;73:119–126. doi: 10.1101/sqb.2008.73.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol Cell Biol. 2005;25:2622–2631. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, Blobel GA. GATA transcription factors and cancer. Genes Cancer. 2010;1:1178–1188. doi: 10.1177/1947601911404223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.