ABSTRACT
Regulation of icaADBC-encoded polysaccharide intercellular adhesin (PIA)/poly-N-acetylglucosasmine (PNAG) production in staphylococci plays an important role in biofilm-associated medical-device-related infections. Here, we report that the AraC-type transcriptional regulator Rbf activates icaADBC operon transcription and PIA production in Staphylococcus epidermidis. Purified recombinant Rbf did not bind to the ica operon promoter region in electrophoretic mobility shift assays (EMSAs), indicating that Rbf regulates ica transcription indirectly. To identify the putative transcription factor(s) involved in Rbf-mediated icaADBC regulation, the ability of recombinant Rbf to interact with the promoter sequences of known icaADBC regulators was investigated. Recombinant Rbf bound to the sarR promoter and not the sarX, sarA, sarZ, spx, and srrA promoters. Reverse transcription (RT)-PCR demonstrated that Rbf acts as a repressor of sarR transcription. PIA expression and biofilm production were restored to wild-type levels in an rbf sarR double mutant grown in brain heart infusion (BHI) medium supplemented with NaCl, which is known to activate the ica locus, but not in BHI medium alone. RT-PCR further demonstrated that although Rbf does not bind the sarX promoter, it nevertheless exerted a negative effect on sarX expression. Apparently, direct downregulation of the SarR repressor by Rbf has a dominant effect over indirect repression of the SarX activator by Rbf in the control of S. epidermidis PIA production and biofilm formation.
IMPORTANCE The importance of Staphylococcus epidermidis as an opportunistic pathogen in hospital patients with implanted medical devices derives largely from its capacity to form biofilm. Expression of the icaADBC-encoded extracellular polysaccharide is the predominant biofilm mechanism in S. epidermidis clinical isolates and is tightly regulated. Here, we report that the transcriptional regulator Rbf promotes icaADBC expression by negatively regulating expression of sarR, which encodes an ica operon repressor. Furthermore, Rbf indirectly represses the ica operon activator, SarX. The data reveal complicated interplay between Rbf and two Sar family proteins in fine-tuning regulation of the biofilm phenotype and indicate that in the hierarchy of biofilm regulators, IcaR is dominant over the Rbf-SarR-SarX axis.
INTRODUCTION
Staphylococci are responsible for the majority of biofilm-mediated device-related infections (1). Biofilms assist in the evasion of the host immune response and offer increased resistance to antimicrobial drugs. Many hospital patients undergo procedures involving the insertion of foreign biomaterials, ranging from simple intravascular catheters to more sophisticated ventricular assist devices. Vulnerable hospital patients with underlying medical conditions are particularly susceptible to device-related infections frequently caused by antibiotic-resistant pathogens. Thus, device-related infections represent a serious clinical problem, which in turn underpins the importance of understanding the mechanisms by which staphylococci form biofilms. Production of the icaADBC operon-encoded polysaccharide intercellular adhesin (PIA) or poly-N-acetylglucosasmine (PNAG) remains the best-understood mechanism of biofilm production in staphylococci (1–3). In Staphylococcus epidermidis, the cell wall-anchored autolysin (4) and the accumulation-associated protein (5–7) have also been implicated in the primary attachment and maturation phases of biofilm formation, respectively.
The contributions of many transcription factors to the regulation of biofilm formation in staphylococci underline the importance of the phenotype. Major regulators of the ica operon include the IcaR repressor (8, 9); a number of members of the 11-member Staphylococcus accessory regulator (Sar) family of proteins (10, 11); and the regulator of biofilm formation, Rbf. The rbf gene was first identified in 2004 in Staphylococcus aureus by transposon mutagenesis (12). Lim et al. reported that rbf was present in 22 of 27 S. aureus clinical isolates tested and that inactivation of the rbf gene in strain 8325-4 resulted in a biofilm-negative phenotype in tryptone soya broth (TSB) medium supplemented with glucose or NaCl (12). Rbf is a member of the AraC/XylS family of transcriptional regulators, defined by a conserved ∼100-amino-acid region comprising a DNA binding domain, which can positively or negatively regulate gene expression (13). Rbf controls the biofilm phenotype in S. aureus by increasing the production of PIA (14). Rbf-mediated activation of the ica operon is indirect and is achieved by activation of sarX, which in turn binds upstream of icaR within the icaA coding region, leading to repression of icaR and concomitant activation of the ica operon (15). The sarX gene is located immediately downstream of rbf on the S. aureus and S. epidermidis chromosomes, and overexpression of rbf in S. aureus UAMS-1 led to a >50-fold increase in sarX expression (14). Unlike in S. aureus, the contribution of sarX to S. epidermidis biofilm is influenced by growth conditions, playing a regulatory role in media supplemented with glucose (11) but not salt (unpublished data), suggesting a different role for SarX in the regulation of biofilm in these two organisms. Here, we report that Rbf-mediated biofilm regulation in S. epidermidis is mediated via SarR, which encodes a transcriptional repressor of the ica locus.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) medium (Sigma) supplemented when required with kanamycin (Kan) (50 μg/ml) or carbenicillin (Car) (50 μg/ml), with the exception of E. coli strain Rosetta when used for protein expression, which was grown in overnight expression medium (Novagen) supplemented with chloramphenicol (Cam) (34 μg/ml) and carbenicillin (50 μg/ml).
TABLE 1.
Strains and plasmids used in the study
| Strain or plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| CSF41498 | Biofilm-positive cerebrospinal fluid isolate (Beaumont Hospital, Dublin, Ireland) | 9 |
| 1457 | Biofilm-positive isolate from infected central venous catheter (Hamburg, Germany) | 36 |
| rbf mutant | CSF41498 derivative; rbf::ermB Emr | This study |
| 1457 sarR | 1457 derivative; ΔsarR::tet Tcr | This study |
| CSF41498 sarR | CSF41498 derivative; ΔsarR::tet transduced from 1457; Tcr | This study |
| rbf sarR mutant | CSF41498 derivative; ΔsarR::tet rbf::ermB Tcr Emr | This study |
| pCR-Blunt II-TOPO | PCR cloning vector; Kmr Apr | Invitrogen |
| pLI50 | Escherichia coli-Staphylococcus cloning vector; Apr (E. coli) Camr (Staphylococcus) | 37 |
| pMal-c2x | E. coli MBP protein expression vector; Camr | NEB |
| pET30 EK/LIC | His tag cloning vector | Novagen |
| pEPSA5 | E. coli-Staphylococcus cloning vector; multiple cloning site downstream of xylose-inducible promoter | 17 |
| pBT2 | Temperature-sensitive E. coli-Staphylococcus shuttle vector; Apr (E. coli) Camr (Staphylococcus) | 16 |
| pUC19 | E. coli cloning vector; Apr | NEB |
| pROJ6448 | pRN5101::pC221 AluI-C; temperature-sensitive staphylococcal origin of replication | 38 |
| pSERB1 | 2,445-bp rbf fragment in pCR-Blunt II-TOPO | This study |
| pSERB2 | rbf gene in pSERB1 interrupted with ermB | This study |
| pSERB3 | rbf::erm from pSERB2 in shuttle vector pBT2 | This study |
| pSERB5 | 2,800-bp fragment containing rbf and its upstream region in pCR-Blunt II-TOPO | This study |
| pSERB6 | rbf fragment from pSERB5 cloned into pLI50 | This study |
| pEPrbf | rbf and its upstream region in pEPSA5 | This study |
| pLIsarR | sarR and its upstream region in pLI50 | This study |
| pTSERM6 | Promoterless rbf in pCR-Blunt II-TOPO | This study |
| pSERM6 | rbf from pTSERM6 downstream of malE in pMAL-c2x | This study |
| psarR-his | sarR gene in pET30 EK/LIC | This study |
S. epidermidis and S. aureus strains were routinely grown at 37°C or 30°C on brain heart infusion (BHI) medium (Oxoid, UK). When required, BHI was supplemented with Cam (10 μg/ml), erythromycin (10 μg/ml), or tetracycline (Tet) (5 μg/ml). BHI was also supplemented with 4% NaCl as required.
Construction of S. epidermidis rbf and sarR mutants.
The CSF41498 rbf mutant was constructed as follows. A 2,445-bp fragment containing the rbf gene from S. epidermidis CSF41498 was amplified by PCR using the primers SErbf1 and SErbf3 and cloned into the pCR-Blunt II-TOPO plasmid (Invitrogen) to create pSERB1. The plasmid was then digested with ClaI and MunI to release a 182-bp fragment, which was replaced with a 1,227-bp EcoRI-ClaI fragment containing the ermB gene from pEC5 (16) to generate pSERB2. In this construct, the rbf gene is disrupted by the ermB gene inserted 937 bp from the rbf start codon. A 3,490-bp BamHI-PstI fragment from pSERB2 containing the rbf::ermB allele was subcloned into the E. coli-Staphylococcus shuttle vector pBT2 (16). The resulting plasmid, designated pSERB3, was transformed into S. epidermidis CSF41498 via RN4220, as described previously (11). The presence of the rbf::ermB allele on the chromosome was confirmed by PCR using primers SErbf1 and SErbf2 (Table 2). For rbf overexpression and complementation experiments, a 2,800-bp fragment was amplified using primers SErbf1 and SErbf2 and cloned into a pCR-Blunt II-TOPO plasmid (Invitrogen) to create pSERB4 before being subcloned into pLI50 to generate pSERB4. A similar fragment was amplified using primers SErbf_rbsEcoR1 and SErbf_Kpn2 and cloned into the EcoRI and KpnI sites of pEPSA5 (17) under the control of a xylose-inducible promoter to generate pEPrbf.
TABLE 2.
Oligonucleotide primers used in the study
| Target | Primer | Sequence (5′–3′) |
|---|---|---|
| gyrB | GYRB1 | TTATGGTGCTGGACAGATACA |
| GYRB2 | CACCGTGAAGACCGCCAGATA | |
| icaA | KCA1 | AACAAGTTGAAGGCATCTCC |
| KCA2 | GATGCTTGTTTGATTCCCT | |
| Pica | SEicaProm1 | TGCGTTATCAATAATCTTATCTTTCAA |
| SEicaProm2 | GAAAAGTAAAAAGTTAAATACATGCAT | |
| PsarR | SEsarR_comp1 | TATAAAACCACTCCTCTGATGCACATCTTG |
| SEsarR_comp2 | ATATACTAGTTTATTATGTGATATTTACAA | |
| PsarX | SEsarXPROMBio1 | TGCAGTATATTTAGTTGAAATATATAAAAA |
| SEsarXPROMBio2 | AATCTGCACCTCCAAATATAAGTAGACAAC | |
| PsrrA | SEsrr_promBio1 | ACACCAAAAAGATGTAAATTACCATTAAGAT |
| SEsrr_promBio2 | ACTTTCTACTACCTCCTACACTTGCTGTTA | |
| PsarA | SEsarA_prom1 | TAATGAAACCTCCCTATTTATATCATA |
| SEsarA_prom2 | AAAATGTTAGTAAAATTCTTTCCAAAA | |
| PsarZ | SEsarZpromBio1 | TTTTCGTACTCCTCCATTTTTTAAAAAATT |
| SEsarZpromBio2 | TTAATCACTCCTTGTTAAGGTAACAATATT | |
| Pspx | SEspx_promBio1 | ATTAGATGCCTACTTTCTAATTAATATTGT |
| SEspx_promBio2 | ACATCTCACTCTCTTATAGAATGAATTTAA | |
| rbf (pLI50 complementation) | SErbf1 | ATCAAAAAGTTGGCGAACCTTTTCA |
| SErbf2 | CAAAAGAGCCTGGAGAAAAGTATCA | |
| rbf (pEPSA5 complementation) | SErbf_rbsEcoR1 | CGAGCTGAATTCGAGGGAAAGAGGTAAAGATA |
| SErbf_Kpn2 | CGAGCTGGTACCTTAAGTTGTGCTACGCCTTTTAT | |
| rbf (protein purification) | SErbf1 | ATGGCAAATTCTTGTTTGCAT |
| SErbf2 | TTAAGTTGTGCTACGCCTTTTATTT | |
| sarR (protein purification) | 1589 | GACGACGACAAGATGGGAAAAATTAAAGACATCAATG |
| 1590 | GAGGAGAAGCCCGGTTATTTGATATAGTTTTCTAATTC | |
| sarR (allele replacement) | 1058 | ATCCTAGGATCGGGTTACTTATCATTAGTG |
| 1059 | ATCCTATCTAGACGCATTAACCAAATCATTG | |
| sarR (allele replacement) | 1060 | ATCCTAGTCGACGCGTGCATGATGAAAGAACAG |
| 1061 | ATCCTACTGCAGCCGTGTCAATGTCAACTTAG | |
| sarR (complementation) | SEsarR1 | TTATTTGATATAGTTTTCTAATTCTAAAATC |
| SEsarR_comp2 | ATATACTAGTTTATTATGTGATATTTACAA | |
| sarX | SE_sarXRT2F | GCAGATTTTGAATGAGCAGAAAT |
| SE_sarXRT2R | ATCTAACTCTCCTGTAGCCA |
To construct an S. epidermidis 1457 sarR mutant, a 1,048-bp sarR gene and flanking DNA fragment were amplified from 1457 chromosomal DNA using primers 1058 (BamHI tailed) and 1059 (XbaI tailed) and ligated into the BamHI and XbaI sites of pUC19. An 851-bp sarR gene and flanking DNA fragment obtained from amplification with primers 1060 (SalI tailed) and 1061 (PstI tailed) were ligated into the corresponding sites. Next, the tetM gene was amplified from pJF12 into the SalI site. Finally, the temperature-sensitive mobilization staphylococcal replicon pR0J6448 was ligated into the PstI site.
The sarR::Tcr allele was transduced into S. epidermidis CSF41498 using phage A6C, as described previously (6). The sarR primers (SEsarR1 and SEsarR_comp2) were used to amplify a fragment containing the sarR gene and upstream promoter sequences with subsequent cloning into pLI50 (via pCR-Blunt II-TOPO) for complementation experiments.
Biofilm assays.
Semiquantitative measurements of biofilm formation under static conditions were determined using Nunclon tissue culture-treated (ΔSurface) 96-well polystyrene plates (Nunc, Denmark), as described previously (9). Each strain was tested at least three times, and average results are presented. A biofilm-positive phenotype was defined as an A490 of ≥0.17. Primary attachment assays were performed as described previously (11).
Biofilm flow cell experiments.
The BioFlux 1000z microfluidic system (Fluxion Biosciences Inc., San Francisco, CA) was used to assess biofilm formation under shear flow conditions. Biofilms were grown in BHI and BHI supplemented with NaCl (4% [wt/vol]). The system was initiated by adding 200 μl of medium to the output wells of a 48-well plate and priming the channels for 5 min at 5.0 dynes/cm2. After priming, the medium was aspirated from the output wells and replaced with a 50-μl suspension of bacteria grown to early exponential growth phase and adjusted to an A600 of 0.8. A further 50 μl of medium was added to the input wells, and the channels were seeded by pumping from the output wells to the input wells for 3 to 5 s at a speed of 3 dynes/cm2. Bacteria were allowed to attach to the surface of the plate for 1 h at 37°C. Excess inoculum solution was aspirated from the output wells, and a further 1 ml of medium was added to the input wells. The flow rate was set at 0.4 dyne/cm2 for 18 h, and bright-field images were captured every 5 min at ×10 magnification.
RNA purification and real-time RT-PCR.
Cultures were grown in BHI medium supplemented with 1% glucose or 4% NaCl where indicated. Cells were harvested following overnight growth (∼16 h) or at an A600 of 1.0 and immediately stored at −20°C in RNAlater (Ambion) to ensure maintenance of RNA integrity prior to purification. RNA was extracted as described previously (6, 11). RNA integrity was examined visually by agarose gel electrophoresis, and the RNA concentration was determined using a NanoDrop spectrophotometer. Reverse transcription (RT)-PCR was performed on a Roche LightCycler using the RNA amplification kit SYBR green I (Roche) as previously described (6, 11). Each experiment was performed at least three times, and means of the data with standard deviations (SD) are presented. RelQuant software (Roche Biochemicals) was used to measure relative expression of target genes. The gyrB gene was used as an internal standard in real-time RT-PCR experiments. Each experiment was performed at least three times, and averages of the data with standard deviations are presented. The primers used for real-time RT-PCR are listed in Table 2.
PIA assays.
PIA immunoblots were performed based on the method of Cramton et al. (18), as described previously (6, 11).
Purification of recombinant Rbf.
To purify recombinant Rbf protein, the rbf gene was amplified by PCR using Phusion high-fidelity DNA polymerase and primers SErbfprot1 and SErbfprot2 and ligated into the pCR-Blunt II-TOPO plasmid (Invitrogen) to create pTSERM6. Plasmid pTSERM6 was digested with EcoRI, and the rbf gene was subcloned into the pMal-c2x vector at an EcoRI site downstream from the malE gene, resulting in plasmid pSERM6. Restriction analysis was used to confirm the correct orientation of the rbf gene. Plasmid pSERM6 was transformed into E. coli strain Rosetta (Novagen), and protein was purified following a previously described protocol (19).
A single colony of E. coli strain Rosetta harboring pSERM6 was used to inoculate 10 ml overnight expression medium (Novagen) containing Car (50 μg/ml) and Cam (34 μg/ml). Cultures were grown overnight at 37°C with shaking and good aeration. The cells were pelleted and resuspended in 5 ml column buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT]). The cells were incubated on ice and lysed by sonication. Samples were centrifuged at 14,000 rpm for 30 min at 4°C. The soluble fraction was diluted in 50 ml column buffer and passed through an amylose resin column at a speed of 1 ml/min. The column was washed and eluted with column buffer containing 10 mM maltose. Purified fractions were collected in 500-μl aliquots and analyzed on a 10% SDS-PAGE gel, and concentrations were determined using a NanoDrop spectrophotometer at A280. The protein was stored at −80°C in 50-μl aliquots.
Purification of recombinant SarR.
The sarR gene was amplified using primers 1589 and 1590 and cloned into pET30 EK/LIC (Novagen) using ligation-independent cloning methodologies. The resulting plasmid, pSarR-his, was electroporated into E. coli BL21(DE3) (Novagen) for protein production. BL21(DE3) containing pSarR-his was grown in 1 liter of 2× YT medium containing 30 mg kanamycin/ml. The culture was grown to an A600 of 0.6, induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and grown for an additional 2 h. The culture was subsequently pelleted by centrifugation and resuspended in 100 ml binding buffer (50 mM Tris, 30 mM imidazole, 500 mM NaCl, pH 7.4). The E. coli cells were then lysed by 4 passages through an EmulsiFlex (Abestin, Inc.). Proteases were inhibited via 0.4 mM phenylmethylsulfonyl fluoride (PMSF). The soluble cell extract was obtained by centrifugation at 12,000 × g for 30 min at 4°C and applied to a HisTrap column (GE Healthcare) at a flow rate of 0.5 ml/min and subsequently washed with 20 column washes of binding buffer. Purified SarR was eluted with elution buffer (50 mM Tris, 500 mM imidazole, 500 mM NaCl, pH 7.4) and then dialyzed against 50 mM Tris (pH 7.5).
Electrophoretic mobility shift assays (EMSAs).
The promoter regions were amplified from S. epidermidis strain CSF41498 genomic DNA using the biotinylated primers listed in Table 2 and Phusion high-fidelity enzyme. The resultant PCR products were first purified from a 2% agarose gel using the QIAquick gel extraction kit (Qiagen) and subsequently purified from a 5% nondenaturing polyacrylamide gel. The DNA concentration was determined using a NanoDrop spectrophotometer.
A biotinylated DNA probe containing the promoter region of a target gene was added to increasing concentrations of recombinant Rbf or SarR protein. A 20-μl binding reaction mixture containing various amounts of protein, 0.2 μg poly(dI-dC) in binding buffer (100 mM Tris, 500 mM KCl, 10 mM DTT, pH 7.5), 5% glycerol, 5 mM MgCl2, 4.5 ng biotinylated probe was used unless otherwise stated. The reaction mixture was incubated at room temperature for 20 min and loaded onto a 5% nondenaturing polyacrylamide gel and electrophoresed at 100 V for 65 min. DNA was transferred onto a Biodyne-B nylon membrane (Pall Corporation) at 4°C in prechilled 0.5% Tris-borate-EDTA (TBE) and electrophoresed at 80 V for 60 min. The membrane was cross-linked under UV light for 10 min. Detection of the bands was performed using a Pierce LightShift chemiluminescent electrophoretic mobility shift assay kit (Pierce Chemicals, Rockford, IL) and a FluorChem FC2 chemiluminescent unit (Alpha Innotech).
Statistical analysis.
Two-tailed, two-sample, equal-variance Student t tests (Microsoft Excel) were used to determine statistically significant differences in assays performed during this study. A significant difference was indicated as a P value of <0.05.
RESULTS AND DISCUSSION
Mutation of rbf impairs the biofilm phenotype of S. epidermidis CSF41498.
An rbf deletion mutation in which 180 bp of the rbf gene was replaced with the erythromycin resistance gene ermB was constructed in CSF41498. No significant difference in the growth rate was observed in the rbf mutant compared to CSF41498 (data not shown). However, biofilm formation by the rbf mutant under static conditions (Fig. 1A) or flow conditions generated using a BioFlux instrument (Fig. 1B) was reduced compared to that of CSF41498 in BHI medium and BHI medium supplemented with 4% NaCl (BHI NaCl) (Fig. 1A). Consistent with this, the rbf mutation was associated with reduced icaA transcription, particularly in BHI NaCl (Fig. 1C), and PIA production (Fig. 1D). However, impaired biofilm production by the rbf mutant did not correlate with impaired primary attachment to polystyrene compared to CSF41498 (data not shown).
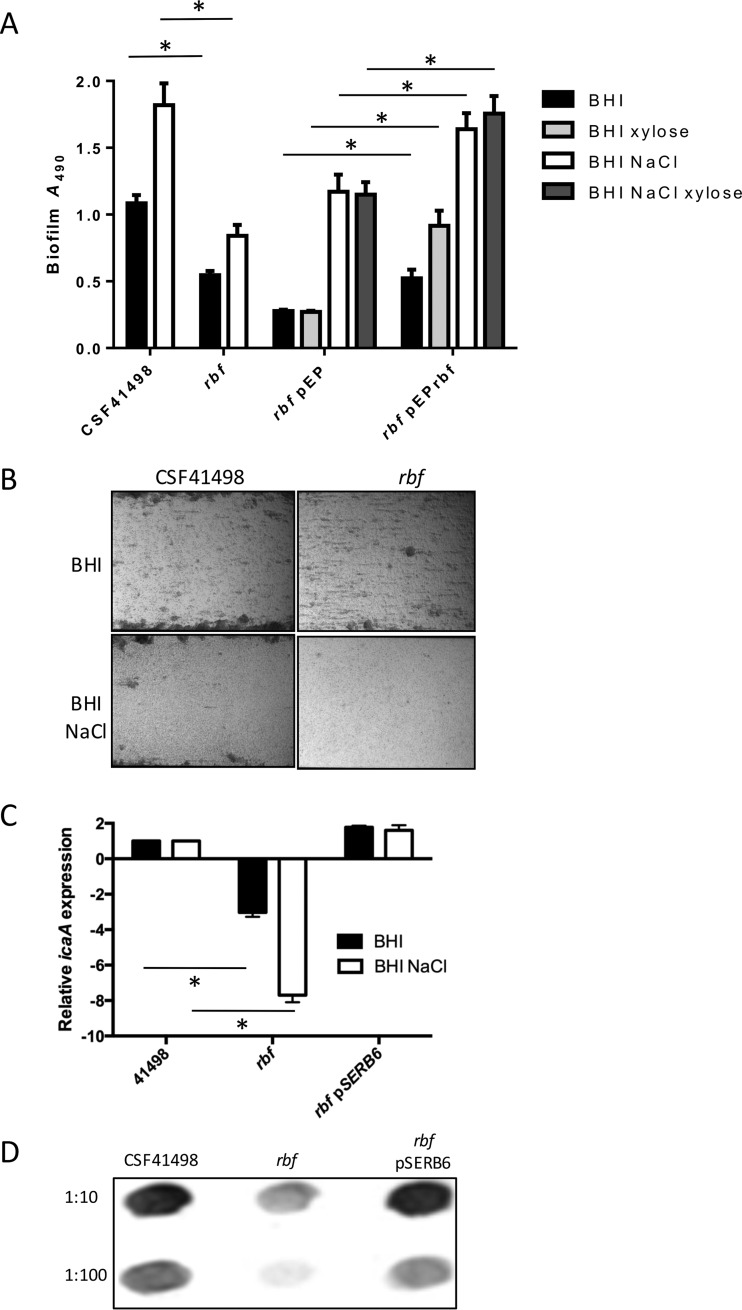
FIG 1.
Rbf regulates icaADBC transcription, PIA expression, and biofilm production in S. epidermidis. (A) Comparative biofilm phenotypes of CSF41498 and its isogenic rbf mutant, complemented where indicated with plasmid pEPSA5 (empty-vector control) or pEPrbf. Semiquantitative measurements of biofilm formation under static conditions were performed in tissue culture-treated 96-well plates. All the strains were grown at 37°C in BHI and BHI NaCl for 24 h. BHI and BHI NaCl were also supplemented with 0.5% xylose for strains carrying pEPrbf. Experiments were repeated at least five times, and average results are presented. Standard errors of the mean are indicated, and the asterisks denote significant differences (P < 0.05) as determined by Student's t test. (B) CSF41498 and its isogenic rbf mutant grown in BHI medium and BHI NaCl in flow cells at 0.6 dyne/cm2 shear using a BioFlux 1000Z instrument. The bright-field images depicting biofilm accumulation after 18 h were captured at ×10 magnification and are representative of the results of the results of three independent experiments. (C) Comparative measurement of icaA transcription by real-time RT-PCR in CSF41498 and its isogenic rbf mutant. Total RNA was extracted from cultures grown at 37°C to an A600 of 1.0. RelQuant software (Roche) was used to measure the relative expression of icaA compared to that of the constitutively expressed gyrB gene. Transcript levels of icaA in the rbf mutant strain were then compared to those in CSF41498 pLI50, which were assigned a value of 1. The data presented are the averages of three separate experiments, and standard deviations are indicated. Statistical significance between the rbf or rbf pEPrbf strains and CSF41498 as determined by Student's t test is indicated by the asterisks (P ≤ 0.05). (D) Comparative immunoblot analysis of PIA production in whole-cell extracts of CSF41498, its isogenic rbf mutant, and the rbf mutant complemented with pSERB6 grown overnight at 37°C in BHI. Cell extracts were diluted 1:10 and 1:100. The blot shown is representative of the results of three independent experiments.
To complement the rbf mutant, a 2,797-bp fragment containing the rbf gene and its upstream regulatory sequences was cloned into the E. coli-Staphylococcus shuttle vector pLI50 to yield pSERB6. icaA transcript levels and PIA production were restored to wild-type levels in the rbf mutant carrying pSERB6 (Fig. 1C and D). However, the rbf gene carried on pSERB6 failed to complement the biofilm effect of the rbf mutant (data not shown). Given that carriage of the pSERB6 plasmid restored PIA production, this observation is difficult to explain but may suggest that inappropriate rbf expression or carriage of pLI50 may impact Rbf-mediated regulation of the ica locus or another factor(s) involved in the biofilm phenotype. Efforts to complement the rbf mutant on the low-copy-number plasmid pRB474 also failed because carriage of the plasmid alone significantly increased CSF41498 biofilm production (data not shown). Next, the rbf gene was placed under the control of a xylose-inducible promoter in pEPSA5 (17). Although supplementation of the growth medium with xylose had a generally positive effect on biofilm production by all the strains, successful complementation of biofilm production by the rbf mutant in BHI medium (0.5% xylose), BHI NaCl (0.5% xylose), and BHI NaCl alone was observed (Fig. 1A). Notably, leaky expression from pEPrbf in the absence of xylose was sufficient to complement the rbf mutant in BHI medium supplemented with NaCl (Fig. 1A). Taken together, these data are suggestive of a complex role of Rbf in controlling the S. epidermidis biofilm phenotype but support the conclusion that Rbf influences biofilm formation in S. epidermidis by positively regulating icaADBC operon expression and PIA production.
Rbf does not bind to the S. epidermidis ica operon promoter.
To investigate how Rbf regulates ica operon transcription, we purified recombinant Rbf protein and performed EMSAs with the ica operon promoter. Rbf is a member of the AraC/XylS family of transcription factors, most of which are highly insoluble and difficult to purify (13). Therefore, the rbf gene was cloned into the pMal-c2x vector (New England Biolabs [NEB]) downstream from the malE gene to take advantage of previous observations that tagging with maltose binding protein (MBP) increases the solubility of recombinant proteins (20). The resulting plasmid, pSERM6, was transformed into E. coli Rosetta, a strain containing pRARE, a plasmid used for expressing rare E. coli codons, and grown overnight in commercial OnEx medium (Novagen) prior to purification of recombinant Rbf. A 218-bp biotinylated oligonucleotide Pica probe was incubated with increasing concentrations of recombinant Rbf, separated on a 5% acrylamide gel, and transferred to a positively charged nylon membrane, and the probe was detected using a Pierce LightShift chemiluminescent EMSA kit (Pierce Chemicals, Rockford, IL). The recombinant Rbf protein did not bind to the ica operon promoter probe (Fig. 2A). Weak interaction between the Pica probe and the highest concentration (20.37 pg) of recombinant Rbf was easily disrupted with poly(dI-dC), indicating that the complex was not specific (data not shown).
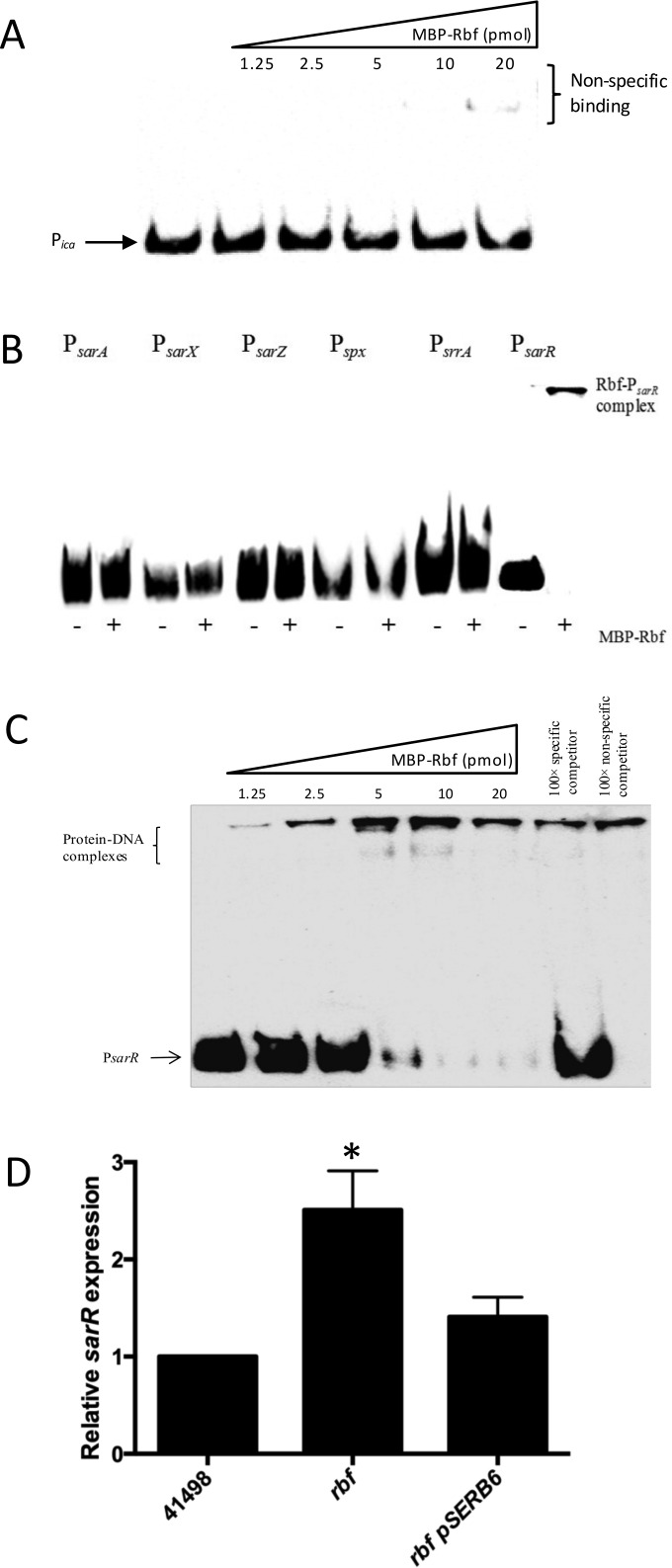
FIG 2.
Recombinant Rbf binds to the sarR promoter and regulates sarR transcription. (A) Recombinant Rbf does not bind to the ica promoter. Increasing concentrations (1.25 to 20 pmol) of recombinant Rbf protein were added to a biotinylated oligonucleotide Pica probe. (B) Recombinant Rbf binding to the sarA, sarX, sarZ, spx, srrA, and sarR promoters. Twenty picomoles of recombinant Rbf protein was added to each biotinylated oligonucleotide probe before being separated on a 5% polyacrylamide gel. (C) Recombinant Rbf binds specifically to the sarR promoter. Increasing concentrations (1.25 to 20 pmol) of recombinant Rbf protein were added to a biotinylated oligonucleotide PsarR probe. The protein-DNA interactions were competed with 100× specific or nonspecific competitor DNA. (D) Comparative measurement of sarR and gyrB (control) transcription in S. epidermidis strains CSF41498 pLI50, rbf pLI50, and rbf pSERB6. RNA was prepared from cultures grown overnight to stationary phase (∼16 h) at 37°C in BHI. RelQuant software (Roche, Switzerland) was used to compare the relative expression of sarR to that of the constitutively expressed gyrB gene. sarR transcript levels in all strains were compared to sarR transcript levels in CSF41498 pLI50, which was assigned a value of 1. The data presented are the averages of three separate experiments. Standard deviations are indicated, and the asterisk denotes a significant difference as determined by Student's t test (P ≤ 0.05).
Rbf binds specifically to the S. epidermidis sarR promoter and regulates sarR transcription.
To investigate the possibility that Rbf indirectly regulates the ica operon by controlling the expression of another biofilm regulator, the ability of recombinant Rbf to interact with biotinylated promoter probes comprising the promoter regions of the sarA (21), sarX (11), sarZ (22), sarR (Fey Laboratory, unpublished data), spx (23), and srrA (24) genes was next investigated. Rbf bound with high specificity to the sarR promoter (Fig. 2B and C) and not to any of the other tested promoters (Fig. 2B). Expression of sarR was increased approximately 2.5-fold in the CSF41498 rbf mutant (Fig. 2D), suggesting that Rbf functions as a repressor of sarR transcription. The sarR gene was first described in 2000 and encodes a 13.6-kDa protein known to bind the sarA promoter and to regulate sarA expression (25). SarR can also regulate the transcriptional activity of the accessory gene regulator (agr) promoter (26), which is also a major target of SarA (27).
SarR is a repressor of ica operon expression and biofilm in S. epidermidis.
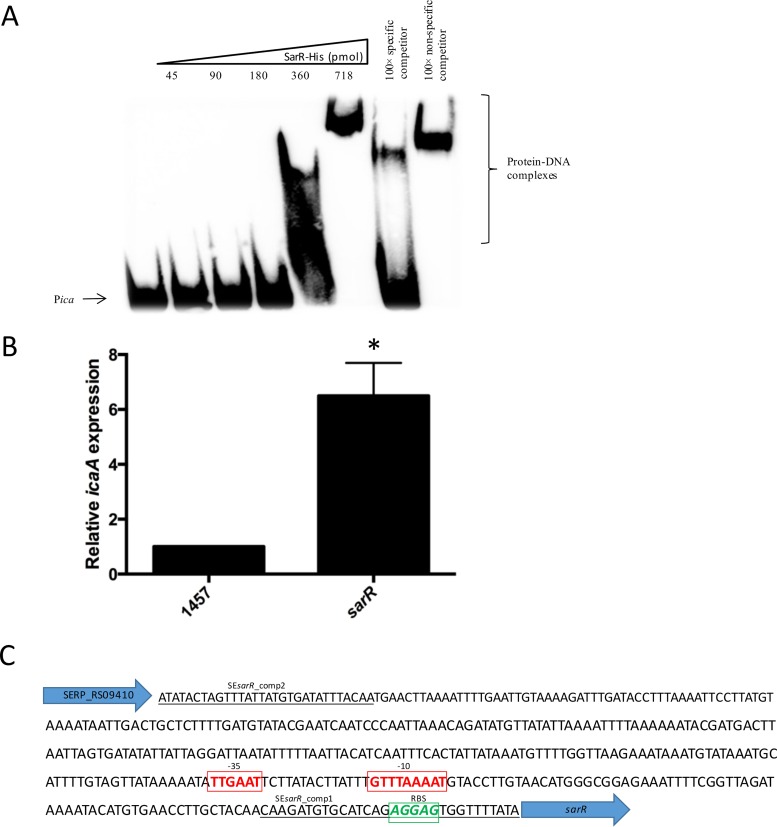
To further investigate the hypothesis that Rbf controls ica operon expression by regulating sarR expression, recombinant SarR was purified and used in EMSAs. Recombinant SarR was relatively soluble, and the pET30 system (Novagen) was employed to express and purify an N-terminal His-tagged SarR recombinant protein, as described in Materials and Methods. These experiments demonstrated that recombinant SarR bound specifically to the ica operon promoter (Fig. 3A). Next, a sarR::tet deletion mutation was constructed in S. epidermidis 1457 using allele replacement, as described in Materials and Methods. Using RT-PCR, icaA transcription was found to be increased 6.5-fold in the sarR mutant compared to 1457 (Fig. 3B). Inspection of the intergenic sequence upstream of the sarR open reading frame and downstream of SERP_RS0910 using the Softberry bacterial promoter prediction software (BPROM) identified a strong candidate promoter based on alignment with canonical −10 and −35 RNA polymerase recognition sequences (Fig. 3C).
FIG 3.
Recombinant SarR binds to the ica operon promoter and regulates ica transcription. (A) Recombinant SarR binds to the ica promoter. Increasing concentrations (45 to 718 pmol) of recombinant SarR protein were added to a biotinylated oligonucleotide PsarR probe. The protein-DNA interactions were competed with 100× specific or nonspecific competitor DNA. (B) Comparative measurement of icaA and gyrB (control) transcription in S. epidermidis strain 1457 and its sarR mutant. RNA was prepared from cultures grown to an A600 of 1.0 at 37°C in BHI NaCl. RelQuant software (Roche, Switzerland) was used to measure the relative expression of icaA against the constitutively expressed gyrB gene. icaA transcript levels in all strains were compared to icaA transcript levels in CSF41498 pLI50, which were assigned a value of 1. The data presented are the averages of three separate experiments. Standard deviations are indicated, and the asterisk denotes a significant difference as determined by Student's t test (P ≤ 0.05). (C) Intergenic nucleotide sequence between SERP_RS09410 and sarR on the chromosome of S. epidermidis RP62A. The binding sites for the SEsarR_comp1 and SEsarR_comp2 oligonucleotide primers are underlined. The predicted sarR ribosome binding site (RBS) (green letters) and promoter based on canonical −10 and −35 RNA polymerase binding sites (red letters) were identified using Softberry bacterial promoter prediction software (BPROM).
Mutation of sarR restores biofilm-forming capacity in an rbf mutant.
Using phage A6C, the sarR mutation was transduced into CSF41498, and its isogenic rbf mutant and transductants were confirmed by PCR of the sarR locus (data not shown). Immunoblots revealed that PIA production was significantly increased in the sarR mutant grown in both BHI and BHI NaCl (Fig. 4A). Consistent with this, the sarR mutation promoted biofilm formation in both BHI and BHI NaCl under shear flow in the BioFlux instrument (Fig. 4B). In contrast, under static growth conditions, the sarR mutation was associated with increased biofilm formation only in BHI NaCl and not in BHI (Fig. 4C).
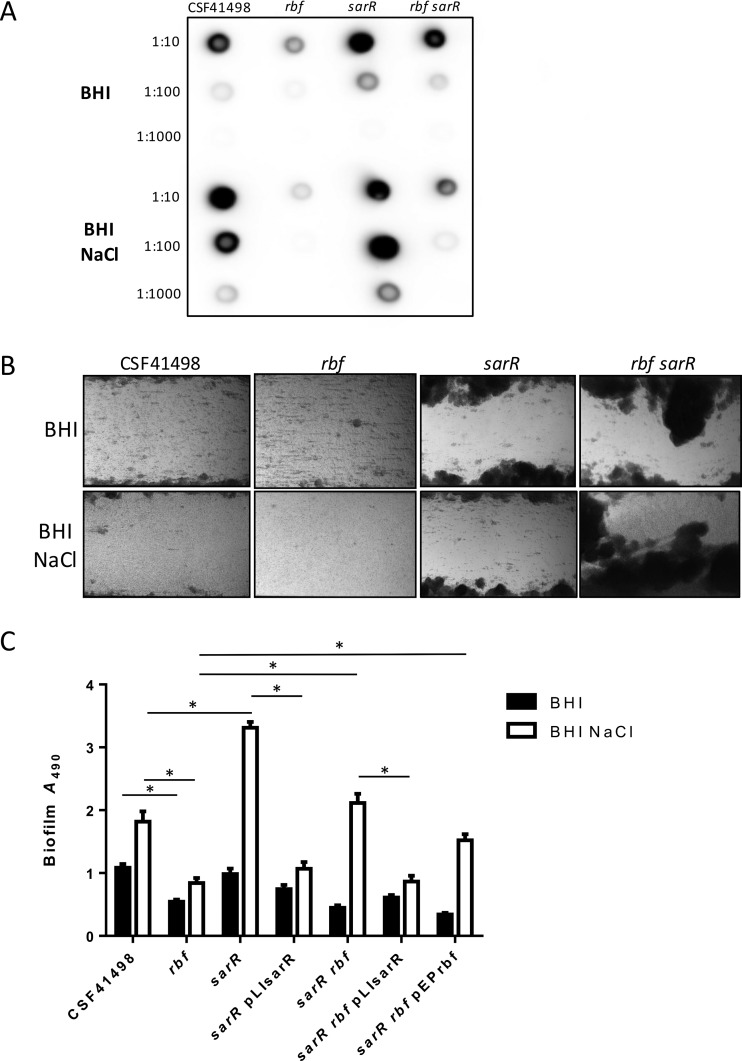
FIG 4.
SarR-regulated PIA production and biofilm development in S. epidermidis. (A) Comparative immunoblot analysis of PIA production in whole-cell extracts of CSF41498 and its isogenic rbf, sarR, and rbf sarR mutants grown overnight at 37°C in BHI medium and BHI NaCl. Cell extracts were diluted 1:10, 1:100, and 1:1,000. The blot shown is representative of the results of three independent experiments. (B) CSF41498 and its isogenic rbf, sarR, and rbf sarR mutants grown in BHI and BHI NaCl in flow cells at 0.4 dyne/cm2 shear using a BioFlux 1000Z instrument. The bright-field images depicting biofilm accumulation after 18 h were captured at ×10 magnification and are representative of the results of three independent experiments. (C) Comparative biofilm phenotypes of CSF41498 and its isogenic rbf, sarR, and rbf sarR mutants, complemented where indicated with pLIsarR or pEPrbf. Semiquantitative measurements of biofilm formation under static conditions were performed in tissue culture-treated 96-well plates. All the strains were grown at 37°C in BHI and BHI NaCl for 24 h. Experiments were repeated at least five times, and average results are presented. Standard errors of the mean are indicated, and the asterisks denote a significant difference as determined by Student's t test (P < 0.05).
In the rbf sarR double mutant, PIA production was restored to wild-type levels in BHI medium (Fig. 4A), whereas in BHI NaCl, PIA expression was increased but remained lower than that of CSF41498 (Fig. 4A). In the BioFlux instrument, the rbf sarR double mutant formed robust biofilms in both BHI and BHI NaCl under shear flow (Fig. 4B), whereas under static growth conditions, biofilm production by the rbf sarR double mutant was restored to wild-type levels only in BHI NaCl and not in BHI medium (Fig. 4C). Complementation of sarR mutants with the sarR gene on plasmid pLI50 significantly reduced biofilm production in BHI NaCl (Fig. 4C).
Taken together, these data reveal that Rbf regulates icaADBC expression and biofilm, at least in part, by controlling sarR expression. The negative impact of the rbf mutation S. epidermidis biofilm production was reversed by a second mutation in sarR via a mechanism that was highly dependent on growth conditions (i.e., osmotic stress and shear flow). The complex relationship between the PIA and biofilm phenotypes expressed by the rbf sarR double mutant indicates that SarR and Rbf may also regulate PIA production at the posttranscriptional level and/or icaADBC-independent biofilm factors. In this context, it is noteworthy that SarR regulates the expression of the two major virulence and biofilm regulators in S. aureus, namely, sarA and agr (25, 26), which influence PIA-independent and PIA-dependent biofilm mechanisms (10, 28, 29). Furthermore, in S. aureus, Rbf regulates >50 genes, including the teichoic acid biosynthetic gene, tagB, and lrgAB and lytS (14), which can impact biofilm formation by influencing the surface charge and cell lysis-mediated release of extracellular DNA, respectively (30, 31).
As noted above, in addition to SarA (10) and SarX (11), SarR is the third member of the Sar family known to bind the ica promoter. Manna and Cheung previously reported that in S. aureus, SarR and SarA bind to similar target DNA sequences within the agr promoter in order to activate transcription (26). Because both SarR and SarA bind to the sarA promoter to repress sarA transcription (25, 32), it is possible that SarR and SarA bind to the same or overlapping target DNA sequences within the ica promoter to negatively or positively regulate transcription, respectively. Indeed, the possibility that SarA and SarR can heterodimerize to regulate target gene expression, which contributes to the complex regulation of the agr locus and may also impact icaADBC expression, has been proposed (26, 33). Our data also show that SarR primarily impacts the S. epidermidis biofilm phenotype in media supplemented with NaCl, which is known to repress transcription of the icaR repressor (8, 9). This observation suggests that IcaR is preeminent in the hierarchy of ica operon regulators and that the major role of SarR, and by extension Rbf, is to repress and fine-tune icaADBC transcription, primarily when IcaR levels are low. Interestingly, to date, SarR has not been implicated in PIA-mediated biofilm formation in S. aureus, although mutation of sarR was shown to increase autolytic activity (34), which may impact biofilm by increasing the release of extracellular DNA (30, 35). However, as observed here, it remains possible that SarR also regulates the ica operon in S. aureus under conditions when IcaR expression is reduced.
Rbf negatively regulates sarX expression in S. epidermidis.
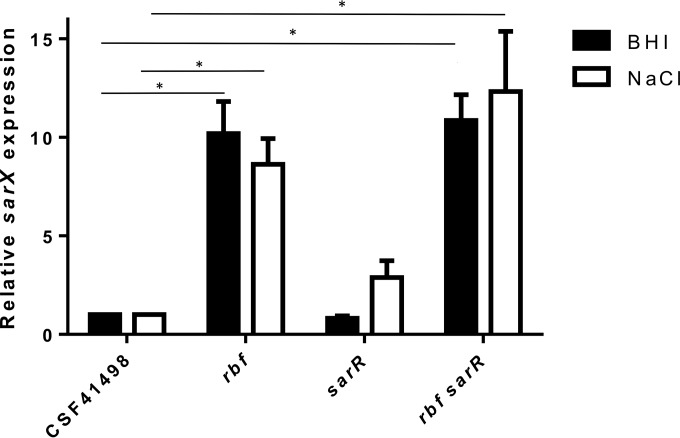
In S. aureus, Rbf activates expression of sarX, and Rbf-mediated regulation of the ica operon is dependent on SarX (15). Recombinant SarX binds to ica operon regulatory sequences in both S. aureus and S. epidermidis (11, 15). LightCycler RT-PCR to measure the effect of the Rbf and SarR mutations on sarX expression revealed that, opposite to what occurs in S. aureus, mutation of rbf was associated with increased sarX expression in both BHI medium and BHI NaCl (Fig. 5). These results were somewhat unexpected, because SarX activates the ica operon in both S. aureus and S. epidermidis. Expression of sarX was also increased in the sarR rbf mutant (Fig. 5). Increased biofilm formation by the sarR and sarR rbf mutants can be attributed to derepression of the ica operon in the absence of the SarR repressor. Mutation of sarR had no effect on sarX expression in BHI medium but was associated with a 2.8-fold increase in BHI NaCl medium, which was not statistically significant (P = 0.8) (Fig. 5).
FIG 5.
Rbf negatively regulates sarX transcription in S. epidermidis. Shown are comparative measurements of sarX and gyrB (control) transcription in S. epidermidis CSF41498, rbf, sarR, and rbf sarR strains. RNA was prepared from cultures grown to an A600 of 1.0 at 37°C in BHI medium and BHI NaCl. RelQuant software (Roche, Switzerland) was used to measure the relative expression of sarX against that of the constitutively expressed gyrB gene. sarX transcript levels in all the strains were compared to sarX transcript levels in CSF41498, which were assigned a value of 1. The data presented are the averages of at least three separate experiments. Standard deviations are indicated, and the asterisks denote a significant difference as determined by Student's t test (P ≤ 0.05).
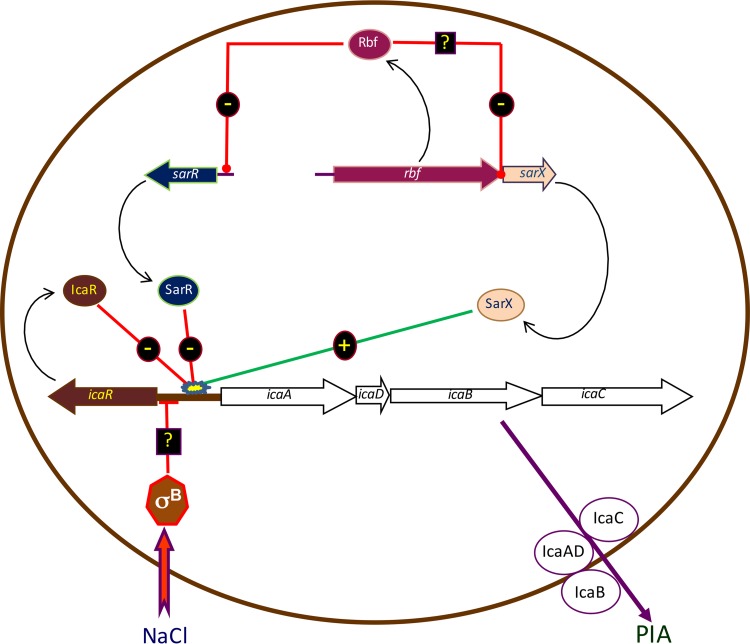
In summary, we propose an updated model for the regulation of icaADBC expression in S. epidermidis (Fig. 6) in which Rbf promotes biofilm formation by directly regulating expression of sarR. SarR and SarX exert opposite effects on icaADBC expression, with SarR acting as a repressor and SarX as an activator. Given that the dominant effect of an Rbf mutation is to reduce icaADBC expression, PIA production, and biofilm (Fig. 1), it appears that Rbf-mediated downregulation of the SarR repressor is dominant over Rbf-mediated repression of the SarX activator. In the hierarchy of biofilm regulators, our data suggest that the Rbf-SarR-SarX axis appears to be subservient to IcaR, as evidenced under osmotic stress induced by NaCl, which is associated with repression of icaR transcription and activation of PIA production.
FIG 6.
Model for S. epidermidis icaADBC regulation by Rbf, SarX, and SarR. Rbf indirectly regulates ica operon expression by binding to the sarR promoter and negatively regulating sarR transcription. SarR binds to the ica operon promoter to repress icaADBC transcription. SarX acts independently of both SarR and Rbf, binding to the ica operon promoter to increase icaADBC transcription (11). Under osmotic stress in medium supplemented with NaCl, SigB-dependent repression of icaR (8, 9) and the concomitant activation of the ica operon are dominant over Rbf, SarX, and SarR.
ACKNOWLEDGMENTS
This study was funded by grants HRA_POR/2012/51 and HRA_POR/2010/90 from the Irish Health Research Board to J.P.O., by grants from the Irish Research Council to C.C. and S.T.O., and by National Institute of Allergy and Infectious Diseases grant R01 AI049311 to P.D.F.
We are grateful to Chia Lee and Brian Conlon for support and advice over the course of the study. The rabbit anti-PIA serum was a gift from T. Maira Litran and G. B. Pier.
REFERENCES
- 1.Hogan S, Stevens NT, Humphreys H, O'Gara JP, O'Neill E. 2015. Current and future approaches to the prevention and treatment of staphylococcal medical device-related infections. Curr Pharm Des 21:100–113. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy H, Rudkin JK, Black NS, Gallagher L, O'Neill E, O'Gara JP. 2015. Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front Cell Infect Microbiol 5:1. doi: 10.3389/fcimb.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zapotoczna M, O'Neill E, O'Gara JP. 2016. Untangling the diverse and redundant mechanisms of Staphylococcus aureus biofilm formation. PLoS Pathog 12:e1005671. doi: 10.1371/journal.ppat.1005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heilmann C, Hussain M, Peters G, Gotz F. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol 24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 5.Schumacher-Perdreau F, Heilmann C, Peters G, Gotz F, Pulverer G. 1994. Comparative analysis of a biofilm-forming Staphylococcus epidermidis strain and its adhesion-positive, accumulation-negative mutant M7. FEMS Microbiol Lett 117:71–78. doi: 10.1111/j.1574-6968.1994.tb06744.x. [DOI] [PubMed] [Google Scholar]
- 6.Conlon BP, Geoghegan JA, Waters EM, McCarthy H, Rowe SE, Davies JR, Schaeffer CR, Foster TJ, Fey PD, O'Gara JP. 2014. Role for the A domain of unprocessed accumulation-associated protein (Aap) in the attachment phase of the Staphylococcus epidermidis biofilm phenotype. J Bacteriol 196:4268–4275. doi: 10.1128/JB.01946-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaeffer CR, Woods KM, Longo GM, Kiedrowski MR, Paharik AE, Büttner H, Christner M, Boissy RJ, Horswill AR, Rohde H, Fey PD. 2015. Accumulation-associated protein (Aap) enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect Immun 83:214–226. doi: 10.1128/IAI.02177-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlon KM, Humphreys H, O'Gara JP. 2002. Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS Microbiol Lett 216:171–177. doi: 10.1111/j.1574-6968.2002.tb11432.x. [DOI] [PubMed] [Google Scholar]
- 9.Conlon KM, Humphreys H, O'Gara JP. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol 184:4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, Penades JR, Lasa I. 2003. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol 48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 11.Rowe SE, Mahon V, Smith SG, O'Gara JP. 2011. A novel role for SarX in Staphylococcus epidermidis biofilm regulation. Microbiology 157:1042–1049. doi: 10.1099/mic.0.046581-0. [DOI] [PubMed] [Google Scholar]
- 12.Lim Y, Jana M, Luong TT, Lee CY. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J Bacteriol 186:722–729. doi: 10.1128/JB.186.3.722-729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. 1997. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev 61:393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cue D, Lei MG, Luong TT, Kuechenmeister L, Dunman PM, O'Donnell S, Rowe S, O'Gara JP, Lee CY. 2009. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J Bacteriol 191:6363–6373. doi: 10.1128/JB.00913-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cue D, Lei MG, Lee CY. 2013. Activation of sarX by Rbf is required for biofilm formation and icaADBC expression in Staphylococcus aureus. J Bacteriol 195:1515–1524. doi: 10.1128/JB.00012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruckner R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett 151:1–8. doi: 10.1016/S0378-1097(97)00116-X. [DOI] [PubMed] [Google Scholar]
- 17.Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, C KG, King P, McCarthy M, Malone C, Misiner B, Robbins D, Tan Z, Zhu Zy ZY, Carr G, Mosca DA, Zamudio C, Foulkes JG, Zyskind JW. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol 43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 18.Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun 67:5427–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowe SE, O'Gara JP. 2016. Electrophoretic mobility shift assays. Methods Mol Biol 1373:155–167. doi: 10.1007/7651_2015_277. [DOI] [PubMed] [Google Scholar]
- 20.Kapust RB, Waugh DS. 1999. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci 8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tormo MA, Knecht E, Gotz F, Lasa I, Penades JR. 2005. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology 151:2465–2475. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 22.Chen PR, Nishida S, Poor CB, Cheng A, Bae T, Kuechenmeister L, Dunman PM, Missiakas D, He C. 2009. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol Microbiol 71:198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pamp SJ, Frees D, Engelmann S, Hecker M, Ingmer H. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J Bacteriol 188:4861–4870. doi: 10.1128/JB.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulrich M, Bastian M, Cramton SE, Ziegler K, Pragman AA, Bragonzi A, Memmi G, Wolz C, Schlievert PM, Cheung A, Doring G. 2007. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol Microbiol 65:1276–1287. doi: 10.1111/j.1365-2958.2007.05863.x. [DOI] [PubMed] [Google Scholar]
- 25.Manna A, Cheung AL. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect Immun 69:885–896. doi: 10.1128/IAI.69.2.885-896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manna AC, Cheung AL. 2006. Transcriptional regulation of the agr locus and the identification of DNA binding residues of the global regulatory protein SarR in Staphylococcus aureus. Mol Microbiol 60:1289–1301. doi: 10.1111/j.1365-2958.2006.05171.x. [DOI] [PubMed] [Google Scholar]
- 27.Chien Y, Manna AC, Cheung AL. 1998. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol Microbiol 30:991–1001. doi: 10.1046/j.1365-2958.1998.01126.x. [DOI] [PubMed] [Google Scholar]
- 28.Fitzpatrick F, Humphreys H, O'Gara JP. 2005. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J Clin Microbiol 43:1973–1976. doi: 10.1128/JCM.43.4.1973-1976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, Loughman A, Foster TJ, O'Gara JP. 2008. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol 190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holland LM, Conlon B, O'Gara JP. 2011. Mutation of tagO reveals an essential role for wall teichoic acids in Staphylococcus epidermidis biofilm development. Microbiology 157:408–418. doi: 10.1099/mic.0.042234-0. [DOI] [PubMed] [Google Scholar]
- 31.Lehman MK, Bose JL, Sharma-Kuinkel BK, Moormeier DE, Endres JL, Sadykov MR, Biswas I, Bayles KW. 2015. Identification of the amino acids essential for LytSR-mediated signal transduction in Staphylococcus aureus and their roles in biofilm-specific gene expression. Mol Microbiol 95:723–737. doi: 10.1111/mmi.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung AL, Nishina K, Manna AC. 2008. SarA of Staphylococcus aureus binds to the sarA promoter to regulate gene expression. J Bacteriol 190:2239–2243. doi: 10.1128/JB.01826-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung AL, Nishina KA, Trotonda MP, Tamber S. 2008. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol 40:355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucas AL, Manna AC. 2013. Phenotypic characterization of sarR mutant in Staphylococcus aureus. Microb Pathog 57:52–61. doi: 10.1016/j.micpath.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Christner M, Heinze C, Busch M, Franke G, Hentschke M, Bayard Duhring S, Buttner H, Kotasinska M, Wischnewski V, Kroll G, Buck F, Molin S, Otto M, Rohde H. 2012. sarA negatively regulates Staphylococcus epidermidis biofilm formation by modulating expression of 1 MDa extracellular matrix binding protein and autolysis-dependent release of eDNA. Mol Microbiol 86:394–410. doi: 10.1111/j.1365-2958.2012.08203.x. [DOI] [PubMed] [Google Scholar]
- 36.Mack D, Siemssen N, Laufs R. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect Immun 60:2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee CY, Buranen SL, Ye ZH. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101–105. doi: 10.1016/0378-1119(91)90399-V. [DOI] [PubMed] [Google Scholar]
- 38.Projan SJ, Archer GL. 1989. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J Bacteriol 171:1841–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]