Abstract
Herpes simplex virus 1 (HSV-1) is a widespread global pathogen, of which the strain KOS is one of the most extensively studied. Previous sequence studies revealed that KOS does not cluster with other strains of North American geographic origin, but instead clustered with Asian strains. We sequenced a historical isolate of the original KOS strain, called KOS63, along with a separately isolated strain attributed to the same source individual, termed KOS79. Genomic analyses revealed that KOS63 closely resembled other recently sequenced isolates of KOS and was of Asian origin, but that KOS79 was a genetically unrelated strain that clustered in genetic distance analyses with HSV-1 strains of North American/European origin. These data suggest that the human source of KOS63 and KOS79 could have been infected with two genetically unrelated strains of disparate geographic origins. A PCR RFLP test was developed for rapid identification of these strains.
Keywords: KOS, KOS63, KOS79, HSV-1, genome, variation, genetic distance, co-infection
Introduction
Infection with human herpesvirus 1, or herpes simplex virus 1 (HSV-1; genus Simplexvirus, subfamily Alphaherpesvirinae, family Herpesviridae, order Herpesvirales) is widespread in humans, and is the causative agent of recurrent herpes labialis or genitalis (1). Initial infection takes place at epithelial or mucosal surfaces, where virus replicates actively to form self-limited lesions that involve interactions with innate and adaptive immunity (1). HSV-1 also enters sensory nerve endings and migrates in axons of the peripheral nervous system (PNS) by retrograde transport to neuronal nuclei located within sensory or sympathetic ganglia (2–4). It is in this cell population that HSV-1 establishes latency. The virus may spontaneously reactivate and produce intermittent shedding and/or clinical diseases throughout life (3, 5). Occasional invasion of the central nervous system (CNS) by HSV-1 can result in rare but severe and sometimes fatal encephalitis (6, 7). While antiviral drugs have been developed for HSV, these are only able to target the lytic replication stages and the intermittent cycles of productive viral replication (8–11). The latent state is largely refractory to current antivirals (8, 12). HSV remains the most studied herpesvirus, and knowledge of HSV has driven key advances in our fundamental knowledge of the mechanisms of herpesvirus entry, replication, gene expression, assembly and egress (1, 12–14).
The vast majority of laboratory and genetic studies have been restricted to a relatively small number of HSV-1 strains and isolates, of which some of the most common are HSV-1 strains F, KOS, 17, SC16, and McKrae (1, 15–21). Strains of HSV differ greatly in phenotype, particularly in in vivo animal model systems of pathogenesis (18, 22–26). Variants of the same name or source are often presumed to be similar, but may show different genetic and phenotypic characterizations, demonstrating that this assumption does not always hold true (22, 27). For example, strains H166 and H166syn are distinctly different strains, both in phenotype and genotype, but arose from the same source patient (28, 29). Two recently published genome sequences of HSV-1 McKrae, which were based on strains propagated in different labs, revealed coding differences in genes such as UL36 and UL56 (30, 31). We recently demonstrated that clonal variants of strains KOS and F could differ in plaque phenotype and harbor numerous genetic differences despite their overall clonal identity (29). The genomes of five variants of KOS have been published (Table 1) (29, 32–34), and many more variants have been utilized in prior studies (15, 22, 34–37), before genome sequencing was a viable option. The history and genome sequences of several of these variants have recently been documented by Colgrove et al (34). These KOS variants have been noted to differ in virulence in animal models, as well as other phenotypes, but the genetic basis of these differences is not yet known (22, 29, 34, 38).
Table 1.
KOS variants and strains used for genome sequence comparisons.
| Abbreviated name | Parent strain | Accession | Source lab (reference) |
|---|---|---|---|
| KOSDavido | KOS | JQ673480 | Davido and colleagues (32) |
| KOSLarge | KOS | KM222721 | Szpara and colleagues (29) |
| KOSKinchington | KOS | JQ780693 | Kinchington and colleagues1 |
| KOSKnipe | KOS | KT899744.1 | Knipe and colleagues (34) |
| KOS1.1 | KOS | KT887225.1 | Knipe and colleagues (34) |
| KOS63 | KOS | TBD | Dix (22), Szpara and colleagues2 |
| KOS79 | KOS79 | TBD | Dix (22), Szpara and colleagues2 |
| 17 | 17 | JN555585 | McGeoch, Davison, and colleagues (20, 21, 76) |
KOSKinchington is described only by GenBank record
KOS63 and KOS79 are presented for the first time here.
Two variants of HSV-1 KOS, called KOS63 and KOS79, were noted relatively early on to differ dramatically in phenotype, both in vitro and in vivo (22). Limited genetic analyses using restriction fragment length polymorphisms (RFLP) and single-gene analysis by PCR suggested several differences. HSV-1 KOS63, which has an orolabial origin, has been passaged numerous times in multiple laboratories (15, 22, 34, 35, 37, 38), and is believed to represent the standard KOS isolates that circulate in many laboratories, but this has not been clearly established before this work. Another orolabial isolate attributed to the same individual, KOS79, has undergone minimal passage in the laboratory (22). In a matched comparison using both peripheral and intracerebral routes of HSV-1 infection, Dix et al. found that KOS79 was highly neurovirulent and neuroinvasive in mice, while KOS63 was much less so (22). KOS63 and KOS79 were suggested to be distinct strains on the basis of polypeptide synthesis and RFLP analyses, although the primary data were not shown (22). KOS79 has also been shown to harbor an extended array of tandem repeats in the neurovirulence protein ICP34.5, relative to a shorter array found in a plaque-purified variant of KOS called KOS321 (36, 39). A number of differences in the ICP0 promoter region of KOS and KOS79 have also been noted (40).
We set out to address the relatedness of the historical isolate KOS63, and the more virulent strain KOS79. We sequenced the full genomes of KOS79 and KOS63 and compared them to five published genome sequences of KOS (29, 32–34). We found that KOS63 and previously sequenced KOS variants are closely related to one another, akin to clonal variants from a parental population (29). However KOS63 and its clonal variants were all genetically distant from KOS79 – differing as much as unrelated strains isolated from individuals in disparate geographic regions. KOS79 clusters most closely with North American and European strains, while KOS63 and its variants cluster with Asian strains. These data provide further evidence that individuals may harbor quite disparate viral strains, and will enable future evaluations of the genetic basis of the neurovirulence of strain KOS79.
Results
Full genome sequencing of KOS79 and comparison to KOS
We used genome-wide comparative sequence analysis to illuminate the genetic relatedness of HSV-1 KOS79 to HSV-1 KOS63 and other previously described isolates of the KOS strain (Table 1). To do this, we first applied Illumina high-throughput sequencing to obtain sequence read data from viral nucleocapsid DNA of strains KOS79 and KOS63. Consensus genomes were generated for each strain using a previously described combination of de novo assembly and reference-guided assembly methods (Table 2; see Methods for details) (29). The genomes of KOS79 and KOS63 do not contain any truncated or missing proteins, with the exception of US9 in KOS63. The presence of a SNP (C>T) in US9, which generates an early stop codon that truncates the protein, has been described in multiple prior sequences of the KOS strain (29, 32, 41). Finding this identical SNP in KOS63 supported the premise that it is indeed closely related to the previously sequenced KOS isolates.
Table 2.
Sequencing statistics for HSV-1 strains KOS79 and KOS63.
To analyze the genetic relatedness of KOS79, KOS63, and the previously sequenced KOS strains from our lab and others, we compared the identity and number of variant proteins among these strains (Table 3 and Supplementary Table 1). As an out-group, we included for comparison the unrelated HSV-1 reference strain 17. We observed that the newly sequenced KOS63 and previously sequenced KOS variants shared an average of 99.2% DNA identity (Table 3). In contrast, comparison of KOS63 or the other KOS variants to either KOS79 or strain 17 resulted in a lower DNA identity of 98.5%. This minor shift in percent identity across the genome translated into a sizable number of proteins harboring coding variations in each pairwise comparison. Between KOS63 and the KOS variants, an average of just 6 proteins differ in any pairwise comparison (Table 3; see Supplementary Table 1 for complete list of variations). This is akin to what was previously observed for sister clones from a common parental virus stock (29). In contrast, pairwise comparison of any of these vs. KOS79 or strain 17 yields a similarly high number of over 60 proteins that harbor coding variations in each pairwise comparison (Table 3). Nine proteins are completely conserved between KOS79 and any of the KOS63-like variants: UL15, UL16, UL20, UL26.5, UL33, VP26 (UL35), UL45, VP22 (UL49A), and UL55, several of which were also highly conserved in our prior global analysis of more than 20 independent HSV-1 strains (42). Overall, the amount of difference observed in these comparisons suggested that KOS79 is as different from KOS63-like strains as the unrelated strain 17.
Table 3.
Pairwise comparisons of genome-wide DNA identity (upper right) and number of proteins with coding variations (lower left) in HSV-1 strains.
| # of proteins with coding variations* | Percent pair-wise DNA identity → | ||||||||
| Strains | 17 | KOS79 | KOS63 | KOSDavido | KOSLarge | KOSKinchington | KOSKnipe | KOS1.1 | |
| 17 | 98.3% | 98.4% | 99.0% | 98.3% | 98.5% | 98.9% | 98.7% | ||
| KOS79 | 57 | 98.4% | 98.1% | 98.6% | 98.4% | 98.1% | 98.1% | ||
| KOS63 | 61 | 65 | 99.2% | 99.3% | 99.2% | 99.1% | 99.1% | ||
| KOSDavido | 61 | 65 | 4 | 99.0% | 99.3% | 99.6% | 99.7% | ||
| KOSLarge | 63 | 65 | 7 | 10 | 99.6% | 99.1% | 99.0% | ||
| KOSKinchington | 61 | 65 | 3 | 2 | 9 | 99.3% | 99.2% | ||
| KOSKnipe | 61 | 65 | 5 | 4 | 11 | 3 | 98.9% | ||
| KOS1.1 | 61 | 65 | 2 | 5 | 8 | 4 | 6 | ||
See Supplementary Table 2 for a list of specific proteins that vary between each pair of strains.
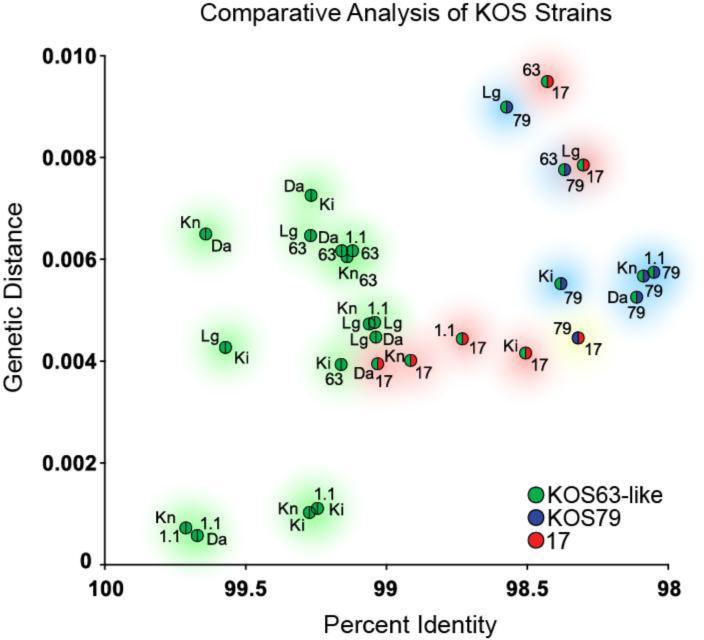
Since percent DNA identity does not reflect the location or distribution of differences between these genomes, we next estimated the evolutionary distance between these strains. As anticipated, the KOS63-like strains had the smallest genetic distance between them in any pairwise comparison, correlating with their high DNA identity (Figure 1). Comparison of either KOS79 or strain 17 to any of the KOS63-like variants resulted in a larger estimate of genetic distance, in keeping with the lower DNA identity (Figure 1). SplitsTree analysis of KOS79, KOS63-like variants, and strain 17 confirmed the close relationship among the KOS63-like variants, and placed KOS79 and strain 17 at nearly equidistant points away from the KOS63-like variants (Supplementary Figure 1). The distance between KOS79 and any of the other KOS variants suggests that it is a distinct and unrelated strain.
Figure 1.
Genome sequence comparisons reveal that KOS63 is as different from KOS79 as from the HSV-1 reference strain 17. The genome sequences of multiple variants of KOS63 (green) were compared to those of KOS79 (blue) and the HSV-1 reference strain 17 (red). For every possible pairwise combination of genomes, we calculated the genetic distance and the percent DNA identity (see Methods for details; see also Table 3). Points are color-coded to reflect the pairs in each comparison, with strain labels next to each point (see below for abbreviations). The x-axis reflects decreasing percent identity, while the y-axis reflects increasing genetic distance. In both cases, more disparate pairwise comparisons will plot further from the origin. Shaded circles provide a visual guide to the groups of pairwise comparisons. Green shading indicates pairwise combinations of KOS63-like strains, blue shading indicates combinations involving strain KOS79, and red shading indicates combinations involving strain 17. Yellow shading indicates the sole pairwise combination of strains 17 and KOS79. Trimmed genome sequences, lacking the terminal copies of the large repeats, were used for all comparisons to avoid giving undue weight to the repeats. Reiteration lengths in several published strains (32, 34) are artificially matched to strain 17, so these regions were removed from all comparisons as well (see Methods for details). Abbreviations are as follows: Da, KOSDavido from Davido and colleagues (32); Lg, KOSLarge from Szpara and colleagues (29); Ki, KOSKinchington from Kinchington and colleagues (33); Kn, KOSKnipe and 1.1, KOS1.1, from Knipe and colleagues (34); 63, KOS63 as described by Dix and colleagues (22), with genome sequenced here; 79, KOS79 as described by Dix and colleagues (22), with genome sequenced here; 17, HSV-1 reference strain 17, as initially published by McGeoch and colleagues (20, 21), and revised by Davison and colleagues (76). All GenBank accessions are listed in Table 1.
Genetic distance analysis and the likely geographic origin of strain KOS79
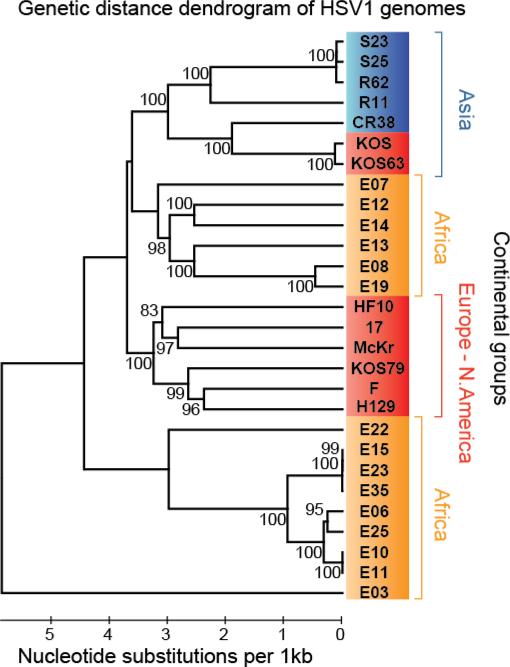
In a previous comparison of HSV-1 genomes from diverse geographic areas, we found that a majority of strains clustered near those from the same geographic origin (42). The KOSDavido isolate included in that publication was the sole exception, where despite its USA origin, KOSDavido clustered with strains of Asian origin (42, 43). To place KOS79 within the context of this study, we integrated KOS79 and KOS63 into the prior 26-genome alignment and computed the genetic distance among these strains. In this analysis, KOS79 clustered with 100% bootstrap confidence with HSV-1 strains F and H129, which were also isolated in the USA (Figure 2). This trio is found within a larger 100% bootstrap confidence cluster, which includes three other European and North American strains, HF10, 17 and McKrae. This contrasts with KOS63 and KOSDavido, which are assigned to the Asian genetic distance cluster with equal confidence. Although attributed to the same individual, KOS63 and KOS79 are fundamentally distinct strains that appear to have distant geographic historical origins.
Figure 2.
Genetic distance reveals that KOS63 and KOSDavido cluster separately from KOS79 in a global collection of HSV-1 strains.
We extended a prior dendrogram analysis of 26 HSV-1 strains of global distribution (42) to include strains KOS63 and KOS79. KOS63 and the originally-included KOSDavido strain both cluster with HSV-1 strains of Asian origin, while KOS79 groups with HSV-1 strains of European and North American origin. Dendrogram was calculated with UPGMA in MEGA (1,000 bootstrap replicates). Branch confidence values are indicated on the tree. Scale bar indicates the number of nucleotide substitutions per kilobase.
The extensive recombination that is postulated to occur in HSV precludes a cladistic analysis using these data on genetic distance (42, 44–48). To investigate the possibility of recombination between KOS63 and KOS79 in the original host, we performed SimPlot analysis using KOS79, KOS63, a consensus of the European cluster (strains HF10, 17, McKrae, F and H129) and a consensus of the Africa-1 cluster (strains E08, E12, E13, E14 and E19) from our prior study (Supplementary Figure 2A) (42). We observed that KOS79 is most closely related to the European consensus over most of its length, except for a few short regions where it is most similar to the Africa-1 consensus (e.g. between kilobases 45-50 and 97-102) or where no predominant background can be identified (e.g. between kilobases 60-65 and 80-85). The corresponding BootScan plot (Supplementary Figure 2B) shows chi-shaped crossover signals supportive of recombination with an African strain at 97-102 kb, although the other two areas of African similarity have less well formed signs of crossover. BootScan analysis suggests potential recombination between KOS79 and KOS63 at 60-65 kb and 80-85kb, though this finding should be viewed with caution because the SimPlot analysis implies genetic equidistance between all strains in these regions. Accurate recombination analysis in HSV-1 will always be complicated by the deep evolutionary history of the species, the extent of recent intercontinental travel with plentiful opportunities for recombination, and the fundamental genetic similarity of all known strains (e.g. more than 98.5% identical over any 10 kb window).
Rapid PCR test to distinguish KOS79 and KOS63
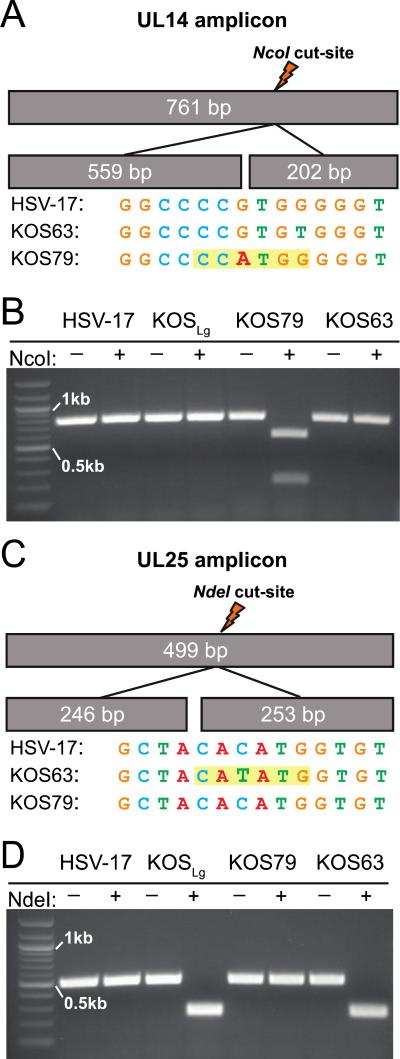
Previous analyses have often grouped KOS63 and KOS79 together (39, 40), although the genomic data now indicate that these viruses are likely to be significantly different. Since the possibility exists that some labs may have initiated studies with a KOS isolate derived from either KOS63 or KOS79, we considered it important to develop an easy assay to distinguish these strains. We therefore designed primers to amplify segments of the tegument proteins UL14 and UL25 (Supplementary Table 2), which each contain a strain-specific restriction digest site for NcoI or NdeI (Figure 3). The KOS79 genome contains an NcoI site in UL14 while KOS63 and its variants do not (Figure 3A). KOS63 and its variants contain an NdeI site in UL25 that is not present in KOS79 (Figure 3C). To confirm these sequencing results, nucleocapsid DNA from HSV-1 strains 17, KOS63, and KOS79 were used to PCR amplify these regions of UL14 and UL25. These amplicons were then subjected to restriction digest and analyzed. Examination of digestion products revealed the expected patterns (Figure 3B,D). Sanger sequencing was used to confirm ten additional SNPs in the UL14 amplicon, and one additional SNP in the UL25 amplicon, that differ between KOS63 and KOS79 (data not shown). This diagnostic tool can be implemented to quickly screen and identify KOS virus stocks or recombinant derivatives within any laboratory as being either KOS63-like or KOS79.
Figure 3.
Restriction digest patterns of HSV-1 UL14 and UL25 amplified regions. Schematic representation of UL14 (A) and UL25 (C) amplicons including predicted size and location of strain-specific NcoI (A) or NdeI (C) sites within each PCR amplicon. The sequence surrounding the distinctive NcoI cut site in KOS79 (A) and NdeI cut site in KOS63 (C) are highlighted in yellow, with other HSV-1 strains shown for comparison. Electrophoretic analyses of UL14 (B) and UL25 (D) amplicons with and without enzyme addition are shown for HSV-1 strains 17, KOSLg, KOS79, and KOS63. Bands at 0.5kb and 1kb are marked on the 100 bp ladder in each gel.
RFLP analysis of KOS strains
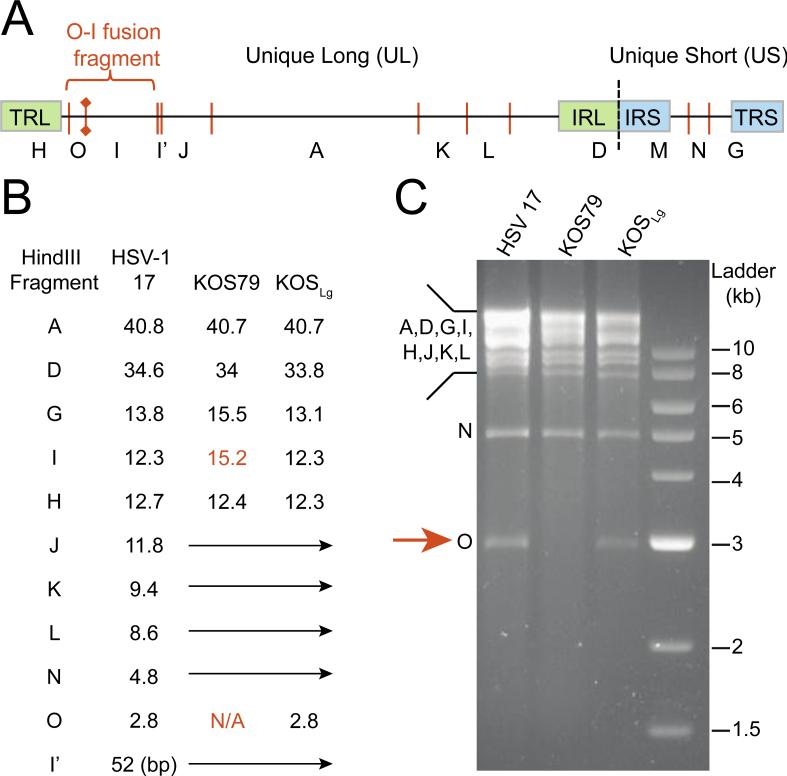
To validate the newly assembled genomes for KOS63-like and KOS79 using an alternative method, actual RFLP patterns were compared to those predicted from each genome sequence. Computational comparison of KOS63 and KOS79 predicted the loss of a HindIII restriction site in glycoprotein M (gM; UL10) in strain KOS79, resulting in the loss of a 2.8 kb band that exists in KOS63-like variants. In the historical lettering system for HSV-1 DNA restriction fragments, this 2.8 kb band was referred to as the “O” fragment (49–51) (Figure 4A). As expected, the 2.8 kb “O” fragment was not present after HindIII digestion of KOS79 DNA, but was present in strains KOS and 17 (Figure 4B-C). This O-I fusion event has been previously noted to occur in African and European strains of HSV-1, but not in Asian strains (52). Finding the fusion in KOS79, but not KOS63, correlates with the geographic history suggested for each strain by genetic distance analysis (Figure 2). Taken together, these results indicate the wide genetic divergence of KOS79 and KOS63, and highlight that genetic comparisons of these strains will reveal many differences regardless of the phenotype under examination (22, 39, 40).
Figure 4.
RFLP analysis of HSV-1 KOS variants.
Diagram of the HSV-1 genome (top) and predicted HindIII restriction sites (bottom). The location (A) and sizes (B) of the historical lettering systems for HindIII fragments (49–51) predicted for these genomes are also illustrated. Fusion of fragment O and I, due to loss of a HindIII site in gM (UL10), has been previously noted in a number of HSV-1 strains. This loss occurs in strain KOS79, both in the genome sequence (B) and in RFLP analysis (C) after HindIII digestion of viral nucleocapsid DNA. Abbreviations in the genome diagram are as follows: TRL/IRL, terminal/internal repeat of the long region; IRS/TRS, internal/terminal repeat of the short region.
Discussion
KOS is one of the most extensively used strains in HSV-1 research, particularly in the United States. It was isolated from a volunteer in the 1960s (15, 43). In addition to its use in many mechanistic and genetic studies, the KOS strain has also been used to develop vaccine antigens and vectors (53–56). The full genome sequence of HSV KOS (here called KOSDavido) was first published in 2012 (32), and later duplicated with a high degree of fidelity by the Kinchington group, using a 1981 master stock obtained from Neal DeLuca at the University of Pittsburgh (JQ780693; here called KOSKinchington) (33), and further detailed by Knipe, Coen and colleagues using a master stock provided by Priscilla Schaffer (here called KOSKnipe and KOS1.1) (34). A plaque-purified isolate of KOS (called KOSLarge) was sequenced by our own group using a stock obtained from Hendricks, Kinchington and colleagues (29, 57). Previous work by Iwasaki and colleagues (37) highlighted potential immune-evasion differences in the isolates of HSV-1 KOS held by different labs, but the genetic basis of these variants has not been investigated. Here we present the first analysis of overall genetic identity among the five publicly available genome sequences of KOS, and we extend this comparison to include newly sequenced genomes of the historically-dated strains KOS63 and KOS79. These additional strains have distinct levels of virulence in animal models, as previously characterized by Dix et al (22), with KOS79 showing significantly higher levels of neurovirulence and neuroinvasiveness than KOS63.
One of the differences between KOS79 and KOS63 that may contribute to the higher virulence of KOS79 in vivo is its intact US9 protein. Previous authors have shown that US9 is involved in anterograde transport of viral capsids and glycoproteins to axonal termini in related alphaherpesviruses, such as pseudorabies virus (PRV) (58–60). However the role of US9 in anterograde transport of HSV-1 is less clear (61–64). KOS63 and its variants have a mutation present in the TATA-box promoter region of the membrane protein US9, as well as a substitution at position 58 in the US9 gene. This substitution introduces an early stop codon, and truncates the last 32 residues of the US9 protein (32, 41). This mutation removes the stop codon and elongates the neighboring open reading frame of US8A, a protein of undetermined function (32, 41). The new availability of complete coding sequences for KOS79 and KOS63 will enable future studies on how the differences in other herpesvirus proteins contribute to their observed differences in virulence phenotypes.
KOS63 and KOS79 were reportedly isolated from the same volunteer on separate occasions (15, 22, 43). The historical record does not allow us to confirm these isolations or to collect a fresh isolate from the same individual, but the frequent usage of KOS63-like variants in labs around the world makes it imperative to clarify their history as much as possible. We present here a rapid PCR and restriction digest test to allow labs to easily distinguish between KOS79 and the classic KOS63-like strains. In past studies, authors did not distinguish whether KOS63 and KOS79 were passage variants—i.e. high- and low-passage variants from a common parental stock—or were unrelated isolates (40, 39). Here we showed that KOS79 is genetically distinct from KOS63-like strains, with sufficient divergence that one could posit independent geographic acquisitions of HSV-1 infection (43). These data remove the possibility that KOS63 represents a passage variant of KOS79, a hypothesis that arose from examples of attenuation by serial passage for related herpesviruses (65–67). If they arose in the same source, the historical dating of strains KOS63 and KOS79 (from 1963 and 1979 respectively; (22)) suggests that these viruses reactivated independently of one another and at different times. It is impossible to determine now whether these historical isolates also arose from different body sites, which emphasizes the importance of recording full clinical details for new isolates. In future studies of extant cases, it will be important to determine how viral reactivation and shedding occur in dual- or multiply-infected individuals, with attention to aspects of space, time, and genetic variation.
Methods
Virus culture and DNA isolation
Stocks of HSV-1 KOS63 and KOS79 were obtained from R. Dix and expanded in adherent MRC-5 (ATCC®, CCL-171) human fetal lung fibroblast cells. MRC-5 cultures grown in Eagle's Minimum Essential Media (EMEM; Sigma-Aldrich) were infected at an MOI of 0.01 and harvested when cultures displayed significant cytopathic-effect (CPE). These master stocks were titered on Vero cells (ATCC®, CCL-81) for easier plaque visualization. For DNA preparation, MRC-5 cultures were infected at an MOI of 5 and viral genomic DNA (gDNA) was isolated via a nucleocapsid preparation assay previously described (68). Similarly prepared gDNA from KOSLarge (29) and from plaque-purified HSV-1 strain 17 were used in the RFLP and PCR studies below.
Next generation sequencing
Viral gDNA was sheared and prepared for sequencing on an Illumina MiSeq. Viral nucleocapsid DNA was sheared using a Covaris M220 sonicator/disruptor, with the following parameters: 60 seconds duration, peak power 50, 10% duty cycle, at 4°C. The Illumina TruSeq DNA sample prep kit was used to prepare barcoded sequencing libraries, according to the manufacturer's low-throughput protocol. Libraries were quantified and assessed by Qubit (Invitrogen, CA), Bioanalyzer (Agilent), and qPCR for library adaptors (KAPA Biosystems). Paired-end sequencing (2 × 300 bp length) was carried out on our lab's Illumina MiSeq instrument, according to manufacturer's recommendations, with 17 pM input. All sequence data has been deposited at the NCBI Short Read Archive under BioProject ID PRJNA295931. Sequence alignments are also openly available from the Lancaster University data archive at http://dx.doi.org/10.17635/lancaster/researchdata/29.
Consensus viral genomes were assembled using a recently described viral genome assembly (VirGA) workflow (29). Briefly, this workflow performs a number of quality control filters, including removal of contaminating host sequences, adaptors from library preparation, or image-acquisition artifacts. It then carries out multiple iterations of de novo assembly using SSAKE, which are combined into longer blocks of sequence (contigs) using Celera and GapFiller. The Mugsy alignment package is used to match these contigs to a reference genome for HSV-1 (strain 17, JN555585). The best matching contigs are stitched into a single consensus genome using a VirGA-specific script called mafnet. Finally, an array of quality-control measures are used to annotate, query, and improve the draft consensus genome. These include comparing the pileup of raw sequence reads to the initial draft genome, detecting polymorphisms within the sequence, identification of sequence features, gaps, and low coverage areas, and validation of selected areas by PCR and Sanger sequencing. Consensus genome sequences for KOS79 and KOS63 have been deposited in GenBank under accessions KT425109 and KT425110.
Pairwise identity and genetic distance comparisons
Trimmed genome sequences, lacking the terminal copies of the large repeats, were used for genetic distance comparisons to avoid giving undue weight to the repeats. In strains KOSDavido, KOSKnipe, and KOS1.1, the lengths of several reiterations (also known as variable number tandem repeats, or VNTRs) have been artificially set to match the reiteration copy number from the reference strain 17 (32, 34). We used the annotation of these VNTRs in the GenBank accession of KOSDavido (JQ673480) to locate and remove these regions from all pairwise calculations of distance and percent identity. ClustalW2 (69) was used to construct pairwise global nucleotide alignments between whole genome sequences and pairwise global amino acid alignments between ORFs. These alignments were utilized by downstream custom Python scripts to calculate percent identity, protein differences, and genetic distance between samples. Further, an all-vs.-all alignment was used to generate a NeighborNet phylogenetic network in SplitsTree (44, 45, 70) with Uncorrected P distances in order to illustrate potential evolutionary relationships between strains.
Global genetic distance and recombination analyses
KOS79 was incorporated into the genome-wide alignment from Szpara et al. (42) using MAFFT with input parameters specified to freeze the prior alignment (71). Addition of KOS79 and KOS63 did not alter the published alignment of any of the other 26 HSV-1 genomes (42). The source alignment is found on the accompanying data website to Szpara et al. (42): http://szparalab.psu.edu/hsv-diversity/. Genetic distances were calculated in MEGA6 (72) using the Maximum Composite Likelihood measure and a dendrogram was produced using the Unweighted Paired Group Mean (Arithmetic) method (UPGMA) with 1000 bootstrap replicates (73, 74). Recombination was analyzed using SimPlot (75).
RFLP analysis
Nucleocapsid DNA was digested overnight at 37°C using the restriction enzymes BamHI, EcoRI, and HindIII (New England BioLabs), according to the company's specifications. 250 ng of DNA was used in each reaction along with the recommended buffer for each restriction enzyme. Samples were visualized on a 1.0% agarose gel with ethidium bromide (Fisher Bio-Reagents) after overnight separation. Digital digests of the corresponding draft genomes were carried out using the software Geneious (version 7.1.7, created by Biomatters, available from http://www.geneious.com). These predicted patterns were compared against actual banding from above digestions, to assess draft genome matching to the observed RFLP banding patterns.
Restriction digest of PCR amplicons
Primers were designed flanking NcoI and NdeI restriction endonuclease cut sites in UL14 and UL25 respectively (Table 1). HSV 17, KOS79, KOSLg and KOS gDNA was boiled for five minutes and then snap-cooled on ice for five minutes. PCR was conducted in 50 μl reaction volumes using 3 μl of template DNA, 1 unit of PrimeStar GXL DNA polymerase (Clontech), 200 mM deoxynucleoside triphosphate, 1 μM primers, and 1X buffer. PCR conditions in an Eppendorf Mastercycler Nexus gradient were as follows: 98°C for 2 minutes, followed by 35 cycles of denaturation at 98°C for 10 seconds, 15 seconds of annealing at 55°C for UL25 or 60°C for UL14, and primer extension at 68°C for 1 minute. Final extension was carried out at 68°C for 2 minutes. PCR amplicons were then subjected to restriction digest using NcoI and NdeI (New England Biosciences). 20 μl of PCR mixture containing the amplicon of interest was mixed with 5 μl of enzyme-appropriate buffer, 24 μl of water, and 1 μl of restriction endonuclease. Samples were incubated at 37°C for 1 hour, then heat inactivated at either 65°C for NdeI or 80°C for NcoI. Digested samples were then purified using a QIAquick PCR purification kit (Qiagen). Standard agarose gel electrophoresis was used to separate and visualize bands. Gels were run at 100 volts for 1 hour at room temperature.
Supplementary Material
Highlights.
Viral forensic genomics reveals historical relationships from current data
Two strains of HSV-1 attributed to the same source are genetically distinct
Acknowledgements
We dedicate this work to Dr. Kendall O. Smith, whose early contributions and mentoring of herpes virologists have led to makny subsequent discoveries. This work was supported by grants to MLS from NIH-NIAID K22 AI095384 and the Virus Pathogens Resource (ViPR) Bioinformatics Resource Center. PRK was supported by NIH EY015291, EY08098 the Eye & Ear Institute of Pittsburgh and Research to Prevent Blindness Inc. The authors thank L.W. Enquist, in whose lab this work was initiated, and members of the Enquist lab for their feedback during early phases of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roizman B, Knipe DM, Whitley R. Fields Virology. 6th ed. Lippincott Williams & Wilkins; Philadelphia, PA.: 2013. Herpes Simplex Viruses; pp. 1823–1897. [Google Scholar]
- 2.Corey L, Spear PG. Infections with Herpes Simplex Viruses. N Engl J Med. 1986;314:686–691. doi: 10.1056/NEJM198603133141105. [DOI] [PubMed] [Google Scholar]
- 3.Croen KD, Ostrove JM, Dragovic LJ, Smialek JE, Straus SE. Latent herpes simplex virus in human trigeminal ganglia. Detection of an immediate early gene “anti-sense” transcript by in situ hybridization. N Engl J Med. 1987;317:1427–1432. doi: 10.1056/NEJM198712033172302. [DOI] [PubMed] [Google Scholar]
- 4.Koyuncu OO, Hogue IB, Enquist LW. Virus infections in the nervous system. Cell Host Microbe. 2013;13:379–93. doi: 10.1016/j.chom.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roizman B, Whitley RJ. An inquiry into the molecular basis of HSV latency and reactivation. Annu Rev Microbiol. 2013;67:355–374. doi: 10.1146/annurev-micro-092412-155654. [DOI] [PubMed] [Google Scholar]
- 6.Haynes RE, Azimi PH, Cramblett HG. Fatal herpesvirus hominis (herpes simplex virus) infections in children. Clinical, pathologic, and virologic characteristics. JAMA. 1968;206:312–319. [PubMed] [Google Scholar]
- 7.Whitley RJ, Soong SJ, Linneman C, Liu C, Pazin G, Alford CA. Herpes simplex encephalitis. Clinical Assessment. JAMA. 1982;247:317–320. [PubMed] [Google Scholar]
- 8.Coen DM, Whitley RJ. Antiviral drugs and antiviral drug resistance. Curr Opin Virol. 2011;1:545–547. doi: 10.1016/j.coviro.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Field HJ, Clercq E de. Effects of Oral Treatment with Acyclovir and Bromovinyldeoxyuridine on the Establishment and Maintenance of Latent Herpes Simplex Virus Infection in Mice. J Gen Virol. 1981;56:259–265. doi: 10.1099/0022-1317-56-2-259. [DOI] [PubMed] [Google Scholar]
- 10.Whitley RJ, Soong SJ, Hirsch MS, Karchmer AW, Dolin R, Galasso G, Dunnick JK, Alford CA. Herpes simplex encephalitis: vidarabine therapy and diagnostic problems. N Engl J Med. 1981;304:313–318. doi: 10.1056/NEJM198102053040602. [DOI] [PubMed] [Google Scholar]
- 11.Zakirova NF, Shipitsyn AV, Jasko MV, Prokofjeva MM, Andronova VL, Galegov GA, Prassolov VS, Kochetkov SN. Phosphoramidate derivatives of acyclovir: Synthesis and antiviral activity in HIV-1 and HSV-1 models in vitro. Bioorg Med Chem. 2012;20:5802–5809. doi: 10.1016/j.bmc.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Johnston C, Koelle DM, Wald A. HSV-2 : in pursuit of a vaccine. J Clin Invest. 2011;121:4600–4609. doi: 10.1172/JCI57148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: an update. Virus Res. 2009;143:222–34. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 14.Pellett PE, Roizman B. Fields Virology. 6th ed. Lippincott Williams & Wilkins; Philadelphia, PA.: 2013. Herpesviridae; pp. 1802–1822. [Google Scholar]
- 15.Smith KO. Relationship Between the Envelope and the Infectivity of Herpes Simplex Virus. Exp Biol Med. 1964;115:814–816. doi: 10.3181/00379727-115-29045. [DOI] [PubMed] [Google Scholar]
- 16.Williams LE, Nesburn AB, Kaufman HE. Experimental Induction of Disciform Keratitis. Arch Ophthalmol. 1965;73:112–114. doi: 10.1001/archopht.1965.00970030114023. [DOI] [PubMed] [Google Scholar]
- 17.Ejercito PM, Kieff ED, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 18.Hill TJ, Field HJ, Blyth WA. Acute and recurrent infection with herpes simplex virus in the mouse: a model for studying latency and recurrent disease. J Gen Virol. 1975;28:341–353. doi: 10.1099/0022-1317-28-3-341. [DOI] [PubMed] [Google Scholar]
- 19.Knipe DM, Ruyechan WT, Roizman B, Halliburtont IANW. Molecular genetics of herpes simplex virus : Demonstration of regions of obligatory and nonobligatory identity within diploid regions of the genome by sequence replacement and insertion. Proc Natl Acad Sci U S A. 1978;75:3896–3900. doi: 10.1073/pnas.75.8.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mcgeoch DJ, Dolan A, Donald S, Street C. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 1986;14:1727–1746. doi: 10.1093/nar/14.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGeoch DJ, Dalrymple MA, Davison AJ, Dolan A, Frame MC, McNab D, Perry LJ, Scott JE, Taylor P. The Complete DNA Sequence of the Long Unique Region in the Genome of Herpes Simplex Virus Type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 22.Dix RD, McKendall RR, Baringer JR. Comparative neurovirulence of herpes simplex virus type 1 strains after peripheral or intracerebral inoculation of BALB/c mice. Infect Immun. 1983;40:103–112. doi: 10.1128/iai.40.1.103-112.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sedarati F, Stevens JG. Biological basis for virulence of three strains of herpes simplex virus type 1. J Gen Virol. 1987;68:2389–2395. doi: 10.1099/0022-1317-68-9-2389. [DOI] [PubMed] [Google Scholar]
- 24.Stevens JG. HSV-1 neuroinvasiveness. Intervirology. 1993;35:152–163. doi: 10.1159/000150306. [DOI] [PubMed] [Google Scholar]
- 25.Goel N, Mao H, Rong Q, Docherty JJ, Zimmerman D, Rosenthal KS. The ability of an HSV strain to initiate zosteriform spread correlates with its neuroinvasive disease potential. Arch Virol. 2002;147:763–73. doi: 10.1007/s007050200024. [DOI] [PubMed] [Google Scholar]
- 26.Brandt CR, Kolb AW, Shah DD, Pumfery AM, Kintner RL, Jaehnig E, Van Gompel JJ. Multiple Determinants Contribute to the Virulence of HSV Ocular and CNS Infection and Identification of Serine 34 of the US1 Gene as an Ocular Disease Determinant. Invest Ophthalmol Vis Sci. 2003;44:2657–2668. doi: 10.1167/iovs.02-1105. [DOI] [PubMed] [Google Scholar]
- 27.Buchman TG, Simpson T, Nosal C, Roizman B. The Stucture of Herpes Simplex Virus DNA and its Application to Molecular Epidemiology. Ann N Y Acad Sci. 1980;354:279–290. doi: 10.1111/j.1749-6632.1980.tb27972.x. [DOI] [PubMed] [Google Scholar]
- 28.Heller M, Dix RD, Baringer JR, Schachter J, Conte JE., Jr Herpetic proctitis and meningitis: recovery of two strains of herpes simplex virus type 1 from cerebrospinal fluid. J Infect Dis. 1982;146:584–588. doi: 10.1093/infdis/146.5.584. [DOI] [PubMed] [Google Scholar]
- 29.Parsons LR, Tafuri YR, Shreve JT, Bowen CD, Shipley MM, Enquist LW, Szpara ML. Rapid Genome Assembly and Comparison Decode Intrastrain Variation in Human Alphaherpesviruses. mBio. 2015;6:e02213–14. doi: 10.1128/mBio.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macdonald SJ, Mostafa HH, Morrison LA, Davido DJ. Genome sequence of herpes simplex virus 1 strain McKrae. J Virol. 2012;86:9540–9541. doi: 10.1128/JVI.01469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watson G, Xu W, Reed A, Babra B, Putman T, Wick E, Wechsler SL, Rohrmann GF, Jin L. Sequence and comparative analysis of the genome of HSV-1 strain McKrae. Virology. 2012;433:528–37. doi: 10.1016/j.virol.2012.08.043. [DOI] [PubMed] [Google Scholar]
- 32.Macdonald SJ, Mostafa HH, Morrison LA, Davido DJ. Genome Sequence of Herpes Simplex Virus 1 Strain KOS. J Virol. 2012;86:6371–6372. doi: 10.1128/JVI.00646-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Payne KM, Russell DA, Kinchington PR. Complete genome sequence of Herpes Simplex Virus Type-1 strain KOS. GenBank Access JQ780693 [Google Scholar]
- 34.Colgrove RC, Liu X, Griffiths A, Raja P, Deluca NA, Newman RM, Coen DM, Knipe DM. History and genomic sequence analysis of the herpes simplex virus 1 KOS and KOS1.1 sub-strains. Virology. 2016;487:215–221. doi: 10.1016/j.virol.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Little SP, Schaffer PA. Expression of the Syncytial Genetic and Phenotypic ( syn ) Phenotype in HSV-1 , Strain KOS : Studies of Mutants in Two syn Loci. Virology. 1981;112:686–702. doi: 10.1016/0042-6822(81)90314-7. [DOI] [PubMed] [Google Scholar]
- 36.Holland TC, Marlin SD, Levine M, Glorioso J. Antigenic variants of herpes simplex virus selected with glycoprotein-specific monoclonal antibodies. J Virol. 1983;45:672–682. doi: 10.1128/jvi.45.2.672-682.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato A, Linehan MM, Iwasaki A. Dual recognition of herpes simplex viruses by TLR2 and TLR9 in dendritic cells. Proc Natl Acad Sci U S A. 2006;103:17343–17348. doi: 10.1073/pnas.0605102103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Davido DJ, Morrison LA. HSV-1 strain McKrae is more neuroinvasive than HSV-1 KOS after corneal or vaginal inoculation in mice. Virus Res. 2013;173:436–40. doi: 10.1016/j.virusres.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao H, Rosenthal KS. An N-terminal Arginine-rich Cluster and a Proline-Alanine-Threonine Repeat Region Determine the Cellular Localization of the Herpes Simplex Virus Type 1 ICP34.5 Protein and Its Ligand, Protein Phosphatase 1. J Biol Chem. 2002;277:11423–11431. doi: 10.1074/jbc.M111553200. [DOI] [PubMed] [Google Scholar]
- 40.Miles DA. The role of ICP0 expression in reactivation of herpes simplex virus type 1 strains KOS-63 and KOS-79. In: Stroop William., editor. PhD dissertation. University of Arkansas at Little Rock; Arkansas, USA.: 2000. [Google Scholar]
- 41.Negatsch A, Mettenleiter TC, Fuchs W. Herpes simplex virus type 1 strain KOS carries a defective US9 and a mutated US8A gene. J Gen Virol. 2011;92:167–72. doi: 10.1099/vir.0.026484-0. [DOI] [PubMed] [Google Scholar]
- 42.Szpara ML, Gatherer D, Ochoa A, Greenbaum B, Dolan A, Bowden RJ, Enquist LW, Legendre M, Davison AJ. Evolution and diversity in human herpes simplex virus genomes. J Virol JVI. 2013:01987–13. doi: 10.1128/JVI.01987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grose C. Korean War and the Origin of Herpes Simplex Virus 1 Strain KOS. J Virol. 2014;88:1–2. doi: 10.1128/JVI.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norberg P, Tyler S, Severini A, Whitley R, Liljeqvist J-A, Bergstrom T. A genome-wide comparative evolutionary analysis of herpes simplex virus type 1 and varicella zoster virus. PloS One. 2011;6:1–8. doi: 10.1371/journal.pone.0022527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norberg P, Kasubi MJ, Haarr L, Bergstrom T, Liljeqvist J-A. Divergence and recombination of clinical herpes simplex virus type 2 isolates. J Virol. 2007;81:13158–13167. doi: 10.1128/JVI.01310-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiry E, Meurens F, Muylkens B, McVoy M, Gogev S, Thiry J, Vanderplasschen A, Epstein A, Keil G, Schynts F. Recombination in alphaherpesviruses. Rev Med Virol. 2005;15:89–103. doi: 10.1002/rmv.451. [DOI] [PubMed] [Google Scholar]
- 47.Bowden R, Sakaoka H, Donnelly P, Ward R. High recombination rate in herpes simplex virus type 1 natural populations suggests significant co-infection. Infect Genet Evol. 2004;4:115–23. doi: 10.1016/j.meegid.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 48.Kolb AW, Ané C, Brandt CR. Using HSV-1 genome phylogenetics to track past human migrations. PloS One. 2013;8:1–9. doi: 10.1371/journal.pone.0076267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skare J, Summers WC. Structure and Function of Herpesvirus Genomes: II. EcoRI, XbaI, and HindIII Endonuclease cleavage sites on Herpes Simplex Virus Type I DNA. Virology. 1977;76:581–595. doi: 10.1016/0042-6822(77)90240-9. [DOI] [PubMed] [Google Scholar]
- 50.Wilkie NM, Cortini R, Clements JB. Structural studies and physical maps for the herpes simplex virus genome. J Antimicrob Chemother. 1977;3(Suppl A):47–62. doi: 10.1093/jac/3.suppl_a.47. [DOI] [PubMed] [Google Scholar]
- 51.Lonsdale DM, Brown SM, Subak-Sharpe JH, Warren KG, Koprowski H. The polypeptide and the DNA restriction enzyme profiles of spontaneous isolates of herpes simplex virus type 1 from explants of human trigeminal, superior cervical and vagus ganglia. J Gen Virol. 1979;43:151–71. doi: 10.1099/0022-1317-43-1-151. [DOI] [PubMed] [Google Scholar]
- 52.Sakaoka H, Aomori T, Saito H, Sato S, Kawana R, Hazlett DT, Fujinaga K. A comparative analysis by restriction endonucleases of Herpes Simplex Virus type 1 isolated in Japan and Kenya. J Infect Dis. 1986;153:612–16. [PubMed] [Google Scholar]
- 53.McCarthy AM, McMahan L, Schaffer PA. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J Virol. 1989;63:18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Osorio Y, Cohen J, Ghiasi H. Improved Protection from Primary Ocular HSV-1 Infection and Establishment of Latency Using Multigenic DNA Vaccines. Investig Opthalmology Vis Sci. 2004;45:506. doi: 10.1167/iovs.03-0828. [DOI] [PubMed] [Google Scholar]
- 55.Kaur A, Sanford HB, Garry D, Lang S, Klumpp SA, Watanabe D, Bronson RT, Lifson JD, Rosati M, Pavlakis GN, Felber BK, Knipe DM, Desrosiers RC. Ability of herpes simplex virus vectors to boost immune responses to DNA vectors and to protect against challenge by simian immunodeficiency virus. Virology. 2007;357:199–214. doi: 10.1016/j.virol.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X, Broberg E, Watanabe D, Dudek T, DeLuca N, Knipe DM. Genetic engineering of a modified herpes simplex virus 1 vaccine vector. Vaccine. 2009;27:2760–2767. doi: 10.1016/j.vaccine.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramachandran S, Knickelbein JE, Ferko C, Hendricks RL, Kinchington PR. Development and pathogenic evaluation of recombinant herpes simplex virus type 1 expressing two fluorescent reporter genes from different lytic promoters. Virology. 2008;378:254–264. doi: 10.1016/j.virol.2008.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lyman MG, Feierbach B, Curanovic D, Bisher M, Enquist LW. Pseudorabies Virus Us9 Directs Axonal Sorting of Viral Capsids. J Virol. 2007;81:11363–11371. doi: 10.1128/JVI.01281-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lyman MG, Kemp CD, Taylor MP, Enquist LW. Comparison of the Pseudorabies Virus Us9 Protein with Homologs from Other Veterinary and Human Alphaherpesviruses. J Virol. 2009;83:6978–6986. doi: 10.1128/JVI.00598-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor MP, Enquist LW. Axonal spread of neuroinvasive viral infections. Trends Microbiol. 2015 doi: 10.1016/j.tim.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snyder A, Polcicova K, Johnson DC. Herpes simplex virus gE/gI and US9 proteins promote transport of both capsids and virion glycoproteins in neuronal axons. J Virol. 2008;82:10613–24. doi: 10.1128/JVI.01241-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGraw HM, Awasthi S, Wojcechowskyj JA, Friedman HM. Anterograde Spread of Herpes Simplex Virus Type 1 Requires Glycoprotein E and Glycoprotein I but Not Us9. J Virol. 2009;83:8315–8326. doi: 10.1128/JVI.00633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Howard PW, Howard TL, Johnson DC. Herpes Simplex Virus Membrane Proteins gE/gI and US9 Act Cooperatively To Promote Transport of Capsids and Glycoproteins from Neuron Cell Bodies into Initial Axon Segments. J Virol. 2013;87:403–414. doi: 10.1128/JVI.02465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Draper JM, Huang G, Stephenson GS, Bertke AS, Cortez D a, LaVail JH. Delivery of herpes simplex virus to retinal ganglion cell axon is dependent on viral protein Us9. Invest Ophthalmol Vis Sci. 2013;54:962–7. doi: 10.1167/iovs.12-11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cha T, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spatz SJ. Accumulation of attenuating mutations in varying proportions within a high passage very virulent plus strain of Gallid herpesvirus type 2. Virus Res. 2010;149:135–142. doi: 10.1016/j.virusres.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 67.Quinlivan M, Breuer J. Clinical and molecular aspects of the live attenuated Oka varicella vaccine: Studies of the Oka varicella vaccine. Rev Med Virol. 2014;24:254–273. doi: 10.1002/rmv.1789. [DOI] [PubMed] [Google Scholar]
- 68.Szpara ML, Tafuri YR, Enquist LW. Preparation of viral DNA from nucleocapsids. J Vis Exp JoVE. 2011:2–7. doi: 10.3791/3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinforma Appl Note. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 70.Strimmer K, Wiuf C, Moulton V. Recombination analysis using directed graphical models. Mol Biol Evol. 2001;18:97–99. doi: 10.1093/oxfordjournals.molbev.a003725. [DOI] [PubMed] [Google Scholar]
- 71.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sneath PHA, Sokal RR. Numerical Taxonomy. W.H. Freeman and Company; San Francisco, California, USA.: 1973. [Google Scholar]
- 74.Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 75.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davison AJ. Evolution of sexually transmitted and sexually transmissible human herpesviruses. Ann N Y Acad Sci. 2012;1230:E37–E49. doi: 10.1111/j.1749-6632.2011.06358.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.