Abstract
Polymorphisms in toll-like receptor (TLR) and β-defensin (DEFB) genes have been recognized as potential genetic factors that can influence susceptibility to and severity of periodontal diseases (PD). However, data regarding associations between these polymorphisms and PD are still scarce in North American populations, and are not available in HIV+ North American populations. In this exploratory study, we analyzed samples from HIV+ adults (n = 115), who received primary HIV care at 3 local outpatient HIV clinics and were monitored for PD status. We genotyped a total of 41 single nucleotide polymorphisms (SNPs) in 8 TLR genes and copy number variation (CNV) in DEFB4/103A. We performed regression analyses for levels of 3 periodontopathogens in subgingival dental plaques (Porphyromonas gingivalis [Pg], Treponema denticola [Td], and Tannerella forsythia [Tf]) and 3 clinical measures of PD (periodontal probing depth [PPD], gingival recession [REC], and bleeding on probing [BOP]). In all subjects combined, 2 SNPs in TLR1 were significantly associated with Td, and one SNP in TLR2 was significantly associated with BOP. One of the 2 SNPs in TLR1 was significantly associated with Td in Caucasians. In addition, another SNP in TLR1 and a SNP in TLR6 were also significantly associated with Td and Pg, respectively, in Caucasians. All 3 periodontopathogen levels were significantly associated with PPD and BOP, but none was associated with REC. Instrumental variable analysis showed that 8 SNPs in 6 TLR genes were significantly associated with the 3 periodontopathogen levels. However, associations between the 3 periodontopathogen levels and PPD or BOP were not driven by associations with these identified SNPs. No association was found between DEFB4/103A CNV and any periodontopathogen level or clinical measure in all samples, Caucasians, or African Americans. Our exploratory study suggests a role of TLR polymorphisms, particularly TLR1 and TLR6 polymorphisms, in PD in HIV+ North Americans.
Introduction
Periodontal diseases (PD) modified by HIV, together with other oral infections, is considered to be among serious complications of HIV infection, and may have implications not only for oral health but possibly for systemic health as well [1–3]. Although the development of PD is generally accepted to depend on the interaction between the host response and the resident oral microbiota [4–6], new vistas into human genomics have opened in an attempt to decipher whether specific genetic polymorphisms can explain host predisposition or susceptibility to PD [6–11]. The components of innate immunity in PD include toll-like receptors (TLRs) [12,13] and β-defensins [14–17]. It is, therefore, not surprising that polymorphisms in these innate immune response genes (TLR and DEFB, respectively) have been extensively studied as potential genetic factors that may influence susceptibility to and severity of PD [12,18–28]. Although, overall, these studies provide valuable information regarding these genetic associations, they also present some important gaps: First, these studies were conducted primarily in European populations [18–21,26–28]. To the best of our knowledge, data regarding associations between these genetic polymorphisms and PD are still scarce in North American populations [22,24], and are not available in HIV+ North American populations. Second, TLR1 and TLR6, singly and as TLR1/2 and TLR2/6 heterodimers, seem to play a role in PD [29–32]. However, single nucleotide polymorphisms (SNPs) in TLR1 and TLR6 were not included in these studies. Third, regarding genetic variation in DEFB and its association with PD, while most studies [24,25,27] focused on specific SNPs in DEFB1, encoding human β-defensin 1 (hBD-1), only one study [20] focused on copy number variation (CNV) in DEFB4, encoding hBD-2. Finally, previous studies have focused on polymorphisms in either TLR [18,19,21–23,26,28] or DEFB [20,24,25,27] genes. Here, it is important to mention that TLRs have been shown to mediate the expression of hBDs in various tissues [33], and, therefore, the TLR-hBD interplay may be one of the critical determinants of HIV-associated oral infections. TLR2 and TLR4 are involved in hBD-2 induction [34–37] and TLR9 may also be involved [38]. TLR2 is also involved in hBD-3 induction [39], and hBD-3 has been shown to regulate myeloid cell activity through interaction with TLR1 and TLR2 [40–42]. Since myeloid cells play an important role in host immune responsiveness in chronic stages of PD, a full understanding of the interplay between TLRs and hBDs is crucial. Moreover, investigating the cumulative effect of polymorphisms in TLR and DEFB on PD will provide a more comprehensive understanding of how these interrelated innate immune components influence the complex outcomes in PD and, maybe, other inflammatory-mediated diseases. The present study takes the first step toward closing these gaps.
Previously, to provide a comprehensive view of PD in the era of highly active antiretroviral therapy (HAART), we characterized an urban, predominantly African American HIV+ cohort according to select immunologic and virologic markers, the presence of subgingival pathogens, dental care utilization, and oral health behaviors [43]. We evaluated PD by using distinct clinical measures, i.e., periodontal probing depth (PPD), gingival recession (REC), clinical attachment level (CAL, i.e., PPD+REC), and bleeding on probing (BOP) [2,43]. In addition, we collected subgingival dental plaque samples and quantified DNA levels of specific pathogens associated with severe PD, i.e., Porphyromonas gingivalis (Pg), Treponema denticola (Td), and Tannerella forsythia (Tf) [43]. Thus, our study provided a detailed and extensive characterization of PD that includes the immunological framework of the cohort [44]. To minimize the potential misclassification from using categories (i.e., mild, moderate, and severe; or chronic vs. aggressive), we characterized the distinct clinical measures of PD as continuous variables. This methodology has allowed us to explore the relationship between HIV and PD more comprehensively [2,43,45]. A potential limitation of our study was that the inclusion of a comparison group was not feasible. However, the focus of our original study was to address the interrelationships among clinical, microbial, and immunological factors, and how they are relevant to studying PD in HIV+ adult subjects. It should be noted that this in-depth, singular focus resulted in important epidemiological and methodological findings that can help frame future HIV-related studies in the post-HAART era [44].
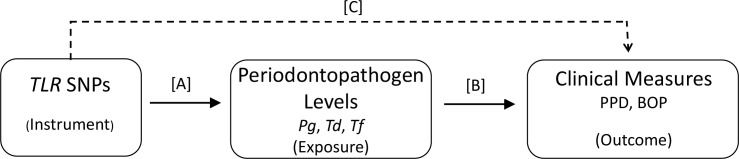
The aim of our present exploratory study was to analyze associations between genetic polymorphisms in both TLR and DEFB genes and periodontopathogen levels and clinical measures of PD in our well-characterized HIV+ cohort [2,43,45]. A long-term goal is to provide an understanding of the biological role of TLR-hBD in PD in HIV+ individuals. For our aim, we evaluated a total of 41 SNPs in 8 TLR genes (TLR1, 2, 3, 4, 6, 7, 8, and 9) [46] and CNV in DEFB4/103A, encoding hBD-2/hBD-3. In addition to linear regression, frequently presented as an optimal method in genetic association studies [47], we employed instrumental variable (IV) analysis [48–50] as a complementary, not comparative, analytic approach. In this approach, a genetic variant is treated as an instrument that is assumed to be associated with an outcome only through its association with an exposure (or risk factor or intermediate variable) (Fig 1). This instrument can then be exploited to obtain causal inferences about the effect of an exposure on an outcome [49,50].
Fig 1. Diagrammatic Representation of Instrumental Variable Analysis.
Instrumental variable models use associations C and A to estimate the relationship between an exposure/risk factor and an outcome (B). Note that the instrument is not supposed to have a direct effect on the outcome, hence this line (C) is dashed. Abbreviations: Pg, Porphyromonas gingivalis; Td, Treponema denticola; Tf, Tannerella forsythia; PPD, periodontal probing depth; BOP, bleeding on probing.
We found that a total of 5 SNPs in TLR1, TLR2, and TLR6 were significantly associated with 2 periodontopathogen levels and one clinical measure of PD.
Materials and Methods
Study design and participants
In our retrospective study, adult subjects receiving primary HIV care from 3 outpatient HIV clinics in Cleveland, OH, were recruited and monitored for PD status in a research study conducted between May 2005 and March 2009 [2]. Subject recruitment process has been described elsewhere [43,45]. Inclusion criteria were: medication-compliant adult subjects, age 18 or older, who were taking HAART for < 2 years at baseline. Exclusion criteria included evidence of a history of cardiovascular disease or Type I or II diabetes mellitus, fewer than 20 teeth, uncontrolled systemic illnesses, diagnosis or treatment of cancer in the past 5 years, pregnancy, and need for antibiotic prophylaxis prior to dental care as per the American Dental Association and other guidelines [43,45]. IRB approval was obtained from University Hospitals Case Medical Center (UHCMC); all subjects signed a written UHCMC IRB-approved informed consent document.
Periodontal disease measurements and definition
PPD, REC, and BOP were determined at 6 sites/tooth by one dentist (LTV) as previously described [2,43]. Consistent with our previous report, PD was defined as the percent of teeth with ≥ 1 of 6 sites meeting or exceeding the following thresholds: PPD ≥ 5 mm and REC ≥ 0 mm [43]. BOP was defined as the percent of teeth with ≥ 4 of 6 sites exhibiting BOP [2]. We did not include CAL as a variable into our analyses, because we were unable to process this data for 28% (32/115) of our subjects due to a lack of data management and budgetary resources. We acknowledge that classification of PD has proven to be problematic. PD is challenging to define clinically, and all classification systems produced to date have their imperfections and their critics [51,52]. The use of different case definitions has a great impact on the prevalence and extent rates of PD [53,54], and thus can influence the results and associations presented in studies, including our own. We also collected and pooled subgingival dental plaque samples from 8 pre-determined sites, based on the study by Fleiss et al. [55], and then quantified DNA levels of Pg, Td, and Tf by real-time PCR in units of log genome copy number/μg total DNA as previously described [43].
Genetic analysis
DNA was extracted from 200 μl of packed blood pellets from study subjects using the QIAamp 96 spin blood kit (QIAGEN, Valencia, CA, USA). A total of 41 SNPs in 8 TLR genes (TLR1, 2, 3, 4, 6, 7, 8 and 9) were genotyped using Illumina’s GoldenGate genotyping assay system combined with VeraCode Technology (Illumina Inc., San Diego, CA, USA). These SNPs were located in promoter regions, 5′-untranslated regions (UTR), exons, introns, and 3′-UTR (S1 Table). A detailed description of how these SNPs were selected has been provided elsewhere [46]. Allelic discrimination was performed using a BeadXpress Reader (Illumina Inc., San Diego, CA, USA) according to the manufacturer’s instructions.
For the determination of DEFB4/DEFB103A CNV, the real-time quantitative PCR assay was used as described [56]. Reference genes TBP (TATA-Box Binding Protein, GenBank accession #AL031259) and DEFB1 (encoding hBD-1, GenBank accession #NT_023736), specific primer sets producing only one specific product of ~150 bp at 54°C annealing temperature, reaction mix, and conditions were used as described [56], and the Bio-Rad CFX96TM system (Bio-Rad Laboratories, Hercules, CA, USA) was used for the PCR analysis. Each sample was run in triplicate. Data were analyzed by the comparative Ct method, and the copy numbers were calculated as described [56]. We have assessed the reliability and validity of this assay using 151 multi-population samples, which included 46 well-characterized samples from 5 diverse populations from the Coriell Cell Repositories [57].
Statistical analysis
Minor allele frequencies (MAF) were calculated using PLINK v1.07. Pairwise linkage disequilibrium (LD) between SNPs of a TLR gene or 2 genes that are nearby (TLR1 and TLR6 [12 kb], TLR7 and TLR8 [10 kb]) was determined for both Caucasians and African Americans using SHEsis (http://analysis.bio-x.cn/myAnalysis.php). Strong LD was defined by high values for both D′ (≥0.8) and r2 (≥0.5) parameters [58].
Linear regression analysis was performed on all 41 SNPs for each of the 3 periodontopathogen levels (Pg, Td, Tf) and the 3 clinical measures (PPD, REC, BOP) using PLINK v1.07. All covariates included in the initial iteration of the regression models are shown in Table 1. Backward stepwise regression was used when selecting significant covariates for each model. Initially, all subjects were included in a single analysis, adjusting for self-identified race. Regression analysis was repeated after stratifying for race, analyzing Caucasians and African Americans separately. SNPs were coded under an additive genetic model, and then under a dominant genetic model [46]. For all SNP association tests, the significance threshold α was determined by using SNPSpDlite [59]. SNPSpDlite calculates a multiple testing correction for SNPs that are in LD with one another, by calculating the LD correlation matrix for given SNPs, then estimating the number of independent tests within the sample. This is an alternative to the more conservative Bonferroni correction, which assumes all tests are independent. Thus, the significance threshold, α, for all SNP association tests was 0.001 (effective number of independent tests = 35). The additive and dominant models were tested separately, with the same significance threshold (0.001) applied to both sets of results.
Table 1. Characteristics of the Study Cohort*.
| Characteristic | n (%) | Mean (SD) |
|---|---|---|
| Age (years)† | 41 (9.6) | |
| Race† | ||
| Caucasian | 38 (33%) | |
| African American | 69 (60%) | |
| Other/Not known | 8 (7%) | |
| Gender† | ||
| Male | 88 (77%) | |
| Female | 27 (23%) | |
| Education† | ||
| High School/GED or more | 55 (48%) | |
| Smoking† | ||
| Ever smoked | 73 (64%) | |
| Number of years smoked | 14 (12.0) | |
| BMI (kg/m2)† | 27 (7.5) | |
| History of HTN† | 31 (27%) | |
| HIV clinical measures | ||
| Baseline CD4+ T-cell count (cells/μl) | 504.6 (324.1) | |
| Baseline viral load (copies/ml)† | 16519 (48717) | |
| Nadir CD4+ T-cell count (cells/μl)† | 159 (133.0) | |
| Time since first seropositive (months)† | 108.2 (83.6) | |
| HAART duration (months)† | 52.5 (55.1) | |
| Periodontal measures | ||
| PPD ≥ 5 mm (% teeth ≥ 1 site/tooth) | 36.1 (24.3) | |
| REC > 0 mm (% teeth ≥ 1 site/tooth) | 55.4 (31.1) | |
| BOP ≥ 4 sites/tooth (% teeth) | 47.4 (20.1) | |
| Microbial measures | ||
| (log genome copy number/μg DNA) | ||
| Porphyromonas gingivalis | 3.69 (2.6) | |
| Treponema denticola | 3.93 (2.3) | |
| Tannerella forsythia | 4.77 (2.1) |
*Abbreviations: GED, general educational development; BMI, body mass index; HTN, hypertension; HAART, highly active antiretroviral therapy; PPD, periodontal probing depth; REC, gingival recession; BOP, bleeding on probing.
†Covariates included in the initial iteration of the regression models.
To determine whether a statistically significant relationship existed between a periodontopathogen level and a clinical measure, preliminary linear regression analysis on all samples was conducted in R (http://www.r-project.org/). Again, backward stepwise regression was used to select only those covariates which significantly contributed to the model. If a significant relationship (p<0.001) was identified between a given periodontopathogen level and clinical measure, only then was the pair included in the IV analysis.
Instrumental variable analysis was used to test the hypothesis that a given TLR SNP may be driving the relationship between each periodontopathogen level and clinical measure pair [60,61]. Each of the 41 SNPs was tested as an IV, while a periodontopathogen level was treated as the exposure and a clinical measure as the outcome. We performed IV analysis using a 2-stage method comprising 2 regression stages: the first-stage regression of the exposure on the IVs, and the second-stage regression of the outcome on the fitted values of the exposure from the first stage. The IV analysis was performed using the ivreg function within the AER package (http://CRAN.R-project.org/package=AER). Two p values were generated: the first p value (pA) describes the relationship between the TLR SNP (IV) and the periodontopathogen level (exposure), and the second p value (pB) describes the mechanistic relationship between the periodontopathogen level (exposure) and clinical measure (outcome) (Fig 1). Both p values <0.05 were considered significant.
To analyze the effect of CNV on each of the 6 measures of PD, linear regression analysis was performed in R. Two different approaches were used to include CNV in the regression model: binary or categorical. Using a cutoff at the median (CNV = 5), CNV was recoded as a binary variable where 1 = CNV ≥ 5, 0 = CNV < 5 (reference group). To create the categorical variable, CNV was divided into CNV < 5, CNV = 5 (reference group), and CNV > 5. For each model, binary or categorical, backward stepwise regression was used as before to select only those covariates which significantly contributed to the model at the α = 0.05 level.
Results
Study subjects, minor allele frequencies, and linkage disequilibrium patterns
The cohort characteristics, including demographics, HIV infection correlates, and PD measurements of the study subjects (n = 115), are presented in Table 1. The majority of the subjects were African Americans, with a predominance of males. The MAF of all 41 TLR SNPs in all subjects combined are presented in S2 Table. MAF ranged from 0.01 to 0.50, which concurred with those reported by our group for another HIV cohort, with similar race and gender distribution, from the same geographic area [46]. The pairwise strong LD patterns of TLR genes for both Caucasians and African Americans are presented in S3 Table. Caucasians showed a higher overall extent of LD than African Americans.
Regression analysis of SNPs
In all subjects combined, 2 SNPs in TLR1 (−2192T>C, 1805G>T) were significantly associated with Td (Table 2), and one SNP in TLR2 (597T>C) was significantly associated with BOP (Table 3). One of the 2 SNPs in TLR1 (1805G>T) was significantly associated with Td in Caucasians (Table 2). In addition, another SNP in TLR1 (−7202G>A) and a SNP in TLR6 (−502T>C) were also significantly associated with Td and Pg, respectively, in Caucasians (Table 2).
Table 2. Association Between Periodontopathogen Levels and TLR SNPs*.
| Group | Periodonto-pathogen | Gene | rs number | SNP | Amino acid | Allele | Test | β | p value |
|---|---|---|---|---|---|---|---|---|---|
| All samples | Td | TLR1 | rs5743595 | −2192T>C | - | C | Add | 1.27 | 0.001a |
| Dom | 1.80 | 0.001a | |||||||
| rs5743551 | −7202G>A | - | A | Add | -0.95 | 0.002 | |||
| rs5743618 | 1805G>T | Ser602Ile | G | Add | -1.11 | 0.001a | |||
| TLR6 | rs5743795 | −1401G>A | - | A | Add | 1.39 | 0.004 | ||
| Dom | 1.48 | 0.004 | |||||||
| Caucasian | Pg | TLR6 | rs1039559 | −502T>C | - | C | Add | -2.10 | 0.001a |
| Td | TLR1 | rs5743551 | −7202G>A | - | A | Add | -1.57 | 0.001a | |
| rs5743618 | 1805G>T | Ser602Ile | G | Add | -1.56 | <0.001a | |||
| African American | Td | TLR1 | rs5743618 | 1805G>T | Ser602Ile | G | Add | -1.72 | 0.009 |
| TLR2 | rs4696480 | −16934T>A | - | T | Dom | -1.37 | 0.006 | ||
| TLR6 | rs5743810 | 745T>C | Ser249Pro | T | Add | -1.81 | 0.003 | ||
| TLR8 | rs5744077 | 28A>G | Met10Val | G | Add | -1.74 | 0.003 | ||
| rs5744080 | 645C>T | His215His | C | Add | -1.37 | 0.008 | |||
| Tf | TLR1 | rs5743618 | 1805G>T | Ser602Ile | G | Dom | -1.51 | 0.002 | |
| TLR8 | rs1548731 | +3121T>C | - | T | Add | 0.86 | 0.004 | ||
| rs5744077 | 28A>G | Met10Val | G | Add | -1.22 | 0.008 |
*Abbreviations: Td, Treponema denticola; Pg, Porphyromonas gingivalis; Tf, Tannerella forsythia; Add, additive genetic model; Dom, dominant genetic model.
aSignificant after the correction for multiple testing (α = 0.001).
Table 3. Association Between Clinical Measures of PD and TLR SNPs*.
| Group | Clinical measure | Gene | rs number | SNP | Amino acid | Allele | Test | β | p value |
|---|---|---|---|---|---|---|---|---|---|
| All samples | REC | TLR6 | rs2381289 | 4224C>T | - | T | Dom | 13.26 | 0.007 |
| BOP | TLR2 | rs3804099 | 597T>C | Asn199Asn | T | Add | 8.89 | 0.001a | |
| Dom | 14.66 | <0.001a | |||||||
| TLR9 | rs187084 | −1486C>T | - | C | Add | -7.31 | 0.010 | ||
| Caucasian | PPD | TLR4 | rs10759932 | −1607T>C | - | C | Dom | 18.12 | 0.005 |
| REC | TLR2 | rs1898830 | −15607A>G | - | G | Add | 12.92 | 0.006 | |
| Dom | 15.87 | 0.006 | |||||||
| BOP | TLR9 | rs352139 | +1174G>A | - | A | Dom | 20.57 | 0.004 | |
| rs352140 | 1635G>A | Pro545Pro | A | Add | -12.98 | 0.006 | |||
| African American | REC | TLR6 | rs2381289 | 4224C>T | - | T | Dom | 19.66 | 0.005 |
| BOP | TLR2 | rs3804099 | 597T>C | Asn199Asn | T | Dom | 14.70 | 0.004 |
*Abbreviations: REC, gingival recession; PPD, periodontal probing depth; BOP, bleeding on probing; Add, additive genetic model; Dom, dominant genetic model.
aSignificant after the correction for multiple testing (α = 0.001).
In addition to the aforementioned TLR SNPs, which were significant after the correction for multiple testing (α = 0.001), there were SNPs that were associated with the periodontopathogen levels (Table 2) and clinical measures (Table 3) at a lower significance level of 0.002–0.01. These p values may be indicative of suggestive associations, which require further validation and/or larger sample sizes. Comparing the results presented in Table 2 and Table 3, no TLR SNP was common to both periodontopathogen levels and clinical measures. The TLR2 SNPs were not in LD, and the TLR6 SNP LD patterns differed between Caucasians and African Americans (S3 Table).
Instrumental variable analysis
Preliminary linear regression analysis on all samples showed that all 3 periodontopathogen levels were significantly associated with PPD and BOP (p<0.001), but none was associated with REC (p≥0.02) (S4 Table).
Instrumental variable analysis using all 41 TLR SNPs (IVs), the 3 periodontopathogen levels (exposures), and PPD and BOP (outcomes) showed that 8 SNPs in TLR1 (n = 2), TLR2 (n = 1), TLR4 (n = 1), TLR6 (n = 1), TLR8 (n = 2), and TLR9 (n = 1) were significantly associated with the 3 periodontopathogen levels (Pg, 2 SNPs; Td, 6 SNPs; Tf, 1 SNP) (pA<0.05) (Fig 1, S5 Table). These pA values suggest that there is a relationship between these 8 IVs and 3 exposures. However, none of the periodontopathogen levels were associated with PPD or BOP after accounting for the effects of these SNPs (pB≥0.1) (Fig 1, S5 Table). These pB values suggest that the relationship between the 3 exposures and 2 outcomes is not driven by these IVs.
Distribution and association of CNV
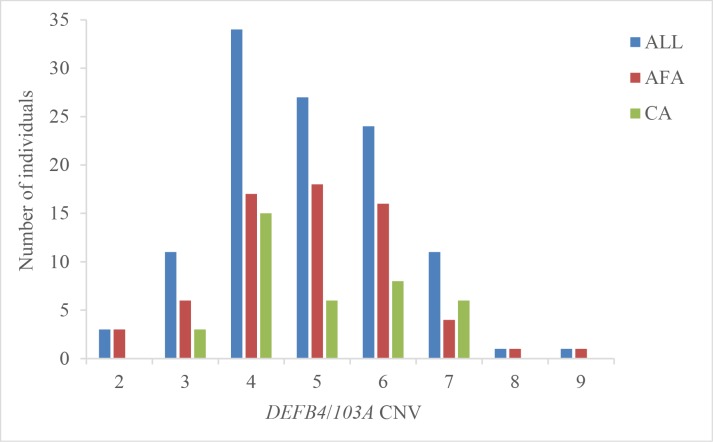
The distribution of integer DEFB4/103A copy numbers in all samples and the 2 racial groups is shown in Fig 2. No difference was observed in the median and mean copy numbers in all samples (5.0 and 4.9, respectively), Caucasians (5.0 and 5.0, respectively), and African Americans (5.0 and 4.9, respectively). Linear regression analysis using a binary (CNV ≥ 5, CNV < 5) or categorical (CNV < 5, CNV = 5, CNV > 5) model did not show any association between CNV and any periodontopathogen level or clinical measure in all samples, Caucasians, or African Americans (p>0.05, data not shown).
Fig 2. Distribution of DEFB4/103A CNV in All Samples, African Americans (AFA), and Caucasians (CA).
Discussion
In this exploratory study, utilizing samples from HIV+ North American subjects, we found that a total of 5 SNPs in TLR1 (n = 3), TLR2 (n = 1), and TLR6 (n = 1) were significantly associated with 2 periodontopathogen levels (Table 2) and one clinical measure of PD (Table 3).
TLR1 gene and/or protein expression may be upregulated in PD [29,32], and TLR1/2 heterodimer signaling may be involved in inflammatory response in PD [30,31]. To date, no report is available regarding the role of TLR1 SNPs in PD. We found that TLR1 −2192C, −7202A, and 1805G alleles were significantly associated with Td (Table 2). The information regarding the functional relevance of −2192T>C is limited. The −2192C allele may be protective for atopic asthma [62]. In that report, PBMCs of atopic asthma patients carrying −2192C showed increased expression of TLR1 mRNA and protein, increased production of proinflammatory cytokine TNF-α and TH1 cytokines IL-12 and IFN-γ, and decreased production of TH2 cytokine IL-4 after stimulation with TLR1/2 ligand Pam3CSK4 [62]. −7202G>A and 1805G>T were functionally relevant in sepsis, tuberculosis, leprosy, and candidemia, where −7202A and 1805G were associated with lower NF-κB activation and signaling, and decreased inflammatory cytokine production, including that of IL-6 (references cited in [46]). The lipooligosaccharide of Td has been shown to bind to gingival fibroblasts, and this binding is mediated by the co-receptor CD14 [63]. Furthermore, stimulating fibroblasts with Td lipooligosaccharide significantly increased the secretion of IL-6 [63]. CD14 and IL-6 are known to be involved in the inflammatory process of PD [64,65]. Thus, our finding that TLR1 −2192C, −7202A, and 1805G alleles are significantly associated with Td is noteworthy, and may be considered as a starting point in identifying the contribution of TLR1 variation to PD.
TLR2 gene and/or protein expression may be upregulated [12,29,32], not affected [66], or downregulated in PD [67]. Soluble salivary TLR2 may be considered a potential prognostic or periodontal health maintenance marker for chronic periodontitis [64]. We found that TLR2 597T allele was associated with BOP (all samples, p≤0.001; African Americans, p = 0.004) (Table 3). BOP has long been considered an indicator of a subgingival inflammatory response to bacterial pathogens; there was a direct relationship between BOP and subgingival endoscopic biofilm and calculus indexes [68]. In our study, the levels of Pg, Td, and Tf DNA in subgingival plaque were highly correlated with BOP (S4 Table). Others have reported that the mRNA levels of TLR2 in human gingival fibroblasts were positively correlated with the number of Pg in subgingival plaque [69]. This Pg-induced TLR2 expression may be partially dependent on TNF-α [69].
Most previous studies did not include 597T>C [12,19,22,23]. One study, conducted in 2 Northwest European populations, did include this SNP [21] but found no association with PD. Being a synonymous SNP (Asn199Asn), the molecular mechanism of 597T>C is not clear. The 597T allele may have a role in protection against Mycobacterium tuberculosis (Mtb), M. leprae [70,71], and the lymphatic filariasis nematode Wuchereria bancrofti [72], which harbors an intracellular symbiotic bacterium Wolbachia. It may be that 597T>C is in LD with a highly polymorphic (GT)n dinucleotide repeat within intron-2 that affects gene regulation [71]. However, the (GT)n repeat polymorphism was not previously associated with PD [18]. In addition, 597T>C is in LD with a 22-bp insertion-deletion polymorphism (Δ22 [−196 to −174]) in the 5′-UTR [72]. Located in close proximity to the NF-κB and SP1 transcription factor binding sites, TLR2 Δ22 is a functional polymorphism [72]. We did not study the (GT)n repeat and Δ22 polymorphisms, but they could form the basis of a future study. Given that HIV/AIDS continues to disproportionately affect African Americans, evaluating the functional and clinical effects of 597T>C in further studies is highly relevant.
TLR6 protein may be upregulated in PD [29], and TLR2/6 heterodimer signaling may be involved in inflammatory responses in PD [31]. However, as is the case for TLR1 SNPs, the role of TLR6 SNPs in PD has not yet been determined. We found that TLR6 −502C allele was significantly associated with Pg in Caucasians (Table 2). The functional relevance of this SNP could be due to the fact that it is in strong LD with non-synonymous 745T>C (−502C-745T) (S3 Table). Others have found 745T protective in tuberculosis patient studies and associated with lower NF-κB signaling, decreased production of IL-6 in whole blood stimulated with TLR2/6 ligand PAM2 and Mtb lysate [73], and increased production of IFN-γ in whole blood stimulated with Bacillus Calmette-Guérin as well as PBMCs stimulated with TLR1/6 lipopeptide ligands [74]. In addition, −502T>C is in strong LD with 4224C>T (S3 Table), located in the 3′-UTR. It is well known that the 3′-UTR significantly determines the stability, localization, translation, and degradation of mRNA [75].
All 3 periodontopathogen levels were significantly associated with PPD and BOP (S4 Table). In our IV analysis, a total of 8 SNPs were significantly associated with the 3 periodontopathogen levels, the majority with Td (pA values, S5 Table). However, the relationship between each periodontopathogen level and clinical measure pair was not driven by associations with these identified SNPs (pB values, S5 Table). Instrumental variable analysis is being increasingly employed in epidemiology to investigate the potential causal effects of an exposure [60,61]. However, major limitations of this analysis result from the strict assumptions that need to be satisfied for the method to be reliable [49,50]: Although TLR SNPs were associated with the periodontopathogen levels (exposures), they may be “weak instruments”–such an IV explains a relatively small proportion of variance in the exposure. It may also be that there is limited statistical power. Insufficient statistical power is a common characteristic of many IV analysis studies and, therefore, a careful selection of instruments plus an adequate sample size are deemed necessary for this method to be able to make reliable conclusions. It is also possible that pleiotropy, where one genetic variant has multiple functions, may be occurring. Among the 5 significant SNPs in this study, 3 (TLR1 −7202G>A, 1805G>T; TLR6 −502T>C) were associated with HIV status in our previous study conducted on another similar HIV cohort [46]. Our study subjects, particularly African Americans, may have population stratification, as the contribution of European ancestry to African-American populations can vary substantially (3% to >30%) [46]. Finally, while we tested for common confounders, it is possible that hidden confounding from unmeasured variables may have affected the analysis. Despite these limitations, 6 of the 8 SNPs from our IV analysis should be investigated further, as they were also significantly or suggestively associated with the periodontopathogen levels or clinical measures of PD (S5 Table).
We did not find an association between DEFB4/103A CNV and any of the measures of PD in any analysis (data not shown). This result seems contrary to the only report available, which found an association between low DEFB4 CNV and severe form of periodontitis [20]. There could be a number of reasons for this difference: First, Jaradat et al. [20] analyzed the association between DEFB4 CNV and generalized chronic periodontitis severity, classified as slight-to-moderate and severe subgroups. We, on the other hand, evaluated PD by its component parts (the 3 selected clinical measures of PD) because they may represent different biological processes contributing to PD [43]. To minimize the possibility of misclassification by categorizing data, we defined and analyzed all PD data as continuous variables. Using this approach, we have uncovered associations between PD measures and markers of cardiovascular disease risk [2] as well as PD measures and HIV-related immunological markers in the era of HAART [45]. Second, using different methods, it has been found that DEFB4/103A genes consistently vary from 2 to 12 copies per diploid genome, with a median or mean copy number of 4 in most populations [57], including the one analyzed by Jaradat et al. [20]. In our study, the median and mean copy number was 5 in all population groups. This could be related to the relatively small sample size (n = 115)–a factor that may have also contributed to the lack of association. Finally, our study was not designed to measure the hBD-2/hBD-3 protein concentrations.
In conclusion, our exploratory study provides new insights into the association of genetic variation in TLR with PD measures in HIV+ North Americans. To the best of our knowledge, this is the first study to analyze the role of TLR1 and TLR6 SNPs in PD; therefore, we cannot directly compare our findings to other studies. The mechanisms by which the aforementioned TLR SNPs, singly, in haplotypes, or in heterodimers, influence PD need to be further elucidated. Analysis of mRNA and protein levels of the TLR variants, and investigation of interactions of the TLR variants with adapter molecules and subsequent recruitment of downstream targets are needed to define the biological mechanisms that underlie these genetic associations.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank all our research participants. We are indebted to Drs. Michael Lederman and Benigno Rodriguez for providing the HIV-infected patient samples from the CFAR specimen repository. We are thankful to Simone Edelheit and Milena Rajak (Genomics Core Facility) for performing the TLR SNP genotyping. We sincerely thank Dr. Carolyn Myers, Dave McNamara, and Dr. Scott Sieg for helpful discussions and critical evaluation of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Institutes of Health DE15746, DE21376, and DE023740 to L.T.V.; National Institutes of Health (through CFAR) AI36219 to L.T.V.; National Institutes of Health 1P01DE019759 to A.W., P.A.Z.; Fogarty International Center D43TW007377 to P.A.Z.; and National Institutes of Health T32HL007567 to C.M.S.
References
- 1.Ryder MI, Nittayananta W, Coogan M, Greenspan D, Greenspan JS. Periodontal disease in HIV/AIDS. Periodontol 2000. 2012;60(1):78–97. 10.1111/j.1600-0757.2012.00445.x . [DOI] [PubMed] [Google Scholar]
- 2.Vernon LT, Babineau DC, Demko CA, Lederman MM, Wang X, Toossi Z, et al. A prospective cohort study of periodontal disease measures and cardiovascular disease markers in HIV-infected adults. AIDS Res Hum Retroviruses. 2011;27(11):1157–66. 10.1089/AID.2010.0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zelkha SA, Freilich RW, Amar S. Periodontal innate immune mechanisms relevant to atherosclerosis and obesity. Periodontol 2000. 2010;54(1):207–21. 10.1111/j.1600-0757.2010.00358.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knight ET, Liu J, Seymour GJ, Faggion CM Jr., Cullinan MP. Risk factors that may modify the innate and adaptive immune responses in periodontal diseases. Periodontol 2000. 2016;71(1):22–51. 10.1111/prd.12110 . [DOI] [PubMed] [Google Scholar]
- 5.Kornman KS. Mapping the pathogenesis of periodontitis: a new look. J Periodontol. 2008;79(8 Suppl):1560–8. 10.1902/jop.2008.080213 . [DOI] [PubMed] [Google Scholar]
- 6.Stabholz A, Soskolne WA, Shapira L. Genetic and environmental risk factors for chronic periodontitis and aggressive periodontitis. Periodontol 2000. 2010;53:138–53. 10.1111/j.1600-0757.2010.00340.x . [DOI] [PubMed] [Google Scholar]
- 7.Laine ML, Crielaard W, Loos BG. Genetic susceptibility to periodontitis. Periodontol 2000. 2012;58(1):37–68. 10.1111/j.1600-0757.2011.00415.x . [DOI] [PubMed] [Google Scholar]
- 8.Feng P, Wang X, Casado PL, Kuchler EC, Deeley K, Noel J, et al. Genome wide association scan for chronic periodontitis implicates novel locus. BMC Oral Health. 2014;14:84 10.1186/1472-6831-14-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimizu S, Momozawa Y, Takahashi A, Nagasawa T, Ashikawa K, Terada Y, et al. A genome-wide association study of periodontitis in a Japanese population. J Dent Res. 2015;94(4):555–61. 10.1177/0022034515570315 . [DOI] [PubMed] [Google Scholar]
- 10.Vieira AR, Albandar JM. Role of genetic factors in the pathogenesis of aggressive periodontitis. Periodontol 2000. 2014;65(1):92–106. 10.1111/prd.12021 . [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Sun X, Xiao L, Xie C, Xuan D, Luo G. Gene polymorphisms and periodontitis. Periodontol 2000. 2011;56(1):102–24. 10.1111/j.1600-0757.2010.00371.x . [DOI] [PubMed] [Google Scholar]
- 12.Ford PJ, Gamonal J, Seymour GJ. Immunological differences and similarities between chronic periodontitis and aggressive periodontitis. Periodontol 2000. 2010;53:111–23. 10.1111/j.1600-0757.2010.00349.x . [DOI] [PubMed] [Google Scholar]
- 13.Mahanonda R, Pichyangkul S. Toll-like receptors and their role in periodontal health and disease. Periodontol 2000. 2007;43:41–55. 10.1111/j.1600-0757.2006.00179.x . [DOI] [PubMed] [Google Scholar]
- 14.Dommisch H, Jepsen S. Diverse functions of defensins and other antimicrobial peptides in periodontal tissues. Periodontol 2000. 2015;69(1):96–110. 10.1111/prd.12093 . [DOI] [PubMed] [Google Scholar]
- 15.Wang P, Duan D, Zhou X, Li X, Yang J, Deng M, et al. Relationship between expression of human gingival beta-defensins and levels of periodontopathogens in subgingival plaque. J Periodontal Res. 2015;50(1):113–22. 10.1111/jre.12187 . [DOI] [PubMed] [Google Scholar]
- 16.Diamond G, Ryan L. Beta-defensins: what are they really doing in the oral cavity? Oral Dis. 2011;17(7):628–35. 10.1111/j.1601-0825.2011.01799.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guncu GN, Yilmaz D, Kononen E, Gursoy UK. Salivary antimicrobial peptides in early detection of periodontitis. Front Cell Infect Microbiol. 2015;5:99 10.3389/fcimb.2015.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folwaczny M, Glas J, Tonenchi L, Torok HP. Microsatellite GT polymorphism in intron 2 of human Toll-like receptor (TLR) 2 gene and susceptibility to periodontitis. Clin Oral Investig. 2011;15(3):435–41. 10.1007/s00784-010-0396-8 . [DOI] [PubMed] [Google Scholar]
- 19.Holla LI, Vokurka J, Hrdlickova B, Augustin P, Fassmann A. Association of Toll-like receptor 9 haplotypes with chronic periodontitis in Czech population. J Clin Periodontol. 2010;37(2):152–9. 10.1111/j.1600-051X.2009.01523.x . [DOI] [PubMed] [Google Scholar]
- 20.Jaradat SW, Hoder-Przyrembel C, Cubillos S, Krieg N, Lehmann K, Piehler S, et al. Beta-defensin-2 genomic copy number variation and chronic periodontitis. J Dent Res. 2013;92(11):1035–40. 10.1177/0022034513504217 . [DOI] [PubMed] [Google Scholar]
- 21.Richter GM, Graetz C, Pohler P, Nothnagel M, Dommisch H, Laine ML, et al. Common genetic risk variants of TLR2 are not associated with periodontitis in large European case-control populations. J Clin Periodontol. 2012;39(4):315–22. 10.1111/j.1600-051X.2011.01846.x . [DOI] [PubMed] [Google Scholar]
- 22.Sahingur SE, Xia XJ, Gunsolley J, Schenkein HA, Genco RJ, De Nardin E. Single nucleotide polymorphisms of pattern recognition receptors and chronic periodontitis. J Periodontal Res. 2011;46(2):184–92. 10.1111/j.1600-0765.2010.01327.x . [DOI] [PubMed] [Google Scholar]
- 23.Song GG, Kim JH, Lee YH. Toll-like receptor (TLR) and matrix metalloproteinase (MMP) polymorphisms and periodontitis susceptibility: a meta-analysis. Mol Biol Rep. 2013;40(8):5129–41. 10.1007/s11033-013-2616-1 . [DOI] [PubMed] [Google Scholar]
- 24.Wohlfahrt JC, Wu T, Hodges JS, Hinrichs JE, Michalowicz BS. No association between selected candidate gene polymorphisms and severe chronic periodontitis. J Periodontol. 2006;77(3):426–36. 10.1902/jop.2006.050058 . [DOI] [PubMed] [Google Scholar]
- 25.Loo WT, Bai LJ, Fan CB, Yue Y, Dou YD, Wang M, et al. Clinical application of human β-defensin and CD14 gene polymorphism in evaluating the status of chronic inflammation. J Transl Med. 2012;10 Suppl 1:S9 10.1186/1479-5876-10-S1-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozturk A, Vieira AR. TLR4 as a risk factor for periodontal disease: a reappraisal. J Clin Periodontol. 2009;36(4):279–86. 10.1111/j.1600-051X.2009.01370.x . [DOI] [PubMed] [Google Scholar]
- 27.Schaefer AS, Richter GM, Nothnagel M, Laine ML, Ruhling A, Schafer C, et al. A 3' UTR transition within DEFB1 is associated with chronic and aggressive periodontitis. Genes Immun. 2010;11(1):45–54. 10.1038/gene.2009.75 . [DOI] [PubMed] [Google Scholar]
- 28.Schroder NW, Meister D, Wolff V, Christan C, Kaner D, Haban V, et al. Chronic periodontal disease is associated with single-nucleotide polymorphisms of the human TLR-4 gene. Genes Immun. 2005;6(5):448–51. 10.1038/sj.gene.6364221 . [DOI] [PubMed] [Google Scholar]
- 29.Beklen A, Hukkanen M, Richardson R, Konttinen YT. Immunohistochemical localization of Toll-like receptors 1–10 in periodontitis. Oral Microbiol Immunol. 2008;23(5):425–31. 10.1111/j.1399-302X.2008.00448.x . [DOI] [PubMed] [Google Scholar]
- 30.Martins MD, Jiao Y, Larsson L, Almeida LO, Garaicoa-Pazmino C, Le JM, et al. Epigenetic modifications of histones in periodontal disease. J Dent Res. 2016;95(2):215–22. 10.1177/0022034515611876 . [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto C, Oda T, Yokoyama S, Tominari T, Hirata M, Miyaura C, et al. Toll-like receptor 2 heterodimers, TLR2/6 and TLR2/1 induce prostaglandin E production by osteoblasts, osteoclast formation and inflammatory periodontitis. Biochem Biophys Res Commun. 2012;428(1):110–5. 10.1016/j.bbrc.2012.10.016 . [DOI] [PubMed] [Google Scholar]
- 32.Scheres N, Laine ML, Sipos PM, Bosch-Tijhof CJ, Crielaard W, de Vries TJ, et al. Periodontal ligament and gingival fibroblasts from periodontitis patients are more active in interaction with Porphyromonas gingivalis. J Periodontal Res. 2011;46(4):407–16. 10.1111/j.1600-0765.2011.01353.x . [DOI] [PubMed] [Google Scholar]
- 33.Froy O. Regulation of mammalian defensin expression by Toll-like receptor-dependent and independent signalling pathways. Cell Microbiol. 2005;7(10):1387–97. 10.1111/j.1462-5822.2005.00590.x . [DOI] [PubMed] [Google Scholar]
- 34.Kumar A, Zhang J, Yu FS. Toll-like receptor 2-mediated expression of β-defensin-2 in human corneal epithelial cells. Microbes and infection / Institut Pasteur. 2006;8(2):380–9. 10.1016/j.micinf.2005.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HY, Takeshita T, Shimada J, Akopyan A, Woo JI, Pan H, et al. Induction of beta defensin 2 by NTHi requires TLR2 mediated MyD88 and IRAK-TRAF6-p38MAPK signaling pathway in human middle ear epithelial cells. BMC Infect Dis. 2008;8:87 10.1186/1471-2334-8-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romano Carratelli C, Mazzola N, Paolillo R, Sorrentino S, Rizzo A. Toll-like receptor-4 (TLR4) mediates human β-defensin-2 (HBD-2) induction in response to Chlamydia pneumoniae in mononuclear cells. FEMS Immunol Med Microbiol. 2009;57(2):116–24. 10.1111/j.1574-695X.2009.00586.x . [DOI] [PubMed] [Google Scholar]
- 37.Vora P, Youdim A, Thomas LS, Fukata M, Tesfay SY, Lukasek K, et al. β-defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. J Immunol. 2004;173(9):5398–405. 10.4049/jimmunol.173.9.5398 . [DOI] [PubMed] [Google Scholar]
- 38.Platz J, Beisswenger C, Dalpke A, Koczulla R, Pinkenburg O, Vogelmeier C, et al. Microbial DNA induces a host defense reaction of human respiratory epithelial cells. J Immunol. 2004;173(2):1219–23. 10.4049/jimmunol.173.2.1219 . [DOI] [PubMed] [Google Scholar]
- 39.Sumikawa Y, Asada H, Hoshino K, Azukizawa H, Katayama I, Akira S, et al. Induction of β-defensin 3 in keratinocytes stimulated by bacterial lipopeptides through toll-like receptor 2. Microbes and infection / Institut Pasteur. 2006;8(6):1513–21. 10.1016/j.micinf.2006.01.008 . [DOI] [PubMed] [Google Scholar]
- 40.Funderburg N, Lederman MM, Feng Z, Drage MG, Jadlowsky J, Harding CV, et al. Human β-defensin-3 activates professional antigen-presenting cells via Toll-like receptors 1 and 2. Proc Natl Acad Sci U S A. 2007;104(47):18631–5. 10.1073/pnas.0702130104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Funderburg NT, Jadlowsky JK, Lederman MM, Feng Z, Weinberg A, Sieg SF. The Toll-like receptor 1/2 agonists Pam(3) CSK(4) and human β-defensin-3 differentially induce interleukin-10 and nuclear factor-kappaB signalling patterns in human monocytes. Immunology. 2011;134(2):151–60. 10.1111/j.1365-2567.2011.03475.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Funderburg NT, Sieg SF. Diminished responsiveness to human β-defensin-3 and decreased TLR1 expression on monocytes and mDCs from HIV-1-infected patients. J Leukoc Biol. 2012;92(5):1103–9. 10.1189/jlb.1111555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vernon LT, Demko CA, Whalen CC, Lederman MM, Toossi Z, Wu M, et al. Characterizing traditionally defined periodontal disease in HIV+ adults. Community Dent Oral Epidemiol. 2009;37(5):427–37. 10.1111/j.1600-0528.2009.00485.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vernon LT, Jayashantha P, Chidzonga MM, Komesu MC, Nair RG, Johnson NW. Comorbidities associated with HIV and antiretroviral therapy (clinical sciences): a workshop report. Oral Dis. 2016;22 Suppl 1:135–48. 10.1111/odi.12412 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vernon LT, Demko CA, Babineau DC, Wang X, Toossi Z, Weinberg A, et al. Effect of Nadir CD4+ T cell count on clinical measures of periodontal disease in HIV+ adults before and during immune reconstitution on HAART. PLoS One. 2013;8(10):e76986 10.1371/journal.pone.0076986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willie B, Hall NB, Stein CM, Jurevic RJ, Weinberg A, Mehlotra RK, et al. Association of Toll-like receptor polymorphisms with HIV status in North Americans. Genes Immun. 2014;15(8):569–77. 10.1038/gene.2014.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buzkova P. Linear regression in genetic association studies. PLoS One. 2013;8(2):e56976 10.1371/journal.pone.0056976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Theodoratou E, Palmer T, Zgaga L, Farrington SM, McKeigue P, Din FV, et al. Instrumental variable estimation of the causal effect of plasma 25-hydroxy-vitamin D on colorectal cancer risk: a mendelian randomization analysis. PLoS One. 2012;7(6):e37662 10.1371/journal.pone.0037662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Hinke S, Davey Smith G, Lawlor DA, Propper C, Windmeijer F. Genetic markers as instrumental variables. J Health Econ. 2016;45:131–48. 10.1016/j.jhealeco.2015.10.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2016;0(0):1–26. 10.1177/0962280215597579 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Highfield J. Diagnosis and classification of periodontal disease. Aust Dent J. 2009;54 Suppl 1:S11–26. 10.1111/j.1834-7819.2009.01140.x . [DOI] [PubMed] [Google Scholar]
- 52.Savage A, Eaton KA, Moles DR, Needleman I. A systematic review of definitions of periodontitis and methods that have been used to identify this disease. J Clin Periodontol. 2009;36(6):458–67. 10.1111/j.1600-051X.2009.01408.x . [DOI] [PubMed] [Google Scholar]
- 53.Costa FO, Guimaraes AN, Cota LO, Pataro AL, Segundo TK, Cortelli SC, et al. Impact of different periodontitis case definitions on periodontal research. J Oral Sci. 2009;51(2):199–206. 10.2334/josnusd.51.199 . [DOI] [PubMed] [Google Scholar]
- 54.Eke PI, Page RC, Wei L, Thornton-Evans G, Genco RJ. Update of the case definitions for population-based surveillance of periodontitis. J Periodontol. 2012;83(12):1449–54. 10.1902/jop.2012.110664 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fleiss JL, Park MH, Chilton NW, Alman JE, Feldman RS, Chauncey HH. Representativeness of the "Ramfjord teeth" for epidemiologic studies of gingivitis and periodontitis. Community Dent Oral Epidemiol. 1987;15(4):221–4. 10.1111/j.1600-0528.1987.tb00525.x . [DOI] [PubMed] [Google Scholar]
- 56.Linzmeier RM, Ganz T. Human defensin gene copy number polymorphisms: comprehensive analysis of independent variation in α- and β-defensin regions at 8p22-p23. Genomics. 2005;86(4):423–30. 10.1016/j.ygeno.2005.06.003 . [DOI] [PubMed] [Google Scholar]
- 57.Mehlotra RK, Zimmerman PA, Weinberg A, Jurevic RJ. Variation in human β-defensin genes: new insights from a multi-population study. Int J Immunogenet. 2013;40(4):261–9. 10.1111/iji.12021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferlin A, Ganz F, Pengo M, Selice R, Frigo AC, Foresta C. Association of testicular germ cell tumor with polymorphisms in estrogen receptor and steroid metabolism genes. Endocr Relat Cancer. 2010;17(1):17–25. 10.1677/ERC-09-0176 . [DOI] [PubMed] [Google Scholar]
- 59.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–9. 10.1086/383251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greenland S. An introduction to instrumental variables for epidemiologists. Int J Epidemiol. 2000;29(4):722–9. 10.1093/ije/29.4.722 . [DOI] [PubMed] [Google Scholar]
- 61.Morris NJ, Gray-McGuire C, Stein CM. Mendelian randomization in family data. BMC Proc. 2009;3 Suppl 7:S45 10.1186/1753-6561-3-s7-s45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kormann MS, Depner M, Hartl D, Klopp N, Illig T, Adamski J, et al. Toll-like receptor heterodimer variants protect from childhood asthma. J Allergy Clin Immunol. 2008;122(1):86–92, e1-8. 10.1016/j.jaci.2008.04.039 . [DOI] [PubMed] [Google Scholar]
- 63.Grenier D. Binding properties of Treponema denticola lipooligosaccharide. J Oral Microbiol. 2013;5:21517 10.3402/jom.v5i0.21517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prakasam S, Srinivasan M. Evaluation of salivary biomarker profiles following non-surgical management of chronic periodontitis. Oral Dis. 2014;20(2):171–7. 10.1111/odi.12085 . [DOI] [PubMed] [Google Scholar]
- 65.Feghali K, Tanabe S, Grenier D. Soluble CD14 induces cytokine release by human oral epithelial cells. J Periodontal Res. 2011;46(1):147–52. 10.1111/j.1600-0765.2010.01311.x . [DOI] [PubMed] [Google Scholar]
- 66.Sahingur SE, Xia XJ, Voth SC, Yeudall WA, Gunsolley JC. Increased nucleic acid receptor expression in chronic periodontitis. J Periodontol. 2013;84(10):e48–57. 10.1902/jop.2013.120739 . [DOI] [PubMed] [Google Scholar]
- 67.Sarah SM, Tamilselvan S, Kamatchiammal S, Suresh R. Expression of Toll-like receptors 2 and 4 in gingivitis and chronic periodontitis. Indian J Dent Res. 2006;17(3):114–6. 10.4103/0970-9290.29879 . [DOI] [PubMed] [Google Scholar]
- 68.Checchi L, Montevecchi M, Checchi V, Zappulla F. The relationship between bleeding on probing and subgingival deposits. An endoscopical evaluation. Open Dent J. 2009;3:154–60. 10.2174/1874210600903010154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wara-aswapati N, Chayasadom A, Surarit R, Pitiphat W, Boch JA, Nagasawa T, et al. Induction of toll-like receptor expression by Porphyromonas gingivalis. J Periodontol. 2013;84(7):1010–8. 10.1902/jop.2012.120362 . [DOI] [PubMed] [Google Scholar]
- 70.Hart BE, Tapping RI. Genetic diversity of toll-like receptors and immunity to M. leprae infection. J Trop Med. 2012;2012:415057 10.1155/2012/415057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thuong NT, Hawn TR, Thwaites GE, Chau TT, Lan NT, Quy HT, et al. A polymorphism in human TLR2 is associated with increased susceptibility to tuberculous meningitis. Genes Immun. 2007;8(5):422–8. 10.1038/sj.gene.6364405 . [DOI] [PubMed] [Google Scholar]
- 72.Junpee A, Tencomnao T, Sanprasert V, Nuchprayoon S. Association between Toll-like receptor 2 (TLR2) polymorphisms and asymptomatic bancroftian filariasis. Parasitol Res. 2010;107(4):807–16. 10.1007/s00436-010-1932-9 . [DOI] [PubMed] [Google Scholar]
- 73.Shey MS, Randhawa AK, Bowmaker M, Smith E, Scriba TJ, de Kock M, et al. Single nucleotide polymorphisms in toll-like receptor 6 are associated with altered lipopeptide- and mycobacteria-induced interleukin-6 secretion. Genes Immun. 2010;11(7):561–72. 10.1038/gene.2010.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Randhawa AK, Shey MS, Keyser A, Peixoto B, Wells RD, de Kock M, et al. Association of human TLR1 and TLR6 deficiency with altered immune responses to BCG vaccination in South African infants. PLoS Pathog. 2011;7(8):e1002174 10.1371/journal.ppat.1002174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3' untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9(5):563–76. 10.4161/rna.20231 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.